The mechanisms by which microtubule dynamics are regulated are not fully understood. In this study, the authors show that five proteins are sufficient to reproduce all three phases of microtubule plus end dynamics in vitro, and, furthermore, a mitotic kinase can modulate the dynamicity.
Abstract
Cytoplasmic microtubules (MTs) undergo growth, shrinkage, and pausing. However, how MT polymerization cycles are produced and spatiotemporally regulated at a molecular level is unclear, as the entire cycle has not been recapitulated in vitro with defined components. In this study, we reconstituted dynamic MT plus end behavior involving all three phases by mixing tubulin with five Drosophila melanogaster proteins (EB1, XMAP215Msps, Sentin, kinesin-13Klp10A, and CLASPMast/Orbit). When singly mixed with tubulin, CLASPMast/Orbit strongly inhibited MT catastrophe and reduced the growth rate. However, in the presence of the other four factors, CLASPMast/Orbit acted as an inducer of pausing. The mitotic kinase Plk1Polo modulated the activity of CLASPMast/Orbit and kinesin-13Klp10A and increased the dynamic instability of MTs, reminiscent of mitotic cells. These results suggest that five conserved proteins constitute the core factors for creating dynamic MTs in cells and that Plk1-dependent phosphorylation is a crucial event for switching from the interphase to mitotic mode.
Introduction
Complementary to genetic analysis, in vitro reconstitution of intracellular processes, using defined factors, is a prerequisite to understand the process at a molecular level. This can determine if a given effect is directly mediated by each factor and defines a minimal set of factors that can execute the process (Loisel et al., 1999; Garner et al., 2007; Shintomi et al., 2015; Yeeles et al., 2015). Dynamic microtubules (MTs) are composed of tubulin proteins and play critical roles in various intracellular processes in eukaryotic cells, such as spindle formation and cell polarization. Tubulin proteins, at particular concentrations, can autonomously and stochastically polymerize and depolymerize in vitro; this is referred to as MT dynamic instability (Mitchison and Kirschner, 1984; Horio and Hotani, 1986; Walker et al., 1988). In cells, free MT plus ends in the interphase cytoplasm repeatedly undergo three phases (growth, shrinkage, and pausing) in a stochastic order (Shelden and Wadsworth, 1993; Rogers et al., 2002). Multiple proteins control cellular MT dynamics, and depletion of these proteins results in alterations to MT dynamicity and consequently a change in the length of MT-based higher-order structures such as cilia or the mitotic spindle (Howard and Hyman, 2007; Akhmanova and Steinmetz, 2008; Goshima and Scholey, 2010; Hu et al., 2015). Most, if not all, of these proteins can individually modulate MT dynamics in vitro. Examples include XMAP215, which can increase MT growth rate, or kinesin-13, which can increase “catastrophe” frequency (frequency of growth-to-shrinkage or pause-to-shrinkage transitions; Howard and Hyman, 2007). When two or more factors are mixed with tubulin, the dynamic MT behavior is better reproduced (Kinoshita et al., 2001; Li et al., 2012; Zanic et al., 2013). However, cellular MT dynamics, with all three phases, have not been successfully reproduced in vitro. Moreover, upon entry into mitosis, MTs become more dynamic, wherein MT growth rate and catastrophe frequency are increased and pausing is suppressed. However, the molecular basis of this cell cycle–dependent switch is also unclear.
The Drosophila melanogaster S2 cell line is a promising model system for the reconstitution of MT plus end dynamics. Parameters of MT dynamics in interphase and mitosis have been obtained through live-cell microscopy of fluorescent markers (Rogers et al., 2002; Brittle and Ohkura, 2005; Sousa et al., 2007; Li et al., 2011; Trogden and Rogers, 2015). Using this cell line, a handful of proteins required for dynamic regulation of MTs have been identified through multiple functional studies, including a large-scale functional genomics screen (Goshima et al., 2007; Hughes et al., 2008; Moutinho-Pereira et al., 2013). For instance, cells depleted of three conserved MT plus end tracking proteins Msps (XMAP215/ch-TOG orthologue), EB1 (a member of the EB protein family), and Sentin (a functional homolog of mammalian SLAIN2) similarly decreased MT dynamicity, wherein MT growth rate was reduced and pausing was drastically elevated (Rogers et al., 2002; Brittle and Ohkura, 2005; Li et al., 2011). To initiate in vitro reconstitution of MT plus end dynamics in S2 cells, we purified EB1, Sentin, and XMAP215Msps proteins, mixed them with pig brain–derived tubulins, and assessed MT plus end polymerization dynamics (Li et al., 2012). Compared with MTs generated solely with tubulin, or with one of these proteins, MTs formed in the presence of all three proteins had faster growth and more frequent catastrophe events, which is consistent with their depletion phenotype in cells (Rogers et al., 2002; Brittle and Ohkura, 2005; Li et al., 2011, 2012). However, even with various concentrations and combinations of these proteins, an in vitro system that closely reproduces the in vivo dynamics has not yet been constructed; most critically, pausing and the “rescue” event (shrinkage-to-growth or shrink-to-pause transitions) were observed with much lower frequency than in cells (Li et al., 2012).
We reasoned that a failure to reconstitute these three phases realistically might be caused by missing factors in the in vitro reaction. In this study, we considered Klp10A (kinesin-13) and CLASP (Drosophila name is Mast or Orbit) as likely candidates for these missing factors, because RNAi depletion in S2 cells affected MT dynamics and mitotic spindle size (Goshima et al., 2005; Mennella et al., 2005; Sousa et al., 2007). Furthermore, orthologous proteins of other species were shown to modulate MT dynamics in vitro (Mennella et al., 2005; Al-Bassam et al., 2010). In addition, with recently established methodology (Widlund et al., 2012), we purified tubulin proteins from S2 cells and used them to replace pig tubulin proteins. In the reaction, in vivo MT dynamics were reproduced qualitatively (and, to some extent, quantitatively) when five factors were mixed with S2 tubulin. Furthermore, phosphorylation by the mitotic kinase Polo (Plk1 in mammals) shifted MT dynamics to the mitotic mode.
Results
CLASPMast/Orbit suppresses MT growth and catastrophe
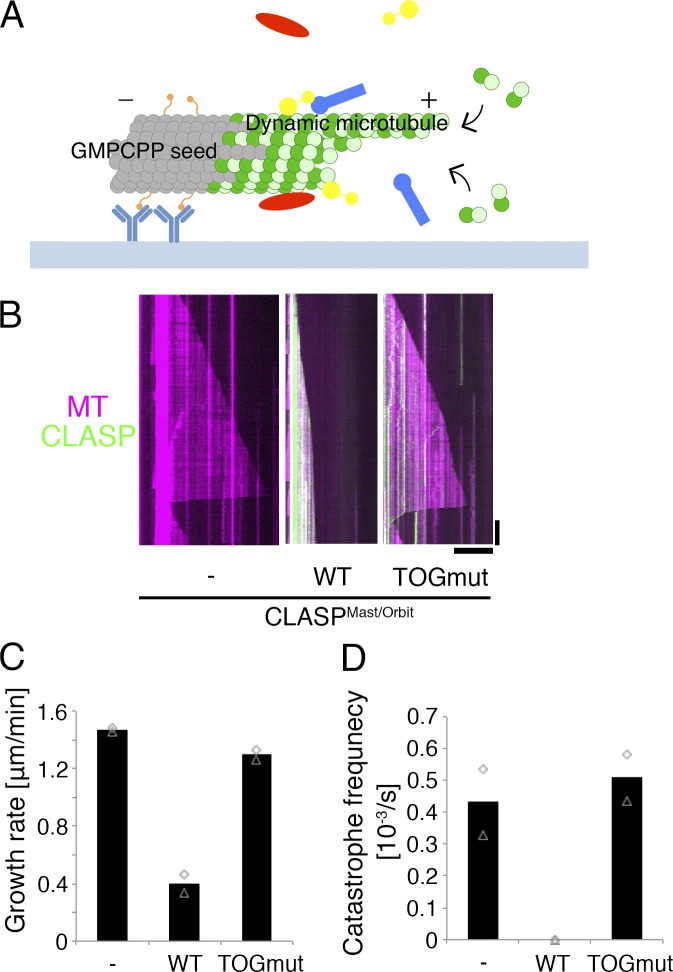
We first characterized the activity of S2 tubulin in the MT plus end polymerization assay, in which dynamic MTs are generated from a stable MT seed made with GMPCPP-tubulin (Bieling et al., 2007; Fig. 1 A). As expected, S2-derived tubulins showed repeated growth-shrinkage cycles, as observed with pig tubulins, although each parameter was quantitatively different (Fig. S1, A, C, and D).
Figure 1.
CLASPMast/Orbit slows growth and inhibits catastrophe in vitro. (A) Schematic of in vitro MT polymerization assay used in this study. Green, tubulin; orange, biotinylation; light blue, anti-biotin antibody; red/yellow/blue, recombinant proteins. (B) CLASPMast/Orbit slows MT growth and inhibits catastrophe. Kymographs show GFP-CLASPMast/Orbit (wild type [WT] and TOG domain mutant [TOGmut]) and MT (10 µM S2 tubulin). (C and D) MT growth rate and catastrophe frequency in the presence of 45 nM CLASPMast/Orbit (wild type and TOG domain mutant) and 10 µM S2 tubulin. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black columns (two independent experiments). Actual values are plotted in these graphs (n = 15–22 MTs). Bars: (horizontal) 10 µm; (vertical) 2 min.
Full-length kinesin-13Klp10A was purified from Escherichia coli, and it showed potent MT depolymerization activity toward both ends of seed MTs (Fig. S1 E), as expected (Rogers et al., 2004; Mennella et al., 2005).
GFP-tagged CLASPMast/Orbit was purified from insect Sf21 cells (Fig. S1 A). Gel filtration chromatography indicated that the protein does not form aggregates (Fig. S1 B). When mixed with S2 tubulin and MT seeds, GFP-CLASPMast/Orbit was localized along the seed and dynamic MTs, and a dramatic reduction in MT growth rate and catastrophe frequency was observed (Fig. 1, B–D). Such activity was not observed, and MT association was reduced, when we used a mutant GFP-CLASPMast/Orbit in which four amino acid residues critical for MT or tubulin binding in the TOG2 and TOG3 domains were mutated (Al-Bassam et al., 2010; Al-Bassam and Chang, 2011; Leano et al., 2013; Fig. 1, B–D). Thus, the observed growth- and catastrophe-suppression activities were derived from CLASPMast/Orbit, and not from contaminated proteins of Sf21 cells. In addition, the result indicated that the effect of GFP-CLASPMast/Orbit (on MT growth rate and catastrophe) requires its binding to tubulin and/or MTs. Previous RNAi experiments suggested that CLASPMast/Orbit induces MT pausing (Sousa et al., 2007; Trogden and Rogers, 2015), whereas purified fission yeast CLASPCls1 protein was shown to have potent rescue (shrinkage-to-growth) activity (Al-Bassam et al., 2010). However, we could not assess if Drosophila CLASPMast/Orbit has these activities in this experiment, because MT shrinkage was never observed (Fig. 1 D).
CLASP has an EB1-binding SxIP motif and abundantly localizes at the growing end of MTs in cells (Sousa et al., 2007; Honnappa et al., 2009). We reconstituted the plus end–tracking behavior of CLASPMast/Orbit by adding EB1 to the reaction (Fig. S1 F). EB1 on its own slightly elevated MT growth rate and catastrophe frequency (Li et al., 2012). However, we could not detect any additional effects of CLASPMast/Orbit on growth rate; in addition, catastrophe could not be observed regardless of the presence or absence of EB1 (Fig. S1 G). These results suggest that EB1-dependent plus end enrichment of CLASPMast/Orbit is not a prerequisite for the two CLASPMast/Orbit-mediated activities toward plus ends.
Reconstitution of three-phase MT dynamics with five factors
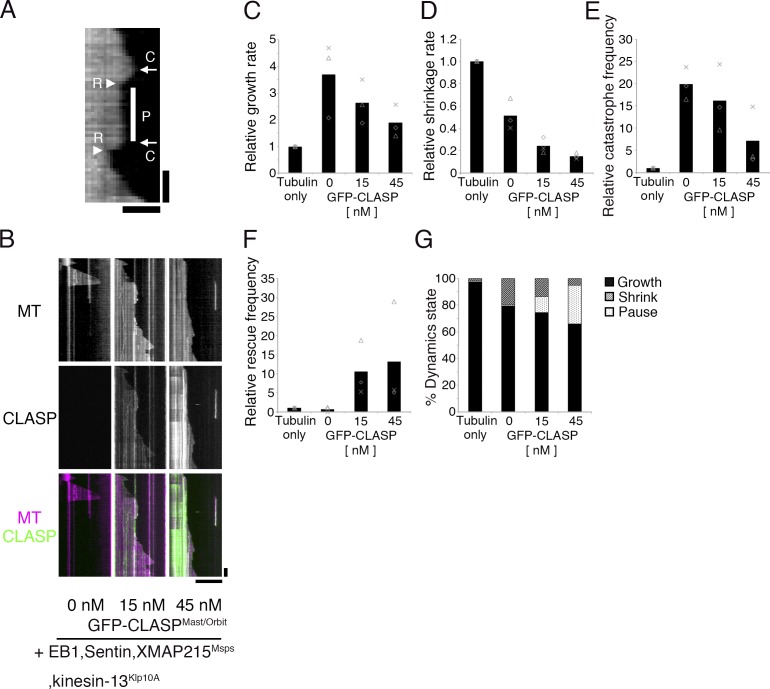
We next mixed all five factors in the reaction with S2-derived tubulin. Interestingly, we observed that MTs repeated cycles of shrinkage and growth, accompanied by frequent rescue and pausing; this behavior had never been observed using just one to three factors (Fig. 2 and Table 1). Each parameter value was within range of the corresponding cellular value, and the overall stochastic behavior was reminiscent of interphase MT dynamics in S2 cells (Fig. S2).
Figure 2.
Reconstitution of dynamic MTs with five factors. (A) An example of pause (P) identified in the kymograph. Catastrophe (C) and rescue (R) events are also marked. Bars: (horizontal) 2 µm; (vertical) 30 s. (B–G) Kymographs and kinetic parameters of MT polymerization dynamics with 0–45 nM GFP-CLASPMast/Orbit in the presence of 15 µM S2 tubulin, XMAP215Msps, EB1, Sentin, and kinesin-13Klp10A. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black columns (three independent experiments). Relative values are plotted in the graphs, whereas the actual values are presented in Table 1. Bars: (horizontal) 10 µm; (vertical) 1 min.
Table 1. Kinetic parameters of in vitro–reconstituted MT polymerization dynamics with the addition of five factors.
| Condition | CLASP concentration | n | Growth rate | Shrinkage rate | Catastrophe frequency | Rescue frequency |
|---|---|---|---|---|---|---|
| nM | µm/min | µm/min | ×10−3 s−1 | ×10−3 s−1 | ||
| Tubulin alone | 0 | 3 | 1.6 ± 0.1 (60) | 37.9 ± 8.2 (30) | 0.9 ± 0.2 (31) | 10.5 ± 6.1 (6) |
| Plus EB1/Sentin/Msps/Klp10A/CLASP | 0 | 3 | 5.8 ± 2.2 (88) | 19.0 ± 3.6 (86) | 17.7 ± 2.0 (86) | 5.3 ± 5.8 (7) |
| 15 | 3 | 4.2 ± 1.3 (260) | 9.1 ± 2.5 (219) | 13.9 ± 3.6 (217) | 81.6 ± 11.2 (211) | |
| 45 | 3 | 3.0 ± 1.0 (166) | 5.6 ± 0.9 (88) | 5.7 ± 3.9 (86) | 86.1 ± 23.9 (87) | |
| P (CLASP 45 nM vs. 0 nM) | 0.116 (n = 3) | 0.003 (n = 3) | 0.009 (n = 3) | 0.005 (n = 3) | ||
| 0.002 (exp1) | <10−4 (exp1) | |||||
| <10−4 (exp2) | <10−4 (exp2) | |||||
| <10−4 (exp3) | <10−4 (exp3) |
Values are mean ± SD of three independent experiments (total number of events). See Fig. 2 for a graphic presentation of these results.
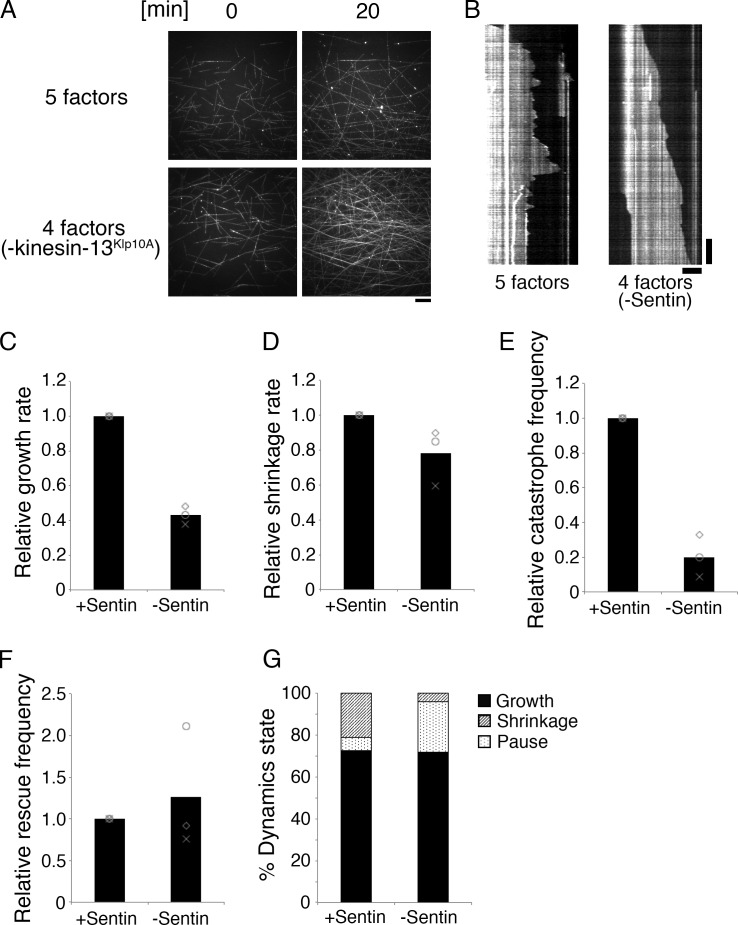
To validate the similarity between in vitro and in vivo dynamics, we omitted kinesin-13Klp10A, Sentin, or CLASPMast/Orbit from the reaction and compared the outcome with in vivo RNAi results. In S2 cells, kinesin-13Klp10A knockdown decreases the catastrophe frequency (Mennella et al., 2005). Sentin knockdown inhibits the dynamic nature of MTs, specifically through increasing the pause duration and decreasing growth and shrinkage rates and catastrophe frequency (Li et al., 2011). CLASPMast/Orbit depletion in S2 cells increased the growth and shrinkage rates but reduced the pause duration (Sousa et al., 2007; Trogden and Rogers, 2015). Interestingly, we generally reproduced these tendencies in our in vitro assays through removal of kinesin-13Klp10A, Sentin, or CLASPMast/Orbit. Without kinesin-13Klp10A, catastrophe was dramatically suppressed and the imaging field was occupied by numerous MTs (Fig. 3 A; only seven catastrophe events were observed during a 413-min growing phase). In the case of CLASPMast/Orbit, all seven parameters were changed in a qualitatively consistent manner (Fig. 2, B–F; “0 nM CLASP” corresponds to CLASP depletion; see also Fig. S3 [A–E], for comparison to published in vivo results). Sentin depletion had a similar impact; changes in six out of seven parameters were consistent in vivo and in vitro (Fig. 3, B–G; Fig. S3, F–J; and Table S1).
Figure 3.
Effect of Sentin or kinesin-13Klp10A removal from the MT polymerization reaction. (A) MTs dramatically increased because of reduced catastrophe frequency in the absence of kinesin-13Klp10A. The other four factors were mixed with 15 µM S2 tubulin for 20 min. Bar, 10 µm. (B–G) MT polymerization dynamics in the presence or absence of Sentin. The other four factors (EB1, XMAP215Msps, kinesin-13Klp10A, and 15 nM CLASPMast/Orbit) were mixed with 15 µM S2 tubulin (three independent experiments). Actual values are presented in Table S1. Bars: (horizontal) 5 µm; (vertical) 2 min.
Lattice-bound CLASPMast/Orbit induces pausing in shrinking MTs
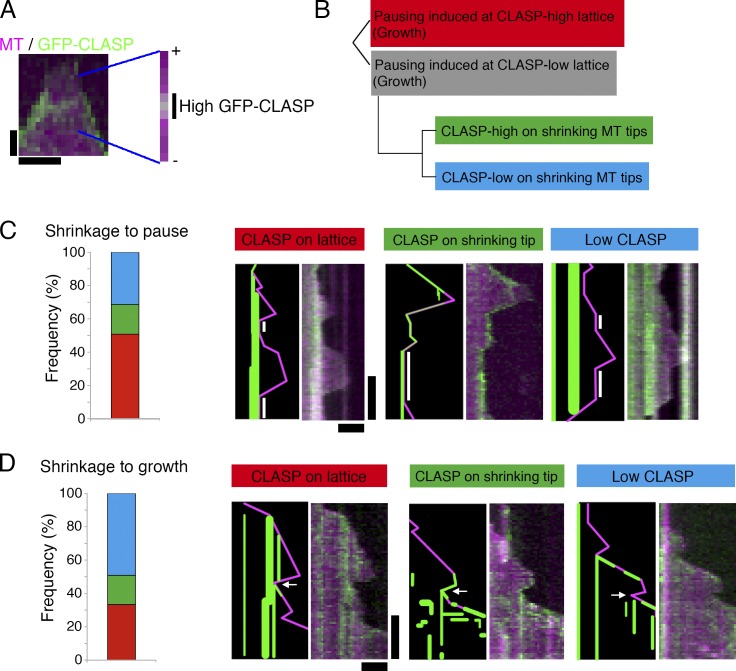
Because pausing was rarely observed in the absence of CLASPMast/Orbit, we investigated whether GFP-CLASPMast/Orbit accumulation is correlated with pause induction. Because CLASPMast/Orbit was often abundantly localized to the growing end of MTs via EB1 binding, we focused our analysis on shrinking MTs. We first surveyed GFP-CLASPMast/Orbit signal distribution along the lattice of shrinking MTs (n = 21) and observed that 24% of the pixels contained a high GFP signal (Fig. 4 A). This result indicated that if pausing is induced at random locations, the event has a 24% chance of occurring at GFP-accumulation sites. However, we identified 51% (n = 51) of shrinkage-to-pause events occurring at GFP-occupied sites (red in Fig. 4, B and C). Furthermore, in 9 out of the 25 events, in which pausing was induced at sites with low GFP-CLASPMast/Orbit abundance, MTs already had strong GFP signals during shrinking (green in Fig. 4, B and C). Thus, GFP-CLASPMast/Orbit accumulation and pause induction were well correlated, albeit not perfectly. To investigate whether CLASPMast/Orbit on the lattice recruits other factors, we mixed MT seeds with TagRFP-CLASPMast/Orbit and GFP-Sentin; Sentin is proposed to act as a bridge between CLASPMast/Orbit and XMAP215Msps (Trogden and Rogers, 2015) but does not localize to the MT lattice on its own (Li et al., 2012). We did not observe lattice localization of the GFP-Sentin signals in the presence of TagRFP-CLASPMast/Orbit, suggesting that prerecruitment of the Sentin–XMAP215Msps complex is not required for CLASPMast/Orbit to induce pausing (Fig. S1 H).
Figure 4.
Pausing was induced at the CLASPMast/Orbit accumulation site with a high probability. (A) GFP-CLASPMast/Orbit distribution along the MT. An MT that just started to shrink is highlighted. Marked are the regions at which GFP-CLASPMast/Orbit was accumulated. Bars: (horizontal) 30 s; (vertical) 1 µm. (B–D) Classification of shrinkage-to-pause (C; n = 51) or shrinkage-to-growth (D; n = 63) events based on GFP-CLASPMast/Orbit accumulation at the turning point. Kymographs and the schematic drawings are displayed. Actual data are derived from the experiments performed in Fig. 2 (shrinkage to growth) or Fig. 5 (shrinkage to pause). Pausing phases are marked with white bars, whereas arrows indicate the rescue points for regrowth. Bars: (horizontal) 2 µm; (vertical) 1 min.
Similar analysis was performed for shrinkage-to-growth events (Fig. 4, B and D). In this case, the correlation was weaker than for the pause but was still higher than expected from random occurrence (33% events occurred at GFP accumulation site [n = 63], which occupied 15% of the pixels [n = 31]). This result indicates that small amounts of CLASPMast/Orbit can also trigger the shrinkage-to-growth rescue, which differs from the results of fission yeast CLASPCls1, wherein enrichment was perfectly correlated with the rescue event (Al-Bassam et al., 2010). We could not rule out the possibility that our microscope was not sensitive enough to detect moderately accumulated GFP-CLASPMast/Orbit. Regardless, the results suggest that fly CLASP is a potent pause/rescue-inducing factor in the presence of four other factors.
Increase in MT dynamicity by Plk1Polo kinase
MT dynamics are subjected to mitotic regulation. During mitosis, the rate of astral MT growth and the frequency of catastrophe events increase, whereas the frequency of rescue events decreases (Belmont et al., 1990; Rusan et al., 2001). It was shown that the addition of active Cdk1 to the interphase frog extracts triggers the interphase-mitosis transition of MT dynamics (Verde et al., 1990). However, the event downstream of Cdk1 remains unclear. We first hypothesized that Cdk1 might directly phosphorylate one or more of the five factors, or tubulin itself, to modulate MT dynamics. Autoradiography experiments indicated that kinesin-13Klp10A and XMAP215Msps are phosphorylated by the recombinant Cdk1–cyclin B complex (Fig. S4 A). Next, we incubated the five proteins and tubulin with Cdk1–cyclin B, followed by a MT polymerization assay. Here, kinetic parameters were not significantly or reproducibly changed in four experiments comparing the effect of Cdk1 (Fig. S4, B–F; and Table S2). These results were not consistent with the idea that Cdk1 phosphorylation directly regulates the activities of these six proteins, although they did not entirely disprove it.
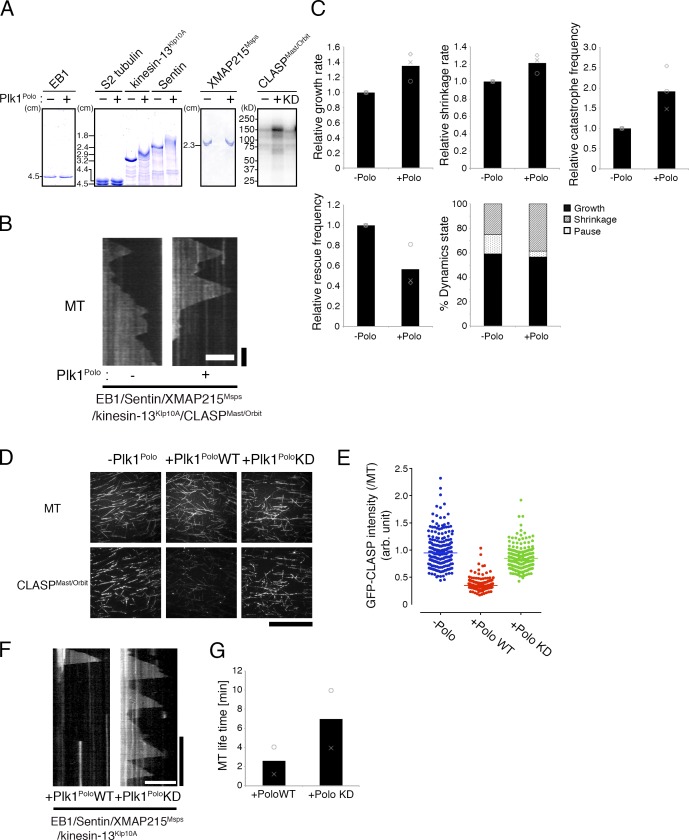
We considered it likely that a factor other than Cdk1 is the direct contributor to the increase in MT dynamicity during mitosis. Polo-like kinase 1 (Plk1) consists of a complex regulatory circuit during mitotic entry (Lindqvist et al., 2009; Zitouni et al., 2014). We therefore next hypothesized that Plk1 may be responsible for the change in dynamics. Interestingly, incubation of five factors and tubulin with recombinant Plk1Polo affected kinetic parameters in a reproducible manner; growth rate and catastrophe increased, whereas rescue and pausing were reduced, making MTs more dynamic (Fig. 5, B and C; and Table S3). Phos-tag and autoradiography analyses showed that kinesin-13Klp10A, Sentin, and CLASPMast/Orbit were phosphorylated by Plk1Polo, whereas no gel shift was observed after Phos-tag analysis for EB1, XMAP215Msps, or S2 tubulin (Fig. 5 A). When GFP-CLASPMast/Orbit was mixed with tubulin, we observed a reduction in MT lattice association in the presence of Plk1Polo (Fig. 5, D and E). This result was consistent with the increase of MT dynamicity conferred by Plk1Polo, as lattice-associated CLASPMast/Orbit frequently induced pausing (Fig. 4 C). This effect was at least in part phosphorylation dependent, because kinase-inactive (kinase-dead [KD]) Plk1Polo, purified in an identical manner to wild type, showed a milder effect (Fig. 5, D and E). Conversely, when active Plk1Polo was mixed with the other four factors and tubulin, we observed rapid depolymerization of MT seeds (Fig. 5, F and G). The seed depolymerization suggested that kinesin-13Klp10A is activated by Plk1Polo. Thus, Plk1Polo-mediated phosphorylation affected the activities of CLASPMast/Orbit and kinesin-13Klp10A. Whether Sentin, which also has catastrophe-inducing activity (Li et al., 2012), is a critical substrate of Plk1Polo remains to be determined.
Figure 5.
Phosphorylation by Plk1Polo elevated MT dynamicity in vitro. (A) Autoradiography (rightmost panel) and Phos-tag gel electrophoresis (others) showed phosphorylation of CLASPMast/Orbit, kinesin-13Klp10A, and Sentin by Plk1Polo. The markers of the Phos-tag gel indicate the distance from the top of the resolving gel. (B and C) Kymographs and kinetic parameters showing that treatment of five MT dynamic-modulating factors with Plk1Polo makes MTs more dynamic (10 µM S2 tubulin; 45 nM CLASPMast/Orbit). In the graph, the mean and relative values of each experiment are plotted (three independent experiments). Actual values are presented in Table S3. Bars: (horizontal) 5 µm; (vertical) 1 min. (D and E) Association of 5 nM GFP-CLASPMast/Orbit with the MT lattice (10 µM S2 tubulin) was drastically reduced by incubation with 100 nM wild-type Plk1Polo (P < 0.0001, Mann–Whitney U test) and more modestly with a kinase-dead (KD) mutant (n = 227 [−Polo], 212 [+PoloWT], and 214 [+PoloKD]). Bar, 50 µm. (F and G) MT depolymerization activity of kinesin-13Klp10A was enhanced by incubation with wild-type Plk1Polo (two independent experiments). Four factors (EB1, XMAP215Msps, Sentin, and 10 nM kinesin-13Klp10A) and 15 µM S2 tubulin were included in this reaction, and seed depolymerization was assessed after generating the kymograph. Note that the lifetime for wild-type Plk1Polo is likely overestimated because some seeds had been already depolymerized when the imaging was started. Bars: (horizontal) 10 µm; (vertical) 5 min.
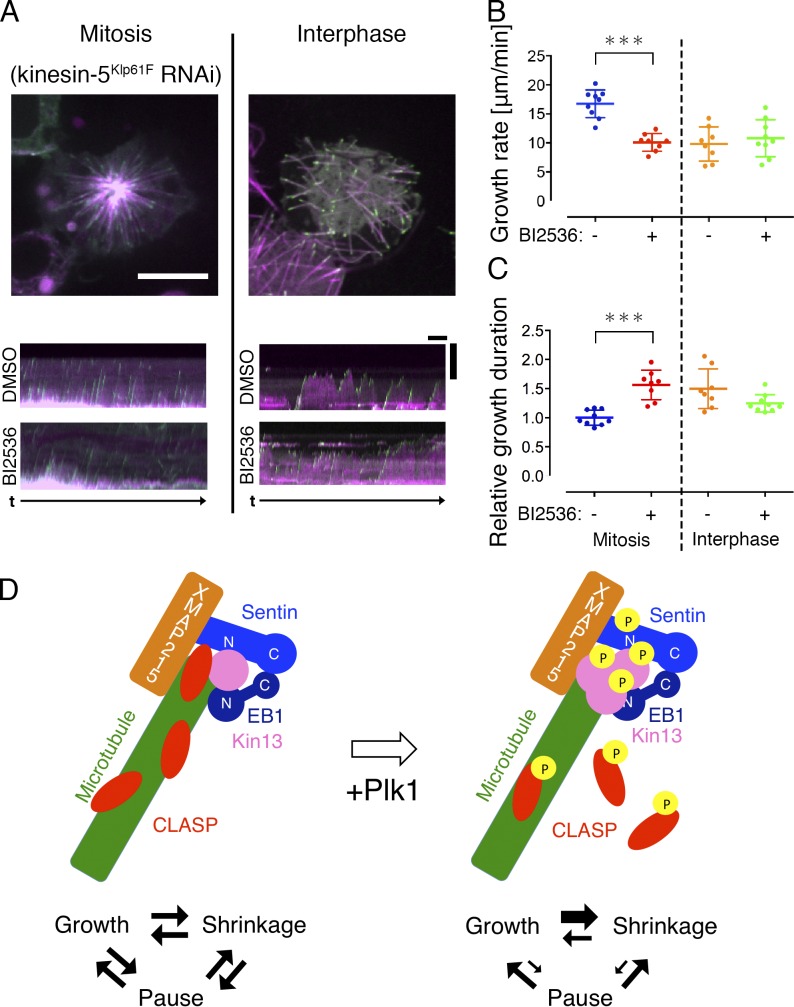
To investigate the involvement of Plk1Polo in MT dynamics in vivo, we measured growth velocity of mitotic astral MTs in S2 cells in the presence or absence of the Plk1Polo inhibitor BI2536. S2 cells have heterogeneity in centrosome numbers, which might alter the parameters associated with MT dynamics of each aster. Therefore, we measured astral MT dynamics in the monopolar spindle that was induced by RNAi knockdown of the kinesin-5 motor Klp61F (Fig. 6 A). Interestingly, mitotic EB1-GFP velocity was decreased by 40% after BI2536 treatment, which matched the velocity in interphase (Fig. 6 B). The decrease was unlikely induced by off-target effect of BI2536 on tubulin, because the MT growth rate during interphase was unaffected by this treatment. We next used the same dataset to compare growth time. Because pausing is a rare event during S2 mitosis (Li et al., 2011), we considered that disappearance of the EB1-GFP signals in the mitotic aster mostly corresponded to catastrophe. Duration of EB1-GFP tracking was 56% longer in the presence of BI2536, and this was similar to interphase values, suggesting that Plk1Polo elevated catastrophe frequency in mitotic cells (Fig. 6 C).
Figure 6.
Plk1Polo inhibition dampens MT dynamicity in vivo. (A–C) Plk1Polo inhibition decreased MT growth rate and catastrophe frequency in mitosis, but not in interphase. Mitotic astral MTs in the monopolar spindle induced by Klp61F (kinesin-5) RNAi were analyzed (100–200 comets per cell, and eight or nine cells in each condition). Each dot in the graph represents the mean value obtained from a cell. Asterisks indicate statistically significant differences (***, P < 0.0001; Turkey’s multiple comparisons test; the normality of data distribution was checked by D’Agostino–Pearson omnibus normality test and Shapiro–Wilk normality test). Error bars indicate SDs. Bar for cell images, 10 µm; horizontal bar for kymographs, 1 min; vertical bar for kymographs, 5 µm. (D) Reconstitution of regulated microtubule polymerization dynamics was achieved in this study. Three-phase polymerization cycles were observed when five factors were mixed with tubulin. Addition of the active Plk1 kinase increased MT dynamicity, wherein pausing became rare.
Discussion
This study aimed to reproduce cellular MT polymerization dynamics in vitro more faithfully than previous studies. Toward this goal, we expanded the materials for reconstitution to seven purified proteins (including tubulin and a kinase). To the best of our knowledge, this presents the most complex reconstitution of MT polymerization dynamics. Our in vitro experiments, together with previous loss-of-function analyses in vivo, suggest that five proteins, XMAP215Msps, EB1, Sentin, kinesin-13Klp10A, and CLASPMast/Orbit, constitute the core machinery that regulates MT dynamics in the S2 cytoplasm (Fig. 6 D, left). In the presence of these five factors, typical MT behavior was reproduced, in which growth, shrinkage, and pausing phases repeat in a stochastic order. The change in dynamics in the absence of CLASPMast/Orbit, Sentin, or kinesin-13Klp10A supports this model. Omission of each factor in vitro resulted in MT dynamics that deviated from that of untreated S2 cytoplasm; dynamics became more similar to that observed upon RNAi depletion of corresponding factors. Moreover, phosphorylation by the mitotic kinase Plk1Polo resulted in more dynamic MTs, reminiscent of what occurs during interphase-to-mitosis transitions (Fig. 6 D, right).
CLASP induces pausing
A previous study using purified yeast CLASPCls1 identified rescue (shrinkage-to-growth) and catastrophe-inhibiting activities, but not pausing or growth-inhibiting activity (Al-Bassam et al., 2010). An intriguing observation in the current study was the emergence of pause when CLASPMast/Orbit was added with the four other factors. This illustrates the value of multifactor reconstitution and suggests that an equilibrium of CLASPMast/Orbit with MT polymerase and depolymerase produces such a state. It is possible that such activity was conferred by physical interaction with other factors; for instance, it was recently shown that XMAP215Msps interacts with CLASPMast/Orbit via Sentin in S2 cells (Trogden and Rogers, 2015). In S2 cells, RNAi depletion of EB1, Sentin, or XMAP215Msps increases the pause duration in interphase, and this phenotype could not be readily explained (Rogers et al., 2002; Brittle and Ohkura, 2005; Li et al., 2011). On the basis of current in vitro results, this phenotype may be interpreted that when EB1, Sentin, or XMAP215 is depleted, CLASPMast/Orbit dominates at the plus end and induces pausing together with the other three factors.
Plk1 elevates MT dynamicity
Another finding in this study was the increase in MT dynamicity by Plk1Polo kinase. The involvement of Plk1Polo is not inconsistent with the classical analysis in which Cdk1 triggers elevation of MT dynamicity in frog egg extracts (Verde et al., 1990). It has been shown that Cdk1 and Plk1 comprise a complex signaling network, and the addition of Cdk1 to egg extracts may well activate the Plk1 kinase. Consistent with the in vitro data, we observed reversion of MT dynamics, from mitotic to interphase mode, by treating prometaphase cells with a Plk1Polo inhibitor. An in vitro kinase assay showed that Sentin, kinesin-13Klp10A, and CLASPMast/Orbit are the substrates of Plk1Polo. The appearance of multiple bands in the electrophoresis gel indicates that multiple residues are potential Plk1Polo phosphorylation sites. Determining the substrate residues that are phosphorylated to influence MT dynamics in vivo is an important future research topic. However, our results do not exclude the possibility that Plk1Polo phosphorylation of other proteins also contributes to changes in MT dynamics in cells.
Comparison between in vitro and in vivo data
Although we believe we have achieved hitherto the most in vivo–relevant reconstitution, we admit that the study is still not a precise reproduction of S2 MT dynamics. First, other factors may also contribute to MT dynamics in cells. S2 cells express additional factors that can modulate MT dynamicity in vitro, such as D-TPX2 and two other kinesin-13 family members (Klp59C and Klp59D); however, RNAi depletion of these factors results in a phenotype that is not as severe as that observed after ablation of the five factors used in this study (Goshima and Vale, 2003; Mennella et al., 2005; Goshima, 2011). Second, our data on MT dynamics are basically limited to free MTs in the cytoplasm. Other types of MTs, such as cortical and peripheral MTs, are often bound to other protein or membranous structures and show different dynamic kinetics (Trogden and Rogers, 2015), and additional factors are likely necessary for reconstitution. For instance, human CLASP depletion decreased catastrophe as well as rescue at the cell cortex, which is not entirely consistent with the consequence we observed here in vitro (Mimori-Kiyosue et al., 2005); GSK3 kinase is known to phosphorylate CLASP and thereby regulate peripheral MT dynamics (Kumar et al., 2009; Trogden and Rogers, 2015). Kinetochore MTs in metaphase are also bound to many other proteins. However, a key feature of kinetochore MT dynamics might have been partially reproduced with one of our assay conditions. In cells, CLASPMast/Orbit is most prevalent at the outer kinetochore region during metaphase and is required for kinetochore MT polymerization (Maiato et al., 2005). The slow but persistent growth of kinetochore MTs has resemblance to that observed when a high concentration of CLASPMast/Orbit was mixed with four other factors; MTs grew slower and rarely shrank even in the presence of catastrophe-promoting factors (Fig. 2; 45 nM CLASPMast/Orbit). Thus, although multiple other MT-regulating proteins are present at the outer kinetochore, it is tempting to speculate that accumulation of CLASPMast/Orbit is a key event that distinguishes kinetochore MTs from highly dynamic astral MTs.
Finally, another difference between our system and the in vivo situation involves the environment and protein concentrations. We used a standard nonviscous buffer in our assay, which would be different from the cytoplasm that is highly viscous, containing many other molecules. Furthermore, our rough estimate indicated that cellular concentrations of CLASPMast/Orbit and kinesin-13Klp10A were 45- to 80-fold higher than what we used in vitro, a level currently unachievable by our purification method (Fig. S5). One possible explanation for this discrepancy is that cellular proteins are regulated spatially or posttranslationally, and the number of actual proteins that can target MTs is small. Alternatively, the antagonistic activity of CLASPMast/Orbit and kinesin-13Klp10A on catastrophe suggests that the molecular ratio of the two proteins rather than the absolute abundance of each protein might be critical. In support of this idea, the ratio of CLASPMast/Orbit and kinesin-13Klp10A was comparable between in vitro (1:6.7) and in vivo (1:3.7) conditions. Regardless, our reconstitution admittedly is qualitative, and quantitative comparison with in vivo kinetic parameters remains difficult. Converting the kinetic parameters and protein concentrations to in vivo values remains a key challenge in the reconstitution of MT dynamics and perhaps also for other complex cellular events (Shintomi et al., 2015).
Toward reconstitution of mammalian MT dynamics
The data obtained in this study might be largely applicable to mammalian cells. EB1, XMAP215Msps, kinesin-13Klp10A, CLASPMast/Orbit, and Plk1Polo have clear orthologues in mammals. Although not conserved in amino acid sequences, Sentin is likely a functional homologue of mammalian SLAIN2, because both proteins bind to EB1 and XMAP215Msps, bridging these two molecules for synergistic MT growth acceleration (Li et al., 2011; van der Vaart et al., 2011). Furthermore, Plk1-dependent activation of kinesin-13 has been demonstrated in vitro (Jang et al., 2009; Zhang et al., 2011). Because multiple paralogous/homologous genes have been identified for CLASP (1 and 2), EB (1, 2, 3), SLAIN (1 and 2), and kinesin-13 (Kif2a, Kif2b and MCAK), mammalian cells might have further layers of complexity compared with Drosophila. In fact, it was recently shown that human CLASP1 and an isoform of CLASP2 have common and also distinct activities (Yu et al., 2016). Regardless, a reasonable first attempt toward reconstitution of mammalian MT dynamics would be to purify and mix homologues of the five proteins identified in this study.
Materials and methods
Cloning
Primers to amplify full-length CLASPMast/Orbit were 5′-CACCATGGCCTATCGGAAGCCCAGCGACCTGG-3′ and 5′-TGACGACGATGCCGCGGAGGAGTTCTTGG-3′. The amplified DNAs were cloned into pENTR-D/TOPO, digested with NotI and AscI, blunted, and cloned into the pFastBac-His-mGFP vector (StuI digested). His-CLASPMast/Orbit was constructed by Gateway cloning into the pDEST10 vector. Kinesin-13Klp10A gene was amplified with the primers 5′-CACCATGGACATGATTACGGTGGGGCA-3′ and 5′-CTAACGCTTGCCATTCGGCGAA-3′. The amplified DNAs were cloned into pENTR-D/TOPO, followed by Gateway cloning into pDEST17. Plk1Polo was amplified with 5′-TTGCGGCCGCCACCATGGCCGCGAAGCCCGAGG-3′ and 5′-TTGGCGCGCCTTATGTGAACATCTTCTCCAGC-3′, and ligated into pENTR-D/TOPO after NotI–AscI digestion. The gene was then cloned into pDEST10 by the Gateway reaction.
Protein purification
S2 tubulin was purified by using a previously described method (Widlund et al., 2012). GST-tagged TOG1 domain (Saccharomyces cerevisiae Stu2, 1–306 aa) expression was induced in E. coli BL21 in 2 liters of Terrific Broth, 0.2 mM IPTG for 18 h at 25°C (TOG1-encoding plasmid was a gift from P. Widlund, Max Planck Institute, Dresden, Germany). Harvested cells were lysed with 120 ml 2×PBS (5.4 mM KCl, 3 mM KH2PO4, 16.2 mM Na2HPO4, and 274 mM NaCl) supplemented with 500 U benzonase (EMD Millipore) and protease inhibitors (0.5 mM PMSF and peptide cocktails [1 µg/ml leupeptin, pepstatin, chymostatin, and aprotinin]) by the Advanced Digital Sonifier D450 (Branson). The lysate was incubated with 5 ml GSTrap (GE Healthcare) for 1 h at 4°C. After washing once, GST-TOG1 were eluted with 20 mM reduced glutathione in 2×PBS, pH 8.0, followed by dialysis using PD-10 desalting column (GE Healthcare) and 3.5 ml coupling buffer (100 mM NaHCO3 with 100 mM NaCl at pH 8.2). After dialysis, 80 mM MgCl2 was added. For conjugation of GST-TOG1, 3.5 ml GST-TOG1 protein solution was applied to 1 ml HiTrap NHS-activated HP columns (GE Healthcare; preactivated by 1 mM HCl) and recirculated at 1.5 ml/min in the column for 20 min. The remaining reactive groups were blocked by a 30-min incubation with 0.5 M ethanolamine and 0.5 M NaCl, pH 8.3. The column was then washed with 1×PBS containing 50% glycerol and was then stored at −20°C. 700 ml S2 cell culture was lysed with 7 ml MRB80 buffer (80 mM K-Pipes, pH 6.8, 4 mM MgCl2, and 1 mM EGTA) supplemented with 125 U benzonase (EMD Millipore) by a Dounce tissue grinder. The lysate was clarified by 9,400 g and 135,000 g centrifugation and applied to a 1-ml TOG1-conjugated column. Tubulin was eluted with the elution buffer (0.5 M (NH4)2SO4 in MRB80, 10 µM GTP), followed by desalting with the PD-10 desalting column (GE Healthcare) and MRB80. The protein solution was concentrated to >40 µM by Amicon Ultra-4 MWCO 30K. The concentrated solution was flash frozen with liquid nitrogen and stored at −80°C. EB1 was purified using E. coli following a protocol described previously (Li et al., 2011, 2012). GST-EB1 expression was induced in BL21-AI with 0.2% arabinose for 16 h at 25°C. Harvested cells were lysed using the Advanced Digital Sonifier D450 (Branson) and the lysis buffer (25 mM Tris-HCl, 1 mM EGTA, 150 mM NaCl, and 1 mM DTT) supplemented with protease inhibitors (0.5 mM PMSF and peptide cocktails). After incubation with glutathione Sepharose beads for 1 h at 4°C, GST was cleaved using HRV 3C protease (Thermo Fisher Scientific), followed by gel filtration chromatography (MRB80, 100 mM KCl, and 1 mM DTT). His-kinesin-13Klp10A was purified using the standard Ni-NTA purification with the buffer identical to that of EB1, except that 1 mM ATP was supplied. His-Sentin, His-GFP-Sentin, and XMAP215Msps-HAHis were purified with insect Sf21 cells essentially as described previously (Li et al., 2012). 1.5 × 106 uninfected Sf21 cells ml–1 in an Erlenmeyer flask were treated with baculovirus and incubated with shaking for 3 d (120 rpm at 27°C). The cells were lysed using 5 ml (per 50-ml cell culture) lysis buffer (50 mM K-Hepes, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 30 mM imidazole, and 1% Triton X-100, pH 7.6, with protease inhibitors). After incubation with nickel-coated beads (0.5-ml bed volume) for 1 h, the proteins were eluted using 250 µl elution buffer (lysis buffer supplemented with 200 mM imidazole without 1% Triton X-100). Elution was repeated five to seven times. Buffer was exchanged to 1× MRB80 containing 100 mM KCl and 1 mM DTT using the desalting column PD MiniTrap G-25 (GE Healthcare). His-GFP-CLASPMast/Orbit (wild type and TOG mutant [W334E, K421A, W852E, K940A, which reside in the TOG2 and TOG3 domains]), His-CLASPMast/Orbit, TagRFP-CLASPMast/Orbit, and His-Plk1Polo (wild type and K54A KD mutant) were purified in a manner identical to that used for Sentin, except that the lysis buffer contained 100 mM KCl and 0.1–0.2% Tween-20 (CLASP), or cells were treated with 100 nM okadaic acid 1 h before collection (Plk1Polo). Purified S2 tubulin, EB1, kinesin-13Klp10A, and Plk1Polo were flash frozen (with 20% glycerol for EB1 and 20% sucrose for kinesin-13Klp10A and Plk1Polo), whereas other proteins were kept on ice and used within 48 h. Recombinant Cdk1–cyclin B complex was purchased from New England Biolabs, Inc. Gel filtration chromatography of GFP-CLASPMast/Orbit was performed with a Superdex 200 increase 10/300 GL column equilibrated with a gel filtration buffer (80 mM K-Pipes, pH 6.8, 100 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 1 mM DTT).
In vitro MT polymerization assay
The in vitro MT polymerization assay was performed basically following the method described in Li et al. (2012), which referred to Bieling et al. (2010) and Gell et al. (2010). Flow chambers (22 mm [width] × 1 mm [height] × 0.15 mm [depth]) were assembled between a coverslip and a precleaned micro slide glass with double-sided tape. Glass was soaked for 3 d in water supplied with the detergent SCAT 20-X, rinsed with fresh water, fired with methanol, and then silanized. The silanized coverslip was coated with antibiotin (1–5% in 1× MRB80; Invitrogen), and the nonspecific surface was blocked with Pluronic F127 (1% in 1× MRB80; Invitrogen). Biotinylated MT seeds (50–100 µM tubulin mix containing 10% biotinylated pig tubulin and 10% Alexa Fluor 568–labeled pig tubulin with 1 mM GMPCPP) were specifically attached to the functionalized surface by biotinylated tubulin–antibiotin links. After the chamber was washed with 1× MRB80, MT growth was initiated by flowing tubulin (containing 3.3% Alexa Fluor 568–labeled pig tubulin), EB1 (400 nM), Sentin (200 nM), GFP-CLASPMast/Orbit (0–45 nM), kinesin-13Klp10A (100 nM, with 1 mM ATP), and/or XMAP215Msps-HA (100 nM) into the assay buffer (1× MRB80, 75 mM KCl, 1 mM GTP, 0.5 mg/ml k-casein, and 0.1% methylcellulose) and an oxygen scavenger system (50 mM glucose, 400 µg/ml glucose-oxidase, 200 µg/ml catalase, and 4 mM DTT). The samples were sealed with candle wax. We used fixed concentrations of EB1, Sentin, and XMAP215Msps based on our previous study, where the effect of these three factors combined has been demonstrated at these concentrations (Li et al., 2012). Kinesin-13Klp10A was used at 100 nM, because increasing this protein further depolymerized virtually all MT seeds in the imaging field. To match the estimated cellular tubulin concentration (Fig. S5), we used 15 µM S2 tubulin for our main reconstitution experiments involving five or four nontubulin proteins (Figs. 2 and 3). However, in some experiments, we used 10 µM S2 tubulin to save the protein whose yield was low compared with yields of pig brain–derived tubulin (Fig. 1 B and Fig. 5, B and D). When the samples were treated with Cdk1–cyclin B or Plk1Polo kinase, 5 or 100 nM recombinant kinase (with 2 mM ATP), respectively, was added to the reaction. During the experiments, the samples were kept at 27°C; images were collected every 3 s for 20 min via total internal reflection fluorescence microscopy with a Ti system (Nikon), EMCCD camera (Evolve; Roper), 100× (1.49 NA) lens, and a 488-nm/561-nm excitation laser. The microscopes were controlled by Micromanager.
Data analysis
All parameters of MT plus end dynamics in vitro, except those related to pause, were determined in accordance with a method described previously (Li et al., 2012). In brief, kymographs were generated for every MT seed identified in the image field, and ∼10 MTs, with a traceable plus end, were randomly selected. The starting and end points of growth and shrinkage were identified in the kymograph. The duration of and change in MT length for each phase were measured, and the rate was determined by calculating the mean velocity between the start and end points of each phase. Catastrophe frequency was determined by dividing the number of shrinkage events by the sum of growth and pause time. The transition from shrinkage to pause or growth was considered a rescue event, and the rescue frequency (for shrinkage time) was calculated. When MTs did not grow or shrink more than two pixels (∼0.35 µm) for five or more frames (15 s), this period was defined as pause; this definition was comparable to that used in the in vivo pause measurement of our previous study (Li et al., 2011). In the analysis presented in Figs. 1 and S1, we did not count the pause phase, because it was impossible to identify the starting point of the pause phase of these slowly and persistently growing MTs; for them, we obtained the growth rate throughout. Catastrophe frequency in the absence of kinesin-13Klp10A was measured by directly tracing the MT plus ends, because MTs were overly crowded and kymographs could not be generated.
Data presentation
We followed Li et al. (2012) for data presentation. As was described previously (Li et al., 2012), some degree of variation in MT behavior was observed on different experiment days, possibly because of differences in protein batches. To adjust for this variation, we have provided most of the graphs for the values for each condition relative to the control data acquired on the same day. In contrast, the mean values of three experiments are provided in the tables. For example, when we obtained growth rates of a1, a2, and a3 (micrometers per minute) in three independent experiments using tubulin alone, as well as b1, b2, and b3 (micrometers per minute) after CLASPMast/Orbit addition, we present the relative values as b1/a1, b2/a2, and b3/a3 in gray and (b1/a1 + b2/a2 + b3/a3)/3 in black in the figures. In the tables, however, we provide the mean rates of (a1 + a2 + a3)/3 and (b1 + b2 + b3)/3 separately with SDs. For all parameters, the p-values based on an unpaired two-sided Student’s t test analysis of three or four experiments were obtained and are presented in Tables 1, S1, S2, and S3. However, the statistical significance of these differences as well as normality of the data distribution could not be properly discussed because sample sizes were too small; nevertheless, the values served as a good indicator of the degree of change. For growth and shrinkage rates where sufficiently large numbers of MTs were analyzed, we additionally provided p-values for each experiment based on a Mann-Whitney U test (therefore, three or four p-values are displayed in each column).
Cell biology
S2 cells that express EB1-GFP and mCherry-tubulin were cultured and RNAi was performed as previously described (Goshima et al., 2007; Bettencourt-Dias and Goshima, 2009). Schneider’s medium (Gibco) supplemented with 10% serum and antibiotics was used for cell culture. For imaging, cells were plated onto a 96-well, glass-bottom plate that was precoated with 5 µl of 0.5 mg/ml concanavalin A solution (1 h at 24°C). For Klp61F RNAi, cells were treated with double-stranded RNA (dsRNA) targeting the 5′ UTR region for 72 h and then plated. The primer sequences for dsRNA were 5′-TATTTGCGCATTATTTTAAAATTG-3′ and 5′-CATATTGATCAATTGAAAC-3′ (Goshima and Vale, 2005), to which the T7 promoter sequences (5′-TAATACGACTCACTATAGGG-3′) were added. For in vitro transcription, the PCR product was mixed with T7 RNA polymerase and 7.5 mM ribonucleotide triphosphates and incubated at 37°C overnight (∼16 h). To complete annealing of dsRNA, the reaction mixture was incubated at 95°C for 5 min using a heat block and then cooled down gradually by leaving the block at room temperature. S2 cells in a 24-well plastic culture plate were treated with 5 µg dsRNA in serum-free medium for 50 min, followed by serum addition. Live imaging of EB1-GFP was performed using spinning-disc confocal microscopy with a 100× 1.40 NA or 100× 1.45 NA objective lens (23–25°C). A CSU-X1 confocal unit (Yokogawa Electric Corporation) attached to a Nikon TE or Ti inverted microscope and EMCCD camera ImagEM (Hamamatsu Photonics) were used for image acquisition. The microscopes and attached devices were controlled using Micromanager. To eliminate actin cytoskeleton that might have physically prevented MT growth (Li et al., 2011), an actin inhibitor (2.5 µM latrunculin A) was added to cells before imaging. In the Plk1Polo inhibition experiment, cells were treated with control DMSO or 1 µM BI 2536, which was shown to be effective in S2 cells (Januschke et al., 2013; Kachaner et al., 2014), and images were acquired at 3-s intervals.
Phos-tag SDS-PAGE, autoradiography, and immunoblotting
Phosphorylation of purified proteins by Plk1Polo, except CLASPMast/Orbit, was evaluated by Phos-tag SDS-PAGE (Kinoshita et al., 2006). 1 mM ATP was added to the reaction. EB1 was run on an 8% acrylamide gel, whereas a 5% gel was used for tubulin, Sentin, and kinesin-13Klp10A. The gel for XMAP215Msps was a mixture of 3% acrylamide and 0.5% agarose, wherein agarose assisted in gel polymerization. Acrylamide conjugated with the Phos-tag molecule at 20 µM. For unknown reasons, the GFP-CLASPMast/Orbit band was not clearly detectable on any type of gel. CLASPMast/Orbit phosphorylation was therefore monitored by autoradiography, where 113 nM Plk1Polo (wild type or KD) was mixed with 1 µM GFP-CLASPMast/Orbit in MRB80/DTT with 100 mM KCl, 0.1 mM ATP (containing 110 nM ATP-P32), and 80 mM β-glycerophosphate for 30 min at 24–25°C, followed by SDS-PAGE and Coomassie staining. Autoradiography was used to test for phosphorylation by Cdk1, where 27 nM recombinant Cdk1–cyclin B was mixed with 2.1 µM EB1, 1.6 µM tubulin, 1.7 µM Sentin, 1.8 µM kinesin-13Klp10A, 1.2 µM CLASPMast/Orbit, or 2.2 µM XMAP215Msps. Immunoblotting of the S2 extract was performed with the following antibodies: anti–α-tubulin (DM1A, 1:1,000, mouse; Sigma-Aldrich), anti-CLASPMast/Orbit (1:300, rabbit; gift from H. Maiato, University of Porto, Porto, Portugal), anti-EB1 (1:1,000, rabbit; gift from S. Rogers, University of North Carolina, Chapel Hill, NC), anti-Klp10A (1:300, rabbit; gift from D. Sharp, Albert Einstein College of Medicine, Bronx, NY), anti-Sentin (1:1,000, rabbit; Li et al., 2011), and anti-XMAP215Msps (1:500, rabbit; gift from H. Ohkura, University of Edinburgh, Edinburgh, Scotland, UK).
Online supplemental material
Fig. S1 shows recombinant proteins used in this study, kinesin-13Klp10A activity and a comparison between S2-derived tubulin and pig brain tubulin. Fig. S2 shows comparison of in vitro and in vivo parameters associated with MT dynamics. Fig. S3 compares the in vitro and in vivo effects of CLASPMast/Orbit and Sentin. Fig. S4 displays the effect of Cdk1–cyclin B in MT dynamics in vitro. In Fig. S5, cellular protein concentration of five factors and tubulin were estimated. Kinetic parameters of in vitro reconstituted MT polymerization dynamics are presented in Tables S1, S2, and S3.
Supplementary Material
Acknowledgments
We wish to thank Tomomi Tani (Marine Biological Laboratory, Woods Hole, MA), Wenjing Li (Tsinghua University, China), Momoko Nishina, Rie Inaba, Tomoya Edzuka, and Tomoko Nishiyama (Nagoya University, Japan) for technical assistance and Wenjing Li and Tomomi Kiyomitsu (Nagoya University) for valuable comments regarding the manuscript.
This work was supported by the Grants-in-Aid for Scientific Research (Japan Society for the Promotion of Science KAKENHI; 15H01317 and 15KT0077). T. Moriwaki is a recipient of a Japan Society for the Promotion of Science predoctoral fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- dsRNA
- double-stranded RNA
- KD
- kinase dead
- MT
- microtubule
References
- Akhmanova A., and Steinmetz M.O.. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., and Chang F.. 2011. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21:604–614. 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., and Chang F.. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 19:245–258. 10.1016/j.devcel.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Hyman A.A., Sawin K.E., and Mitchison T.J.. 1990. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 62:579–589. 10.1016/0092-8674(90)90022-7 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., and Goshima G.. 2009. RNAi in Drosophila S2 cells as a tool for studying cell cycle progression. Methods Mol. Biol. 545:39–62. 10.1007/978-1-60327-993-2_3 [DOI] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., and Surrey T.. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., Hentrich C., Piehler J., and Surrey T.. 2010. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 95:555–580. 10.1016/S0091-679X(10)95028-0 [DOI] [PubMed] [Google Scholar]
- Brittle A.L., and Ohkura H.. 2005. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 24:1387–1396. 10.1038/sj.emboj.7600629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E.C., Campbell C.S., Weibel D.B., and Mullins R.D.. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 315:1270–1274. 10.1126/science.1138527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C., Bormuth V., Brouhard G.J., Cohen D.N., Diez S., Friel C.T., Helenius J., Nitzsche B., Petzold H., Ribbe J., et al. . 2010. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 95:221–245. 10.1016/S0091-679X(10)95013-9 [DOI] [PubMed] [Google Scholar]
- Goshima G. 2011. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS One. 6:e28120 10.1371/journal.pone.0028120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Scholey J.M.. 2010. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26:21–57. 10.1146/annurev-cellbio-100109-104006 [DOI] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. 10.1091/mbc.E05-02-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., and Vale R.D.. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. . 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Horio T., and Hotani H.. 1986. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 321:605–607. 10.1038/321605a0 [DOI] [PubMed] [Google Scholar]
- Howard J., and Hyman A.A.. 2007. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19:31–35. 10.1016/j.ceb.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Hu Z., Liang Y., Meng D., Wang L., and Pan J.. 2015. Microtubule-depolymerizing kinesins in the regulation of assembly, disassembly, and length of cilia and flagella. Int. Rev. Cell Mol. Biol. 317:241–265. 10.1016/bs.ircmb.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Hughes J.R., Meireles A.M., Fisher K.H., Garcia A., Antrobus P.R., Wainman A., Zitzmann N., Deane C., Ohkura H., and Wakefield J.G.. 2008. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 6:e98 10.1371/journal.pbio.0060098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.Y., Coppinger J.A., Seki A., Yates J.R. III, and Fang G.. 2009. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 122:1334–1341. 10.1242/jcs.044321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., and Gonzalez C.. 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Kachaner D., Pinson X., El Kadhi K.B., Normandin K., Talje L., Lavoie H., Lépine G., Carréno S., Kwok B.H., Hickson G.R., and Archambault V.. 2014. Interdomain allosteric regulation of Polo kinase by Aurora B and Map205 is required for cytokinesis. J. Cell Biol. 207:201–211. 10.1083/jcb.201408081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., and Koike T.. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 5:749–757. 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Arnal I., Desai A., Drechsel D.N., and Hyman A.A.. 2001. Reconstitution of physiological microtubule dynamics using purified components. Science. 294:1340–1343. 10.1126/science.1064629 [DOI] [PubMed] [Google Scholar]
- Kumar P., Lyle K.S., Gierke S., Matov A., Danuser G., and Wittmann T.. 2009. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell Biol. 184:895–908. 10.1083/jcb.200901042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leano J.B., Rogers S.L., and Slep K.C.. 2013. A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure. 21:939–950. 10.1016/j.str.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Miki T., Watanabe T., Kakeno M., Sugiyama I., Kaibuchi K., and Goshima G.. 2011. EB1 promotes microtubule dynamics by recruiting Sentin in Drosophila cells. J. Cell Biol. 193:973–983. 10.1083/jcb.201101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., and Goshima G.. 2012. Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 199:849–862. 10.1083/jcb.201206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., Rodríguez-Bravo V., and Medema R.H.. 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185:193–202. 10.1083/jcb.200812045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa R., Pantaloni D., and Carlier M.F.. 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 401:613–616. 10.1038/44183 [DOI] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., and Rieder C.L.. 2005. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47. 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Rogers G.C., Rogers S.L., Buster D.W., Vale R.D., and Sharp D.J.. 2005. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7:235–245. 10.1038/ncb1222 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., and Akhmanova A.. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153. 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S., Stuurman N., Afonso O., Hornsveld M., Aguiar P., Goshima G., Vale R.D., and Maiato H.. 2013. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc. Natl. Acad. Sci. USA. 110:19808–19813. 10.1073/pnas.1320013110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., and Sharp D.J.. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., and Vale R.D.. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884. 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N.M., Fagerstrom C.J., Yvon A.M., and Wadsworth P.. 2001. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell. 12:971–980. 10.1091/mbc.12.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden E., and Wadsworth P.. 1993. Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J. Cell Biol. 120:935–945. 10.1083/jcb.120.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Takahashi T.S., and Hirano T.. 2015. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol. 17:1014–1023. 10.1038/ncb3187 [DOI] [PubMed] [Google Scholar]
- Sousa A., Reis R., Sampaio P., and Sunkel C.E.. 2007. The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil. Cytoskeleton. 64:605–620. 10.1002/cm.20208 [DOI] [PubMed] [Google Scholar]
- Trogden K.P., and Rogers S.L.. 2015. TOG proteins are spatially regulated by Rac-GSK3beta to control interphase microtubule dynamics. PLoS One. 10:e0138966 10.1371/journal.pone.0138966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S.M., Bjelic S., Demmers J., Vorobjev I., Hoogenraad C.C., Steinmetz M.O., and Akhmanova A.. 2011. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193:1083–1099. 10.1083/jcb.201012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Labbé J.C., Dorée M., and Karsenti E.. 1990. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 343:233–238. 10.1038/343233a0 [DOI] [PubMed] [Google Scholar]
- Walker R.A., O’Brien E.T., Pryer N.K., Soboeiro M.F., Voter W.A., Erickson H.P., and Salmon E.D.. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. 10.1083/jcb.107.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P.O., Podolski M., Reber S., Alper J., Storch M., Hyman A.A., Howard J., and Drechsel D.N.. 2012. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell. 23:4393–4401. 10.1091/mbc.E12-06-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles J.T., Deegan T.D., Janska A., Early A., and Diffley J.F.. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 519:431–435. 10.1038/nature14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Signorile L., Basu S., Ottema S., Lebbink J.H., Leslie K., Smal I., Dekkers D., Demmers J., and Galjart N.. 2016. Isolation of functional tubulin dimers and of tubulin-associated proteins from mammalian cells. Curr. Biol. 26:1728–1736. 10.1016/j.cub.2016.04.069 [DOI] [PubMed] [Google Scholar]
- Zanic M., Widlund P.O., Hyman A.A., and Howard J.. 2013. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15:688–693. 10.1038/ncb2744 [DOI] [PubMed] [Google Scholar]
- Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G., et al. . 2011. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286:3033–3046. 10.1074/jbc.M110.165340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S., Nabais C., Jana S.C., Guerrero A., and Bettencourt-Dias M.. 2014. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15:433–452. 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.