Abstract
Acidic mammalian chitinase (AMCase) is implicated in asthma, allergic inflammation, and food processing. Little is known about genetic and evolutional regulation of chitinolytic activity of AMCase. Here, we relate human AMCase polymorphisms to the mouse AMCase, and show that the highly active variants encoded by nonsynonymous single-nucleotide polymorphisms (nsSNPs) are consistent with the mouse AMCase sequence. The chitinolytic activity of the recombinant human AMCase was significantly lower than that of the mouse counterpart. By creating mouse-human chimeric AMCase protein we found that the presence of the N-terminal region of human AMCase containing conserved active site residues reduced the enzymatic activity of the molecule. We were able to significantly increase the activity of human AMCase by amino acid substitutions encoded by nsSNPs (N45, D47, and R61) with those conserved in the mouse homologue (D45, N47, and M61). For abolition of the mouse AMCase activity, introduction of M61R mutation was sufficient. M61 is conserved in most of primates other than human and orangutan as well as in other mammals. Orangutan has I61 substitution, which also markedly reduced the activity of the mouse AMCase, indicating that the M61 is a crucial residue for the chitinolytic activity. Altogether, our data suggest that human AMCase has lost its chitinolytic activity by integration of nsSNPs during evolution and that the enzyme can be reactivated by introducing amino acids conserved in the mouse counterpart.
Keywords: acidic mammalian chitinase, amino acid substitutions, chitinolytic activity, pseudogene, reactivation, single-nucleotide polymorphisms
Introduction
Chitin, a linear β-1, 4-linked polymer of N-acetyl-D-glucosamine (GlcNAc), is one of the most abundant carbohydrate polymers in nature and it is a major component of fungal cell walls and exoskeletons of invertebrates (Aam et al. 2010; Khoushab and Yamabhai 2010).
Despite the absence of endogenous chitin, humans and mice express two active chitinases, chitotriosidase (Chit1) and acidic mammalian chitinase (AMCase) (Renkema et al. 1995; Boot et al. 2001; Lee et al. 2011; Bueter et al. 2013). First human chitinase, Chit1, was identified in Gaucher disease patients (Hollak et al. 1994; Renkema et al. 1995). AMCase, encoded by the CHIA gene, was the second discovered mammalian chitinase and was named for its acidic isoelectric point (Boot et al. 2001).
AMCase has attracted considerable attention due to its increased expression under specific pathological conditions. For example, significant increases in AMCase mRNA and protein levels were detected in an induced asthma mouse model (Zhu et al. 2004) as well as in antigen-induced mouse models of allergic lung inflammation (Reese et al. 2007). In humans, AMCase has been shown to be increased in lungs of allergen-exposed patients with asthma and in alveolar macrophages in cases of fatal asthma (Zhu et al. 2004). AMCase is highly expressed in mouse stomach. A robust peak of activity was observed at pH 2.0, suggesting that AMCase can function as a digestive enzyme that breaks down chitin also as part of the host defense against chitin-containing pathogens in the gastric contents (Boot et al. 2001; Ohno et al. 2012, 2013; Kashimura et al. 2015).
Multiple AMCase variants have been identified based on single nucleotide polymorphisms (SNPs) and associated with asthma risk in humans (Bierbaum et al. 2005; Chatterjee et al. 2008). Seibold et al. (2009) described nonsynonymous SNPs (nsSNPs) that variably endowed isoforms of human AMCase with differential enzymatic activity and showed that a haplotype encoding an isoform of AMCase with heightened enzyme activity was associated with protection from asthma in humans. No detailed knowledge is available regarding genetic and evolutional regulation of chitinolytic activity of AMCase.
In this study, we show that chitinolytic activity of human AMCase is significantly lower than that of the mouse counterpart and investigate the chitinolytic activity of naturally occurring human AMCase variants encoded by nsSNPs. We relate these polymorphisms to the active mouse AMCase, and show that the highly active variants contain SNPs consistent with the mouse AMCase sequence.
Results
Chitinolytic Activity of Human AMCase Is Significantly Lower than That of the Mouse AMCase
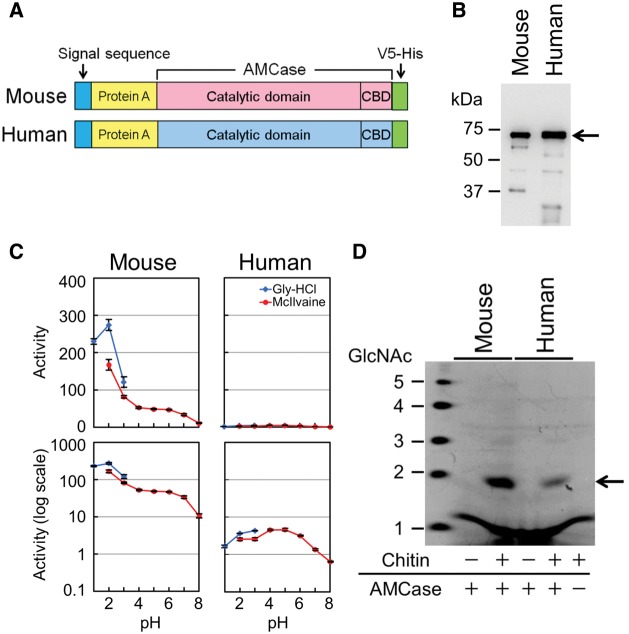
We expressed mouse and human AMCase in the periplasmic space of E. coli as a fusion protein containing Staphylococcus aureus Protein A and V5-His tag (Kashimura et al. 2013) as described in the “Materials and Methods” section (fig. 1A and B; supplementary figure S1, Supplementary Material online). To compare the chitin hydrolytic activities of these recombinant proteins, we used 4-methylumbelliferyl (4-MU) N,N’-diacetyl-β-D-chitobioside and colloidal chitin, a fluorogenic and a high molecular weight substrate, respectively.
Fig. 1.
Chitinolytic activity of human AMCase is significantly lower as compared with mouse AMCase. (A) Schematic representation of E. coli-expressed mouse and human AMCase fusion proteins. (B) Western blot analysis of the recombinant proteins using anti-V5 antibody. Arrow highlights the positions of the fusion proteins (Protein A-AMCase-V5-His). (C) Comparison of the chitinolytic activities of mouse and human AMCase proteins using fluorogenic substrate. The values represent actual scale (upper panels) and log scale (lower panels). Error bars represent the mean ± standard deviation from a single experiment conducted in triplicate. (D) Degradation products of colloidal chitin by mouse and human AMCase proteins. Chitin fragments generated by the recombinant proteins were analyzed by fluorophore-assisted carbohydrate electrophoresis. Chitin oligomers are shown in the left margin as standards. Both recombinant proteins released mainly (GlcNAc)2 fragments (highlighted with the arrow).
Consistently with previous reports, recombinant mouse AMCase had the highest activity at pH 2.0 (Boot et al. 2001; Kashimura et al. 2013), whereas the recombinant human AMCase was most active at around pH 4.0–5.0 (Chou et al. 2006; Seibold et al. 2009; Goedken et al. 2011) with the human enzyme being significantly less active as compared with the mouse AMCase (fig. 1C; supplementary table S1_1 and S1_2, Supplementary Material online; P < 0.01). The chitinolytic activity of the human AMCase was 1/75 and 1/11 of that of the mouse AMCase at pH 2.0 and at pH 4.0, respectively (supplementary table S1_1, Supplementary Material online), which were essentially consistent with previous reports (Goedken et al. 2011). Human AMCase was able to degrade the colloidal chitin producing primarily N,N′-diacetylchitobiose [(GlcNAc)2] fragments at pH 2.0, however its hydrolyzing activity was significantly lower than that of the mouse AMCase (fig. 1D; supplementary table S1_3, Supplementary Material online). There was no significant difference in the chitinolytic activities of the recombinant proteins analyzed by a fluorogenic substrate of 4-MU N,N′-diacetyl-β-D-chitobioside and colloidal chitin using fluorophore-assisted carbohydrate electrophoresis (supplementary table S1_2 and S1_3, Supplementary Material online).
N-Terminal Region in Human AMCase Reduces Chitinolytic Activity
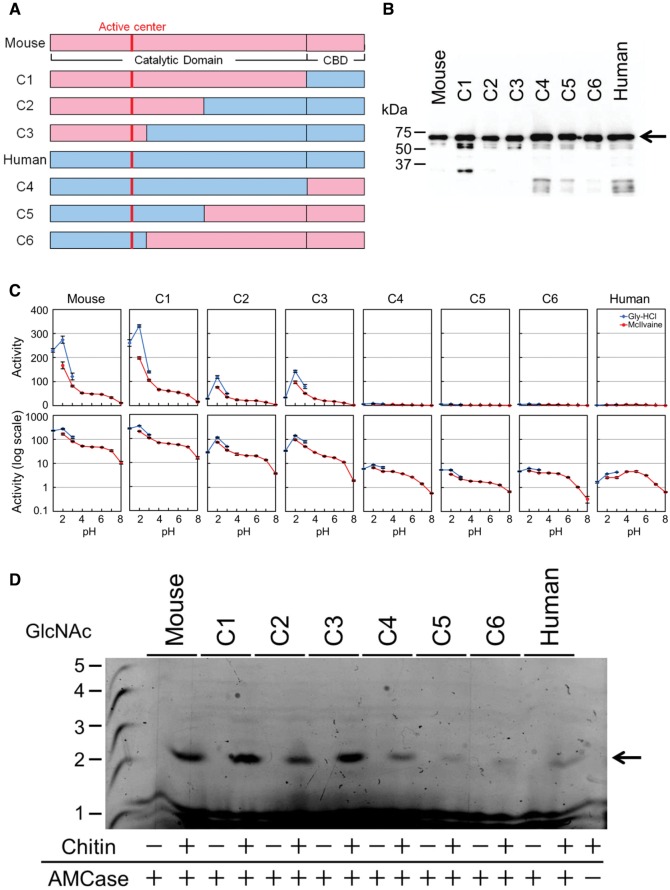
Although human and mouse AMCase proteins share 82% sequence identity and 86% sequence similarity (Boot et al. 2001), these two homologues significantly differ with respect to their enzymatic activities (fig. 1C and D; supplementary table S1, Supplementary Material online). To determine regions responsible for the reduced chitinolytic activity in human AMCase, we expressed chimeric human–mouse AMCase proteins in E. coli (fig. 2A and B; supplementary fig. S2 and supplementary table S2, Supplementary Material online) and examined the influence of the catalytic domain (CatD) and the chitin-binding domain (CBD). Chimeras C1, C2, and C3, which all contained mouse N-terminal region, exhibited strong chitinolytic activity toward the fluorogenic substrate comparable to mouse AMCase, however the activity was dropping with increasing content of human AMCase sequence (fig. 2C; supplementary table S2_1, Supplementary Material online; P < 0.01). In contrast, the chitinolytic activities were very low in chimeras C4, C5, and C6 as compared with mouse AMCase (fig. 2C; supplementary table S2_1, Supplementary Material online; P < 0.01) and similar to that of the human enzyme (fig. 2C; supplementary table S2_1, Supplementary Material online; P < 0.01). Although each of the chimeric proteins released mainly (GlcNAc)2 fragments from colloidal chitin at pH 2.0, chimeras C4, C5, and C6 and human AMCase produced these fragments at very low levels (fig. 2D; supplementary table S2_2, Supplementary Material online). Thus, the N-terminal region with conserved active site amino acid residues in human AMCase significantly reduced chitinolytic activity.
Fig. 2.
Presence of N-terminal region derived from human AMCase containing active center significantly reduced the chitinolytic activity of chimeric enzymes. (A) Schematic representation of E. coli-expressed AMCase chimeric proteins. The amino acid sequences are color coded as follows: pink, mouse sequence; blue, human sequence. (B) Western blot analysis of the expressed AMCase chimeric proteins as described in figure 1B. (C) Comparison of the chitinolytic properties of chimeric proteins with mouse and human AMCase in figure 1C. (D) Degradation products of colloidal chitin by mouse AMCase, human AMCase, and six AMCase chimeric proteins.
Chitinolytic Activity of Human AMCase Is Controlled by Amino Acid Substitutions
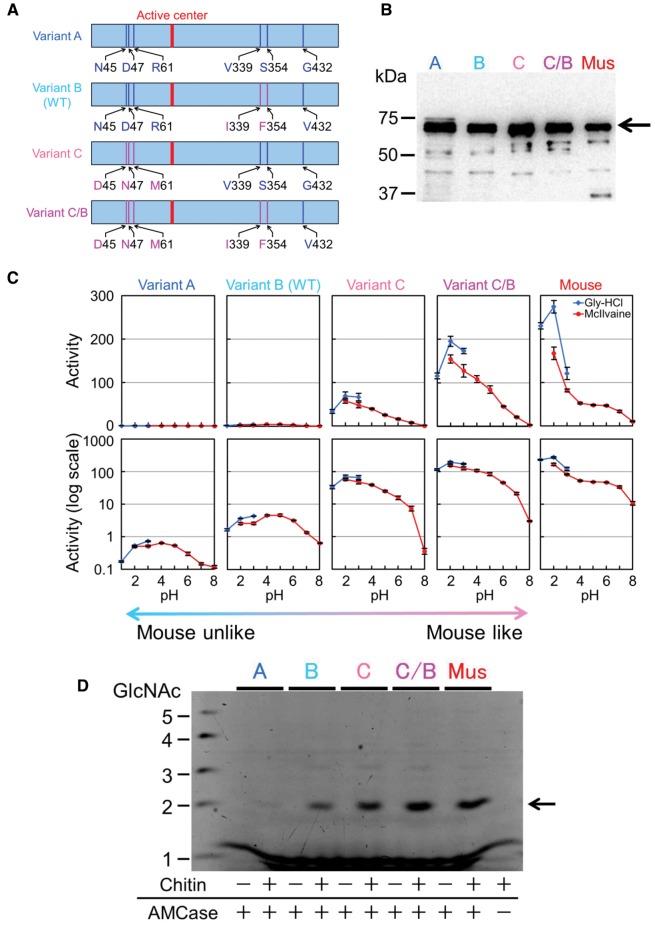
Eight nsSNPs including three novel variants [A290G (N45D), G296A (D47N), and G339T (R61M)] near the area coding for the enzyme active site have been identified (Seibold et al. 2009). We constructed three naturally occurring variants (A, B, and C) and one artificial protein variant (C/B) to examine the effect of six of these polymorphisms present in the catalytic domain and/or hinge region on the chitinolytic activity of the human AMCase (fig. 3A; supplementary fig. S3A and B, Supplementary Material online). In variant A, all substituted amino acids differ from those in mouse AMCase (fig. 3A; supplementary fig. S3A and B, Supplementary Material online). Variant B corresponds to the so-called “wild-type” (WT) human AMCase described in previous reports (Boot et al. 2001; Chou et al. 2006; Seibold et al. 2009). In contrast to variants A and B, three amino acid substitutions of the N-terminal region in variant C are in agreement with the mouse homologue (fig. 3A; supplementary fig. S3A and B, Supplementary Material online). Additionally, we constructed variant C/B with the N- and C-terminal regions consistent with variants C and B, respectively (fig. 3A and B; supplementary fig. S3A and B, Supplementary Material online).
Fig. 3.
Chitinolytic activity of human AMCase can be controlled by amino acid substitutions. (A) Schematic representation of E. coli-expressed human AMCase variants. Human AMCase amino acid substitutions, which are not conserved in the mouse counterpart are shown in blue. The amino acid substitutions conserved in the mouse counterpart are shown in pink. (B) Western blot analysis of tested recombinant AMCase variants. (C) Comparison of the chitinolytic activities of human AMCase variants with mouse AMCase as described in figure 1C. (D) Degradation products of colloidal chitin by human AMCase variants and mouse AMCase. A, variant A; B, variant B; C, variant C; C/B, variant C/B; Mus, mouse.
Chitinolytic activity of variant A was much lower than that of the mouse AMCase (fig. 3C; supplementary table S3_1, Supplementary Material online; P < 0.01). The activity of variant B (WT human AMCase) was approximately 5-fold higher than that of variant A and variant C had about 100- and 20-fold higher activity than variant A and B, respectively (fig. 3C; supplementary table S3_1, Supplementary Material online; P < 0.01). In addition, optimal pH for variants A and B was at around pH 4.0–5.0, whereas variant C showed highest chitinolytic activity at pH 2.0 (fig. 3C; supplementary table S3_1, Supplementary Material online). Surprisingly, the activity of variant C/B was substantially higher and reached the level of the mouse AMCase at pH 2.0 (fig. 3C; supplementary table S3_1, Supplementary Material online). Although each of the human variants released primarily (GlcNAc)2 fragments from the colloidal chitin at pH 2.0, variant A produced very low amounts of these fragments whereas variant C/B had equal activity as the mouse AMCase (fig. 3D; supplementary table S3_2, Supplementary Material online). Thus, amino acid substitutions encoded by nsSNPs at the N-terminal region in human AMCase had distinct effects on the chitinolytic activity and we were able to reactivate the human AMCase by introducing three residues (N45D, D47N, and R61M), which were conserved in the mouse homologue (fig. 3A and B).
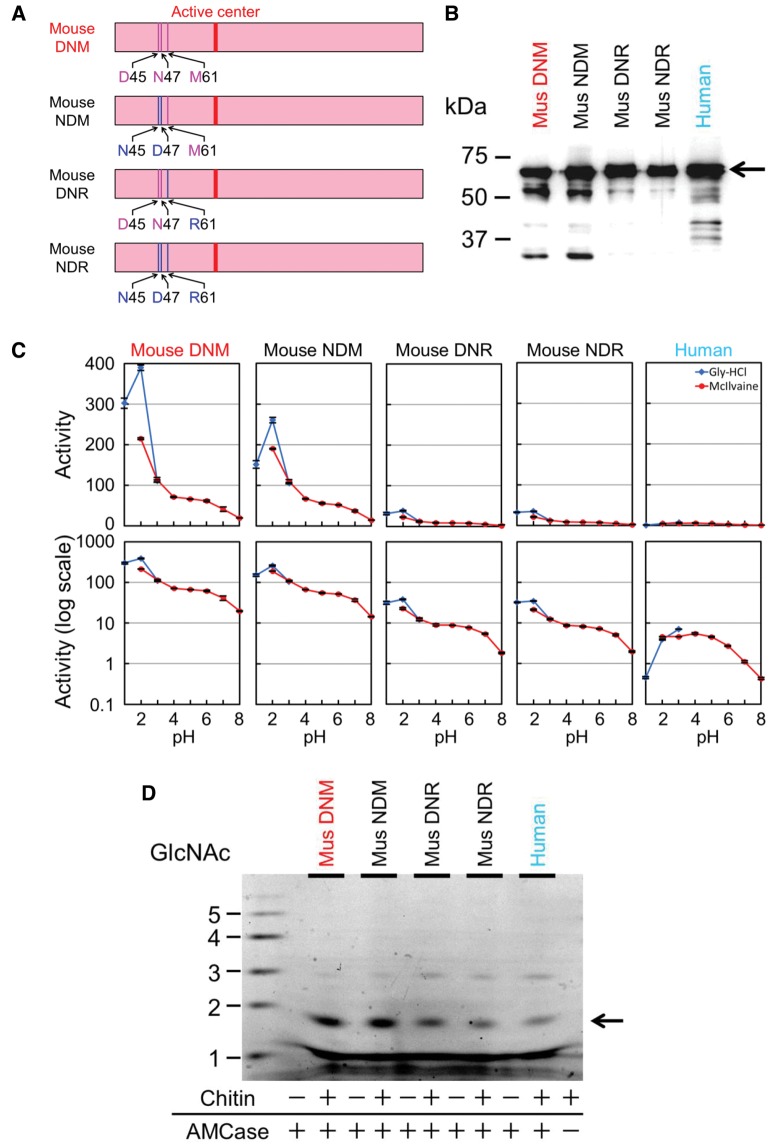
Inactivation of Mouse AMCase by Introducing M61R Mutation
We next attempted to dissect the effect of three amino acid substitutions at the N-terminal region on the chitinolytic activity of mouse AMCase. Amino acid residues in WT mouse AMCase are D45, N47, and M61 (we named this type DNM) in the N-terminal region (fig. 4A). We constructed mutant mouse AMCase proteins named NDM (N45, D47, and M61), DNR (D45, N47, and R61), and NDR (N45, D47, and R61) (underlined are mutated amino acids; fig. 4A and B). The chitinolytic activity of NDM was at the same level as that of the WT mouse AMCase (DNM) (fig. 4C; supplementary table S4_1, Supplementary Material online). In contrast, the activities of DNR and NDR were significantly lower than that of the WT mouse AMCase and comparable to the WT (variant B) human enzyme (fig. 4C; supplementary table S4_1, Supplementary Material online; P < 0.01). Both DNR and NDR mutants produced very low amounts of (GlcNAc)2 fragments whereas the NDM mutant had equal activity as the WT mouse AMCase (fig. 4D; supplementary table S4_2, Supplementary Material online). Thus, D45N and N47D substitution have low effects, whereas M61R substitution found in human AMCase has a crucial role in the activity of the enzyme.
Fig. 4.
Inactivation of mouse AMCase by introducing M61R mutation. (A) Schematic representation of WT (DNM) and mutant proteins (NDM, DNR, and NDR) mouse AMCase. Western blot analysis of the recombinant AMCase variants (B), comparison of the chitinolytic activities of mouse AMCase mutants (C) and degradation products of colloidal chitin by mouse AMCase mutants (D) were carried out as described in figure 1B–D.
M61 Is the Crucial Residue for High Chitinolytic Activity
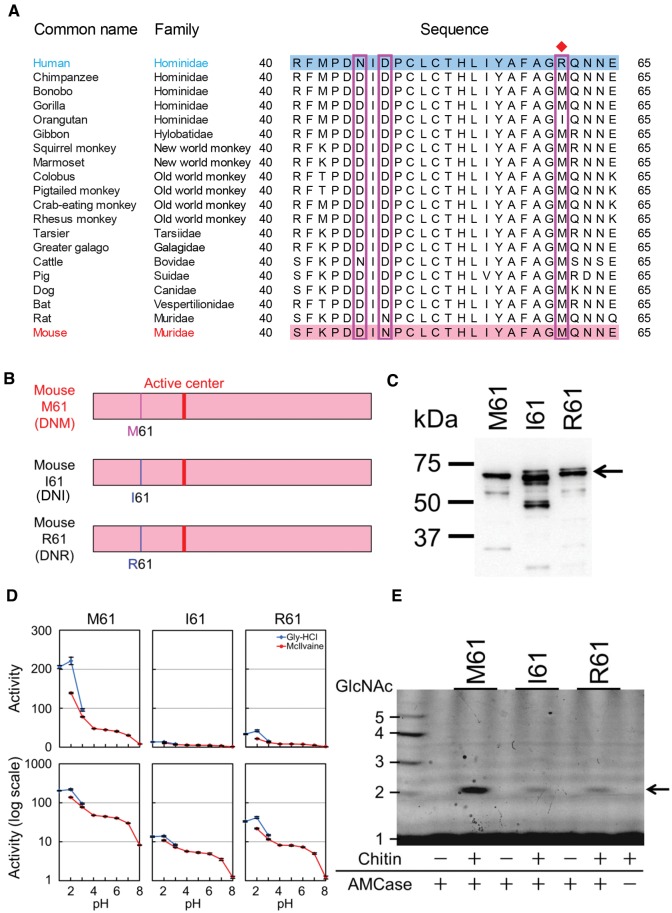
We further examine the amino acid at the position 61 at the N-terminal region of AMCase in New World monkeys, Old World monkeys, and hominoids as well as distantly related mammals. M61 is conserved in mammals as well as in most primates other than orangutan (Pongo abelii) and human (fig. 5A; supplementary table S5_1, Supplementary Material online). Orangutan and human have M61I and M61R substitutions, respectively.
Fig. 5.
M61 is a crucial residue for high chitinolytic activity. (A) Alignments of the amino acid at position 61 in New World monkeys, Old World monkeys, and hominoids as well as distal mammals. M61 is conserved in mammals as well as in most primates other than Orangutan and human. Orangutan has M61I substitution, whereas human WT AMCase is M61R. Red diamond indicates the amino acid at the position 61. (B) Schematic representation of WT (M61) and mutant (I61 and R61) mouse AMCase proteins. Western blot analysis of the recombinant mouse AMCase proteins (C), comparison of the chitinolytic activities of mouse WT and mutant AMCase proteins (D), degradation products of colloidal chitin by mouse AMCase mutants (E) were carried out as described in figure 1B–D.
We next examined the effect of the M61I substitution on the chitinolytic activity of WT mouse AMCase. We constructed mutant mouse AMCase proteins named I61 (DNI, orangutan) and R61 (DNR, human) as well as M61 (DNM, WT mouse AMCase). Chitinolytic activities of I61 and R61 were significantly lower when compared with that of the WT mouse AMCase (fig. 5D and E; supplementary table S5_2 and S5_3, Supplementary Material online). Thus, both M61R and M61I substitutions have major effects on the whole enzyme activity. Based on these results, we concluded that the M61R mutation is specific for humans and that Methionine at this position (M61) is the crucial residue for high chitinolytic activity.
Evolutionary Analysis of AMCase in Hominoidea
As shown above, M61R nonsynonymous substitution resulted in a significant reduction of chitinolytic activity of the human enzyme. As the WT human AMCase has lost the function, an excess of nonsynonymous mutations in the gene is anticipated due to the relaxation of functional constraint. We compared the ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS) in humans and other hominoidea.
To examine the evolutionary history of AMCase genes, we constructed a phylogeny and calculated dN/dS ratio using calculator PAML (Xu and Yang 2013). When we compared the dN/dS ratio in hominoidea, the value of human AMCase was 3.04, indicating that human AMCase underwent natural selection (Darwinian selection) (fig. 6A). It is interesting to note that AMCase is undergoing purifying selection in other hominoidea.
Fig. 6.
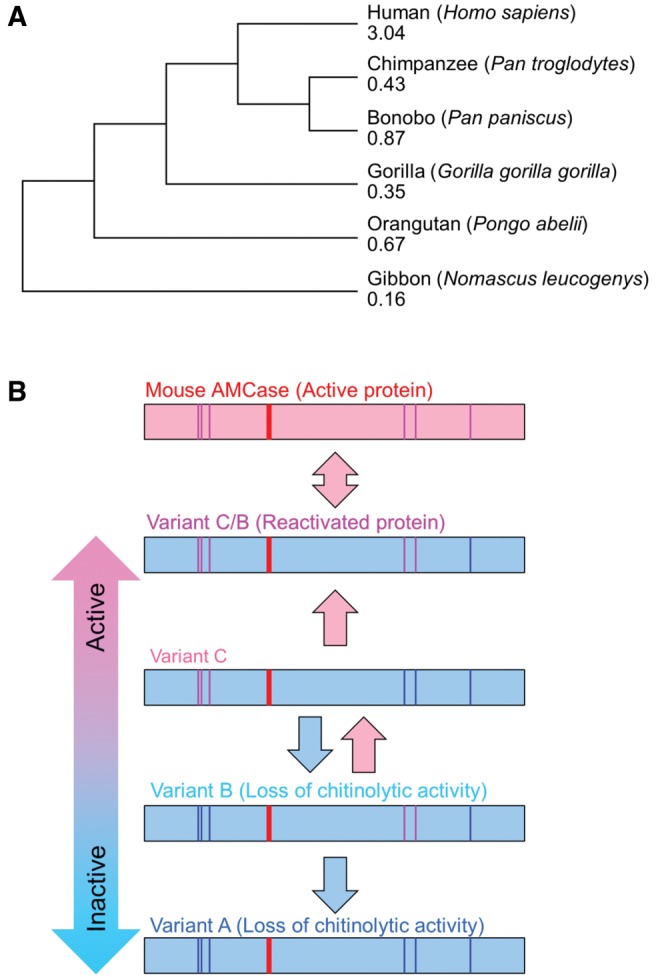
Evolutionary analysis of AMCase. (A) Human AMCase is undergoing natural selection. Phylogenetic tree was constructed by the unweighted pair group method with arithmetic mean (UPGMA). Rate ratios of nonsynonymous-to-synonymous substitutions dN/dS (ω) were calculated by PAMLX and indicated (shown below the species name). (B) Loss and gain of chitinolytic activity by amino acid substitutions in human AMCase. Human AMCase variants were defined by single nucleotide polymorphisms. The chitinolytic activity of these variants depended on location and pattern of the amino acid substitutions. The chitinolytic activity in human AMCase can be recovered by the combination of the N-terminal sequence of variant C and the C-terminal region of variant B. Resulting novel variant C/B had high specific activity comparable with the mouse homologue.
Discussion
In this study, we investigated chitinolytic activity of naturally occurring human AMCase variants encoded by nsSNPs. The activity of the human enzyme was significantly reduced by specific amino acids that are not present in the mouse AMCase. Variants A and B (fig. 3A) could be considered as the enzyme isoforms whose activities have been lost by the amino acid substitutions. Thus, one could hypothesize that human AMCase became an inactive pseudogene during the course of evolution (fig. 6B, lower part). This assumption is supported by a comparison with primates as well as distantly related mammals (fig. 5A; supplementary table S5_1, Supplementary Material online). Pseudogenization is a genomic evolutionary phenomenon which has been thought to provide opportunities for phenotypic adaptations by gene function loss (Wang et al. 2006).
As shown in fig. 6A, only human AMCase showed the high dN/dS ratio, whereas the evolutionary event of the AMCase gene in other hominoidea is close to be neutral. Thus, human AMCase is likely to be a novel gene undergoing loss of function during the course of evolution, providing an opportunity for some unknown phenotypic adaptation. In addition to the reduced chitinolytic activity of human AMCase, the expression level of AMCase mRNA and its product were relatively low in the human stomach when compared with that in mouse (Ohno et al. 2013). Since modern humans do not consume a significant amount of chitin-containing foods, this is not unexpected. Many stomach diseases are associated with infection by exogenous organisms. The severity of gastritis-associated infections such as Helicobacter pylori may correlate with the activity of endogenous enzymes (Cozzarini et al. 2009; Nookaew et al. 2013). It thus remains to be determined whether the low level of chitinolytic activity of AMCase in the human stomach participates in the response to gastric disorders.
Biochemical data presented in this report for WT human AMCase expressed in E. coli was essentially similar to the previous work by Seibold et al. expressed in insect cells (Seibold et al. 2009) and Goedken et al. (2011) in COS cells. These results indicate that our recombinant proteins recapitulate key biochemical features of AMCase that were obtained from different sources in the previous studies and would validate the use of E coli-expressed AMCase proteins in the subsequent chimeric and mutational analyses.
Seibold et al. (2009) already identified eight nonsynonymous single nucleotide polymorphisms including three novel variants (A290G, G296A, and G339T) near the gene area coding for the enzyme active site and showed that the AMCase variant G339T (R61M) is associated with asthma protection in some populations. Here we showed that M at the position 61 is highly conserved in the many mammals other than orangutan and humans (fig. 5A). In addition, introduction of M61R and M61I to WT mouse AMCase lead to a significant reduction of its chitinolytic activity (figs. 4 and 5). Taken together, our present and previous data by Seibold et al. (2009) indicate that G339T (R61M) in humans is associated with asthma protection in the fashion of gain of function.
Since the allele frequency of G339T is 26% in African Americans (Seibold et al. 2009), G339T (R61M) is known to be a nsSNP found in the modern human populations (rs41282496). Recently, the genomes of Neandertals and Denisovans have been sequenced (Green et al. 2010; Meyer et al. 2012; Prufer et al. 2014). It will be interesting to determine whether they carry M61 and/or R61 in their AMCase genome.
We were able to reactivate the human AMCase by combining two relatively inactive isoforms present in humans. Resulting new variant, C/B (fig. 3A), containing five amino acids conserved in mouse homologue, had high chitinolytic activity and pH profile comparable to the mouse AMCase (fig. 3C and D; fig. 6B, upper part). These data indicated that specific amino acids introduced to the enzyme can reduce its activity (particularly M61R and M61I substitutions are critical for the chitinolytic activity), whereas presence of amino acids conserved in active enzymes reactivate the human enzyme to the level of the mouse homologue. In addition, we determined specific amino acids by nsSNPs which are responsible for reducing chitinolytic activity of human AMCase. The concept outlined here could be utilized for enhancement of the enzyme activity by minimal genetic manipulation, e.g., by genome-editing technologies, in the future (fig. 6B, upper part).
Elevated or reduced AMCase levels have been reported in numerous diseases such as asthma, allergic inflammation, ocular allergy, dry eye syndrome, stomach cancer, adenoid hypertrophy, conjunctivitis, neuromyelitis, gastritis, or nasal polyp formation (Zhu et al. 2004; Reese et al. 2007; Bucolo et al. 2008; Seibold et al. 2008; Cozzarini et al. 2009; Musumeci et al. 2009; Park et al. 2009; Bucolo et al. 2011; Correale and Fiol 2011; Heo et al. 2011; Nookaew et al. 2013). In addition, AMCase is an important downstream effector of interleukin-13 stimulation in Th2 helper cell-mediated immune responses to pathogens, parasites and ovalbumin (Zhu et al. 2004; Hartl et al. 2009). Although the chitinolytic activity of the human AMCase was significantly lower than that of the mouse AMCase, our novel C/B variant containing five amino acids conserved in mouse AMCase had high chitinolytic activity and pH profile comparable to the mouse AMCase. Thus, our recombinant proteins could be useful for understanding pathophysiological roles of AMCase in humans. This research could potentially lead to development of AMCase-related therapies as well as for the treatment of specific human diseases resulting from low levels and/or activity of AMCase, e.g., by supplementation therapy.
Materials and Methods
RNA, AMCase cDNA Preparation, and E. coli Expression Vectors
We used human stomach total RNA from the Human Total RNA Master Panel II (Clontech Laboratories) and reverse transcribed the RNA into cDNA, as previously described (Ohno et al. 2012). Mature Human AMCase was amplified from the human stomach cDNA by PCR using KOD Plus DNA polymerase (Toyobo) and oligonucleotide primers (Sigma-Aldrich Life Science Japan) anchored with the restriction sites for EcoRI and XhoI (supplementary data file S1, Supplementary Material online) as described previously (Kashimura et al. 2013). Amplified cDNA was digested with EcoRI and XhoI and cloned into the same sites of the pEZZ18/pre-Protein A-AMCase-V5-His (mouse version) (Kashimura et al. 2013). The entire nucleotide sequence of the resulting plasmid DNA (pEZZ18/human AMCase variant/V5-His) was confirmed by sequencing (Eurofins Genomics). Our initial human AMCase cDNA corresponds to variant C (see fig. 3A and supplementary fig. S3A, Supplementary Material online).
Site-Directed Mutagenesis by Primer Extension
We constructed human variant A, B, and C/B as well as chimeric and mutant proteins essentially by the method of site-directed mutagenesis by primer extension (Reikofski and Tao 1992). Detailed methods are described in supplementary text and supplementary data file S1, Supplementary Material online.
Preparation of the Recombinant Human and Mouse AMCase Proteins Expressed in E. coli
Using the plasmid DNAs (the pEZZ18/pre-Protein A-AMCase-V5-His), we transformed E. coli BL21 (DE3) (Novagen) to express pre-Protein A-AMCase-V5-His proteins (figs. 1A, 2A, 3A, 4A, and 5B). Transformed E. coli BL21 (DE3) strains were grown in 1 L LB medium containing 100 µg/mL ampicillin at 37 °C for 18 h. Cells were harvested by centrifugation at 5,000 x g for 20 min at 4 °C. The recombinant protein was prepared from the periplasmic fraction of E. coli and purified by IgG Sepharose chromatography as described previously (Kashimura et al. 2013). The protein-containing fractions were desalted using PD MidiTrap G-25 (GE Healthcare) equilibrated with the TS buffer [20 mM Tris-HCl (pH 7.6), 150 mM NaCl and a protease inhibitor (Complete, Roche)]. We also produced Protein A-WT mouse AMCase-V5-His and Protein A-V5-His as described previously (Kashimura et al. 2013).
SDS-Polyacrylamide Gel Electrophoresis and Western Blot
The obtained protein fractions were analyzed using standard SDS-polyacrylamide gel electrophoresis (PAGE), followed by Western blot using anti-V5-HRP monoclonal antibody (Invitrogen). The immunoblots were analyzed and quantified using the Luminescent Image Analyzer (ImageQuant LAS 4000, GE Healthcare) according to the manufacturer’s instructions.
Chitinase Enzymatic Assays
Chitinase enzyme activity was determined with the fluorogenic substrate 4-MU β-D- N, N′-diacetylchitobioside hydrate (Sigma-Aldrich) as a substrate in McIlvaine’s buffer (0.1 M citric acid and 0.2 M Na2HPO4; pH 2.0 to pH 8.0) or 0.1 M Gly-HCl buffer (pH 1.0 to pH 3.0) at 37 °C for 30 min as described previously (Ohno et al. 2013). The fluorescence of released 4-MU was measured using spectrofluorophotometer RF-5300PC (Shimadzu) with excitation at 360 nm and emission at 450 nm. The amount of product (4-MU) was estimated using a standard curve based on 4-MU (Sigma-Aldrich).
Degradation of Colloidal Chitin by AMCase
Colloidal chitin was prepared from shrimp shell chitin (Sigma-Aldrich) as described previously and used as a substrate to determine the chitinase activity (Kashimura et al. 2013). All enzymatic reactions using colloidal chitin (at a final concentration of 1 mg/mL) as substrates were carried out in a volume of 50 μL containing E. coli-expressed proteins in McIlvaine’s buffer (pH 2.0). The reactions were incubated for 16 h at 37 °C. The chitin fragments generated from these reactions were labeled covalently at their reducing end groups with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid (ANTS, Invitrogen), and the resulting fluorescent derivatives were separated by high-resolution PAGE, as described by Jackson (1990) and quantified using the Luminescent Image Analyzer as described above. N-acetyl chitooligosaccharides (Seikagaku Corporation) were used as standards.
Phylogenetic Analysis
cDNA sequences of hominoidea AMCase were retrieved from the NCBI GenBank (supplementary data file S2, Supplementary Material online). A phylogenetic tree was constructed by the unweighted pair group method with arithmetic mean (UPGMA) method with MEGA 7 (http://www.megasoftware.net) (Kumar et al. 2016). Bootstrap analyses were conducted with 1,000 replicates. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are expressed in units of the number of base substitutions per site.
Calculation of Substitution Rates
Rate ratios of nonsynonymous-to-synonymous substitutions dN/dS (ω) were calculated by PAMLX software, version 1.3.1 (Xu and Yang 2013). We employed a free-ratio model to allow the dN/dS ratios to vary for each branch.
Statistical Analyses
Data are shown as mean with standard deviation (SD). Chitinolytic activities were compared by Student’s t-test.
Supplementary Material
Supplementary figures S1–S3, tables S1–S5, text and data files S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Haruko Miyazaki and Nobuyuki Nukina for their encouragement, to Shotaro Honda, Satoshi Wakita, Daisuke Mizutani, Eri Tabata and Yasutada Imamura for valuable suggestions. This work was supported by a Grant from the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan (to F.O.); by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (grant numbers 15J10960, 16K07699 to M.O. and F.O., respectively); by the Project Research Grant from the Research Institute of Science and Technology, Kogakuin University (to F.O.) and in part by a grant of the Strategic Research Foundation Grant-aided Project for Private Universities (S1411005) from the Ministry of Education, Culture, Sport, Science and Technology, Japan (to M.S. and F.O.). P.O.B. received support from ALS Association and Mayo Clinic Center for Regenerative Medicine.
References
- Aam BB, Heggset EB, Norberg AL, Sorlie M, Varum KM, Eijsink VG. 2010. Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs. 8:1482–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A. 2005. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med. 172:1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. 2001. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 276:6770–6778. [DOI] [PubMed] [Google Scholar]
- Bucolo C, Musumeci M, Maltese A, Drago F, Musumeci S. 2008. Effect of chitinase inhibitors on endotoxin-induced uveitis (EIU) in rabbits. Pharmacol Res. 57:247–252. [DOI] [PubMed] [Google Scholar]
- Bucolo C, Musumeci M, Musumeci S, Drago F. 2011. Acidic Mammalian chitinase and the eye: implications for ocular inflammatory diseases. Front Pharmacol. 2:43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueter CL, Specht CA, Levitz SM. 2013. Innate sensing of chitin and chitosan. PLoS Pathogens 9:e1003080.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee R, Batra J, Das S, Sharma SK, Ghosh B. 2008. Genetic association of acidic mammalian chitinase with atopic asthma and serum total IgE levels. J Allergy Clin Immunol. 122:202–208, 208.e1–208.e7. [DOI] [PubMed] [Google Scholar]
- Chou YT, Yao S, Czerwinski R, Fleming M, Krykbaev R, Xuan D, Zhou H, Brooks J, Fitz L, Strand J, et al. 2006. Kinetic characterization of recombinant human acidic mammalian chitinase. Biochemistry 45:4444–4454. [DOI] [PubMed] [Google Scholar]
- Correale J, Fiol M. 2011. Chitinase effects on immune cell response in neuromyelitis optica and multiple sclerosis. Multiple Scler. 17:521–531. [DOI] [PubMed] [Google Scholar]
- Cozzarini E, Bellin M, Norberto L, Polese L, Musumeci S, Lanfranchi G, Paoletti MG. 2009. CHIT1 and AMCase expression in human gastric mucosa: correlation with inflammation and Helicobacter pylori infection. Eur J Gastroenterol Hepatol. 21:1119–1126. [DOI] [PubMed] [Google Scholar]
- Goedken ER, O'Brien RF, Xiang T, Banach DL, Marchie SC, Barlow EH, Hubbard S, Mankovich JA, Jiang J, Richardson PL, et al. 2011. Functional comparison of recombinant acidic mammalian chitinase with enzyme from murine bronchoalveolar lavage. Protein Express Purif. 75:55–62. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D, He CH, Koller B, Da Silva CA, Kobayashi Y, Lee CG, Flavell RA, Elias JA. 2009. Acidic mammalian chitinase regulates epithelial cell apoptosis via a chitinolytic-independent mechanism. J Immunol. 182:5098–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo KW, Hur DY, Park SK, Yang YI, Kwak HH, Kim TY. 2011. Expression of chitinases in hypertrophied adenoids of children. Otolaryngol Head Neck Surg. 145:660–665. [DOI] [PubMed] [Google Scholar]
- Hollak CE, van Weely S, van Oers MH, Aerts JM. 1994. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J Clin Investig. 93:1288–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. 1990. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Detection of picomolar quantities by an imaging system based on a cooled charge-coupled device. Biochem J. 270:705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashimura A, Kimura M, Okawa K, Suzuki H, Ukita A, Wakita S, Okazaki K, Ohno M, Bauer PO, Sakaguchi M, et al. 2015. Functional properties of the catalytic domain of mouse acidic mammalian chitinase expressed in Escherichia coli. Int J Mol Sci. 16:4028–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashimura A, Okawa K, Ishikawa K, Kida Y, Iwabuchi K, Matsushima Y, Sakaguchi M, Sugahara Y, Oyama F. 2013. Protein A-mouse acidic mammalian chitinase-V5-His expressed in periplasmic space of Escherichia coli possesses chitinase functions comparable to CHO-expressed protein. PLoS One 8:e78669.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoushab F, Yamabhai M. 2010. Chitin research revisited. Mar Drugs. 8:1988–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. 2011. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 73:479–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci M, Aragona P, Bellin M, Maugeri F, Rania L, Bucolo C, Musumeci S. 2009. Acidic mammalian chitinase in dry eye conditions. Cornea 28:667–672. [DOI] [PubMed] [Google Scholar]
- Nookaew I, Thorell K, Worah K, Wang S, Hibberd ML, Sjovall H, Pettersson S, Nielsen J, Lundin SB. 2013. Transcriptome signatures in Helicobacter pylori-infected mucosa identifies acidic mammalian chitinase loss as a corpus atrophy marker. BMC Med Genomics.6:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Togashi Y, Tsuda K, Okawa K, Kamaya M, Sakaguchi M, Sugahara Y, Oyama F. 2013. Quantification of chitinase mRNA levels in human and mouse tissues by real-time PCR: species-specific expression of acidic mammalian chitinase in stomach tissues. PLoS One 8:e67399.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Tsuda K, Sakaguchi M, Sugahara Y, Oyama F. 2012. Chitinase mRNA levels by quantitative PCR using the single standard DNA: acidic mammalian chitinase is a major transcript in the mouse stomach. PLoS One 7:e50381.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Cho HW, Heo KW, Hur DY, Lee HK. 2009. Role of acidic mammalian chitinase and chitotriosidase in nasal polyps. Otolaryngol Head Neck Surg. 141:462–466. [DOI] [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM. 2007. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikofski J, Tao BY. 1992. Polymerase chain reaction (PCR) techniques for site-directed mutagenesis. Biotechnol Adv. 10:535–547. [DOI] [PubMed] [Google Scholar]
- Renkema GH, Boot RG, Muijsers AO, Donker-Koopman WE, Aerts JM. 1995. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J Biol Chem. 270:2198–2202. [DOI] [PubMed] [Google Scholar]
- Seibold MA, Donnelly S, Solon M, Innes A, Woodruff PG, Boot RG, Burchard EG, Fahy JV. 2008. Chitotriosidase is the primary active chitinase in the human lung and is modulated by genotype and smoking habit. J Allergy Clin Immunol. 122:944–950.e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold MA, Reese TA, Choudhry S, Salam MT, Beckman K, Eng C, Atakilit A, Meade K, Lenoir M, Watson HG, et al. 2009. Differential enzymatic activity of common haplotypic versions of the human acidic Mammalian chitinase protein. J Biol Chem. 284:19650–19658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 101:11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. 2006. Gene losses during human origins. PLoS Biol. 4:e52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang Z. 2013. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 30:2723–2724. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. 2004. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 304:1678–1682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.