Highlight
Transcriptional analysis (mRNA and small RNA) of rice flower organs demonstrates correlations of 24-nt phased small RNAs with anther maturation and with three small RNA-binding Argonautes.
Key words: Anther, Argonaute, microRNA, phasiRNA, rice, spikelet.
Abstract
Dissection of the genetic pathways and mechanisms by which anther development occurs in grasses is crucial for both a basic understanding of plant development and for examining traits of agronomic importance such as male sterility. In rice, MULTIPLE SPOROCYTES1 (MSP1), a leucine-rich-repeat receptor kinase, plays an important role in anther development by limiting the number of sporocytes. OsTDL1a (a TPD1-like gene in rice) encodes a small protein that acts as a cofactor of MSP1 in the same regulatory pathway. In this study, we analyzed small RNA and mRNA changes in different stages of spikelets from wild-type rice, and from msp1 and ostdl1a mutants. Analysis of the small RNA data identified miRNAs demonstrating differential abundances. miR2275 was depleted in the two rice mutants; this miRNA is specifically enriched in anthers and functions to trigger the production of 24-nt phased secondary siRNAs (phasiRNAs) from PHAS loci. We observed that the 24-nt phasiRNAs as well as their precursor PHAS mRNAs were also depleted in the two mutants. An analysis of co-expression identified three Argonaute-encoding genes (OsAGO1d, OsAGO2b, and OsAGO18) that accumulate transcripts coordinately with phasiRNAs, suggesting a functional relationship. By mRNA in situ analysis, we demonstrated a strong correlation between the spatiotemporal pattern of these OsAGO transcripts and phasiRNA accumulations.
Introduction
Rice (Oryza sativa), as a major crop, has been widely used as a monocot model species to explore the genetic basis of flower development in higher plants (Yoshida and Nagato, 2011). Rice anther development is one of the major topics studied in rice flower development, and changes in the cytological morphology in different developmental stages of rice anthers have been well-described (Zhang and Wilson, 2009; Zhang et al., 2011). Tapetum and microsporocyte specification is a crucial event in male fertility, occurring at early stages of anther development in plants; a number of genes have been discovered as regulators of cell fate specification (Zhang and Yang, 2014). For example, EXCESS MICROSPOROCYTES 1 (EMS1, or EXTRA SPOROGENOUS CELLS, EXS), a member of the leucine-rich repeat receptor-like kinase (LRR-RLK) family, specifies tapetal identity and limits the number of pollen mother cells (PMCs) in Arabidopsis (Canales et al., 2002; Zhao et al., 2002). The small secreted protein, TAPETAL DETERMINANT 1 (TPD1), has been reported as a ligand of EMS1/EXS with a deterministic role in the cell fate of the tapetum (Jia et al., 2008).
The EMS1/EXS ortholog in rice is MULTIPLE SPOROCYTE (MSP1) (Nonomura et al., 2003), while the ligand protein TPD1 has two TPD1-like orthologs in rice, including OsTDL1A and OsTDL1B, among which OsTDL1A (also known as MICROSPORELESS2, MIL2) may interact with MSP1 (Zhao et al., 2008; Hong et al., 2012). Although msp1 and ostdl1a mutants display defects in anther and ovule development, both show a phenotype of complete male sterility, while partially maintaining female fertility (Nonomura et al., 2003; Hong et al., 2012; Yang et al., 2016). MSP1, as a receptor-like kinase in an upstream signaling pathway, affects many other downstream genes involved in rice anther development. For example, a loss of function of MSP1 will largely down-regulate the expression of other genes involved in rice anther development, such as Undeveloped Tapetum1 (UDT1) and Tapetum Degeneration Retardation (TDR) (Jung et al., 2005; Li et al., 2006). Maize MAC1 is the ortholog of OsTDL1A, having similar functions in limiting archesporial cell proliferation in maize anthers (Wang et al., 2012). Therefore, the OsTDL1A-MSP1 pathway plays a central role in early stages of rice anther development to simultaneously specify the tapetum and limit the number of pollen mother cells (Zhang and Yang, 2014).
Small RNA pathways play roles in both flower development and gametogenesis in plants. Some conserved miRNAs appear to function similarly in flower development across different plant species, such as Arabidopsis, tomato, petunia, rice, and maize (Luo et al., 2013). In rice, miR172 targets APETALA2 (AP2) genes controlling inflorescence architecture and spikelet meristem identity (Zhu et al., 2009; Lee and An, 2012). A number of rice SQUAMOSA Promoter Binding Protein-Like (OsSPL) genes, including OsSPL14, are targeted by miR156; this pathway has a role in flowering time, panicle architecture, grain yield, and other developmental phenotypes (Xie et al., 2006; Jiao et al., 2010; Miura et al., 2010). Other miRNAs, such as miR159 and miR164, are also reported to be involved in rice floral development (Tsuji et al., 2006; Adam et al., 2011). In addition to miRNAs, trans-acting siRNAs (tasiRNAs), dependent on the activities of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and DICER-LIKE 4 (DCL4), generated from non-coding transcripts such as TAS3, play roles in both vegetative and reproductive development in both rice and maize by targeting genes encoding auxin response factors (ARFs). For example, the rice mutant of SHOOTLESS2 (SHL2), the ortholog of Arabidopsis RDR6, displays a severe phenotype of misregulation of adaxial-abaxial polarity patterning in both the lemma and anther (Toriba et al., 2010), while the maize mutant leafbladeless1 (lbl1, a loss of function of an SGS3 ortholog, SUPPRESSOR OF GENE SILENCING 3) is defective in tasiRNA biogenesis and shows a pleotropic phenotype, including sterile male inflorescences (Nogueira et al., 2007). The RDR6-dependent small RNA biogenesis pathway not only produces tasiRNAs, but also yields two large populations of phased secondary siRNAs (phasiRNAs) in the reproductive tissues of monocots (reviewed in Fei et al., 2013). Data have suggested that these 21- and 24-nt reproductive phasiRNAs, triggered by miR2118 and miR2275, respectively, may play crucial roles in microgametogenesis in maize, because the accumulation of phasiRNAs shows highly stage-specific patterns in maize anther development (Zhai et al., 2015). In addition, the Argonaute (AGO) protein MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1), previously demonstrated to be essential for sporogenesis in rice anthers (Nonomura et al., 2007), has recently been shown to recruit 21-nt phasiRNAs (Komiya et al., 2014).
In a recent study, we showed that the MSP1-OsTDL1A partners are master regulators of downstream transcription factors that are involved in plant anther development (Yang et al., 2016). Therefore, we hypothesized that loss-of-function of MSP1-OsTDL1A may also cause great downstream changes in non-coding RNAs and small RNAs. Here, we systematically characterized changes in small RNA and mRNA, especially non-coding PHAS transcripts, across early developmental stages of rice spikelets in wild-type, msp1 and ostdl1a backgrounds using deep sequencing data. We found comprehensive changes of miRNAs, phasiRNAs, and PHAS transcripts in early stages of rice spikelet development. Importantly, the reproductive phasiRNAs displayed stage-specific expression patterns during early stages of anther development, suggesting that the timing of phasiRNA biogenesis is crucial in rice microsporogenesis. Furthermore, phasiRNA and mRNA changes in different developmental stages and mutant backgrounds facilitated the identification of several rice AGOs, in addition to MEL1, that potentially load phasiRNAs.
Materials and methods
Plant materials and growth conditions
All the rice plants used in this study were in a genetic background of variety 9522, a japonica rice. The two male-sterile mutants, ostdl1a and msp1-4, are from a rice mutant library made by 60Co γ-ray radiation; the molecular details of these mutants are described in Yang et al. (2016). Plants were grown in the paddy field of Shanghai Jiao Tong University in China.
Small RNA and RNA-seq library construction
For small RNA library construction, total RNA enriched for small RNA was extracted. The small RNA faction between 18 to 30 nt in length was collected by gel separation, then ligated to 5′ and 3′ adaptors and purified. These small RNAs were reverse transcribed by RT-PCR and finally amplified via PCR. For RNA-seq libraries, after the total RNA extraction and DNase I treatment, magnetic beads with oligo(dT) were used to isolate mRNA. Mixed with the fragmentation buffer, the mRNA was fragmented into short fragments. The cDNA was synthesized using the mRNA fragments as templates. Short fragments were purified for end repair and single nucleotide A (adenine) addition, ligated to adapters, and then the second strand was degraded using UNG (Uracil-N-Glycosylase). After agarose gel electrophoresis, the suitable fragments were selected for PCR amplification as templates. All the small RNA and RNA-seq libraries were sequenced on an Illumina HiSeq 2000 platform by BGI (BGI-Shenzhen, China).
Small RNA data analysis
Small RNA sequencing data were preprocessed by removing adapters, and then mapped to the version 7.0 of the rice genome assembly from the Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu, accessed 23 September 2016) using the program Bowtie (Langmead et al., 2009). Small RNA reads that were mapped to the tRNAs and rRNAs were filtered, and reads mapped to the rice genome from different libraries were normalized to reads per 5 million reads (RP5M) for comparisons. The Bioconductor (www.bioconductor.org) package ‘edgeR’ was used for small RNA differential analysis (P<0.05, FDR<0.05). Rice miRNA sequences were downloaded from miRBase (Release 21; http://www.mirbase.org/, accessed 23 September 2016) (Kozomara and Griffiths-Jones, 2014). The R (www.r-project.org, accessed 23 September 2016) package ‘pheatmap’ was used to represent the average abundance of miRNAs from three biological replicates. PHAS locus identification was performed using the same method described previously by Zhai et al. (2011). Briefly, small RNA sequencing data from different libraries were combined together to increase the sequencing depth for PHAS loci identification. A phasing score of 25 was used as a stringent cut-off, followed by a manual check to remove loci producing highly abundant small RNAs in other sizes, which are most likely degradation products from t/rRNAs. The overall phasiRNA abundance for each PHAS locus was calculated by summing up the normalized abundance of 21- or 24-nt small RNAs generated from each corresponding 21-PHAS and 24-PHAS locus.
RNA-seq data analysis
Paired-end strand-specific RNA-seq reads (90bp × 2) were mapped to the rice genome sequences allowing no more than two mismatches using ‘Tophat’ (Trapnell et al., 2009). The BAM files generated by ‘Tophat’ were sorted and indexed using ‘SAMtools’ (Li et al., 2009), and then visualized via Integrative Genomics Viewer (IGV) (Robinson et al., 2011). The program ‘Cufflinks’ (Trapnell et al., 2012) was used for transcriptome assembly, differential analysis of gene expression, and calculation of the FPKM value (fragments per kilobase of transcript per million mapped reads). For differential analysis of gene expression, we used ‘q-value<0.01’ and ‘fold change > 2’ as cut-offs. Bar graphs and line charts representing FPKM values of gene expression were plotted using the Bioconductor package ‘cummeRbund’ (Trapnell et al., 2012).
Microarray data analysis
Gene lists were inputted into the webserver Rice Oligonucleotide Array Database (http://www.ricearray.org/, accessed 23 September 2016; Cao et al., 2012). Specific public microarray datasets were selected to acquire the abundance values of each gene. The normalized gene expression values were further visualized as heatmaps using the R package ‘pheatmap’.
In situ hybridizations
Freshly collected samples were fixed in formalin-acetic acid-alcohol (FAA) and dehydrated in a series of graded ethanol concentrations; these samples were then infiltrated with Histo-clear II, embedded in Paraplast Plus, and subsequently processed into 6-μm thick sections using a Leica RM2245 rotary microtome. Templates for RNA probe synthesis were amplified by PCR from the cDNA. Probes were transcribed in vitro under the T7 promoter with RNA polymerase, using the DIG RNA labeling kit (Roche). The RNA in situ hybridizations were carried out as described by Kouchi and Hata (1993) and Li et al. (2006). The forward and reverse RT-PCR primers were as follows: OsAGO1d, 5′-GCAATACCACCCACAAGGAC-3′ and 5′-GGTTCCAATACTCCCACTTCC-3′; OsAGO18, 5′-CAGTAT AACAGTACGGAACGC-3′ and 5′-TGTCATTACAACAAGTAG GAGG-3′.
Accession numbers
RNA-seq and small RNA data are available from Genbank, under GEO accession number GSE77300.
Results
Comparative analysis of small RNAs in spikelets of wild-type and mutant rice
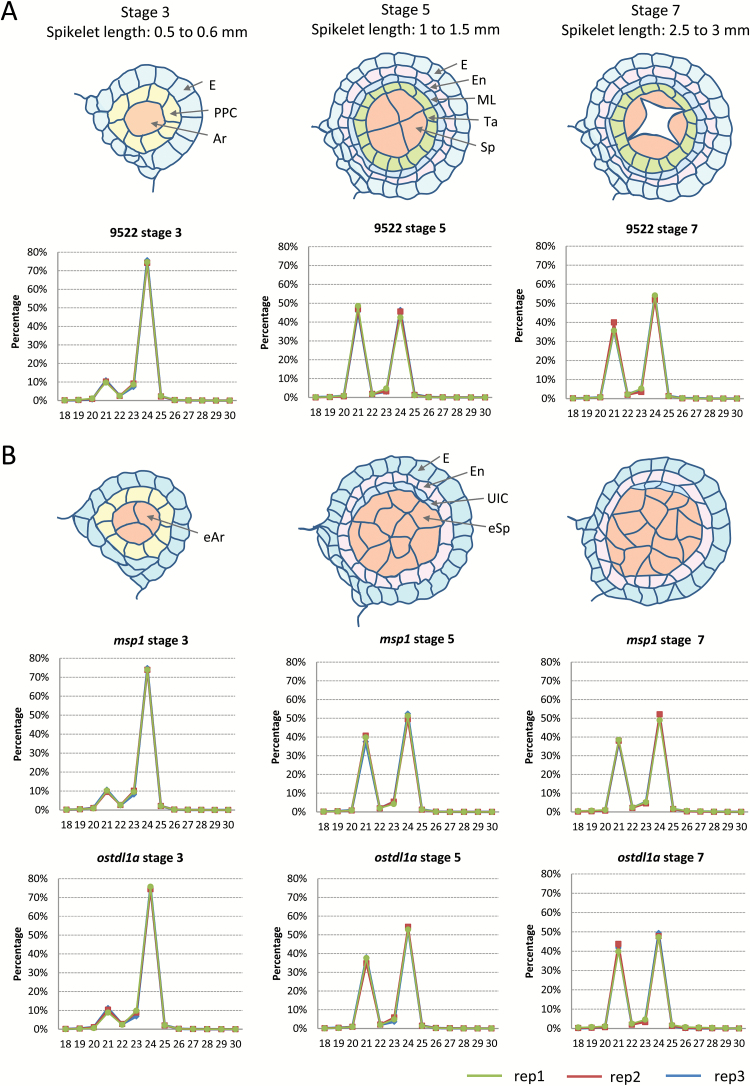
Small RNAs play crucial roles in mediating both transcription and translation, with different classes distinguishable by their distinct biogenesis pathways (Axtell, 2013). The recent discovery of reproductive phasiRNAs in monocots indicates that this special class of small RNAs may be important for male reproduction, although the underlying mechanism remains to be elucidated (Johnson et al., 2009; Song et al., 2012; Zhai et al., 2015). To assess small RNA and mRNA changes across different stages of rice spikelet development and to understand how they are impacted by perturbation of the OsTDL1A-MSP1 pathway, we prepared small RNA and RNA-seq libraries from spikelets of wild-type rice cultivar 9522, and the mutants msp1-4 (‘msp1’ hereafter) and ostdl1a [library information is listed in Supplementary Table S1 at JXB online; mutant information is described in Yang et al. (2016), and is also shown in Supplementary Fig. S1]. We performed three biological replicates for each genotype and stage. The lengths of rice spikelets correspond to different anther developmental stages (Fig. 1). Specifically, stage 3 (0.15–0.2mm), stage 5 (0.25–0.3mm), and stage 7 (0.4–0.45mm) of rice anthers correspond to 0.5–0.6mm, 1.0–1.5mm, and 2.5–3.0mm rice spikelets, respectively (Zhang et al., 2011) (Fig. 1A); hereafter, we will refer to these sizes of rice spikelets as stage 3, stage 5, and stage 7 spikelets, respectively. Samples were collected at these three stages because MSP1 and OsTDL1A mainly function at early stages (stage 3 to stage 5) of rice anther development (Yang et al., 2016), while stage 5–7 is an important stage at which meiocytes start meiosis (Zhang et al., 2011).
Fig. 1.
Small RNA size distribution in different developmental stages and backgrounds of rice spikelets. (A) Schematic representation of rice anther structures in different stages of spiklets of wild-type rice. Each layer of cells is indicated by an arrow: epidermis (E); primary parietal cell (PPC); archesporial cell (Ar); endothecium (En); middle layer (ML); tapetum (Ta); sporogenous cell (Sp). Small RNA size distributions in the different stages of wild-type rice spikelets are shown below. (B) Schematic representation of rice anther structures in different stages of spiklets of the msp1 and ostdl1a mutants. Excessive archesporial cell (eAr); unknown identity cell (UIC); excessive sporogenous cell (eSp). Small RNA size distributions in the different stages of the mutant spikelets are shown below.
Sequencing reads of small RNAs were aligned to the rice genome and normalized to 5 million (5M), and the distribution of lengths in different stages was analyzed. The three replicates were nearly identical (Fig.1). The 24-nt small RNAs account for ~75% of all small RNAs at stage 3 in wild-type rice. Interestingly, with the development of spikelets, there was a shift in the predominant size class, with 21-nt small RNAs the largest proportion (nearly 50%) in the profile at stage 5. In stage 7, the proportion (~55%) of 24-nt small RNAs was once again larger than the 21-nt counterpart (<40%), but was far smaller than that in stage 3. The fluctuation of small RNA percentages in different sizes that accompanied spikelet development may represent shifts in small RNA biogenesis and transcriptome changes in rice reproductive tissues, consistent with prior reports in maize (Zhai et al., 2015). Similar to the wild-type spikelets, in the two mutants changes in small RNA size proportions occurred more significantly in the transition from stage 3 to stage 5 than from stage 5 to stage 7. The only apparent difference was a reduction in the proportion of 21-nt small RNAs in the mutants in stage 5 spikelets (Fig. 1B). To investigate this difference, and others less readily apparent, we checked the levels of small RNAs that are often 21 nt in length, miRNAs and phasiRNAs.
miRNA expression patterns in different developmental stages of rice spikelets
A number of miRNAs have characterized roles in plant development, targeting several families of transcription factors or other development-related genes (Jones-Rhoades et al., 2006; Chen, 2009). miRNAs, such as miR156 and miR172, have been proven to control flower development at a post-transcriptional level in both Arabidopsis and rice (Aukerman and Sakai, 2003; Chen, 2004; Xie et al., 2006; Wu et al., 2009a; Zhu et al., 2009; Jiao et al., 2010; Lee and An, 2012). Therefore, the expression patterns of miRNAs are important to our understanding of rice spikelet development.
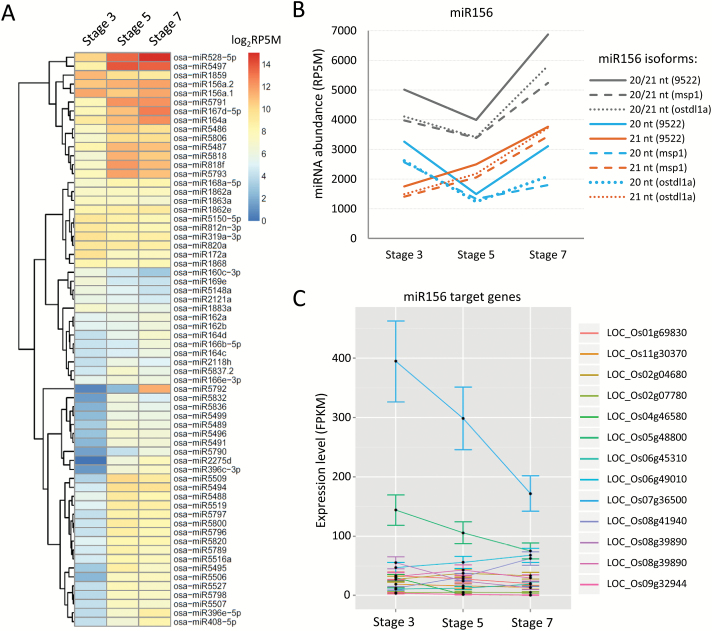
Genome-wide differential analysis of miRNA expression was performed in wild-type rice across different stages of spikelet development (see Supplementary Tables S2 and S3). From stage 3 to stage 5, >60 miRNAs increased significantly, while only 18 miRNAs decreased during the same time period; from stage 5 to stage 7, the numbers of miRNAs with significantly different levels (up or down) were both fewer than 20 (Fig. 2A; Table 1). Comparing different stages in the msp1 and ostdl1a mutant backgrounds, similar sets (both qualitatively and quantitatively) of differentially accumulating miRNAs were observed, suggesting that the male sterile phenotype of these mutants has a limited impact on miRNA levels, relative to the wild-type. This was further confirmed by the observation that very few miRNAs were identified as significantly different in their levels when comparing the two mutants with the wild-type (Table 1). Interestingly, among the few impacted miRNAs in the two mutants, miR2275, which triggers 24-nt phasiRNA production, was totally abolished (Supplementary Table S3, and see below).
Fig. 2.
miRNA expression in different developmental stages of spikelets in wild-type rice cultivar 9522. (A) Differentially expressed miRNAs in different developmental stages of spikelets. Only miRNAs with abundance greater than 50 RP5M are included in the heatmap. (B) 20-nt miR156 and 21-nt isoforms display distinct expression patterns; levels indicated as ‘20/21’ show the sum of abundance of the 20 and 21 nt isoforms. (C) Expression levels of miR156 target genes in different stages of rice spikelet development.
Table 1.
Numbers of differentially abundant miRNAs, identified by pairwise comparisons of rice spikelets in different stages and backgrounds.
Numbers of significantly up- and down-regulated miRNAs, as indicated by the arrows (P<0.05, FDR<0.05). miRNAs are from the miRBase release 21. ‘S3’, ‘S5’, and ‘S7’ refer to stages 3, 5, and 7, respectively.
Among the miRNAs differentially expressed in wild-type rice across the developmental stages, the level of miR164 showed a ~7-fold increase from stage 3 to stage 7, whereas miR172 showed a dramatic decline of ~4-fold from stage 3 to stage 5, then remained relatively steady to stage 7 (see Supplementary Fig. S2A). Prior work using PARE/degradome data confirmed the targets of a number of miRNAs in rice (Li et al., 2010; Zhou et al., 2010). From RNA-seq data, we obtained the expression levels of both miR164 and miR172 target genes. Transcript levels of the most abundant miR164 target (LOC_Os12g05260) decreased from 180 FPKM (stage 3) to 125 FPKM (stage 5), and the expression level increased slightly in stage 7 compared to stage 5, and a similar trend was observed for the target gene LOC_Os06g23650 (Supplementary Fig. S2B); these were the only two targets that showed an inverse relationship with miR164 abundance. Three miR172 targets showed an inverse correlation with miR172 levels (Supplementary Fig. S2C). miR156 showed an interesting pattern across the developmental stages of rice spikelets: a 21-nt miR156 isoform increased gradually from stage 3 to stage 7, while the 20-nt miR156 firstly decreased from stage 3 to stage 5, and then increased at stage 7 (Fig. 2B). The underlying mechanism is unknown, but it is possible that these two isoforms of miR156 are generated from different MIR156 genes, and are differentially regulated with distinct targets or cellular expression patterns, exerting distinct patterns of control on the expression of OsSPLs. Therefore, studies of individual MIR156 genes would be important in the future to demonstrate how the miR156 family members differentially control spikelet or panicle development. Similar to miR164 and miR172, a subset of miR156 targets showed an inverse correlation with miR156 (Fig. 2C). Overall, the abundance levels of miRNA target transcripts in the two mutants were quite similar to those in wild-type, indicating that mutations in the OsTDL1A-MSP1 pathway had a limited impact on rice miRNAs and their targets in spikelets (Table 1; Supplementary Fig. S2B–D). Among the miRNA targets validated from prior work, only a subset had the expected inverse correlation with miRNA levels; thus, other factors may impact target transcript levels, including both transcriptional (e.g. transcription factors and epigenetic regulation) and post-transcriptional (e.g. mRNA turnover or sequestration) factors. Taken together, these data indicate that miRNAs together with their target genes are dynamically modulated during early stages of rice spikelet development, and are largely independent of the OsTDL1A-MSP1 pathway.
Timing of reproductive phasiRNA biogenesis
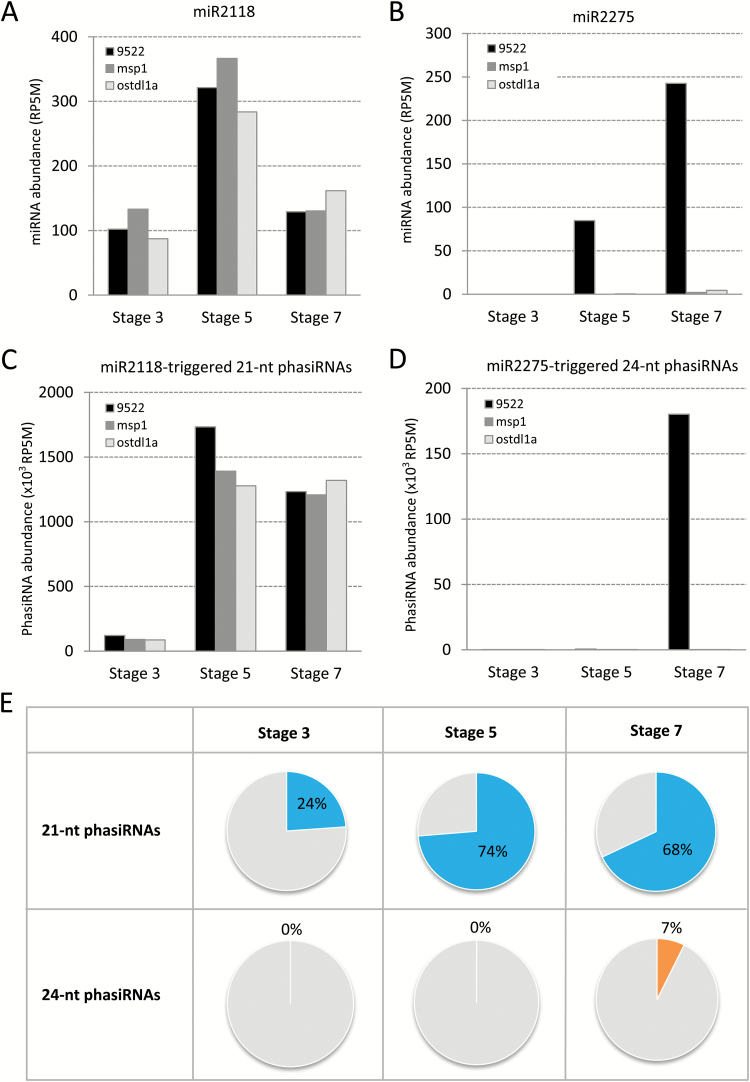
PhasiRNAs, in addition to miRNAs, represent another class of small RNAs of great interest, because two populations are exclusively abundant in the reproductive tissues of monocots (Arikit et al., 2013). More specifically, miR2118 and miR2275 are triggers of 21-nt and 24-nt reproductive phasiRNAs, respectively, in both rice and maize (Song et al., 2012; Zhai et al., 2015). We checked the abundance levels of both miRNAs in different stages and backgrounds; miR2118 abundance peaked at stage 5 in wild-type spikelet libraries, reaching 320 reads per 5 million reads (RP5M), and then dropped to 130 RP5M at stage 7 (Fig. 3A). Similar abundances were observed in msp1 and ostdl1a mutants, indicating that miR2118 is not impacted in both mutants, possibly explained by miR2118 accumulation in the epidermis of anthers (Zhai et al., 2015), a cell layer apparently not defective in the msp1 and ostdl1a mutants or in the mac1 maize mutant. In contrast to miR2118, in wild-type spikelets, miR2275 was not expressed in stage 3, and then increased in stage 5 (~85 RP5M) and stage 7 (~240 RP5M) (Fig. 3B); miR2275, as mentioned above, is essentially absent in both msp1 and ostdl1a mutants, at all stages.
Fig. 3.
miR2118, miR2275, and phasiRNA abundances in rice spikelets. Levels of miR2118 (A), miR2275 (B), miR2118-triggered 21-nt phasiRNAs (C), and miR2275-triggered 24-nt phasiRNAs (D) in different stages and backgrounds of rice spikelets. (E) The percentage of 21-nt (top, blue slices with percentage numbers) and 24-nt phasiRNAs (bottom, orange slices with percentage numbers) out of the total population of 21-nt and 24-nt genome-matched small RNAs in wild-type rice spikelets.
We calculated both 21- and 24-nt phasiRNA abundances to see how phasiRNAs change during rice spikelet development and whether they are affected in the two mutants. Phasing analysis resulted in 1843 21-PHAS loci and 50 24-PHAS loci (see Supplementary Table S4). By summing phasiRNA abundances from each PHAS locus, we obtained the overall phasiRNA abundances for both 21- and 24-nt phasiRNAs. In wild-type rice spikelets, the overall abundance of 21-nt phasiRNAs was ~120 000 RP5M in stage 3, increasing by >10-fold in stage 5, and then decreasing by <30% in stage 7 (Fig. 3C). Compared to the wild-type, 21-nt phasiRNAs had a very similar pattern in the two mutants, although with a slightly lower total abundance in stage 5. The 24-nt counterparts had only a few hundred reads in stage 3 and stage 5, but then increased to more than 180 000 RP5M in stage 7 of the wild-type spikelets (Fig. 3D). Consistent with the observation that miR2275 is absent in msp1 and ostdl1a mutants at all stages, 24-nt phasiRNAs were also diminished in both mutants. Considering the developmental defect in these mutants is largely in the anthers and not other tissues in the spikelets (Yang et al., 2016), we infer that the loss of 24-nt phasiRNAs is due to the defective anther development. Furthermore, as in maize (Zhai et al., 2015), rice 21-nt phasiRNAs initiated at an early stage in anther development, the 24-nt phasiRNAs appeared later, coincident with or just before meiosis, and the absence of miR2275 and 24-nt phasiRNAs in msp1 and ostdl1a mutants is similar to maize mac1 (the ortholog of ostdl1a in maize).
We next calculated the proportion of 21- and 24-nt phasiRNAs in the entire genome-matched populations of the 21- and 24-nt small RNAs in rice spikelets (Fig. 3E). We found that 21-nt phasiRNAs accounted for 24% of total 21-nt siRNAs in wild-type rice spikelets, increased to a remarkable 74% of the total in stage 5, and then reduced slightly to 68% at stage 7. This compares to 60% of all 21-mers at the peak in maize (Zhai et al., 2015); this higher proportion in rice spikelets versus isolated maize anthers may reflect the fact that there are ~4-fold as many genomic loci generating 21-nt phasiRNAs in rice compared to maize. The 24-nt phasiRNAs were almost absent at stages 3 and 5, followed by a substantial increase to 7% of the total at stage 7 (Fig. 3E). This compares to 64% of the total 24-mers in isolated maize anthers (Zhai et al., 2015); this much lower proportion in rice may reflect the fact that there are fewer loci, or that the peak abundance of 24-nt phasiRNAs is later than stage 7. Overall, these results reveal that both miRNA triggers and phasiRNAs are largely up-regulated at specific stages in rice anthers.
Interconnected PHAS locus transcription and phasiRNA bursts
We next examined the phasiRNA precursors (PHAS transcripts) in the RNA-seq data to assess the correlation of phasiRNA and PHAS mRNA levels. As many 21-PHAS loci are found in large clusters in the rice genome (Johnson et al., 2009) (see Supplementary Table S4), we selected a representative cluster on chromosome 3 for this analysis. Our earlier observations in wild-type rice showed a stage 5 peak for 21-nt phasiRNAs, which reduced slightly at stage 7 (Fig. 3C); in the RNA-seq data at stage 3 and stage 5, 21-PHAS transcripts levels were generally consistent with phasiRNA production; however, at stage 7, 21-PHAS transcripts levels were very low (Fig. 4) while phasiRNA levels were still high. This is an indication that 21-nt phasiRNAs persist longer than their precursor transcripts, possibly for a prolonged role in later stages of spikelet development. As for msp1 and ostdl1a mutants, levels of neither 21-nt phasiRNAs nor 21-PHAS transcripts were impacted (stage 5 for both mutants is shown in Supplementary Fig. S3; stage 7 data are not shown but were not appreciably different from the wild-type). Consistent with the precursor molecules consisting of a polyadenylated mRNA paired with an RDR6-derived antisense strand, the strand-specific RNA-seq reads were mapped to only one strand at a given PHAS locus (Fig. 4). An examination of the RNA-seq data for 24-PHAS loci showed a strong boost in transcript levels from stage 5 to stage 7 in wild-type rice anthers (Supplementary Fig. S4A). However, unlike 21-PHAS loci, both 24-nt phasiRNAs and 24-PHAS mRNAs were absent in the RNA-seq data from msp1 and ostdl1a mutants (Supplementary Fig. S4A, B). In summary, sequencing data revealed the developmental modulation of phasiRNA biogenesis in rice anthers, with 24-phasiRNAs and their precursors disrupted in the msp1 and ostdl1a mutants, reflecting an intimate association of these transcripts with anther development and microsporogenesis in rice.
Fig. 4.
21-nt PHAS precursor transcripts peak coincidentally with their phasiRNA products. We examined a randomly selected 21-PHAS locus on rice chromosome 3 in wild-type (cultivar 9522) rice to assess the peak of abundance relative to the 21-phasiRNAs that peak at stage 5. Because PHAS loci are highly clustered in the rice genome, we selected a region of ~24kb as an example; in this case, the PHAS loci are interlaced with repetitive sequences. Each dot is a small RNA; light blue represent 21-nt sRNAs, green represent 22-nt, and orange represent 24-nt. Yellow shaded regions are predicted DNA transposons; pink shaded regions are predicted retrotransposons; orange shaded regions are inverted repeats. The small pink box is an annotated miRNA. The phasiRNA loci are essentially the distinct blocks of 21-nt sRNAs (light blue dots). The strand-specific RNA-seq data is represented as an IGV screenshot; blue bars are top-strand reads, and red bars are bottom-strand reads. There was a paucity of RNA-seq reads from these PHAS loci data in stage 7 at this cluster of loci, whereas 21-phasiRNAs were still abundant at stage 7.
Identification of specialized Argonautes that load reproductive phasiRNAs
Argonautes (AGOs) are core effector proteins in small RNA-mediated silencing pathways. Different small RNAs are preferentially recruited into specific AGOs, mainly determined by the 5′-terminal nucleotide of the small RNA (Mi et al., 2008). The ten AGO proteins of Arabidopsis are reasonably well studied, but the functional roles of the ~17 to 19 AGOs in grass genomes are less well described (Zhang et al., 2015). The naming for OsAGOs in this study is consistent with the description by Zhang et al. (2015). Moreover, it is still unclear which AGOs recruit the abundant reproductive phasiRNAs of grasses. A recent study in rice showed that the germline-specific AGO protein MEL1 associates with 21-nt phasiRNAs that have 5′-terminal cytosine (Komiya et al., 2014). In addition, the AGO(s) that recruits 24-nt phasiRNAs is still unknown, although ZmAGO18b is enriched in tapetum and germ cells in maize anthers (Zhai et al., 2014), where 24-nt phasiRNAs accumulate (Zhai et al., 2015). Considering the dramatic changes of phasiRNA abundances across different stages of rice spikelet development, we hypothesized that AGO proteins that recruit phasiRNAs may show a gene expression pattern correlated with phasiRNA abundances. Therefore, we examined the expression of all AGOs in rice from stages 3 to 7 (Fig. 5A). We found that MEL1 peaked at stage 5, the same stage as the peak of accumulation of 21-nt phasiRNAs; therefore, MEL1 expression was indeed correlated with phasiRNA production. In the RNA-seq data, MEL1 levels were slightly higher in both mutants (see Supplementary Fig. S5), perhaps because of the excessive number of sporocytes, where MEL1 is expressed, in mutants (Nonomura et al., 2007). Intriguingly, we found that OsAGO1d showed the same pattern as MEL1, suggesting a possible functional connection between the OsAGO1d protein and reproductive phasiRNAs (Fig. 5A). Similar to MEL1, OsAGO1d expression was barely impacted in msp1 and ostdl1a mutants (Supplementary Fig. S5).
Fig. 5.
Abundance of OsAGO transcripts in different stages and backgrounds of anther development. (A) mRNA levels of all nineteen OsAGOs in wild-type rice. OsAGO1d and MEL1 are highlighted because only these two AGO transcripts peaked at stage 5. (B) OsAGO2b, OsAGO18, and OsAGO5b displayed substantial up-regulation at stage 7 compared to stages 3 and 5. OsAGO2b and OsAGO18, but not OsAGO5b, is defective in the msp1 and ostdl1a mutants at stage 7. Significant differences (Student’s t-test) are indicated: *P<0.01 and **P<0.05; ‘ns’ indicates no significant difference.
We speculated that AGOs that accumulate in stage 7 could function with 24-nt phasiRNAs. Gene expression analysis showed that only three OsAGOs, namely OsAGO2b, OsAGO5b, and OsAGO18, displayed a substantial up-regulation at stage 7 compared to stages 3 and 5 (Fig. 5B). Since 24-nt phasiRNA accumulation was deficient in msp1 and ostdl1a mutants, the result of defects in cell layers important for their biogenesis, it is possible that the OsAGOs that load 24-nt phasiRNAs are similarly impacted in both mutants. We found that the levels of OsAGO2b and OsAGO18, but not OsAGO5b, were partially reduced in the msp1 and ostdl1a mutants (Fig. 5B), suggesting that OsAGO2b and OsAGO18 are candidates for roles with 24-nt phasiRNAs. To further confirm their specificity to anthers, we checked their expression patterns using published rice anther microarray datasets. Consistent with our RNA-seq data, OsAGO2b and OsAGO18, but not OsAGO5b, showed a meiosis-specific expression pattern in rice anthers (see Supplementary Fig. S6).
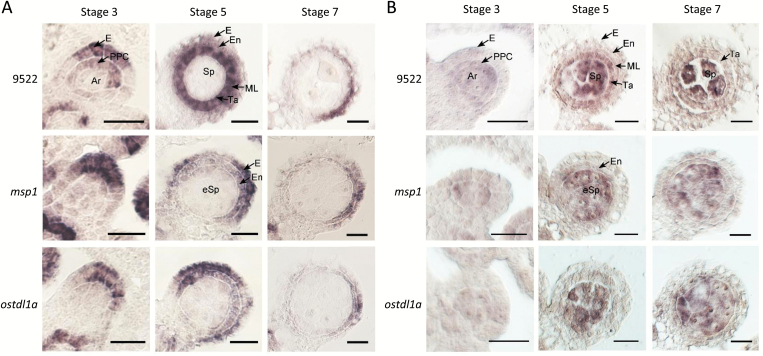
Temporal-spatial expression of phasiRNA-associated AGOs revealed by in situ hybridizations
To connect the temporal specificity of transcript accumulation with spatial patterns, we performed in situ hybridizations or examined published images for selected rice AGO genes. Published images for OsAGO2b demonstrate its accumulation in sporocytes and wall layers of rice anthers at pre-meiotic stages, and in later meiosis stage the transcript is restricted predominantly to the tapetum layer and microspores (Deveshwar et al., 2011). We performed in situ hybridizations in rice anthers to examine the transcript accumulation patterns of OsAGO1d and OsAGO18 (Fig. 6). These results showed that OsAGO1d accumulates highly in the distal epidermis and primary parietal cells of the anther lobe at stage 3. This OsAGO1d pattern is reminiscent of miR2118, which accumulates in the distal cells of the epidermis in maize anthers (Zhai et al., 2015). In stage 5, OsAGO1d accumulates highly in the middle layer and the tapetal layer, which are both differentiated from the primary parietal cells. In stage 7, the level of OsAGO1d is much lower than that of stage 5 (Fig. 6A). As for OsAGO18, its abundance was enriched in tapetal and sporogenous cells at stages 5 and 7 (Fig. 6B). Therefore, the patterns of OsAGO1d and OsAGO18 across different developmental stages of rice anthers are consistent with the results of RNA-seq data from rice spikelets, and these OsAGO transcripts are highly correlated both spatially and temporally with reproductive phasiRNAs (comparing our in situs with those of the phasiRNAs in maize anthers in Zhai et al., 2015). Taken together, these analyses suggest a possible functional connection between grass reproductive phasiRNAs and the three AGO proteins encoded by OsAGO1d, OsAGO2b, and OsAGO18.
Fig. 6.
RNA in situ hybridization of OsAGO1d and OsAGO18 in different stages and backgrounds of rice anthers. Expression patterns of OsAGO1d (A) and OsAGO18 (B), performed in stages 3, 5, and 7 of anthers, from either wild-type cultivar 9522 or the msp1 or ostdl1a mutants, as indicated. Each image shows one anther lobe. Cell layers are labeled as in Fig. 1. Scale bars indicate 20 μm.
Discussion
Like most complex developmental changes, rice spikelet development entails widespread changes in mRNAs and small RNAs. Similar data for mutants in MSP1 and OsTDL1A, which encode interacting proteins with crucial roles in initiating early stage reproductive development, demonstrate the impact of disordered cell specification in rice reproductive tissues (Yang et al., 2016).
miRNAs play important roles in plant development. We identified miRNAs with differential accumulation patterns during rice spikelet development, including conserved miRNAs (miR156, miR172, and miR164). Among these, the role of miR164 in development is not well studied, although it accumulates in spikelet and floral meristems (Adam et al., 2011). While variation in miRNA levels in different stages of rice spikelets suggests that miRNAs are active, one interesting observation from the msp1 and ostdl1a mutants is that levels of most miRNAs and their targets were largely not impacted in the mutants, despite the block in development (Fig. 2B, C; Supplementary Fig. S2). This suggests that these miRNA-involved gene silencing pathways are genetically independent or upstream of the OsTDL1A-MSP1 pathway.
We also assessed phasiRNA production in rice spikelets. Two classes of phasiRNAs have distinct accumulation patterns in grass anthers (Zhai et al., 2015); as in maize, we found that rice phasiRNAs peak at specific stages during spikelet development. We showed that miR2118, the trigger of 21-nt phasiRNAs, accumulates to the highest level at stage 5 and drops severely in stage 7, while 21-nt phasiRNAs are relatively slightly retarded, peaking at stage 5 but decreasing only slightly at stage 7. As in maize, miR2275 peaks later (stage 7), at the stage at which 24-nt phasiRNAs reached the highest abundance that we measured. In the msp1 and ostdl1a mutants, 24- but not 21-nt phasiRNAs were depleted – consistent with data from the maize mutant mac1 (the ortholog of rice OsTDL1A) (Wang et al., 2012). Therefore, the timing of phasiRNA biogenesis is conserved in rice and maize, two Poaceae evolutionarily separated by ~50 million years (Wolfe et al., 1989).
AGO proteins are key catalytic components that associate with small RNAs, and different AGOs function as either RNA binders/slicers or chromatin modifiers by loading different classes of small RNAs (Waterhouse, 2016). Therefore, to know which AGOs load these reproductive phasiRNAs would greatly help elucidate their functions. Considering that in the mutants 24-nt phasiRNAs were impacted but miRNAs were largely not, we may be able to infer AGOs with roles in phasiRNA function. Compared to 10 AGOs in Arabidopsis, the rice and maize genomes encode more, 19 and 17 respectively (Zhang et al., 2015). AGO5 expression in Arabidopsis is specific to somatic ovule tissues, with a role in megagametogenesis (Tucker et al., 2012). A potentially conserved function of AGO5 in plant gametogenesis has been shown in rice, as the AGO5 relative MEL1 (OsAGO5c) binds 21-nt reproductive phasiRNAs with 5′ C (Komiya et al., 2014). AGO1 in rice has four homologs (OsAGO1a, OsAGO1b, OsAGO1c, and OsAGO1d); OsAGO1a/b/c predominantly recruit miRNAs and other small RNAs with 5′-terminal uridine (Wu et al., 2009b). Our RNA-seq data showed that rice OsAGO1d accumulates in spikelets, synchronous with MEL1, making OsAGO1d a strong candidate for further functional analysis. In rice, OsAGO18 is induced upon viral infection, and has been shown to confer resistance to viruses by sequestering miR168, suppressing OsAGO1 expression (Wu et al., 2015). Maize has two homologs of AGO18; ZmAGO18b is specific to the tapetum and germ cells (Zhai et al., 2014). Our data showed that OsAGO18 and OsAGO2b transcripts increase substantially at stage 7, coincident with 24-nt phasiRNA accumulation. Moreover, a recent study on Arabidopsis AGO3 showed that this poorly characterized Argonaute mediates RdDM by binding 24-nt siRNAs (Zhang et al., 2016). Phylogenetic analysis showed that Arabidopsis AGO3 and rice OsAGO2b are close to each other (Zhang et al., 2015), suggesting that OsAGO2b could also recruit 24-nt small RNAs, such as the reproductive phasiRNAs, to regulate epigenetic modifications. Furthermore, in situ hybridization results for OsAGO1d, OsAGO18, and OsAGO2b showed that these OsAGOs have distinct expression patterns during rice anther development, suggesting that they may associate with either 21- or 24-nt phasiRNAs in a stage-specific manner. In summary, our results suggest a functional relevance between OsAGO1d, OsAGO18, and OsAGO2b and grass reproductive phasiRNAs.
What might be the function of these AGO proteins loaded with phasiRNAs? Prior work on MEL1 (associated with 21-nt phasiRNAs) suggests a role in histone modifications; histone H3 lysine 9 dimethylation (H3K9me2) is decreased in the mel1 mutant (Nonomura et al., 2007). Yet the reduced H3K9me2 in mel1 could be an indirect effect of other epigenetic changes, as many chromatin modifications in plants and other organisms are intricately linked (Castel and Martienssen, 2013). Considering the lack of sequence complementarity of phasiRNAs and other regions in the genome (Zhai et al., 2015), it is possible that reproductive phasiRNAs act primarily in cis or impact cis-adjacent regions by a spreading mechanism. Indeed, a recent study on maize reported that both 21- and 24-PHAS loci showed higher levels of CHH methylation in meiocytes than other tissues, such as seedlings (Dukowic-Schulze et al., 2016). This finding suggests that reproductive phasiRNAs may play an important role in chromatin remodeling in cis around the stage of meiosis. Overall, the coordinated accumulation of 21- and 24-nt phasiRNAs and several AGO transcripts during rice reproductive development suggests that more detailed and comprehensive analyses of DNA methylation and histone modifications are needed, particularly when coupled with mutants in phasiRNA pathways.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. MSP1 and OsTDL1A transcript levels in stage 3 of rice spikelets in the wild-type cultivar 9522 and two mutants.
Figure S2. miRNA and target transcript levels in different stages and backgrounds of rice spikelets.
Figure S3. 21-nt phasiRNAs and precursor transcripts were unaffected in both msp1 and ostdl1a mutants at stage 5.
Figure S4. 24-nt phasiRNAs are strongly impacted in stage 7 spikelets of the two rice mutants.
Figure S5. mRNA levels of MEL1 and OsAGO1d in different stages and backgrounds of rice spikelets.
Figure S6. Expression of AGOs in different tissues and developmental stages of rice anthers in public microarray datasets.
Table S1. Summary information for small RNA and RNA-seq libraries prepared in this study.
Table S2. miRNA levels in different libraries.
Table S3. Differentially expressed miRNAs in different stages and backgrounds of rice spikelet development.
Table S4. 21- and 24-nt phasiRNA abundance from each PHAS locus.
Acknowledgements
Research in the Meyers lab is supported by the US National Science Foundation Plant Genome Research Program (award #1339229). Collaborative research and travel between the Zhang and Meyers lab is supported by the British Council’s Global Innovation Initiative, which funds the Global Innovations Network (GII) (see https://www.cpib.ac.uk/GII/ for more details, accessed 23 September 2016). Research in the Zhang lab is supported by National Natural Science Foundation of China (31430009; 31110103915; 32322040; 31271698); China Innovative Research Team, Ministry of Education, and the Program of Introducing Talents of Discipline to Universities (111 Project, B14016); National Key Basic Research Developments Program, Ministry of Science and Technology, China (2013CB126902); the Science and Technology Commission of Shanghai Municipality (grant no. 13JC1408200); Leading Scientist in Agriculture of Shanghai Municipality. We gratefully acknowledge the assistance of Mayumi Nakano with data handling, and Jixian Zhai for helpful discussions.
References
- Adam H, Marguerettaz M, Qadri R, et al. 2011. Divergent expression patterns of miR164 and CUP-SHAPED COTYLEDON genes in palms and other monocots: implication for the evolution of meristem function in angiosperms. Molecular Biology & Evolution 28, 1439–1454. [DOI] [PubMed] [Google Scholar]
- Arikit S, Zhai J, Meyers BC. 2013. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Current Opinion in Plant Biology 16, 170–179. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annual Review of Plant Biology 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. 2002. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Current Biology 12, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC. 2012. The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature Reviews Genetics 14, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cellular and Developmental Biology 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveshwar P, Bovill WD, Sharma R, Able JA, Kapoor S. 2011. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biology 11, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze S, Sundararajan A, Ramaraj T, Kianian S, Pawlowski WP, Mudge J, Chen C. 2016. Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Frontiers in Plant Science 7, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. The Plant Cell 25, 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Tang D, Shen Y, Hu Q, Wang K, Li M, Lu T, Cheng Z. 2012. MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytologist 196, 402–413. [DOI] [PubMed] [Google Scholar]
- Jia G, Liu X, Owen HA, Zhao D. 2008. Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proceedings of the National Academy of Sciences, USA 105, 2220–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Research 19, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G. 2005. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. The Plant Cell 17, 2705–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura K. 2014. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant Journal 78, 385–397. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. 1993. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Molecular & General Genetics 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, An G. 2012. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant Journal 69, 445–461. [DOI] [PubMed] [Google Scholar]
- Li H Handsaker B Wysoker A Fennell T Ruan J Homer N Marth G Abecasis G Durbin R, and 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, et al. 2006. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. The Plant Cell 18, 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. 2010. Transcriptome-wide identification of microRNA targets in rice. Plant Journal 62, 742–759. [DOI] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L. 2013. Evolutionary conservation of microRNA regulatory programs in plant flower development. Developmental Biology 380, 133–144. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. 2007. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Development 21, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N. 2003. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. The Plant Cell 15, 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2007. A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. The Plant Cell 19, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nature Biotechnology 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, et al. 2012. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant Journal 69, 462–474. [DOI] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, Hirano HY. 2010. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. The Plant Cell 22, 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, et al. 2006. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant Journal 47, 427–444. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. 2012. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ. 2012. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P. 2016. RNA interference: A dark horse in the AGO stable. Nature Plants 2, 16059. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH. 1989. Date of the monocot–dicot divergence estimated from chloroplast DNA sequence data. Proceedings of the National Academy of Sciences, USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009a The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang Z, Wang Y, et al. 2015. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. 2009b Rice microRNA effector complexes and targets. The Plant Cell 21, 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. 2006. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiology 142, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Qian X, Chen M, Fei Q, Meyers BC, Liang W, Zhang D. 2016. Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiology 171, 2085–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y. 2011. Flower development in rice. Journal of Experimental Botany 62, 4719–4730. [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes & Development 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V, Meyers BC. 2015. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proceedings of the National Academy of Sciences, USA 112, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z. 2014. Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. Journal of Integrative Plant Biology 56, 1042–1052. [DOI] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L. 2011. Cytological analysis and genetic control of rice anther development. Journal of Genetics & Genomics 38, 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA. 2009. Stamen specification and anther development in rice. Chinese Science Bulletin 54, 2342–2353. [Google Scholar]
- Zhang D, Yang L. 2014. Specification of tapetum and microsporocyte cells within the anther. Current Opinion in Plant Biology 17, 49–55. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xia R, Meyers BC, Walbot V. 2015. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Current Opinion in Plant Biology 27, 84–90. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Guo X, Wang X-J, Zhang X. 2016. Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nature Plants 2, 16049. [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H. 2002. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes & Development 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Herve P, Xue Q, Bennett J. 2008. OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant Journal 54, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Gu L, Li P, Song X, Wei L, CHen Z, Cao X. 2010. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Frontiers in Biology 5, 67–90. [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. 2009. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biology 9, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.