Abstract
Ohno proposed that the expression levels of X-linked genes have been doubled to compensate the degeneration of Y-linked homologs during the evolution of mammalian sex chromosomes, but RNA sequencing in human somatic tissues showed no such upregulation for the vast majority of X-linked genes. Here we report that the X to autosome expression ratio equals ∼1 in haploid human parthenogenetic embryonic stem (pES) cells and ∼0.5 in diploidized pES cells, both with one active X chromosome. Although we confirmed the upregulation of ∼5% of X-linked genes encoding members of large protein complexes in diploids, these genes are also upregulated in haploids, breaking the otherwise balanced dosage. These findings argue against Ohno’s hypothesis for both haploid and diploid cells and demonstrate that, at least in humans, precise gene regulation for dosage balance, even for members of large protein complexes, is much less critical than is commonly thought.
Keywords: dosage compensation, sex chromosome, evolution, dosage balance, mammal.
Nearly 50 years ago, Ohno famously hypothesized that, during the origin of the mammalian sex chromosomes from a pair of autosomes, the per-allele expression levels of X-linked genes in males should be doubled to compensate the degeneration of the Y-linked homologs (Ohno 1967). This proposed 2-fold upregulation of X-linked genes, presumably also present in females, could explain why one of females' two X chromosomes needs to be inactivated. Thus, Ohno's hypothesis forms the theoretical foundation of the current understanding of mammalian sex chromosome evolution and sex chromosome dosage compensation (Charlesworth 1996; Payer and Lee 2008). The empirical test of Ohno's hypothesis, however, has had a bumpy history (He and Zhang 2016). Early microarray-based gene expression profiling found an expression ratio between one active X (Xa) and two autosomes (AA) in human somatic tissues to be ∼1, or Xa:AA ∼1, supporting Ohno's hypothesis (Nguyen and Disteche 2006). RNA sequencing (RNA-seq)-based expression profiling, however, found Xa:AA ∼0.5, refuting Ohno's hypothesis (Xiong et al. 2010). A subsequent analysis of human proteomic data also found Xa:AA ∼0.5 (Chen and Zhang 2015). More importantly, RNA-seq expression comparisons between human X-linked genes and their chicken autosomal orthologs, which represent proto-X genes, confirmed the lack of the hypothesized 2-fold upregulation in mammalian X chromosome evolution (Julien et al. 2012; Lin et al. 2012). Intriguingly, however, for ∼5% of X-linked genes that encode components of large protein complexes, a 2-fold upregulation was detected in human RNA-seq data (Pessia et al. 2012) and confirmed in the comparison with chicken RNA-seq data (Lin et al. 2012), suggesting that these genes are dosage compensated, probably because they are dosage-sensitive due to the stoichiometric requirement for members of a large protein complex and thus are subject to relatively strong natural selection for dosage balance. It is worth noting that Ohno's hypothesis concerns diploid cells. For haploid cells, the number of X chromosomes, relative to the number of sets of autosomes is 0, 1, and 1, for Y-carrying sperm, X-carrying sperm, and eggs, respectively. Apparently, X-chromosome dosage compensation is impossible for Y-carrying sperm and not needed for eggs and X-carrying sperm. Note that, although X is silenced during male meiosis, it is reactivated in spermatids (Daish and Grutzner 2010; Turner 2015). Early microarray data suggested an Xa:A expression ratio of ∼1 in X-carrying spermatids and ∼0.9 in secondary oocytes (i.e., post-meiosis I) (Nguyen and Disteche 2006). But given the poor performance of microarray in quantifying between-chromosome expression ratios (Xiong et al. 2010), these results require reexamination. It is especially interesting to examine the ∼5% of X-linked genes that are dosage-sensitive, because their 2-fold upregulation observed in diploid cells would lead to an Xa:A expression ratio of 2 and thus dosage imbalance if it persists in haploid cells. If dosage balance in haploid cells is important to these dosage-sensitive genes, one should expect no upregulation of these genes in haploids. In other words, the dosage compensation hypothesis predicts a diploid-specific 2-fold upregulation of these genes. Here we address the above questions using RNA-seq data generated from haploid human parthenogenetic embryonic stem (pES) cell lines originating from haploid oocytes (Sagi et al. 2016).
The data are from five cell lines, including two pES cell lines (pES10 and pES12), two differentiated embryoid bodies − EB1 and EB2 − that were, respectively, from pES10 and pES12, and neural progenitor cells (NPCs) differentiated from pES10 (Sagi et al. 2016). Because haploid cells spontaneously and irreversibly diploidize over passages, Sagi et al. (2016) used fluorescence-activated cell sorting (FACS) to separate haploids from diploids, followed by validation via visualization of ploidy by DNA fluorescence in situ hybridization (FISH) and quantification of centromere protein foci. To analyze haploid cells and diploidized cells in the same cell cycle phase, the authors used FACS to separately isolate G1-phase cells (before DNA replication) and G2/M-phase cells (after DNA replication). They confirmed on the basis of low Xist expression level and low H3K27me3 staining that the single X chromosome in haploid cells is transcriptionally active in both phases. By contrast, only one of the two X chromosomes is active in both phases in diploid cells (Sagi et al. 2016).
Because Ohno’s hypothesis is about genes that existed before the origin of the mammalian X, we followed previous studies (Xiong et al. 2010; Lin et al. 2012) to focus on human genes having one-to-one orthologs in chicken, and acquired 11,861 autosomal genes and 360 X-linked genes for analysis. We computed the ratio in median mRNA expression level between X-link genes and autosomal genes in haploid cells, and referred to it as the Xa:A expression ratio, because the comparison is between one active X and one set of autosomes. Similarly, we computed the expression ratio between one active X and two sets of autosomes in diploids and referred to it as the Xa:AA expression ratio. An expression ratio of 1 means dosage balance.
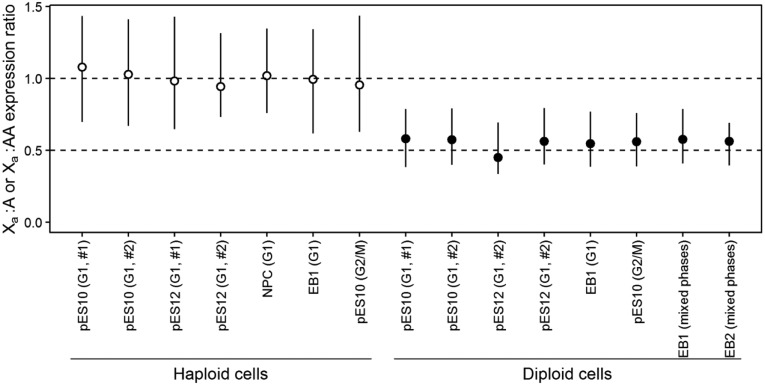
We found the Xa:A expression ratio to be ∼1 in all haploid cell lines studied (fig. 1). Specifically, the 95% confidence interval of the estimated ratio overlaps with 1 but not 0.5 or 2 for each haploid cell line (fig. 1). This result is similar to the previous microarray-based finding in X-carrying spermatids and secondary oocytes (Nguyen and Disteche 2006). By contrast, the Xa:AA expression ratio is ∼0.5, with its 95% confidence interval overlapping with 0.5 but not 1 for each diploid cell line (fig. 1). The Xa:AA ratio in diploids remains close to 0.5 (supplementary fig. S1, Supplementary Material online) when we removed genes known to escape X-inactivation in diploid cells; these genes are expressed from both X chromosomes and therefore are irrelevant to Ohno's hypothesis. Together, these results demonstrate an overall lack of X upregulation at the mRNA level in both haploid and diploid cells, which results in dosage balance in haploids but imbalance in diploids. It should be mentioned that Sagi et al. also observed higher expressions of X-linked genes (relative to all genes) in haploids than diploids using the data analyzed here, but because they assumed dosage balance in diploids, they misinterpreted their results as demonstrating dosage imbalance in haploids (Sagi et al. 2016).
Fig. 1.
General absence of X-chromosome upregulation in human haploid and diploid cells. Shown are the median Xa:A expression ratio in haploid cells (open circles) and median Xa:AA expression ratio in diploid cells (closed circles). Error bars show 95% confidence intervals of the medians, estimated by, respectively, bootstrapping X-linked and autosomal genes 1,000 times. Cell cycle phases (G1 or G2/M) are indicated. #1 and #2 indicate biological replicates. See main text for the meanings of the cell line names.
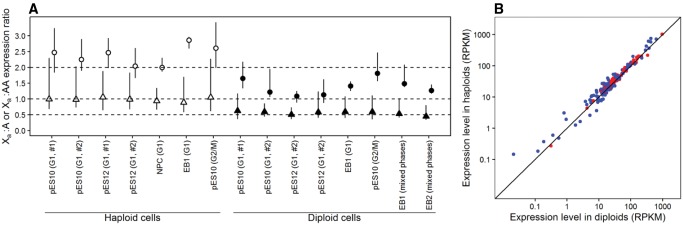
We next examined the X-linked genes that encode members of small and large protein complexes, respectively. For genes encoding members of small complexes, Xa:A is ∼1 in haploid cells and Xa:AA is ∼0.5 in diploid cells (triangles in fig. 2A), similar to the overall pattern of all genes in figure 1. For genes encoding members of large complexes, Xa:AA is ≥1 in diploid cells (circles in fig. 2A). Specifically, in all eight cell lines, the 95% confidence interval of the estimated median Xa:AA ratio surpasses 0.5; it further surpasses 1 in six cases and overlaps with 1 in two cases. Thus, diploid cells show strong upregulation of X-linked genes encoding components of large protein complexes, to a degree that exceeds Ohno's prediction in most of the cell lines examined here. The corresponding Xa:A ratio for these genes significantly surpasses 1 in all haploid cell lines and is close to 2 (fig. 2A). In one case, it is even significantly >2. Thus, these X-linked genes are upregulated in haploids where dosage balance requires no upregulation. As a result, dosage balance is broken in haploids for these presumably dosage-sensitive genes. One potential explanation for the lack of dosage balance for members of large protein complexes in haploid cells is that these complexes are not functionally needed in haploids. However, this hypothesis is unlikely to be true because many complexes play house-keeping functions that should be required by all cells. Indeed, the expression levels of genes encoding components of large complexes are overall not lower in haploids than in diploids. For instance, for the cell line pES10 in G1-phase (replicate #1), 52 large complexes show higher median expressions in haploid than diploid cells, significantly more than that (19) exhibiting the opposite pattern (fig. 2B and supplementary table S1, Supplementary Material online). Of the six cell lines for which matched haploid and diploid expression data are available, only replicate #2 of pES10 in G1-phase has significantly >50% of large complexes with lower median expressions in haploids than diploids (supplementary fig. S2 and table S1, Supplementary Material online). When the data from the six cell lines are combined, >50% of large complexes have higher median expressions in haploids than in diploids (supplementary table S1, Supplementary Material online). In other words, there is no evidence that large protein complexes are less needed in haploids than in diploids.
Fig. 2.
Upregulation of X-linked genes encoding members of large protein complexes in haploid and diploid cells. (A) Median Xa:A expression ratio in haploid cells (open symbols) and median Xa:AA expression ratio in diploid cells (closed symbols) for members of small (triangles) and large (circles) protein complexes. Error bars show 95% confidence intervals estimated by bootstrapping the relevant protein complexes 1,000 times. (B) Median expression levels of protein complex components are not lower in haploids than diploids. Each dot represents either a small (blue) or a large (red) protein complex, showing the median expression levels of genes encoding all components of the complex in pES10 (G1, #1) haploid cells and diploid cells, respectively. The diagonal line shows equal expressions in haploid and diploid cells. The number of circles above the diagonal is significantly greater than that below the diagonal for both blue (P = 7.6 × 10 − 4, two-tailed binominal test) and red (P = 1.1 × 10 − 4) circles. RPKM, number of RNA-seq reads per kilobase of exon per million reads mapped.
In summary, for the vast majority of genes on X, there is no hypothesized 2-fold upregulation in either haploid or diploid cells. Consequently, their expression levels are imbalanced with those of autosomal genes in diploids but are naturally balanced in haploids. For ∼5% of X-linked genes encoding members of large protein complexes, upregulation is observed in both haploid and diploid cells, resulting in an approximate dosage balance in diploids but imbalance in haploids. It may be that, because the human genome spends more time in diploids than in haploids, natural selection for dosage balance of these dosage-sensitive genes is stronger in diploids than in haploids, resulting in the upregulation of these genes in diploids. But the fact that ploidy-specific expression regulation of these genes did not evolve suggests that even for these dosage-sensitive genes, dosage imbalance is apparently tolerated (in haploids). Taken together, our results argue against Ohno's hypothesis for both haploid and diploid cells and demonstrate that, at least in humans, precise gene regulation for dosage balance, even for members of large protein complexes, are much less critical than is commonly thought (Veitia et al. 2015). In the future, it would be interesting to generate proteomic data from human haploid cells to allow validating the above findings at the proteomic level, as was recently done in human diploid cells (Chen and Zhang 2015). It would also be interesting to confirm our results by comparing human haploid transcriptomic or proteomic data with the corresponding data from a bird, as was previously done regarding diploid transcriptomic results (Julien et al. 2012; Lin et al. 2012). Finally, the upregulation of the X-linked genes encoding large protein complexes awaits a mechanistic understanding.
Materials and Methods
Gene models as well as mapping of EnsEMBL gene IDs to UniProt/SwissProt accessions in human were downloaded from EnsEMBL (release 80) (Cunningham et al. 2015). Human and chicken one-to-one orthologs were also downloaded from the same release of EnsEMBL. The number of RNA-seq reads per kilobase of exon per million reads mapped (RPKM), supplied by the original authors of the study (Sagi et al. 2016), was used to measure the gene expression level in a sample. X inactivation was previously assessed by analyzing the expressions of 401 X-linked genes in a panel of rodent/human somatic cell hybrids using reverse transcription polymerase chain reaction (Carrel and Willard 2005). Following previously set criteria, we considered a gene in Carrel and Willard's data to escape from X inactivation if the expression of the gene from an inactive X exceeds 10% of the corresponding expression from an active X in at least eight of nine biological replicates (Sharp et al. 2011). The gene models from EnsEMBL were used to transform the gene names in Carrel and Willard’s data to EnsEMBL Gene IDs. In the end, 58 genes were identified to have escaped from X inactivation, of which 25 have one-to-one orthologs in chicken. Following a previous study (Pessia et al. 2012), we obtained the list of members of human protein complexes from HPRD release 9 (www.hprd.org). The gene models from EnsEMBL were used to transform protein complex members in HPRD to EnsEMBL Gene IDs. Following previous studies (Lin et al. 2012; Pessia et al. 2012), we designated human protein complexes with seven or more members as large complexes and those with six or fewer members as small complexes. This resulted in 552 autosomal and 39 X-linked genes encoding members of 72 large complexes, and 285 autosomal and 71 X-linked genes encoding members of 133 small complexes. Similar to the main analysis, we then focused on genes that have one-to-one orthologs in chicken. For each protein complex that is encoded by at least one X-linked gene with gene expression data and one autosomal gene with gene expression data, we computed the ratio between the median gene expression level for the X-linked genes and that for the autosomal genes. We then computed the median of the above ratio for all protein complexes for each cell line.
Supplementary Material
Supplementary figures S1–S2 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Dr Soochin Cho for valuable comments. This work was supported in part by U.S. National Institutes of Health grant R01GM103232 to J.Z. and by Sun Yat-Sen University and the Program of Recruitment of Global Experts to X.C.
References
- Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1996. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 6:149–162. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J. 2015. No X-chromosome dosage compensation in human proteomes. Mol Biol Evol. 32:1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daish TJ, Grutzner F. 2010. Meiotic sex chromosome inactivation In: Encyclopedia of life sciences. Chichester: John Wiley & Sons; p. 1–9. [Google Scholar]
- He X, Zhang J. 2016. X-chromosome dosage compensation In: Encyclopedia of life sciences. Chichester: John Wiley & Sons; p. 1–7. [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, Daish T, Grutzner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10:e1001328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. 2012. Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 109:11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet. 38:47–53. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. New York: Springer-Verlag. [Google Scholar]
- Payer B, Lee JT. 2008. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 42:733–772. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. 2012. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 109:5346–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi I, Chia G, Golan-Lev T, Peretz M, Weissbein U, Sui L, Sauer MV, Yanuka O, Egli D, Benvenisty N. 2016. Derivation and differentiation of haploid human embryonic stem cells. Nature 532:107–111. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. 2011. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 21:1592–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. 2015. Meiotic silencing in mammals. Annu Rev Genet. 49:395–412. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Veyrunes F, Bottani S, Birchler JA. 2015. X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J Mol Cell Biol. 7:2–11. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Chen X, Chen Z, Wang X, Shi S, Wang X, Zhang J, He X. 2010. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 42:1043–1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.