Murugesan et al. report that actomyosin arcs at the T cell synapse are formin-generated structures that directly propel T cell receptor cluster movement. The authors reveal the origin, organization, and functions of a major cytoskeletal network during synapse maturation.
Abstract
Actin assembly and inward flow in the plane of the immunological synapse (IS) drives the centralization of T cell receptor microclusters (TCR MCs) and the integrin leukocyte functional antigen 1 (LFA-1). Using structured-illumination microscopy (SIM), we show that actin arcs populating the medial, lamella-like region of the IS arise from linear actin filaments generated by one or more formins present at the IS distal edge. After traversing the outer, Arp2/3-generated, lamellipodia-like region of the IS, these linear filaments are organized by myosin II into antiparallel concentric arcs. Three-dimensional SIM shows that active LFA-1 often aligns with arcs, whereas TCR MCs commonly reside between arcs, and total internal reflection fluorescence SIM shows TCR MCs being swept inward by arcs. Consistently, disrupting actin arc formation via formin inhibition results in less centralized TCR MCs, missegregated integrin clusters, decreased T–B cell adhesion, and diminished TCR signaling. Together, our results define the origin, organization, and functional significance of a major actomyosin contractile structure at the IS that directly propels TCR MC transport.
Introduction
Recognition of antigen on the surface of an antigen-presenting cell (APC) initiates signaling cascades within the T cell that drive large-scale reorganization of its actin cytoskeleton (Beemiller and Krummel, 2013; Yu et al., 2013; Kumari et al., 2014). This reorganization is essential for the formation of the immunological synapse (IS), the specialized interface between the two cells (Monks et al., 1998; Grakoui et al., 1999). Initially, activation of actin polymerization within the T cell at the periphery of its contact with the APC drives the spreading of the T cell across the surface of the APC. Once spreading is complete, continued actin polymerization begins to drive an inward flow of actin toward the center of the contact site and in the plane of the IS. By coupling this inward flow with depolymerization at the center of the IS, the T cell creates an ongoing centripetal flow of actin that is thought to be a major driving force for the inward movement of T cell receptor microclusters (TCR MCs) and integrin clusters in the T cell’s plasma membrane (Bunnell et al., 2001; Varma et al., 2006; Kaizuka et al., 2007; Babich et al., 2012; Beemiller et al., 2012; Smoligovets et al., 2012; Yi et al., 2012). Over the next 5–10 min, the inward movement of receptor clusters culminates in the formation of a mature IS, in which TCR MCs are concentrated at the center of the IS (the central supramolecular activation cluster [cSMAC]), and leukocyte functional antigen 1 (LFA-1), the T cell’s major integrin, is concentrated in a surrounding ring (the peripheral SMAC [pSMAC]). Importantly, actin assembly and dynamics are intimately linked not just to TCR MC movement, but to virtually every key event during IS formation, including signalosome assembly and tuning (Mattila et al., 2016), integrin activation (Comrie et al., 2015a,b), the mechanical regulation of T cell signaling (Chen and Zhu, 2013), and effector functions such as lytic granule release (Brown et al., 2011; Mace et al., 2012; Basu et al., 2016). Clearly, a full understanding of how actin cytoskeletal forces are created and organized at the IS is required to define the mechanisms by which they drive T cell function.
Numerous laboratories have used diffraction-limited imaging of T cells engaged with planar lipid bilayers containing freely diffusing activators (e.g., anti-CD3 and intercellular adhesion molecule 1 [ICAM-1]) to correlate the dynamics of actin flow and receptor cluster movement in an ideal imaging plane (Dustin, 2009). Importantly, these studies revealed robust, polymerization-driven, actin retrograde flow in a ring surrounding the pSMAC now known as the distal SMAC (dSMAC; Kaizuka et al., 2007; Babich et al., 2012; Beemiller et al., 2012; Yi et al., 2012). Moreover, the rate of centripetal TCR MC movement in this radially symmetric dSMAC roughly correlated with the rate of inward actin flow (Kaizuka et al., 2007), and elegant biophysical studies demonstrated frictional coupling between the TCR MCs and actin flow (DeMond et al., 2008; Yu et al., 2010). Less clear, however, is what propels TCR MC movement across the pSMAC, especially as GFP-actin, the reporter typically used to image actin dynamics at the IS, does not reveal obvious actin organization there (Kaizuka et al., 2007). Using F-Tractin, an indirect reporter for F-actin, we, in contrast, identified concentric actin arcs in the pSMAC that are decorated with myosin II (Yi et al., 2012). Additionally, we showed that the lamellipodial-like dSMAC and lamella-like pSMAC exhibit distinct rates of inward actin flow and that the rates of centripetal TCR MC movement across these two zones matched their distinct actin flow rates (Yi et al., 2012). Nevertheless, the existence of these arcs has been questioned (Beemiller and Krummel, 2013; Le Floc’h and Huse, 2015), and they have never been observed in primary T cells. Moreover, an alternate mechanism to drive TCR MC movement across the pSMAC has been proposed that involves dynein-dependent, microtubule-based transport (Hashimoto-Tane et al., 2011). Finally, the role of myosin II in TCR MC movement has been controversial (Hammer and Burkhardt, 2013).
In this study, we sought to determine how the actin arcs are created and how they become organized into concentric structures. Beyond that, we sought to define the spatial relationship between the arcs and receptor clusters during IS maturation and to quantitate the contributions made by arcs to receptor cluster distribution, T cell–APC adhesion, and proximal TCR signaling. Finally, we sought to directly image the arc-dependent translocation of TCR MCs. Key to accomplishing these goals was the use of total internal reflection fluorescence–structured-illumination microscopy (TIRF-SIM), which, by virtue of its high spatiotemporal resolution (Kner et al., 2009; Beach et al., 2014; Li et al., 2015), proved pivotal in reaching several of our main conclusions.
Results
SIM reveals the architecture and dynamics of F-actin at the IS
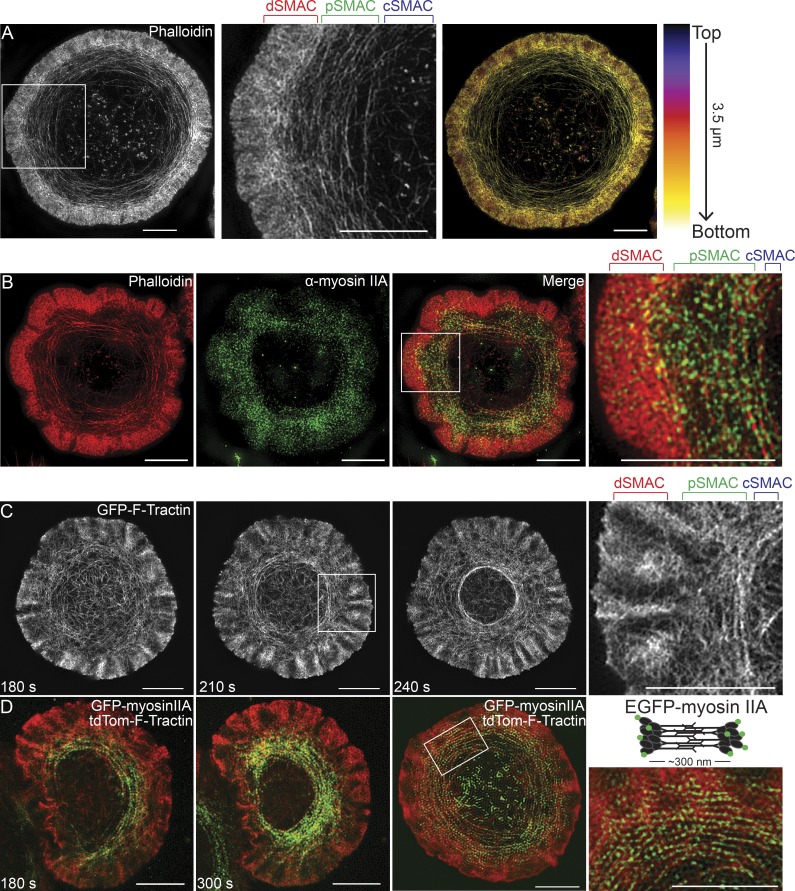
To characterize the architecture of actin networks at the IS at higher resolution, we imaged activated, phalloidin-stained Jurkat T cells with 3D-SIM to reveal endogenous F-actin structures at ∼120-nm lateral and ∼250-nm axial resolution. Consistent with our previous work at lower resolution (Yi et al., 2012), 3D-SIM demonstrated that the IS is comprised of three distinct zones of F-actin: an outer, dense ring corresponding to the branched actin network in the dSMAC, a middle ring comprised of concentric actin arcs corresponding to the pSMAC, and a central, relatively actin-depleted zone corresponding to the cSMAC (Fig. 1 A, first and second panels). Color coding the 3D projection of this cell according to z position (Fig. 1 A, third panel) shows that these actin structures are largely confined to the plane of the IS, indicating that their dynamics will occur in close association with the plasma membrane at the IS. Also consistent with previous work, 3D-SIM images of Jurkats stained for actin and endogenous myosin IIA, the dominant myosin II isoform in T cells, showed that the concentric actin arcs in the pSMAC are highly enriched for this contractile motor (Fig. 1 B).
Figure 1.
SIM imaging reveals concentric actomyosin arcs in the pSMAC region of the Jurkat T cell IS. (A, first and second panel) 3D-SIM image of an activated Jurkat T cell stained with phalloidin. (A, third panel) Cell in first panel color-coded by z position. Lighter colors are closer to the coverslip. (B) 3D-SIM image of a Jurkat T cell stained with phalloidin (red) and anti–myosin IIA antibody (green). Individual channels and the merged image are shown. (C) Still images from a TIRF-SIM video of a Jurkat T cell expressing GFP–F-Tractin (Video 1). (D) Still images from two color TIRF-SIM videos of two different Jurkat T cells expressing GFP–myosin IIA and tdTomato–F-Tractin (Video 2, corresponding to third panel). SMAC zones bracketed in A–C at top. Bars, 5 µm.
We next sought to visualize the dynamics of IS actin networks using a live-cell compatible SIM technique. Given that these networks are close to the plasma membrane, we used TIRF-SIM, which merges the power of SIM (in this study, ∼100-nm xy resolution) with TIRF (enhanced signal-to-noise ratio for structures close to the coverslip) in a format that displays little bleaching or phototoxicity while providing high temporal resolution (Kner et al., 2009; Li et al., 2015). Fig. 1 C (first three panels) shows still images taken from a video of a Jurkat T cell expressing GFP–F-Tractin, an indirect, dynamic reporter for F-actin (Johnson and Schell, 2009), 180, 210, and 240 s (Fig. 1 C) after engagement with an activating surface (Video 1). It is immediately apparent that F-Tractin faithfully reports the two actin networks and three IS zones identified in fixed images of phalloidin-stained cells. This is most clearly seen in Fig. 1 C (fourth panel), as well as in the corresponding video. Additionally, TIRF-SIM imaging of Jurkats expressing GFP–myosin IIA and tdTomato–F-Tractin revealed in dynamic fashion the enrichment of myosin on the actin arcs in the pSMAC (Fig. 1 D, first and second panels). Importantly, close examination of these images, stills from Video 2 (Fig. 1 D, third and fourth panels), and Video 2 itself revealed the presence of individual, 300-nm-long myosin IIA bipolar filaments decorating the actin arcs and moving inward with them. Together, these observations confirm and enhance our previous understanding of actin architecture at the Jurkat T cell IS and provide additional support for a radially symmetric contractile actomyosin network within the pSMAC.
Actomyosin arcs are also prominent structures at the IS of primary mouse T cells
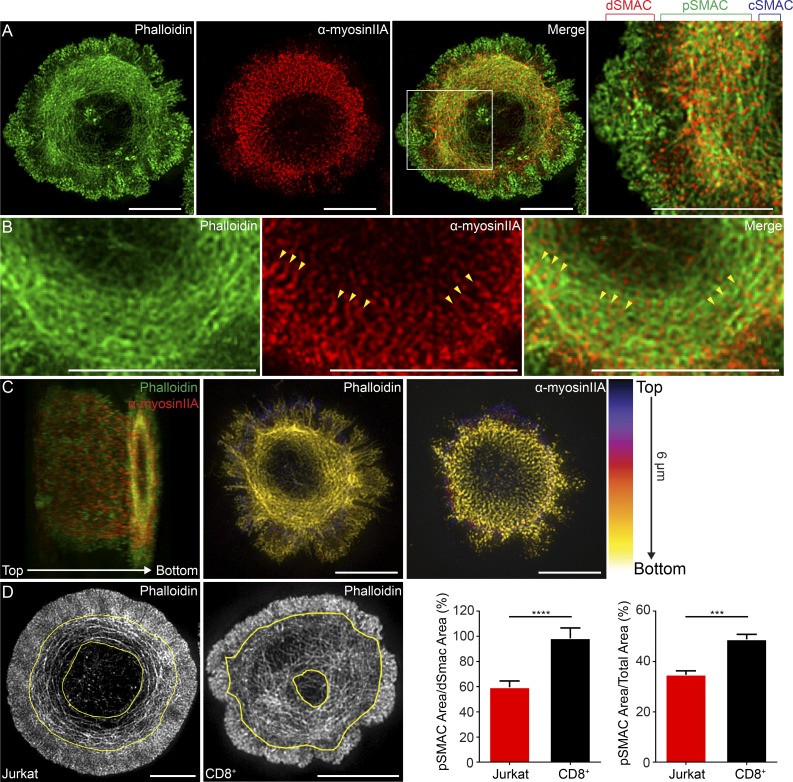
We performed 3D-SIM on activated primary mouse CD8+ T cells stained with phalloidin and an antibody to myosin IIA to reveal endogenous structures. Fig. 2 A shows that the IS of a CD8+ T cell is essentially indistinguishable from that of a Jurkat T cell. Also, like Jurkats, the two major F-actin networks at the CD8+ T cell IS are very close to the IS membrane (Fig. 2 C). Strikingly, enlarged images of typical CD8+ T cell synapses reveal an almost sarcomere-like pattern for myosin IIA embedded in the actin arcs comprising their pSMAC (Fig. 2 B, arrowheads). Perhaps most importantly, rather than being difficult to detect or absent, actomyosin arcs are an even more prominent component of the mouse CD8+ T cell IS than the Jurkat T cell IS (Fig. 2 D). Finally, still images from TIRF-SIM imaging of primary mouse CD8+ T cells expressing GFP–myosin IIA and tdTomato–F-Tractin demonstrated the formation of the actomyosin arcs in the pSMAC (Fig. S1, A and B). Together, these results demonstrate in unequivocal fashion the existence of actomyosin arcs at the IS of primary mouse T cells.
Figure 2.
Actomyosin arcs are also a very prominent feature of the primary mouse CD8+ T cell IS. (A) 3D-SIM image of a primary mouse CD8+ T cell stained with phalloidin (green) and anti–myosin IIA antibody (red). (B) Magnified views of actomyosin arcs in a separate CD8+ T cell stained as in A. Yellow arrowheads mark sarcomere-like pattern of myosin IIA. (C, left) Cell in B color-coded by z position; each channel is separated in middle and right panels. Lighter colors are closer to the coverslip. (D) Area occupied by pSMAC actin arcs (bracketed by yellow traces) in typical Jurkat IS (first panel) and primary mouse CD8+ T cell IS (second panel). The ratio of pSMAC to dSMAC area (third panel) and pSMAC to total IS area (fourth panel) for Jurkat and mouse CD8+ T cells. n = 17–24 cells/condition. Data are means ± SEM. SMAC zones bracketed in A at top. Bars, 5 µm. ***, P < 0.001; ****, P < 0.0001.
3D-SIM and TIRF-SIM reveal linear actin filaments/bundles embedded within the dSMAC that appear to give rise to the actin arcs in the pSMAC
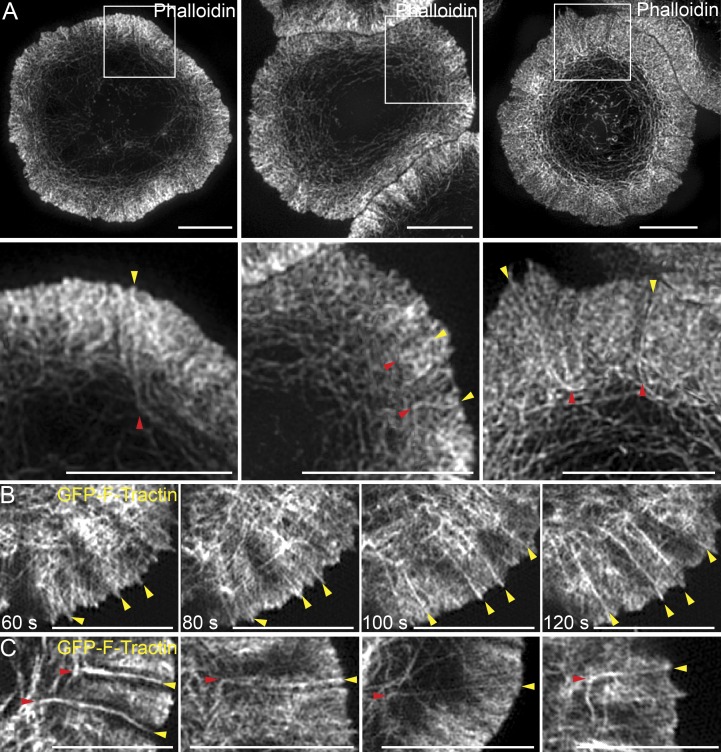
Closer examination of fixed, phalloidin-stained Jurkat synapses using 3D-SIM revealed linear actin filaments/bundles embedded within the branched actin network of the dSMAC. These structures appear to originate at the distal edge of the IS, traverse the dSMAC, and bend upon exiting the dSMAC to give rise to the actin arcs of the pSMAC (Fig. 3 A, yellow arrowheads in the bottom panels mark the origin of these structures and red arrowheads mark their exit from the dSMAC). Further support for their existence came from live-cell TIRF-SIM imaging of Jurkat T cells expressing GFP–F-Tractin. Specifically, Fig. 3 B shows still images taken from Video 4 of a quadrant of a Jurkat T cell synapse 180, 240, 300, and 360 s after engagement with an activating surface. Close examination reveals the progressive formation of several linear actin filaments/bundles (Fig. 3 B, arrowheads) at the distal edge of the IS that are oriented perpendicular to the plasma membrane, traverse the dSMAC, and then bend and splay upon exit from the dSMAC to form arcs (Video 3). Fig. 3 C shows four clear examples of such bending upon exit from the inner aspect of the dSMAC (see red arrowheads), and Video 4 shows all of the events described in this section occurring across a broad section of a Jurkat IS.
Figure 3.
SIM imaging reveals linear actin filaments/bundles embedded in the branched actin network of dSMAC. (A) 3D-SIM images of three representative phalloidin-stained Jurkat T cells. (B) Successive still images from a TIRF-SIM video of a Jurkat expressing GFP–F-Tractin (Video 3). (C) Four still images from TIRF-SIM videos of Jurkats expressing GFP–F-Tractin. Yellow arrowheads mark the origin of linear actin filaments/bundles embedded in the branched actin network of the dSMAC, whereas red arrowheads mark where they bend upon exit from the dSMAC. See also Video 4. Bars, 5 µm.
The linear actin filaments/bundles, and the actin arcs they give rise to, are formin generated
From a topological standpoint, the linear actin filaments/bundles observed in Fig. 3 are consistent with actin structures built by formins (Kovar, 2006; Breitsprecher and Goode, 2013). Specifically, they originate at the plasma membrane, the site where formins are typically active, and project inwards, one of two fates typically exhibited by formin-generated filaments (the other being projection outward to make structures like filopodia). Additional indirect evidence that these structures are formin generated stems from the observation that the arcs are very poorly labeled by GFP-actin (Kaizuka et al., 2007) because formins are known to incorporate modified actins into filaments very ineffectively (Chen et al., 2012). This is completely consistent with our previous observations (Yi et al., 2012), which we confirmed and extended in this study using colocalization analyses (Fig. S1, C–E).
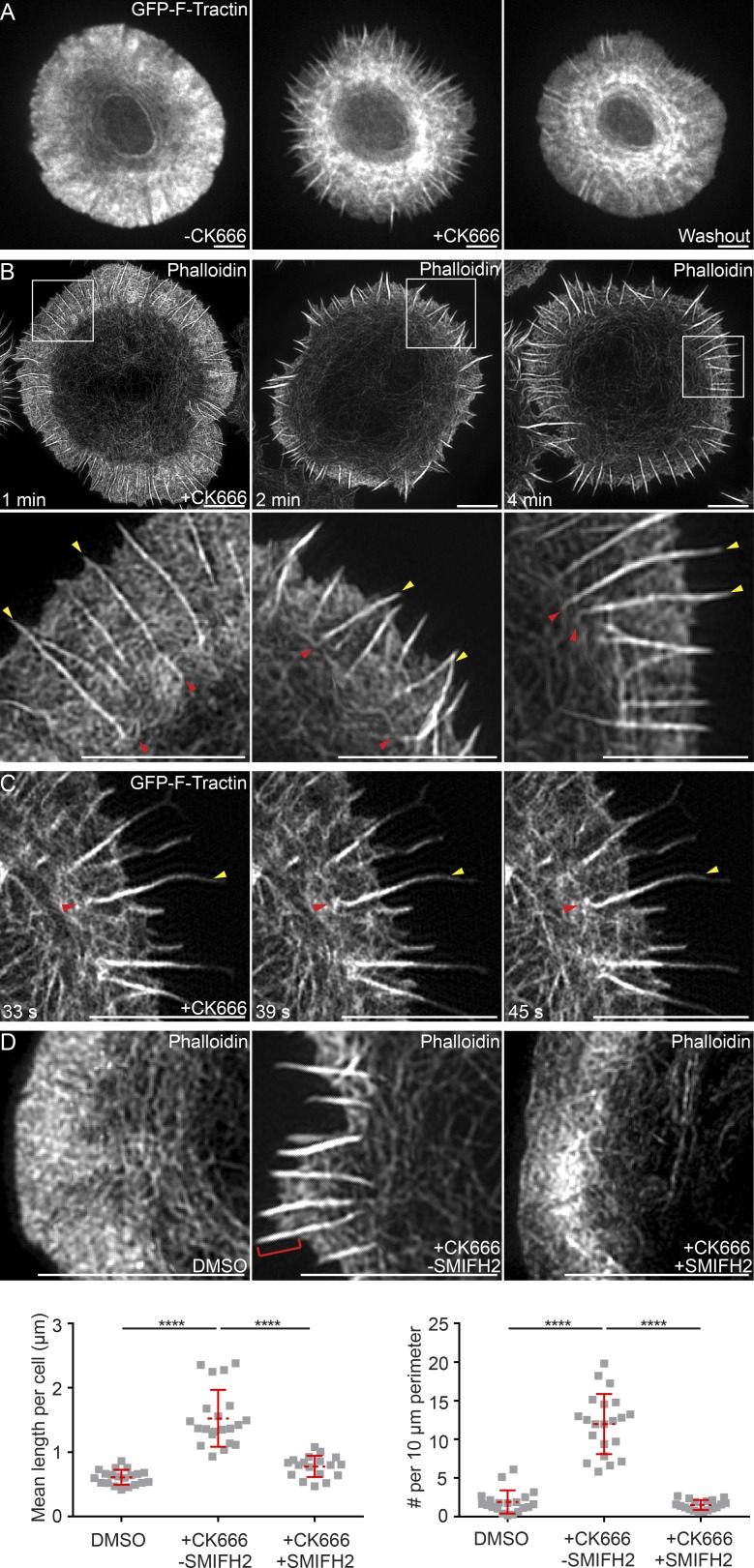
Recent, paradigm-shifting studies in both yeast (Burke et al., 2014) and mammalian cells (Lomakin et al., 2015; Rotty et al., 2015) have demonstrated that the Arp2/3 complex and formins are competing for a limiting pool of actin monomer, such that when one actin nucleator is inhibited, the network created by the other becomes more robust. We reasoned, therefore, that if the linear actin filaments/bundles embedded within the dSMAC are formin generated, they should be augmented upon Arp2/3 inhibition. Consistently, Video 5 and the still images taken from it (Fig. 4 A) show that the addition of the Arp2/3 inhibitor CK666 to activated Jurkat T cells expressing GFP–F-Tractin resulted in the dramatic collapse of the Arp2/3-dependent, branched actin network comprising the dSMAC and the progressive enhancement of the linear actin filaments/bundles, which took on the appearance of radial actin “spikes” in which actin polymerization and retrograde flow continued (Fig. 4 A, compare second panel [+CK666] to first panel [control]; note that this effect was completely reversible in third panel [washout]; see also Video 5 and Gomez et al., 2007). 3D-SIM imaging of fixed, phalloidin-stained Jurkat T cells treated with CK666 for 1, 2, and 4 min revealed these spikes in greater detail as they emerged from the shrinking lamellipodial network and increased in robustness over time (Fig. 4 B, top). Importantly, actin arcs were still present in these Arp2/3-inhibited cells. Moreover, the corresponding insets in Fig. 4 B (bottom) show that the CK666-enhanced actin filaments/bundles in the dSMAC bend and reorient into actin arcs at the dSMAC/pSMAC boundary (the red arrowheads mark bend points where the spikes marked by yellow arrowheads give rise to arcs). This bending and reorientation process could be observed in real time in Jurkats expressing GFP–F-Tractin and imaged using TIRF-SIM (Video 6 and the still images taken from it in Fig. 4 C). Together, these observations indicate that arc assembly is largely Arp2/3 independent and argue that the arcs are created by one or more formins acting at the distal edge of the IS.
Figure 4.
Arp2/3 inhibition augments the assembly of the formin-nucleated linear actin filaments in the dSMAC. (A) Still images from a confocal video of a Jurkat T cell expressing GFP–F-Tractin before (left) and 3 min after (middle) treatment with 25 µM CK666 and after drug washout (right; and Video 5). (B) 3D-SIM images of phalloidin-stained Jurkat T cells treated with 25 µM CK666 for the indicated times. (C) Still images from a TIRF-SIM video of a Jurkat T cell expressing GFP–F-Tractin treated with CK666 as in B (Video 6). (D, top) 3D-SIM images of Jurkats treated with DMSO, 25 µM CK666, or 25 µM CK666 plus 10 µM SMIFH2 and stained with phalloidin. The red bracket in the second panel marks a spike. (D, bottom) Quantitation of mean length of linear actin spikes per cell (left) and their surface density (right). n = 16–22 cells/condition. In B and C, yellow arrowheads mark linear actin spikes, and red arrowheads mark their bend points. Data are means ± SD. Bars, 5 µm. ****, P < 0.0001.
To provide more direct support for the hypothesis that the actin spikes observed upon Arp2/3 inhibition are formin generated, we compared cells treated with CK666 alone to cells treated with both CK666 and the pan-formin inhibitor small-molecule inhibitor of formin homology 2 domain (SMIFH2; Rizvi et al., 2009). As before, 3-min incubation in the presence of CK666 resulted in a considerable diminution in the size of the branched actin array comprising most of the dSMAC and the appearance of actin spikes at the cell edge (Fig. 4 D, compare second panel [+CK666 −SMIFH2] to first [DMSO control] panel; the red bracket in the second panel marks a typical actin spike). Importantly, addition of CK666 to cells that had been pretreated with SMIFH2 resulted in a dramatic reduction in the formation of spikes (Fig. 4 D, compare third panel [+CK666 +SMIFH2] to second panel [+CK666 −SMIFH2]). This reduction is indicated by significant decreases in both the mean length of spikes (Fig. 4 D, bottom left) and the number of spikes per unit of cell perimeter (Fig. 4 D, bottom right) for cells treated with both inhibitors relative to cells treated with CK666 alone. These results provide additional support for the idea that the linear actin filaments/bundles embedded within the dSMAC and contiguous with the actin arcs in the pSMAC are formin-generated structures.
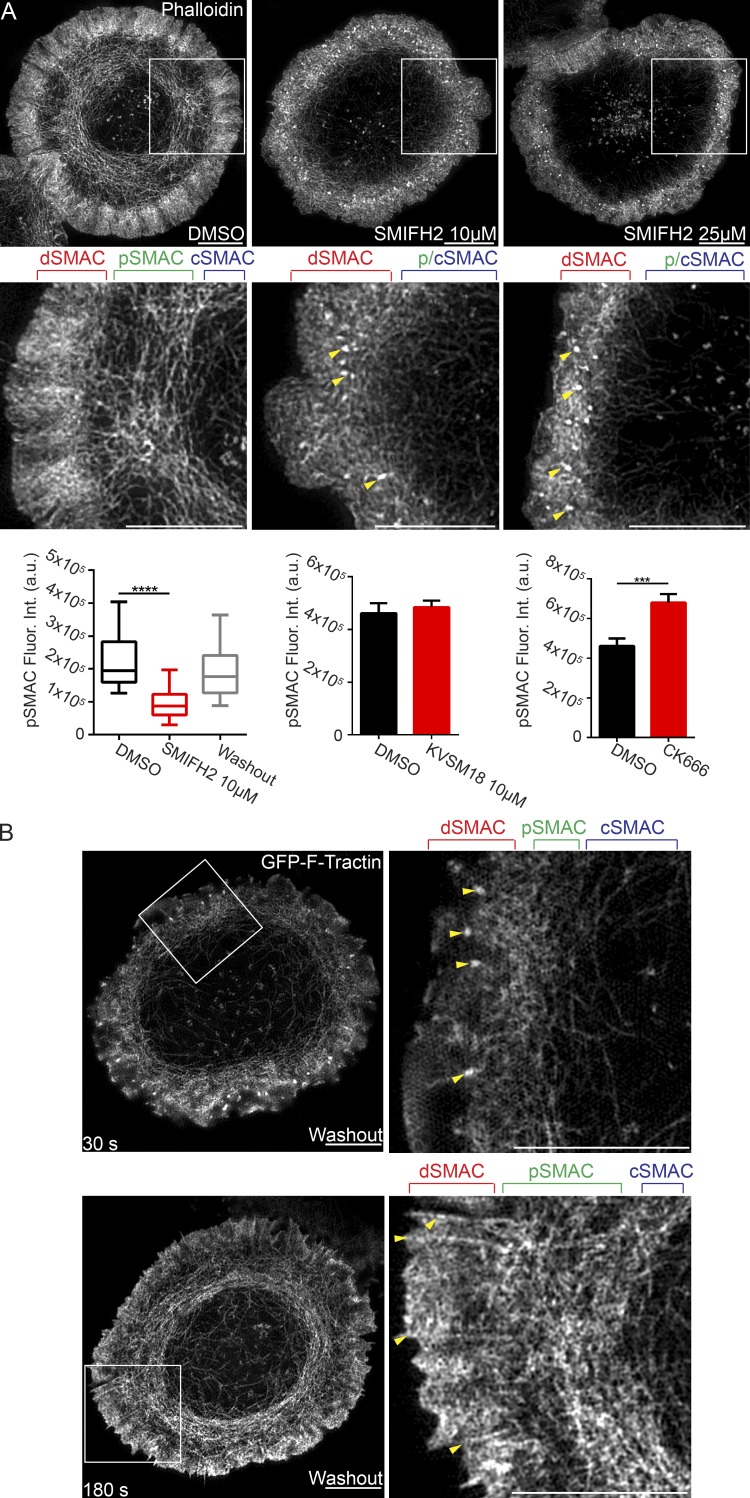
As a more direct test for the role of formins in actin arc creation, we imaged arc formation with and without SMIFH2. To accomplish this, we allowed Jurkats to activate for 2 min before SMIFH2 addition and then asked if its addition prevented further actin arc formation. We observed that 5-min incubation in the presence of SMIFH2 (both 10 and 25 µM) resulted in a dramatic decrease in the presence of actin arcs within the pSMAC, as revealed by 3D-SIM imaging of fixed, phalloidin-stained cells (Fig. 5 A, compare second and third panels and the corresponding insets in the fifth and sixth panels to the DMSO control in the first and fourth panels). To quantitate arc loss, we measured total phalloidin fluorescence in the pSMAC area of control cells and cells treated with 10 µM SMIFH2. Fig. 5 A (bottom left) shows that SMIFH2-treated cells exhibited significantly less F-actin in their pSMACs, consistent with the loss of actin arcs after formin inhibition. Importantly, SMIFH2-induced arc loss is likely attributable to formin inhibition rather than off-target effects because treatment with 10 µM KVSM18, a closely related but inactive analogue of SMIFH2 (Rizvi et al., 2009), does not cause arc loss (Fig. 5 A, bottom middle). Interestingly, formin-inhibited cells also exhibited a dramatic increase in the number of F-actin foci in the dSMAC (Fig. 5 A, arrowheads). We believe these foci correspond to the Arp2/3-dependent F-actin foci described recently (Kumari et al., 2015) and that they become more robust upon formin inhibition as a result of an increase in actin monomer available for Arp2/3-dependent nucleation. On the flip side, the inhibition of Arp2/3-dependent nucleation using CK666 increased the content of arcs in the pSMAC (Fig. 5 A, bottom right), presumably because of an increase in actin monomer available for formin-dependent nucleation.
Figure 5.
Formin inhibition blocks actin arc formation. (A, micrographs) 3D-SIM images of Jurkat T cells treated with DMSO, 10 µM SMIFH2, or 25 µM SMIFH2 and stained with phalloidin. Yellow arrowheads mark actin foci. (A, bottom) Mean phalloidin fluorescence in the pSMAC. n = 21–30 cells/condition. Box plots are centered on means and display upper and lower quartile ranges and min to max values. Bar graphs are means ± SEM. (B, top) Still image from a TIRF-SIM video of a Jurkat T cell expressing GFP–F-Tractin 30 s after SMIFH2 washout. Yellow arrowheads mark actin foci. (B, bottom) Still image from the same TIRF-SIM video 180 s after SMIFH2 washout. Yellow arrowheads mark linear actin filaments. SMAC zones are bracketed in A and B at top. See also Video 7. Bars, 5 µm. ***, P < 0.001; ****, P < 0.0001. a.u., arbitrary units; Fluor. Int., fluorescence intensity.
Importantly, the SMIFH2-mediated inhibition of actin arc formation was reversible, as washout of SMIFH2 resulted in the rapid resumption of actin arc formation. This is apparent in still images taken from a TIRF-SIM video (Video 7) 30 (Fig. 5 B, top) and 180 s (Fig. 5 B, bottom) after SMIFH2 washout. Careful examination of the magnified insets in Fig. 5 B (right panels) show that SMIFH2 washout was accompanied not only by the appearance of newly formed arcs in the pSMAC (Fig. 5 B, bottom right), but also by the disappearance of the actin foci (Fig. 5 B, yellow arrowheads in top right), and the reappearance of linear actin filaments/bundles embedded in the dSMAC (yellow arrowheads in bottom right). Within 5 min of washout, the content of F-actin in the pSMAC was restored to control levels (Fig. 5 A, bottom left).
We next tested if one or more formins could be detected at the tips of CK666-enhanced actin spikes by immunofluorescence staining for endogenous proteins. Previous studies (Gomez et al., 2007; Ramabhadran et al., 2011) have provided evidence for the expression of four formins in Jurkat T cells: mDia1, mDia2, formin-like protein 1 (FMNL1), and the non-CAAX version of inverted formin-2 (INF2). Of these, we were able to detect clear signals for mDia1, FMNL1, and non-CAAX INF2 at the tips of CK666-enhanced actin spikes (Fig. S2 A, micrographs). Moreover, the fraction of spike tips that stained for these three formins, which ranged from ∼35 to ∼55%, was significantly higher than the fraction that stained for mDia2 or with preimmune sera (Fig. S2 A, right panel), arguing that the tip staining seen for mDia1, FMNL1, and non-CAAX INF2 is specific. Of these three formins, mDia1 appears to be the most highly expressed in Jurkats (Gomez et al., 2007). Importantly, Jurkat T cells in which we achieved near-complete knockdown of mDia1 using shRNA (Fig. S2 B, top left) exhibited a dramatic decrease in the content of arcs in their pSMAC relative to control cells expressing a scrambled shRNA plasmid (Fig. S2 B, micrographs and bottom left). Also like SMIFH2-treated cells, mDia1 knockdown cells exhibited a proliferation of Arp2/3-dependent actin foci (Fig. S2 B, bottom right, arrowheads). Together, these data argue that the formin mDia1 plays a major role in arc formation. Consistent with this conclusion, Jurkats treated with a membrane-permeant pan-activator of Rho GTPases (CN01), which activate most formins including mDia1, exhibited accentuated linear actin structures embedded in their dSMAC, in the amount of arcs in their pSMAC, and in the overall size of their pSMAC (Fig. S3, A–C). Finally, we found that tropomyosin, a universal marker of formin-generated structures (Tojkander et al., 2012), strongly decorates actin arcs. Specifically, Fig. S3 (D–F) show that GFP-tagged nonmuscle tropomyosin TM4 is highly enriched on the arcs in the pSMAC, whereas α-actinin, which typically cross-links Arp2/3-generated branched actin networks, is largely restricted to the dSMAC. Together, the results in this section provide compelling evidence that arcs arise largely, if not entirely, from formin-dependent nucleation at the outer edge of the dSMAC.
Myosin II organizes the linear actin filaments/bundles emanating from the dSMAC into the concentric actin arcs of the pSMAC
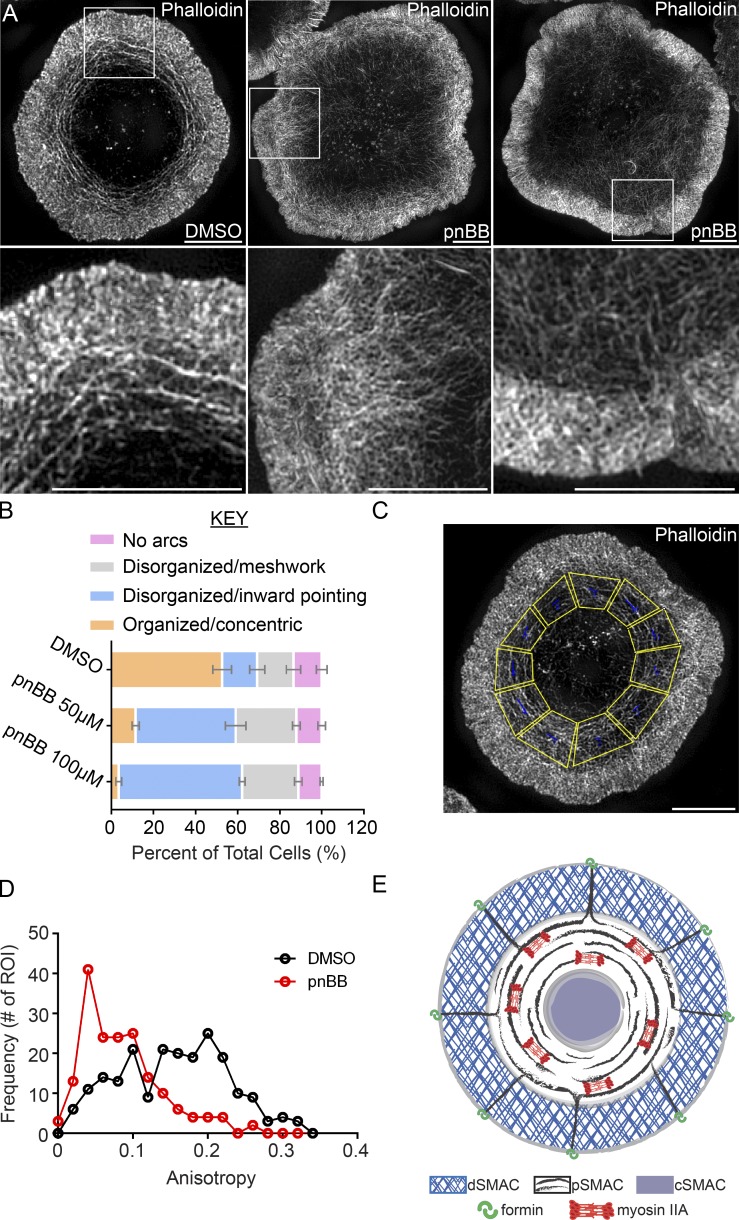
Given the enrichment of myosin IIA at the pSMAC and the inherent ability of bipolar myosin II filaments to organize actin networks, we asked if myosin II is required to organize the linear actin filaments/bundles emanating from the dSMAC into the concentric actin arcs of the pSMAC. To selectively and conditionally inhibit myosin IIA, we used para-nitro-blebbistatin (pnBB), a new variant of blebbistatin that lacks the original molecule’s phototoxicity (Képiró et al., 2014). In all experiments, Jurkats were pretreated with pnBB for 30 min followed by activation on antibody-coated coverslips for 10 min. 3D-SIM images of fixed, phalloidin-stained cells show that preincubation with 50 µM pnBB resulted in large-scale disruption of actin arc organization in the pSMAC (Fig. 6 A, compare second and third panels, and the corresponding insets in fifth and sixth panels to the DMSO control in first and fourth panels). This result was borne out by visual scoring of arc organization (Fig. 6 B), as only ∼12% of cells treated with 50 µM pnBB and ∼5% of cells treated with 100 µM pnBB have highly organized concentric actin arcs, as compared with ∼52% of control, DMSO-treated Jurkats (Fig. 6 B, orange bars). In place of organized arcs, the pSMAC region of pnBB-treated cells exhibited either: (1) actin filaments/bundles that projected inward toward the center of the IS (Fig. 6 A, middle panels; and Fig. 6 B, blue bars); or (2) a highly disorganized, meshlike organization (Fig. 6 A, right panels; Fig. 6 B, gray bars; and Video 8).
Figure 6.
Myosin IIA inhibition disrupts actin arc organization. (A) 3D-SIM images of a Jurkat pretreated with DMSO and two Jurkats pretreated with 50 µM pnBB for 30 min and stained with phalloidin (Video 9). (B) Cells were scored for actin arc morphology as indicated. n = 100 cells/condition; three experiments per condition; data are means ± SD (see Materials and methods for details). (C) Boxed regions covering the pSMAC (yellow) were quantitated for actin arc anisotropy in D using FibrilTool. n = 22–26 cells/condition. (E) Model depicting formin-dependent actomyosin arc formation at the IS. Bars, 5 µm.
To further quantify the defects in actin arc organization seen upon myosin II inhibition, we determined the anisotropy of actin structures in the pSMAC using FibrilTool, which measures how well a structure of interest (in this study, actin arcs) in a given region of interest (ROI; in this study, the pSMAC) is organized in parallel arrays. The values obtained range from 0, when the orientation of the structures in the ROI is random, to 1, when all of the structures are oriented in the same direction. To deal with the fact that the IS is radially symmetric, we divided the pSMAC region in 3D-SIM images of fixed, phalloidin-stained Jurkats into 10–12 trapezoid-shaped ROIs of similar size (Fig. 6 C). Fig. 6 D shows that the anisotropy of pSMAC actin structures is significantly lower in cells treated with 50 µM pnBB (red line) than in control, DMSO-treated cells (black line), corroborating the qualitative assessments described in Fig. 6 (A and B). Together, these data argue strongly that bipolar filaments of myosin IIA organize formin-generated, linear actin filaments/bundles emanating from the dSMAC into concentric actin arcs populating the pSMAC. Importantly, stochastic variation in the direction the linear filaments bend as they exit the dSMAC should ensure that bipolar myosin II filaments are routinely engaged with actin filaments of opposing orientation, a prerequisite for myosin II–dependent contraction and inward movement of arcs (Fig. 6 E). This self-organizing property should propel the formation of the radially symmetric contractile actomyosin network in the pSMAC.
Actomyosin arcs facilitate the accumulation of TCR and LFA-1 clusters at the cSMAC and inner aspect of the pSMAC, respectively
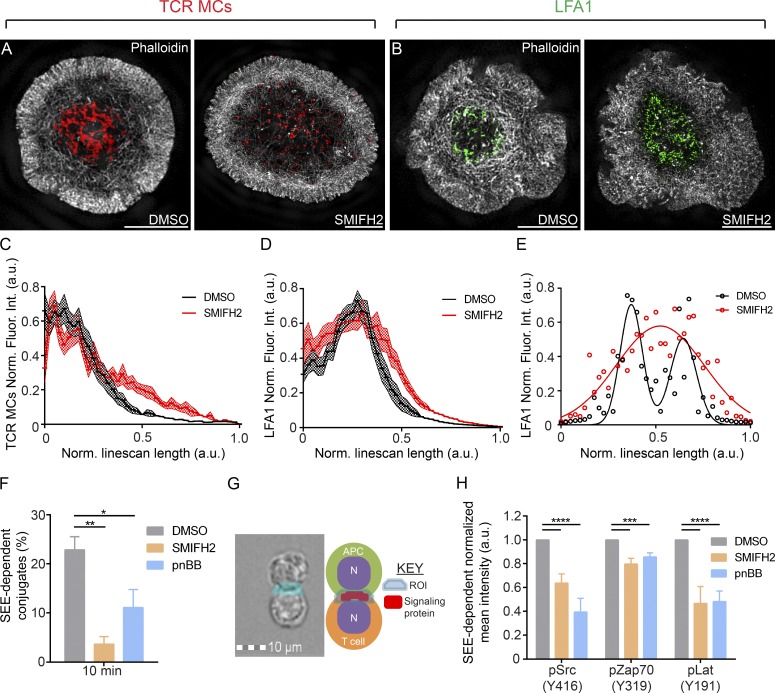
Glass-supported planar lipid bilayers displaying stimulatory molecules to mimic the APC surface have been used extensively to correlate cytoskeletal dynamics in the plane of the IS with TCR MC and integrin cluster position and dynamics in T cells (Kaizuka et al., 2007; Hartman et al., 2009). As a first test for the role of actin arcs in IS formation, we determined the distributions of F-actin, TCR MCs, and the open, active conformation of LFA-1’s β2 subunit in control, DMSO-, and SMIFH2-treated Jurkats after a 10-min engagement with bilayers containing fluorescent anti-CD3 antibody and unlabeled ICAM (and where antibody M24 against the fixation-sensitive, open, active conformation of the β2 integrin was present for the last 4 min). At 10 min, the cells were fixed, stained with phalloidin, and the distributions of F-actin (Fig. 7, A and B, white), TCR MCs (red), and open, active LFA-1 (green) imaged with 3D-SIM. As expected, control, DMSO-treated Jurkats exhibited a strong accumulation of TCR MCs in their cSMAC (i.e., inside the arcs populating their pSMAC; Fig. 7 A, left), and the accumulation of active LFA-1 in a ring along the inner aspect of their pSMAC/actin arcs (Fig. 7 B, left). Strikingly, SMIFH2 treatment disrupted the centralization of TCR MCs (Fig. 7 A, right) and the formation of the ring of active LFA-1 (Fig. 7 B, right), accompanied by the loss of actin arcs (Fig. 7, A and B, right). These conclusions are supported by radial plot profiles, which report the mean distribution of TCR MCs (Fig. 7 C) and active LFA-1 clusters (Fig. 7 D) between the cell center (0 on the x axis) and the cell edge (1.0 on the x axis). Relative to the DMSO control (black lines), SMIFH2 addition (red lines) reduced the centralization of TCR MCs (Fig. 7 C, as indicated by a flattening and rightward shift in the red line) and broadened the distribution of active LFA-1 clusters (as indicated by a broadening of the red line on both sides of the black peak in Fig. 7 D; P < 0.0001). The effect of SMIFH2 on the distribution of the open, active integrin becomes especially clear when we fit line scans across the IS to Gaussian distributions (Fig. 7 E), in which the normal bimodal distribution of active LFA-1 across the IS that corresponds to the pSMAC ring (black circles and black fitted line) is lost upon SMIFH2 treatment (red circles and red fitted line). Finally, consistent with the fact that arcs require myosin II to assemble into radially symmetric contractile structures, we observed similar defects in the distributions of TCR MCs and active LFA-1 in Jurkats treated with pnBB (Fig. S4 A). Collectively, these results argue strongly that actin arcs, in conjunction with myosin II bipolar filaments, make a major contribution to the centripetal movement and distribution of both TCR MCs and integrin clusters at the IS.
Figure 7.
Functional consequences of formin inhibition. (A) 3D-SIM images of phalloidin-stained (grayscale) DMSO-treated or 10 µM SMIFH2-treated Jurkat T cells stimulated on planar lipid bilayers containing Alexa Fluor 647–labeled anti-CD3 (red) to report TCR MC distribution. (B) Same as in A, except cells were stained with M24 antibody (green) during IS formation to determine the distribution of open, active LFA-1. Radial plot profiles of TCR MCs (C) and LFA-1 clusters (D) after DMSO or SMIFH2 treatment. n = 14–16 cells/condition; Data are means ± SEM; P < 0.0001, two-way ANOVA. (E) Line scans across cells in B fit to Gaussian distributions. (F) Adhesion conjugate assay after DMSO, SMIFH2, or pnBB treatment. n ≥ 3 independent experiments; data are means ± SEM. (G) Combination masking strategy to quantitate fluorescence at the T cell IS. (H) Imaging flow cytometry of Jurkat T–B cell conjugates fixed, immunostained, and analyzed on a flow cytometer for background-corrected MFI (Norm. Fluor. Int.) of the indicated signaling proteins after DMSO, SMIFH2, or pnBB treatment. n ≥ 3 independent experiments; data are means ± SEM. Note that the differences remain statistically significant even when background correction is omitted (DNS). Bars, 5 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. a.u., arbitrary units; Norm. Fluor. Int., normalized fluorescence intensity.
Actomyosin arcs promote robust T cell–APC adhesion and TCR proximal signaling
To further gauge the physiological significance of actin arcs at the IS, we measured adhesion and TCR proximal signaling in control and formin-inhibited Jurkats present in conjugates with an APC. For the adhesion assay, Jurkats were mixed with staphylococcal enterotoxin E (SEE)–loaded Raji B cells in the presence or absence of SMIFH2, incubated for 10 min, fixed, immunostained with fluorescent antibodies against OKT3 to mark Jurkats and human leukocyte antigen (HLA) to mark Raji B cells, and the fraction of total Jurkat cells present in OKT3 and HLA double-positive conjugates was determined by flow cytometry analysis. Fig. 7 F shows that inhibition of arc formation with SMIFH2 results in an approximately fivefold reduction in cell adhesion relative to the DMSO control.
To test for possible defects in TCR proximal signaling after the loss of arcs, Jurkat T cells and Raji B cells expressing plasma membrane markers farnesylated GFP and farnesylated RFP, respectively, were mixed together in the absence or presence of SMIFH2, incubated for 10 min, fixed, and immunostained with fluorescent antibodies specific for the tyrosine phosphorylated versions of Src (Y416), Zap70 (Y319), or Lat (Y191) as proxies for the extent of proximal TCR signaling. To quantitate the signals for pSrc, pZap70, and pLat at the IS, we used an Amnis ImageStream flow cytometer, which combines the sample-size advantages of flow cytometry with the sensitivity of fluorescence microscopy. Specifically, we identified GFP and RFP double-positive T–B cell conjugates and measured the mean fluorescence intensity (MFI) of the three phosphorylated signaling molecules on the T cell side of the IS using a masking strategy to identify the contact area between the two cells (Fig. 7 G and Materials and methods). Fig. 7 H shows that disruption of arc formation using SMIFH2 resulted in marked reductions relative to the DMSO control in the content of pSrc (Y416) and pLAT (Y191) at the IS of Jurkat–Raji cell conjugates and a small but significant reduction in the content of pZAP70 (Y319). Of note, significant reductions in the phosphorylation of these signaling molecules were also seen in Western blots of SMIFH2-treated Jurkats activated using soluble anti-CD3 (Fig. S5, A and B; C shows that KVSM18-treated Jurkats did not show a drop in Lat phosphorylation). Finally, consistent with the data in preceding figures and the work of others (Ilani et al., 2009; Kumari et al., 2012; Yu et al., 2012), Jurkats in which myosin II was inhibited using pnBB phenocopied SMIFH2-treated cells as regards defects in adhesion (Fig. 7 F) and signaling (Fig. 7 H). Together, these results argue that formin-generated actomyosin arcs are required for robust adhesion and TCR proximal signaling in T cell–APC conjugates.
TCR MCs reside between arcs, whereas active LFA-1 coincides with arcs
To further define the physical relationship among actomyosin arcs, TCR MCs, and active LFA-1 in maturing synapses, we engaged Jurkats with bilayers as in Fig. 7. In this case, however, the anti–active LFA-1 antibody M24 was added 2 min after engagement, and cells were fixed at 6 min instead of 10 min so as to capture IS organization during maturation. After staining with phalloidin, the distributions of F-actin (Fig. 8 A, white), TCR MCs (Fig. 8 A, red), and open, active LFA-1 (Fig. 8 A, green) were imaged by 3D-SIM to precisely define the spatial relationships among them. Fig. 8 A (first panel) shows a SIM image of a typical Jurkat 6 min after engagement. Fig. 8 A (second panel) shows an enlargement of a region in this cell’s pSMAC. Fig. 8 A (bottom nine panels) shows the signals in this boxed region as a series of three pairwise comparisons. We made three observations from these comparisons. First, the individual signals for TCR MCs (Fig. 8 A, third panel), F-actin (Fig. 8 A, fourth panel), and their overlay (Fig. 8 A, fifth panel) indicate that TCR MCs (red) tend to reside between actin arcs (white). Second, the individual signals for TCR MCs (Fig. 8 A, sixth panel), active LFA-1 (Fig. 8 A, seventh panel), and their overlay (Fig. 8 A, eighth panel) indicate that TCR MCs (red) also tend to reside between LFA-1–rich structures (green). Finally, the individual signals for F-actin (Fig. 8 A, ninth panel), active LFA-1 (Fig. 8 A, tenth panel), and their overlay (Fig. 8 A, eleventh panel) indicate that active LFA-1 (green) tends to coincide with actin arcs (white). Importantly, these observations were borne out by quantitation. First, Pearson’s coefficient analyses of the entire pSMAC region show that the colocalization of F-actin and active LFA-1 is much more significant than the colocalization of F-actin and TCR MCs, LFA-1 and TCR MCs, and a randomized control in which images were rotated 90° relative to each other (Fig. 8 B). Second, line scans and radial plot profiles across population average projections of individual TCR MCs (Fig. 8, C and D) show overlapping intensity profiles for actin arcs (black line) and active LFA-1 (green line) that bracketed and were largely excluded from the peak of TCR MC intensity (red line). Finally, analysis of samples stained for endogenous myosin II show that, as expected from the preceding data, the position of myosin II correlated well with that of LFA-1 and F-actin (Fig. 8 B and Fig. S4 B). Together, these highly resolved static SIM images argue that, on average, TCR MCs reside between actin arcs, whereas active LFA-1 (and myosin II) coincide with actin arcs during IS maturation.
Figure 8.
On average, TCR MCs reside between arcs, whereas active LFA-1 coincides with arcs in maturing synapses. (A) Three-color 3D-SIM imaging of endogenous actin (grayscale), TCR MCs (red), and open, active LFA-1 (green) at a Jurkat T cell IS 6 min after bilayer engagement. (A, bottom nine panels) Pairwise comparisons of the actin, LFA-1, and TCR MC channels and overlays. (B) Pearson’s correlation coefficient; n = 20–40 cells/correlation. Box plots are centered on means and display upper and lower quartile ranges and min to max values. (C) Line scans across population-averaged TCR MCs. n = 110. (D) Radial plot profiles of population-averaged TCR MCs. n = 11 cells; 10–15 TCR MCs/cell. Data are means ± SEM. Note that TCR MCs overlap considerably with F-actin during initial cell spreading (Lam Hui et al., 2014). Bars, 1 µm. ****, P < 0.0001. a.u., arbitrary units; MyoIIA, myosin IIA; Norm. Fluor. Int., normalized fluorescence intensity.
TCR MCs are swept inward by actin arcs
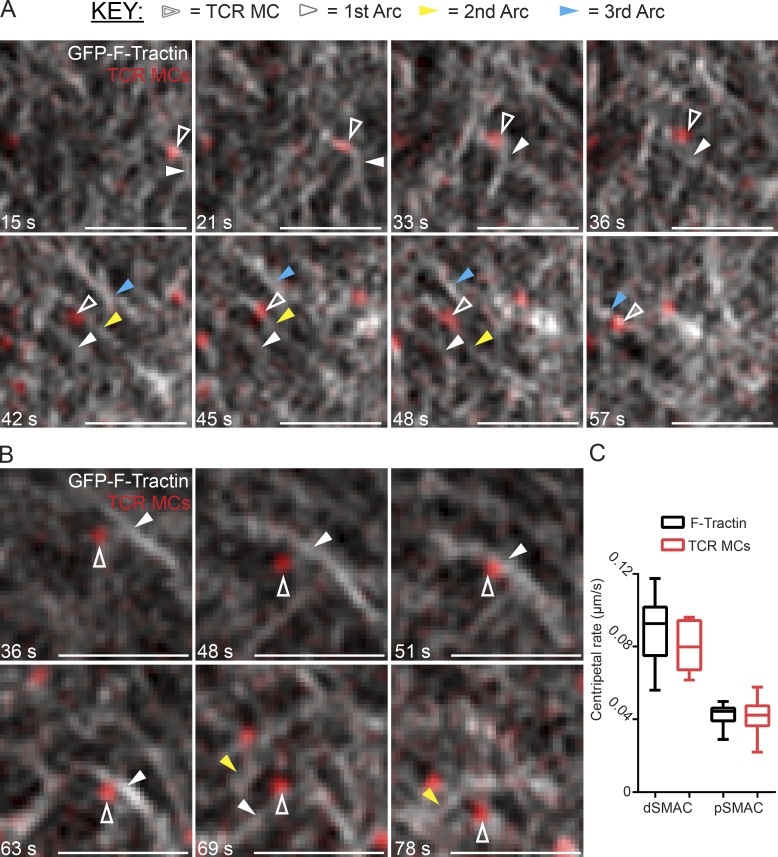
The static SIM images in Fig. 8 demonstrating that TCR MCs commonly reside between actin arcs suggest that TCR MCs may be “swept” inward by these arcs. To look for such sweeping behavior, we used live TIRF-SIM to image Jurkat T cells expressing GFP–F-Tractin and engaged with bilayers containing Alexa Fluor 548–labeled anti-CD3 antibody to report the position of TCR MCs. Video 9 and still images taken from it (Fig. 9 A) show a clear example of this sweeping behavior, in which a single TCR MC (Fig. 9 A, white open arrowheads) is swept inward for a time by one arc (Fig. 9 A, first through third panels, white closed arrowheads), slips past it (Fig. 9 A, fourth panel) to be immediately picked up and swept inward by a second arc (Fig. 9 A, fifth through seventh panels, yellow arrowheads), and then slips again to be picked up by a third arc (Fig. 9 A, eighth panel, blue arrowheads). Video 10 and the still images taken from it in Fig. 9 B show a second example of this sweeping behavior, in which a TCR MC (Fig. 9 B, open arrowheads) is swept inward successively by two arcs (Fig. 9 B, first through fourth panels, white closed arrowheads, and fifth and sixth panels, yellow closed arrowheads). Consistent with our previous work (Yi et al., 2012), the rates of inward arc flow and TCR MC movement are not statistically different in these videos (Fig. 9 C), suggesting that the immobile phase arising from slippage is not large enough to significantly uncouple MC movement from arc flow, at least in a bilayer context. Collectively, these findings provide strong evidence that formin-generated, LFA-1– and myosin II–rich actin arcs in the pSMAC sweep TCR MCs inward to facilitate their robust accumulation in the cSMAC.
Figure 9.
Actin arcs propel the centripetal transport of TCR MCs across the pSMAC. (A and B) Still images from two separate TIRF-SIM videos of Jurkat T cells expressing GFP–F-Tractin (grayscale) stimulated on planar lipid bilayers containing Alexa Fluor 568–labeled anti-CD3 antibody (red) to report TCR MC movement (Videos 9 and 10). (C) Quantitation of rates of centripetal movement of actin and TCR MCs across the dSMAC and pSMAC obtained from Videos 9 and 10. n = 15–19 measurements/condition. Box plots are centered on means and display upper and lower quartile ranges and min to max values. Bars, 1 µm.
Discussion
In this study, we used SIM to define the organization of the Jurkat T cell’s actin cytoskeleton in unprecedented clarity and demonstrate in unequivocal fashion the existence of actomyosin arcs in the pSMAC region of their IS. By combining SIM imaging with the use of specific inhibitors, we showed how formins and myosin II cooperate to create these actin arcs, and we demonstrated their functional importance. We also used TIRF-SIM to provide the first subdiffraction-limited imaging of actin-driven TCR MC transport. Importantly, we showed that actomyosin arcs are also major components of primary mouse T cell synapses, arguing that they may play significant roles in promoting T cell effector functions in vivo.
Although we presented many lines of evidence that the arcs are formin generated, current models for the formation of anisotropic lamellar actin structures like the arcs described in this study involve not only formin-dependent nucleation, but also the repurposing of the branched actin network in the lamellipodium after debranching and cross-linking (Medeiros et al., 2006; Burnette et al., 2011; Tojkander et al., 2012; Henson et al., 2015; Tee et al., 2015). Although we cannot rule out some contribution to arc formation from this pathway, it appears from our imaging and inhibitor studies that the linear filaments made by formins at the edge of the dSMAC are primarily responsible for arc formation. Regarding the fate of the branched actin network in the dSMAC, the addition of Jasplakinolide, a drug that prevents the depolymerization of actin filaments, causes the rapid formation of a large F-actin ring at the dSMAC/pSMAC boundary (Yi et al., 2012). We think, therefore, that the majority of the dSMAC is disassembled at this boundary, with the ensuing release of monomer supporting continued Arp2/3- and formin-dependent actin nucleation at the outer edge of the dSMAC.
With regard to the functional significance of pSMAC arcs, we show in Jurkat T cells that disruption of their formation via formin inhibition results in numerous functional deficiencies, including less centralized TCR MCs, missegregated integrin clusters, decreased T–B cell adhesion frequency, and diminished TCR proximal signaling in conjugates. Moreover, myosin II inhibition phenocopies formin inhibition, consistent with the fact that arcs require myosin II for their organization and contraction. This latter finding is consistent with a subset of previous studies of myosin II function at the IS (Hammer and Burkhardt, 2013). Given that centrosome repositioning is facilitated by formins (Gomez et al., 2007) and helps sustain T cell signaling (Martín-Cófreces et al., 2008), it remains possible that some portion of the defects seen upon formin inhibition are downstream of reduced centrosome repositioning.
Our demonstration that actomyosin arcs are commonly decorated with the open, active conformation of LFA-1 during IS maturation diverges from the recent results of Comrie et al. (2015a), who reported essentially no overlap between active LFA-1 and myosin II at the IS. Although part of this discrepancy could be because of the different T cells studied (Jurkat in this study versus human CD4+), it may have more to do with the time points chosen for imaging (6–10 min in this study versus 30 min). Interestingly, Comrie et al. (2015a) attributed LFA-1 activation primarily to polymerization-driven actin retrograde flow and minimally to myosin II contractility. This conclusion was based in part on their observation that activation was strongly inhibited when retrograde flow was blocked using Jasplakinolide, but not when myosin II contraction was inhibited using blebbistatin. We note, however, that using Jasplakinolide to impair actin retrograde flow may also impair actomyosin arc function by rapidly depleting the actin monomer pool, based on our demonstration in this study that this pool is normally limiting for nucleation in T cells. We also note that their use of blebbistatin (no preincubation and addition 5 min after activation) may have underestimated myosin II’s role in LFA-1 activation. That said, we also see evidence that actin retrograde flow can promote LFA-1 activation, because open, active LFA-1 was still present at the IS when arc formation had been inhibited (Fig. 7). Nevertheless, our demonstration that the open, active conformation of LFA-1 commonly colocalizes with actomyosin arcs in maturing synapses argues that actomyosin arcs probably play a significant role in integrin activation in T cells, as in mesenchymal cells.
Perhaps our most remarkable observation, made possible by the enhanced spatiotemporal resolution provided by 3D-SIM and TIRF-SIM, was that TCR MCs in the pSMAC appear to be swept inward by the actomyosin arcs. This conclusion has relevance for the longstanding question of how inward actin flow propels the centripetal transport of MCs. Studies by DeMond et al. (2008) and Yu et al. (2010) showed that thwarting the inward movement of MCs using barriers placed in the bilayer causes actin flow to slow transiently at the site of the thwarted MC. This seminal observation argues for a frictional coupling between TCR MCs and a viscoelastic actin cytoskeleton. Our TIRF-SIM videos showing that arcs sweep MCs inward and that slippage events can occur are consistent with this conclusion. Whether the basis for this frictional coupling is simply physical collision between the actin and the MC or involves transient physical tethering between them (or some combination of both) remains to be seen. The imaging described in this study could serve useful in efforts to resolve this latter issue. Finally, these SIM modalities could prove valuable in defining the contributions made by actomyosin force generation to phase-separated cluster formation, ligand discrimination, and mechanotransduction at the IS (Chen and Zhu, 2013; Depoil and Dustin, 2014; Su et al., 2016).
Materials and methods
Cell culture and transfection
E6.1 Jurkat T cells were maintained at 37°C in Iscove’s modified Dulbecco’s media (Thermo Fisher Scientific) supplemented with 10% FBS (Sigma-Aldrich), sodium pyruvate, l-glutamine, penicillin-streptomycin, and MEM nonessential amino acids solution (Thermo Fisher Scientific). Transfections were performed with 1.0 × 106 cells/ml and 2 to 3 µg plasmid DNA by nucleofection using Amaxa kit V (Lonza). Primary mouse T cells were obtained via dissection of mouse inguinal, axillary, brachial, and mesenteric lymph nodes. CD8+ T cells were isolated via negative selection using a CD8a+ T cell isolation kit (Kit II, MACS; Miltenyi Biotec) and grown in DMEM supplemented with 5% FBS, l-glutamine, and penicillin-streptomycin. The cells were then activated for 72 h with plate-bound 2.5 µg/ml anti-mouse CD3 and 1 µg/ml anti-mouse CD28 antibodies (BD), followed by stimulation with 50 U/ml recombinant interleukin-2 (R&D Systems) every 48 h.
Plasmids and reagents
F-Tractin tagged with mGFP or tdTomato were gifts from M. Schell (Uniformed Services University, Bethesda, MD). Anti–myosin IIA antibody was purchased from Sigma-Aldrich. shRNA plasmids for control and mDia1 knockdown were gifts from D. Billadeau (Mayo Clinic, Rochester, MN). Alexa-conjugated secondary antibodies and phalloidin were purchased from Thermo Fisher Scientific. Alexa Fluor 488–conjugated anti–human LFA-1 (M24 clone) antibody was purchased from BioLegend. Anti–INF2 non-CAAX antibody was a gift from H. Higgs (Dartmouth University, Hanover, NH). Anti-mDia1 antibody was purchased from Bethyl Laboratories, Inc. Anti–FMNL1 and anti–α-actinin antibodies were purchased from Sigma-Aldrich. GFP-Tm4 was a gift from P. Gunning (University of New South Wales, Sydney, New South Wales, Australia). pnBB was purchased from Octopharma. CN01 was purchased from Cytoskeleton, Inc. The inactive SMIFH2 analogue KVSM18 was a gift from D. Kovar (University of Chicago, Chicago, IL). CK666, SMIFH2, ML-7, Y27632, and DMSO for vehicle control were purchased from Sigma-Aldrich.
Immunofluorescence
Jurkat T cells were allowed to adhere to the substrate for 7 min at 37°C and then fixed for 15 min in 4% (wt/vol) PFA solution (Electron Microscopy Sciences) in cytoskeletal buffer (300 mM NaCl, 10 mM EGTA, 10 mM glucose, 10 mM MgCl2, 20 mM Pipes, pH 7.4, and 0.4% sucrose). Samples were then incubated in a blocking solution consisting of 10% FBS (Sigma-Aldrich), 0.01% sodium azide (Sigma-Aldrich), 1× PBS, and 0.2% saponin (Sigma-Aldrich) for 15 min at room temperature (RT). After three washes in 1× PBS, the cells were stained with primary antibody (1:100- to 500-fold dilution in 1× PBS) for 1 h at RT, followed by secondary antibody (1:1,000-fold dilution in 1× PBS) or phalloidin (1:200-fold dilution) for 1 h at RT.
Preparation of supported planar lipid bilayers
Liposomes and glass-supported planar lipid bilayers were prepared as described previously (Varma et al., 2006; Yi et al., 2012; Crites et al., 2015). All lipids were purchased from Avanti Polar Lipids, Inc. Liposomes were created using a mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine, biotin–CAP-PE (1% molar ratio), and 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (DGS)–NTA (1% molar ratio) lipids. To generate unilamellar liposomes, lipids were lyophilized overnight, reconstituted in nitrogen-purged Tris-saline buffer, sonicated in a water bath, and passed through an extrusion membrane (∼50-nm pore size) using a mini-extruder kit (Avanti Polar Lipids, Inc.). Bilayers were created in flow chambers assembled using glass coverslips that had been cleaned using Piranha solution (70% [vol/vol] sulfuric acid [Sigma-Aldrich]; and 30% [vol/vol] hydrogen peroxide [Thermo Fisher Scientific]). After depositing liposomes onto the flow chambers, the chambers were washed with HBS buffer (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer saline) and 5% (wt/vol) casein (Sigma-Aldrich) to block nonspecific sites (Yi et al., 2012). Next, a 1:2 ratio of monobiotinylated Alexa Fluor 568/647– or rhodamine X–labeled (10 µg/ml) anti-CD3ε antibody and streptavidin (85878; Sigma-Aldrich) were added to the flow chambers along with histidine-tagged ICAM-1, either unlabeled or labeled with Alexa Fluor 647 (0.5 µg/ml). Anti-CD3ε antibody (OKT3; Biovest International Inc.) was monobiotinylated and labeled with fluorescent dyes as described previously (Carrasco et al., 2004). The uniformity and lateral mobility of lipids in the bilayers was assessed by FRAP.
Preparation of stimulatory glass coverslips
Coverslip substrates coated with immobilized antibodies to stimulate Jurkat T cells were prepared as described previously (Bunnell et al., 2003). In brief, eight-well coverglass chamber slides (Lab-Tek; Thermo Fisher Scientific) were washed in HCL and 70% (vol/vol) ethanol. After three washes in 1× PBS, each well was incubated for 30 min at RT with 500 µl of 0.01% (vol/vol) poly-l-lysine (Sigma-Aldrich). Wells were then incubated for 1 h at 37°C with 500 µl of a solution containing 20 µg/µl anti-CD3ε antibody and 20 µg/µl anti-CD28 antibody (BD) diluted in 1× PBS. For primary mouse CD4+/CD8+ cells, the same procedure was followed except that mouse anti-CD3/CD28 antibodies (BD) were used instead.
Microscopy
3D-SIM was performed on a 3D-SIM Imaging System (GE DeltaVision OMX; Applied Precision Ltd.) equipped with a 60× 1.42 NA objective (Olympus). Raw data were reconstructed using Softworx (Applied Precision Ltd.; GE Healthcare) and 0.003 for the wiener filter constant. TIRF-SIM imaging was performed as described previously (Beach et al., 2014; Li et al., 2015). In brief, images were acquired using an inverted microscope (Axio Observer; ZEISS) fitted with a 100× 1.49 NA objective (Olympus) and a spatial light modulator to provide phase grating for structured illumination (Li et al., 2015). Confocal imaging was performed on an IX81 with a 150× 1.4 NA objective (Olympus) fitted with a spinning disk confocal unit (Yokogawa Electric Corporation). Linear adjustments to images were made using ImageJ software (National Institutes of Health).
Adhesion conjugate assay
The adhesion conjugate assay was performed as previously described (Cannons et al., 2010; Zhao et al., 2012). In brief, Raji B cells were pulsed with 2 µg/ml SEE for 1 h at 37°C and washed three times in medium. Jurkat T cells were mixed with SEE-pulsed Raji B cells at a ratio of 1:4 and incubated for 10 min at 37°C. For myosin II inhibition, Jurkat T cells were preincubated with 50 µM pnBB or DMSO control. For formin inhibition, 25 µM SMIFH2 was added to the Jurkat T cell and Raji B cell mixture. Samples were fixed using 4% PFA and stained with FITC-conjugated anti-human CD3 antibody (OKT3; eBioscience) and PE-conjugated anti–human HLA–antigen D related (DR) antibody (LN3; eBioscience). Conjugate frequency was analyzed using flow cytometry (LSR II; BD) after collecting a minimum of 10,000 events, with the OKT3+ HLA-DR+ double-positive population representing Jurkat T–Raji B cell conjugates.
Amnis ImageStream flow cytometry assay
Imaging flow cytometry was performed as previously described (Markey et al., 2015). In brief, Jurkat T and Raji B cells were transfected with GFP- and RFP-farnesyl plasmids, respectively, using a nucleofector kit (Amaxa; Lonza). Raji B cells were pulsed with 2 µg/ml SEE for 1 h at 37°C and washed three times in medium. Jurkat T cells were mixed with SEE-pulsed Raji B cells at a ratio of 1:1 and incubated with NucBlue Live Ready Probes Reagent (Thermo Fisher Scientific) for 10 min at 37°C. Samples were fixed using 4% PFA, permeabilized using 0.2% saponin, and stained for phosphorylated signaling molecules. Anti–phosphorylated Src (Y416) antibody, anti–phosphorylated Zap70 (Y319)/Syk (Y352) antibody, and anti–phosphorylated Lat (Y191; Cell Signaling Technology) were used as primary antibodies, followed by an Alexa Fluor 647 secondary antibody (Thermo Fisher Scientific). For data acquisition, a flow cytometer (Amnis ImageStream Gen-X Mark II; EMD Millipore) with four lasers (405, 488, 561, and 642 nm) was used. All acquisition was performed at a magnification of 40 collecting a minimum of 30,000 events with a negative control (SEE-null) and compensation controls. Focused cells based on the gradient root mean square feature were analyzed. Doublet events were gated from the aspect ratio analysis and then refined further using GFP and RFP double-positive cells. Jurkat T cell and Raji B cell doublets were successfully identified using this strategy, and the selection of high image quality, focused doublets within the viewing window allowed the refining of final gating (n = 25–225 cells). A combination mask was then used to identify pixels at the interface (valley mask) and overlap with the T cell (intensity mask). Background-corrected MFIs at the masked region were calculated by subtracting the MFI of the control (SEE-null) from the MFI of each phosphoprotein in different inhibition conditions. These values were then used for statistical analyses (Prism 6; GraphPad Software).
In vitro T cell activation assay
To activate Jurkats using soluble anti-CD3 antibody (Balagopalan et al., 2011), cells were washed with serum-free RPMI 1640, resuspended to a concentration of 3 × 107 cells/ml, and activated with anti-CD3 (1 µg/ml) for the indicated time points. Activation was halted by lysing the cells using 95°C 2× sample buffer and boiling for 3 min. Samples were then passed through a small-gauge syringe needle to shear the DNA and loaded onto a 10% precast polyacrylamide gel (Thermo Fisher Scientific). The separated proteins were transferred to nitrocellulose membrane that was then blocked overnight at 4°C in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% Tween 20) supplemented with 5% milk. All antibodies were diluted in TBST containing 5% milk. Primary antibodies were added to the membrane for 2 h at RT, followed by three 10-min washes in TBST and a 1-h incubation at RT with the appropriate secondary antibody. The proteins were visualized by chemiluminescence. Quantitation of the phosphorylation status of the signaling proteins was performed by measuring the integrated density of the bands on ImageJ (National Institutes of Health) followed by background correction and normalization to an anti–actin antibody (Sigma-Aldrich) loading control.
Image and statistical analyses
All statistical analyses were performed using Prism 6 (GraphPad Software). Unless otherwise indicated, all statistical comparisons were performed using unpaired, two-tailed t tests. Box and whiskers plots are centered on the mean and display upper and lower quartile ranges and min to max values. All data represent mean ± SEM except Figs. 4 D and 6 B, which are mean ± SD. To compare the relative pSMAC footprint at the IS of Jurkat T cells versus primary CD8+ T cells (Fig. 2 D), the total IS area was determined based on thresholds for F-actin intensity. Outlines for the dSMAC and pSMAC were manually determined using the F-actin channel and superimposed on the thresholded image. The area within the identified regions was measured using ImageJ (National Institutes of Health). To compare the actin spike morphology in Fig. 4 D (bottom panels), the Filotracker script from MATLAB application CellGeo was used on max projections of phalloidin-stained 3D-SIM images as previously described (Tsygankov et al., 2014). To qualitatively assess actin arc morphology, maximum projections of 3D-SIM images of phalloidin-stained Jurkat T cells were scored manually into four categories: Organized/concentric, Disorganized/inward pointing, Disorganized/meshwork, and No arcs. Note that Organized/concentric was used to describe cells with robust actin arcs, similar to representative images of control cells. To quantify the content of actin arcs in the pSMAC, maximum projections of phalloidin-stained 3D-SIM images of Jurkat T cells were used to capture the interior of the IS (i.e., everything except the dSMAC). Note that we chose to measure all regions of the IS except the dSMAC because formin inhibited cells do not display enough actin arcs to accurately define a pSMAC/cSMAC boundary. Note also that formin-inhibited cells display an increase in cSMAC phalloidin signal because of an increase in the frequency of F-actin foci. Therefore, our results likely underestimate the loss of pSMAC actin arc fluorescence upon SMIFH2 treatment. Imaging parameters across conditions were kept identical. The integrated pixel intensities of the outlined regions of interest for each condition were measured using ImageJ (National Institutes of Health). The FibrilTool plugin for ImageJ was used to measure actin arc morphology as described previously (Boudaoud et al., 2014). In brief, the pSMAC regions in maximum projections of phalloidin-stained 3D-SIM images were divided into 10–12 trapezoid shaped ROIs of similar size to measure the anisotropy of arcs in the radial symmetric pSMAC. To measure the distribution of TCR MCs and LFA-1 at the IS, we used the radial plot profile plugin for ImageJ. The plugin approximated the outline of the cell based on manual drawing of a quadrangle that best fit the cellular dimensions (identified by F-actin staining). This outline was superimposed on the channel displaying TCR MCs or LFA-1, followed by radial plot profile generation. Radial plot profile intensities were background subtracted and normalized for intensity. To account for cell size variance, cell radii were normalized to 1, and intensities were binned 41 times along the normalized radii. The two-way analysis of variance (ANOVA) was used to test for statistical significance between control and experimental condition datasets. The distribution of TCR MCs and LFA-1 at the IS was also measured by drawing line scans across the IS. After background subtraction, the line scans were normalized as described above for radial plot profiles, but fit to Gaussian distributions and tested for single versus two-Gaussian fit in Prism (GraphPad Software). To compare the spatial associations between F-actin, TCR MCs, LFA-1, and myosin IIA, 3D-SIM images were thresholded using the F-actin channel to find the center of the cell (xy coordinates). The TCR MC channel was then thresholded to identify the MCs and find their centers (xy coordinates). A 40 × 40-pixel box was drawn to crop 10–15 pSMAC TCR MCs per cell. These images were aligned and rotated by measuring the angles between the MC and the center of the cell in Excel using the ArcTan function (Microsoft). The images from each channel were then superimposed to obtain population averaged images for radial plot profiles and line scan analyses as described earlier in this section. To assess colocalizations, Pearson’s coefficient analyses were performed using the JaCop plugin in ImageJ. To measure the speed of centripetal actin and TCR MC movement, kymographs were made from TIRF-SIM videos using the MultipleKymograph plugin from ImageJ. The dSMAC and pSMAC regions were identified by the relatively abrupt change in slope for F-actin flow, and rates were determined by measuring slope angles.
Online supplemental material
Fig. S1 shows that actomyosin arcs exist in primary mouse CD8+ T cells and that td-Tomato–F-Tractin, but not RFP-actin, labels actin arcs. Fig. S2 shows that several formins (including mDia1) are enriched at the tips of actin spikes and that mDia1 knockdown inhibits arc formation. Fig. S3 shows that an activator of Rho GTPases enhances arcs and that arcs label robustly with TM4 but not α-actinin. Fig. S4 shows that myosin II inhibition disrupts receptor cluster distribution and that myosin II coincides with actin and open, active LFA-1 in the pSMAC. Fig. S5 shows that SMIFH2 inhibits signaling in T cells stimulated with soluble antigen and that KVSM18 does not inhibit Lat phosphorylation. Video 1 shows the dynamics of F-actin networks at the IS using TIRF-SIM. Video 2 shows the dynamics of actomyosin arcs using two-color TIRF-SIM in a Jurkat. Video 3 shows the formation of linear actin filaments/bundles at the distal edge of the IS. Video 4 shows linear actin filaments/bundles, embedded in the dSMAC, flowing inward and giving rise to pSMAC actin arcs. Video 5 shows the reversible augmentation of linear actin filaments/bundles upon Arp2/3 inhibition. Video 6 shows augmented actin filaments/bundles that flow inward and bend to give rise to actin arcs. Video 7 shows the recovery of actin arc formation upon washout of the formin inhibitor. Video 8 shows that myosin II inhibition disrupts the organization of actin arcs. Videos 9 and 10 show TCR MCs being swept inward by actin arcs. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201603080.dv.
Supplementary Material
Acknowledgments
The authors thank the National Heart, Lung, and Blood Institute FACS Core for help with Amnis ImageStream flow cytometry; Rajat Varma for help with liposome preparations; Henry Higgs, Dan Billadeau, Peter Gunning, David Kovar, and Michael Schell for reagents; and Travis Crites, Alex Ritter, Connie Sommers, Senta Kapnick, Jennifer Cannons, Lakshmi Balagopalan, Sarah Hitchcock, Larry Samelson, Pam Schwartzberg, and Clare Waterman for helpful discussions.
This work was supported by Intramural National Heart, Lung, and Blood Institute grant 1ZIAHL006121-04 to J.A. Hammer.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- APC
- antigen-presenting cell
- cSMAC
- central supramolecular activation cluster
- dSMAC
- distal supramolecular activation cluster
- FMNL1
- formin-like protein 1
- HLA
- human leukocyte antigen
- ICAM
- intercellular adhesion molecule
- INF2
- inverted formin-2
- IS
- immunological synapse
- LFA-1
- leukocyte functional antigen 1
- MFI
- mean fluorescence intensity
- pnBB
- para-nitro-blebbistatin
- pSMAC
- peripheral supramolecular activation cluster
- ROI
- region of interest
- SEE
- staphylococcal enterotoxin E
- SIM
- structured-illumination microscopy
- SMAC
- supramolecular activation cluster
- SMIFH2
- small-molecule inhibitor of formin homology 2 domain
- TCR MC
- T cell receptor microcluster
- TIRF
- total internal reflection fluorescence
References
- Babich A., Li S., O’Connor R.S., Milone M.C., Freedman B.D., and Burkhardt J.K.. 2012. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J. Cell Biol. 197:775–787. 10.1083/jcb.201201018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L., Ashwell B.A., Bernot K.M., Akpan I.O., Quasba N., Barr V.A., and Samelson L.E.. 2011. Enhanced T-cell signaling in cells bearing linker for activation of T-cell (LAT) molecules resistant to ubiquitylation. Proc. Natl. Acad. Sci. USA. 108:2885–2890. 10.1073/pnas.1007098108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Whitlock B.M., Husson J., Le Floc’h A., Jin W., Oyler-Yaniv A., Dotiwala F., Giannone G., Hivroz C., Biais N., et al. . 2016. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 165:100–110. 10.1016/j.cell.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach J.R., Shao L., Remmert K., Li D., Betzig E., and Hammer J.A. III. 2014. Nonmuscle myosin II isoforms coassemble in living cells. Curr. Biol. 24:1160–1166. (published erratum appears in Curr. Biol. 2015. 25:402) 10.1016/j.cub.2014.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemiller P., and Krummel M.F.. 2013. Regulation of T-cell receptor signaling by the actin cytoskeleton and poroelastic cytoplasm. Immunol. Rev. 256:148–159. 10.1111/imr.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemiller P., Jacobelli J., and Krummel M.F.. 2012. Integration of the movement of signaling microclusters with cellular motility in immunological synapses. Nat. Immunol. 13:787–795. 10.1038/ni.2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaoud A., Burian A., Borowska-Wykręt D., Uyttewaal M., Wrzalik R., Kwiatkowska D., and Hamant O.. 2014. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 9:457–463. 10.1038/nprot.2014.024 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D., and Goode B.L.. 2013. Formins at a glance. J. Cell Sci. 126:1–7. 10.1242/jcs.107250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.C., Oddos S., Dobbie I.M., Alakoskela J.M., Parton R.M., Eissmann P., Neil M.A., Dunsby C., French P.M., Davis I., and Davis D.M.. 2011. Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super-resolution microscopy. PloS Biol. 9:e1001152 (published erratum appears in PLoS Biol. 2012. 10:8) 10.1371/journal.pbio.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell S.C., Kapoor V., Trible R.P., Zhang W., and Samelson L.E.. 2001. Dynamic actin polymerization drives T cell receptor-induced spreading: A role for the signal transduction adaptor LAT. Immunity. 14:315–329. 10.1016/S1074-7613(01)00112-1 [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Barr V.A., Fuller C.L., and Samelson L.E.. 2003. High-resolution multicolor imaging of dynamic signaling complexes in T cells stimulated by planar substrates. Sci. STKE. 2003:PL8. [DOI] [PubMed] [Google Scholar]
- Burke T.A., Christensen J.R., Barone E., Suarez C., Sirotkin V., and Kovar D.R.. 2014. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol. 24:579–585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D.T., Manley S., Sengupta P., Sougrat R., Davidson M.W., Kachar B., and Lippincott-Schwartz J.. 2011. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 13:371–381. 10.1038/ncb2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons J.L., Qi H., Lu K.T., Dutta M., Gomez-Rodriguez J., Cheng J., Wakeland E.K., Germain R.N., and Schwartzberg P.L.. 2010. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 32:253–265. 10.1016/j.immuni.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Y.R., Fleire S.J., Cameron T., Dustin M.L., and Batista F.D.. 2004. LFA-1/ICAM-1 interaction lowers the threshold of B cell activation by facilitating B cell adhesion and synapse formation. Immunity. 20:589–599. 10.1016/S1074-7613(04)00105-0 [DOI] [PubMed] [Google Scholar]
- Chen W., and Zhu C.. 2013. Mechanical regulation of T-cell functions. Immunol. Rev. 256:160–176. 10.1111/imr.12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Nag S., and Pollard T.D.. 2012. Formins filter modified actin subunits during processive elongation. J. Struct. Biol. 177:32–39. 10.1016/j.jsb.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie W.A., Babich A., and Burkhardt J.K.. 2015a F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J. Cell Biol. 208:475–491. 10.1083/jcb.201406121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie W.A., Li S., Boyle S., and Burkhardt J.K.. 2015b The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J. Cell Biol. 208:457–473. 10.1083/jcb.201406120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crites T.J., Maddox M., Padhan K., Muller J., Eigsti C., and Varma R.. 2015. Supported lipid bilayer technology for the study of cellular interfaces. Curr. Protoc. Cell Biol. 68:24.5.1–24.5.31. 10.1002/0471143030.cb2405s68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond A.L., Mossman K.D., Starr T., Dustin M.L., and Groves J.T.. 2008. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys. J. 94:3286–3292. 10.1529/biophysj.107.119099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depoil D., and Dustin M.L.. 2014. Force and affinity in ligand discrimination by the TCR. Trends Immunol. 35:597–603. 10.1016/j.it.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L. 2009. Supported bilayers at the vanguard of immune cell activation studies. J. Struct. Biol. 168:152–160. 10.1016/j.jsb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T.S., Kumar K., Medeiros R.B., Shimizu Y., Leibson P.J., and Billadeau D.D.. 2007. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 26:177–190. 10.1016/j.immuni.2007.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., and Dustin M.L.. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227. 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Hammer J.A. III, and Burkhardt J.K.. 2013. Controversy and consensus regarding myosin II function at the immunological synapse. Curr. Opin. Immunol. 25:300–306. 10.1016/j.coi.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman N.C., Nye J.A., and Groves J.T.. 2009. Cluster size regulates protein sorting in the immunological synapse. Proc. Natl. Acad. Sci. USA. 106:12729–12734. 10.1073/pnas.0902621106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Tane A., Yokosuka T., Sakata-Sogawa K., Sakuma M., Ishihara C., Tokunaga M., and Saito T.. 2011. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity. 34:919–931. 10.1016/j.immuni.2011.05.012 [DOI] [PubMed] [Google Scholar]
- Henson J.H., Yeterian M., Weeks R.M., Medrano A.E., Brown B.L., Geist H.L., Pais M.D., Oldenbourg R., and Shuster C.B.. 2015. Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol. Biol. Cell. 26:887–900. 10.1091/mbc.E14-07-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilani T., Vasiliver-Shamis G., Vardhana S., Bretscher A., and Dustin M.L.. 2009. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat. Immunol. 10:531–539. 10.1038/ni.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.W., and Schell M.J.. 2009. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell. 20:5166–5180. 10.1091/mbc.E09-01-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y., Douglass A.D., Varma R., Dustin M.L., and Vale R.D.. 2007. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl. Acad. Sci. USA. 104:20296–20301. 10.1073/pnas.0710258105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Képiró M., Várkuti B.H., Végner L., Vörös G., Hegyi G., Varga M., and Málnási-Csizmadia A.. 2014. para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor. Angew. Chem. Int. Ed. Engl. 53:8211–8215. 10.1002/anie.201403540 [DOI] [PubMed] [Google Scholar]
- Kner P., Chhun B.B., Griffis E.R., Winoto L., and Gustafsson M.G.. 2009. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods. 6:339–342. 10.1038/nmeth.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R. 2006. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18:11–17. 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kumari S., Vardhana S., Cammer M., Curado S., Santos L., Sheetz M.P., and Dustin M.L.. 2012. T lymphocyte myosin IIA is required for maturation of the immunological synapse. Front. Immunol. 3:230 10.3389/fimmu.2012.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Curado S., Mayya V., and Dustin M.L.. 2014. T cell antigen receptor activation and actin cytoskeleton remodeling. Biochim. Biophys. Acta. 1838:546–556. 10.1016/j.bbamem.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Depoil D., Martinelli R., Judokusumo E., Carmona G., Gertler F.B., Kam L.C., Carman C.V., Burkhardt J.K., Irvine D.J., and Dustin M.L.. 2015. Actin foci facilitate activation of the phospholipase C-γ in primary T lymphocytes via the WASP pathway. eLife. 4:e04953 10.7554/eLife.04953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Hui K., Kwak S.I., and Upadhyaya A.. 2014. Adhesion-dependent modulation of actin dynamics in Jurkat T cells. Cytoskeleton (Hoboken). 71:119–135. 10.1002/cm.21156 [DOI] [PubMed] [Google Scholar]
- Le Floc’h A., and Huse M.. 2015. Molecular mechanisms and functional implications of polarized actin remodeling at the T cell immunological synapse. Cell. Mol. Life Sci. 72:537–556. 10.1007/s00018-014-1760-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Shao L., Chen B.C., Zhang X., Zhang M., Moses B., Milkie D.E., Beach J.R., Hammer J.A. III, Pasham M., et al. . 2015. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 349:aab3500 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin A.J., Lee K.C., Han S.J., Bui D.A., Davidson M., Mogilner A., and Danuser G.. 2015. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 17:1435–1445. 10.1038/ncb3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace E.M., Wu W.W., Ho T., Mann S.S., Hsu H.T., and Orange J.S.. 2012. NK cell lytic granules are highly motile at the immunological synapse and require F-actin for post-degranulation persistence. J. Immunol. 189:4870–4880. 10.4049/jimmunol.1201296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey K.A., Gartlan K.H., Kuns R.D., MacDonald K.P., and Hill G.R.. 2015. Imaging the immunological synapse between dendritic cells and T cells. J. Immunol. Methods. 423:40–44. 10.1016/j.jim.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Martín-Cófreces N.B., Robles-Valero J., Cabrero J.R., Mittelbrunn M., Gordón-Alonso M., Sung C.H., Alarcón B., Vázquez J., and Sánchez-Madrid F.. 2008. MTOC translocation modulates IS formation and controls sustained T cell signaling. J. Cell Biol. 182:951–962. 10.1083/jcb.200801014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P.K., Batista F.D., and Treanor B.. 2016. Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling. J. Cell Biol. 212:267–280. 10.1083/jcb.201504137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N.A., Burnette D.T., and Forscher P.. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:215–226. 10.1038/ncb1367 [DOI] [PubMed] [Google Scholar]
- Monks C.R., Freiberg B.A., Kupfer H., Sciaky N., and Kupfer A.. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86. 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Ramabhadran V., Korobova F., Rahme G.J., and Higgs H.N.. 2011. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell. 22:4822–4833. 10.1091/mbc.E11-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., Haynes E.M., Suarez C., Winkelman J.D., Johnson H.E., Haugh J.M., Kovar D.R., and Bear J.E.. 2015. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell. 32:54–67. 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoligovets A.A., Smith A.W., Wu H.J., Petit R.S., and Groves J.T.. 2012. Characterization of dynamic actin associations with T-cell receptor microclusters in primary T cells. J. Cell Sci. 125:735–742. 10.1242/jcs.092825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., and Vale R.D.. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 352:595–599. 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee Y.H., Shemesh T., Thiagarajan V., Hariadi R.F., Anderson K.L., Page C., Volkmann N., Hanein D., Sivaramakrishnan S., Kozlov M.M., and Bershadsky A.D.. 2015. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 17:445–457. 10.1038/ncb3137 [DOI] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., and Lappalainen P.. 2012. Actin stress fibers--assembly, dynamics and biological roles. J. Cell Sci. 125:1855–1864. 10.1242/jcs.098087 [DOI] [PubMed] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460. 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R., Campi G., Yokosuka T., Saito T., and Dustin M.L.. 2006. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 25:117–127. 10.1016/j.immuni.2006.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Wu X.S., Crites T., and Hammer J.A. III. 2012. Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol. Biol. Cell. 23:834–852. 10.1091/mbc.E11-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Wu H.J., Kaizuka Y., Vale R.D., and Groves J.T.. 2010. Altered actin centripetal retrograde flow in physically restricted immunological synapses. PLoS One. 5:e11878 10.1371/journal.pone.0011878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Fay N.C., Smoligovets A.A., Wu H.J., and Groves J.T.. 2012. Myosin IIA modulates T cell receptor transport and CasL phosphorylation during early immunological synapse formation. PLoS One. 7:e30704 10.1371/journal.pone.0030704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Smoligovets A.A., and Groves J.T.. 2013. Modulation of T cell signaling by the actin cytoskeleton. J. Cell Sci. 126:1049–1058. 10.1242/jcs.098210 [DOI] [PubMed] [Google Scholar]
- Zhao F., Cannons J.L., Dutta M., Griffiths G.M., and Schwartzberg P.L.. 2012. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 36:1003–1016. 10.1016/j.immuni.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.