Abstract
Classical zinc fingers (ZFs) are traditionally considered to act as sequence-specific DNA-binding domains. More recently, classical ZFs have been recognised as potential RNA-binding modules, raising the intriguing possibility that classical-ZF transcription factors are involved in post-transcriptional gene regulation via direct RNA binding. To date, however, only one classical ZF-RNA complex, that involving TFIIIA, has been structurally characterised. Yin Yang-1 (YY1) is a multi-functional transcription factor involved in many regulatory processes, and binds DNA via four classical ZFs. Recent evidence suggests that YY1 also interacts with RNA, but the molecular nature of the interaction remains unknown. In the present work, we directly assess the ability of YY1 to bind RNA using in vitro assays. Systematic Evolution of Ligands by EXponential enrichment (SELEX) was used to identify preferred RNA sequences bound by the YY1 ZFs from a randomised library over multiple rounds of selection. However, a strong motif was not consistently recovered, suggesting that the RNA sequence selectivity of these domains is modest. YY1 ZF residues involved in binding to single-stranded RNA were identified by NMR spectroscopy and found to be largely distinct from the set of residues involved in DNA binding, suggesting that interactions between YY1 and ssRNA constitute a separate mode of nucleic acid binding. Our data are consistent with recent reports that YY1 can bind to RNA in a low-specificity, yet physiologically relevant manner.
INTRODUCTION
Classical zinc fingers (ZFs) are ∼30-residue protein domains that are extremely abundant in complex organisms and have a well-established role as a DNA-binding module in transcription factors (TFs, reviewed in (1,2)). In recent years, however, there has been increasing recognition that classical ZFs, which often occur in tandem arrays, can participate in diverse protein–RNA and protein–protein interactions (reviewed in (2,3)). The canonical example of this phenomenon is the TF TFIIIA, which contains nine classical ZFs. Fingers 4–7 bind both the internal control region of the 5S RNA gene, promoting transcription, and also the 5S rRNA message itself (4–6). Transport of this ribonucleoprotein complex out of the nucleus prevents DNA binding by TFIIIA, negatively regulating 5S rRNA transcription. Another well-studied case is the Wilms’ tumour suppressor protein (WT1), a transcription factor containing four classical ZFs. Isoforms lacking a three amino acid (KTS) insertion between the third and fourth fingers bind Igf-2 exon 2 DNA, whilst those bearing the KTS insertion preferentially bind the corresponding RNA; this latter activity is proposed to regulate levels of alternatively spliced Igf-2 transcripts (7,8). Similarly, the classical ZFs of human ZNF74 have been suggested to be involved in both transcriptional regulation and mRNA metabolism depending on splicing context (9,10).
Despite the implication that non-DNA-binding functions might be more common for classical ZFs than previously recognised, experimental evidence for dual DNA/RNA interactions is still relatively limited. To date, TFIIIA remains the only classical-ZF protein for which structural data on both DNA and RNA complexes are available (4,5). Nevertheless, there are other RNA-binding classical-ZF proteins that have been shown to display specificity for RNA over DNA. For example, the ZFs of ZFa, JAZ-1 and wig-1, which are characterised by elaborations on the normal classical ZF fold and also long non-canonical inter-ZF linkers, preferentially bind double-stranded (ds) RNA (11–13). A number of classical-ZF proteins are also known to bind single-stranded (ss) RNA from competition assays using homopolymeric RNA, although their in vivo RNA targets are unknown (9,14,15).
Yin-Yang 1 (YY1) is a TF involved in diverse regulatory pathways, including many implicated in oncogenesis, and the protein is proposed to function as a transcriptional repressor, activator and initiator in the context of different promoters (reviewed in (16–18)). YY1 comprises (i) a large and disordered N-terminal region that contains transcriptional activation and repression domains, and (ii) four C-terminal classical ZFs (separated by canonical five-residue linkers) that bind a defined DNA sequence motif (19,20). The YY1–DNA interaction has been extensively characterised, both biochemically and structurally (21,22). YY1 ZFs are also thought to interact with various protein partners (reviewed in (3,17)), although no biophysical data on purified proteins is currently available.
Several studies also support a functional role for YY1 as an RNA-binding protein. YY1 has been suggested to be required for assembly of Xenopus oocyte messenger ribonucleoprotein particles (mRNPs) through a direct interaction with various mRNAs (23,24). It has also been reported that both DNA and RNA binding by YY1 are important for the physical association of the long non-coding RNA X-inactivation specific transcript (Xist) with the Xist locus during X-inactivation in mouse embryonic fibroblasts (25). Xist RNA is transcribed from and remains localised to the X chromosome chosen to be inactivated in each mammalian female somatic cell. This localisation of Xist initiates a cascade of events that leads to silencing of genes in cis. However, the mechanisms by which Xist is localised and the inactivation signal is spread along the chromosome have not been delineated (reviewed in (26,27)). Jeon and Lee proposed a model in which YY1 acts as a protein tether linking the Xist locus and its transcript, thereby requiring both modes of nucleic acid binding by YY1 for effective Xist localisation (25). Most recently, it has been proposed that the ability of YY1 to bind nascent mRNA transcripts is partially responsible for the retention of YY1 at actively transcribed promoters (28). Given the abundance of ZF transcription factors in mammalian genomes (∼800 proteins out of the ∼1400 transcription factors), the scope for an additional layer of regulatory control of gene expression through transcription factor-RNA interactions is enormous.
Currently, however, little is known about the molecular details underlying YY1-RNA interactions. YY1 does not bind the RNA equivalent of its cognate DNA sequence (23), and indeed it has never been determined whether YY1 recognises a specific motif in its proposed target RNAs. In vitro binding assays indicate a preference for polyU and AU-rich duplexes (23), whereas pulldowns of Xist RNA fragments implicate the ∼2-kb B/C repeat regions of Xist as the YY1 target (25). It has been suggested that the N-terminal domain of YY1 mediates RNA binding (28); however, this region has been shown to be largely disordered (29). Given the precedent of nucleic acid binding by ZFs and the higher level of conservation of the ZFs (compared to the N-terminal region) between Xenopus, mouse and human, we judged that this latter region was a more likely candidate to display RNA-binding properties.
In this study, we have directly assessed the interaction of purified recombinant YY1 ZFs with RNA, using SELEX to interrogate the RNA-binding preferences of YY1 ZFs; we have then used mutagenesis, microscale thermophoresis (MST) assays, gel shifts and NMR spectroscopy to identify residues involved in RNA binding. We show that the YY1 ZF cluster is able to bind many different RNAs including but not limited to Xist/XIST with micromolar affinity, and that the interaction is distinct from the DNA-binding activity of YY1. These results are consistent with the hypothesis that classical ZF TFs might play a role in RNA biology and begin to provide biochemical details of their interactions with RNA.
Abbreviations: ds, double-stranded; EMSA, electrophoretic mobility shift assay; MST, microscale thermophoresis; NMR, nuclear magnetic resonance; RBP, RNA-binding protein; SELEX, Systematic Evolution of Ligands by Exponential Enrichment; ss, single-stranded; ZF, zinc finger.
MATERIALS AND METHODS
Plasmid constructs
Human YY1FL (1–414) and YY1D (298–404) in pGEX-2T were gifts from Levon Khachigian (University of New South Wales). YY1(F1–4) (292–414), YY1(F1–3) (292–381), YY1(F2–4) (324–414), YY1(F12) (292–351), YY1F23 (324–381) and YY1F34 (352–414) were amplified by PCR from YY1FL and cloned into a modified pGEX-6P vector using BamHI/EcoRI restriction sites. Mutants of YY1(F1–4) (pGEX-6P) were generated using the QuickChange method (30). The numbering of YY1 constructs is that of the full-length human protein. ZRANB2(F12), consisting of two RanBP2-type ZFs (1–95) joined by a shortened linker (Δ45-64), was a gift from Mitchell O'Connell.
Human XIST RepC and mouse Xist RepC were cloned into pcDNA3.1. The following ssRNA Pentaprobes were used.
| Pentaprobe | Sequence |
|---|---|
| 2 | TATCTTACTTTAGTTTCATTTAATTGTGTTGTAC TCTCCTCTGCGTTCACTTAGCTTAACTTGGTTTG GCTTGATTTGACTTCAGTTGCGCTCTATTCTA |
| 7 | TAAAGTAAGATAAGGCAAGACAAGGTAAAACG GAACAGAACCGAAGGGAAGAGAAGCAAAGCGA AAGGAAATAAACAAAAGAAAAATTCGTAGAATTCCG |
| 9 | ATGGGTAGGATAGGCGAGGCCAGACCAGCGGA GCAGGGGAGGGTTAGACGAGACTAGATCGAAC TCAACGATTGAATGCACAGGACAGTAGAATAG AGCG |
Protein purification
All proteins were expressed as N-terminal fusions with glutathione-S-transferase (GST). Escherichia coli Rosetta (DE3) pLysS cells transformed with each construct were grown at 37°C to an OD of ∼0.5 before induction with IPTG (isopropyl-β-d-thiogalactopyranoside) and overexpression at 25°C for 18 h. For isotopically labeled samples, the cells were washed and resuspended in M9 minimal media containing 15NH4Cl, 13C-glucose (for double-labeled YY1(F1–4)) and 15N or 15N/13C Celtone media (Cambridge Isotope Laboratories, Tewksbury, MA, USA) before induction.
All GST-fusion constructs were purified by GSH affinity chromatography in lysis buffer (50 mM Tris pH 7.5, 500 mM NaCl, 100 μM ZnCl2, 0.5 μM PMSF, 20 μg/mL DNAse I, 1 mM DTT). For EMSAs, the GST-fusion proteins were dialysed into buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT prior to use in binding assays. For MST and NMR, YY1(F1–4) was cleaved from GST using HRV-3C protease and purified by cation exchange (UNO S1, in 50 mM Tris pH 7.5, 50–1000 mM NaCl, 1 mM DTT). Protein concentration was determined by UV absorbance at 280 nm.
Preparation of nucleic acid probes
Unlabeled and 5′ fluorescein-labeled m2 RNA (AGGCGAUGGUGAGC), poly(A) RNA and forward (AGGGTCTCCATTTTGAAGCG) and reverse (CGCTTCAAAATGGAGACCCT) strands of the AAV P5 DNA site (21) were purchased as synthetic oligonucleotides from IDT (Integrated DNA Technologies). Linear transcription templates for Pentaprobes 2, 7 and 9 were prepared by digesting pcDNA3 carrying the Pentaprobe sequences with ApaI, followed by end-filling with Klenow fragment. The RNA Pentaprobes were 32P-labeled by in vitro transcription with T7 RNA polymerase using α-32P-UTP. The forward strand of AAV P5 DNA was 32P-labeled using γ-32P-ATP and T4 polynucleotide kinase, then annealed with an excess of unlabeled reverse strand by heating at 95°C followed by slow cooling. 32P-labeled probes were purified using spin columns and ethanol precipitation, and were resuspended in water prior to use.
SELEX
The SELEX library was purchased as synthetic ssDNA oligonucleotides (IDT), consisting of a T7 promoter, followed by a constant region, a central randomised region of 25 or 50 nt, and a second constant region. The sequence of this ssDNA library is as follows: GAAATTAATACGACTCACTATAGGGAGGACGATGCGG(N25 or 50)AGACGACGAGCGGGA. The library was amplified by PCR (5 cycles) using primers to the constant regions to produce template for the initial transcription of the ssRNA library. The ssRNA library was generated by in vitro transcription of this template using T7 polymerase, with the resultant ssRNA bearing the randomised region flanked by two 15-nt constant regions. The transcribed RNA was treated with RQ1 DNAse (Promega) to remove any remaining dsDNA template.
Purified GST-fusion proteins were immobilised on glutathione beads and the loading quantified by SDS-PAGE with BSA standards. Beads loaded with 10–300 pmol of protein were incubated with an excess of RNA library, then washed with five column volumes of SELEX buffer (20 mM MOPS pH 7, 100 mM KCl, 5 mM MgCl2, 5% [v/v] glycerol, 0.1% [v/v] Triton X-100, 0.1 mM PMSF, 1 mM DTT). The protein and any bound RNA were eluted using 100 mM reduced glutathione in SELEX buffer, and the RNA recovered by ethanol precipitation and resuspension of the pellet in water. The library for the subsequent SELEX round was prepared by reverse transcription of the recovered RNA, followed by PCR (6 or 10 cycles) for amplification and re-introduction of the T7 promoter.
In total, six rounds of selection were conducted. Stringency was increased over successive rounds of selection by increasing the RNA molar excess 10–60-fold for GST/ZRANB2(F12)/YY1D, and 2–15-fold for GST/ZRANB2(F12)/YY1(F1–4). The absolute quantity of protein in each round of selection was either kept the same or decreased from the previous round.
EMSAs
Varying quantities of protein (0–10 μM) were incubated with a constant trace quantity of 32P-labeled DNA or RNA probe in gel shift buffer (10 mM MOPS pH 7, 50 mM KCl, 5 mM MgCl2, 10% glycerol [v/v]) on ice for 30 min before loading onto a 6% or 8% polyacrylamide gel cast in 1× TB buffer (90 mM Tris, 90 mM boric acid, 5 mM MgCl2). Gels were run at 200 V for 1–2 h in 0.5× TB buffer and exposed on a phosphor screen overnight. The screen was imaged on a Typhoon FLA9000 scanner.
MST
Protein samples and 5′ fluorescein-labeled m2 RNA were dialysed into the same buffer (20 mM phosphate pH 7, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT). Varying quantities of protein were incubated with 200 nM m2 RNA at room temperature for 15 min, before aspiration into standard-treated glass capillaries. Fluorescence and thermophoresis were measured using a Monolith NT.115 instrument (NanoTemper Technologies GmbH, München, Germany).
NMR spectroscopy
15N- and 13C/15N-labeled YY1(F1–4) samples were dialysed into 20 mM sodium phosphate (pH 7), 100 mM NaCl, 5 mM MgCl2, 1 mM DTT before concentrating to 150–300 μM. For 15N-HSQC titration, m2 RNA was dialysed into the same buffer as the protein. Protein samples were filtered (0.22 μm) prior to addition of D2O (5–10% [v/v]) and DSS (2,2-dimethyl-2-silapentane-5-sulfonic acid). Two molar equivalents of m2 RNA was titrated in increments of 0.25 eq. into the 15N YY1(F1–4) sample. All spectra were acquired at 298 K on Bruker 600 MHz or 800 MHz NMR spectrometers, and the data processed using TOPSPIN3 (Bruker, Karlsruhe, Germany) and SPARKY ((31)). Chemical shift perturbation values were calculated as a weighted average of changes in 1H and 15N chemical shift, as follows:
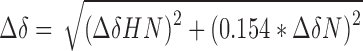 |
TALOS+ (32) was used to calculate phi and psi angles for YY1(F1–4).
RESULTS
The zinc fingers of YY1 bind RNA
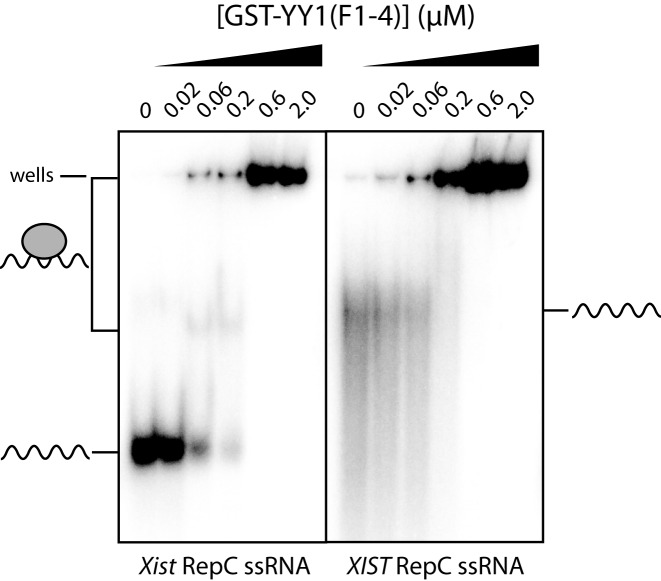
As an initial corroboration of the interaction observed by Jeon and Lee, we conducted electrophoretic mobility shift assays (EMSAs) with the YY1 ZFs using 32P-labeled mouse Xist/human XIST repeat C (RepC) RNA probes. The RepC region consists of 14 approximately 120-nucleotide repeat units in mouse Xist, whilst human XIST has a single unit with ∼50% sequence identity to the murine RepC repeats (26). The human XIST RepC sequence and one repeat unit from the mouse Xist RepC were transcribed in vitro, and the four ZFs of YY1 were expressed in and purified from E. coli as a GST-fusion protein (GST-YY1(F1–4)). In EMSAs, a clear, shifted band is formed by GST-YY1(F1–4) with the mouse Xist probe at 60 nM protein, whereas such a band is not apparent with the human XIST probe despite the reduction in free RNA probe (Figure 1). At higher protein concentrations, complexes that do not move into the gel form in both cases.
Figure 1.
EMSAs of 32P-labeled Xist/XIST RNA probes with GST-YY1F1–4. A constant amount of the RepC region of either mouse Xist (left) or human XIST (right) RNA probe (wavy line) was incubated with the indicated concentrations of protein (0–2 μM, grey oval) and analysed on 6% polyacrylamide native gels. A shifted band or disappearance of RNA probe indicates interaction with the protein. Note that at higher protein concentrations the YY1-RNA complexes are found in the wells of the gel, indicating that they are very large and/or low in solubility.
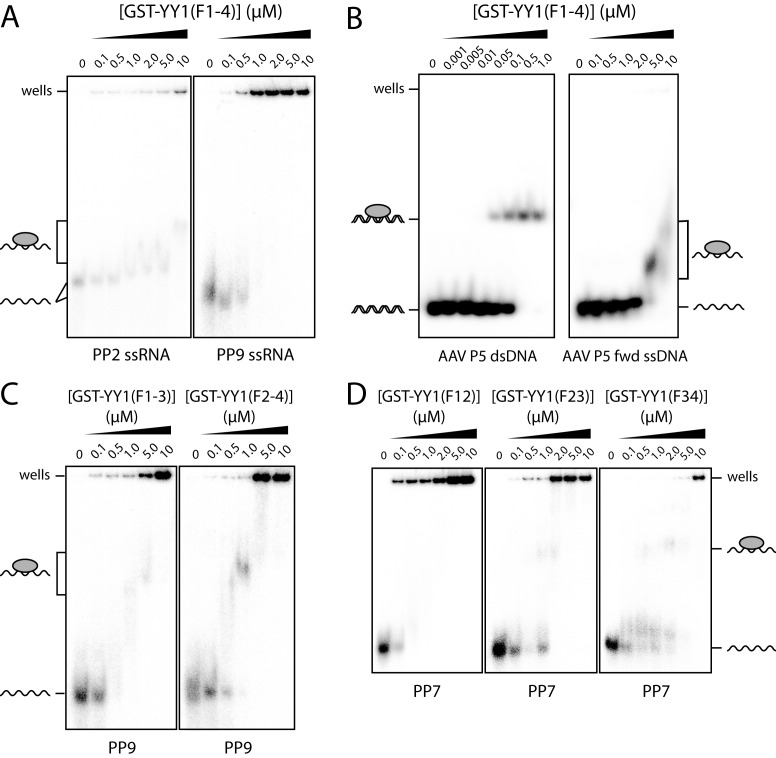
To assess more broadly the ability of YY1(F1–4) to bind to RNA, we tested this construct in EMSAs with Pentaprobes (PPs), a set of twelve 100-bp, single-stranded RNA sequences that collectively represent all possible 5-mers (33,34). The diverse set of binding sites available on these probes makes them a good screen for nucleic acid binding activity, and it is likely that they contain both single- and double-stranded elements. We chose two representative Pentaprobes of the 12, namely PP2 and PP9, for the EMSAs. As shown in Figure 2A, YY1(F1–4) begins to interact with both PP2 and PP9 at a protein concentration of ∼0.5 μM (judging from the intensity change in the probe-only band). In the case of PP2, a clear band is observed at 10 μM YY1(F1–4), whereas for PP9, the complex formed is very large and/or has low solubility and remains in the gel wells. This differential interaction is suggestive of some level of selectivity for different RNA sequences. For comparison, we carried out an EMSA using a dsDNA probe derived from the initiator element of adeno-associated virus P5 (AAV P5) promoter, a known target site for YY1 (35). Previous isothermal titration calorimetery data showed that YY1 can bind this site with nanomolar affinity (22). Our EMSA shows the formation of a single well-defined band consistent with the formation of a protein–DNA complex. An EMSA with a single-stranded version of this DNA probe also shows an interaction, albeit one with substantially lower affinity (Figure 2B). It is notable that the RNA Pentaprobes interact with YY1(F1–4) at somewhat lower concentrations than the ssDNA, suggesting some preference for ssRNA over ssDNA.
Figure 2.
YY1 binds a range of nucleic acids in vitro using multiple ZFs. A constant amount of each probe was incubated with increasing quantities of protein (0–10 μM) and analysed on 6% polyacrylamide native gels. All polypeptides are GST-fusion proteins and are represented as grey ovals. (A) YY1(F1–4) with ssRNA Pentaprobes 2 and 9. (B) YY1(F1–4) with the adeno-associated virus P5 promoter, a cognate dsDNA site, and the coding strand only of the same site (AAV P5 fwd ssDNA). (C) YY1 3-ZF constructs with Pentaprobe 9 ssRNA. (D) YY1 2-ZF constructs with Pentaprobe 7 ssRNA.
It has been shown previously that not all of the TFIIIA and WT1 ZFs are required for those proteins to interact with RNA (7,36). In order to isolate the minimal RNA-binding domain of YY1, the 4-ZF unit was subdivided into two- and three-ZF constructs, which were expressed as GST-fusion proteins. EMSAs with PP7 and PP9 showed that both three-ZF constructs (YY1(F1–3) and YY1(F2–4)) gave rise to discrete shifted bands at protein concentrations of ∼1 μM (Figure 2C). Similarly, all two-ZF constructs shifted the RNA probes (Figure 2D), indicating that none of the ZFs are indispensable for RNA binding. Nevertheless, RNA-binding capability is not equally distributed over all the fingers; some differences were observed in the concentrations of protein required to shift the free RNA probe. The removal of ZFs also appears, for all of the proteins except YY1(F12), to increase the tendency for the complex to run into the gel (Figure 2C and D). This observation might suggest that the overall interaction strength is diminished in proteins with fewer ZFs or that they are able to bind to fewer sites on the Pentaprobes, reducing the size of the complex and/or the likelihood of aggregation or precipitation.
Probing the RNA sequence specificity of YY1 ZFs
In order to interrogate the RNA-binding preferences of YY1, we used systematic evolution of ligands by exponential enrichment (SELEX). In this approach, preferred RNA sequences are selected from an initial randomised library through multiple rounds of binding and amplification.
SELEX was conducted on GST-YY1(F1–4) and on GST alone. As a positive control, a selection was also carried out on GST-ZRANB2(F12), a GST-fusion of the two RanBP2-type ZFs of ZRANB2; this double-ZF domain binds ssRNA containing the sequence GGU(N)xGGU with high specificity and a high nanomolar affinity (37,38). DNA oligonucleotide libraries containing a randomised region of 50 nt [GST-YY1(F1–4) and GST alone] or 25 nt [GST-ZRANB2(F12)] were transcribed into RNA libraries for selection. A selection was also performed using a mutant of GST-YY1(F1–4) in which ZF4 was crippled by removal of a zinc-ligating histidine (YY1D), which allowed partial assessment of the importance for RNA binding of having all four ZFs intact.
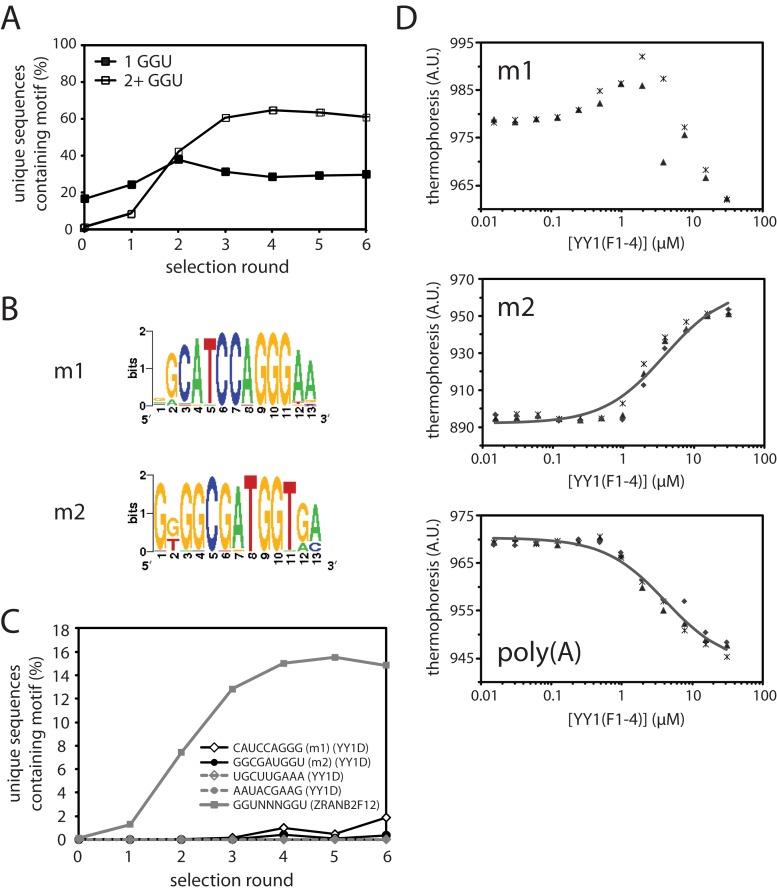
The proteins were immobilised on glutathione-Sepharose beads and used to pull down RNA, which was reverse-transcribed and amplified to generate the library for the subsequent round of selection. Stringency was increased in each round of selection by increasing the RNA:protein molar ratio. After six rounds of selection, the cDNA libraries recovered from each round were deep-sequenced on the Illumina HiSeq platform. Between 46K and 620K unique sequences were recovered per barcode from the sequencing, with minimum Phred quality scores of 24 and mean scores >30 across all base positions. To search for enriched sequence motifs in these libraries, we used the program MERMADE (adapted from http://korflab.ucdavis.edu/BnS/ (39)). No motifs were detected for GST, providing confidence that we did not select in our experiments for aptamers that recognize GST or glutathione-Sepharose beads. Examination of the selected ZRANB2(F12) library revealed significant enrichment of double-GGU motifs (Figure 3A), as expected. The proportion of unique sequences containing two or more GGU sites increased in the ZRANB2 libraries over rounds 1–4, indicating that the number of rounds and the stringency conditions used in our SELEX experiments are appropriate for selecting RNA sequences specifically bound by a protein with high nanomolar affinity. Previous measurements in literature suggest YY1 binds RNA with an affinity in this range (23), although our EMSAs suggest that the interaction for the ZFs in isolation is somewhat weaker. However, because selection for double-GGU motifs by ZRANB2(F12) saturates after 3–4 rounds of SELEX, we reasoned that even for a weaker binder, motifs should be detectable by round 6.
Figure 3.
SELEX analysis of YY1 and ZRANB2(F12). (A) Enrichment of sequences containing one (solid squares) or two or more (open squares) GGU sites in the ZRANB2(F12) SELEX. (B) Weblogos of the two most highly enriched motifs identified by MERMADE from round 6 of the YY1D SELEX dataset. (C) Enrichment over successive rounds of selection of motifs m1 and m2 in YY1D SELEX libraries, and also of a representative double-GGU motif in ZRANB2(F12) libraries, as a percentage of the number of unique sequences recovered in each round. For comparison, the percentage of two other 9-mers in each round of YY1D are also shown (dashed lines). (D) Binding of YY1(F1–4) to fluorescent m2/polyA RNA in MST assays. Data points from independent titrations are shown, fitted to a 1:1 binding model.
MERMADE was not able to identify any enriched motifs in the YY1(F1–4) SELEX experiment. However, two motifs were identified in round 6 for YY1D (Figure 3B). These sequences, m1 and m2, were the most highly enriched sequences over other 9-mers over successive rounds of selection (Figure 3C), although the level of selection was much lower than for the GGU motifs observed in the ZRANB2(F12) experiment. Although these sequences are not found within the C-repeat region of Xist/XIST, we tested their ability to bind to YY1(F1–4) in microscale thermophoresis (MST) binding assays, using chemically synthesized, fluorescein-labeled RNAs (Figure 3D). YY1(F1–4) appeared to bind both m1 and m2 with roughly micromolar affinity. The interaction between YY1(F1–4) and m1 was clearly complex; the binding isotherm was biphasic in nature, perhaps indicating the presence of a second, non-specific binding event. Consequently, only the m2 data could be readily fitted to a 1:1 binding model (Kd = 3.8 ± 0.6 μM). The binding of either m1 or m2 was not significantly tighter than the interaction of YY1(F1–4) with a poly(A) sequence of the same length (Kd = 4.2 ± 0.5 μM, Figure 3D). Thus, although YY1 clearly displays a degree of selectivity for some RNAs (judging from our EMSAs), it was not sufficiently pronounced for a preference to be readily detectable in our SELEX experiment.
It is possible that YY1 recognises an RNA structural motif rather than a specific sequence. The 50-nt randomized region used in the YY1(F1–4) selection should allow for a reasonable degree of structure formation within the randomized region. However, the detection of consensus structural motifs from a dataset of the present size is currently not computationally tractable (40,41). Furthermore, we would also envisage being able to at least detect some sort of sequence signature that would manifest as a motif in the MERMADE analysis if a conserved structural element did exist. It is also notable that both earlier work (23) and our MST data show that YY1(F1–4) can bind both poly(A) and poly(U) sequences (which are known to display little or no stable structure), suggesting that RNA secondary structure elements are not critical for the interaction.
YY1 zinc fingers interact with RNA and DNA via distinct subsets of residues.
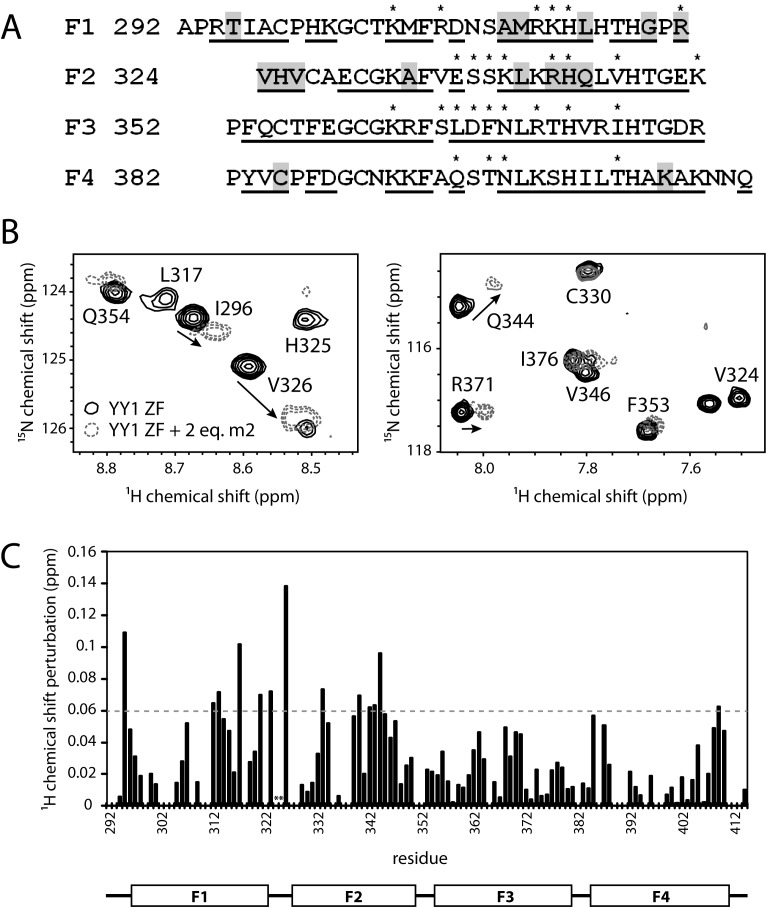
To probe the molecular details of RNA binding by YY1, we conducted 15N-heteronuclear single-quantum-coherence (15N-HSQC) NMR experiments. We expressed 13C- and 15N-labeled YY1(F1–4) and assigned resonances in the 15N-HSQC spectrum using the standard CBCA(CO)NH/HNCACB approach. 76% of backbone amide resonances were able to be assigned (Figure 4A and Supporting Figure S1) and the distribution of backbone phi and psi angles predicted from these chemical shifts (using TALOS+) were in close agreement with the values observed in the X-ray crystal structure of YY1 bound to DNA (PDB code: 1UBD). 15N-HSQC spectra of 15N-labeled YY1(F1–4) alone and in the presence of up to two molar equivalents of the SELEX-derived RNA motif m2 (AGGCGAUGGUGAGC) were then acquired (Figure 4B and Supporting Figure S1). The m2 sequence was used for this experiment due to its more straightforward binding behaviour in MST assays with YY1 compared with m1. Significant chemical shift changes were observed for a distinct subset of the resonances. The largest chemical shift perturbations occur mostly in the first two ZFs (Figure 4C), consistent with m2 being derived from SELEX using YY1D, in which only the N-terminal three fingers are intact. Within the first two ZFs, 14 residues have perturbations of at least 1 S.D. greater than the mean, of which only three (specifically, R323, R342 and H343) overlap with the 14 residues known to make direct contacts with DNA in the YY1–DNA crystal structure (21) (Figure 4A). This difference suggests that the mode of interaction with RNA is distinct from that with DNA, and that RNA binding is not simply an ‘off-target’ nucleic acid interaction.
Figure 4.
Mapping of the YY1-RNA interaction by chemical shift perturbation experiments. (A) Amino acid sequence of YY1(F1–4) showing DNA contact residues (indicated by asterisks) and residues with chemical shift perturbations greater than 1 S.D. from the mean, following the addition of m2 RNA (highlighted in grey; see also (C)). Residues that could be assigned from 3D triple-resonance NMR spectra are underlined. (B) Partial 15N-HSQC spectra of YY1(F1–4) alone (black, solid lines) and in the presence of two molar equivalents of m2 RNA (grey dashed lines). The direction of peak shifting is indicated by arrows. (C) Plot of chemical shift perturbations along the primary sequence of YY1(F1–4) in the presence of two molar equivalents of m2 RNA. The dashed grey line represents the mean + 1 S.D. from the mean chemical shift change across all measured residues. Asterisks indicate peaks no longer present in the spectrum after addition of 0.25 molar equivalents of RNA.
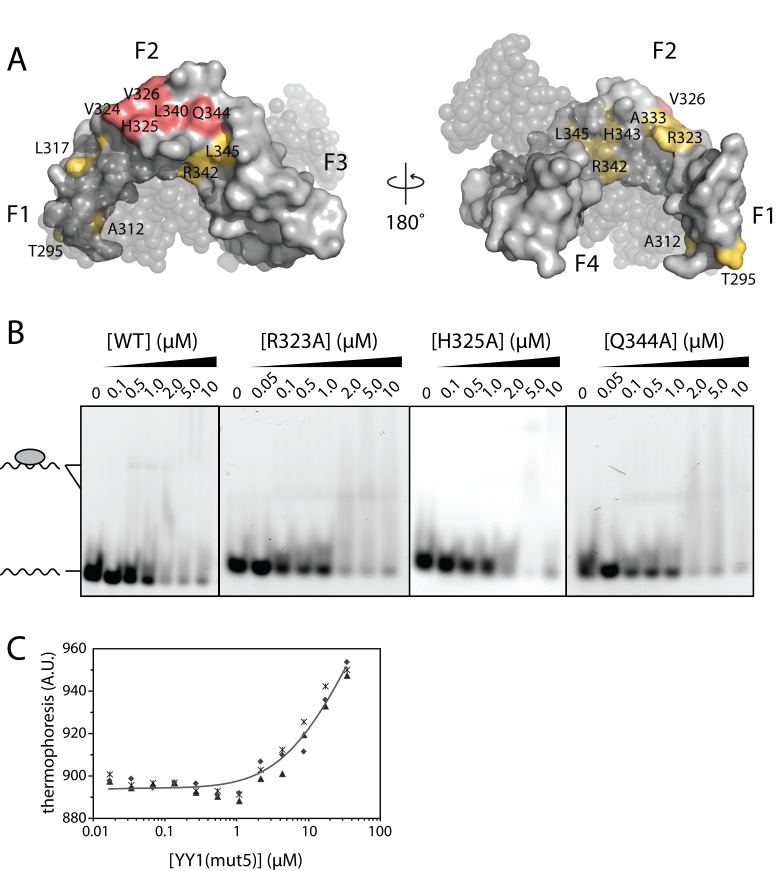
Mapping of the residues with largest chemical shift perturbations onto the YY1–DNA crystal structure identified clusters of residues that most likely comprise the RNA interaction surface (Figure 5A). We chose three residues that underwent the largest chemical shift perturbation (R323, H325, Q344) and individually mutated these to alanine. 1D 1H NMR spectra of these mutants showed that folding was not disrupted by the mutations (Supporting Figure S2). However, none of these individual point mutations significantly affected the affinity for m2 RNA in EMSAs (Figure 5B). We next selected five m2-perturbed residues (V324, H325, V326, L340, Q344) that constituted a contiguous surface on YY1 in the YY1–DNA structure (and are not involved in DNA binding), and simultaneously mutated these to alanine, creating YY1(mut5). These mutations give rise to an at least eight-fold decrease in m2 affinity, from Kd = 3.8 ± 0.6 μM (wild-type) to Kd = 31 ± 9 μM (mut5) as measured by MST assays (Figure 5C). Note that our inability to access the end of the binding isotherm means that the calculated Kd represents an upper limit in affinity.
Figure 5.
Mutagenesis of YY1(F1–4). (A) Mapping of YY1(F1–4) resonances most perturbed by m2 RNA (>1 S.D. greater than the mean chemical shift perturbation, see Figure 4C) onto the crystal structure of the YY1(F1–4)+DNA complex, in yellow or pink (PDB ID: 1UBD). Residues simultaneously mutated to create YY1(mut5) are indicated in pink. DNA is shown as gray, translucent spheres. (B) EMSAs of fluorescent m2 RNA probe with YY1(F1–4) point mutants R323A, H325A and Q344A. (C) Binding of YY1(mut5) to fluorescent m2 RNA in MST assays. Data points from independent titrations are shown, fitted to a 1:1 binding model.
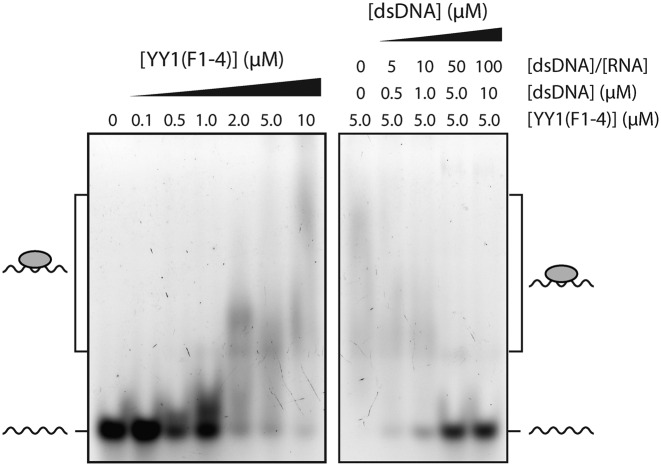
The NMR data suggest that it might be possible for both DNA and RNA to bind to the YY1 ZFs simultaneously. Such an arrangement could constitute a mechanism to explain the model of Xist tethering proposed by Jeon and Lee, which posits that YY1 acts as a bridge between Xist RNA and the inactive X chromosome (25). In order to directly test this hypothesis, we conducted a competition EMSA by titrating the YY1(F1–4)-m2 RNA complex with an increasing molar excess of dsDNA containing the cognate YY1 binding site (AAV P5 dsDNA, used in Figure 2B). Figure 6 shows the titration of 100 nM fluorescent m2 RNA probe with YY1(F1–4). At a YY1(F1–4) concentration of 5 μM, the band corresponding to m2 free probe (100 nM) has largely disappeared, indicating that the majority of the RNA is bound. Incubation of 100 nM m2 with 5 μM YY1(F1–4) in the presence of excess dsDNA inhibits m2 binding to YY1, as evidenced by reappearance of free m2 probe in the lanes containing 5 and 10 μM dsDNA.
Figure 6.
DNA and RNA compete for overlapping binding sites on YY1. Competition EMSA of YY1(F1–4)-m2 RNA complex with dsDNA. All samples were analysed on an 8% polyacrylamide native gel. Left: 100 nM fluorescent m2 RNA probe was incubated with increasing quantities of YY1(F1–4) (0–10 μM). Right: 5 μM YY1(F1–4) and 100 nM fluorescent m2 RNA were incubated with an increasing molar excess of unlabeled AAV P5 dsDNA.
DISCUSSION
YY1 zinc fingers bind ssRNA with low specificity
YY1 has been suggested to bind RNA in the contexts of Xenopus oocyte development and murine X-inactivation. The present study shows that ZFs of YY1 can bind with micromolar affinity to ssRNA and might therefore be responsible for these observed activities. Our EMSA and MST data indicate that YY1(F1–4) binds directly to Xist RNA and displays at least some ability to distinguish different RNA sequences. However, this level of selectivity was not sufficient to consistently select a sequence motif in YY1 SELEX experiments. In general, we observe that YY1 does not appear to require RNA secondary structure for binding, as indicated by its affinity for poly(A), though we cannot rule out that there may be a structural element to recognition of its native RNA targets. A recent CLIP-Seq study suggests that YY1 binds the 5′ end of nascent transcripts (28), although no preferred sequence motif was identified, perhaps consistent with the idea that RNA sequence specificity is not required in its biological function.
Although the YY1 ZFs are clearly sufficient for RNA binding, it is possible that the N-terminal region of YY1 might modulate the RNA-binding properties of the ZFs, either in its own right or by recruitment of protein partners. Sigova et al. (28) ascribe the ability of YY1 to bind nascent transcripts to the N-terminal half of the protein rather than the ZF domain, although the EMSAs supporting this conclusion are not unequivocal. The affinity of the YY1 ZFs in our EMSAs does appear to be somewhat lower than in EMSAs of the full-length protein to RNA sequences of comparable length (23). Additionally, it has been suggested that full-length YY1 forms a homo-oligomer, allowing YY1 to bind DNA sequences lacking the cognate recognition site (42), and it is possible that YY1 oligomerisation also influences RNA specificity.
The molecular nature of the YY1-RNA interaction
Our NMR experiments identify residues likely to be involved in the YY1–RNA interaction and suggest that YY1 ZFs bind RNA in a manner distinct from its DNA binding. Changes in chemical shift are caused by changes in the chemical environment experienced by each residue, which may be due to either direct ligand contacts or changes in local conformation. Consequently, the precise nature of each residue's contribution to the interaction requires corroboration. Our mutagenesis data indicate either that the RNA-contact surface of the protein is quite extensive, or that a significant number of the RNA contacts might be made by backbone atoms. NMR studies have shown that the linker between ZFs in TFIIIA becomes ordered on binding to both DNA (43,44) and RNA (45). Furthermore, the TFIIIA ZFs are able to reorient substantially in order to switch from DNA to RNA binding (46). The large chemical shift perturbations around the linker between fingers 1 and 2 (G321–V326; Figure 4C) might be indicative of similar changes induced by RNA binding to YY1. Nevertheless, mutation of residues R323, V324, H325 and V326 does not cause the same losses of DNA-binding affinity observed for TFIIIA linker mutants (47), suggesting that the role of the linker residues may be less important, or at least less sequence-dependent, in the YY1-RNA interaction.
The chemical shift perturbations induced by RNA binding to YY1 are modest compared with those observed in similar experiments with other RNA-binding proteins. Sequence-specific, single-stranded RNA binding domains such as the FUS RRM (48), TIA-1 RRM (49) and ZRANB2 RanBP2-type ZF (37) display amide proton chemical shift changes in the order of 0.15–1.2 ppm at the threshold of 1 S.D. above than the mean. In comparison, individual classical ZFs of the double-stranded RNA-binding proteins JAZ and ZFa exhibit perturbations of less than 0.6 ppm (12). This disparity is attributable to hydrogen bonding to RNA bases by sequence-specific ssRNA-binding proteins, which induces larger changes in environment for participating amide groups, and also to closer proximity of the protein to the aromatic ring currents of the bases.
The interaction of YY1 with RNA produces even smaller chemical shift changes—we observe a maximum HN chemical shift change of 0.14 ppm in the fully bound state, although V324 and H325 disappeared during the titration (and therefore could not be tracked) and most likely undergo somewhat larger shifts. One possible explanation for these small chemical shift changes is that the interaction involves rapid exchange between multiple binding conformations/orientations (of the protein and/or the RNA), resulting in averaging of the chemical shift changes from each distinct binding event. If these distinct binding events induce shift changes that are relatively uncorrelated, the overall observed changes will be small. Consistent with this idea, we recently observed substantially smaller chemical shift changes when the ZF protein ZNF217 bound to a non-specific DNA target than when it bound a sequence for which it had specificity (50). This explanation would also partially account for our mutagenesis data, which indicate that the contribution of individual residues to overall affinity is small, perhaps due to the existence of a number of binding modes.
Whilst chemical shift perturbations indicate that the DNA and m2-binding surfaces on YY1(F1–4) are distinct, dsDNA competes efficiently with m2 RNA for YY1(F1–4) binding in an EMSA, suggesting that m2 RNA and dsDNA cannot bind simultaneously. These data are also consistent with independent affinity measurements, which suggest dsDNA binds YY1 zinc fingers at least an order of magnitude more tightly than m2 RNA. Therefore, if YY1 bridges Xist RNA and DNA, it is unlikely that this occurs via the zinc fingers.
Relationship between YY1 and other RNA-binding classical-ZF proteins
YY1 does not appear to interact with RNA in the same way as known RNA-binding classical ZF proteins. In contrast with TFIIIA, which specifically recognises the structure formed by a 61-nt 5S RNA construct (5,51), YY1 is able to interact with shorter sequences that lack secondary structure entirely (e.g. poly(A)). Furthermore, sequence and structural comparison suggests that YY1 does not belong to the class of specialised dsRNA-binding classical ZF proteins typified by ZFa, including JAZ and wig-1, which have long linkers between fingers instead of the conserved TGEKP classical ZF linker (11).
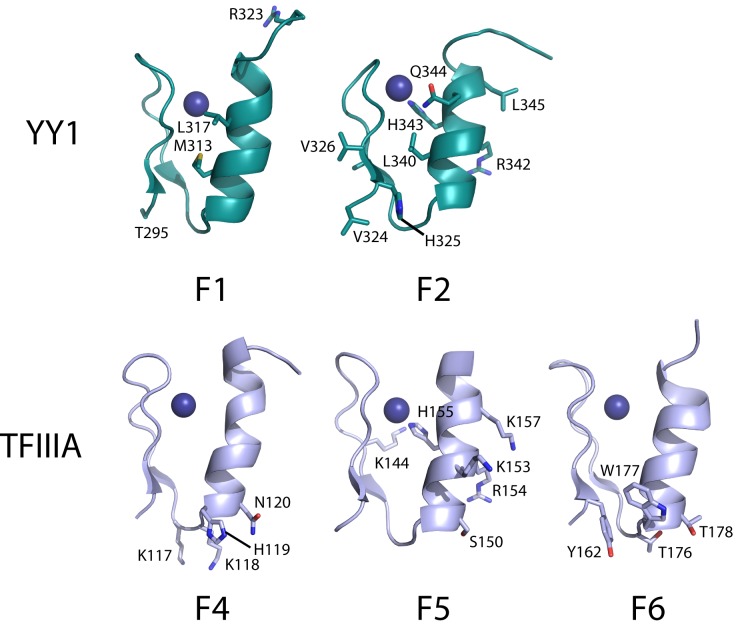
Mapping of the YY1 residues perturbed in 15N-HSQC titrations with m2 RNA onto the structure of YY1 in complex with DNA (PDB: 1UBD) suggests a contiguous RNA-binding surface of ∼1600 Å2 (assuming all perturbed residues directly contact RNA), comparable to that of TFIIIA with 5S RNA. Fourteen residues distributed across TFIIIA F4–6 make crystal contacts with RNA, and the same number of residues in YY1 fingers 1 and 2 have m2-induced chemical shift perturbations greater than 1 S.D. above the mean. However, there is no clear similarity between TFIIIA and YY1 in the arrangement or identity of these residues. This disparity contrasts with the DNA-binding situation, which involves residues at positions –1, +1, 2, 3 and 6 of the α-helix and is conserved across all classical ZF-DNA interactions. Only three RNA-binding residues occupy equivalent positions in TFIIIA finger 5 (R154, H155, K157) and YY1 finger 2 (R342, H343, L345); of these, the Arg and His residues are also involved in DNA binding (Figure 7). Notably, TFIIIA makes just two base-specific contacts with 5S RNA, involving residues at positions –2, –1, +1 and +2 of the α-helices of fingers 4 and 6 (5). The equivalent residues in YY1 fingers 1 and 2 were not perturbed in our NMR titration experiments (although it should be noted that these experiments rarely identify all residues involved in an interaction as only the environment of the backbone amide group is being assessed).
Figure 7.
Comparison of YY1 and TFIIIA ZFs involved in RNA binding. Fingers 1 and 2 of YY1 (from PDB ID: 1UBD) and fingers 4–6 of TFIIIA (from PDB ID: 1UN6) are shown as ribbon diagrams. Residues involved in RNA binding are labeled and displayed as sticks.
From these observations, a mode of RNA-binding by classical ZFs may be hypothesised in which some DNA-binding residues are repurposed for RNA binding whilst peripheral residues unrelated to DNA binding impart specificity for RNA to the interaction. Differences in RNA interaction modes by classical ZFs might reflect differences in the nature of the RNA targets of each protein. The inherent flexibility of RNA and its ability to adopt heterogeneous structures compared with dsDNA suggests that modes of recognition even within a single class of protein domains may be equally diverse; indeed, this has already been observed in the well-characterised RNA recognition motifs (RRMs) (reviewed in (52,53)).
Does YY1 bind to Xist/XIST?
Our data show that human YY1(F1–4) (100% sequence identity with mouse and Xenopus YY1(F1–4)) is clearly able to bind both mouse and human Xist/XIST RNA. A lack of conservation in the C-repeat region of Xist/XIST may indicate a divergence between mouse and human in its requirement for Xist/XIST localisation. In addition to differences in copy number, the human C-repeat unit only shares ∼50% sequence identity with the murine counterpart. It has been noted that the mechanism of X-inactivation is likely to differ between mouse and human, in particular the effectors associated with the Xist transcript (26). Taken in this context, the differences in the EMSAs suggest that the interaction of YY1 with RepC RNA in X-inactivation may be species-specific. Indeed, the function of YY1 as an RNA-binding protein certainly appears to extend beyond its role in X-inactivation, as exemplified by its involvement in mRNP formation in Xenopus (23), an organism that does not employ an XY sex determination system.
It has been proposed that YY1 binds RNA transcripts from the promoters it occupies (28), and it is possible that the binding of YY1 to Xist/XIST RNA represents a specific case of this phenomenon. However, recent studies have identified proteins bound to mouse Xist lncRNA both at the onset of X-inactivation and in fully differentiated cell lines (54,55). Notably, these sets of proteins did not include YY1, but did include RYBP, a protein that interacts with YY1 (56). Other studies have suggested that the primary role of YY1 in both mouse and human X-inactivation lies in the transcriptional activation of Xist/XIST (57,58). It is therefore worth considering that the association of YY1 with Xist RNA might in fact be indirect.
Is RNA-binding activity likely to be a general feature of classical zinc-finger proteins?
Recent high-throughput RNA-binding protein discovery experiments have employed an oligo-dT pulldown and mass spectrometry approach to interrogate the mRNA-bound proteome in HeLa (59) and HEK cells (60). Classical ZFs are under-represented in these ‘mRNA interactomes’; less than 1% of known human classical ZF proteins were observed, compared to 60% and 40%, respectively, of the CCHC- and CCCH-type ZFs (classes of ZF that are known to bind RNA from previous work (61,62)). These data suggest that classical ZFs might not, in general, have an RNA-binding function, although there are several reasons why such interactions might not have been observed. For example, the involvement of classical ZFs in RNA metabolism might occur primarily in development (which will not be reflected in differentiated cancer cell lines). Indeed the RNA-related functions of TFIIIA (63), WT1 (9) and ZNF74 (10) do appear to play developmental roles. It is also possible that classical ZF proteins that play dual roles as DNA- and RNA-binding proteins (again, such as TFIIIA and WT1) are not easily recovered in pulldown assays of this type if they partition mostly or partly with DNA. Conversely, a recent survey of DNA-binding preferences of single classical ZF domains using bacterial one-hybrid screening and protein binding microarrays suggests that up to 40% of human classical ZFs do not bind DNA (64). Whether these represent RNA- or protein-binding ZFs remains to be determined.
In summary, we have shown that the ZFs of YY1 are able to interact with multiple RNA sequences, and that their specificity is not limited to putative endogenous RNA target Xist. The mode of interaction appears to be distinct from both the DNA binding of YY1 and the RNA binding of other classical ZF proteins. The possibility that RNA interaction may form part of the functional repertoire of this multifunctional classical ZF transcription factor is tantalising and further studies to investigate in vivo RNA targets of YY1 will yield insight into the role of classical ZF proteins in RNA biology.
FUNDING
National Health and Medical Research Council of Australia [1063188, 1058916]. Funding for open access charge: University of Sydney research funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Iuchi S. Three classes of C2H2 zinc finger proteins. CMLS-Cell. Mol. Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razin S.V., Borunova V.V., Maksimenko O.G., Kantidze O.L. Cys2His2 zinc finger protein family: Classification, functions, and major members. Biochemistry-Moscow. 2012;77:217–226. doi: 10.1134/S0006297912030017. [DOI] [PubMed] [Google Scholar]
- 3.Brayer K., Segal D. Keep your fingers Off My DNA: protein–protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 2008;50:111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 4.Nolte R.T., Conlin R.M., Harrison S.C., Brown R.S. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu D., Alexandra Searles M., Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 6.Pelham H.R., Brown D.D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl. Acad. Sci. U.S.A. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caricasole A., Duarte A., Larsson S.H., Hastie N.D., Little M., Holmes G., Todorov I., Ward A. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laity J.H., Chung J., Dyson H.J., Wright P.E. Alternative splicing of Wilms' tumor suppressor protein modulates DNA binding activity through isoform-specific DNA-induced conformational changes. Biochemistry. 2000;39:5341–5348. doi: 10.1021/bi9926678. [DOI] [PubMed] [Google Scholar]
- 9.Grondin B., Bazinet M., Aubry M. The KRAB zinc finger gene ZNF74 encodes an RNA-binding protein tightly associated with the nuclear matrix. J. Biol. Chem. 1996;271:15458–15467. doi: 10.1074/jbc.271.26.15458. [DOI] [PubMed] [Google Scholar]
- 10.Côté F., Boisvert F.-M., Grondin B., Bazinet M., Goodyer C.G., Bazett-Jones D.P., Aubry M. Alternative promoter usage and splicing of ZNF74 multifinger gene produce protein isoforms with a different repressor activity and nuclear partitioning. DNA Cell. Biol. 2001;20:159–173. doi: 10.1089/104454901300069004. [DOI] [PubMed] [Google Scholar]
- 11.Möller H.M., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Solution structure of the N-terminal zinc fingers of the Xenopus laevis double-stranded RNA-binding protein ZFa. J. Mol. Biol. 2005;351:718–730. doi: 10.1016/j.jmb.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Burge R.G., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Structural characterization of interactions between the double-stranded RNA-binding zinc finger protein JAZ and nucleic acids. Biochemistry. 2014;53:1495–1510. doi: 10.1021/bi401675h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Méndez-Vidal C., Wilhelm M.T., Hellborg F., Qian W., Wiman K.G. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 2002;30:1991–1996. doi: 10.1093/nar/30.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köster M., Kühn U., Bouwmeester T., Nietfeld W., El-Baradi T., Knöchel W., Pieler T. Structure, expression and in vitro functional characterization of a novel RNA binding zinc finger protein from Xenopus. EMBO J. 1991;10:3087–3093. doi: 10.1002/j.1460-2075.1991.tb07861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreazzoli M., De Lucchini S., Costa M., Barsacchi G. RNA binding properties and evolutionary conservation of the Xenopus multifinger protein Xfin. Nucleic Acids Res. 1993;21:4218–4225. doi: 10.1093/nar/21.18.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Lee J.-S., Galvin K.M. Everything you have ever wanted to know about Yin Yang 1. BBA-Rev. Cancer. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 17.Thomas M.J., Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 18.Gordon S., Akopyan G., Garban H., Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2005;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 19.Hyde-DeRuyscher R.P., Jennings E., Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yant S.R., Zhu W., Millinoff D., Slightom J.L., Goodman M., Gumucio D.L. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human β globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houbaviy H.B., Usheva A., Shenk T., Burley S.K. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houbaviy H.B., Burley S.K. Thermodynamic analysis of the interaction between YY1 and the AAV P5 promoter initiator element. Chem. Biol. 2001;8:179–187. doi: 10.1016/s1074-5521(00)90066-8. [DOI] [PubMed] [Google Scholar]
- 23.Belak Z.R., Ovsenek N. Assembly of the Yin Yang 1 transcription factor into messenger ribonucleoprotein particles requires direct RNA binding activity. J. Biol. Chem. 2007;282:37913–37920. doi: 10.1074/jbc.M708057200. [DOI] [PubMed] [Google Scholar]
- 24.Belak Z.R., Ficzycz A., Ovsenek N. Biochemical characterization of Yin Yang 1 - RNA complexes. Biochem. Cell Biol. 2008;86:31–36. doi: 10.1139/o07-155. [DOI] [PubMed] [Google Scholar]
- 25.Jeon Y., Lee J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plath K., Mlynarczyk-Evans S., Nusinow D.A., Panning B. XIST RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 27.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 28.Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Górecki A., Bonarek P., Górka A.K., Figiel M., Wilamowski M., Dziedzicka-Wasylewska M. Intrinsic disorder of human Yin Yang 1 protein. Proteins. 2015;83:1284–1296. doi: 10.1002/prot.24822. [DOI] [PubMed] [Google Scholar]
- 30.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91–91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goddard T.D., Kneller D.G. SPARKY 3. San Francisco: University of California; 2006. [Google Scholar]
- 32.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan A.H.Y., Czolij R., Mackay J.P., Crossley M. Pentaprobe: a comprehensive sequence for the one-step detection of DNA-binding activities. Nucleic Acids Res. 2003;31:e124. doi: 10.1093/nar/gng124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendak K., Loughlin F.E., Cheung V., O'Connell M.R., Crossley M., Mackay J.P. A rapid method for assessing the RNA-binding potential of a protein. Nucleic Acids Res. 2012;40:e105–e105. doi: 10.1093/nar/gks285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y., Seto E., Chang L.-S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüippel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 36.Theunissen O., Rudt F., Guddat U., Mentzel H., Pieler T. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- 37.Loughlin F.E., Mansfield R.E., Vaz P.M., McGrath A.P., Setiyaputra S., Gamsjaeger R., Chen E.S., Morris B.J., Guss J.M., Mackay J.P. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5 ' splice site-like sequences. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5581–5586. doi: 10.1073/pnas.0802466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell M.R., Vandevenne M., Nguyen C.D., Matthews J.M., Gamsjaeger R., Segal D.J., Mackay J.P. Modular assembly of RanBP2-type zinc finger domains to target single-stranded RNA. Angew. Chem. Int. Ed. 2012;51:5371–5375. doi: 10.1002/anie.201200866. [DOI] [PubMed] [Google Scholar]
- 39.Zykovich A., Korf I., Segal D.J. Bind-n-Seq: high-throughput analysis of in vitro protein–DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro B.A., Yingling Y.G., Kasprzak W., Bindewald E. Bridging the gap in RNA structure prediction. Curr. Opin. Struct. Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Capriotti E., Marti-Renom M.A. Computational RNA structure prediction. Curr. Bioinform. 2008;3:32–45. [Google Scholar]
- 42.López-Perrote A., Alatwi H.E., Torreira E., Ismail A., Ayora S., Downs J.A., Llorca O. Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases. J. Biol. Chem. 2014;289:22614–22629. doi: 10.1074/jbc.M114.567040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wuttke D.S., Foster M.P., Case D.A., Gottesfeld J.M., Wright P.E. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity1. J. Mol. Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 44.Foster M., Wuttke D., Clemens K., Jahnke W., Radhakrishnan I., Tennant L., Reymond M., Chung J., Wright P. Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA. J. Biomol. NMR. 1998;12:51–71. doi: 10.1023/a:1008290631575. [DOI] [PubMed] [Google Scholar]
- 45.Lee B.M., Xu J., Clarkson B.K., Martinez-Yamout M.A., Dyson H.J., Case D.A., Gottesfeld J.M., Wright P.E. Induced fit and ‘Lock and Key’ recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 2006;357:275–291. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Brown R.S. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Choo Y., Klug A. A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X., Niu C., Ren J., Zhang J., Xie X., Zhu H., Feng W., Gong W. The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim. Biophys. Acta. 2013;1832:375–385. doi: 10.1016/j.bbadis.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang I., Hennig J., Jagtap P.K.A., Sonntag M., Valcárcel J., Sattler M. Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res. 2014;42:5949–5966. doi: 10.1093/nar/gku193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandevenne M., Jacques D.A., Artuz C., Nguyen C.D., Kwan A.H.Y., Segal D.J., Matthews J.M., Crossley M., Guss J.M., Mackay J.P. New Insights into DNA Recognition by Zinc Fingers Revealed by Structural Analysis of the Oncoprotein ZNF217. J. Biol. Chem. 2013;288:10616–10627. doi: 10.1074/jbc.M112.441451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neely L.S., Lee B.M., Xu J., Wright P.E., Gottesfeld J.M. Identification of a minimal domain of 5 S ribosomal RNA sufficient for high affinity interactions with the RNA-specific zinc fingers of transcription factor IIIA1. J. Mol. Biol. 1999;291:549–560. doi: 10.1006/jmbi.1999.2985. [DOI] [PubMed] [Google Scholar]
- 52.Auweter S.D., Oberstrass F.C., Allain F.H.T. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mackay J.P., Font J., Segal D.J. The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 2011;18:256–261. doi: 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- 54.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minajigi A., Froberg J.E., Wei C., Sunwoo H., Kesner B., Colognori D., Lessing D., Payer B., Boukhali M., Haas W., et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349:aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García E., Marcos‐Gutiérrez C., del Mar Lorente M., Moreno J.C., Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makhlouf M., Ouimette J.-F., Oldfield A., Navarro P., Neuillet D., Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapman A.G., Cotton A.M., Kelsey A.D., Brown C.J. Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 2014;15:89–89. doi: 10.1186/s12863-014-0089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 60.Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 61.Lee S.J., Michel S.L.J. Structural metal sites in nonclassical zinc finger proteins involved in transcriptional and translational regulation. Accounts Chem. Res. 2014;47:2643–2650. doi: 10.1021/ar500182d. [DOI] [PubMed] [Google Scholar]
- 62.Muriaux D., Darlix J.-L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honda B.M., Roeder R.G. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980;22:119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- 64.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]