Highlight
Osmotic stress rapidly induces strong transcriptome responses within the root apex and interferes with growth through a deep remodeling of hormonal status rather than by repressing the machinery of expansive growth.
Key words: Division, elongation, hormonal signaling, osmotic stress, Populus nigra, root apical meristem, root growth, transcriptome
Abstract
Molecular regulation of growth must include spatial and temporal coupling of cell production and cell expansion. The underlying mechanisms, especially under environmental challenge, remain obscure. Spatial patterns of cell processes make the root apex well suited to deciphering stress signaling pathways, and to investigating both processes. Kinematics and RNA-sequencing were used to analyze the immediate growth response of hydroponically grown Populus nigra cuttings submitted to osmotic stress. About 7400 genes and unannotated transcriptionally active regions were differentially expressed between the division and elongation zones. Following the onset of stress, growth decreased sharply, probably due to mechanical effects, before recovering partially. Stress impaired cell expansion over the apex, progressively shortened the elongation zone, and reduced the cell production rate. Changes in gene expression revealed that growth reduction was mediated by a shift in hormone homeostasis. Osmotic stress rapidly elicited auxin, ethylene, and abscisic acid. When growth restabilized, transcriptome remodeling became complex and zone specific, with the deployment of hormone signaling cascades, transcriptional regulators, and stress-responsive genes. Most transcriptional regulations fit growth reduction, but stress also promoted expression of some growth effectors, including aquaporins and expansins. Together, osmotic stress interfered with growth by activating regulatory proteins rather than by repressing the machinery of expansive growth.
Introduction
Root growth is responsive to changes in the environment (Walter et al., 2009). The biophysics of root growth has been studied in response to different environmental cues, such as low temperature (Pritchard et al., 1990), carbon (Muller et al., 1998), or water (Sharp et al., 1988; Spollen et al., 2008). Following a sudden change in water availability, growth shows a two-phased response: an acute response followed by a new, usually lower, steady-state growth rate (Skirycz and Inzé, 2010). Most studies have focused on comparing root growth in the two successive steady states (before stress versus the new steady growth rate, e.g. Spollen et al., 2008), while the responses occurring during the transition period remain largely undocumented. Pritchard et al. (1991) and Frensch and Hsiao (1994) showed that cell turgor pressure decreased very rapidly following the onset of osmotic stress, stopping growth, but was restored in growing cells in at most a couple of hours. Growth was then only partially restored, indicating that molecular and physiological rewiring had taken place in addition to physical responses.
Aiming to discover key regulators of drought acclimation, drought-induced molecular responses have been studied extensively (Lawlor, 2013). Altering the expression of regulators of drought responses has enhanced drought tolerance but often at the cost of growth, suggesting that the molecular pathways controlling growth and drought response are intertwined. Some studies have specifically focused on transcriptome remodeling in growing organs such as the root apex (e.g. Cohen et al., 2010). Others have gone further, depicting drought-induced transcriptional changes in differentially responding sections along the root apex (Poroyko et al., 2007; Opitz et al., 2016). Indeed, since root growth involves both cell proliferation and cell expansion, which occur sequentially along the root apex, harmonious and sustainable growth requires that the activities of these two processes are coupled (Silk, 1992; Baskin, 2013). Sensitivity of cell elongation to water deficit has been shown in several species using kinematics (Sharp et al., 1988; Triboulot et al., 1995). The importance of apical meristem activity in determining growth rate has been highlighted (Beemster and Baskin, 1998), and its sensitivity to water deficit has been shown in maize and poplar roots (Fraser et al., 1990; Sacks et al., 1997; Bizet et al., 2015b). The balance between proliferation and elongation processes, as well as their underlying molecular control, is a current research focus (Tardieu et al., 2011; Baskin, 2013; De Vos et al., 2014). A gene expression map at high spatial resolution has led to better knowledge of the transcriptional circuits linked to root development in Arabidopsis thaliana (Birnbaum et al., 2003; Brady et al., 2007; Iyer-Pascuzzi et al., 2009). Takatsuka and Umeda (2014) pointed out the importance of considering the roots of plant species other than Arabidopsis in order to gain further insights into the networks involved in growth regulation. RNA-sequencing now allows a holistic view of the root transcriptome in any species. Relative to the small roots of Arabidopsis, the large adventious roots of poplar cuttings are easier to dissect and allow continuous monitoring of growth.
To better understand how proliferation and elongation responses are coupled in a growing organ, attention must be paid to the dynamics of environmentally induced changes in growth rate (Silk, 1992; Sharp et al., 2004; Beemster et al., 2005). In this study, we analyzed the immediate growth response to osmotic stress within the root apex of poplar, using kinematics and RNA-sequencing. We addressed the question of the timing of transcriptome remodeling (by sampling roots both 30min and 3h after stress onset) considering both proliferation and elongation processes (by analyzing the transcriptome within the division zone and the elongation zone separately). In the whole root system of Arabidopsis, gene expression was rapidly altered after osmotic stress onset, triggering a continuous response which increased with time (Kilian et al., 2007). We thus hypothesized that the zone specificity of the responses would build up over these early steps. We aimed to assess the molecular events involved in the establishment of growth inhibition.
Osmotic stress reduced both cell expansion and cell production in Populus root. As early as 30min after stress onset, signaling pathways were activated, with the earliest steps involving hormone metabolism in particular. Our work highlights the canonical function of abscisic acid (ABA) in the growth response to osmotic stress, as well as its interplay with other hormones. Transcriptome rewiring became more complex as growth reached a new steady state. Distinct groups of genes were identified in each zone. Comparison with transcriptome responses to drought in soil suggested that some of these molecular events were not transient, but steadily activated for a long-term acclimation.
Materials and methods
Plant material, growth conditions, and stress application
Populus nigra cuttings (genotype 6J29) were grown as in Bizet et al. (2015b). The hydroponic tanks were randomly assigned to control or stress conditions. Osmotic stress was applied by replacing the nutrient solution with one supplemented with 160g l−1 polyethylene glycol (PEG 4000, Merck Chemicals, Darmstadt, Germany), generating an osmotic potential of −0.35MPa (Wescor 5500, Logan, UT, USA), and without lowering the oxygen level (oxymeter HQ40D, Hach Lange, Noisy-le-grand, France) (Supplementary Fig. S1 at JXB online). In controls, the nutrient solution was replaced by fresh solution. Solutions were fully replaced in <10min.
Control and osmotic stress treatments were performed in parallel, with two batches of eight cuttings per treatment. The experiment was run twice, independently. Apices of roots longer than 2cm were collected 0.5h and 3h after the change of nutrient solution and stored in RNAlater® (Ambion, Austin, TX, USA).
Root growth monitoring, growth parameter determination, and microscopy
Once a root was longer than 2cm, the cutting was transferred to a transparent tank filled with circulating nutrient solution. Root growth was monitored in the dark under near-infrared illumination as in Bizet et al. (2015b). In this device, the PEG-added solution fully replaced the control solution in 3min, without any manipulation of the root or pause in the growth monitoring. Images were taken every 6min (Supplementary Fig. S2A). Raw velocity profiles were obtained by particle image velocimetry, using Kineroot (Basu et al., 2007). The velocity profile along the root apex was interpolated with a cubic smoothing spline (smooth.spline, spar 0.5, R Development Core Team, 2014). The elemental elongation rate (EER) profile was calculated by spatially differentiating the velocity profile (Erickson, 1976). Root growth rate was determined as the maximum velocity (Supplementary Fig. S2B). Growth zone length was measured as the distance between the quiescent center and the position where EER fell below 3%. EERmax was defined as the maximum value of EER (Supplementary Fig. S2C). The growth zone includes the so-called division zone (DZ), where cells divide and slowly expand, and the elongation zone (EZ) where cells rapidly elongate. The length of the DZ was determined using infrared brightness as in Bizet et al. (2015b). Briefly, the drop of brightness below 70% of its maximum, probably due to lower cell wall density, corresponded to the end of DZ (Supplementary Fig. S2D). EZ length was calculated as growth zone length minus DZ length.
Following growth monitoring, root apices were fixed and sectioned longitudinally to determine the cell length profile (Bizet et al., 2015b; Supplementary Fig. S2E). Cell production rate was computed as the ratio between velocity at the end of the DZ and mean cell length in the apical meristem.
RNA isolation, library preparation, and Hiseq 2500 sequencing
Root apices were dissected under a microscope in RNAlater®. After removal of the 0.5mm apical part of the root (corresponding to the root cap), two consecutive segments were collected: (i) a 1.5mm long segment corresponding to the DZ; and (ii) a 4.5mm long segment corresponding to the EZ. Four replicates were built by pooling eight root segments per sampling unit (zone×treatment×time). Root segments in DZ and EZ were paired-distributed (DZ and EZ libraries with the same index were constructed from the same pool of roots). The two experiments were randomly represented among replicates.
RNA was extracted using a Spectrum Plant Total RNA kit (Sigma, St Louis, MO, USA) with a DNAse I treatment. Absence of genomic DNA was confirmed by PCR with intron-flanking primers. RNA quality was assessed using an Experion RNA StdSens Analysis kit (Bio-Rad, Hercules, CA, USA).
Real-time PCRs were performed with 250ng of RNA, as described in Bizet et al. (2015a). Amplicons were sequenced (Supplementary Fig. S3).
RNA were checked on Caliper GX (PerkinElmer, Waltham, MA, USA). Libraries were prepared with 1.5 µg of RNA using a ‘TruSeq Stranded mRNA Sample Prep kit’ (Illumina, San Diego, CA, USA) with an 8-fold multiplexing level. mRNA was fragmented for 3min at 94 °C. Purification was performed using 0.6X Agencourt AMPure XP beads (Beckman Coulter, Villepinte, France). Libraries were quantified using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and quality tested by Agilent 2100 Bioanalyzer High Sensitivity (Agilent Technologies, Santa Clara, CA, USA). RIN values of samples ranged from 7.3 to 9.4 (median=9). Libraries were processed with Illumina cBot for cluster generation on the flow cell, and sequenced in paired-end mode on HiSeq2500 (Illumina). Raw data processing was performed using CASAVA v1.8.2 of the Illumina pipeline for both format conversion and de-multiplexing.
Reads were deposited in the Sequence Read Archive (SRA) under study accession number SRP067564.
RNA-sequencing processing
Quality filtering and trimming of paired-end reads were performed using the erne-filter command (Erne v1.3, default parameters except --min-size=70; Del Fabbro et al., 2013). For each sample, reads were mapped to the Populus trichocarpa genome sequence (unmasked v3.0 Phytozome v10) using TopHat2 v2.12 (Trapnell et al., 2009; D. Kim et al., 2013). The P. trichocarpa genome was indexed with the ‘bowtie-build’ command from the genome sequence fasta file. TopHat2 default parameters were used except: -r 235, --mate-std-dev 55, -g 10, --read-mismatche 10, --read-edit-dist 10, and --read-gap-length 10. Gene features were provided with the –G option (Ptrichocarpa_210_v3.0.gene_exons.gff3 file). Reads that mapped once were extracted from BAM files (accepted_hits.bam, TopHat2 output) using the NH:i:1 tag as described by Loraine et al. (2013). Further analyses were performed on single-mapping reads. A reference annotation-based transcript assembly was constructed (RABT; Roberts et al., 2011). Assemblies were performed using Cufflinks v2.2.1 (Trapnell et al., 2010), which was supplied with the Ptrichocarpa_210_v3.0.gene_exons.gff3 file. A merged.gtf file was created from the 32 transcripts.gtf output files using Cuffmerge.
Based on their mapping, the 41 226 loci were assigned to three categories: unique predicted gene, putative uTAR (unannotated transcriptionally active regions), or chimera (spanning over several predicted genes or tandem repeats). Ambiguous contigs due to assembly artifacts were removed.
Expression analysis and detection of differentially expressed genes
Cuffdiff2, supplied with the merged.gtf file, was used with default parameters (Trapnell et al., 2012). For each locus, the number of single-mapping reads gave access to a normalized count-based expression that was used to perform a hierarchical clustering of the 32 libraries (Euclidean distance using R package Stats). Mean and normalized expression values were computed as FPKM (fragments per kilobase of transcript per million mapped reads). Eight pairwise comparisons were designed on the basis of their biological relevance (differential expression between zones: EZ-CTL-0.5h versus DZ-CTL-0.5h, EZ-CTL-3h versus DZ-CTL-3h, EZ-PEG-0.5h versus DZ-PEG-0.5h, and EZ-PEG-3h versus DZ-PEG-3h; stress response: DZ-PEG-0.5h versus DZ-CTL-0.5h, DZ-PEG-3h versus DZ-CTL-3h, EZ-PEG-0.5h versus EZ-CTL-0.5h, and EZ-PEG-3h versus EZ-CTL-3h). A locus was denoted ‘expressed’ or ‘not expressed’ from its status in Cuffdiff2. No significant expression was found for 13 933 loci (call ‘Notest’), and checked on normalized count and FPKM. Differential gene expression was expressed as the Log2 of the fold change (FC). Differentially expressed genes (DEGs) were identified by Cuffdiff2 (Student t-test, P-value <0.05) after correction for multiple testing [false discovery rate (FDR), P-value <0.05] and applying the threshold (|Log2(FC)|≥2).
Identification, in silico validation, and functional annotation of uTARs
The P. trichocarpa genomic sequence at the uTAR location was used as the query for a homology search in plant databases. BLASTN was performed against the expressed sequence tag (EST) database with default parameters and 10–5 as the maximal e-value (NCBI, http://www.ncbi.nlm.nih.gov), and the best matched poplar EST was reverse-blasted on the P. trichocarpa genome v3.0 (Suppplementary Table S3). NCBI BLAST+ executable (v2.2.8) was used on a local platform against the Non-Redundant Protein Sequence database (April 2014). The command line ‘blastx’ was executed with default parameters except that the e-value threshold was set to 1. BLAST results and the JBrowse interface were used to discard uTARs mapped on repeats.
Gene annotation, ontology, and enrichment analysis
Gene ontology (GO) assignment, GO-Slim summarization, and enrichment analysis were performed using Blast2Go (v2.8; Conesa et al., 2005). The 41 335 P. trichocarpa primary transcripts were blasted against A. thaliana (‘blastx’, default parameters, except e-value threshold=0.01). Only the best hit was imported, linking poplar gene models to the GO identifiers of their closest Arabidopsis orthologs. GO was summarized as plant GO-Slim terms. Enrichment analysis consisted of a Fisher’s exact test combined with an FDR correction for multiple testing. A corrected P-value of 0.05 was used to identify enriched GO terms.
Arabidopsis microarrays
Three Arabidopsis studies were selected for a pair-wise comparison of DZ and EZ (Birnbaum et al., 2003; De Rybel et al., 2010; Tsukagoshi et al., 2010). Affymetrix ATH1 Arrays were normalized using R (gcrma 2.40, default parameters; Irizarry et al., 2003). Differential expression was computed for Columbia wild-type as Log2(FC), moderated t-tests implemented in the eBayes function (limma 3.24.12; Smyth, 2004), and FDR corrections for multiple testing were employed. A zone-preferred expression [corrected P-value <0.05, Log2(FC)≥2] was detected for 1173, 2972, and 4349 AGI in GSE21876, E-MEXP-2912, and GSE5749, respectively.
Results
The dynamics of root growth in response to osmotic stress
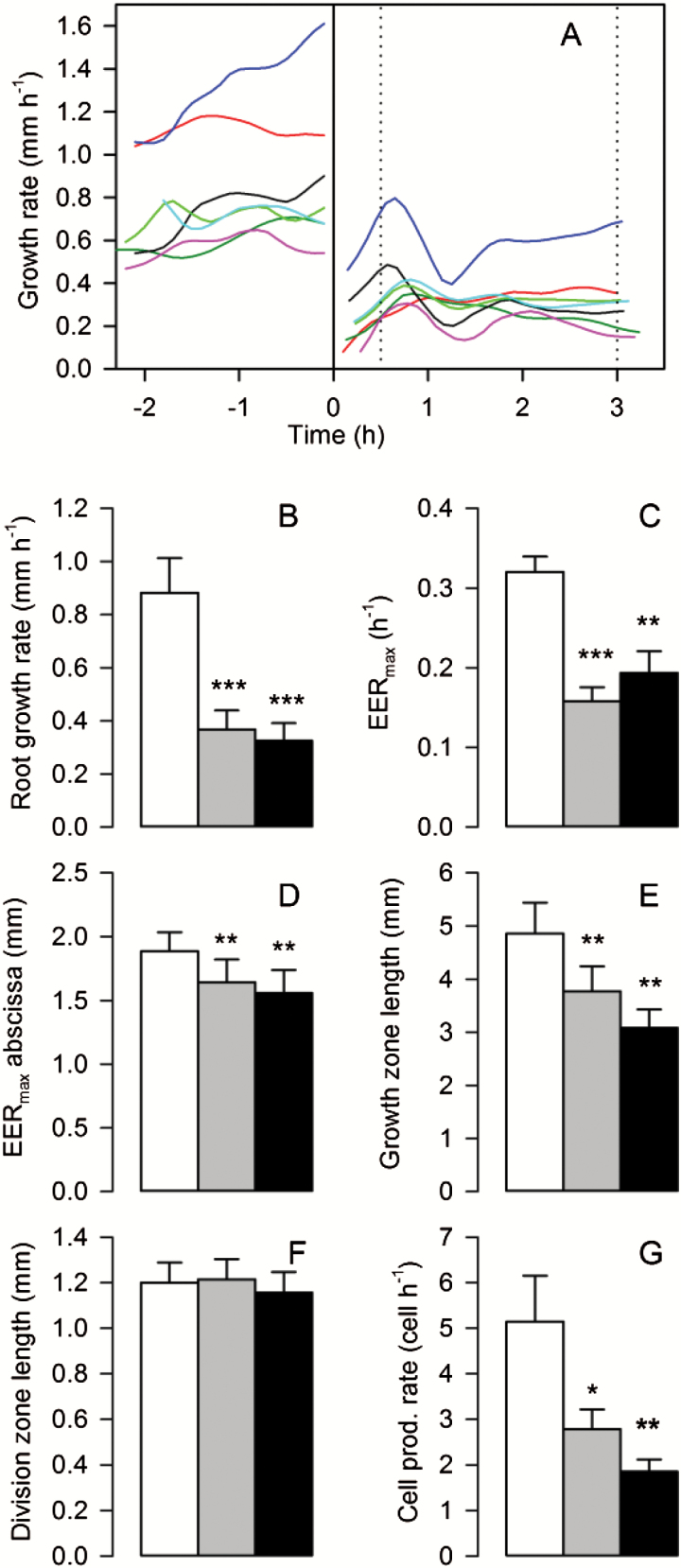
As the growth of poplar root is relatively sensitive to osmotic stress, a moderate osmotic stress (−0.35MPa) was applied by adding PEG to the nutrient solution. The growth response to osmotic stress of poplar roots was determined under infrared illumination. Our experimental set-up enabled the growth of individual roots to be monitored without any root manipulation. Before stress, the mean growth rate was 0.83mm h−1 but with a large range (Fig. 1A). Immediately following the onset of stress, root contraction and movements prevented focus and growth monitoring for a few minutes, probably due to changes in cell turgor. After 10min under stress, root growth was strongly reduced but was already recovering. During this transition period, all roots showed a common pattern of growth rate, with smooth oscillations passing through a maximum within 45min of stress application. Growth rate reached a new steady state after 2h under stress.
Fig. 1.
Root growth response to osmotic stress. (A) Individual root growth rate versus time. The solid line indicates the time of stress onset, the dotted lines the harvest time points. (B–G) Growth parameters measured before stress, and after 0.5h and 3h under stress (white, gray, and black, respectively): (B) root growth rate, (C) maximal elemental elongation rate (EERmax), (D) EERmax abscissa, (E) growth zone length, (F) division zone length, and (G) cell production rate. Mean ±SE, n=7, asterisks denote paired t-tests P-value (***<0.001, **<0.01, and *<0.05) after ANOVA.
Kinematic analyses were performed during the transition period (after 0.5h under stress) and during the steady states (before stress and after 3h under stress). Osmotic stress significantly decreased the root growth rate to the same extent at both time points (Fig. 1B). The maximal elemental elongation rate (EERmax) was decreased by 50%, and its position on the root axis was shifted towards the quiescent center (Fig. 1C, D). The growth zone was reduced according to stress duration (Fig. 1E). In contrast, the length of the division zone remained unchanged throughout the experiment (Fig. 1F). The rate of cell production by the meristem was also reduced (Fig. 1G). Since osmotic stress did not impact cell length within the division zone (Supplementary Fig. S2F), the reduced cell production rate reflected a reduced cell division rate. Taken together, osmotic stress reduced both cell processes (production and expansion), as well as the length of the elongation zone, but not that of the division zone.
Quality of RNA-sequencing raw data
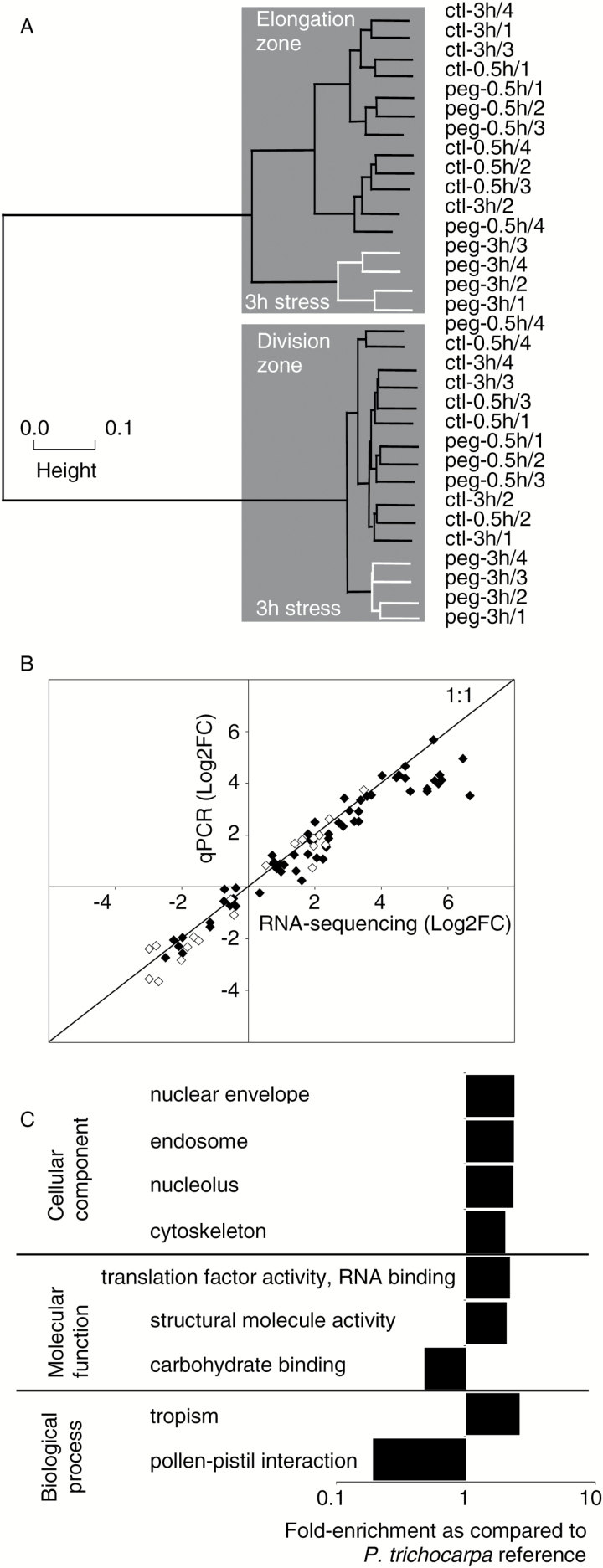
The dynamics of transcriptome remodeling under osmotic stress were investigated in the DZ and the EZ using 32 cDNA libraries. RNA-sequencing was performed using Hiseq2500 technology, generating ~30 million pair-end reads of 100bp per library (Supplementary Table S1). Following quality filtering, reads were mapped onto the P. trichocarpa genome using a reference annotation-based transcript assembly. Only reads that mapped exactly once onto the genome were considered (~95% of mapped reads; Supplementary Table S1). Overlapping reads per locus were counted and normalized using Cuffdiff2 (~13 million counts per library). Transcriptomes were strongly structured according to zone (Fig. 2A). Within the DZ and EZ clusters, the libraries corresponding to roots collected after 3h under stress were gathered together, indicating not only the impact of this treatment on the transcriptome but also its reproducibility. The remaining libraries were close, indicating a temporal stability of gene expression under control conditions and only minor changes during the transition period. Normalized expression was computed per sampling unit (zone×treatment×time) as the mean of four independent biological replicates and expressed in FPKM. RNA-sequencing results were confirmed by qPCR for 23 loci, with both methods being highly consistent (y=0.85x, R2=0.92, n=132; Fig. 2B; Supplementary Fig. S3).
Fig. 2.
The root apex transcriptome. (A) Hierarchical clustering performed on normalized count-based expression. Gray boxes highlight the main clusters gathering libraries from the division zone and from the elongation zone. The second-order clustering (white) highlights 3h long osmotic stress libraries. (B) Fold changes in gene expression assessed by RNA-sequencing and qPCR. Black and white symbols correspond to annotated genes and uTARs, respectively (Supplementary Fig. S3). (C) Highest GO enrichments in the root apex transcriptome as compared with the P. trichocarpa genome (|Fold-change enrichment| ≥2, corrected P-value <0.05, Blast2GO; Supplementary Table S2).
The root apex transcriptome
The transcriptome of the P. nigra root apex consisted of 25 262 expressed genes, covering ~62% of the 41 335 loci containing protein-coding transcripts of the P. trichocarpa v3 genome. Using Blast2go, 92% of these transcripts were associated with a GO. As compared with the P. trichocarpa genome, the root apex transcriptome was significantly enriched in 96 GOs while 41 GOs were significantly under-represented (Fig. 2C; Supplementary Table S2A). For cellular component, the most significant enrichments were ‘nuclear envelope’, ‘endosome’, ‘nucleolus’, and ‘cytoskeleton’, while ‘extracellular space’ was the most strongly depleted term. In terms of molecular function, ‘structural molecule activity’ and ‘translation factor activity, RNA binding’ were over-represented, while ‘carbohydrate binding’ was under-represented. The main enrichments for biological process were ‘tropism’ and ‘translation’.
RNA-sequencing revealed some putative novel transcribed regions (uTARs), which were inspected manually. Based on homology searches and JBrowse inspection, 258 uTARs were cross-validated. They were not only expressed in P. nigra libraries and mapped onto the P. trichocarpa genome but were also supported by a poplar mRNA and/or matched a predicted protein motif (Supplementary Table S3). About 20% of the cross-validated uTARs overlapped a predicted transcript in the previous version of the P. trichocarpa genome.
Expression patterns within the growth zone
A large compendium of genes exhibited differential expression in the DZ and EZ under at least one condition (time×treatment) (Supplementary Table S4; 7341 genes), and, for >70% of them, consistently under the four conditions (Table 1).
Table 1.
Number of differentially expressed genes
| Differential expressiona in the division zone (DZ) and the elongation zone (EZ) | Under at least one condition (time×treatment) | 7341 DEGs |
| Under the four conditions (time×treatment) | 5152 DEGs | |
| DZ-preferred expression | 1953 DEGs | |
| EZ-preferred expression | 3199 DEGs | |
| Differential expressiona in response to stress | Under at least one comparison (zone×time) | 1162 DEGs |
| In the DZ In the EZ |
216 DEGs 393 DEGs |
a Student t-test P-value <0.05; FDR P-value <0.05 and threshold, |Log2(FC)| ≥2.
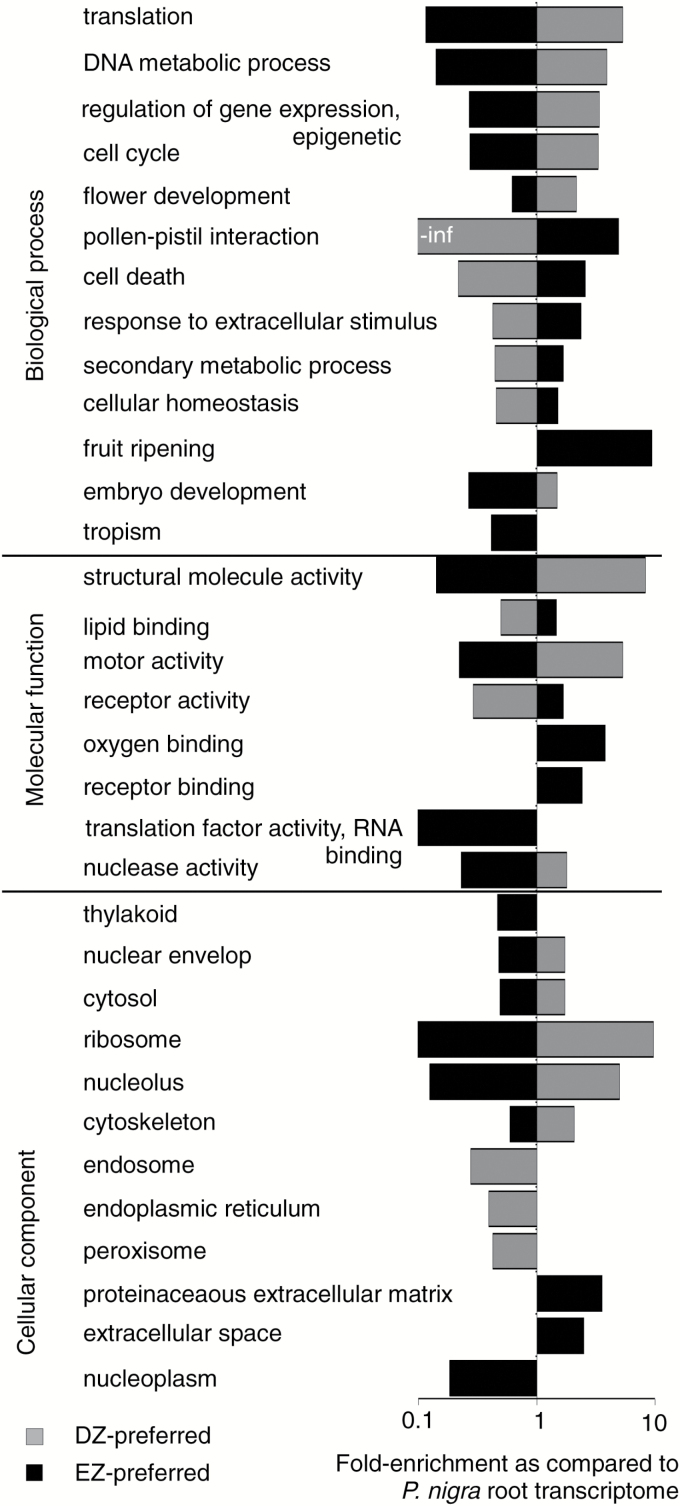
A total of 1953 genes and 18 uTARs were preferentially expressed in the DZ (‘DZ-preferred’; Supplementary Tables S3, S4). Among them, 910 genes and five uTARs exhibited at least 4-fold higher expression in the DZ than in the EZ, regardless of growth conditions. Comparing the DZ-preferred genes with the root apex transcriptome revealed significant functional enrichments (Fig. 3; Supplementary Table S2B). As expected for actively dividing cells, the most enriched GO terms were ‘translation’, ‘DNA metabolic process’, ‘regulation of gene expression’, and ‘cell cycle’, and encompassed a large set of genes associated with ‘ribosome’, ‘nucleolus’, and ‘cytoskeleton’. On the other hand, 3199 genes and 24 uTARs were expressed preferentially in the EZ, including 2011 genes and 14 uTARs showing at least 4-fold higher expression in the EZ than in the DZ (‘EZ-preferred’; Supplementary Tables S3, S4). The most enriched GO term was ‘fruit ripening’, gathering genes involved in the synthesis of ethylene (Fig. 3; Supplementary Table S2B). For cellular component, ‘proteinaceous extracellular matrix’ and ‘extracellular space’ were over-represented.
Fig. 3.
Transcriptomes of the division and elongation zone. Highest GO enrichments among zone-preferred expressed Populus genes in the division zone (gray bars) and in the elongation zone (black bars) as compared with P. nigra root transcriptome (|fold change enrichment| >2, corrected P-value <0.05; Supplementary Table S2).
With the exception of ‘cell wall’ (Supplementary Table S2B), functional enrichments were zone specific. For instance, ‘receptor binding’ or ‘oxygen binding’ were over-represented among the EZ-preferred genes. Similarly ‘flower development’ was over-represented among the DZ-preferred genes, due to the association of this ontology with genes related to cell cycle and chromatin structure. Notably, most of the GO terms over-represented among DZ-preferred genes were significantly depleted among EZ-preferred genes, and vice versa. Such over- and under-representations of functional groups in the two core sets corroborated the proper sampling of the DZ and EZ during root dissection.
A cross-species comparison was carried out using the AGI of the closest Arabidopsis homolog. Agreeing with the well-known amplification of gene number in the Populus genome, the 7341 Populus genes that showed a differential expression in the DZ and EZ (Table 1) were related to 4752 AGI (Supplementary Table S4). Among them, 1730 AGI exhibited a zone-preferred expression in the Arabidopsis root apex, thus consolidating the expression patterns of 2410 Populus genes. Such a strong overlap was unexpected, especially considering the highly duplicated poplar genome.
The overview of gene functions confirmed that genes related to cell division and cell elongation were expressed in the expected zones (Supplementary Table S4). Most cyclin and cyclin-dependent kinase genes showed a DZ-preferred expression, as did the 86 Populus genes for which the closest Arabidopsis gene was expressed preferentially in the quiescent center (Nawy et al., 2005). Genes related to cell wall modification (such as xyloglucan endotransglucosylase / hydrolase or expansin) exhibited mostly EZ-preferred expression. WRKY transcription factor genes were expressed preferentially in the EZ. Concerning the 41 aquaporins expressed in the root apex, transcripts accumulated preferentially in the EZ, with the exception of PtPIP1;4, which was expressed preferentially in the DZ (Supplementary Table S4).
Differential gene expression during the osmotic stress response
Regardless of zone and time, 1162 genes and 53 uTARs were responsive to osmotic stress, with a change in expression above the threshold |Log2(FC)|≥ 2 as compared with the respective control (‘PEG-responsive genes’; Supplementary Tables S3, S4). The stress response covered 26 Mapman bins (Fig. 4; Supplementary Table S4). About one-third of DEGs corresponded to genes with unknown function (bin 35). The stress response covered distinct metabolic pathways (related to cell wall or lipid, amino acid and sugar metabolism, or encoding large enzyme families, such as cytochrome P450) and transporters (including aquaporin). However, the key components triggered by osmotic stress were regulatory processes (~330 mapped genes). Several organization levels were involved, as showed by the numerous transcription factors (bin 27.3), as well as genes related to protein modification and degradation (bin 29.4 and 29.5), signaling (bin 30), and hormones (bin 17). Mapping highlighted the fact that the stress-responsive genes were distributed unevenly among the DZ and EZ, the DZ transcriptome being less responsive than the EZ transcriptome. Moreover the number of DEGs increased with time under stress in both zones.
Fig. 4.
Transcriptional remodeling under osmotic stress. Mapman visualization of PEG-responsive genes retrieved for the division zone (DZ) and the elongation zone (EZ) after 0.5h or 3h under stress (1189 mapped of 1162 data points). Numbers and names refer to bins (http://mapman.gabipd.org/).
GO analyses were used to test whether the response encompassed functional enrichments. Half an hour after stress onset, 216 DEGs were detected in the DZ (53 down-regulations and 163 up-regulations; Fig. 4), leading to a few functional enrichments (Fig. 5; Supplementary Table S2C). For cellular component, ‘extracellular space’ and ‘cell wall’ were the most enriched terms. The most enriched biological processes were ‘response to endogenous stimulus’, ‘lipid metabolic process’, and ‘signal transduction’, confirming that the osmotic stress was indeed sensed. Meanwhile, 393 DEGs were detected in the EZ (including 302 up-regulations; Fig. 4). Some biological processes were strongly enriched (‘fruit-ripening’, ‘response to endogenous stimulus’, and ‘cell communication’; Fig. 5; Supplementary Table S2D), as well as the molecular function’s ‘sequence-specific DNA binding transcription factor activity’, indicating that signaling and regulatory pathways were activated. The most enriched term for cellular component was ‘extracellular space’ (Fig. 5; Supplementary Table S2D). Three hours after stress onset, enlarged sets of DEGs were retrieved in both the DZ and the EZ (272 and 588 DEGs, respectively), leading to numerous functional enrichments. The temporal deployment of the transcriptional responses was thus converted into physiologically relevant processes. Concerning the DZ (Fig. 5; Supplementary Table S2E), ‘pollen–pistil interaction’, ‘response to abiotic stimulus’, and ‘response to stress’, together with ‘oxygen binding’ and ‘sequence-specific DNA binding transcription factor activity’ were among the most enriched GO terms, whereas they were absent in the early response. For the EZ (Fig. 5; Supplementary Table S2F), enrichments concerned ‘oxygen binding’, ‘DNA binding’, ‘response to abiotic stimulus’, ‘response to stress’, ‘response to external stimulus’, and ‘signal transduction’. For GO terms enriched at both time points, transcriptional responses strengthened with time. Fold-enrichment values were similar or increased, while DEG numbers increased strongly. For instance, the number of DEGs in ‘response to endogenous stimulus’ almost doubled with time.
Fig. 5.
Functional enrichments of the response. Gene ontology fold-enrichments among PEG-responsive genes retrieved for the DZ and EZ after 0.5h or 3h under stress as compared with P. nigra root transcriptome. Only non-redundant GO terms are shown (|fold change enrichment| >2, corrected P-value <0.05; Supplementary Table S2).
Our transcriptome analysis revealed that regulations were highly structured by time, highlighting a rapid but labile response (268 DEGs only at 0.5h; Supplementary Table S4). Other DEGs contributed to sustained or late responses, their expression being regulated at 3h. This schedule was consistent with responses to longer stress in poplar and maize apex (Cohen et al., 2010; Opitz et al., 2016; Supplementary Table S4). Using poplar to maize orthology and a stringent threshold for differential expression, a weak overlap was found with maize roots experiencing a 6h long PEG stress (Supplementary Table S4). Most of the overlapping genes were involved in sustained or late responses. In two other poplar species experiencing 36h or 10 d long soil water deprivation (Cohen et al., 2010), most DEGs were common again, with sustained or late responses. Long water deprivation provoked an energy deficit transcriptional response (Cohen et al., 2010), which did not occur here, as expected from a short-term response (Supplementary Table S4). Species, growth conditions, plant stature, stress application and duration, and even internal cell status distinguished the poplar studies, and 190 highly responsive genes constituted an unexpectedly high overlap. For ~80% of them, the direction of transcriptional regulation was identical, suggesting their key roles for stress acclimation. DEGs strongly responsive to both PEG and drought might be constitutive components of the water deficit response in poplar roots.
Discussion
Plant growth is intimately intertwined with environmental conditions, and growth plasticity is a crucial property that allows sessile organisms to withstand stress. Water distribution in soil, as well as water availability, fluctuate in both time and space. Local water depletion acts both as a stress and as a positional cue shaping root systems (Robbins and Dinneny, 2015). Elucidating how changes in water availability are transmitted to changes in root growth remains a current focus. In the root axis, cells coming from the DZ rapidly enlarged within the EZ, making the root apex relevant to dissecting the cellular components of stress-inducible responses (Baskin, 2013). The spatial patterning of these responses has been documented at several scales, increasing our knowledge of the complexity of controls underlying root growth variation (reviewed in Yamaguchi and Sharp, 2010). While most of these studies focused on responses in the mid-term, here we documented the earliest impacts of osmotic stress on the growing root of Populus, focusing on the dynamics of cell processes and the establishment of underlying controls.
Kinetics and kinematics reveal the sensitivity of cell division and cell elongation in Populus root
Following osmotic stress onset, root growth was strongly reduced but recovered rapidly (Fig. 1). After some oscillation, the growth rate stabilized at a lower level than in controls. While not mentioned by the few studies that have monitored growth following osmotic stress onset (Pritchard et al., 1991; Frensch and Hsiao, 1994), such oscillations could result from the different time courses of the processes underlying growth recovery, such as osmoregulation, cell wall relaxation, or water transport. Osmotic stress shortened the growth zone and reduced the cell elongation rate (Fig. 1), as previously shown for other species (Sharp et al., 1988; Pritchard et al., 1991; Triboulot et al., 1995; Sacks et al., 1997). A paradigm arose from these latter studies, stating that cell elongation is insensitive to water stress in the apical part of the root. In contrast, we recently showed that stress-driven impairment of cell elongation encompassed this apical region (corresponding to the DZ) in a euramerican poplar hybrid (Bizet et al., 2015b). Consistently, P. nigra exhibited a significant reduction in cell expansion rate within the DZ, indicating that cytoplasmic growth was also impacted by osmotic stress. In maize, water stress caused a large change in the cell production rate and local cell division rate profile (Fraser et al., 1990; Sacks et al., 1997). Here, osmotic stress rapidly impaired the cell production rate without affecting DZ length (Fig. 1). Since stress duration was very short compared with the ~20h cell cycle (Ivanov, 1997; Bizet et al., 2015b), a shortening of the DZ would have required numerous precocious exits of the cell cycle. Similarly, 10h of osmotic stress was too short to induce mitotic exit in Arabidopsis leaves (Skirycz et al., 2011).
Stress-induced transcriptional remodeling was surveyed separately in the DZ and EZ. The DZ encompasses dividing and slowly expanding cells, while the EZ has rapidly elongating cells (Supplementary Fig. S2). The EZ was shortened but the sampling size was kept identical regardless of growing conditions. Stressed EZ samples thus included a small quantity of mature tissue, but contamination was negligible since RNA concentration decreases strongly moving away from the quiescent center as a result of dilution by cell expansion (Merret et al., 2010). Functional enrichments, clustering, and cross-comparison with gene profiling in Arabidopsis confirmed that zones were properly separated from each other. The DZ transcriptome was consistent with dividing and slowly expanding cells, while the EZ transcriptome was associated with rapidly elongating cells.
The build-up of hormonal control underlying growth inhibition
Our experimental set-up combined a proper application of osmotic stress—that controls for hypoxia and prevents root manipulation—with a sampling strategy based on fine analysis of root growth. In this context, gene expression profiling was a powerful tool to reveal physiologically relevant processes with regards to growth inhibition. Depicting the early molecular events triggered by osmotic stress, we showed that genes related to hormones (including metabolism, primary targets, and signaling cascades) encompassed 10% of DEGs after 0.5h under stress, and up to 20% after 3h. Hormone networks have been dissected in A. thaliana roots (Takatsuka and Umeda, 2014; De Smet et al., 2015). Meanwhile most knowledge about hormonal control of root growth has been generated from artificial modulation of their levels—by hormone applications and/or targeted mutants. Here, the build-up of hormonal interplays that accompanied root growth inhibition was dissected under physiological conditions and is summarized in Fig. 6.
Fig. 6.
Hormone responses during osmotic stress root growth inhibition. The growth response is summarized by typical profiles of the elemental elongation rate before stress, and after 0.5h and 3h under stress. Tables summarized PEG-responsive genes related to hormone metabolism and signaling, as well as hormone-responsive genes (Supplementary Table S4). Down-regulated expression was indicated with an arrow. Metabolic genes are in bold and hormone carriers are underlined. Yellow, blue, and green backgrounds corresponded to the spatial patterns of gene regulation (DZ, EZ, and both zones, respectively).
Ethylene and auxin pathways were rapidly elicited (Fig. 6). The expression of auxin biosynthetic genes increased (flavin monooxygenase, YUCCAs and the tryptophan aminotransferase gene, TAA1), suggesting de novo auxin biosynthesis. Consistently, auxin signaling and response were activated (AUX/IAA, GH3, LBD16, SAUR; Fig. 6; Okushima et al., 2005). Cell-to-cell transport of auxin is mediated by efflux and influx carriers (ABCB, PIN, and AUX families) and influenced by APYRASEs (Kaneda et al., 2011, Liu et al., 2012). A localized activation of these genes occurred after 3h under stress (Fig. 6), in accordance with their drought responsiveness in other species (OsPin3t, Zhang et al., 2012; APY2 and ABCB15, Cohen et al., 2010). Local auxin maxima and auxin gradient might thus be reshaped along stressed roots. Transcript accumulation of 1-Amino-Cyclopropane-1-Carboxylate Synthase and Oxidase and Ethylene Response Factor supported stress-driven accumulation of ethylene (Vandenbussche et al., 2012). Together, our results fit a model of ethylene-mediated inhibition of root growth by a modulation of auxin biosynthesis and transport.
As classically reported under osmotic stress, the ABA pathway was activated (Fig. 6). In Arabidopsis, AtNCED3 controls the level of endogenous ABA under drought (Iuchi et al., 2001). Here, sustained activation of poplar orthologs suggested an enhanced ABA biosynthesis together with a decreased catabolism (ABA 8'-hydroxylase genes, CYP707A1). During the early response, ABA increase was supported by transcriptional regulation of an ABA receptor (PYL6) and an ABA-dependent transcriptional activator (VND-INTERACTING2; Yang et al., 2011). However, key ABA-responsive genes were not yet activated, indicating that signaling was only primed. Among the ABA-dependent pathway (Finkelstein, 2013), ABI5, AFP, ABF, and RD26 were activated after 3h under stress. Additionally, PYL expression was repressed while expression of PP2C genes and HB7/12 was increased (Fig. 6). In Arabidopsis, ATHB12 and ATHB7 transcription factor genes mediated a negative feedback by up-regulating the expression of PP2C and down-regulating the expression of PYL (Valdès et al., 2012). An ABA exporter-encoding gene, localized in the plasma membrane in Arabidopsis (ABCG25; Kuromori et al., 2010), was transcriptionally activated in the DZ. As in droughted roots (Cohen et al., 2010), genes related to ABA catabolism (CYP707A2 and CYP707A4) were activated in the EZ. Together, ABA appeared finely tuned in stressed roots via metabolic and transport feedback. ABA-mediated transcriptional responses occurred in both zones, but required some time to establish fully. Such dynamics support the canonical role of ABA accumulation under drought, counteracting the growth inhibition due to ethylene accumulation (Sharp, 2002).
Levels of the growth-promoting hormones brassinosteroids and gibberellins were decreased after 3h under stress (Fig. 6). Among brassinosteroid primary targets (Coll-Garcia et al., 2004), expression of BRH1 and EXO genes suggests that brassinosteroids were lowered rapidly (Fig. 6). The transient repression of several EXO genes drove functional enrichments in ‘extracellular space’ in both zones (Fig. 5). Beyond being a hallmark of brassinosteroid status, EXO was proposed as a potential mediator of the brassinosteroid-promoted growth necessary for cell expansion (Schröder et al., 2009; Aya et al., 2014). Later, repression of brassinosteroid biosynthetic gene expression (SQE1 and DWF genes) occurred in a zone-preferred manner (Fig. 6). DWF4 repression could reflect negative feedback (Zhang et al., 2009), but the decrease in brassinosteroids was confirmed by PP2A repression (Tang et al., 2011). Gibberellins have been proposed to mediate growth inhibition and physiological responses to drought (Zawaski and Busov, 2014). Here, early responsive DEGs included transcriptional factors (MYB62 and GIS3), DELLA protein (RGL2), and F-Box protein (SLY2), which are all hallmarks of a decrease in gibberellin. However, the regulation of bHLH transcription factor (bHLH137, BNQ3, PRE5, HBI1, and IBL1) involved in gibberellin and brassinosteroid signaling pathways (Ikeda et al., 2012) precluded any firm conclusions. Homeostasis of bioactive gibberellins results from their production via the action of GA-20-oxidases, and their inactivation by catabolic GA-2-oxidases (Hauvermale et al., 2012). Thus, transcriptional regulation of GA2OX4, GA2OX8, and GA20OX1 was expected to reduce gibberellin (Fig. 6). The signaling cascade, responses, and feedback were consistent with lowered gibberellin levels (Zhang et al., 2009; Feurtado et al., 2011). ABA antagonized gibberellin and brassinosteroid pathways (Zentella et al., 2007; Zhang et al., 2009). Consistent with our data, ATHB12 was reported to decrease GA20OX1 expression and growth (Son et al., 2010). Overall, growth-promoting hormones were reduced consistently with stress inhibition of root growth.
Artificial increases in cytokinins negatively impacted root growth and DZ length in Arabidopsis (Werner et al., 2003). Cytokinin signals depend on its transmission through a multistep phosphorelay [RESPONSE REGULATORS (RRs) Müller, 2011] and its modulation by KISS ME DEADLY F-box proteins (H.J. Kim et al., 2013). As shown in Fig. 6, accumulation of RR transcripts suggested that cytokinin levels increased in stressed roots. Bioactive cytokinins depend on de novo synthesis (IPTs, isopentenyl transferases), catabolism (CKX, cytokinin oxidase/dehydrogenase), and activation (LOG, lonely guy) (Kuroha et al., 2009). In Arabidopsis, dehydration or ABA treatments reduced IPT expression but also, as a secondary response, repressed CKX expression (Nishiyama et al., 2011). Here, osmotic stress repressed IPT5 expression while increasing CKX3 expression in the EZ. These responses mimicked the negative feedback provoked by cytokinin treatment (Brenner and Schmülling, 2012). In the DZ, the levels of bioactive cytokinins appeared tuned through regulatory rather than metabolic processes (LOG and KMD; Fig. 6). Alteration of cytokinin status and signaling thus plays a role in the inhibition of root growth.
Defense hormones also participate in stress responses (Fig. 6). Osmotic stress provoked a rapid activation of jasmonate metabolism in the EZ. ST2A, LOX2, and OPR2 were drought responsive in poplar roots (Cohen et al., 2010), and LOX5 and OPR3 in the EZ of maize roots (Opitz et al., 2016). Jasmonates were then perceived and JAZ activated. Consistent with our results, jasmonates inhibited root growth through interaction with auxin (Chen et al., 2011). Salicylic acid (SA) fluctuated during the stress response (Figs 4–6). In the DZ, early repression of SA-inducible PR1 was followed by the up-regulation of SA-inducible NHL25. In the EZ, the pathway of SA conversion into volatile methylsalicylate was activated, suggesting a systemic signaling (Chen et al., 2003).
Distinct sets of genes were recruited in the DZ and the EZ (Fig. 6). Osmotic stress reshaped hormonal status and probably modified local hormone maxima, controlling cell proliferation and expansion.
Impairment of growth machinery
In addition to changes in hormonal status, growth response was accompanied by a strong remodeling of transcription factors, kinases, and regulators of protein turnover, lipid metabolism, cell wall properties, and transporters (Fig. 4). Growth was restricted, but stress promoted the expression of growth effectors. As found in maize (Yamaguchi and Sharp, 2010; Opitz et al., 2016), transcriptional regulation occurred for genes related to cell wall properties, including EXPANSIN, XTH, PECTIN LYASE, PECTIN METHYL ESTERASE, and CELLULOSE SYNTHASE (Cosgrove, 2005). The cell wall-strengthening XTHs were down-regulated, and the wall-loosening EXPs (especially EXPA1 and EXPA8) and PECTINE LYASE genes were up-regulated. In the root of stressed maize, the pattern of EXPA1 accumulation fitted expansion maintenance, contributing to growth maintenance of turgor-reduced cells (Wu et al., 2001). Its ortholog accumulated over the whole poplar growth zone, but the cell expansion rate was not maintained, suggesting that facilitation of expansion via EXPA expression might not be sufficient to counteract stress impact. Osmotic stress also elicited expression of aquaporin, PtPIP2;7, PtPIP2;10, PtTIP1;1, PtTIP1;2, PtTIP2;2, PtTIP2;3, and PtTIP2;4 (Supplementary Table S4). Although members of this multigene family show redundancy and high stress responsiveness (Cohen et al., 2013), these transcriptional regulations suggested an increase in membrane hydraulic conductivity (Maurel, 2007). As cell expansion rate depends on wall extensibility as well as on membrane hydraulic conductivity (Lockhart, 1965), the up-regulation of aquaporin also pointed to facilitation of cell expansion.
The cell production rate was reduced by osmotic stress but none of the core cell cycle genes was retrieved among the most stress-responsive genes in the DZ, even if they were detected as preferentially DZ-expressed. In this short-term response, post-transcriptional regulation of cell cycle components could be responsible for the reduction of the cell division rate, as demonstrated in Arabidopsis leaves (Skirycz et al., 2011). On the other hand, the reduced growth rate may underlie this response. Cells have to reach a size threshold before dividing, so lowered cytoplasmic growth may participate in control of the cell cycle, as previously proposed (Bizet et al., 2015b). These working hypotheses are not exclusive and both require further examination.
RNA-sequencing provided a genome-wide diagnostic of the molecular regulation underlying stress responses. Many DEGs were also drought-responsive genes that contributed to acclimation to soil water deprivation (Cohen et al., 2010), suggesting that drought and osmotic stress, as well as short- and long-term responses, share common components. In conclusion, our work revealed that osmotic stress interfered with growth by changing hormonal status and activating regulatory proteins rather than by repressing the underlying machinery of expansive growth.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Number of sequenced and single-mapped reads in the 32 libraries.
Table S2. Gene ontology analyses reduced to most specific terms.
Table S3. In silico validation of unannotated transcriptionally active regions (uTARs).
Table S4. Differentially expressed genes.
Figure S1. Osmotic pressure and oxygen saturation of nutrient solutions.
Figure S2. Growth traits within P. nigra root.
Figure S3. qPCR validation: primer sequences and expression patterns.
Acknowledgements
The research has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under the grant agreement no. FP7-311929 (WATBIO), and from the French National Research Agency through the Laboratory of Excellence ARBRE (ANR-12- LABXARBRE-01). MR was supported by WATBIO and Region Lorraine (no. 12000425). The authors thank the facility of Functional Ecology (PTEF OC081) at the research center INRA Nancy-Lorraine. The authors thank Cyril Buré for his daily excellent technical support, Maryline Harroué and Sarah Liéger for histological analyses, Allyson Hollander for qPCR analysis, and Emmanuelle Morin for her help in bioinformatics analysis. The authors acknowledge Marc Villar and Catherine Bastien from the UR0588-INRA Unit for access to the referenced Populus nigra clone 6-J29, and Olivier Forestier from Guéméné-Penfao ONF Nursery for preparation of the cuttings.
Glossary
Abbreviations:
- DEG
differentially expressed gene
- EER
elemental elongation rate
- uTAR
unannotated transcriptionally active region.
References
- Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. 2014. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant and Cell Physiology 55, 897–912. [DOI] [PubMed] [Google Scholar]
- Baskin TI. 2013. Patterns of root growth acclimation: constant processes, changing boundaries. Wiley Interdisciplinary Reviews: Developmental Biology 2, 65–73. [DOI] [PubMed] [Google Scholar]
- Basu P, Pal A, Lynch JP, Brown KM. 2007. A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiology 145, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI. 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiology 116, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inze D, Vuylsteke M. 2005. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiology 138, 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. 2003. A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Bizet F, Bogeat-Triboulot M-B, Montpied P, Christophe A, Ningre N, Cohen D, Hummel I. 2015a Phenotypic plasticity toward water regime: response of leaf growth and underlying candidate genes in Populus. Physiologia Plantarum 154, 39–53. [DOI] [PubMed] [Google Scholar]
- Bizet F, Hummel I, Bogeat-Triboulot M-B. 2015b Length and activity of the root apical meristem revealed in vivo by infrared imaging. Journal of Experimental Botany 66, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Brenner W, Schmülling T. 2012. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biology 12, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, D’Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. 2003. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. The Plant Journal 36, 577–588. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, et al. 2011. The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. The Plant Cell 23, 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Bogeat-Triboulot M-B, Tisserant E, et al. 2010. Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Bogeat-Triboulot M-B, Vialet-Chabrand S, Merret R, Courty P-E, Moretti S, Bizet F, Guilliot A, Hummel I. 2013. Developmental and environmental regulation of Aquaporin gene expression across Populus species: divergence or redundancy? PLoS One 8, e55506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll-Garcia D, Mazuch J, Altmann T, Müssig C. 2004. EXORDIUM regulates brassinosteroid-responsive genes. FEBS Letters 563, 82–86. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6, 850–861. [DOI] [PubMed] [Google Scholar]
- Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8, e85024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet S, Cuypers A, Vangronsveld J, Remans T. 2015. Networks involved in hormonal control of root development in Arabidopsis thaliana: a framework for studying its disturbance by metal stress. International Journal of Molecular Sciences 16, 19195–19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos D, Vissenberg K, Broeckhove J, Beemster GTS. 2014. Putting theory to the test: which regulatory mechanisms can drive realistic growth of a root? PLoS Computational Biology 10, e1003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RO. 1976. Modeling of plant growth. Annual Review of Plant Physiology 27, 407–434. [Google Scholar]
- Feurtado JA, Huang D, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EW, Cutler AJ. 2011. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. The Plant Cell 23, 1772–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. 2013. Abscisic acid synthesis and response. Arabidopsis Book 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser TE, Silk WK, Rost TL. 1990. Effects of low water potential on cortical cell length in growing regions of maize roots. Plant Physiology 93, 648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frensch J, Hsiao TC. 1994. Transient responses of cell turgor and growth of maize roots as affected by changes in water potential. Plant Physiology 104, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM. 2012. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiology 160, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. 2012. A triantagonistic basic helix–loop–helix system regulates cell elongation in Arabidopsis. The Plant Cell 24, 4483–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. 2001. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. The Plant Journal 27, 325–333. [DOI] [PubMed] [Google Scholar]
- Ivanov VB. 1997. Relationship between cell proliferation and transition to elongation in plant roots. International Journal of Developmental Biology 41, 907–915. [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Simpson J, Herrera-Estrella L, Benfey PN. 2009. Functional genomics of root growth and development in Arabidopsis. Current Opinion in Plant Biology 12, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Schuetz M, Lin BS, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. 2011. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. Journal of Experimental Botany 62, 2063–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2 accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Chiang YH, Kieber JJ, Schaller GE. 2013. SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proceedings of the National Academy of Sciences, USA 110, 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. 2009. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. The Plant Cell 21, 3152–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. 2010. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proceedings of the National Academy of Sciences, USA 107, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW. 2013. Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. Journal of Experimental Botany 64, 83–108. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu J, Clark G, et al. 2012. Role for apyrases in polar auxin transport in Arabidopsis. Plant Physiology 160, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart JA. 1965. An analysis of irreversible plant cell elongation. Journal of Theoretical Biology 8, 264–275. [DOI] [PubMed] [Google Scholar]
- Loraine AE, McCormick S, Estrada A, Patel K, Qin P. 2013. RNA-Sequencing of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiology 162, 1092–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C. 2007. Plant aquaporins: novel functions and regulation properties. FEBS Letters 581, 2227–2236. [DOI] [PubMed] [Google Scholar]
- Merret R, Moulia B, Hummel I, Cohen D, Dreyer E, Bogeat-Triboulot M-B. 2010. Monitoring the regulation of gene expression in a growing organ using a fluid mechanics formalism. BMC Biology 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B. 2011. Generic signal-specific responses: cytokinin and context-dependent cellular responses. Journal of Experimental Botany 62, 3273–3288. [DOI] [PubMed] [Google Scholar]
- Muller B, Stosser M, Tardieu F. 1998. Spatial distributions of tissue expansion and cell division rates are related to irradiance and to sugar content in the growing zone of maize roots. Plant, Cell and Environment 21, 149–158. [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. 2005. Transcriptional profile of the Arabidopsis root quiescent center. The Plant Cell 17, 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, et al. 2011. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. The Plant Cell 23, 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz N, Marcon C, Paschold A, Malik WA, Lithio A, Brandt R, Piepho H-P, Nettleton D, Hochholdinger F. 2016. Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. Journal of Experimental Botany 67, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poroyko V, Spollen WG, Hejlek LG, Hernandez AG, LeNoble ME, Davis G, Nguyen HT, Springer GK, Sharp RE, Bohnert HJ. 2007. Comparing regional transcript profiles from maize primary roots under well-watered and low water potential conditions. Journal of Experimental Botany 58, 279–289. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Jones RGW, Tomos AD. 1990. Measurement of yield threshold and cell wall extensibility of intact wheat roots under different ionic, osmotic and temperature treatments. Journal of Experimental Botany 41, 669–675. [Google Scholar]
- Pritchard J, Jones RGW, Tomos AD. 1991. Turgor, growth and rheological gradients of wheat roots following osmotic stress. Journal of Experimental Botany 42, 1043–1049. [Google Scholar]
- R Development Core Team. . 2014. R: a language and environment for statistical computing. http://www.R-project.org Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Robbins NE, II, Dinneny JR. 2015. The divining root: moisture-driven responses of roots at the micro- and macro-scale. Journal of Experimental Botany 66, 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Sequencing. Bioinformatics 27, 2325–2329. [DOI] [PubMed] [Google Scholar]
- Sacks MM, Silk WK, Burman P. 1997. Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Physiology 114, 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder F, Lisso J, Lange P, Müssig C. 2009. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biology 13, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE. 2002. Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant, Cell and Environment 25, 211–222. [DOI] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. 2004. Root growth maintenance during water deficits: physiology to functional genomics. Journal of Experimental Botany 55, 2343–2351. [DOI] [PubMed] [Google Scholar]
- Sharp RE, Silk WK, Hsiao TC. 1988. Growth of the maize primary root at low water potentials I. Spatial distribution of expansive growth. Plant Physiology 87, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk WK. 1992. Steady form from changing cells. International Journal of Plant Science 153, S49–S58. [Google Scholar]
- Skirycz A, Inzé D. 2010. More from less: plant growth under limited water. Current Opinion in Biotechnology 21, 197–203. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, et al. 2011. Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. The Plant Cell 23, 1876–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3, 1–25. [DOI] [PubMed] [Google Scholar]
- Son O, Hur YS, Kim YK, et al. 2010. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant and Cell Physiology 51, 1537–1547. [DOI] [PubMed] [Google Scholar]
- Spollen WG, Tao W, Valliyodan B, et al. 2008. Spatial distribution of transcript changes in the maize primary root elongation zone at low water potential. BMC Plant Biology 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M. 2014. Hormonal control of cell division and elongation along differentiation trajectories in roots. Journal of Experimental Botany 65, 2633–2643. [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, et al. 2011. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nature Cell Biology 13, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Granier C, Muller B. 2011. Water deficit and growth. Co-ordinating processes without an orchestrator? Current Opinion in Plant Biology 14, 283–289. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Sequencing. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-Sequencing experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Sequencing reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulot M-B, Pritchard J, Tomos D. 1995. Stimulation and inhibition of pine root growth by osmotic stress. New Phytologist 130, 169–175. [Google Scholar]
- Takatsuka H, Umeda M. 2014. Hormonal control of cell division and elongation along differentiation trajectories in roots. Journal of Experimental Botany 10, 2633–2643. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. [DOI] [PubMed] [Google Scholar]
- Valdés AE, Overnäs E, Johansson H, Rada-Iglesias A, Engström P. 2012. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Molecular Biology 80, 405–418. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Vaseva I, Vissenberg K, Van Der Straeten D. 2012. Ethylene in vegetative development: a tale with a riddle. New Phytologist 194, 895–909. [DOI] [PubMed] [Google Scholar]
- Walter A, Silk WK, Schurr U. 2009. Environmental effects on spatial and temporal patterns of leaf and root growth. Annual Review of Plant Biology 60, 279–304. [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Thorne ET, Sharp RE, Cosgrove DJ. 2001. Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiology 126, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Sharp RE. 2010. Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant, Cell and Environment 33, 590–603. [DOI] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. 2011. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. The Plant Cell 23, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawaski C, Busov VB. 2014. Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PLoS One 20, e86217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, et al. 2007. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. The Plant Cell 10, 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li J, Zhang W, Yan S, Wang R, Zhao J, Li Y, Qi Z, Sun Z, Zhu Z. 2012. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. The Plant Journal 72, 805–816. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X. 2009. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proceedings of the National Academy of Sciences, USA 106, 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.