Abstract
Nemaline myopathy (NM) is a congenital muscle disorder associated with muscle weakness, hypotonia, and rod bodies in the skeletal muscle fibers. Mutations in 10 genes have been implicated in human NM, but spontaneous cases in dogs have not been genetically characterized. We identified a novel recessive myopathy in a family of line-bred American bulldogs (ABDs); rod bodies in muscle biopsies established this as NM. Using SNP profiles from the nuclear family, we evaluated inheritance patterns at candidate loci and prioritized TNNT1 and NEB for further investigation. Whole exome sequencing of the dam, two affected littermates, and an unaffected littermate revealed a nonsense mutation in NEB (g.52734272 C>A, S8042X). Whole tissue gel electrophoresis and western blots confirmed a lack of full-length NEB in affected tissues, suggesting nonsense-mediated decay. The pathogenic variant was absent from 120 dogs of 24 other breeds and 100 unrelated ABDs, suggesting that it occurred recently and may be private to the family. This study presents the first molecularly characterized large animal model of NM, which could provide new opportunities for therapeutic approaches.
Introduction
One of the most common human congenital myopathies, accounting for 17 % of cases, nemaline myopathy (NM) is characterized by the presence of rod bodies in the skeletal muscle fibers, muscle weakness, and hypotonia (Romero et al. 2013). Proximal limb muscles, facial muscles, and neck flexors are most frequently affected by muscle weakness, while respiratory insufficiency is the primary cause of death (Romero et al. 2013). NM is clinically heterogeneous and is classified into six subtypes based on severity, pattern of muscle weakness, and age of onset (Wallgren-Pettersson and Laing 2011). Ten genes have been implicated in NM: ACTA1, CFL2, KBTBD13, KLHL40, KHL41, LMOD3, NEB, TNNT1, TPM2, and TPM3 (Nowak et al. 2015). These genes encode proteins associated with the skeletal muscle sarcomere thin filament or Kelch domain proteins (Malfatti et al. 2014).
While founder mutations have been reported in Ashkenazi Jewish (NEB) (Anderson et al. 2004), Old Order Amish (TNNT1) (Johnston et al. 2000), and Turkish (TPM3) (Lehtokari et al. 2008) populations, most NM cases involve de novo mutations. Among cases explained at the genetic level, most are attributed to recessive mutations in NEB (usually in compound heterozygosity) and dominant mutations in ACTA1 (Romero et al. 2013). It is often infeasible to identify de novo mutations through Sanger sequencing due to numerous candidate genes and the prohibitive size of the NEB transcript (~26 kb, 183 exons). In recent years, next generation sequencing technologies that allow for simultaneous capture of candidate genes have expedited the genetic characterization of individual NM cases (Chen et al. 2013; Scoto et al. 2013; Güttsches et al. 2015; Marra et al. 2015).
Mouse models harboring mutations in ACTA1 (Nowak et al. 2009; Nguyen et al. 2011; Ravenscroft et al. 2011), LMOD3 (Tian et al. 2015), KLHL40 (Garg et al. 2014), NEB (Bang et al. 2006; Witt et al. 2006; Li et al. 2015), and TPM3 (Corbett et al. 2001; de Haan et al. 2002) are available for study of NM subtypes, but no large animal models have been described. Dog models have been instrumental in advancing therapeutic strategies for hereditary muscle disorders such as Duchenne muscular dystrophy and myotubular myopathy (Kornegay et al. 2012; Childers et al. 2014). While spontaneously occurring forms of NM have been reported in a Border collie and a Schipperke, they were not characterized at the molecular level (Shelton and Engvall 2005). Herein, we describe a novel NM in a family of American bulldogs (ABDs) and determine the pathogenic variant through genome-wide SNP profiling and whole exome sequencing (WES).
Materials and methods
Animals
A 5-month-old male ABD was evaluated at the University of Guelph Veterinary Teaching Hospital for non-progressive generalized muscle weakness, exercise intolerance, and tremors beginning at approximately 2 months of age. A female littermate had similar clinical signs, while another female littermate was reported to be clinically unaffected. Two other littermates could not be located. The sire and dam of the litter were cousins, and there was no prior history of muscle disease in the family. Samples from the family were collected for diagnostic procedures and submitted with owner consent.
Histopathology and immunohistochemistry
Biopsies from the triceps, biceps femoris, and suprascapular muscles were collected from both affected dogs under general inhalational anesthesia following electrophysiological examination. The biopsy specimens were either snap frozen in isopentane (pre-cooled in liquid nitrogen) or immersion fixed in 2.5 % glutaraldehyde for electron microscopy. Sections (8 µm) were further processed using a standard panel of histological and histochemical stains and reactions (Dubowitz et al. 2013). Similar staining and reactions were performed on age-matched control muscles from the tissue archives of the Comparative Neuromuscular Laboratory.
Electron microscopy
Glutaraldehyde-fixed muscle specimens were post-fixed in 1 % aqueous osmium tetroxide prior to dehydration and embedding in araldite resin. Thick sections (1 µm) were stained with toluidine blue-basic fuchsin prior to light microscopic examinations, while thin sections (60–90 nm) were stained with uranyl acetate and lead citrate prior to examination in a Zeiss 10 electron microscope.
DNA preparation
DNA was extracted from muscles of both affected dogs and blood from the sire, dam, and unaffected littermate using the DNeasy extraction kit (Qiagen, Hilden, Germany). Whole blood samples from ABDs recruited for an unrelated study were obtained as controls (Mauldin et al. 2014). DNA was isolated following the Gentra PureGene protocol (Qiagen). Genomic DNAs from purebred dogs of other breeds were available from our private DNA bank at Clemson University.
Illumina SNP arrays
To date, 10 genes have been identified in human cases of NM; their canine counterparts lie on 10 different chromosomes. To investigate inheritance patterns at each gene, SNP profiles were generated for the nuclear family using the Illumina CanineHD BeadChip, containing 173,662 SNPs (Illumina, San Diego, CA, USA). Genotypes at polymorphic SNPs encompassing the candidate genes (flanking the genes within 1 Mb) were examined for consistency with an autosomal recessive mode of inheritance.
Whole exome sequencing
Whole exome sequencing (WES) data were generated for the dam, two affected siblings, and an unaffected sibling at CeGaT GmbH (Tübingen, Germany). Genomic DNA (1 µg) from each sample was mechanically sheared to fragments of approximately 180–250 bp (Covaris LE220, Woburn, MA, USA). Fragment sizes were verified for quality control (Fragment Analyzer, Advanced Analytical, Ankeny, IA, USA). The fragment library was hybridized with 120-mer biotinylated RNA baits from the SureSelect XT Canine All Exon kit (Agilent, Santa Clara, CA, USA), which was designed based on the UCSC CanFam2 Ensembl and Refseq tracks as well as human protein alignments. Magnetic streptavidin beads were used for purification according to the manufacturer’s instructions. Library DNA was amplified, sequencing barcodes and adapters were added (Illumina), and equimolar amounts of each sample were pooled. The pool was sequenced on both lanes of a Rapid Flowcell on a HiSeq 2500 instrument (Illumina), generating paired-end 2 × 100 bp sequences, comprising on average 6 GB per sample. Sequencing data were demultiplexed (Illumina bcl2fastq 1.8.2), sequencing adapters were trimmed (skewer 0.1.116), and the resulting sequence mapped to the canine genome (CanFam3.1) using the Burrows-Wheeler Aligner (bwa 0.7.2-r351). PCR duplicates and low-quality alignments were removed (samtools 0.1.18 and internal software). Variant calling was performed using a dual pipeline of bcftools (0.1.17) and varscan (2.3.5), and internal software was used to combine these files into a single variant call file (VCF) per sample.
Reads were visualized using integrated genomics viewer (IGV) (Thorvaldsdóttir et al. 2013). Known variants from Ensembl dbSNP (Can Fam3.1) and from private whole genome sequence databases were excluded. NEB was manually screened for variants predicted to have an effect on the protein.
Genotyping of g.52734272
Primers for PCR were designed to capture a 489 bp region encompassing g.52734272 (Forward 5′-AAGTCCCAGCAGCAACATAA-3′, Reverse 5′-GTCCAAAGTGGTCGGTCCT-3′). Products were amplified using ReddyMix master mix (Thermo Scientific, Waltham, MA, USA) with 0.4 µM primers, 50 ng DNA, and water for a total volume of 25 µL. Thermal cycling conditions were as follows: 95 °C for 5 min; five touchdown cycles of 95 °C for 30 s, 58 °C for 30 s reducing 1 °C per cycle, and 72 °C for 1 min; 31 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; and a 10-min final elongation at 72 °C. Direct sequencing was carried out with BigDye Terminator using an ABI 3730xl Genetic Analyzer to validate g.52734272.
Using the same primers and thermal cycling conditions described above, genotyping of g.52734272 in unrelated ABDs and other breeds was carried out through a restriction enzyme digest with 5 µL PCR product and either BfaI CutSmart (New England Biolabs, Ipswich, MA, USA) or FspBI FastDigest (Thermo Scientific) for total reaction volumes of 25 and 15 µL, respectively. BfaI (FspBI) recognizes and cleaves (^) the following sequence: 5′-C^TAG-3′. Digests were visualized on a 1.2 % agarose gel, where wild-type alleles are uncut (489 bp) and mutant alleles are cut once (248 and 241 bp).
Gel electrophoresis and western blotting
Frozen tissue samples were ground to a fine powder using a glass mortar and pestle chilled in liquid nitrogen. After 20 min of priming at −20 °C, tissue was resuspended in a 1:1 mixture of an 8 M urea buffer (8 M urea, 2 M thiourea, 0.05 M Tris–HCl, 0.075 M dithiothreitol, as well as 3 % SDS and 0.03 % bromophenol blue, pH 6.8) and 50 % glycerol with protease inhibitors (0.04 mM E-64, 0.16 mM leupeptin, and 0.2 mM PMSF). The solutions were mixed in a 60 °C water bath for 4 min, followed by a 10 min incubation at the same temperature. Residual debris was removed via centrifugation at 13,000 rpm, and the supernatant was flash frozen for storage at −80 °C. Initial protein analysis was done via a 2–7 % gradient acrylamide gel. Western blot was performed on the samples using 0.8 % agarose gels run at 15 mA per gel for 2 h and 50 min. Following this, they were transferred onto Immobilon-P PVDF membranes (Millipore, Billerica, MA, USA) using a semi-dry transfer unit (Bio-Rad, Hercules, CA, USA) for 2.5 h at 164 mA. Membranes were briefly stained with Ponceau S to check for transferred proteins. Following removal of the stain, membranes were incubated with primary antibody at 4 °C overnight. Nebulin N-terminal and C-terminal expression was quantified using primary antibodies specific to those regions (Myomedix #6969 and #6964, respectively, http://www.myomedix.com). Expression was normalized to the integrated optical density of myoglobin heavy chain (MHC) obtained from the Ponceau S staining. The fluorescence of the western blots was analyzed using Odyssey Infrared Imaging System. Ponceau S images were analyzed with One-D scan EX (Scanalytics Inc., Rockville, MD, USA).
Results
Affected dogs could independently ambulate, had generalized atrophy, and the myopathy was relatively non-progressive (Supplemental video). Atrophy of the cervical and dorsal thoracic limb muscles was noted with bilateral hypertrophy of the triceps muscles. Serum creatine kinase (CK) activities were mildly elevated. Electromyography (EMG) revealed spontaneous electrical activity, consisting mainly of fibrillation potentials, within the proximal appendicular muscles of the thoracic limbs and the cervical paraspinal musculature. Motor nerve conduction velocity (MNCV) testing showed a mild decrease in the latency of the tibial and ulnar nerves. Respiratory difficulties were not present.
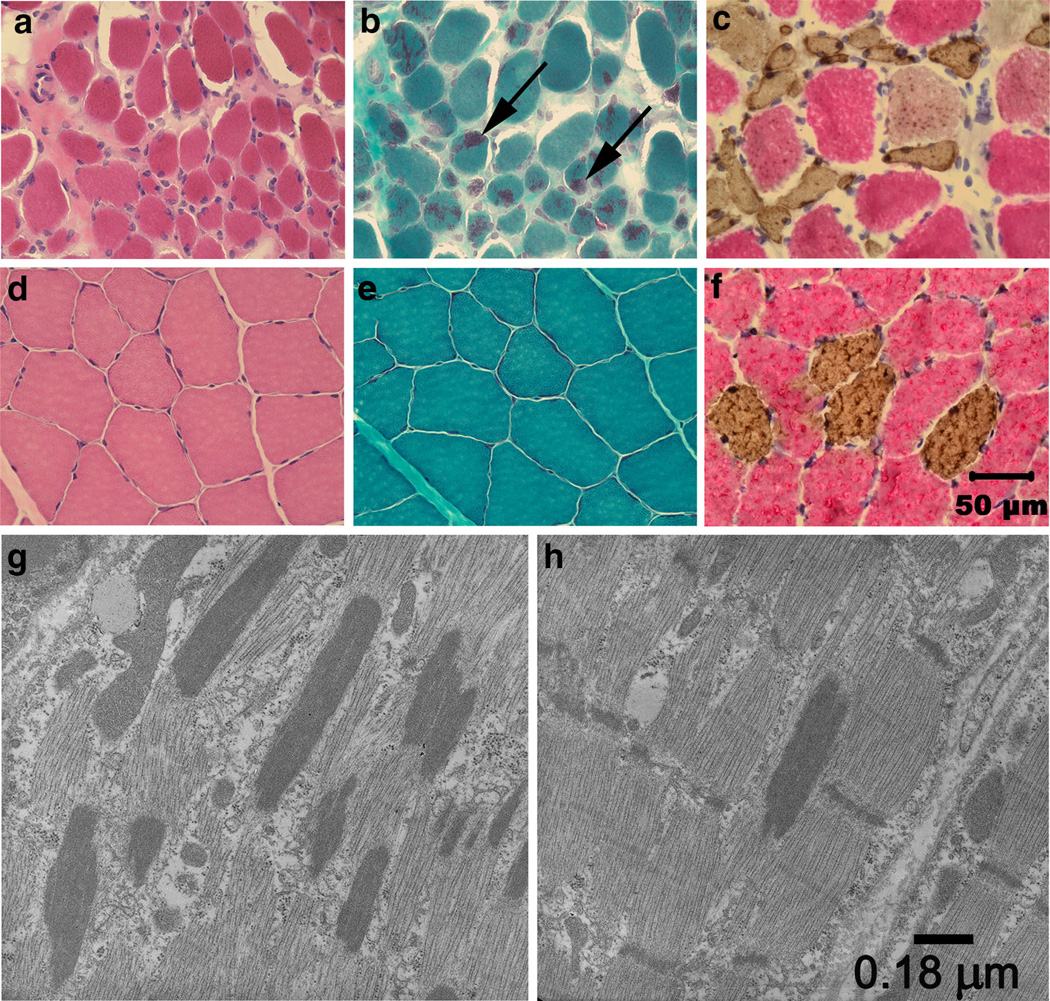
A marked variability in myofiber size and generalized atrophy was present in muscles from the affected ABDs (Fig. 1a) compared to control muscle (Fig. 1d). The predominant abnormality found in >50 % of the muscle fibers from all affected muscles examined was rod-like inclusion bodies highlighted with the modified Gomori trichrome stain (Fig. 1b); these structures were not observed in control muscles (Fig. 1e) or with H&E staining (Fig. 1a, d). Rod-like structures tended to be centralized or peripherally distributed in the muscle fibers and were present in both slow twitch type 1 and fast twitch type 2 muscle fibers. Atrophic fibers were also of both fiber types (Fig. 1c, f). Numerous rods were apparent along the long axis parallel to that of the muscle filaments (Fig. 1g). The rods were in structural continuity with Z disks (Fig. 1h), had the same electron density as the Z lines of adjacent sarcomeres, and had a similar lattice pattern of periodic cross striations.
Fig. 1.
Histopathology establishes NM. Cryosections from the triceps muscle of an affected pup (a–c) and archived control triceps muscle (d–f) were stained with H&E (a, d), modified Gomori trichrome (b, e) and following incubation with monoclonal antibodies against type 1 and type 2 myosin heavy chains (c, f). Excessive variability in myofiber size and atrophy were observed in the affected muscle with the H&E stain (a) compared to control muscle (d). Numerous myofibers in the affected muscle contained rod bodies (b, arrows) not evident in control muscle (e). Both type 1 and type 2 fibers were atrophic (c) with fibers of both fiber types similar in size in control muscle (f). Bar 50 µm for images a–f. By electron microscopy, numerous electron dense rods were apparent along the long axis parallel to that of the muscle fiber (g). The rods were in structural continuity with Z disks (h), had the same electron density as the Z lines of adjacent sarcomeres, and had a similar lattice pattern of periodic cross striations. Bar 0.18 µm for images g and h
The absence of disease in the sire and dam indicates that transmission is likely autosomal recessive. We hypothesized that the parents were heterozygous for a deleterious allele inherited identical-by-descent through a common grandparent and that their affected progeny were homozygous. Allelic inheritance was manually evaluated for 28 polymorphic SNPs within or flanking the 10 candidate genes; only markers representing TNNT1 (CFA1) and NEB (CFA19) were consistent with a simple autosomal recessive pattern (Table 1).
Table 1.
Genotypes for the nuclear family are reported for polymorphic SNPs (CanFam2) representing each of the 10 candidate genes
| Gene (Chr) | SNP position |  |
||||
|---|---|---|---|---|---|---|
| TNNT1 (1) | 105264488 | AG | AG | GG | GG | AG |
| 105432142 | GG | AG | AG | AG | GG | |
| 105598975 | GG | CG | CG | CG | GG | |
| 106497529 | AG | AG | AA | AA | AG | |
| ACTA1 (4) | 12713391 | AG | AA | AG | AG | AA |
| 12875911 | AG | AA | AG | AG | AA | |
| TPM3 (7) | 45997866 | CC | AC | CC | CC | CC |
| 46022207* | AT | AA | AA | AT | AT | |
| 46034262* | AC | AA | AA | AC | AC | |
| 46049585* | AG | AG | AA | AG | AG | |
| 46303991 | CC | AC | CC | CC | CC | |
| CFL2 (8) | 16507437 | CC | AC | CC | AC | CC |
| 16587360 | GG | AG | GG | AG | GG | |
| TPM2 (11) | 55241525* | AC | CC | CC | AC | AC |
| 55248320* | AA | AG | AA | AG | AA | |
| NEB (19) | 55761948* | GG | AG | GG | GG | GG |
| 55777049* | GG | AG | GG | GG | GG | |
| 55824838* | AG | AG | AA | AA | AG | |
| LMOD3 (20) | 25536336 | AG | AA | AA | AA | AG |
| 25605665 | AA | GG | AG | AG | AG | |
| 25625642 | AG | AA | AA | AA | AG | |
| KLHL40 (23) | 14709678 | AC | AA | AC | AC | AC |
| 14867876 | AG | GG | AG | AG | AG | |
| KBTBD13 (30) | 32370798 | AG | GG | GG | GG | AG |
| 32443189 | AG | GG | GG | GG | AG | |
| 32458624 | AC | AA | AA | AA | AC | |
| KLHL41 (36) | 17244342 | AG | AA | AA | AA | AG |
| 17280474 | AG | GG | AG | AG | GG | |
When possible, SNPs lying within the gene are used (asterisks); otherwise, the most proximal flanking SNPs are reported. When present, flanking SNPs (within 1 Mb) for which the affected dogs are homozygous are also shown. SNPs fitting a recessive pattern of inheritance are highlighted in bold
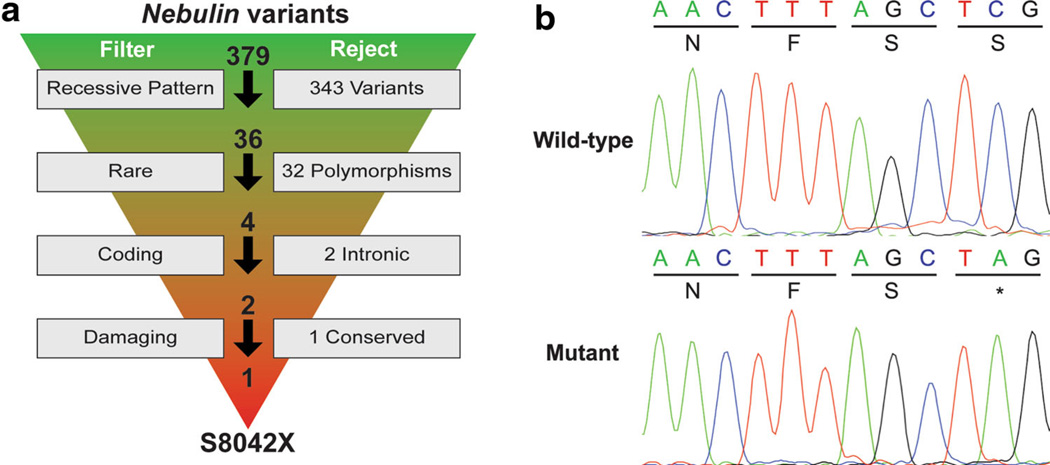
We conducted WES of four family members because of the large size of NEB. Across the samples, exome coverage, mappable reads, yields, and mean quality scores averaged 30×, 59 million, 6 Mb, and 37.1, respectively. VCFs were used to filter variants within the two candidate genes for those fitting recessive transmission (homozygous alternate in both affected dogs, heterozygous in the dam, and heterozygous or homozygous reference in the unaffected sibling). This step eliminated all 11 variants identified within TNNT1 and 343 variants in NEB. Thirty-six remaining variants in NEB were filtered to remove common polymorphisms, non-coding variants, and non-damaging variants (Fig. 2a). Only one overtly deleterious variant was identified in NEB, a nonsense mutation g.52734272 C>A, corresponding to human exon 169 (S8042X; NP_001258137.1). In addition, we confirmed that no variants in the other eight candidate genes segregated with the phenotype. The NEB nonsense mutation was verified by Sanger sequencing (Fig. 2b) and was absent in 100 unrelated ABDs and five dogs each from 24 other breeds, determined by restriction digest genotyping.
Fig. 2.
Filtering of NEB variants reveals S8042X. a Filtering parameters are shown to the left, with the number of rejected variants to the right. Total variants prior to each filtering parameter are shown above the arrows, beginning with the total number of variants. b Chromatograms from Sanger sequencing show the wild-type and mutant alleles
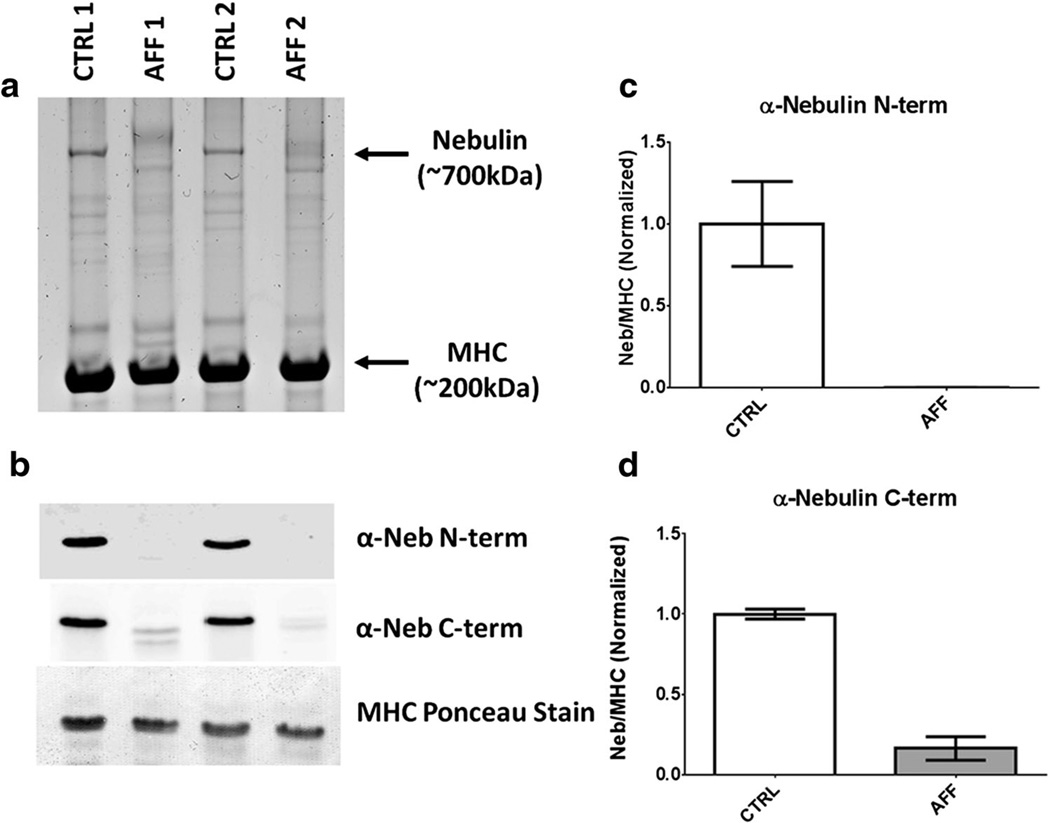
Protein analysis of the triceps biopsies from the affected dogs suggested a loss of NEB, which was then confirmed via western blotting (Fig. 3a, b). These values were quantified, revealing NEB expression at 0.3 and 16.2 % of their wild-type counterparts for the N-terminus and C-terminus, respectively (Fig. 3c, d).
Fig. 3.
Nebulin is drastically reduced in skeletal muscles with S8042X. a Protein analysis on a 2–7 % gradient acrylamide gel reveals a clear loss of nebulin in muscles from affected dogs compared to healthy controls. b Western blots for the same samples using antibodies to nebulin’s N- and C-terminus. c and d Average quantification (CTRL 1 and CTRL 2; AFF 1 and AFF 2) of western blots, indicating a severe loss of nebulin protein in affected dogs.
Discussion
Histological and clinical findings from the affected ABDs are consistent with primary NM. While the nemaline rod bodies are hallmarks of the disease, these morphological features provide little indication of subtype or genetic etiology. The most common of the six subtypes is typical congenital NM, which is most often attributed to mutations in NEB (Romero et al. 2013, Wallgren-Pettersson et al. 2011). The clinical presentation of independent ambulation and relatively non-progressive muscle weakness at 2 months of age in the ABDs is consistent with typical congenital NM (North and Ryan 2002).
We used a bipartite approach to uncover a nonsense mutation in NEB causing ABD NM. To refine the list of 10 known candidate genes, we generated genome-wide SNP profiles. Affected dogs inherited identical SNP haplotypes spanning TNNT1 and NEB; therefore, these genes were prioritized for further study. Together, they comprise nearly 200 exons, rendering Sanger sequencing impractical. Instead, we utilized WES, an approach that provides high exon read coverage and the data to parse the rest of the exome if no mutations are found in candidate genes.
NEB, or nebulin, is a giant protein (600–900 kDa) that stabilizes the actin thin filament of skeletal muscle sarcomeres and is critical for proper thin filament formation and muscle contraction (Labeit et al. 2011). NEB mutations account for up to 50 % of molecularly characterized NM cases (North 2011); most are truncating (Lehtokari et al. 2014). NEB has a multitude of isoforms achieved primarily through alternative splicing of three sets of exons (63–66, 143–144, and 167–177); truncating NEB mutations are frequently found in the latter set. In general, homozygous truncating mutations may be less tolerated in ubiquitously expressed exons (Lehtokari et al. 2014). Consistent with these trends observed in human myopathies, S8042X lies in the alternatively spliced exons 167–177.
We elected to investigate NEB protein levels, rather than transcript presence, in cases and controls due to limited tissue quantities. Protein gels show little banding in the NEB region, a finding consistent with the absence of protein detected with the N-terminus anti-nebulin antibody. The C-terminus antibody showed a low level of protein (~16 % of the levels in the control tissues) but with a mobility slightly larger than that of NEB in the control samples. It is possible that this is a unique splice isoform of nebulin that has low abundance and excludes the N-terminus. Alternatively, the C-terminus antibody may be cross-reacting with another protein similar in size or a degradation product of a larger protein. We consider the cross-reactivity hypothesis more likely because we have detected titin degradation products of a size similar to NEB that cross-react with NEB C-terminus antibodies in previous work. Regardless, our data suggest that S8042X results in very low levels of NEB, probably due to nonsense-mediated decay. The inability to successfully express normal levels of full-length nebulin likely underlies the pathology of the affected ABDs.
While we report the first mutation causing NM in dogs, this is not the first pathogenic variant described in canine NEB. Ahram et al. (2015) identified NEB missense mutations associated with primary angle-closure glaucoma (PACG) in Basset hounds. Interestingly, affected Basset hounds display no clinical signs of NM (Ahram et al. 2015). Ocular abnormalities were not identified in either of the ABDs with NM, nor are they associated with human NM attributed to NEB.
The recent development of WES enrichment kits for canines (Broeckx et al. 2014, 2015) will facilitate faster discovery of pathogenic variants, particularly when candidate genes are sizable or numerous. Massive parallel sequencing of several exome libraries in a single hi-seq lane is an economical approach for sequencing of multiple individuals, relative to whole genome resequencing, and permits simultaneous identification and filtering of variants. To date, this report is one of few to use WES in a nuclear family to identify a pathogenic variant in dogs (Ahonen et al. 2013; Ahram et al. 2015). Rather than requiring the widespread distribution of the ABD mutation throughout the population in order to gather sufficient cases for analysis with traditional methods, WES allowed us to detect the mutation directly in the two probands.
Although no carriers were found in a survey of unrelated ABDs (extended family members were unavailable for study), a genetic test now exists for S8042X should NM arise again in the breed. The identification of carriers holds potential for the development of a colony for testing of therapeutic approaches for humans.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R15AR062868 and R01AR053897. The authors wish to thank the dog owners for contributing samples to this study, and James R. Clark for helping prepare Fig. 2.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-016-9644-9) contains supplementary material, which is available to authorized users.
Contributor Information
G. Diane Shelton, Email: gshelton@ucsd.edu.
Leigh Anne Clark, Email: lclark4@clemson.edu.
References
- Ahonen SJ, Arumilli M, Lohi H. A CNGB1 frameshift mutation in Papillon and Phalene dogs with progressive retinal atrophy. PLoS One. 2013;8:e72122. doi: 10.1371/journal.pone.0072122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahram DF, Grozdanic SD, Kecova H, Henkes A, Collin RW, Kuehn MH. Variants in nebulin (NEB) are linked to the development of familial primary angle closure glaucoma in Basset hounds. PLoS One. 2015;10:e0126660. doi: 10.1371/journal.pone.0126660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Ekstein J, Donnelly MC, Keefe EM, Toto NR, LeVoci LA, Rubin BY. Nemaline myopathy in the Ashkenazi Jewish population is caused by a deletion in the nebulin gene. Hum Genet. 2004;115:185–190. doi: 10.1007/s00439-004-1140-8. [DOI] [PubMed] [Google Scholar]
- Bang ML, Li X, Littlefield R, Bremner S, Thor A, Knowlton KU, Lieber RL, Chen J. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckx BJG, Coopman F, Verhoeven GEC, Bavegems V, De Keulenaer S, De Meester E, Van Niewerburgh F, Deforce D. Development and performance of a targeted whole exome sequencing enrichment kit for the dog (Canis Familiaris Build 31) Sci Rep. 2014;4:5597. doi: 10.1038/srep05597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckx BJG, Hitte C, Coopman F, Verhoeven GEC, De Keulenaer S, De Meester E, Derrien T, Alfoldi J, Lindblad-Toh K, Bosmans T, Gielen I, Van Bree H, Van Ryssen B, Saunders JH, Van Niewerburgh F, Deforce D. Improved canine exome designs, featuring ncRNAs and increased coverage of protein coding genes. Sci Rep. 2015;5:12810. doi: 10.1038/srep12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang JL, Tang BS, Sun ZF, Shi YT, Shen L, Lei LF, Wei XM, Xiao JJ, Hu ZM, Pan Q, Xia K, Zhang QY, Dai MZ, Liu Y, Ashizawa T, Jiang H. Using next-generation sequencing as a genetic diagnostic tool in rare autosomal recessive neurologic Mendelian disorders. Neurobiol Aging. 2013;34:2442.e11–2442.e17. doi: 10.1016/j.neurobiolaging.2013.04.029. [DOI] [PubMed] [Google Scholar]
- Childers MK, Joubert R, Poulard K, Moal C, Grange RW, Doering JA, Lawlor MW, Rider BE, Jamet T, Daniéle N, Martin S, Riviére C, Soker T, Hammer C, Van Wittenberghe L, Lockard M, Guan X, Goddard M, Mitchell E, Barber J, Williams JK, Mack DL, Furth ME, Vignaud A, Masurier C, Mavilio F, Moullier P, Beggs AH, Buj-Bello A. Gene therapy prolongs survival and restores function in murine and canine models of myotubular myopathy. Sci Trans Med. 2014;6:220ra10. doi: 10.1126/scitranslmed.3007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett MA, Robinson CS, Dunglison GF, Yang N, Joya JE, Stewart AW, Schnell C, Gunning PW, North KN, Hardeman EC. A mutation in alpha-tropomyosin (slow) affects muscle strength, maturation and hypertrophy in a mouse model for nemaline myopathy. Hum Mol Genet. 2001;10:317–328. doi: 10.1093/hmg/10.4.317. [DOI] [PubMed] [Google Scholar]
- de Haan A, van der Vliet MR, Gommans IM, Hardeman EC, van Engelen BG. Skeletal muscle of mice with a mutation in slow alpha-tropomyosin is weaker at lower lengths. Neuromuscul Disord. 2002;12:952–957. doi: 10.1016/s0960-8966(02)00138-4. [DOI] [PubMed] [Google Scholar]
- Dubowitz V, Sewry CA, Oldfors A. Muscle biopsy: a practical approach. 4th. Oxford: Saunders Elsevier; 2013. [Google Scholar]
- Garg A, O’Rourke J, Long C, Doering J, Ravenscroft G, Bezprozvannaya S, Nelson BR, Beetz N, Li L, Chen S, Laing NG, Grange RW, Bassel-Duby R, Olson EN. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J Clin Invest. 2014;124:3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttsches AK, Dekomien G, Claeys KG, von der Hagen M, Huebner A, Kley RA, Kirschner J, Vorgerd M. Two novel nebulin variants in an adult patient with congenital nemaline myopathy. Neuromuscul Disord. 2015;25:392–396. doi: 10.1016/j.nmd.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Kelley RI, Crawford TO, Morton DH, Agarwala R, Koch T, Schäffer AA, Francomano CA, Biesecker LG. A novel nemaline myopathy in the Amish caused by a mutation in troponin T1. Am J Hum Genet. 2000;67:814–821. doi: 10.1086/303089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay JN, Bogan JR, Bogan DJ, Childers MK, Li J, Nghiem P, Detwiler DA, Larsen CA, Grange RW, Bhavaraju-Sanka RK, Tou S, Keene BP, Howard JF, Jr, Wang J, Fan Z, Schatzberg SJ, Styner MA, Flanigan KM, Xiao X, Hoffman EP. Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm Genome. 2012;2:85–108. doi: 10.1007/s00335-011-9382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Ottenheijm CA, Granzier H. Nebulin, a major player in muscle health and disease. FASEB J. 2011;25:822–829. doi: 10.1096/fj.10-157412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtokari VL, Pelin K, Donner K, Voit T, Rudnik-Schöneborn S, Stoetter M, Talim B, Topaloglu H, Laing NG, Wallgren-Pettersson C. Identification of a founder mutation in TPM3 in nemaline myopathy patients of Turkish origin. Eur J Hum Genet. 2008;16:1055–1061. doi: 10.1038/ejhg.2008.60. [DOI] [PubMed] [Google Scholar]
- Lehtokari VL, Kiiski K, Sandaradura SA, Laporte J, Repo P, Frey JA, Donner K, Marttila M, Saunders C, Barth PG, den Dunnen JT, Beggs AH, Clarke NF, North KN, Laing NG, Romero NB, Winder TL, Pelin K, Wallgren-Pettersson C. Mutation update: the spectra of nebulin variants and associated myopathies. Hum Mutat. 2014;35:1418–1426. doi: 10.1002/humu.22693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Buck D, De Winter J, Kolb J, Meng H, Birch C, Slater R, Escobar YN, Smith JE, III, Yang L, Konhilas J, Lawlor MW, Ottenheijm C, Granzier HL. Nebulin deficiency in adult muscle causes sarcomere defects and muscle-type-dependent changes in trophicity: novel insights in nemaline myopathy. Hum Mol Genet. 2015;24:5219–5233. doi: 10.1093/hmg/ddv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfatti E, Lehtokari VL, Böhm J, De Winter JM, Schäffer U, Estournet B, Quijano-Roy S, Monges S, Lubieniecki F, Bellance R, Viou MT, Madelaine A, Wu B, Taratuto AL, Eymard B, Pelin K, Fardeau M, Ottenheijm CA, Wallgren-Pettersson C, Laporte J, Romero NB. Muscle histopathology in nebulin-related nemaline myopathy: ultrastrastructural findings correlated to disease severity and genotype. Acta Neuropathol Commun. 2014;2:44. doi: 10.1186/2051-5960-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra JD, Engelstad KE, Ankala A, Tanji K, Dastgir J, De Vivo DC, Coffee B, Chiriboga CA. Identification of a novel nemaline myopathy-causing mutation in the troponin T1 (TNNT1) gene: a case outside of the old order Amish. Muscle Nerve. 2015;51:767–772. doi: 10.1002/mus.24528. [DOI] [PubMed] [Google Scholar]
- Mauldin EA, Wang P, Evans E, Cantner CA, Ferracone JD, Credille KM, Casal ML. Autosomal recessive congenital ichthyosis in American bulldogs is associated with NIPAL4 (ICHTHYIN) deficiency. Vet Pathol. 2014;52:654–662. doi: 10.1177/0300985814551425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MA, Joya JE, Kee AJ, Domazetovska A, Yang N, Hook JW, Lemckert FA, Kettle E, Valova VA, Robinson PJ, North KN, Gunning PW, Mitchell CA, Hardeman EC. Hypertrophy and dietary tyrosine ameliorate the phenotypes of a mouse model of severe nemaline myopathy. Brain. 2011;134:3516–3529. doi: 10.1093/brain/awr274. [DOI] [PubMed] [Google Scholar]
- North KN. Clinical approach to the diagnosis of congenital myopathies. Semin Pediatr Neurol. 2011;18:216–220. doi: 10.1016/j.spen.2011.10.002. [DOI] [PubMed] [Google Scholar]
- North KN, Ryan MM. Nemaline Myopathy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews. Seattle: University of Washington; 2002. [Google Scholar]
- Nowak KJ, Ravenscroft G, Jackaman C, Filipovska A, Davies SM, Lim EM, Squire SE, Potter AC, Baker E, Clément S, Sewry CA, Fabian V, Crawford K, Lessard JL, Griffiths LM, Papadimitriou JM, Shen Y, Morahan G, Bakker AJ, Davies KE, Laing NG. Rescue of skeletal muscle alpha-actin-null mice by cardiac (fetal) alpha-actin. J Cell Biol. 2009;185:903–915. doi: 10.1083/jcb.200812132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak KJ, Davis MR, Wallgren-Pettersson C, Lamont PJ, Laing NG. Clinical utility gene card for: nemaline myopathy. Eur J Hum Genet. 2015;23 doi: 10.1038/ejhg.2015.12. doi: 10.1038/ejhg.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft G, Jackaman C, Bringans S, Papadimitriou JM, Griffiths LM, McNamara E, Bakker AJ, Davies KE, Laing NG, Nowak KJ. Mouse models of dominant ACTA1 disease recapitulate human disease and provide insight into therapies. Brain. 2011;134:1101–1115. doi: 10.1093/brain/awr004. [DOI] [PubMed] [Google Scholar]
- Romero NB, Clarke NF. Congenital myopathies. Handb Clin Neurol. 2013;113:1321–1336. doi: 10.1016/B978-0-444-59565-2.00004-6. [DOI] [PubMed] [Google Scholar]
- Scoto M, Cullup T, Cirak S, Yau S, Manzur AY, Feng L, Jacques TS, Anderson G, Abbs S, Sewry C, Jungbluth H, Muntoni F. Nebulin (NEB) mutations in a childhood onset distal myopathy with rods and cores uncovered by next generation sequencing. Eur J Hum Genet. 2013;21:1249–1252. doi: 10.1038/ejhg.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton GD, Engvall E. Canine and feline models of human inherited muscle diseases. Neuromuscul Disord. 2005;15:127–138. doi: 10.1016/j.nmd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Ding S, You Y, Li TR, Liu Y, Wu X, Sun L, Xu T. Leiomodin-3-deficient mice display nemaline myopathy with fast-myofiber atrophy. Dis Model Mech. 2015;8:635–641. doi: 10.1242/dmm.019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallgren-Pettersson C, Sewry CA, Nowak KJ, Laing NG. Nemaline myopathies. Semin Pediatr Neurol. 2011;18:230–238. doi: 10.1016/j.spen.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.