Highlight
In vitro study shows that AtCCD4 claves all-trans-bicyclic-carotenoids, excludes its direct involvement in generating plastid retrograde signals supposedly derived from cis-desaturation intermediates, and demonstrates that AtCCD7 converts 9-cis-acylic carotenes.
Key words: Apocarotenoids, carotenoids, carotenoid cleavage dioxygenase, CCD4, CCD7, retrograde signaling
Abstract
The Arabidopsis carotenoid cleavage dioxygenase 4 (AtCCD4) is a negative regulator of the carotenoid content of seeds and has recently been suggested as a candidate for the generation of retrograde signals that are thought to derive from the cleavage of poly-cis-configured carotene desaturation intermediates. In this work, we investigated the activity of AtCCD4 in vitro and used dynamic modeling to determine its substrate preference. Our results document strict regional specificity for cleavage at the C9–C10 double bond in carotenoids and apocarotenoids, with preference for carotenoid substrates and an obstructing effect on hydroxyl functions, and demonstrate the specificity for all-trans-configured carotenes and xanthophylls. AtCCD4 cleaved substrates with at least one ionone ring and did not convert acyclic carotene desaturation intermediates, independent of their isomeric states. These results do not support a direct involvement of AtCCD4 in generating the supposed regulatory metabolites. In contrast, the strigolactone biosynthetic enzyme AtCCD7 converted 9-cis-configured acyclic carotenes, such as 9-cis-ζ-carotene, 9'-cis-neurosporene, and 9-cis-lycopene, yielding 9-cis-configured products and indicating that AtCCD7, rather than AtCCD4, is the candidate for forming acyclic retrograde signals.
Introduction
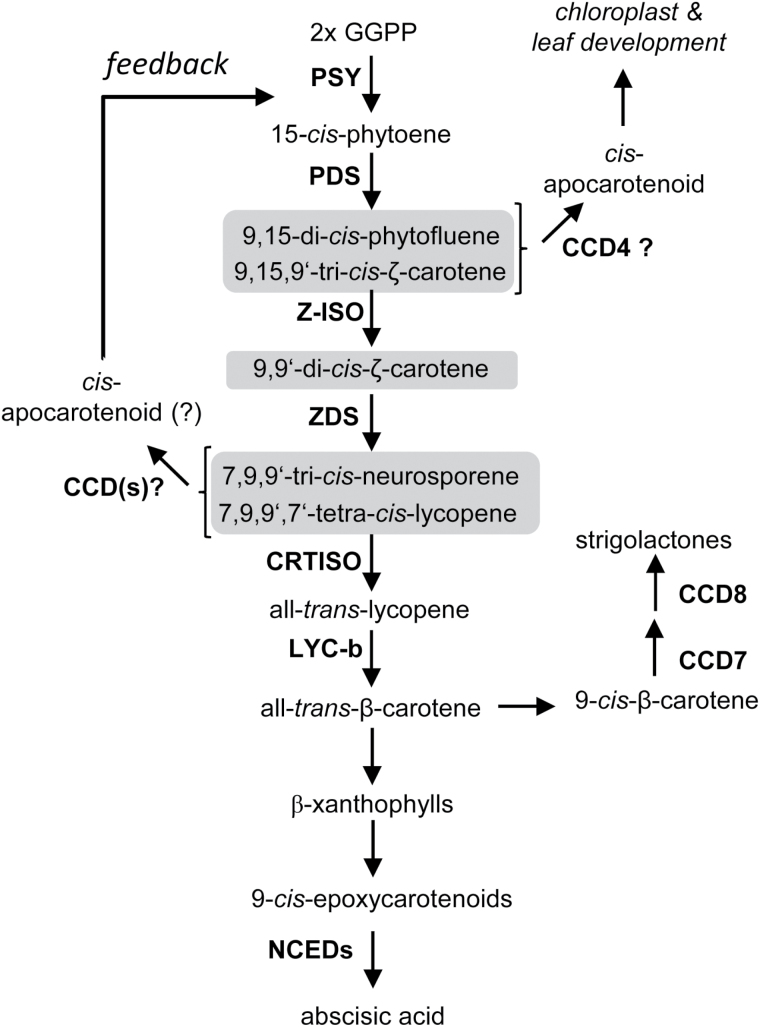
Apocarotenoids are carotenoid cleavage products. They are generally synthesized by members of the carotenoid cleavage oxygenase (CCO) family, which are present in animals, plants, fungi, and eubacteria (von Lintig and Vogt, 2000; Ruch et al., 2005; Auldridge et al., 2006; Prado-Cabrero et al., 2007; Walter and Strack, 2011; Sui et al., 2013; Nisar et al., 2015). Whether these enzymes utilize a monooxygenase or a dioxygenase mechanism remains debated; however, the plant enzymes are commonly referred to as carotenoid dioxygenases (CCDs). The formation of apocarotenoids in plants can be the starting point for biosynthetic pathway branches leading to important regulatory compounds (Fig. 1), such as the phytohormones abscisic acid (ABA) (Schwartz et al., 1997; Zhu, 2002; Seo, 2002) and the strigolactones (SLs; Alder et al., 2012; Al-Babili and Bouwmeester, 2015).
Fig. 1.
Carotenoid biosynthesis and formation of carotenoid-derived signaling molecules. Desaturation intermediates are shaded. The scheme shows established and hypothetical signals generated by carotenoid cleavage dioxygenases (CCDs). The formation of abscisic acid and SLs is initiated by 9-cis-epoxycarotenoid cleavage dioxygenases (NCEDs) and CCD7, respectively. Hypothetical signals are indicated by a question mark. PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, ζ-carotene isomerase; CRTISO, carotenoid cis-trans isomerase; LCY-b, lycopene β-cyclase; D27 (DWARF27), all-trans/9-cis-β-carotene isomerase. See text for further explanations.
The Arabidopsis CCD family includes nine enzymes classified as either 9-cis-epoxycarotenoid dioxygenases (NCEDs), NCED2, 3, 5, 6, and 9, or CCDs, CCD1, 4, 7, and 8. NCEDs are involved in ABA biosynthesis and catalyze the stereospecific cleavage of the C11–C12 double bond in 9-cis-configured epoxycarotenoids, to yield the ABA precursor xanthoxin (Schwartz et al., 1997). CCD1 is the only plant CCD that is localized in the cytosol. Arabidopsis AtCCD1 (Schwartz et al., 2001) and homologs from several plant species catalyze symmetrical double cleavage at the C9–C10 and C9'–C10' double bonds in cyclic and acyclic all-trans-configured carotenoids. CCD1 enzymes also target other double bonds in linear carotenes, yielding flavors, such as 6-methyl-5-hepten-2-one, geranial, and farnesyl acetone (Vogel et al., 2008; Ilg et al., 2009, 2014). Additional cleavage sites in apocarotenoids have also been documented (Ilg et al., 2014). Moreover, it was assumed that CCD1-type enzymes convert apocarotenoids rather than carotenoids in planta and that they may act as a scavenger for oxidatively damaged carotenoids (Scherzinger and Al-Babili, 2008; Ilg et al., 2010). In contrast, CCD7 and CCD8 are devoted to the biosynthesis of SLs (Gomez-Roldan et al., 2008; Umehara et al., 2008; Al-Babili and Bouwmeester, 2015). Here, CCD7 catalyzes the stereospecific cleavage of the trans-configured C9'–C10' double bond in 9-cis-β-carotene produced by the all-trans/9-cis-β-carotene isomerase DWARF27 (Alder et al., 2012; Bruno and Al-Babili, 2016), yielding β-ionone and 9-cis-β-apo-10'-carotenal (Alder et al., 2012; Bruno et al., 2014). The latter is then converted by CCD8, which catalyzes combination of repeated oxygenation and intramolecular rearrangements, into carlactone (Alder et al., 2012). However, CCD8 enzymes also catalyze a common CCD reaction by cleaving all-trans-β-apo-10'-carotenal/ol (Alder et al., 2008) at the C13–C14 double bond, yielding β-apo-13-carotenone (d'orenone). D'orenone is a natural metabolite that can inhibit root hair growth by interfering with auxin transport (Schlicht et al., 2008).
CCD4 enzymes, the subject of this study, regulate carotenoid homeostasis in different tissues, such as Arabidopsis seeds (Gonzalez-Jorge et al., 2013), chrysanthemum petals (Ohmiya et al., 2006), and peach fruit pulp (Brandi et al., 2011; Falchi et al., 2013). CCD4 enzymes from Crocus, Chrysanthemum, and apple produce β-ionone, indicating a single or double cleavage activity at the C9–C10 and/or the C9'–C10' double bond in β-carotene (Rubio et al., 2008; Huang et al., 2009). It was recently demonstrated that the potato CCD4b cleaves the C9–C10 and/or the C9'–C10' double bond in β-carotene (C40), by identifying all-trans-β-apo-10'-carotenal (C27) as the second product formed together with β-ionone (C13) (Bruno et al., 2015). Though with much lower efficiency, the potato enzyme also cleaved α-carotene and unepoxydated xanthophylls, pointing to β-carotene as the main substrate of CCD4 in planta (Bruno et al., 2015). However, other members of the CCD4 subfamily show different regional and substrate specificity. For instance, Citrus CCD4 enzymes cleave the C7'–C8' double bond in β-carotene, β,β-cryptoxanthin, and zeaxanthin, leading to the fruit peel pigments, β-apo-8'-carotenal and β-citraurin, and the volatiles, cyclocitral and 3-OH-cyclocitral (Rodrigo et al., 2013; Ma et al., 2013).
CCD enzymes might also form hitherto unknown regulatory compounds. Apart from ABA and SLs, additional carotenoid-derived regulatory molecules have been postulated and thought to arise through cleavage of desaturation intermediates of the carotenoid biosynthesis pathway (Fig. 1) (Moise et al., 2014). For instance, the tomato tangerine mutant is impaired in the activity of carotenoid cis-trans isomerase (CRTISO; Fig. 1), which results in 7,9,7',9'-tetra-cis-lycopene (prolycopene) accumulation in fruits, while the r(2997) yellow-flesh does not express the fruit-specific phytoene synthase gene (PSY1), leading to a lack of fruit carotenoids. It was shown that the accumulation of prolycopene and its poly-cis-configured precursors in the yellow-flesh background is accompanied by a partial restoration of PSY1 transcription. This epistatic behavior of the downstream over the upstream pathway activity is thought to be mediated by carotene cleavage product(s) derived from prolycopene or its less desaturated precursors (Kachanovsky et al., 2012). Moreover, the use of Arabidopsis mutants and carotene desaturation inhibitors pointed to the presence of apocarotenoid(s) that modulates leaf and chloroplast development and which is thought to derive from phytofluene and/or ζ-carotene. It has been suggested that AtCCD4 is involved in the formation of this regulatory signal(s) (Avendaño-Vázquez et al., 2014). Furthermore, potato ccd4 knock-down lines develop tubers with elevated carotenoid content, which also show changed shape and premature sprouting (Campbell et al., 2010). In saffron, CCD4 transcript levels increase under abiotic stress and during senescence (Rubio et al., 2008; Rubio-Moraga et al., 2014).
In the current work, we investigated the cleavage activity of AtCCD4 in vitro, using analytical and kinetic approaches. In addition, we evaluated the potential of this enzyme in generating the signals proposed to derive from cis-configured carotene desaturation intermediates, in comparison with the homolog, SL biosynthetic enzyme AtCCD7. Our data indicate that AtCCD7 rather than AtCCD4 may be a candidate capable of forming these signal molecule(s).
Materials and methods
Cloning and in vitro assays
pThio-AtCCD4: RNA was isolated from 4-week-old Arabidopsis seedlings using Plant RNA Purification Reagent (Invitrogen®) and cDNA was synthesized using SuperScript™ RNase H reverse transcriptase (Invitrogen, Paisley, UK). AtCCD4 full-length cDNA was amplified by PCR with the primers FP 5'-CCGGAGCTCCGGTTATGCCTAACGTG-3' and RP 5'-AGTGAGCTCTATATTGTTAAAGCTTATTAAGGT-3', both containing SacI restriction sites. The digested and purified PCR product was ligated into SacI-digested and dephosphorylated pThio®-vector. The pThio-AtCCD7 was described previously (Alder et al., 2012).
pThio-AtCCD4 was transformed into BL21 (DE3) Escherichia coli cells harboring the plasmid pGro7 (Takara Bio Inc.). Cells were grown at 37 °C in 50 ml of 2 YT growth medium supplemented with chloramphenicol and ampicillin. Protein expression was induced with 0.2% (w/v) arabinose at an OD600 of 0.5. Cells were grown for 4 h at 28 °C and harvested. Cell pellets were re-suspended in 1 ml of modified LEW buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mg ml−1 lysozyme, 1 mM dithiothreitol; pH 8.0), passed twice through a French pressure cell at 10 000 psi, and centrifuged at 20 000 g for 5 min. Protein was quantified using the Quick Start™ Bradford Protein Assay (Bio-Rad Laboratories) and adjusted to 20 µg µl−1.
Assays were performed in a total volume of 200 µl. Purified substrates (30 µM in CHCl3) were mixed with 20 µl of ethanolic Triton X-100 (2%, v/v; Sigma), dried using a vacuum evaporator, and dissolved in 100 µl of 2× incubation buffer [2 mM TCEP, 0.4 mM FeSO4, 200 mM HEPES-NaOH pH 7.8, and 2 mg ml−1 catalase (Sigma)]. Assays were started by the addition of each 50 µl lysate and H2O, and then incubated for 1 h, if not stated otherwise, under shaking (200 rpm) at 30 °C in darkness. For extraction, 2 vols of acetone were added, followed by a short sonication and the addition of 3 vols of light petroleum/diethylether (2:1, v/v). After centrifugation, the epiphase was dried and redissolved in 40 µl of CHCl3. A 5 µl aliquot of the extract was subjected to HPLC analysis with system 1, using tocopherol acetate (0.1 mg ml−1) as an internal standard.
Substrate preparation and identification
Substrates were purified using thin-layer silica gel plates (Merck). Synthetic apolycopenals and apocarotenals were kindly provided by BASF (Germany). β-Carotene was obtained from Roth, β,β-cryptoxanthin was from Sigma, and neoxanthin, violaxanthin, and α-carotene were from CaroteNature. Lutein was isolated from Narcissus pseudonarcissus petals, and 9-cis-violaxanthin from fresh spinach leaves. Zeaxanthin, lycopene, and ζ-carotene isomers were extracted from E. coli strains expressing the respectively mutagenized carotenoid gene cluster from Pantoea ananatis (Prado-Cabrero et al., 2007). 9,15-di-cis-Phytofluene, prolycopene, and proneurosporene were isolated from fruits of the tangerine tomato mutant and identified with the aid of published data (Clough and Pattenden, 1979; Carey et al., 1983). 9'-cis-Phytofluene was isolated from Dunaliella salina. 15-cis- and all-trans-Phytofluene were purchased from CaroteNature. 9-cis-β-Carotene was obtained by iodine-isomerization of β-carotene (Zechmeister, 1962) and subsequent HPLC purification. ζ-Carotene isomers were purified using a YMC C30 column and MeOH:TBME (3:1, v/v) as the eluent (HPLC system 4). The identification of isomers was carried out according to Carey et al. (1983) and Breitenbach and Sandmann (2005). All-trans-neurosporene was obtained from Rhodovulum sulfidophilum (Hagemann et al., 1996). 9'-cis-Neurosporene was prepared enzymatically by CRTI according to Yu et al. (2011) and using the substrate 7,9,9'-tri-cis-neurosporene isolated from tangerine fruit (Carey, et al., 1983). Synthetic 9-cis-lycopene was obtained from Buchem (The Netherlands). Carotenoid substrates were quantified photometrically according to their molar extinction coefficients (Britton et al, 1995).
Dynamic modeling and data processing
The decay of β-carotene (β), β-cryptoxanthin (cry), and zeaxanthin (zea) to β-ionone (β-io) and 3-OH-β-ionone (OH-β-io) via the intermediates β-apo-10-carotenal (β-10) and 3-OH-β-apo-10-carotenal (OH-β-10 is modeled by a set of ordinary differential equations (ODEs) following mass-action kinetics:
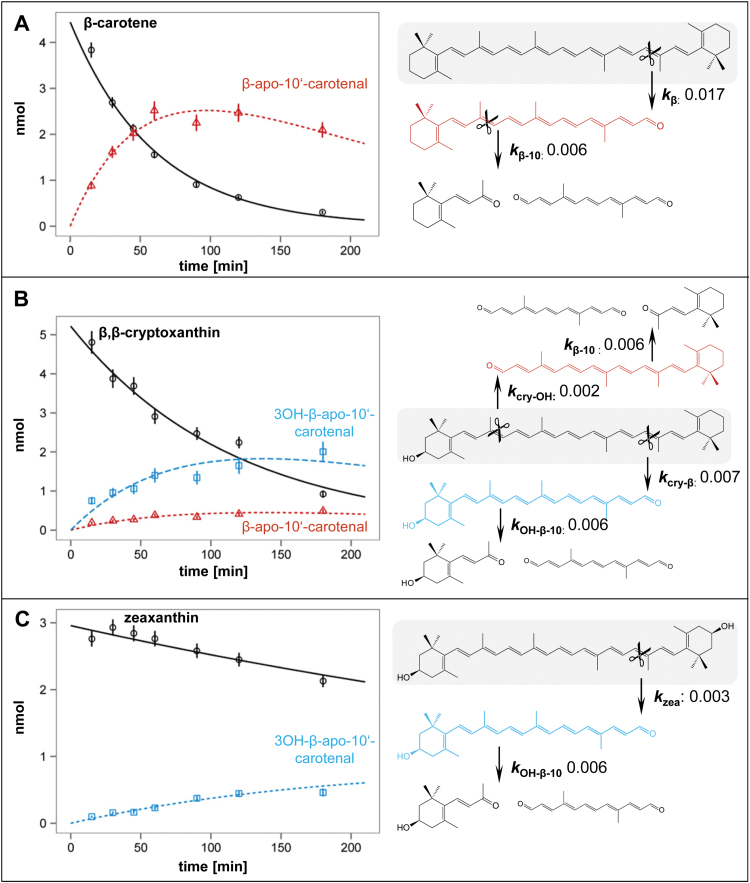
In total, the model contains 13 free parameters, denoted by θ, which are determined from data. The parameters of interest are the rate constants ki, i = β, cry-OH, cry-β, zea, β-10,OH- β-10. Other parameters are the initial concentrations of the states at time point zero and one conversion factor needed to account for a general change in the enzyme activity between datasets shown in Fig. 3A–C).
Fig. 3.
Time course and dynamic modeling of the AtCCD4 reaction. The conversion of three bicyclic carotenoids (shaded in gray) into the corresponding monocyclic C27 apocarotenoids and the subsequent cleavage of the latter are shown in (A–C). Note that the final products (3-OH)-β-ionone and the C14-dialdehyde were detected but could not be accurately quantified because of volatility, partition behavior, and instability. The symbols denote experimental data points and represent the average (± SEM) of 3–4 replicates. The line through the data points represents the model fit.
The parameters θ are estimated via the maximum likelihood method assuming normally distributed noise. The maximization of the likelihood translates into minimizing the cost function
where x i denotes the measured value of data point i with uncertainty σi, and x(t i, θ) gives the model prediction at time t i. The ODEs together with their sensitivity equations are integrated with the lsodes solver (Soetaert et al., 2010). Non-linear derivative-based optimization of the cost function is performed by a trust region optimizer (Nocedal and Wright, 1999). Parameter identifiability and confidence intervals are computed by the profile likelihood method (Raue et al., 2009). All analyses have been performed with the packages cOde/dMod for dynamic modeling in R. The detailed model specification encompassing alle experimental conditions can be found in Supplementary dataset S1 at JXB online.
Data pre-processing
Each experiment was repeated at least three times; some were repeated four times. Mean values of the experiments were taken as input for the modeling. Measurement uncertainties were estimated by the error model which accounts for an absolute and a relative contribution. The variance parameters s 0 and s rel are determined via maximum-likelihood estimation from the negative log-likelihood:
where ϕni–1 is the probability density of the χ2 distribution with n i–1 degrees of freedom and the index corresponds to data point i with mean value x i and variance v i determined from n i replicates. Standard errors of the mean were computed from the estimated variance and the number of replicates .
HPLC analysis and purification
Carotenoids were analyzed using a Shimadzu UFLC XR separation module equipped with an SPD-M20A photodiode array detector (Shimadzu). The column temperature was held at 40 °C. A YMC-pack-C30 reversed phase column (150 × 3 mm i.d., 5 µm; YMC Europe) was used throughout. The separation system 1 solvent system consisted of A: MeOH/TBME (1:1, v/v) and B: MeOH/H2O/TBME (30:10:1, v/v/v). The flow rate was 0.6 ml min−1 with a gradient from 100% B to 0% B within 20 min and maintenance of the final conditions for 4 min. The separation system 2 solvent system utilized A: MeOH/TBME (1:1, v/v) and B: MeOH/H2O/TBME (30:10:1, v/v/v). The flow rate was 0.6 ml min−1 with a gradient from 100% B to 0% B within 24 min and maintenance of the final conditions for 4 min. Separation system 3 employed the solvent system A: MeOH/TBME (4:1, v/v) and B: MeOH/H2O/TBME (30:10:1, v/v/v). The flow rate was 0.6 ml min−1 with a gradient from 100% B to 40% B within 20 min, to 0% in 5 min and maintenance of the final conditions for 10 min. Separation system 4 was isocratic with the solvent MeOH/TBME (3:1, v/v) at a flow rate of 2 ml min−1.
Mass spectrometry
Volatile cleavage products such as β-ionone, 6-methyl-5-hepten-2-one (MHO), and geranylacetone were collected by solid phase micro extraction (SPME; PDMS, 100 µm; Supelco). The fiber was exposed to the assay gas phase for 15 min and desorbed in the injector of the Trace GC coupled to a Trace DSQ II mass spectrometer (Thermo Fisher Scientific). Separation was achieved on a 30 m Zebron ZB-5 column 0.25 mm i.d., 0.25 µm film thickness (Phenomenex). The initial temperature of 50 °C was held constant for 5 min, followed by a ramp of 25 °C min−1 to a final temperature of 280 °C which was maintained for 5 min. The helium carrier gas flow rate was 1 ml min−1 and the injector temperature was set to 220 °C. Electron impact ionization (EI) was used at an ion source potential of 70 eV and a source temperature of 200 °C. Spectra were matched to the NIST (2.0) database using the Excalibur software. Additionally, standards of β-ionone and MHO (Sigma) were used.
Non-volatile reaction products were identified by LC-MS using a Dionex UltiMate 3000 UPLC coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Sample separation was achieved with a Hypersil Gold C18 UPLC column (150 × 2.1 mm i.d., 1.9 µm) and the solvent system A, 0.05% (v/v) formic acid in H2O, and B, 0.05% (v/v) formic acid in acetonitrile. Initial conditions were 70% B for 1 min, followed by a gradient to 100% B within 4 min. The final conditions were maintained for 10 min, all at a flow rate of 0.5 ml min−1. Ionization of apocarotenoids was achieved with atmospheric pressure chemical ionization (APCI) and analyzed in the positive mode. Nitrogen was used as sheath and auxiliary gas, set to 20 and 10 arbitrary units, respectively. The vaporizer temperature was set to 350 °C and the capillary temperature was 320 °C. The spray voltage was set to 5 kV and the normalized collision energy (NCE) to 35 arbitrary units. For data analysis, the TraceFinder (3.1) software and authentic apocarotenoid standards were used.
Results
AtCCD4 cleaves cyclic carotenoids and apocarotenoids
Carotenoid-accumulating E. coli strains have frequently been used to test carotenoid-cleaving enzymes in vivo (von Lintig and Vogt, 2000; Booker et al., 2004; Prado-Cabrero et al., 2007; Frusciante et al., 2014). However, only a limited number of carotenoid species can be produced in this system, and the stereospecificity of the reactions cannot be determined. We therefore resorted to in vitro assays utilizing an N-terminal thioredoxin–AtCCD4 fusion expressed in BL21 E. coli cells harboring the groEL-ES chaperone system (Alder et al., 2008; Ilg et al., 2009; Bruno et al., 2014). Experiments were carried out with particle-free cell lysates and substrate-containing detergent micelles.
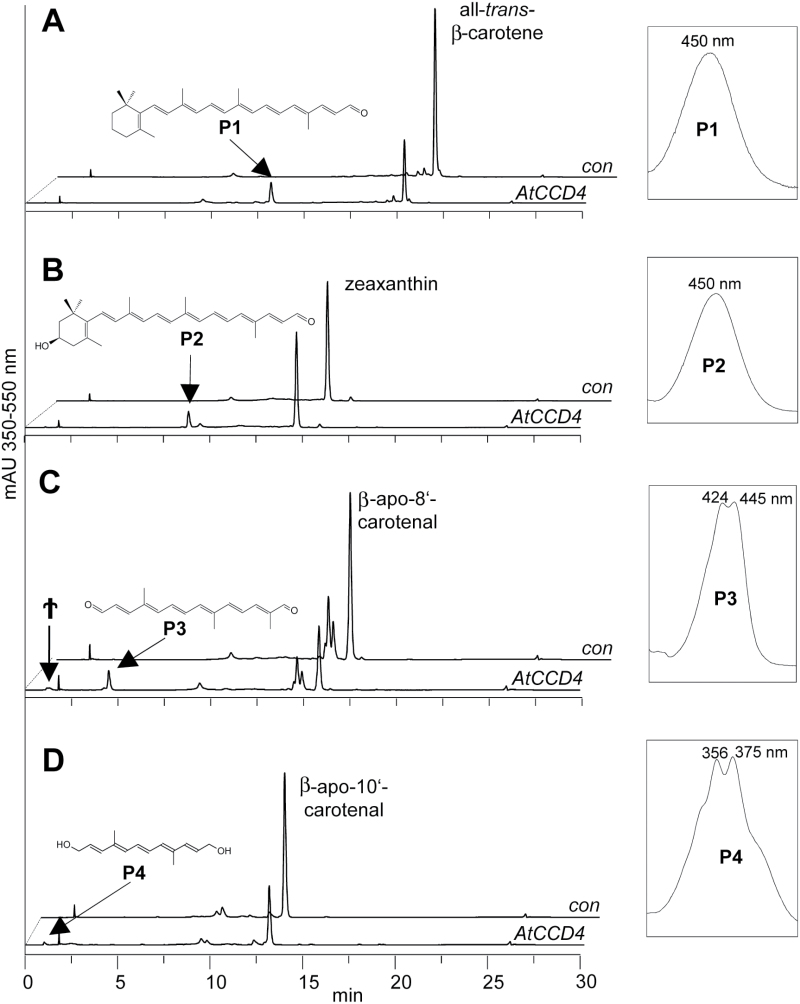
First, we tested the cleavage of all-trans-β-carotene. The enzyme converted this substrate into product P1 that was identified as all-trans-β-apo-10'-carotenal by chromatographic and UV/VIS spectral comparison with the authentic reference (Fig. 2A). This product is consistent with a single cleavage, at either the C9–C10 or the C9'–C10' double bond. β-Ionone, the second cleavage product expected to arise upon the cleavage at this site, was identified by GC-MS analysis (Supplementary Fig. S1). Next, we investigated the stereospecificity of AtCCD4 by testing the conversion of 9-cis-β-carotene whose cleavage could liberate the SL precursor 9-cis-β-apo-10'-carotenal, which could indicate a contribution of AtCCD4 to SL biosynthesis. However, only marginal activity was observed, as witnessed by barely detectable traces of 9-cis-β-apo-10'-carotenal (Supplementary Fig. S2F). We also tested the cleavage of the asymmetric α-carotene that contains an ε-ionone and a β-ionone ring. This incubation led to a lower proportion of all-trans-β-apo-10'-carotenal (P1; Supplementary Fig. S2A) compared with all-trans-ε-apo-10'-carotenal (P5; Supplementary Fig. S2A), indicating preferred cleavage at the C9'–C10' double bond adjacent to the β-ionone ring. The enzyme also cleaved the asymmetric β,β-cryptoxanthin (carrying β-ionone and the 3-OH-β-ionone ring) at either the C9–C10 or the C9'–C10' double bond yielding traces of β-apo-10'-carotenal (P1) and higher amounts of 3-OH-β-apo-10'-carotenal (P2), indicating a preference for the double bond in the unhydroxylated moiety (Supplementary Fig. S2B). Though a previous report excluded the conversion of the dihydroxylated substrate zeaxanthin (Huang et al., 2009), we detected a cleavage at the C9–C10 or the C9'–C10' double bond, yielding 3-OH-β-apo-10'-carotenal (P2; Fig. 2B). The dihydroxylated lutein, which carries a 3-OH-β- and -ε-ionone ring was converted—with a low efficiency—into 3-OH-β-apo-10'-carotenal and 3-OH-ε-apo-10'-carotenal (P2 and P6; Supplementary Fig. S2C), which arise by cleaving the C9–C10 or the C9'–C10' double bond. We also observed very low cleavage activity with all-trans-violaxanthin, which led to 5,6-epoxy-3-OH-β-apo-10'-carotenal (Supplementary Fig. S2D) tentatively identified based on the literature (Schwartz et al., 2001). The 9-cis isomer of violaxanthin and all-trans-neoxanthin were not converted (Supplementary Fig. S2E, G). Taken together, AtCCD4 cleaved most of the C40-bicyclic carotenoids at the C9–C10 or the C9'–C10' double bond, with a preference for the double bond adjacent to unsubstituted β-ionone rings. Hydroxylation of both ionone rings led to a decrease in activity but did not affect the position of the cleavage site.
Fig. 2.
HPLC analysis of AtCCD4 activity. (A) Incubation of the enzyme with all-trans-β-carotene yielded β-apo-10'-carotenal (P1). (B) Cleavage of zeaxanthin yielded 3-OH-β-apo-10'-carotenal (P2). (C) Incubation with β-apo-8'-carotenal led to a C17 dialdehyde (P3) and traces of the corresponding C17 dialcohol (†). (D) β-Apo-10'-carotenal yielded the C14 dialcohol rosafluene (P4). UV/VIS spectra of the products are depicted in insets. The structures of the products are given. For structures of the substrates, see Supplementary Fig. 5. Analysis was performed using HPLC system 1.
Next, we tested apocarotenoids with different chain lengths. AtCCD4 cleaved β-apo-8'-carotenal (C30) at the C9–C10 double bond, yielding a C17 dialdehyde (P3; Fig. 2C) and the corresponding C17 dialcohol (†; Fig. 2C) that probably derived from the dialdehyde product. The reduction of dialdehydes is frequently observed in assays utilizing E. coli lysate and is probably caused by an unspecific aldehyde dehydrogenase activity (Rodrigo et al., 2013; Bruno et al., 2015). GC-MS analysis confirmed the presence of the volatile β-ionone in these assays, which is expected to arise upon the formation of the C17-dialdehyde (Supplementary Fig. S1B). Consistently, the C14 dialcohol rosafluene was detected (P4; Fig. 2D) upon incubation with either β-apo-10'-carotenal (C27) or 3-OH-β-apo-10'-carotenal. Incubation with the shorter apocarotenal β-apo-12'-carotenal (C25) led only to traces of β-ionone. In conclusion, all apocarotenoids regardless of their chain length were cleaved at the C9–C10 double bond.
As shown above, AtCCD4 converted the C40 carotenoids β-carotene, β-cryptoxanthin, and zeaxanthin into β-apo-10'-carotenal and/or 3-OH-β-apo-10'-carotenal. However, this enzyme also cleaved its own products, β-apo-10'-carotenal and/or 3-OH-β-apo-10'-carotenal. To understand the biological function of AtCCD4, it is crucial to determine the preferred substrate, i.e. whether the enzyme favors C40-carotenoids or apocarotenoids. For this purpose, the assessment of standard parameters, K M and V max, may be flawed due to the biphasic system used and to uncertainties concerning the equivalent micellar packing of the different substrate species in an enzyme-accessible form. Therefore, we have resorted to modeling of time course experiments in one homogeneous assay system in which bicyclic carotenoids are converted into monocyclic apocarotenoids that are again subjected to a further cleavage.
Time course and modelling
Dynamic modeling of AtCCD4 activity in time course experiments was carried out to determine rate constants for the primary (cleavage of bicyclic C40 carotenoids) and secondary cleavage reactions (cleavage of apocarotenoids) depicted in Fig. 3. Based on this, the rank order of bicyclic substrate decay is determined by the number of OH functions present, namely β-carotene (k β) >β-cryptoxanthin (k cry-OH+k cry-β) >zeaxanthin (k zea), the k-ratios being 1.0:1.9:5.7 (Fig. 3A–C). The asymmetric β-cryptoxanthin is preferentially cleaved at the C9–C10 site next to the unhydroxylated ionone (k cry-β=3.5 k cry-OH; Fig. 3B) corroborating that hydroxylation hinders cleavage.
It is conceivable that the two equivalent half sides of the symmetric β-carotene substrate can be cleaved by AtCCD4 with the same probability in terms of ‘half-side substrate recognition’. Instead of the expected k β≈2k β-10 (reflecting the double concentration of cleavable end-groups), the model supports k β=2.8 k β-10, indicating discrimination between the uncleaved bicyclic and the pre-cleaved monocyclic substrate. Furthermore, along these lines of thinking, β-ionone-containing substrate half-sides should be equivalent in bicyclic substrates, namely k β≈k cry-β. However, this is not the case either, so that the binding of the entire carotenoid molecule and the simultaneous recognition of both rings must be assumed. This notion is further supported by the fact that the model is consistent with a completely different behavior of the monocyclic C27 apocarotenoid substrates. Here, the preference for cleavage at unhydroxylated ionone rings is lost, the rate constants for the cleavage of β-apo-10'-carotenal and 3-OH-β-apo-10'-carotenal being very similar (k β-0≈k OH-β-10). This was experimentally verified by time course experiments using both apocarotenoids as substrates (Supplementary dataset S1). Consequently, losing one of the two binding determinants seemingly abolishes the differentiation observed with the bicyclic substrates.
As a result of hydroxylation, rate constants decreasd in bicylic substrates, whereas they remained relatively constant in monocyclic apocarotenoids. Opposing relative substrate preferences are consequently found. The absence of one ring can lower (k β/k β-10=2.8) or increase (k zea/k OH-β-10=0.4) the rate constants relative to those obtained for bicyclic symmetric substrates. This is further exemplified by the asymmetric (monohydroxylated) β-cryptoxanthin, where simultaneously rate constants relatively increase (k cry-OH/k β-10=0.3) or remain unchanged (k cry-β/k OH-β-10=1.0).
CCD4 and/or CCD7 as potential generators of regulatory metabolites
AtCCD4 is thought to cleave the cis-configured phytoene desaturase (PDS) products phytofluene and/or ζ-carotene to generate regulatory molecules involved in leaf development (Avendaño-Vázquez et al., 2014). Furthermore, genetic evidence indicated a role for an undefined CCD in cleaving cis-configured ζ-carotene desaturase (ZDS) desaturation intermediates to generate a feedback signaling compound regulating the transcription of the tomato PSY1 (Kachanovsky et al., 2012). Therefore, we examined four ζ-carotene isomers as potential AtCCD4 substrates. The canonical PDS product 9,15,9'-tri-cis-ζ-carotene as well as 9,9'-di-cis-ζ-carotene were used, the latter assuming a relevant contribution of the C15,C15' double bond-specific ζ-carotene-cis-trans-isomerase (Z-ISO) in planta. However, none of these substrates was cleaved (Supplementary Fig. S3A–D). Likewise, the enzyme did not convert the canonical PDS intermediate 9,15-di-cis-phytofluene (Supplementary Fig. S3E) or the isomers 15-cis-phytofluene and all-trans-phytofluene (Supplementary Fig. S3E). Thus, our in vitro data do not support the proposed involvement of CCD4 in leaf development.
To account also for a possible role in generating ζ-carotene- and lycopene-derived signals, we investigated AtCCD4 cleavage activity with the ZDS desaturation intermediate 7,9,9'-tri-cis-neurosporene (proneurosporene) and product 7,9,9',7'-tetra-cis-lycopene (prolycopene; Supplementary Fig. S3F, G). Assuming a relevant contribution of CRTISO, which can generate partially trans-isomerized neurosporene and lycopene (Isaacson et al., 2004; Yu et al., 2011), we also assayed 9'-cis-neurosporene (Supplementary Fig. S3H). However, the enzyme did not cleave any of these linear substrates (Supplementary Fig. 3A–H). Thus, our data are not in favor of an involvement of CCD4 in the regulation of PSY expression.
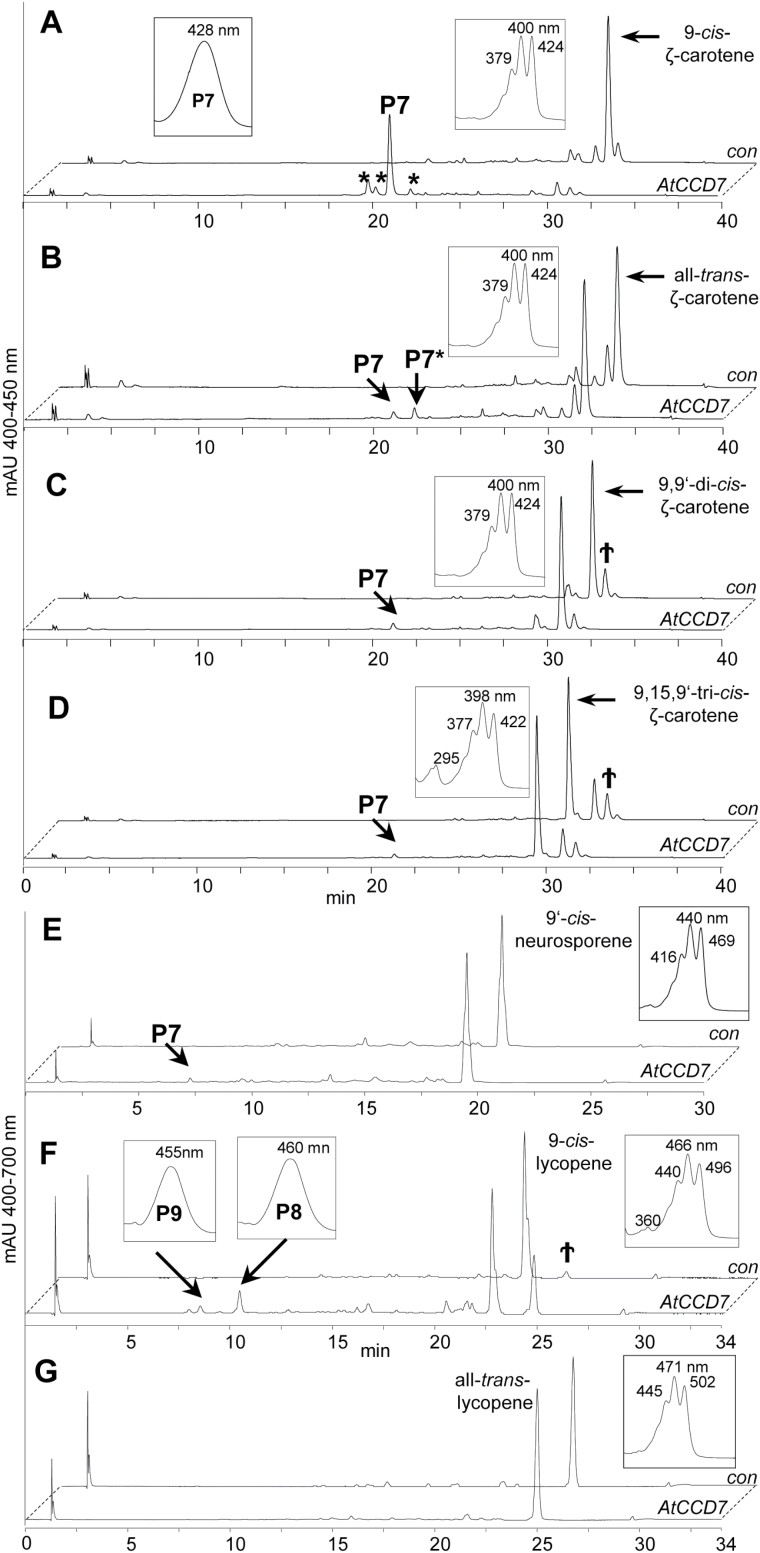
CCD7 enzymes are highly stereospecific, cleaving only 9-cis-configured substrates, such as 9-cis-β-carotene (Bruno et al., 2014), which is produced from the all-trans species by the isomerase DWARF27 (Alder et al., 2012; Bruno and Al-Babili, 2016). We therefore tested AtCCD7 as a candidate for the cleavage of cis-configured carotene desaturation intermediates. However, AtCCD7 did not convert any of the tri-cis-configured PDS or ZDS intermediates and products—9,15-di-cis-phytofluene, 9,15,9'-tri-cis-ζ-carotene, 7,9,9'-tri-cis-neurosporene, and 7,9,9',7-tetra-cis-lycopene. (Fig. 4D; Supplementary Fig. S4). Furthermore, the non-canonical isomers of phytofluene (9-cis, 15-cis, all-trans; Supplementary Fig. S4) were also not converted.
Fig. 4.
HPLC analysis of AtCCD7 activity with cis-configured carotene desaturation intermediates. (A) AtCCD7 cleaved 9-cis-ζ-carotene yielding P7, tentatively identified as 9-cis-ζ-apo-10'-carotenal. The minor peaks (asterisks) represent unspecific ζ-apo-10'-carotenal isomers—confirmed by isobaric masses (see Fig. 5A)—that arise upon sample processing. (B–D) Formation of traces of P7 upon incubation with other ζ-carotene isomers was due to minor cross-contamination with the 9-cis isomer (indicated by †). Product P7* corresponding to all-trans- ζ-apo-10'-carotenal was probably formed from P7 by unspecific isomerization. (E) AtCCD7 showed low cleavage activity with 9'-cis-neurosporene yielding P7, and (F) with 9-cis-lycopene yielding P8 and P9. P8 was identified by LC-MS analysis as all-trans-apo-10'-lycopenal, P9 as the putative 9-cis-apo-10'-lycopenal (see Fig. 6). The appearance of the all-trans-lycopene isomer upon incubation (indicated by †) and the formation of P8 are probably due to spontaneous cis-to-trans isomerization of 9-cis-lycopene and P8, respectively. (G) AtCCD7 did not convert all-trans-lycopene. UV/VIS spectra are shown as insets. For structures of the substrates, see Supplementary Figure 5. HPLC system 3 (A–D), HPLC system 1 (E), and HPLC system 2 (F, G) were used.
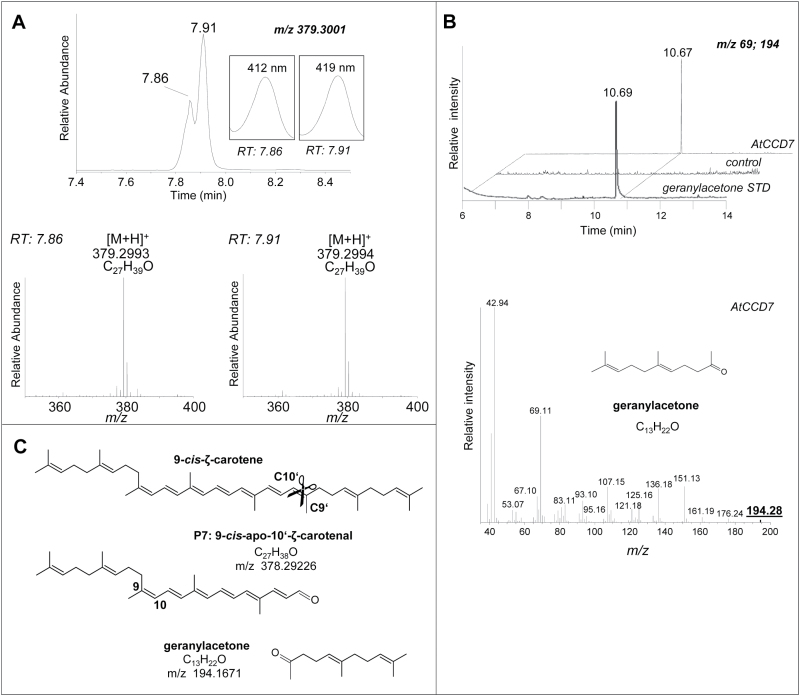
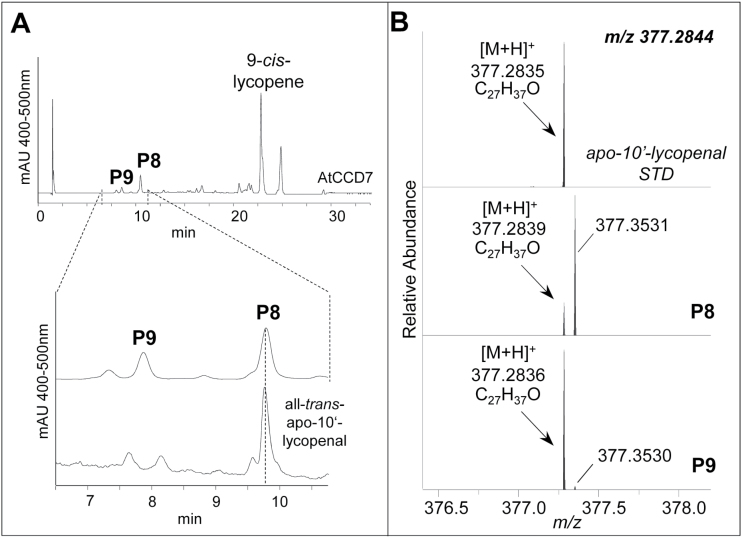
However, AtCCD7 cleaved the 9-cis-configured isomer of ζ-carotene (Fig. 4A–D) yielding the product P7 and small amounts of compounds with similar chromatographic and spectral characteristics (marked by asterisks in Fig. 4A). Subsequent LC-MS and GC-MS analyses revealed the formation of tentatively 9-cis-configured ζ-apo-10'-carotenal (P7, C27; Fig. 5A) and the volatile geranylacetone (C13; Fig. 5B). The other minor products detected in this incubation are probably cis/trans-isomers of P7, which can be formed by non-enzymatic isomerization of P7. Incubation with a ζ-carotene sample, which contained all-trans-ζ-carotene as the major isomer, led to traces of P7. We also detected a minor peak (P7*) tentatively identified as the all-trans-isomer of ζ-apo-10'-carotenal. It can be assumed that P7* was formed from P7 by non-enzymatic isomerization. The formation of P7 trace amounts from the all-trans-ζ-carotene substrate is most probably due to unavoidable minor cross-contamination with the 9-cis species (indicated by † in Fig. 4C, D). Cleavage of 9-cis-ζ-carotene indicated an activity with 9'-cis-neurosporene that is half identical with ζ-carotene, while the second, more desaturated half corresponds to lycopene. To obtain this substrate, we used recombinant CRTISO that can produce the corresponding 9'-mono-cis isomer from the poly-cis-compounds proneurosporene and prolycopene (Isaacson et al., 2004; Yu et al., 2011). Incubation with AtCCD7 led to a single detectable product, most probably ζ-apo-10'-carotenal (P7), as verified by UV/VIS spectroscopic properties (Fig. 4E). This product suggests a cleavage in the more desaturated half side of neurosporene (Fig. 4E; Supplementary Fig. S5 XVIII). This half side should be trans-configured, based on CCD7’s strict stereospecificity (Bruno et al., 2014), and on the preference of CRTISO (Yu et al., 2011). Consequently the ζ-apo-10-carotenal yielded from 9'-cis-neurosporene is expected to carry a 9'-cis configuration, as shown in Fig. 5C. It needs to be noted that the conversion was much lower as compared with that obtained with 9-cis-ζ-carotene. (Fig. 4A). AtCCD7 showed weak activity with 9-cis-lycopene, yielding product P8 accompanied by traces of P9 (Fig. 4F), while all-trans-lycopene was not converted (Fig. 4G). LC-MS analysis and comparison with an all-trans-apo-10'-lycopenal standard identified both products as apo-10'-lycopenals, of which P8 represents the all-trans-species and P9 the 9-cis species (Fig. 6A, B). As shown in Fig. 4F, unspecific isomerization of 9-cis- to all-trans-lycopene in the presence of detergents and cleared cell lysates was commonly observed (indicated by † in Fig. 4F). In accordance with this observation and due to AtCCD7’s specificity for cleavage of the trans-configured C9–C10 double bond, we assume that P8 was formed from the sole cleavage product P9 by non-enzymatic isomerization.
Fig. 5.
Identification of the 9-cis-ζ-carotene cleavage product. (A) The 9-cis-ζ-carotene cleavage product P7 (see Fig. 4A) was analyzed by LC-MS. AtCCD7 produced a putatively 9-cis-configured ζ-apo-10'-carotenal (7.91) accompanied by variable amounts of unspecific cis-trans-isomers (7.86), as shown by their isobaric masses. UV/VIS spectra of the products are depicted in insets. (B) GC-MS co-elution with the authentic reference (trace extracted at the indicated masses) and spectral comparison with the NIST 2.0 database identified geranylacetone as the volatile second cleavage product. (C) Schematic cleavage pattern of 9-cis-ζ-carotene. m/z denotes calculated isobaric masses.
Fig. 6.
Identification of the 9-cis-lycopene AtCCD7 cleavage products. (A) The 9-cis-lycopene cleavage products P8 and P9 (see Fig. 4F) were analyzed by LC-MS. AtCCD7 produced a putatively 9-cis-configured apo-10'-lycopenal (P9) that was accompanied by the all-trans species identified based on an authentic reference (lower trace). (B) The isomeric nature is shown by the isobaric masses. For structures of the substrates, see Supplementary Fig. S5. HPLC system 2 was used for separation.
Discussion
AtCCD4 cleavage site specificity and substrate preference
We have characterized the substrate, stereospecificity, and the cleavage pattern of AtCCD4. The results presented document AtCCD4 as an enzyme strictly cleaving the C9–C10 double bond. This regional specificity of cleavage was observed with bicyclic C40 carotenes and maintained with standard plant xanthophylls and derived (hydroxy-) apocarotenoids (Fig. 2A, B; Supplementary Fig. S2). The cleavage site is thus independent of hydroxyl functions in the C3 and C3' positions or on the presence of a second ionone ring. Cleavage site specificity was also unaffected by the chain length in (monocyclic) apocarotenoids (Fig. 2C, D). However, AtCCD4 requires the presence of at least one ionone ring for activity, since we did not observe any conversion of linear carotenes (Supplementary Fig. S3). Moreover, AtCCD4 is specific for all-trans-configured substrates and did not convert any mono- or poly-cis-configured substrates. This rules out a hypothetically possible direct contribution to ABA and SL biosynthesis (see AtCCD4 and regulatory molecules). Thus, AtCCD4 substrates need to be bicyclic or monocyclic and in the all-trans configuration to be converted.
Dynamic modeling of AtCCD4 time course experiments was carried out using different types of substrates, namely unhydroxylated (β-carotene), monohydroxylated (β-cryptoxanthin), and dihydroxylated (zeaxanthin) carotenoids, and the derived mono- and unhydroxylated apocarotenoids arising as secondary substrates upon primary cleavage. The resulting rate constants demonstrate that (i) unhydroxylated bicyclic C40 carotenes are preferred over C27 apocarotenoids; (ii) the presence of one or more OH groups in the C40 substrate reduces cleavage activity; and (iii) the latter does not apply to C27 apocarotenoids which, hydroxylated or not, were converted with very similar albeit low rate constants. It is worth noting that AtCCD1, an enzyme also targeting the C9–C10 double bond, prefers apocarotenoids over bicyclic carotenoids (Schmidt et al., 2006; Ilg et al., 2010). This difference is probably due to different biological functions and might be reflected by the cytoplasmatic localization of CCD1 that may contribute to carotenoid degradation in planta (Gonzalez-Jorge et al., 2013; see the Introduction).
Upon incubation with violaxanthin, AtCCD4 showed only a marginal cleavage activity. However, two earlier publications on CCD4 mutants from potato (RNAi) and Arabidopsis (T-DNA insertion) reported on elevated total carotenoid levels, mainly of violaxanthin followed by antheraxanthin, and lutein (Campbell et al., 2010; Gonzalez-Jorge et al., 2013). This apparent contradiction can be explained by different substrate specificity in planta or by end-product accumulation that is caused by reducing the cleavage rate of the β-carotene precursor. Our in vitro data are consistent with the second option and, hence, they are in line with the in vivo data. However, it cannot be excluded that the enzyme’s preference is affected by the microenvironment in planta.
As deduced from the corresponding rate constants, AtCCD4 showed higher preference for C40 carotenoids than for apocarotenoids, indicating that the enzyme recognizes the entire C40 substrate and not only half sides. To gain insight into possible underlying structural elements allowing this discrimination, AtCCD4 structure predictions were carried out with I-TASSER (Zhang, 2008). The maize enzyme VP14 (NCED 3 in Arabidopsis), whose crystal structure has been elucidated (Messing et al., 2010), was used as a comparator. AtCCD4 and VP14 share higher amino acid sequence homology (40% sequence identity) and structural similarity (TM score of 0.901) as compared with the other two CCDs for which structural information is available, SynACO (Kloer et al., 2005) and RPE65 (Kiser et al., 2009).
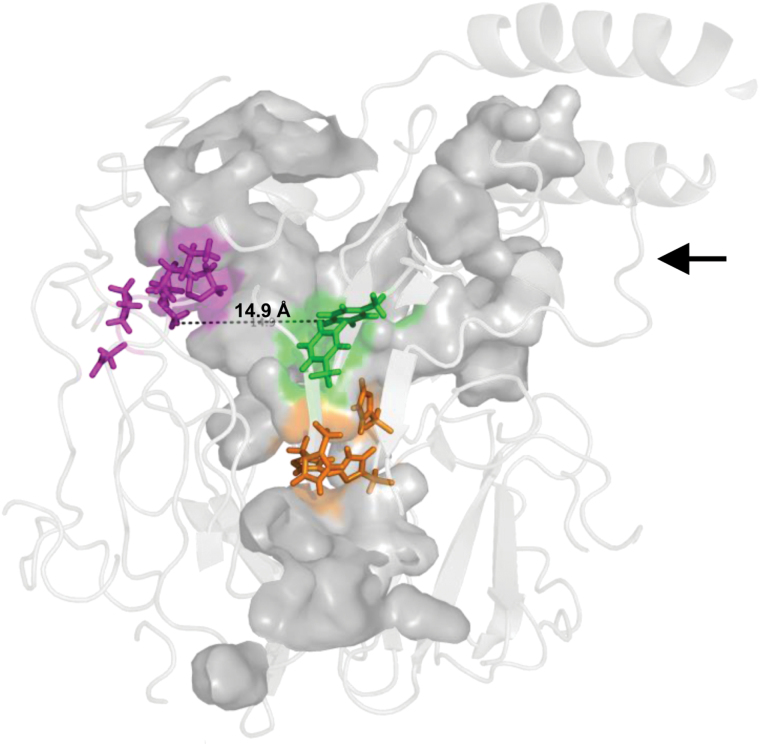
Common CCD structural features (Sui et al., 2013), including a hydrophobic surface patch to mediate association with membranes, are conserved (Supplementary Fig. S7A, B). Here, a cavity extends (arrowhead in Supplementary Fig. S7B) towards the active center lined with mainly hydrophobic residues. It is about 40 Å in length and hence well capable of accommodating entire C40 carotenoid substrates, as suggested with VP14 (Messing et al., 2010). This supports our observations that are in favor of a recognition of the entire substrate molecule by AtCCD4. The cavity features a conserved motif at its back end (Messing et al., 2010) consisting of two charged and three hydrophobic residues (D499PMPK503 in AtCCD4; pink in Fig. 7 and Supplementary Fig. S6) that might act as a ‘bumper’ for one ionone ring to restrict substrate penetration. These residues thus contribute to the positioning of the C9–C10 site relative to the active center that is formed by the conserved His298, His347, His412, and His590 (orange in Fig. 7 and Supplementary Fig. S6) co-ordinating Fe2+ on the central axis of the seven-blade β-propeller domain (blue in Supplementary Fig. S6A). Substrate positioning in VP14 is thought also to be aided by the three phenylalanine residues Phe171, Phe411, and Phe589, of which Phe411 is substituted by Ile411 in AtCCD4 (green in Fig. 7 and Supplementary Fig. S6) (Messing et al., 2010). The position of the second ring appears less defined, at loop structures situated at the tunnel entrance. The structural identity of the aliphatic proportion in bicyclic substrates would consequently place a high selective significance on the ‘bumper’ site capable of accommodating unsubstituted β-ionone functions but also C3' hydroxylated functionss albeit with lower effectiveness. The distance to the reactive center of ~15 Å (dotted line, Fig. 7) might consequently determine regional specificity of cleavage.
Fig. 7.
Predicted substrate cavity of AtCCD4. The substrate cavity is highlighted in gray and the conserved histidine residues co-ordinating Fe2+ are in orange. The conserved DPMPK motif on the back of the substrate cavity thought to restrict substrate penetration is shown in magenta. The ‘caging’ phenylalanine residues are highlighted in green. The distance from the rear of the cavity to the active center is ~15 Å, approximately the size of a β-ionone moiety, thus positioning the C9–C10 double bond for cleavage. For further explanations, see text.
VP14 is specific for 9-cis-configured epoxidated xanthophyll. This stereospecificity has been attributed to the presence of Val478, Ala478, or Ile478 in NCEDs, while CCDs carry phenylalanine or methionine in this position. With phenylalanine in this position, this would define AtCCD4 as a trans-specific CCD, a condition that is met by our data.
AtCCD4 and regulatory molecules
For decades, evidence for retrograde regulation of carotenoid biosynthesis has been put forward, for instance in tomato (Giuliano et al., 1993), maize (Bai et al., 2009), daffodil (Al-Babili et al., 1999), potato (Diretto et al., 2010), and carrot (Arango et al., 2014). The underlying mechanisms are far from being understood. However, it can be assumed that less lipophilic apocarotenoids that can be transported outside of the plastid play the role of signaling molecules, as known for ABA and SLs (Nambara and Marion-Poll, 2005; Raghavendra et al., 2010; Al-Babili and Bouwmeester, 2015). As outlined (see the Introduction), data by Kachanovsky et al. (2012) and Avendaño-Vazquez et al. (2014) point to poly-cis-configured carotene desaturation intermediates as signal precursors, and the latter publication suggested a participation of AtCCD4 in the generation of such signals.
However, our in vitro studies show that AtCCD4 is incapable of cleaving any carotene desaturation intermediates. AtCCD4 cleaved neither the cis-ζ-carotene isomers 9,15,9'-tri-cis-, 9,9'-di-cis-, and 9-cis-ζ-carotene nor all-trans-ζ-carotene (Supplementary Fig. S3). Moreover, it did not convert any other linear carotene, such as phytofluene, neurosporene, or lycopene, irrespective of their stereo-configuration (Supplementary Fig. S3E–H). Our data are consistent with the report of Huang et al. (2009), which showed that AtCCD4 does not cleave ζ-carotene or lycopene in carotenoid-accumulating E. coli cells.
Our data do not support a contribution of AtCCD4 to the biosynthesis of the two known carotenoid-derived hormones ABA and SLs. In the case of SLs, this assumption is based on the stereospecificity of this enzyme that did not cleave 9-cis-β-carotene (Supplementary Fig. 2F), which would lead to 9-cis-apo-10'-carotenal, the SL biosynthesis intermediate formed by AtCCD7. Similarly, we did not observe any conversion of 9-cis-violaxanthin (Supplementary Fig. 2G), indicating that AtCCD4 neither contributes to nor directly interferes with ABA biosynthesis. This corroborates the previous observation that ABA levels in ccd4 knock-down potatoes remain unaffected (Campbell et al., 2010).
The enzyme produces all-trans-β-apo-10'-carotenal and -3-OH-β-apo-10'-carotenal from bicyclic carotenoids. Given that AtCCD4 mediates the synthesis of a signaling molecule, it might be speculated that this molecule is a derivative of these apocarotenoids. Indeed, all-trans-β-apo-10'-carotenal in vitro is a precursor of β-apo-13-carotenone (d'orenone) that is formed by CCD8 enzymes (Alder et al., 2008). D'orenone has been shown to exert a regulatory function upon external application, affecting root hair growth in Arabidopsis (Schlicht et al., 2008). Moreover, this compound triggers indole-3-acetic acid (IAA) synthesis in the ectomycorrhizal basidiomycete Tricholoma vaccinum and promotes lateral root growth in the host tree spruce (Wagner et al., 2016). Taken together, our data do not support the hypothesis that AtCCD4 mediates the formation of signaling molecules from linear intermediates of the carotenoid biosynthesis pathway. However, it cannot be excluded that this enzyme plays a role in leaf morphogenesis or other developmental processes, for instance as a structural component of a complex regulating such processes. This assumption is based on the report of Naested et al. (2004) that shows the association of AtCCD4 with the protein VAR3 in a complex required for plastid development.
A possible role for AtCCD7 in forming linear retrograde signals
Intriguingly, both known carotenoid-derived hormones, ABA and SLs, originate from 9-cis-configured carotenoids (Al-Babili and Bouwmeester, 2015). The 9-cis configuration is also present in the acyclic carotenes that are thought to be the precursor of new signaling molecule(s). All desaturation intermediates have a 9-cis configuration (as in phytofluene) or are cis-configured at both the C9–C10 and C9'–C10' double bond (Fig. 1; for structures, see Supplementary Fig. S5). Moreover, it can be assumed that plants also contain other 9-cis-configured carotenes, such as 9'-cis-neurosporene and 9-cis-lycopene, which do not act as pathway intermediates. 9'-cis-Neurosporene and 9-cis-lycopene can be enzymatically formed in vitro by the carotene isomerase CRTISO (Isaacson et al., 2004; Yu et al., 2011), a property that has been exploited in this study to generate these substrates.
Based on the reported specificity of AtCCD7 for 9-cis-configured bicyclic carotenoids (Bruno et al., 2014), we tested the capability of this enzyme in cleaving desaturation intermediates and 9-cis-configured acyclic carotenes. The enzyme cleaved 9-cis-ζ-carotene, but did not show activity with any of the other ζ-carotene isomers tested. Thus, besides the strict specificity for 9-cis-configured carotenoids, AtCCD7 does not require the presence of an ionone ring structure for activity, in contrast to AtCCD4. Though to a much smaller extent, AtCCD7 also cleaved 9'-cis-neurosporene.
AtCCD7 maintained the regional specificity of the cleavage when incubated with acyclic carotenes. Taking the synthetic NMR-confirmed 9-cis-lycopene substrate as an example, cleavage of the trans-configured C9'–C10' double bond yielded 9-cis-apo-10'-lycopenal. The stereo-configuration of the product was demonstrated by its isobaric mass combined with different chromatographic behavior, compared with authentic all-trans-apo-10'-lycopenal (Fig. 6A, B). We conclude that AtCCD7 consequently formed 9-cis-ζ-apo-10'-carotenal from 9-cis-ζ-carotene and from 9'-cis-neurosporene. It remains to be shown whether the two AtCCD7 products 9-cis-ζ-apo-10'-carotenal and 9-cis-apo-10'-lycopenal are precursors of the signaling compounds that are thought to arise from carotene desaturation intermediates.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. GC-MS analysis of AtCCD4 volatile products.
Figure S2. HPLC analysis of AtCCD4 activity with different C40 xanthophylls and carotenes.
Figure S3. HPLC analysis of AtCCD4 activity with carotene desaturation intermediates.
Figure S4. HPLC analysis of AtCCD7 activity with carotene desaturation intermediates.
Figure S5. Structures of substrates cleaved by AtCCD4 and AtCCD7.
Figure S6. Sequence alignment of VP14, AtCCD4, and AtNCED3.
Figure S7. AtCCD4 3D model prediction.
Supplementary dataset S1. Modeling of AtCCD4 activity.
Acknowledgements
This work was supported by the European Union (METAPRO; FP7 KBBE-2009-3-1-01) and the King Abdullah University of Science and Technology (KAUST).
References
- Al-Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Al-Babili S, Hartung W, Kleinig H, Beyer P. 1999. CPTA modulates levels of carotenogenic proteins and their mRNAs and affects carotenoid and ABA content as well as chromoplast structure in Narcissus pseudonarcissus flowers. Plant Biology 1, 607–612. [Google Scholar]
- Alder A, Holdermann I, Beyer P, Al-Babili S. 2008. Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochemical Journal 416, 289–96. [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arango J, Jourdan M, Geoffriau E, Beyer P, Welsch R. 2014. Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. The Plant Cell 26, 2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ. 2006. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Current Opinion in Plant Biology 9, 315–321. [DOI] [PubMed] [Google Scholar]
- Avendaño-Vázquez A-O, Cordoba E, Llamas E, et al. 2014. An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. The Plant Cell 26, 2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Kim E-H, DellaPenna D, Brutnell TP. 2009. Novel lycopene epsilon cyclase activities in maize revealed through perturbation of carotenoid biosynthesis. The Plant Journal 59, 588–599. [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, Mccarty D, Klee H, Leyser O. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Brandi F, Bar E, Mourgues F, Horváth G, Turcsi E, Giuliano G, Liverani A, Tartarini S, Lewinsohn E, Rosati C. 2011. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biology 11, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach J, Sandmann G. 2005. ζ-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220, 785–793. [DOI] [PubMed] [Google Scholar]
- Britton G, Liaaen-Jensen S, Pfander H, eds. 1995. Carotenoids, Vol 1B: Spectroscopy. Basel: Birkhäuser Verlag. [Google Scholar]
- Bruno M, Al-Babili S. 2016. On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27. Planta 243, 1429–1440. [DOI] [PubMed] [Google Scholar]
- Bruno M, Beyer P, Al-Babili S. 2015. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Archives of Biochemistry and Biophysics 572, 126–133. [DOI] [PubMed] [Google Scholar]
- Bruno M, Hofmann M, Vermathen M, Alder A, Beyer P, Al-Babili S. 2014. On the substrate- and stereospecificity of the plant carotenoid cleavage dioxygenase 7. FEBS Letters 588, 1802–1807. [DOI] [PubMed] [Google Scholar]
- Campbell R, Ducreux LJM, Morris WL, Morris J, Suttle JC, Ramsay G, Bryan GJ, Hedley PE, Taylor M. 2010. The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiology 154, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L, Clough JM, Pattenden G. 1983. Application of nuclear magnetic resonance spectroscopy in the stereochemical assignment of poly-Z-isomeric conjugated polyene isoprenoids. Journal of the Chemical Society, Perkin Transactions 1, 3005–3009. [Google Scholar]
- Clough JM, Pattenden G. 1979. Naturally occurring poly-cis carotenoids. Stereochemistry of poly-cis lycopene and its congeners in ‘Tangerine’ tomato fruits. Journal of the Chemical Society, Chemical Communications, 616–619. [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, Beyer P, Giuliano G. 2010. Transcriptional–metabolic networks in beta-carotene-enriched potato tubers: the long and winding road to the Golden phenotype. Plant Physiology 154, 899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi R, Vendramin E, Zanon L, Scalabrin S, Cipriani G, Verde I, Vizzotto G, Morgante M. 2013. Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. The Plant Journal 76, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, et al. 2014. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proceedings of the National Academy of Sciences, USA 111, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. 1993. Regulation of carotenoid biosynthesis during tomato development. The Plant Cell 5, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–195. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Ha S-H, Magallanes-Lundback M, et al. 2013. Carotenoid cleavage dioxygenase 4 is a negative regulator of β-carotene content in Arabidopsis seeds. The Plant Cell 25, 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann GE, Sturgis JN, Wagner E, Robert B, Beyer P, Tadros MH. 1996. The effects of the detergent LDAO on the carotenoid metabolism and growth of Rhodovulum sulfidophilum. Microbiological Research 151, 421–426. [Google Scholar]
- Huang F-C, Molnár P, Schwab W. 2009. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. Journal of Experimental Botany 60, 3011–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilg A, Beyer P, Al-Babili S. 2009. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS Journal 276, 736–747. [DOI] [PubMed] [Google Scholar]
- Ilg A, Yu Q, Schaub P, Beyer P, Al-Babili S. 2010. Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta . Planta 232, 691–699. [DOI] [PubMed] [Google Scholar]
- Ilg A, Bruno M, Beyer P, Al-Babili S. 2014. Tomato carotenoid cleavage dioxygenases 1A and 1B: relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio 4, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Ohad I, Beyer P, Hirschberg J. 2004. Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiology 136, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachanovsky DE, Filler S, Isaacson T, Hirschberg J. 2012. Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proceedings of the National Academy of Sciences, USA 109, 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. 2009. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proceedings of the National Academy of Sciences, USA 106, 17325–17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. 2005. The structure of a carotenoid oxygenase. Science 308, 267–269. [DOI] [PubMed] [Google Scholar]
- Ma G, Zhang L, Matsuta A, Matsutani K, Yamawaki K, Yahata M, Wahyudi A, Motohashi R, Kato M. 2013. Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage cioxygenase 4 in the flavedo of citrus fruit. Plant Physiology 163, 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing SA, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM. 2010. Structural insights into maize viviparous 14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. The Plant Cell 22, 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise AR, Al-Babili S, Wurtzel ET. 2014. Mechanistic aspects of carotenoid biosynthesis. Chemical Reviews 114, 164–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naested H, Holm A, Jenkins T, et al. 2004. Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. Journal of Cell Science 117, 4807–4818. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Nisar N, Li L, Shan L, et al. 2015. Carotenoid metabolism in plants. Molecular Plant 8, 68–82. [DOI] [PubMed] [Google Scholar]
- Nocedal J, Wright SJ. 1999. Numerical optimization. Berlin: Springer Science + Business Media, LLC. [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. 2006. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiology 142, 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Cabrero A, Estrada AF, Al-Babili S, Avalos J. 2007. Identification and biochemical characterization of a novel carotenoid oxygenase: elucidation of the cleavage step in the Fusarium carotenoid pathway. Molecular Microbiology 64, 448–460. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. 2010. ABA perception and signalling. Trends in Plant Science 15, 395–401. [DOI] [PubMed] [Google Scholar]
- Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, Timmer J. 2009. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25, 1923–1929. [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Alquézar B, Alós E, Medina V, Carmona L, Bruno M, Al-Babili S, Zacarías L. 2013. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. Journal of Experimental Botany 64, 4461–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Rambla JL, Santaella M, Gómez MD, Orzaez D, Granell A, Gómez-Gómez L. 2008. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. Journal of Biological Chemistry 283, 24816–24825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Moraga A, Rambla JL, Fernández-de-Carmen A, Trapero-Mozos A, Ahrazem O, Orzáez D, Granell A, Gómez-Gómez L. 2014. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Molecular Biology 86, 555–569. [DOI] [PubMed] [Google Scholar]
- Ruch S, Beyer P, Ernst H, Al-Babili S. 2005. Retinal biosynthesis in Eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Molecular Microbiology 55, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Scherzinger D, Al-Babili S. 2008. In vitro characterization of a carotenoid cleavage dioxygenase from Nostoc sp. PCC 7120 reveals a novel cleavage pattern, cytosolic localization and induction by highlight. Molecular Microbiology 69, 231–244. [DOI] [PubMed] [Google Scholar]
- Schlicht M, Samajová O, Schachtschabel D, Mancuso S, Menzel D, Boland W, Baluska F. 2008. D'orenone blocks polarized tip growth of root hairs by interfering with the PIN2-mediated auxin transport network in the root apex. The Plant Journal 55, 709–717. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Kurtzer R, Eisenreich W, Schwab W. 2006. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. Journal of Biological Chemistry 281, 9845–9851. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart J. 2001. Characterization of a novel carotenoid cleavage dioxygenase from plants. Journal of Biological Chemistry 276, 25208–25211. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, Mccarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Seo M. 2002. Complex regulation of ABA biosynthesis in plants. Trends in Plant Science 7, 41–48. [DOI] [PubMed] [Google Scholar]
- Soetaert K, Petzoldt T, Setzer RW. 2010. Solving differential equations in R: package deSolve. Journal of Statistical Software 33, 1–25.20808728 [Google Scholar]
- Sui X, Kiser PD, von Lintig J, Palczewski K. 2013. Structural basis of carotenoid cleavage: from bacteria to mammals. Archives of Biochemistry and Biophysics 539, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Tan B, Mccarty DR, Klee HJ. 2008. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. Journal of Biological Chemistry 283, 11364–11373. [DOI] [PubMed] [Google Scholar]
- von Lintig J, Vogt K. 2000. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. Journal of Biological Chemistry 275, 11915–11920. [DOI] [PubMed] [Google Scholar]
- Wagner K, Krause K, David A, Kai M, Jung E-M, Sammer D, Kniemeyer O, Boland W, Kothe E. 2016. Influence of zygomycete-derived D'orenone on IAA signaling in Tricholoma–spruce ectomycorrhiza. Environmental Microbiology 18, 2470–2480. [DOI] [PubMed] [Google Scholar]
- Walter MH, Strack D. 2011. Carotenoids and their cleavage products: biosynthesis and functions. Natural Product Reports 28, 663–692. [DOI] [PubMed] [Google Scholar]
- Yu Q, Ghisla S, Hirschberg J, Mann V, Beyer P. 2011. Plant carotene cis-trans isomerase CRTISO: a new member of the FAD(RED)-dependent flavoproteins catalyzing non-redox reactions. Journal of Biological Chemistry 286, 8666–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmeister L. 1962. Cis trans isomeric carotenoids, vitamins A and arylpolyenes. New York: Academic Press. [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.