Abstract
Linker histones are an integral component of chromatin but how these proteins promote assembly of chromatin fibers and higher order structures and regulate gene expression remains an open question. Using Förster resonance energy transfer (FRET) approaches we find that association of a linker histone with oligonucleosomal arrays induces condensation of the intrinsically disordered H1 CTD in a manner consistent with adoption of a defined fold or ensemble of folds in the bound state. However, H1 CTD structure when bound to nucleosomes in arrays is distinct from that induced upon H1 association with mononucleosomes or bare double stranded DNA. Moreover, the H1 CTD becomes more condensed upon condensation of extended nucleosome arrays to the contacting zig-zag form found in moderate salts, but does not detectably change during folding to fully compacted chromatin fibers. We provide evidence that linker DNA conformation is a key determinant of H1 CTD structure and that constraints imposed by neighboring nucleosomes cause linker DNAs to adopt distinct trajectories in oligonucleosomes compared to H1-bound mononucleosomes. Finally, inter-molecular FRET between H1s within fully condensed nucleosome arrays suggests a regular spatial arrangement for the H1 CTD within the 30 nm chromatin fiber.
INTRODUCTION
The hierarchical packaging of the eukaryotic genome into chromatin plays a fundamental role in regulation of gene expression and other DNA-dependent processes. The basic repeating subunit of chromatin, the nucleosome, is comprised of 147 bp segments of DNA wound around an octamer of core histone proteins to form the nucleosome core and ∼50 bp of ‘linker’ DNA that connects nucleosome cores to form vast oligonucleosome arrays (1). Linker histones (H1s) bind to the exterior of nucleosomes and linker DNA and promote folding of nucleosome arrays into chromatin fibers and other high order structures (2–4). Transcription-related acetylation and other chromatin remodeling activities can destabilize higher-order chromatin structures, resulting in the displacement of H1s from transcriptionally active regions (5–9). However, the mechanism(s) by which linker histones stabilize higher chromatin structures remain undefined.
Despite their ability to stabilized condensed states of chromatin, deletion of H1s does not lead to a global deregulation of gene expression. Rather, specific subsets of genes appear to be regulated in a manner linked to H1s by multiple mechanisms (10,11). Nevertheless, for most H1 subtypes a direct correlation exists between gene activity and depletion of H1 from promoter regions, especially for the variant H1.2. (8,9). Mapping of H1 distribution across the genome shows a lower H1 occupancy at the transcription start site of active genes and indicates displacement of H1 from cis-regulatory elements is critical to their function (8,9,12). Moreover, Drosophila H1 has been shown to repress transposable elements and repetitive sequences by facilitating recruitment of the H3 K9 methyltransferase Su(var)3-9 (13). Overall linker histones are essential proteins in mammals as inactivation of three mouse H1 variants (H1.c, H1.d and H1.e) results in a decreased H1/core nucleosome ratio and embryonic lethality (11,14).
Metazoan linker histones (H1s) have a tripartite structure that includes a short 25–30 amino acid residue N-terminal domain, ∼80 residue globular domain containing a ‘winged-helix’ fold found in the FOX/Forkhead-family of DNA-binding factors, and an extended ∼100 residue C-terminal domain (CTD) (1,15). While the globular domain directs structure-specific recognition and binding of H1s to nucleosomes, both in vitro and in vivo experiments show that the chromatin condensing function is primarily provided by the CTD (16–18). This ∼100 residue domain typically contains ∼40 basic residues, nearly all lysines, with few, if any, acidic residues. Analysis of salt-dependent chromatin condensation indicates that the CTD stabilizes higher-order structure via an electrostatic mechanism in which nearly all positively charged residues in the CTD are involved in neutralization of the polyanionic backbone of DNA (19). The CTD does not exhibit defined structure when H1 is free in aqueous solutions of physiological pH and ionic strengths (20–22). However, peptides derived from H1 CTDs have been shown to adopt defined structural elements in structure-promoting solvents, and in the presence of DNA fragments or alkaline pH (19,22,23). Moreover, both the overall H1 CTD amino acid residue content and evidence that residue composition rather than primary sequence is more important for chromatin condensation in vitro suggest that the CTD functions as an intrinsically disordered domain (17,24). Such domains exhibit disorder when free in solution but adopt ordered structure(s) when bound to macromolecular partners. In support of this idea, recent work demonstrates that an H1 CTD undergoes a drastic condensation consistent with a disorder→order transition and adoption of defined structure(s) upon binding to both DNA and mononucleosomes (25,26). However, how such transitions in CTD structure are related to its chromatin condensation function remains undefined.
To better understand how H1 promotes formation of higher order chromatin structures we investigated structural changes within the H1 CTD upon binding of a linker histone to nucleosomal arrays using fluorescence and förster resonance energy transfer (FRET) approaches. We find that, similar to binding to individual nucleosomes, association of H1 with nucleosomes in arrays induces condensation of the H1 CTD in a manner consistent with adoption of defined structure(s). However oligonucleosome-induced H1 CTD structure is distinct compared to that found when H1 is bound to mononucleosomes or bare double stranded DNA. In addition, we find that the CTD structure changes during the initial stages of folding of the chromatin fiber and analysis of inter-molecular FRET implies a regular arrangement for H1 within condensed fibers. Finally we provide evidence that linker DNA conformation is a key molecular determinant of the H1 CTD structure within nucleosome arrays.
MATERIALS AND METHODS
Expression and purification of core histones and H1s
H1°a from Xenopus laevis (27), hereafter referred to as H1, was expressed in bacterial cells using the plasmid pET3aH1(0)a (15). The coding sequences for H1 CTD mutants H1 K195C, H1 G101CG195C, H1 T173C and H1 G101CT173C were generated by the QuickChange site-directed mutagenesis kit (Stratagene) from this plasmid using the top-strand primer GAAATCTGGACGGTGTA AGTAAGGAT, GCCAGTGAGGGCATGCAAGGTA A AG and bottom-strand primers CTTTAGACCTGCCA CATTCATTCCTA, and GGCCTTCTTTACCTTGCAT GCCCTCA. Proteins were expressed and purified as described previously (25). The concentrations of the purified protein stocks were determined by quantitative comparison to H1 standards on SDS-PAGE (25). H2A and H2B were expressed and purified as preformed dimer as described previously (28). Coding sequences for H3 containing a cysteine to alanine substitution at position 110 (H3C110A) and H4 harboring an alanine to cysteine substitution at position 15 (H4A15C) were subcloned into the pET3d expression plasmid. The proteins were expressed and purified as preformed tetramers as described (29).
Nucleosome and nucleosomal array reconstitution
The plasmid p207-12 (30) was digested with EcoRV to release 207-bp DNA fragments containing the 601-nucleosome positioning sequence for nucleosome reconstitution centrally located in a 30-N-30 template where N = the 147 bp nucleosome core DNA region. A fragment containing 12 × 207-bp tandem repeats of the 601-nucleosome positioning sequence, 12 × 207-601 DNA, was generated by digesting the same plasmid with XbaI, HindIII and DraI. DNA fragments used in nucleosomal array reconstitution were generated by digesting plasmid pXP-10 with EcoRI to release a 215-bp fragment containing nucleotides −78 to +137 of a Xenopus borealis somatic-type 5S RNA gene (28). Digest products were isolated from 0.8% polyacrylamide gels as described (28).
Nucleosome reconstitution conditions were empirically optimized by titrating H3/H4 and H2A/H2B to DNA. Typical reconstitution conditions were 10 μg of 207 bp 601 DNA fragment and saturating amounts (∼5.0 μg of each) of H3/H4 and 5 μg H2A/H2B in the reconstitution buffer (10 mM Tris, pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol (DTT) and 2 M NaCl) of a 200 μl total volume. Nucleosomes were reconstituted via standard salt dialysis (28). Two reconstitution reactions were combined (400 μl total) and nucleosomes were purified by sedimentation through 10.0 ml 7–20% sucrose gradients (10 mM Tris– HCl, pH 8.0 and 3 mM EDTA) with ultracentrifugation at 34 000 g for 18 h in a Beckmann SW41 rotor at 4°C. Nucleosome fractions (500 μl) were collected in 0.6-ml siliconized tubes pretreated with bovine serum albumin (BSA) (0.3 mg/ml) in TE overnight at 4°C. Fractions were analyzed by electrophoresis on nucleoprotein gels [5% acrylamide, 0.5 TBE (1 90 mM Tris base, 90 mM boric acid, 2.5 mM EDTA)], stained with ethidium bromide. BSA was added to peak fractions to 0.15 mg/ml and the samples were stored at 4°C to prevent dissociation. The purified nucleosomes were found to be stable for several weeks.
Nucleosome arrays were reconstituted with the 12 × 207-601 DNA and histone octamers in the presence of monomer 5S DNA as competitor to buffer histone concentration (31). The 5S DNA was added to the reconstitution reaction at an equal mass to the 12 × 207-601 DNA. Typical reconstitution conditions were 4.3 μg of H3/H4 tetramer, 5 μg of 12 × 207-601 DNA fragment, 5 μg of 5S DNA fragment and 5.8 μg of H2A/H2B dimer in the reconstitution buffer of a 200 μl total volume. Core histone proteins were mixed with DNA in 2 M NaCl/TE, followed by dialysis against 1.5 M NaCl/TE for 1 h, 1 M NaCl/TE for 3 h, 0.7 M NaCl/TE for 3 h and finally 10 mM Tris, 0.1 mM EDTA overnight. Nucleosomal arrays were purified by sedimentation through 10.0 ml 7–30% sucrose gradients (10 mM Tris– HCl, pH 8.0 and 3 mM EDTA) with ultracentrifugation at 28 000 g for 14 h in a Beckmann SW41 rotor at 4°C. Nucleosomal array fractions were collected in 0.6-ml siliconized tubes pretreated with BSA (0.3 mg/ml) in TE overnight at 4°C. Fractions were examined by electrophoresis on nucleoprotein gels. Nucleosome saturation of arrays was assessed by EcoRI digestion into monosomes and MgCl2-dependent self-association (32). Digestion of oligonucleosome species digestion yields ∼99% mono nucleosomes, as expected for saturated templates. BSA was added to peak fractions to 0.15 mg/ml and samples stored at 4°C to prevent dissociation. Di-nucleosomes and tri-nucleosomes were reconstituted on 601 DNA templates containing two or three copies of tandemly repeated 207 bp fragments via standard salt dialysis. The level of nucleosome saturation was determined by electrophoretic mobility shift assays (EMSAs) and AvaI digestion to release monosomes. EMSAs shows a two- and three-step up-shift for H1s titrated into di- and tri-nucleosomes, respectively.
FRET and smFRET
FRET analysis with labeled H1 was performed with the H1 double mutant H1G101C/K195C, H1G101C/T173C and the combination of single mutants H1 G101C, H1 K195C and H1 T173C. These proteins were prepared, expressed and purified as described above and the cysteines were subsequently reduced in 50 mM DTT for 1 h. Upon reduction, DTT was removed and the protein was purified by ion-exchange chromatography and quick-frozen (26). The protein was labeled with maleimido-Cy3, maleimido-Cy5 or a 50/50 mixture of the two. Emission spectra were recorded and FRET efficiency was calculated as described previously (26). For FRET analysis of salt-dependent folding the mutant H4 A15C was prepared, expressed, purified and reduced in the H3/H4 tetramer form as described above. The tetramer was labeled with maleimido-Cy3, Maleimido-Cy5 or a 50/50 mixture. The labeled proteins were reconstituted to 12 × 207-601 nucleosomal arrays and placed in a siliconized glass cuvette. Emission spectra were recorded with excitation at 515- and 610-nm excitation wavelengths on a Horiba Jobin Yvon FluoroMax-4 spectrofluorometer with 5-nm slit widths in both excitation and emission channels. Spectra were recorded in presence of increasing concentrations of NaCl or MgCl2 as indicated in the Figure 1 legend. We confirmed that the anisotropies for labeled H1 within nucleosomes, DNA and nucleosomal array complexes are not significantly altered compared to free H1, or compared to free dyes, consequently a kappa-squared value of two-third was used for FRET efficiency calculations. Spectra for labeled H1s were recorded in the absence or presence of mono-nucleosomes or nucleosome arrays as indicated in the figure legends. FRET efficiency was calculated as described (33) using maximum peak heights. For the Cy3-Cy5 pair, we used a base Förster radius (R0) of 5.4 nm (34), which we employed for calculations related to free H1. Due to fluorophore quenching in the nucleosome and nucleosomal array, we calculated an adjusted R0 of 5.01, by estimation of the change in the quantum efficiency of donor (Cy3) fluorescence and the resulting effect on the overlap integral. Note that changes in quantum yield of fluorescence of acceptor (Cy5) are taken into account in the calculation of FRET efficiency (26).
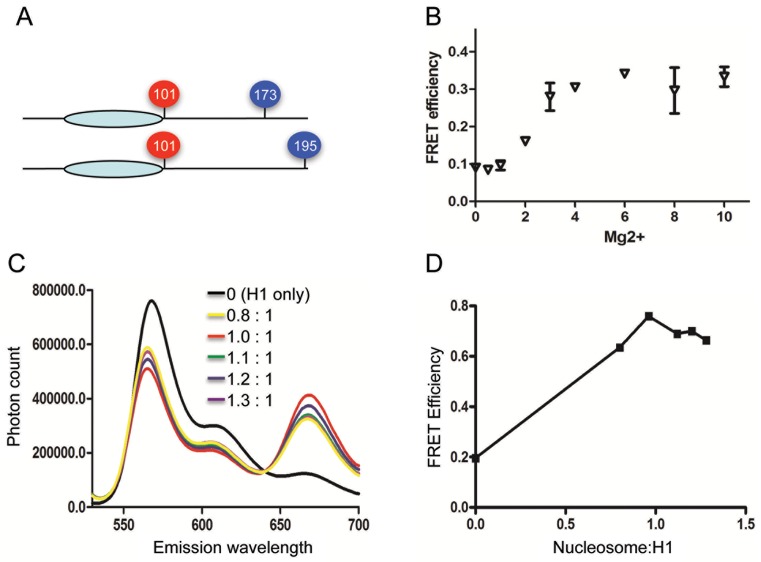
Figure 1.
H1 association with nucleosome arrays induces condensation of the CTD. (A) Position of Cy3/Cy5 attachment sites in H1 G101C/T173C and H1 G101C/K195C. (B) Reconstituted arrays containing exhibit Mg2+-dependent folding consistent with nucleosome saturation. Arrays containing Cy3 and Cy5 labeled H4 A14C were mixed with a 10-fold excess of arrays containing unlabeled histones to eliminate inter-array FRET. Plot of Intra-array FRET efficiency versus MgCl2 concentration. (C) Fluorescence emission spectra of Cy3/Cy5-labeled H1 G101C/K195C bound to arrays at increasing nucleosome:H1 ratios. Excitation of samples at 515 nm. (D) Plot of FRET efficiencies calculated from data shown in (C).
Single molecule FRET measurements were carried out as previously described (35) on a Nikon TE2000 microscope (Tokyo Japan) with a customized total internal reflection (TIR) illumination setup for fluorophore excitation at 532 nm and an EMCCD camera (Andor IXON DU-897, Belfast Ireland) for signal collection. The microscope slide surface was passivated with a lipid bilayer as we published elsewhere (36). The surface also contained a sub-monolayer of biotinylated polyethylene glycol (PEG) for nucleosome immobilization. The preparation of the biotinylated nucleosome was as described above. H1 at 50 nM was mixed with the nucleosome at 3 nM and subsequently injected onto the surface-passivated slide that had been pre-treated with streptavidin. Upon injection, a total of 50 movies each lasting ∼30 s were taken within 30 min total. No oxygen scavenging system or triplet state quencher was used in order to avoid any artifacts due to these chemicals. The emission was first filtered with a long-pass filter (HQ550LP, Chroma Technology, Bellows Falls VT) and divided into Cy3 and Cy5 spectral regions with a dichroic mirror (650DCXL, Chroma Technology). The TIR illuminated area of 50 × 100 μm2 that was spectrally separated into the two spectral regions was projected on the EMCCD camera. The fluorescence intensities of individual Cy3 spots and the corresponding Cy5 spots were obtained from each movie frame (50 ms/frame). The approximate smFRET efficiencies from single FRET pairs in each movie frame were computed based on the fluorescence intensities with a formula ICy5/(ICy3 + ICy5) where I is single fluorophore intensity. Each H1 molecule was categorized into either the lower (<0.5) or higher (>0.5) FRET population according to its time-averaged FRET efficiency. The smFRET efficiencies from each population were combined together to construct the corresponding FRET histogram.
RESULTS
H1CTD exhibits distinct extents of condensation upon binding to nucleosome arrays compared to mononucleosomes or bare DNA
Previously we documented that the H1 CTD undergoes a drastic condensation upon binding to nucleosomes or naked DNA fragments, consistent with a transition from an unstructured, disordered conformation to a defined structure or ensemble of structures (22,25,26). To further understand the behavior of H1 in a more physiological context, and how the H1 CTD promotes folding and condensation of chromatin, we used FRET to monitor structural changes within this domain upon binding to a 12mer-nucleosome array. The H1 mutants G101C/K195C and G101C/T173C were modified with Cy3- and Cy5-maleimide to place fluorophore pairs at either end of the CTD, or one fluorophore at an interior position within the CTD (Figure 1A). Nucleosomal arrays were reconstituted on a DNA template consisting of 12 × 207 bp tandem repeats of DNA segments containing the 601 nucleosome positioning sequence (37). The reconstituted arrays showed ∼50% self-association at ∼3 mM MgCl2, while EcoRI digestion yields almost exclusively mononucleosomes, indicating that the array templates were saturated with nucleosomes (Supplementary Figure S1). Moreover, inter-nucleosome, intra-array FRET between fluorophores attached to the core histone surface (see ‘Materials and Methods’ section) exhibited a MgCl2-dependent transition consistent with salt-dependent folding of saturated arrays lacking H1 (Figure 1B).
As observed previously, Cy5 emission (peak at ∼670) from Cy3/Cy5-labeled H1 G101C/K195C in the absence of nucleosomes was low upon excitation of Cy3 (at 515 nm), yielding calculated FRET efficiencies consistent with an unstructured CTD (Figure 1C) (26). However, upon addition of nucleosomal arrays, significant increases in Cy5 emission were observed, indicating increased FRET and decreased distance between the fluorophore-labeled positions (Figure 1C). Determination of relative FRET efficiencies over a range of H1 concentrations shows the efficiency is maximal at a stoichiometry of ∼1 H1 per nucleosome (Figure 1D). Similar results were observed for Cy3/Cy5 labeled G101C/T173C (see below). Interestingly, the FRET response for the free H1s did not change as salt concentrations were raised from 5 to 90 mM NaCl (Figure 2A), indicating that general charge shielding alone is insufficient to cause detectable condensation of the CTD.
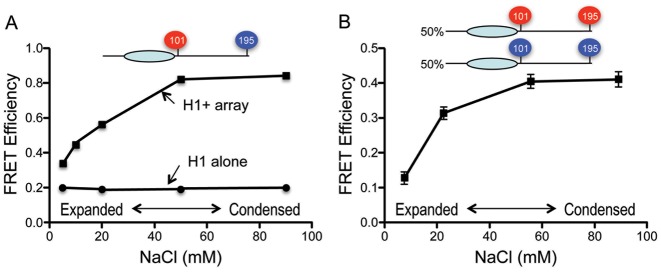
Figure 2.
H1 CTD FRET during salt-dependent folding of nucleosome arrays. (A) Total FRET increases upon salt-dependent folding of 12-mer nucleosome arrays containing Cy3/Cy5-labeled H1 G101C/K195C (squares). FRET response of the labeled H1 in the absence of arrays over the same range of salt concentrations is shown (circles). (B) Inter-molecular FRET between labeled H1s increases upon salt-dependent folding of nucleosome arrays. A 50/50 mixture of Cy3-labeled and Cy5 labeled H1 G101C/K195C was bound to 12-mer nucleosome arrays and FRET efficiency determined over the range of salt concentrations shown.
Nucleosome arrays containing H1 adopt an extended ‘beads-on-string’ conformation in low ionic strength buffers, but form condensed 30-nm fiber-like structures and higher order structures at NaCl concentrations ≥50 mM (7). We found that Cy3/Cy5-labeled H1 G101C/K195C bound to 207 bp × 12 nucleosome arrays exhibited increases in FRET as the NaCl concentration was raised over the range known to cause folding of the array (Figure 2A). The central globular domain of the linker histone directs structure-specific nucleosome binding via contacts with exiting/enter linker DNA and the DNA at the nucleosome dyad (38,39). Therefore H1 CTDs bound to adjacent nucleosomes may be positioned close enough to allow inter-molecular FRET within condensed arrays. Indeed, nucleosomal arrays bound with a 50:50% mixture of Cy3-only and Cy5-only labeled H1 G101C/K195C exhibit significant intermolecular FRET that increases as the salt concentration is raised to induce folding of the array (Figure 2B). This indicates that at least some of the FRET increase observed with the double-labeled H1 (Figure 2A) is due to inter-molecular FRET.
We wished to assess the extent of CTD folding when H1 was bound to nucleosomes within the arrays. However, the total FRET component observed in Figure 2A is a sum of intra- and inter-molecular FRET occurring in the chromatin. To isolate intra-molecular FRET, we diluted the labeled H1 in the sample with unlabeled protein until the inter-molecular FRET contribution was reduced to background levels (26). We found that intermolecular FRET from 207 bp × 12 arrays bound by a 50:50 mixture of Cy3-only and Cy5-only labeled H1 G101C/K195C reaches background levels after ∼5-fold dilution with unlabeled protein (Figure 3, blue triangles). Of note, this result indicates that the number of labeled H1s within FRET distance within the folded/condensed arrays is far less than that found in H1–DNA complexes (26) (see ‘Discussion’ section). A similar result was observed using Cy3-only or Cy5-only labeled H1G101C/T173C (Figure 3, red triangles).
Figure 3.
H1 CTD conformation in nucleosome arrays is distinct from that in mononucleosomes. Intramolecular FRET for Cy3/Cy5 labeled H1 G101C/K195C or H1 G101C/T173C (squares as indicated) bound to arrays was obtained by increasing dilution of labeled protein with unlabeled H1 (Fang et al., (26)). A 50/50 mix of Cy3-labeled and Cy5-labeled H1 G101C/K195C or H1 G101C/T173C bound to arrays (inverted or regular triangles), with increasing dilution by unlabeled H1. Dotted lines indicate intra-molecular FRET values. Inset: table of intra-molecular FRET efficiencies for H1s bound to mononucleosomes (H-Nuc) or arrays (H1-Array).
We then determined intra-molecular FRET representing CTD condensation by similarly diluting the Cy3-/Cy5 double-labeled H1s with unlabeled H1. Increasing the ratio of unlabeled/labeled H1 reduced total FRET efficiency in samples of fully condensed arrays until reaching a constant value of ∼0.42 for H1G101C/K195C and ∼0.52 for H1G101C/T173C (Figure 3). Since we have established that the inter-molecular FRET contribution has been eliminated under such conditions, these values represents the intrinsic amount of intra-molecular FRET for the H1 CTD bound to nucleosomes within the arrays. In contrast, FRET efficiencies of 0.79 and 0.61 respectively, were observed for double labeled H1G101C/K195C and H1G101C/T173C are bound to mononucleosomes (25,26). Thus the extent of CTD condensation upon H1 binding to nucleosomes within arrays is distinct compared to H1 bound to isolated mono-nucleosomes. Moreover, the relative distance between residues 101–173 is less than that between 101–195 when H1 is bound to arrays (Figure 3), while the opposite relationship was found when H1 is bound to mononucleosomes (26).
Relative orientation of H1 CTDs within the chromatin fiber
Based on above experiments demonstrating significant inter-molecular FRET between neighboring H1s (Figure 2B), we wondered whether the specific labeled sites on the H1 CTD occupied defined distances from each other in nucleosomal arrays and how these distances might vary during chromatin fiber compaction. We ascertained the relative distances between the specific labeled sites within the H1 CTD when bound to nucleosomal arrays by determining inter-molecular FRET for pairwise 1:1 combinations of single-site labeled H1s (Cy3-H1G101C/Cy5-H1G101C, Cy3-H1G101C/Cy5-H1K195C and Cy3-H1K195C/Cy5-H1K195C) bound to nucleosome arrays, in 5, 20, 50 and 80 mM NaCl, corresponding to extended, partially folded, maximally folded, and maximally folded/self-associated arrays, respectively (7). In 5 mM NaCl the three combinations each yielded small amounts of FRET, with a calculated FRET efficiency of about ∼0.13 (Figure 4). This result indicates that in the extended nucleosomal arrays, all three labeled sites are barely within FRET distance and the relative average distances between all combinations are approximately the same. Upon raising the salt concentration, the FRET response increased and differences between pairs of sites became evident, with FRET response peaking at 50 mM NaCl (Figure 4), where the arrays are fully folded into chromatin fiber structures (7). The highest FRET efficiency is observed with the Cy3-H1K195C/Cy5-H1K195C pair, followed by Cy3-H1G101C/Cy5-H1K195C, while the lowest response was observed with Cy3-H1G101C/Cy5-H1G101C. Thus within the compacted chromatin fiber, H1 residue 195 is closest in space to the same residue on adjacent H1s, with larger distances between residue 101 and 195 and the greatest distance between the N-terminal regions of the CTDs on adjacent H1s. Raising the NaCl concentration further, to 80 mM NaCl, resulted in a slight dampening of the highest FRET efficiencies, likely due to accumulation of self-associated arrays, which also contribute to the intermolecular FRET signal (unpublished results). Interestingly, these spatial relationships are distinct from those observed within H1–DNA complexes (26), suggesting that the relative arrangement of H1s within arrays does not resemble that found within H1–DNA aggregates.
Figure 4.
H1 CTD C-termini of neighboring H1s are closer in space than N-termini in condensed chromatin. H1 G0101C and H1 K195C were specifically modified with either Cy3 (red) or Cy5 (blue) and FRET pairs bound to 12-mer nucleosome arrays in equimolar ratios. Intermolecular FRET efficiencies were determined at various salt concentrations, and plotted. See text for description.
H1CTD folding depends on chromatin fiber conformation
The increase in FRET response from Cy3/Cy5-labeled H1 G101C/K195C upon array folding (Figure 2A) and differences with that observed upon binding to mononucleosomes raised the possibility that the CTD structure may be linked to the folding state of the chromatin. To assess whether the conformation of the H1 CTD is related to folding of the nucleosome array, we measured intra-molecular FRET for Cy3-/Cy5-labeled H1 G101CK195C and H1G101CT173C bound to nucleosomal arrays in solutions containing increasing concentrations of NaCl. As above, intra-molecular FRET was determined using the labeled H1 dilution method to eliminate inter-molecular FRET contributions (26).
Arrays bound by H1 in 10 mM NaCl exist in an uncompact conformation known as a loose zig-zag structure (2,40). Labeled H1 exhibited a FRET efficiency of ∼0.45 that was diminished by dilution of the labeled H1 with unlabeled protein to about 0.33 (Figure 5A, black line). Thus the total FRET response is comprised largely by intra-molecular FRET, with little inter-molecular FRET between H1s on these arrays. This is expected as nucleosome centers on fully expanded arrays are separated by distances greater than the maximum range for efficient inter-molecular FRET (Figure 4).
Figure 5.
H1 CTD condensation increases during the first stage of nucleosome array folding. Nucleosome arrays were bound by either Cy3/Cy5 labeled H1 G101C/K195C (A) or H1 G101C/T173C (B) with increasing dilution with unlableled H1 to eliminate inter-molecular FRET. Arrays were adjusted to 10, 20, 50, and 90 mM NaCl and FRET efficiencies determined. (C) Schematic of array folding in 10, 20 and >50 mM NaCl and corresponding FRET efficiencies for Cy3/Cy5 labeled H1 G101C/K195C bound to arrays with different degrees of salt-dependent folding.
We found that when the NaCl concentration is raised to 20 mM, there is a significant increase in FRET from the labeled H1s within the arrays, to an efficiency of ∼0.55 (Figure 5A, green line). At this salt concentration, H1-containing arrays adopt a moderately condensed zig-zag conformation (7,40). Similar to the fully less compact arrays, dilution of labeled H1 with unlabeled protein on arrays in 20 mM NaCl indicates the majority (∼80%) of the total FRET response is intra-molecular, with a FRET efficiency of 0.42. A similar result was obtained at 10 and 20 mM NaCl with arrays containing Cy3/Cy5 labeled H1 G101C/T173C (Figure 5B, black and green lines). In contrast, the FRET response from labeled H1 bound to mononucleosomes remains constant over a range from 10 to 50 mM NaCl (Supplementary Figure S2). Thus our results indicate that the transition from a loose to a moderately condensed zig-zag conformation is accompanied by a conversion to a more condensed CTD in which the labeled pairs in both H1 G101C/K195C and H1 G101C/T173C are closer in space.
We next investigated the H1 CTD conformation in fully condensed arrays in either 50 or 90 mM NaCl (7). Importantly, we observed a significant increase in FRET for arrays containing either Cy3/Cy5 labeled G101C/K195C or H1 G101C/T173C in these elevated salt conditions (Figure 5, red and purple lines) compared to that observed in 20 mM NaCl. However, upon dilution with unlabeled H1, the FRET efficiencies for both 50 and 90 mM samples were reduced ∼40% to values identical to those observed for arrays in 20 mM NaCl, indicating that the entirety of the increase was due to inter-molecular FRET. This result is consistent with the expected transition of moderately condensed arrays in 20 mM NaCl to fully condensed structures in 50 and 90 mM NaCl, and the closer apposition of labeled H1s within these arrays. Importantly upon elimination of inter-molecular FRET, the identical values of intra-molecular FRET are observed for arrays in 20, 50 and 90 mM NaCl indicates that the H1 CTD adopts a similar structure in moderately folded and fully condensed arrays.
Molecular determinants of H1CTD condensation
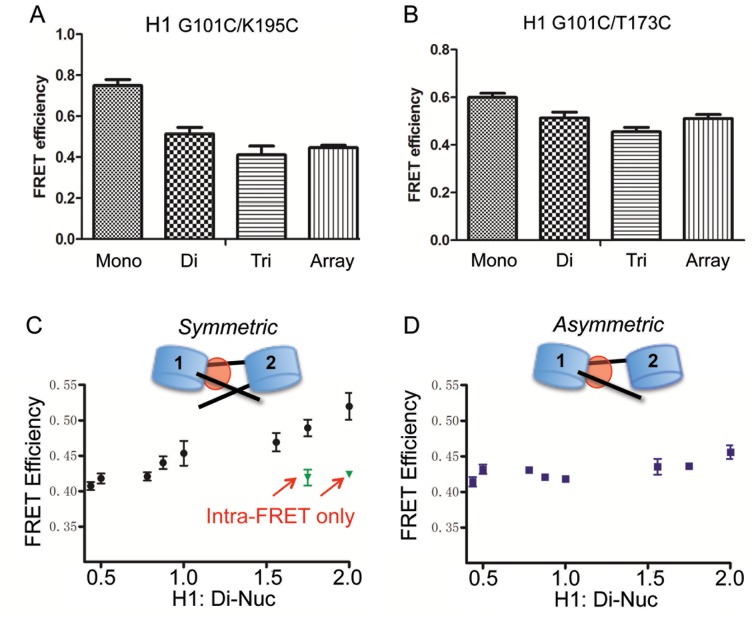
To further investigate why the H1 CTD adopts distinct structure(s) when bound to nucleosomes within arrays compared to mononucleosomes, we compared the condensation state of the CTD when H1 is associated with mononucleosomes, dinucleosomes and trinucleosomes and 207 bp × 12 nucleosome arrays. All chromatin complexes were prepared with a 1:1 stoichiometry of H1:nucleosomes. We found that H1 bound to all oligonucleosomes exhibited similar, reduced FRET efficiencies compared to the H1-mononucleosome complex (Figure 6A). For H1 G101C/T173C, a similar trend was observed with all oligonucleosomes having similar, lower total FRET response compared to mononucleosomes (Figure 6B)
Figure 6.
H1 CTD condensation in mononucleosomes is distinct compared to oligonucleosomes. Cy3/Cy5 labeled H1 G101C/K195C (A) or H1 G101C/T173C (B) was bound to mononucleosomes (30-N-30), dinucleosomes (30-N-60-N-30), or tri-nucleosomes (30-N-60-N-60-N-30) or 207 bp × 12-mer arrays (with 60 bp linker DNA) and FRET efficiencies compared. (C and D) Linker DNA from the distal nucleosome does not influence H1 CTD structure in dinucleosomes. Increasing amounts of H1 were incubated with either a symmetric (30-N-60-N-30) (C) or asymmetric (30-N-60-N-0) (D) dinucleosome containing or lacking terminal linker DNA segment extending from nucleosome 2. Black circles and squares indicate total FRET measured while green triangles depict intra-molecular FRET signal in the presence of an excess of unlabeled H1 (see text). Note that symmetric and asymmetric disomes bind 2 and 1 H1s, respectively.
It is possible that within the array, neighboring H1s affect each other's environment and thus the extent of CTD folding. To test this possibility, we prepared arrays in which the H1:nucleosome stoichiometry was reduced so that on average only one H1 would be bound to each array. We find that even at a low stoichiometry, the intra-molecular FRET response from Cy3/Cy5-labeleld H1 G101C/K195C was similar to that found in saturated arrays (results not shown). Thus, neighboring H1s apparently do not significantly influence each other's structure and such and effect does not account for the differences observed in H1 structure in mono- versus dinucleosomes.
A popular class of models for the folded chromatin fiber posits that linker DNA is relatively straight between adjacent nucleosomes, positioned on opposite sides of the fiber (3). To account for observed differences in H1 structure in mononucleosomes vs arrays, we considered the possibility that an H1 CTD bound to nucleosome N might interact with the distal linker DNA of neighboring nucleosome N + 1 within arrays (Figure 6C and D). To test this, we prepared symmetric and asymmetric di-nucleosomes (30-N-60-N-30 and 30-N-60-N-0, respectively), in which the free linker DNA on nucleosome 2 was present or deleted, and assessed whether the deletion caused the H1 structure to resemble that found in mononucleosomes. Binding of H1 exhibited a similar total FRET response for both symmetric and asymmetric dinucleosomes at substoichiometric ratios of H1 per dinucleosome (1:1). However, at a ratio of 2 H1s per dinucleosome the FRET response was increased for the symmetric but not for the asymmetric dinucleosome (Figure 6C and D). Dilution with unlabeled H1 showed that increase in FRET was entirely due to an intermolecular FRET component for the symmetric dinucleosome upon binding of 2 H1s, while no intermolecular FRET was observed for the asymmetric construct at the same H1 concentration (Supplementary Figure S3). This result is consistent with the binding of only 1 H1 to the asymmetric disome as the lack of the linker DNA on nucleosome 2 reduces H1 binding affinity about 5-fold (41). Therefore, H1 bound to either dinucleosome construct exhibits equivalent intra-molecular FRET efficiencies, indicating that the DNA from an adjacent (N+1) nucleosome does not alter the folding of the H1 CTD bound to nucleosome N. Thus structural features of the N + 1 nucleosome other than the distal linker DNA must be responsible for influencing the H1 CTD structure bound to nucleosome N.
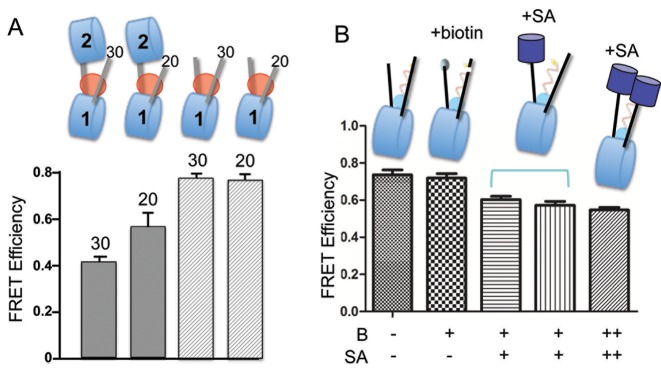
To further dissect the molecular determinants influencing H1 CTD folding in oligonucleosomes, we hypothesized that neighboring nucleosomes might sterically constrain linker DNA trajectory, thereby altering the H1 CTD's environment. We therefore prepared asymmetric dinucleosomes in which the free linker DNA arm of nucleosome 1 was progressively shortened to test whether a steric constraint with nucleosome 2, if present, might be reduced (Figure 7A, top). Indeed, we observed that reducing the linker DNA length to 20 bp from 30 bp results in a significant increase in the FRET response observed with the asymmetric di-nucleosome. A similar change in the linker DNA length associated with a monosome does not result in a similar change in FRET (Figure 7A). This result suggests that the linker DNA trajectory is altered by the presence of a neighboring nucleosome, which, in turn influences H1 CTD structure. To further test this idea we replaced the neighboring nucleosome with streptavidin, to test whether any bulky substituent might influence the linker DNA path and alter CTD structure. Indeed, binding of streptavidin to mononucleosomes biotinylated at either one or both ends of the linkers caused a detectable decrease in FRET, resembling that observed in the oligonucleosomes (Figure 7B). These results indicate that the trajectory of the linker DNA and the structure of the H1 CTD are coupled.
Figure 7.
Neighboring nucleosomes constrain trajectory of linker DNA and affect H1 CTD conformation. (A) Decreasing linker DNA length increases CTD condensation in dinucleosomes but not in mononucleosomes. Asymmetric dinucleosomes with 30 and 20 bp linker DNAs 30-N-60-N-0 and 20-N-60-N-0, respectively were prepared, along with monosomes controls (30-N-30 and 30-N-20), bound by Cy3/Cy5 labeled H1 G101C/K195C and FRET efficiencies determined, as indicated. (B) Attachment of streptavidin to the end of linker DNA alters CTD conformation. Linker DNA ends in 30-N-30 mononucleosomes were modified with biotin (B) and streptavidin added (SA) as indicated. Bracket indicates samples where the order of addition of H1 and SA were reversed; ++ indicates nucleosomes in which both ends of the linker DNAs were biotinylated.
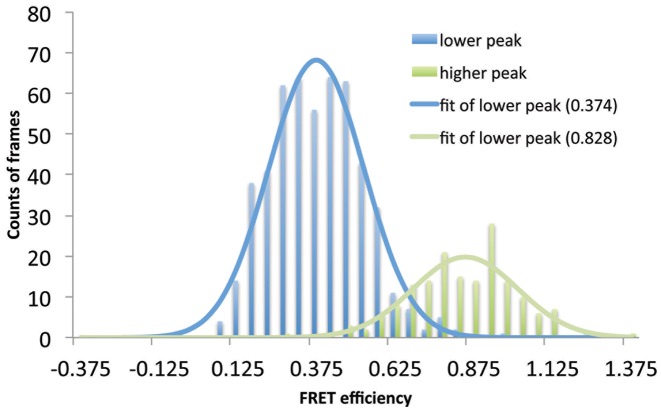
To substantiate the above results obtained with bulk FRET techniques we determined FRET of Cy3/Cy5-labeled H1 G101C/K195C bound to nucleosomes using a single-molecule approach. Nucleosomes in which one of the linker DNA ends was biotinylated (30-N-30-B) were tethered to a passivated surface in which a lipid bilayer is combined with a biotinylated PEG sub-monolayer (36). Labeled H1 was flown into the chamber as described in the ‘Materials and Methods’ section and 50 movies of the surface were taken within the first 30 min after H1 injection. Three fluorescent H1 species were observed; a species exhibiting little or no FRET response and two species exhibiting FRET responses of ∼0.4 and ∼0.8 (Figure 8). A control with only the naked DNA template resulted in about a 10-fold lower percentage of H1 showing FRET. Gratifyingly, the FRET efficiencies of the two nucleosome-bound species corresponded well to the efficiencies measured for H1 bound to monosomes (∼0.8) and nucleosome arrays (∼0.4) in our bulk FRET experiments. We hypothesize that attachment of the nucleosomes to the surface may slow equilibration of nucleosome and H1 CTD conformations, resulting in the heterogeneous distribution of FRET states. Thus we interpret the two states observed in the single molecule measurements as representative of the distinct H1-monosome and H1-oligonucleosome structures detected in bulk experiments, suggesting a bimodal nature of linker DNA/CTD conformations.
Figure 8.
Single molecule FRET histograms of H1 bound to 30-N-30 mononucleosomes reveals two major populations. The H1 molecules observed with smFRET were categorized into two populations (lower- and high-FRET) according to their time-averaged FRET efficiencies. The smFRET efficiencies from each population were combined together to construct the corresponding FRET histogram. The lower peak (∼0.4) corresponds to the FRET histogram of the lower-FRET population and the high peak (∼0.8) corresponds to the FRET histogram of the higher-FRET population. Each histogram was fit to a Gaussian distribution to yield the average FRET efficiency of 0.374 (lower-FRET) or 0.828 (higher-FRET), revealing at least two distinct native conformations of the H1 CTD. Nearly identical values were obtained when the histogram was analyzed directly without categorization of values (Supplementary Figure S4)
DISCUSSION
The H1 CTD is critical for stabilizing native chromatin structures and numerous post-translational modifications within this domain are correlated with alterations in chromatin structure associated with gene transcription and other nuclear processes. We previously demonstrated that that the H1 CTD undergoes a drastic conformational change, from an unfolded, random coil to a condensed structure or ensemble of structures upon binding to mononucleosomes. In this work we discovered that binding of H1 to oligonucleosome arrays similarly induced folding of the CTD but that the ultimate conformation(s) is (are) distinct compared to that observed when H1 is bound to mononucleosomes and dependent on array structure. The distinct H1 CTD conformations are not due to the influence of nearby H1s bound to neighboring nucleosomes as similar results are obtained with arrays sub-saturated with H1. Moreover, we found that a single neighboring nucleosome is sufficient to induce distinct H1 CTD folding, and that the distal linker DNA of a neighboring nucleosome in a dinucleosome construct does not play a role as might be predicted from models for the condensed chromatin fiber. Our results suggest that neighboring nucleosomes impose constraints on linker DNA linker trajectory and support a model in which linker DNA conformation and the structure of the H1 CTD are coupled.
Our observation the H1 CTD transitions from a disordered chain to a condensed structure, with a probable unique fold or ensemble of folds, is consistent with data indicating that the CTD has the amino acid residue content and biochemical properties of an intrinsically disordered protein (IDP) domain (17). Moreover, H1 CTD peptides in the presence of DNA, detergents or structure stabilizing solvents have been shown to exhibit secondary structural elements (21–23,42) and modeling studies indicate a linker-DNA dependent condensation of the H1 CTD (43). In the current work find that the H1 CTD maintains this disordered state over a wide range of salt concentrations, from 10 mM to 2M NaCl (Figure 2A and unpublished results), indicating that homogenous charge neutralization is insufficient to recapitulate the nucleosomal environment experienced by H1. Moreover, a hallmark of IDPs is the ability to adopt different structures dependent upon specific macromolecular binding partners. We find that the CTD exhibits unique conformations when bound to naked DNA, mono-nucleosomes (26), and nucleosome arrays (this work), suggesting that the CTD senses a distinct environment in each. Our data indicate that the CTD adopts a less compact overall structure when bound to nucleosomes within arrays than with mono-nucleosomes.
We find that the CTD exhibits the lowest apparent extent of condensation when the array is maximally uncondensed in low salt (10 mM NaCl) buffers, but undergoes additional condensation when the arrays are more condensed in buffer containing 20 mM NaCl. This transition in CTD structure coincides with a transition in array structure from a loose zig-zag arrangement to a more compact folding intermediate known as the contacting zig-zag (40,44) (Figure 9). This result suggests that the CTD is sensitive to changes in the in array structure and that factors that alter the propensity of the CTD to fold may influence and be a means of regulation of oligonucleosome folding. For example, certain phosphorylation events reduce the propensity of the CTD to fold and may be a means of regulating structure (our unpublished results). Given that the coordination is at the earliest extents of array folding there may be specific phosphorylation events that promote unfolding of chromatin fibers to allow access of trans-acting factors.
Figure 9.
Scheme showing states of CTD folding associated with free H1 and H1 bound to expanded arrays, condensed arrays and mono-nucleosomes (bottom). Cartoon of CTD structures (top) are meant to depict random coil for the free protein (–), and increasing states of compaction upon binding to expanded arrays, condensed arrays, and mononucleosomes, respectively. Note that the extent of CTD compaction is grossly inversely correlated with the separation between linker DNA in each chromatin species.
Interestingly, beyond the initial stage of salt-dependent folding, further compaction of the array in 50 and 90 mM NaCl does not result in significant changes in the H1 CTD conformation, suggesting that salient aspects of array structure and linker DNA conformation relevant to CTD environment are unchanged beyond formation of the contacting zig-zag conformation in 20 mM NaCl. These results are consistent with a recent study indicating that the contacting zig-zag is a persistent feature of higher order chromatin structure in both interphase and mitotic chromatin (45). Clearly, array condensation in buffers containing 20–50 mM NaCl results in increased H1–H1 intermolecular FRET (Figures 4 and 5). In this case the closer apposition of H1s its likely due to further condensation of individual arrays, perhaps to canonical 30 nm fiber-like structures (7,46) rather than inter-array interactions, as little self-association is expected at 50 mM NaCl (7). Interestingly, the H1 CTD structure appears unchanged even upon increasing the NaCl concentration to 90 mM, where array–array self-association is likely to occur, with conversion of folded arrays to interdigitated fibers (47). Thus the environment of H1 appears to remain relatively constant during formation of multiple higher order chromatin structures beyond the contacting zig-zag arrangement.
Our data indicates that a critical factor contributing to the distinct structures observed in mononucleosomes and arrays is linker DNA conformation. Addition of a single nucleosome adjacent to a nucleosome-bound H1 (i.e. as in the asymmetric dinucleosome) results in H1-CTD conformation similar to that found in arrays (Figure 6A and B). This effect appears to be due to steric constraints imposed by neighboring nucleosomes as shortening the free linker DNA arm in an asymmetric dinucleosome to be more ‘monosome-like’ while addition of a bulky streptavidin to the free linkers in a mononucleosomes alters CTD conformation to be more ‘oligonucleosome-like’ (Figure 7). Moreover the changes in CTD structure we observed during the earliest stages of salt-dependent array folding may also be linked to changes in linker DNA conformation. Cy3/Cy5 H1 G101-K195C within the low salt (10 mM NaCl) loose zig-zag arrays, exhibits a FRET efficiency of 0.32, where linker DNA entry/exit angles are expected to be greatest. Upon folding to the contacting zig-zag form in 20 mM NaCl, the entry/exit angle is expected to decrease, while the FRET efficiency increases to 0.42. Thus the folding of the nucleosome array appears coupled to H1 CTD conformation via alterations in linker DNA conformation.
We also find that the CTD exhibits a unique orientation within the folded array. Intermolecular FRET indicates that the C-terminal end of each H1 CTD is closer in space than the labeled site (G101C) on the N-terminus. Thus our data are consistent with model exhibiting radial symmetry, such that the CTD would be arrayed about a central axis with the C-terminal end of the protein oriented toward the center of the fiber. We note that the spatial relationships between sites within the CTD observed for H1 bound to nucleosomes arrays is exactly opposite that observed for H1 bound to naked DNA fragments. In the latter case, residue 101, nearest the globular domain, was closest in space to residue 101 on other H1s. It is also interesting to note that dilution of inter-molecular in H1–DNA complexes required 20-30-fold excess of unlabeled protein, in contrast to the ∼5-fold excess required for nucleosome arrays. This implies that each H1 within the condensed array has only a few other H1s within FRET distance, while many H1s within the DNA aggregates are close in space. Overall this observation is consistent with an extended and ordered structure in which each H1 is bound to a nucleosome within the array. It will be interesting in future experiments to compare relative distances between other positions within H1 within condensed nucleosome arrays.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM52426 to J.J.H., GM097286 to T.-H.L.]. Funding for open access charge: NIH GM52426.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cutter A.R., Hayes J.J. A brief review of nucleosome structure. FEBS Lett. 2015;589:2914–2922. doi: 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bian Q., Belmont A.S. Revisiting higher-order and large-scale chromatin organization. Curr. Opin. Cell Biol. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pepenella S., Murphy K.J., Hayes J.J. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnick E.H., Bustin M., Marsaud V., Richard-Foy H., Hager G.L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annunziato A.T., Hansen J.C. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carruthers L.M., Bednar J., Woodcock C.L., Hansen J.C. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 8.Cao K., Lailler N., Zhang Y., Kumar A., Uppal K., Liu Z., Lee E.K., Wu H., Medrzycki M., Pan C., et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millan-Arino L., Islam A.B., Izquierdo-Bouldstridge A., Mayor R., Terme J.M., Luque N., Sancho M., Lopez-Bigas N., Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen X., Gorovsky M.A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 11.Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Izzo A., Kamieniarz-Gdula K., Ramirez F., Noureen N., Kind J., Manke T., van Steensel B., Schneider R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Lu X., Wontakal S.N., Kavi H., Kim B.J., Guzzardo P.M., Emelyanov A.V., Xu N., Hannon G.J., Zavadil J., Fyodorov D.V., et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Y., Nikitina T., Morin-Kensicki E.M., Zhao J., Magnuson T.R., Woodcock C.L., Skoultchi A.I. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caterino T.L., Hayes J.J. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 17.Lu X., Hamkalo B., Parseghian M.H., Hansen J.C. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 2009;48:164–172. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X., Hansen J.C. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 19.Clark D.J., Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 20.Bradbury E.M., Cary P.D., Chapman G.E., Crane-Robinson C., Danby S.E., Rattle H.W., Boublik M., Palau J., Aviles F.J. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The conformation of histone H1. Eur. J. Biochem. 1975;52:605–613. doi: 10.1111/j.1432-1033.1975.tb04032.x. [DOI] [PubMed] [Google Scholar]
- 21.Clark D.J., Hill C.S., Martin S.R., Thomas J.O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roque A., Iloro I., Ponte I., Arrondo J.L., Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol. Chem. 2005;280:32141–32147. doi: 10.1074/jbc.M505636200. [DOI] [PubMed] [Google Scholar]
- 23.Roque A., Ponte I., Suau P. Role of charge neutralization in the folding of the carboxy-terminal domain of histone H1. J. Phys. Chem. B. 2009;113:12061–12066. doi: 10.1021/jp9022579. [DOI] [PubMed] [Google Scholar]
- 24.Hansen J.C., Lu X., Ross E.D., Woody R.W. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 25.Caterino T.L., Fang H., Hayes J.J. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered h1 carboxyl-terminal domain. Mol. Cell. Biol. 2011;31:2341–2348. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang H., Clark D.J., Hayes J.J. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2011;40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutledge R.G., Neelin J.M., Seligy V.L. Isolation and expression of cDNA clones coding for two sequence variants of Xenopus laevis histone H5. Gene. 1988;70:117–126. doi: 10.1016/0378-1119(88)90110-2. [DOI] [PubMed] [Google Scholar]
- 28.Hayes J.J., Lee K.M. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods. 1997;12:2–9. doi: 10.1006/meth.1997.0441. [DOI] [PubMed] [Google Scholar]
- 29.Kan P.Y., Caterino T.L., Hayes J.J. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol. Cell. Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikitina T., Ghosh R.P., Horowitz-Scherer R.A., Hansen J.C., Grigoryev S.A., Woodcock C.L. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 31.Huynh V.A., Robinson P.J., Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Hayes J.J. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell. Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirier M.G., Oh E., Tims H.S., Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii Y., Yoshida T., Funatsu T., Wazawa T., Yanagida T.Y. Fluorescence resonance energy transfer between single fluorophores attached to a coiled-coil protein in aqueous solution. Chem. Phys. 1999;277:163–173. [Google Scholar]
- 35.Lee J.Y., Wei S., Lee T.H. Effects of histone acetylation by Piccolo NuA4 on the structure of a nucleosome and the interactions between two nucleosomes. J. Biol. Chem. 2011;286:11099–11109. doi: 10.1074/jbc.M110.192047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue H., Fang H., Wei S., Hayes J.J., Lee T.H. Single-Molecule Studies of the Linker Histone H1 Binding to DNA and the Nucleosome. Biochemistry. 2016;55:2069–2077. doi: 10.1021/acs.biochem.5b01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogge R.A., Kalashnikova A.A., Muthurajan U.M., Porter-Goff M.E., Luger K., Hansen J.C. Assembly of nucleosomal arrays from recombinant core histones and nucleosome positioning DNA. J. Vis. Exper. 2013 doi: 10.3791/50354. doi:10.3791/50354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syed S.H., Goutte-Gattat D., Becker N., Meyer S., Shukla M.S., Hayes J.J., Everaers R., Angelov D., Bednar J., Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou B.R., Jiang J., Feng H., Ghirlando R., Xiao T.S., Bai Y. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bednar J., Horowitz R.A., Grigoryev S.A., Carruthers L.M., Hansen J.C., Koster A.J., Woodcock C.L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White A.E., Hieb A.R., Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 2016;6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roque A., Teruel N., Lopez R., Ponte I., Suau P. Contribution of hydrophobic interactions to the folding and fibrillation of histone H1 and its carboxy-terminal domain. J. Struct. Biol. 2012;180:101–109. doi: 10.1016/j.jsb.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Luque A., Collepardo-Guevara R., Grigoryev S., Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furrer P., Bednar J., Dubochet J., Hamiche A., Prunell A. DNA at the entry-exit of the nucleosome observed by cryoelectron microscopy. J. Struct. Biol. 1995;114:177–183. doi: 10.1006/jsbi.1995.1017. [DOI] [PubMed] [Google Scholar]
- 45.Grigoryev S.A., Bascom G., Buckwalter J.M., Schubert M.B., Woodcock C.L., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson P.J., Fairall L., Huynh V.A., Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeshima K., Rogge R., Tamura S., Joti Y., Hikima T., Szerlong H., Krause C., Herman J., Seidel E., DeLuca J., et al. Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 2016;35:1115–1132. doi: 10.15252/embj.201592660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.