Abstract
Neisseria meningitidis causes bacterial meningitis and septicemia. It evades the host complement system by upregulating expression of immune evasion factors in response to changes in temperature. RNA thermometers within mRNAs control expression of bacterial immune evasion factors, including CssA, in the 5′-untranslated region of the operon for capsule biosynthesis. We dissect the molecular mechanisms of thermoregulation and report the structure of the CssA thermometer. We show that the RNA thermometer acts as a rheostat, whose stability is optimized to respond in a small temperature range around 37°C as occur within the upper airways during infection. Small increases in temperature gradually open up the structure to allow progressively increased access to the ribosome binding site. Even small changes in stability induced by mutations of imperfect base pairs, as in naturally occurring polymorphisms, shift the thermometer response outside of the desired temperature range, suggesting that its activity could be modulated by pharmacological intervention.
INTRODUCTION
Pathogenic microbes precisely control the expression of virulence factors in response to the host microenvironments. Neisseria meningitidis is a Gram-negative diplococcus and an obligate human pathogen, which is an important cause of septic shock and meningitis (1). Infection with this bacterium usually leads to asymptomatic colonization of the human nasopharynx (2), the site of several thermal gradients (3), but the meningococcus has evolved elaborate molecular strategies to evade host immunity, especially the complement system (4). Virtually all strains recovered from the blood or cerebrospinal fluid of patients with meningococcal disease express a polysaccharide capsule, an essential virulence determinant that promotes resistance against complement-mediated bacteriolysis (4), while around 60% of isolates carried in the nasopharynx are acapsulate. Strains which express sialic-acid containing capsules (belonging to serogroups B, C, Y and W) account for >95% of all cases in Europe and USA (5). For most isolates causing disease, N. meningitidis expresses a capsule composed of α2-8 linked sialic acid (serogroup B) which is a molecular mimic of a human molecule, and which provokes little if any immune response. For other serogroups, the capsule is a key antigen in several successful vaccines against the meningococcus (6). Polysaccharide-only and protein:polysaccharide-conjugate vaccines have proven effective at preventing meningococcal disease and are in routine use worldwide.
We have shown that the bacterium enhances expression of its immune evasion mechanisms as the temperature rises from those found in the nasopharynx (32°C and upward) to those in the systemic circulation (37°C) (7). Levels of the capsule, factor H binding protein (fHbp) (8) and the lipopolysaccharide-specific sialyl transferase (LST), all of which contribute to subversion of the complement system, increase in N. meningitidis over this narrow physiological range (9) to enable the meningococcus to sense local inflammation; upregulate its immune evasion mechanisms, and avoid elimination by host effectors, such as complement and phagocytes (10). Furthermore, thermal regulation by the meningococcus might provide an advantage to bacteria as they migrate to systemic circulation, which is at higher temperature than the nasopharynx (11).
The biosynthesis of sialic acid in the meningococcus, which is incorporated into lipopolysaccharide (LPS) and capsule, is encoded by the capsule biosynthesis operon, css. The thermal response in capsule production is mediated by an RNA thermometer located within 110 nucleotides of the 5′-untranslated region (5′-UTRs) of the css operon. Typically RNA thermometers located in the 5′-UTRs of a transcript form secondary structures at lower temperatures, which sequester the ribosome binding site (RBS). This prevents ribosomes accessing the mRNA, thereby preventing protein translation. As the temperature increases, the secondary structure is lost, allowing translation to proceed (12,13). Unlike other RNA thermometers studied so far (12), however, the css thermometer precisely regulates a single biochemical pathway, rather than affecting global responses to temperature. Furthermore, it responds to relatively small changes in temperature associated with the nasal cavity (14,15) and the host response (7,16), raising the mechanistic question of how this activity is structurally encoded within this RNA element.
In this study, we dissect the molecular mechanisms governing the activation of the cssA thermometer from N. meningitidis and report its three-dimensional structure. To date, there are remarkably few reports of the structure of RNA thermometers (17,18). The Cyanobacterial thermometer, one of the smallest naturally occurring RNA thermometers (18), consists of only 24 nucleotides, while a 34 nucleotide microdomain of a larger RNA structure has also been characterized (17). The cssA RNA thermometer usually contains two copies of an 8 nucleotide sequence (TATACTTA) located just upstream of the RBS and 15 nucleotides upstream of the cssA start codon (Figure 1) (7,19); with this 8 nucleotide repeat, the sequence of the 5′-UTR is remarkably conserved. However, some clinical isolates of N. meningitidis have lost one copy of the 8 nucleotide sequence and have enhanced capsule expression. Under these circumstances, polymorphisms in the 5′-UTR restores CssA thermoregulation and capsule expression (7).
Figure 1.
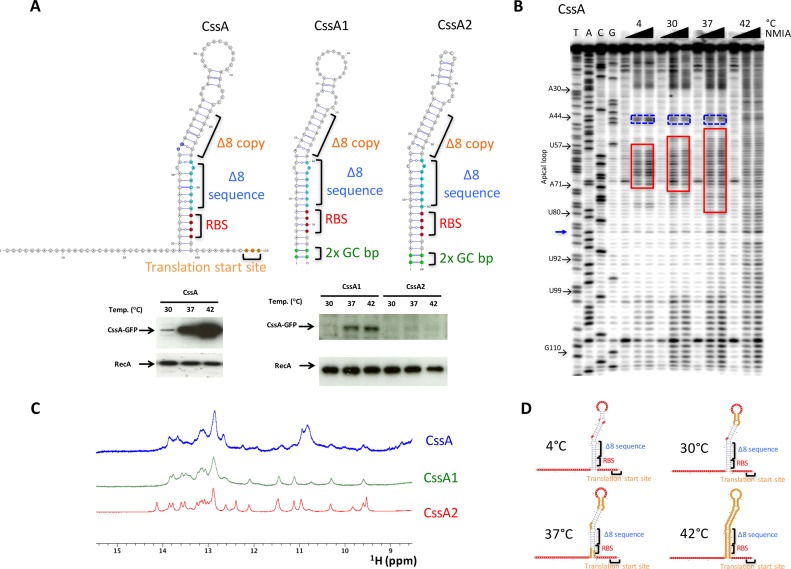
Thermosensing activity of CssA thermometer. (A) Proposed RNA secondary structure for the CssA RNA thermometers and for two variants (CssA1 and CssA2) prepared to perform nuclear magnetic resonance (NMR) studies; predicted structures were generated with VARNA (22). The lower panel shows the thermoregulated activity of the corresponding RNA constructs as assayed in Escherichia coli by western blot analysis of the protein products. The A44 and G45 nucleotides are highlighted with blue fill color on the CssA RNA secondary structure. (B) Selective 2′-OH acylation analyzed by primer extension (SHAPE) (21) for the CssA RNA establishes the RNA secondary structure at different temperatures, well below and just above activation of protein synthesis (4–42°C). (C) An overlay of the 1D imino region for the CssA RNA and for the two modified variants (CssA1 and CssA2). The considerable similarities in the imino resonances peak demonstrate the presence of very similar structures across all three RNAs. (D) Temperature-dependent SHAPE data summarized on the secondary structure of the CssA RNA; the 5′- and 3′-tails are reactive at all temperatures.
Using chemical probing and nuclear magnetic resonance (NMR), we show that, as the temperature is raised, the RNA thermometer unfolds, making the RBS accessible for efficient translation; these changes occur over temperatures that are relevant to the bacterium in the human nasopharynx. By applying selective 2′-OH acylation analyzed by primer extension (SHAPE) chemistry to map the RNA structure as a function of temperature, we demonstrate that the opening mechanism is not an ‘on-off’ process, but proceeds progressively, much like a rheostat. Even single nucleotide changes that increase the stability of the structure by a few degrees shift the temperature response outside of the physiological range, suggesting that this mechanism has exquisitely adapted to facilitate translation at temperatures that trigger immune evasion by N. meningitidis. Therefore our work demonstrates that every base counts in the capsule thermometer. Furthermore, the data provide insights into the impact of naturally occurring polymorphisms in the thermometer, and indicate that this structure can be modified to respond to different temperature changes and be exploited in bioengineering.
MATERIALS AND METHODS
RNA preparation
All RNAs studied during this work were prepared by in vitro transcription using T7 polymerase and ‘ultramer’ oligonucleotides (purchased from IDT) or regular DNA templates, depending on the length of the RNA (Supplementary Table S1). ‘Ultramer’ oligonucleotides were specifically used for RNAs longer >75 nucleotides. Additional details are provided in the Supplementary sections. The RNAs were purified by anion exchange chromatography with a salt gradient of 0.1–2M NaCl. The RNAs (which eluted at 200–600 mM salt concentration) were run on denaturing PAGE to analytically confirm size and purity. The 13C/15N- uniformly labeled RNA samples were also aligned in the presence of 10–15 mg/ml bacteriophage Pf1 (ASLA biotech) and residual dipolar couplings (RDCs) were measured using ARTSY (20).
Selective 2′-OH acylation analyzed by primer extension (SHAPE)
We performed SHAPE (21) chemical probing on the wild-type CssA thermometer, and on the CssA1 and CssA2 sequences over the temperature range of 4–42°C. The resulting chemical modifications were analyzed by primer extension with a radiolabeled DNA oligonucleotide and separated by gel electrophoresis. In order to accommodate primer binding, CssA1 and CssA2 were transcribed with a 3′-extension while for the wild-type RNA a primer complementary to the 3′-single stranded region was sufficient. Additional details are provided in the Supplementary Data.
The predicted RNA structures were generated with VARNA (22).
Mutagenesis studies and thermosensing activity
In order to drive the structure-based mutagenesis to alter the thermosensing activity, six different mutants were created (Supplementary Tables S1 and 2; Figure 2). Three variants (CssA6, CssA71, CssA72 and CssA75) were tested for thermometer activity as well (Supplementary Table S2 and Figure 3), while others had already been previously examined.
Figure 2.
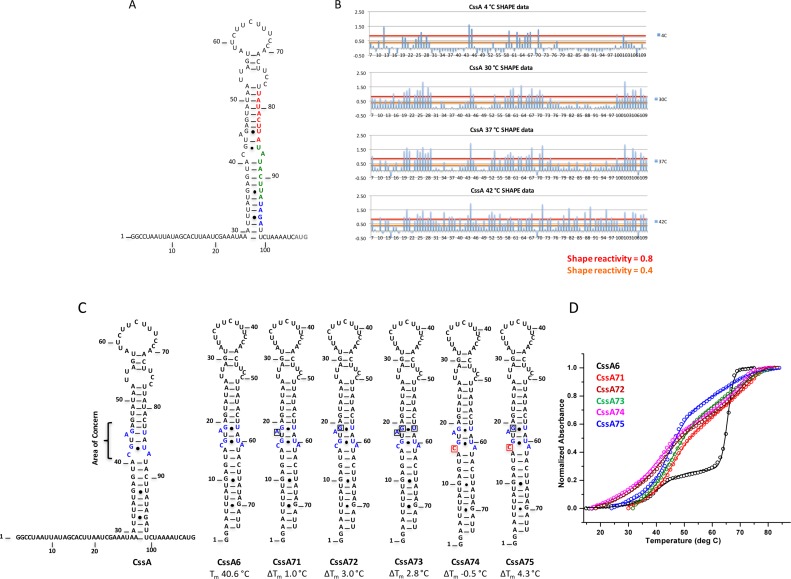
Per residue SHAPE reactivity and mutations introduced to stabilize the structure of the CssA thermometer. (A) Secondary structure of the CssA thermometer as predicted by mfold (35). Red and green colored nucleotides correspond to the two copies of the 8 bp sequence (TATACTTA), whereas blue colored nucleotides correspond to the RBS. The dark gray colored AUG represents the translation start site. (B) Quantification of SHAPE reactivity over the temperature range 4–42°C. Reactivity is classified as follows: 0.4–0.8 and >0.8 correspond to reactive and highly reactive sites, respectively. At 4°C, nucleotides at the 5′ and 3′-single stranded regions, the apical loop and the internal AG-U are chemically modified, indicative of a flexible, open structure; as the temperature increases, the flexibility spreads from the apical loop further down the structure; by 42°C, the entire RNA structure becomes reactive. (C) Secondary structure of various mutated thermometers (CssA6, 71, 72, 73, 74 and 75), where single nucleotide substitutions were introduced to alter the thermodynamic stability of the thermometer. Except for CssA74, all mutations generated a more stable RNA. The area highlighted in blue within the secondary structure is the region where mutations were introduced. Red colored nucleotides represent the mutated residue. (D) Thermal melting results for CssA6, CssA71, CssA72, CssA73, CssA74 and CssA75, shown in black, red, wine, olive-green, magenta and blue colors, respectively The CssA thermometer and its variants have two unfolding transitions around 39–45°C and 69°C, as is most evident in the black melting curve corresponding to CssA. This behavior is due to the formation of a dimeric, more stable RNA structures at the concentrations required for optical melting studies, which are not physiologically relevant.
Figure 3.
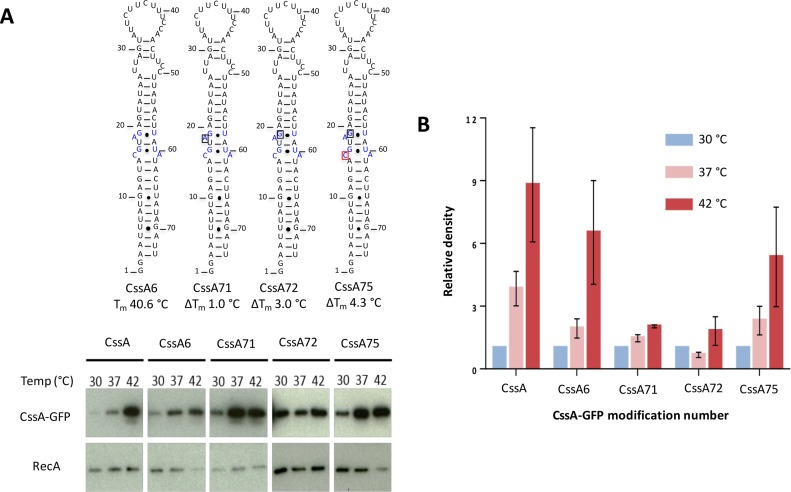
Correlation of thermosensing activity in Escherichia coli with thermodynamic stability. (A) Western blot analysis of CssA-GFP expression. Total protein isolated from indicated strains were subject to western blot analysis. Membranes were probed with antibodies recognizing GFP or RecA. Nucleotide shown in black box is removed in the respective sequence whereas the nucleotide in red box is mutated to U. Their respective melting temperature is shown (in°C) at the bottom of secondary structure. The blue colored residues in CssA6 are mutated/deleted in CssA71, CssA72 and CssA75. (B) Thermosensing properties of the CssA RNA constructs averaged over three biological replicates. The bar chart shows average expression of GFP relative to the loading control RecA, together with their standard deviations.
UV melting studies
Ultraviolet (UV) melting experiments were conducted on an Agilent 8453 diode array UV-Visible spectrophotometer by recording the absorbance verses temperature at 260 nM (Supplementary Table S3). The spectrophotometer has an external temperature controller which drives a Peltier heat pump attached to the sample holder to control the temperature in the range 10–90°C in 1 cm path length quartz cuvettes with a volume of 1400 μl. The rate of heating was kept at 1°C/min and each experiment was done at least twice to estimate reproducibility. The concentration of the RNAs was maintained at 1–2 μM in the NMR buffer. All the RNA samples were subjected to snap cooling at 90°C followed by degassing using a high-speed centrifuge before performing the thermal melting experiment to remove any bubbles. The melting temperature (Tm) was obtained using a two or multi-state equation (23) within Origin 8 (www.originlab.com).
Small angle X-ray scattering (SAXS)
RNA samples for small angle X-ray scattering (SAXS) were prepared similarly to samples made for NMR measurements, but in the lower concentration range of 1–2 mg/ml. SAXS experiments were recorded on beamline 4–2 at the Stanford Synchrotron Radiation Laboratory implementing inline FPLC-SAXS set-up (24,25). Frames with consistent Rg-values were merged in Primus (26). The particle distance distribution function P(r) plots were calculated using GNOM (27) and used for low resolution ab initio shape reconstruction with DAMMIN (28). A total of 16 models were generated with DAMMIN using in-house written scripts. The best and final model was selected with a suite of softwares (DAMSEL, DAMSUP, DAMAVER (29) and DAMFILT) (http://www.embl-hamburg.de/biosaxs) for comparison and fitting to the NMR structure. DAMSEL compares the models, finds the most probable model and identifies the outliers; DAMSUP aligns all models with the most probable model; DAMAVER averages these aligned models and computes a probability map; and DAMFILT filters the average model at a given default cut-off volume which is the expected volume of the generated PDB. DAMFILT removes loosely defined and lower occupancy atoms and generates a most probable compact model. Chi-square (χ2) values were determined by fitting the lowest energy NMR structure to the SAXS scattering profile. CRYSOL (30) was used to back calculate the Scattering profile from the pdb structure and determine the Chi-square from fit of the back-calculated scattering profile with the experimental SAXS data (Supplementary Tables S4 and 5).
NMR experiments
All NMR experiments were executed on Bruker Avance 800 or 600 MHz spectrometers equipped with triple resonance cryogenic probes. Exchangeable 2D 1H-1H NOESY and 2D [1H-15N] HSQC spectra were recorded in 95% H2O/5% 2H2O including 10 mM NaCl, 10 mM Phosphate pH 6.0 and 0.1 μM ethylenediaminetetraacetic acid at 7°C. The non-exchangeable 2D 1H-1H NOESY, 1H-1H TOCSY, DQF-COSY, 2D [1H-13C] HSQC, 1H-13C NOESY-HSQC and HCCH-TOCSY spectra were recorded in 99.9% 2H2O buffer at 25°C. The exchangeable and non-exchangeable NOESY spectra were recorded with various mixing times (50–250 ms) to facilitate spectral assignments and quantification of cross peak intensities by comparison of peak intensities. HNN-COSY experiments were recorded to obtain H-bonding constraints for the RNAs (31). Temperature dependent 1D imino spectra were recorded in the range of 5–45°C. All NMR data were processed either with Topspin (www.bruker.com) or NMRPipe (32) and analyzed using Sparky (33). For RDCs, ARTSY experiments (20) were recorded on (13C, 15N)-labeled RNA on an Agilent 900 MHz NMR system at NMRFAM equipped with a 5 mm 1H(13C/15N) cryogenic probe, oriented in the presence of 10–15 mg/ml bacteriophage Pf1 (ASLA biotech). Dephasing delays of 11.6, 5.0 and 5.5 ms were used for NH's, H6/H8/H2 and H1′/H5, respectively. The processing of ARTSY experiments recorded for isotropic and anisotropic samples was done with NMRPipe. Analysis was done using scripts kindly provided by Dr Ad Bax.
NMR spectral assignments
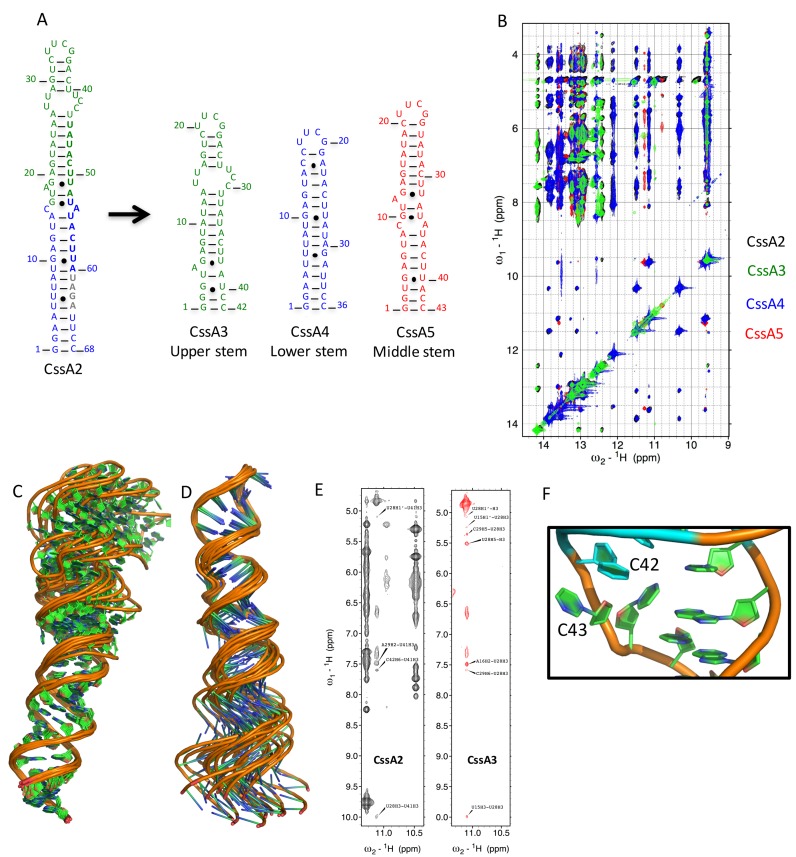
In order to calculate the RNA structure, we divided it into three segments, which overlap to generate the complete thermometer. Overlay of the CssA3, Cssa4 and CssA5 2D 1H-1H NOESY spectra with those of CssA2 showed remarkable similarities in the chemical shifts and allowed the direct transfer of assignments from the smaller RNAs to the complete thermometer (Supplementary Figures S1, 2, 3 and 4). Thus, analysis of CssA3, CssA4 and CssA5 RNA samples enabled the bulk of the assignments to be obtained. Details of the experiments used are provided in the supplementary section.
Experimental constraints
We generated restraint tables for CssA2 by combining CssA3, CssA4 and CssA5 distance restraints and torsion angles. Overlaying CssA3, CssA4 and CssA5 H2O and D2O 1H-1H NOESY spectra on the equivalent CssA2 spectra revealed a highly transferable pattern of chemical shifts and NOEs for the H1′-H6/H8/H2 walk and for the base pairs, including all of CssA4 base pairs (except for the A-C mismatch) and all of CssA3, including the unusual U-U mismatch (but excluding the A-U base pair below the bulged GA). Restraints from CssA3 spanned nucleotides G16-U53; those obtained with CssA4 spanned G1-A14 and U55-C68, while CssA5 contributed information at and around the A-C mismatch, namely A14-U17 and A52-U55. These restraints were used in the final structure calculation starting from simulated annealing through the addition of RDCs for final refinement. The NMR experimental constraints are summarized in Supplementary Table S4 and described in the Supplementary Data.
Structure determination
Xplor-NIH (34,35) was used for all structure calculations with a simulated annealing protocol. The target function used in simulated annealing included NOE derived and hydrogen bond distance constraints (final force constant = 50 kcal mol−1 Å2) as well as dihedral torsion angle (final force constant = 200 kcal mol−1 rad−2) restraints. Non-bonded interactions are described by a quartic van der Waals repulsion term (final force constant of 4 kcal mol−1 Å2 and a van der Waals radius scale factor of 0.8) and a torsion angle database potential (RAMA; with final weighting of 1). The simulated annealing procedure starts from randomized coordinates and initially undergoes high temperature torsion angle dynamics (8000 steps at 3500 K). The bath temperature is gradually cooled from 3500 to 298 K introducing the van der Waals terms and incrementally raising the force constants for a number of terms (angles, impropers, dihedral angles, NOEs, van der Waals repulsion and RAMA). Following the final cooling step, the molecule underwent two sequential final Powell minimizations, first in torsion angle space and then in Cartesian space. The calculations are repeated multiple times with different random number seeds for the assignment of initial velocities; the lowest energy structures without violations of distance (>0.3Å) or torsion angle (>5°) restraints were used for further refinement. Additional details are provided in the supplementary section.
RESULTS
The expression of three different genes involved in immune evasion by Neisseria is activated by temperature-sensing RNA thermometers within the 5′-UTR of the corresponding mRNAs. Unlike other RNA thermometers studied so far (12,17), which respond in a sigmoidal fashion to changes in temperature, the CssA thermometer responds in a graded manner within a narrow range corresponding to differences in the nasal cavity and central body temperature. According to mfold (36), the CssA thermometer forms a long imperfect stem-loop structure flanked by unfolded tails and capped by a 13 nucleotides large apical loop (Figure 1A). We used SHAPE chemistry and NMR to define the structure of this RNA and understand the mechanism of regulation.
Secondary structure of the CssA thermometer
SHAPE (21) chemical probing was performed to establish the RNA secondary structure between 4 and 42°C (Figure 1B), well below and just above the temperature of activation of protein synthesis. At 4°C, a temperature much lower than the bacterium encounters in the nasopharynx, chemical reactivity is observed only for flexible nucleotides within the 5′- and 3′-end single stranded regions, the large apical loop and the internal A/G-U bulge.
NMR spectroscopy was used to establish the base pairing unambiguously. We prepared two additional constructs, CssA1 and CssA2 (Figure 1A), to improve the spectral quality, which is compromised by the long unstructured tails and the large apical loop in the wild-type CssA RNA. In CssA1, both flanking tails were removed (two terminal GC base-pairs were added to facilitate transcription); for CssA2, the apical loop was exchanged with a stable UUCG tetraloop. CssA2 is not a good model for temperature regulation. For this, we employ other RNAs that are not as stabilized. However, all such constructs are dimeric at the NMR concentration. In contrast the UUCG containing construct is not dimeric and that allows structure determination. Thus, it is an excellent model for the structure, as demonstrate by the chemical shift superposition (see below).
Western blots demonstrate that CssA1 retains the behavior observed for CssA (albeit less efficiently as a result of the two extra base pairs), whereas the UUCG tetraloop abolishes activity over the temperature range (Figure 1A), by providing thermodynamic stability (37). However, the NH region of these three RNAs shows strong similarities indicating that the base pairing is the same as observed for the wild-type sequence (Figure 1C), a result confirmed by a thorough analysis of two dimensional spectra. As demonstrated later, the wild-type CssA sequence forms a duplex structure. The doubling of its effective molecular size leads to much broader NMR spectra (Figure 1C) and prevents high quality NMR analysis. Once this was done, spectral assignments confirmed the secondary structure generated by SHAPE, as shown in Figure 1A.
Temperature-dependent opening of the CssA thermometer
We used SHAPE and NMR to probe how temperature affects the RNA secondary structure. As shown by the SHAPE data (Figure 1B) and summarized in the site-dependent accessibility plots (Figures 1D and 2), the CssA thermometer starts unraveling from the apical loop (the unpaired A58-A70 region) followed by the A71-U78 region. Intense SHAPE reactivity is observed at all temperatures for A44 and G45 (as indicated by blue boxed on the gels; these nucleotides are located at the ‘inflection’ point of the structures of Figure 1A and are labeled in dark blue), suggesting that they might provide breathing points. All nucleotides become reactive at 42°C (Figure 2B), over a narrow temperature range, as the thermometer opens up from the top down. At and above 42°C, base pairing of the ribosome binding sequence is completely disrupted (Figure 1A).
Effect of single nucleotide changes on stability and activity
The results of Figure 1 suggest that the activity of the thermometer reflects its dynamic accessibility, and that temperature-induced opening of the secondary structure simply provides access to the RBS for activation of protein synthesis. To test this hypothesis, different mutations were introduced within the RNA (Figure 2C; Supplementary Tables S1 and 2), either around the internal A44G45-U84 bulge highlighted by the SHAPE data or by modifying the nucleotides of the G-U or C-A pairs to alter the RNA stability only subtly. The resulting melting curves report on the fraction of the RNA that is open (single stranded) or closed (base paired) at any given temperature for each RNA, because the change in absorption at 260 nM depends on whether a base pair is stacked on another or not (single stranded).
The starting structure for introducing mutations is called CssA6, which is identical to the CssA thermometer but lacks the flexible 5′-and 3′-end tails that complicate thermodynamic analysis. The mutations increased the thermostability by only 2–6°C (Figure 2C and Supplementary Table S2); larger changes would prevent correlating chemical physical stability with translational activation. For example, CssA71 (where the Adenine in the bulge is removed) is stabilized by ∼3°C while CssA75 (where the Adenine in the bulge is removed and the Guanine mutated to Adenine, respectively, to generate a UA base pair in place of a C-A mismatch) is stabilized by almost 6°C.
The melting curves for these RNAs (Figure 2D) are biphasic, but the only relevant transition occurs around 40°C; the second transition corresponds to melting of a dimeric structure, which is not physiologically relevant and is just a consequence of the concentrations required for biophysical studies. These dimeric structures are observed most clearly in SAXS that reveal a molecule about twice the size of the expected stem loop (Supplementary Figure S5A and B). Once we introduced a UUCG tetraloop, as in CssA2, the RNA adopted a single stem-loop conformation, as defined both by the molecule size and shape in SAXS experiments and by the thermodynamic melting experiments. We only consider the lower temperature transition in the remainder of this analysis.
The introduction of these mutations has a decisive effect on the thermometer activity (Figure 3). To study these mutations in a biological background, we fused these RNA constructs upstream of a GFP reporter system in pEGFP-N2 and transformed them into Escherichia coli. Their thermosensing properties were observed by western blotting using a GFP antibody in three biological replicates (Figure 3A). Expression of GFP (relative to loading control RecA), is shown in Figure 3B. We observed strong reduction in thermosensing ability for CssA71 and CssA72, which have increased thermodynamic stability created by deleting A18 (CssA71) or by the A18-G transversion (CssA72) (Figure 3). These effects parallel the increased thermodynamic stability of these two mutants of 1 and 3°C, respectively. Thus, the data indicate that the thermosensing activity over the physiological temperature range reflects the thermodynamic stability of the RNA, and that even small changes in temperature are sufficient to shift the response. However, stability alone is not sufficient to explain the entire spectrum of activity. When we further stabilized the structure by introducing a C15-A mutation (CssA75), partial thermosensing activity was restored.
The thermosensing activity of CssA was evaluated for a few additional mutations introduced in the background of a deletion of 8 nucleotides (instead of two copies of UAUACUUA, a single copy is retained) observed in polymorphic bacterial strains6. The first two mutants (Δ8 bp (AU/GC) where UAUACUUA becomes UAUGCCUA) and Δ8 bp (A/G) where UAUACUUA becomes UAUGCUUA) have similar thermal melting profile to CssA and partially restore thermosensing activity, whereas the Δ8bp (U/C) (UAUACUUA becomes UAUACCUA) mutation induces a thermal behavior as seen for CssA-Δ8bp and demonstrates unregulated CssA expression at all temperatures6 (Supplementary Tables S1, 2 and 3). Thus, mutations generated within the 8 bp sequence have variable effect in E. coli plasmid reporters. The SHAPE results for the Δ8 bp construct suggests that A81 (mutated in the AU/GC or A/G changes) is more reactive whereas U83 (for U/C change) is comparatively less reactive (Supplementary Figure S6). Changing A81 to G81 would stabilize the helix and restore thermosensing activity to wild-type levels in the presence of a single 8 bp sequence, while mutating the relatively stable nucleotide U83 to C83 would lead to additional disruption of the stem structure and would completely abolish the thermosensing activity and upregulate CssA production at every temperature.
Altogether, these data demonstrate that the thermosensing activity of the CssA thermometer responds to even small changes in temperature and, less strongly, to mutations of certain nucleotides, suggesting that the thermometer acts as a bona fide molecular rheostat whose optimal temperature response can be optimized by simple mutations.
Naturally occurring polymorphisms in the CssA thermometer
The sequence of the CssA thermometer is remarkably conserved among clinical N. meningitidis isolates. However, in some strains, one copy of the 8 base repeat is lost, and this change leads to upregulation of protein production at every temperature (7). In order to understand the mechanistic basis of this behavior, we performed thermal melting for several Δ8 bp constructs (referred to as CssA76, CssA77 and CssA78; Supplementary Tables S2 and 3). In CssA76, the first 8 bp adjacent to the RBS were switched with nucleotides present in the opposite strand, whereas the second 8 bp region was interchanged with the opposite strand in CssA77; both 8 bp regions were switched with nucleotides from the other strand in CssA78. As a result of these changes, CssA77 has a similar thermal profile to CssA6, while CssA76 and CssA78, have lower melting points, around 35°C (7). These results are consistent with the dysregulation of thermosensing activity observed in strains that carry these polymorphic variations (7).
NMR analysis of the CssA thermometer
The wild-type CssA sequence is not suitable for structural investigation. The relatively poor quality of the imino section of the NMR spectra is reflected in even broader spectra for the non-exchangeable region of the spectrum. The same was also found for CssA1 or CssA6, and is due to the formation of a dimeric structure that cannot be avoided at NMR concentrations but is irrelevant to the biology of the thermometers. Only when we introduced a UUCG tetraloop, as in CssA2, the RNA adopted only a stem-loop conformation, as defined both by the molecule size and shape in SAXS experiment and by the behavior in thermodynamic melting experiments. When we compared imino spectra for CssA1 and CssA2, they looked nearly identical, with the few extra peaks in CssA2 originating from the UUCG-tetraloop (Supplementary Figure S5D). These results, and the monomeric structure of the RNA, demonstrate that CssA2 is an accurate model to probe the structure of the CssA thermometer.
We assigned the NMR spectra (Figure 4A) and recorded 1D imino proton spectra for both CssA1 and CssA2 to assess the thermal response at the nucleotide level at temperatures used for the SHAPE analysis (Figure 4B). We found that the upper helix is more thermolabile than the lower stem, as in SHAPE studies. However, imino peaks corresponding to the lower helix are still intact in both CssA1 and CssA2 even at 45°C. This indicates that, as the temperature increases, the RNA unfolds from the apical loop and this destabilization spreads down toward A30-U39, the region of the thermometer containing the RBS, thus making it accessible for ribosome binding.
Figure 4.
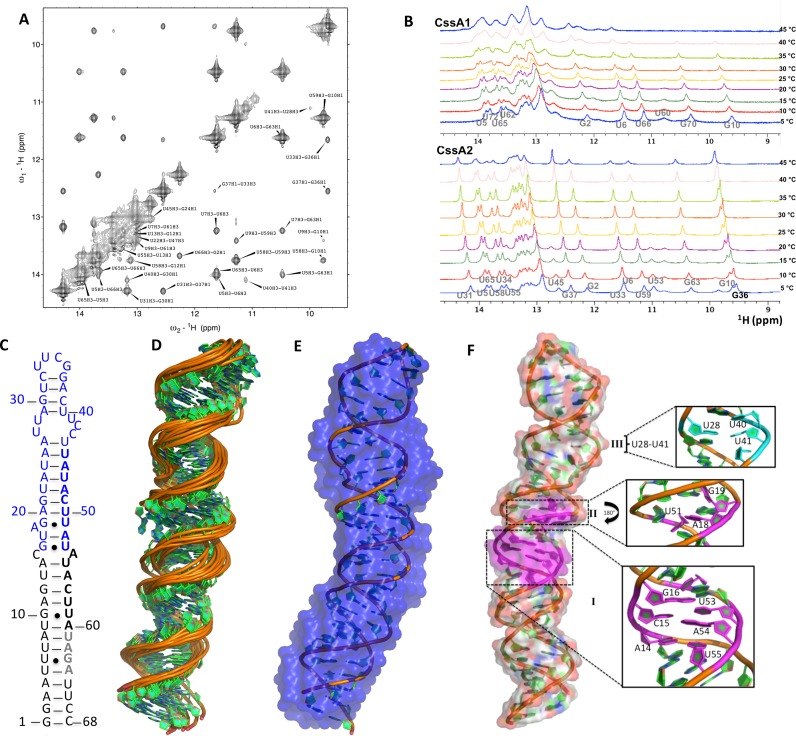
Structural analysis of the CssA thermometer. (A) Imino-region of the NOESY spectrum of CssA2, which allowed assignments of the base pair resonances. (B) Temperature dependence of the imino spectra for CssA1 (top) and CssA2 (bottom) reveals opening of the structure at the level of individual base pairs. (C) Schematic representation of the secondary structure of CssA2, based on the NMR structure. Residues corresponding to CssA3 and CssA4 are highlighted in blue and black, respectively, to show how the complete structure was constructed by splicing together the two segments. (D) NMR ensemble of 10 refined structures of CssA2 (residues 1–64). (E) The lowest energy structure of CssA2 in cartoon representation superposed on the SAXS model shown in blue color; the SAXS results were not used for refinement, they therefore provide independent validation of the structure. (F) Complete structure of CssA2, highlighting local structural features of the CssA core structure: (I) A14-U55, C15*A54, G16-U53; (II) A18G19-U51 in magenta and (III) U28-U41 in light green color.
NMR structure of the CssA thermometer
We used a ‘divide and conquer’ approach to obtain a much larger number of constraints for structure determination than would be possible if we just studied the complete RNA, whose large and asymmetric structure leads to rapid relaxation of the NMR signal. We divided CssA2 into three partially overlapping segments, designated CssA3, CssA4 and CssA5, which, when the common base pairs are superimposed, generate CssA2. CssA3 spans the upper helix whereas CssA4 spans the lower stem; CssA5 consists of middle stem of CssA2 (Figures 4C–F and 5A). Overlaying the NOESY NMR spectra of the three segments on the CssA2 spectra revealed a highly transferable pattern of chemical shifts and NOESY cross-peaks for both the exchangeable and non-exchangeable protons (Figure 5B). Thus, we generated a restraint table for CssA2 by combining distance and torsion angle restraints from each segment to generate a structure for the complete RNA. These restraints were used in the final structure calculation starting from simulated annealing through the addition of RDCs for final refinement (38,39) (Supplementary Table S4). We calculated the overall CssA2 structure (Figure 4) starting from simulated annealing and subsequent refinement using first local RDCs for the upper and lower stems to refine the local geometry, then introduced global RDCs obtained for the CssA2 NH's to orient the upper and lower stems along with the NOEs. The top 10 structures without violations in distance (>0.3Å) or torsion angle (>5°) restraints were analyzed (Supplementary Table S4). For further validation, we used the SAXS-generated envelope; we found that the NMR-generated structure fits very well within the SAXS-model, which was not used in any way in the refinement (Figure 4E).
Figure 5.
NMR Assignment strategy and structural insight into CssA2 thermometer. (A) Three smaller constructs (CssA3, CssA4 and CssA5) were prepared to help structure determination of CssA2. Green and blue bold letters correspond to the 8 base sequence whereas gray colored bold nucleotides are the RBS. CssA3 corresponds to the upper stem of CssA2, CssA4 overlaps with the lower stem while CssA5 is representative of middle region of CssA. CssA5 was only used to refine the relative orientation of the upper and lower helices. (B) Overlay of the imino regions of 2D 1H-1H NOESY spectra recorded in 93% H2O and 7% 2H2O on Bruker Avance 800 MHz spectrometer at 7°C for CssA2, CssA3, CssA4 and CssA5. The black color corresponds to CssA2, green color is for CssA3, blue color for CssA4 and red color is used for CssA5. An excellent match is seen between each of the fragments (CssA3/4/5) and CssA2. (C) 1H-15N HSQC spectrum recorded in 95% H2O/5% 2H2O for CssA2 labeled with assigned peaks. The assignments were based on the spectra recorded for CssA2, CssA3, CssA4 and CssA5. Structural superposition of CssA2 RNA overlayed on (C) the lower helix (corresponding to CssA4) or (D) the upper helix (corresponding to CssA3). (E) Direct NMR evidence to support the U28*U41 base pair observed in NOESY spectra of CssA2 and CssA3, to demonstrate the similarities between the two constructs. (F) Orientation of C42 and C43 shown on the structure. The C43 base and ribose are projected toward the major groove, while the C42 ribose points inward, but its base has moved outside the helix, exposing both bases.
The CssA thermometer forms an extended helix of ∼100 Å in length with a twist of around 30 deg around the A18G19-U51 bulge, which forms a locally flexible hinge. The structure has three local distortions: the wobble pair G16-U53 (I), an A18G19U51 bulge (II) and a U-C-rich internal loop (III) (Figure 4F). The lower helix (corresponding to CssA4) is well defined, with a local rmsd of only 0.68 Å (Figure 5C and Supplementary Figure S3) whereas the upper helix (corresponding to CssA3) showed some flexibility in the regions A26-U28, C42-U44 and converges with a higher backbone rmsd of 1.1 Å (Figure 5D). These observations suggest that the apical helix is only partially stable because of the presence of irregular structure whereas the lower helix is more rigid, a result consistent with the SHAPE and NMR data that indicate top-down melting of the secondary structure. The U28-U41 base pair in region III is only observed below 30°C (Figures 4F and 5E). The A18*U51 base pair in region II also forms transiently; U51(H3) is near A18 (H2), indicating formation of a base pair with A18, while G19 is outside of the A18-U51 plane (Figure 4F). In region I, A14 is perfectly aligned toward U55 (H3, O4) while the C15 base moves away from A54 and makes no contact with either base. G16 (H1/O6) forms a wobble base pair with U53 which opens up at 37°C (Figure 4B and F). Near region III, C42 and C43 remain very flexible in the core structure (Figures 4F and 5F) as supported by strong SHAPE reactivity for the CssA thermometer (Figure 1B and Supplementary Figure S5C).
DISCUSSION
RNA thermometers provide an elegant mechanism for microbial adaption to different temperatures (40). Their advantage is that they function independently of a response regulator, and do not require de novo transcription. They are energetically economical and offer rapid activation and reversibility and are therefore ideally suited for responding to abrupt changes in the environment.
Several enteric pathogens control the expression of key transcription factors or cellular heat shock responses through RNA thermometers. For example, Salmonella (41), Yersina pestis (42), E. coli, Vibrio cholerae (16) and Listeria monocytogenes (43) employ RNA thermometers to re-program gene expression during transition from the external environment to the higher temperatures found in the mammalian gastrointestinal tract (44). In these examples, RNA thermometers sense a rapid and large fluctuation in temperature as the key cue for adaptation to distinct setting within and outside their host. Most offer a binary response to the ambient environment, with translation either being completely stalled by a stable secondary RNA structure preventing access of ribosomes to the nascent mRNA, or being active when the RNA structure melts, allowing protein synthesis. There are other emerging examples of thermometers which promote translation at lower temperatures in microbes as divergent as Thermus thermophilus (45) and E. coli to mediated cold shock or adaptive responses (46); this post-transcriptional response is mediated by secondary structures in the 5′ UTR of the mRNA which form at lower temperatures to either protect the transcript from degradation, or enable more efficient access of ribosomes to the RBS.
In contrast, the CssA thermometer from N. meningitidis regulates bacterial evasion from the host immune responses by controlling the biosynthesis of a single molecule, rather than mediating pleiotropic responses in the bacteria. Subtle increases in temperature, as observed in the air cavities during inflammation, activate protein synthesis downstream of this RNA structure (7). Thus, the CssA thermometer responds in a gradual manner, like a rheostat, over what in physical chemical terms is a very modest temperature change. We think of this structure as a rheostat because the thermometer activity increases in a more or less constant manner over the temperature range, consistent with the progressive melting of the structure, leading to progressive increase in protein expression, with a maximum temperature that can also be gradually moved with small changes in stability.
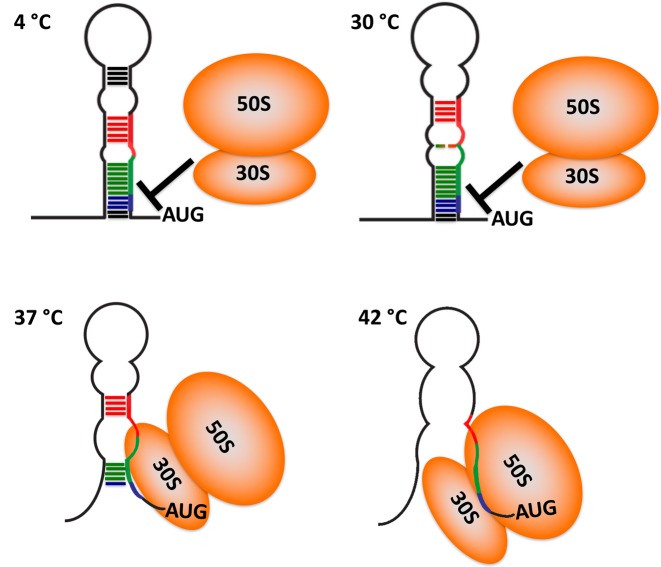
The NMR structure, SHAPE analysis and the correlation between thermodynamic stability and activity, lead to the model of thermosensing illustrated in Figure 6. Although the structure of the thermometer is ‘closed’ at temperatures below activation, in the narrow transition temperature range around 37°C, the thermometer exists in equilibrium between closed and open states. The ribosome samples the population of open thermometers to recognize the RBS and initiate translation. The structure begins to ‘melt’, from the top of the helix downward, starting from the A18G19-U51 internal loop, while the RBS sequence remains occluded by complementary base pairing; around 37°C, the lower helix becomes partially open making the RBS available for the ribosome to initiate translation (7). As the temperature rises to 42°C, as occurs during infection, the thermometer becomes completely destabilized, liberating the RBS and permitting ribosome entry (Figure 6 and Supplementary Video S1).
Figure 6.
Model for the thermosensing activity of the CssA thermometer. Cartoon representation of temperature induced unzipping the thermometer, as revealed by NMR spectroscopy and SHAPE assays. Dark colored lines represent stable base pairs, whereas dotted lines correspond to weaker base-pairs which open up at the higher temperatures. At 4°C, all base pair regions are stably formed, and the ribosome cannot access the RBS which is fully occluded. As the temperature is raised toward 30°C, the RNA structure opens up, starting near the apical loop and the A44G45-U84 bulge in center of the repeated 8 bp elements, shown in red and green, respectively. At this temperature, the RBS, shown in blue color, is still inaccessible and protein synthesis inhibited. By 42°C, the thermometer structure is fully open, leading to efficient translation. Around the narrow transition temperature range near 37°C, the thermometer exists in equilibrium between closed and open states. The ribosome (depicted in orange) samples the population of open thermometers to recognize the RBS and initiate translation.
The architecture of the CssA thermometer is reminiscent of the cyanobacterial hsp17 thermometer (18). In the cyanobacterial structure (one of the smallest thermometers) the 3 nucleotide bulge separates the start codon from the RBS. This is distinct from the CssA thermometer, where the bulge is larger and located on one side of the RBS and translation start site. Similar to N. meningitidis, however, the hsp17 structure responds gradually to temperatures, albeit over a larger temperature range in vitro, achieved via a zippering mechanism, which enables the translation of chaperones, involved in the heat shock response.
We previously identified naturally occurring polymorphisms in the 5′ UTR of the CssA mRNA, which lead to dysregulated production of CssA (7). These changes distort the structure in a manner that is amplified when the temperature increases. Single nucleotide substitutions that stabilize the structure by only a few degrees, lead to a corresponding decrease in activation of protein synthesis and a shift of activation to higher temperature. This result suggests that small molecules that stabilize the RNA would prevent evasion of the host immune response. The NMR structure of the thermometer identifies distortions in the long base paired helix that could be used for targeting the RNA with small molecules within the upper helix, where the structure begins to melt. Furthermore, the substitutions demonstrate that the optimal activity of the thermometer itself can be optimized by simple engineering of its sequence, providing an opportunity to regulate protein synthesis at different temperatures.
RNA thermometers provide a quick, reversible, and energetically efficient strategy for a bacterium to respond to changes in temperature. As our findings demonstrate, the atomic structure of the thermometer and its response can be exquisitely adjusted to suit the requirements of the bacterium; among disease isolates, even the loss of an 8 nucleotide repeat sequence can be counterbalanced by the acquisition of compensatory mutations that precisely restore the function of the thermometer. Therefore, pathogenic strains of N. meningitidis retain the capacity to upregulate the expression of capsule as they transit from the cooler environment of the nasopharynx to the temperature in the systemic circulation, following invasion of the epithelial barrier. Our study provides the molecular mechanism underlying this key step in virulence, and for understanding the effects of polymorphisms which are more prevalent in carried strains, and thence the transition from commensalism to disease in this important human pathogen.
Supplementary Material
Acknowledgments
We are grateful to Drs Mike Guttman and Marco Tonelli for help with the collection of SAXS data at Stanford Synchrotron Radiation Lightsource (SSRL) and ARTSY experiments at National Magnetic Resonance Facility at Madison (NMRFAM), respectively.
ACCESSION NUMBERS
NMR assignments of CssA2, CssA3, CssA4 and CssA5 were submitted to BMRB under submission ID 25784, 25781, 25780 and 25785, respectively. The structure of CssA2, CssA3, CssA4 and CssA5 were deposited with PDB IDs: 2n6w, 2n6t, 2n6s and 2n6x, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [5RO1 GM110569 to G.V.]; Wellcome Trust and Medical Research Council (to C.M.T.); Federal Contract from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [HHSN272201200025C]; Swedish Research Council [Dnr: 2014–2050 to E.L.]; NIH [P41GM103399] (NIGMS), old number [P41RR002301] to National Magnetic Resonance Facility at Madison; University of Wisconsin-Madison; NIH [P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220]; NSF [DMB-8415048, OIA-9977486, BIR-9214394]; USDA. Funding for open access charge: NIH [5RO1 GM110569].
Conflict of interest statement. None declared.
REFERENCES
- 1.Pizza M., Rappuoli R. Neisseria meningitidis: pathogenesis and immunity. Curr. Opin. Microbiol. 2015;23:68–72. doi: 10.1016/j.mib.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Yazdankhah S.P., Caugant D.A. Neisseria meningitidis: an overview of the carriage state. J. Med. Microbiol. 2004;53:821–832. doi: 10.1099/jmm.0.45529-0. [DOI] [PubMed] [Google Scholar]
- 3.Keck T., Leiacker R., Riechelmann H., Rettinger G. Temperature profile in the nasal cavity. Laryngoscope. 2000;110:651–654. doi: 10.1097/00005537-200004000-00021. [DOI] [PubMed] [Google Scholar]
- 4.Schneider M.C., Exley R.M., Ram S., Sim R.B., Tang C.M. Interactions between Neisseria meningitidis and the complement system. Trends Microbiol. 2007;15:233–240. doi: 10.1016/j.tim.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Harrison O.B., Brueggemann A.B., Caugant D.A., van der Ende A., Frosch M., Gray S., Heuberger S., Krizova P., Olcen P., Slack M., et al. Molecular typing methods for outbreak detection and surveillance of invasive disease caused by Neisseria meningitidis, Haemophilus influenzae and Streptococcus pneumoniae, a review. Microbiology. 2011;157:2181–2195. doi: 10.1099/mic.0.050518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L.K., Carlone G.M., Borrow R. Advances in the development of vaccines against Neisseria meningitidis. N. Engl. J. Med. 2010;362:1511–1520. doi: 10.1056/NEJMra0906357. [DOI] [PubMed] [Google Scholar]
- 7.Loh E., Kugelberg E., Tracy A., Zhang Q., Gollan B., Ewles H., Chalmers R., Pelicic V., Tang C.M. Temperature triggers immune evasion by Neisseria meningitidis. Nature. 2013;502:237–240. doi: 10.1038/nature12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider M.C., Prosser B.E., Caesar J.J., Kugelberg E., Li S., Zhang Q., Quoraishi S., Lovett J.E., Deane J.E., Sim R.B., et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swartley J.S., Marfin A.A., Edupuganti S., Liu L.J., Cieslak P., Perkins B., Wenger J.D., Stephens D.S. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quagliarello V. Dissemination of Neisseria meningitidis. N. Engl. J. Med. 2011;364:1573–1575. doi: 10.1056/NEJMcibr1101564. [DOI] [PubMed] [Google Scholar]
- 11.Cole P. Recordings of respiratory air temperature. J. Laryngol. Otol. 1954;68:295–307. doi: 10.1017/s0022215100049690. [DOI] [PubMed] [Google Scholar]
- 12.Grosso-Becera M.V., Servin-Gonzalez L., Soberon-Chavez G. RNA structures are involved in the thermoregulation of bacterial virulence-associated traits. Trends Microbiol. 2015;23:509–518. doi: 10.1016/j.tim.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Narberhaus F., Waldminghaus T., Chowdhury S. RNA thermometers. FEMS Microbiol. Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 14.McFadden E.R., Jr, Pichurko B.M. Intraairway thermal profiles during exercise and hyperventilation in normal man. J. Clin. Investig. 1985;76:1007–1010. doi: 10.1172/JCI112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole P. Some aspects of temperature, moisture and heat relationships in the upper respiratory tract. J. Laryngol. Otol. 1953;67:449–456. doi: 10.1017/s0022215100048908. [DOI] [PubMed] [Google Scholar]
- 16.Weber G.G., Kortmann J., Narberhaus F., Klose K.E. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14241–14246. doi: 10.1073/pnas.1411570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury S., Maris C., Allain F.H., Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner D., Rinnenthal J., Narberhaus F., Schwalbe H. Mechanistic insights into temperature-dependent regulation of the simple cyanobacterial hsp17 RNA thermometer at base-pair resolution. Nucleic Acids Res. 2015;43:5572–5585. doi: 10.1093/nar/gkv414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo H., Tang C.M., Exley R.M. Mechanisms of avoidance of host immunity by Neisseria meningitidis and its effect on vaccine development. Lancet Infect. Dis. 2009;9:418–427. doi: 10.1016/S1473-3099(09)70132-X. [DOI] [PubMed] [Google Scholar]
- 20.Ying J., Wang J., Grishaev A., Yu P., Wang Y.X., Bax A. Measurement of (1)H-(15)N and (1)H-(13)C residual dipolar couplings in nucleic acids from TROSY intensities. J. Biomol. NMR. 2011;51:89–103. doi: 10.1007/s10858-011-9544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson K.A., Merino E.J., Weeks K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 22.Darty K., Denise A., Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnwal R.P., Jobby M.K., Devi K.M., Sharma Y., Chary K.V. Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea. J. Mol. Biol. 2009;386:675–689. doi: 10.1016/j.jmb.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 24.Guttman M., Lee K.K. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J. Virol. 2013;87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M., Weinkam P., Sali A., Lee K.K. All-atom ensemble modeling to analyze small-angle x-ray scattering of glycosylated proteins. Structure. 2013;21:321–331. doi: 10.1016/j.str.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 27.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 28.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing (vol 76, pg 2879, 1999) Biophys. J. 1999;77:2896–2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svergun D., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 31.Dingley A.J., Nisius L., Cordier F., Grzesiek S. Direct detection of N-H[...]N hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008;3:242–248. doi: 10.1038/nprot.2007.497. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Goddard T.D., Kneller D.G. SPARKY 3. San Francisco: University of California; 2002. [Google Scholar]
- 34.Schwieters C.D., Clore G.M. Using small angle solution scattering data in Xplor-NIH structure calculations. Prog. Nucl. Magn. Reson. Spectrosc. 2014;80:1–11. doi: 10.1016/j.pnmrs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varani G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 38.Lipsitz R.S., Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu. Rev. Biophys. Biomol. Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 39.Prestegard J.H., Al-Hashimi H.M., Tolman J.R. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 40.Johansson J. RNA thermosensors in bacterial pathogens. Contrib. Microbiol. 2009;16:150–160. doi: 10.1159/000219378. [DOI] [PubMed] [Google Scholar]
- 41.Waldminghaus T., Heidrich N., Brantl S., Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007;65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 42.Hoe N.P., Goguen J.D. Temperature sensing in yersinia-pestis—translation of the Lcrf activator protein is thermally regulated. J. Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 44.Morita M.T., Tanaka Y., Kodama T.S., Kyogoku Y., Yanagi H., Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mega R., Manzoku M., Shinkai A., Nakagawa N., Kuramitsu S., Masui R. Very rapid induction of a cold shock protein by temperature downshift in Thermus thermophilus. Biochem. Biophys. Res. Commun. 2010;399:336–340. doi: 10.1016/j.bbrc.2010.07.065. [DOI] [PubMed] [Google Scholar]
- 46.Giuliodori A.M., Di Pietro F., Marzi S., Masquida B., Wagner R., Romby P., Gualerzi C.O., Pon C.L. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.