Abstract
The flagellum is a key innovation linked to eukaryogenesis. It provides motility by regulated cycles of bending and bend propagation, which are thought to be controlled by a complex arrangement of seven distinct dyneins in repeated patterns of outer- (OAD) and inner-arm dynein (IAD) complexes. Electron tomography showed high similarity of this axonemal repeat pattern across ciliates, algae, and animals, but the diversity of dynein sequences across the eukaryotes has not yet comprehensively been resolved and correlated with structural data. To shed light on the evolution of the axoneme I performed an exhaustive analysis of dyneins using the available sequenced genome data. Evidence from motor domain phylogeny allowed expanding the current set of nine dynein subtypes by eight additional isoforms with, however, restricted taxonomic distributions. I confirmed the presence of the nine dyneins in all eukaryotic super-groups indicating their origin predating the last eukaryotic common ancestor. The comparison of the N-terminal tail domains revealed a most likely axonemal dynein origin of the new classes, a group of chimeric dyneins in plants/algae and Stramenopiles, and the unique domain architecture and origin of the outermost OADs present in green algae and ciliates but not animals. The correlation of sequence and structural data suggests the single-headed class-8 and class-9 dyneins to localize to the distal end of the axonemal repeat and the class-7 dyneins filling the region up to the proximal heterodimeric IAD. Tracing dynein gene duplications across the eukaryotes indicated ongoing diversification and fine-tuning of flagellar functions in extant taxa and species.
Keywords: axoneme, cilium, flagellum, dynein, last eukaryotic common ancestor.
Introduction
Flagella (and cilia) belong to the major innovations of the early eukaryote (Mitchell 2007; Carvalho-Santos et al. 2011; Katz 2012; Koumandou et al. 2013). They produce motility by bending and function in cell movement, feeding, morphogenesis, and cell division (Roy 2009; Lindemann and Lesich 2010; Langousis and Hill 2014). Flagella are built on a canonical 9 + 2 axoneme, which consists of nine microtubule doublets symmetrically arranged around a central pair of two singlet microtubules (Inaba 2007; Lindemann and Lesich 2010; Ishikawa 2015). The outer doublet microtubules provide the scaffold for the attachment of dynein complexes, which produce sliding of the microtubules by using ATP. The dyneins are arranged in complex arrays of single-headed, heterodimeric and heterotrimeric inner- (IDA) and outer-dynein arm (ODA) complexes (fig. 1A) and regulate flagellar wave forms and force production. The functional specialization of the axonemal dyneins in conjunction with a multitude of dynein genes conserved across several extant eukaryotes tempted David Asai almost 20 years ago to propose the multi-dynein hypothesis (Asai 1995). It states, that multiple dynein isoforms exist that each perform specific tasks in the cell. Each dynein is encoded by a separate gene, is located at a specific place, produces a unique range of forces, and thus the isoforms are not functionally interchangeable. Also, the sequence, and thus the cellular function, of each isoform is conserved across eukaryotes.
Fig. 1.
Phylogenetic tree of the dynein motor domains. (A) Schematic representation of the various cytoplasmic and axonemal dynein complexes. Axonemal dyneins are plotted onto the axonemal repeat as far as their exact positions were known. For better orientation, the composition and naming of the ODAs of four model organisms are given. (B) Unrooted maximum-likelihood topology generated under the JTT + Γ model as implemented in FastTree. The tree is based on all dynein motor domains excluding fragmented sequences, and excluding the divergent and long-branching cytoplasmic dyneins from Microsporidia and Piroplasmida for better presentation, resulting in 3021 dynein motor domain sequences. Branch support values are given for the major class-defining nodes. The positions of the DHCs of Homo sapiens and Chlamydomonas reinhardtii are given using their old nomenclature for better orientation (see also table 1 and supplementary tables S1 and S5, Supplementary Material online, for name translation schemes). Orphan DHCs are shown in black. Alternative class names for the cytoplasmic dyneins (Pfister et al. 2005) and the outer-arm, OADs (Asai and Wilkes 2004), and inner-arm dyneins, IADs (Asai and Wilkes 2004; Morris et al. 2006; Wickstead and Gull 2007), are given for comparison. The abbreviations ODA (outer dynein arm) and IDA (inner dynein arm) are usually used for describing the entire complexes and functional entities, while OAD (outer-arm dynein) and IAD (inner-arm dynein) are usually used to denote only the respective DHCs. The scale bar corresponds to estimated amino acid substitutions per site. The same tree with focus on the orphan dyneins is shown in supplementary fig. S4, Supplementary Material online. The full tree is available as supplementary data S1, Supplementary Material online. A maximum-likelihood tree generated under the WAG + Γ model and including the divergent cytoplasmic dyneins is presented in supplementary fig. S3, Supplementary Material online, for comparison. (C) Number of species within selected major taxa. Only taxa with more than five species in subtaxa were selected. The remaining species were grouped as “others”. Although the analyzed species are dominated by metazoans and fungi, dynein repertoires were also identified for 130 species from other taxa. (D) Number of annotated dyneins per taxon. (E) Number of “orphans” (currently unclassified dyneins) per taxon. Most of the orphans belong to taxa with low taxonomic sampling with genomes of only one or two species of the respective taxon available.
Subsequently obtained structural, biochemical and sequence data seemed to support this hypothesis. The best-analyzed examples in this respect are the axonemal dyneins from Chlamydomonas reinhardtii (DiBella and King 2001; Kamiya 2002; Bui et al. 2008; Kamiya and Yagi 2014). Chlamydomonas encodes three dynein heavy chains [DHCs, often also abbreviated as DYH (Wilkes et al. 2008) or DYN (Pfister et al. 2005)] that together with multiple smaller subunits form the heterotrimeric outer-dynein arm complex (Mitchell and Brown 1994; Wilkerson et al. 1994), which repeats every 24 nm along the axoneme (Huang et al. 1979; Goodenough and Heuser 1982; Ishikawa et al. 2007) (fig. 1A). The inner-dynein arms are part of a complex repeat structure based on a heterodimeric DHC complex and six single-headed DHCs (Kagami and Kamiya 1992; Bui et al. 2008). Analyses of dynein-deficient mutants have shown each of these IDAs lacking at a specific position in the repeat structure indicating that the IDAs of Chlamydomonas are specifically targeted to a certain position in the axoneme, and that they are not substituted by other homologs (Bui et al. 2008; Bui et al. 2012). In contrast, there are only two types of cytoplasmic DHCs. One of them is responsible for the specific task of the retrograde intraflagellar transport and accordingly referred to as IFT-dynein (Kozminski et al. 1993; Hou and Witman 2015). The other cytoplasmic dynein is responsible for many different cytoplasmic motor activities such as the steady state localization of the Golgi complex and endosomal membranes near the cell center (Allan et al. 2002; Vaughan 2005), the movement of ER-Golgi transport complexes (Murshid and Presley 2004), and the organization of the mitotic spindle during interphase and mitosis (Compton 1998).
Dyneins are large multi-subunit motor protein complexes that use ATP as energy for the retrograde movement along microtubule filaments (Kikkawa 2013; Roberts et al. 2013; Carter et al. 2016). The DHC is not homologous to the proteins of the other two eukaryotic motor protein families, the kinesins and the myosins, that share a common ancestor (Kull et al. 1998). Instead, the DHCs belong to the AAA+ superfamily (ATPases associated with various cellular activities) (Neuwald et al. 1999). Most AAA+ proteins form hexamers that are organized in a ring structure. In contrast, the six AAA+ modules of the DHC are consecutively encoded on the same gene interrupted by the so-called stalk- and strut-domains that mediate and regulate the binding of dynein to the microtubules (Carter et al. 2011; Kon et al. 2012). The linker-domain N-terminal to the motor domain and the “C sequence” termed region C-terminal to AAA6 are located on the front and back face of the AAA+ ring (Roberts et al. 2009; Carter et al. 2011; Kon et al. 2012). In contrast to myosins and kinesins, phylogenetic analyses of dyneins based on deep sequence and taxonomic sampling are still missing. Published studies are based on 50 to 150 dynein sequences from ten to 25 species (Asai and Wilkes 2004; Höök and Vallee 2006; Morris et al. 2006; Wickstead and Gull 2007; Wilkes et al. 2008; Hartman and Smith 2009; Wickstead and Gull 2012). The common scheme that crystallized from these analyses is nine major dynein subfamilies present in all major eukaryotic taxa: a cytoplasmic dynein, an IFT-dynein and seven axonemal dyneins. Accordingly, the last eukaryotic common ancestor (LECA) is supposed to have already encoded this diversified set of dynein genes.
In this study, I have manually annotated 3,272 DHC sequences from 636 species to perform a comprehensive and exhaustive analysis of the evolution of DHC isoforms in eukaryotes. This analysis revealed the origin and identity of the outermost outer-arm dynein (OAD) indicating why this third OAD is only present in plants and SAR (Stramenopiles, Alveolata, and Rhizaria), and not in metazoans and excavates. To advance our understanding of cilium evolution in the early eukaryote, I correlated the sequence data with available structural data from axonemes suggesting that the axonemal repeat was built in four distinct steps of dynein gene duplications. The deep sequence sampling indicates conservation of a core axoneme consisting of the heteromeric dyneins, and species-specific fine-tuning of axonemal motility by a variable part comprising the single-headed dyneins.
Results
Comparative Genomics Resolves Problems with Reconstructing DHC Genes
Using comparative genomics strategies I generated a dataset of 3,272 DHC sequences from 636 eukaryotes (fig. 1). Because automatic gene predictions are error-prone (especially all the gene predictions of the very long and exon-rich DHC genes contain many mispredicted exons/introns and unrecognized N-terminal/C-terminal parts), and because even those gene predictions are available for only a small subset of all sequenced eukaryotic genomes, I manually assembled and annotated all DHC sequences used in this study. Due to their size, only a few full-length cDNA sequences of DHCs are available that could be and were used as guidance for the manual assembly of further sequences. Additional difficulties for the manual DHC gene assemblies arose from the many problems in the available contig assemblies that only become evident when analyzing large genes such as the DHCs, which are encoded on dozens to several hundreds of kbp. For example, all versions prior to the most recent human reference genome sequence (versions prior to GRCh38) contain a gap of about 56 kbp within the DHC7C gene corresponding to about 860 residues. The eukaryotic genomes were analyzed in an iterative search process over many years. On the one hand, reanalysis of improved genome assemblies often helped to fill sequence gaps. On the other hand, the availability of a newly sequenced related species allowed revising the annotation of divergent regions (e.g. the N-terminal ends or loop regions) by manual comparative analysis of the respective genomic regions. The genome assemblies of closely related species were also reanalyzed, if further orthologs or paralogs were identified in new species, to reveal potential dyneins missed in earlier searches. For example, the newly identified metazoan-specific class-11, class-12, and class-13 DHCs show very low sequence homology in TBLASTN searches when searched with one of the well-known DHCs (supplementary fig. S1, Supplementary Material online). As soon as I was able to reconstruct these genes in one metazoan species, I reanalyzed all previously analyzed metazoans. A somewhat extreme case for the gene prediction effort was the Symbiodinium minutum genome, for which TBLASTN suggests four to five dyneins (supplementary fig. S2, Supplementary Material online) but that contains at least 21 dynein homologs (up to 272 exons/per gene, up to 65 of the usually rare GA−AG introns/per gene). With this iterative approach, the search for divergent DHC homologs and the accuracy of the DHC gene annotations have continuously been re-evaluated and improved. The N-terminal ends (N-terminal 100–200 residues) are the most divergent parts of the dynein sequences and might, despite all efforts, still contain mispredicted sequences. The N-terminal tails are responsible for dynein localization and their sequences were supposed to reveal common schemes apart from the force-producing motor domains. The main classification of the dyneins is based on the phylogenetic analysis of the head and linker domains (together referred to as the motor domain).
High Sequence and Taxonomic Sampling Permits Unambiguous Classification
Phylogenetic tree reconstructions of various motor domain datasets, generated by including and excluding divergent and long branching dyneins, revealed 100% support for 17 groups of dyneins that were subsequently termed classes (fig. 1B, supplementary data S1, Supplementary Material online). The support for the grouping of the 17 classes is independent of the amino acid substitution model used for tree reconstruction (compare fig. 1B and supplementary fig. S3 and data S2, Supplementary Material online). 75 Dyneins from taxonomically unrelated species did not group to any of the 17 classes and remained unclassified (termed “orphans”, supplementary fig. S4, Supplementary Material online). All designated subtypes have distinct domain architectures (see below) supporting that appropriate nodes had been taken for classification. Key to this unambiguous classification is the extremely high sequence and taxonomic sampling. In previous analyses based on only a few, taxonomically very divergent species, support for most subtypes was low, many dyneins were misplaced, and representative sequences for the dynein subtypes, which I identified here as new classes, were missing at all (Asai and Wilkes 2004; Höök and Vallee 2006; Morris et al. 2006; Wickstead and Gull 2007; Wilkes et al. 2008; Hartman and Smith 2009; Wickstead and Gull 2012). The misplaced dyneins together with historically grown species-specific nomenclatures and multiple overlapping and contradicting naming schemes in public sequence databases resulted in highly confusing descriptions in the literature with respect to dynein subtypes, orthologs, and paralogs. For example, the orthologs of the Chlamydomonas OAD dyneins γ, β, and α (innermost, central, outermost OAD) were termed α, β, γ for Tetrahymena thermophila, α and β in Strongylocentrotus purpuratus, and γ and β for human.
Suggestion for an Extensible and Unifying Nomenclature
Recently, a unifying taxonomy has been proposed for ciliary dyneins based on Chlamydomonas genes (Hom et al. 2011). Although the Chlamydomonas axoneme is the most and best studied model system in flagellum research, the proposed taxonomy is not able to reflect the complex history of dynein sequence evolution with its many taxon-specific gene duplications. Numbering dyneins consecutively (Hom et al. 2011) results in unrelated numbers for orthologs and paralogs within and across species. The terms DYNC1H1/DYNC2H1 and OAD/IAD are in use to designate the cytoplasmic and axonemal dynein classes, respectively, but the DHC genes/proteins have never been renamed accordingly. Such a nomenclature is also not extensible as it always requires determining a function before naming a new class. To allow immediate identification and comparison of subtypes and variant isoforms, I therefore suggest a nomenclature for the DHC genes/proteins similar to that used for myosins and kinesins. I suggest giving each subtype a certain num1ber and naming subtype variants by letters (fig. 1B, table 1, supplementary text and Supplementary tables S1–S6, Supplementary Material online). Variant designations were harmonized to reflect phylogeny-based sequence homology and species relationships as far as possible. Although another nomenclature might cause some controversy in the first instance, a consistent and extensible taxonomy for the dyneins of all eukaryotes will have considerable advantages in the long term. There are mainly four model organisms, which are extensively studied, Chlamydomonas reinhardtii, Homo sapiens, Strongylocentrotus purpuratus (and other sea urchins), and Tetrahymena thermophila, and their species-specific nomenclature might be used in parallel (table 1; more detailed name translation tables are provided by supplementary tables S1–S6, Supplementary Material online).
Table 1.
Name translating schemes for major model organisms.
| Class | Proposed name | Gene/protein names used in the literature | Function | |||
|---|---|---|---|---|---|---|
| Tetrahymena | Chlamydomonas | Sea urchin | Human | |||
| 1 | DHC1 | DHC1/DHC1a | DYNC1H1 | DYNC1H1 | cytoplasmic 1 | |
| 2 | DHC2 | DHC2/DHC1b | DHC1b | DYNC2H1 | DYNC2H1 | cytoplasmic 2, IFT |
| 3 |
|
DHC3 | DHC15 |
|
|
OAD gammainnermost OAD |
| 4α | DHC4A | DHC5 | DHC13 | OAD alphaoutermost OAD | ||
| 4β |
|
DHC4 | DHC14 | DNAH9 |
|
OAD betacentral OAD |
| 5 | DHC5 | DHC6 | DHC1, 1α I1/f | DNAH10 | DNAH10 | IAD1 alpha |
| 6 | DHC6 | DHC7 | DHC10, 1β I1/f | DNAH2 | DNAH2 | IAD1 beta |
| 7 |
|
|
|
|
|
IAD3 |
| 8 |
|
|
DHC2, d | DNAH1 | DNAH1 | IAD4 |
| 9α |
|
|
|
|
|
IAD5 |
| 9β | DHC9B | DHC15 | IAD5 | |||
| 11 | DHC11 | — | HD1 | |||
| 12 | DHC12 | — | ||||
| 13 | DHC13 | — | ||||
| orphan | pcr4, DHC12 | |||||
The Number of Dynein Subfamilies Extends to 17
It has been noted that many eukaryotes possess supposed to be highly aberrant forms of DHCs, including the human DYNHD1 protein (table 1), which were suggested to be remnants of early dynein duplications and, therefore, to represent “ghost genes” (Gibbons 2012). However, if such “ghost genes” were remnants they would be expected to be present in only a few extant species and to be considerably disturbed (e.g. frame-shifts and in-frame stop codons) or completely disbanded in closely related species. While some highly disturbed and fragmented dyneins were found in fish and bird genomes (accordingly called “pseudogenes”), all dyneins of the newly defined subfamilies and also the unclassified dyneins do represent bona fide dynein homologs. In total, these dyneins comprise 216 sequences (6.6% of the dataset) from 110 (17.3%) species (fig. 1). In contrast to the cytoplasmic, IFT and axonemal dyneins, the new classes are currently taxonomically restricted. The class-10 and class-17 dyneins are specific to Chytridiomycota and Neocallimastigomycota fungi, class-12 first appeared in the ancestor of the Holozoa (Choanoflagellida and Metazoa), and class-11 and class-13 are currently specific to metazoans. Dyneins of the apusozoan Thecamonas trahens and the cryptophyte Guillardia theta consistently group with high support basal to the class-12 and class-13 dyneins (supplementary fig. S4, Supplementary Material online), and the origin of these classes might therefore date back to the ancient Opisthokont or even the LECA. Because of their restricted distribution, I would suggest these Thecamonas and Guillardia dyneins to be treated as unclassified (“orphans”) as long as further supporting data are not available. The classes 14 to 16 are currently specific to Oomycota, a subgroup of the Stramenopiles.
The Motor Domains Dominate the Overall Domain Architecture of the Dyneins
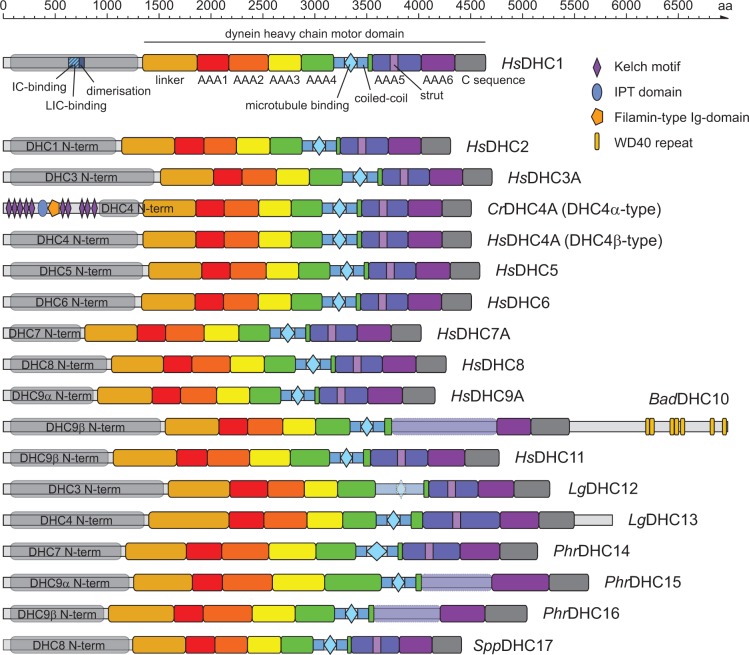
The overall domain organizations of the various dyneins are very similar (fig. 2). The motor domains consist of an N-terminal linker domain followed by six consecutive AAA+ domains, which are interrupted by the stalk and strut (fig. 2). The N-terminal 800–1,500 amino acids encode oligomerization sites and contain regions for the assembly with class-specific components. Some of the new classes have alterations to this general scheme supporting their assignment as distinct classes. The class-10 DHCs have a very long unique region at the position, where usually the AAA5 domain is found, and encode a 1,500 amino acid C-terminal tail region containing seven WD40 domains. The WD40 domains might form a seven-bladed propeller-like structure (most beta-sheet propellers have seven blades) or higher-bladed propellers with noncanonical further blades. In all class-10 dyneins the motor domain from AAA5 to the C-terminus is encoded in a single exon thus excluding potential gene prediction and annotation artifacts with respect to the C-terminal tails (supplementary fig. S5, Supplementary Material online). The class-10 domain architecture seemed suggestive of a dynein fused to its intermediate chain. However, the WD40 region does not show any homology to known intermediate chain proteins. Class-12 dyneins have an extremely unusual stalk domain that does not contain a typical microtubule-binding site nor coiled-coil regions. Class-13 dyneins have longer AAA5 domains and unique C-terminal extensions of low complexity (fig. 2). Except for their amino acid composition these extensions do not show any conservation across homologs. Class-14 dyneins have specific extended loops in the microtubule-binding domain. Class-15 and class-16 dyneins have several class-specific extensions to AAA+ domains and unique regions, where AAA5 domains are usually found. It has been noted based on sequence alignments that the “C sequence” is absent in “fungal” DHCs (Mocz and Gibbons 2001). The present data show that the “C sequence” is absent in DHCs from Ascomycota, Microsporida and Fungi incertae sedis, but present in Basidiomycota, Chytridiomycota, Blastocladiomycota, Entomophthoromycota, Monoblepharidomycota, and Neocallimastigomycota. The “C sequence” is also absent in DHC1 proteins from Bacillariophyta (diatoms) and DHC14 proteins from Albuginales, indicating that the “C sequence” has been lost independently in different dynein subtypes and in different taxa. In all other dyneins, the “C sequence” is present. Given the deep taxonomic sampling and broad coverage of dynein subtypes of this study, these few exceptions rather suggest that the “C sequence” is in principle an essential, nonvariable part of the motor domain. Although the sequences of the ATP hydrolysis sites and the microtubule binding sites have not been analyzed in detail here, it seems reasonable to propose similar characteristics for force generation and transduction for the DHCs of the same class.
Fig. 2.
Domain organization of representative dyneins. The dyneins are characterized by a large motor domain of about 3300 residues that consists of a so-called linker region connecting the N-terminal tail to the motor domain, six AAA+ domains forming a ring-shaped structure, a coiled-coil based stalk domain containing the microtubule-binding region, and a so-called C-sequence. Sequence homology between the N-terminal tails of previously existing classes and new classes is indicated. Subunit binding-sites (IC, intermediate chain; LIC, light-intermediate chain) and the region responsible for dimerization are known for the cytosplasmic class-1 dyneins and indicated. Domains shown in transparent indicate sequence regions that could not unambiguously be aligned to the respective regions of the other dynein classes. For example, the strut regions within the AAA5 domains could not be identified in class-10, class-15 and class-16 dyneins. All domain schemes are drawn to scale. Species abbreviations are: Bad, Batrachochytrium dendrobatidis (Chytridiomycota); Cr, Chlamydomonas reinhardtii; Hs, Homo sapiens; Lg, Lottia gigantean (mollusc); Phr, Phytophthora ramorum (Stramenopiles); Spp, Spizellomyces punctatus (Chytridiomycota).
Support for the Monophyly of Each of the New Classes and Suggestions for Their Localization and Function
Given their restricted phylogenetic distribution, are the newly defined classes really distinct dynein isoforms, should not they instead be combined into a common further major supergroup, or should not they be better defined as divergent subtypes of the nine ubiquitous classes? The newly defined classes grouped together in most phylogenetic trees, which might point to an overlooked common origin or be the result of tree reconstruction problems such as long-branch attraction artifacts. However, the bootstraps support for a monophyletic group is very low (fig. 1B, supplementary fig. S4, Supplementary Material online) and several classes and orphans often group outside this supergroup (supplementary fig. S3, Supplementary Material online). Thus, by comparing the N-terminal tail regions I suspected to identify remnants of the origin of the newly identified classes not observable in the motor domains. Because the motor domain phylogeny did not reveal common origins with the nine known classes, I aligned the N-terminal tails of each new class with the tails of each of the known classes to identify the closest relative, and finally generated a phylogenetic tree of the N-terminal tail domains (fig. 3).
Fig. 3.
Phylogenetic tree of the dynein tail domains. Unrooted phylogenetic tree of the dynein’s N-terminal tails generated with the maximum-likelihood method under the JTT + Γ model as implemented in FastTree. The tree is based on all dynein N-terminal tail domains excluding fragmented and partial sequences, resulting in 2738 dynein tail domain sequences. Branch support values are given for the major class-defining nodes. Orphan DHCs are shown in dark gray. The scale bar corresponds to estimated amino acid substitutions per site. Species abbreviations are: Apas, Aphanomyces astaci (oomycete); Aua, Aureococcus anophagefferens (Stramenopiles); Bds, Bodo saltans (kinetoplastid); Cr, Chlamydomonas reinhardtii; Dap, Daphnia magna (arthropod); Gil, Giardia lamblia (diplomonad); Gt, Guillardia theta (cryptophyte); Klf, Klebsormidium flaccidum (streptophyte); Pf, Plasmodium falciparum (alveolate); Ssa, Spironucleus salmonicida (diplomonad); Sysa, Symbiodinium sp A1 (dinoflagellate); Tct, Thecamonas trahens (Apusozoa); Tv, Trichomonas vaginalis (parabasalid).
Interestingly, the N-terminal tails of the newly defined dynein classes and all orphan dyneins (with the single exception of an orphan diplomonad dynein) could well be aligned across their entire lengths to one of the nine known classes (fig. 3). These findings suggest that the eight new classes each originated from one of the nine ancient classes, retained the N-terminal tail regions with their oligomerization domains and intermediate and light chain binding regions, but evolved their motor domains to derive new force production, force transduction and microtubule-binding characteristics. The sequence homology and phylogenetic grouping of the N-termini of the new classes and the orphan dyneins (except one of the diplomonad orphans) suggests that all these dyneins are localized to axonemes (fig. 3). Because the N-termini encode the binding-sites to axonemal substructures, it is most likely that these dyneins will occupy similar positions in the axonemal repeat as the major dyneins with the same N-termini. For example, the class-12 and class-13 dyneins have N-terminal tails similar to the outer-arm class-3 and class-4 dyneins, respectively (fig. 3), suggesting that the class-12 and class-13 dyneins might form a similar heterodimeric dynein complex localizing to the outer dynein arm positions. However, the dynein head domains are different and will therefore cause different axonemal bending characteristics. Experimental data is needed to determine whether these divergent dyneins are distributed randomly across the entire flagellum, whether these dyneins localize to certain flagellar substructures (e.g. to the posterior or distal part of the flagellum), or whether these dyneins are restricted to tissue-, cell-type, or developmental stage-specific flagella. In accordance with the phylogeny of the motor domains and the domain architecture analysis, the different origins of the N-terminal tails also support the assignment of the new classes as separate, distinct new classes.
Tail Domain Divergence and Homology within and across Dynein Subtypes
The N-terminal tail regions are, with some notable exceptions, conserved within classes but different between the groups of homodimeric, heteromeric, and single-headed dyneins. Accordingly, the complete tail regions of the OAD (DHC3 and DHC4) could be aligned, and also parts of the tails of the cytoplasmic (DHC1) and IFT (DHC2) dyneins, and parts of the heterodimeric inner-arm dyneins (DHC5 and DHC6). Also, there is some homology between the heterodimeric inner-arm and OAD (fig. 3).
Surprisingly, there were two groups of N-terminal tails that could not be aligned to the tails of the other respective class members: A group of outer-arm DHC4 dyneins including the OADα from Chlamydomonas (see below) and a group of single-headed DHC7 dyneins including the Chlamydomonas IAD-b (also termed “DHC5”), IAD-e (“DHC8”) and “DHC4” dyneins (table 1). This group of DHC7 dyneins is restricted to Viridiplantae and two subbranches of Stramenopiles (Labrinthulomycetes and Pelagophyceae) and their tails are similar to tails of a subgroup of class-9 dyneins (hereafter termed DHC9β; fig. 3 and supplementary fig. S6, Supplementary Material online). DHC9β dyneins are present in all major taxa except Holozoa, Fungi and plants suggesting origin of such a DHC9β prototype in the LECA and later taxon-specific loss. The ancestors of the Viridiplantae and the two Stramenopiles subbranches most likely obtained these DHC7(head)/DHC9β(tail) chimera independently of each other. The identification of these chimeric dyneins immediately explains why these DHC7 dyneins share identical smaller subunits (actin and centrin) with the DHC9A (IAD5 group, “DHC7”, IAD-g) (Kamiya and Yagi 2014). The regular DHC7 dyneins share the actin and p28 subunits, and the DHC8 (“DHC2”, IAD-d) dynein binds p44, actin, p38, and p28. In contrast to a previous assumption (Kamiya and Yagi 2014), homology of the N-terminus of the orphan dynein DHC_A (pcr4, “DHC12”) with DHC8 dyneins suggests binding of the DHC8 and not the DHC7 subunits.
An Early Eukaryotic Gene Duplication Led to a Distinct Outermost Outer-Arm Dynein
Here, for the first time, I was able to distinguish two subgroups of the outer dynein arm β (DHC4) family: the central (termed DHC4β following the Chlamydomonas naming) and the outermost (termed DHC4α) dyneins of the heterotrimeric outer dynein arm complex (supplementary fig. S7, Supplementary Material online). However, these subfamilies are not sister groups but the DHC4α are a monophyletic group within the DHC4β. Given its phylogenetic distribution it is most likely that the DHC4 dynein duplicated in the LECA. Subsequently, the last common ancestor of the Diaphoretickes, which comprise the Archaeplastida (plants, green and red algae), the SAR, the Haptophyceae and the Cryptophyceae (Adl et al. 2012), exchanged the N-terminal tail of one of the DHC4 dyneins resulting in the DHC4α subgroup. The DHC4α dyneins have a very specific, but conserved, N-terminal tail consisting of multiple Kelch motifs separated by a tandem arrangement of an IPT (Ig-like, plexins, transcription factors) and a Filamin-type immunoglobulin domain that both are supposed to have immunoglobulin like folds (fig. 2).
Duplications of Partial Genes Enable Outer Dynein Arm Diversity in the Diplomonad Spironucleus salmonicida
The flagellated protozoan parasite Giardia lamblia is known to encode the OADγ GilDHC3 and the OADα GilDHC4 genes in two and four genes, respectively (supplementary fig. S8, Supplementary Material online), which are located on different chromosomes/supercontigs and are transcribed into independent mRNAs (Kamikawa et al. 2011; Roy et al. 2012). The two genes of GilDHC3 are trans-spliced and translated into a single polypeptide chain (Kamikawa et al. 2011). Of the partial GilDHC4 genes the three C-terminal parts are trans-spliced into one major mRNA. The N-terminal part and the combined C-terminal parts are translated independently and subsequently combined into the OAD complex (Roy et al. 2012). These data are supported by independent genome assemblies of seven Giardia strains. The recently completed genome of Spironucleus salmonicida (Xu et al. 2014) contains four DHC3 genes, a single exon DHC3 gene (termed DHC3A) and three partial genes similar to the Giardia split DHC3 gene (supplementary fig. S8, Supplementary Material online). The two copies of the N-terminus encoding part of the split DHC3 gene could independently trans-splice to the C-terminus encoding gene resulting in altogether three different OADγ transcripts. The three trans-spliced parts of the Giardia DHC4 gene are present as a single exon gene in Spironucleus. The N-terminus encoding DHC4 gene is duplicated in Spironucleus enabling the formation of two different ODAα protein complexes (supplementary fig. S8, Supplementary Material online). Unlike Giardia, Spironucleus contains promoter-like motifs that might regulate the temporal transcription of the duplicated genes. Also, the different outer-arm isoforms could show distinct spatial distributions similar to the major and minor isoforms of the Chlamydomonas single-headed IADs (Yagi et al. 2009).
Axonemal Dyneins Are Usually Either All Present or All Absent
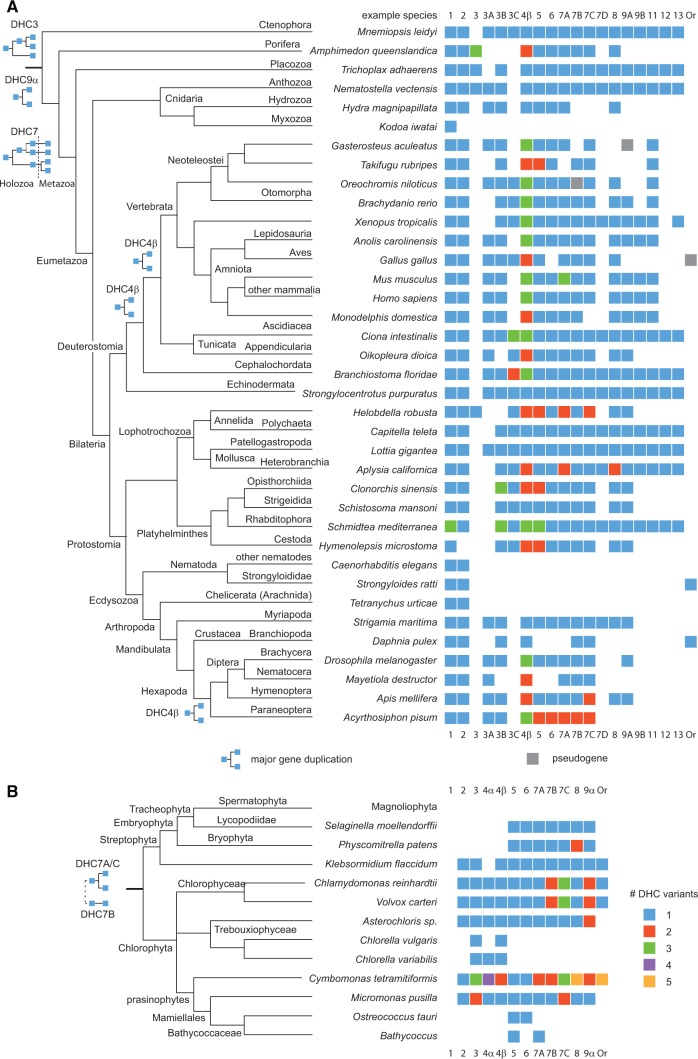
In general, the full set of eight (or ten for species also encoding DHC4α and DHC9β) axonemal dyneins is either present or lost completely (fig. 4, supplementary figs S9 and S10, Supplementary Material online). There are very few exceptions: (1) As noted before, the Coccidia (e.g. Toxoplasma gondii), Haemosporida (e.g. Plasmodium species), and some diatoms (e.g. Thalassiosira species) do not contain IFT dyneins (supplementary fig. S10, Supplementary Material online) (Wickstead and Gull 2007; Wickstead and Gull 2012). In addition, IFT dynein has been lost in the Eustigmatophyceae branch of the Stramenopiles (e.g. Nannochloropsis species), some green algae (Chlorella species and Bathycoccaceae), and Cestoda (e.g. Hymenolepsis species) (fig. 4 and supplementary fig. S10, Supplementary Material online). (2) The outer-arm dyeins have been lost in Bathycoccaceae, Embryophyta, and Mastigamoeba balamuthi (Amoebozoa), and only the DHC4α dynein in all Rhizaria, in Thalassiosira oceanica and Chlorella variabilis (fig. 4 and supplementary fig. S10, Supplementary Material online). (3) All inner-arm dyneins have been lost in Chlorella, Thalassiosira and Nannochloropsis species. The two-headed inner-arm dyneins DHC5 (IAD-1fα) and DHC6 (IAD-1fβ) are absent in Nematocera (insects, e.g. Mayetiola destructor) and Branchiopoda (crustaceans, e.g. Daphnia species). DHC5 dyneins are absent in Plasmodium species (Wickstead and Gull 2012), and DHC6 dyneins are absent in Bathycoccus species and birds (fig. 4 and supplementary fig. S10, Supplementary Material online). Single-headed inner-arm dyneins (DHC7, DHC8, and DHC9; fig. 1) have been lost in Ostreococcus species, DHC7 and DHC9 in Plasmodium species, and DHC8 and DHC9 in Bathycoccus species, Paraneoptera (insects, e.g. Acyrthosiphon pisum), Nematocera, Branchiopoda, and Neoteleostei (fish, e.g. Takifugu rubripes). DHC8 has been lost in Brachycera, and DHC9 dyneins have been lost in birds, Hydrozoa (e.g. Hydra magnipapillata), and Porifera (e.g. Amphimedon queenslandica). The presence of only three fragmented dynein genes in Emiliania huxleyi was attributed to the loss of most flagellar genes in the sequenced strain and is not a characteristic of the species itself (von Dassow et al. 2015). Thus, given the deep taxonomic sampling of this study, loosing a single axonemal dynein is a rare event.
Fig. 4.
Evolution of dynein diversity within metazoans and plants. (A) Dynein inventories were compiled for representative metazoans and plotted onto the most widely accepted phylogenetic tree of the Metazoa. Early metazoan evolution has been adapted from (Ryan et al. 2013). Dynein class variants are listed separately whenever their origin dates back to the last metazoan common ancestor. Gene duplications are indicated at branches where they most likely happened. The “DHC3” group contains DHC3 dyneins that do not group consistently to one of the designated subtypes. “Or” is the abbreviation for “orphan”, which means unclassified DHC sequence. Daphnia species contain a so far unclassified dynein, whose tail is similar to DHC7A dyneins but whose motor domain is extremely divergent and does not group to the DHC7 dyneins. In addition to the nematode-typical DHC1 and DHC2 dyneins, Strongyloides species contain a third dynein, which is to some extent fragmented, does not group to any known dynein class, and might be a pseudogene. (B) Dynein inventories have been compiled for all sequenced genomes in the taxon Viridiplantae and were plotted onto the most widely accepted phylogenetic tree of the green algae and plants. A more detailed tree including data from transcriptome shotgun assemblies is shown in supplementary fig. S9, Supplementary Material online.
Resolving Controversial Assignments of Dyneins to Subfamilies
At the dynein subfamily level, the new data allow revising some previous claims: (A) In contrast to previous analyses (Wickstead and Gull 2007, 2012), cytoplasmic DHC1 dyneins were identified in Aureococcus anophagefferens (Stramenopiles) and Batrachochytrium dendrobatidis (Chytridiomyceta), and also in Glaucocystophyceae (Cyanophora paradoxa) and Rhodophyta (Chondrus crispus) (supplementary figs S9 and S10, Supplementary Material online). This demonstrates, that DHC1 has not been lost at the origin of the Archaeplastida (Glaucocystophyceae, Rhodophyta, Viridiplantae), but at the origin of the Viridiplantae and by species-specific loss events in the other branches. (B) Giardia does not contain a cytoplasmic DHC1, as proposed before (Wickstead and Gull 2007). Such a dynein is not present in any of seven sequenced Giardia species, but encoded in Spironucleus salmonicida, another recently sequenced diplomonad (supplementary fig. S10, Supplementary Material online) (Xu et al. 2014). (C) In contrast to others (Wickstead and Gull 2007), I was not able to identify a DHC8 dynein in Drosophila melanogaster and any of the >80 sequenced Brachycera (fig. 4) suggesting that the previous assignment resulted from a misgrouping due to the low taxonomic sampling in the study.
Phylogenetic Distribution of Cytoplasmic Dynein and Independent Evolution of Differently Split Dynein Genes Within Basidiomycota
As has been noted before (Wickstead and Gull 2012), the cytoplasmic DHC1 dynein is ubiquitously present in eukaryotes and has rarely been lost or duplicated (fig. 4 and supplementary figs S9 and S10, Supplementary Material online). DHC1 has been lost in Viridiplantae, Haptophyceae, Giardia, and all species that do not contain any dynein (see below) and was duplicated in Choanoflagellata, Bigelowiella natans (Rhizaria), Trichomonas vaginalis (Parabasalia) and Thecamonas trahens (Apusozoa), and triplicated in Schmidtea mediterranea.
A split dynein gene has been reported for the Basidiomycota Ustilago maydis (Straube et al. 2001). The two genes of the cytoplasmic dynein UmDHC1 are translated into separate polypeptides, which subsequently assemble into the DHC1 complex (Straube et al. 2001). By deep taxonomic sampling of available Basidiomycota genomes I have identified further split dynein genes resulting from multiple independent gene split events (supplementary fig. S11, Supplementary Material online). While the Ustilaginomycotina (including U. maydis) have the DHC1 genes split within the AAA4 domain before the start of the stalk region, the Agaricomycota (for example Phanerochaete chrysosporum) have the DHC1 genes separated before the motor domain, and the Basidiomycota incertae sedis species (including Wallemia sebi) have the DHC1 gene split into two parts within AAA1 (supplementary fig. S11, Supplementary Material online). The available EST/cDNA data cover the termini of the split genes but do not include gene bridging reads, similar to the split genes of UmDHC1. This strongly supports independent transcription and translation of the genes and disfavors potential trans-splicing. Some Agaricomycota encode alternative versions of the N-terminus encoding genes as consequence of gene duplications, which could result in two or more different DHC1 complexes if all alternative versions were translated genes and not pseudogenes. Analysis of the gene structures of all Basidiomycota DHC1 genes did not reveal any correlation between the three positions for splitting the DHC1 gene and intron positions, which could have provided an explanation for the predisposition of certain regions for generating split genes. However, corresponding introns could have been lost in the analyzed extant Basidiomycota. Not all Basidiomycota encode split DHC1 genes. Some sub-branches like the Tremellomycetes (e.g. Filobasidiella neoformans) and the Pucciniomycotina (e.g. Puccinia graminis) species have single full-length DHC1 genes.
Species Can Live without Dyneins
Although high quality genome assemblies are available, DHC genes could not be identified in several genomes. Among these are the red algae (Rhodophyta: Bangiophyceae) Cyanidioschyzon merolae, Galdieria sulphuraria and Porphyridium purpureum, the yeasts Vanderwaltozyma polyspora and Hanseniaspora valbyensis, the microsporidians Anncaliia algerae and Enterocytozoon bieneusi, the Entamoeba species (taxon Amoebozoa), the apicomplexans Babesia microti and Theileria orientalis, and the flowering plants. The absence of DHCs in flowering plants can be explained by multiple gene loss events in the last common ancestor of the Magnoliophyta, as has been proposed before (Wickstead and Gull 2012). The other examples are members of taxa lacking cilia in all stages of their life cycle and therefore represent cases of species-specific cytoplasmic dynein loss events. These species also do not contain any subunit of the dynactin complex (Hammesfahr and Kollmar 2012) supporting that dynein genes have not been overlooked or missed in assembly gaps. The aforementioned yeasts, microsporidians and apicomplexans have particularly small genomes compared with closely related species providing a possible explanation for the loss of all dynein genes including the smaller subunits.
Discussion
Dynein Evolution Is a Tale of Diversification and Gain of Function, Not a History of Loss
If considering only the presence or absence of the cytoplasmic and axonemal dynein subtypes in extant species, the evolution of the DHC gene family would be dominated by gene loss events, as has been suggested earlier (Wickstead and Gull 2012). This is the simple consequence of the presence of the respective nine dynein subfamilies in the LECA. However, the identification of eight further subfamilies appearing in holozoa/metazoa, Chytridiomycota/Neocallimastigomycota and oomycetes, and the resolution of orthologs and paralogs within subfamilies demonstrate that dynein diversification did not stop at the beginning of eukaryotic radiation (fig. 4). The taxon- and species-specific duplications of the cytoplasmic and axonemal dyneins clearly outnumber the loss events that are mainly restricted to loosing the entire set of axonemal dyneins in certain taxa and species. Similar to the cytoplasmic dynein DHC1, the IFT dynein and the heterodimeric inner-arm dyein complex (DHC5 and DHC6) were rarely duplicated (fig. 4, supplementary figs S9 and S10, Supplementary Material online). The IFT dynein is duplicated in kinetoplastids, Mastigamoeba, Sprionucleus, Trichomonas, and Ichthyophthirius multifiliis (ciliate). The IAD-1β (DHC6) has only been duplicated in Guillardia and Acyrthosiphon pisum (pea aphid), and IAD-1α (DHC5) duplicates were so far only found in Guillardia, Platyhelminthes, Acyrthosiphon, Helobdella robusta (clitellate), and Takifugu rubripes. In contrast, the OADs and the single-headed IADs have independently been duplicated and multiplicated across all taxa (fig. 4, supplementary figs S9 and S10, Supplementary Material online). Because of the lower species sampling in most taxa, duplication events could only be assigned to taxonomic splits in Holozoa/Metazoa, Stramenopiles, and plants (fig. 4). All other duplications have currently to be regarded as species-specific or specific to restricted taxa. In contrast to many cytoskeleton-associated protein families including, for example, coronins (Eckert et al. 2011), dynactin subunits (Hammesfahr and Kollmar 2012), and myosins (Mühlhausen and Kollmar 2013), none of the dynein duplications is related to a known whole-genome duplication event. The OADα (DHC3) split twice and the single-headed dynein DHC9α once at the beginning of the Metazoa (fig. 4, supplementary figs S6 and S12, Supplementary Material online). The OADβ (DHC4β) duplicated independently at the origin of the Chordata and after split of the Cephalochordata, and at the origin of the hexapods (fig. 4, supplementary fig. S7, Supplementary Material online). The single-headed DHC7 dynein subfamily (IAD5) duplicated twice in the ancestor of the Holozoa, and one of the DHC7 orthologs duplicated again after split of the Choanoflagellida (fig. 4, supplementary fig. S13, Supplementary Material online). The DHC7 subfamily duplicated two times in plants, although one of the duplications might even date back to the last common ancestor of the Diaphoretickes or even the LECA (fig. 4, supplementary fig. S13, Supplementary Material online). The Stramenopiles independently duplicated all three OAD subfamilies and all single-headed IADs (supplementary fig. S10, Supplementary Material online). Further species-specific dynein duplications as well as independent losses of certain dynein variants led to distinct sets of dyneins in extant species or closely related extant species (fig. 4).
Mapping Dynein Classes onto the Axonemal Repeat Structure
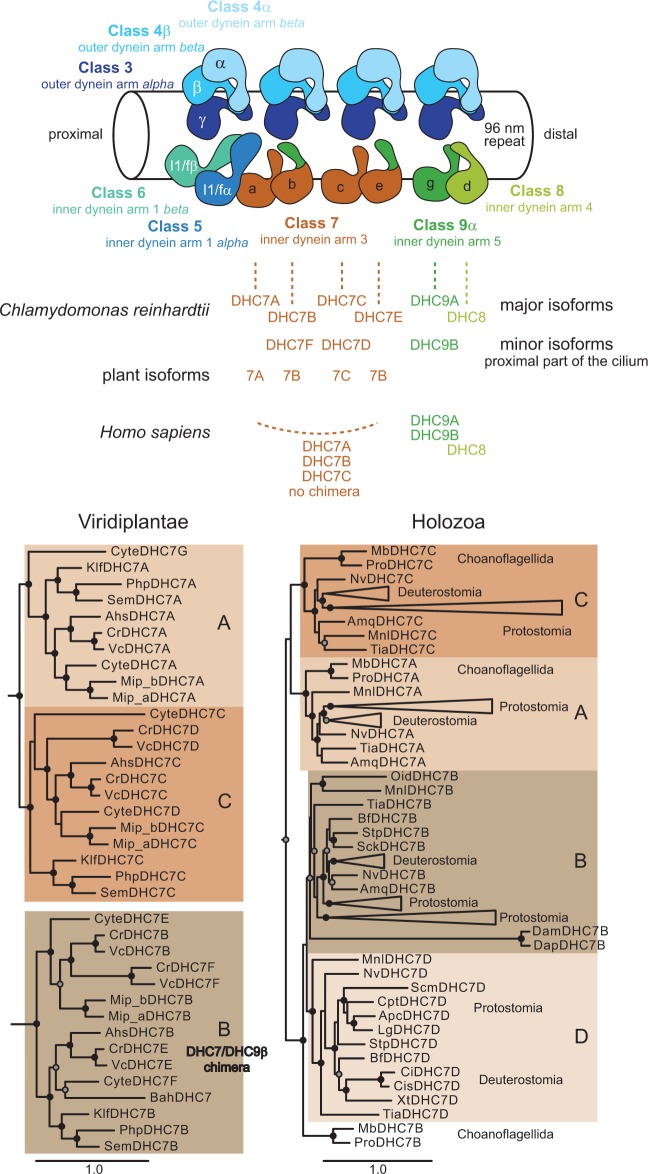
By analyzing Chlamyodomonas dynein-deficient mutants each lacking one or more of the OAD or IAD subsets it has been shown that these are not functionally interchangeable but bind to specific locations in the axonemal repeat structure with their N-terminal tails. Accordingly, the position of each dynein within the repeat is known (fig. 5) (Bui et al. 2012). In addition, electron tomography images of axonemes from Chlamydomonas, the ciliate Tetrahymena, Homo sapiens, and the sea urchin Strongyylocentrotus purpuratus showed compelling structural conservation of the axonemal repeat regions (Pigino et al. 2012; Lin et al. 2014). The deep taxonomic sampling of the sequence data now allows resolving the placing of inner-arm dyneins across species (fig. 5). While the heterodimeric inner-arm dynein complex (DHC5/DHC6) is at the posterior end of the 96 nm axonemal repeat, the two single-headed DHC8 and DHC9 dyneins are at the distal end. The various DHC7 homologs occupy the four positions of single-headed dyneins in-between. Assuming similar overall structures of the axonemes, species with single DHC7 genes, therefore, need 4-fold expression levels of their single DHC7 compared with the other inner-arm dyneins.
Fig. 5.
Structural organization of the dyneins within the axoneme. The structural conservation of axonemes from Chlamydomonas, Tetrahymena, human, and sea urchin at electron-tomography resolution (Pigino et al. 2012; Lin et al. 2014) suggests similar organization of the dynein subfamilies in the axonemal repeat. The positions of the major variants of the Chlamydomonas axonemal dyneins within the axoneme are known and were used to map dynein subfamilies. For better orientation, the names of the corresponding Chlamydomonas dyneins are printed within each schematic dynein. The exact positions within the axonemal repeat of the minor variants of the single-headed Chlamydomonas dyneins are not known, only that these minor isoforms are present in the proximal part of the axoneme (Yagi et al. 2009). Therefore, the minor isoforms are indicated at the position of their closest homolog from the major isoforms. Plants contain three different types of single-headed DHC7 dyneins, of which the DHC7B variant represents the chimeric dynein with DHC7B motor domain and DHC9β N-terminal tail. Below the scheme of the axoneme, the sections of the phylogenetic tree containing the plant and holozoan DHC7 dyneins are shown (the full tree of the DHC7 dyneins is shown in supplementary fig. S13, Supplementary Material online). The ancient holozoan DHC7 dynein has been duplicated twice, independently of the plant DHC7 dyneins. Accordingly, distinct axonemal positions of the human DHC7 dyneins cannot be inferred. The four positions occupied by DHC7 dyneins might be filled by a single human DHC7 dynein (with the three DHC7 dyneins expressed in a tissue- and/or developmental stage-specific pattern) or a mixture of two or three DHC7 orthologs.
The Sequence Data Suggest Considerable Differences of the Axonemal Repeats across Species at High Resolution
The DHC8 dyneins have only been duplicated in restricted branches or specific species (supplementary figs. S9 and S10, Supplementary Material online). Therefore, the DHC8 dyneins of all species can be assumed to occupy the same position in the axonemal repeat. In contrast, the DHC9 dyneins contain two distinct subgroups that most likely date back to the LECA. Holozoa and Viridiplantae encode homologs of only the DHC9α subgroup, which were shown to be located at the distal end of the axonemal repeat. However, the Viridiplantae contain chimeric dyneins each consisting of a DHC9β-N-terminus and a DHC7-head (DHC7B, DHC7E, and DHC7F; fig. 5). Because the dynein N-termini are thought to guide association of the respective dyneins to axonemal positions, DHC9β-type dyneins of other species might be located at similar positions as these chimeric Viridiplantae dyneins. Holozoa do not encode any dynein with DHC9β-type N-terminus but the respective single-headed dynein positions in the axonemal repeats are occupied (Pigino et al. 2012; Lin et al. 2014). These findings indicate that Viridiplantae and other species encoding DHC9β-type dyneins (or dynein tails) might contain additional doublet microtubule-associated proteins guiding the correct localization of the respective single-headed inner-arm dyneins. In contrast, species with single-copy inner-arm dyneins such as Heterolobosea and fungi (supplementary figs S9 and S10, Supplementary Material online) and species with overall very similar single-headed dyneins such as Metazoa do not encode enough single-headed dyneins for the six distinct positions in the axonemal repeat, which must accordingly be filled with multiple identical dyneins.
Although there is strong structural homology at the resolution of electron tomography, these findings indicate, that the axonemal substructures are most likely very different between species of different major eukaryotic taxa at high resolution. As important as studies of Chlamydomonas and Tetrahymena flagella are, only macroscopic and low-resolution characteristics might be representative for all species. The multi-dynein hypothesis states that axonemal dynein isoforms have specific localizations and are not functional interchangeable (Asai 1995). While this hypothesis holds true for the Chlamydomonas and maybe also the Tetrahymena dyneins, it is not true for the single-headed inner-arm dyneins of most other species simply because of too low a number of orthologs (supplementary fig. S10, Supplementary Material online). Already other green algae and all Streptophyta, as well as other Alveolata (Chromerida and Apicomplexa) have reduced sets of single-headed dyneins. The observation of chimeric dyneins also contradicts the general statements of the multi-dynein hypothesis.
The New Data Support the Hypothesis of Independent Evolution of Cytoplasmic andAxonemal Dyneins
The LECA most likely contained dyneins from nine different subgroups (a cytoplasmic dynein, the IFT dynein, and seven axonemal dyneins) and two copies of each of the outer dynein arm beta DHC4β and the single-headed DHC9 inner-arm dynein, combining to at least eleven dynein genes. Mainly two scenarios have been proposed for the evolution of flagellar motility in the LECA before eukaryotic radiation started. In the first scenario, the cytoplasmic dynein was the most ancient dynein and evolved by gene duplication into the ancestor of the IFT and all axonemal dyneins (Hartman and Smith 2009). The most ancient axonemal dynein was supposed to be a homodimeric inner-arm dynein, which lost its dimerization characteristics and subsequently evolved by further gene duplications into the set of seven axonemal dyneins, of which some later established heterodimers (Hartman and Smith 2009). In the second scenario, the first dynein evolved into the ancestor of the seven axonemal dyneins and a prototype cytoplasmic dynein that later split into the cytoplasmic DHC1 dynein and the DHC2 IFT dynein (Wickstead and Gull 2012). In the first scenario, the motile cilium would have evolved from an immotile cilium with sensory functions, while in the second scenario a rudimentary motile cilium was established first, to which in a later step the IFT motor and sensory functions were added. The new data strongly support the independent evolution of cytoplasmic and axonemal dyneins (fig. 6A). Although I did not root the tree of the dynein motor domains (fig. 1, supplementary fig. S3, Supplementary Material online), the inclusion of hundreds of divergent dyneins clearly supports a common origin of the cytoplasmic DHC1 dynein and the DHC2 IFT dynein, and a common origin of the seven axonemal dyneins. Rooting the dynein tree by midasin, another eukaryotic protein with six tandem AAA+ domains, also results in these branchings (Wickstead and Gull 2012) although dynein and midasin are clearly the products of independent AAA+ domain fusions/duplications (Garbarino and Gibbons 2002) and midasin’s inclusion into dynein phylogenies might therefore cause tree reconstruction artifacts. The phylogenetic tree of the N-terminal tails also shows that the DHC2 dyneins are more closely related to DHC1 than to axonemal dyneins. However, the dynein data are not suitable to distinguish between a sensory or motile first scenario for early cilium evolution.
Fig. 6.
Stepwise evolution of the axonemal repeat in the LECA. (A) The phylogeny of the dynein subfamilies suggests independent evolution of the cytoplasmic and axonemal dyneins from a common monomeric ancestor. Subsequent to the emergence of a prototype heterodimer the basic axonemal repeat, as it most likely was present in the LECA, evolved in three steps: First, either (A) the heterodimeric inner dynein arm complex (DHC5/6) is a result of a duplication of the heterodimeric outer dynein arm complex (DHC3/4), or (B) the outer dynein arm complex originated from the inner dynein arm complex. Second, the single-headed dyneins DHC8 and DHC9 appeared by duplication of the heterodimeric inner dynein arm complex and subsequent loss of dimerization capability. Alternatively, the DHC9 dynein could have appeared first by duplication of the DHC6 dynein and evolution into a monomer, which then duplicated resulting in DHC8. Third, the single-headed DHC7 dyneins emerged by gene duplication from a DHC8 dynein. The heterotrimeric outer dynein arm complex evolved after eukaryotic radiation started. The outer dynein arm beta dynein DHC4β still became duplicated in the LECA, but established a new subtype DHC4α dynein by replacing most of the N-terminal tail region at the origin of the Diaphoretickes (SAR + Archaeplastida). B) Topology of the dynein classes in the phylogenetic tree (fig. 1). The topology of the dynein intermediate chains was derived from a tree of more than 1000 DIC sequences and is almost identical to that of previously published trees (Wickstead and Gull 2007, 2012). (C) Schematic tree of several major eukaryotic super-groups. The topology has been derived by combining data from several studies (Parfrey et al. 2011; Adl et al. 2012; Derelle and Lang 2012). Duplication of DHC4β and DHC9 in the LECA and subsequent loss of DHC4α and DHC9β variants in major eukaryotic super-groups are indicated.
Evolution of the Dynein Repeat Structure in the Axoneme of the Early Eukaryote
The topology of the dynein classes in the phylogenetic tree suggests a scenario for the early evolution of the seven axonemal dyneins and the stepwise build-up of the axonemal repeat (fig. 6). The earliest building block of the repeat was a heterodimeric dynein. The ancestral heterodimer could have been derived from a prototype dynein monomer that established a heterodimer after duplication in the axonemal dynein branch and a homodimer in the cytoplasmic branch, or from a prototype heterodimer that split in the cytoplasmic branch to subsequently form two independent dyneins (DHC1 and DHC2). The initial heterodimer could have been an ODA complex (DHC3 + DHC4β) or the IDA complex (DHC5 + DHC6). Whichever was last emerged by duplication of the entire respective other heterodimer. A series of independent duplications from single-headed ancestral dyneins followed by independent heterodimerization as suggested by others (Hartman and Smith 2009) seems highly unlikely. Evidence for a concerted duplication are the close phylogenetic grouping of the DHC4β (OADβ) and the DHC5 (IAD-1fα) dyneins and the pairwise phylogenetic grouping of outer- and inner-am dynein intermediate chains (fig. 6B) (Wickstead and Gull 2007, 2012). Although the current data do not unravel the identity of the ancestral complex, the closer relationship of the OADs to the cytoplasmic dyneins suggests the ODA complex to be the ancestral. This seems to be supported by the finding of many extant eukaryotes in different major lineages having only OADs (fig. 4, supplementary fig. S10, Supplementary Material online). Thus far, the only species with only heterodimeric inner-arm dyneins are the Ostreococcus green algae (fig. 4), which are, however, not thought to build axonemes at all (Henderson et al. 2007). Nevertheless, flagella with absent outer dynein arms are still able to generate motion (Mitchell and Rosenbaum 1985). Next, the first single-headed inner-arm dyneins emerged by duplication of the entire heterodimeric IDA complex or by two subsequent duplications of the DHC6 (IAD-1fβ) dynein (fig. 6A). The topology of the single-headed dyneins in the phylogenetic tree and the absence of dynein intermediate chains associated with single-headed dyneins strongly favor the stepwise evolution by first the emergence of the DHC9 prototype dynein and subsequent loss of intermediate chain-binding and heterodimerisation domains. The DHC9 dynein was then duplicated to establish the DHC8 dynein. With these gene duplications the dynein repeat structure became doubled. Subsequently, the DHC8 dynein underwent a gene duplication resulting in the DHC7 dynein that filled up the 96 nm axonemal repeat (fig. 6) (Oda et al. 2014). Still in the LECA, the OAD DHC4β and the inner-arm dynein DHC9β became duplicated.
Evolution of Axonemal Dyneins since Divergence of Eukaryotic Super-Groups
The last step in the evolution of dynein complexity at a macroscopic scale is the extension of the outer dynein arm complex (fig. 6A). Instead of remaining a DHC4β alternative, the N-terminal tail of the DHC4α duplicate became exchanged in the ancestor of the Diaphoretickes so that the new DHC4α variant could be added as additional component to the ODA complex (fig. 6A). The original DHC4α duplicate was lost in Kinetoplastida, Parabasalia, Diplomonada, and Amorphea, but is still retained in Heterolobosea, demonstrating that dynein diversification did not stop with the LECA starting to diverge into the major lineages that survive to this day (fig. 6C).
The part of the axnomal repeat, which has been established last, is also the part that has underwent the most innovations and divergence until today’s species: multiple independent gene duplications of the single-headed dyneins followed by generation of highly divergent N-termini, and formation of chimeric single-headed dyneins. Duplicates of other dynein genes retained their overall structure and are most likely the result of life-cycle dependent and cell type-specific specialization. While flagella might have similar characteristics in size and shape of the bending at a macroscopic scale, the presented sequence data show that we are just at the beginning of understanding axonemal and dynein diversity at high resolution.
Materials and Methods
Identification and Annotation of DHC Genes
DHC genes have been identified in iterated TBLASTN searches of the completed or almost completed genomes of 636 species starting with using the protein sequence of HsDHC1 as query and proceeding with divergent DHCs. The respective genomic regions covering the search hits were submitted to AUGUSTUS (Stanke and Morgenstern 2005) to obtain gene predictions. However, feature sets are only available for a few species and therefore almost all predictions contained wrong sequences and/or missed exons. Wrong and missing sequence regions became apparent when comparing the predicted protein sequences to other, already corrected dynein sequences in the multiple sequence alignment. Missing exons were manually added by inspecting the three-reading-frame translations of the respective genomic DNA regions, and sequences wrongly predicted as exonic were identified and manually removed. Divergent regions were reconstructed by simultaneously manually comparing the three-reading-frame translations of the respective genomic DNA regions of homologous dyneins of related species. Translations conserved in all respective species were considered as exonic. Where possible, EST and TSA data have been analyzed to help in the annotation process. Protein sequences from closely related species have been obtained using the cross-species gene reconstruction functionality of WebScipio (Hatje et al. 2013) using the dyneins of the closest related species as query sequences and adjusting the search parameters to allow the correct or almost correct reconstruction of protein homologs down to about 80% sequence identity. Nevertheless, also for all these genomes TBLASTN searches have been performed. With this strategy, I tried to minimize the risk to miss more divergent dynein homologs, which might have been derived by species-specific inventions, or dyneins, which are not present in the query species' dynein repertoire due to species-specific gene loss events or due to species-specific assembly gaps. Although I spent considerable efforts in manually completing the entire DHC sequences (and not only the motor domains), the N-terminal ends (the N-terminal 100–200 amino acids) might still contain wrongly predicted exons and miss sequences, because sufficient comparative genomic data are not yet available for all species.
Some of the genes contain alternative splice forms for the motor domain [for an example of a mutually exclusive spliced exon in a DHC1 gene see (Pillmann et al. 2011)]. The different splice forms were not considered independently in the analysis but in all cases the same splice forms were taken for homologous DHCs.
Generating the Multiple Sequence Alignment
The DHC sequence alignment in its current stage was created over years of assembling dynein sequences. The initial alignment was created in 2005 based on the few full-length sequences available at that time. Newly identified DHC sequences were added to this alignment and validated/corrected immediately. To this end, every newly predicted sequence was aligned to its supposed closest relative using ClustalW (Thompson et al. 2002) and this “pre-aligned” sequence added to the multiple sequence alignment. By inspecting the “pre-aligned” sequence in comparison to the already corrected sequences gene prediction errors immediately became obvious. During the subsequent sequence validation process, I manually removed wrongly predicted sequence regions and filled gaps where the automatic gene prediction missed to identify exons. The alignment was then manually adjusted where necessary. Still, gaps remained in many sequences derived from low-coverage genomes. In these cases the integrity of the coding regions before and after the gaps was carefully maintained by clearly separating the coding regions and adding gaps reflecting missing parts of the supposed protein sequences to the multiple sequence alignment.
Incomplete Genes and Pseudogenes
Sequences were termed “Partials” if a small part was missing (up to 5% of the average protein length). Sequences with gaps accounting for more than 5% of the expected sequence length were termed “Fragments”. Note, although some dyneins were missing more than 5% of the sequence and accordingly termed “Fragments”, almost all of these sequences are longer than 1,000 amino acids providing enough data for unambiguous classification. The “Partials” and “Fragments” status was assigned to the dynein motor domains and the N-terminal tail regions separately because of their independent use in the phylogenetic analyses. For the phylogenetic tree reconstructions I excluded all fragmented sequences, because the phylogeny might be affected by incomplete sequences. However, “Partials” and “Fragments” are important for the qualitative analysis to denote the presence of the specific dynein subtype in the respective species.
Fragments have mainly been classified and named based on their obvious homology at the amino acid level. While the class assignment is pretty obvious in most cases, the correct variant designation often needs more careful analyses. Those fragments that did not obviously group to one of the assigned classes or subclasses have sequentially been added to the dataset used to construct the major tree, and trees were calculated for every of these alignments. Some of these fragments could subsequently be classified while a few fragmented dyneins still have to be considered orphans (e.g. there are three fragmented dyneins of unknown classification in the amoebae Mastigamoeba balamuthi). Given the deep sequence and taxonomic sampling of the dataset, even very short fragments of only 100 amino acids should be sufficient now for correctly grouping the dynein fragment into one of the established classes using the BLASTP service at CyMoBase. Thus, it is very unlikely that the orphan “Fragments” will group to one of the established 17 classes if their full-length sequences become available. Seven of the DHCs were termed pseudogenes because they contain far more frame shifts and missing sequences than expected from sequencing or assembly errors.
Preparation of the Datasets for the Phylogenetic Analyses
When I stopped identifying and annotating dyneins to start the final analyses, the dataset consisted of 3,272 dyneins from 636 species. Subsequently, the genomes of several taxonomically important dinoflagellates became available that I analyzed to include in the qualitative analysis. These dyneins are not included in the phylogenetic analyses. These species comprise Vitrella brassicaforis, Symbiodinium sp. A1, Symbiodinium minutum, and Symbiodinium kawagutii. Their dynein repertoires were annotated by BLASTP searches against all other dyneins. The total dataset thus contains 3,351 dyneins from 640 species.
The motor domain is the largest part that is conserved in all DHCs and comprises aa 1,409–4,646 from Homo sapiens DHC1 (aa 4,646 is the C-terminus). The two N-terminal helices present in the high-resolution crystal structure of the Dictyostelium discoideum DHC1 (Kon et al. 2012) are not conserved beyond the class-1 DHCs and this region is therefore not included in the motor domain as defined here. To generate datasets for phylogenetic tree reconstructions, sequences designated “Fragment” or “Pseudogene” were removed from the multiple sequence alignment resulting in 3,082 dyneins. The alignment of the full-length dynein sequences was split into the part comprising the N-terminal tails and the part containing the motor domains (used to generate the tree shown in supplementary fig. S3, Supplementary Material online). The motor domain alignment contains 12,405 alignment positions. The dataset of the N-terminal tails was further reduced by removing all sequences designated “Partials” (tree shown in supplementary fig. S7, Supplementary Material online). To generate a tree for representation purposes, the divergent, long-branching DHC1 sequences from Theileria, Babesia and Microsporidian species were removed from the motor domain alignment (dataset used to generate the tree shown in fig. 1).
Computing and Visualizing Phylogenetic Trees
Phylogenetic trees were generated for all datasets using the maximum-likelihood method with estimated proportion of invariable sites and bootstrapping (1,000 replicates) as implemented in FastTree v.2 (Price et al. 2010). ProtTest v.3.2 failed to run on the datasets. Therefore, I generated phylogenetic trees for every dataset using the JTT + Γ, the WAG + Γ, and the LG + Γ amino acid substitution models as implemented in FastTree. The generated trees were usually identical except for some branchings within classes (e.g. the Placozoa, Cnidaria, Porifera and Ctenophora dyneins show varying topologies) and the placing of a few very divergent dyneins as exemplified in the trees shown in fig. 1 and supplementary fig. S3, Supplementary Material online. Including or excluding sets of DHCs (e.g. the divergent microsporidian class-1 DHCs or all orphan DHCs) did not change the phylogeny of the other classes. Also, including or excluding regions in the alignment, that represent large insertions/loops present in only a few of the DHCs, did not change the tree. For example, the Ascomycota and diatom class-1 DHCs, which do not contain the “C-sequence”, always group correctly to the cytoplasmic dyneins. To exclude bias introduced by the algorithm I computed a phylogenetic tree with RAxML-HPC-Hybrid v. 8.2.8 (Stamatakis 2014) using the high-performance parallel computing implementation at CIPRES (Miller et al. 2011). To generate a small enough but still representative dataset, I reduced the redundancy within the main dataset (supplementary dataset S1, Supplementary Material online) to 70% with CD-Hit (Li and Godzik 2006) and selected conserved alignment blocks with gblocks v. 0.91b and relaxed parameters allowing many contiguous nonconserved positions (Talavera and Castresana 2007). The resultant alignment had 1,202 dynein sequences and 1,710 alignment positions. The amino acid substitution model was LG + Γ + F and the option to halt bootstrapping automatically when certain criteria are met was set (RAxML stopped after 156 bootstrap replicates). The resultant phylogenetic tree showed almost the same topology of the classes as the FastTree generated tree shown in supplementary fig. S3, Supplementary Material online, indicating that the tree topology is independent of the algorithm and independent of reducing redundancy within and removing divergent regions from the alignment. Although I tried various Bayesian tree reconstruction approaches, these failed to converge on the basic dataset within months of computation time. Phylogenetic trees were visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/; last accessed October 8, 2016).
Data Availability
Sequences, domain and motif predictions, and gene structure reconstructions are available at CyMoBase [http://www.cymobase.org (last accessed October 8, 2016), (Odronitz and Kollmar 2006)]. CyMoBase allows searching the data for specific dynein sequences, entire classes, individual species, or taxa, as single selectors or in combinations. In adddition, CyMoBase provides a BLASTP server allowing searching sequences by sequence homology. The results-view also lists references to genome sequencing centers and citations of genome sequence analyses for every matching species. Gene structure visualizations are provided for each sequence, including a reference to the genome assembly used for reconstruction. Each gene structure is linked to WebScipio for in-depth inspection at the nucleotide level.
Supplementary Material
Supplementary data S1 and S2, figures S1–S13 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work has been funded by grants KO 2251/3-1, KO 2251/3-2 and KO 2251/3-3 of the Deutsche Forschungsgemeinschaft.
References
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan VJ, Thompson HM, McNiven MA. 2002. Motoring around the Golgi. Nat Cell Biol. 4:E236–E242. [DOI] [PubMed] [Google Scholar]
- Asai DJ. 1995. Multi-dynein hypothesis. Cell Motil Cytoskeleton. 32:129–132. [DOI] [PubMed] [Google Scholar]
- Asai DJ, Wilkes DE. 2004. The dynein heavy chain family. J Eukaryot Microbiol. 51:23–29. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 183:923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. 2012. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 198:913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. 2011. Crystal structure of the dynein motor domain. Science 331:1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Diamant AG, Urnavicius L. 2016. How dynein and dynactin transport cargos: a structural perspective. Curr Opin Struct Biol. 37:62–70. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 194:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA. 1998. Production of M-phase and I-phase extracts from mammalian cells. Methods Enzymol. 298:331–339. [DOI] [PubMed] [Google Scholar]
- von Dassow P, John U, Ogata H, Probert I, Bendif EM, Kegel JU, Audic S, Wincker P, Da Silva C, Claverie J-M, et al. 2015. Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 9:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 29:1277–1289. [DOI] [PubMed] [Google Scholar]
- DiBella LM, King SM. 2001. Dynein motors of the Chlamydomonas flagellum. Int Rev Cytol. 210:227–268. [DOI] [PubMed] [Google Scholar]
- Eckert C, Hammesfahr B, Kollmar M. 2011. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evol Biol. 11:268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino JE, Gibbons IR. 2002. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics 3:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. 2012. 1 - Discovery of dynein and its properties: a personal account In: King SM, editor. Dyneins. Boston (MA: ): Academic Press; p. 2–87. [Google Scholar]
- Goodenough UW, Heuser JE. 1982. Substructure of the outer dynein arm. J Cell Biol. 95:798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammesfahr B, Kollmar M. 2012. Evolution of the eukaryotic dynactin complex, the activator of cytoplasmic dynein. BMC Evol Biol. 12:95.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H, Smith TF. 2009. The evolution of the cilium and the eukaryotic cell. Cell Motil Cytoskeleton 66:215–219. [DOI] [PubMed] [Google Scholar]
- Hatje K, Hammesfahr B, Kollmar M. 2013. WebScipio: reconstructing alternative splice variants of eukaryotic proteins. Nucleic Acids Res. 41:W504–W509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GP, Gan L, Jensen GJ. 2007. 3-D ultrastructure of O. tauri: electron cryotomography of an entire eukaryotic cell. PLoS ONE 2:e749.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom EFY, Witman GB, Harris EH, Dutcher SK, Kamiya R, Mitchell DR, Pazour GJ, Porter ME, Sale WS, Wirschell M, et al. 2011. A unified taxonomy for ciliary dyneins. Cytoskeleton 68:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höök P, Vallee RB. 2006. The dynein family at a glance. J Cell Sci. 119:4369–4371. [DOI] [PubMed] [Google Scholar]
- Hou Y, Witman GB. 2015. Dynein and intraflagellar transport. Exp Cell Res. 334:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Luck DJ. 1979. Paralyzed flagella mutants of Chlamydomonas reinhardtii. Defective for axonemal doublet microtubule arms. J Biol Chem. 254:3091–3099. [PubMed] [Google Scholar]
- Inaba K. 2007. Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann NY Acad Sci. 1101:506–526. [DOI] [PubMed] [Google Scholar]
- Ishikawa T. 2015. Cryo-electron tomography of motile cilia and flagella. Cilia 4:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Sakakibara H, Oiwa K. 2007. The architecture of outer dynein arms in situ. J Mol Biol. 368:1249–1258. [DOI] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. 1992. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci. 103:653–664. [Google Scholar]
- Kamikawa R, Inagaki Y, Tokoro M, Roger AJ, Hashimoto T. 2011. Split introns in the genome of Giardia intestinalis are excised by spliceosome-mediated trans-splicing. Curr Biol. 21:311–315. [DOI] [PubMed] [Google Scholar]
- Kamiya R. 2002. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 219:115–155. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Yagi T. 2014. Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zool Sci. 31:633–644. [DOI] [PubMed] [Google Scholar]
- Katz LA. 2012. Origin and diversification of eukaryotes. Annu Rev Microbiol. 66:411–427. [DOI] [PubMed] [Google Scholar]
- Kikkawa M. 2013. Big steps toward understanding dynein. J Cell Biol. 202:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature 484:345–350. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 48:373–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 90:5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Vale RD, Fletterick RJ. 1998. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil. 19:877–886. [DOI] [PubMed] [Google Scholar]
- Langousis G, Hill KL. 2014. Motility and more: the flagellum of Trypanosoma brucei. Nat Rev Microbiol. 12:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. 2014. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun. 5:5727.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB, Lesich KA. 2010. Flagellar and ciliary beating: the proven and the possible. J Cell Sci. 123:519–528. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2011. The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery. TG’11. New York (NY): ACM. p. 41:1–41:8.
- Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol. 607:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Brown KS. 1994. Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes. J Cell Sci. 107:635–644. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. 1985. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 100:1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocz G, Gibbons IR. 2001. Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure 9:93–103. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hoffman MP, Obar RA, McCafferty SS, Gibbons IR, Leone AD, Cool J, Allgood EL, Musante AM, Judkins KM, et al. 2006. Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol. 300:219–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S, Kollmar M. 2013. Whole genome duplication events in plant evolution reconstructed and predicted using myosin motor proteins. BMC Evol Biol. 13:202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshid A, Presley JF. 2004. ER-to-Golgi transport and cytoskeletal interactions in animal cells. Cell Mol Life Sci. 61:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43. [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. 2014. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346:857–860. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. 2006. Pfarao: a web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase). BMC Genomics 7:300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 108:13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Fisher EMC, Gibbons IR, Hays TS, Holzbaur ELF, McIntosh JR, Porter ME, Schroer TA, Vaughan KT, Witman GB, et al. 2005. Cytoplasmic dynein nomenclature. J Cell Biol. 171:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T. 2012. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol. 178:199–206. [DOI] [PubMed] [Google Scholar]
- Pillmann H, Hatje K, Odronitz F, Hammesfahr B, Kollmar M. 2011. Predicting mutually exclusive spliced exons based on exon length, splice site and reading frame conservation, and exon sequence homology. BMC Bioinformatics 12:270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. 2013. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 14:713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, et al. 2009. AAA+ Ring and linker swing mechanism in the dynein motor. Cell 136:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. 2009. The motile cilium in development and disease: emerging new insights. Bioessays 31:694–699. [DOI] [PubMed] [Google Scholar]
- Roy SW, Hudson AJ, Joseph J, Yee J, Russell AG. 2012. Numerous fragmented spliceosomal introns, AT-AC splicing, and an unusual dynein gene expression pathway in Giardia lamblia. Mol Biol Evol. 29:43–49. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, NISC Comparative Sequencing Program, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342:1242592.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich-Söldner R, Kahmann R, Steinberg G. 2001. A split motor domain in a cytoplasmic dynein. EMBO J. 20:5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc. Bioinformatics. 00:2.3:2.3.1–2.3.22. [DOI] [PubMed] [Google Scholar]
- Vaughan KT. 2005. Microtubule plus ends, motors, and traffic of Golgi membranes. Biochim Biophys Acta. 1744:316–324. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. 2007. Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. 2012. 2 - Evolutionary Biology of Dyneins A2 In: King SM, editor. Dyneins. Boston (MA: ): Academic Press; p. 88–121. [Google Scholar]
- Wilkerson CG, King SM, Witman GB. 1994. Molecular analysis of the gamma heavy chain of Chlamydomonas flagellar outer-arm dynein. J Cell Sci. 107:497–506. [DOI] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. 2008. Twenty-five dyneins in Tetrahymena: a re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton. 65:342–351. [DOI] [PubMed] [Google Scholar]
- Xu F, Jerlström-Hultqvist J, Einarsson E, Astvaldsson A, Svärd SG, Andersson JO. 2014. Thegenome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet. 10:e1004053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, Kamiya R. 2009. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci. 122:1306–1314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences, domain and motif predictions, and gene structure reconstructions are available at CyMoBase [http://www.cymobase.org (last accessed October 8, 2016), (Odronitz and Kollmar 2006)]. CyMoBase allows searching the data for specific dynein sequences, entire classes, individual species, or taxa, as single selectors or in combinations. In adddition, CyMoBase provides a BLASTP server allowing searching sequences by sequence homology. The results-view also lists references to genome sequencing centers and citations of genome sequence analyses for every matching species. Gene structure visualizations are provided for each sequence, including a reference to the genome assembly used for reconstruction. Each gene structure is linked to WebScipio for in-depth inspection at the nucleotide level.