The mechanisms mediating actomyosin ring contraction during Drosophila cellularization, a developmental division that resembles cytokinesis, are unclear. Xue and Sokac delineate the contribution of cytoskeletal motors and actin-binding proteins to actomyosin ring constriction as Drosophila embryos undergo cleavage.
Abstract
Contraction of actomyosin rings during cytokinesis is typically attributed to actin filaments sliding toward each other via Myosin-2 motor activity. However, rings constrict in some cells in the absence of Myosin-2 activity. Thus, ring closure uses Myosin-2–dependent and –independent mechanisms. But what the Myosin-2–independent mechanisms are, and to what extent they are sufficient to drive closure, remains unclear. During cleavage in Drosophila melanogaster embryos, actomyosin rings constrict in two sequential and mechanistically distinct phases. We show that these phases differ in constriction speed and are genetically and pharmacologically separable. Further, Myosin-2 activity is required for slow constriction in “phase 1” but is largely dispensable for fast constriction in “phase 2,” and F-actin disassembly is only required for fast constriction in phase 2. Switching from phase 1 to phase 2 seemingly relies on the spatial organization of F-actin as controlled by Cofilin, Anillin, and Septin. Our work shows that fly embryos present a singular opportunity to compare separable ring constriction mechanisms, with varying Myosin-2 dependencies, in one cell type and in vivo.
Introduction
Morphogenesis, cell division, wound healing, and cell motility all require the physical contraction of cell surfaces by arrays of actin and Myosin-2 (actomyosin; Munjal and Lecuit, 2014). We often equate these contractions with muscle sarcomeres, where antiparallel actin filaments (F-actin) slide toward each other via Myosin-2 motor activity. However, the contractions that power shape change in many cells defy a sarcomere-like mechanism. For example, in the actomyosin rings that divide cells during cytokinesis, F-actin is not organized in antiparallel bundles but rather forms parallel and mixed polarity bundles and even disordered networks (Fishkind and Wang, 1993; Kamasaki et al., 2007; Reichl et al., 2008). Also, unlike sarcomeres, ring closure in cytokinesis has varying dependencies on Myosin-2. In some cells, reduced Myosin-2 function blocks cytokinesis (Zang et al., 1997; Lord et al., 2005; Mishra et al., 2013; Davies et al., 2014). In mouse cardiomyocytes, engineered Myosin-2 that binds F-actin but lacks motor activity is sufficient to drive ring closure (Ma et al., 2012). Finally, in budding yeast, Myosin-2 is dispensable for cytokinesis, and ring constriction is instead driven by F-actin disassembly (Lord et al., 2005; Mendes Pinto et al., 2012). Thus, ring closure during cytokinesis involves mechanisms that extend beyond the sarcomere model. However, the molecular details of these mechanisms are poorly understood. Also, it is not clear if individual mechanisms, like Myosin-2–independent and F-actin disassembly–dependent contraction, can each stand on their own or always need to be simultaneously coupled for efficient ring closure (Mendes Pinto et al., 2013).
Here, we investigate how actomyosin rings constrict during Drosophila melanogaster cellularization. The fly embryo first develops as a multinucleated cell, passing through 13 nuclear divisions with no intervening cytokinesis. Finally, in cell cycle 14, the embryo undergoes cellularization, whereby ∼6,000 peripherally anchored nuclei are simultaneously packaged into a sheet of cells that goes on to gastrulate and form the hatching larva (Schejter and Wieschaus, 1993). During cellularization, plasma membrane furrows invaginate from the embryo surface to build the lateral sides of cells. In three dimensions, these furrows are hollow columns of membrane that extend to enclose each nucleus (Fig. 1 A). An actomyosin ring forms at the leading edge of each membrane column and constricts perpendicular to furrows to build the bottom of each cell. The molecular composition of these rings is conserved with the contractile rings that drive cytokinesis, and cellularization resembles cytokinesis (Mazumdar and Mazumdar, 2002; Field et al., 2005; Mavrakis et al., 2014; Reversi et al., 2014; He et al., 2016). However, ring constriction in cellularization has not been described at high temporal or spatial resolution.
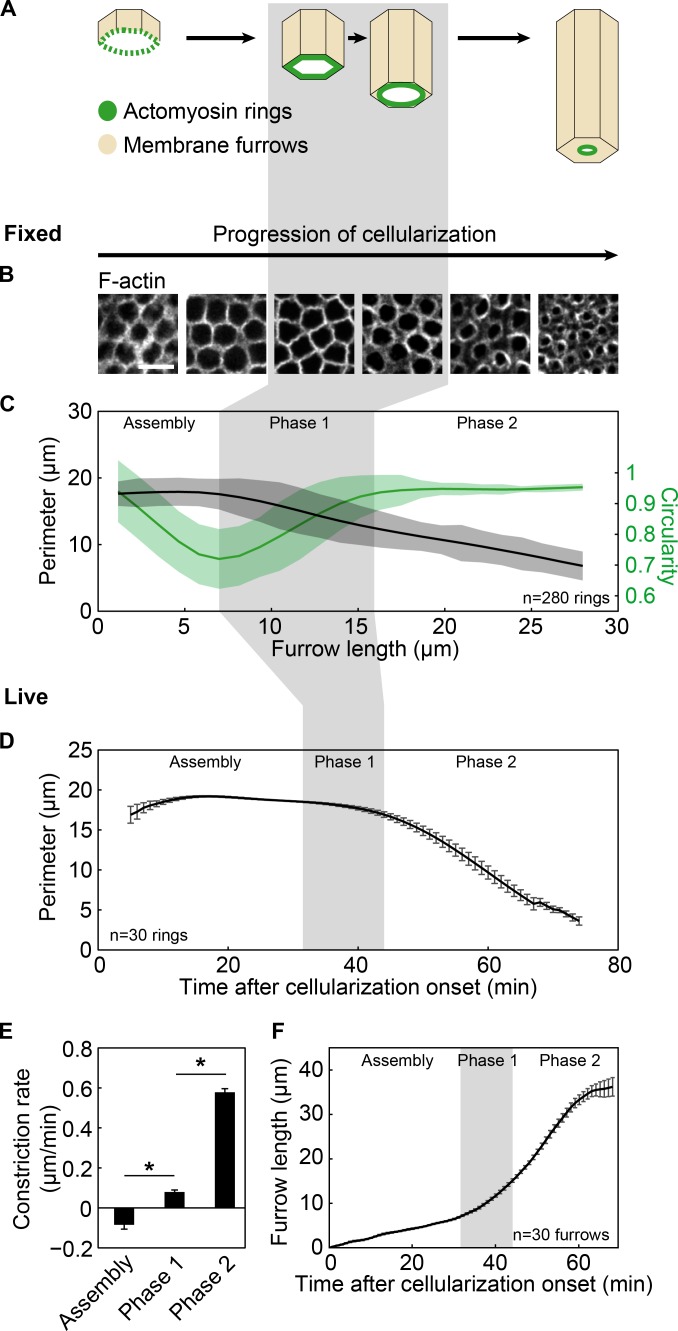
Figure 1.
Rings constrict in distinct morphological and kinetic phases. (A) Membrane furrows ingress during cellularization. An actomyosin ring (green) constricts to close the cell bottom. (B and C) Fixed wild-type embryos. (B) Rings in surface views (F-actin; phalloidin) at furrow lengths of 1, 4, 7, 16, 24, and 28 µm (left to right). Bar, 10 µm. (C) Ring perimeter (black) and circularity (green) versus furrow length (n = 56 embryos, five rings per embryo; the same trends were found in three independent experiments). Splines show trend of change for data points. Clouds show one SD from the moving mean. (D–F) Live wild-type embryos. (D) Ring perimeter versus time. Rings visualized with G-actinRed; see Video 1. (E) Constriction rate per phase. (F) Furrow length versus time. (A–D and F) Gray shading highlights phase 1. (D–F) n = 6 embryos, five rings or furrows per embryo; mean ± SE. *, P < 0.05.
Results and discussion
Rings constrict in morphologically and kinetically distinct phases
We tracked ring closure during cellularization in fixed wild-type embryos using phalloidin to visualize F-actin in rings (Fig. 1 B). We used increasing furrow length to order fixed embryos in developmental time because furrows ingress continuously and with highly reproducible kinetics throughout cellularization (Figard et al., 2013). Surface views of rings revealed changes in ring perimeter and shape as embryos cellularized (Fig. 1, B and C). Shape was measured as circularity (C), where C = 1 for a circle and C < 1 for angular shapes (e.g., C = 0.6 for a triangle; C = 4 πA/P2 [A, area; P, perimeter]; Thomas and Wieschaus, 2004). Plotting ring perimeter or circularity versus furrow length per embryo revealed distinct morphological phases of constriction (Fig. 1, B and C). In an initial “assembly” phase, ring boundaries transitioned from fuzzy to sharp, and ring perimeter did not change, although ring shape morphed from circular to more angular. In constriction “phase 1”, ring perimeter decreased and angular rings rounded into circles again. Finally, in constriction “phase 2”, circular rings shrunk.
We next asked how rates of ring closure in live embryos map to the morphological phases seen in fixed embryos. Rings were visualized in live wild-type embryos by incorporation of injected Rhodamine G-actin (G-actinRed; Fig. 1, D and E; and Video 1). G-actinRed injection did not obviously impede cellularization, as furrows ingressed at normal speed in these embryos (Fig. 1 F). To relate ring closure data from fixed and live imaging, we measured furrow length per embryo or furrow length per time point, respectively (Fig. 1 F). Furrow length in fixed embryos was then approximated to time within 1–2 min of error (Fig. 1, C and D; Lim et al., 2015). Based on this analysis, the transition from phase 1 to phase 2 was seen in live data as a switch from slow to fast constriction. We calculated constriction rates in a 10-min window in the middle of each phase, avoiding data points close to the transitions. During assembly, rings constricted at −0.09 ± 0.02 µm/min (mean ± standard error [SE]; Fig. 1 E), consistent with little perimeter change seen in fixed data. For phase 1, rings rounded up slowly at 0.08 ± 0.01 µm/min, followed by phase 2, when circular rings constricted faster at 0.58 ± 0.02 µm/min (mean ± SE; Fig. 1 E). Thus, rings constrict during cellularization in distinct morphological and kinetic phases.
Constriction phases correlate with distinct dynamics of ring components
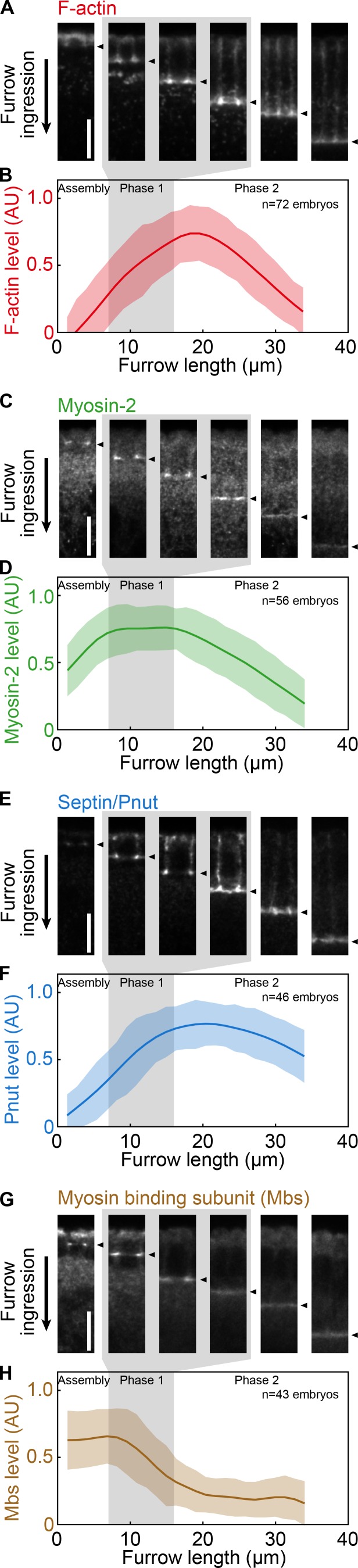
We wondered if distinct machinery drives each phase of ring constriction. In support of this possibility, we found differences in ring composition in phase 1 versus phase 2. In cross sections from fixed embryos (Fig. 2, A, C, E, and G), we measured protein levels in rings for F-actin, Myosin-2, the F-actin bundling and bending protein Septin (Drosophila Peanut [Pnut]; Adam et al., 2000; Mavrakis et al., 2014), and the Myosin-2 binding subunit of myosin phosphatase (Drosophila Mbs/MYPT1; Ong et al., 2010; Vasquez et al., 2014). Each protein displayed a unique profile of change in ring density with increasing furrow length, undergoing a switch in accumulation, maintenance, or depletion coincident with the onset of phase 1 and/or 2 (Fig. 2, A–H). These protein dynamics support that rings close in distinct phases. Because stoichiometric relationships differ between proteins per phase, the mechanisms that drive the phases may differ.
Figure 2.
Constriction phases correlate with distinct dynamics of ring components. (A–H) Fixed wild-type embryos. Gray shading highlights phase 1. (A, C, E, and G) Rings in cross section at increasing furrow lengths, stained for F-actin (phalloidin; A), Myosin-2 (C), Pnut (E), and Mbs (G). Arrowheads show ring position at furrow tips. Bars, 10 µm. (B, D, F, and H) Fluorescence intensity (arbitrary units [AU]) versus furrow length for F-actin (B), Myosin-2 (D), Pnut (F), and Mbs (H; n ≥ 43 embryos per staining and 10 ring profiles per embryo). Splines show trend of change for data points. Clouds show one SD from the moving mean.
Myosin-2 activity is important for ring constriction in phase 1 but is largely dispensable in phase 2
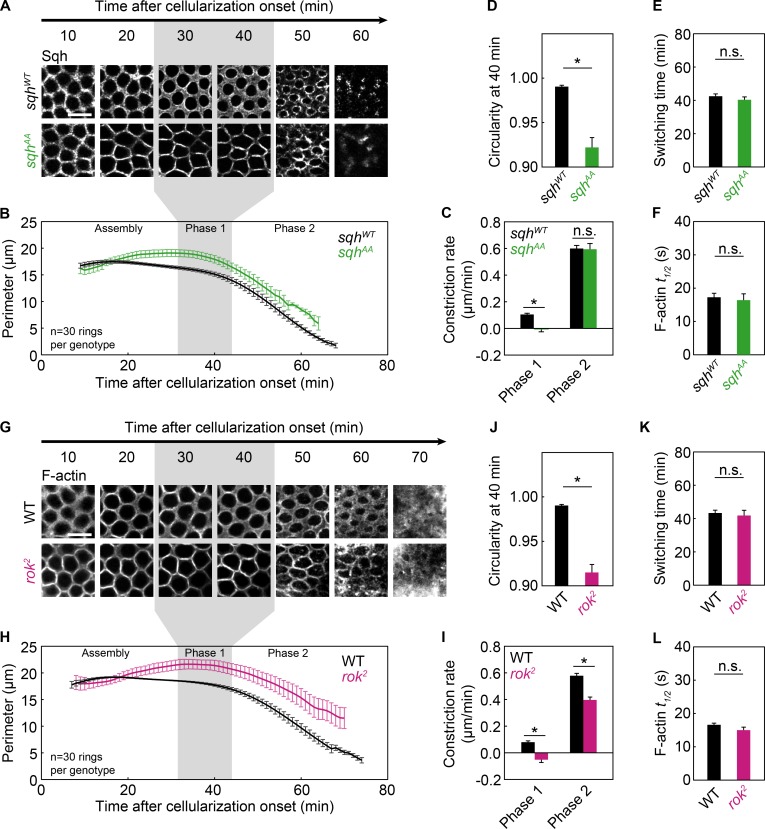
To determine the mechanisms of ring closure, we asked to what extent each phase depends on Myosin-2. To do so, we specifically inhibited Myosin-2. Normally, Rho kinase (Rok) activates Myosin-2 by phosphorylating myosin regulatory light chain (MRLC; Drosophila Spaghetti Squash [Sqh]) at threonine 20 (T20) and serine 21 (S21; Amano et al., 1996; Jordan and Karess, 1997; Ikebe, 2008). Phosphorylated MRLC/Sqh then stimulates motor ATPase activity and Myosin-2 bipolar filament formation (Sellers, 1991; Kamisoyama et al., 1994). So, to block Myosin-2 activity, we used flies expressing a mutated sqh transgene, with T20 and S21 replaced with nonphosphorylatable alanine residues (sqhAA; Vasquez et al., 2014). Control flies expressed wild-type sqh transgene (sqhWT). Mutant or wild-type transgenes were expressed in embryos where endogenous sqh levels are reduced by ∼90% (sqh1 germline clones), and sqhAA embryos display embryonic phenotypes that mimic sqh nulls (Vasquez et al., 2014).
Because the encoded SqhAA and SqhWT proteins are GFP tagged, we imaged rings in live embryos (Fig. 3 A). In sqhAA mutants, rings failed to round up in phase 1 (Fig. 3 A). The constriction rate, calculated in a 10-min window in the middle of the sqhAA phase 1, was negative, and circularity was reduced (Fig. 3, C and D). In contrast, sqhAA rings constricted at the sqhWT rate in phase 2 (Fig. 3, A–D). The switching times from phase 1 to phase 2 for sqhAA and sqhWT embryos, defined as the point of intersection between linear fits for phase 1 and 2 perimeter versus time data, were not significantly different (Fig. 3 E). To ask if perturbed Myosin-2 activity influenced F-actin disassembly, which is required for ring closure in some cells (Mendes Pinto et al., 2012), we measured F-actin turnover in G-actinRed–labeled rings by FRAP (Fig. S1, A–D; and Table S1). However, we found no change in the half-time to recovery (t1/2) or percent mobile fraction of F-actin in sqhAA rings (Fig. 3 F and Table S1). Although this sqhAA approach does not fully remove all Myosin-2 from embryos, our results suggest that Myosin-2 motor activity and/or F-actin cross-linking are important for constriction in phase 1 but are largely dispensable in phase 2.
Figure 3.
Myosin-2 is important for constriction in phase 1, but not in phase 2. (A–F) Live sqhWT (black) and sqhAA (green) embryos. (G–L) Live wild-type (WT; black) and rok2 (pink) embryos. (A and G) Rings visualized with Sqh-GFP (A) or G-actinRed (G) constricting over time (minutes). Bars, 10 µm. (B and H) Ring perimeter versus time. (C and I) Constriction rate per phase. (D and J) Circularity 40 min after cellularization onset. (E and K) Switching time from phase 1 to phase 2. (F and L) t1/2 for F-actin in photobleached rings (G-actinRed; F, n ≥ 10 rings from four or more embryos per genotype; L, n ≥ 26 rings from ≥12 embryos per genotype; mean ± SE; see Fig. S1 and Table S1). (A, B, G, and H) Gray shading highlights phase 1 in wild-type embryos. (G) Wild-type images are the same as in Fig. 4 A and Fig. 5 (A and G). (B–E and H–K) n = 6 embryos per genotype, 5 rings per embryo; mean ± SE. (C–F and I–L) *, P < 0.05; n.s., not significant.
To confirm the varying dependencies on Myosin-2 per phase, we inhibited Myosin-2 in another way. Although less direct than targeting Myosin-2 itself, we disrupted Rok. Rok localizes to rings during cellularization (Vasquez et al., 2014) and is required for Myosin-2 localization to cortical structures (Royou et al., 2004; Dawes-Hoang et al., 2005; Fernandez-Gonzalez and Zallen, 2013; Mason et al., 2013; Vasquez et al., 2014). Accordingly, Myosin-2 was undetectable in rings in rok mutants (rok2 germline clones; Fig. S2, A–D). To visualize rings in live rok2 embryos, we injected G-actinRed. Similar to sqhAA results, constriction in rok2 rings was inhibited in phase 1 but occurred at a rate approaching wild-type in phase 2 (Fig. 3, G–I). Because constriction in phase 1 failed, circularity remained low after phase 1 (Fig. 3 J). Consistent with sqhAA, the switching time to phase 2 and the rate of F-actin turnover did not change significantly in rok2 mutants (Fig. 3, K and L). Thus, rok inhibition and coincident loss of Myosin-2 from rings blocked constriction in phase 1, but not phase 2.
We noted, however, that unlike sqhAA mutants, rok2 embryos showed reduced ring closure speed in phase 2. We wondered if this phenotype might stem from Rok’s action on downstream targets besides MRLC/Sqh, including several F-actin regulators (Amano et al., 2010; Mason et al., 2013). Indeed, F-actin levels in rok2 mutants were reduced in phase 2 (Fig. S2 E), suggesting that the constriction mechanism in phase 2 has specific F-actin requirements.
Cofilin, but not F-actin disassembly, promotes timely switching from phase 1 to phase 2
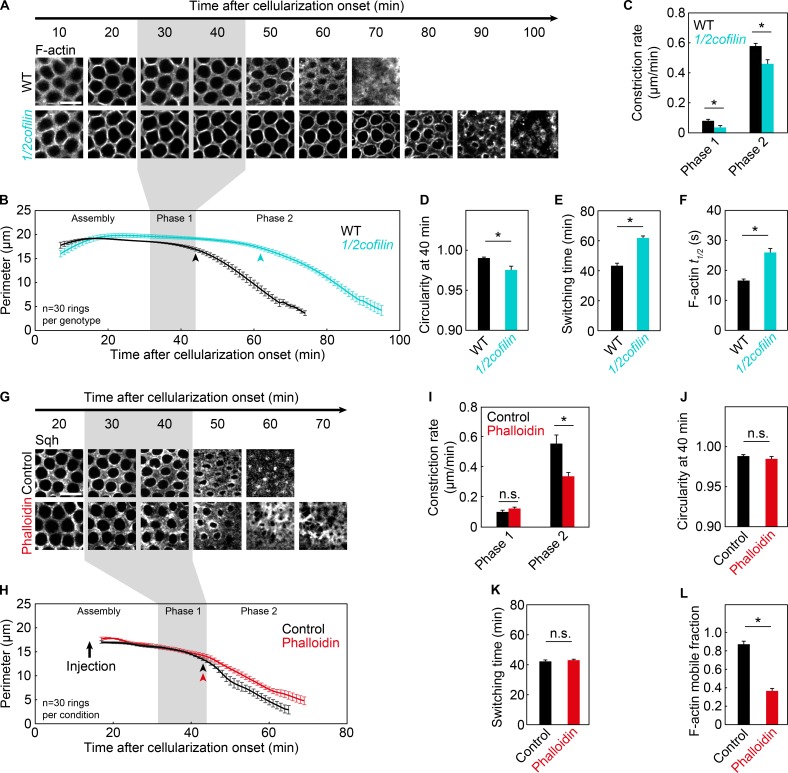
To identify these F-actin requirements, we focused on the role of Cofilin. Cofilin binds and severs F-actin to promote filament disassembly and turnover (Andrianantoandro and Pollard, 2006; McCullough et al., 2008). Cofilin-mediated F-actin disassembly is necessary for Myosin-2–independent ring constriction in budding yeast, and Cofilin modifies the spatial organization and connectivity of F-actin in rings to speed up contractility in vitro (Mendes Pinto et al., 2012, 2013; Ennomani et al., 2016). Thus, we assayed ring closure in embryos with reduced Cofilin. We collected embryos from mothers heterozygous for a loss-of-function allele of cofilin (Drosophila twinstar [tsr1]; Gunsalus et al., 1995), thus reducing maternal Cofilin dosage by half (1/2cofilin). By FRAP, 1/2cofilin embryos showed reduced F-actin disassembly in rings (Fig. 4 F; Fig. S1, E and F; and Table S1).
Figure 4.
Cofilin, but not F-actin disassembly, controls timely switching to phase 2. (A–F) Live wild-type (WT; black) and 1/2cofilin (turquoise) embryos. (G–L) Live DMSO (control; black) and phalloidin-injected (phalloidin; red) embryos. (A and G) Rings visualized with G-actinRed (A) or Sqh-GFP (G), constricting over time (minutes). Bars, 10 µm. (B and H) Ring perimeter versus time. (C and I) Constriction rate per phase. (D and J) Circularity 40 min after cellularization onset. (E and K) Switching time from phase 1 to phase 2. (F and L) t1/2 (F) or percent mobile fraction (L) for F-actin in photobleached rings (G-actinRed; F, n ≥ 26 rings from ≥13 embryos per genotype; L, n ≥ 7 rings from ≥7 embryos per condition; mean ± SE; see Fig. S1, E–H; and Table S1). (A, B, G, and H) Gray shading highlights phase 1 in wild-type embryos. (B and H) Arrowheads indicate switching time. (H) Arrow indicates injection time. (A) Wild-type images are the same as in Fig. 3 G and Fig. 5 (A and G). (B–E and H–K) n = 6 embryos per genotype or condition, five rings per embryo; mean ± SE. (C–F and I–L) *, P < 0.05; n.s., not significant.
To visualize rings in 1/2cofilin embryos, we used G-actinRed (Fig. 4 A). We saw rings constrict in phases 1 and 2 in these embryos (Fig. 4, B and C). However, circularity was low after phase 1, reflecting reduced constriction in phase 1 (Fig. 4, C and D). In addition, fast constriction in phase 2 was significantly slower (Fig. 4 C), and switching from phase 1 to phase 2 was delayed by ∼20 min (Fig. 4 E). We interpret the latter change as a delay rather than no phase 1, because we did see slow constriction in phase 1. Interestingly, this was the first genotype to show delayed switching between constriction phases.
Because Cofilin severs and disassembles F-actin, we hypothesized that phase 1 and 2 constriction, as well as switching between them, depends on F-actin disassembly. To test this, we blocked F-actin disassembly with phalloidin. To visualize rings, embryos expressing SqhWT-GFP were used and injected during ring assembly (Fig. 4, G–L). By FRAP, we confirmed that phalloidin injection stabilized F-actin (Fig. 4 L; Fig. S1, G and H; and Table S1). In fact, the percent mobile fraction of F-actin in these rings was so low that we could not reliably measure t1/2 (Fig. 4 L and Table S1). However, ring constriction in phase 1 was indistinguishable from controls, and switching from phase 1 to phase 2 happened at the normal time (Fig. 4, H–K). The only change was reduced phase 2 constriction speed (Fig. 4, H and I). To make sure our results were not simply caused by a lag between phalloidin injection and its effect on constriction, we repeated the analysis, injecting phalloidin at the onset of phase 2. Again, constriction speed in phase 2 was reduced (Fig. S3, A–C). These results suggest that F-actin disassembly is not required for Myosin-2–dependent constriction in phase 1 or for timely switching from phase 1 to phase 2. However, F-actin disassembly is required for normal constriction in phase 2.
F-actin architecture may control timely switching from phase 1 to phase 2
A molecular activity of Cofilin besides F-actin disassembly seems to promote switching to the phase 2 constriction mechanism. In fact, Cofilin localization to rings is most prominent in phase 1, when F-actin is still accumulating in rings (Fig. S3, D and E). Cofilin debranches F-actin in networks to promote bundling (Chan et al., 2009; Ennomani et al., 2016), makes F-actin mechanically compliant to bending and twisting (Prochniewicz et al., 2005; McCullough et al., 2008; De La Cruz, 2009), influences filament length via severing, and may even bundle F-actin itself (Bamburg and Bernstein, 2016). These activities could cumulatively impact F-actin packing and bending in rings. Thus, we hypothesized that switching from slow to fast constriction depends on the structure and organization of F-actin in rings.
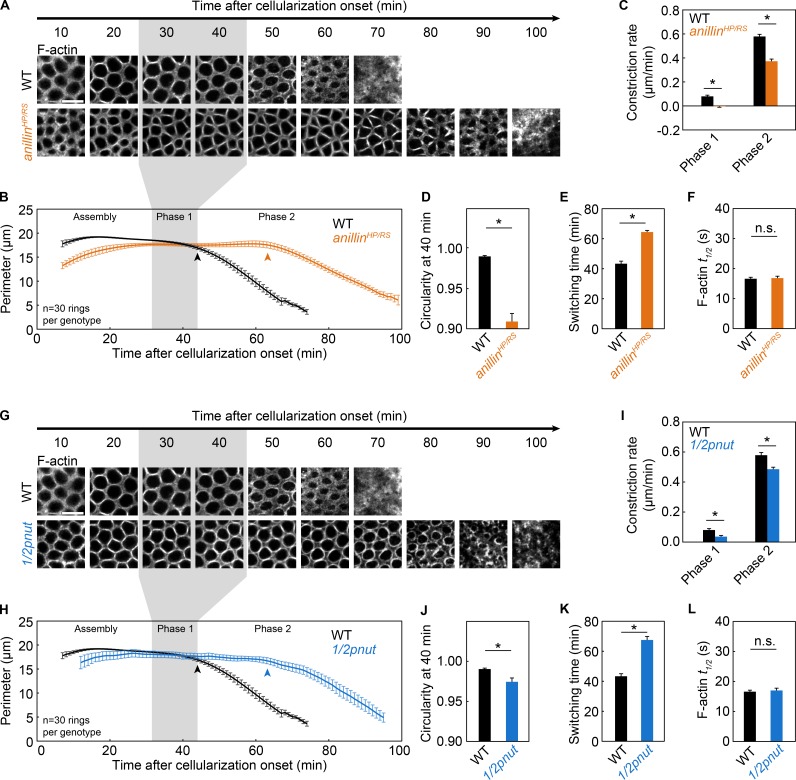
We assayed switching times in mutants where F-actin organization is known to be perturbed. Anillin (Drosophila Scraps/Anillin) is an F-actin cross-linking protein that localizes to rings during cellularization and cytokinesis (Field and Alberts, 1995; Field et al., 2005; Piekny and Maddox, 2010). For cellularization, Anillin recruits Septin/Pnut to rings, and in anillin mutants, F-actin forms aberrant bar-like structures in rings (Field et al., 2005). We prepared mutants transheterozygous for loss-of-function alleles of anillin (anillinHP/RS; Field et al., 2005), and confirmed that Septin/Pnut failed to accumulate in these rings (Fig. S2, G and H). To visualize rings in live anillinHP/RS embryos, we injected G-actinRed (Fig. 5 A). Ring constriction was perturbed in both phase 1 and phase 2 in the anillinHP/RS embryos (Fig. 5 C). Most notably, switching to phase 2 was delayed by ∼20 min (Fig. 5, B and E). In contrast to 1/2cofilin embryos, however, the delay in anillinHP/RS embryos was accompanied by little change in the rate of F-actin turnover (Fig. 5 F and Table S1). Thus, as with phalloidin, there was no correlation between reduced F-actin disassembly and delayed switching. Instead, Anillin may impact the switch via its influence over some aspect of F-actin organization.
Figure 5.
Anillin and Septin control timely switching to phase 2. (A–F) Live wild-type (WT; black) and anillinHP/RS (orange) embryos. (G–L) Live wild-type (WT; black) and 1/2pnut (blue) embryos. (A and G) Rings visualized with G-actinRed constricting over time (minutes). Bars, 10 µm. (B and H) Ring perimeter versus time. (C and I) Constriction rate per phase. (D and J) Circularity 40 min after cellularization onset. (E and K) Switching time from phase 1 to phase 2. (F and L) t1/2 for F-actin in photobleached rings (G-actinRed; F, n ≥ 35 rings from ≥14 embryos per genotype; L, n ≥ 16 rings from ≥6 embryos per genotype; mean ± SE; see Table S1). (A, B, G, and H) Gray shading highlights phase 1 in wild-type embryos. (B and H) Arrowheads indicate switching time. (A and G) Wild-type images are the same as in Figs. 3 G and 4 A. (B–E and H–K) n = 6 embryos per genotype and five rings were measured per embryo; mean ± SE. (C–F and I–L) *, P < 0.05; n.s., not significant.
During cellularization, Septin/Pnut cross-links F-actin in rings into tight parallel bundles, and in vitro, Septin/Pnut promotes curvature of F-actin bundles (Mavrakis et al., 2014). Given that Septin/Pnut fails to localize to rings in anillinHP/RS embryos (Fig. S2, G and H; Field et al., 2005), we directly tested the role of Septin/Pnut in switching from phase 1 to phase 2. We collected embryos from mothers heterozygous for a loss-of-function allele of pnut (pnut02502; Schnorr et al., 2001), reducing the maternal dose of pnut by half (1/2pnut; Fig. S2, I and J). To visualize rings in live 1/2pnut embryos, we injected G-actinRed (Fig. 5 G). Like anillinHP/RS embryos, constriction was disrupted in phases 1 and 2 in 1/2pnut embryos (Fig. 5, I and J), and switching was delayed by ∼20 min (Fig. 5, H and K). We found no change in F-actin turnover (Fig. 5 L and Table S1). Thus, the delay is not associated with a change in F-actin disassembly. Instead, we suggest that the spatial organization of F-actin (e.g., bundling or bending) is critical for timely switching from slow to fast constriction at the transition from phase 1 to phase 2. We envision a model whereby Cofilin, Anillin, and Septin/Pnut control switching time, because they promote this optimal F-actin organization within rings.
In summary, distinct mechanisms drive back-to-back phases of actomyosin ring constriction in cellularization. These mechanisms do not require coupling, as we found genetic and pharmacological perturbations that selectively disrupted one constriction phase while leaving the other intact with its normal wild-type kinetics. To our knowledge, this is the first example where sequential and separable mechanisms of varying Myosin-2 dependencies drive one ring closure event. Adding to what is already known from other cell types, actomyosin rings seem to combine mechanisms in all possible ways, where constriction may be powered by a single mechanism or by multiple mechanisms that either overlap at the same time or occur in sequence (Neujahr et al., 1997; Zang et al., 1997; Lord et al., 2005; Reichl et al., 2008; Burkel et al., 2012; Ma et al., 2012; Mendes Pinto et al., 2012, 2013; Mishra et al., 2013; Davies et al., 2014; Oelz and Mogilner, 2016). Per contractile event, different combinations of mechanisms may have evolved to meet specific mechanical requirements, to increase robustness, or, as suggested here, to tune the kinetics of ring closure. For cellularization, the juxtaposition of phases 1 and 2 will now allow a unique viewing of contraction mechanisms. Our data suggest that Myosin-2–dependent contraction in phase 1 and F-actin disassembly–dependent contraction in phase 2 are distinct mechanisms that can stand alone and need not overlap, as suggested by prior experiments and theory. In addition, during cellularization, F-actin disassembly–dependent contraction proceeds with little or no Myosin-2 cross-linking activity, reinforcing the need to consider roles for alternative F-actin cross-linkers in this mechanism.
Materials and methods
Fly stocks
The wild-type stock was OreR. The sqhAA, sqhWT, and rok2 germline clones were generated by standard Flipase-dominant female sterile methods with published stocks (Chou and Perrimon, 1992; Winter et al., 2001; Verdier et al., 2006; Vasquez et al., 2014). For sqh germline clones, sqh1, FRT101/FM7; sqhAA-gfp/CyO or sqh1, FRT101/FM7; sqhWT-gfp/CyO (provided by A. Martin, Massachusetts Institute of Technology, Cambridge, MA) were crossed with w* ovoD1 v24 P{FRT(whs)}101/C(1)DX, y1 f1/Y; P{hsFLP}38 (FBst0001813; Vasquez et al., 2014). For rok2 germline clones, y1 w1118 Rok2 P{neoFRT}19A/FM7c (FBst0006666) was crossed with P{ovoD1-18}P4.1, P{hsFLP}12, y1 w1118 sn3 P{neoFRT}19A/C(1)DX, y1 w1 f1 (FBst0023880; Winter et al., 2001; Verdier et al., 2006). For phalloidin injections, embryos were collected from sqhAX3; sqh-gfp, which expresses GFP-tagged wild-type Sqh in a sqh mutant background (Royou et al., 2004). For manipulation of maternal dosage of Cofilin and Pnut, 1/2cofilin and 1/2pnut embryos were collected from y1 w*; P{FRT(whs)}G13 P{A92}tsr1/CyO, P{sevRas1.V12}FK1 (FBst0009107) and cn1 P{PZ}pnut02502/CyO; ry506 (FBst0011194) mothers, respectively. This loss-of-function strategy was previously established for F-actin regulators that are maternally provided for cellularization (Zheng et al., 2013). For anillin maternal effect mutants, anillinHP/CyO males and anillinRS/CyO females were crossed to generate anillinHP/RS transheterozygous females, which were crossed with anillinHP/CyO males (Field et al., 2005).
Embryo fixation and staining
For the staining of Mbs, embryos were fixed in 16% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4)/heptane (1:1) for 20 min at room temperature. For all other stainings, embryos were fixed in 16% paraformaldehyde in PBS/heptane (1:1) for 20 min at room temperature. After both fixation methods, embryos were hand-peeled. For primary antibodies, secondary antibodies, and phalloidin, staining conditions are listed in Table S2. The Mbs antibody was a gift from C. Tan (University of Missouri, Columbia, MO).
Microinjection
Embryos were dechorionated in 50% bleach for 1.5 min, washed with water, and mounted on their lateral side with embryo glue (heptane incubated overnight with double-stick tape). Embryos were then desiccated for 6 min, covered with halocarbon oil 700:27 (3:1), and injected at the midpoint of their ventral side as follows. For phalloidin and DMSO injection during assembly or in phase 2, the injection times were 10 min and 40 min after cellularization onset, respectively. For G-actinRed injection, the injection time was 1 h before cellularization onset to allow complete diffusion throughout the embryo. Concentrations were 150 µg/ml phalloidin (EMD Millipore) in 5 mM KCl, 0.1 mM NaP, pH 7, 2% DMSO, and 5 mg/ml G-actinRed (Cytoskeleton, Inc.) in 5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, and 0.2 mM ATP.
Image acquisition
For fixed embryo imaging, single-plane images were collected on a confocal microscope (LSM710; ZEISS) using a 40× water-immersion objective, NA 1.2, with the pinhole set to one airy unit, and resolution of 1,024 × 1,024 pixels. For live-embryo imaging, the microscope and settings were the same except imaging was done on a 25°C heated stage. Mounting media was Aqua-Poly/Mount (Polysciences) for fixed embryos and halocarbon oil (Sigma-Aldrich) for live embryos. To follow constriction rates, time-lapse images were collected en face (i.e., in surface views) at 2.5-min intervals. This interval allowed refocusing of the imaging plane as the rings moved progressively deeper into the embryo. To follow FRAP, time-lapse images were collected in cross section at 1-s intervals. A 1-µm × 1.5-µm box, encompassing a ring in profile, was photobleached with a 561-nm laser at 100% power for 2 s, to reach ∼50% of the original fluorescence intensity. Recovery of the bleached ring was monitored for the next 150 s, until the fluorescence intensity stabilized at a plateau. Three unbleached rings were simultaneously monitored.
Image analysis
Quantification of fluorescence intensity for ring components in fixed embryos was done with custom MATLAB code (Sokac and Wieschaus, 2008), which uses thresholding to identify rings in cross section images. Rings were then hand-selected to avoid falsely identified objects, and their mean intensity was calculated. For each embryo, ∼10 rings were measured. Quantification of fluorescence intensity for FRAP was done using ImageJ.
Measurements of furrow length, ring perimeter, and ring circularity were done manually using ImageJ, except where furrow ingression rate was measured and analyzed with custom MATLAB code as previously described (Figard et al., 2013). For fixed embryos, furrow length was measured in a cross section collected at the embryo midplane by drawing a straight line from the embryo surface to furrow tip, and the mean length was calculated from five furrows per embryo. For fixed and live embryos, perimeter and circularity were measured from surface view images by tracing the inner rim of each ring with the Polygon Tool in ImageJ, and mean values were calculated from five rings per embryo. Because fixed embryos shrink, the measurements of furrow length and perimeter were multiplied by a shrinkage factor of 1.36 for fixed embryos, which was determined by comparing the maximum furrow lengths observed from fixed and live embryos. Note that the circularity versus time profile for live wild-type embryos does not completely recapitulate the fixed circularity data. Although the overall trends of circularity change are the same for fixed and live data (i.e., rings become more circular as they constrict), the live circularity measurement does not readily detect the transitions from assembly to phase 1 or from phase 1 to phase 2. The circularity measurement displays limited sensitivity in detecting small changes in shapes that are close to circular, and rings in wild-type embryos approximate circles throughout cellularization. For live wild-types embryos, circularity values are close to one and deviate by only 2%, over all of cellularization, making it difficult to detect meaningful changes (C = 0.970 ± 0.002 or C = 0.988 ± 0.002 at the lowest or highest value near the beginning or end of cellularization, respectively; mean ± SE; n = 30 rings from six embryos). In addition, circularity differences are accentuated in the fixed imaging. We suggest this is because embryos shrink when fixed, and the ring morphologies seen after shrinkage reveal some underlying property of the rings. This property is likely mechanical, and may even reflect differences in the levels of tension in the rings (Thomas and Wieschaus, 2004).
Data analysis
For ring constriction rate, perimeter versus time measurements per ring were used to generate a continuous curve. Five rings were measured per embryo. All five curves from one embryo were averaged. To find the switching time between phase 1 and phase 2 per live embryo, the mean curve was fitted with a two-line model. The switching point was determined to be the point on the curve closest to the intersection of the two lines. Once the switching time was found for wild-type, sqhAA-gfp, sqhWT-gfp, rok2, phalloidin-injected, and DMSO-injected embryos, the ring constriction rate of each phase was calculated by averaging the derivative of the perimeter curves within a 10-min time window per phase. The phase 1 window was set to end 5 min before the switching point, and the phase 2 window was set to start 8 min after the switching point. For 1/2cofilin, anillinHP/RS, and 1/2pnut embryos, where switching was delayed, the phase 2 window was set the same way as for all other genotypes; however, the phase 1 window was set from 30 to 40 min after cellularization onset, which is the wild-type phase 1 time range.
For change in ring size, shape, and fluorescence intensity versus furrow length for fixed embryos, the mean perimeter, circularity, or intensity value for each embryo was plotted against the mean furrow length of that embryo. Plots were generated using the “curve fitting tool” in MATLAB, according to the “smoothing spline” method with the smoothing parameter P = 0.05. The shaded region around the spline was generated by calculating the standard deviation of all data points within a 10-µm-wide bin centered on any given furrow length, with ∼30 overlapping bins moving along the x axis.
For FRAP, the fluorescence intensity of the bleached ring (Ifrap) and the mean intensity of three reference unbleached rings (Iref) were normalized by using the Phair’s double normalization (Phair et al., 2004): Inorm = (Ifrap/Ifrap-pre)/(Iref/Iref-pre), where Ifrap-pre and Iref-pre are two constants, representing the prebleach intensity of Ifrap and Iref, respectively, and Inorm is the normalized intensity of the bleached ring. The Inorm versus time curve was then fitted with a single exponential equation: Inorm = I∞ − (I∞ − I0) × e(−k × t), where t represents time and t = 0 is the end of bleaching, I0 = Inorm (t = 0), I∞ = Inorm (t = ∞), and k is the reaction rate constant of the intensity recovery process. Therefore, the half-time to recovery was calculated by t1/2 = ln(2)/k, and the mobile fraction was calculated by mobile fraction = (I∞ − I0)/(1 − I0). A small number of rings were discarded from the analysis if the fit had an r2 < 0.6.
Statistics
P-values were calculated using the two-tailed unpaired Student’s t test, with equal variances. All error bars are mean ± SE.
Online supplemental material
Fig. S1 shows that F-actin in rings turns over throughout cellularization, but turnover (i.e., disassembly) is reduced in 1/2cofilin and phalloidin-injected embryos. Fig. S2 shows disruption of various ring components in rok2, anillinHP/RS, and 1/2pnut embryos. Fig. S3 shows that phalloidin injection at the onset of phase 2 slows down ring constriction in phase 2 and that Cofilin localization to rings is most prominent in phase 1. Video 1 shows ring constriction in a wild-type embryo, visualized by injection of G-actinRed. Table S1 contains the t1/2 values and percent mobile fraction for F-actin in rings from all FRAP experiments. Table S2 contains the staining conditions for antibodies and probes.
Supplementary Material
Acknowledgments
We thank I. Golding, H. Xu, and L. Sepúlveda (Baylor College of Medicine) for help with data analysis; W. Bement (University of Wisconsin) for discussing results; and A. Martin and C. Tan for reagents. The Baylor College of Medicine Computational and Integrative Biomedical Research Center provided computing resources.
The work was supported by National Institutes of Health grant R01 GM115111 (A.M. Sokac).
The authors declare no competing financial interests.
Footnotes
Abbreviation used:
- SE
- standard error
References
- Adam J.C., Pringle J.R., and Peifer M.. 2000. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol. Biol. Cell. 11:3123–3135. 10.1091/mbc.11.9.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., and Kaibuchi K.. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249. 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- Amano M., Nakayama M., and Kaibuchi K.. 2010. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken). 67:545–554. 10.1002/cm.20472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., and Pollard T.D.. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23. 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R., and Bernstein B.W.. 2016. Actin dynamics and cofilin-actin rods in alzheimer disease. Cytoskeleton (Hoboken). 73:477–497. 10.1002/cm.21282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel B.M., Benink H.A., Vaughan E.M., von Dassow G., and Bement W.M.. 2012. A Rho GTPase signal treadmill backs a contractile array. Dev. Cell. 23:384–396. 10.1016/j.devcel.2012.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Beltzner C.C., and Pollard T.D.. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19:537–545. 10.1016/j.cub.2009.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.B., and Perrimon N.. 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 131:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T., Jordan S.N., Chand V., Sees J.A., Laband K., Carvalho A.X., Shirasu-Hiza M., Kovar D.R., Dumont J., and Canman J.C.. 2014. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev. Cell. 30:209–223. 10.1016/j.devcel.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes-Hoang R.E., Parmar K.M., Christiansen A.E., Phelps C.B., Brand A.H., and Wieschaus E.F.. 2005. folded gastrulation, cell shape change and the control of myosin localization. Development. 132:4165–4178. 10.1242/dev.01938 [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M. 2009. How cofilin severs an actin filament. Biophys. Rev. 1:51–59. 10.1007/s12551-009-0008-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennomani H., Letort G., Guérin C., Martiel J.L., Cao W., Nédélec F., De La Cruz E.M., Théry M., and Blanchoin L.. 2016. Architecture and connectivity govern actin network contractility. Curr. Biol. 26:616–626. 10.1016/j.cub.2015.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., and Zallen J.A.. 2013. Wounded cells drive rapid epidermal repair in the early Drosophila embryo. Mol. Biol. Cell. 24:3227–3237. 10.1091/mbc.E13-05-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C.M., and Alberts B.M.. 1995. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 131:165–178. 10.1083/jcb.131.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C.M., Coughlin M., Doberstein S., Marty T., and Sullivan W.. 2005. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development. 132:2849–2860. 10.1242/dev.01843 [DOI] [PubMed] [Google Scholar]
- Figard L., Xu H., Garcia H.G., Golding I., and Sokac A.M.. 2013. The plasma membrane flattens out to fuel cell-surface growth during Drosophila cellularization. Dev. Cell. 27:648–655. 10.1016/j.devcel.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkind D.J., and Wang Y.L.. 1993. Orientation and three-dimensional organization of actin filaments in dividing cultured cells. J. Cell Biol. 123:837–848. 10.1083/jcb.123.4.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K.C., Bonaccorsi S., Williams E., Verni F., Gatti M., and Goldberg M.L.. 1995. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131:1243–1259. 10.1083/jcb.131.5.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Martin A., and Wieschaus E.. 2016. Flow-dependent myosin recruitment during Drosophila cellularization requires zygotic dunk activity. Development. 143:2417–2430. 10.1242/dev.131334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe M. 2008. Regulation of the function of mammalian myosin and its conformational change. Biochem. Biophys. Res. Commun. 369:157–164. 10.1016/j.bbrc.2008.01.057 [DOI] [PubMed] [Google Scholar]
- Jordan P., and Karess R.. 1997. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J. Cell Biol. 139:1805–1819. 10.1083/jcb.139.7.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., and Mabuchi I.. 2007. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 178:765–771. 10.1083/jcb.200612018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisoyama H., Araki Y., and Ikebe M.. 1994. Mutagenesis of the phosphorylation site (serine 19) of smooth muscle myosin regulatory light chain and its effects on the properties of myosin. Biochemistry. 33:840–847. 10.1021/bi00169a027 [DOI] [PubMed] [Google Scholar]
- Lim B., Dsilva C.J., Levario T.J., Lu H., Schüpbach T., Kevrekidis I.G., and Shvartsman S.Y.. 2015. Dynamics of inductive ERK signaling in the Drosophila embryo. Curr. Biol. 25:1784–1790. 10.1016/j.cub.2015.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Laves E., and Pollard T.D.. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 16:5346–5355. 10.1091/mbc.E05-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Kovács M., Conti M.A., Wang A., Zhang Y., Sellers J.R., and Adelstein R.S.. 2012. Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc. Natl. Acad. Sci. USA. 109:4509–4514. 10.1073/pnas.1116268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason F.M., Tworoger M., and Martin A.C.. 2013. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol. 15:926–936. 10.1038/ncb2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M., Azou-Gros Y., Tsai F.C., Alvarado J., Bertin A., Iv F., Kress A., Brasselet S., Koenderink G.H., and Lecuit T.. 2014. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat. Cell Biol. 16:322–334. 10.1038/ncb2921 [DOI] [PubMed] [Google Scholar]
- Mazumdar A., and Mazumdar M.. 2002. How one becomes many: blastoderm cellularization in Drosophila melanogaster. BioEssays. 24:1012–1022. 10.1002/bies.10184 [DOI] [PubMed] [Google Scholar]
- McCullough B.R., Blanchoin L., Martiel J.L., and De La Cruz E.M.. 2008. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J. Mol. Biol. 381:550–558. 10.1016/j.jmb.2008.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes Pinto I., Rubinstein B., Kucharavy A., Unruh J.R., and Li R.. 2012. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell. 22:1247–1260. 10.1016/j.devcel.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes Pinto I., Rubinstein B., and Li R.. 2013. Force to divide: structural and mechanical requirements for actomyosin ring contraction. Biophys. J. 105:547–554. 10.1016/j.bpj.2013.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Kashiwazaki J., Takagi T., Srinivasan R., Huang Y., Balasubramanian M.K., and Mabuchi I.. 2013. In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat. Cell Biol. 15:853–859. 10.1038/ncb2781 [DOI] [PubMed] [Google Scholar]
- Munjal A., and Lecuit T.. 2014. Actomyosin networks and tissue morphogenesis. Development. 141:1789–1793. 10.1242/dev.091645 [DOI] [PubMed] [Google Scholar]
- Neujahr R., Heizer C., and Gerisch G.. 1997. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 110:123–137. [DOI] [PubMed] [Google Scholar]
- Oelz D., and Mogilner A.. 2016. Actomyosin contraction, aggregation and traveling waves in a treadmilling actin array. Physica D. 318-319:70–83. 10.1016/j.physd.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S., Foote C., and Tan C.. 2010. Mutations of DMYPT cause over constriction of contractile rings and ring canals during Drosophila germline cyst formation. Dev. Biol. 346:161–169. 10.1016/j.ydbio.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Gorski S.A., and Misteli T.. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414. 10.1016/S0076-6879(03)75025-3 [DOI] [PubMed] [Google Scholar]
- Piekny A.J., and Maddox A.S.. 2010. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 21:881–891. 10.1016/j.semcdb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Prochniewicz E., Janson N., Thomas D.D., and De La Cruz E.M.. 2005. Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 353:990–1000. 10.1016/j.jmb.2005.09.021 [DOI] [PubMed] [Google Scholar]
- Reichl E.M., Ren Y., Morphew M.K., Delannoy M., Effler J.C., Girard K.D., Divi S., Iglesias P.A., Kuo S.C., and Robinson D.N.. 2008. Interactions between myosin and actin crosslinkers control cytokinesis contractility dynamics and mechanics. Curr. Biol. 18:471–480. 10.1016/j.cub.2008.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversi A., Loeser E., Subramanian D., Schultz C., and De Renzis S.. 2014. Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J. Cell Biol. 205:395–408. 10.1083/jcb.201309079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schejter E.D., and Wieschaus E.. 1993. Functional elements of the cytoskeleton in the early Drosophila embryo. Annu. Rev. Cell Biol. 9:67–99. 10.1146/annurev.cb.09.110193.000435 [DOI] [PubMed] [Google Scholar]
- Schnorr J.D., Holdcraft R., Chevalier B., and Berg C.A.. 2001. Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics. 159:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers J.R. 1991. Regulation of cytoplasmic and smooth muscle myosin. Curr. Opin. Cell Biol. 3:98–104. 10.1016/0955-0674(91)90171-T [DOI] [PubMed] [Google Scholar]
- Sokac A.M., and Wieschaus E.. 2008. Local actin-dependent endocytosis is zygotically controlled to initiate Drosophila cellularization. Dev. Cell. 14:775–786. 10.1016/j.devcel.2008.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.H., and Wieschaus E.. 2004. src64 and tec29 are required for microfilament contraction during Drosophila cellularization. Development. 131:863–871. 10.1242/dev.00989 [DOI] [PubMed] [Google Scholar]
- Vasquez C.G., Tworoger M., and Martin A.C.. 2014. Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J. Cell Biol. 206:435–450. 10.1083/jcb.201402004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier V., Johndrow J.E., Betson M., Chen G.C., Hughes D.A., Parkhurst S.M., and Settleman J.. 2006. Drosophila Rho-kinase (DRok) is required for tissue morphogenesis in diverse compartments of the egg chamber during oogenesis. Dev. Biol. 297:417–432. 10.1016/j.ydbio.2006.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C.G., Wang B., Ballew A., Royou A., Karess R., Axelrod J.D., and Luo L.. 2001. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 105:81–91. 10.1016/S0092-8674(01)00298-7 [DOI] [PubMed] [Google Scholar]
- Zang J.H., Cavet G., Sabry J.H., Wagner P., Moores S.L., and Spudich J.A.. 1997. On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell. 8:2617–2629. 10.1091/mbc.8.12.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Sepúlveda L.A., Lua R.C., Lichtarge O., Golding I., and Sokac A.M.. 2013. The maternal-to-zygotic transition targets actin to promote robustness during morphogenesis. PLoS Genet. 9:e1003901 10.1371/journal.pgen.1003901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.