Abstract
Whole slide imaging (WSI) has the potential to be utilized in telepathology, teleconsultation, quality assurance, clinical education, and digital image analysis to aid pathologists. In this paper, the potential added benefits of computer-assisted image analysis in breast pathology are reviewed and discussed. One of the major advantages of WSI systems is the possibility of doing computer-based image analysis on the digital slides. The purpose of computer-assisted analysis of breast virtual slides can be (i) segmentation of desired regions or objects such as diagnostically relevant areas, epithelial nuclei, lymphocyte cells, tubules, and mitotic figures, (ii) classification of breast slides based on breast cancer (BCa) grades, the invasive potential of tumors, or cancer subtypes, (iii) prognosis of BCa, or (iv) immunohistochemical quantification. While encouraging results have been achieved in this area, further progress is still required to make computer-based image analysis of breast virtual slides acceptable for clinical practice.
Keywords: Breast pathology, breast virtual slides, image analysis, whole slide imaging
INTRODUCTION
Whole slide imaging (WSI) has the potential to be utilized in telepathology, clinical education, and digital image analysis to aid pathologists. As different types of specimen have different specifications, comprehensive studies should be carried out in each pathology subspecialty to assess the extent of added benefits of WSI in that field. A large proportion of the pathology slides are related to breast tissue; for example, in the United States, 1.6 million breast biopsies are assessed by the pathologists each year.[1] Recent studies suggested that WSI can be adopted in breast pathology as pathologists’ performance in reading breast slides while using WSI platforms was comparable to conventional microscopy in breast pathology.[2]
One of the major advantages of WSI systems compared to conventional microscopy is the possibility of doing computer-based image analysis on the digital slides. Recently, many researchers and slide scanner vendors have started developing automated methods to facilitate pathologists’ tasks in breast pathology. This review is restricted to the computer-based image analysis in breast pathology and aimed at discussing the previous studies, summarizing their results, and identifying the remaining challenges and areas where further studies are required.
It should be noted that image analysis done in other pathology subspecialties or animals’ tissue might be extendable to breast pathology; however, discussion about the potentials for extending these ideas to breast pathology is out of the scope of this review. For a broader review on digital pathology in general, please refer to the studies by Pantanowitz et al. and Ghaznavi et al.[3,4]
SEARCH STRATEGY
Three different databases, namely Scopus, PubMed, and IEEEXplore, were searched to find relevant studies published after 1995. Our overall search strategy included terms for digital slides (e.g., whole slide, digital pathology, virtual slide) and breast and was limited to English-language, original, human studies. We also searched references of the retrieved articles. The exact search statement for each database can be found in Appendix 1. For studies where the methodology evolved in two or more papers with considerable amount of overlap, only the most expanded version was included in the review.
Studies focusing on the application of WSI or computer-aided analysis to multispectral images or quantification of biomarkers other than four clinically important immunohistochemical (IHC) stains (i.e., estrogen receptor (ER), progesterone receptor (PR), Ki-67, and human epidermal growth factor receptor [HER2]) have been excluded as they are not currently widely used in the clinical practice.
CLASSIFICATION OF THE REVIEWED STUDIES
The primary purpose of the reviewed studies can be classified into four categories: (i) segmentation of desired regions or objects in the slide, (ii) classification of breast slides, (iii) prognosis of breast cancer (BCa), and (iv) IHC quantification. Most of the reviewed studies had a block diagram similar to the one shown in Figure 1. Some of the methods may not include one or more of the steps illustrated in Figure 1. Moreover, multiple steps may have been merged in some studies. As shown, features could be extracted from a segmented object or tissue texture. Each group of studies is discussed in this section.
Figure 1.
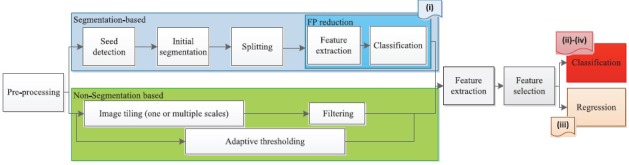
The common steps in the reviewed studies
Before processing the slides, preprocessing steps can be performed to eliminate the background,[5] segment diagnostically relevant area (DRA),[6,7] standardize the color,[6] or separate stain.[8] Color deconvolution is a commonly used preprocessing step to separate the H channel. Ruifrok and Johnston[9] proposed a formulation based on the Beer–Lambert law to map the red, green, and blue (RGB) color space to a set of three stains using color deconvolution. It should be noted that color deconvolution needs prior knowledge about the color vector of each stain (stain matrix). Standard stain matrix for a wide range of stain combinations is provided in a study by Ruifrok and Johnston.[9] However, use of image-specific stain matrix is more accurate. This motivated the development of an image-specific stain normalization algorithm to automatically estimate stain matrix for each slide, such as the one presented in a study by Khan et al.[10] As an alternative solution to overcome color variations due to dyeing, in a study by Ali et al.,[11] stain separation was done adaptively in cyan, magenta, and yellow color space rather than RGB. In addition, the RGB color space is not perceptually uniform. To overcome this problem, in the studies by Dundar et al.[12] and Basavanhally et al.,[13] lab color space which is a perceptually uniform color space was used. Finally, the color deconvolution assumes that the relation between spectral absorbance of a stain mixture and the concentrations of the pure stains is linear. This assumption is valid under monochromatic conditions; however, it introduces an error under nonmonochromatic conditions.[14]
Segmentation of Desired Regions or Objects in the Slide
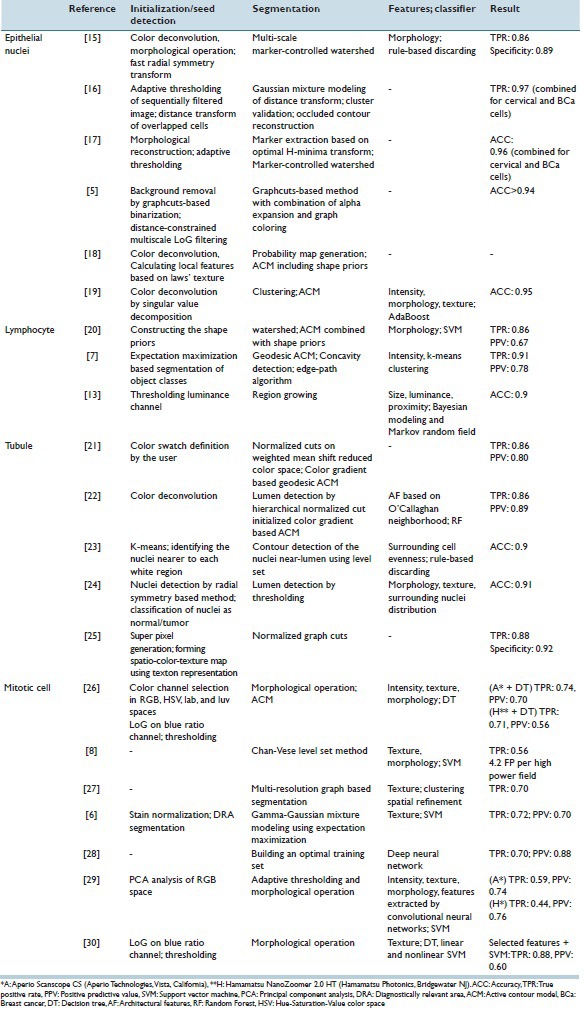
A wide range of image processing methods has been used for segmenting objects in breast virtual slides. Accurate segmentation is important as it is an intermediate step of studies with various purposes. Because of its importance, only proposing a segmentation method to handle difficulties in breast slides has been main subject of 25 reviewed studies. The studies are summarized in Table 1. In this section, methods proposed for segmenting DRAs,[31,32,33,34] epithelial nuclei,[5,15,16,17,18,19,35] lymphocyte cells,[7,13,20] tubule,[21,22,23,24,25] and mitotic figures[6,8,26,27,28,29,30] are discussed.
Table 1.
Summary of the studies aimed at segmentation of structures in breast virtual slides
Diagnostically relevant areas
In pathology slides, large areas are empty. As stated, segmenting DRAs could be used as a preprocessing step to reduce the computational cost[6,7] or to avoid storage of non-DRAs with high magnification.[36] Due to its significance, there are studies aimed only at improving the accuracy of DRAs segmentation.
In the earliest method,[34] thresholding of gray-level image was used to segment DRAs. However, many important features of breast tissues are coded in the color and texture. Therefore, in a study by Mercan et al.,[32] a texton-based approach was proposed to distinguish between DRAs and irrelevant patches. In another study by Khan et al.,[31] Gabor-based texture features were used to differentiate hypocellular from hypercellular stroma. Gabor filters are extensively used in image analysis as they resemble the human visual system.
In a study by Peikari et al.,[33] the areas that attracted pathologists’ attention were found using eye tracking data obtained from pathologists while assessing digital breast slides. The visual bag-of-words model with texture and color features was used to describe DRAs and train a logistic regression and a support vector machine (SVM) to predict DRAs in testing slides.
Epithelial cells
As shown in Figure 1, the segmentation procedure of epithelial cells included seed detection, initial segmentation, splitting, and false positive (FP) reduction. In one of the earliest studies by Dalle et al.,[37] gamma-corrected red channel was used to segment the epithelial cells. K-means clustering in RGB space was also utilized to detect cells.[38] However, in more recent studies, epithelial cells were mostly segmented from the H channel.[15,35,39,40] Thresholding the H channel followed by morphological operation is a low-computational cost approach to detect epithelial cells. Nonetheless, thresholding is sensitive to variations of stain and cannot handle overlapping cells. Hough transform as well as Laplacian of Gaussian (LoG) filtering[41] and its approximation, which is difference of Gaussian (DoG), are also popular tools to detect blob-like objects and are utilized to detect nuclei in breast tissue. They are more robust to the staining variations; however, the Hough transform is a computationally expensive approach and DoG should be deployed in a multi-resolution scheme to address cells with different sizes. The fast radial symmetry transform, which is a computationally efficient, noniterative procedure for localizing radial symmetry objects, has also been utilized for candidate nuclei locations detection.[15]
The initial seeds could be used for further fine segmentation using active contour models (ACMs),[18,19,37,39,40] Graphcuts,[5] or marker-controlled watershed.[15,35] Conventionally, the ACM relies on gray-level image, but breast slides are colored. To address this issue, in a study by Basavanhally et al.,[39] color gradient-based ACM was used. In addition, ACM cannot handle overlapping cells as they rely only on intensity information and do not incorporate knowledge about the nucleus shape. In a study by Veillard et al.,[18] a nucleic shape prior was included to deal with this issue. In a study by Al-Kofahi et al.,[5] Graphcuts could partially handle the segmentation the overlapping cells when combined with distance map constrained multi-scale LoG for initial seed detection. However, Graphcuts led to over-segmentation in enlarged highly textured nuclei and under-segmentation in partially broken or weakly stained touching cells. Marker-controlled watershed is a robust approach for separating the overlapping cells when the initial seeds are correctly localized. However, in case of severely overlapping cells, spurious initial seeds are inevitable. The adaptive H-minima transform[17] and Bayesian classification[16] scheme were proposed to handle severely touching cells.
In the studies by Veta et al.[35] and Vink et al.,[19] an extra FP reduction step was added to improve positive prediction value (PPV). In a study by Veta et al.,[35] morphological features were extracted from each segmented area and rule-based discarding was used to eliminate FPs. In a study by Vink et al.,[19] a more comprehensive feature set including intensity-based features, morphological and textural features was extracted from each segmented area and modified AdaBoost was used for classification of areas as true nuclei or FPs. Further investigations are still required to eliminate FPs and handle overlapping, enlarged, and broken cells.
Lymphocytes
Lymphocytic infiltration is a prognostic indicator; therefore, recently, researchers worked on the automatic segmentation of lymphocytes. In a study by Basavanhally et al.,[13] lymphocytes were initially segmented using region growing, which resulted in a large number of epithelial nuclei being detected as well. Bayesian modeling of size and luminance of lymphocytes and proximity modeling using Markov random field were used to eliminate nuclei. However, region growing cannot handle overlapping cells. To address this, a concavity detection scheme was proposed in a study by Fatakdawala.[7] The expectation maximization-based method was used to initialize the ACM and then overlapping cells were split. Despite achieving a true positive rate (TPR) of 86%, PPV was only 64%. In a study by Ali and Madabhushi,[20] shape priors were incorporated in ACM to handle overlapped cells and the watershed algorithm was used to initialize the ACM. Similarly, high TPR (86%) and low PPV (67%) were achieved. Therefore, it seems that adding an FP reduction module is required to eliminate epithelial nuclei.
Tubules
Tubules are characterized by a white region called lumen, surrounded by a single layer of nuclei in normal breast histopathology. Dalle et al.[42] utilized thresholding followed by morphological operations to segment lumen areas. However, thresholding was not robust to stain variation. Xu et al.[21] proposed a color gradient-based geodesic ACM which was initialized by weighted mean shift clustering and normalized cuts for lumen segmentation. Later, in a study by Basavanhally et al.,[22] domain knowledge was incorporated into method proposed in a study by Xu et al.[21] and each segmented area was classified either as true or false based on architectural features and an accuracy of 86% in detection of true lumen was achieved. Maqlin et al.[23] used heuristic rules based on evenness and closeness of strings of surrounding nuclei to eliminate lumen-like areas and achieved an accuracy of 90%.
The above-mentioned methods associated only the closest nuclei to the lumen. However, it could be surrounded by multiple layers of nuclei. Therefore, in a study by Nguyen et al.,[24] the global distribution of the nuclei and lumina were considered. Furthermore, a comprehensive feature set containing architectural, morphological, intensity-based, and textural features were used to distinguish true lumina from artifacts. Finally, the discussed methods focus on lumen detection; however, nuclear arrangement in tubules could be with a lumen or in solid islands without a lumen. Belsare et al.[25] proposed a novel integrated spatio-color-texture-based graph partitioning method to address this issue and achieved a correct classification rate (CCR) of 92% for segmentation.
Mitotic figures
Mitosis counting is tedious and subject to inter-observer variation. The automatic detection of mitosis could potentially address these problems. Roullier et al.[27] proposed a multi-resolution image analysis strategy for detection of mitotic figures based on Graph-based regularization. The method was analyzed WSI at different levels and segmented the relevant areas and detected the mitoses in the highest magnification, and it was completely unsupervised. However, the detection rate was 70% and no FP reduction step was adopted; hence, further improvement was required for deploying the method in a clinical setting.
In a study by Khan et al.,[6] the pixel intensities of mitotic and nonmitotic areas were modeled by a Gamma-Gaussian mixture. A set of textural and intensity-based features were extracted from each region labeled as mitosis by the first module. The features fed into an SVM classifier which detects FP instances. The obtained TPR and PPV were 72% and 70%. Irshad et al.[30] detected the candidate region in the blue ratio image using thresholding followed by morphological operations and extracted a patch of size 80 pixel × 80 pixel from blue ratio and red and blue channels of RGB color space. A wide range of textural features including Haralick textural, gray-level run length, scale invariant feature transform, and Gabor-based features was extracted from each patch. The principal component analysis was used for dimension reduction. The features were fed into decision tree, linear SVM, and nonlinear SVM. It was shown that a decision tree achieved to the highest performance with a TPR of 76% and a PPV of 75%. Later, Irshad[26] investigated the added value of morphological features and features from other color spaces to the FP reduction step, but the result did not improve.
One of the difficulties in the detection of mitotic figure is its wide range of appearances. To handle this issue, in the studies by Cireşan et al. and Malon and Cosatto,[28,29] the learned features extracted by convolutional neural networks were utilized. Malon and Cosatto[29] achieved a TPR of 59% and a PPV of 75% while in a study by Cireşan et al.,[28] the PPV and TPR were improved to 80% and 70% using an optimized approach for sampling nonmitosis pixels in the training set. Further investigations are still required in mitotic detection filed to improve TPR and PVP and also deal with the wide range of variability in the appearance of mitotic figures.
Classification
Computer-based image analysis of breast slides may aim at classifying the virtual slide into different categories. The classification could be done based on the grade of BCa, the invasive potential of tumors, or cancer subtypes. Table 2 summarizes the purposes and methods of the reviewed studies aimed at classification of breast slides.
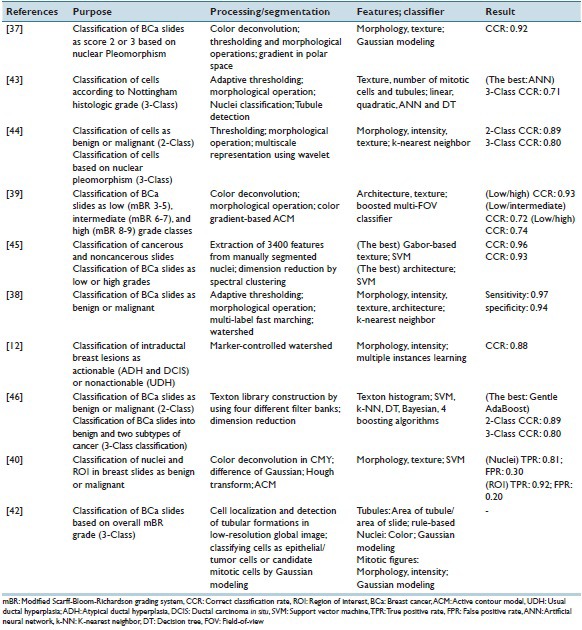
Table 2.
Summary of the studies aimed at breast histopathology slides classification
Cancer grading
Scarff-Bloom-Richardson grading system is a well-known grading system relying on magnitude of tubule formation, nuclear pleomorphism, and mitotic count. Segmenting the mitotic figures (which leads to the mitotic count) has been discussed in sections 1–5. The discussed studies in this section aimed at classifying the slides according to nuclear pleomorphism[37,43,44] or all three factors.[39,45]
The earliest reviewed study by Weyn et al.[44] applied wavelet transform in four levels on the segmented nuclei and the energy of filtered images in each scale was calculated. In addition to wavelet-based, Haralick, intensity-based, and morphological features were extracted and fed into a K-nearest neighbor classifier to separate individual nuclei and also each case in four categories (normal, nuclear atypia Grade I, II, III). A CCR of 64% for classification of individual nuclei and a CCR of 79% for case-based classification were observed. It was shown that textural features (wavelet-based and Haralick features) had a high additive value to intensity-based features. The dataset used in the study was highly imbalance (21 normal vs. eight Grade III cases) and the segmentation method was required further refinement.
In a study by Doyle et al.,[45] the centers of nuclei were manually segmented and a range of intensity-based and textural (Gabor-based and Haralick) features were extracted from each nucleus. The mean, standard deviation, minimum-to-maximum ratio, and mode of these features over all cells in each slide were calculated and formed the feature vector. The architectural features were also extracted from Voronoi diagram, Delaunay triangulation, minimum spanning tree, and nuclei density function. The architectural features resulted in the highest CCR for low- versus high-grade classification while the textural features resulted in a significantly lower CCR (73 vs. 93%). Despite the encouraging CCR, the fact that the segmentation was done manually limits generalizability of the study. In a study by Dalle et al.,[37] the nuclei segmentation was done automatically using polar transform, and area, compactness, and mean intensity were extracted from each segmented nuclei. A high value for CCR (92%) was achieved for scoring nuclear pleomorphism of 2396 region of interests (ROIs). However, the result could be biased as the dataset contained images from only six patients and did not include any patient with Grade I.
Pathologists implicitly integrate features from multiple field-of-views (FOVs) of different sizes when grading BCa. However, automatically selecting an optimal FOV size is not straightforward. In a study by Basavanhally et al.,[39] architectural and textural features were extracted from a multi-FOV of varying sizes and important features at different FOV sizes were identified to distinguish low/high-, low/intermediate-, and intermediate/high-grade patients. Unsurprisingly, the highest performance was obtained when distinguishing low from high-grade patients. Similar to results obtained in a study by Doyle et al.,[45] architectural features performed better than textural ones. It was also observed that the most discriminating architectural features were different in FOVs with various sizes while contrast played a dominant role among textural features. It was also shown that the multi-FOV classifier outperformed multi-scale classifier.
All of the above-mentioned methods extracted features from segmented nuclei only while pathologists rely on features from other structures such as tubules. To overcome this limitation, in a study by Petushi et al.,[43] features were also extracted from the tubule and showed that the density of tubule and number of Grade III would be useful parameters for BCa grading.
Benign versus malignant classification and distinguishing lesion subtypes
A pathologist usually inspects the breast tissue to determine if it is a benign or malignant lesion is present and also to identify the cancer type (if appropriate). Computer-aided detection (CAD) tools could help the pathologists in this task and make the results less susceptible to observer variation.
In the earliest CAD system,[44] it was shown that textural features (wavelet-based and Haralick) outperformed morphological and intensity-based features in differentiating benign from malignant cells. Later, in a study by Doyle et al.,[45] Gabor-based features, which are also a textural feature, achieved higher CCR compared to architectural features, and the diagnostic importance of nuclear texture in differentiating normal from cancerous tissue has been shown. However, the wavelet-based, Haralick, and Gabor-based features are not easily interpretable to pathologists. Furthermore, the high dimension of feature vector when low number of training instances is available increases the chance of overfitting of the classifier to in hand data. Therefore, in a study by Cosatto et al.,[40] only the median nuclear area over an ROI and the number of large well-formed nuclei were utilized to train a linear SVM with a labeled dataset of 335 hand-picked ROIs. A sensitivity of 92% and a specificity of 80% were obtained. However, no information was provided about number of the patients from whom ROIs were picked.
In the real clinical practice, the ultimate goal is distinguishing patients with malignancy and not the individual ROIs. Pathologists judge each case based on multiple ROIs and label it accordingly. In a study by Filipczuk et al.,[38] a larger set from fifty patients (nine ROIs per patient) were classified as either benign or malignant using 84 features (morphological, intensity-based, and textural) extracted from isolated nuclei in each ROI. Sequential forward feature selection was used to reduce number of features, and a k-nearest neighbor was used as a classifier. The final diagnosis for each patient was obtained by a majority voting of the classification of all nine ROIs belonging to the same patient. A CCR of 100% was achieved. Considering the high CCR obtained in the study, a further investigation on this method on a larger data set is useful as no information about number of borderline cases was provided in the paper. Moreover, using majority voting could be questionable as pathologists consider a case malignant when at least one of the ROIs in the slide is positive. To address this issue, in a study by Dundar et al.,[12] learning with multiple instances was used to train an SVM classifier. According to the proposed classifier, a benign case was misclassified when at least one of the ROIs in a slide was classified as malignant, and a malignant case was misclassified when all of the ROIs in a slide were classified as benign. The method has been tested on a dataset of 20 well-defined ductal carcinoma in situ (DCIS), 12 borderline DCSI, 24 atypical ductal hyperplasia (ADH), and 39 usual ductal hyperplasia (UDH). DCSI and ADH cases were grouped as actionable (malignant) while UDH cases were considered nonactionable (benign). An overall accuracy of 87.9% was obtained while the accuracy on the borderline cases was 84.6%, comparable to that of nine pathologists on the same set (81.2% average). Despite encouraging result, for deploying such a system in clinical practice as an aid to pathologists, its additive value to a pathologist's diagnosis should also be assessed. Moreover, the proposed method (classification rule and features) is not easily interpretable to pathologists.
Unlike the above-mentioned studies, pathologists do not segment each individual nucleus within a slide; however, they analyze the scene holistically. In a study by Yang et al.,[46] textural features based on texton-based method were extracted without segmenting the structures in slides. CCRs of 89% and 80% were achieved in benign/malignant and multi-class (benign and two major cancer subtypes) classification.
Prognosis of Breast Cancer
The advent of WSI allows extracting quantitative features which could be helpful in predicting prognosis of BCa. In a study by Veta et al.,[47] an automatic nuclei segmentation algorithm[15] was utilized to extract size-related nuclear morphometric features and their prognostic value in male BCa was investigated. The results demonstrated that mean nuclear area has a significant prognostic value. In another study, Beck et al.[48] showed that quantitative stromal features are associated with survival. A comprehensive set of quantitative features from the BCa epithelium and stroma was extracted by utilizing a machine learning method called computational pathologist. The prognostic model was based on the extracted features and it was shown that the score from the model was strongly associated with overall survival. In addition, assessing significance of features revealed that survival was strongly related to three of the stromal features and the magnitude of association was stronger than the association of survival with epithelial features.
The presence of lymphocytic infiltration is also a prognostic indicator for in HER2 + BCa patients. Currently, pathologists do not routinely report the presence of LI as quantifying it is a tedious job. As discussed in 1–3, recently researchers worked on automatic detection of lymphocytes. However, a further step should be added to grade the extent of lymphocytic infiltration. In a study by Basavanhally et al.,[13] architectural features were extracted from Voronoi diagram, Delaunay triangulation, and minimum spanning tree using the centers of individually detected lymphocytes as vertices. A CCR of 90% was achieved in differentiating patients with high and low lymphocytic infiltration level. However, the dataset contained ROIs only from 12 patients. To assess the generalizability of the method, further analysis on a larger dataset is recommended.
Immunohistochemical Quantification
Currently, the standard procedure in pathology laboratories for assessment of IHC is visual examination of samples by a pathologist. The pathologist determines the status of receptors by counting positively stained cells. Hence, the procedure is tedious and prone to inter-observer variability due to subjectiveness. Computer-assisted methods in the field of IHC quantification aim at quantification of information extracted from IHC-stained samples to reduce inter-observer variability and assessment time.
HER2 receptors typically express on the cell membrane. Therefore, membrane segmentation is one of the main steps of automated methods for quantification of HER2. A color-based approach,[49] water shedding,[50] and skeletonization[51] were used for membrane segmentation. After the segmentation stage, a group of features was extracted from the membrane and then utilized for prediction of HER2 score. The extracted features were based on membrane staining intensity,[49,51,52,53] membrane completeness,[49,53] or membrane color properties.[54,55] Instead of restricting the area for feature selection to the segmented membrane, Ali et al. utilized an algorithm which was previously used for analysis of astronomical images and extracted intensity-based features from the entire image without segmentation.[11,56] The agreement of the reviewed automated methods with the expert scoring is listed in Table 3.
Table 3.
Automatic and semi-automatic methods for immunohistochemical quantification
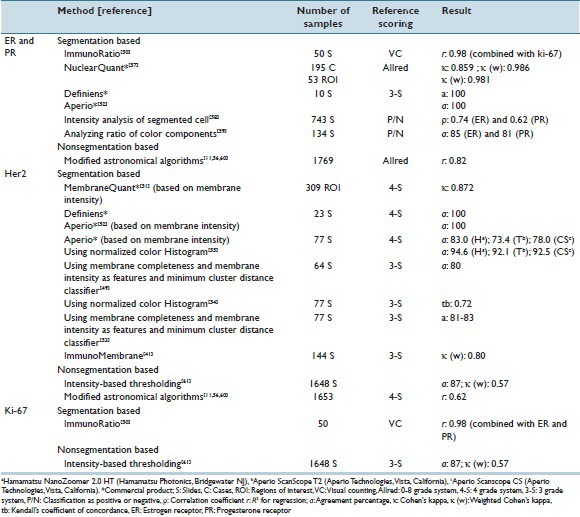
In contrast to HER2 receptors, ER and PR overexpression typically results in nuclear immunoreactivity, and hence, nuclei segmentation is the first step of some of the reviewed methods listed in Table 3. The extracted features from segmented nuclei in the reviewed studies were based on nuclei staining intensity,[57,58] nuclei shape,[52,57] or nuclei color properties.[50,59] Rather than segmenting the nuclei, Amaral et al. proposed a method for predicting quick score values for receptor assessment based on color- and intensity-based features extracted from each pixel of test images without segmentation of nuclei.[60] As shown in Table 3, the reviewed automated methods for ER and PR assessment showed high agreement with the expert scoring.
Similar to the methods for ER and PR assessment, quantitative assessment of Ki-67 could be done by extracting features based on either the segmented cell nuclei[50] or the percentage of stained area.[61] The agreement between the automated methods and the visual examination done by pathologist is reported in Table 3.
ImmunoRatio[50] and ImmunoMembrane[41] are two publicly available web-based applications. ImmunoRatio is a tool for quantitative assessment of ER, PR, and Ki-67 while ImmunoMembrane is an HER2 IHC analysis software. Both applications were tested and matched well with the pathologist's visual examination.[41,50]
DISCUSSION
The emphasis of this review was discussing the computer-based image analysis in breast pathology. In spite of encouraging results achieved by the reviewed studies, further progress is still required to make the CAD tools acceptable for clinical practice. For example, segmentation of severely overlapping and broken cells has not been fully addressed yet. Moreover, the low PPV of segmentation methods suggests that an FP reduction step should follow the initial segmentation. In addition, only a few studies focused on automatic segmentation and grading of tubule formation as well as distinguishing cancer subtypes; hence, further studies in these fields are required. Moreover, most of the studies attempted to extract features from the segmented objects. Further investigation of nonsegmentation-based methods in breast pathology is required as these methods avoid error propagation from the segmentation step and also mimic the human visual system which captures textural features.
Pathologists extract information from multiple ROIs and scales. Using multi-ROI and multi-scale approach to mimic the perception of pathologists could be a potential direction for future studies. Furthermore, clinicians prefer a CAD which provides physically interpretable features and classification rules; however, most of the current tools are “black box” systems.
One of the other major challenges of CAD is variability of breast tissue. Although standardization of slide preparation protocols, color normalization, noise reduction, and quality assurance programs will tackle the tissue variability problems to some extent, there is an inherent variability in the appearance of the objects within the breast tissue, which cannot be compensated. For example, the shape of epithelial cancerous nuclei may vary from almost normal-like round structure to highly irregularly shaped and enlarged nuclei with coarse and marginalized chromatin and prominent nucleoli. Moreover, the fact that different structures in breast histopathology slides may look similar decreases the specificity of CAD in detection of certain features. Another difficulty for segmentation-based CAD is separating clustered or overlapping cells. All these factors that affect adversely on the performance of CAD systems should be addressed to obtain a CAD which is robust enough to be used in the clinical practice of pathology.
In evaluation of CAD studies, inherent inter-pathologist variations should be considered. For example, in a study by Shaw et al.,[2] it was shown that intra- and inter-pathologist agreement for detection of pleomorphism is lower than that of IHC quantification. Similarly, automatic IHC quantification tools usually achieved higher agreement with pathologists’ assessment in comparison with CADs aimed BCa grading [Tables 2 and 3].
Moreover, the additive value of CAD to pathologist's opinion should be investigated as CAD could be potentially used as “second reader.” Finally, one of the major obstacles for researchers working on BCa digital slides is lack of publicly available data sets which enable them to evaluate the performance and robustness of their proposed algorithms. Having such reference databases whose ground truth was built based on a panel of expert pathologists would provide a unique opportunity for comparing different algorithms’ performance against each other. Recently, two publicly available databases for mitosis detection have been introduced;[62] however, more databases containing virtual slides of different BCa types, different grades of BCa, and so on from different scanners are still required.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
APPENDIX
Appendix 1
The advanced search option of the databases was used to find the articles. The search was limited to human studies. Endnote was used for reference management. After combining all references and omitting non-English references, duplicated studies were omitted by using a built-in function in Endnote. Nonoriginal studies (e.g., review paper, abstract paper or report), undetected duplicates, and nonrelevant studies were excluded based on scanning the article's title and abstracts. Then, included papers are downloaded and fully studied and a few of them were further excluded in case they were not original or relevant to the topic of the review.
The following statement was used to search Scopus:
TITLE-ABS-KEY (“breast”) AND (TITLE-ABS-KEY [“virtual slide”] OR TITLE-ABS-KEY [“whole slide”] OR TITLE-ABS-KEY [“digital pathology”] OR TITLE-ABS-KEY [“digital histopathology”] OR TITLE-ABS-KEY [“whole-slide”] OR TITLE-ABS-KEY [“digitized histopathology”] OR TITLE-ABS-KEY [“digital slide”] OR TITLE-ABS-KEY [“digitized slide”] OR TITLE-ABS-KEY [“digitized cytology”] OR TITLE-ABS-KEY [“digital cytology”] OR TITLE-ABS-KEY [“digital cytopathology”] OR TITLE-ABS-KEY [“digitized cytopathology”] OR TITLE-ABS-KEY [“cell segmentation”] OR TITLE-ABS-KEY [“nuclei segmentation”] OR TITLE-ABS-KEY [“nucleus segmentation”]).
The following statement was used to search IEEEXplore
(QT breast QT) AND ([“QT virtual slide QT”] OR [“QT whole slide”] OR [“QT digital pathology”] OR [“QT digital histopathology QT”] OR [“QT whole-slide”] OR [“digital slide”] OR [“QT digital cytology QT”] OR [“QT cell segmentation QT”] OR [“QT nucleus segmentation QT] OR [“QT nuclei segmentation QT”] OR [“QT histometry QT”] OR [“QT histology image QT”] OR [“QT histopathology image QT”] OR [“QT mitotic QT]).
The following statement was used to search PubMed
(“Breast”) AND ([“virtual slide”] OR [“whole slide”] OR [“digital pathology”] OR [“digital histopathology”] OR [“whole-slide”] OR [“digital slide”] OR [“digital cytology”] OR [“cell segmentation”] OR [“nucleus segmentation”] OR [“nuclei segmentation”] OR [“histometry”] OR [“histology image”] OR [“histopathology image”]).
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2016/7/1/43/192814
REFERENCES
- 1.Silverstein M. Where's the outrage? J Am Coll Surg. 2009;208:78–9. doi: 10.1016/j.jamcollsurg.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Shaw EC, Hanby AM, Wheeler K, Shaaban AM, Poller D, Barton S, et al. Observer agreement comparing the use of virtual slides with glass slides in the pathology review component of the POSH breast cancer cohort study. J Clin Pathol. 2012;65:403–8. doi: 10.1136/jclinpath-2011-200369. [DOI] [PubMed] [Google Scholar]
- 3.Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghaznavi F, Evans A, Madabhushi A, Feldman M. Digital imaging in pathology: Whole-slide imaging and beyond. Annu Rev Pathol. 2013;8:331–59. doi: 10.1146/annurev-pathol-011811-120902. [DOI] [PubMed] [Google Scholar]
- 5.Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010;57:841–52. doi: 10.1109/TBME.2009.2035102. [DOI] [PubMed] [Google Scholar]
- 6.Khan AM, Eldaly H, Rajpoot NM. A gamma-gaussian mixture model for detection of mitotic cells in breast cancer histopathology images. J Pathol Inform. 2013;4:11. doi: 10.4103/2153-3539.112696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): Application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:1676–89. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- 8.Veta M, Van Diest PJ, Pluim JP. Detecting Mitotic Figures in Breast Cancer Histopathology Images. In Progress in Biomedical Optics and Imaging – Proceedings of SPIE. 2013 [Google Scholar]
- 9.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–9. [PubMed] [Google Scholar]
- 10.Khan AM, Rajpoot N, Treanor D, Magee D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng. 2014;61:1729–38. doi: 10.1109/TBME.2014.2303294. [DOI] [PubMed] [Google Scholar]
- 11.Ali HR, Irwin M, Morris L, Dawson SJ, Blows FM, Provenzano E, et al. Astronomical algorithms for automated analysis of tissue protein expression in breast cancer. Br J Cancer. 2013;108:602–12. doi: 10.1038/bjc.2012.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundar MM, Badve S, Bilgin G, Raykar V, Jain R, Sertel O, et al. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans Biomed Eng. 2011;58:1977–84. doi: 10.1109/TBME.2011.2110648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:642–53. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 14.Haub P, Meckel T. A model based survey of colour deconvolution in diagnostic brightfield microscopy: Error estimation and spectral consideration. Sci Rep. 2015;5:12096. doi: 10.1038/srep12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veta M, van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JP. Automatic nuclei segmentation in H & E stained breast cancer histopathology images. PLoS One. 2013;8:e70221. doi: 10.1371/journal.pone.0070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung C, Kim C, Chae SW, Oh S. Unsupervised segmentation of overlapped nuclei using Bayesian classification. IEEE Trans Biomed Eng. 2010;57:2825–32. doi: 10.1109/TBME.2010.2060486. [DOI] [PubMed] [Google Scholar]
- 17.Jung C, Kim C. Segmenting clustered nuclei using H-minima transform-based marker extraction and contour parameterization. IEEE Trans Biomed Eng. 2010;57:2600–4. doi: 10.1109/TBME.2010.2060336. [DOI] [PubMed] [Google Scholar]
- 18.Veillard A, Kulikova MS, Racoceanu D. Cell nuclei extraction from breast cancer histopathology images using colour, texture, scale and shape information. Diagn Pathol. 2013;8:1. [Google Scholar]
- 19.Vink JP, Van Leeuwen MB, Van Deurzen CH, De Haan G. Efficient nucleus detector in histopathology images. J Microsc. 2013;249:124–35. doi: 10.1111/jmi.12001. [DOI] [PubMed] [Google Scholar]
- 20.Ali S, Madabhushi A. An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging. 2012;31:1448–60. doi: 10.1109/TMI.2012.2190089. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Janowczyk A, Chandran S, Madabhushi A. A Weighted Mean Shift, Normalized Cuts Initialized Color Gradient Based Geodesic Active Contour Model: Applications to Histopathology Image Segmentation. In SPIE Medical Imaging. International Society for Optics and Photonics. 2010 [Google Scholar]
- 22.Basavanhally A, Yu E, Xu J, Ganesan S, Feldman M, Tomaszewski J, et al. Incorporating Domain Knowledge for Tubule Detection in Breast Histopathology Using O’Callaghan Neighborhoods. In SPIE Medical Imaging. International Society for Optics and Photonics. 2011 [Google Scholar]
- 23.Maqlin P, Thamburaj R, Mammen JJ, Nagar AK. Springer; 2013. Automatic Detection of Tubules in Breast Histopathological Images. In Proceedings of Seventh International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA 2012) [Google Scholar]
- 24.Nguyen K, Barnes M, Srinivas C, Chefd’hotel C. Automatic Glandular and Tubule Region Segmentation in Histological Grading of Breast Cancer. In SPIE Medical Imaging. International Society for Optics and Photonics. 2015 [Google Scholar]
- 25.Belsare AD, Mushrif MM, Pangarkar MA, Meshram N. Breast histopathology image segmentation using spatio-colour-texture based graph partition method. J Microsc. 2016;262:260–73. doi: 10.1111/jmi.12361. [DOI] [PubMed] [Google Scholar]
- 26.Irshad H. Automated mitosis detection in histopathology using morphological and multi-channel statistics features. J Pathol Inform. 2013;4:10. doi: 10.4103/2153-3539.112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roullier V, Lézoray O, Ta VT, Elmoataz A. Multi-resolution graph-based analysis of histopathological whole slide images: Application to mitotic cell extraction and visualization. Comput Med Imaging Graph. 2011;35:603–15. doi: 10.1016/j.compmedimag.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Cireºan DC, Giusti A, Gambardella LM, Schmidhuber J. Springer; 2013. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks. In International Conference on Medical Image Computing and Computer-assisted Intervention. [DOI] [PubMed] [Google Scholar]
- 29.Malon CD, Cosatto E. Classification of mitotic figures with convolutional neural networks and seeded blob features. J Pathol Inform. 2013;4:9. doi: 10.4103/2153-3539.112694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irshad H, Jalali S, Roux L, Racoceanu D, Hwee LJ, Naour GL, Capron F. Automated mitosis detection using texture, SIFT features and HMAX biologically inspired approach. J Pathol Inform. 2013;4:12. doi: 10.4103/2153-3539.109870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan AM, El-Daly H, Simmons E, Rajpoot NM. HyMaP: A hybrid magnitude-phase approach to unsupervised segmentation of tumor areas in breast cancer histology images. J Pathol Inform. 2013;4:1. doi: 10.4103/2153-3539.109802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercan E, Aksoy S, Shapiro LG, Weaver DL, Brunye T, Elmore JG. Localization of Diagnostically Relevant Regions of Interest in Whole Slide Images. In Pattern Recognition (ICPR), 2014 22nd International Conference on. 2014 [Google Scholar]
- 33.Peikari M, Gangeh MJ, Zubovits J, Clarke G, Martel AL. Triaging diagnostically relevant regions from pathology whole slides of breast cancer: A texture based approach. IEEE Trans Med Imaging. 2016;35:307–15. doi: 10.1109/TMI.2015.2470529. [DOI] [PubMed] [Google Scholar]
- 34.Petushi S, Katsinis C, Coward C, Garcia F, Tozeren A. Automated Identification of Microstructures on Histology Slides. In Biomedical Imaging: Nano to Macro, 2004. IEEE International Symposium on 2004. IEEE [Google Scholar]
- 35.Veta M, Huisman A, Viergever MA, van Diest PJ, Pluim JP. Marker-Controlled Watershed Segmentation of Nuclei in H & E Stained Breast Cancer Biopsy Images. In Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on 2011. IEEE [Google Scholar]
- 36.Racoceanu D, Capron F. Towards semantic-driven high-content image analysis: An operational instantiation for mitosis detection in digital histopathology. Comput Med Imaging Graph. 2015;42:2–15. doi: 10.1016/j.compmedimag.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Dalle JR, Li H, Huang CH, Leow WK, Racoceanu D, Putti TC. Nuclear pleomorphism scoring by selective cell nuclei detection. In WACV: IEEE. 2009 [Google Scholar]
- 38.Filipczuk P, Kowal M, Obuchowicz A. Multi-label fast marching and seeded watershed segmentation methods for diagnosis of breast cancer cytology. Conf Proc IEEE Eng Med Biol Soc. 2013;2013:7368–71. doi: 10.1109/EMBC.2013.6611260. [DOI] [PubMed] [Google Scholar]
- 39.Basavanhally A, Ganesan S, Feldman M, Shih N, Mies C, Tomaszewski J, et al. Multi-field-of-view framework for distinguishing tumor grade in ER+breast cancer from entire histopathology slides. IEEE Trans Biomed Eng. 2013;60:2089–99. doi: 10.1109/TBME.2013.2245129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosatto E, Miller M, Graf HP, Meyer JS. Grading Nuclear Pleomorphism on Histological Micrographs. In Pattern Recognition, 2008. ICPR 2008 19th International Conference on 2008 IEEE [Google Scholar]
- 41.Tuominen VJ, Tolonen TT, Isola J. ImmunoMembrane: A publicly available web application for digital image analysis of HER2 immunohistochemistry. Histopathology. 2012;60:758–67. doi: 10.1111/j.1365-2559.2011.04142.x. [DOI] [PubMed] [Google Scholar]
- 42.Dalle JR, Leow WK, Racoceanu D, Tutac AE, Putti TC. Automatic Breast Cancer Grading of Histopathological Images. In 2008 30 th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE. 2008 doi: 10.1109/IEMBS.2008.4649847. [DOI] [PubMed] [Google Scholar]
- 43.Petushi S, Garcia FU, Haber MM, Katsinis C, Tozeren A. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med Imaging. 2006;6:14. doi: 10.1186/1471-2342-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weyn B, van de Wouwer G, van Daele A, Scheunders P, van Dyck D, van Marck E, et al. Automated breast tumor diagnosis and grading based on wavelet chromatin texture description. Cytometry. 1998;33:32–40. doi: 10.1002/(sici)1097-0320(19980901)33:1<32::aid-cyto4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 45.Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated Grading of Breast Cancer Histopathology Using Spectral Clustering with Textural and Architectural Image Features. In Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008 5th IEEE International Symposium on 2008. IEEE [Google Scholar]
- 46.Yang L, Chen W, Meer P, Salaru G, Goodell LA, Berstis V, et al. Virtual microscopy and grid-enabled decision support for large-scale analysis of imaged pathology specimens. IEEE Trans Inf Technol Biomed. 2009;13:636–44. doi: 10.1109/TITB.2009.2020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veta M, Kornegoor R, Huisman A, Verschuur-Maes AH, Viergever MA, Pluim JP, et al. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol. 2012;25:1559–65. doi: 10.1038/modpathol.2012.126. [DOI] [PubMed] [Google Scholar]
- 48.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 49.Gavrielides MA, Masmoudi H, Petrick N, Myers KJ, Hewitt SM. Automated Evaluation of HER-2/neu Immunohistochemical Expression in Breast Cancer Using Digital Microscopy. In 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Proceedings, ISBI. 2008 [Google Scholar]
- 50.Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: A publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Micsik T, Kiszler G, Szabó D, Krecsák L, Hegedus C, Tibor K, et al. Computer aided semi-automated evaluation of HER2 immunodetection – A robust solution for supporting the accuracy of anti HER2 therapy. Pathol Oncol Res. 2015;21:1005–11. doi: 10.1007/s12253-015-9927-6. [DOI] [PubMed] [Google Scholar]
- 52.Lloyd MC, Allam-Nandyala P, Purohit CN, Burke N, Coppola D, Bui MM. Using image analysis as a tool for assessment of prognostic and predictive biomarkers for breast cancer: How reliable is it? J Pathol Inform. 2010;1:29. doi: 10.4103/2153-3539.74186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masmoudi H, Hewitt SM, Petrick N, Myers KJ, Gavrielides MA. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans Med Imaging. 2009;28:916–25. doi: 10.1109/TMI.2009.2012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keller B, Chen W, Gavrielides MA. Quantitative assessment and classification of tissue-based biomarker expression with color content analysis. Arch Pathol Lab Med. 2012;136:539–50. doi: 10.5858/arpa.2011-0195-OA. [DOI] [PubMed] [Google Scholar]
- 55.Keay T, Conway CM, O’Flaherty N, Hewitt SM, Shea K, Gavrielides MA. Reproducibility in the automated quantitative assessment of HER2/neu for breast cancer. J Pathol Inform. 2013;4:19. doi: 10.4103/2153-3539.115879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gertych A, Mohan S, Maclary S, Mohanty S, Wawrowsky K, Mirocha J, et al. Effects of tissue decalcification on the quantification of breast cancer biomarkers by digital image analysis. Diagn Pathol. 2014;9:213. doi: 10.1186/s13000-014-0213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krecsák L, Micsik T, Kiszler G, Krenács T, Szabó D, Jónás V, et al. Technical note on the validation of a semi-automated image analysis software application for estrogen and progesterone receptor detection in breast cancer. Diagn Pathol. 2011;6:6. doi: 10.1186/1746-1596-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, et al. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: Application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10:R89. doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharangpani GM, Joshi AS, Porter K, Deshpande AS, Keyhani S, Naik GA, et al. Semi-automated imaging system to quantitate estrogen and progesterone receptor immunoreactivity in human breast cancer. J Microsc. 2007;226(Pt 3):244–55. doi: 10.1111/j.1365-2818.2007.01772.x. [DOI] [PubMed] [Google Scholar]
- 60.Amaral T, McKenna SJ, Robertson K, Thompson A. Classification and immunohistochemical scoring of breast tissue microarray spots. IEEE Trans Biomed Eng. 2013;60:2806–14. doi: 10.1109/TBME.2013.2264871. [DOI] [PubMed] [Google Scholar]
- 61.Konsti J, Lundin M, Joensuu H, Lehtimäki T, Sihto H, Holli K, et al. Development and evaluation of a virtual microscopy application for automated assessment of Ki-67 expression in breast cancer. BMC Clin Pathol. 2011;11:3. doi: 10.1186/1472-6890-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–48. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]


