Ulmschneider et al. demonstrate that intracellular pH increases during differentiation of Drosophila ovarian epithelial stem cells and mouse embryonic stem cells, that blocking this increase impairs differentiation, and that intracellular pH may regulate the strength of Hedgehog signaling in epithelial stem cells.
Abstract
Despite extensive knowledge about the transcriptional regulation of stem cell differentiation, less is known about the role of dynamic cytosolic cues. We report that an increase in intracellular pH (pHi) is necessary for the efficient differentiation of Drosophila adult follicle stem cells (FSCs) and mouse embryonic stem cells (mESCs). We show that pHi increases with differentiation from FSCs to prefollicle cells (pFCs) and follicle cells. Loss of the Drosophila Na+–H+ exchanger DNhe2 lowers pHi in differentiating cells, impairs pFC differentiation, disrupts germarium morphology, and decreases fecundity. In contrast, increasing pHi promotes excess pFC cell differentiation toward a polar/stalk cell fate through suppressing Hedgehog pathway activity. Increased pHi also occurs with mESC differentiation and, when prevented, attenuates spontaneous differentiation of naive cells, as determined by expression of microRNA clusters and stage-specific markers. Our findings reveal a previously unrecognized role of pHi dynamics for the differentiation of two distinct types of stem cell lineages, which opens new directions for understanding conserved regulatory mechanisms.
Introduction
Cellular differentiation is a central feature of metazoan biology, driving tissue development, homeostasis, and repair. This process is often studied in the context of adult and embryonic stem cell (ESC) biology, where individual steps in the transition from a multipotent progenitor to a differentiated cell type can be carefully monitored. In both cell types, multiple regulatory mechanisms operate in concert to ensure that each step of differentiation occurs in a robust and precise manner. The role of developmental cues, transcription factors, and chromatin state in cellular differentiation has been the focus of intense investigation, but we know much less about the contributions of dynamic cytosolic signals.
In this study, we investigated how changes in intracellular pH (pHi) promote differentiation in the follicle stem cells (FSCs) of the Drosophila melanogaster ovary and mouse ESCs (mESCs). pHi dynamics are known to act as a cytosolic signal that contributes to the regulation of multiple cell processes, including cell cycle progression (Putney and Barber, 2003; Schreiber, 2005), membrane trafficking (Mukherjee et al., 2006; Brown et al., 2009; Kojima et al., 2012), and cell-substrate adhesion (Srivastava et al., 2008; Choi et al., 2013), and is dysregulated in some diseases, such as cancer (Webb et al., 2011; Parks et al., 2013) and neurodegenerative disorders (Harguindey et al., 2007; Wolfe et al., 2013). However, a role for pHi dynamics in metazoan development remains understudied. Here, we show that pHi increases during the differentiation of FSCs and mESCs and is necessary for the efficient initial differentiation of both cell types. In addition, our data suggest a specific role for pHi dynamics in the regulation of Hedgehog (Hh) signaling in the FSC lineage.
Results and discussion
We previously reported that a null allele of DNhe2, an orthologue of the mammalian plasma membrane Na+–H+ exchanger that catalyzes an influx of extracellular Na+ and an efflux of intracellular H+, suppresses dysplasia with expression of activated oncogenes in Drosophila imaginal disks (Grillo-Hill et al., 2015). Through these studies, we noticed that flies homozygous for DNhe2null have reduced fertility. Thus, we performed an egg-laying assay and found that DNhe2null flies laid significantly fewer eggs per day compared with wild-type flies (Fig. 1 A). To investigate further, we searched for defects in oogenesis. The formation of new follicles during early oogenesis requires proper differentiation in the FSC lineage. This begins in the germarium (Fig. 1 B) with a pair of FSCs at the region 2a/2b border (Margolis and Spradling, 1995; Nystul and Spradling, 2007) that divide regularly to self-renew and produce prefollicle cell (pFC) daughters. Upon exiting the niche, a subset of pFCs begin to differentiate into polar and stalk cells (Larkin et al., 1996; Besse et al., 2002; Nystul and Spradling, 2010), which facilitate follicle budding, while the remaining pFCs differentiate into main body follicle cells (FCs) that surround the developing germline cyst. This well-defined lineage makes it possible to identify the stem cell and distinct stages of differentiation in vivo with single-cell resolution.
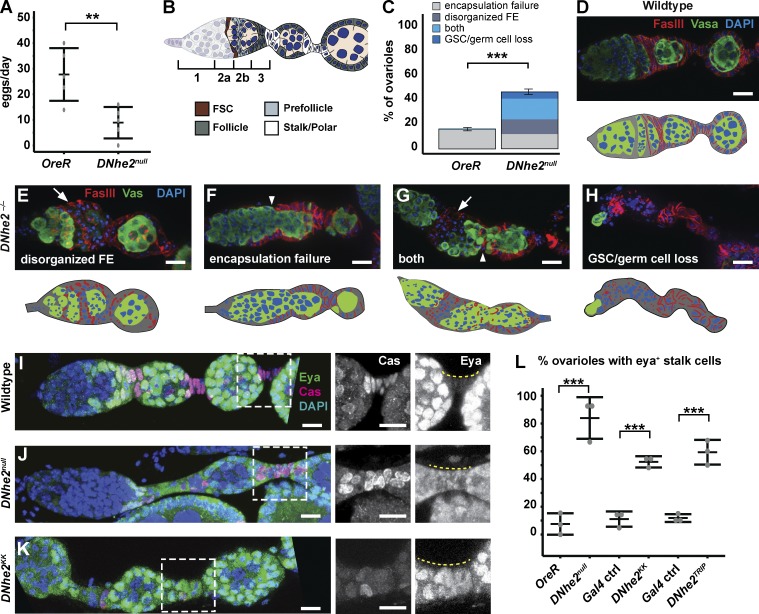
Figure 1.
DNhe2 is necessary for differentiation in the FSC lineage. (A) DNhe2null flies have significantly reduced egg laying. Graph depicts the mean number of eggs laid per female per day. n = 5 independent replicates. **, P < 0.01. (B) Diagram of the germarium showing the four regions, regions 1, 2a, 2b, and 3. Two FSCs (brown) are located in the middle of the germarium, at the region 2a/2b border. Cells that exit the FSC niche become pFCs (light gray) and then differentiate into main body FCs (dark gray), polar cells, or stalk cells (white). (C–H) DNhe2null germaria have morphological defects. (C) The frequency of each type of morphological defect. n = 3 independent replicates; n > 50 ovarioles. ***, P < 0.001. (D) Wild-type germarium stained with FasIII to label FCs, vasa to label germline, and DAPI. (E–H) Examples of germaria from DNhe2null flies. (E) A disorganized follicle epithelium (FE; arrow). (F) A failure of FCs to properly encapsulate the germline, resulting in fused cysts (arrowhead). (G) Both a disorganized follicle epithelium (arrow) and encapsulation defect (arrowhead). (H) A lack of germ cells and germline stem cells (GSCs). (I–L) Decreased DNhe2 impairs stalk cell differentiation. Stalk cells (yellow dashed lines in I–K [right]) are Cas+, Eya– in wild-type ovarioles (I) but are Cas+, Eya+ in DNhe2null (J) or DNhe2KK RNAi ovarioles (K). (L) Penetrance of Eya misexpression in stalks. n > 40 ovarioles for all genotypes; n = 3 independent replicates. ***, P < 0.001. Bars, 10 µm. P-values were determined with a t test in B and a χ2 test in C and L. Error bars represent SEM.
We observed clear morphological phenotypes in 45.2% of the germaria from DNhe2null flies (Fig. 1 C). In most cases, the pFCs either accumulated into disorganized clusters (Fig. 1 E) rather than forming into a single layered epithelium (Fig. 1 D); failed to completely encapsulate a germ cell cyst, resulting in a fusion of two adjacent cysts (Fig. 1 F); or both (Fig. 1 G). In some germaria, there were few or no germ cell cysts, indicating a loss of germ cell production (Fig. 1 H). When stalks were present in DNhe2null ovarioles, stalk cells mostly remained rounded (Fig. 1 J) rather than acquiring their typical flattened shape (Fig. 1 I). In wild-type tissue, pFCs express both Castor (Cas) and Eyes absent (Eya), whereas stalk and polar cells are Cas+, Eya– (Fig. 1 I; Chang et al., 2013). However, in 83 ± 9% (n = 42) of DNhe2null ovarioles, cells in the first stalk downstream from the germarium remained Cas+, Eya+ (Fig. 1 J) compared with 9 ± 4% (n = 86) of wild-type ovarioles (Figs. 1 L and S1, A and B). Knockdown of DNhe2 in FSCs and early FCs using two separate RNAi lines driven by 109–30-Gal4 (FC-Gal4) produced phenotypes similar to DNhe2null flies, including the morphological phenotypes (Fig. S1, D–F) and Cas+, Eya+ cells in the stalks (Fig. 1, K and L; and Fig. S1 C). However, the penetrance was lower, suggesting that there are some noncell autonomous effects on FC differentiation in DNhe2null flies or that knockdown with these RNAi lines is incomplete.
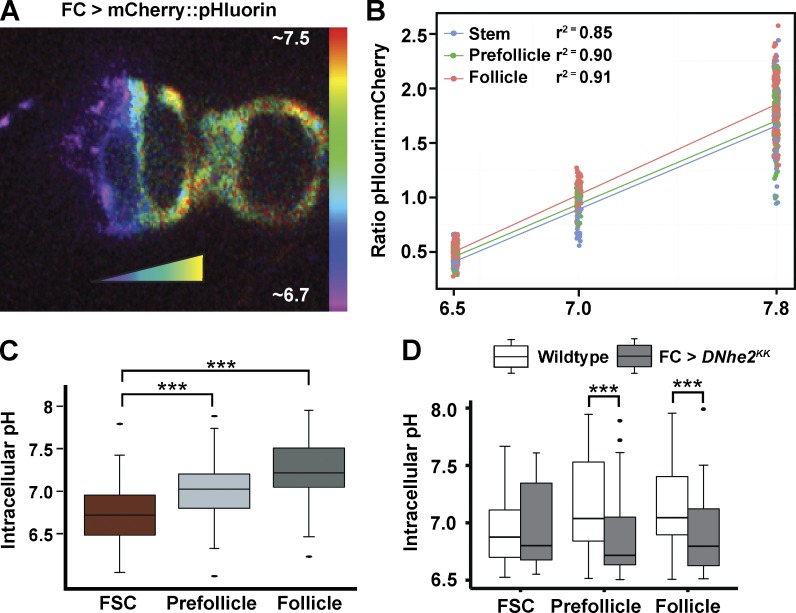
We previously reported that pHi in eye epithelia is regulated by changes in DNhe2 expression (Grillo-Hill et al., 2015) and hence investigated whether there are also pHi changes in ovarioles, focusing on cells in the FSC lineage. We generated FSC clones expressing mCherry::pHluorin (Fig. 2 A), a genetically encoded pH biosensor (Koivusalo et al., 2010; Choi et al., 2013; Rossano et al., 2013; Grillo-Hill et al., 2015). The fluorescence intensity of pHluorin increases with increased pHi within the physiological range (Miesenböck et al., 1998), whereas the fluorescence of mCherry is insensitive to physiological changes in pHi and thus indicates biosensor abundance. Fluorescence ratios were converted to pH values by using the well-characterized nigericin calibration method (Fig. 2 B; Fig. S2, A–C; and Table S1; Meima et al., 2009; Choi et al., 2013; Grillo-Hill et al., 2014, 2015). We found significant differences in pHi between each cell type in wild-type flies, with values of 6.8 ± 0.07 for FSCs, 7.0 ± 0.06 for pFCs, and 7.3 ± 05 for the FCs (P < 0.001; n = 36 germaria; Fig. 2 C and Table S2). Hence, pHi is higher at progressively later stages of differentiation. RNAi knockdown of DNhe2 significantly lowered the pHi of pFCs and FCs by 0.2–0.25 pH units (Fig. 2 D), indicating that the FC differentiation phenotypes we observed are associated with significant reductions in pHi.
Figure 2.
pHi increases during pFC differentiation in a DNhe2-dependent manner. (A) A germarium expressing UAS-mCherry::pHluorin in an FSC clone generated with the FLPout system. The pseudocoloring indicates the ratio of the fluorescence intensities of pHluorin to mCherry, which correlates with pHi. Bar indicates pHi estimates. (B) Three-point measurement of FSC clones expressing mCherry::pHluorin in freshly dissected ovarioles equilibrated to a pH of 6.5, 7.0, or 7.8, with nigericin buffers. The mean ratio of pHluorin to mCherry fluorescence increased linearly with increasing pH for each cell type (r2 = 0.85 for FSC, 0.90 for pFC, and 0.91 for FCs), confirming that the biosensor is responsive to pH changes as predicted when expressed in the follicle epithelium. (C) pHi increases as cells transition from an FSC to an FC. n = 36 germaria; n = 5 independent replicates. (D) Nhe2KK RNAi driven in FCs with FC-Gal4 (gray boxes) causes a significant decrease in pHi compared with control (white boxes) in pFC and FCs. n = 16–17 germaria; n = 3 independent replicates. ***, P < 0.001 (t test).
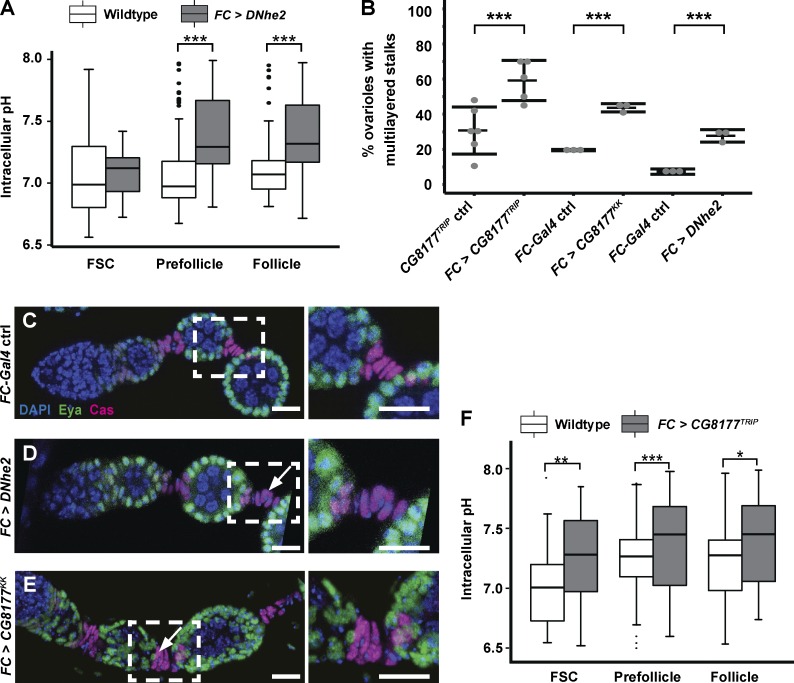
To determine how increased pHi affects FC differentiation, we investigated the phenotypes caused by overexpression of DNhe2. Upon overexpression of DNhe2, pHi in pFCs and FCs increased significantly (0.3–0.35 pH units, P < 0.001; Fig. 3 A), and the stalks between budded follicles were multilayered in 29 ± 4% (n = 113; Fig. 3, B–D; and Fig. S2, D and E). To confirm that the stalk cell differentiation phenotype is caused by changes in pHi, we performed an RNAi screen of 20 genes that are predicted to regulate pH homeostasis and identified CG8177. CG8177 is a putative Cl−/HCO3 exchanger (an acid loader) with the highest similarity to SLC4A1 and SLC4A2. Knockdown of an acid loader is predicted to increase pHi, and, indeed, we found that RNAi knockdown of CG8177 using FC-Gal4 significantly increased pHi by 0.4 pH units. In addition, CG8177 knockdown significantly increased the frequency of multilayered stalks (Fig. 3, B, E, and F; and Fig. S2 F).
Figure 3.
Increasing pHi promotes excessive FC differentiation. (A) Overexpression of DNhe2 (gray boxes) increases pHi relative to controls (white boxes) in pFCs and FCs. n = 15–17 germaria; n = 3 independent replicates. (B) Penetrance of the stalk phenotypes with overexpression of DNhe2 or RNAi knockdown of CG8177. n > 100 for all genotypes; n > 3 independent replicates. (C–E) Overexpression of DNhe2 or knockdown of CG8177 causes multilayered stalks. Stalk cells in a wild-type ovariole (C) form a single row, whereas overexpression of DNhe2 (D) or knockdown of CG8177 (E) causes the formation of multilayered stalks (white arrows). Insets are enlarged in C–E (right). (F) RNAi knockdown of CG8177 (gray boxes) increases pHi relative to controls (white boxes). n = 17–19 germaria; n = 3 independent replicates. Bars, 10 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P-values were determined with a t test in A and F and a χ2 test in B.
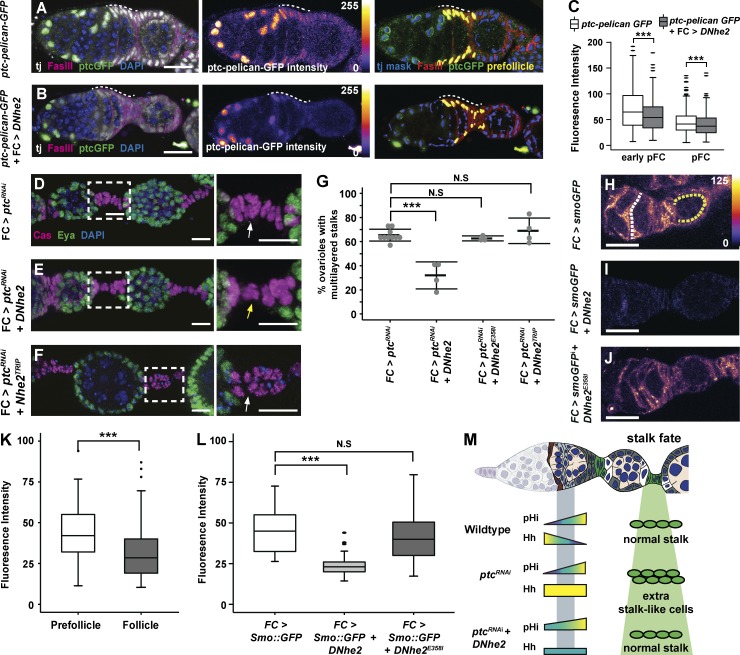
Polar and stalk cell differentiation are regulated by multiple signaling pathways, including Notch and Hh. Notch signaling is activated in a small subset of pFCs and is required specifically for polar cell differentiation, whereas Hh signaling is required in pFCs for differentiation toward the polar and stalk cell fates, but then must be down-regulated during the differentiation process for mature polar and stalk cells to form (Forbes et al., 1996a,b; Tworoger et al., 1999; Zhang and Kalderon, 2000; Besse et al., 2002). To test whether increased pHi promotes polar and stalk cell differentiation by modulating the response to these differentiation cues, we examined the expression of Notch and Hh pathway activity reporters in pFCs overexpressing DNhe2. Overexpression of DNhe2 did not affect the pattern or level of expression of the Notch pathway reporter, notch response element (NRE)–GFP (Fig. S2, G and H). However, we observed a significant decrease in the Hh pathway reporter, Ptc-pelican-GFP, in both early pFCs, just downstream from the FSC niche where Hh signaling is high, as well as in later pFCs, where Hh signaling is lower (Fig. 4, A–C). In addition, overexpression of DNhe2 suppressed the stalk cell phenotype caused by overactivation of Hh signaling. Specifically, we found that knockdown of ptc by RNAi in FCs with FC-Gal4 caused the development of multilayered stalks in 70 ± 2% (n = 467) of ovarioles (Fig. 4, D and G), which is consistent with published results (Forbes et al., 1996a; Tworoger et al., 1999; Chang et al., 2013), whereas overexpression of DNhe2 along with ptc RNAi reduced the frequency of this phenotype to 30 ± 5% (n = 279; Fig. 4, E and G). In contrast, overexpression of DNhe2E358I, an allele with a point mutation that prevents ion translocation (Denker et al., 2000; Grillo-Hill et al., 2015), did not increase pHi (Fig. S2 I) and did not suppress the ptc RNAi phenotype (Fig. 4 G). In addition, RNAi knockdown of DNhe2 did not enhance the ptc RNAi phenotype (Fig. 4, F and G). These observations indicate that increased pHi attenuates wild-type and constitutively active Hh signaling.
Figure 4.
DNhe2 overexpression attenuates Hh signaling. (A and B) Compared with control (A), overexpression of DNhe2 decreases Ptc-pelican-GFP expression. Pseudocolor of GFP channel in A and B reflects fluorescence intensity. Nuclear volumes selected for intensity measurements (yellow) are shown (right). pFC regions are indicated with white dashed lines. (C) Quantification of Ptc-pelican-GFP fluorescence intensities in control germaria (white boxes) with DNhe2 overexpression (gray boxes) in early pFCs or pFCs. n > 450 cells for all genotypes; n = 3 independent replicates. (D–F) Multilayered stalk phenotype in FC > ptcRNAi (D) is less penetrant in FC > ptcRNAi + DNhe2 (E), but not in FC > ptcRNAi + DNhe2TRiP RNAi (F). Insets enlarged in D–F (right). White and yellow arrows indicate stalks with double or single rows of cells, respectively. (G) The frequency of ovarioles with multilayered stalks in each genotype. n > 100 ovarioles for all genotypes; n = 3–6 independent replicates. (H–J) In ovarioles expressing Smo::GFP only (H), the fluorescence intensity is higher in pFCs (yellow dotted line) than in FCs (white dotted line). Coexpression of wild-type DNhe2 (I) but not DNhe2E358I (J) significantly reduces GFP fluorescence throughout the germarium. (K) Quantifications of the fluorescence intensity in germaria with FC-Gal4 driving Smo::GFP expression. n = 31 germaria. (L) Expression of DNhe2 but not DNhe2E358I decreases the fluorescence intensity of Smo::GFP. n > 29 germaria for all genotypes. (M) Summary of the genetic interaction between RNAi knockdown of ptc and overexpression of DNhe2. In wild-type germaria, pHi increases in pFCs as Hh pathway activity decreases, and a normal stalk forms. RNAi knockdown of ptc results in excessive Hh signaling and multilayered stalks. Overexpression of DNhe2 increases pHi, attenuates Hh signaling, and suppresses the stalk cell phenotype caused by RNAi knockdown of ptc. Bars: (A, B, and H–J) 10 µM; (D–F) 20 µm. ***, P < 0.001. P-values were determined with a t test in C, K, and l and a χ2 test in G.
To investigate how increased pHi attenuates Hh signaling, we examined the levels of a Smoothened::GFP (Smo::GFP) fusion protein expressed with FC-Gal4. We found that Smo::GFP levels were highest in region 2b, where pFC pHi is low (Fig. 2 A), and decreased progressively toward region 3, where pHi is higher (Fig. 4, H and K). In contrast, the level of an unrelated fusion protein, CD8::GFP, expressed with the same driver was uniform throughout the germarium (Fig. S2 J), indicating that the decrease in Smo::GFP is not caused by differences in FC-Gal4 expression levels or GFP stability in these regions. In addition, we found that overexpression of wild-type DNhe2, but not DNhe2E358I, significantly decreased Smo::GFP levels throughout the germarium (Fig. 4, I, J, and L). Collectively, our findings suggest a model in which an increase in pHi contributes to the patterning of pFC differentiation, at least in part, by down-regulating Hh pathway activity (Fig. 4 M).
To determine the role of pHi dynamics in the differentiation of another stem cell type, we investigated mESCs (Murry and Keller, 2008; Young, 2011). Naive mESCs are maintained in a pluripotent state when cultured with leukemia inhibitory factor (LIF) and inhibitors of mitogen-activated protein kinase kinase (MEK) and glycogen synthase kinase-3β (2i). Upon removal of LIF2i, naive cells spontaneously differentiate to a primed state by 3–5 d (Ying et al., 2008). We measured pHi in cells loaded with the fluorescent H+-sensitive dye 2,7-biscarboxyethyl-5(6)-carboxyfluorescein (BCECF), calibrating ratios of pH-sensitive and pH-insensitive BCECF signals to pHi values with nigericin buffers as described previously (Meima et al., 2009; Grillo-Hill et al., 2015). Naive mESCs maintained a near-constant pHi between 7.40 ± 0.06 and 7.46 ± 0.07 over 6 d. However, in differentiating cells there was a transient increase in pHi to 7.65 ± 0.06 at 48 h and 7.57 ± 0.05 at 72 h (P < 0.001; Fig. 5 A), with pHi returning to values similar to naive cells at 96 h and 6 d. Because a MEK inhibitor is included in the LIF2i-containing medium for naive mESCs and MEK increases the activity of NHE1 and pHi (Malo et al., 2007), we adapted naive cells to LIF without 2i. In this condition, we observed a similar increase in pHi upon differentiation, indicating that the increased pHi does not merely reflect removal of MEK inhibition of NHE1 (Fig. S3 A).
Figure 5.
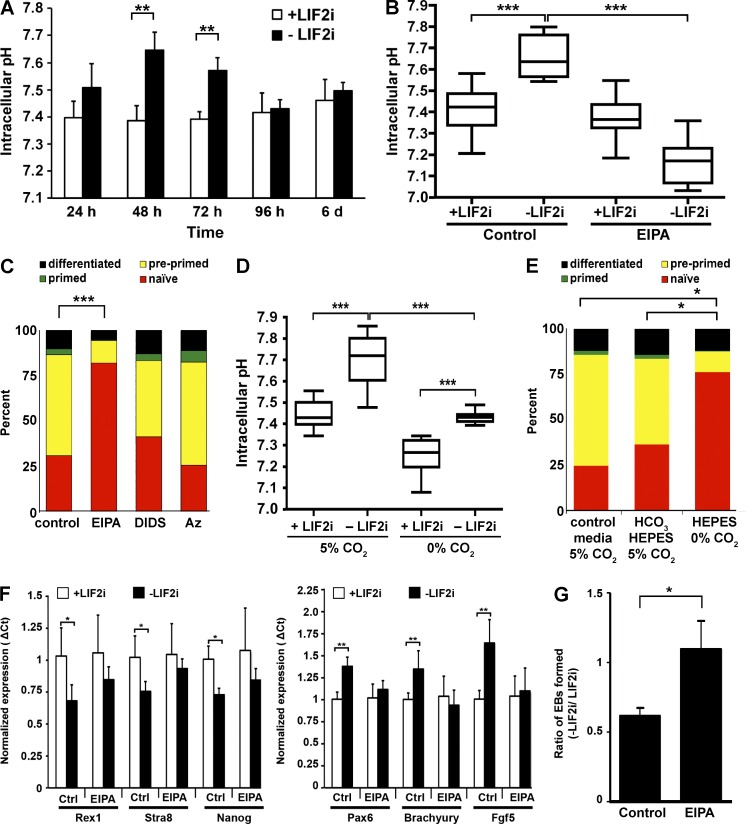
Increased pHi during mESC differentiation is necessary for progression to primed fate. (A) pHi measurements revealed no significant change over 6 d with LIF2i but significant increases at 48 and 72 h after removing LIF2i. n = 3 cell preparations. (B) EIPA blocked the increase in pHi seen at 72 h after removal of LIF2i. n = 5 cell preparations. (C) In the dual-reporter mESC line, naive mESCs expressed mir-290-mCherry; preprimed cells expressed both mir-290-mCherry and mir-302-eGFP; primed cells expressed mir-302-eGFP; and further differentiated cells have silenced both reporters. After 72 h without LIF2i, 30.9% of control cells and 82.0% of EIPA-treated cells expressed mir-290-mCherry alone. n = 6 cell preparations. (D) Cells maintained for 72 h in Hepes-buffered DMEM in the absence of CO2 had lower pHi in the presence and absence of LIF2i compared with cells maintained at 5% CO2. n = 4 cell preparations. (E) In the dual-reporter mESC line, more cells maintained in Hepes-buffered DMEM in the absence of CO2 expressed in the naive cell miRNA mir-290-mCherry (75.4%) compared with cells maintained at 5% CO2 in either a predominantly Hepes-buffered DMEM (36.4%) or control DMEM (24.6%). n = 3 cell preparations. (F) Naive markers (Rex1, Stra8, and Nanog) were expressed at significantly lower levels, and differentiation markers (Pax6, T, and Fgf5) were expressed at significantly higher levels in control cells maintained without LIF2i for 72 h compared with naive cells maintained with LIF2i. In contrast, in the presence of EIPA, naive marker expression did not decrease, nor did differentiation marker expression increase upon removal of LIF2i. (G) Functional pluripotency, as measured by EB formation, is maintained in the presence of EIPA. The number of EBs formed after 72 h without LIF2i was normalized to the number of EBs formed in naive mESCs for each experiment. Control mESCs form fewer EBs than naive mESCs (61 ± 5%). However, mESCs grown without LIF2i but with EIPA for 72 h form EBs as well as naive mESCs (109 ± 20%; P < 0.05). Error bars are mean ± standard deviation; n = 130–150 droplets; n = 3 independent experiments. Box plots display mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t test).
To determine whether increased pHi is necessary for the spontaneous differentiation of naive mESCs, we first changed pHi using the NHE1 selective pharmacological inhibitor 5-(N-ethyl-N-isopropyl) amiloride (EIPA; Putney et al., 2002). In naive cells maintained with LIF2i, pHi was not significantly different in the absence and presence of EIPA (Fig. 5 B). However, with EIPA, pHi did not increase with removal of LIF2i for 72 h and was significantly lower than in control naive cells, possibly reflecting loss of efflux of metabolic acids generated by increased glycolysis that occurs with differentiation (Ge et al., 2010). These data suggest that NHE1 activity is necessary for the increased pHi in primed cells but not for steady-state pHi in naive mESCs.
We determined the differentiation state of mESCs in the absence and presence of EIPA using several approaches. We first used a dual-reporter mESC line expressing developmentally regulated miRNA clusters that are tagged with distinct fluorophores (Parchem et al., 2014). In this reporter line, naive mESCs express mir-290-mCherry, but not mir-302-eGFP; preprimed cells in intermediate stages express both reporters; primed cells express mir-302-eGFP, but not mir-290-mCherry; and both reporters are silenced in cells that have differentiated further. EIPA had no effect on the proportion of naive cells in self-renewing +LIF2i medium (Fig. S3 B). However, after 72 h without LIF2i, there was a significantly higher percentage (82.0%) of naive cells with EIPA compared with controls (30.9%; P < 0.001; Fig. 5 C). After 5 d without LIF2i, there were also more mir-290-mCherry+ cells in the presence of EIPA compared with controls (Fig. S3 C). We found no increase in cell death with EIPA treatment (Fig. S3 D), indicating that attenuated differentiation is not caused by selective elimination of preprimed and/or primed cells. In contrast to attenuated differentiation with EIPA, differentiation with the pharmacological inhibitors 4,4'-diisothiocyano-2,2'-stilbenedisulfonic acid (DIDS), which blocks activity of anion exchangers, or acetazolamide (Az), which blocks carbonic anhydrases, was not different than the control (Fig. 5 C). Moreover, despite the efficacy of both inhibitors in attenuating pHi recovery, when cells were rapidly switched from a HCO3-free Hepes buffer at 0% CO2 to a Hepes-free buffer containing 25 mM NaHCO3 superfused with 5% CO2 (Fig. S3 E), neither inhibitor prevented the increase in pHi with differentiation (Fig. S3 F). Although NHE1 activity was previously shown to be necessary for differentiation of mESCs to cardiomyocytes (Li et al., 2009), to our knowledge, a role for increased pHi in the spontaneous differentiation of mESCs toward primed has not been reported.
To confirm that attenuated differentiation was caused by decreased pHi rather than by inhibiting a possible function of NHE1 independent of H+ efflux, we used another approach to change pHi by maintaining cells in a predominantly Hepes-buffered medium at atmospheric 5% compared with 0% CO2. The pHi and differentiation of cells maintained at 5% CO2 in DMEM containing 5 mM NaHCO3 and 30 mM Hepes was not different than cells maintained at 5% CO2 in control medium containing 44 mM NaHCO3 and no Hepes (Fig. 5, D and E). In contrast, cells maintained at 0% CO2 in a Hepes-buffered medium lacking NaHCO3 had a lower pHi with LIF2i, and although pHi was increased after 72 h without LIF2i, it was significantly lower than controls without LIF2i and similar to controls with LIF2i (Fig. 5 D). Additionally, differentiation of dual-reporter cells maintained at 0% CO2 was significantly attenuated, as indicated by the abundance of mir-290-mCherry+ cells (Fig. 5 E). Collectively, these data indicate that an increase in pHi, likely to a pHi >7.5, is necessary for mESCs to differentiate toward the primed fate.
For a second method to determine the differentiation state of mESCs in the absence and presence of EIPA, we used quantitative RT-PCR to quantify expression of naive and primed markers. For control cells, we found that removal of LIF2i for 72 h decreased the expression of mESC markers Rex1, Stra8, and Nanog and increased the expression of primed cell markers Fgf5, brachyury (T), and Pax6, as expected (Parchem et al., 2014). However, with EIPA, there was no change in the expression of these genes upon removal of LIF2i in cultures (Fig. 5 F), indicating that the increase in pHi is required for the expression of genes that drive differentiation. As a third metric for the differentiation state, we determined functional pluripotency in EIPA-treated cells cultured without LIF2i by testing for the formation of embryoid bodies (EBs) that form spontaneously when mESCs are grown in suspension (Kurosawa, 2007). We grew mESCs with or without LIF2i and EIPA for 72 h and then plated cells in suspension as described previously (Kurosawa, 2007). We found that control cells grown without LIF2i for 72 h formed EBs at a lower frequency than control cells grown with LIF2i (61 ± 5%; Fig. 5 G). However, after 72 h without LIF2i, cells treated with EIPA formed EBs at a significantly higher frequency than control cells (109 ± 20%; P < 0.05), suggesting that mESCs treated with EIPA maintain functional pluripotency even in the absence of LIF2i.
Differentiation of adult and ESCs is regulated by distinct cues and is generally studied independently. Our study reveals that increased pHi contributes to the regulation of differentiation in both cell types. These findings open new directions to determine whether increased pHi is a conserved differentiation signal for other stem cell types and to resolve the molecular mechanisms mediating self-renewal and differentiation by pHi dynamics. As we described previously (Schönichen et al., 2013), regulation of cell processes by pHi dynamics is mediated by protonation of selective pH sensors, which acts as a posttranslational modification to change activity or ligand binding within the narrow physiological pH range. Our findings suggest that, in the FSC lineage, either Smo or a protein that regulates Smo stability may be a pH sensor. However, changes in pHi may also influence Hh signaling through effects on other cellular processes, such as vesicle trafficking, or by changing extracellular pH, neither of which can be assessed with our current tools. In addition, pHi dynamics could converge with Hh signaling and other cues (Simons et al., 2009) to regulate differentiation through a common effector such as the chromatin state, which is responsive to changes in pHi (McBrian et al., 2013). Although the pH sensors mediating mESC differentiation remain to be determined, several pH sensors we recently identified, including cofilin (Frantz et al., 2008), talin (Srivastava et al., 2008), and focal adhesion kinase (Choi et al., 2013), are required for mESC function or differentiation (Lee et al., 2011; Liu et al., 2011; Yun et al., 2012) and therefore may be components of the pH-responsive differentiation program. In summary, our findings show the critical importance of pHi dynamics in two distinct stem cell lineages, which advances our understanding of the cytosolic signals enabling stem cell differentiation.
Materials and methods
pHi measurements in Drosophila
FLPout clones were generated with a 15–30-min heat shock at 37°C and dissected 7–10 d after clone induction. In FSC clones, FSCs can be identified as the most anterior cell in the clone (Margolis and Spradling, 1995; Nystul and Spradling, 2007), pFCs are found just downstream from the FSCs, in region 2b, and more mature FCs are located further downstream, in region 3 of the germarium. For expressing UAS-mCherry::pHluorin with an FC-specific (FC) driver (109–30-Gal4), flies were dissected 7 d after eclosion. For pHi imaging, ovaries were dissected into bicarbonate buffer as described previously (Grillo-Hill et al., 2014, 2015) with 0.25 mg/ml concanavalin A conjugated to Alexa Fluor 647 and incubated for 15 min before imaging to visualize cell membranes (C21421; Thermo Fisher Scientific). Calibration curves were generated to calibrate mCherry::pHluroin ratios to pHi values for each experiment and genotype. Although two-point calibration curves are commonly used, we tested whether the addition of a third point would improve pHi estimates. However, the addition of a third point did not change the slope, Y intercept, or r2 value of the linear regression by >1% (Table S1). Thus, we used two-point calibration curves for all pHi estimates. To generate the calibration curves, fresh ovaries were equilibrated into nigericin buffer, pH 6.5 or 7.5, for 15 min before imaging as described previously (Grillo-Hill et al., 2014, 2015) using 40 µM nigericin (N1495; Invitrogen). 16-bit images in 1,024 × 1,024 format were acquired with Leica Biosystems software on an SP5 or SP8 laser-scanning confocal microscope with a 40× objective with a numerical aperture of 1.3 (Leica Biosystems), with gain, laser power, pinhole size, and other image acquisition settings consistent across matched control and experimental conditions. All imaging was done at room temperature. To obtain the fluorescence intensity measurement, we first subtracted background using a large region of interest in a black area of the image. Then, regions of interest were drawn around individual cells using the concanavalin A–Alexa Fluor 647 dye as a guide to outline individual cells. We measured one to two stem cells and three to five pFCs of bright and uniform intensity and selected the most highly expressed cells for measurement. Each region of interest was measured in one or two slices of 1 µM. If two slices were used, fluorescence intensity was averaged over two slices. Calibration curves for each experiment, genotype, and cell type were generated, and linear regression analysis yielding r2 values of at least 0.70 were obtained and used to calculate pHi values from ratios.
Drosophila tissue immunofluorescence staining
Unless otherwise noted, ovaries were fixed and stained according to previously published immunostaining protocols (Castanieto et al., 2014). To reduce background in ovarioles stained for Cas and Eya, ovaries were blocked in 0.5% BSA (A7979; Sigma-Aldrich) in PBST (PBS + 0.2% Triton X-100) for at least 1 h before addition of both primary and secondary antibodies.
The following antibodies were used: from the Developmental Studies Hybidoma Bank, we purchased mouse anti-Eya (Eya-10H6, 1:50) and mouse anti–Fasciclin III (7G10, 1:100); guinea pig anti–traffic jam (1:1,000) was a gift from A. Spradling (Carnegie Institute of Washington, Baltimore, MD); from Thermo Fisher Scientific, we purchased rabbit anti-GFP (A11122, 1:1,000); from Santa Cruz Biotechnology, Inc., we purchased rabbit anti-vasa (1:1,000); and rabbit anti-castor (1:4,000 or 1:5,000) was a gift from W. Odenwald (National Institutes of Health, Bethesda, MD). The following secondary antibodies were used at 1:1,000: anti–mouse Alexa Fluor 488 and 555 (A11001 and A21422; Invitrogen), anti–rabbit Alexa Fluor 488 and 555 (A11008 and A21428; Invitrogen), and anti–guinea pig Alexa Fluor 633 (A21105; Invitrogen). Images were acquired on an SP5 or SP8 line-scanning confocal microscope for all experiments in which quantitative fluorescence comparisons were made and in experiments in which we scored for the presence of Eya. An M2 Axioimager microscope with an Apotome unit with a 63× Apochromat objective with a numerical aperture of 1.4 (ZEISS) was used for images in which comparisons of morphological features were made. Images were cropped, rotated, contrast adjusted in Photoshop (Adobe), and saved as TIFF files.
Egg-laying assays
Flies closed within 48 h were moved into egg-laying vessels. Egg-laying vessels consist of standard plastic polypropylene fly bottles turned upside down onto standard fly food mixed with one to three drops of methylene blue dye with a small amount of wet yeast. One to three females per bottle were left overnight, up to 20 h, and then eggs were counted. The rate of eggs per fly per day was calculated as the hourly per fly egg-laying rate multiplied by 24 h.
Ptc-pelican-GFP quantitative analysis
The middle section of a germarium varying in thickness from 1 to 5 µM stained with traffic jam, FasIII, GFP, and DAPI were imaged on an SP5 inverted confocal microscope. Imaris 3D analysis software (Bitplane) was used to render 3D surfaces over traffic jam expressing in all somatic cell nuclei. pFCs were then manually identified and selected based on the position and intensity of FasIII staining. Mean Ptc-pelican-GFP intensity per nuclear volume was measured with the Imaris software. Three independent experiments all had similar distributions, means, and standard deviations, so data were aggregated.
Smo::GFP quantitative analysis
The middle section (one to five slices 1 µM thick) of a germarium stained with GFP and FasIII was imaged on an SP8 inverted confocal microscope. Two different types of quantitative analyses were performed. First, for control (109–30-gal4 > Smo::GFP), we quantified the background-subtracted fluorescence intensity for pFCs and FCs. For each of these regions, two or three lines were drawn through the brightest portion of the region, and the mean intensity of the line was measured. For the second quantification, we used the FasIII channel to mask the GFP channel and then took a measurement of the mean fluorescence intensity within each germaria to get a measurement of the total fluorescence intensity in every germaria.
Plotting and statistics for Drosophila data
We used RStudio statistical analysis software to generate plots. For the boxplots displaying the pH data and the quantitative fluorescence measurements, the box represents the interquartile range, which is the middle 50% of the data. The black line is the median, and the whiskers show the range of the data, with outliers of 1.5× the interquartile range plotted as dots. For the dot plots, the lines indicate the median and the standard deviation. We believe that displaying our data thus gives the reader the most accurate picture of the full variation within our data. Unless otherwise noted, we used RStudio to perform statistical comparisons and generate plots.
Stocks
Stocks were generated with standard crossing schemes, maintained on standard molasses food, and cultured at 25°C unless otherwise noted, and all stocks were acquired from the Bloomington Drosophila Stock Center unless otherwise noted. For mCherry::pHluorin clones using the FLPout system, y[1] w[*]; P{w[+mC]=GAL4-Act5C(FRT.CD2).P}S (Bloomington ID 4780). For ptcRNAi experiments, ptcRNAi is a transgenic RNAi line from the Transgenic RNAi Project collection (y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03223}attP2; Bloomington ID 28795). All experiments using FC> refers to y1 w*; P{GawB}109-30 /CyO (Bloomington ID 7023). For DNhe2 knockdown via RNAi, DNhe2KK102518 is a transgenic line containing P{KK102518} (Vienna Drosophila Resource Center, FlyBase ID FBti0117476). DNhe2HMC03243 is a transgenic line from the Transgenic RNAi Project collection (y1 v1; P{TRiP.HMC03243}attP2; Bloomington ID 51491). For DNhe2 and UAS-mCherry::pHluorin, UAS-DNhe2, UAS-DNhe2E358I, DNhe2null, and UAS-mCherry::pHluorin were as described by Grillo-Hill et al. (2015). For CG8177 knockdown via RNAi, CG8177TRIP refers to y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMC03399}attP40 (Bloomington ID 51827). CG8177KK refers to P{KK100095}VIE-260B (Bloomington ID v109594, obtained from the Vienna Drosophila Resource Center). For Smo::GFP analysis, y[1] w[*]; P{w[+mC]=UAS-smo.GFP}2 (Bloomington ID 44624). For Ptc-pelican-GFP analysis, yw*; Ptc-pelican-GFP (a gift from T. Kornberg, University of California, San Francisco, San Francisco, CA). NRE-GFP refers to w[1118]; P{w[+m*]=NRE-EGFP.S}5A (Bloomington ID 30727). CD8 GFP refers to yw; UAS-CD8::GFP/cyo (a gift from B. Ohlstein, Columbia University, New York, NY).
mESC culture
Wild-type v6.5 and dual-reporter mESCs were a gift from R. Blelloch (University of California, San Francisco, San Francisco, CA). Cells were maintained as reported previously (Parchem et al., 2014). For differentiation assays, cells were plated in DMEM (BRL 0569; Gibco) containing LIF (ESGRO; EMD Millipore) and inhibitors for MEK (PD0325901, 04–0006-10; Stemgent) and glycogen synthase kinase-3β (CHIR99021, 04–0004-10; Stemgent), termed LIF2i. After plating for 24 h, spontaneous differentiation was initiated by washing cells three times in PBS and then maintaining cells for the indicated times in media without LIF2i. FACS analysis of dual-reporter mESCs was performed at the University of California, San Francisco, Flow Cytometry Core on an LSR II flow cytometer (BD), and analysis was performed using FACSDiva software (BD).
To make EBs, cells were plated and differentiated as described previously (Kurosawa, 2007) with or without EIPA for 72 h and then plated into hanging droplets (104 cells per droplet) in media without LIF2i on the lids of 10-cm Petri dishes. Hanging droplets were cultured for 72 h. Droplets were washed in media without LIF2i to low-adhesion 6-well cell culture dishes, and the number of EBs was counted the next day. Data are reported as the percentage of EBs formed, calculated by dividing the number of EBs formed by cells cultured without LIF2i by the number of EBs formed in control cells maintained in media with LIF2i. EB data reported are from three independent cell preparations with the following numbers of droplets per condition: control +LIF2i (150), control −LIF2i (150), EIPA +LIF2i (132), and EIPA −LIF2i (141). Data were analyzed, and statistical analysis was performed in Excel (Microsoft) using a paired t test.
pHi measurements of mESCs
pHi was measured as described previously (Meima et al., 2009; Grillo-Hill et al., 2015). In brief, cells maintained in 24-well plates were loaded with 1 µM fluorescent H+-sensitive dye BCECF in an HCO3-containing buffer (110 mM NaCl, 5 mM KCl, 10 mM glucose, 25 mM NaHCO3, 1 mM MgSO4, 1 mM KPO4, and 2 mM CaCl2, pH 7.4) and washed, and ratios of BCECF fluorescence at Ex490/Em530 and Ex440/Em530 were acquired every 15 s for 5 min using a SpectraMax M5 plate reader (Molecular Dynamics). The fluorescence ratios were converted to pHi by calibrating each well with 10 µM nigericin (Invitrogen) in 105 mM KCl. Pharmacological inhibitors were added 24 h after plating and included EIPA (ALX-550-266-M005; final concentration of 10 µM; Enzo), DIDS (309795; final concentration of 1 µM; EMD Millipore), and Az (A6011; final concentration of 200 µM; Sigma-Aldrich). The efficacy of DIDS and Az was confirmed by their ability to inhibit pHi recovery after rapidly switching cells from a nominally HCO3-free Hepes buffer at 0% CO2 to a Hepes-free buffer containing 25 mM NaHCO3 superfused with 5% CO2, as previously described (Hulikova et al., 2014). For experiments with altered CO2 and NaHCO3 medium, cells were plated and maintained in control DMEM for 24 h, washed, and them maintained for 72 h in the absence or presence of LIF2i in NaHCO3-free DMEM (D5030; Sigma-Aldrich) supplemented with 4.5 g/liter glucose, 30 mM Hepes, pH 7.4, 0.11 mg/liter Na-pyruvate, 1× Glutamax (35050–061; Gibco), 1× nonessential amino acids, 2 mM glutamine, 100 mg/liter penicillin/streptomycin, and 15% fetal bovine serum. Medium for cells maintained at 5% CO2 but not for cells maintained at 0% CO2 was also supplemented with 5 mM NaHCO3.
Quantitative RT-PCR experiments
RNA was extracted from mESCs maintained in the absence or presence of LIF2i for 72 h using TRIzol (Ambion) according to the manufacturer’s protocol with the following modifications: 800 µl TRIzol was added to cells, 2 µl glycoblue (AM9515; Ambion) was added to visualize the RNA pellet, and the pellet was rinsed in 75% EtOH. RNA purity was assessed on a Nanodrop spectrometer. cDNA was synthesized using a Superscript III kit (18080093; Invitrogen) from 2 mg RNA and diluted 1:1 before being added to the PCR reaction. Primers were as described in Parchem et al. (2014). Quantitative PCR was performed with SYBR Greener (quantitative PCR SuperMix for ABI prism; 11760; Invitrogen) according to the manufacturer’s protocol on a Real-Time PCR Viaa7 instrument (Applied Biosystems). Quantitative PCR analysis was done in the open source RStudio software using the ReadqPCR and NormqPCR (Perkins et al., 2012). Open source packages are available on the Bioconductor website. For each gene, three to five independent cell preparations were used.
Online supplemental material
Fig. S1 shows additional images of the Eya channel for images stained with Cas and Eya. The images were background subtracted in ImageJ and then displayed as either pseudocolored images to reflect fluorescence intensity or as grayscale. Additional morphological features of FC > DNhe2KK stained with FasIII and Vasa are also shown. Fig. S2 shows example images of calibration curves, example calibrations for quantitative pH imaging, and images of the Notch reporter, NRE-GFP, as well as quantitative pH measurements for FC > DNhe2E358I. Fig. S3 shows additional aspects of the interaction between changes in pHi and mESC differentiation, including the lack of an effect of EIPA on cell survival, dual-reporter expression indicating EIPA maintains more cells in a naive state, and increased pHi in differentiating cells being independent of relieving MEK inhibition. Table S1 shows the differences in the slope and intercept of the linear regression model using two or three calibration points. Table S2 shows the summary of the pHi estimates for the wild-type controls in all experiments.
Supplementary Material
Acknowledgments
We thank Sumitra Tatapudy for assistance with image acquisition of mCherry::pHluorin, Bradley Webb for assistance with quantitative pHi imaging analysis, and Torsten Wittmann and members of the Wittmann, Barber, and Nystul laboratories for helpful discussions. We also thank Robert Blelloch, Tom Kornberg, and Ophir Klein for suggestions on the manuscript.
This work was funded by the California Institute of Regenerative Medicine fellowship TG2-01153 to B. Ulmschneider and National Institutes of Health grants GM097158 to T.G. Nystul, GM47413 to D.L. Barber, and GM116384 to D.L. Barber and T.G. Nystul.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Az
- acetazolamide
- EB
- embryoid body
- ESC
- embryonic stem cell
- FC
- follicle cell
- FSC
- follicle stem cell
- Hh
- Hedgehog
- LIF
- leukemia inhibitory factor
- mESC
- mouse ESC
- NRE
- notch response element
- pFC
- prefollicle cell
- pHi
- intracellular pH
References
- Besse F., Busson D., and Pret A.-M.. 2002. Fused-dependent Hedgehog signal transduction is required for somatic cell differentiation during Drosophila egg chamber formation. Development. 129:4111–4124. [DOI] [PubMed] [Google Scholar]
- Brown D., Paunescu T.G., Breton S., and Marshansky V.. 2009. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid–base homeostasis and vesicle trafficking. J. Exp. Biol. 212:1762–1772. 10.1242/jeb.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanieto A., Johnston M.J., and Nystul T.G.. 2014. EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. eLife. 3 10.7554/eLife.04437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-C., Jang A.C.-C., Lin C.-H., and Montell D.J.. 2013. Castor is required for Hedgehog-dependent cell-fate specification and follicle stem cell maintenance in Drosophila oogenesis. Proc. Natl. Acad. Sci. USA. 110:E1734–E1742. 10.1073/pnas.1300725110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.-H., Webb B.A., Chimenti M.S., Jacobson M.P., and Barber D.L.. 2013. pH sensing by FAK-His58 regulates focal adhesion remodeling. J. Cell Biol. 202:849–859. 10.1083/jcb.201302131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker S.P., Huang D.C., Orlowski J., Furthmayr H., and Barber D.L.. 2000. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell. 6:1425–1436. 10.1016/S1097-2765(00)00139-8 [DOI] [PubMed] [Google Scholar]
- Forbes A.J., Lin H., Ingham P.W., and Spradling A.C.. 1996a hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 122:1125–1135. [DOI] [PubMed] [Google Scholar]
- Forbes A.J., Spradling A.C., Ingham P.W., and Lin H.. 1996b The role of segment polarity genes during early oogenesis in Drosophila. Development. 122:3283–3294. [DOI] [PubMed] [Google Scholar]
- Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M.J.S., Jacobson M.P., and Barber D.L.. 2008. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183:865–879. 10.1083/jcb.200804161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Meng W., Zhou H., and Bhowmick N.. 2010. Could stroma contribute to field cancerization? Med. Hypotheses. 75:26–31. 10.1016/j.mehy.2010.01.019 [DOI] [PubMed] [Google Scholar]
- Grillo-Hill B.K., Webb B.A., and Barber D.L.. 2014. Ratiometric imaging of pH probes. Methods Cell Biol. 123:429–448. 10.1016/B978-0-12-420138-5.00023-9 [DOI] [PubMed] [Google Scholar]
- Grillo-Hill B.K., Choi C., Jimenez-Vidal M., and Barber D.L.. 2015. Increased H+ efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. eLife. 4 10.7554/eLife.03270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harguindey S., Reshkin S.J., Orive G., Arranz J.L., and Anitua E.. 2007. Growth and trophic factors, pH and the Na+/H+ exchanger in Alzheimer’s disease, other neurodegenerative diseases and cancer: new therapeutic possibilities and potential dangers. Curr. Alzheimer Res. 4:53–65. 10.2174/156720507779939841 [DOI] [PubMed] [Google Scholar]
- Hulikova A., Aveyard N., Harris A.L., Vaughan-Jones R.D., and Swietach P.. 2014. Intracellular carbonic anhydrase activity sensitizes cancer cell pH signaling to dynamic changes in CO2 partial pressure. J. Biol. Chem. 289:25418–25430. 10.1074/jbc.M114.547844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M., Welch C., Hayashi H., Scott C.C., Kim M., Alexander T., Touret N., Hahn K.M., and Grinstein S.. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188:547–563. (published erratum appears in J. Cell Biol. 2010. 189:385) 10.1083/jcb.200908086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A., Toshima J.Y., Kanno C., Kawata C., and Toshima J.. 2012. Localization and functional requirement of yeast Na+/H+ exchanger, Nhx1p, in the endocytic and protein recycling pathway. Biochim. Biophys. Acta. 1823:534–543. 10.1016/j.bbamcr.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Kurosawa H. 2007. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 103:389–398. 10.1263/jbb.103.389 [DOI] [PubMed] [Google Scholar]
- Larkin M.K., Holder K., Yost C., Giniger E., and Ruohola-Baker H.. 1996. Expression of constitutively active Notch arrests follicle cells at a precursor stage during Drosophila oogenesis and disrupts the anterior–posterior axis of the oocyte. Development. 122:3639–3650. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Lee Y.J., and Han H.J.. 2011. Role of hypoxia-induced fibronectin–integrin β1 expression in embryonic stem cell proliferation and migration: involvement of PI3K/Akt and FAK. J. Cell. Physiol. 226:484–493. 10.1002/jcp.22358 [DOI] [PubMed] [Google Scholar]
- Li X., Karki P., Lei L., Wang H., and Fliegel L.. 2009. Na+/H+ exchanger isoform 1 facilitates cardiomyocyte embryonic stem cell differentiation. Am. J. Physiol. Heart Circ. Physiol. 296:H159–H170. 10.1152/ajpheart.00375.2008 [DOI] [PubMed] [Google Scholar]
- Liu J., He X., Qi Y., Tian X., Monkley S.J., Critchley D.R., Corbett S.A., Lowry S.F., Graham A.M., and Li S.. 2011. Talin1 regulates integrin turnover to promote embryonic epithelial morphogenesis. Mol. Cell. Biol. 31:3366–3377. 10.1128/MCB.01403-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo M.E., Li L., and Fliegel L.. 2007. Mitogen-activated protein kinase–dependent activation of the Na+/H+ exchanger is mediated through phosphorylation of amino acids Ser770 and Ser771. J. Biol. Chem. 282:6292–6299. 10.1074/jbc.M611073200 [DOI] [PubMed] [Google Scholar]
- Margolis J., and Spradling A.. 1995. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 121:3797–3807. [DOI] [PubMed] [Google Scholar]
- McBrian M.A., Behbahan I.S., Ferrari R., Su T., Huang T.-W., Li K., Hong C.S., Christofk H.R., Vogelauer M., Seligson D.B., and Kurdistani S.K.. 2013. Histone acetylation regulates intracellular pH. Mol. Cell. 49:310–321. 10.1016/j.molcel.2012.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meima M.E., Webb B.A., Witkowska H.E., and Barber D.L.. 2009. The sodium-hydrogen exchanger NHE1 is an Akt substrate necessary for actin filament reorganization by growth factors. J. Biol. Chem. 284:26666–26675. 10.1074/jbc.M109.019448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G., De Angelis D.A., and Rothman J.E.. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394:192–195. 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Kallay L., Brett C.L., and Rao R.. 2006. Mutational analysis of the intramembranous H10 loop of yeast Nhx1 reveals a critical role in ion homoeostasis and vesicle trafficking. Biochem. J. 398:97–105. 10.1042/BJ20060388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C.E., and Keller G.. 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 132:661–680. 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Nystul T., and Spradling A.. 2007. An epithelial niche in the Drosophila ovary undergoes long-range stem cell replacement. Cell Stem Cell. 1:277–285. 10.1016/j.stem.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Nystul T., and Spradling A.. 2010. Regulation of epithelial stem cell replacement and follicle formation in the Drosophila ovary. Genetics. 184:503–515. 10.1534/genetics.109.109538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., and Blelloch R.. 2014. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 14:617–631. 10.1016/j.stem.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks S.K., Chiche J., and Pouysségur J.. 2013. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat. Rev. Cancer. 13:611–623. 10.1038/nrc3579 [DOI] [PubMed] [Google Scholar]
- Perkins J.R., Dawes J.M., McMahon S.B., Bennett D.L.H., Orengo C., and Kohl M.. 2012. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics. 13 10.1186/1471-2164-13-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney L.K., and Barber D.L.. 2003. Na–H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem. 278:44645–44649. 10.1074/jbc.M308099200 [DOI] [PubMed] [Google Scholar]
- Putney L.K., Denker S.P., and Barber D.L.. 2002. The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42:527–552. 10.1146/annurev.pharmtox.42.092001.143801 [DOI] [PubMed] [Google Scholar]
- Rossano A.J., Chouhan A.K., and Macleod G.T.. 2013. Genetically encoded pH-indicators reveal activity-dependent cytosolic acidification of Drosophila motor nerve termini in vivo. J. Physiol. 591:1691–1706. 10.1113/jphysiol.2012.248377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönichen A., Webb B.A., Jacobson M.P., and Barber D.L.. 2013. Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 42:289–314. 10.1146/annurev-biophys-050511-102349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R. 2005. Ca2+ signaling, intracellular pH and cell volume in cell proliferation. J. Membr. Biol. 205:129–137. 10.1007/s00232-005-0778-z [DOI] [PubMed] [Google Scholar]
- Simons M., Gault W.J., Gotthardt D., Rohatgi R., Klein T.J., Shao Y., Lee H.-J., Wu A.-L., Fang Y., Satlin L.M., et al. 2009. Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat. Cell Biol. 11:286–294. 10.1038/ncb1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava J., Barreiro G., Groscurth S., Gingras A.R., Goult B.T., Critchley D.R., Kelly M.J.S., Jacobson M.P., and Barber D.L.. 2008. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc. Natl. Acad. Sci. USA. 105:14436–14441. 10.1073/pnas.0805163105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworoger M., Larkin M.K., Bryant Z., and Ruohola-Baker H.. 1999. Mosaic analysis in the Drosophila ovary reveals a common hedgehog-inducible precursor stage for stalk and polar cells. Genetics. 151:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.A., Chimenti M., Jacobson M.P., and Barber D.L.. 2011. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 11:671–677. 10.1038/nrc3110 [DOI] [PubMed] [Google Scholar]
- Wolfe D.M., Lee J.-H., Kumar A., Lee S., Orenstein S.J., and Nixon R.A.. 2013. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur. J. Neurosci. 37:1949–1961. 10.1111/ejn.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A.. 2008. The ground state of embryonic stem cell self-renewal. Nature. 453:519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. 2011. Control of the embryonic stem cell state. Cell. 144:940–954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.P., Ryu J.M., Kim M.O., Park J.H., and Han H.J.. 2012. Rapid actions of plasma membrane estrogen receptors regulate motility of mouse embryonic stem cells through a profilin-1/cofilin-1-directed kinase signaling pathway. Mol. Endocrinol. 26:1291–1303. 10.1210/me.2012-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Kalderon D.. 2000. Regulation of cell proliferation and patterning in Drosophila oogenesis by Hedgehog signaling. Development. 127:2165–2176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.