Abstract
Many studies show that short non-coding sequences are widely conserved among regulatory elements. More and more conserved sequences are being discovered since the development of next generation sequencing technology. A common approach to identify conserved sequences with regulatory roles relies on topological changes such as hairpin formation at the DNA or RNA level. G-quadruplexes, non-canonical nucleic acid topologies with little established biological roles, are increasingly considered for conserved regulatory element discovery. Since the tertiary structure of G-quadruplexes is strongly dependent on the loop sequence which is disregarded by the generally accepted algorithm, we hypothesized that G-quadruplexes with similar topology and, indirectly, similar interaction patterns, can be determined using phylogenetic clustering based on differences in the loop sequences. Phylogenetic analysis of 52 G-quadruplex forming sequences in the Escherichia coli genome revealed two conserved G-quadruplex motifs with a potential regulatory role. Further analysis revealed that both motifs tend to form hairpins and G quadruplexes, as supported by circular dichroism studies. The phylogenetic analysis as described in this work can greatly improve the discovery of functional G-quadruplex structures and may explain unknown regulatory patterns.
INTRODUCTION
Short, non-coding and recurring sequences, referred as sequence motifs, often indicate protein binding sites or motifs that confer structural modifications that alter interaction with proteins. The latter, also referred to as structural nucleic acid motifs may be detected using mathematical predictions of putative alternate topologies instead of the common B-DNA (1,2). Since the development of next generation sequencing technologies, an unprecedented amount of sequence data is available to scan for such structure motifs. Whilst most structural motif prediction algorithms rely on Watson–Crick base-pairing and associated topologies, only a few tools are available that focus on scanning for alternate nucleic acid structures.
The G-quadruplexes (GQs) are a large group of evolutionarily conserved higher order structures present in both lower (bacteria) (3) and higher organisms (worms and humans) (4,5). G-quadruplexes consist of guanine-rich sequences which form a core of stacked G-tetrads, plenary aligned guanines held by Hoogsteen H-bonding, and connected by loops arising from intervening mixed sequences that are not usually involved in tetrads themselves (6). The combination of the number of stacked tetrads, the polarity of the strands, the location and length of the loops as well as the syn versus anti conformation of the guanine bases leads to a vast variety of G-quadruplex topologies.
The GQ DNA structures were first discovered in vitro and later it was demonstrated that the telomeric G-rich single-stranded DNA sequences fold into a GQ structure under near-physiological in vitro conditions. Subsequently human telomeric repetitive sequences (dTAAGGG) have been shown to form GQ structures in vivo (6–9). While telomeric GQs are believed to be responsible for maintaining telomere length and stability, emerging evidence of non-telomeric GQs and their non-random distribution along the genome indicates alternative biological roles. GQs are enriched in certain regulatory elements such as CpG islands, enhancers, insulators and promoters, suggesting their involvement in gene regulation (8,10). Indeed, it was demonstrated that promoters of proto-oncogenes, c-Myc and KRAS, contain nuclease hypersensitive elements (NHEs) that form GQs that are interchangeable between a double-stranded form and GQs form (11–16). It was demonstrated that these interchangeable states of NHEs regulate recruitment of transcription factors to the promoters, thus regulating gene transcriptions. Small molecules selectively stabilizing GQ structures (c-Myc) or oligonucleotides mimicking a GQ DNA structure (KRAS) effectively manipulate gene transcriptions, thus the GQ structures have emerged as a considerable target for drug design because of their potential in combating cancer (12–16).
While several studies have focused on identifying GQ motifs and their roles in eukaryotes, there has not been nearly enough study on GQs in prokaryotes. Though many features of prokaryotic DNA organizations differ from eukaryotes (circular versus linear, absence of a chromatin-like nucleosome structure, etc.), it was not until recently that GQ DNA structures were found to be enriched in the genomes of lower organisms including bacteria (3,17–21). Computational interrogation of 140 different bacterial genomes revealed that potential GQ-forming sequences are highly enriched in regulatory regions (−200 to −1 bp upstream from the transcription start site) of many prokaryotic genes (3). Indeed, similar to eukaryotes, where the effects of GQs on gene expression were already known, a recently published elegant work provided evidence that GQ sequences are critical for gene expressions in bacteria, strongly indicating that regulatory roles of GQ sequences are highly conserved between eukaryotes and bacteria (21). Additionally, it was also reported that, similar to specific binding of human MSH2/MSH6 (bacterial homolog of MutSα) to GQs located in the immunoglobulin switch regions, bacterial E. coli MutSα was found to bind to GQ-forming sequences (19,21). In addition to MutS, Henderson et al. identified many non-telomeric GQ DNA binding proteins in human, yeast, Escherichia coli and Arabidopsis. The GQ binding proteins showed different affinities toward different G-quadruplex-forming sequences, indicating a relation between topology and function (22). Strikingly, the occurrence of non-homologous recombination between the pilin gene (pilE) locus and silent pilS loci was found to be caused by GQs in the upstream of the pilE gene in the pathogen Neisseria gonorrhoeae, thus leading to pilin antigenic variation, which is essential for the pathogenic bacterium to evade the host's defense response (23,24).
Here, we have applied a rather restrict algorithm to identify 52 highly putative G-quadruplex forming (HPGQ) motifs in E. coli. Phylogenetic classification of these sequences revealed two groups of highly conserved HPGQs with 16 and 7 members. To our surprise all members of both groups were located only within intergenic regions and especially close to the 3′UTR and mostly between operons, indicating a regulatory role. Both mathematical and biophysical studies indicated that these sequences can be found in three different structural variations, duplex, hairpin and GQ, indicating a regulatory switch.
MATERIALS AND METHODS
Prediction of putative G-quadruplex groups
The E. coli reference genome, K12 MG1655 (NC_000913.3), was obtained from NCBI database and scanned for putative G-quadruplexes using quadparser software (3,25) with the standard pattern GSNL1GSNL2GSNL3GS where G refers to guanine and N refers to any nucleotide including guanines. While S is set to be equal to or bigger than 3, and L1-3 are set to be between 1 and 7. The program was run using both strands. The sequences and their starting positions were listed and confirmed using NCBI sequence viewer (25,26). The G-quadruplexes predicted by this software may differ from other G-quadruplex finding software due to algorithm differences (25).
The sequence list was processed using Clustal omega tool to develop a multiple sequence alignment with a minimum of three iterations. In Clustal omega, the following parameters were used: –output-order = tree-order –percent-id –dealign –iter = 3 (27–29). Alignment results were used for the phylogenetic tree generation using ClustalW version 2. ClustalW, where the alignment file was used as the input, was employed to generate the phylogenetic tree with UPGMA as the clustering method (30,31). For large data sets, the neighbor-joining method is preferred for better accuracy (31,32). Grouping was performed as followed; a minimum of three identical HPGQs in a cluster was accepted as a new group. The groups were then expanded by inclusion of other members while moving along the phylogenetic branch until a sequence with the second mutation was encountered. The ratio of the number of mutations (n) to the total alignment length of the shortest GQ-forming sequence (N) is referred to as the degree of divergence.
Motif discovery and enrichment
The locations of the two main HPGQ groups were mapped according to the gene open reading frames (ORFs) on the genome as defined on the NCBI database. The distance to the genes and their directions were listed. The order of the genes in the operon was taken from RegulonDB (33).
Selected HPGQ sequences and their flanking regions (±200 bp) were used for motif finding studies. In order to reveal other conserved motifs in the flanking regions we used the GLAM2 web tool (34). We chose GLAM2 over MEME because such a motif does not necessarily have an identical distance to the G quadruplex forming sequence and GLAM2 is capable of finding ‘gapped’ motifs. HPG1 and HPG2 were scanned separately using only the G quadruplex forming strand and the maximum number of columns to be aligned was set to 80, instead of the default value, 50. Because GLAM2 returns a non-compatible motif format to be used for the motif enrichment tool, GOMo, we also applied HPG1 and HPG2 to the MEME motif finder tool, both separately and combined (common motif) using the same parameters as mentioned for GLAM2. Motifs discovered from the MEME analysis were scanned to reveal gene ontology (GO) terminology of associated genes using GOMo with default parameters (34).
In order to discover any degenerate HPGQs with similar sequences that were not detected due to a restriction of the G-quadruplex algorithm, we scanned the E. coli K12 MG1655 uid57779 upstream sequences for the common motif using the FIMO web tool. A large number of hits were identified by FIMO and these were limited by designating a P-value cut-off. The cut-off value was determined by testing the motifs in evolutionarily distant species. For the conservation study reference bacteria species were selected from the NCBI database and scanned for the motif in their upstream sequences.
Centrimo was used to discover associated motifs and transcription factor binding sites in the flanking regions in order to find any transcription binding factors associated with the HPGQs. The HPGQ sequences and their flanking regions (±200 bp) were scanned for motifs from the CollecTF (bacterial TF motifs) database using default options (35).
Secondary structure prediction
Secondary structural modeling was performed using the ViennaRNA package (36). Initially, two sequences were obtained to represent HPG1 motif and HPG2 motifs using RNAalifold (37) and the sequence alignment was performed using GLAM2 yielding two sequences that were 63 and 54 bases long, respectively. Next, the sequences were scanned for hairpin and G-quadruplex formation by RNAsubopt using ‘no lonely pair’ (–noLP), and ‘scan for g quadruplex’ (-g) parameters. It is important to note that the default parameter file is for RNA secondary structures. For that reason, we specified the parameter file obtained from RNAalifold Webserver modified for DNA structures as defined by Reuter and Mathews (37–40). Chosen secondary structures and their free energies were listed to represent various topologies.
Circular dichroism and gel retardation analysis
The oligonucleotide sequences, dTTT TCT CCC TCT CCC TTT GGG AGA GGG CCG GGG TGA GGG CAA AAA CGC GCA C and dGTG CGC GTT TTT GCC CTC ACC CCG GCC CTC TCC CAA AGG GAG AGG GAG AAA A, were obtained commercially (Sentegen Biotech, Ankara, Turkey) and 100 μM stock solutions were prepared in a 10 mM TE buffer at pH 7.2. Oligonucleotides were diluted to 5 μM in a 2.5 mM Hepes buffer at pH 7.2 and appropriate salt concentrations unless otherwise is indicated. Samples were heated at a temperature of 90°C for 30 min before being allowed to cool down to room temperature over 16 h. Circular dichroism (CD) measurements were conducted using a Jasco J-810 CD spectrophotometer with a 0.1 cm cell path length. The measurements were taken between 200 and 300 nm at 100 nm/min speed and 1 nm bandwidth. Melting profiles were formed from CD measurements of the samples at stepwise increasing temperatures and allowing to equilibrate for ∼5 min. The CD signal strengths at 260 nm were then plotted and 4-parameter sigmoidal curves were fitted using SigmaPlot. A gel retardation study was performed in 15% polyacrylamide gel in TBE buffer using the samples prepared for CD measurements.
RESULTS
Identification of highly putative G-quadruplexes
A G-quadruplex may refer to a large group of topologies with a common feature of the G-tetrad, a non-canonical assembly of four guanines on a plane. Because the stacks of G-tetrads are required to stabilize the structure, four repeats or tracts, of guanines are necessary in any sequence to form an intramolecular GQ. While the length of these repeats has the biggest impact on stability, so do the sequences linking the guanine tracts (G-tracts). A generally accepted algorithm of GSNL1GSNL2GSNL3GS is used to determine the putative G-quadruplex forming sequences, where S refers to the length of the GQ-stem, length of the guanine tract and L1-3 defines the lengths of the loops, independent from each other. While the effect of the loop lengths on GQ formation is highly debatable, the repeat length or stem length were, as expected, highly influential due to cumulative π–π interactions. In most cases a tract length of 3 is considered a stringent rule for the formation of GQ in vivo. In order to capture only highly putative G-quadruplexes (HPGQs) we restricted S to be above or equal to 3 while L1-3 were set to be between 1 and 7. A whole genome scan using this algorithm yielded only 52 sequences within the reference E. coli genome (K12 MG1655, NC_000913.3). These highly putative G-quadruplexes are abbreviated as HPGQs and listed in Supplementary Table S1. In the cases of sequences with overlapping patterns due to the presence of more than four guanine tracts, we selected the largest patterns to include all G-tracts. It is important to note that such patterns may adopt multiple topologies depending on the G-tracts that take part in the G-tetrad formations.
Phylogenetic classification of HPGQs
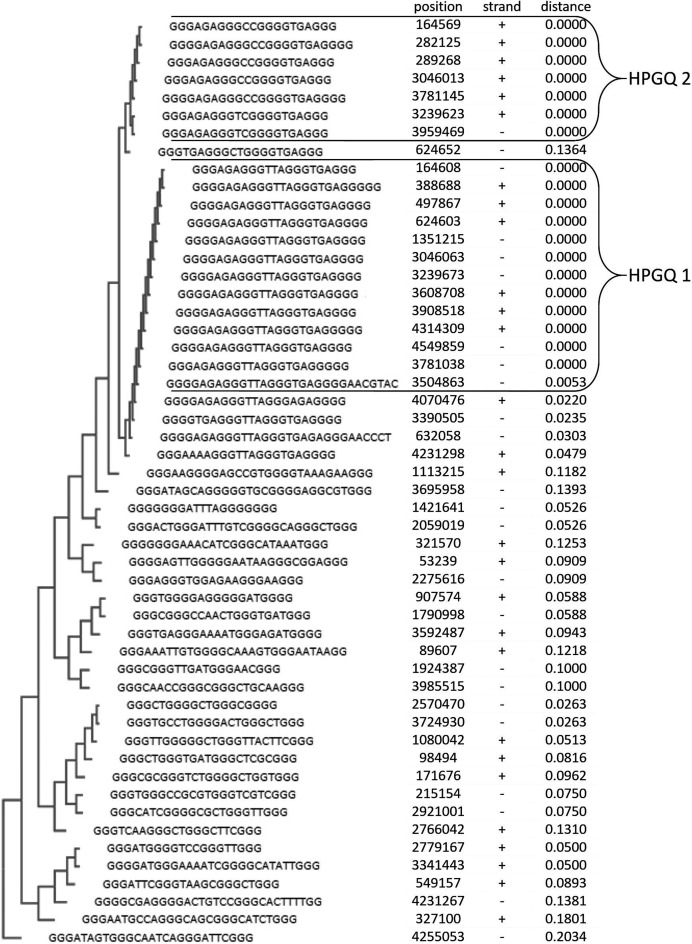
As previously mentioned, a G-quadruplex may have a very large variety of topologies. When investigating the role of a GQ, this topological variety should be taken into account because any protein–GQ interaction would be strongly dependent on the topology of the GQ (9,41). Since topology is strongly controlled by sequence, it is possible to relate the sequence of a GQ to its function. Keeping that in mind, biologically relevant sequences are often conserved. This is also applicable for GQs just as it is for any biomolecule. If a particular topology is expected to have a specific biological role, its sequences are highly likely to be conserved. Such GQ elements may be revealed when the predicted GQ-forming sequences are clustered and classified. In order to classify the GQs, we decided to use phylogenetic analysis. The sequences of E. coli HPGQs were aligned using Clustal omega (42,43). The alignment was sent to ClustalW for developing an unrooted phylogenetic tree to find similar and therefore evolutionarily and functionally connected sequences. Each group is initiated when at least three HPGQs with identical shortest G-quadruplex-forming sequences are found within. This excludes any extensions at the flanks if present. The groups were then expanded by including neighboring branch members until a second mutation was detected within the shared shortest G-quadruplex-forming sequence. In other words, every group consisted of sequences containing a GQ-forming sequence identical to each other or different with a single mismatch, with insertions/deletions counting as a mismatch. For the two HPGQ groups we have detected, this corresponded to a maximum degree of divergence of 0.048. This value is calculated from n/N where n equals the number of mismatches in the alignment (n = 1) and N is the length of the shared shortest G-quadruplex-forming sequence at the initiation of the groups (N = 21 for HPG1 and HPG2). With a degree of divergence threshold of 0.048 the sequences were grouped into two distinct HPGQ groups with 16 and 7 members, HPG1 and HPG2, respectively (Figure 1). None of the unclustered HPGQs were found to have a lower degree of divergence from HPG1 or HPG2 and left ungrouped. It should be noted that some of these sequences may not be evolutionarily related and thus may be placed at outlier branches by ClustalW. Such sequences are disregarded as they do not cluster. Since we are interested in the vicinity of each of the clusters, the relationship between evolutionarily unrelated G-quadruplex groups are irrelevant for this study. Our research continued with the two groups identified at this point.
Figure 1.
Phylogenetic tree of Escherichia coli HPGQs. The phylogenetic tree was constructed using ClustalW tool implementing the UPGMA method. The common predictive GQ motifs called HPG1 and HPG2 are exclusively clustered together. The first column represents the position of the HPGQs on the genome while the second column refers to genes located on the strand. The third column displays their phylogenetic distances to the nearest node as provided by ClustalW web tool and the degree of divergence from the closest HPGQ group is shown in the last column.
Interestingly, the alignment of the motifs revealed that 13 members of HPG1 and another 5 members of HPG2 had the identical HPGQ sequences within each group; dGGGAGAGGGTTAGGGTGAGGG and dGGGAGAGGGCCGGGGTGAGGG, respectively. Moreover, these sequences showed a striking similarity with a difference only in the second loop (dTTA versus dCC). Because the difference is restricted to the middle loop, it is expected to have a distinguishable topological difference mainly occurs on a single side of the G-quadruplex (i.e. top or bottom), therefore both structures may still share a similar topology.
Genomic distribution of HPGQs in E. coli
The high conservation of the HPGQs indicates a biological role for the motif. In order to investigate the biological function, we found locations of HPGQ motifs in relation to the nearest ORFs and operons within the E. coli genome. Surprisingly, all motifs were located at sites flanking ORFs and none within. Their mid-point relative to the nearest genes, the directions of the genes and the strand of the motifs are listed in Table 1.
Table 1. List of HPGQs positions and relative positions of the related genes. Shown is the landscape for each member of HPG1 and HPG2 groups and their neighboring genes. While the ‘HPGQ no’ refers to the order of HPGQ motifs in Supplementary Table S1, the ‘midpoint’ and ‘strand’ indicates the position of the motif in the genome and the strand in which the motif is located, respectively. The ‘gene names’ refer to the genes located on either sides of the HPGQ according to ncbi annotations. ‘Direction’ indicates the transcription direction of the gene given in the neighboring column in accordance with the HPGQ (towards or outwards). The distance of the HPGQs to the associated ORF is given under ‘distance to next ORF’. The orders of corresponding genes in its operon and the total gene count in the operon according to RegulonDB database are shown in the ‘operon order/gene count’, respectively.
| HPGQ no | Operon Order/ Gene Count | Gene Name | Direction | Distance to next ORF | Midpoint | Strand | Distance to next ORF | Direction | Gene Name | Operon Order/ Gene Count | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HPG1 | 6 | 1/1 | hrpB | >>> | 63 | 164608 | - | 111 | >>> | mrcB | 1/1 |
| 13 | 4/4 | tauD | >>> | 31 | 388688 | + | 54 | <<< | hemB | 1/1 | |
| 15 | 1/1 | adk | >>> | 38 | 497867 | + | 177 | >>> | hemH | 1/1 | |
| 17 | 1/1 | fepB | <<< | 82 | 624603 | + | 271 | >>> | entC | 1/5 | |
| 22 | 1/1 | fabI | <<< | 165 | 1351215 | - | 181 | <<< | yjcD | 1/1 | |
| 33 | 1/1 | ygfF | <<< | 151 | 3046063 | - | 94 | <<< | gcvP | 3/3 | |
| 35 | 1/1 | alx | >>> | 117 | 3239673 | - | 260 | >>> | sstT | 1/1 | |
| 38 | 5/5 | frlR | >>> | 74 | 3504863 | - | 56 | <<< | yhfS | 2/2 | |
| 40 | 1/1 | zntA | >>> | 48 | 3608708 | + | 32 | <<< | tusA | 1/1 | |
| 43 | 3/3 | lldD | >>> | 11 | 3781038 | - | 165 | >>> | trmL | 1/1 | |
| 45 | 4/5 | pstB | <<< | 142 | 3908519 | + | 19 | <<< | pstA | 3/5 | |
| 51 | 1/1 | yjdP | >>> | 101 | 4314309 | + | 24 | <<< | phnP | 11/11 | |
| 52 | 6/6 | fimH | >>> | 138 | 4549859 | - | 83 | <<< | gntP | 1/1 | |
| 48 | 4/4 | yihR | <<< | 64 | 4070476 | + | 28 | <<< | yihS | 3/4 | |
| 37 | 1/1 | aaeR | >>> | 45 | 3390505 | - | 67 | <<< | tldD | 1/1 | |
| 19 | 1/1 | cstA | >>> | 51 | 632058 | - | 108 | >>> | ybdD | 1/1 | |
| HPG2 | 5 | 1/1 | hrpB | >>> | 24 | 164569 | + | 150 | >>> | mrcB | 1/1 |
| 9 | 1/1 | yagA | <<< | 131 | 282125 | + | 142 | >>> | yagE | 1/2 | |
| 10 | 2/2 | yagI | <<< | 95 | 289268 | + | 22 | <<< | argF | 1/2 | |
| 32 | 1/1 | ygfF | <<< | 101 | 3046013 | + | 144 | <<< | gcvP | 3/3 | |
| 44 | 3/3 | lldD | >>> | 117 | 3781145 | + | 59 | >>> | trmL | 1/1 | |
| 34 | 1/1 | alx | >>> | 67 | 3239623 | + | 310 | >>> | sstT | 1/1 | |
| 46 | 1/1 | ilvC | >>> | 13 | 3959469 | - | 52 | <<< | ppiC | 1/1 |
Since all HPGQs are located between genes, we decided to investigate the directionality of the neighboring genes. Most of the HPGQs are found to be between tandem gene pairs and the distances between ORFs were found to be ranging between 377 bp and 65 bp. Such small distances between ORFs complicates locating the putative regulatory regions between tandem gene pairs since 200 bp upstream of the transcription start site is often regarded as regulatory (3). Furthermore, while most of the HPGQs we identified are present between tandem gene pairs, 8 out of 16 HPG1 members were upstream of operons, indicating a role in the initiation of transcription.
For the HPGQs found between genes of the same operon a terminator role is plausible by the blocking of RNA polymerase by GQs and the prevention of further transcription. This is also supported by previous studies indicating that polymerases ‘fall off’ during the transcription of GQ-induced DNA sequences (11,13). For HPG1, six of the members were found between convergent gene pairs while only a single member is located between divergent gene pairs. The fact that HPGQs were closer to the ORFs at 3′ the end may suggest this hypothesis further, however, it should be noted that previous studies have shown that non-B-motif formations may have regulatory effect even at a greater distance to the ORF (2).
The genomic distribution of HPGQs relative to each other were also analyzed. This revealed a surprising pattern. Six HPG1 and HPG2 members (HPGQs 5, 6, 32, 33, 34 and 35) were found to be located in close proximity (<50 bp) and in pairs (Supplementary Figure S1). Curiously, these pairs were also located on the opposite strands. In addition, an HPG1 member, HPGQ 17 was also found 50 nt upstream of HPGQ 18, which is a degenerate form of the common sequence of HPG2. Considering that in most cases they were present in the opposite strands, HPG1 and HPG2 motifs may not only have similar function but also function together.
Search for conserved pattern in flanking regions
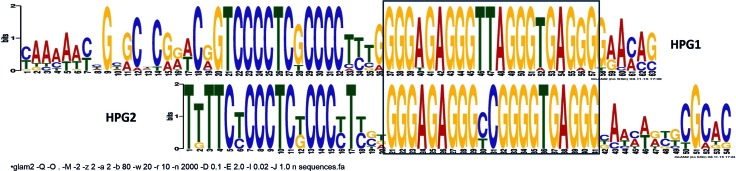
The close proximity of HPGQs made us wonder if there is a conserved sequence pattern in the flanking regions. To reveal any conservation in the flanks we used the GLAM2 tool from the MEME suite (34,44) to scan sequences from 200 bp upstream to 200 bp downstream of the middle of the each HPGQ (401 bp in total). The results show that both HPGQ classes possess a common C-rich feature ending ∼3 bases upstream of the HPGQs. The difference occurs after the C-rich region – a highly conserved 4 nt long T-rich region precedes C-rich region for HPG2s. The alignment also reveals the similarity of the HPG1 and HPG2 sequences. The difference of these groups peak at the composition of the second loop (Figure 2).
Figure 2.
Nucleotide alignment of HPG1 (bottom) and HPG2 (top) ‘gapped’ motifs as obtained from GLAM2. The motifs discovered using GLAM2 were aligned and GQ-forming G-rich regions are framed within the box.
The close proximity of the members of HPG1 and HPG2s, as well as similar extended motifs is a strong indication that HPG1 and HPG2 are associated to the same biological processes, which suggests that a higher degree of divergence threshold could be acceptable during phylogenetic classification. For that reason, we decided to find a common motif for further analysis. Unfortunately, the motif output of GLAM2 tool is incompatible with other motif scanning and enrichment tools. This is because, unlike MEME, GLAM2 finds ‘gapped’ motifs which reveals motifs even if there are insertions or deletions creating gaps in the motif. For that reason, a common motif was found for HPG1 and HPG2 using the MEME tool. MEME analysis resulted in a 41 bp long motif with well-conserved C-rich and G-rich regions (Table 2).
Table 2. GOMo analysis for prediction of associated GO terms linked to ‘ungapped’ HPGQ motifs. WebLogos of consensus HPG1 and HPG2 motifs were generated by the MEME motif analysis tool. Statistical significance of the discovered motifs are represented as E-value for each motif. These motifs were associated to molecular function and biological process GO terms by GOMO tool. The motifs were mainly related to iron–sulfur cluster binding, maltose-transporting ATPase activity, anaerobic respiration and maltose transport. The scores of the associated GO terms are given in brackets as calculated by the geometric mean of rank-sum tests for the particular GO term.

|
Gene ontology analysis for the common motif of clustered members
The motif obtained from MEME analysis employing both HPG1 and HPG2 flanking sequences was subsequently used to find associated GO terms in E. coli using GOMo tool (45) by scanning between 1000 bp upstream and 200 bp downstream of the first genes in operons. Among 208 predictions, a significant abundance in the molecular function domain is observed. Three of the top five predictions with 100% specificity were related to iron and sulfur cluster binding, indicating a regulatory role of HPGQs (Table 2).
Interestingly, when HPG1 and HPG2 sequences were processed separately by MEME and the identified motifs were subsequently used for the search of associated GO terms, a HPG1-based motif was associated with iron–sulfur cluster binding as well, whereas a HPG2-based motif was found to be related to maltose transport, both with 100% specificity. As a control, the common motif was used to scan Saccharomyces genus, resulting in only a single prediction with a specificity of 75% along with two predictions with negligible (0%) specificity. It is important to note that a higher discrepancy can be observed between motifs obtained by GLAM2 and MEME due to gaps present in the C-rich region. This is especially noticeable for a motif obtained from HPG2 sequences with their flanking regions.
Search for degenerate HPGQs
G-quadruplex algorithm is capable of finding most of the G-quadruplex forming sequences, however, the genome may also have degenerate forms of HPGQs which did not fit our algorithm. These sequences may not be able to be detected due to a mutation at guanine sites. In order to reveal such sequences, the FIMO tool (46) was used with the common motif in E. coli upstream sequences. The FIMO tool output is an array of sequences with increasing q-value, defined as false discovery rate. In order to filter false positive predictions, evolutionarily distant prokaryotic upstream sequences were scanned for the motif within. The decimal fraction of the lowest q-value obtained from the distant prokaryotic species was chosen as the q-value cut-off. With a cut-off of 0.01, an additional 3 motifs were found in E. coli upstream sequences, indicating that a motif scan may reveal additional but degenerate HPGQs (Supplementary Table S2). It is also important to note that the degenerate HPGQs may still form GQ with the assistance of single guanines in the flanks or the loops if present. For that reason, any degenerate member should be approached cautiously and not be included in the group without additional analysis.
HPGQ is not associated with any known motifs
The mode of mechanism of a regulatory motif may be dependent on the transcription factor (TF) binding ability of HPGQs in double stranded, hairpin or G-quadruplex state. It is expected that if the duplex formation is required for a TF to bind, for such a duplex-motif TF-binding would not depend on or require the preservation of G-quadruplex formation and may only be an additional control mechanism to render the duplex-motif out of reach. In such case, we would expect to find a TF-binding motif preserved over a G-quadruplex forming sequence. In other words, if such a motif is found, it can be suggested that the G-quadruplex formation may inhibit TF-binding rather than activation, given that the motif is preserved within G-rich region. We scanned for known motifs listed in CollecTF database (35) within the common HPGQ motif using the Centrimo tool (44). Employing an E-cut-off value of 0.1 the scan yielded no motifs and increasing the E-cut-off to 1 returned only a single E. coli motif, binding site for Macrodomain Ter protein (MatP), with low probability, indicating that HPGQ motif is not associated with known motifs. However, that does not mean that there is not a TF-binding site in the vicinity of the HPGQ that is affected by G-quadruplex formation. Similarly, no TF-binding motif was detected within 200 bp range of HPGQs when scanned for CollecTF motifs.
HPG1 motif is conserved throughout bacteria
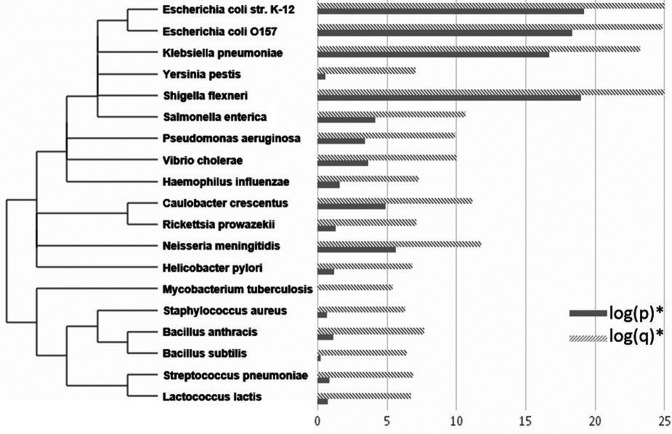
It is generally accepted that highly conserved sequences also have a functional value, so it is expected that HPGQs should also be conserved among other bacteria if they have biological relevance and function. In order to investigate the evolutionary conservation of the motifs with respect to other bacteria, FIMO tool was used to scan for a common motif in the upstream sequences of other bacterial species. We have compared the best hits with the lowest P-value, the probability of a random sequence with as good or better score as well as q-value, the false discovery rates as described by the FIMO website. We found that the common motif was highly conserved for bacteria that are closely related to E. coli and less conserved with increasing distance from E. coli in the phylogenetic tree, since closely related species scored better with the motif than distant species (Figure 3). This indicates the motif may have a biological role conserved in other bacteria as well as E. coli. Unexpectedly, somewhat more related species, Y. pestis and S. enterica, showed an exception to this trend with little conservation of the motif in their upstream sequences.
Figure 3.
The conservation of the common motif among bacteria. The phylogenetic tree based on ncbi taxonomy database (left) and the negative logarithm of P-value (solid bars) and q-values (striped bars) of the best FIMO analysis matches using the common motif in Table 2 within given prokaryotic upstream sequences (right) are presented.
Topological variations of HPGQs
If a structural motif is to have a direct influence on regulatory mechanisms (e.g. TF interaction), it is expected to be able to switch between alternate states and topologies. In other words, the structure should not be rigid or inert toward other factors and be able to switch between states. For GQs located on duplex DNA, the B-form can be considered as an alternate state in most cases. Also, the additional C-rich region conserved in the 5′ flank of HPGQs indicates the formation of a hairpin together with two of the G-tracts suggesting that the HPGQ motifs may adopt the native B-form, G-quadruplex or hairpin like structure. The plausibility of these states can be seen by the comparison of free energies for each possible topology. In order to identify the specific nucleotides involved in the GQ or hairpin structures, free energies were calculated using the ViennaRNA package (36) for both HPG1 and HPG2 motifs (Table 3). The free energies indicate close thermodynamic properties between hairpin and GQ structures demonstrating that presence of a structural switch is possible. However, it should be remembered that the topologies are associated strongly with intracellular conditions, ligands, ions etc. Moreover, a structural switch between alternate GQ topologies (i.e. parallel versus anti parallel) is also a possibility that may be revealed with further analysis.
Table 3. Optimal and suboptimal secondary structures of common HPGQ motifs and the free energies of thermodynamic ensembles. The sequences were assembled according to the most abundant nucleotide in each position of the motif and aligned. The folding patterns of each sequence and the free energies of proposed structures were calculated using ViennaRNA suite and ‘RNAsubopt –g –P dna_mathews.par –e 2 –s ––noLP’ command line. While ‘+’ represents Hoogsteen hydrogen bonding under G-quadruplex formation, (‘&’) represents Watson–Crick base-pairing.

|
Structural studies confirm presence of the switch
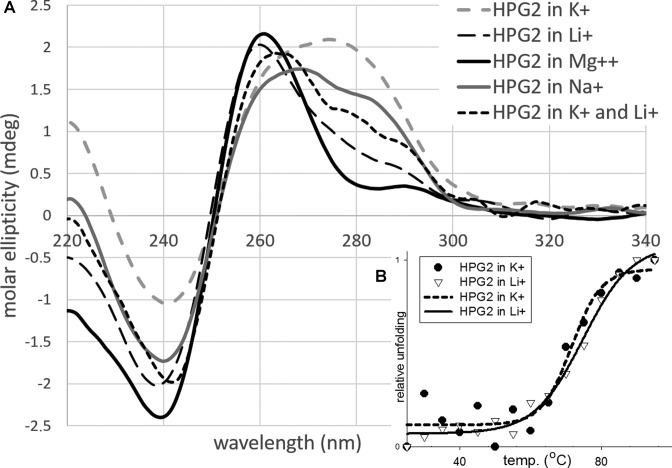
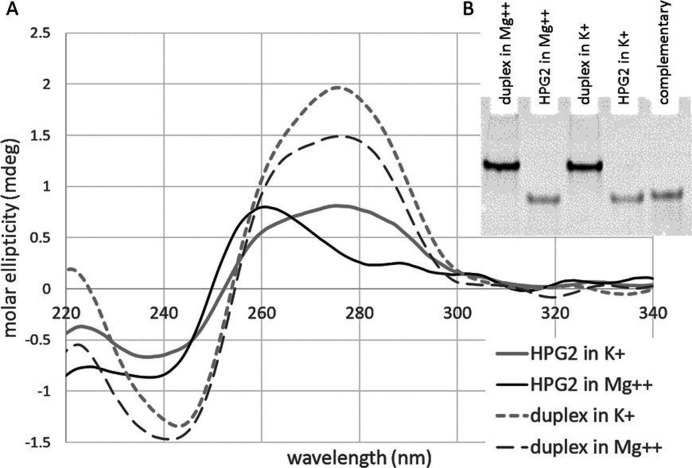
In order to show the structural variation of the HPGQs and to investigate a putative switch, the topology of putative G-quadruplex-forming HPG2 sequence as listed in Table 3 and its complementary sequence (cHPG2) were determined using CD spectrophotometry in the presence of various ions (K+, Li+, Na+ and Mg++). The use of CD spectrophotometry is a well-established technique that determines structural transitions including sub-types of G-quadruplexes (47). Generally, a positive band at 265 nm paired with a negative band at 240 nm indicates a parallel G4 structure whilst the antiparallel G4 structure produces two positive bands at 295 and 240 nm (48). Indeed, our CD measurements revealed a structural transition in the presence of different ions under in vitro conditions. In the presence of 100 mM K+, a monovalent cation well-known for its G-quadruplex stabilizing effect, a dip at 240 nm and a wide peak at 275 nm was observed for the GQ-forming strand of HPG2 (Figure 4A). This CD signature is found similar to CD signatures detected for the human telomere sequence, d[T2G3(T2AG3)3A] (49). The latter peak was replaced with another at 260 nm in the presence of either 10 mM Mg++ or 100mM Li+. It is important to note that the presence of Li+, which is known to greatly destabilize G-quadruplexes (50,51), is expected to promote the formation of a hairpin duplex structure which is also promoted in the presence of Mg++. Unlike Li+, free Mg++ ions are present in the E. coli cytosol up to 3 mM (52) which suggests that the structure promoted by Li+ is also stimulated by Mg++ and for that reason, may have biological relevance. Replacing the ions with 100 mM Na+ or a mixture of 50 mM Li+ and 50 mM K+ corresponded to a transition between the G-quadruplex and hairpin.
Figure 4.
(A) Circular dichroism (CD) and (B) melting analysis of HPG2 structures in the presence of various ions. The oligonucleotides were dissolved in a 2.5 mM Hepes buffer at pH 7.2 in the presence of either 100 mM KCl (dashed grey line), 100 mM LiCl (dashed thin black line), 10 mM MgSO4 (thick black line), 100 mM NaCl (thick grey line) or 50 mM KCl and 50 mM LiCl (dashed thick black line). Samples were heated up to 90°C for 30 min and allowed to cool down to room temperature for 16 h. The melting curves of the HPG2 motif sequence as presented in Table 2 in the presence of K+ and Li+ according to the change at 260 nm using CD spectrophotometer indicates similar melting temperatures for both structures (73°C and 71°C, respectively).
When incubated together with the complementary sequence the positive peak shifted towards 280 nm in the presence of either ion (Mg++ or K+) indicating a similar topology for both ions (Figure 5A). Indeed, gel retardation analysis also indicated that in the presence of the complementary sequence the duplex formation was preferred regardless of the ion present (Figure 5B).
Figure 5.
(A) Circular dichroism and (B) gel retardation analysis of HPG2 structures in the absence (solid lines) and presence (dashed lines) of complementary sequence in the presence of K+ (grey lines) or Mg++ (black lines). The oligonucleotides were dissolved in a 2.5 mM Hepes buffer at pH 7.2 in the presence of either 100 mM KCl or 10 mM MgSO4. Duplex formation was achieved by incubation equal amounts of GQ-forming strand and its complementary. Samples were heated up to 90°C for 30 min and allowed to cool down to room temperature for 16 h before measurement. The oligonucleotide concentrations were set to 1 μM for CD experiment and 5 μM for gel retardation.
A melting study of the structures of HPG2 was also performed to compare the structural stability of the two states in the presence of 100 mM K+ versus 100 mM Li+ (Figure 4B). The signal strength at 260 nm was used for the melting experiments because it was the most significantly changing wavelength when CD signatures were compared at different temperatures in the presence of K+ or Li+. The melting temperatures of these two structures were found to be relatively close in the presence of either ion (73°C and 71°C, respectively) which is in accordance with the mathematical predictions made previously (Table 3). It is important to note that such high melting temperatures require additional chaperons for the occurrence of a switch at physiological temperatures; however, the similar melting temperatures and similar free energies of both states suggests that members of HPG1 and HPG2 may be found in either state in vivo.
Identification of G-quadruplexes with shorter stem G-tract lengths
It is well-established that K+ has a strong stabilizing effect on G-quadruplexes (53–55). Considering the high K+ content in the E. coli cytosol, up to 200 mM (56), together with previous studies indicating a role of G-quadruplexes with shorter stem length (S) in prokaryotic gene regulation, we decided to perform the G-quadruplex prediction analysis with a new set of parameters. S was set to be equal or higher than 2 while L1-3 were set to be between 1 and 5 rather than 1 and 7. The shorter stem length (S) compromises the GQ stability. Since longer loop lengths result in a significant destabilization as well, (57–59) the maximum loop lengths (L1-3) was limited to 5 nt, so that the discovered GQs would not be too unstable.
The phylogenetic tree analysis and classification returned a large number of intragenic G-quadruplex groups, both within non-coding and coding regions (see Supplementary Data). We adopted the neighbor-joining method instead of UPGMA to obtain a more sensitive phylogenetic classification.
Surprisingly, the most of the clustered putative G-quadruplex (pGQ) groups were present in intragenic regions. pGQs within protein-coding regions were not processed further since these sequences may be conserved for their amino acid code rather than the G-quadruplexes they might form. We also detected pGQs groups in non-protein coding regions which consisted of tRNA, sRNA and rRNA genes. It is not surprising to observe several G-quadruplexes in non-protein coding RNA genes since the RNA-based G-quadruplexes are known to be much more stable in comparison to DNA-based ones and may not have to compete with duplex formation. Among pGQs in the intergenic regions, the majority of the sequences were present in repeat regions. However, groups found in the repeating regions were also disregarded since these sequences were present in very large and highly identical regions. The conservation was not only for a small region within the vicinity of GQ forming sequence, instead the region was conserved as a whole and showed no indication of conservation due to G-quadruplex formation.
To our amazement, after intense analysis the only G-quadruplex groups located between gene pairs and not associated with repeat regions were HPG1 and HPG2. Only a single degenerate was included in HPG1 when the group was discovered using an altered algorithm indicating that the motif enrichment can still be used to find other degenerate members (Supplementary Figure S2). This was mainly due to the fact that the other degenerate members were filtered by the degree of divergence threshold. In other words, they could be found by quadparser using the pattern designed for 2-tetrad GQs. However, even then they would be filtered out as they consisted of more than two nucleotide difference from the GQ-forming sequence of HPG1s. When a looser threshold was adopted these sequences can be included. Briefly, the use of the alternate algorithm enabled us to discover additional degenerate members of HPG1 and HPG2 without additional motif scanning however, the phylogenetic analysis became a much more rigorous work.
Only one pGQ group was found to be conserved among various regions; three members in tRNA-coding, three members in intergenic, one in sRNA and one in protein-coding regions. We disregarded the single members found in protein coding region mentioned above. Motif discovery using Glam2 yielded a motif with conserved G-quadruplex forming region only, indicating no alternative structure (Supplementary Figure S3).
DISCUSSION
Any intramolecular G-quadruplex forming sequence requires loop sequences between the G-tracts. Each following G-tract is required to fold onto itself in intramolecular GQs in order to align with other G-tracts. The necessary flexibility for the alignment is provided by this loop. Generally, the loop requires a greater length than a single nucleotide and is usually no longer than 7. Because the role of the loop is mainly steric, it is expected that the sequence of the loops are less effective than the stem on the formation of GQs. Indeed, most studies show that the loop composition has a marginal effect on the stability of the structures, while the length is highly influential on topology as well as stability. However, when considering a biological function of a GQ, we need to take into account the entire topology, not only the ability to form a G-quadruplex. In this case, the loop compositions may have a direct role through interactions with other biological molecules. For that reason, we hypothesized that if a G-quadruplex has a direct interaction with proteins, its topology and, consequently, its loop sequences should be conserved between GQ motifs of similar origin or function and thus, may be discovered through phylogenetic classification.
With this aim, for the first time, we have applied a classification for putative G-quadruplex forming sequences based on phylogenetic analysis to put weight on the loop sequences when studying GQs. Our analysis of the E. coli genome revealed two groups of putative G-quadruplex forming motifs by scanning for putative GQs using a generally accepted algorithm that allows freedom of the loop sequence. Sequences are then grouped using a phylogenetic clustering web tool revealing two major groups with very high sequence similarity. Interestingly these two groups are located between prokaryotic open reading frames and most of them are between operons indicating a regulatory role. Since the loop sequence influences the final topology of the G-quadruplex and its ability to form H-bonds with other biomolecules, the strong conservation of the loops clearly indicates that biological significance of the G quadruplexes not only comes from the formation of stacked G-tetrads but the formation of a specific G-quadruplex topology. For that reason, the investigation of any putative G-quadruplex without classification overlooks the potentially different roles of G-quadruplexes.
It should be remembered that the G-quadruplex algorithm did not allow any flexibility on the unity of the guanine-tracts; in other words, any sequences similar to HPGQ except an insertion/deletion mutation inside the guanine tract would not have been found. In order to include these, we have scanned the whole genome using the motif revealing three additional degenerate HPGQs. It should be remembered that the tool used to discover degenerate GQs has no regard for the preservation of the G-quadruplex, and the process gives equal weight to the mutations in any position of the motif. Coincidentally, the degenerate sequences could have fit our algorithm after a single substitution to extend the G-rich region. Moreover, it is not possible to eliminate G-quadruplex formation for degenerate forms since it is known that singled-out guanines in the flanks may take part in the formation of G-tetrads.
Genomic distribution analysis of these sequences showed strong correlation with each other, since they are often found in pairs and on opposite strands. In order to reveal a common motif including both HPG1 and HPG2 members an alignment was performed. Surprisingly, the alignment of all HPGQs together with their flanking sequences revealed the presence of a conserved C-rich region at the 5′ flank of each HPGQ and made us suspect the formation of non-GQ topologies. This indicated that HPGQs may have the ability to switch, not only between GQ and duplex DNA but also, a hairpin-like topology and even an antiparallel triplex. Thermodynamic calculations showed that both GQ and hairpin structures are possible indicating that these structures may switch between the two topologies. This transition would be an indication of an active role in the regulatory mechanism. We also showed using CD spectrophotometer measurements that such a structural transition occurs for HPGQ motifs depending on the associated ion. Whilst G-quadruplex formation was expected in the presence of K+, our findings clearly indicated that an alternative topology occurs in the presence of Mg++. The latter is similar to the structure established in the presence of Li+, which is a G-quadruplex destabilizing ion. Since both Mg++ and K+ are present in the E. coli cytosol, a regulatory switch based on both states is possible. Similar transitions between GQ and hairpins have previously been shown to have biological relevance (60,61). However it cannot be conclusive as the CD signatures of G-quadruplexes were shown to vary and the CD signature we obtained in Mg++ resembles to that of an alternate GQ-forming sequence (62). Further laboratory work will clarify potential topologies as well as the regulatory mechanism of HPGQs found in this study.
Briefly, the term G-quadruplex represents a large variety of topologies with one common feature, a non-canonical guanine tetrad. Besides this feature the tertiary structures may greatly vary depending on the tetrad count, loops or the buffer conditions. These directly influence the potential interactions of these molecules and thus their biological role. For this reason, it is essential to keep in mind that the generally accepted algorithms detects with no regard to the topology, stability or the tendency to prefer double stranded helix over G quadruplex. In order to eliminate the sequences fitting to the algorithm only coincidentally but which has no relevant G quadruplex formation, we applied a phylogenetic analysis to the output. Such analysis clusters the sequences depending on the loop sequences, and indirectly, the topological similarities of G-quadruplexes if formed. Our analysis of the E. coli genome revealed that two major groups of putative G-quadruplexes have highly similar loop sequences. Unexpectedly, 13 and 7 out of 52 were identical. A common motif was used to find three similar motifs in the genome but was missed by the algorithm due to a mutation inside the guanine tracts. It can be argued that mutations in the G-tract would prevent G-quadruplex formation; however, this is dependent on several factors since alternate nucleobases may replace a single guanine of a G-tetrad and still establish stable G-quadruplex. Alternatively, we found that looser rules may include these degenerates in the groups. Motif discovery studies showed that the predicted GQ-forming sequences were actually part of a larger motif which proposed formation of a hairpin as well as a GQ topology. Structural studies supported existence of a transition between hairpin and GQ by these motifs. Further structural and functional studies may reveal the biological role of these G-quadruplex motifs represented here where we are not inclined to speculate.
In conclusion, this work explains the use of well-established tools to cluster and enrich motifs as a follow-up to G-quadruplex prediction and in the process discovers a conserved bacterial motif conserved in various bacterial species and capable of switching between topological states suggesting to a regulatory role. Analysis of more complex model organism genomes is necessary for the evaluation of this methodology and better understanding of the biological roles of G-quadruplexes.
Supplementary Material
Acknowledgments
The authors are grateful to Zehra Sayers for providing access to CD spectrophotometer, Ines Karmous for her assistance during the measurements and Rose Doluca for the proofreading of this work. The authors also thank Oliver E. Blacque for critical reading of the manuscript and his valuable suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for Open Access charge: Dr Nezih Hekim.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pérez-Martín J., de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Hatfield G.W., Benham C.J. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 3.Rawal P., Kummarasetti V.B.R., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S.K., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paeschke K., Katrin P., Tomas S., Jan P., Daniela R., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 8.Du Z., Zhao Y., Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy J., Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2007;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cogoi S., Xodo L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S., Paramasivam M., Filichev V., Géci I., Pedersen E.B., Xodo L.E. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009;52:564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 17.Yadav V.K., Abraham J.K., Mani P., Kulshrestha R., Chowdhury S. QuadBase: Genome-wide database of G4 DNA occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2007;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frees S., Menendez C., Crum M., Bagga P.S. QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs. Hum. Genomics. 2014;8:8. doi: 10.1186/1479-7364-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrat E.A., Johnson B.R., Williams J.D., Borchert G.M., Larson E.D. G-quadruplex recognition activities of E. Coli MutS. BMC Mol. Biol. 2012;13:23. doi: 10.1186/1471-2199-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du X., Wojtowicz D., Bowers A.A., Levens D., Benham C.J., Przytycka T.M. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 2013;41:5965–5977. doi: 10.1093/nar/gkt308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holder I.T., Hartig J.S. A matter of location: Influence of G-Quadruplexes on Escherichia coli gene expression. Chem. Biol. 2014;21:1511–1521. doi: 10.1016/j.chembiol.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Kang S.-G., Henderson E. Identification of non-telomeric G4-DNA binding proteins in human, E. coli, yeast, and Arabidopsis. Mol. Cells. 2002;14:404–410. [PubMed] [Google Scholar]
- 23.Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahoon L.A., Seifert H.S. Focusing homologous recombination: Pilin antigenic variation in the pathogenic Neisseria. Mol. Microbiol. 2011;81:1136–1143. doi: 10.1111/j.1365-2958.2011.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong H.M., Stegle O., Rodgers S., Huppert J.L. A toolbox for predicting g-quadruplex formation and stability. J. Nucleic Acids. 2010;2010:564946. doi: 10.4061/2010/564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfsberg T.G. Using the NCBI map viewer to browse genomic sequence data. Curr. Protoc. Bioinform. 2010;29:1–25. doi: 10.1002/0471250953.bi0105s29. [DOI] [PubMed] [Google Scholar]
- 27.Aiyar A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 2000;132:221–241. doi: 10.1385/1-59259-192-2:221. [DOI] [PubMed] [Google Scholar]
- 28.Higgins D.G., Sharp P.M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 29.Sievers F., Higgins D.G. Clustal omega. Curr. Protoc. Bioinform. 2014;48:1–16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 30.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W., Sun Z. Random local neighbor joining: a new method for reconstructing phylogenetic trees. Mol. Phylogenet. Evol. 2008;47:117–128. doi: 10.1016/j.ympev.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Gascuel O., Steel M. Neighbor-joining revealed. Mol. Bio. Evol. 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- 33.Salgado H., Peralta-Gil M., Gama-Castro S., Santos-Zavaleta A., Muniz-Rascado L., Garcia-Sotelo J.S., Weiss V., Solano-Lira H., Martinez-Flores I., Medina-Rivera A., et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2012;41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frith M.C., Saunders N.F.W., Kobe B., Bailey T.L. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiliç S., White E.R., Sagitova D.M., Cornish J.P., Erill I. CollecTF: a database of experimentally validated transcription factor-binding sites in Bacteria. Nucleic Acids Res. 2014;42:D156–D160. doi: 10.1093/nar/gkt1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenz R., Ronny L., Bernhart S.H., Siederdissen C.H. zu, Hakim T., Christoph F., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernhart S.H., Hofacker I.L., Will S., Gruber A.R., Stadler P.F. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruber A.R., Bernhart S.H., Lorenz R. The ViennaRNA web services. Methods Mol. Biol. 2015;1269:307–326. doi: 10.1007/978-1-4939-2291-8_19. [DOI] [PubMed] [Google Scholar]
- 39.Mathews D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brázda V., Václav B., Lucia H., Jack L., Miroslav F. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002 doi: 10.1002/0471250953.bi0203s00. doi:10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 44.Bailey T.L., Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buske F.A., Bodén M., Bauer D.C., Bailey T.L. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 2010;26:860–866. doi: 10.1093/bioinformatics/btq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kypr J., Kejnovska I., Renciuk D., Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaushik M., Kaushik S., Bansal A., Saxena S., Kukreti S. Structural diversity and specific recognition of four stranded G-quadruplex DNA. Curr. Mol. Med. 2011;11:744–769. doi: 10.2174/156652411798062421. [DOI] [PubMed] [Google Scholar]
- 49.Randazzo A., Antonio R., Spada G.P., da Silva M.W. Circular dichroism of quadruplex structures. Topics Curr. Chem. 2012;330:67–86. doi: 10.1007/128_2012_331. [DOI] [PubMed] [Google Scholar]
- 50.Davis J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 51.Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 52.Alatossava T., Jütte H., Kuhn A., Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Włodarczyk A., Grzybowski P., Patkowski A., Dobek A. Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA. Photon correlation spectroscopy and circular dichroism studies. J. Phys. Chem. B. 2005;109:3594–3605. doi: 10.1021/jp045274d. [DOI] [PubMed] [Google Scholar]
- 54.Kim B.G., Long J., Dubins D.N., Chalikian T.V. Ionic Effects on VEGF G-Quadruplex Stability. J. Phys. Chem. B. 2016;120:4963–4971. doi: 10.1021/acs.jpcb.6b03731. [DOI] [PubMed] [Google Scholar]
- 55.Kim B., Byul K. Effects of salt on the stability of a G-Quadruplex from the human c-MYC promoter. Biophys. J. 2016;110:405. doi: 10.1021/acs.biochem.5b00097. [DOI] [PubMed] [Google Scholar]
- 56.Shabala L., Bowman J., Brown J., Ross T., McMeekin T., Shabala S. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol. 2009;11:137–148. doi: 10.1111/j.1462-2920.2008.01748.x. [DOI] [PubMed] [Google Scholar]
- 57.Pandey S., Agarwala P., Maiti S. Effect of loops and G-quartets on the stability of RNA G-quadruplexes. J. Phys. Chem. B. 2013;117:6896–6905. doi: 10.1021/jp401739m. [DOI] [PubMed] [Google Scholar]
- 58.Guedin A., Gros J., Alberti P., Mergny J.-L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnov I., Ivan S., Shafer R.H. Effect of Loop Sequence and Size on DNA Aptamer Stability †. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 60.Kuo M.H.-J., Zi-Fu W., Ting-Yuan T., Ming-Hao L., Hsu S.-T.D., Jing-Jer L., Ta-Chau C. Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 2015;137:210–218. doi: 10.1021/ja5089327. [DOI] [PubMed] [Google Scholar]
- 61.Romanucci V., Gaglione M., Messere A., Potenza N., Zarrelli A., Noppen S., Liekens S., Balzarini J., Di Fabio G. Hairpin oligonucleotides forming G-quadruplexes: new aptamers with anti-HIV activity. Eur. J. Med. Chem. 2015;89:51–58. doi: 10.1016/j.ejmech.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 62.Lam E.Y.N., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.