Abstract
Histone proteins are synthesized in large amounts during S-phase to package the newly replicated DNA, and are among the most stable proteins in the cell. The replication-dependent (RD)-histone mRNAs expressed during S-phase end in a conserved stem-loop rather than a polyA tail. In addition, there are replication-independent (RI)-histone genes that encode histone variants as polyadenylated mRNAs. Most variants have specific functions in chromatin, but H3.3 also serves as a replacement histone for damaged histones in long-lived terminally differentiated cells. There are no reported replacement histone genes for histones H2A, H2B or H4. We report that a subset of RD-histone genes are expressed in terminally differentiated tissues as polyadenylated mRNAs, likely serving as replacement histone genes in long-lived non-dividing cells. Expression of two genes, HIST2H2AA3 and HIST1H2BC, is conserved in mammals. They are expressed as polyadenylated mRNAs in fibroblasts differentiated in vitro, but not in serum starved fibroblasts, suggesting that their expression is part of the terminal differentiation program. There are two histone H4 genes and an H3 gene that encode mRNAs that are polyadenylated and expressed at 5- to 10-fold lower levels than the mRNAs from H2A and H2B genes, which may be replacement genes for the H3.1 and H4 proteins.
INTRODUCTION
The bulk of the histone proteins are synthesized coordinately with DNA during S-phase and are very stable after incorporation into chromatin. These proteins are encoded by replication-dependent (RD)-histone mRNAs which are the only known cellular eukaryotic mRNAs that do not end in a polyA tail but end instead in a conserved stem-loop. The protein that binds the stem-loop, SLBP, is required for processing of the RD-histone pre-mRNAs and also functions during the entire histone mRNA life cycle including transport from the nucleus, translation and mRNA degradation (1). Canonical RD-histone processing occurs by endonucleolytic cleavage following SLBP binding to the stem-loop (2). This cleavage is directed by the U7 snRNP (3–5) that interacts with a second histone specific sequence called the histone downstream element (HDE) (6). These histone mRNAs are expressed at high levels during S-phase increasing 35-fold as cells enter S-phase and decreasing rapidly at the end of S-phase (7). SLBP levels are also cell cycle regulated and the increased levels of SLBP as cells enter S-phase allows increased processing of histone mRNA. The degradation of SLBP at the end of S-phase prevents further accumulation of histone mRNAs or protein (8,9).
Most studies of histone mRNAs have focused on their expression and regulation in growing cells. An unresolved question is how histone proteins are synthesized after cells have terminally differentiated and no longer re-enter S-phase. This is a particularly pertinent question for long-lived cells. In dividing cultured cells, histones are stable with <5% turnover of labelled histones over 8 generations (10). The half-life of histone protein in chicken brain tissue has been estimated at 19 days (11). In a recent proteomics study, histones were identified as among the most stable proteins in mammalian cells (12), with histone H3.1 and H4 significantly more stable than histones H2A and H2B. Histone turnover likely occurs as a result of damage to existing histone proteins. However, many mammalian cells have life spans substantially longer than even the slowly turning over histone proteins (13). How, then, do non-dividing cells such as these maintain proper levels of histone proteins that are critical for genome stability and the regulation of gene expression?
One possibility is that there are distinct genes that are expressed constitutively and encoded by polyadenylated mRNAs. The histone H3F3 genes found in all multicellular organisms encode the variant histone H3.3, which is synthesized constitutively, and plays an important role in gene regulation. It also serves as a replacement histone H3 variant. In mice, histone H3.3 becomes the predominant histone H3 protein in non-dividing cells over the lifespan of the mouse (14). Similar to H3.3, histone H1O is encoded by a polyadenylated RNA and the amount of H1O protein increases after terminal differentiation (15). There are several variants of histone H2A (e.g. H2A.Z, macroH2A), which are expressed from polyadenylated mRNAs, but these have specific functions, and likely do not serve as ‘replacement’ variants. For example, the histone H2A.X protein is expressed from a single gene and is involved primarily in DNA repair (16). In S-phase cells, the H2A.X mRNA ends in a stem-loop and is cell cycle regulated. Outside of S-phase the same gene expresses a longer polyadenylated mRNA (17,18). There are no reports of variant histone H2B genes or histone H4 genes that express only polyadenylated mRNAs (e.g. like H1O or H3.3).
Some polyadenylated core histone mRNAs are produced in small amounts in cultured cells as a result of knockdown of a number of factors (NELF (19), ARS2 (20), chromatin modifiers (21), P-TEFb (21), Y-RNAs (22) and SLBP (23)). These treatments likely result in a perturbation of canonical histone pre-mRNA processing after the stem-loop. In addition, Kari et al. have shown that there is production of some polyadenylated and spliced histone H2B mRNAs when human fibroblasts are differentiated into adipocytes in vitro (24). In some of these studies, the relative proportion of polyadenylated mRNAs and properly processed mRNAs were determined and the amount of polyA+ RNA was very small (<5%) (19,22). In other studies, only increases in polyadenylated histone mRNAs over the very small amounts of polyadenylated histone mRNAs present in control cells was described (21).
Here, we show that in terminally differentiated tissues, a subset of histone genes in the two RD-histone mRNA clusters remain active and encode polyadenylated mRNAs. These include one histone H1 gene, H1C, previously shown to be expressed in many adult mouse tissues (25), as well as genes for the four core histone proteins. All these mRNAs are polyadenylated and, in most cases, the polyA signal is 3′ of the stem-loop and extends the 3′ UTR of the histone mRNA. One or more histone H2B mRNAs (depending on the species) from the HIST1 cluster are formed by splicing around the histone stem-loop, resulting in a polyadenylated histone mRNA lacking the stem-loop. These mRNAs likely encode the ‘replacement’ H2A, H2B and H4 proteins in non-dividing cells. Surprisingly, terminally differentiated cells also express a number of genes thought to be required only for RD-histone gene expression, including most of the genes required for processing histone mRNA.
MATERIALS AND METHODS
RNA extraction from mouse tissue
Post-natal day 1 (P1 mice) and dissected mouse liver and brain were quick-frozen with liquid nitrogen and stored 80°C until use. Sections of tissue (100–200 mg) were weighed and placed in a ceramic mortar filled will liquid nitrogen and ground by hand. The resulting tissue powder was transferred to a 15-ml tube and 1 ml of Trizol (Invitrogen) was added per 50 mg of tissue, and processed as recommended by the manufacturer. The RNA pellet was air dried and resuspended in 300 μl of 0.3 M sodium acetate (pH 5.6), extracted with phenol/chloroform and ethanol precipitated.
Preparation of protein lysate from mouse tissues
The tissue powder was prepared as described above and was resuspended in 1 ml of NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 0.5% NP-40) per 100 mg of tissue, rotated at 4°C for 20 min, and the cell lysates clarified by centrifugation at 16 000 xg. The supernatant was transferred to new tubes and the protein concentration was determined by Bradford Assay.
S1 Nuclease protection assays
The S1 nuclease assays were performed as previously described (26), using either total cell RNA or RNA fractionated on oligo(dT) cellulose into polyA+ and polyA− fractions. Each gene was cloned into pUC19 and 5 μg of each plasmid digested at restriction site in the ORF resulting in the generation of a 5′ overhang that can be labelled by incorporation of α-[32P]-dCTP (3000 Ci/mmol) using the Klenow fragment of DNA pol I. After labelling the plasmid was then digested with a 2nd restriction site downstream of the 3′ end. Description of restriction site usage and probe size is found in Supplementary Table S1. After hybridization and digestion with S1 nuclease as described previously, the protected fragments were analysed by electrophoresis on a 6% polyacrylamide gel containing 8M urea followed by autoradiography.
3T3-L1 differentiation
3T3-L1 pre-adipocytes were cultured in DMEM supplemented with 1X penicillin-streptomycin solution (Corning) and 10% (v/v) fetal calf serum and differentiated with modifications to protocol described previously (27). To induce differentiation, 2-day post confluent pre-adipocytes (day 0) were treated with an induction media containing 1 mM dexamethasone, 0.5 mM isobutylmethylxanthine and 5 μg/ml insulin (all from Sigma) in 10% fetal bovine serum (FBS) supplemented DMEM. Two days later, induction media was replaced with 5 μg/ml insulin only until fully differentiated (day 12) with media changed every 2 days. Differentiation was confirmed by OilRed O stain and qRT-PCR quantification of pre-adipocyte and adipocyte marker gene expression (28).
Northern blotting for U7 snRNA
U7 snRNA was detected by Northern blotting as previously described using 5′ end labelled antisense oligonucleotides and 25 μg of total cell RNA (29). As an internal control the gel was stained prior to blotting and the intensity of 5S rRNA staining determined.
Biotinylated RNA pulldown
Cell lysates were diluted to 1 mg/ml protein in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 0.5% NP-40) and final concentration of EDTA was brought to 20 mM. For each experiment, 100 μl lysate was incubated with 10 μl of 10 μM biotinylated 30 nt stem-loop RNA (Dharmacon) for 1 h as previously described (30). The biotinylated RNA was recovered by binding to streptavidin-agarose beads followed by centrifugation. The unbound protein was saved for Western blot analysis. Beads were washed 4 times with 1 ml of NP-40 lysis buffer. Bound proteins were recovered in 25 μl of SDS loading buffer and resolved on an 8% SDS-PAGE gel for Western blot analysis.
Analysis of RNA-seq data
Reads from Illumina Human BodyMap Project were aligned to hg19 using MapSplice2 with default settings. UCSC gene annotations were downloaded and used to create transcriptome annotations for RSEM. We manually scanned the histone gene cluster to identify any genes expressed in the tissues, and confirmed that there were reads that included the stem-loop and downstream sequences, as well as junction spanning reads that defined the splice sites for all 10 histone genes in each of the human samples. We computed gene expression levels for UCSC genes using RSEM with the settings –estimate-rspd and –paired-end. RSEM aligns reads to a reference transcriptome in a way designed to find all possible genes that each read could have originated from and then uses a Bayesian network model to estimate the abundance of all genes simultaneously. This strategy is ideal for dealing with genes that may have many multimapped reads, as is the case with histone genes. For duplicated genes, RSEM computes a separate expression level for each annotated locus. The expression levels that RSEM reports are in units of FPKM (fragments per kilobase per million reads). Data from the cow and mouse tissues were provided by Jason Merkin and Chris Burge (31).
RESULTS
Histone genes express polyadenylated mRNAs in human tissues
There are two major histone gene clusters in mammals (32). The large HIST1 cluster contains more than 50 genes, including all of the RD-H1 proteins and the H2A.1, H2B.1 and H2B.2, H3.1 and H4 isoforms of core histone genes. The smaller cluster, HIST2, contains about 10 genes for core histones, including a central pair of duplicated histone H2A and H3 genes that encode the H2A.2 and H3.2 protein variants, and depending on the species, at least one H4 gene and 2–3 other H2A and H2B genes. The structure and gene order in both clusters is conserved in mammals, although there are examples of small differences due to creation of pseudogenes and differing extents of the duplication of the central gene pair in HIST2 in some species. In addition, there are three histone genes in the small HIST3 cluster, which are not expressed in somatic cells and a lone H4 gene, HIST4H4, expressed coordinately with the genes in HIST1 and HIST2, all of which have been conserved in mammals. All of these histone genes encode mRNAs that end in the stem-loop. Here, we analysed expression of histone mRNAs in human tissues and in tissues of two other mammals, mouse and cow, with sequencing data provided by Jason Merkin and Chris Burge (31).
The non-polyadenylated histone mRNAs are typically not present in the RNAs sequenced in high throughput sequencing projects as part of the analysis of gene expression. However, Illumina provided human sequences derived from total cell RNA (ribominus) from 15 adult tissues. The mouse and cow sequences were all polyA+ mRNA from terminally differentiated tissues. We also analysed expression of histone genes in human breast tumour tissue (both ribominus and polyA+) and normal adjacent breast tissue (poly A+) from the TCGA project at UNC.
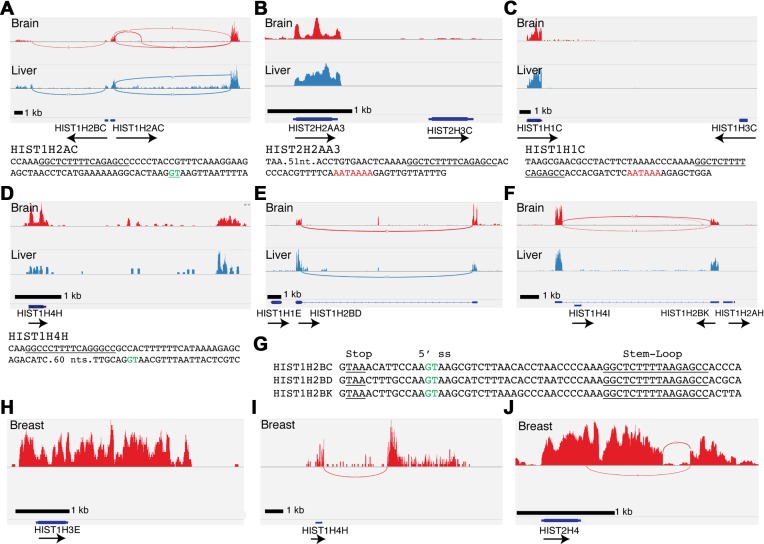
The histone mRNA expression pattern was similar in all 15 human tissues in the Illumina data set. There was essentially no histone mRNA expressed in tissues from most histone genes, as expected, since most of the tissues consisted largely of non-dividing cells. The ‘replication-independent’ histone genes, H3.3, H2A.Z, the polyadenylated form of histone H2A.X, histone H1O, and macroH2A, were expressed in all the tissues (Supplementary Figure S1). We were surprised there was expression of the same subset of RD-histone genes in all 15 terminally differentiated human tissues as polyadenylated mRNAs (Table 1). The ten expressed histone mRNAs were the HIST1H2AC, HIST1H2BC (Figure 1A), HIST2H2AA3 (Figure 1B), HIST1H1C (Figure 1 C), HIST1H4H (Figure 1D), HIST1H2BD (Figure 1E) and HIST1H2BK (Figure 1F), HIST1H3E (Figure 1H), HIST2H4 (Figure 1J) and HIST1H2BE (data not shown). Of these, HIST1H2BC, HIST1H2AC, HIST1H2BD, HIST1H2BK, HIST1H4H and HIST2H4 were expressed as spliced and polyadenylated mRNAs (Table 1). The HIST1H1C, HIST1H3E, HIST1H2BE gene and the duplicated HIST2H2AA3 genes were expressed as mRNAs that were polyadenylated, but not spliced. Screenshots of the expression of these genes are shown in Figure 1, together with examples of genes adjacent to HIST2H2AA3 (Figure 1B), HIST1H2BD (Figure 1E) and HIST1H2BK (Figure 1F) that were not expressed. In each case, there were multiple sequence reads extending through the stem-loop into the adjacent DNA, and they terminated following a polyadenylation signal downstream of histone stem-loop (Figure 1A–F, lower panels). This is in contrast to the polyA− histone RNAs expressed in HeLa cells (33,34) where the reads terminated before the stem-loop (the random primer used to prepare cDNA is not able to prime cDNA within the stem-loop).
Table 1. Polyadenylated Human histone mRNAs and their mouse orthologs.
| Human Gene | Is the mRNA spliced? | Does the mature mRNA retain its stem-loop? | FPKM | Mouse Ortholog | Is the mouse ortholog PolyA+? |
|---|---|---|---|---|---|
| Hist1H1C | No | Yes | 14.9 ± 3.6 | Hist1H1c | Yes |
| Hist1H2AC | Yes | Yes | 21.1 ± 5.5 | Hist1H2ac | No |
| Hist2H2AA3 | No | Yes | 9.2 ± 3.2 | Hist2H2aa1 | Yes |
| Hist1H2BC | Yes | No | 1.0 ± 0.5 | Hist1H2bc | Yes |
| Hist1H2BD | Yes | No | 8.6 ± 2.7 | Pseudogene | N.A. |
| Hist1H2BK | Yes | No | 29.5 ± 4.7 | Hist1H2bk | No |
| Hist2H2BE | No | Yes | 10.9 ± 3.7 | Hist2H2be | N.D. |
| Hist1H3E | No | Yes | 1.1 ± 0.3 | Hist1H3e | N.D. |
| Hist1H4H | Yes | Yes | 1.3 ± 0.2 | Hist1H4h | N.D. |
| Hist2H4 | Yes | Yes | 2.9 ± 0.9 | Hist2H4 | Yes |
For each human gene, table indicates whether or not mature polyA mRNA is spliced or whether the histone stem-loop is retained in the mature polyA mRNA. Mouse orthologs of human genes that produce polyadenylated mRNAs were identified and analyzed as to whether or not they produce polyadenylated mRNAs. *Two isoforms present in Human Hist1H2AC. FPKM ± Standard Deviation. N.D.= Not detected, N.A. = Not applicable.
Figure 1.
A subset of replication-dependent histone mRNAs are expressed in terminally differentiated tissues. (A–F) Sashimi plots RNA-Seq (ribominus) of total RNA from human tissues in the Illumina BodyMap project showing expression of the indicated histone genes in brain (Red) and liver (Blue) for (A) HIST1H2AC and HIST1H2BC; (B) HIST2H3C (not expressed) and HIST2H2AA3; (C) HIST1H3C (not expressed) and Hist1H1C; (D) HIST1H4H; (E) HIST1H1E (not expressed) and Hist1H2BD; (F) Hist1H4I (not expressed), Hist1H2BK and Hist1H2AH (not expressed). Note that there is no expression of adjacent histone genes. Sequences below sashimi plots indicate important sequence motifs; stem-loops(underlined), 5′ splice sites (green) and polyadenylation signals (red). (G) Sequence comparison of HIST1H2B genes whose mRNAs are spliced and polyadenylated. Conserved 5′ splice site (green) and stem-loop (underlined) are indicated. Note that the location of the 5′ splice site results in removal of the stem-loop from the polyadenylated mRNA. (H–J) Sashimi plots of normal human breast tissue from a representative TCGA sample for (H) HIST1H3E, (I) HIST1H4H and (J) HIST2H4.
The HIST1H2AC, HIST1H4H and HIST2H4 genes were expressed as polyadenylated mRNAs that were spliced after the normal 3′ end of the histone mRNA. Note the green highlighted sequence indicating the 5′ splice donor. This processing reaction results in mature mRNA that contains both a stem-loop and a polyA tail, similar to the structure of the polyadenylated histone mRNAs that were not spliced. The HIST1H2AC gene shares a promoter with the adjacent HIST1H2BC, which is also expressed as a spliced and polyadenylated mRNA (Figure 1A), albeit at lower levels than HIST1H2AC. Two other H2B genes, HIST1H2BD and HIST1H2BK, are also expressed as spliced and polyadenylated mRNAs (Figure 1E–F). The HIST1H2BK gene was adjacent to the HIST1H2AH gene and the HIST1H2BD was adjacent to the HIST1H1E gene, neither of which was expressed (Figure 1E–F). In contrast to HIST1H2AC, the spliced and polyadenylated H2B genes produced mature mRNAs that lacked the stem-loop, because the 5′ splice donor site is located between the stop codon and stem-loop (Figure 1G, green highlighted sequence). Note that the H2B mRNAs will not be targets for nonsense-mediated decay (NMD). Efficient NMD requires that the stop codon be >55 nts from the exon-junction and that the efficiency of NMD increases only as the distance between the stop codon and multiple exon junctions increase (35,36). The distance between the stop codon and 5′ splice site is >55 nts for HIST1H4H (Figure 1D) and Hist2H4 and this may contribute to the low levels of expression of these genes among the spliced histone mRNAs (Table 1, c.f. HIST1H4H and HIST1H2AC, HIST1H2BC, HIST1H2BK, HIST1H2BD).
To further confirm the expression of these genes in normal human tissue, we analysed sequencing data from the TCGA project at UNC-Chapel Hill, where adjacent normal breast tissue as well as breast tumour tissue was sequenced, and where the sequencing was done to a deeper depth. The data from normal tissue confirmed the results from the data obtained from Illumina, and similar levels of expression of the polyadenylated histone mRNAs were found in each sample (Table 1). In this data set, we identified a single histone H3 gene, HIST1H3E, which was expressed at a similar level as the HIST1H4H gene (Figure 1H), each with an FPKM of about 1, compared to the level of 10–30 FPKM found for the H2A, H2B and HIST1H1C genes. Analysis of this data set also demonstrated sequencing reads that span splice junctions for the two expressed H4 genes, HIST1H4H and HIST2H4 (Figure 1I and J). The histone H2A and H2B mRNAs were expressed at a significantly higher level than the histone H4 or H3 mRNAs. The H3/H4 protein tetramer is much more stable than the H2A/H2B protein dimer (12); therefore, it is likely that it does not require as high expression to maintain proper levels within chromatin. In total, 10 of the 65 histone genes in the HIST1 and HIST2 clusters were expressed in tissues as polyadenylated mRNAs (Table 1).
Similar sets of histone mRNAs are expressed in tissues of other mammals
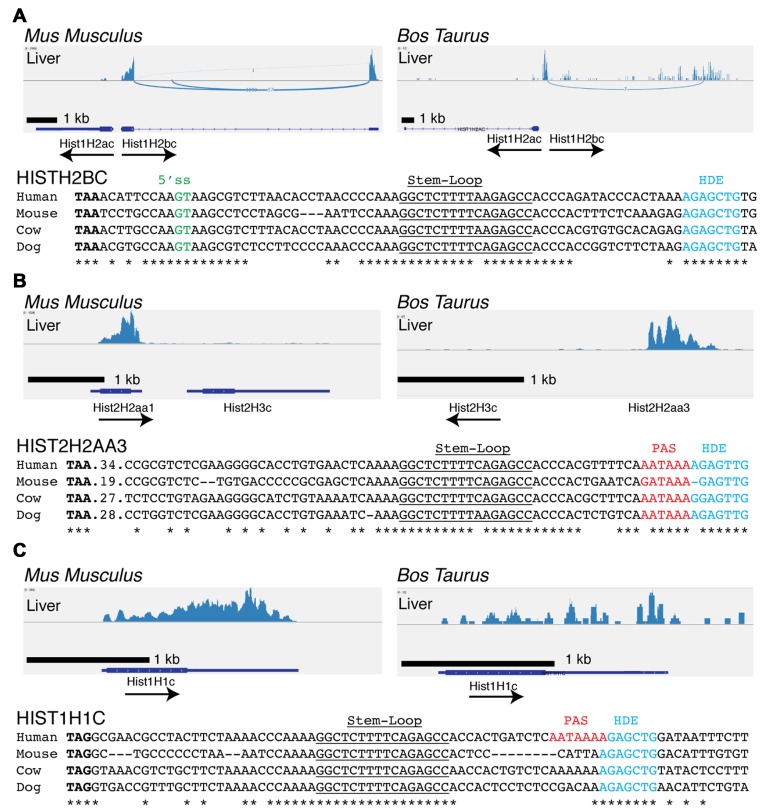
We examined the expression of histone genes from the tissues of two other mammals, mouse and cow, using sequencing data obtained from Jason Merkin and Chris Burge (31) (GSE41637). These data contained only sequences from polyadenylated mRNAs, but were sufficient for us to identify the replication-dependent histone mRNAs that were expressed as polyadenylated mRNAs in these non-dividing tissues (Figure 2). Since the mouse core histone genes are over 95% conserved at the nucleotide level (37,38), it was not possible to unambiguously map reads containing only coding region sequences to individual genes. However, the polyadenylated mRNAs could be mapped because of the unique sequences in the 3′ UTR before and after the stem-loop. Like the human histone genes, the cow histone genes have enough differences in the coding regions to assign sequencing reads to individual histone genes. Although the histone gene cluster was not completely annotated in the cow genome, we determined that its organization was the same as the mouse and human cluster and identified the orthologues of each of the genes expressed in humans. Note that the naming convention for all human genes has the human genes capitalized (e.g. HIST1H2BC) and the orthologous genes in other mammals (i.e. mouse and cow) genes in lower case (e.g. Hist1H2bc) (32).
Figure 2.
Polyadenylation of a subset of histone mRNAs is conserved in mouse and cow. (A) Sashimi plots of the expression of the Hist1H2bc gene and Hist1H2ac gene from the mouse (left) and cow (right) liver. Below are the sequences of the Hist1H2bc 3′ end from four mammals showing conserved 5′ splice sites (green), stem-loop (underlined), HDE (blue) and stop codon (Bold). *'s mark invariant nts. (B) Sashimi plots of the expression of the Hist2H2aa1 and Hist2H3c gene from the mouse (left) and cow (right) liver. Below is the sequence comparison of the HIST1H2AA3 3′ end showing conserved polyadenylation signal (red), the stem-loop (underlined) and the HDE (Blue). (C) Sashimi plots of the expression of the HIST1H1C gene from the mouse (left) and cow (right). Below is the sequence comparison of the HIST1H1C 3′ end. Note that the polyadenylation signal used in human is not conserved in mouse, cow or dog but the gene is expressed from a poly(A) signal further downstream resulting in a longer 3′UTR (41).
The orthologues of the HIST1H2BC (Figure 2A), HIST2H2AA3 (Figure 2B) and HIST1H1C (Figure 2C) genes were expressed in both cow and mouse brain (data not shown) and liver (Figure 2). The Hist1H2bd gene was also expressed in cow, but not in mouse where it is a pseudogene (32). The HIST1H2BK gene is the most highly expressed H2B gene in human tissues (Table 1), but it's orthologue, Hist1H2bk, is not expressed in mouse or cow tissues as the 5′ splice site in the Hist1H2bk gene is less well conserved throughout evolution and is not conserved in mouse or cow (Supplementary Figure S2). As in humans, the Hist1H2bc gene is found in a gene pair with Hist1H2ac in mouse and cow. In humans, both mRNAs are spliced and polyadenylated, but expression of the Hist1H2ac gene was not detected in either mouse or cow (Figure 2A). The sequencing depth for the mouse and cow tissues was lower than that for the human tissues. If the mouse and cow Hist1H2ac genes were expressed at similar levels to the human orthologue, we would have detected that expression, since the human HIST2H2AC gene expression was similar to the HIST2H2AA3 gene whose expression was readily detected in the mouse and cow samples. We certainly cannot rule out low levels of expression of the mouse and cow Hist1H2ac genes. Note that the histone HIST1H3E and HIST1H4H and HIST2H4 genes are expressed at very low levels in the human tissues (∼10% of the H2a and H2b genes, Table 1). The sequencing depth in the cow and mouse tissues was not sufficient to determine whether or not they were expressed in these tissues, and only the much deeper TCGA sequencing data compared to the Illumina data, allowed us to quantify their expression.
The splice site conserved in the human HIST1H2B genes (Figure 1G) is also conserved in the two cow genes (Hist1H2bc and Hist1H2bd) and in the mouse Hist1H2bc gene (Figure 2A). Examination of the sequenced mammalian genomes revealed that the 10 nts surrounding the 5′ splice site in the 3′ UTR in the HIST1H2BC and HIST1H2BD sequences were highly conserved across mammalian species (Supplementary Figure S2A and B). The sequence of the HIST1H2BK 3′UTR in primates was similar to the human HIST1H2BC and HIST1H2BD genes, but was totally dissimilar in other mammals (Supplementary Figure S2C).
The mouse Hist2h2aa1 and cow Hist2H2aa3 genes are orthologues of the human HIST2H2AA3 gene, and have a conserved polyadenylation site in the HDE (Figure 2B, red). This gene is also expressed as a polyadenylated mRNA in mouse round spermatids (39). The HIST1H1C gene was also expressed in tissues from all three species (Figures 1C and 2C). In humans, the polyA signal is immediately after the stem-loop, within the HDE (Figures 1C and 2C). However, in mice the polyA site is more than 1 kB 3′ of the stem-loop (25,40,41). Thus, the expression of the HIST1H2BC, HIST1H2BD, HIST2H2AA3 and HIST1H1C genes has been conserved throughout mammalian evolution, and they function in mammals to provide histone mRNAs expressed in terminally differentiated cells in addition to expressing non-polyadenylated mRNAs that are cell-cycle regulated in growing cells.
Validation of polyadenylated histone mRNA expression in adult mouse liver
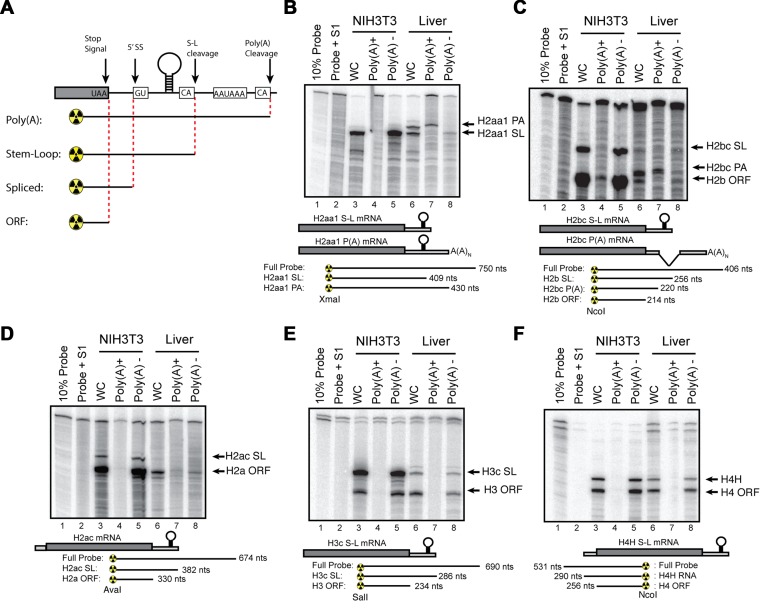
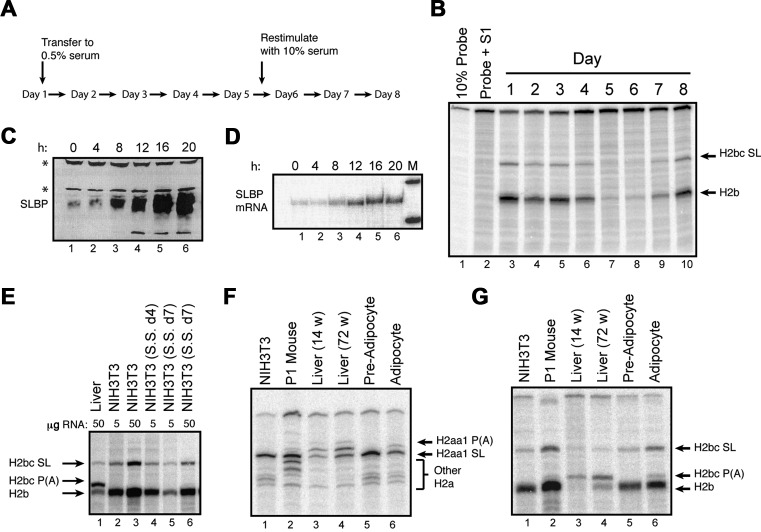
To confirm the expression of the mouse histone mRNAs detected by high-throughput sequencing we analysed histone gene expression in actively growing cultured fibroblasts (NIH3T3 cells) and liver tissue from 72-week-old mice. The liver is a relatively homogenous tissue with hepatocytes making up more than 70% of the cells. In healthy adult livers, the vast majority of these hepatocytes are quiescent, only re-entering the cell cycle after injury. Unlike the human histone genes, the coding regions of the mouse histone genes are highly conserved (>95%) at the nucleotide level (37). Because of the conserved ORF sequences, we can use an S1 nuclease mapping assay to simultaneously analyse both the mRNA expressed from a specific gene, as well as the mRNAs from all the other histone genes encoding that histone protein (26,42). A probe labelled in the coding region will protect all the histone mRNAs to a region close to the stop codon or initiation codon, as well as a band from the specific gene extending to the 3′ end or the 5′ end of the transcript, depending on whether one is probing the 3′ or the 5′ end of the mRNA (Figure 3A). This highly sensitive method allows for mapping of multiple isoforms of mRNA from a single sample. Further, since all of the radioactivity is at the end of the probe, the relative intensity of the bands gives the relative amounts of the transcripts. A generalized schematic is shown in Figure 3A. In each experiment below, the different protected fragments for a given probe are diagrammed below the autoradiogram.
Figure 3.
Expression of histone mRNAs is mouse fibroblasts cells and mouse liver. (A) Schematic of generalized S1 nuclease protection assay performed on mouse histone mRNA. A generalized histone mRNA is depicted indicating possible 5′splice sites (5'ss), stem-loop cleavage sites (S-L cleavage) or poly(A) cleavage sites. S1 nuclease protection probes will protect a given mRNA to the point that cleavage or splicing occurs (indicated below). Note that due to the high degree of conservation in mice, all of the mRNAs for a specific histone class (i.e. H2B.1 or H2A.2) will be protected up to the stop codon. (B–F) Total cell RNA from NIH3T3 cells and mouse liver was fractionated into polyA+ and polyA− fractions. Total RNA (whole cell (WC)) and poly(A)+ and poly(A−) RNA were probed in an S1 nuclease protection assay. Five micrograms of total 3T3 cell RNA and 50 μg of liver RNA was analysed using the S1 nuclease protection assay of a 3′ labelled fragment at the indicated restriction enzyme site for (B) Hist2H2aa1, (C) Hist1H2bc, (D) Hist1H2ac, (E) Hist2H3c or (F) Hist2H4H. The probe used for histone H4H was labelled at the 5′ end of the indicated restriction site. For each experiment, below autoradiogram, possible mRNA isoforms are indicated as well as protected fragments and sizes for S1 nuclease probes.
Total RNA from NIH3T3 cells and mouse liver was fractionated into polyA− and polyA+ RNA using oligo-d(T) cellulose. Orthologues of human histone HIST1H2BC (Hist1H2bc), HIST2H2AA3 (Hist2H2aa1) and HIST1H4H (Hist1H4h), which were expressed in human tissues, and the HIST2H3C (Hist2H3c) gene, adjacent to the Hist2H2AA3 gene, which was not expressed in human tissues, were analysed by S1 nuclease mapping, using 5 μg of total RNA from exponentially growing NIH3T3 cells and 50 μg of total liver RNA (Figure 3). In actively proliferating NIH3T3 cells, most of the 3′ S1 probes map two protected fragments: one extending to the 3′ end of the mRNA ending in the stem-loop and a second fragment, mapping near the stop codon, due to the high level of conservation between the open reading frames of all mouse histone genes of a given class (26). This band is indicated by the name of that class of histone protein (e.g. H3.1 or H2a.1). If the histone mRNA is spliced or polyadenylated then the probe is protected up to the point where the mRNA sequence diverges from the probe (Figure 3A; e.g. at the 5′ splice site in Hist1H2bc, before the 3′ end of the mRNA or at the polyA addition site in Hist2H2aa1).
The S1 nuclease protection assays confirmed the low levels of histone mRNAs in liver and there were easily detectable amounts of the polyadenylated forms of Hist2H2aa1 (Figure 3B, lane 7) and Hist1H2bc (Figure 3C, lane 7) mRNAs. There were no polyadenylated mRNAs detected in the NIH3T3 cells. There is some non-polyadenylated histone mRNA detected in the liver that gives the same pattern of S1 nuclease protected fragments as the NIH3T3 cell mRNA. This likely arises from a low level of expression of the histone mRNAs expressed in growing cells. Note that the non-polyA mRNA expressed in mouse liver is greatly reduced compared to NIH3T3 cells, and we used 10 times more RNA (50 μg versus 5 μg) for assay of liver RNA, compared to neonatal mice or growing 3T3 cells. We assayed the same RNAs with probes for Hist1H2ac (Figure 3D), Hist2H3c (Figure 3F) and Hist1H4h (Figure 3E) genes. There was no polyadenylated mRNAs detected from these genes in mouse, consistent with the sequencing data.
Differential transcription of individual histone genes in the adult mouse liver
Persistent expression of this subset of histone genes might be due to continued transcription of only these specific genes following exit from cell cycle. To investigate this possibility, we analysed GRO-Seq data sets performed by Fang et al. on adult mouse liver (43) and by Min et al. on mouse embryonic stem cells (mESCs) and mouse embryonic fibroblasts (MEFs) (44). In GRO-Seq, the position of RNA polymerases capable of elongating already initiated transcripts is determined. Many genes have RNA polymerase II bound and paused at the promoter after synthesis of a short (∼50 nt RNA). These RNAs are elongated during the GRO-Seq experiment as are any RNA polymerases that were elongating transcripts when the nuclei were isolated (45,46). A major regulatory point in transcription is the transition to processive transcription when P-TEFb action releases the stalled RNA polymerase. For genes transcribed at a low rate, there is a prominent GRO-Seq peak where the polymerase pauses, and for genes transcribed at a high rate there is a smaller peak and transcription throughout the gene (45,47). There are also genes that have a paused polymerase which do not express RNA. The level of expression of the histone genes encoding polyadenylated mRNAs is low in mouse liver compared to their rate of expression in growing cells, and one would expect this to be reflected in the GRO-Seq data.
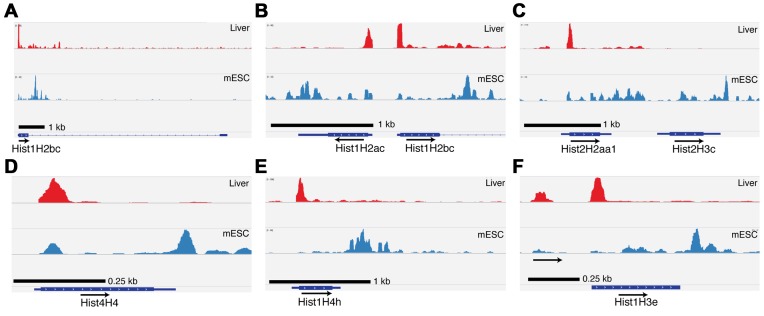
In mouse liver, all the histone genes that express polyadenylated mRNAs (Table 1) contained sequence reads corresponding to active transcription (e.g. Hist1H2bc (Figure 4A) and Hist2h2aa1 (Figure 4C)) with prominent peaks near the start site of transcription, and small amounts of transcription detected in the gene body, characteristic of relatively lowly expressed genes (46). In contrast, these genes were highly expressed in actively growing mESC (Figure 4, lower blue panels) or MEFs (data not shown), with transcribing polymerases detected throughout the gene characteristic of highly active genes, as well as a peak of activity near the 3′ end where the polymerases would be terminating. In the liver there were genes that were not active as detected by GRO-Seq, (e.g. Hist2H3c), while the adjacent gene was transcribed (e.g. Hist1H2aa1, Figure 4C). The Hist1H2ac gene gave a GRO-Seq signal similar to the Hist1H2bc gene (Figure 4B), even though mRNA was only produced from the Hist1H2bc gene. Note that it is not possible using GRO-Seq to distinguished a gene expressed at a low level, from a gene that has a stalled polymerase and does not express mature mRNAs, since in a low expressed gene, the polymerase is stalled at the promoter most of the time. Surprisingly, there were many genes that were active based on the GRO-Seq signal, but for which we did not detect as accumulating histone mRNAs by RNA-Seq or the S1 nuclease protection assay, e.g. Hist1H2ac, (Figure 4B) and Hist4H4, Hist1H4h and Hist1H3e (Figure 4D–F). Overall 80% of the histone genes had an active GRO-Seq signal at the promoter, while only 20% had no GRO-Seq signal. Thus, surprisingly the promoters of most of the histone genes in non-dividing tissues were ‘active’ but did not accumulate RNA. Whether this is due to a lack of processing signals, resulting in rapid degradation of the transcript, or failure of the polymerase to enter productive elongation is not clear. Release of paused polymerases is clearly a major regulated event on many genes in mammalian cells (45,47). We verified these findings by analysing RNA Polymerase II ChIP-Seq data sets performed by Sun et al. on mouse liver and brain (not shown) (48). These data confirmed data found by GRO-Seq; however, experimental limitations of their ChIP-Seq only define a region ∼400 nts at which RNA Pol II was engaged. Therefore, these data were not as precise, but were consistent with GRO-Seq data.
Figure 4.
Paused polymerases are present on many histone genes in mouse liver. We analysed the GRO-Seq data for mouse liver (43) and from mouse embryonic stem cells (mESC) and mouse embryo fibroblasts (44)(not shown). The data for selected histone genes are shown in panels A–F for liver (top) and mESC (bottom). (A) GRO-Seq reads for Hist1H2bc demonstrate a low level of transcription in mature mouse liver and in mESC. In the liver there is a large peak of paused polymerase and a low level of transcription within the gene. Note that characteristic of the highly expressed genes in the mESC, there is a large peak at the 3′ end of the gene, but smaller peak at the 5′ end of the gene. (B) GRO-Seq reads at the 5′ end of the Hist1H2ac suggest engagement of polymerase at the promoter in both liver and mESC. However, Hist1H2ac mRNA does not accumulate in mouse liver (Table 1). Note that reads throughout the ORF and 3′UTR of the gene are absent in liver in contrast to mESC where it is actively transcribed. (C) GRO-Seq reads for the Hist2H2aa1 gene shows a low level of expression in the liver, and a high level of expression in the mESC. In contrast Hist2H3c is not expressed in the liver (there is no paused polymerase) but shows active transcription in the mESC cells. (D–F) (D) Hist4H4, (E) Hist1H4h and (F) Hist1H3e have a paused polymerase in the liver and active transcription in the mESC cells.
Differentiation, but not exit from the cell cycle, results in expression of polyadenylated histone mRNAs
The switch to production of polyadenylated histone mRNAs derived from a few genes could result from exiting the cell cycle, or could be part of the process of terminal differentiation. To distinguish between these two processes mouse NIH3T3 fibroblasts were arrested either by serum starvation or 3T3-L1 fibroblasts were arrested and induced to terminally differentiate into adipocytes. We arrested NIH3T3 cells by serum starvation (Figure 5A), causing them to enter a ‘G0’ state. Arrested cells can be stimulated to re-enter the cell cycle by addition of 10% FBS, and they enter S-phase 12–16 h later. There is a large change in histone mRNA levels between cells stimulated to enter S-phase and the arrested cells (49). Upon arrest by serum starvation, levels of Hist1H2bc and other H2B.1 mRNAs were reduced as has been previously reported (Figure 5B) (49). SLBP protein is barely detectable (Figure 5C, Lane 1), while SLBP mRNA is reduced only 4- to 5-fold (Figure 5D), characteristic of the post-transcriptional regulation of SLBP during the cell cycle (9). Serum starvation of NIH3T3 cells leads to a 90% reduction of Hist1H2bc levels as determined by comparing the intensity of the protected bands from 5 micrograms of RNA from growing cells with 50 micrograms of RNA from the arrested cells (see Figure 5E, cf. lanes 2 and 6). There was no detectable polyadenylated mRNA expressed from the H2bc gene in the arrested cells (Figure 5E, compared to the abundant polyadenylated H2bc mRNA in the same amount of liver mRNA (Figure 5E, lane 1). Note that the ratio of the other H2b.1 mRNAs (the protected fragment at the termination codon) and the specific Hist1H2bc mRNA ending at the stem-loop did not change upon serum starvation. SLBP protein levels rapidly increase as cells re-enter S-phase (Figure 5D, Lanes 2–6) with the maximum number of cells in S-phase 16–20 h after serum stimulation. Therefore, the reduction of SLBP levels is not itself a determinant for the switch from histone mRNAs ending in a stem-loop to those ending in a polyA tail.
Figure 5.
Differentiation but not cell cycle arrest activates polyadenylation of histone mRNA. (A) NIH3T3 cells were arrested in G0 by serum starvation and stimulated to reenter the cell cycle by the addition of 10% FBS as previously described (7). (B) Five micrograms of total cell RNA from the indicated time points was analysed by S1 nuclease mapping for the Hist1H2bc mRNA. Lane 7 is 12 h after addition of serum and lane 8 is 30 h after addition of serum. Serum starvation did not result in production of spliced Hist1H2bc mRNA (see also panel E). (C and D) Levels of SLBP protein (panel C) and mRNA (panel D) were determined in cells serum starved for 6 days (lanes 1) and over 20 h after restimulation with serum (lanes 2–6) by Western blotting and RNase protection assays, respectively. Levels of SLBP protein reduced >20-fold in cells serum-starved for 6 days, while levels of SLBP mRNA (panel D) were only reduced 4-fold. The levels of both SLBP protein and SLBP mRNA increased starting 8 h after re-addition of 10% serum. The * (panel C) indicate cross-reacting proteins whose levels don't change. (D) The RNase protection assay protects a 276 nt fragment of SLBP mRNA. The markers are 350 nt and 242 nt DNA fragments. (E) S1 nuclease protection assays of Hist1H2bc mRNA in 50 μgs of mouse liver RNA (lane 1), 5 or 50 μgs of RNA from growing NIH 3T3 cells (lanes 2,3), 5 μgs of RNA from cells starved for 4 days (lane 4) and 5 μgs or 50 μgs of RNA from cells serum starved for 7 days (lanes 5 and 6). Note that the most abundant H2b mRNA in liver is the polyadenylated H2b mRNA. (E and F) Hist2Haa1 mRNA (panel E) and Hist1H2bc mRNA (panel F) were analysed by S1 nuclease protection assay in actively growing pre-adipocyte 3T3-L1 cells and differentiated adipocyte 3T3-L1 cells. Lane 1: 5 μg 3T3-L1 RNA; Lane 2: 5 μg neonatal (P1) mouse RNA; Lane 3, 50 μg 14 week liver RNA; Lane 4: 50 μg 72 week liver RNA; Lane 5: 5 μg pre-adipocyte RNA; lane 6. 50 μg adipocyte RNA. Results demonstrate the production of polyadenylated (E) or spliced and polyadenylated (F) mRNA after differentiation.
An alternative possibility is that terminal differentiation, rather than exiting the cell cycle, triggers the switch from the utilization of canonical histone 3′ end processing signals to polyadenylation signals. Kari et al. demonstrated that when human fibroblasts are differentiated into adipocytes in vitro, there is polyadenylation of the human HIST1H2BC, D and K mRNAs and the HIST1H2AC mRNA, although they did not determine the fraction of these mRNAs that were polyadenylated (24), or whether there was also polyadenylation of these RNAs in fibroblasts that were arrested but not differentiated. To test whether growth arrest or differentiation of the cells led to polyadenylation of histone mRNA, we compared histone gene expression in serum-starved 3T3 fibroblasts and 3T3-L1 fibroblasts differentiated into adipocytes (50). Like the NIH3T3 cells, the pre-adipocyte cells expressed solely stem-loop isoforms of Hist2H2aa1 and Hist1H2bc (Figure 5F and G, lane 5). After seven days of differentiation, the cells had exited the cell cycle and differentiated into adipocytes as indicated by the presence of oilRED droplets in the cells (Supplementary Figure S3A), decrease of expression of Pref-1, and induction of expression of Ppar2γ (51)(Supplementary Figure S3B). There was a large reduction in histone mRNA levels in the differentiated cell cultures compared to the pre-adipocytes (Supplementary Figure S3C, cf. lane 1 and 3). We used 10 times more RNA when probing adipocyte versus pre-adipocytes. As reported by Kari et al. (24), after differentiation, adipocytes began producing polyadenylated isoforms of these two mRNAs (Figure 5F and G, cf. lanes 5 and 6). The detection of this band was not due to the increased loading of mRNA, as we did not detect it in pre-adipocyte RNA when 50 μg was analysed (Supplementary Figure S3C, lane 2). Less polyadenylated mRNA was produced compared to the mRNA from the same genes in mouse liver, and there was still some normally processed Hist2H2aa1 and Hist1H2bc mRNA produced, likely as a result of not all cells entering terminal differentiation. Thus, the process of terminal differentiation may result in altering the pathway of histone mRNA expression, to activate expression of H2A and H2B mRNA isoforms that are normally expressed in terminally differentiated adult tissues.
Adult tissues express specific proteins and mRNAs required for histone mRNA biosynthesis
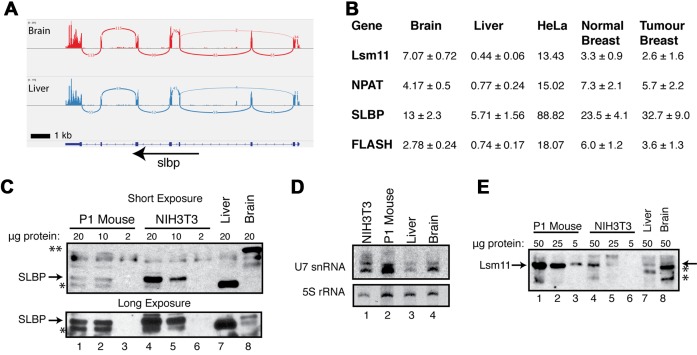
There are several genes involved specifically in histone mRNA metabolism and the expression of these genes has not been studied in terminally differentiated tissues. They include SLBP, which binds the stem-loop and is associated with histone mRNA in all steps of histone mRNA metabolism; Lsm10 and Lsm11, proteins found only in the Sm ring of U7 snRNP (52,53); NPAT, a factor required for histone gene expression (54–56) that is concentrated in the Histone Locus Body (HLB), and FLASH, which binds to the Lsm11 protein and is essential for histone pre-mRNA processing (57). FLASH is also involved in FAS-casp8 dependent apoptosis, but this may be connected to its role in the HLB, since an aspect of this process is re-localization of FLASH from the HLB to the mitochondria (58). Previously, the Schümperli lab showed that human tissues expressed SLBP mRNA based on a tissue Northern blot, but did not determine if SLBP protein was present (59). We confirmed that SLBP mRNA is expressed in mouse (Figure 6A) and in human (Supplementary Figure S4E) liver and brain. In addition Lsm10, Lsm11, FLASH and NPAT mRNAs are expressed in both human (Supplementary Figure S4) and mouse (Supplementary Figure S5) liver and brain, as well as all the other human tissues in the Illumina BodyMap Project.
Figure 6.
Expression of Histone Specific factors in mammalian tissues. (A) Sashimi plot demonstrating continued expression of SLBP mRNA in mouse brain (top) and liver (bottom) from (31) (B) RNA-Seq data from 3 replicates of mouse liver and mouse brain (31) and 4 sets of paired human normal breast and breast tumour tissue were analysed for levels of expression of Lsm11, NPAT, SLBP and FLASH. For the 4 human and 3 mouse samples, the average plus standard deviation is indicated. Note the expression levels of all the factors except SLBP were slightly higher in normal tissue. The expression levels of SLBP were higher in the tumour tissue (P = 0.06) in a Student's t-test. (C) Analysis of SLBP by Western blotting in cells and tissues from neonatal (P1) mouse (lanes 1–3), 3T3 cells (lanes 4–6) and mouse liver (lane 7) and brain (Lane 8). The amount of protein loaded is indicated above each lane. Tissue specific cross-reacting bands are indicated (*, liver and **, brain) (D) Analysis of U7 snRNA levels in cells and tissues. Twenty five micrograms of total cell RNA from 3T3 cells, a neonatal (P1) mouse, liver and brain were resolved on a 15% urea-polyacrylamide gel and transferred to a nylon membrane. Membranes were probed for U7 snRNA and visualized by exposure to phosphor screen. The gel was stained with ethidium bromide and photographed to determine the relative amounts 5S rRNA. (E) Analysis for Lsm11 by Western blotting in cells and tissues. The indicated amounts of the same extracts used in panel C are shown. Note the high levels of Lsm11 in the P1 mouse (arrow) consistent with the high amount of U7 snRNA in the P1 mouse.
We also quantified the expression data of these factors in mouse liver and brain and in the TCGA data from human breast cancer and the adjacent normal tissue (Figure 6B). Both the normal and tumour tissues expressed the mRNAs for all the factors involved in histone mRNA biosynthesis. There were increased levels of SLBP mRNA relative to the expression of the other factors in the tumour compared to the normal tissue. Lsm11, NPAT and FLASH mRNAs were also expressed in normal tissue, at a similar level in both normal and tumour tissue, with the normal tissue slightly higher. The mRNAs for all the factors were expressed at higher levels in cultured HeLa cells.
The expression of many proteins, including SLBP, is regulated post-transcriptionally (9), and the levels of mRNA do not necessarily reflect the amount of protein present. In cultured cells, SLBP protein is tightly cell-cycle regulated by proteolysis and translational regulation, and there are only small changes in SLBP mRNA level, while U7 snRNP is present throughout the cell cycle (9). We analysed the amounts of SLBP protein, U7 snRNA and Lsm11 protein in mouse liver. SLBP was readily detected in lysates from exponentially growing NIH3T3 fibroblasts and from neonatal (P1) mice (Figure 6C, lanes 1–6). Consistent with SLBP being present in high levels only in S-phase cells, SLBP was not detected in the liver lysate by Western analysis, and was present at very low levels, if at all, in the brain lysate (long exposure; Figure 6C, lanes 7,8, lower panel). There was a prominent band detected by the α-SLBP antibody in the liver and in the NIH3T3 lysate which migrated slightly faster than SLBP (indicated by (*) in Figure 6C) and another band in brain lysate migrating much slower (indicated by **). However, these cross-reacting bands do not bind the histone stem-loop in an RNA affinity purification assay, while SLBP does (Supplementary Figure S6), indicating that they are cross-reacting proteins.
We also assayed for both U7 snRNA by Northern blotting (Figure 6D) and the U7 snRNP specific protein, Lsm11, by Western blotting (Figure 6E), in mouse liver, brain, NIH3T3 cells and the neonatal (P1) mouse. The levels of U7 snRNA and the U7 core protein Lsm11 in the P1 mouse were similar to those in NIH3T3 cells, consistent with many of the cells in the mouse rapidly proliferating. The amount of U7 snRNA in mouse liver and brain was lower than the P1 mouse (Figure 6D, lanes 2–4), but, unlike SLBP, was still readily detectable and Lsm11 was also detectable at low levels (Figure 6E, lanes 7 and 8). The mRNAs for U7 snRNP components Lsm10 and Lsm11 were also present in adult tissues (Figure 6B; Supplementary Figures S4 and S5). These results strongly suggest that there is U7 snRNP present in terminally differentiated cells.
DISCUSSION
Polyadenylation of histone mRNAs allows for expression of histones outside of S-phase
Following terminal differentiation, it is necessary to replace histones as they are lost, since the proper ratio of DNA and histone proteins is critical for genomic stability (60,61). The average age of many epithelial cells in the human body is several years with neuronal cells living even longer (62). In these cells, there must be a mechanism to replace lost histones that have a half-life of weeks to months (11,12). In continuously growing cells, there are small amounts of replication-dependent histone mRNAs expressed in G1 cells (7) and in fibroblasts arrested in G0 there are also small amounts of histone mRNA processed at the stem-loop (Figure 5B) (49).
In the data from the Illumina BodyMap project, in which total RNA (ribominus) was sequenced and hence both polyA+ histone mRNA as well as any normally processed histone mRNAs were sequenced, we did not detect histone mRNA processed at the stem-loop from histone genes that are normally actively expressed (e.g. HIST2H3C or HIST1H1E, Figure 1), although a subset of histone genes expressed polyadenylated RNAs. In contrast, ENCODE data from ribominus RNA samples from growing cells there are large amounts of replication dependent histone mRNAs expressed and very small amounts of polyadenylated histone mRNAs were expressed (33,63).
A variety of mechanisms might be utilized for synthesis of different histone proteins in terminally differentiated tissues. One obvious mechanism to provide replacement histone is to utilize different genes to encode the replacement histones. This is the case for the histone H3 genes, but not the other core histone genes. Previously, H3.3 (H3F3A, H3F3B) had been identified as a replacement histone that accumulates as a large fraction of the histone H3 in non-dividing cells over an extended period of time (months to years) (64), as well as playing important functions in gene regulation in growing cells. The H3.3 protein is expressed from genes that produce a spliced polyadenylated mRNA that is not linked to the replication dependent histone gene clusters (65,66). Histone H1O has been suggested to function as a replacement histone for H1 proteins, and again, this gene is expressed as a polyadenylated mRNA lacking a stem-loop and is not linked with the replication-dependent histone gene cluster (67,68).
For the other 3 classes of core histone genes, there are no obvious replacement histone genes, although there are variant proteins for histone H2A. All of the polyadenylated H2A variants (H2A.X, H2A.Z, macroH2A) have distinct functions. No H2B or H4 genes have been identified outside of those in the replication-dependent clusters. We propose that the small subset of the replication-dependent genes that are expressed as polyadenylated mRNAs in terminally differentiated cells encode the replacement H2A, H2B and H4 histone proteins. We found both RD-histone H2A and RD-histone H2B genes expressed in all tissues. Most of these mRNAs contain both a stem-loop and a polyA tail, similar to the H2A.X mRNA expressed outside of S-phase. The exceptions are the HIST1H2BC, HIST1H2BD and HIST1H2BK mRNAs, in humans, and Hist1H2bc and Hist1H2bd mRNAs in other mammals. These spliced H2B mRNAs have a conserved structure, using a 5′ donor site before the stem-loop, resulting in an mRNA that does not contain the stem-loop and is not a target for nonsense mediated decay. The 3′ UTR and 5′ splice site are conserved among all these H2B genes, even though they encode slightly different proteins. The other expressed human H2B gene, HIST2H2BE, is found in the HIST2 cluster, and this gene encodes a polyadenylated mRNA that still contains the stem-loop, similar to HIST1H1C or HIST2H2AA3.
Recently, it has been shown that over a six-month period in mouse brain, that histone H3.1, H3.2 and H4 are significantly more stable than histone H2A and H2B (12). This observation suggests that the H2A-H2B protein dimer has a shorter half-life than the H3-H4 tetramer (12). Therefore, H2A and H2B genes would need to be expressed at higher levels to compensate for the increased turnover. There is an isolated RD-H4 gene on human chromosome 11, but this HIST4H4 gene although conserved as an isolated gene in mammals, is expressed only as a stem-loop RNA (32), and is not expressed in non-dividing tissues. We identified two possible replacement histone H4 genes, which were expressed at very low levels (∼5- to 10-fold less than the H2A and H2B genes), in all human tissues we analysed. Since there have been no other histone H4 genes or cDNAs identified other than the RD-histone H4 genes, these two genes likely provide the replacement histone H4 protein. The depth of sequencing in the cow and mouse samples was not sufficient to detect a similar level of expression from the H4 genes in these species.
Histone H3.3 has long been recognized as a replacement histone, which replaces most of the histone H3 in terminally differentiated cells. However, a significant amount of the H3.2 protein remains in non-dividing cells in the mouse (14) (see Figure 7). We identified one RD-histone H3 gene, HIST1H3E, which is expressed at low levels in terminally differentiated human cells, similar to the level of the HIST1H4H gene. This is consistent with the possibility that there is a small amount of ‘replacement’ histone H3 encoded by this replication-dependent gene.
Figure 7.
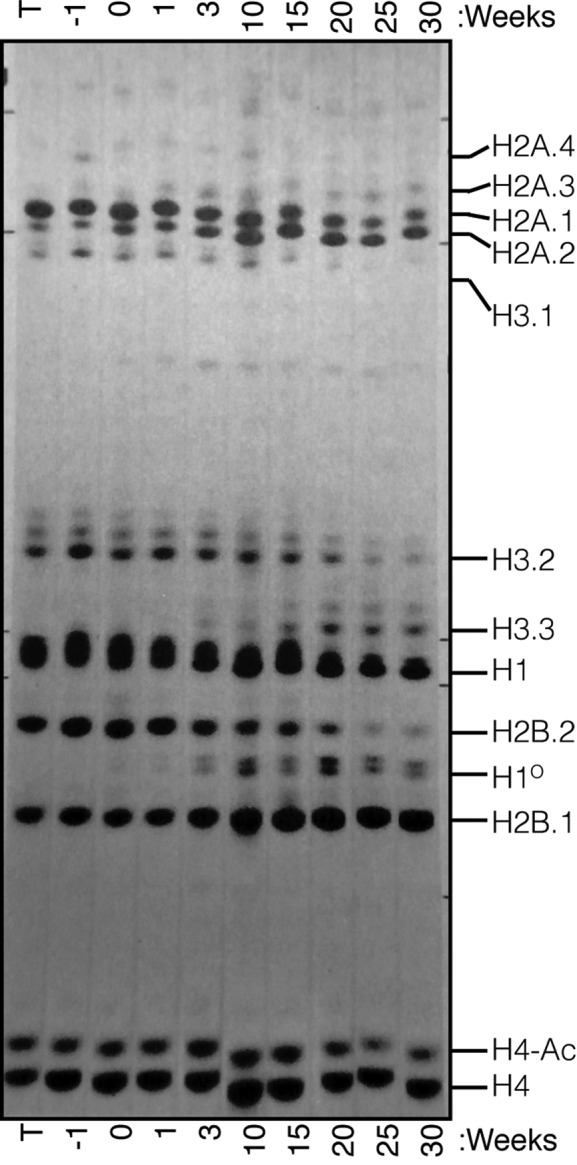
Expression of Histone proteins in mouse liver. Total histones were isolated from purified nuclei were prepared from the livers of mice from 1 week before birth (−1) to 30 weeks of age. The histones were resolved on Triton-X/Acid/Urea Polyacrylamide gels (6 mM Triton-X 100, 5% Acetic Acid, 6M Urea) and stained with Amido Black. ‘T’ is histones from mouse thymus that shows a similar pattern as mouse embryos. The H2A.1, H2B.2 and H3.1 and H3.2 proteins decrease over time, while the H2A.2, H2B.1, H3.3 and H10 proteins accumulate with time. The multiple bands for H3.2, H3.3 and H4 proteins are acetylated forms of histones. These changes in protein levels are consistent with the core histone genes that continue to be expressed in tissues. Reproduced from Zweidler, A. (1984) In: Stein, G., Stein, W. and Marzluff, W. F. (eds.), Histone genes: structure, organization and regulation. John Wiley and Sons, New York, pp. 373–395 with permission of John Wiley & Sons Ltd. Copyright © 1984 by John Wiley & Sons, Inc.
The pattern of histone protein changes that occur in mouse liver as a function of age was determined by A. Zweidler (14) over 30 years ago, and a figure showing the changes in liver histones from birth to 30 weeks of age is reproduced in Figure 7. There are changes in the subtypes of 3 core histones, H2A, H2B and H3, encoded by RD-histone genes (all the histone H4 genes encode the same protein). There are increases in the relative amounts of the H2A.2 protein (encoded by the Hist2H2aa1 gene), the H2B.1 protein (encoded by the Hist1H2bc gene), and the H3.2 protein, (encoded by the Hist1H3h gene) in mouse liver during the first 30 weeks. At the same time the H3.3 and H1O proteins increase. Thus, the RD-histone genes we have identified encode the proteins that accumulate in adult mouse non-dividing tissues.
Polyadenylation of histone mRNAs may be a feature of terminal differentiation
Knockdown of a number of factors in RNA biosynthesis by RNAi in cultured human cells leads to polyadenylation of a small percentage of histone mRNA. These include knockdown of NELF, a transcription elongation inhibitor (19); ARS2, a factor required for efficient cell cycle progression (20) that is also present in the NEXT complex that binds to nascent capped transcripts (69); CDK9 (21), a transcription elongation factor; and RNF20 and RNF40, which are involved in H2B ubiquitination (21). In many of these experiments the primary assay is measurement of changes of polyadenylated histone mRNA by qRT-PCR of oligo(dT) primed cDNA. Results are often reported as ‘fold-increase’ and large fold-increases in polyadenylated histone mRNAs are often observed using this technique. In the cases where the expression of the polyadenylated histone mRNAs relative to normally processed histone mRNAs have been measured (19,20,22), the amount of poly(A)+ histone mRNA is in the range of 2–3% of the properly processed RNA. The large increases reported reflect the fact that the amount of polyadenylated histone mRNA under normal conditions is extremely low. It is likely that all of these treatments disrupt the coupling between transcription and histone 3′ end processing, resulting in a small amount of RNA, which is not processed normally and may then be polyadenylated. The HIST2H2AA3 mRNA and several histone H2B mRNAs which are expressed in tissues are among those affected. In at least one case, the polyadenylated histone mRNA formed is not the same as the polyadenylated histone mRNAs expressed in tissues. Knockdown of SLBP resulted in expression of a novel histone H4 mRNA detected by Northern blotting (23). This RNA was derived from the HIST1H4J gene, which is not expressed in human tissues.
The switch in histone gene expression is likely a consequence of terminal differentiation, and not simply a consequence of cells stopping cell division. When 3T3 fibroblasts are arrested cells in G0 by serum starvation, we do not detect expression of polyadenylated histone mRNAs over the 7 day serum starvation. When we differentiated the mouse 3T3-L1 fibroblasts over 7 days, polyadenylated Hist1h2bc and Hist2h2aa1 mRNAs were expressed, consistent with activation of this pathway in terminally differentiated cells, and not simply as a result of serum starvation. Our data contrasts with the report of Pirngruber et al. that serum starvation of HTC116 cells resulted in polyadenylation of HIST1H2BC mRNA (70), and other pharmacological treatments that also arrest cells show a similar result. In their study, they reported a 3-fold increase in polyadenylated HIST1H2BC in response to serum starvation, but they did not measure the absolute amount of polyadenylated mRNAs expressed. In subsequent experiments, Kari et al. (24) reported that there are increases in the same polyadenylated H2B and H2A mRNAs that we observed are expressed in human tissues, when human fibroblasts differentiate into adipocytes in vitro. They did not compare these results with the effect of serum starving these fibroblasts.
Rai et al. have identified some histone genes that are expressed as polyadenylated mRNAs in response to DNA damage or senescence (71). In their study, they propose the concept of ‘chromostasis,’ through which DNA replication independent chromatin regulators maintain dynamic chromatin in senescent cells in culture. They demonstrate that HIRA is required for efficient deposition of histones during senescence. Importantly, using affymetrix microarrays, they identify some of the genes identified in this study as expressed (HIST1H2AC, HIST1H2BC and HIST1H2BK) in senescent cells, which may represent another type of terminal differentiation.
How does the switch to polyadenylated histone mRNAs occur?
A combination of factors likely determines how histone mRNAs switch their 3′ end formation and which genes remain expressed. In actively growing cells, at the G1/S-phase transition, CycE/Cdk2 phosphorylates NPAT (54,55,72,73) resulting in a high rate of expression of processed histone mRNAs. In cultured mammalian cells, there is a significant rate of histone gene transcription outside of S-phase (49) or when histone RNA levels are rapidly reduced by inhibiting DNA replication (74,75). Surprisingly, the available GRO-Seq data from Fang et al. (43) show that in mouse liver most histone genes (about 80%) have stalled RNA polymerase II at their promoters including the genes that express polyadenylated histone mRNAs. Many of these genes do not express histone mRNA in tissues.
One could imagine two different extremes for the state of these genes. First, only the genes expressed in tissues release RNA polymerases at a low rate from their promoters into productive elongation or second, many of the RD-histone genes with stalled polymerases at their promoters are transcribed at a low level but do not accumulate any mRNA because there are not effective splicing/polyadenylation signals. Comparing the GRO-Seq data from growing cells with mouse liver and brain, it is clear that a major regulatory step is at the level of transition of the RNA polymerase into processive elongation after synthesis of the short transcript. Genes that are expressed as polyadenylated mRNAs also have a stalled polymerase at their promoter, and the release of this polymerase into productive elongation is likely tightly controlled resulting in production of low levels of polyA mRNA. The importance of regulation at this step has been emphasized recently by studies of Adelman et al. (47,76). The levels of some of these mRNAs (HIST1H3E, HIST2H4, HIST1H4H and HIST1H2AC) may be further reduced by NMD, since they are all spliced with an intron in the 3′ UTR more than 100 nts 3′ of the stop codon. Both the HIST1H2B mRNAs, and the HIST2H2AA3 mRNAs that are expressed are not sensitive to NMD and accumulate at significantly higher levels that the other transcripts that are spliced.
Some of the histone genes, e.g. the Hist2H3c gene adjacent to the expressed Hist2H2aa1 gene, are not expressed in tissues (do not have a stalled polymerase at the promoter), although the Hist2H3c gene is expressed at high levels in cultured mouse cells (75). In growing cultured cells, the histone genes do not show a strong GRO-Seq peak at the promoter, but show transcription throughout the gene with a major peak 3′ of the processing site, consistent with the high level of RNA polymerase present at histone genes (77,78).
The expression of components of the histone locus body, NPAT and FLASH, in terminally differentiated cells suggest that they play a role in the expression of histone genes in tissues, supporting a basal level of transcription of the histone genes, likely by forming a histone locus body. In normally growing fibroblasts (54,56) and ES cells (79,80), the HLB persists throughout the cell cycle, as it does in Drosophila, both in replicating and non-replicating cells (81–83). SLBP mRNA was also expressed in the tissues. It was expressed at higher levels in cultured cells compared to the other mRNAs, and at higher levels in human breast tumours relative to the adjacent normal tissue. In contrast to the components of U7 snRNP, the level of SLBP protein in the liver and brain is dramatically reduced despite expression of the SLBP mRNA, as it is in cultured cells in G1. The reduction of SLBP protein is unlikely to be the determinant for the switch to polyadenylated histone mRNA in differentiated cells. This is most clearly demonstrated by arrest of fibroblasts in a G0 state, which reduces the levels of SLBP protein, but does not cause result in the accumulation of polyadenylated histone mRNA, and the failure of the HIST1H4J gene to be expressed in tissues, although it is expressed as a polyadenylated mRNA in HeLa cells when SLBP is knocked down (23).
In summary, we report a subset of histone genes express polyadenylated histone mRNAs (many of which are also spliced) in terminally differentiated tissues. These mRNAs likely encode the replacement core histones which turn over very slowly in these cells, which are very long-lived. The proteins expressed from these genes encode the same histone variants proteins shown to accumulate in mouse tissues by Fred Zweidler years ago, replacing some of the histone proteins originally present in these tissues.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Jessica Sorrentino and Dr Norman Sharpless for providing frozen dissected mouse liver. We would like to thank Dr Jason Merkin and Dr Chris Burge for sharing data prior to publication and Dr Nancy Kedersha for critical reading of the manuscript.
Footnotes
Present address: Shawn M. Lyons, Department of Medicine, Harvard Medical School, Division of Rheumatology, Immunology and Allergy, Brigham and Women's Hospital, Boston, MA 02115, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM29832 to W.F.M., GM67113 to Y.X., CA130795 to M.L.W.]; Summer Undergraduate Research fellowships from the UNC Office of Undergraduate Research [to C.H.C., A.G. and B.W.]; NSF Graduate Research Fellowship [DGE-1144081] and NIH BD2K Fellowship [T32 CA201159] to J.D.W. Funding for open access charge: National Institutes of Health (NIH) [GM29832 to W.F.M., GM67113 to Y.X., CA130795 to M.L.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Marzluff W.F., Wagner E.J., Duronio R.J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Mowry K.L., Steitz J.A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 4.Gick O., Krämer A., Keller W., Birnstiel M.L. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotten M., Gick O., Vasserot A., Schaffner G., Birnstiel M.L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 1988;7:801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaufele F., Gilmartin G.M., Bannwarth W., Birnstiel M.L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- 7.Harris M.E., Böhni R., Schneiderman M.H., Ramamurthy L., Schümperli D., Marzluff W.F. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng L.X., Dominski Z., Yang X., Elms P., Raska C.S., Borchers C.H., Marzluff W.F. Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol. Cell. Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitfield M.L., Zheng L.X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R. Conservation of histones in chromatin during growth and mitosis in vitro. J. Mol. Biol. 1969;40:457–466. doi: 10.1016/0022-2836(69)90165-x. [DOI] [PubMed] [Google Scholar]
- 11.Bondy S.C. The synthesis and decay of histone fractions and of deoxyribonucleic acid in the developing avian brain. Biochem. J. 1971;123:465–469. doi: 10.1042/bj1230465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyama B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R., 3rd, Hetzer M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler S.J., Williams N.I., Stanton G.B., Cameron J.L., Greenough W.T. Maturation time of new granule cells in the dentate gyrus of adult macaque monkeys exceeds six months. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10326–10331. doi: 10.1073/pnas.1017099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zweidler A. In: Histone genes: structure, organization and regulation. Stein G, Stein W, Marzluff WF, editors. NY: John Wiley and Sons; 1984. pp. 373–395. [Google Scholar]
- 15.Panyim S., Chalkley R. A new histone found only in mammalian tissues with little cell division. Biochem. Biophys. Res. Commun. 1969;37:1042–1049. doi: 10.1016/0006-291x(69)90237-x. [DOI] [PubMed] [Google Scholar]
- 16.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 17.Bonner W.M., Mannironi C., Orr A., Pilch D.R., Hatch C.L. Histone H2A.X gene transcription is regulated differently than transcription of other replication-linked histone genes. Mol. Cell. Biol. 1993;13:984–992. doi: 10.1128/mcb.13.2.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannironi C., Bonner W.M., Hatch C.L. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Gruber J.J., Olejniczak S.H., Yong J., La Rocca G., Dreyfuss G., Thompson C.B. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirngruber J., Shchebet A., Schreiber L., Shema E., Minsky N., Chapman R.D., Eick D., Aylon Y., Oren M., Johnsen S.A. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohn M., Ihling C., Sinz A., Krohn K., Huttelmaier S. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 2015;29:1998–2003. doi: 10.1101/gad.266486.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan K.D., Mullen T.E., Marzluff W.F., Wagner E.J. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kari V., Karpiuk O., Tieg B., Kriegs M., Dikomey E., Krebber H., Begus-Nahrmann Y., Johnsen S.A. A subset of histone H2B genes produces polyadenylated mRNAs under a variety of cellular conditions. PLoS One. 2013;8:e63745. doi: 10.1371/journal.pone.0063745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z.F., Sirotkin A.M., Buchold G.M., Skoultchi A.I., Marzluff W.F. The mouse histone H1 genes: Gene organization and differential regulation. J. Mol. Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- 26.Graves R.A., Wellman S.E., Chiu I.M., Marzluff W.F. Differential expression of two clusters of mouse histone genes. J. Mol. Biol. 1985;183:179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- 27.Student A.K., Hsu R.Y., Lane M.D. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J. Biol. Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 28.Gerin I., Bommer G.T., Lidell M.E., Cederberg A., Enerback S., Macdougald O.A. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J. Biol. Chem. 2009;284:10755–10763. doi: 10.1074/jbc.M809115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominski Z., Yang X.C., Purdy M., Marzluff W.F. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9422–9427. doi: 10.1073/pnas.1533509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominski Z., Yang X., Kaygun H., Marzluff W.F. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 31.Merkin J.J., Chen P., Alexis M.S., Hautaniemi S.K., Burge C.B. Origins and impacts of new mammalian exons. Cell Rep. 2015;10:1992–2005. doi: 10.1016/j.celrep.2015.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzluff W.F., Gongidi P., Woods K.R., Jin J., Maltais L.J. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 33.Brooks L., 3rd, Lyons S.M., Mahoney J.M., Welch J.D., Liu Z., Marzluff W.F., Whitfield M.L. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA. 2015;21:1943–1965. doi: 10.1261/rna.053389.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Sun X., Qian Y., LaDuca J.P., Maquat L.E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 37.DeBry R.W., Marzluff W.F. Selection on silent sites in the rodent H3 histone gene family. Genetics. 1994;138:191–202. doi: 10.1093/genetics/138.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor J.D., Wellman S.E., Marzluff W.F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J. Mol. Evol. 1986;23:242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- 39.Moss S.B., Ferry R.A., Groudine M. An alternative pathway of histone mRNA 3′ end formation in mouse round spermatids. Nucleic Acids Res. 1994;22:3160–3166. doi: 10.1093/nar/22.15.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng G.H., Skoultchi A.I. Rapid induction of polyadenylated H1 histone mRNAs in mouse erythroleukemia cells is regulated by c-myc. Mol. Cell. Biol. 1989;9:2332–2340. doi: 10.1128/mcb.9.6.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng G.H., Nandi A., Clerk S., Skoultchi A.I. Different 3′-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu T.J., Liu L., Marzluff W.F. Mouse histone H2A and H2B genes: four functional genes and a pseudogene undergoing gene conversion with a closely linked functional gene. Nucleic Acids Res. 1987;15:3023–3039. doi: 10.1093/nar/15.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang B., Everett L.J., Jager J., Briggs E., Armour S.M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M.A. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min I.M., Waterfall J.J., Core L.J., Munroe R.J., Schimenti J., Lis J.T. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Core L.J., Waterfall J.J., Gilchrist D.A., Fargo D.C., Kwak H., Adelman K., Lis J.T. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henriques T., Gilchrist D.A., Nechaev S., Bern M., Muse G.W., Burkholder A., Fargo D.C., Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H., Wu J., Wickramasinghe P., Pal S., Gupta R., Bhattacharyya A., Agosto-Perez F.J., Showe L.C., Huang T.H., Davuluri R.V. Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res. 2011;39:190–201. doi: 10.1093/nar/gkq775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLisle A.J., Graves R.A., Marzluff W.F., Johnson L.F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol. Cell. Biol. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green H., Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 51.Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 52.Pillai R.S., Grimmler M., Meister G., Will C.L., Luhrmann R., Fischer U., Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma T.L., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., Harper J.W. Cell cycle-regulated phosphorylation of p220 NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye X., Wei Y., Nalepa G., Harper J.W. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J.Y., Kennedy B.K., Lawrence B.D., Barbie D.A., Matera A.G., Fletcher J.A., Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X.C., Burch B.D., Yan Y., Marzluff W.F., Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milovic-Holm K., Krieghoff E., Jensen K., Will H., Hofmann T.G. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007;26:391–401. doi: 10.1038/sj.emboj.7601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin F., Schaller A., Eglite S., Schümperli D., Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meeks-Wagner D., Hartwell L.H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 61.Salzler H.R., Davidson J.M., Montgomery N.D., Duronio R.J. Loss of the histone pre-mRNA processing factor stem-loop binding protein in Drosophila causes genomic instability and impaired cellular proliferation. PLoS One. 2009;4:e8168. doi: 10.1371/journal.pone.0008168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spalding K.L., Bhardwaj R.D., Buchholz B.A., Druid H., Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 63.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franklin S.G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 65.Brush D., Dodgson J.B., Choi O.R., Stevens P.W., Engel J.D. Replacement variant histone genes contain intervening sequences. Mol. Cell. Biol. 1985;5:1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pehrson J., Cole R.D. Histone H10 accumulates in growth-inhibited cultured cells. Nature. 1980;285:43–44. doi: 10.1038/285043a0. [DOI] [PubMed] [Google Scholar]
- 68.Pehrson J.R., Cole R.D. Histone H1 subfractions and H10 turnover at different rates in nondividing cells. Biochemistry. 1982;21:456–460. doi: 10.1021/bi00532a006. [DOI] [PubMed] [Google Scholar]
- 69.Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche Nel H., Gostan T., Vandermoere F., Robert M.C., Cusack S., et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013;20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- 70.Pirngruber J., Johnsen S.A. Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3′ end processing through p21, NPAT and CDK9. Oncogene. 2010;29:2853–2863. doi: 10.1038/onc.2010.42. [DOI] [PubMed] [Google Scholar]
- 71.Rai T.S., Cole J.J., Nelson D.M., Dikovskaya D., Faller W.J., Vizioli M.G., Hewitt R.N., Anannya O., McBryan T., Manoharan I., et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014;28:2712–2725. doi: 10.1101/gad.247528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao G., Bracken A.P., Burkard K., Pasini D., Classon M., Attwooll C., Sagara M., Imai T., Helin K., Zhao J. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 2003;23:2821–2833. doi: 10.1128/MCB.23.8.2821-2833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao J.Y., Dynlacht B., Imai T., Hori T., Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heintz N., Sive H.L., Roeder R.G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graves R.A., Marzluff W.F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol. Cell. Biol. 1984;4:351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams L.H., Fromm G., Gokey N.G., Henriques T., Muse G.W., Burkholder A., Fargo D.C., Hu G., Adelman K. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol. Cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng B., Li T., Rahl P.B., Adamson T.E., Loudas N.B., Guo J., Varzavand K., Cooper J.J., Hu X., Gnatt A., et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anamika K., Gyenis A., Poidevin L., Poch O., Tora L. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS One. 2012;7:e38769. doi: 10.1371/journal.pone.0038769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghule P.N., Becker K.A., Harper J.W., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220(NPAT) in human embryonic stem cells. J. Cell. Physiol. 2007;213:9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- 80.Ghule P.N., Dominski Z., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J. Cell. Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tatomer D.C., Rizzardi L.F., Curry K.P., Witkowski A.M., Marzluff W.F., Duronio R.J. Drosophila Symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus. 2014;5:613–625. doi: 10.4161/19491034.2014.990860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White A.E., Burch B.D., Yang X.C., Gasdaska P.Y., Dominski Z., Marzluff W.F., Duronio R.J. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White A.E., Leslie M.E., Calvi B.R., Marzluff W.F., Duronio R.J. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.