Abstract
Histone methylation by lysine methyltransferase enzymes regulate the expression of genes implicated in lineage specificity and cellular differentiation. While it is known that Set7 catalyzes mono-methylation of histone and non-histone proteins, the functional importance of this enzyme in stem cell differentiation remains poorly understood. We show Set7 expression is increased during mouse embryonic stem cell (mESC) differentiation and is regulated by the pluripotency factors, Oct4 and Sox2. Transcriptional network analyses reveal smooth muscle (SM) associated genes are subject to Set7-mediated regulation. Furthermore, pharmacological inhibition of Set7 activity confirms this regulation. We observe Set7-mediated modification of serum response factor (SRF) and mono-methylation of histone H4 lysine 4 (H3K4me1) regulate gene expression. We conclude the broad substrate specificity of Set7 serves to control key transcriptional networks in embryonic stem cells.
INTRODUCTION
Together with transcription factors, histone-modifying complexes supervise the expression of genes that are critical for cell fate decisions and differentiation (1,2). Intrinsic to gene regulation are a handful of critical signaling molecules – Oct4 (octamer-binding transcriptional factor 4), Sox2 (Sry box-containing gene 2) and Nanog regulate the expression of genes in embryonic stem cells (ESCs) whilst maintaining pluripotent cell fate (3). The promoters in ESCs are distinctively modified by tri-methylation of histone H3 lysine 4 (H3K4me3) and histone H3 lysine 27 (H3K27me3) (4,5) that form bivalent domains critical for developmental genes (6). During ESC differentiation chromatin is subject to dynamic histone modification that precisely regulate gene expression (7). Specific signaling stimuli regulate bivalent chromatin domains critical for ESC differentiation. However, despite these recent advances, the epigenetics field remains largely uncharted with respect to understanding the physiological function of histone modifications (8). Uncovering their function in vivo remains challenging and to this end the extensive mapping of chromatin modifications has identified critical domains implicated in the regulation of developmental genes (9).
Many of the mechanistic insights into transcriptional regulation are derived from studies that have used pathway-specific readouts to induce different cell fates in cultured cells and other model systems subject to extracellular stimulation (7,10). Of particular relevance is phenotypic switching using mESCs have led to important mechanistic insights (11) and fundamental discoveries in cell differentiation (6,12). While lineage specificity is directly under the control of precise programs of chromatin modification (13), many of the original insights into transcriptional control have come from models of smooth muscle cell (SMC) differentiation. SMCs do not undergo terminal differentiation and maintain remarkable plasticity that can switch between phenotypic states in response to extracellular cues (14). Markers of SMC differentiation include contractile protein expression of smooth muscle alpha actin (ACTA2), smoothelin, calponin (CNN1), transgelin (TAGLN) and smooth muscle myosin heavy chain (MYH11) (15). During de-differentiation to the ‘synthetic’ phenotype the expression of contractile proteins is significantly reduced with re-entry into the cell-cycle, with SMCs becoming more proliferative with increased protein synthesis and migration accompanied with extracellular matrix secretion (16).
Since the original findings that CArG elements and SRF binding regulate the expression of smooth muscle (SM) genes, our understanding of phenotypic plasticity has increased significantly (17–19). SM-associated gene expression depends on combinatorial interactions of cis-regulatory elements and trans-regulatory binding factors. Myocardin (MYOCD) is a co-activator that binds SRF at CArG sequence elements to promote RNA polymerase II interaction and regulation of the contractile phenotype (20). Considered one of the best-characterized mechanisms of SMC differentiation and plasticity, CArG-SRF dependent transcriptional control serves as a paradigm mechanism of gene activation. Indeed, much of what we have learned about phenotypic switching has come indirectly from studying transcription factor binding that have defined the opposing actions of MYOCD that promote de-differentiation by trans binding factors such as KLF4 and FoxO4 (21,22). Phenotypic switching is primarily attained at the transcriptional level, typically, SRF binds CArG elements as a dimer to regulate the expression of genes implicated in SMC differentiation (23). Interestingly, the CArG-SRF interaction also participates in the regulation of skeletal- and cardiac-muscle specific genes (24,25). A better understanding of how SRF-CArG dependent transcriptional control is necessary to clarify how this regulatory complex is coordinated in response to upstream signaling and the expression of cell-specific SM-associated genes. Of major interest is context-dependent gene regulation which are coordinated by chromatin modification and the interactions of DNA bound transcriptional factors that function together with activators and repressors to control differentiation and phenotypic switching.
SMCs derived from mouse embryoid bodies display epigenetic changes that serve to alter chromatin structure, not surprisingly, when assayed for transcriptional competence, the local chromatin structure is more open at CArG-box sequences (26). Moreover, the characteristic binding of SRF together with histone H3K4 dimethylation (H3K4me2) and H3K9 acetylation (H3K9ac) enrichment are not observed in non-SMCs. These results highlight a potential molecular basis for cell type-specific and promoter-specific patterns of gene regulation (13). Phenotypic switching is defined by a series of histone modifications that serve as a paradigmatic model of SRF binding to the CArG box (27). The de-differentiated phenotype is distinguished by compacted CArG box chromatin that signal HDAC interaction and the loss of SRF which is responsible for the suppression of MYOCD transcription. This fine balance in transcriptional control shifts to a more open chromatin structure associated with permissive histone modifications; namely, SRF interaction and p300 binding that promotes differentiation. Furthermore, the effectiveness of specific signaling cues can tightly control SMC gene expression which has been shown to rely on the capacity of transcriptional activators such as PDGF-BB stimulation or vascular injury to regulate gene expression (22,28). Once again, the common principle here is context-specific and signaling-dependent transcriptional control.
While the regulatory mechanisms implicated in cell plasticity involve the coordinated actions of specific histone tail modifications, the precise role of histone modifying enzymes remains largely uncharted. A major gap in our knowledge of ESC differentiation relates to mechanisms of epigenetic control. Clearly, the identification of the enzymes that confer specific histone determinants that signal the events that promote permissive and more open chromatin structures are essential to dissect the role of epigenetic signaling as a cause or consequence of ESC differentiation. Furthermore, while the mechanisms that regulate SMC-specific transcription involve ubiquitous transcription factors such as p300 and SRF, the functional importance of histone methyltransferase enzymes during ESC differentiation remain unclear. Recent experimental studies have shown Set7-mediated mono-methylation of Sox2 suppresses transcriptional activity and induces degradation of this pluripotency factor in mouse ESCs (29). The functional importance of this repressive epigenetic mark is the control of Sox2 stability and represents a regulatory switch that determines ESC fate. Yet an important unanswered question remains, does Set7 regulate the expression of genes by histone or non-histone protein modification upon ESC differentiation?
In this article, we characterize the role of the Set7 methyltransferase using Stem cell antigen-1 (Sca1+) cells as a model of differentiation. We observe elevated Set7 expression and the regulation of SM-associated genes upon Sca1+ cell differentiation. We show the pluripotent transcription factors, Sox2 and Oct4 regulate Set7 expression during mESC differentiation. Transcriptional network analysis shows the expression of genes associated with differentiation are broadly regulated. More specifically, we define the expression of SM-associated genes are significantly reduced in Set7 deficient cells. SM-associated gene regulation is coordinated by the actions of the Set7 enzyme modifying H3K4me1 as well as interactions with the SRF protein. Furthermore, pharmacological inhibition of Set7 methyltransferase activity reduces the expression of SM-associated genes that closely corresponds with gene expression patterns derived from Set7 deficient cells. The experimental results indicate that Set7 is required for the regulation of SM-associated gene expression. Our studies provide a framework to understand stem cell commitment by the Set7 enzyme during ESC differentiation.
MATERIALS AND METHODS
The expanded Methods section is available in the Online Data Supplement.
Cell culture
Mouse Embryonic Stem cells (mESCs; a gift from Dr Mark Denham, University of Melbourne) were maintained in stem cell medium containing Dulbecco's modified Eagles medium (DMEM) supplemented with ES qualified fetal bovine serum (FBS), knockout serum replacement (KSR), non-essential amino acids (NEAA), antibiotics (penicillin/streptomycin), β-mercaptoethanol (Gibco) and 103units/ml LIF (ESGRO, Millipore). Mouse smooth muscle cells (mSMCs) isolated from aortas were cultured in DMEM/ Nutrient Mixture F-12 (DMEM/F12) supplemented with FBS, NEAA and antibiotics. 293FT cells (Invitrogen) were cultured in DMEM supplemented with FBS, L-glutamine and antibiotics. All cells were maintained at 37°C with 5% CO2 incubator. (R)-PFI-2 (a kind gift by Dr P. J. Brown, University of Toronto, Canada) was used for inhibition of Set7 methyltransferase activity. Cell differentiation and transfection experiments are described in the Online Data Supplement.
RNA analysis
RNA isolation and gene expression analyses were performed according to protocols previously described (30–32).
Protein analysis
Protein extraction, immunoprecipitation, immunoblotting and methyltransferase activity assay analysis were performed according to protocols previously described (31,32).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to protocols previously described (31,33). In brief, cells were cross-linked with 1% formaldehyde for 10 min and then quenched with 0.125M glycine for 10 min. Cell pellets were resuspended in SDS Lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0 and proteinase inhibitor cocktail) and sonicated for 30 min at 30 s intervals using a Bioruptor (Diagenode). A total of 4 µg of pre-cleared chromatin with Dynabeads protein A (Invitrogen) was immunoprecipitated using anti-OCT4, anti-H3K4me1, anti-H3K4me3 (Abcam), anti-H3K4me2, anti-H3K27me3, anti-H3K9/K14ac, anti-H4ac, anti-CTCF, anti-rabbit IgG (Millipore), anti-SOX2, anti-STAT3 (Cell Signaling Technology) or anti-SRF (Santa Cruz) antibodies. Quantification of the immunoprecipitated DNA was performed by qPCR and data are calculated as relative to the IgG-negative control and represented as enrichment of immunoprecipitated DNA samples corrected against the input DNA.
Statistical analysis
Statistical analyses was performed using GraphPad Prism 6.0 (San Diego, CA, USA). Statistical significance was evaluated by Student's t-test. A P-value less than 0.05 were considered significant. Error bars on all figures are represented as a standard error of the mean (SEM).
RESULTS
Set7 activation in Sca1+ cell differentiation from mESCs
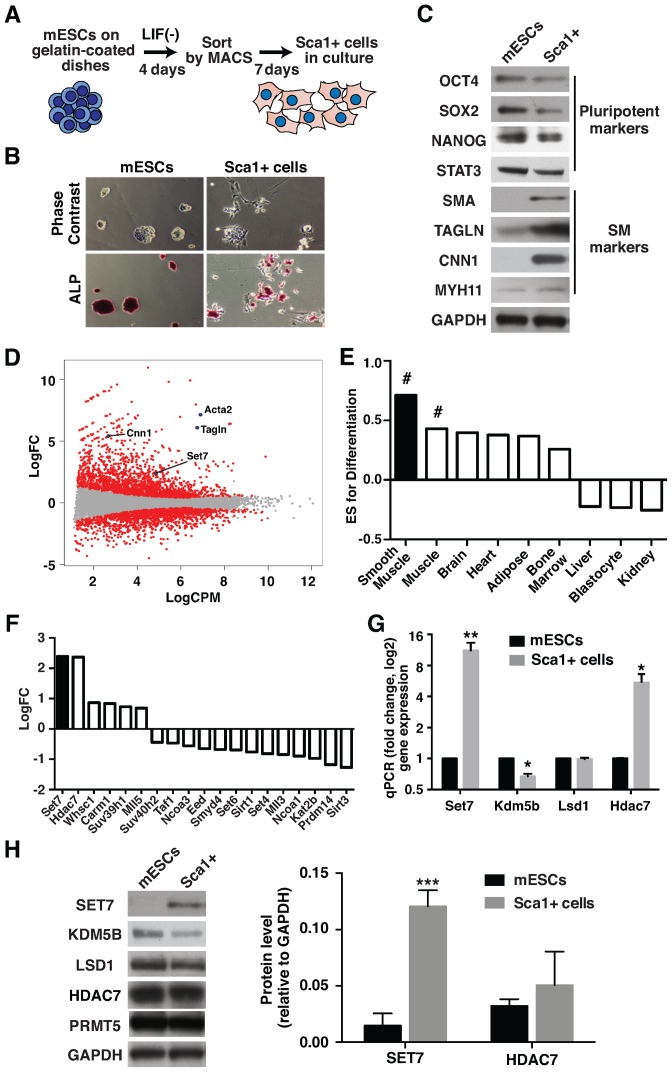
Mouse embryonic stem cells (mESCs) were differentiated in culture by removal of leukemia inhibitory factor (LIF). Stem cell antigen-1 positive (Sca1+) progenitor cells were isolated by cell sorting (34) (Figure 1A). We observed distinct morphological changes to mESCs with reduced alkaline phosphatase (ALP) staining consistent with Sca1+ cell differentiation (Figure 1B). The expression of pluripotency genes OCT4, SOX2 and NANOG were reduced and the expression of SM-associated genes increased in differentiated Sca1+ cells (Supplementary Figure S1 and Figure 1C). Gene expression changes in mESC and Sca1+ cells were determined by RNA-Sequencing (RNA-Seq). We observed 1307 up-regulated and 997 down-regulated genes upon Sca1+ cell differentiation (Figure 1D). Gene set enrichment analysis (GSEA) showed increased expression of genes related to extracellular matrix organization and muscle contraction (Supplementary Figure S2). We also observed activation of SM-associated genes upon Sca1+ cell differentiation (Figure 1E). Consistent with the observed changes in gene expression, immunoblotting experiments confirm increased expression of SMA, TAGLN and CNN1 proteins (Figure 1C). These results suggest that Sca1+ cells are committed toward the SM lineage. Because histone modifying enzymes play a critical role in lineage differentiation we examined their expression. Set7 was upregulated upon Sca1+ cell differentiation (Figure 1F). Consistent with RNA-Seq analysis, the expression of Set7 mRNA and protein were also elevated (Figure 1G and H). The expression of HDAC7, which is a marker of SMC differentiation (35), was also increased in Sca1+ cells.
Figure 1.
Set7 is up-regulated during Sca1+ cell differentiation. (A) Schematic representation of the experimental procedure for Sca1+ cell differentiation. Mouse ESCs were cultured without LIF on gelatin-coated dishes for 4 days. Sca1+ cells were isolated by magnetic activated cell sorting (MACS) using anti-Sca1 immunomagnetic beads, followed by incubation with growth medium for 7 days. (B) Morphological changes of mESC differentiation to Sca1+ cells. Upper panel indicates the representative phase contrast images of mESC and Sca1+ cells. Loss of alkaline phosphatase (ALP) staining was observed during differentiation (Bottom panel). Original magnification, 40X. (C) Protein expression profiles derived from mESC and Sca1+ cells. Immunoblot analysis of cell lysates prepared from mESC and Sca1+ cells was performed using antibodies specific to OCT4, SOX2, NANOG, STAT3, SMA, Transgelin (TAGLN), Calponin (CNN1) and MYH11. GAPDH was used for protein loading control. The results shown represent three independent experiments. (D) Genome-wide expression analysis for mESC differentiation to Sca1+ cells. RNA-Sequencing (RNA-Seq) was performed from mESC and Sca1+ cells. MA plots show the Log2 fold change (LogFC) and Log2 counts per million (LogCPM) derived from mESCs and Sca1+ cells. Significant changes in expression (up- and down-regulated genes with a P-value < 0.05) are highlighted in red. (E) Gene set enrichment analysis (GSEA) of Sca1+ cell differentiation based on tissue type (smooth muscle, adipose, heart, muscle, brain, kidney, liver, blastocyte and bone marrow). A positive Enrichment Score (ES) indicates gene sets up-regulated in Sca1+ cells. # FDR q-value = 0.01. (F) RNA-Seq identifies differentially expressed genes in mESC and Sca1+ cells that are implicated in histone tail modification. Signal shown represents the LogFC of normalized read counts of genes in Sca1+ cells. (G) Increased Set7 and Hdac7 expression in Sca1+ cells. mRNA levels of the histone methyltransferase (Set7), demethylase (Lsd1, Kdm5b) and deacetylase (Hdac7) were assessed by quantitative RT-PCR (qRT-PCR) using Gapdh as the internal control. Gene expression was normalized against mESCs. (H) Protein expression of histone modifiers during Sca1+ cell differentiation. Immunoblot analysis of cell lysates prepared from mESC and Sca1+ cells was performed using antibodies specific to SET7, KDM5B, LSD1, HDAC7 and PRMT5 (left panel). GAPDH was used as a loading control. The results shown are a representative of three independent experiments. Quantification of SET7 and HDAC7 protein levels in mESC and Sca1+ cells (right panel). Protein blotting signals were quantified by an infrared imaging system. Data are presented as a ratio SET7 and HDAC7 relative to GAPDH signal. Error bars represent the standard error of the mean of at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Oct4 and Sox2 regulate Set7 expression in stem cells
LIF signaling maintains mESC pluripotency. The removal of LIF from the media pushes mESC differentiation forward and reduces ALP positive cells (Supplementary Figure S3A). Reduced Oct4 and Sox2 gene expression was inversely correlated with Set7 gene expression (Supplementary Figure S3B). Conversely, Sca1+ cells stimulated by LIF for 4 days increased expression of Oct4, Sox2, Nanog and Stat3 and suppressed Set7 expression (Supplementary Figure S3C). We observed no significant change in Set7 expression by LIF stimulation in SMCs (Supplementary Figure S3C). Based on these results, we postulate Set7 expression is subject to regulation by Oct4, Sox2 and Nanog upon mESC differentiation.
Next, we analyzed transcription factor binding sites (TFBS) in the Set7 promoter using the JASPAR CORE database (http.//jaspar.genereg.net). We identified three putative binding sites in the Set7 promoter located at R1 for CTCF (−11 to −132), R2 for OCT4/SOX2 (−149 to −303) and R3 for STAT3 (−277 to −381) (Figure 2A). These sites are highly conserved with 95% homology between mouse and human sequences (Supplementary Figure S4). A control region R4 (−880 to −1018) was also assessed that does not contain these binding sites.
Figure 2.
Pluripotent factors Oct4 and Sox2 regulate Set7 expression in mESCs. (A) Schematic illustration of the Set7 proximal promoter region. Mouse Set7 gene is located on chromosome 3qC. Putative binding sites of three transcription factors; CTCF (R1 at −11 to −132 bp), OCT4/SOX2 (R2 at −149 to −303 bp), STAT3 (R3 at −277 to −381 bp) and negative control region (R4 at −880 to −1018 bp) are located upstream of the transcriptional start site (TSS) of the Set7 (Setd7) gene. (B) Identification of the Oct4 and Sox2 binding element necessary for Set7 transcription in mESCs. The reporter constructs (left panel) were generated and transfected into mESCs. Luciferase activity was measured using the FLUOstar Omega (BMG Labtech). Data are expressed as the relative response ratio (RRR%) that quantifies the ratio of the Luciferase firefly luminescence/Renilla luminescence of the experimental reporter activity relative to the control reporter activity. (C) Deletion of the Oct4/Sox2 binding site activates Set7 transcription. Reporter constructs (1.3, 1.0, 0.7 and 0.3) were deleted for the Oct4/Sox2 binding sites and transfected into mESCs. Luciferase activity was normalized against cells transfected with the reporter constructs with wild-type (WT) Set7 promoter. (D) Reduced binding of OCT4 and SOX2 on the Set7 gene promoter following LIF removal. Soluble chromatin was isolated from mESCs stimulated with (black) or without LIF (white) for 4 days, and immunopurified with antibodies that recognize CTCF, OCT4, SOX2, STAT3, H3K4me3 and H3K27me3. qPCR was performed to assay quantitative binding relative to the IgG-negative control. The level of enrichment is shown as the percentage of corrected input. (E) Knockdown of OCT4 reactivates Set7 expression. mESCs were transfected with siRNA targeted to the Oct4 transcript. Immunoblot analysis was performed using anti-OCT4, SOX2, SET7 and GAPDH antibodies (left panel). mRNA levels of Set7, Oct4, Sox2 and Nanog genes were quantified by qRT-PCR using Gapdh as the internal control (right panel). Gene expression was normalized against cells transfected with non-target control (NTC). (F) Overexpression of OCT4 and SOX2 down-regulates Set7 gene expression. Sca1+ cells were transfected with HA-tagged OCT4 (HA-OCT4) and SOX2 (HA-SOX2) constructs followed by protein and RNA isolation after 48 h. Immunoblot analysis was performed using anti-HA, OCT4, SOX2, SET7 and GAPDH antibodies (left panel). Set7 gene expression was assessed by qRT-PCR using Gapdh as the internal control (right panel). Gene expression was normalized against the vector control. Error bars represent the standard error of the mean from three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
To identify important regulatory regions, we cloned the Set7 promoter (1.3 kb upstream from the transcriptional start site) into the luciferase vector and created the deletion derivatives by site-directed mutagenesis (Figure 2B, left). These vectors were transfected into mESCs and assayed for transcription by assessing luciferase activity. We identified the 80-bp region with the OCT4/SOX2 binding site at R2 is an essential element for Set7 expression (Figure 2B, right). Furthermore, we showed that deletion of the OCT4/SOX2 binding site increased luciferase activity in mESCs (Figure 2C). These results imply that Set7 expression is subject to regulation by promoter elements that recognize the pluripotent factors Oct4 and Sox2 during mESC differentiation.
Because specific deletion mutants regulate Set7 transcription we assessed binding of STAT3, OCT4, SOX2 and CTCF at R1, R2, R3 and R4 sites by chromatin immunoprecipitation (ChIP) assays. Soluble chromatin derived from mESCs show reduced OCT4 and SOX2 binding at the R2 site of the Set7 promoter following LIF removal (Figure 2D). No significant changes were observed for STAT3 and CTCF binding (R1 for CTCF and R3 for STAT3). Interestingly, R2 and R3 are enriched with both H3K4me3 and H3K27me3, which are though to participate in forming a bivalent chromatin domain (6). We observed increased H3K4me3 and reduced H3K27me3 at the Set7 promoter following LIF removal (Figure 2D). Having shown OCT4 and SOX2 bind the Set7 proximal promoter, we investigated functional activity in mESCs using Oct4 siRNA, because Oct4 and Sox2 cooperatively interact to regulate gene expression (36). We confirm Set7 activation is associated with suppressed Sox2 and Nanog expression in Oct4KD (knockdown) cells (Figure 2E). Next, we used transient over expression experiments in Sca1+ cells to determine whether OCT4 and SOX2 suppresses Set7 gene expression. While the expression of Nanog and Rex1 genes were increased (data not shown), transient overexpression of OCT4 and SOX2 reduced Set7 expression in Sca1+ cells (Figure 2F). Taken together, the results of these experiments using loss or gain of function cell models suggest Set7 expression is subject to regulation by OCT4 and SOX2.
Set7 dependent transcriptional regulation in a model of mESC differentiation
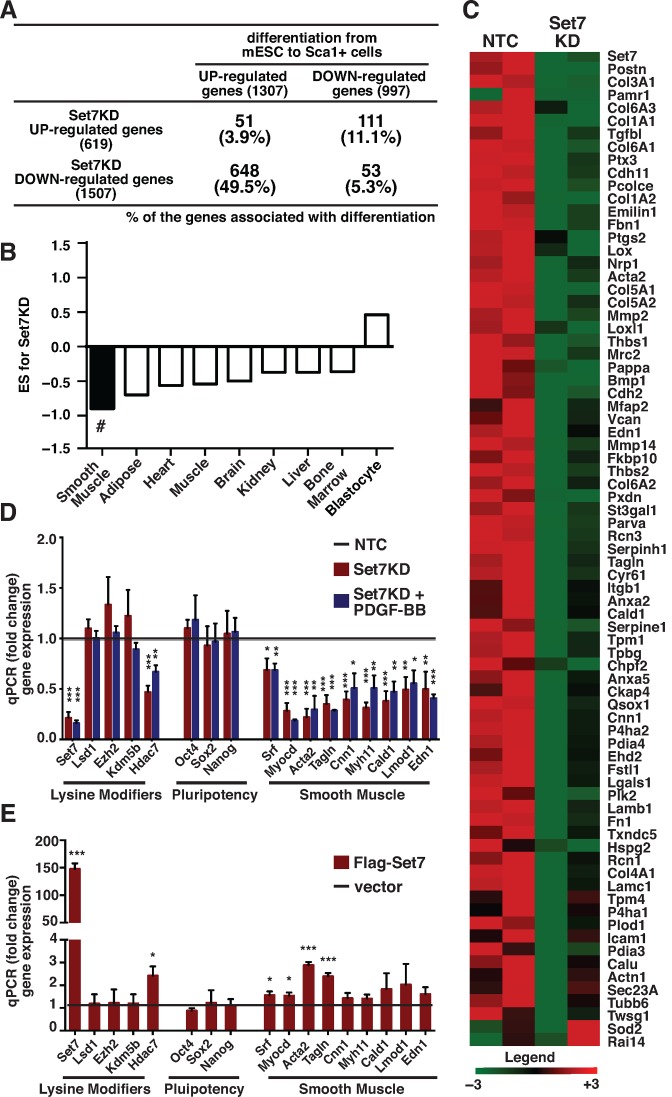
Because precisely regulated patterns of transcription underlie cell differentiation, we investigated the functional role of Set7 using lentivirus-mediated shRNA knockdown (31) (Set7KD, Supplementary Figure S5A). We selected the most effective shRNA target for Set7 protein knockdown from five shRNA constructs (Supplementary Figure S6). RNA-Seq identified 1507 down-regulated and 619 up-regulated genes subject to regulation by Set7 (Figure 3A). We compiled a list of genes implicated in differentiation by profiling transcripts derived from mESC and Sca1+ cells. The expression of differentiation-associated genes in Sca1+ cells was reduced by almost half (49.5%) when compared to Set7KD cells (Figure 3A). GSEA identified differentiation-associated genes were broadly regulated in Set7 deficient cells (Figure 3B). More specifically, the expression of SM-associated genes was significantly down-regulated in Set7KD cells (Supplementary Figure S5B). We also observe a subset of canonical SM-associated genes suppressed in Set7KD cells (Figure 3C). Consistent with RNA-Seq data we confirm reduced expression of Acta2, Cnn1 and Tagln in Set7KD cells by qRT-PCR (Figure 3D and Supplementary Figure S5C and S5D). Conversely, Set7 over-expression in Sca1+ cells increased SM-associated gene and protein expression (Figure 3E and Supplementary Figure S5E). While previous reports have shown that PDGF stimulation induces SMC differentiation (34,35) we found no association with the activation of SM-associated genes in Set7KD Sca1+ cells (Figure 3D). Finally, we show a direct role for Set7 using shRNA in regulating the expression of SM-associated genes in mouse aortic SMCs (Supplementary Figure S7).
Figure 3.
Genome-wide expression analysis shows Set7 as a positive regulator for smooth muscle associated genes. (A) Table summarising the expression of differential mRNA transcripts derived from mESC and Sca1+ cells in response to differentiation (adjusted P-value < 0.01) versus mRNA transcripts that were differentially expressed in Set7KD Sca1+ cells (adjusted P-value < 0.01). The percentages shown are genes associated in Sca1+ cell differentiation. (B) GSEA was used to determine transcriptional changes in Set7KD Sca1+ cells highlighting different tissue types (smooth muscle, adipose, heart, muscle, brain, kidney, liver, bone marrow and blastocytes). A negative Enrichment Score (ES) indicates down-regulated gene sets in Set7KD Sca1+ cells. # FDR q-value = 0.01. (C) Heatmap of SM-associated genes differentially expressed in Set7KD Sca1+ cells. Signal shown represents the Log2 of normalized values of all samples. Genes in red are up-regulated and those shown in green are down-regulated. (D) Gene expression changes observed in Set7KD Sca1+ cells as a positive regulator of SM gene expression independent of PDGF-BB stimulation. Set7KD Sca1+ cells were stimulated with PDGF-BB in culture for 14 days. Assessment of gene expression was performed using qRT-PCR for lysine modifiers, pluripotency markers and SM-associated genes in Set7KD Sca1+ cells (red bar) and Set7KD + PDGF-BB (blue bar) normalized against non-target control cells (black line). Gapdh was used as the internal control. (E) Overexpression of Set7 induces the expression of SM-associated genes in Sca1+ cells. Gene expression was assessed by qRT-PCR for histone modifiers, pluripotency markers and SM-associated genes in Sca1+ cells overexpressing FLAG-tagged Set7 (red bar) normalized against the empty vector (black line). Gapdh was used as the internal control. *P < 0.05, **P < 0.01 and ***P < 0.001.
SM-associated genes are subject to Set7-mediated methyltransferase activity
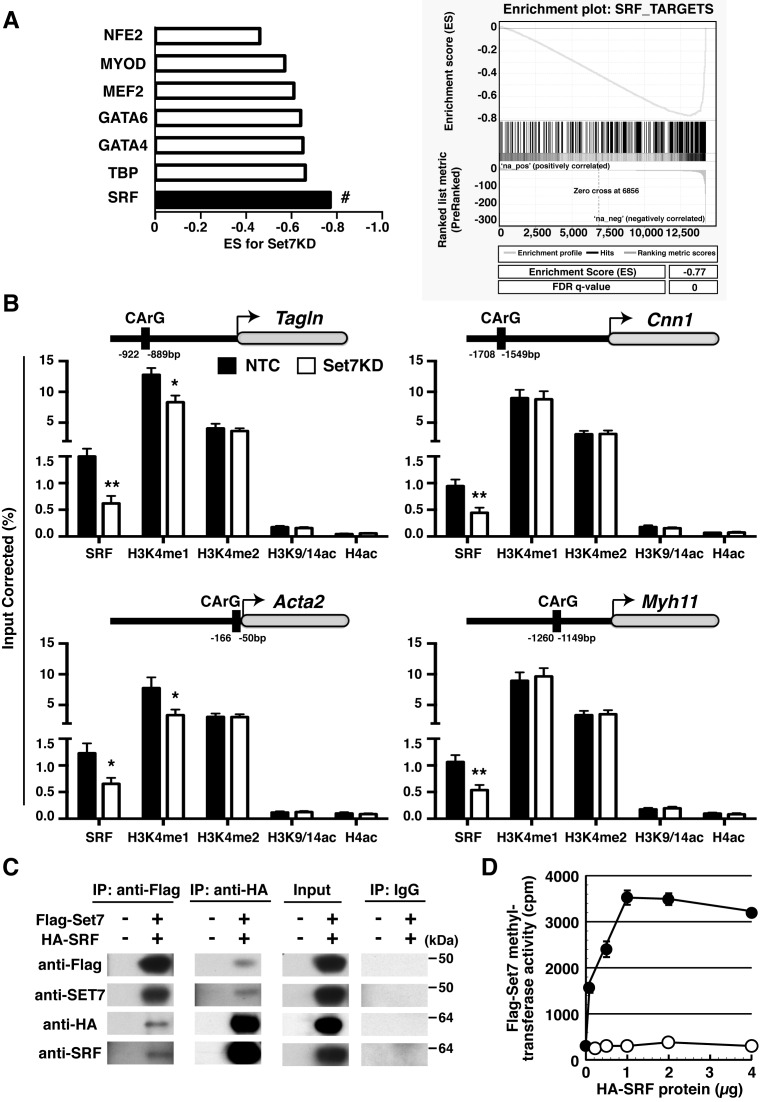
Serum response factor (SRF) regulates smooth muscle gene expression by interacting with myocardin at the conserved CArG (CC(A/T)6GG) binding motif (19,26). We assessed known CArG elements involved with SMC differentiation in Set7KD cells using GSEA (37). We found a strong association for CArG-containing SRF binding sites (FDR q-value = 0.01) (Figure 4A).
Figure 4.
Set7 is required for H3K4me1 and the recruitment of SRF at the promoters of SM-associated genes. (A) SRF gene targets are down-regulated in Set7KD Sca1+ cells. GSEA identifies regulatory targets implicated in SM-associated gene expression in Set7KD cells (Left panel). A negative ES indicates down-regulated transcription factor gene sets by Set7KD. # FDR q-value = 0.01. GSEA identifies SRF gene targets in Set7KD Sca1+ cells (Right panel). SM-associated transcription factors were derived from mSigDB. (B) SM-associated gene expression is subject to SRF and H3K4me1 dependent mechanisms regulated by Set7 methyltransferase. Soluble chromatin was isolated from the Sca1+ cells transduced with shRNA for non-target control (NTC; black) or Set7KD (white) and immunoprecipitated with antibodies against SRF, H3K4me1, H3K4me2, H3K9/14ac and H4ac. Enrichment at CArG sites were assessed at Cnn1, Tagln, Acta2 and Myh11 genes by qPCR. Data are calculated as relative to the IgG-negative control and represented as ChIP DNA normalized against input DNA. Error bars represent the standard error of the mean of at least five independent experiments. *P < 0.05 and **P < 0.01. (C) Set7 interacts with SRF. 293FT cells were co-transfected with Flag-tagged Set7 (Flag-Set7) and HA-tagged SRF (HA-SRF) expression plasmids. Reciprocal immunoprecipitaion assays were performed with anti-Flag or anti-HA beads. Immunoprecipitations were detected by immuno-blotting using anti-FLAG, HA, SET7 and SRF antibodies. The data presented are a representative of four independent experiments. (D) Set7 methylates recombinant SRF protein. Immunopurified Flag-Set7 (black circle) was incubated with (H3)-S-adenosyl-methione and HA-SRF protein for 1 h at 30°C. Tritiated HA-SRF protein was measured by liquid scintillation. Tritiated HA-SRF proteins without Flag-Set7 (white circle) represent background activity. Data are presented as mean ± SD, n = 3.
Studies show di-methylation of H3K4 (H3K4me2) at CArG-containing promoters of SM-associated genes regulate SRF complexes during development (13,26). Furthermore, SRF binding at CArG sites is mediated by the MADS box-domain that contains the target amino acid motif (GRHKPST) (K > R) (S > KYARTPN) K (lysine for methylation is underlined) as a putative target for Set7 enzyme (30,38). We hypothesized the regulation of SM-associated genes was subject to Set7-mediated modification of histone and non-histone proteins, specifically, H3K4 and SRF.
ChIP assays were performed for SRF, H3K4me1, H3K4me2, H3K9/14ac and H4ac (pan-acetylation) at the CArG-containing sites of the Cnn1, Tagln, Acta2 and Myh11 genes. Chromatin derived from Set7KD Sca1+ cells was significantly reduced for SRF binding at CArG-containing sites (Figure 4B). We also observed reduced H3K4me1 at CArG-containing sites for the Tagln and Acta2 genes in Set7KD cells (Figure 4B). While H3K4me2 is as an epigenetic mark found on the promoters of SM-associated genes (26), we observe no significant change at CArG-containing sites. This is also the case for H3K9/14ac and H4ac in Set7KD cells. Taken together, these results suggest Set7 may regulate H3K4me1 and SRF cooperatively on the Tagln and Acta2 genes. Whereas, Set7-mediated SRF binding on the Cnn1 and Myh11 genes is independent of H3K4me1 modification.
Having demonstrated recruitment of SRF at CArG-containing elements is subject to Set7 mediated regulation, we investigated direct protein-interaction by immunoprecipitation using Flag-tagged Set7 (Flag-Set7) and HA-tagged SRF (HA-SRF). Reciprocal immunoprecipitation (Re-IP) experiments demonstrated Set7 interacts with SRF (Figure 4C). We show the specificity of our optimized Re-IP assays for the epitopes using IgG controls. We also investigated whether SRF protein was subject to the methyl-writing ability of the Set7 enzyme using specific methyltransferase activity assay (31). Consistent with the mechanism of SRF recruitment at SM-associated genes (Figure 4B), we confirm the methyltransferase activity derived from immunopurified Set7 can methylate SRF substrate protein (Figure 4D). Taken together, our data indicate Set7 regulates histone and non-histone proteins implicated in the regulation of SM-associated gene expression.
Pharmacological Set7 inhibition reduces the expression of SM-associated genes
Since (R)-PFI-2 is a selective inhibitor of Set7 methyltransferase activity (39), we investigated the effect of enzymatic inhibition by this compound on SM-associated gene expression (Figure 5A). We demonstrate significant inhibition of Set7 enzymatic activity using the (R)-PFI-2 compound on SRF recombinant protein as well as the H3K4 peptide substrates (Figure 5B). To show Set7-mediated regulation of SM-associated gene expression, we exposed Sca1+ cells and mouse aortic SMCs with (R)-PFI-2 (20μM) for 2 days. Sca1+ cell viability and cytotoxicity did not significantly change with (R)-PFI-2 exposure (Supplementary Figure S8). The Set7 inhibitor reduced the expression of SM-associated genes such as Acta2, Tagln, Cnn1 and Myh11 in Sca1+ cells (Figure 5C). In parallel experiments, we confirmed Set7-mediated regulation of SM-associated genes in mouse aortic SMCs using (R)-PFI-2. The experimental results show the pharmacological Set7 inhibitor regulates the expression of SM-associated genes.
Figure 5.
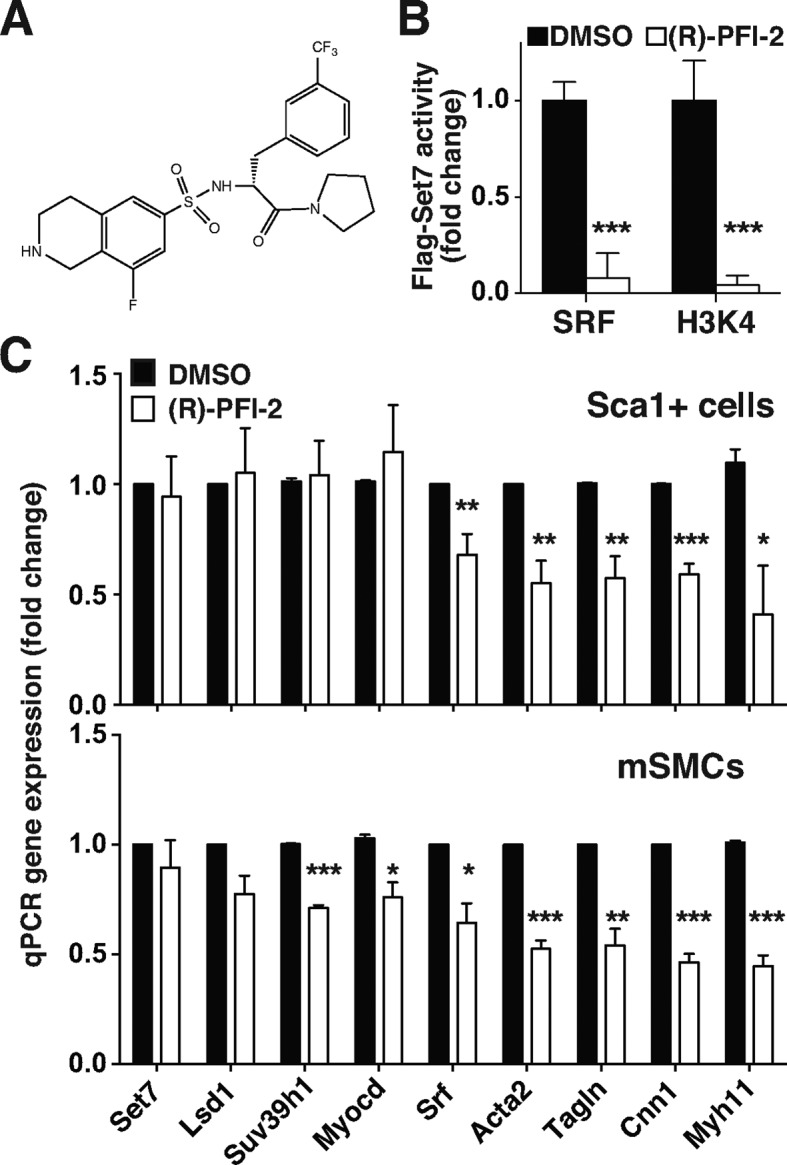
Pharmacological inhibition of Set7 methyltransferase activity attenuates the expression of SM-associated genes. (A) Chemical structure of the Set7 inhibitor, (R)-PFI-2. (B) Inhibition of SRF and H3K4 methylation by the (R)-PFI-2 compound. A total of 5 μM of (R)-PFI-2 was added in each reaction and the activity assessed. Flag-Set7 activity is presented as fold-change normalized to DMSO control. A total of 2 nmol of H3K4 peptide was used as a control substrate. (C) Set7 inhibitor (R)-PFI-2 attenuates expression of SM-associated genes. Sca1+ cells (upper panel) and mouse aortic SMCs (lower panel) were incubated with 20 μM (R)-PFI-2 for 2 days. DMSO was used as a vehicle control. mRNA levels of histone modifiers and SM-associated markers were assessed by qRT-PCR using Gapdh as the internal control. Gene expression was normalized against DMSO control. Error bars represent the standard error of the mean of at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
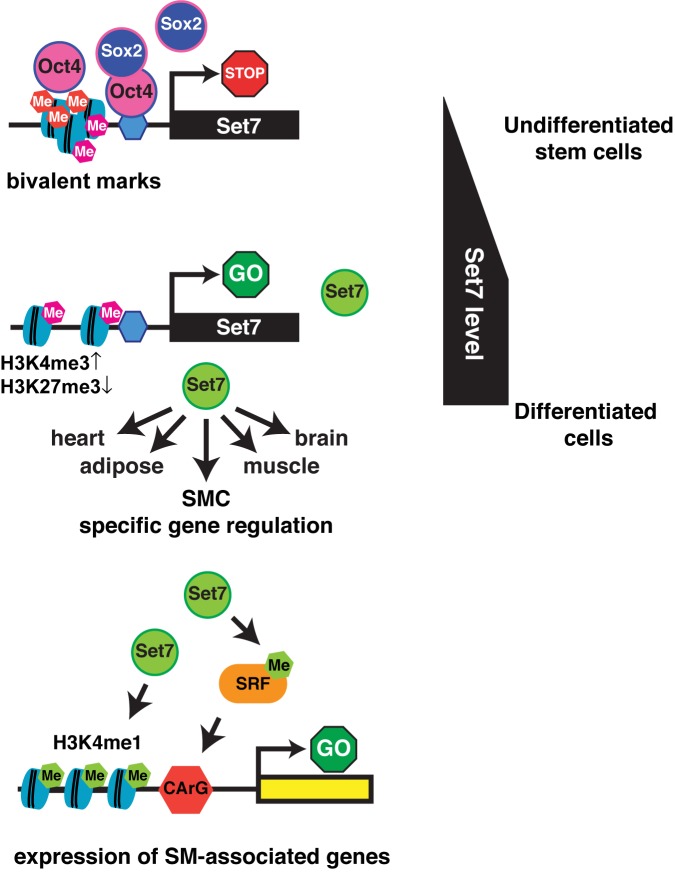
Figure 6 illustrates the proposed mechanistic events associated with the regulation of Set7 by OCT4 and SOX2 upon ESC differentiation. Our study highlights the importance of Set7 methyltransferase activity regulating SM-associated genes by histone and non-histone substrates.
Figure 6.
Schematic illustration of Set7 dependent transcriptional network in mESC differentiation. In its repressed and undifferentiated state, the Set7 promoter is characterized with a bivalent domain and enriched with OCT4 and SOX2. Following ESC differentiation OCT4 and SOX2 are released from the Set7 promoter for transcriptional activation associated with H3K4me3. Increased Set7 protein promotes the broad expression of differentiation-associated genes (SMC, Heart, Adipose, Muscle and Brain). The results of experimental studies performed here indicate Set7-dependent control of SM-associated genes by SRF methylation and H3K4me1.
DISCUSSION
In the present study, we have shown that Set7 is regulated by the pluripotent transcription factors, Oct4 and Sox2 during mESC differentiation. In contrast to other histone modifiers, the expression of Set7 is significantly up-regulated in Sca1+ cells as well as embryoid bodies, myoblast and human ESC differentiation (29,40,41). While these results suggest that Set7 has a role in differentiation we still do not know the regulatory machineries and epigenetic determinants that segregate and define different cell fates. Because the Set7 promoter has a very high CpG rich region we did investigate regulation by DNA methylation. However, we did not detect changes in the methylation status of the Set7 promoter during mESC differentiation (data not shown). Oct4 and Sox2 are critical for stem cell differentiation (3) and Set7-mediated methylation regulates the Sox2 switch during early embryonic development (29). We show the binding of Oct4/Sox2 is critical for Set7 gene regulation. Our studies suggest gene expression controlled by H3K4me1, as well as Sox2, might be important regulatory events during ESC differentiation. In a separate publication that compliments the study results presented here, Oct4 knockdown dramatically up-regulates Set7 expression in human ESCs (42). We speculate the inactivation of Set7 by Oct4 and Sox2 is important for ESC maintenance. Whether Set7 coordinates the expression of other downstream factors necessary for differentiation remains unknown and an area for future investigation.
While specific signaling cues can tightly stimulate transcription, reciprocal regulation has been shown to involve lysine specific methyl-writing and methyl-erasing activities. Infact, this robustness represents a unique balance of co-regulators that extend the functional cooperativity such as those mediated by Set7 and the lysine-specific demethylase 1, LSD1, in regulating HIF-1α transcription (43). Furthermore, the post-translational modification of histone H3 lysine 4 by Set7 methyltransferase regulates myogenic promoters during skeletal muscle differentiation (40). The expression of skeletal muscle gene targets is coordinated by cooperative competition of Set7 and Suv39h1 for MyoD regulation. This is primarily attained at the transcriptional level with Set7 competing with Suv39h1 for the binding of target sequences. In this study, we show the expression of SM-associated genes is segregated by the modification of histone and non-histone substrates conferred by Set7 methyltransferase activity.
The broad substrate specificity of Set7 to regulate the stability of non-histone proteins also serves to control nuclear processes during embryonic development. Indeed, this is highlighted in mice lacking Set7, with early embryonic lethality (44). While that study went onto to show p53 is subject to Set7 mediated methylation, the role of the lysine methyltransferase during embryogenesis remain poorly understood. A key challenge to understand the physiological role of Set7 is characterizing the regulatory function during development: in the skeletal muscle example described previously, differentiation occurs by Set7 mediated activation (40). Other studies have shown tissue-specific activation of the Set7 gene during β-cell development is dependent on the (pancreatic and duodenal homeobox 1) PDX1 transcription factor (45) with the capacity to reprogram endocrine cells (46). Studies investigating p53-dependant transcriptional regulation show the homozygous knockout of the Set7 allele in mice are viable, which is at odds with an earlier study (44), without causing marked consequences on tumor suppressor activity (47,48). While explanations for the discordant experimental results remain unclear, further studies are required to characterize the role of the lysine methyltransferase during embryogenesis to investigate the mechanistic link between the Set7 enzyme with differentiation and smooth muscle function.
While our network analysis suggests SM-associated genes are under Set7 regulation, a previous study showed that CArG-box containing SM-associated genes are enriched for H3K4me2 and regulated directly by SRF (26). Given the requirement for SRF in regulating skeletal muscle growth and maturation (49), we speculate that Set7 cooperatively regulates transcription by context-dependent mechanisms. For example, we observe asymmetric modification of H3K4me1 and H3K4me2 at CArG-box chromatin. This is primarily observed on Tagln and Acta2 promoters with reduced H3K4me1 and SRF binding in Set7KD Sca1+ cells. Whereas, Cnn1 and Myh11 genes show indistinguishable H3K4me1 and H3K4me2 at CArG box-sequences, these genes are dependent on SRF interaction for transcriptional control. This unique interplay between SRF at CArG-box chromatin and the asymmetric enrichment of histone modifications highlight distinguishable regulatory events mediated by Set7 at SM-associated genes.
Despite some of the fundamental advances already discussed, a major gap in our knowledge relates to the dynamic yet subtle sequestration of Set7 between cellular compartments. Not surprisingly, cellular position is central to lysine substrate activity, which is subject to specific extracellular signals. The effectiveness of these signaling cues relies on their capacity to robustly control Set7-mediated methylation. With little known of the subcellular localization of specific macromolecular complexes, Set7 mono-methylates Yap at K494 (Yes-associated protein) to promote cytoplasmic localization by the Hippo pathway (50). Interestingly, in high-density embryonic fibroblasts, Set7 is predominantly cytoplasmic, which is consistent with other studies (31). This is intriguing because human endothelial cells stimulated by hyperglycemia alters the subcellular locality and this is associated with distinguishable histone and non-histone regulatory events. While these (33,51) and other studies indicate biochemical properties as a glycemic sensor (52), nuclear accumulation with specific nucleosomal preferences highlights dynamic Set7 activity. This fascinating link with extracellular signaling also carries with it precise instructions for Set7 activity that include broad substrate specificity (30). While H3 is mono-methylated at K4 to regulate the expression of some SM-associated genes, understanding how SRF methylation is regulated by Set7 is an area of future investigation. The MADS box-domain of SRF which recognizes CArG binding sites, contains the F161-SKRK-T166 motif and is implicated in the regulation of SM-associated genes (26). We have shown that (R)-PFI-2 inhibits Set7 methyltransferase activity against the SRF protein substrate and suppresses SM-associated gene expression. Since, a subset of genes is associated with asymmetric methylation, this suggests methylation of H3K4 and SRF is a dynamic process and studies examining aspects of demethylation may provide clues to the control of cytoplasmic and nuclear location.
Supplementary Material
Acknowledgments
The authors sincerely thank Dr Mark Denham for mESCs, Dr P. J Brown for (R)-PFI-2, Monash Micro Imaging-AMREP and Christian Orlowski and Antony Kaspi for their assistance with RNA sequencing and bioinformatic analyses.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [NHMRC GNT0526681, GNT1048377]; Juvenile Diabetes Research Foundation [JDRF 5-2008-298, 27-2012-451]; Diabetes Australia Research Trust [DART]; Victorian Government's Operational Infrastructure Support program [in part]. Funding for open access charge: Baker IDI Heart and Diabetes Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gan Q., Yoshida T., McDonald O.G., Owens G.K. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- 2.Morey L., Santanach A., Di Croce L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol. Cell. Biol. 2015;35:2716–2728. doi: 10.1128/MCB.00266-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Sims R.J., 3rd, Nishioka K., Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 7.Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 9.Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Kumar B., Parrish M.E., Slaughter B.D., Unruh J.R., Gogol M., Seidel C., Paulson A., Li H., Gaudenz K., Peak A., et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015;25:1229–1243. doi: 10.1101/gr.184978.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H., Shukla A., Wang X., Chen W.Y., Bernstein B.E., Roeder R.G. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Alexander M.R., Owens G.K. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 14.Liu R., Leslie K.L., Martin K.A. Epigenetic regulation of smooth muscle cell plasticity. Biochim. Biophys. Acta. 2015;1849:448–453. doi: 10.1016/j.bbagrm.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owens G.K. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 16.Sobue K., Hayashi K., Nishida W. Expressional regulation of smooth muscle cell-specific genes in association with phenotypic modulation. Mol. Cell Biochem. 1999;190:105–118. [PubMed] [Google Scholar]
- 17.Chen J., Kitchen C.M., Streb J.W., Miano J.M. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 18.Kim S., Ip H.S., Lu M.M., Clendenin C., Parmacek M.S. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manabe I., Owens G.K. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J. Clin. Invest. 2001;107:823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long X., Bell R.D., Gerthoffer W.T., Zlokovic B.V., Miano J.M. Myocardin is sufficient for a smooth muscle-like contractile phenotype. Arterioscler. Thromb. Vasc. Biol. 2008;28:1505–1510. doi: 10.1161/ATVBAHA.108.166066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z.P., Wang Z., Yanagisawa H., Olson E.N. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Sinha S., McDonald O.G., Shang Y., Hoofnagle M.H., Owens G.K. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 23.Shore P., Sharrocks A.D. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartorelli V., Kurabayashi M., Kedes L. Muscle-specific gene expression. A comparison of cardiac and skeletal muscle transcription strategies. Circ. Res. 1993;72:925–931. doi: 10.1161/01.res.72.5.925. [DOI] [PubMed] [Google Scholar]
- 25.Du K.L., Ip H.S., Li J., Chen M., Dandre F., Yu W., Lu M.M., Owens G.K., Parmacek M.S. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald O.G., Wamhoff B.R., Hoofnagle M.H., Owens G.K. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J. Clin. Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manabe I., Owens G.K. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ. Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 28.Hendrix J.A., Wamhoff B.R., McDonald O.G., Sinha S., Yoshida T., Owens G.K. 5′ CArG degeneracy in smooth muscle alpha-actin is required for injury-induced gene suppression in vivo. J. Clin. Invest. 2005;115:418–427. doi: 10.1172/JCI22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J.X., Wong J. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Keating S.T., Ziemann M., Okabe J., Khan A.W., Balcerczyk A., El-Osta A. Deep sequencing reveals novel Set7 networks. Cell. Mol. Life Sci. 2014;71:4471–4486. doi: 10.1007/s00018-014-1651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okabe J., Orlowski C., Balcerczyk A., Tikellis C., Thomas M.C., Cooper M.E., El-Osta A. Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circ. Res. 2012;110:1067–1076. doi: 10.1161/CIRCRESAHA.112.266171. [DOI] [PubMed] [Google Scholar]
- 32.Okabe J., Fernandez A.Z., Ziemann M., Keating S.T., Balcerczyk A., El-Osta A. Endothelial transcriptome in response to pharmacological methyltransferase inhibition. ChemMedChem. 2014;9:1755–1762. doi: 10.1002/cmdc.201402091. [DOI] [PubMed] [Google Scholar]
- 33.El-Osta A., Brasacchio D., Yao D., Pocai A., Jones P.L., Roeder R.G., Cooper M.E., Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Q., Zeng L., Zhang Z., Hu Y., Xu Q. Stem cell-derived Sca-1+ progenitors differentiate into smooth muscle cells, which is mediated by collagen IV-integrin alpha1/beta1/alphav and PDGF receptor pathways. Am. J. Physiol. Cell Physiol. 2007;292:C342–352. doi: 10.1152/ajpcell.00341.2006. [DOI] [PubMed] [Google Scholar]
- 35.Margariti A., Xiao Q., Zampetaki A., Zhang Z., Li H., Martin D., Hu Y., Zeng L., Xu Q. Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J. Cell Sci. 2009;122:460–470. doi: 10.1242/jcs.034850. [DOI] [PubMed] [Google Scholar]
- 36.Okumura-Nakanishi S., Saito M., Niwa H., Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 37.Rafehi H., Balcerczyk A., Lunke S., Kaspi A., Ziemann M., Kn H., Okabe J., Khurana I., Ooi J., Khan A.W., et al. Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res. 2014;24:1271–1284. doi: 10.1101/gr.168781.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhayalan A., Kudithipudi S., Rathert P., Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Barsyte-Lovejoy D., Li F., Oudhoff M.J., Tatlock J.H., Dong A., Zeng H., Wu H., Freeman S.A., Schapira M., Senisterra G.A., et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12853–12858. doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao Y., Neppl R.L., Huang Z.P., Chen J., Tang R.H., Cao R., Zhang Y., Jin S.W., Wang D.Z. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castano J., Morera C., Sese B., Boue S., Bonet-Costa C., Marti M., Roque A., Jordan A., Barrero M.J. SETD7 regulates the differentiation of human embryonic stem cells. PLoS One. 2016;11:e0149502. doi: 10.1371/journal.pone.0149502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babaie Y., Herwig R., Greber B., Brink T.C., Wruck W., Groth D., Lehrach H., Burdon T., Adjaye J. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25:500–510. doi: 10.1634/stemcells.2006-0426. [DOI] [PubMed] [Google Scholar]
- 43.Kim Y., Nam H.J., Lee J., Park do Y., Kim C., Yu Y.S., Kim D., Park S.W., Bhin J., Hwang D., et al. Methylation-dependent regulation of HIF-1alpha stability restricts retinal and tumour angiogenesis. Nat. Commun. 2016;7:10347. doi: 10.1038/ncomms10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurash J.K., Lei H., Shen Q., Marston W.L., Granda B.W., Fan H., Wall D., Li E., Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol. Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 45.Deering T.G., Ogihara T., Trace A.P., Maier B., Mirmira R.G. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathiyalagan P., Keating S.T., Al-Hasani K., El-Osta A. Epigenetic-mediated reprogramming of pancreatic endocrine cells. Antioxid. Redox Signal. 2015;22:1483–1495. doi: 10.1089/ars.2014.6103. [DOI] [PubMed] [Google Scholar]
- 47.Campaner S., Spreafico F., Burgold T., Doni M., Rosato U., Amati B., Testa G. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol. Cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Lehnertz B., Rogalski J.C., Schulze F.M., Yi L., Lin S., Kast J., Rossi F.M. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol. Cell. 2011;43:673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Li S., Czubryt M.P., McAnally J., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oudhoff M.J., Freeman S.A., Couzens A.L., Antignano F., Kuznetsova E., Min P.H., Northrop J.P., Lehnertz B., Barsyte-Lovejoy D., Vedadi M., et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 51.Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E.K., Calkin A.C., Brownlee M., Cooper M.E., El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciccarelli M., Vastolo V., Albano L., Lecce M., Cabaro S., Liotti A., Longo M., Oriente F., Russo G.L., Macchia P.E., et al. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia. 2016;59:176–186. doi: 10.1007/s00125-015-3774-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.