ABSTRACT
RNA uridylation is a significant transcriptome-shaping factor in protists, fungi, metazoans, and plants. The 3′ U-additions are catalyzed by terminal uridyltransferases (TUTases), a diverse group of enzymes that along with non-canonical poly(A) polymerases form a distinct group in the superfamily of DNA polymerase β-like nucleotidyl transferases. Within and across studied organisms and subcellular compartments, TUTases differ in nucleotide triphosphate selectivity, interacting partners, and RNA targets. A general premise linking RNA uridylation to 3′–5′ degradation received support from several studies of small RNAs and mRNA turnover. However, recent work on kinetoplastid protists typified by Trypanosoma brucei provides evidence that RNA uridylation may play a more nuanced role in generating functional small RNAs. In this pathogen's mitochondrion, most mRNAs are internally edited by U-insertions and deletions, and subjected to 3′ adenylation/uridylation; guide RNAs (gRNAs) required for editing are U-tailed. The prominent role of uridylation in mitochondrial RNA metabolism stimulated identification of the first TUTase, RNA editing TUTase 1 (RET1). Here we discuss functional studies of mitochondrial uridylation in trypanosomes that have revealed an unorthodox pathway of small RNA biogenesis. The current model accentuates physical coupling of RET1 and 3′–5′ RNase II/RNB-type exonuclease DSS1 within a stable complex termed the mitochondrial 3′ processome (MPsome). In the confines of this complex, RET1 initially uridylates a long precursor to activate its 3′–5′ degradation by DSS1, and then uridylates trimmed guide RNA to disengage the processing complex from the mature molecule. We also discuss a potential role of antisense transcription in the MPsome pausing at a fixed distance from gRNA's 5′ end. This step likely defines the mature 3′ end by enabling kinetic competition between TUTase and exonuclease activities.
KEYWORDS: exonuclease, guide RNA, mitochondria, RNA decay, RNA editing, Trypanosoma, TUTase, uridylation
Introduction
Post-transcriptional 3′ end nucleotide additions wield profound influence on RNA biogenesis, trafficking, function and turnover. With the advent of deep sequencing and the influx of new genomes, it is becoming evident that few such modifications have retained universal functions across various phyla, or even within the same organelle. Eukaryotic mRNA polyadenylation has long been held as an essential stabilizing process, although A-tailing by non-canonical poly(A) polymerases may also induce exosome-dependent decay in the nucleus.1,2 Indeed, mitochondrial mRNA adenylation by homologous poly(A) polymerases may exert opposite effects: degradation in plants3 and stabilization in trypanosomes.4,5 It appears that the 3′ processing history “written” by spatially and temporally resolved dynamic interactions with protein complexes, and chemical nature and extent of modification represent the major determinants of mRNA stability. RNA uridylation activity was described nearly a half-century ago in mammalian tissues and later detected in plants along with stretches of non-encoded uridines in cellular RNAs. Review by Rissland and Norbury provides an excellent historical account leading up to more recent developments.6 Although rigorous biochemical studies have established the presence of terminal uridyltransferase (TUTase) activities in many organisms, S. cerevisiae being a notable exception, the respective enzymes remained unidentified until 2002. The prominent role of RNA uridylation in U-insertion/deletion mRNA editing typical of kinetoplastid protozoans necessitated a tour de force biochemical purification of the major TUTase activity from mitochondria of Leishmania tarentolae. An orthologous enzyme from Trypanosoma brucei was identified in the same work.7 This protein was subsequently shown to uridylate guide RNAs and was termed RNA editing TUTase 1 (RET1) for chronological reasons, and to distinguish it from RNA editing TUTase 2 (RET2). The latter enzyme functions as an integral subunit of the RNA editing core complex in internal mRNA U-insertion editing.8,9 Divergent protein sequences of 2 editing TUTases have been used to define a family of alike enzymes in trypanosomes10 that includes another mitochondrial TUTase MEAT1,11 mitochondrial poly(A) polymerases KPAP14 and KPAP2,12 cytoplasmic TUT313 and TUT414,15 TUTases, and nuclear non-canonical poly(A) polymerases ncPAP1 and ncPAP2.16 Pioneering work from the Norbury laboratory has identified the non-canonical poly(A) polymerase CID1 as cytosolic TUTase linked to deadenylation-independent mRNA decay 17,18 in S. pombe. Although nucleotide triphosphate selectivity often cannot be inferred from primary sequences, X-ray crystallography has established U-specific contacts characteristic for exclusively U-specific RET219 and MEAT1,20 and the basis for a more relaxed A/U-specificity of CID1.21 In mammalian cells, the TUTase/non-canonical PAP family consists of 7 proteins.22 The rapidly growing list of their functions includes regulation of microRNA decay23-25 and processing,26 snRNA processing,27,28 histone mRNA cycling,29 global mRNA decay30 and mitochondrial mRNA adenylation.31 Although the field is still young, some general concepts are already emerging. It appears that uridylation provides chemical means of distinction from adenylation, thereby creating an alternative binding platform for RNA decay and processing effectors. In this Point-of-View, we elaborate on recent advances in mitochondrial RNA processing in trypanosomes and propose an unorthodox model of small RNA biogenesis of which antisense RNA-controlled uridylation-induced 3′–5′ degradation is the key. Although at this time such a mechanism has been described for guide RNAs involved in mRNA editing, an evolutionary conservation of the TUTase-RNase II-like exonuclease coupling and existence of closely-spaced convergent promoters in many organisms32 are indicative of a potentially more general mechanism.
Much ado about U
The mitochondrial genome of Trypanosoma brucei, the causative agent of African sleeping sickness, is composed of a few 25-kb maxicircles and approximately 10,000 of 1-kb minicircles that are catenated and densely packed into a nucleoprotein structure called kinetoplast. Maxicircles encode genes typically found in mitochondrial genomes (rRNAs, subunits of respiratory complexes and a single ribosomal protein) while minicircles produce a diverse population of small RNAs.33,34 Six of the 18 annotated pre-mRNAs contain open reading frames and are referred to as unedited; the remaining transcripts must undergo U-insertion/deletion editing to acquire a protein coding sequence and, sometimes, start and stop codons. The extent of editing varies from insertion of 4 nucleotides into CO2 mRNA to hundreds of Us in pan-edited transcripts. Short (50–60 nt) guide RNAs (gRNAs), transcribed predominantly from minicircles have been recognized as carriers of editing information by their complementarity to fully-edited mRNAs.35 The postulated mechanism by which gRNA directs specific endonucleolytic cleavage of pre-mRNA, subsequent insertion or deletion of a defined number of uridines, and re-ligation of mRNA fragments35 has been confirmed by in vitro assays with mitochondrial extracts36,37 and purified enzymatic editing complexes.38-40 The ensuing work by several groups detailed the composition of the enzymatic RNA editing core complex (RECC,41 20S editosome) responsible for the elemental editing reaction and assigned specific functions to individual endonucleases (REN1, REN2 and REN3), exonucleases REX1 and REX2, RNA ligases REL1 and REL2, and RET2 TUTase.33,34 42 More recently, exploratory proteomics43 and the identification of 2 homologous RNA binding proteins essential for gRNA stability, GRBC1 and GRBC2,44 led to the discovery of the guide RNA binding complex, also referred to as the mitochondrial RNA binding complex 1 (MRB1). Although still an evolving concept,45 this trimodular assembly of ∼20 polypeptides binds RNA editing substrates (pre-edited mRNAs and gRNAs), intermediates (partially edited mRNAs) and products,46 and coordinates interactions with the polyadenylation complex. The RNA-mediated assembly of the RNA editing core and RNA editing substrate binding complexes appears to constitute an ∼2 MDa RNA editing holoenzyme (40S editosome).
Impressive progress in understanding mRNA editing often overshadows the fact that editing is an essential, but only one of several RNA processing steps in mitochondrial gene expression. Except for the identity of mitochondrial RNA polymerase,47,48 we know little about transcription from maxicircle and minicircle templates. A “cryptic” endonuclease that presumably partitions multicistronic precursors prior to editing remains a hypothetical explanation for the observed tight packing of pre-mRNAs49 within the conserved region of the minicircle, yet no such enzyme has been identified. The chemical nature of RNA termini is often instructive about the processing history; we keep in mind that maxicircle-encoded rRNAs and mRNAs bear 5′ monophosphate, but are uridylated and adenylated, respectively.50,51 In contrast, guide RNAs52 and guide RNA-like molecules46 start with 5′ triphosphate indicative of the transcription initiation site, and end with an oligo(U) tail.
Master of many tails
RET1-catalyzed uridylation targets all classes of mitochondrial RNAs,53 but the resultant 3′ extensions can be divided into 2 distinct categories: approximately 13 nt-long continuous U-tails found in mature rRNAs and gRNAs, and 200-300 nt-long A/U-heteropolymers that are added to fully-edited and unedited mRNAs.54 The A/U-tailing is accomplished by a concerted action of KPAP1 poly(A) polymerase and RET1 TUTase, and is aided by kinetoplast polyadenylation factors 1 and 2 (KPAF1/2 complex). It has been proposed that the pre-editing addition of a short A-tail by KPAP1 stabilizes the partially-edited transcript4,5 while the A/U-tailing commits the fully-edited and unedited mRNA to translation by recruiting the small ribosomal subunit.55
Initial cues that 3′ uridylation and nucleolytic processing may be coupled came from observation of 800-1000 nt-long guide RNA precursors accumulating in a T. brucei cell line with silenced endogenous RET1, or in cells overexpressing catalytically inactive enzyme.53 These results were puzzling because RET1 TUTase lacks nuclease activity,56 yet its TUTase activity is apparently required for nucleolytic precursor processing. Furthermore, RET1 knockdown also triggers buildup of rRNA and mRNA precursors along with an increase in the abundance of a few mature unedited mRNAs.53 Conversely, decay rates of de-uridylated gRNAs in RET1 RNAi cells were similar to those of U-tailed molecules, suggesting that gRNA stabilization rests with the GRBC complex and does not depend on the U-tail's presence. To resolve this convoluted picture, Suematsu et al took an immunoaffinity purification approach to define putative components of the RET1 complex.57 Remarkably, RET1, RNase II-like 3′–5′ exonuclease DSS1, and 3 large proteins lacking discernible motifs were identified as nearly stoichiometric components of this complex. Cross-tagging and in vitro reconstitution confirmed that RET1, DSS1 and associated MPSS1-3 proteins form a stable ∼900 kDa particle termed the mitochondrial 3′ processome (MPsome). Genetic knockdowns of verified MPsome components lead to RET1 RNAi-like defects in gRNA processing and arrested cell growth phenotype. The Read laboratory previously identified DSS1 exonuclease58 by similarity to a hydrolytic subunit of the mitochondrial degradosome from S. cerevisiae, which is composed of DSS1 and SUV3 helicase, a member of a conserved Ski2 family of DExH box RNA helicases.59 However, extensive proteomic investigation of the MPsome-associated proteins failed to verify the proposed interaction60 between DSS1 and a putative SUV3 ortholog in trypanosomes.57 Unlike most members of the RNase II/RNR family, DSS1 proteins lack RNA binding motifs and are inactive in autonomous form. In yeast and trypanosomes DSS1 gains exonuclease activity by associating with a cognate complex, albeit by different means. In yeast, DSS1 exonuclease relies on SUV3 helicase for RNA binding and for ATP hydrolysis-driven unwinding of structured RNAs. In trypanosomes, the MPsome-embedded DSS1 likely utilizes RET1 for RNA binding and accumulates the energy of RNA hydrolysis61 to propel the MPsome through long gRNA precursors.
How to start and when to stop?
Historically, guide RNAs were identified as short patches of complementarity between minicircle or maxicircle DNA and fully-edited mRNAs while allowing G-T base-pairing and 2–3 mismatches.62 The limited number of available minicircle sequences, however, precluded comprehensive gRNA annotation, leaving significant coverage gaps for known mRNA editing sites. Initial RNA-Seq studies of small (40–65 nt) mitochondrial RNAs have uncovered an extremely complex transcriptome and highly redundant guide RNA coverage of edited mRNAs.34,63 Although specific values vary depending on assembly and alignment criteria, 60,441 unique species have been identified as potential guide RNAs while the remaining 73,360 small RNA assemblies were designated as non-guide RNAs (ngRNAs).57 Considering the difficulties of accounting for 5′ and 3′ heterogeneities and gRNA redundancy, these values probably overestimate the number of functional gRNA families. In any event, the 5′ triphosphates and 3′ U-tails found in most gRNAs and ngRNAs46 indicate a lack of 5′ processing while the 3′ processing/modification pathway appears to be similar. Mapping sequenced small RNAs to available minicircles has revealed an extremely complex transcriptional profile of these 1 kb-long molecules and immediately suggested an unorthodox mechanism of small RNA biogenesis (Fig. 1). The remarkable juxtaposition of small RNAs that map as sense and antisense to multiple loci suggests that guide RNA genes are transcribed bi-directionally giving rise to long sense and antisense precursors which overlap head-to-head. Similar data were obtained in a related organism Leishmania tarentolae.64 In agreement with this scenario, accumulation of both sense and antisense precursors, along with the loss of their respective gRNA and antisense ngRNA, occurs upon knockdown of either DSS1 or RET1 activities.57 Furthermore, sequencing of gRNA precursor termini has revealed that most of the 800–1000 nt-long primary transcripts also contain 3′ U-tails. The in vitro reconstitution experiments featuring affinity purified MPsome and a series of model RNA substrates have confirmed the main postulates of the gRNA processing model presented in Fig. 1. First, it has been demonstrated that the U-tail's presence facilitates MPsome-catalyzed degradation of a long gRNA precursor; this explains the inhibition of processing by a point mutation that inactivates TUTase activity.53 Second, the MPsome's highly processive 3′–5′ exonucleolytic activity appears to be fine-tuned to degrade structured RNAs, but to pause 11–13 nucleotides from the stable 30–40 nt double-stranded region formed by overlapping sense and antisense precursors. The pausing seems to be stochastic as the MPsome eventually moves through the double-stranded RNA until the substrate is degraded to 5–7 nucleotides. Third, the pausing creates a double-stranded processing intermediate with a single-stranded 3′ overhang, which incidentally corresponds to the length of a minimal RNA substrate for RET1 TUTase.56 Although the secondary uridylation was not captured in reconstitution experiments, it seems plausible that the MPsome pausing generates a “window of opportunity” for RET1 TUTase to add an oligo(U) tail thereby disengaging the MPsome from mature gRNA. Although this model provides a conceivable explanation for the existence of the U-tail in small RNAs, the following steps leading to double-stranded region separation and loading of functional gRNAs onto the guide RNA binding complex44 remain unclear. This process is likely to be accompanied by the asymmetric degradation of anti-sense ngRNAs; indeed, these molecules are typically present at much lower steady-state levels than cognate gRNAs are. Likewise, one would expect guide RNA to be maintained in a single-stranded state favorable for hybridization with pre-edited mRNA during editing.
Figure 1.
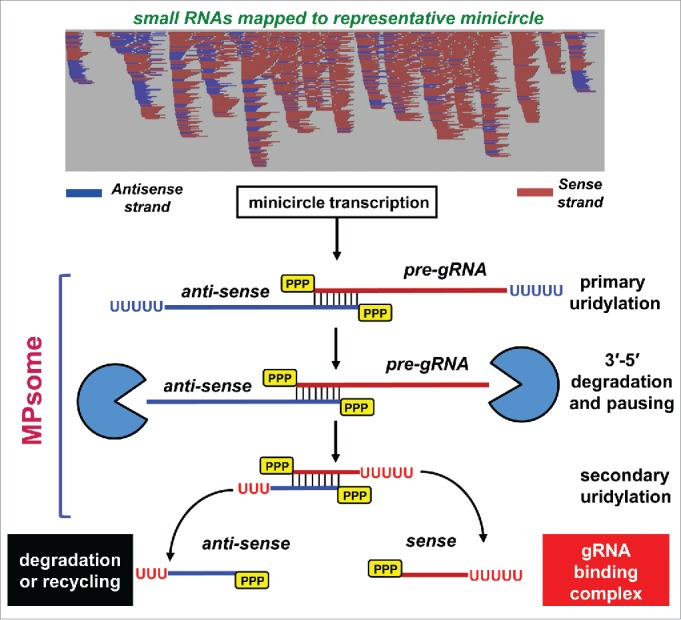
Guide RNA Biogenesis Model. Bidirectional transcription of gRNA gene generates sense and antisense precursors with overlapping 5′ regions. Mitochondrial 3′ processome-embedded RET1 TUTase and DSS1 3′–5′ exonuclease catalyze 3 coupled reactions: primary precursor uridylation, processive precursor degradation and secondary uridylation of trimmed gRNA. Primary uridylation stimulates DSS1hydrolytic activity, which provides energy for unwinding the secondary structures along gRNA precursor. The MPsome stochastically pauses at 10–12 nt from stable duplex regions formed by head-to-head hybridization of sense and antisense primary transcripts. The MPsome pausing allows RET1 TUTase to engage the single-stranded 3′ overhang and perform secondary uridylation. This step may disengage the MPsome from the duplex intermediate. Double-stranded RNA intermediate likely undergoes unwinding before mature gRNA can be sequestered by the gRNA binding complex and delivered into the editing pathway.
Although the mechanisms of microRNAs, siRNAs, and most other classes of small RNA processing vary to some extent, an underlying mechanism invokes multiple cleavages of a partially double-stranded precursor by RNase III-like endonucleases.65 In contrast, trypanosomal mitochondrial guide RNAs are derived from the 5′ extremity of a primary molecule by uridylation-induced, antisense transcription-controlled 3′–5′ exonucleolytic degradation.
Outlook
As debate on the evolutionary origins of RNA editing continues,66,67 the experimental focus is on molecular mechanisms that adapted ancient catalytic modules (RNase III, nucleotidyl transferase, ligase etc.) to function in protein assemblies capable of introducing RNA-programmed sequence changes. To some extent, the elemental editing reactions still resemble the pathways of their origin. For example, subunits of the core editing complex RET2 TUTase and U-specific 3′–5′ REX1 exonuclease are reminiscent of Pol β DNA polymerase and AP exonuclease, respectively, acting in concert on abasic DNA lesions. By the same token, guide RNA biogenesis reflects an evolutionarily conserved RNA decay pathway involving functional coupling between TUTase and RNase-II-like exonuclease, wherein uridylation stimulates RNA decay.30,68,69 However, sequestering the enzymes with seemingly opposing nucleotidyl transferase and hydrolase activities into the stable protein complex (MPsome) probably serves a more nuanced purpose of antisense transcription-dependent 3′end definition. It seems likely that such an antisense transcription-dependent mechanism is more universal and eventually will be discovered in other organisms. Indeed, there is ample evidence of closely-spaced convergent promoters in mammalian and other cells.32 The current model of gRNA biogenesis highlights the lack of knowledge about the nature of mitochondrial promoters, transcription factors and coupling between transcription and guide RNA processing. For example, the notion of guide RNA genes being flanked by imperfect inverted repeats70 may need to be re-examined in the light of precursor processing mechanisms. Based on a RET1 knockdown study,53 one could envisage that the MPsome is also responsible for processing maxicircle-encoded rRNA and mRNA precursors. These findings position controlled the 3′–5′ degradation as the main nucleolytic processing pathway in mitochondria of trypanosomes, which may also function in bulk RNA decay. With that, however, comes an inherent challenge to the decade-old concept of endonucleolytic partitioning of multicistronic maxicircle precursors that originate outside of the gene-packed conserved region. In T. brucei, minicircles typically encode several gRNAs so each precursor is bound to contain several gRNAs, of which all but the most 5′ unit are degraded. If the same concept is applicable to maxicircle precursor processing, then one must assume the existence of gene-specific promoters and a source of antisense transcripts to delimit the 3′–5′ degradation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of our laboratories for fruitful discussions. This work was supported by NIH grants AI091914 and AI101057 to RA, and AI11315 to IA.
ORCID
Takuma Suematsu http://orcid.org/0000-0003-1430-6580
Liye Zhang http://orcid.org/0000-0003-2104-6828
References
- 1.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005; 121:713-24; PMID:15935758; http://dx.doi.org/ 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 2.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al.. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005; 121:725-37; PMID:15935759; http://dx.doi.org/ 10.1016/j.cell.2005.04.030 [DOI] [PubMed] [Google Scholar]
- 3.Lange H, Sement FM, Canaday J, Gagliardi D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci 2009; 14:497-504; PMID:19716749; http://dx.doi.org/ 10.1016/j.tplants.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 4.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J 2008; 27:1596-608; PMID:18464794; http://dx.doi.org/ 10.1038/emboj.2008.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol Cell Biol 2005; 25:1634-44; PMID:15713623; http://dx.doi.org/ 10.1128/MCB.25.5.1634-1644.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rissland OS, Norbury CJ. The Cid1 poly(U) polymerase. Biochim Biophys Acta 2008; 1779:286-94; PMID:18371314; http://dx.doi.org/ 10.1016/j.bbagrm.2008.03.003 [DOI] [PubMed] [Google Scholar]
- 7.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell 2002; 108:637-48; PMID:11893335; http://dx.doi.org/ 10.1016/S0092-8674(02)00647-5 [DOI] [PubMed] [Google Scholar]
- 8.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci U S A 2003; 100:10617-22; PMID:12954983; http://dx.doi.org/ 10.1073/pnas.1833120100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst NL, Panicucci B, Igo RP Jr., Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell 2003; 11:1525-36; PMID:12820966; http://dx.doi.org/ 10.1016/S1097-2765(03)00185-0 [DOI] [PubMed] [Google Scholar]
- 10.Aphasizhev R. RNA uridylyltransferases. Cell Mol Life Sci 2005; 62:2194-203; PMID:16158189; http://dx.doi.org/ 10.1007/s00018-005-5198-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA 2009; 15:1322-37; PMID:19465686; http://dx.doi.org/ 10.1261/rna.1538809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao CY, Read LK. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol Biochem Parasitol 2007; 154:158-69; PMID:17543398; http://dx.doi.org/ 10.1016/j.molbiopara.2007.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett 2004; 572:15-8; PMID:15304317; http://dx.doi.org/ 10.1016/j.febslet.2004.07.004 [DOI] [PubMed] [Google Scholar]
- 14.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol 2007; 366:882-99; PMID:17189640; http://dx.doi.org/ 10.1016/j.jmb.2006.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual Role of the RNA Substrate in Selectivity and Catalysis by Terminal Uridylyl Transferases. Proc Natl Acad Sci U S A 2007; 104:14634-9; PMID:17785418; http://dx.doi.org/ 10.1073/pnas.0704259104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etheridge RD, Clemens DM, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol 2009; 164:66-73; PMID:19070634; http://dx.doi.org/ 10.1016/j.molbiopara.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 2009; 16(6):616-23; PMID:19430462; http://dx.doi.org/ 10.1038/nsmb.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(a) polymerases. Mol Cell Biol 2007; 27:3612-24; PMID:17353264; http://dx.doi.org/ 10.1128/MCB.02209-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J 2005; 24:4007-17; PMID:16281058; http://dx.doi.org/ 10.1038/sj.emboj.7600861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J Mol Biol 2010; 399:464-75; PMID:20403364; http://dx.doi.org/ 10.1016/j.jmb.2010.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol 2012; 19:782-7; PMID:22751018; http://dx.doi.org/ 10.1038/nsmb.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell 2014; 158:980-7; PMID:25171402; http://dx.doi.org/ 10.1016/j.cell.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 23.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 2009; 16:1021-5; PMID:19713958; http://dx.doi.org/ 10.1038/nsmb.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009; 138:696-708; PMID:19703396; http://dx.doi.org/ 10.1016/j.cell.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 25.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008; 32:276-84; PMID:18951094; http://dx.doi.org/ 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 26.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012; 151:521-32; PMID:23063654; http://dx.doi.org/ 10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- 27.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 2006; 12:1494-504; PMID:16790842; http://dx.doi.org/ 10.1261/rna.87706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trippe R, Richly H, Benecke BJ. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur J Biochem 2003; 270:971-80; PMID:12603330; http://dx.doi.org/ 10.1046/j.1432-1033.2003.03466.x [DOI] [PubMed] [Google Scholar]
- 29.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 2008; 22:50-65; PMID:18172165; http://dx.doi.org/ 10.1101/gad.1622708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014; 159:1365-76; PMID:25480299; http://dx.doi.org/ 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang JH, Tong L. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta 2012; 1819:992-7; PMID:22172994; http://dx.doi.org/ 10.1016/j.bbagrm.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013; 14:880-93; PMID:24217315; http://dx.doi.org/ 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- 33.Aphasizheva I, Aphasizhev R. U-Insertion/Deletion mRNA-Editing Holoenzyme: Definition in Sight. Trends Parasitol 2015; 32(2):144-56; PMID:26572691; http://dx.doi.org/ 10.1016/j.pt.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aphasizhev R, Aphasizheva I. Mitochondrial RNA editing in trypanosomes: Small RNAs in control. Biochimie 2014; 100:125-31; PMID:24440637; http://dx.doi.org/ 10.1016/j.biochi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 1990; 60:189-98; PMID:1688737; http://dx.doi.org/ 10.1016/0092-8674(90)90735-W [DOI] [PubMed] [Google Scholar]
- 36.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 1996; 84:831-41; PMID:8601307; http://dx.doi.org/ 10.1016/S0092-8674(00)81062-4 [DOI] [PubMed] [Google Scholar]
- 37.Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA [see comments]. Science 1996; 273:1189-95; http://dx.doi.org/ 10.1126/science.273.5279.1189 [DOI] [PubMed] [Google Scholar]
- 38.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 1997; 16:4069-81; PMID:9233816; http://dx.doi.org/ 10.1093/emboj/16.13.4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol 2000; 20:8447-57; PMID:11046141; http://dx.doi.org/ 10.1128/MCB.20.22.8447-8457.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 2003; 22:913-24; PMID:12574127; http://dx.doi.org/ 10.1093/emboj/cdg083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist 2010; 161:2-6; PMID:19945343; http://dx.doi.org/ 10.1016/j.protis.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Read LK, Lukes J, Hashimi H. Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip Rev RNA 2016; 7:33-51; PMID:26522170; http://dx.doi.org/ 10.1002/wrna.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics 2007; 7:534-45; PMID:18073385; http://dx.doi.org/ 10.1074/mcp.M700430-MCP200 [DOI] [PubMed] [Google Scholar]
- 44.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-Binding Complex from Mitochondria of Trypanosomatids. Molecular Cell 2008; 32:198-209; PMID:18951088; http://dx.doi.org/ 10.1016/j.molcel.2008.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukes J. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol 2013; 29:91-9; PMID:23305619; http://dx.doi.org/ 10.1016/j.pt.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 2014; 34:4329-42; PMID:25225332; http://dx.doi.org/ 10.1128/MCB.01075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clement SL, Koslowsky DJ. Unusual organization of a developmentally regulated mitochondrial RNA polymerase (TBMTRNAP) gene in Trypanosoma brucei. Gene 2001; 272:209-18; PMID:11470527; http://dx.doi.org/ 10.1016/S0378-1119(01)00538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grams J, Morris JC, Drew ME, Wang ZF, Englund PT, Hajduk SL. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. Journal of Biological Chemistry 2002; 277:16952-9; PMID:11859084; http://dx.doi.org/ 10.1074/jbc.M200662200 [DOI] [PubMed] [Google Scholar]
- 49.Koslowsky DJ, Yahampath G. Mitochondrial mRNA 3′ cleavage polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol Biochem Parasitol 1997; 90:81-94; PMID:9497034; http://dx.doi.org/ 10.1016/S0166-6851(97)00133-3 [DOI] [PubMed] [Google Scholar]
- 50.Adler BK, Harris ME, Bertrand KI, Hajduk SL. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol Cell Biol 1991; 11:5878-84; PMID:1719373; http://dx.doi.org/ 10.1128/MCB.11.12.5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol 1992; 52:231-40; PMID:1352374; http://dx.doi.org/ 10.1016/0166-6851(92)90055-O [DOI] [PubMed] [Google Scholar]
- 52.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell 1990; 62:391-7; PMID:1695552; http://dx.doi.org/ 10.1016/0092-8674(90)90375-O [DOI] [PubMed] [Google Scholar]
- 53.Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Molecular and Cellular Biology 2010; 30:1555-67; PMID:20086102; http://dx.doi.org/ 10.1128/MCB.01281-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide Repeat Proteins Stimulate mRNA Adenylation/Uridylation to Activate Mitochondrial Translation in Trypanosomes. Molecular Cell 2011; 42:106-17; PMID:21474072; http://dx.doi.org/ 10.1016/j.molcel.2011.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aphasizhev R, Aphasizheva I. Emerging roles of PPR proteins in trypanosomes: Switches, blocks, and triggers. RNA Biol 2013; 10:1495-500; PMID:24055869; http://dx.doi.org/15060068 10.4161/rna.26215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J Biol Chem 2004; 279:24123-30; PMID:15060068; http://dx.doi.org/ 10.1074/jbc.M401234200 [DOI] [PubMed] [Google Scholar]
- 57.Suematsu T, Zhang L, Aphasizheva I, Monti S, Huang L, Wang Q, Costello CE, Aphasizhev R. Antisense Transcripts Delimit Exonucleolytic Activity of the Mitochondrial 3′ Processome to Generate Guide RNAs. Mol Cell 2016; 61:364-78; PMID:26833087; http://dx.doi.org/ 10.1016/j.molcel.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattiacio JL, Read LK. Roles for TbDSS-1 in RNA surveillance and decay of maturation by-products from the 12S rRNA locus. Nucleic Acids Res 2008; 36:319-29; PMID:18032430; http://dx.doi.org/ 10.1093/nar/gkm690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dziembowski A, Malewicz M, Minczuk M, Golik P, Dmochowska A, Stepien PP. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol Gen Genet 1998; 260:108-14; PMID:9829834; http://dx.doi.org/ 10.1007/s004380050876 [DOI] [PubMed] [Google Scholar]
- 60.Mattiacio JL, Read LK. Evidence for a degradosome-like complex in the mitochondria of Trypanosoma brucei. FEBS Lett 2009; 583:2333-8; PMID:19540236; http://dx.doi.org/ 10.1016/j.febslet.2009.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science 2012; 336:1726-9; PMID:22745434; http://dx.doi.org/ 10.1126/science.1216848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maslov DA, Simpson L. Strategies of kinetoplastid cryptogene discovery and analysis. Methods Enzymol 2007; 424:127-39; PMID:17662839; http://dx.doi.org/ 10.1016/S0076-6879(07)24006-6 [DOI] [PubMed] [Google Scholar]
- 63.Koslowsky D, Sun Y, Hindenach J, Theisen T, Lucas J. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res 2013; 42(3):1873-86; PMID:24174546; http://dx.doi.org/ 10.1093/nar/gkt973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simpson L, Douglass SM, Lake JA, Pellegrini M, Li F. Comparison of the Mitochondrial Genomes and Steady State Transcriptomes of Two Strains of the Trypanosomatid Parasite, Leishmania tarentolae. Plos Neglect Trop D 2015; 9:e0003841; PMID:26204118; http://dx.doi.org/22952398 10.1371/journal.pntd.0003841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Bio 2014; 15:509-24; PMID:25027649; http://dx.doi.org/22952398 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 66.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol 2012; 4:a011403; PMID:22952398; http://dx.doi.org/ 10.1101/cshperspect.a011403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science 2010; 330:920-1; PMID:21071654; http://dx.doi.org/ 10.1126/science.1198594 [DOI] [PubMed] [Google Scholar]
- 68.Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013; 497:244-8; PMID:23594738; http://dx.doi.org/ 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 2013; 32:1842-54; PMID:23503588; http://dx.doi.org/ 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jasmer D, Stuart K. Sequence organization in African trypanosome minicircles is defined by 18 base pair inverted repeats. Mol Biochem Parasitol 1986; 18:321-32; PMID:3960057; http://dx.doi.org/ 10.1016/0166-6851(86)90089-7 [DOI] [PubMed] [Google Scholar]


