Panas et al. review the mechanisms that control the dynamic formation and disassembly of RNA stress granules.
Abstract
The accumulation of stalled translation preinitiation complexes (PICs) mediates the condensation of stress granules (SGs). Interactions between prion-related domains and intrinsically disordered protein regions found in SG-nucleating proteins promote the condensation of ribonucleoproteins into SGs. We propose that PIC components, especially 40S ribosomes and mRNA, recruit nucleators that trigger SG condensation. With resolution of stress, translation reinitiation reverses this process and SGs disassemble. By cooperatively modulating the assembly and disassembly of SGs, ribonucleoprotein condensation can influence the survival and recovery of cells exposed to unfavorable environmental conditions.
Introduction
Regulation of mRNA stability and translation plays an important role in controlling gene expression. Newly transcribed mRNAs bind to a plethora of RNA-binding proteins (RNA-BPs) to assemble RNP particles, the protein components of which influence the mRNP subcellular localization, as well as their rates of translation and decay. In cells subjected to adverse conditions, inhibition of translation initiation leads to the accumulation of stalled translation preinitiation complexes (PICs) that condense to form non–membrane-enclosed foci known as stress granules (SGs; Kedersha et al., 1999; Buchan and Parker, 2009; Kedersha and Anderson, 2009). The corecruitment of proteins that regulate cell signaling helps cells to survive exposure to unfavorable environmental conditions (Kedersha et al., 2013).
SGs belong to a class of diverse subcellular entities known as RNA granules (Anderson and Kedersha, 2006). Examples of RNA granules connected to mRNA metabolism include SGs, processing bodies (P bodies), and neuronal granules. Their classification is mostly based on their localization, composition, and proposed functions. SGs, perhaps the most well-characterized members of the family, have been implicated in the pathogenesis of many diseases including cancer, neurodegeneration, inflammatory disorders, and viral infections (Wolozin, 2012, 2014; Lloyd, 2013; Anderson et al., 2014; Buchan, 2014). Thus mechanistic details of SG assembly/disassembly and resulting effects on cell signaling and survival programs will lead to a better understanding of the underlying disease pathology.
SGs are cytoplasmic foci assembled when untranslated mRNPs accumulate in cells subjected to biotic stress (e.g., viral infections; McInerney et al., 2005; White et al., 2007; White and Lloyd, 2012) or environmental stress (e.g., oxidation, heat, and starvation; Anderson and Kedersha, 2002, 2006, 2009; Buchan and Parker, 2009). Their formation is tightly connected to the disassembly of translating polysomes: an increase in the fraction of untranslated mRNAs favors SG assembly, whereas an increase in the fraction of translated mRNAs (in polysomes) favors SG disassembly (Kedersha et al., 2000). Mechanistically, the polysome/SG dynamics is achieved by regulating the activities of eukaryotic initiator factor 2 (eIF2), the cap-binding eIF4F complex (consisting of eIF4E, eIF4A, and eIF4G), or both. These complexes serve as early checkpoints in the regulation of translation initiation and concomitantly modulate SG formation.
During translation, most mRNAs are circularized by interactions between eIF4G at its 5′ end and poly(A)-binding protein (PABP) at its 3′ poly(A) tail (Fig. 1 A; Tarun and Sachs, 1995; Kahvejian et al., 2005). Recruitment of eIF4F to the 5′ cap of mRNA is a translational checkpoint under stringent control of mechanistic target of rapamycin (mTOR), a serine/threonine kinase that couples cellular metabolism to protein synthesis. Under optimal growth conditions, mTOR constitutively phosphorylates eIF4E-binding protein (4E-BP) to prevent it from binding to eIF4E and inhibiting translation. In cells subjected to metabolic stress (e.g., amino acid starvation), inactivation of mTOR results in the accumulation of hypophosphorylated 4E-BP, which binds eIF4E and inhibits translation (Sonenberg and Hinnebusch, 2009). By blocking eIF4F assembly, eIF4E:4E-BP complexes inhibit translation initiation and prevent polysome assembly. The resulting noncanonical PICs that lack factors required to initiate translation seed the assembly of SGs (Fig. 1 B1).
Figure 1.
Model of canonical and noncanonical PIC formation under various conditions. Note that the diagram is not time resolved, and hence some depicted interactions may not occur simultaneously or be competitive. (A) Assembly of the 48S PIC during normal conditions. The ternary complex joins the 40S ribosomal subunit and forms a 43S preinitiation complex. The eIF4F complex (eIF4E, eIF4A, and eIF4G) binds together with eIF4B to the 5′ cap of the mRNA. The eIF4F-bound mRNA associates with the 43S PIC, and then scans to the AUG start codon, where 48S PIC formation occurs. (B) Formation of noncanonical PICs during various stress conditions. Here, we depict three different signaling pathways that assemble noncanonical PICs: (1) mTOR inhibition by sodium selenite, amino acid (AA) starvation, rapamycin, or oxidative stress (H2O2), which leads to hypophosphorylation of 4E-BP, which then interacts with eIF4E and blocks translation initiation; (2) various stresses activate distinct eIF2α kinases that phosphorylate eIF2α, deplete the ternary complex, and promote the assembly of a noncanonical PIC; and (3) compounds such as pateamine A, 15-deoxy-Δ(12,14)-prostaglandin J2 (15d PGJ2), hippuristanol, silvestrol, or tiRNA target the eIF4F complex, also creating a noncanonical PIC. These noncanonical PICs differ from canonical PICs in composition and exposure of the 40S subunit interface and mRNA. These interfaces recruit RNA-BPs such as G3BP and TIA-1/R, increasing the local concentration of these proteins to promote LLPS and assembly of SG seeds. Figure modified from Jackson et al. (2010).
Assembly of the eIF2/GTP/tRNAiMet ternary complex that delivers initiator tRNAiMet to the 40S ribosomal subunit is another checkpoint of translation initiation. The ternary complex and the mRNA/eIF4F complex combine with the 40S ribosomal subunit before the recruitment of the 60S ribosomal subunit to initiate protein synthesis. Under stress conditions, eIF2 is a substrate for one of four stress-sensing serine/threonine kinases (heme-regulated eIF2α kinase [HRI], general control nonderepressible 2 [GCN2], protein kinase R [PKR], and PKR-like ER kinase [PERK]) that phosphorylate serine residue 51 of eIF2α (Wek et al., 1995; Srivastava et al., 1998; Harding et al., 2000; McEwen et al., 2005). Phosphorylated eIF2α (ph-eIF2α) inhibits efficient GDP-GTP exchange, leading to a decrease in levels of translationally competent ternary complexes, and inhibits translation initiation. Because eIF2α phosphorylation does not affect translation elongation (Shenton et al., 2006), ribosomes already engaged in translation “run off” the mRNA, leading to polysome disassembly and accumulation of noncanonical 48S PICs that lack charged tRNAiMet and early initiation factors eIF2 and eIF5. The influx of stalled 48S complexes leads to SG assembly (Fig. 1 B2; Kedersha et al., 2002).
Although phosphorylation of eIF2α is a major trigger of SG formation, ph-eIF2α–independent triggers of SG assembly have also been described. These triggers can be divided into two classes. Examples of the first class, which inhibits the function of the RNA helicase eIF4A, include (a) 15-deoxy-Δ(12,14)-prostaglandin J2, a natural lipid inflammatory mediator belonging to a class of prostaglandins that bind to eIF4A and block its interaction with eIF4G (Kim et al., 2007); (b) pateamine A, a metabolite isolated from the sea sponge Mycale sp. that dissociates eIF4A from eIF4G by inhibiting eIF4A helicase or ATPase activities (Bordeleau et al., 2006a; Dang et al., 2006); (c) hippuristanol, a steroid isolated from the coral Isis hippuris, that allosterically inhibits eIF4A and reduces ATPase, helicase, and RNA-binding activities (Bordeleau et al., 2006b; Tsumuraya et al., 2011; Cencic et al., 2013); and (d) silvestrol, a rocaglate isolated from the plant genus Aglaia, that stimulates the RNA-binding and helicase activities of eIF4A, resulting in sequestration of eIF4A from the eIF4F complex (Fig. 1 B3; Bordeleau et al., 2008; Cencic et al., 2009; Kogure et al., 2013). Examples of compounds in the second class, which disrupt eIF4E interaction with eIF4G, include (a) sodium selenite, a compound that inactivates mTOR to promote the assembly of inhibitory eIF4E:4E-BP1 complexes (Fujimura et al., 2012); (b) hydrogen peroxide, an oxidative agent that indirectly promotes binding of 4E-BP1 to eIF4E by promoting 4E-BP hypophosphorylation (Fig. 1 B1; Emara et al., 2012); and (c) tRNA-derived stress-induced RNAs (tiRNAs) produced by angiogenin-induced cleavage of mature tRNAs that displace eIF4G/eIF4A from cap-bound eIF4E (Emara et al., 2010; Ivanov et al., 2011a). tiRNA-repressed PICs are then assembled into SGs with help from the RNA chaperone YB-1 (Fig. 1 B3; Lyons et al., 2016). In some cases (e.g., treatment with antineoplastic vinca alkaloid drugs), both eIF4F complex– and ph-eIF2α–mediated translation inhibition contribute to SG assembly (Szaflarski et al., 2016). Similarly, under conditions of nutrient starvation, both eIF2α phosphorylation (via GCN2) and inactivation of mTOR signaling are required for SG formation, although this mechanism selectively packages mRNAs bearing 5′-terminal oligopyrimidine tracts (5′TOPs) into SGs (Damgaard and Lykke-Andersen, 2011; Ivanov et al., 2011b). As a common theme, each of these mechanisms inhibits the assembly of eIF4F and promotes the assembly of noncanonical PICs and SGs. Finally, it is important to emphasize that untranslated mRNAs are essential and defining components of SGs. As such, pharmacologic interventions (e.g., cycloheximide and emetine) that trap mRNAs at polysomes prevent SG assembly. In contrast, treatment with puromycin, an antibiotic that promotes polysome disassembly, potently stimulates SG assembly (Kedersha et al., 2000).
SGs are compositionally and functionally diverse
Proteomic and genetic screens have identified hundreds of proteins that either are components of SGs or modulate their assembly (Ohn et al., 2008; Jain et al., 2016). A stalled PIC including 40S ribosomal subunits and diverse RNA-BPs is included in a hypothesized, relatively stable SG “core” that may be biochemically purified or visualized using electron or superresolution fluorescence microscopy (Souquere et al., 2009; Jain et al., 2016). One study showed that ∼50% of SG proteins lack annotated RNA-binding domains, suggesting that protein–protein interactions play important roles in SG assembly. Such non-mRNP components include various adaptor/scaffolding proteins and diverse enzymes such as protein/lipid kinases and phosphatases; methyl-, glucosyl-, and ribosyltransferases; GTPases; ATPases; ubiquitin-modifying enzymes; and RNA or DNA nucleases and helicases (Jain et al., 2016). SGs also contain molecules involved in diverse signaling pathways, allowing them to function as RNA-centric signaling hubs that communicate a “state of emergency” to other signaling and metabolic pathways (Kedersha et al., 2013). Importantly, the composition of SGs is different in cells subjected to different types of stress, as seen during treatment with the chemotherapy drugs sodium selenite and vinca alkyloids. Whereas vinca alkaloid–induced SGs are similar to canonical SGs induced by sodium arsenite (Szaflarski et al., 2016), selenite-induced SGs lack the SG marker eIF3 and exhibit reduced recruitment of 40S ribosomal subunit proteins (Fujimura et al., 2012). Remarkably, whereas vinca alkaloid– or sodium arsenite–induced SGs are anti-apoptotic and promote cell survival, selenite-induced SGs inhibit cell survival (although effects of these drugs on cellular metabolism may be pleiotropic and also act through SG-independent mechanisms). Thus different SG subtypes with different composition (e.g., lacking canonical components such as eIF3) may have opposite effects on cellular metabolism and survival.
Intrinsically disordered protein regions
To organize complex biochemical reactions, eukaryotic cells assemble membrane-enclosed and non–membrane-enclosed compartments that have specialized functions. Membrane-enclosed compartments include the nucleus, mitochondria, Golgi complex, and lysosomes. Non–membrane-enclosed compartments include nucleoli (Hernandez-Verdun, 2011), Cajal bodies (Cioce and Lamond, 2005), P bodies, and SGs (Buchan, 2014). Until recently, the physical forces that maintain non–membrane-enclosed entities as stable structures within the cell have been mysterious. Recent studies have shown that weak interactions between low-complexity and intrinsically disordered protein regions (IDPRs) can facilitate a phase separation that concentrates these proteins into discrete subcellular domains (Tompa, 2005; Keating, 2012; Brangwynne, 2013; Nott et al., 2015). IDPRs differ from structured proteins in amino acid composition, complexity, charge, flexibility, and hydrophobicity: they are deficient in order-promoting amino acids, such as Ile, Leu, Val, Trp, Tyr, Phe, Cys, and Asn and enriched in disorder-promoting amino acids such as Ala, Arg, Gly, Gln, Ser, Pro, Glu, and Lys. IDPRs are highly dynamic in that they can rapidly move from compact to extended conformations (Wright and Dyson, 1999; Tompa, 2002; Uversky and Dunker, 2010; Uversky, 2013).
The abundance of IDPRs increases with organism complexity: eukaryotic proteins typically have more IDPRs than bacteria or archaea to fulfill more complex tasks such as signaling or intercellular communication (Ward et al., 2004; Peng et al., 2014). The lack of a specific tertiary structure gives IDPRs the ability to serve as hub proteins that bring together proteins and nucleic acids required for a specific function (Kriwacki et al., 1996; Singh et al., 2007). Binding to specific substrates can determine the structural conformation that they adopt (Kriwacki et al., 1996; Uversky, 2003). Their conformation can also be modulated by post-translational modifications (PTMs) found within or in close proximity to IDPRs, allowing context-dependent modulation of their structure (Iakoucheva et al., 2004; Pejaver et al., 2014). Indeed, some PTMs regulate not only the conformation of SG-associated proteins but also the assembly of SGs. Phosphorylation of the SG-nucleating protein G3BP within its IDPR (at serine 149) impairs its ability to induce SG formation (Tourrière et al., 2003; Kedersha et al., 2016). Similarly, MK2-induced phosphorylation of the SG-nucleating protein tristetraprolin on serine residues 52 and 178 triggers its egress from SGs (Stoecklin et al., 2004; Kedersha et al., 2005). Because PTM sites have been linked to specific disease phenotypes (Li et al., 2010), IDPR structural alterations and their effects on SG assembly may contribute to the pathogenesis of cancer, cardiovascular disease, diabetes, and neurodegenerative disease (Uversky, 2014).
Liquid–liquid phase separation
In some cases (e.g., P granules found in one-cell-stage Caenorhabditis elegans embryos or Xenopus laevis extrachromosomal nucleoli), IDPR-induced liquid–liquid phase separation (LLPS) produces spherical liquid droplets, a morphology that minimizes the interface with adjacent cytoplasm. These liquid droplets fuse with each other, exhibit “dripping” phenomena, rapidly (within seconds) rearrange their contents (Brangwynne et al., 2009, 2011; Weber and Brangwynne, 2012; Brangwynne, 2013; Lee et al., 2013; Patel et al., 2015), and reassume a spherical morphology after shear stress (Brangwynne et al., 2009). Although SGs are rarely spherical, several observations suggest that IDPR-induced LLPS may contribute to some aspects of their properties. IDPRs are found in many SG proteins, including TIA-1/R and G3BP, which play important roles in SG condensation (Han et al., 2012; Kato et al., 2012; Kedersha et al., 2013, 2016).
TIA-1 is composed of three N-terminal RNA recognition motifs (RRMs), an IDPR, and a glutamine-rich prion-related domain (PRD; Kedersha et al., 2000; Gilks et al., 2004). Its PRD, which is essential for SG assembly, forms small spherical cytoplasmic bodies when overexpressed. Like amyloid proteins, the PRD can assume a protease-resistant conformation indicative of highly stable protein–protein interactions. Remarkably, the prion domain of yeast Sup35 can substitute for the TIA-1-PRD in SG condensation. These results suggest that a conformational change within the PRD can contribute to the nucleation of SGs. In support of this conclusion, an IDPR within Pub1, a yeast homolog of TIA-1, also promotes the assembly of liquid droplets in a salt- and temperature-dependent manner. Over time, these droplets are transformed into more rigid and less dynamic hydrogels, as revealed by FRAP analysis (Lin et al., 2015). This transformation from liquid droplets to hydrogels and even insoluble amyloids was also observed using recombinant FUS, a protein involved in the pathogenesis of amyotrophic lateral sclerosis (ALS; Burke et al., 2015; Murakami et al., 2015; Patel et al., 2015). Furthermore, heterogeneous nuclear RNP A1 (hnRNPA1) is an SG protein (Kim et al., 2013) that also promotes LLPS that depends on a C-terminal IDPR. Molecular crowding, electrostatic interactions, and RNA enhance hnRNPA1-induced phase transition (Molliex et al., 2015). Additionally, hyperosmotically induced stress can induce the formation of SGs via molecular crowding (Bounedjah et al., 2012; Kedersha et al., 2016), which suggests a connection between LLPS and SGs.
The SG nucleator G3BP contains a structured NTF2-like domain and an RRM domain; IDPRs are found in an acidic region, a PxxP domain, and an RGG domain. G3BP has not yet been shown to promote LLPS, and the mechanism by which G3BP nucleates SG assembly is still elusive. Knockout of G3BP renders cells unable to form SGs in response to ph-eIF2α stresses or eIF4A inhibition, showing the importance of G3BP in SG assembly (Kedersha et al., 2016). The RGG motif of G3BP interacts with 40S ribosomal subunits, connecting the translation machinery with SGs, and loss of this motif prevents G3BP from promoting SG assembly (Kedersha et al., 2016). The NTF2-like domain oligomerizes (Tourrière et al., 2003; Vognsen and Kristensen, 2012; Schulte et al., 2016) and interacts with Caprin1 and USP10 in a mutually exclusive manner (Kedersha et al., 2016). USP10 overexpression prevents SG formation, whereas Caprin1 promotes it, suggesting that this competition for G3BP includes a regulatory mechanism that modulates SG assembly. USP10 may prevent G3BP from nucleating SG assembly by locking G3BP in a conformation that influences the NTF2-like domain and mediates or prevents G3BP oligomerization. It is also possible that USP10 blocks the helicase function of G3BP (Costa et al., 1999), as helicase activity of some proteins, e.g., DDX6/RCK, has been shown to be required for RNA granule assembly (Ohn et al., 2008).
The observation that SG proteins can promote the formation of liquid droplets that can transition into insoluble aggregates suggests that RNP granules may be linked to the pathological inclusions associated with neurodegenerative diseases such as ALS, frontotemporal lobar degeneration, and Alzheimer’s disease (Wolozin, 2012; Vanderweyde et al., 2013). The recent discovery that 1,6-hexanediol, an aliphatic alcohol that disrupts weak hydrophobic interactions, dissolves liquid droplets without affecting insoluble aggregates may provide a tool to test this hypothesis. This reagent dissolves yeast P bodies but not low glucose–induced yeast SGs, indicating that the physical nature of these particular RNP granules is different. Because 1,6-hexanediol dissolves both mammalian P bodies and arsenite-induced mammalian SGs (Kroschwald et al., 2015), the relative contribution of low-affinity IDPR-induced LLPS and high-affinity amyloid-like aggregation to the assembly of different types of RNP granules is not obvious. It should be noted that low glucose–induced yeast SGs and arsenite-induced mammalian SGs have markedly different compositions and functions, making it unclear whether they are even orthologous granules (Ohn et al., 2008; Kedersha et al., 2013; Mitchell et al., 2013). Nevertheless, these experiments suggest that LLPS plays a role in some aspect of the assembly or maintenance of arsenite-induced nonspherical SGs in mammalian cells. Like liquid droplets, some components of these SGs are highly dynamic: FRAP analysis reveals that G3BP1, TIA-1/R, and PABP rapidly shuttle in and out of these granules (Kedersha et al., 2000, 2005). In contrast, FMRP (Gareau et al., 2013), FASTK (Kedersha et al., 2005), IGF2BP1 and HuR (Bley et al., 2015), and CPEB (Mollet et al., 2008) are relatively fixed in these granules, suggesting a possible scaffolding function (Kedersha et al., 2005). In stressed cells, mRNA transit studies revealed that roughly one third of endogenous mRNAs is diffusely scattered throughout the cytoplasm, one third shuttles in and out of SGs with a time constant of 300 s, and one third is immobile within SGs (Zhang et al., 2011).
The similarities and differences between SGs and liquid droplets might be reflected in the heterogeneity within the SG itself. Indeed, recent studies suggest that SGs are composed of stable cores surrounded by a more dynamic shell (Jain et al., 2016; Protter and Parker, 2016). Protter and Parker (2016) propose two models that could lead to the assembly of these two-component granules. The first model posits that untranslated mRNPs promote the condensation of SG cores that then recruit a dynamic, morphologically amorphous shell. The cores may form more stable protein–protein aggregates over time, similar to the in vitro transition from liquid droplets to hydrogels to insoluble aggregates. The second model suggests that untranslated mRNPs promote LLPS to form liquid droplets held together by weak interactions. Core formation then results from higher-affinity interactions that form during a structural transition at the center of the droplet (Protter and Parker, 2016). To test these models, a 1,6-hexanediol treatment should dissolve the shell component while leaving the core remaining. A related study investigating the biochemical and cellular behavior of engineered multivalent proteins found that scaffold proteins form phase-separated liquids. In this context, scaffold proteins recruit client proteins possessing similar interaction elements, provided that free scaffold binding sites are available. These studies suggest that RNP granule assembly is regulated by the availability of scaffold protein-binding sites (Banani et al., 2016) and may explain how USP10 and Caprin1 regulate the ability of G3BP to mediate SG formation (Kedersha et al., 2016).
Ribosomal proteins and SG assembly: A speculative model
In both models, untranslated mRNPs are nucleators of SG cores; here, we propose that stalled 48S PICs can serve as a seed for SG condensation. As depicted in Fig. 1 B, there are at least three ways to form noncanonical PICs that stall translation initiation and nucleate SG assembly. Under these conditions, translating ribosomes run off the mRNA, and polysomes are disassembled. This results in long stretches of exposed mRNA, which might attract nucleating RNA-BPs such as TIA-1, by binding to 5′TOP motifs (Damgaard and Lykke-Andersen, 2011), and G3BP, by interacting with the 40S ribosome (Kedersha et al., 2016), thus increasing their local concentration and favoring LLPS, liquid droplet formation, and nucleation of SGs. At the same time, the noncanonical PIC is likely to expose the 40S ribosomal subunit interface, which would normally be covered by the 60S subunit, to recruit RNA-BPs, to either exposed 18S rRNA or ribosomal proteins, and nucleate SGs.
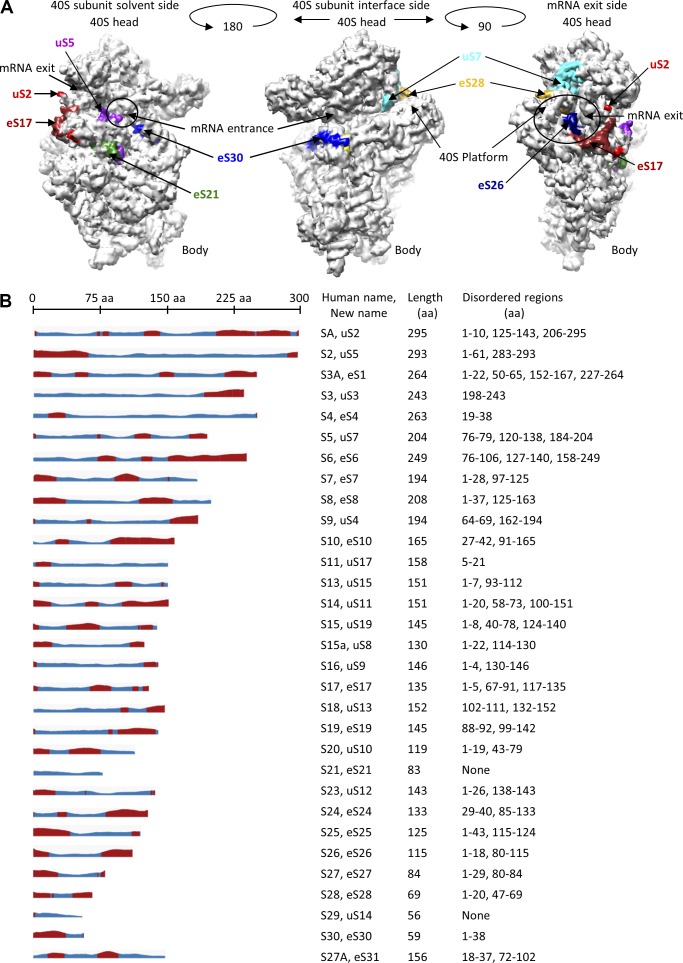
An analysis of the disordered regions within 40S ribosomal subunit proteins identified several candidates with IDPRs that might contribute to the condensation event. Fig. 2 B catalogs 40S ribosomal proteins with IDPRs that may mediate LLPS to nucleate SG assembly. Specific examples include (a) uS5 and eS30 proteins found at the mRNA entry site, which were identified in a functional RNAi screen as essential for SG assembly (Ohn et al., 2008); (b) exposed 40S ribosomal proteins known to tether the 40S head to the 40S platform region of neighboring ribosomes, leading to the formation of a condensed structure, in which mRNA cannot loop outside the polysome, hindering the ability of RNA-BPs to interact with mRNA (Myasnikov et al., 2014); and (c) ribosomal proteins residing at the 40S subunit solvent site or mRNA exit, including uS2, eS17, eS21, uS7, eS28, and eS26, that could recruit RNA-BP nucleators. The ability of G3BP to interact with isolated 40S ribosomal subunits, but not 80S ribosomes, allows it to mediate LLPS after the assembly of stalled PICs (Kedersha et al., 2016).
Figure 2.
Human 40S ribosomal proteins containing disordered regions. (A) Images based on the crystal structure of the human 40S ribosomal subunit (protein database: 4UG0; Khatter et al., 2015). Views from the 40S subunit solvent site (left), 40S subunit interface site (middle), and mRNA exit site (right). Disordered 40S ribosomal proteins at the mRNA entry sites (left and middle) are uS2 (red), uS5 (violet), eS21 (green), and eS30 (blue). Disordered 40S ribosomal proteins, which are located at the platform and close to the mRNA exit site, include uS7 (light blue), eS17 (dark red), eS26 (dark blue), and eS28 (orange). Ribosomal RNA and other 40S ribosomal proteins are shown in light gray. (B) Schematic representation of IDPRs in 40S ribosomal subunit proteins. The disordered regions are from the protein database website. The disorder calculations are based on the JRONN method (Yang et al., 2005). Red, potentially disordered; blue, potentially ordered. Shown are the nomenclature of small ribosomal subunit names, the amino acid length, and the potentially disordered regions.
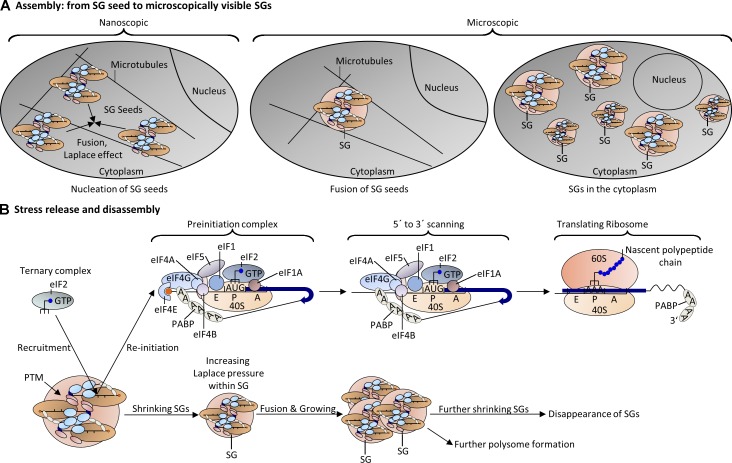
Once the ribosomal SG seed is formed, the increased local concentration of SG nucleators and the stalled noncanonical PICs could act as a sink to recruit more SGs nucleators via IDPRs. Furthermore, the negatively charged and accessible mRNA stretches might also recruit more RNA-BPs via electrostatic interactions. Over time, proximal ribosomal seeds can fuse to form larger structures and become visible in the microscope. This event might give SGs their characteristic irregular structure (Fig. 3 A), a result of steric hindrance between the stalled 48S PICs.
Figure 3.
Model of SG assembly and disassembly. (A) Once nanoscopic SG seeds are formed (Fig. 1 B), nearby seeds attract each other via weak electrostatic interactions, interact with neighboring SG seeds, and coalescence to form irregular microscopically visible SGs. Microscopically visible SGs can fuse to produce larger SGs. (B) After stress release, events that promote SG disassembly may include increase in concentrations of ternary complexes; phosphorylation of 4E-BP by mTOR, releasing the eIF4E block; and reactivation of eIF4A activities. These events might trigger the formation of translationally competent PICs to reinitiate translation at the surface of SGs. Because translating ribosomes displace mRNA-bound RNA-BPs, those complexes are then detached from SGs. As SGs shrink, the Laplace pressure increases to promote further fusion with adjacent SGs. Over time, fewer and larger SGs appear before they eventually disappear from the cytoplasm. Other SG proteins such as USP10 can potentiate disassembly by maintaining G3BP in a soluble conformation. PTM of ribosomal and SG proteins might also contribute to the disassembly.
Various forces can influence the size and conformation of condensing SGs. Because small SGs have a higher Laplace pressure (the pressure difference between the inside and outside of a curved surface) than larger SGs, they will tend to fuse with adjacent SGs, an effect known as Ostwald ripening (Mendoza et al., 2004). Their final size may be controlled by the number of molecules used to build up the droplet (Decker et al., 2011). The connection between molecule number and droplet size has been discussed elsewhere (Goehring and Hyman, 2012; Brangwynne, 2013). The finding that overexpression of G3BP or TIA-1 leads to the formation of considerably larger SGs than those induced by environmental stress (Tourrière et al., 2003; Gilks et al., 2004) suggests that the composition of SGs can also influence the ultimate size of the granule.
Live cell imaging has revealed that SGs move in the cytoplasm (Kedersha et al., 2005). Ribosomes (Suprenant et al., 1989; Hamill et al., 1994) as well as SGs interact with microtubules, and disruption of the microtubule network with nocodazole or vinblastine disrupts the formation of SGs (Ivanov et al., 2003; Chernov et al., 2009). The attachment of ribosomes and SGs to microtubules suggests that nanoscopic SG seeds can be actively transported in the crowded cytoplasm. Furthermore, microtubule motor proteins have been shown to influence the assembly and movement of SGs (Loschi et al., 2009), supporting the notion of active transport. These mobile SG seeds may serve as concentration sinks in the cytoplasm that are affected by concentration differences, composition differences, electrostatic interactions, and Laplace pressures to grow into typical irregular SGs (Fig. 3 A).
Once stress is released, SGs disassemble and translation resumes. In addition, recent articles describe a dynamic remodeling process in yeast SGs that is dependent on ATP-dependent helicases and remodelers. ATP is required for SG assembly and dynamics (Jain et al., 2016). Although PTM of SG proteins can promote SG disassembly, the detailed mechanism by which SGs are disassembled is not known. In mammalian SGs, disassembly occurs when the inciting stress is relieved, allowing resumption of protein synthesis, or in the case of cold shock SGs, when temperature is raised but before translation is resumed (Hofmann et al., 2012). In the first case, it is likely that replenishment of translation initiation factors (e.g., ternary complex) allows translation to resume at the surface of the SG as stalled PICs are converted into polysomes (Fig. 3 B). This process would displace SG-nucleating proteins such as TIA-1/R and G3BP, shrinking the SG and increasing its Laplace pressure. This should promote fusion with adjacent SGs, a phenomenon that has been observed in cells allowed to recover from stress (Anderson and Kedersha, 2002; Kedersha et al., 2002). In the second case, the rapid temperature-dependent disassembly of cold-shock SGs upon warming is evidence for LLPS (Hofmann et al., 2012). Disassembly can also be influenced by activities of heat shock proteins, such as HSP70. Pharmacological inhibition of HSP70 delays SG disassembly (Ganassi et al., 2016), and the concentration-dependent aggregation of the PRD domain of TIA-1 is inhibited by HSP70 overexpression (Gilks et al., 2004). It suggests that HSP70 can act as a disaggregase and play a role in SG surveillance and dynamics. Interestingly, the overexpression of USP10 prevents the formation of SGs by directly binding G3BP via an FGDF motif (Kedersha et al., 2016). This motif is also found in the nonstructural protein 3 (nsP3) of Semliki Forest virus (Panas et al., 2012, 2015; Schulte et al., 2016), suggesting that USP10 and nsP3 may influence the physical state of G3BP to influence SG formation. Last, PTMs of ribosomal proteins and SG proteins may cooperatively influence SG dynamics. Methylation of G3BP at R447 has been proposed to affect interactions with the 40S ribosomal subunit and repress SG formation (Tsai et al., 2016). In addition, ER stress–induced ubiquitination within IDPRs of uS5 and uS3 has been proposed to contribute to reprogramming of protein translation (Higgins et al., 2015).
Concluding remarks, problems, and perspectives
Experiments performed in cell-free systems have nicely revealed how individual proteins bearing IDPRs can promote LLPS to produce non–membrane-enclosed bodies. Just as environmental conditions (e.g., ionic composition and strength, pH, temperature) can coax individual proteins to aggregate into crystals in vitro, specific environmental conditions can also coax many proteins bearing IDPRs to undergo LLPS to assemble droplets, hydrogels, and aggregates. In cells, the assembly of multicomponent RNA granules is vastly more complex. This is further complicated by the ability of PTM of individual proteins and RNAs to influence granule assembly. Thus, our current state of knowledge is not sufficient to fully understand how RNA granules are assembled and disassembled within living cells.
Despite our incomplete understanding of granule assembly, the link between RNA granules and aggregation events that contribute to neurodegenerative diseases such as ALS, frontotemporal lobar degeneration, and Alzheimer’s disease makes it imperative that we learn more about these mechanisms. It is clear that the large-scale rearrangements of molecules accompanying SG assembly and disassembly are tightly regulated processes, and defects in their organization may influence cellular metabolism and survival under stress. Because dysregulated aggregation of proteins or mRNPs can impair cellular functions, redundant mechanisms that disassemble or remove aggregates have evolved (Tyedmers et al., 2010; Doyle et al., 2013). In addition, PTMs such as phosphorylation, ubiquitination, and polyuridylation can be used to tag proteins and RNAs for removal (Wippich et al., 2013). Studying the molecular mechanisms by which LLPS leads to droplet formation and subsequent aggregation, as well as the relationship between physiological RNA granules and pathological aggregates, could improve our understanding of the pathogenesis of neurodegenerative disease.
Finally, it is important to note that most of what we know about SGs was discerned from traditional structural and microscopy techniques. Development of new approaches that allow study of such heterogeneous, dynamic, and amorphous entities as SGs is a major challenge for the future. Recent progress in superresolution microscopy, such as development of lattice light-sheet microscopy, may provide new insights into SG biology. These methods have already advanced studies of complex biological processes such as embryogenesis and entry of viruses into living cells/organisms. It will be exciting to apply these and other new technologies to studies of SG assembly and disassembly.
Acknowledgments
We thank Alexander G. Myasnikov for help in the preparation of the 40S ribosomal crystal structure images used in Fig. 2 and the Anderson laboratory for helpful discussions and feedback on the manuscript.
This work is supported by the National Institutes of Health (GM111700 and CA168872 to P. Anderson and NS094918 to P. Ivanov).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ALS
- amyotrophic lateral sclerosis
- BP
- binding protein
- eIF
- eukaryotic initiator factor
- IDPR
- intrinsically disordered protein region
- LLPS
- liquid–liquid phase separation
- mTOR
- mechanistic target of rapamycin
- P body
- processing body
- PIC
- preinitiation complex
- PRD
- prion-related domain
- PTM
- post-translational modification
- SG
- stress granule
References
- Anderson P., and Kedersha N.. 2002. Stressful initiations. J. Cell Sci. 115:3227–3234. [DOI] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2006. RNA granules. J. Cell Biol. 172:803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2009. Stress granules. Curr. Biol. 19:R397–R398. 10.1016/j.cub.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., and Ivanov P.. 2014. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta. 1849:861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., and Rosen M.K.. 2016. Compositional control of phase-separated cellular bodies. Cell. 166:651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glaß M., Möller B., and Hüttelmaier S.. 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 43:e26 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M.E., Cencic R., Lindqvist L., Oberer M., Northcote P., Wagner G., and Pelletier J.. 2006a RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 13:1287–1295. 10.1016/j.chembiol.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., and Pelletier J.. 2006b Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220. 10.1038/nchembio776 [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Robert F., Gerard B., Lindqvist L., Chen S.M., Wendel H.G., Brem B., Greger H., Lowe S.W., Porco J.A. Jr., and Pelletier J.. 2008. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118:2651–2660. 10.1172/JCI34753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Hamon L., Savarin P., Desforges B., Curmi P.A., and Pastré D.. 2012. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J. Biol. Chem. 287:2446–2458. 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P. 2013. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203:875–881. 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., and Hyman A.A.. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 108:4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11:1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell. 36:932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K.A., Janke A.M., Rhine C.L., and Fawzi N.L.. 2015. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 60:231–241. 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Carrier M., Galicia-Vázquez G., Bordeleau M.E., Sukarieh R., Bourdeau A., Brem B., Teodoro J.G., Greger H., Tremblay M.L., et al. . 2009. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 4:e5223 10.1371/journal.pone.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Robert F., Galicia-Vázquez G., Malina A., Ravindar K., Somaiah R., Pierre P., Tanaka J., Deslongchamps P., and Pelletier J.. 2013. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood Cancer J. 3:e128 10.1038/bcj.2013.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov K.G., Barbet A., Hamon L., Ovchinnikov L.P., Curmi P.A., and Pastré D.. 2009. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284:36569–36580. 10.1074/jbc.M109.042879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M., and Lamond A.I.. 2005. Cajal bodies: A long history of discovery. Annu. Rev. Cell Dev. Biol. 21:105–131. 10.1146/annurev.cellbio.20.010403.103738 [DOI] [PubMed] [Google Scholar]
- Costa M., Ochem A., Staub A., and Falaschi A.. 1999. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 27:817–821. 10.1093/nar/27.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C.K., and Lykke-Andersen J.. 2011. Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev. 25:2057–2068. 10.1101/gad.17355911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., and Liu J.O.. 2006. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- Decker M., Jaensch S., Pozniakovsky A., Zinke A., O’Connell K.F., Zachariae W., Myers E., and Hyman A.A.. 2011. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21:1259–1267. 10.1016/j.cub.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Doyle S.M., Genest O., and Wickner S.. 2013. Protein rescue from aggregates by powerful molecular chaperone machines. Nat. Rev. Mol. Cell Biol. 14:617–629. 10.1038/nrm3660 [DOI] [PubMed] [Google Scholar]
- Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.F., and Anderson P.. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285:10959–10968. 10.1074/jbc.M109.077560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M.M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., and Anderson P.. 2012. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423:763–769. 10.1016/j.bbrc.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K., Sasaki A.T., and Anderson P.. 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40:8099–8110. 10.1093/nar/gks566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I., Mediani L., Poser I., Lee H.O., Seguin S.J., Morelli F.F., Vinet J., Leo G., et al. . 2016. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell. 63:796–810. 10.1016/j.molcel.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Gareau C., Martel D., Coudert L., Mellaoui S., and Mazroui R.. 2013. Characterization of fragile X mental retardation protein granules formation and dynamics in Drosophila. Biol. Open. 2:68–81. 10.1242/bio.20123012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., and Anderson P.. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398. 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring N.W., and Hyman A.A.. 2012. Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol. 22:R330–R339. 10.1016/j.cub.2012.03.046 [DOI] [PubMed] [Google Scholar]
- Hamill D., Davis J., Drawbridge J., and Suprenant K.A.. 1994. Polyribosome targeting to microtubules: Enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J. Cell Biol. 127:973–984. 10.1083/jcb.127.4.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S.L.. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., and Ron D.. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. 2011. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2:189–194. 10.4161/nucl.2.3.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R., Gendron J.M., Rising L., Mak R., Webb K., Kaiser S.E., Zuzow N., Riviere P., Yang B., Fenech E., et al. . 2015. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell. 59:35–49. 10.1016/j.molcel.2015.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S., Cherkasova V., Bankhead P., Bukau B., and Stoecklin G.. 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell. 23:3786–3800. 10.1091/mbc.E12-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., and Dunker A.K.. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32:1037–1049. 10.1093/nar/gkh253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P.A., Chudinova E.M., and Nadezhdina E.S.. 2003. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290:227–233. 10.1016/S0014-4827(03)00290-8 [DOI] [PubMed] [Google Scholar]
- Ivanov P., Emara M.M., Villen J., Gygi S.P., and Anderson P.. 2011a Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 43:613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Kedersha N., and Anderson P.. 2011b Stress puts TIA on TOP. Genes Dev. 25:2119–2124. 10.1101/gad.17838411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., and Pestova T.V.. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., and Sonenberg N.. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104–113. 10.1101/gad.1262905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. . 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating C.D. 2012. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res. 45:2114–2124. 10.1021/ar200294y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90:155–185. 10.1016/S1877-1173(09)90004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., and Anderson P.. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., and Anderson P.. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., and Anderson P.. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 38:494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., and Anderson P.. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212:845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H., Myasnikov A.G., Natchiar S.K., and Klaholz B.P.. 2015. Structure of the human 80S ribosome. Nature. 520:640–645. 10.1038/nature14427 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A., et al. . 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495:467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.J., Kim J.H., and Jang S.K.. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 26:5020–5032. 10.1038/sj.emboj.7601920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T., Kinghorn A.D., Yan I., Bolon B., Lucas D.M., Grever M.R., and Patel T.. 2013. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS One. 8:e76136 10.1371/journal.pone.0076136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriwacki R.W., Hengst L., Tennant L., Reed S.I., and Wright P.E.. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA. 93:11504–11509. 10.1073/pnas.93.21.11504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S.. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 4:e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.F., Brangwynne C.P., Gharakhani J., Hyman A.A., and Jülicher F.. 2013. Spatial organization of the cell cytoplasm by position-dependent phase separation. Phys. Rev. Lett. 111:088101 10.1103/PhysRevLett.111.088101 [DOI] [PubMed] [Google Scholar]
- Li S., Iakoucheva L.M., Mooney S.D., and Radivojac P.. 2010. Loss of post-translational modification sites in disease. Pac. Symp. Biocomput. 2010:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., and Parker R.. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 60:208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.E. 2013. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip. Rev. RNA. 4:317–331. 10.1002/wrna.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M., Leishman C.C., Berardone N., and Boccaccio G.L.. 2009. Dynein and kinesin regulate stress-granule and P-body dynamics. J. Cell Sci. 122:3973–3982. 10.1242/jcs.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons S.M., Achorn C., Kedersha N.L., Anderson P.J., and Ivanov P.. 2016. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 44:6949–6960. 10.1093/nar/gkw418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.J., Anderson P., and Kaufman R.J.. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- McInerney G.M., Kedersha N.L., Kaufman R.J., Anderson P., and Liljeström P.. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 16:3753–3763. 10.1091/mbc.E05-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza R., Savin I., Thornton K., and Voorhees P.W.. 2004. Topological complexity and the dynamics of coarsening. Nat. Mater. 3:385–388. 10.1038/nmat1138 [DOI] [PubMed] [Google Scholar]
- Mitchell S.F., Jain S., She M., and Parker R.. 2013. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20:127–133. 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., and Weil D.. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell. 19:4469–4479. 10.1091/mbc.E08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T., Michel C.H., et al. . 2015. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 88:678–690. 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikov A.G., Afonina Z.A., Ménétret J.F., Shirokov V.A., Spirin A.S., and Klaholz B.P.. 2014. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat. Commun. 5:5294 10.1038/ncomms6294 [DOI] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., and Baldwin A.J.. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 57:936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P.. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10:1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Varjak M., Lulla A., Eng K.E., Merits A., Karlsson Hedestam G.B., and McInerney G.M.. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 23:4701–4712. 10.1091/mbc.E12-08-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Schulte T., Thaa B., Sandalova T., Kedersha N., Achour A., and McInerney G.M.. 2015. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog. 11:e1004659 10.1371/journal.ppat.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. . 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Pejaver V., Hsu W.L., Xin F., Dunker A.K., Uversky V.N., and Radivojac P.. 2014. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 23:1077–1093. 10.1002/pro.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Oldfield C.J., Xue B., Mizianty M.J., Dunker A.K., Kurgan L., and Uversky V.N.. 2014. A creature with a hundred waggly tails: Intrinsically disordered proteins in the ribosome. Cell. Mol. Life Sci. 71:1477–1504. 10.1007/s00018-013-1446-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S., and Parker R.. 2016. Principles and properties of stress granules. Trends Cell Biol. 26:668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T., Liu L., Panas M.D., Thaa B., Dickson N., Götte B., Achour A., and McInerney G.M.. 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol. 6:6 10.1098/rsob.160078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D., Smirnova J.B., Selley J.N., Carroll K., Hubbard S.J., Pavitt G.D., Ashe M.P., and Grant C.M.. 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281:29011–29021. 10.1074/jbc.M601545200 [DOI] [PubMed] [Google Scholar]
- Singh G.P., Ganapathi M., and Dash D.. 2007. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 66:761–765. 10.1002/prot.21281 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G., and Weil D.. 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122:3619–3626. 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- Srivastava S.P., Kumar K.U., and Kaufman R.J.. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273:2416–2423. 10.1074/jbc.273.4.2416 [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W.F., Blackwell T.K., and Anderson P.. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313–1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprenant K.A., Tempero L.B., and Hammer L.E.. 1989. Association of ribosomes with in vitro assembled microtubules. Cell Motil. Cytoskeleton. 14:401–415. 10.1002/cm.970140310 [DOI] [PubMed] [Google Scholar]
- Szaflarski W., Fay M.M., Kedersha N., Zabel M., Anderson P., and Ivanov P.. 2016. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget. 7:30307–30322. 10.18632/oncotarget.8728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. Jr., and Sachs A.B.. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997–3007. 10.1101/gad.9.23.2997 [DOI] [PubMed] [Google Scholar]
- Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27:527–533. 10.1016/S0968-0004(02)02169-2 [DOI] [PubMed] [Google Scholar]
- Tompa P. 2005. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579:3346–3354. 10.1016/j.febslet.2005.03.072 [DOI] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., and Tazi J.. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai W.C., Gayatri S., Reineke L.C., Sbardella G., Bedford M.T., and Lloyd R.E.. 2016. Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumuraya T., Ishikawa C., Machijima Y., Nakachi S., Senba M., Tanaka J., and Mori N.. 2011. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem. Pharmacol. 81:713–722. 10.1016/j.bcp.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Tyedmers J., Mogk A., and Bukau B.. 2010. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11:777–788. 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. 2003. A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 21:211–234. 10.1080/07391102.2003.10506918 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. 2013. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta. 1834:932–951. 10.1016/j.bbapap.2012.12.008 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. 2014. Introduction to intrinsically disordered proteins (IDPs). Chem. Rev. 114:6557–6560. 10.1021/cr500288y [DOI] [PubMed] [Google Scholar]
- Uversky V.N., and Dunker A.K.. 2010. Understanding protein non-folding. Biochim. Biophys. Acta. 1804:1231–1264. 10.1016/j.bbapap.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T., Youmans K., Liu-Yesucevitz L., and Wolozin B.. 2013. Role of stress granules and RNA-binding proteins in neurodegeneration: A mini-review. Gerontology. 59:524–533. 10.1159/000354170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vognsen T., and Kristensen O.. 2012. Crystal structure of the Rasputin NTF2-like domain from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 420:188–192. 10.1016/j.bbrc.2012.02.140 [DOI] [PubMed] [Google Scholar]
- Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., and Jones D.T.. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337:635–645. 10.1016/j.jmb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Weber S.C., and Brangwynne C.P.. 2012. Getting RNA and protein in phase. Cell. 149:1188–1191. 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Wek S.A., Zhu S., and Wek R.C.. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497–4506. 10.1128/MCB.15.8.4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., and Lloyd R.E.. 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20:175–183. 10.1016/j.tim.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., and Lloyd R.E.. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2:295–305. 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., and Pelkmans L.. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 152:791–805. 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- Wolozin B. 2012. Regulated protein aggregation: Stress granules and neurodegeneration. Mol. Neurodegener. 7:56 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. 2014. Physiological protein aggregation run amuck: Stress granules and the genesis of neurodegenerative disease. Discov. Med. 17:47–52. [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., and Dyson H.J.. 1999. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321–331. 10.1006/jmbi.1999.3110 [DOI] [PubMed] [Google Scholar]
- Yang Z.R., Thomson R., McNeil P., and Esnouf R.M.. 2005. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 21:3369–3376. 10.1093/bioinformatics/bti534 [DOI] [PubMed] [Google Scholar]
- Zhang J., Okabe K., Tani T., and Funatsu T.. 2011. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J. Cell Sci. 124:4087–4095. 10.1242/jcs.090951 [DOI] [PubMed] [Google Scholar]