Abstract
Regulation of translation plays a critical role in determining mRNA fate. A new role was recently reported for a subset of RGG-motif proteins in repressing translation initiation by binding eIF4G1. However the signaling mechanism(s) that leads to spatial and temporal regulation of repression activity of RGG-motif proteins remains unknown. Here we report the role of arginine methylation in regulation of repression activity of Scd6, a conserved RGG-motif protein. We demonstrate that Scd6 gets arginine methylated at its RGG-motif and Hmt1 plays an important role in its methylation. We identify specific methylated arginine residues in the Scd6 RGG-motif in vivo. We provide evidence that methylation augments Scd6 repression activity. Arginine methylation defective (AMD) mutant of Scd6 rescues the growth defect caused by overexpression of Scd6, a feature of translation repressors in general. Live-cell imaging of the AMD mutant revealed that it is defective in inducing formation of stress granules. Live-cell imaging and pull-down results indicate that it fails to bind eIF4G1 efficiently. Consistent with these results, a strain lacking Hmt1 is also defective in Scd6-eIF4G1 interaction. Our results establish that arginine methylation augments Scd6 repression activity by promoting eIF4G1-binding. We propose that arginine methylation of translation repressors with RGG-motif could be a general modulator of their repression activity.
INTRODUCTION
Messenger RNAs in cytoplasm can exist in different functional states. They can get translated, degraded or remain stored in a translationally repressed state depending on the physiological state of the cell (1). The storage of mRNAs in a translationally repressed state can allow them to return back to translation (2,3). Repression of translation and subsequent storage of mRNAs play an important role in regulating gene expression to affect various cellular processes (4). Despite its importance, the molecular basis of movement of mRNAs between a translationally active and a repressed state have been elucidated only for a handful of mRNAs (5–7).
Protein factors that aid in the movement of mRNAs out of translating pool are in general referred to as translation repressors. Recently, a subset of RGG-motif containing proteins in yeast was shown to have translation repression activity (8). These proteins repress translation initiation by binding eIF4G1 through their RGG-motifs. Temporal and spatial regulation of translation repressors in general and specifically of the RGG-motif containing repressors remains poorly explored.
To address the above issue we focused on Scd6, an RGG-motif containing protein. Scd6 is a conserved translation repressor that binds eIF4G1 through its RGG-motif and inhibits the formation of 48S complex (8,9). The Scd6 repression mechanism could lead to stabilization of target mRNAs because it preserves the integrity of cap-binding complex on repressed mRNAs which would be expected to result in protection from decapping complex (10). However, Scd6 can also act as a decapping activator. In Saccharomyces cerevisiae, Δscd6 has synthetic decapping defect with Δedc3 (Edc3 is another related decapping activator) (11). Consistent with this, Scd6 and its ortholog from Arabidopsis and Schizosaccharomyces pombe physically interact with Dcp2 (9,12,13). The S. pombe ortholog also directly stimulates decapping in vitro, albeit weakly (12). Thus Scd6 could be an important mRNA fate determinant that might act by differentially affecting translation and mRNA stability.
The roles of Scd6 ortholog in other model system supports the Scd6 repression mechanism identified in yeast (8). The Arabidopsis ortholog, DCP5, represses translation of mRNAs encoding seed storage proteins (13). hRAP55 in humans has been shown to localize to stress granules (SGs) and P-bodies (PBs), both of which are markers of translation repression (14). The Scd6 ortholog in Xenopus (xRAP55) represses translation upon tethering to mRNA (15). Both xRAP55 and CAR-1 (Caenorhabditis elegans ortholog) have been shown to associate with respective Dhh1 (a DEAD-box RNA helicase and translation repressor) orthologs (16,17). A conserved role of Scd6 in translation repression argues that the mechanism of Scd6 repression identified in yeast might also be conserved. Interestingly xRAPB, which is a variant of xRAP55, has recently been shown to associate with translating mRNAs upon overexpression (18). The mechanism and significance of this observation remains to be assessed.
RGG-motif proteins are characterized by the presence of single or multiple RGG- and/or RGX-motifs. Scd6 contains a single RGG- and seven RGX- motifs at its C-terminus. RGG-/RGX-motifs are sites of arginine methylation in other RGG-motif proteins (19). Hmt1 (human PRMT1 homolog) is the predominant methyltransferase in yeast that has been shown to catalyze both mono- and asymmetric dimethylation reactions (20). RNA metabolism related functions are anticipated to be strongly affected by arginine methylation since RNA-binding proteins are the largest group of arginine-methylated proteins (21). Arginine methylation has been shown to affect protein–protein and protein–RNA interactions in both positive and negative manner suggesting that a diversity of molecular interactions will be affected by arginine modification (22,23).
The role of arginine methylation in modulating the function of RGG-motif containing translation repressors has not been addressed. The RGG-motif of Scd6 is important for its repression activity (8,9). We hypothesized that arginine methylation of Scd6 at its C-terminal RGG-motif could play an important role in modulating this repression activity. We have tested the above hypothesis and report here that arginine methylation indeed promotes Scd6 repression activity by augmenting its interaction with eIF4G1.
MATERIALS AND METHODS
Yeast strains and plasmids
All plasmids and strains used in this study are listed in Supplementary Tables S1 and S2 respectively. Yeast strains used in this study are BY4741 (wild-type) or its derivatives. Strains were grown on either standard yeast extract/peptone medium or synthetic medium supplemented with the appropriate amino acids and 2% glucose or galactose (when required). For galactose induction, strains were grown in presence of glucose at 30°C until OD600 reached 0.3–0.4. Cells were then pelleted and washed with 2% galactose containing media followed by induction for 2 h (in case of microscopy experiments) or 12 h (in case of pull-down experiments from yeast cells).
In vitro methylation assay
His-Scd6-FLAG, His-Scd6ΔRGG-FLAG, His-Npl3 and His-Hmt1 were purified in recombinant form by Ni-NTA chromatography (Thermo Fisher Scientific, catalog no. 88222). Glutathione S-transferase (GST) was purified using glutathione sepharose (GE Healthcare, catalog no. 17075605). The methylation buffer (24) contained 100 mM Tris–Cl pH8, 200 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM Dithiothreitol (DTT). A total of 0.5 uCi of 3H S-adenosyl methionine (SAM) (AdoMet, specific activity 55–85 Ci/mmole; PerkinElmer, catalog no. NET155H001MC) was used in the reaction along with 20 µM unlabeled SAM ((New England Biolabs) NEB; catalog no. B9003S). A total of 10 µg of purified His-Scd6-FLAG and His-Npl3 were used in 50 µl reaction along with 7.5 µg of Hmt1. The reaction mixture was incubated at 37°C for 1 h following which reaction was stopped by addition of sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer. The entire reaction was analyzed by SDS-PAGE followed by staining with Coomassie Brilliant Blue. The gel was then soaked in En3hance solution (PerkinElmer, Catalog no. 6NE9701), dried and subjected to fluorography. For detecting arginine methylation using arginine methylation specific antibodies, recombinant His-Scd6-FLAG/His-Scd6ΔRGG-FLAG (100 µg) was incubated with recombinant His-Hmt1 (100 µg) in the presence of methylation buffer (100 mM Tris pH8, 200 mM NaCl, 2 mM EDTA, 1 mM DTT) in a 250-µl reaction, with or without 1 mM cold SAM at 37°C for 2 h. A total of 15% of reaction was loaded and analyzed by SDS-PAGE followed by western blotting using mono methyl arginine (MMA) antibody ((Cell Signaling Technology) CST, catalog no. 8711; 1:1000 dilution).
Protein purification, pull-downs and western blotting
Proteins were purified from Escherichia coli according to standard protocols using glutathione sepharose (GE, catalog no. 17075605) or Ni-NTA agarose (Thermo Fisher Scientific, catalog no. 88222). To remove RNA that might provide bridging interactions, extracts were treated for 20 min with RNase A (1 mg/ml) from Qiagen (catalog no. 19101). Purified protein was concentrated and dialyzed into 20 mM Tris–Cl pH7.5, 100 mM NaCl, 10% glycerol and 1 mM DTT. Western analysis was performed using anti-GST (CST, catalog no. 2624; 1:1000 dilution), anti-His (CST, catalog no. 2366; 1:1000 dilution), Peroxidase anti-peroxidase (PAP) (Sigma, catalog no. P1291; 1:500 dilution), anti-GFP (Santa Cruz, catalog no. sc-9996; 1:1000 dilution), anti-PGK1 (Abcam, catalog no. ab113687; dilution 1:1000) and anti-eIF4G1 (Cocalico Biologicals; 1:1000 dilution).
For performing pull-downs from yeast, cells were grown and induced as indicated above. Cells from a 15 ml galactose induced culture were broken open in 200 µl lysis buffer containing 50 mM Tris–Cl pH7.5, 50 mM NaCl, 2 mM MgCl2, 0.1% Triton-X100, 1 mM β-Mercaptoethanol, 1× Complete mini-EDTA-free tablet (Roche, catalog no. 04693132001) and lysed by vortexing at 4°C in bead-beater with glass beads. Unbroken cells and debris were removed by centrifugation at 5500 rpm for 5 min at 4°C, followed by a 2 min spin at 14000 rpm to remove any protein aggregates. A total of 500 µg of total protein was used for the pull-down reactions in 1 ml buffer having 50 mM NaH2PO4 pH8, 300 mM NaCl, 10 mM Imidazole and 50 µl of Ni-NTA beads. The reaction mix was nutated at 4°C for 2 h. Following this, beads were washed thrice (10 min each) with buffer having 50 mM NaH2PO4 pH8, 300 mM NaCl and 40 mM Imidazole in all cases except in Figure 1A where 100 mM imidazole washes were performed to get rid of the arginine methylated band running at same position as Scd6ΔRGG. After washing, 100 µl of SDS-PAGE loading dye was added to beads. About 5% of input and 30% of pellet was analyzed by SDS-PAGE followed by western blotting.
Figure 1.
Scd6 gets arginine methylated in vivo. (A) Galactose-inducible Scd6 and its RGG-deletion variant was pulled down from wild-type yeast cells. The blots were probed with mono-methyl arginine (MMA) specific antibody and Peroxidase-anti peroxidase (PAP) antibody, in that order after stripping the blot. * Represents another arginine-methylated protein. (B) Quantitation of three independent experiments (n = 3) that were performed as in A. (C) Galactose-inducible His-Scd6 was pulled down from wild-type and Δhmt1 cells followed by probing as explained in A. * Represents another arginine-methylated protein. (D) Quantitation of three independent experiments (n = 3) that were performed as mentioned in C.
For glutathione pull-downs, cells were lysed and spun as above. After removing the input sample, the supernatant was nutated for 2 h at 4°C with 30 µl of Glutathione Sepharose-4B (GE Healthcare) in 1 ml reaction mix with buffer containing 50 mM Tris–Cl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.25 µg RNase A. Beads were washed three times (10 min each) with wash buffer containing 50 mM Tris–Cl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100. A total of 40 µl of SDS-PAGE loading dye was added to beads and analyzed by SDS-PAGE. About 5% input and 40% pellet was loaded followed by western blotting. Glutathione pull-downs using recombinant proteins were set as described earlier (8).
Tandem mass spectrometry coupled to liquid chromatography
Tandem mass spectrometry (MS/MS) coupled to liquid chromatography analysis of trypsin-digested gel bands as described (25) was carried out using a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with an Advion nanomate ESI source (Advion, Ithaca, NY, USA), following ZipTip (Millipore, Billerica, MA, USA) C18 sample clean-up according to the manufacturer's instructions. Peptides were eluted from a C18 precolumn (100-μm id × 2 cm, Thermo Fisher Scientific) onto an analytical column (75-μm ID × 10 cm, C18, Thermo Fisher Scientific) using a 5% hold of solvent B (acetonitrile, 0.1% formic acid) for 5 min, followed by a 5–7% gradient of solvent B over 5 min, 7–15% gradient of solvent B over 45 min, 15–35% gradient of solvent B over 60 min, 35–40% gradient of solvent B over 28 min, 40–85% gradient of solvent B over 5 min, 85% hold of solvent B for 10 min and finally a return to 5% in 1 min and another 10 min hold of 5% solvent B. All flow rates were at 400 nl/min. Solvent A consisted of water and 0.1% formic acid. Data dependent scanning was performed by the Xcalibur v 2.1.0 software (26) using a survey mass scan at 30 000 resolution in the Orbitrap analyzer scanning m/z 350–2000, followed by alternating collision-induced dissociation and higher collision dissociation MS/MS of the five most intense ions measured in the Orbitrap at 7500 resolution.
Precursor ions were selected by the monoisotopic precursor selection setting with selection or rejection of ions held to a +/− 10 ppm window. Dynamic exclusion was set to place any selected m/z on an exclusion list for 45 s after a single MS/MS. All MS/MS spectra were searched against a combined protein database of S. cerevisiae and E. coli proteins downloaded from UniproKB, and also included the YPR129W primary sequence, using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific) considering fully or semi tryptic peptides with up to two missed cleavages. Iodoacetamide derivatives of cysteines, oxidation of methionines, methylation and dimethylation of arginines were all specified as variable modifications. This database contained 30 213 protein entries. Proteins were identified at 99% confidence with XCorr score cut-offs (27) as determined by a reversed database search using the Percolator algorithm (http://per-colator.com) (28). Identified modified peptides were considered with a q-value < 0.01 (29). The protein and peptide identification results were also visualized with Scaffold v 3.6.1 (Proteome Software Inc., Portland OR, USA), a program that relies on various search engine results (i.e.: Sequest, X! Tandem, MASCOT) and which uses Bayesian statistics to reliably identify more spectra (30,31). Proteins were accepted that passed a minimum of two peptides identified at 95% peptide confidence and 99.9% protein confidence by the Peptide and Protein Profit algorithms, respectively, within Scaffold.
Microscopy
For all experiments, yeast cultures were grown to OD600 of 0.3-0.5 in the appropriate synthetic drop-out media at 30°C. Galactose inductions were performed as described above for 2 h. After induction, cells were pelleted and spotted on coverslips for immediate microscopic examination at room temperature. All images were acquired using a Deltavision RT microscope system running softWoRx 3.5.1 software (Applied Precision, LLC), using an Olympus 100×, oil-immersion 1.4 NA objective. Exposure time and transmittance settings for Green Fluorescent Protein (GFP) channel were 0.2 s and 32%, respectively. Images were collected as 512 × 512 pixel files with a CoolSnapHQ camera (Photometrics) using 1 × 1 binning for yeast. All yeast images were deconvolved using standard softWoRx deconvolution algorithms. ImageJ (32) was used to adjust all images to equal contrast ranges according to the experiment conducted or protein examined. For each experiment, 150–180 cells were counted. Data from three independent experiments was used for quantitation and statistical significance was calculated using t-test.
RESULTS
Scd6 is arginine methylated in RGG-motif dependent manner
RNA binding proteins are the largest group of arginine methylated proteins (22). Several RNA binding proteins with RGG-motifs such as Sbp1 (33,34), Npl3 (35), Gbp2 (36) have been shown to be substrates for arginine methylation. Because Scd6 is an RNA-binding protein with RGG-motif, we hypothesized that it might be a substrate for arginine methylation.
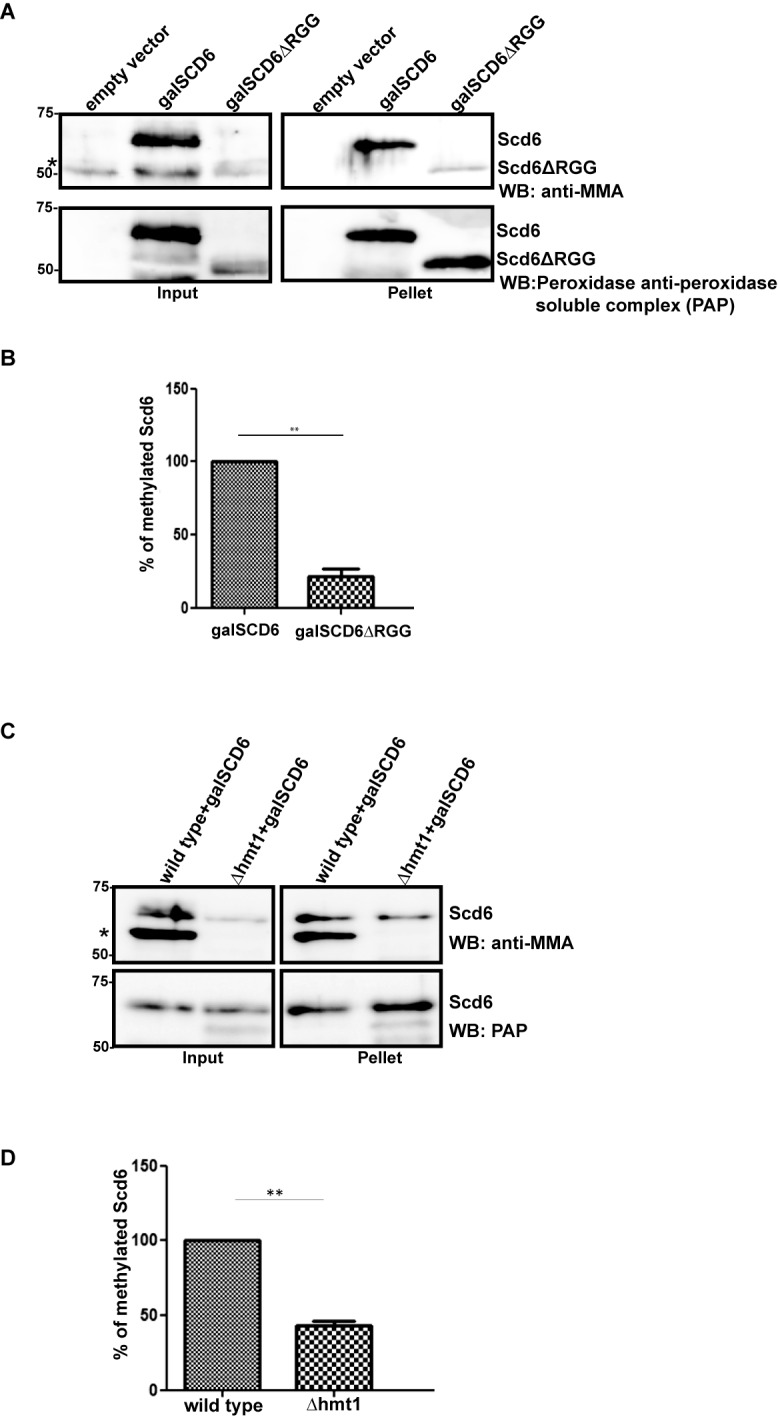
To test this hypothesis, we first examined if Scd6 gets methylated in vivo. We performed His-pull-downs from BY4741 yeast strain expressing either full length Scd6 or a variant lacking the C-terminal RGG domain, Scd6ΔRGG from a galactose inducible promoter. MMA specific antibody (CST, catalog no. 8711) was used to assess the methylation status of Scd6. We observed that full length Scd6 is methylated whereas Scd6ΔRGG shows very weak cross-reactivity with MMA antibody (Figure 1A and B), despite full length and the ΔRGG variant being expressed at similar levels as detected by western analysis using PAP antibody, which specifically detects the ZZ-tag at Scd6 C-terminal. Similar results were observed with another mono-methylation specific antibody from Abcam (catalog no. ab414; data not shown). We interpret these results to indicate that Scd6 is arginine methylated, with the RGG domain being the primary site of modification.
Since Hmt1 is the predominant arginine methyltransferase in yeast (20), we checked if Hmt1 was required for methylation of Scd6 using the same approach. We observed that the MMA antibody cross-reacted with Scd6 to a significantly (2-fold) reduced level in Δhmt1 strain (Figure 1C and D). Put together, these results indicate that Scd6 is arginine methylated, at least partially in an Hmt1-dependent manner and the RGG-motif is required for its efficient methylation.
To determine if Scd6 was a direct substrate of Hmt1, we tested the ability of Hmt1 and Scd6 to physically interact. We performed glutathione pull-downs with lysates from yeast cells expressing GST-Scd6 under the control of galactose inducible promoter. We observed that Hmt1 was present in the pellet fraction of strain expressing GST-Scd6 but not in strain containing empty vector (Figure 2A), indicating that Scd6 binds Hmt1 in vivo.
Figure 2.
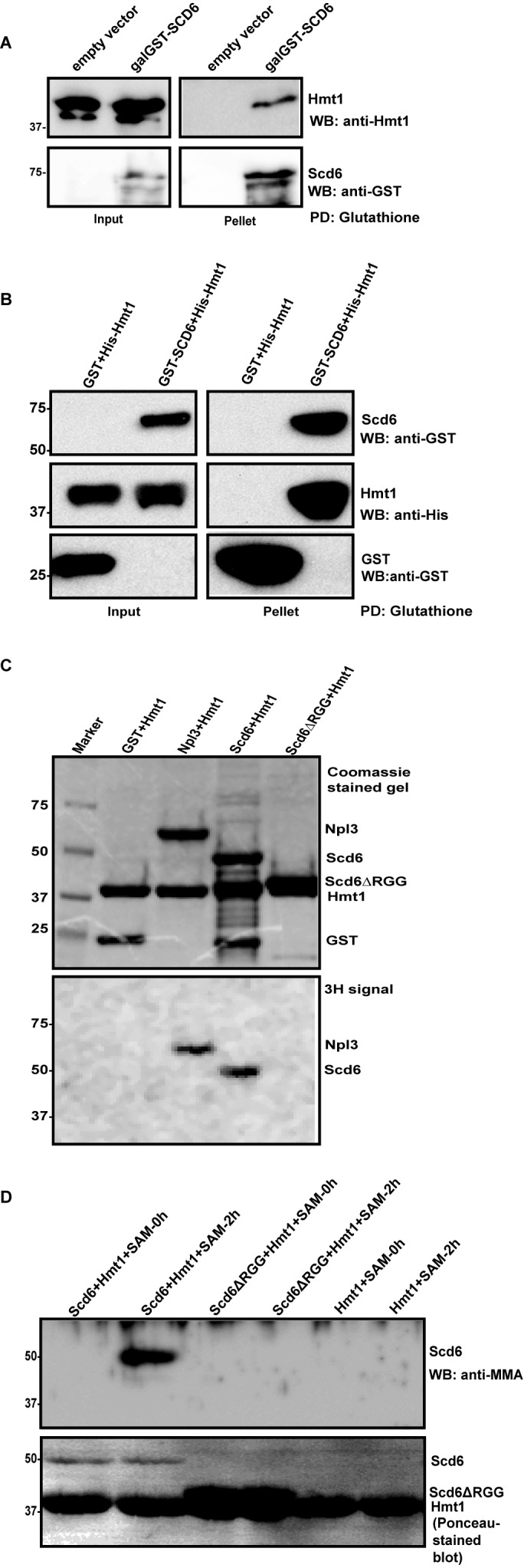
Scd6 is a direct target of Hmt1. (A) Interaction of Hmt1 with GST-Scd6. Glutathione pull-down was performed from cells expressing GST-Scd6 under galactose-inducible promoter. Hmt1 was probed with anti-Hmt1 antibody (a kind gift from Michael Yu). (B) Recombinant purified His-Hmt1 was incubated with recombinant GST-Scd6 followed by glutathione pull-down. (C) Purified GST (negative control), Npl3 (positive control), full-length Scd6 and Scd6ΔRGG mutant were in vitro methylated by incubating with purified Hmt1 in presence of 3H-labeled S-adenosyl methionine (SAM). Top panel shows Coomassie Brilliant Blue-stained gel and bottom panel shows fluorograph of the same gel. (D) Recombinant purified Scd6 and Scd6ΔRGG were incubated with recombinant purified Hmt1 in presence or absence of SAM followed by probing with MMA antibody. Lower panel shows the Ponceau-S stained blot which indicates the amount of each protein present.
To test if this binding was direct, we purified GST-Scd6 and His-Hmt1 in recombinant form. Glutathione pull-downs were performed with GST-Scd6. We observed that Hmt1 was present in pellet fraction along with GST-Scd6 but not with GST alone (Figure 2B) indicating that Scd6 and Hmt1 can bind each other, which would be consistent with Hmt1 binding and directly methylating Scd6.
To examine if Hmt1 methylates Scd6 we performed in vitro methylation assay with recombinant proteins in two different ways. In first approach, purified recombinant Scd6 was methylated with purified recombinant Hmt1 in presence of 3H-labeled SAM (used as a methyl-donor in the methylation reaction). We observed that Scd6 but not the RGG-deletion mutant incorporated tritium (Figure 2C). Purified Npl3 and GST served as positive and negative controls respectively. The doublet visible in the lane with Scd6ΔRGG in Figure 2C, top panel and Figure 2D, lower panel consists of the Scd6ΔRGG running at position similar to that of Hmt1 due to their similar sizes. In the second approach, recombinant purified Scd6 was in vitro methylated using purified Hmt1 and cold SAM for 2 h followed by western analysis with MMA antibody. Methylation of Scd6 was evident based on its cross-reactivity with MMA antibody (Figure 2D). The RGG-deletion mutant of Scd6 did not cross-react with antibody suggesting that the RGG-motif was important for its methylation. We conclude based on above results that Scd6 is a substrate of Hmt1 and its RGG-motif is required for methylation.
The in vivo and in vitro results clearly indicate that Scd6 gets arginine methylated at its RGG-motif and Hmt1 plays an important role in its methylation.
Mapping methylation sites on Scd6
Scd6 contains eight -RGG/-RGX repeats and three arginines in its C-terminal RGG domain (residues 283–349, Figure 3A) (8). Identifying specific residues that get methylated in vivo would allow us to make mutations that specifically test the role of methylation in affecting Scd6 repression activity. It is possible that all of the arginines in RGG-motif get methylated or perhaps there could be preferred sites of methylation under different conditions.
Figure 3.
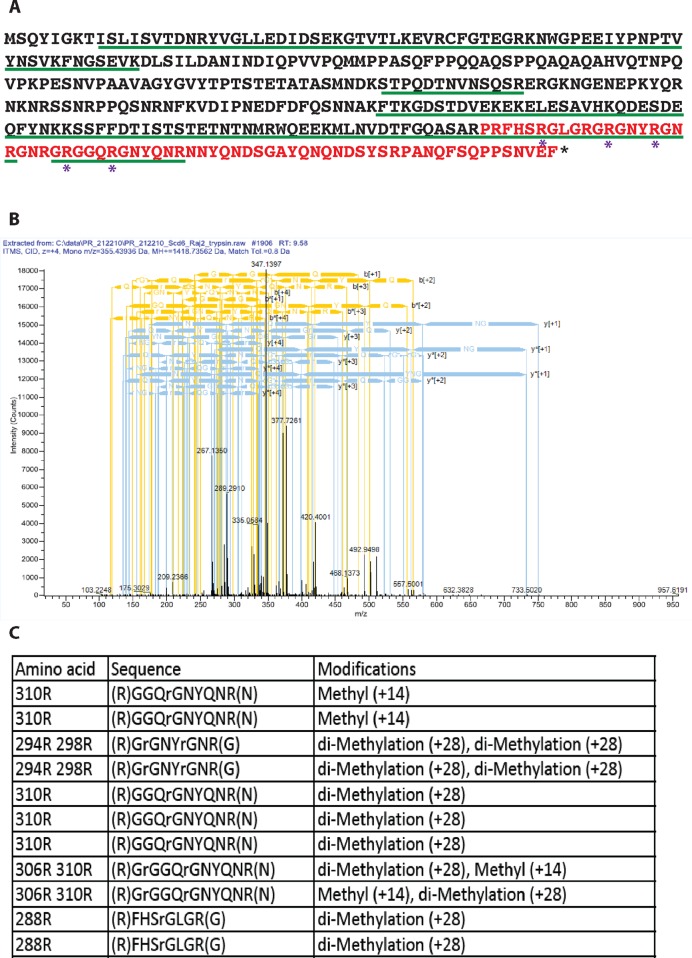
Identification of methylation sites in Scd6 by mass spectrometry. (A) Scd6 protein sequence showing arginine residues (marked with *) observed to be methylated in vivo by mass spectrometry analysis. Residues marked in red represent the RGG-motif (283–349). The green underlined residues represent the extent of protein coverage. (B) A representative mass spectrometry chart of GRGGQRGNYQNR peptide. (C) Table listing methylated peptides that were detected by mass spectrometry along with their methylation status.
In order to identify the arginine residues that get methylated in vivo, we purified GST-Scd6 from yeast cells and submitted it for mass spectrometry analysis. Of the eleven arginines present in the RGG-motif, peptides containing nine arginines were detected and we observed that five of those arginines were methylated (Figure 3A; methylated arginines are marked with *). A representative mass spectrum of one of the methylated RGG-motif peptides has been shown (Figure 3B). Both mono- and di-methylated arginines were detected for 306R and 310R (Figure 3C) whereas only di-methylated peptides were identified for 288R, 294R and 298R. The relative significance of mono- and di-methylation in general remains to be established. Mass spectrometry results identified specific methylated arginine residues of Scd6 in vivo suggesting that multiple arginines in the RGG-motif are methylated. It is possible that in the Scd6 RGG-motif, arginine residues besides the five detected by mass spectrometry also get methylated. We may not have detected such methylated residues in our mass spectrometry analysis either due to the absence of trypsin-cleaved peptide fragments containing these residues (304R and 335R) or due to very low levels of methylated arginine containing peptides for other residues.
Mutation of multiple arginines is required to create an arginine methylation defective (AMD) mutant
To test the significance of arginine methylation in Scd6 repression activity, it was imperative to create a mutant that was methylation defective. Since Hmt1 has many substrates in yeast, phenotypes of the Δhmt1 mutant may not be due to defects in Scd6 methylation only. We began with sequentially creating arginine to alanine mutations of modified residues identified in the mass spectrometry in Scd6 RGG-motif. We observed that increasing the number of arginine to alanine mutations lead to concomitant decrease in methylation as observed using MMA antibody (Figure 4B and C). A mutant with nine arginines converted to alanine was the most defective in methylation (henceforth referred to as arginine methylation defective [AMD] mutant, Figure 4A–C). Mut3 has all five arginines detected by mass spectrometry analysis converted to alanine in addition to R292, R301 and R304. However it still has significantly higher methylation levels than AMD (Figure 4C) suggesting that arginine residues other than the five detected by mass spectrometry get methylated. We observed that the expression level of the AMD mutant was comparable to that of wild-type protein indicating that the decreased signal with the MMA antibody is not due to lower expression of this variant (Figure 4D). Based on this result, we have used the AMD mutant as a tool for assessing the role of arginine methylation in regulating ability of Scd6 to repress translation.
Figure 4.
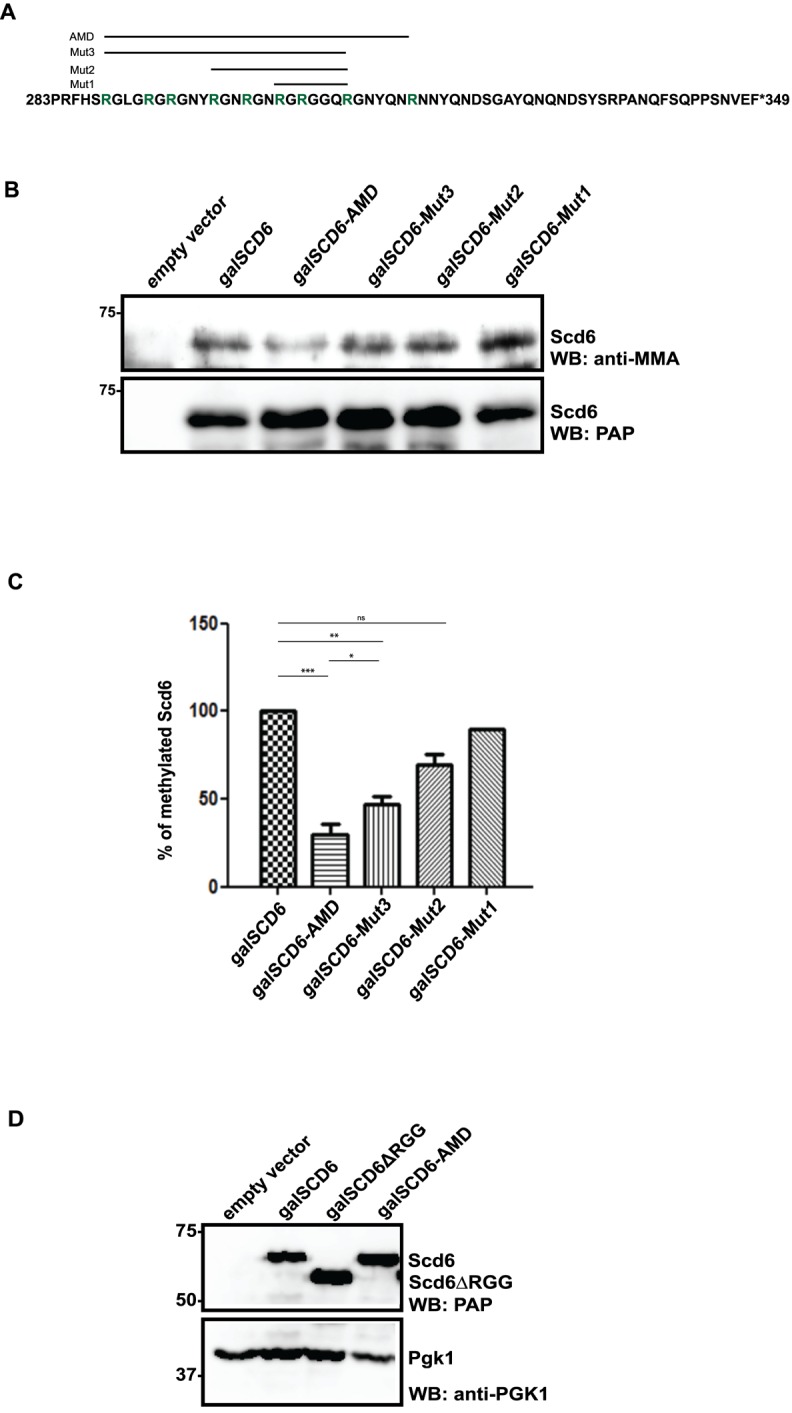
Arginine methylation defective (AMD) mutant of Scd6. (A) Arginines (green) mutated to alanine in different mutants have been shown. Three, five, eight and nine arginines were mutated in Mut1, Mut2, Mut3 and AMD mutant, respectively. Only the RGG-motif sequence (283–349) of Scd6 has been shown. (B) His pull-down was performed from cells expressing galactose-induced mutants. Western analysis was performed with MMA antibody followed by probing with Peroxidase anti-peroxidase (PAP). (C) Quantitation of three independent experiments (n = 3) that were performed as described in B. (D) Expression level analysis of AMD mutant in yeast cells using PAP antibody.
Scd6 AMD mutant is defective in repressing translation
Overexpression of Scd6 (and other translation repressors such as Dhh1 and Ded1) leads to a growth defect (9,17,37) due to global translation repression. Translation repression defective mutants of above proteins are compromised in causing overexpression mediated growth defect. We tested the ability of AMD mutant to repress translation using this type of growth assay. We observed that the AMD mutant rescues the growth defect of overexpressed wild-type Scd6 (Figure 5A) arguing that the methylation defect in the AMD mutant hampers its ability to repress translation. It is noteworthy that the growth defect rescue by AMD mutant was similar to the one observed for RGG-deletion mutant of Scd6 (Figure 5A) suggesting that the translation repression defect in AMD mutant is comparable to the one in RGG-deletion mutant.
Figure 5.
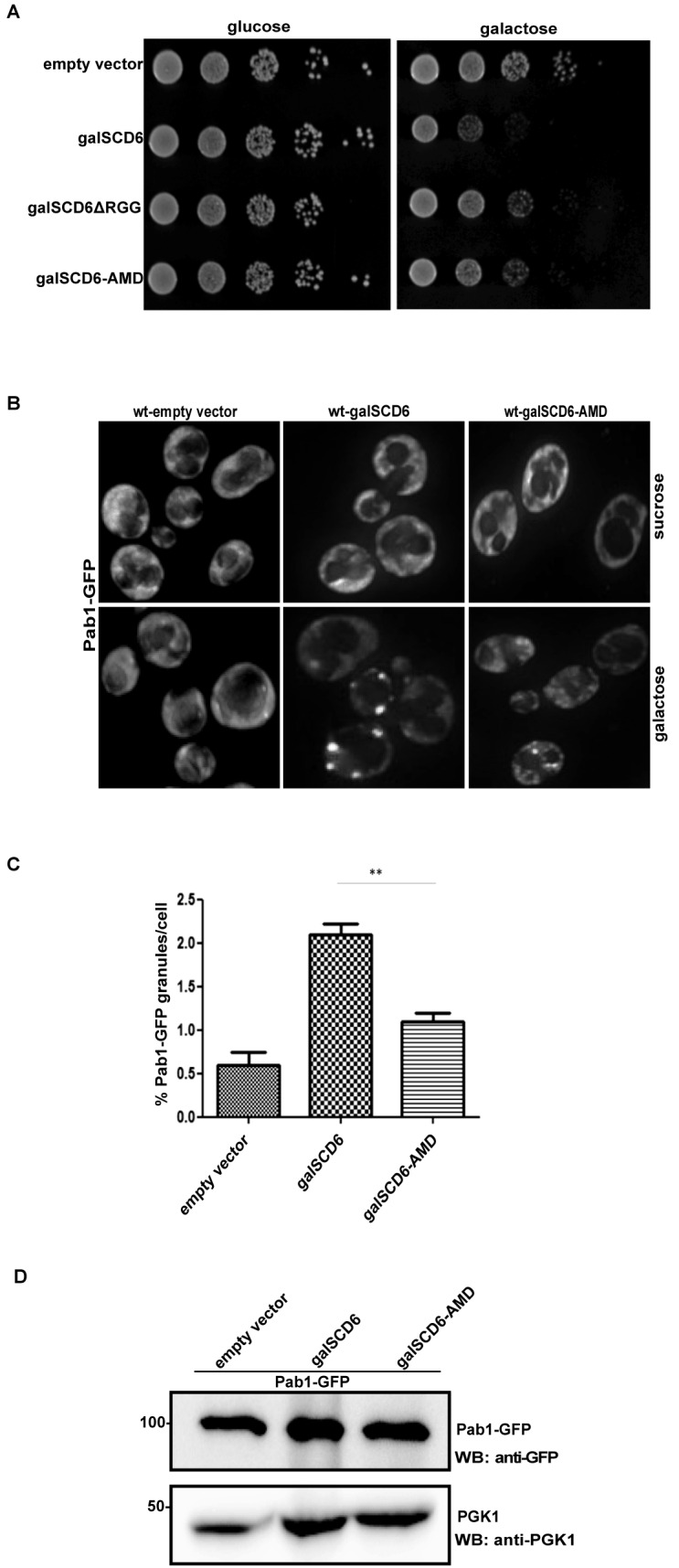
AMD mutant of Scd6 is defective in repressing translation. (A) AMD mutant rescues growth defect caused by overexpression of SCD6. Five independent growth assay experiments (n = 5) were performed. (B) Live cell imaging of stress granule formation upon overexpression of wild-type and AMD mutant in Pab1-GFP strain. (C) Quantitation of Pab1-GFP granule formation by AMD mutant. Three independent experiments (n = 3) were considered for quantitation. (D) Western analysis to compare Pab1-GFP levels in cell lysates from strains expressing wild-type Scd6 and AMD mutant. Blots were probed with anti-GFP (upper panel) followed by probing with anti-PGK1 (lower panel).
Translationally repressed mRNAs accumulate in RNA granules such as mRNA processing-bodies and SG. The formation of granules in response to translation repression is evolutionarily conserved (38). Translation repressors like Scd6 and Ded1 upon overexpression induce the formation of RNA granules, which harbour translationally repressed mRNPs (8,37). We next tested the ability of AMD mutant to induce formation of RNA granules using Pab1-GFP strain (39) since Pab1 is a core SG marker (40). We observed that the AMD mutant induced significantly fewer SG than the wild-type Scd6 protein (Figure 5B and C). We confirmed that decreased granule formation was not due to decrease in expression of Pab1-GFP upon AMD mutant overexpression (Figure 5D). This observation is consistent with the growth assay result where AMD is defective in translation repression. Based on the results of growth and RNA granule assay presented in Figure 5, we conclude that the AMD mutant is defective in repressing translation.
AMD mutant fails to induce eIF4G1-foci formation in a manner comparable to wild-type
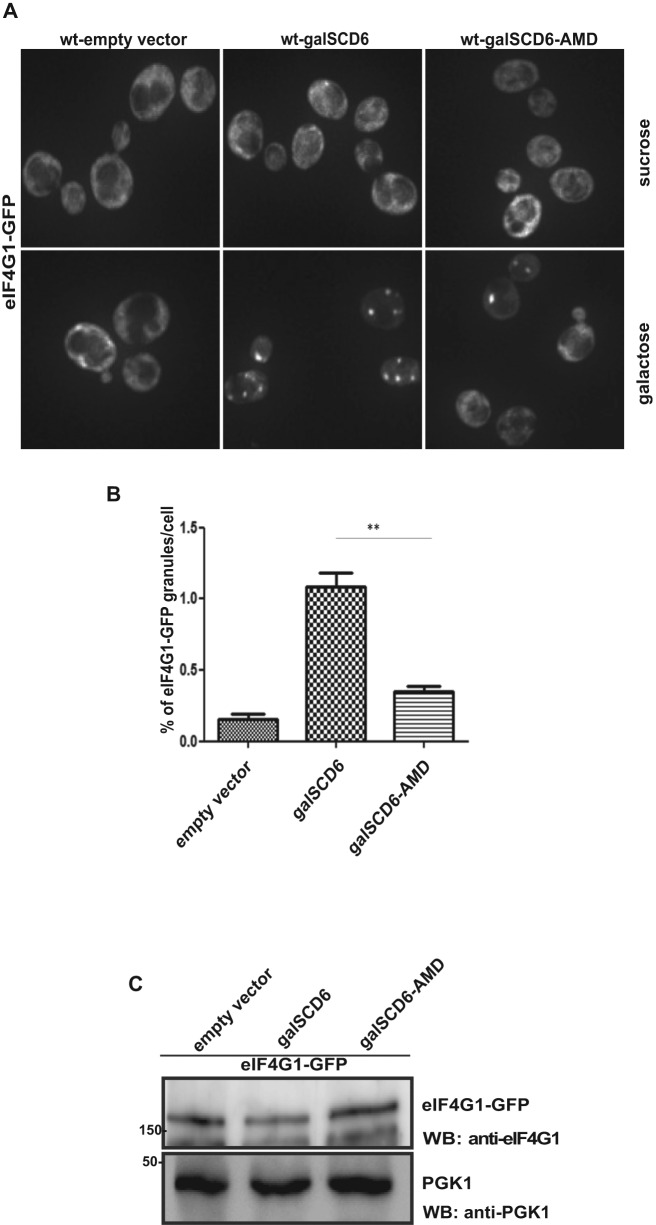
Scd6 binds eIF4G1 leading to its accumulation in RNA granules along with the repressed mRNAs (8). Since the AMD mutant is defective in repressing translation, we wanted to check if it was proficient in inducing formation of eIF4G1 containing repression mRNPs. We tested this by overexpressing wild-type Scd6 and the AMD mutant in an eIF4G1-GFP (39) strain and observed that overexpression of wild-type Scd6 induced formation of eIF4G1-foci as reported earlier (Figure 6A and B). However, the AMD mutant induced fewer eIF4G1-foci as compared to wild-type Scd6 (Figure 6A and B), although the expression level of eIF4G1-GFP was comparable in each case (Figure 6C). We conclude that arginine methylation of Scd6 is important for driving cytoplasmic eIF4G1-GFP into repression foci.
Figure 6.
AMD mutant is defective in inducing eIF4G1-granule formation. (A) Live-cell imaging of eIF4G1-GFP strain upon overexpression of wild-type and AMD mutant of Scd6. (B) Quantitation of eIF4G1-GFP granule formation. Three independent experiments (n = 3) were considered for quantitation. (C) Western analysis to compare eIF4G1-GFP levels in cell lysates from strains expressing wild-type Scd6 and AMD mutant. Blots were probed with anti-eIF4G1 (upper panel) followed by probing with anti-PGK1 (lower panel).
Arginine methylation of Scd6 promotes its interaction with eIF4G1
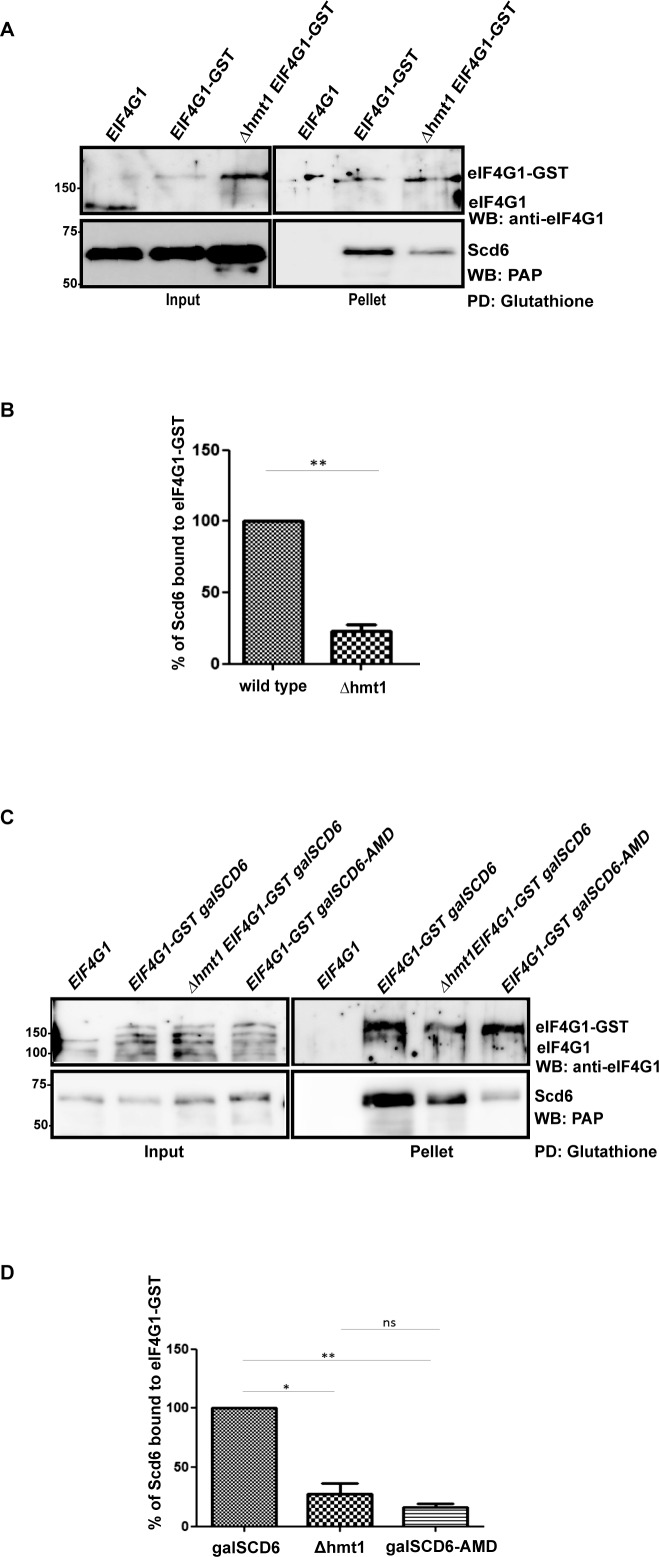
Scd6 has been reported to repress translation by binding eIF4G1 through its RGG-motif (8). Based on the observation that Scd6 RGG-motif gets methylated at multiple arginine residues and the Scd6 AMD mutant is also defective in repressing translation, we hypothesized that arginine methylation of Scd6 could be important for its ability to bind eIF4G1. Our hypothesis predicted compromised Scd6–eIF4G1 interaction in absence of Hmt1 because Scd6 methylation is defective in this background (Figure 1C and D). To test this, we pulled down eIF4G1-GST (genomically tagged) from wild-type and Δhmt1 cells and observed that a significantly reduced amount of Scd6 came down from Δhmt1 cells as compared to wild-type cells (Figure 7A and B). To further establish the role of arginine methylation in promoting Scd6–eIF4G1 interaction, binding of eIF4G1 with AMD mutant was tested. We observed that the AMD mutant was significantly defective in binding eIF4G1 as compared to wild-type Scd6 (Figure 7C and D) in yeast cells. Based on the results presented here for eIF4G1–Scd6 interaction in Δhmt1 background and for eIF4G1–AMD mutant interaction, we conclude that arginine methylation of Scd6 promotes its interaction with eIF4G1 to augment its repression activity.
Figure 7.
Arginine methylation promotes Scd6-eIF4G1 interaction. (A) Glutathione pull-downs from yeast cells expressing eIF4G1-GST in wild-type and Δhmt1 background. Scd6 was expressed from galactose-inducible plasmid. (B) Quantitation of three independent experiments (n = 3) that were performed as explained in A. (C) Glutathione pull-down from eIF4G1-GST strain expressing wild-type Scd6 or AMD mutant under galactose-inducible promoter. (D) Quantitation of three independent glutathione pull-downs (n = 3) that were performed as described in C.
DISCUSSION
We demonstrate in this work that conserved translation repressor Scd6 gets arginine methylated at its RGG motif and provide several pieces of evidence to support this conclusion. First, Scd6 gets methylated in vivo and methylation is defective in absence of RGG motif and Hmt1 based on western analysis performed using anti-mono methyl arginine antibody (Figure 1). Second, purified Hmt1 methylates purified Scd6 in vitro, but not its RGG-motif lacking variant (Figure 2). Third, mass spectrometry analysis identified multiple methylated arginine residues in Scd6 (Figure 3). These observations identify Scd6 as an arginine methylated translation repressor in yeast.
The ability of Scd6 to get methylated depends on Hmt1 enzyme (Figure 1), although some methylation occurs in absence of Hmt1. Nevertheless, this is consistent with the observation that Hmt1 is the predominant methyltransferases in yeast and has been shown to methylate other RGG-motif proteins (20,41). It has been recently reported that hRAP55 (human ortholog of Scd6) also gets arginine methylated and interacts with PRMT1, which is the human homolog of Hmt1 (42). However the functional significance of hRAP55 methylation has not been demonstrated.
We also provide evidence that methylation promotes the repression activity of Scd6 using the AMD mutant and Δhmt1 strain. First, the AMD mutant of Scd6 rescued the over expression growth defect compared to wild-type Scd6 (Figure 5A). Second, the AMD mutant did not efficiently induce the formation of SG, which arise as a result of translation repression (Figure 5B and C). Third, Scd6-eIF4G1 binding was compromised in Δhmt1 mutant (Figure 7A and B). Fourth, the AMD mutant was defective both in binding eIF4G1 and in inducing its localization to RNA granules (Figures 6A–C, 7C and D). These results argue that arginine methylation of Scd6 modulates its repression activity, at least in part, by promoting interaction with eIF4G1. To our knowledge this is the first example of a direct role of arginine methylation in modulating activity of a translation repressor.
Interestingly, Scd6 overexpression growth defect was not rescued in absence of Hmt1 (data not shown). We think that residual methylation occurring in absence of Hmt1 (Figure 1C and D) could allow Scd6 overexpression mediated growth arrest. The methyltransferase(s) that might methylate Scd6 in absence of Hmt1 remains to be identified.
The evolutionary conservation of Scd6 arginine methylation combined with results presented in this work argue for a role of methylation in modulating translation repression activity of Scd6 orthologs including hRAP55. Understanding the basis of how methylated arginines promote Scd6–eIF4G1 interaction will be an important future direction.
Our work raises the possibility that arginine methylation could be a general regulator of RGG-motif containing proteins affecting translation. Sbp1, Npl3 and Ded1 are other proteins shown to repress translation by binding eIF4G1 (10,37). All the three proteins have been shown to get arginine methylated in vivo (33,35,43) although the contribution of this modification to their repression activity has not been determined. It is interesting to note that numerous proteins involved in neurological disorders (e.g. FUS, FMRP, ATXN-2 and hnRNPA1) contain RGG-motifs and have roles in RNA biogenesis/function including the regulation of translation (44,45). As such, a comprehensive understanding of the impact of arginine methylation on global translation would impinge on studying regulation of other RGG-motif proteins involved in translation by arginine methylation. One interesting possibility is that eIF4G1 might function as a ‘reader’ of methylated arginines, a role reminiscent of Tudor proteins, and thereby integrate the methylation status of various translational repressors into a biological outcome. However, since arginine methylation can affect interactions with protein/RNA in either positive or negative manners, its effects are likely to be differential on proteins involved in translation.
Understanding the diversity and specificity of methyltransferases will be an important future challenge. Although Hmt1 has been reported to be the predominant methyltransferase (20), it is likely that other methyltransferases could be playing a back-up role. A recent report indicates that methylation of many yeast proteins remain unaffected in a triple mutant lacking Hmt1, Rmt2 and Hsl7 (43), which suggests there are unidentified arginine methyltransferases. Thus identifying the functions and targets of numerous putative methyltransferases (46) and proteins that can modify the function of these enzymes (47) would be important for elucidating mechanisms that ensure specificity of arginine methylation of proteins in general and specifically RNA-binding proteins.
Supplementary Material
Footnotes
Present address: Shanaya Shah, Department of Microbiology, University of California Davis, Davis, CA 95616, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome-DBT Intermediate Fellowship [IA/I/12/2/500625 to P.I.R.]; Wellcome-DBT Intermediate fellowship [IA/I/12/2/500625] project assistantship (to S.S., V.V.); Council for Scientific and Industrial Research Junior Research Fellowship (to G.P.); Mass spectrometry data acquired by Arizona Proteomics Consortium supported by NIEHS [ES06694 to the SWEHSC], NIH/NCI [CA023074 to AZCC] and BIO5 Institute of the University of Arizona; Thermo Fisher LTQ Orbitrap Velos mass spectrometer [1S10 RR028868-01]; NIH [GM045443]; Howard Hughes Medical Institute (to R.P.); HHMI Postdoctoral Fellowship (to P.I.R.); DBT-Wellcome Trust India Alliance [IA/I/12/2/500625]. Funding for open access charge: DBT-Wellcome Trust India Alliance [IA/I/12/2/500625].
Conflict of interest statement. None declared.
REFERENCES
- 1.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 4.Sonenberg N., Hinnebusch A.G. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann K., Grskovic M., Gebauer F., Hentze M.W. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell. 2005;122:529–540. doi: 10.1016/j.cell.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Berchowitz L.E., Kabachinski G., Walker M.R., Carlile T.M., Gilbert W.V., Schwartz T.U., Amon A. Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanenbaum M.E., Stern-Ginossar N., Weissman J.S., Vale R.D. Regulation of mRNA translation during mitosis. eLife. 2015;4:e07957. doi: 10.7554/eLife.07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajyaguru P., She M., Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissan T., Rajyaguru P., She M., Song H., Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajyaguru P., Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11:2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decourty L., Saveanu C., Zemam K., Hantraye F., Frachon E., Rousselle J.C., Fromont-Racine M., Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromm S.A., Truffault V., Kamenz J., Braun J.E., Hoffmann N.A., Izaurralde E., Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012;31:279–290. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J., Chua N.H. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W.H., Yu J.H., Gulick T., Bloch K.D., Bloch D.B. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K.J., Ogawa K., Takagi M., Imamoto N., Matsumoto K., Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- 16.Boag P.R., Nakamura A., Blackwell T.K. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- 17.Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladomery M., Sommerville J. The Scd6/Lsm14 protein xRAPB has properties different from RAP55 in selecting mRNA for early translation or intracellular distribution in Xenopus oocytes. Biochim. Biophys. Acta. 2015;1849:1363–1373. doi: 10.1016/j.bbagrm.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Thandapani P., O'Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Gary J.D., Lin W.J., Yang M.C., Herschman H.R., Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 21.Chen C., Nott T.J., Jin J., Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev. Mol. Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 22.Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Cote J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 23.Calvanese V., Lara E., Suarez-Alvarez B., Abu Dawud R., Vazquez-Chantada M., Martinez-Chantar M.L., Embade N., Lopez-Nieva P., Horrillo A., Hmadcha A., et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13736–13741. doi: 10.1073/pnas.1001399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green D.M., Marfatia K.A., Crafton E.B., Zhang X., Cheng X., Corbett A.H. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 25.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 26.Andon N.L., Hollingworth S., Koller A., Greenland A.J., Yates J.R., 3rd, Haynes P.A. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Qian W.J., Liu T., Monroe M.E., Strittmatter E.F., Jacobs J.M., Kangas L.J., Petritis K., Camp D.G. 2nd, Smith R.D. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Prot. Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 28.Spivak M., Weston J., Bottou L., Kall L., Noble W.S. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J. Prot. Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kall L., Storey J.D., MacCoss M.J., Noble W.S. Posterior error probabilities and false discovery rates: two sides of the same coin. J. Prot. Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 30.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 31.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 32.Abramoff M, M.P. Ram S. Image processing with ImageJ. Biophotonics Int. 2009;11:36–42. [Google Scholar]
- 33.Frankel A., Clarke S. RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem. Biophys. Res. Commun. 1999;259:391–400. doi: 10.1006/bbrc.1999.0779. [DOI] [PubMed] [Google Scholar]
- 34.Plank M., Fischer R., Geoghegan V., Charles P.D., Konietzny R., Acuto O., Pears C., Schofield C.J., Kessler B.M. Expanding the yeast protein arginine methylome. Proteomics. 2015;15:3232–3243. doi: 10.1002/pmic.201500032. [DOI] [PubMed] [Google Scholar]
- 35.Henry M., Borland C.Z., Bossie M., Silver P.A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erce M.A., Abeygunawardena D., Low J.K., Hart-Smith G., Wilkins M.R. Interactions affected by arginine methylation in the yeast protein-protein interaction network. Mol. Cell. Proteomics. 2013;12:3184–3198. doi: 10.1074/mcp.M113.031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilliker A., Gao Z., Jankowsky E., Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchan J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 40.Buchan J.R., Muhlrad D., Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yagoub D., Hart-Smith G., Moecking J., Erce M.A., Wilkins M.R. Yeast proteins Gar1p, Nop1p, Npl3p, Nsr1p, and Rps2p are natively methylated and are substrates of the arginine methyltransferase Hmt1p. Proteomics. 2015;15:3209–3218. doi: 10.1002/pmic.201500075. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto K., Nakayama H., Yoshimura M., Masuda A., Dohmae N., Matsumoto S., Tsujimoto M. PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol. 2012;9:610–623. doi: 10.4161/rna.19527. [DOI] [PubMed] [Google Scholar]
- 43.Low J.K., Hart-Smith G., Erce M.A., Wilkins M.R. Analysis of the proteome of Saccharomyces cerevisiae for methylarginine. J. Proteome Res. 2013;12:3884–3899. doi: 10.1021/pr400556c. [DOI] [PubMed] [Google Scholar]
- 44.Blackwell E., Ceman S. Arginine methylation of RNA-binding proteins regulates cell function and differentiation. Mol. Rep. Dev. 2012;79:163–175. doi: 10.1002/mrd.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneb H.M., Dion P.A., Rouleau G.A. The FUS about arginine methylation in ALS and FTLD. EMBO J. 2012;31:4249–4251. doi: 10.1038/emboj.2012.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrossian T., Clarke S. Bioinformatic Identification of Novel Methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue K., Mizuno T., Wada K., Hagiwara M. Novel RING finger proteins, Air1p and Air2p, interact with Hmt1p and inhibit the arginine methylation of Npl3p. J. Biol. Chem. 2000;275:32793–32799. doi: 10.1074/jbc.M004560200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.