Abstract
Rec8 is a prominent component of the meiotic prophase chromosome axis that mediates sister chromatid cohesion, homologous recombination and chromosome synapsis. Here, we explore the prophase roles of Rec8. (i) During the meiotic divisions, Rec8 phosphorylation mediates its separase-mediated cleavage. We show here that such cleavage plays no detectable role for chromosomal events of prophase. (ii) We have analyzed in detail three rec8 phospho-mutants, with 6, 24 or 29 alanine substitutions. A distinct ‘separation of function’ phenotype is revealed. In the mutants, axis formation and recombination initiation are normal, as is non-crossover recombination; in contrast, crossover (CO)-related events are defective. Moreover, the severities of these defects increase coordinately with the number of substitution mutations, consistent with the possibility that global phosphorylation of Rec8 is important for these effects. (iii) We have analyzed the roles of three kinases that phosphorylate Rec8 during prophase. Timed inhibition of Dbf4-dependent Cdc7 kinase confers defects concordant with rec8 phospho-mutant phenotypes. Inhibition of Hrr25 or Cdc5/polo-like kinase does not. Our results suggest that Rec8's prophase function, independently of cohesin cleavage, contributes to CO-specific events in conjunction with the maintenance of homolog bias at the leptotene/zygotene transition of meiotic prophase.
INTRODUCTION
Meiosis involves a complex succession of chromosomal events that result in the physical connection of homologous chromosomes. These events occur during a prolonged prophase period that precedes the two meiotic divisions. Connections between homologs are established during prophase and then ensure that maternal and paternal homologous chromosomes segregate to opposite poles later, during the first meiotic division (1,2). These connections, seen cytologically as ‘chiasmata’, result from the combined effects of reciprocal crossovers (COs) between non-sister chromatids of homologs and sister connections along the chromosome arms.
Sister chromatid cohesion, mediated by ring-shaped cohesin complexes, is crucial for meiotic chromosomal events. The meiotic cohesin complex, formed in the presence of the meiosis-specific α-kleisin subunit Rec8, abundantly occupies the conjoined sister structural axes that form during prophase (3–6). Rec8 is required for axis formation and, thus, ensuing formation of synaptonemal complexes (SCs) that join the axes of homologs at mid-prophase (5).
Meiotic recombination occurs in direct physical and functional association with chromosome axes at all stages (7). In a mutant devoid of Rec8 (rec8Δ), recombination still occurs relatively efficiently, with nearly-normal levels of recombination-initiating double-strand breaks (DSBs) and high levels of interhomolog recombination products. However, significant aberrancies are also apparent. DSB levels are modestly reduced and their processing is defective, as seen by hyper-resection of their single-strand DNA tails. In meiotic recombination, recombination occurs preferentially between homologs rather than sister chromatids (‘homolog bias’). In wild-type (WT) meiosis, homolog bias is established when a DSB engages a partner duplex. The recombination pathway then bifurcates into crossover (CO) and noncrossover (NCO) branches. In the absence of Rec8, establishment of homolog bias and pathway bifurcation are normal and NCO products appear at relatively high levels (4,8). However, the crossover branch of the pathway exhibits significant defects: progression through the several steps of CO recombination is significantly delayed; homolog bias is lost at a key intermediate stage and CO levels are reduced.
Rec8 is prominently subject to multiple phosphorylations, most of which cluster within a sequence-non-conserved central region of the molecule (3,9). Such phosphorylation is known to be essential for removal of Rec8 immediately prior to and during the two divisions (9–11; below). Importantly, it is the overall level of phosphorylation that is important for this effect, not simply phosphorylation of certain particular sites (9). However, Rec8 is also known to be abundantly phosphorylated not only during the divisions but throughout meiosis, from very early stages onward (9,11; this work). Moreover, mutant Rec8 proteins lacking phosphorylation at multiple different positions (rec8 ‘phospho-mutations’) confer significant defects during prophase. Together these findings have led to the suggestion that phosphorylation might have significant interesting prophase roles (3,12).
Existing data suggest that recombination is defective in rec8 phospho-mutants. All the analyzed phospho-alleles exhibit delayed exit from prophase (3,12). Where tested (3), these delays dependent on recombination initiation by the dedicated DSB trans-esterase Spo11, implying that they result from defective recombinational progression. Further, for one tested allele, a recombination defect was revealed by direct analysis (12). Interestingly, in analyzed cases, axis formation was seen to be normal while SC formation is defective (12; below). Interestingly, some rec8 phospho-alleles, including the three examined in detail in the current study, are normal for prophase sister chromatid cohesion. Phospho-mutant defects can thus identify important roles for Rec8 that go beyond its basic function in global sister cohesion.
The present study was initiated to further investigate the roles of Rec8 for meiotic prophase events, with further attention to the possibility that phosphorylation might be involved. Four types of studies were carried out.
First, we asked whether Rec8's (phosphorylation-dependent) separase-mediated cleavage is, or is not, also involved in Rec8's prophase roles. It is well-established that after prophase I, the programmed stepwise removal of Rec8 is essential for regular chromosome segregation over two meiotic divisions (13–15). Segregation of homologous chromosomes during meiosis I requires the release of arm cohesion between the sister chromatids while centromeric sister connections are maintained; segregation of sisters during meiosis II then requires the release of centromeric sister connections (14,16–18). Moreover, at both stages, cohesin removal involves programmed cleavage by separase, which is activated by the anaphase-promoting complex/cyclosome (APC/C) (19). There are hints that cleavage might not be involved. Appearance of detectable Rec8 cleavage products parallels the occurrence of meiosis I division, suggesting that cleavage might not be possible as early as prophase (3,20). Moreover, there are precedents for significant roles of Rec8/cohesin phosphorylation that do not involve cleavage. In mitotically dividing mammalian cells, cohesin is removed by a phosphorylation-dependent cleavage-independent process at the prophase-to-metaphase transition, phosphorylation-dependent cleavage occurring only later, during mitosis per se (21–23). An analogous situation pertains in budding yeast meiosis: cohesin is partially released from chromosomes at the prophase/metaphase transition by a separase-independent mechanism that is dependent on Cdc5 kinase, and thus presumably phosphorylation (11). Nonetheless, despite these considerations, roles for a low level of cohesin cleavage during meitoic prophase are not excluded and the possible involvement of such cleavage has never previously been directly examined.
Second, we wished to precisely define the prophase phenotypes conferred by three previously-characterized rec8 phospho-mutations: rec8-6A, rec8-24A and rec8-29A. We thus performed physical analysis of DNA recombination synchronous meiotic time courses, including identification of intermediates and CO and NCO products, in direct comparison with wild-type and a rec8Δ null mutant as described previously (4,8). We also further examined the effects of rec8 phospho-mutations on development of prophase axes and ensuing formation of synaptonemal complex.
Third, three different kinases have been implicated in phosphorylation of Rec8, at different stages and for different purposes. We were interested to examine the roles of these kinases for meiotic recombination. Casein kinase 1δ/ϵ (CK1δ/ϵ, Hrr25 in yeast) and Dbf4-dependent Cdc7 kinase (DDK) collaboratively account for Rec8 phosphorylation in early/mid-prophase (9). Polo-like kinase (‘PLK’; Cdc5 in yeast) can also phosphorylate Rec8 (24–26). Cdc5 is activated in mid/late prophase and has roles in the late prophase stages of recombination (27) and in cohesin removal at the prophase-to-metaphase transition (11). However, Cdc5 is not required for SC formation (27) suggesting that it is likely not involved in early/mid-prophase functions of Rec8; moreover, its late-stage recombination role is unaffected by a rec8 phospho-mutation, suggesting that it may also play no role in Rec8 phosphorylation at that stage.
Finally, the intimate physical and functional interplay between structural components and biochemical components of the recombination complex are established prior to DSB formation and continues throughout the entire recombination process (7). We have further explored this interplay for rec8 phospho-mutant phenotypes in relation to the roles of two other key types of molecules: Mek1, without which early inter-homolog recombination complexes are not properly assembled and/or activated (4,8), and two members of the ZMM ensemble, which forms at sites of CO-designations to nucleate SC formation and shepherd ensuing SC-associated biochemical steps of CO formation. Two ZMM components were considered: SUMO ligase Zip3 and SC component Zip1.
The findings presented provide new information with respect to all four lines of investigation. More speculatively, our results are consistent with a role for Rec8 modification in a specific constellation of effects related to CO formation and CO-dependent SC formation.
MATERIALS AND METHODS
Yeast strains
We used diploid Saccharomyces cerevisiae SK1 strains. The details regarding strain constructions are shown in Supplementary Table S1. Deletion mutant strains were constructed by polymerase chain reaction (PCR)-based 1-step gene disruption and confirmed by yeast colony PCR.
Meiotic time courses
Synchronous meiosis was achieved as described previously (4,8). Cells were first patched onto YPG plates (1% yeast extract, 2% bacto-peptone, 2% bacto-agar and 3% glycerol) and grown overnight, and single colonies were then picked from YPD plates (1% yeast extract, 2% bacto-peptone, 2% bacto-agar and 2% glucose) after 2 days of growth. To synchronize supplemented pre-sporulation (SPS) cultures at the G1 phase, a 1/500 dilution of the culture was grown at 30°C in SPS medium (0.5% yeast extract, 1% bacto-peptone, 0.67% yeast nitrogen base without amino acids, 1% potassium acetate and 0.05 M potassium biphthalate, pH 5.5). Meiosis was initiated by transferring cells to sporulation medium (SPM; 1% potassium acetate, 0.02% raffinose and 0.01% antifoam) pre-warmed to 30°C. For physical analysis of zmm mutants at 33°C (Figure 8), the cells were maintained at 30°C and then shifted to 33°C after 2.5 h growth in SPM (4). For analysis of Mek1-dependent meiotic recombination in rec8-6A, rec8-29A and rec8Δ mutants, a single culture was synchronized and divided into four identical sporulation cultures. Then, to inhibit Mek1 kinase activity in the mek1as strain, fresh 1-NA-PP1 (1 μM, final concentration) was added to three identical cultures after 3 h, 5 h and 7 h. Meiotic division was monitored by fluorescence microscopy after 4′,6-diamidino-2-phenylindole (DAPI) staining (4). The events of meiosis I and II were calculated from nuclear DAPI staining results (n = 200). Esp1 expression was induced by the addition of 50 μM CuSO4, at 2 h, 5 h or 7 h after transferring the PCUP1-ESP1 strain to SPM. hrr25-as and cdc7-as3 were inhibited with chemical inhibitors (5 μM 1NM-PP1 for hrr25-as and 15 μM PP1 for cdc7-as3) at 2 h after transferring the hrr25-as and cdc7-as3 strains to SPM medium. For analysis of the ndt80Δ PCUP1-CDC5 mutant, a single SPS culture was synchronized and divided into two identical SPM cultures. In one of these cultures, the Cdc5 protein was induced by the addition of 50 μM CuSO4 at 7 h after the induction of meiosis.
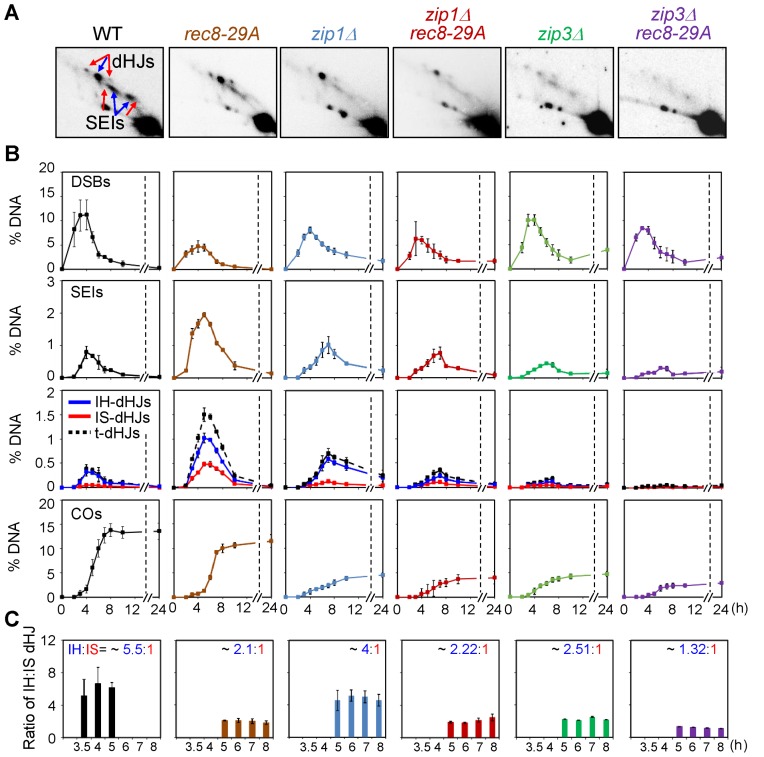
Figure 8.
Effects of rec8 phospho-mutant alleles on JM formation and homolog bias in zip1Δ and zip3Δ. For analysis at 33°C, meiotic cultures were maintained at 30°C and then shifted to 33°C after 2.5 h growth in SPM. (A) Representative 2D-gel images of JMs in various strains. (B) Quantification of DSBs, SEIs, dHJs and COs in WT, rec8-29A, zip1Δ, zip1Δ rec8-29A, zip3Δ and zip3Δ rec8-29A (mean ± SEM for three cultures). (C) IH:IS dHJ ratio in WT, rec8-29A, zip1Δ, zip1Δ rec8-29A, zip3Δ and zip3Δ rec8-29A (mean ± SEM for three cultures).
Physical analysis
Genomic DNA preparation and physical analysis was performed as described (4,8). For physical analysis of meiotic recombination, genomic DNA was treated with psoralen and DNA inter-strand crosslinks were induced using 365-nm ultraviolet light. DNA from each time point was extracted using a guanidine and phenol extraction method (4,8). Genomic DNA (2 μg) was digested with 80 units XhoI and dissolved in DNA loading buffer after precipitation with sodium acetate and ethanol. Electrophoresis in 1D gels (0.6% SeaKem LE agarose in Tris-borate-ethylenediaminetetra acetic acid (TBE)) was performed in TBE buffer at 2 V/cM for 24 h. For two-dimensional (2D) gel analysis, 2.5 μg of XhoI-digested DNA was loaded onto 0.4% SeaKem gold agarose gels (lacking ethidium bromide), and whole genomic DNA was separated. Gels were stained with 0.5 μg/ml ethidium bromide in TBE, and slices of each DNA lane containing DNA of interest were cut from the gel. Gel slices were placed into a 2D-gel tray for subsequent electrophoresis. DNA was positioned so that the higher molecular weights were to the left side. Two-dimensional gels (0.8% SeaKem LE agarose gel in TBE containing 0.5 μg/ml ethidium bromide) were poured onto the gel slices. Electrophoresis was performed in pre-chilled TBE containing 0.5 μg/ml ethidium bromide at 6 V/cm for 6 h at 4°C. For IH-CO and IH-NCO assays, 2 μg of DNA was digested with XhoI and NgoMIV and analyzed by 1D gel electrophoresis, as described above. Gels were subjected to Southern blot analysis with ‘Probe A’ after transferring onto nylon membranes (Bio-Rad). Probes were radiolabeled with 32P-dCTP using a Random Priming Kit (Agilent Technologies). The URA3-arg4 physical recombination reporter system and the CTAB/CoHex/Mg2+ based procedure to stabilize HJ containing recombination intermediates have been described (28). Hybridizing DNA signals were visualized and quantified using a phosphoimager with Quantity One software (Bio-Rad).
Chromosome spreading and immunofluorescence for yeast cytology
Fixation of yeast cells and chromosomes spreading for immunofluorescence assays were performed as described previously (4). Briefly, cells were spheroplasted in ZK buffer (25 mM Tris-Cl, 0.8 M KCl; pH 7.5,) with dithiothreitol (DTT; final concentration = 50 mM) for 2 min at room temperature with gentle mixing. The 100T zymolyase was added to reaction samples at an optimal concentration, and incubated for 30 min. Pellets were obtained by centrifugation, washed once and resuspended in cold MES buffer. Fixation and lysis were conducted using 3% paraformaldehyde and 3.4% sucrose, and 1% lipsol detergent, respectively, after which the samples were placed on clean slides. After drying, the slides were dipped in 0.2% Photo-Flo (Kodak Ltd.) for 30 s and transferred to TBS buffer (25 mM Tris-Cl, pH 8.0, 136 mM NaCl, 3 mM KCl), followed by blocking TBS buffer with 1% bovine serum albumin (BSA). A primary mouse monoclonal HA antibody (Santa Cruz Biotechnology, Catalog # sc-7392) was diluted 1:1000 and used to detect a recombinant 3HA-tagged Rec8 protein. A rabbit polyclonal anti-Zip1 antibody (Santa Cruz Biotechnology, Catalog # sc-33733) and a mouse monoclonal anti-Myc antibody (Santa Cruz Biotechnology, Catalog # sc-40) were used to detect Zip1 and Zip3-13myc, respectively. Secondary antibodies for recognizing the primary antibodies were Alexa-fluor-488-conjugated, goat anti-mouse IgG (Jackson ImmnoResearch, Catalog # 115-545-003, diluted 1:500) and a Cy3-conjugated goat, anti-rabbit IgG (Jackson ImmnoResearch, Catalog # 111-165-003, diluted 1:500). Images were acquired using a fluorescence microscope with 100x magnification. The contrast and brightness of images were adjusted using Adobe Photoshop software. The intensities of chromosomal arrays from WT and mutant Rec8 strains were measured and quantified using ImageJ software (National Institutes of Health). The number of Zip3 foci was quantified with GraphPad Prism Software.
Western blot analysis
Yeast cells were washed in distilled water and resuspended in 0.3 M NaOH. After a 5-min incubation at room temperature, samples were centrifuged at 5000 rpm for 1 min. After discarding the supernatant, 1x protein loading buffer was added to each pellet, mixed well and boiled for 5 min. Equal amounts of cell lysates were analyzed by sodium dodecyl sulfide-polyacrylamide gel electrophoresis and Western blotting was performed using standard procedures. To analyze Rec8 dephosphorylation, yeast cell extracts were incubated in a dephosphorylation buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 0.01% NP40, 2 mM MnCl2, 2 mM DTT) at 30°C for 3 h with or without lambda phosphatase while agitating. WT and mutant Rec8 proteins were detected with an HA antibody (Santa Cruz Biotechnology, Catalog # sc-7392, diluted 1:1000). An anti-Pgk1 antibody (Invitrogen, Catalog # 459250, diluted 1:5000) was used to detect the Pgk1 protein as a loading control.
Analysis of sister cohesion
We used PCUP1-ESP1 diploid cells carrying the Lac operator and a GFP-tagged Lac repressor on chromosome XV to analyze sister cohesion. Esp1 expression was induced with 50 μM CuSO4, added at 2 h after transferring cells to SPM medium. Samples were harvested at the indicated time points and fixed in 40% ethanol plus 0.1 M sorbitol, then stored at −20°C (4). For sister chromatid cohesion analysis, fixed cells were spun down and then resuspended in 10 mM Tris (pH 8.0) and 1 mM EDTA containing 1 μg/ml DAPI. Sister association was visualized immediately using an Olympus BX53 fluorescence microscope and analysis software, using filters for measuring GFP and DAPI fluorescence.
Spore viability tests
Single diploid colonies from WT and mutant strains were inoculated in SPM medium, grown overnight at 30°C and then >100 tetrads were dissected for each strain onto YPD plates. The plates were incubated at 30°C for 2 days.
RESULTS
Physical recombination analysis system
Meiotic recombination initiates by programmed DSBs. Each DSB engages a partner duplex via a presumptive nascent D-loop interaction. This interaction usually involves a homolog chromatid, rather than the sister, reflecting the strong tendency for meiotic recombination to exhibit ‘homolog bias’ (Introduction). At the nascent D-loop stage, the recombination pathway bifurcates, with some intermediates yielding ‘CO’ products and others yielding products lacking reciprocal exchange of flanking regions (‘NCOs’) (29). CO-fated interactions progress via two long-lived intermediates that are detectable by physical analysis – single-end invasions and double Holliday junctions (SEIs and dHJs, collectively referred to as ‘joint molecules’ or ‘JMs’), finally yielding COs at the end of pachytene (30). Particular features are required to maintain homolog bias during these transitions, specifically at the transition from SEIs to dHJs (4,8; below). NCO-fated interactions progress by a different sequence of events, likely synthesis-dependent strand annealing, for which physical intermediates have not yet been detected.
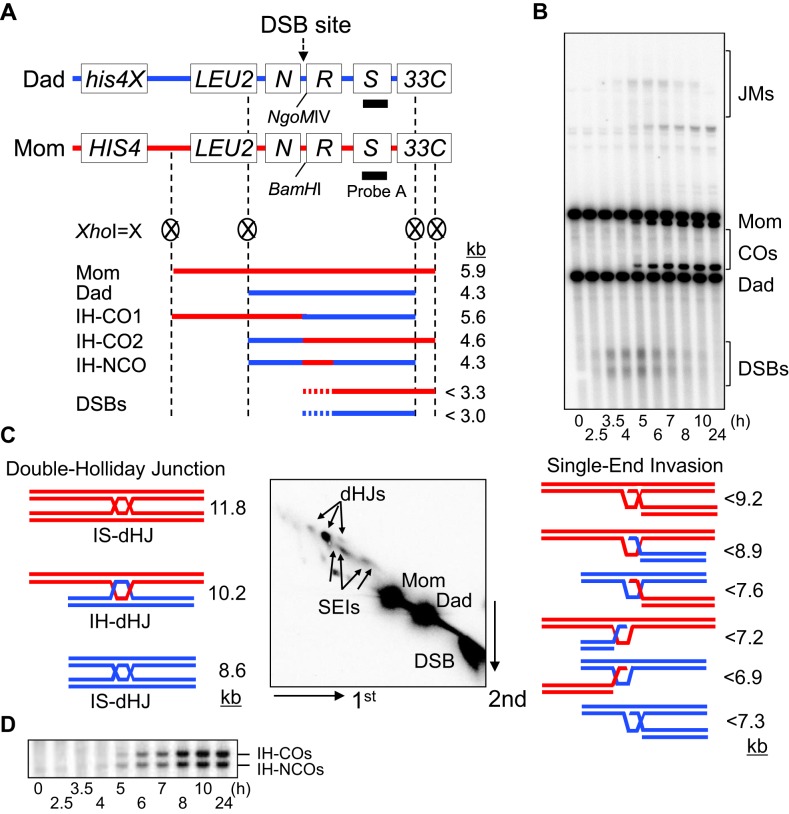
Here, DNA events of meiotic recombination were monitored using the HIS4LEU2 hotspot assay system for chromosome III (4,8,31–34). Cell cultures were synchronized in the G1 stage, and then transferred to fresh SPM medium to induce meiosis. Meiotic genomic DNA samples were prepared using an optimized guanidine-phenol extraction method (4,8,31–34). XhoI restriction site polymorphisms between ‘Mom’ and ‘Dad’ parental alleles produce diagnostic restriction species for DSBs, JMs and recombinant chromosomes (Figure 1). Each enzyme-digested DNA sample from the time course was subjected to one-dimensional (1D) gel electrophoresis to analyze DSBs and COs (Figure 1A and B). To analyze JM intermediates, cell samples from the meiotic time course were treated with psoralen and exposed to UV light (365 nm) to generate inter-strand crosslinks for stabilizing recombination intermediates (Figure 1C). Native/native 2D gel electrophoresis reveals the formation of distinct JM branched structures and the progression of meiotic recombination (4,8,31–34). The various species of SEIs and dHJs exhibit distinct migration mobilities, thus allowing characterization via their distinguishable shapes and sizes in 2D-gel analysis (Figure 1C). An important feature of JM analysis is that it permits the detection and evaluation of homolog bias. The species corresponding to double Holliday junctions between homologs and between sisters are readily distinguishable from one another, and the tendency for meiotic recombination to occur between homologs manifested in a 5:1 ratio of inter-homolog to inter-sister dHJs (i.e. IH:IS dHJ ratio = ∼5:1) (4). Physical analysis can also distinguish IH–CO and IH–NCO products. Digestion of genomic DNA with XhoI and NgoMIV, yields diagnostic species of 4.6 and 4.3 kb, respectively, as defined in 1D-gels (Figure 1A and D). For all analyses, the radiolabeled ‘Probe A’ was used for Southern hybridization, and the hybridized DNA signals were quantified (Figure 1A).
Figure 1.
Physical analysis of meiotic recombination. (A) Physical map of the HIS4LEU2 hotspot showing diagnostic restriction sites and the position of the probe. Fragments diagnostic for recombinant intermediates digested with XhoI or XhoI + NgoMIV and position of ‘Probe A’ are shown. N, NFS1; R, RRP7; S, STE50. Mom, mom species; Dad, dad species; IH-CO, interhomolog-crossover; IH-NCO, interhomolog-noncrossover; DSBs, double-strand breaks. (B) One-dimensional (1D) gel analysis showing wild-type (WT) double-strand breaks (DSBs) and crossover (CO) species. Mom, mom species; Dad, dad species; JMs, joint molecules. (C) Two-dimensional (2D) gel analysis of JM species. SEI, single end invasion; IS-dHJ, intersister double-Holliday Junction; IH-dHJ, interhomolog double-Holliday Junction. (D) IH-CO and IH-NCO fragments representing each set of recombinants in the WT strain.
Lack of Rec8 cleavage confers no defect in recombination
Rec8 is phosphorylated at multiple sites during prophase (3,15) (confirmed in Supplementary Figure S1). Phosphorylation is canonically known to be required for separase-mediated cleavage of cohesin during the metaphase-to-anaphase transitions of mitosis and, analogously, during the two meiotic divisions (19,20,35,36). To ensure appropriate timing, separase is kept inactive by inhibitory binding of securin Pds1 (35–37) and then activated by APC/C mediated proteolysis of Pds1 (37). We were interested to know whether cohesin cleavage might also occur during prophase, albeit at a low level, but sufficient to influence the recombination process.
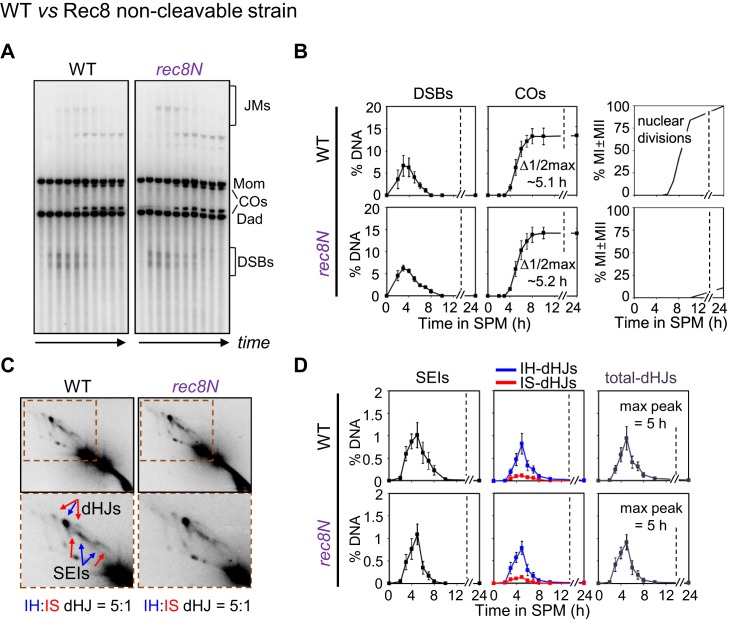
To investigate this possibility, we analyzed the progression of recombination in time-course experiments using an allele of REC8, rec8N, which is known to severely reduce or eliminate Rec8 cleavage (19). The rec8N allele comprises three mutations (E428R, R431E and R453E). A strain carrying this allele is completely blocked for occurrence of the meiotic divisions due to lack of separase-mediated cleavage of Rec8 (Figure 2, Supplementary Figure S2) (19). Analysis of recombination in the rec8N background revealed no detectable change in recombination. The rec8N mutant efficiently formed recombination intermediates at quantities similar to wild-type, with a normal IH:IS dHJ ratio of ∼5:1, and with timely turnover of intermediates into meiotic recombination products that occurred at normal levels (Figure 2, Supplementary Figure S2). We conclude that meiotic recombination progresses normally, independently of Rec8 cleavage.
Figure 2.
Rec8 cleavage is not required for meiotic recombination in prophase I. (A) Representative 1D gel images for WT and rec8N (non-cleavable Rec8). See also Supplementary Figure S2. Time points are 0, 2.5, 3.5, 4, 5, 6, 7, 8, 10 and 24 h. (B) Quantitative analysis of DSBs, total COs and nuclear divisions during meiosis (mean ± SEM for three cultures). (C) Representative 2D gel images for WT and rec8N. (D) Quantitative analysis of SEIs and dHJs (mean ± SEM for three cultures).
Analysis of SC formation in a rec8N mutant further reveals no obvious difference from wild-type meiosis, with well-formed SCs appearing prominently at the appropriate time (19). These findings confirm that chromosome axes assemble, and SCs form, independent of Rec8 cleavage.
rec8 phospho-mutants exhibit a ‘separation of function’ phenotype: normal DSB formation and normal NCO formation plus a specific constellation of CO-related defects
Our previous analyses have characterized in detail the nature of meiotic recombination in a rec8Δ null mutant (4). We have now analogously analyzed recombination in three rec8 mutants, rec8-6A, rec8-24A and rec8-29A. In these three alleles, the indicated number of multiple phosphorylation sites have been mutated so that they encode alanine rather than the original amino acid, with concomitant reductions in phosphorylation (3,12). For this study, we have confirmed that phosphorylation is strongly reduced in rec8-29A, although a low level of residual phosphorylation can still be detected (Supplementary Figure S1B). For convenience, we refer to these alleles as rec8 ‘phospho-mutations’. Detailed analysis reveals a common, specific constellation of defects in the three mutants that comprise a subset of those conferred by rec8Δ, thus defining a ‘separation of function’ phenotype. This phenotype pertains specifically to events related to CO formation. Interestingly, also, the severity of this phenotype increased in relation to the number of mutated residues.
rec8 phospho-mutants exhibit normal DSB formation and resection
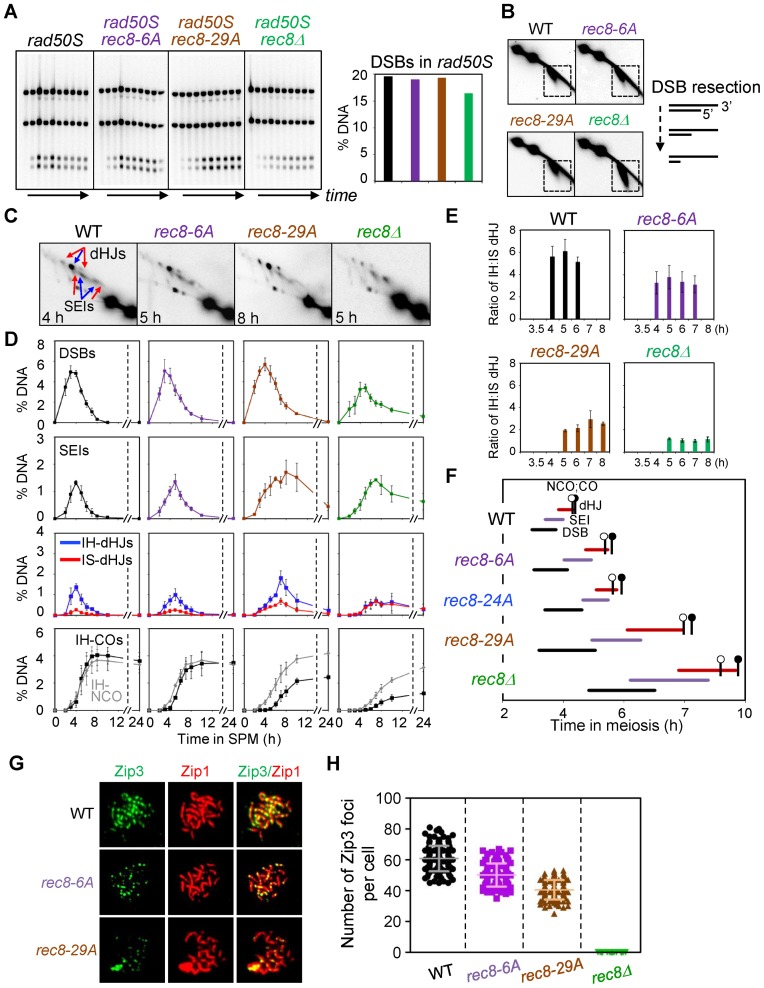
In rec8Δ, DSB formation is modestly reduced, as defined by 1D gel analysis in a strain background (rad50S) where DSBs accumulate rather than turning over to later forms (4). In the present study, WT, rec8-6A and rec8-29A yeast strains both exhibited similar levels of DSBs at four loci as assayed in a rad50S background (Figure 3A, Supplementary Figure S3). In contrast, the rad50S rec8Δ mutant, analyzed in parallel, exhibited a reduction of DSB levels, e.g. by ∼20% compared to the other rad50S strains at the HIS4LEU2 hotspot (Figure 3A, Supplementary Figure S3). The rec8-24A mutant was not examined by rad50S analysis but likely also exhibits normal DSB levels as inferred from standard time course analysis.
Figure 3.
Analysis of meiotic recombination in rec8 phospho-mutant strains. (A) Analysis of DSB formation in rad50S strains. (Left) 1D gel analysis of rad50S DSBs. (Right) Quantification of DSB levels in the left gel analysis. (B) 2D-gel analysis of DSB resection in WT and indicated single mutants. Illustration shows DSB resection of 5′-termini to generate 3′-single-strand DNA tail. (C) Representative 2D gel images for WT and mutant strains. (D) Quantitative analysis of DSBs, SEIs, dHJs, IH-COs and IH-NCOs from meiotic time courses (mean ± SEM for three cultures). ‘% DNA’ in Y-axis of the plot indicates the percentage of total hybridizing signals. (E) Ratio of IH/IS dHJ over time plotted as percentage maximum level (mean ± SEM for three cultures). (F) Timing and kinetics of recombination events in each of the strains. Relative timing of DSBs, SEIs, dHJs, IH-COs and IH-NCOs were analyzed as described (4,30). The lifespan of each recombination event was measured by the timing of the length, beginning and end of recombinant DNA formation. IH-CO and IH-NCO products are marked by black and blank circles, respectively, to denote their appearance and disappearance in approximately 50% of cells. (G) Representative images showing Zip3 foci and Zip1 array of spread chromosome in each indicated strains immunostained for Zip3-13myc and Zip1. (H) Quantification of the number of Zip3 foci in WT (sampled at 5 h), Rec8-phospho mutants (sampled at 6 h for rec8-6A and at 7 h for rec8-29A) and rec8Δ (sampled at 7 h). Scatter plots show the number of Zip3 foci at pachytene chromosomes (mean ± SD, n = 100–150 nuclei). Each symbol indicates a numerical value for individual nuclei.
In a WT (RAD50) setting, DSB ends are resected at their 5′ termini, with >90% of DSBs being resected by ≥500 nucleotides (4,38). In 2D gels, DSBs are detected as a downward diagonal spike due to decreased molecular weight plus accelerated mobility of ssDNA-containing species in the second dimension (Figure 3B). The absence of Rec8 confers a DSB hyper-resection phenotype, detected as somewhat elongated signals as compared with those of the WT (4; Figure 3B). However, all three Rec8 phosphorylation-defective strains exhibit the behavior of the WT strain in respect to DSB resection (Figure 3B, Supplementary Figure S4).
rec8 phospho-mutants normal NCO formation but reduced formation of COs
In WT cells, the final levels of diagnostic IH-CO and IH-NCO species were ∼4.1% and ∼3.7%, respectively, with IH-NCOs appearing slightly earlier than IH-COs. By comparison, the rec8Δ strain shows reduced formation of both types of products; however, CO levels are differentially reduced as compared to NCOs (Figure 3D). All three phospho-mutant strains exhibit overall levels of IH-NCO products that are relatively similar to those observed for WT strains and a significant defect in CO formation: the levels of IH-CO products were progressively reduced in the three cases, to 86%, 79% and 60% of the WT level in rec8-6A, rec8-24A and rec8-29A, respectively (Figure 3D, Supplementary Figure S4). As a result of these effects, the CO:NCO ratio was reduced from 1.11:1 in WT to 0.93:1, 0.75:1, 0.57:1 and 0.42:1 in the three rec8 phospho-mutants and rec8Δ, respectively (Supplementary Table S2). We have also confirmed that effects on CO formation are not specific to the HIS4LEU2 hot spot: rec8 phospho-mutants rec8-6A and rec8-29A exhibit delayed and reduced formation of CO products at the ARG4 DSB hot spot region on chromosome VIII (Supplementary Figure S5).
rec8 phospho-mutants exhibit defective homolog bias
In WT cells, dHJs occur peak at ∼4 h and exhibit a IH:IS-dHJ ratio of 5:1, defining the preference for use of the homolog versus the sister during CO formation (Figure 3C–E). Previous analysis has shown that the rec8Δ mutant is defective in maintenance of homolog bias. In the absence of Rec8, homolog bias is established relatively normally (as implied by relatively efficient IH-NCO formation); however, along the CO branch of the pathway, this bias is lost, resulting in a 1:1 IH:IS dHJ ratio (4) (Figure 3C–E). This 1:1 ratio is also observed in other types of mutants and is interpreted to be specifically diagnostic of a defect in maintenance of homolog bias at the SEI-to-dHJ transition (4). Moreover, for rec8Δ, this defect can explain much of the differential reduction of CO levels relative to NCO levels as seen in rec8Δ, due to the fact that many CO-fated inter-homolog interactions are diverted to an inter-sister fate (4). The rec8-6A, rec8-24A and rec8-29A mutants exhibit progressively decreasing IH:IS dHJ ratios of 4.4:1 and 4.1:1 and 2.5:1, which thus approach but do not reach the 1:1 ratio observed in rec8Δ (Figure 3E, Supplementary Figure S5, Table S2). Since the three mutants exhibit normal NCO formation and a progressively increasing differential reduction in CO:NCO ratio, an plausible interpretation is that the three rec8 phospho-mutants confer progressively increasing defects in maintenance of homolog bias.
rec8 phospho-mutants exhibit reduced formation of CO-correlated Zip3 foci
The SUMO E3 ligase Zip3 is a component of the ZMM complex that specific implements formation of COs (versus NCOs) and nucleates synapsis initiation. Zip3 localizes to prominent foci that correspond to the sites of progressing CO-fated recombinational interactions (39). These foci are cleanly detected at mid-pachytene (39–42), approximately the time of the SEI-to-dHJ transition when homolog bias is lost in rec8Δ. We have analyzed Zip3 foci in rec8 phospho-mutants and rec8Δ with a strain expressing a Myc tag inserted at the C-terminus of Zip3. The number of Zip3-Myc foci observed in WT cells was 59.4 ± 8.21 and was progressively reduced to 48.4 ± 6.44 and 39.3 ± 5.02 in rec8-6A and rec8-29A, respectively, with no Zip3 foci detected in the rec8Δ mutant (Figure 3G and H). These data provide another indication that events along the meiotic CO recombination pathway are defective in rec8 phospho-mutants, with progressive severity in relation to the number of mutations.
rec8 phospho-mutants exhibit delayed progression along the CO pathway
In WT cells, DSBs, SEIs and dHJs appear and disappear sequentially during meiosis (Figure 3C and D). In rec8Δ, progression through these stages is significantly delayed. This is shown qualitatively by prolonged presence of all three species (Figure 3C and D) and quantitatively by ‘lifespan analysis’ (Figure 3F). The three rec8 phospho-mutants exhibit analogous delays, with the rec8-6A, rec8-24A and rec8-29A mutants exhibiting progressively stronger delays which, in rec8-29A, approach that observed in rec8Δ.
rec8 phospho-mutant strains exhibit normal axis formation but defective SC formation
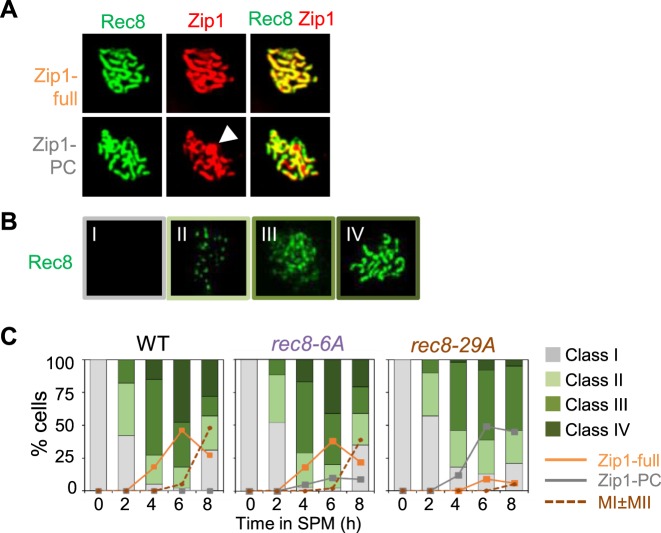
Prophase axes, and thus SCs, are undetectable in a rec8Δ mutant (4,5). We have now characterized meiotic chromosome axis morphogenesis in rec8 phospho-mutant cells as compared to WT (Figure 4A and B). Surface spread nuclei were scored for detectable Rec8 signals according to four categories as defined previously (4): category I (no Rec8 staining), category II (modest numbers of foci), category III (larger numbers of foci with a clear tendency for forming linear arrays, leptotene/zygotene) and category IV (strongly staining lines, zygotene/pachytene) (4; Figure 4B). In both WT and rec8 phospho-mutants, nuclei staining positive for Rec8 progressively disappeared (category I) or appeared (category II–IV), implying that the phospho-mutations do not abrogate Rec8 loading onto chromosomes. rec8-6A cells exhibit no significant defect in the temporal progression of Rec8 staining. In contrast, rec8-29A cells exhibit a defect in SC formation at leptotene/zygotene, by two criteria. First, rec8-6A and rec8-29A cells exhibit an elevation in the level of SC-related polycomplexes, aberrant assemblies that are diagnostic of delayed or defective installation of SC along the chromosomes. Second, rec8-29A cells progressed from category I to III at a normal rate and then exhibit a significant delay in the progression specifically at this stage, as seen by delayed progression from category III to category IV as compared to WT cells (Figure 4). Additionally the rec8-29A mutant exhibits decreased numbers of fully synapsed chromosomes, and a dramatically elevated frequency of Zip1 polycomplex (PC) formation, implying a severe defect in SC installation at leptotene/zygotene (Figure 4C).
Figure 4.
Chromosome analysis of WT, rec8-6A and rec8-29A cells immunostained for Rec8 and Zip1. (A) Chromosome spreads of meiotic cells immunostained for Rec8-3HA and Zip1. Representative images of spread nuclei immunostained for Rec8-3HA in green and Zip1 in red. Nuclei were immunostained with anti-HA (for Rec8-3HA) and anti-Zip1 at pachytene in spread chromosomes. Zip1-full, Zip1 full assembly; Zip1-PC, Zip1 polycomplex. Arrowhead indicates a Zip1-PC. (B) Chromosome spreads of meiotic cells immunostained for Rec8-3HA. Nuclear spreads were categorized to four classes: Category I, no staining; Category II, modest numbers of foci; Category III: larger numbers of foci with a clear tendency for linear arrays; Category IV: strongly staining lines. Category III appears after DSB formation and Category IV appears contemporaneously with SC formation in advance of CO formation, analyzed in the previous study (4). (C) Quantification of appearance and disappearance for Rec8-3HA staining category in (B) over time in meiosis. The formation of Zip1-PC is also quantified. Over 150 nuclei were analyzed for each time point.
These structural defects of rec8 phospho-mutants correspond to, and further extend, the ‘separation of function’ phenotype defined by analysis of DNA events. DSBs occur in the context of developing prophase chromosome axes. Both of these aspects are defective in rec8Δ and normal in the rec8 phopho-mutants. Thereafter, SC formation and CO-designation are concomitant events and are positionally and functionally linked. SC specifically nucleated at CO sites (43,44) and SC nucleation and the first steps of CO-formation (formation of SEIs) are coordinately mediated by a common set of functions, i.e. the ZMMs, with CO recombination proceeding in SC-associated complexes. rec8 phosphomutants are significantly defective in SC formation as well as CO-related SC-localized events of recombination.
We further find that rec8 phospho-mutants exhibit a reduction in the frequency of viable spores (Supplementary Figure S6), in accord with diverse defects in recombination and chromosome morphogenesis during prophase and ensuing delays and defects in the meiotic divisions.
Premature induction of separase confers a rec8Δ null phenotype for chromosomal and DNA events
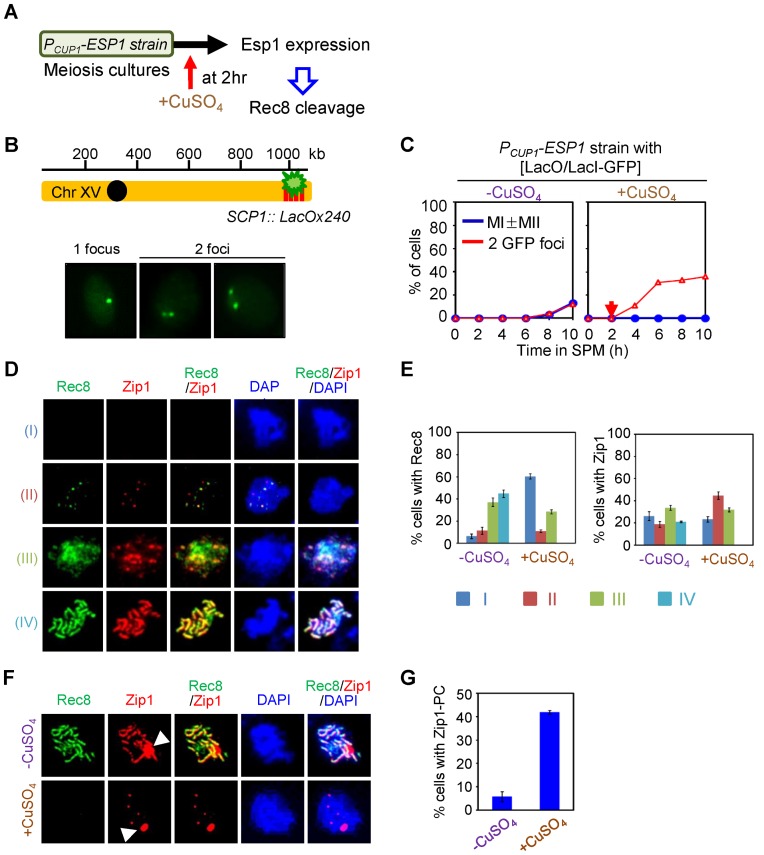
Evidence presented above suggests that cleavage of Rec8 plays no detectable role in meiotic recombination. However, Rec8 is phosphorylated during prophase, and phosphorylation is required for separase-mediated cleavage during the meiotic divisions. We were therefore interested to confirm that Rec8 remains susceptible to cleavage during prophase. Such susceptibility has been shown in a mutant condition where the APC/C is prematurely activated (45) but has never been examined in an otherwise WT strain background. For this purpose, we constructed a copper-inducible allele of esp1 by replacing the native ESP1 promoter with the CUP1 promoter, which is repressed in the absence of CuSO4 and can be specifically induced by addition of CuSO4 (Figure 5, Supplementary Figures S7 and S8). We then analyzed events of interest in the PCUP1-ESP1 strain both without CuSO4 addition and after the addition of CuSO4 to synchronous meiotic cultures at appropriate time points.
Figure 5.
Rec8 cleavage at early prophase I exhibits defects in sister chromatid cohesion and synaptonemal complex formation. (A) Experimental regimen to express Esp1 protein during meiotic prophase I. (B and C) Analysis of sister chromatid cohesion. Strains carrying LacO array at chromosome XV and expressing Lac repressor conjugated with green fluorescence protein were analyzed for sister association in fixed whole cells (the diagram showing the location of Lac operator array at chromosome XV in B). Single culture was synchronized and divided into two SPM cultures; then Esp1 expression was induced by addition of 50 μM CuSO4 at 2 h (red arrow) in one of the two cultures. One focus indicates unreplicated chromatid or unseparated sister chromatids. Two foci indicate replicated and separated sister chromatids. (D) Esp1 overexpression in the early stages of meiotic prophase I defects in SC formation and Rec8 assembly onto chromosomes. Nuclei were immunostained with anti-HA (for Rec8-3HA) and anti-Zip1 in spread chromosomes. Representative images showing spread nuclei immunostained for Rec8-3HA and Zip1 according to four categories (category I, no signals; category II, modest number of foci; category III, large number of foci with fuzzy pattern; category IV, fully assembled lines). (E) Chromosome spreads were analyzed for the percentage of cells in Zip1 and Rec8 staining in PCup1-ESP1 strain in the presence or absence of CuSO4, according to the categories as shown in (D) (mean ± SD, n = 150–200 nuclei). (F) Representative images showing the formation of Zip1-polycomplex (PC) in PCup1-ESP1 strain in the presence or absence of CuSO4. White arrowheads indicate Zip1-PCs. (G) Quantitative analysis showing the percentages of Zip1-PCs shown in (F) (mean ± SD, n = 150–200 nuclei).
In the absence of Esp1 induction, Rec8 cleavage was undetectable throughout the period of meiosis corresponding to stages through and well beyond prophase. Furthermore, sister separation at the time of the two divisions was substantially reduced and, correspondingly, occurrence of the first meiotic division was strongly delayed (Figure 5A–C, Supplementary Figure S8).
In contrast, when expression of Esp1 activated at early prophase I (t = 2 h) the level of full-length Rec8 proteins began to decline and Rec8 cleavage products accumulated rapidly (Supplementary Figure S8). This same effect was observed in an otherwise WT strain background and also in an ndt80Δ background where meiosis arrests at the late prophase stage of pachytene, confirming that the observed effects are occurring before exit from this stage, as expected. Moreover, in this regime, sister chromatid separation increases immediately upon induction, rather than after prophase as in the normal case; Rec8 association with prophase chromosomes is dramatically reduced to nearly undetectable level; and prophase axis and SC formation is severely compromised (Figure 5B–G). These findings imply that Rec8 is susceptible to Esp1-mediated cleavage during early/mid-meiotic prophase. By implication: (i) prophase phosphorylation is sufficient to enable efficient cleavage; and (ii) absence of detectable Rec8 cleavage during prophase in WT meiosis is due to absence of separase, not to insensitivity of prophase Rec8 to cleavage.
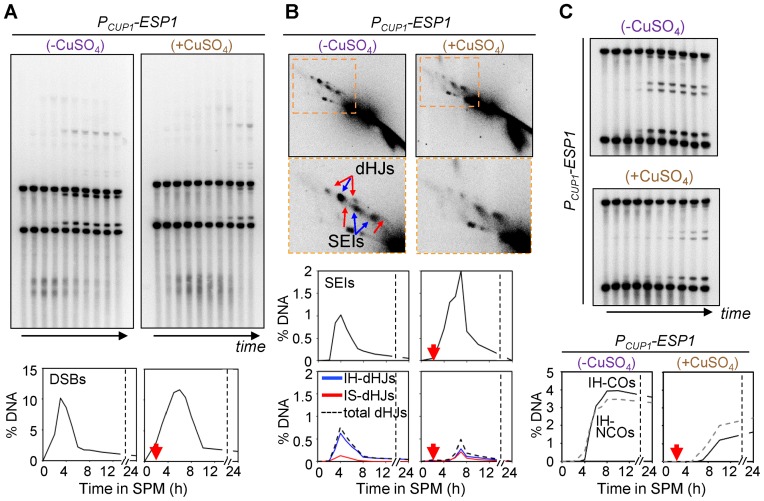
Recombination phenotypes of PCUP1-ESP1 cells were also defined physical analysis in the presence or absence of CuSO4 (Figure 5). In the absence of CuSO4, meiotic recombination proceeded exactly as in a WT strain: DSBs occur at ∼2 h; SEI and dHJ intermediates subsequently formed at normal levels, and recombination products appear with the same timing and levels as in WT (Figure 6). These findings provide a second line of evidence that separase-mediated cleavage of Rec8 is not involved in meiotic recombination. In contrast, when Esp1 was expressed at t = 2 h, when DSB formation is still in progress in most cells, dramatic effects were observed, with the observed recombination phenotypes perfectly resembling those observed in a compete rec8Δ null mutant described above, including hyper-resection of DSBs, delayed formation and turnover of JMs, an IH:IS dHJ ratio of ∼1:1 and a reduction in both CO and NCO products, with a disproportionate reduction of COs versus NCOs (Figure 6).
Figure 6.
Rec8 cleavage at early prophase I exhibits defects in meiotic recombination. (A) 1D gel analysis and quantification of DSB. Time points are 0, 2.5, 3.5, 4, 5, 6, 7, 8, 10 and 24 h. (B) Representative Southern blot images of 2D gel analysis and quantification of JM levels. (C) Analysis of IH-CO and IH-NCO.
Addition of CuSO4 at t = 5 h, when SEIs and dHJs are both present at high levels, conferred detectably delayed turnover of DSBs, SEIs and dHJs (Supplementary Figure S9). Interestingly, we also reproducibly observe an usual ‘second peak’ of DSBs, SEIs and dHJs, suggesting that this condition enables a second round of DSB formation. This effect can be explained by the observation that extra DSBs occur late in meiosis in situations where SC formation is defective (46), a condition that could be expected in the current protocol. We further found that Esp1 expression at t = 7 h has no detectable effect on the assayed events of recombination (Supplementary Figure S9). Since most or all cells have reached the dHJ or later stages by this point, this result provides further evidence Rec8's critical roles for recombination are implemented during earlier events of the recombination process.
Roles of protein kinases Hrr25, Cdc7 and Cdc5 in meiotic recombination
The observations presented above show that rec8 mutations that eliminate phosphorylation confer prominent defects in prophase, including in meiotic recombination. Moreover, the strengths of the defects increases in relation to the number of mutated sites. These phenomena support notion that the mutant phenotypes do, in fact, result from a paucity of phosphorylation, as has been prominently assumed in previous studies of such mutants (3,12). To further investigate this possibility, we asked whether the appropriate diagnostic recombination defects observed in phospho-mutants are also conferred by direct elimination of any of the three kinases known to mediate Rec8 phosphorylation.
Cdc7, but not Hrr25, might mediate Rec8 phosphorylation for recombination
Hrr25 and DDK, together, account for Rec8 phosphorylation at early/mid-prophase (9). We thus first investigated whether/how inactivation of the Hrr25 and/or DDK kinases affects meiotic prophase recombination. We constructed hrr25-as and cdc7-as3 alleles which render the corresponding kinase activities sensitive to chemical inhibitors (9,47). We then analyzed hrr25-as, cdc7-as3 and hrr25-as cdc7-as3 double mutant strains to determine the effects of inhibitor addition at appropriate times in meiosis (Figure 7, Supplementary Figure S10).
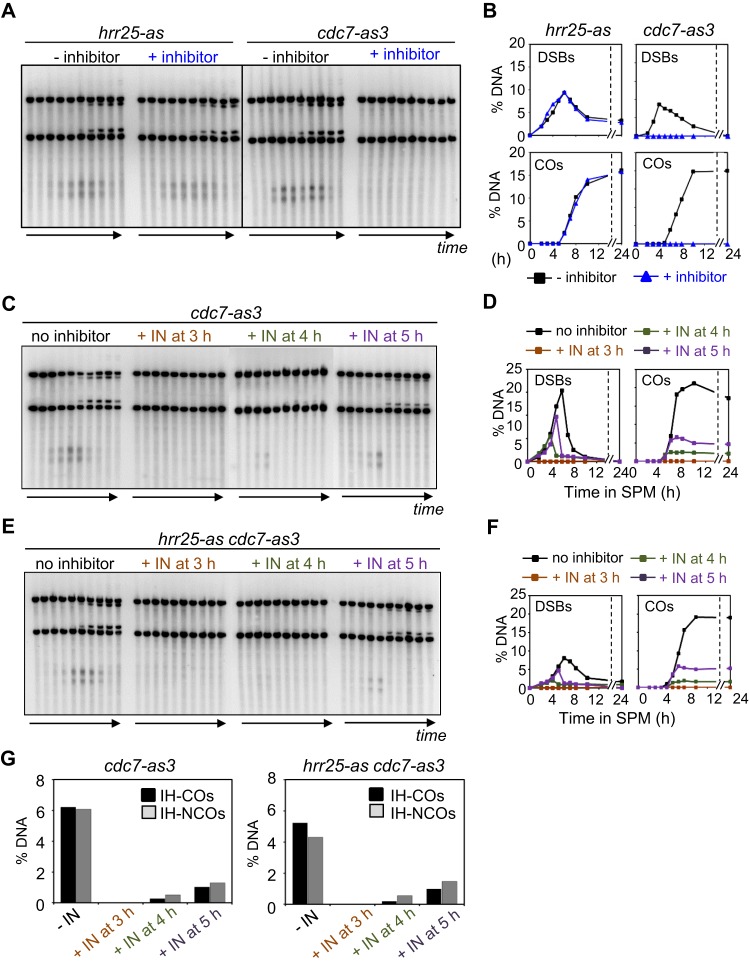
Figure 7.
Meiotic recombination in the absence of kinase activity of Hrr25 and Cdc7. (A) 1D Southern blot analysis of meiotic recombination in hrr25-as and cdc7-as3 strain. (B) Quantification of DSB and CO species in the presence or absence of chemical inhibitor. (C) 1D Southern blot analysis of meiotic recombination in cdc7-as3 mutant. (D) Quantification of DSB and CO species in the presence or absence of chemical inhibitor. (E) 1D Southern blot analysis of meiotic recombination in hrr25-as cdc7-as3 double mutant. (F) Quantification of DSB and CO species in the presence or absence of chemical inhibitor. (G) Quantification of IH-CO and IH-NCO in cdc7-as3 and hrr25-as cdc7-as3 strains. See also Supplementary Figure S10C.
Since Rec8 is phosphorylated from pre-meiotic S phase onward, we first explored the effects of adding inhibitor at 2 h. In accord with known early roles of Cdc7/Dbf4, cdc7-as3 cells exhibited no detectable DSBs or COs and meiotic progression through the two divisions was completely blocked (Figure 7A and B, Supplementary Figure S10). Flow cytometry profiles of cdc7-as3 also showed that inactivation of Cdc7 induced a pre-meiotic S phase delay (Supplementary Figure S10). Given the early role of Cdc7 revealed by these studies, we appreciated that detection of a specific effect of kinase inhibition on post-DSB stages of recombination defective in rec8 phospho-mutants would require addition of inhibitor at a later time point, after execution of early Cdc7 functions. We therefore next examined recombination in a cdc7-as3 single mutant strain, in the absence of inhibitor or upon addition of inhibitor at t = 3, 4 or 5 h (Figure 7C and D). In the absence of inhibitor, DSBs and COs appear and disappear normally (Figure 7C and D). In contrast, addition of inhibitor at the three time points permits progressively higher levels of DSB formation and corresponding progressively higher levels of CO products and, analogously NCO products (Figure 7C, D and G), implying progressively reduced effects as more and more cells progress past critical Cdc7-dependent step(s). These patterns suggest specifically that inactivation of Cdc7 blocks DSB formation but that already-present DSBs can progress relatively efficiently to mature products. There also is no obvious delay in DSB turnover or the time of appearance of products. Interestingly, however, there is a decrease in the CO:NCO ratio to 0.78, a value that is lower than the value of ∼1.11 observed in wild type and in the same strain in the absence of inhibitor.
In contrast to these results, addition of inhibitor to hrr25-as cells at 2 h had no effect on DSB or CO levels, which occurred normally as seen in untreated cells, although the meiotic cell cycle progression was defective (Figure 7A and B, Supplementary Figure S10A and S10B). Furthermore, an hrr25-as cdc7-as3 double mutant exhibits the same phenotype as the cdc7-as3 single mutant, including a reduced CO:NCO ratio of 0.67, regardless of the time of inhibitor addition (Figure 7C–G, Supplementary Figure S10C, Table S2).
The resulting phenotype of recombination observed in cdc7-as3 with late inhibitor addition corresponds to that observed in the rec8 phospho-mutants: the observed CO:NCO ratios (0.78 and 0.67) are very similar to that observed in rec8-24A and rec8-29A mutants (0.84 and 0.58, respectively). However, we have not yet determined whether the Cdc7 role is eliminated by rec8 phospho-mutations, as expected in such a case. In contrast, phosphorylation of Rec8 by Hrr25 plays is not relevant to the prophase roles of Rec8 for recombination.
Cdc5-mediated phosphorylation is not required for normal (Rec8-mediated) homolog bias
We also probed for any possible role of Cdc5-dependent Rec8 phosphorylation in recombination. Previous work has shown that when meiosis is arrested at mid-pachytene in an ndt80Δ mutant, dHJs tend to accumulate and CO formation is defective while NCO formation proceeds to completion. Starting from this condition, forced expression of Cdc5 provokes disassembly of pachytene chromosome structure, disappearance of dHJs and appearance of CO products (27). These effects are not altered in a rec8 phospho-mutant, suggesting that Rec8 is not the target for these later effects (27). Nonetheless, we were interested to examine this sequence of events in more detail. We were interested to know, e.g. whether the dHJs that accumulate in the absence of Cdc5 are normal or are defective for homolog bias (as expected if Cdc5-mediated phosphorylation of Rec8 at an earlier stage were required for homolog bias maintenance).
For this purpose, we used strains in which the CDC5 promoter was replaced with the CUP1 promoter. In the absence of CuSO4, PCUP1-CDC5 ndt80Δ cells exhibited pachytene arrest, and the total levels of JMs accumulated to high levels, as in previously studied Cdc5-depleted conditions. We observe strong accumulation of SEIs, as well as of dHJs, to ∼7% and ∼6.2% of total DNA, respectively (Supplementary Figure S11). However, the IH:IS dHJ ratio observed in arrested cells is ∼5.65:1, essentially the same as that observed in WT (∼5.5:1). These findings imply that Cdc5 is not required for establishment or maintenance of homolog bias and thus cannot be not required for any potential role of Rec8 in the latter process.
We further find that, as in previous studies (above), upon Cdc5 induction in arrested cells, at t = 7 h after initiation of meiosis, all JMs were efficiently resolved to IH-COs and recombination products very rapidly reached plateaus levels, with WT levels of COs (Supplementary Figure S11; 4.71% in COs). This pattern suggests that, prior to Cdc5 induction, a normal number of DSBs have been committed to the CO fate, further implying normal establishment and maintenance homolog bias. We note, however, that the level of NCOs rises to a higher than normal levels. While the basis for this effect is not known, it does not match the effects observed in rec8 mutants and thus is unlikely to be related to a role of Cdc5 in Rec8 phosphorylation.
Taken together, these findings support the view that, as expected from previous work, Cdc5-mediated phosphorylation of Rec8 does not play a role for homolog bias during meiotic CO recombination.
Rec8 is required for robust homolog bias in the absence of ZMM functions Zip1 and Zip3
ZMM proteins mediate progression of CO-fated recombination intermediates and nucleation of SC formation (29). Since Rec8 also plays significant roles in CO recombination and SC formation, it was of interest to understand the dependencies between the two sets of functions, in particular with respect to recombination. Such analysis is made possible by the rec8 phospho-mutants that form axes and are then specifically defective in SC formation. For this purpose, we carried out DNA analyses in rec8-29A zmm double mutants, in comparison with single mutants. Experiments were performed at 33°C, a temperature at which CO recombination is most tightly controlled (Figure 8, See also 4,29).
Analysis of zip1Δ and zip3Δ mutants confirms previous findings that IH-NCOs form normally, while the formation of CO-specific post-DSB recombination intermediates is specifically and severely abrogated (29) (Figure 8, Supplementary Figure S12). 2D-gel analysis of meiotic recombination events further shows, however, that a significant number of SEIs and dHJs can be detected (Figure 8A and B). This is likely because as very small subset of DSBs progress to these stages and then remain there for a very long time (29). Zip1-deleted and Zip3-deleted cells exhibited modestly lower levels of homolog bias than WT (IH:IS dHJ ratios of ∼4:1 and 2.5:1 in each mutant respectively versus ∼5.5:1 in WT; Figure 8C).
Analysis of zip1Δ rec8-29A and zip3Δ rec8-29A double mutants reveals that this rec8 phospho-mutant still confers important defects even in the absence of ZMM functions (Figures 3 and 8). The presence of rec8-29A significantly reduces IH-COs and IH-NCOs in both the zip1Δ and zip3Δ backgrounds (Supplementary Figure S12) and also reduces dHJ ratios to ∼2.22:1 in zip1Δ rec8-29A (Student's t-test, P < 0.0148) and ∼1.32:1 in zip3Δ rec8-29A (Student's t-test, P < 0.0102) (Figure 8C). These values are similar to, or lower than, the IH:IS dHJ ratio in the rec8-29A single mutant, (2.5:1). These results suggest that, the function(s) abrogated by rec8-29A is(are) distinct from the role(s) of ZMM proteins Zip1 and Zip3.
Absence of Mek1 ameliorates delayed recombination in the rec8-29A mutant
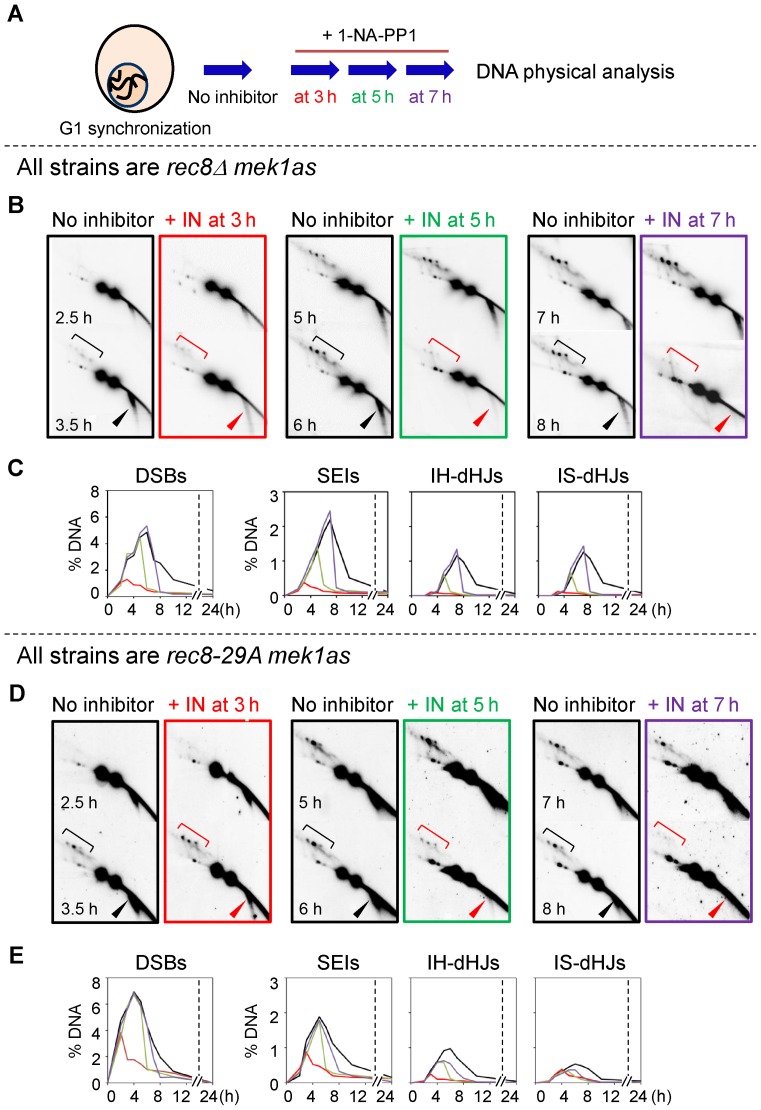
Mek1 is required for setting up a meiosis-specific recombination ensemble (8). In the absence of Mek1, homolog bias is not established and meiosis-specific recombination fails (4,8,48,49). It could be expected that this early and primary role would be upstream of the roles of Rec8 affected by rec8 phospho-mutations that involve later steps. To ask if this is true, we utilized a mek1as strain, which encodes a mutant Mek1 kinase protein in which ATP-binding, and thus kinase activity, can be inhibited using the compound 1-NA-PP1 (4,8,48,49). Hereafter, the designations ‘mek1as(+IN)’ and ‘mek1as(-IN)’ signify mek1as strains grown in the presence and absence of 1-NA-PP1, respectively.
Our previous study shows that meiotic recombination in mek1as(-IN) exhibits the same pattern as was observed in WT cells (4) and that addition of inhibitor at the time of meiotic DNA replication stage, and thus prior to assembly of recombination complexes and initiation of recombination, results in a rapid progression of meiotic recombination. Moreover, recombination tends to exhibit strong sister bias and a resultant paucity of IH-COs and IH-NCOs (4). We have also previously shown that Mek1 kinase inactivation relieves all of the recombination timing observed in a rec8Δ mutant (4,8). We further confirmed this finding in the present study, showing alleviation of delay even at a low temperature, not previously examined (Supplementary Figure S13).
We next asked whether Mek1 kinase inactivation could overcome either the timing delays and/or the residual homolog bias observed rec8 phospho-mutant strains. We therefore examined recombination after inactivating Mek1 kinase at various time points during meiosis in the rec8Δ mek1as, rec8-29A mek1as and rec8-6A mek1as strains in parallel analyses (Figure 9 and Supplementary Figures S14 and S15). In the rec8Δ mek1as(-IN) mutant, the progression of recombination was delayed compared to that in the mek1as(-IN) strain (Figure 9). In the absence of Mek1 activity, the rec8Δ strain exhibited accelerated DSB turnover and JM formation (4). These results were obtained regardless of whether the inhibitor was added at 3 h, 5 h or 7 h (Figure 9B and C). All DSBs and recombination intermediates in the rec8Δ mek1as strain were immediately processed upon elimination of Mek1 kinase activity (Figure 9B and C; Supplementary Figure S14 and S15). Thus, in rec8Δ mek1as(+IN), the dramatic delays in progression detected in rec8Δ were no longer observed: DSBs, SEIs and dHJs progressed rapidly and both IH-COs and IH-NCOs appeared (Figure 9B and C; Supplementary Figures S14 and S15). The rec8-29A mek1as and rec8-6A mek1as mutants also exhibited rapid turnover of DSBs, SEIs and dHJs, with very prompt formation of IH-COs and IH-NCOs as soon as Mek1 kinase was inactivated, as also observed for the rec8Δ mutant (Figure 9D and E; Supplementary Figures S14–S16). These results suggest that the regulation of meiotic progression via Mek1 is independent of Rec8 in general and Rec8 phospho-mutant functions in particular. Conversely, the defects associated with rec8 phospho-mutants are limited to normal IH-directed recombination, as the IS-biased recombination observed upon Mek1 inactivation proceeds efficiently in those mutants.
Figure 9.
Role of Mek1 kinase activity in meiotic recombination of rec8Δ and rec8 phospho-mutant. (A) Schematic representation of experimental procedures (see Materials and Methods). (B) Representative images of 2D-gels of rec8Δ mek1as in the absence or presence of 1-NA-PP1 at the indicated time points. Square brackets and arrowheads denote JMs and DSBs, respectively. (C) Quantification of recombination intermediates in the rec8Δ mek1as strain shown in (B). (D) Representative images of 2D-gels of rec8-29A mek1as in the absence or presence of 1-NA-PP1 at the indicated time points. Square brackets and arrowheads denote JMs and DSBs, respectively. (E) Quantification of recombination intermediates in the rec8-29A mek1as strain shown in (D).
DISCUSSION
The presented results provide four new pieces of information regarding the roles of meiotic cohesin subunit Rec8 for prophase recombination and structure.
Rec8 prophase roles do not involve cleavage by separase
No detectable change meiotic recombination is observed in a rec8N mutant where Rec8 is resistant to phosphorylation-dependent separase-mediated cleavage. Moreover, Rec8 remains fully sensitive to separase during prophase, as shown here by the effects of artificial Esp1 induction. Thus, if Rec8 phosphorylation plays a role for meiotic recombination, that role does not involve separase-mediated cleavage.
rec8 phospho-mutations confer a ‘separation of function’ defect that specifically affects CO-related events
Recombination-initiating DSBs occur in axis-associated recombination complexes and are significantly dependent on the presence of several axis components. Correspondingly, aberrant DSB formation/resection in rec8Δ is attributable to the fact that absence of Rec8 also results in a complete absence of detectable chromosome axes. The results presented here further demonstrate that three non-null rec8 alleles carrying mutations in multiple phosphorylation sites exhibit normal axis formation and normal DSB formation, further documenting the link between these two processes.
All analyzed rec8 phospho-mutant alleles exhibit four defects specifically related CO recombination: (i) normal formation of inter-homolog NCOs but defective formation of COs; (ii) defective homolog bias during CO recombination, presumably due to defective bias maintenance; (iii) delayed progression from one stage to another along the CO pathway; and (iv) defective SC formation. The first three phenotypes correspond to a subset of the defects described previously for rec8Δ (4). SC formation in rec8Δ could not be specifically analyzed due to the prior defect in axis formation. Thus, the rec8 phospho-mutations comprise ‘separation of function’ alleles that define a specific constellation of sub-functions of Rec8 related to CO formation. At least two of these effects are implemented at mid-prophase. SC formation and SEI formation occur at zygotene/early pachytene and Rec8's maintenance of homolog bias function comes into play at the SEI-to-dHJ transition, which occurs at early/mid- pachytene. For rec8Δ, and potentially for the phospho-alleles, homolog bias defects can largely account for the deficit of COs; however, modest defect(s) at later stages of CO formation are not totally excluded, either as downstream consequences of earlier defects and/or due to later Rec8 roles.
Strikingly, the three rec8 phospho-alleles exhibit progressively severe defects in all four of affected processes, in correlation with the number of mutated phosphorylation sites, from very modest defects in rec8-6A to a nearly complete rec8Δ phenotype in rec8-29A. These mutations were created specifically to abrogate phosphorylation at the indicated number of positions. Indeed, the rec8-24A mutant has been shown to completely block phosphorylation-dependent cleavage when separase is prematurely-induced at prophase. Moreover, it is generally considered that phosphorylation of multiple residues has a cumulative global effect on Rec8 function, aside from any unique roles for individual phosphorylations at specific sites (3,12). We cannot yet exclude the possibility that the observed phenotypes reflect effects of the alanine substitution mutations that are unrelated to the absence of phosphorylation. Nonetheless, it is tempting to think that a global effect of phosphorylation could underlie the rec8 phospho-mutant separation of function role(s), as has been widely assumed in previous studies (3,12).
DDK, but not HRR25 or Polo-like kinase, has a differential role in CO formation that is concordant with the rec8 phospho-mutant defect
At the time of rec8 CO-related defects (zygotene/early pachytene), Rec8 phosphorylation appears to be mediated essentially by Hrr25 and Cdc7/DDK (9). We find that Hrr25 plays no role for meiotic recombination. In contrast, by using timed inactivation of Cdc7, we can show that Cdc7 inactivation during mid-prophase provokes a differential reduction in CO recombination as compared to NCO recombination. Thus Cdc7 is differentially required for CO recombination versus NCO formation. This is the same type of defect seen for rec8 phospho-mutants. Thus, if phosphorylation of Rec8 is indeed important for its prophase functions, Cdc7 could be the responsible kinase. This notion can be tested in future studies by asking whether absence of Cdc7 does or does not have an effect on recombination in rec8Δ or rec8 phospho-mutants.
Interestingly, previous studies have shown that absence of Cdc7 reduces, but does not eliminate, phosphorylation of Rec8 during early/mid-prophase (9), with Hrr25 responsible for the remaining level of modification. Moreover, in the present study, the hrr25 cdc7 double mutant condition confers the same recombination phenotype as the cdc7 single mutant condition whereas the hrr25 single mutant condition shows no significant defect. Thus, whatever is the DDK substrate relevant for its role in prophase recombination, the effects of DDK and Hrr25 on that substrate are significantly different.
Rec8 is also phosphorylated by Polo-like kinase/Cdc5. Several lines of evidence suggest that Cdc5 is unlikely to be responsible for any putative phosphorylation-dependent roles of Rec8 in prophase events. First, Cdc5 is induced later, at early/mid-pachytene (9). Second, we show here Cdc5, is not required for the signature role of Rec8, maintenance of homolog bias. Third, a previous study shows that Cdc5 is not required for normal SC formation, another signature Rec8 role (3,27). Fourth, while Cdc5 is known to be required for late stages of CO formation, a rec8 phospho-mutant exhibited the same sensitivity to Cdc5's effects as wild type, implying that Rec8 is likely not the relevant target at that stage.
Functions abrogated in rec8 phospho-mutants are downstream of Mek1 and independent of ZMM proteins Zip1 and Zip3
Mek1 activity is required for assembling the normal meiotic interhomolog-directed recombination ensemble. Accordingly, we find that the rec8 phospho-mutant defects, like that those of rec8Δ (4), are downstream of Mek1. We further find that the rec8 phospho-mutant defect in homolog bias is still observed in the absence of either of two ZMM components, Zip3 and Zip3, which play important roles in mediating progression of CO recombination. This result is significant because it implies that that Rec8's role in homolog bais is distinct from that of ZMM proteins. One way of understanding this distinction is to consider that ZMM proteins are the primary mediators of the first step of CO formation, formation of SEIs from nascent DSB/partner interactions (29) whereas the role of Rec8 in maintenance of homolog bias occurs at the next step, at the transition from SEIs to dHJs (4). Further, ZMM's main role is to properly execute SEI formation, with loss of homolog bias as a secondary consequence of this execution failure; in contrast, for Rec8, the primary role is to actively maintain homolog bias during a critical ensuing transition. And finally, the role of ZMMs for SEI formation is exerted at the leading DSB end, which makes the initial contact with a partner duplex; the role of Rec8, in contrast, is likely mediated at the other, lagging DSB end, which is released from the ‘donor’ axis and incorporated into the emerging CO recombination complex at the SEI-to-dHJ transition. Rec8's role in maintenance of homolog bias is exerted at this stage and acts somehow to ensure appropriate differentiation between the leading and lagging ends during this critical transition.
Synthesis and Speculation
The current studies provide further circumstantial evidence in favor of the hypothesis that prophase phosphorylation of Rec8 plays important role(s) specifically for CO formation. If this hypothesis is correct, our data further imply that such a role(s) are independent of phosphorylation-dependent Rec8 cleavage by separase. Phosphorylation is known to promote separase-independent release of cohesin from chromosomes in several situations, most notably the prophase I/metaphase I transition of budding yeast meiosis (11) and the analogous prophase-to-metaphase transition in mitotically cycling mammalian cells (50). Cytological studies show that sister cohesin/cohesion/Rec8 is specifically absent at sites of chiasmata (i.e. COs) along diplotene chromosomes, as illustrated for grasshopper and mouse (14,51,52). Moreover, a recent study using super-resolution microscopy in mouse meiocytes suggests that Rec8 prevents local separation of sister chromatid axes and illegitimate SC assembly (53,54). Studies of Sordaria prophase chromosomes further show that WT chromosomes exhibit axis splitting and loss of axis integrity specifically at CO sites and that this tendency is exaggerated when Rec8 is absent (14). Taken together, we propose that Rec8-cohesin loading is critical for the function and shape of the chromosome axis, which might also include dynamic adjustments of recombination associated axis features via the degree of local Rec8 phosphorylation, as well as subtle, targeted modulations of sister chromatid cohesion by mechanisms independent of Rec8 cleavage. Collectively, these data indicate that Rec8 in prophase regulates faithful homolog segregation by guiding the outcomes of meiotic recombination.
Supplementary Material
Acknowledgments
The authors are grateful to Nancy Hollingsworth, Michael Lichten, Wolfgang Zachariae, Angelika Amon and Kim Nasmyth for strains and plasmids used in this study; Franz Klein and Akira Shinohara for helpful discussions and comments on the manuscript; and anonymous four reviewers for comments and suggestions on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning [2012M3A9C6050367; 2014R1A2A1A11051584; 2015R1A2A1A15054378 to K.P.K, FWF SFB F34, I031-B to M.X and NIH RO1 GM-044794 to N.K.]. Funding for open access charge: National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kleckner N., Zickler D., Jones G.H., Dekker J., Padmore R., Henle J., Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 3.Brar G.A., Kiburz B.M., Zhang Y., Kim J.E., White F., Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 4.Kim K.P., Weiner B.M., Zhang L., Jordan A., Dekker J., Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 6.Lin W., Wang M., Jin H., Yu H.G. Cohesin plays a dual role in gene regulation and sister-chromatid cohesion during meiosis in Sacchromyces cerevisiae. Genetics. 2011;187:1041–1051. doi: 10.1534/genetics.110.122358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Har.b Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong S., Sung Y., Yu M., Lee M., Kleckner N., Kim K.P. The logic and mechanism of homologous recombination partner choice. Mol. Cell. 2013;51:440–453. doi: 10.1016/j.molcel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katis V.L., Lipp J.J., Imre R., Bogdanova A., Okaz E., Habermann B., Mechtler K., Nasmyth K., Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambon J.P., Touati S.A., Berneau S., Cladière D., Hebras C., Groeme R., McDougall A., Wassmann K. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr. Biol. 2013;23:485–490. doi: 10.1016/j.cub.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Yu H.G., Koshland D. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell. 2005;123:397–407. doi: 10.1016/j.cell.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Brar G.A., Hochwagen A., Ee L.S., Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol. Biol. Cell. 2009;20:1030–1047. doi: 10.1091/mbc.E08-06-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Gálová M., Petronczki M., Gregan J., Cetin B., et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 14.Storlazzi A., Tesse S., Ruprich-Robert G., Gargano S., Pöggeler S., Kleckner N., Zickler D. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 2008;22:796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y. A one-sided view of kinetochore attachment in meiosis. Cell. 2006;126:1030–1032. doi: 10.1016/j.cell.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Lee J., Okada K., Ogushi S., Miyano T., Miyake M., Yamashita M. Loss of Rec8 from chromosome arm and centromere region is required for homologous chromosome segregation and sister chromatid separation, respectively, in mammalian meiosis. Cell Cycle. 2006;5:1488–1455. doi: 10.4161/cc.5.13.2903. [DOI] [PubMed] [Google Scholar]
- 17.Kitajima T.S., Miyazaki Y., Yamamoto M., Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokobayashi S., Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Buonomo S.B., Clyne R.K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 20.Kitajima T.S., Miyazaki Y., Yamamoto M., Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauf S., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumara I., Vorlaufer E., Stukenberg P.T., Kelm O., Redemann N., Nigg E.A., Peters J.M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 23.Liang Z., Zickler D., Prentiss M., Chang F.S., Witz G., Maeshima K., Kleckner N. Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion cycles. Cell. 2015;161:1124–1137. doi: 10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attner M.A., Miller M.P., Ee L.S., Elkin S.K., Amon A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14278–14283. doi: 10.1073/pnas.1311845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiamata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell. Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- 26.Lee B.H., Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- 27.Sourirajan A., Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allers T., Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell. 2001;8:225–231. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- 29.Börner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 30.Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 31.Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N. Sgs1, BLM ortholog, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi N., Brown M.S., Bishop D.K., Börner G.V. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol. Cell. 2015;57:797–811. doi: 10.1016/j.molcel.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martini E., Diaz R.L., Hunter N., Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:282–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasanuma H., Tawaramoto M.S., Lao J.P., Hosaka H., Sanda E., Suzuki M., Yamashita E., Hunter N., Shinohara M., Nakagawa A., et al. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasymth K. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 36.Salah S.M., Nasymth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 2000;109:27–34. doi: 10.1007/s004120050409. [DOI] [PubMed] [Google Scholar]
- 37.Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae in controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 38.Zakharyevich K., Ma Y., Tang S., Hwang P.Y., Boiteux S., Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L., Wang S., Yin S., Hong S., Kim K.P., Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature. 2014;511:551–556. doi: 10.1038/nature13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphryes N., Leung W.K., Argunhan B., Terentyev Y., Dvorackova M., Tsubouchi H. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 2013;9:e1003194. doi: 10.1371/journal.pgen.1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinohara M., Oh S.D., Hunter N., Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 42.Shinohara M., Hayashihara K., Grubb J.T., Bishop D.K., Shinohara A. DNA damage response clamp 9-1-1 promotes assembly of ZMM proteins for formation of crossovers and synaptonemal complex. J. Cell. Sci. 2015;128:1494–1506. doi: 10.1242/jcs.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson K.A., Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Espagne E., de Muyt A., Zickler D., Kleckner N.E. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E5059–E5068. doi: 10.1073/pnas.1416411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oelschlaegel T., Schwickart M., Matos J., Bogdanova A., Camasses A., Havlis J., Shevchenko A., Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 46.Thacker D., Mohibullah N., Zhu X., Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan L., Zhang C., Shokat KM., Hollingsworth N.M. Chemical inactivation of Cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–1774. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan L., de los Santos T., Zhang C., Shokat K., Hollingsworth N.M. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terentyev Y., Johnson R., Neale M.J., Khisroon M., Bishop-Bailey A., Goldman A.S. Evidence that MEK1 positively promotes interhomologue double-strand break repair. Nucleic Acids Res. 2010;38:4349–4360. doi: 10.1093/nar/gkq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hauf S., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valdeolmillos A.M., Viera A., Page J., Prieto I., Santos J.L., Parra M.T., Heck M.M., Martínez-A C., Barbero J.L., Suja J.A., et al. Sequential loading of cohesin subunits during the first meiotic prophase of grasshoppers. PLoS Genet. 2007;3:e28. doi: 10.1371/journal.pgen.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agostinho A., Mannebergm O., van Schendel R., Hernández-Hernández A., Kouznetsova A., Blom H., Brismar H., Höög C. High density of REC8 constrains sister chromatid axes and prevents illegitimate synaptonemal complex formation. EMBO Rep. 2016;17:901–913. doi: 10.15252/embr.201642030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishiguro K., Watanabe Y. The cohesin REC8 prevents illegitimate inter-sister synaptonemal complex assembly. EMBO Rep. 2016;17:783–784. doi: 10.15252/embr.201642544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.