Abstract
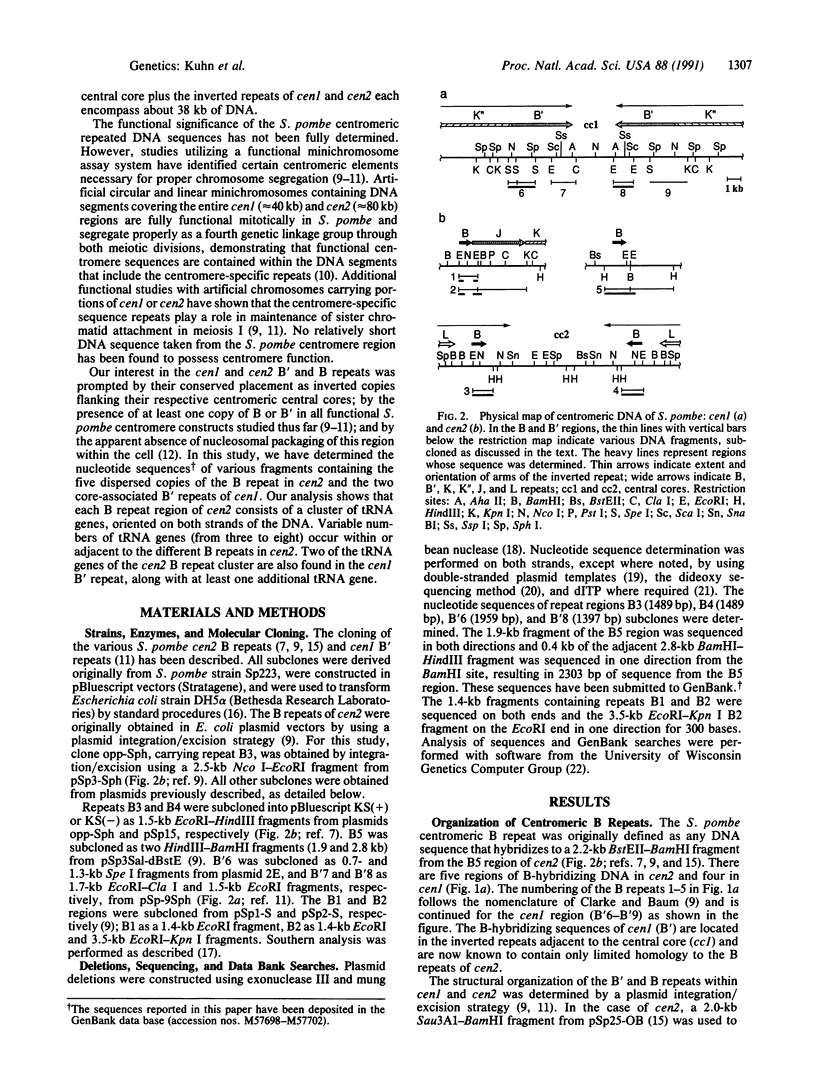
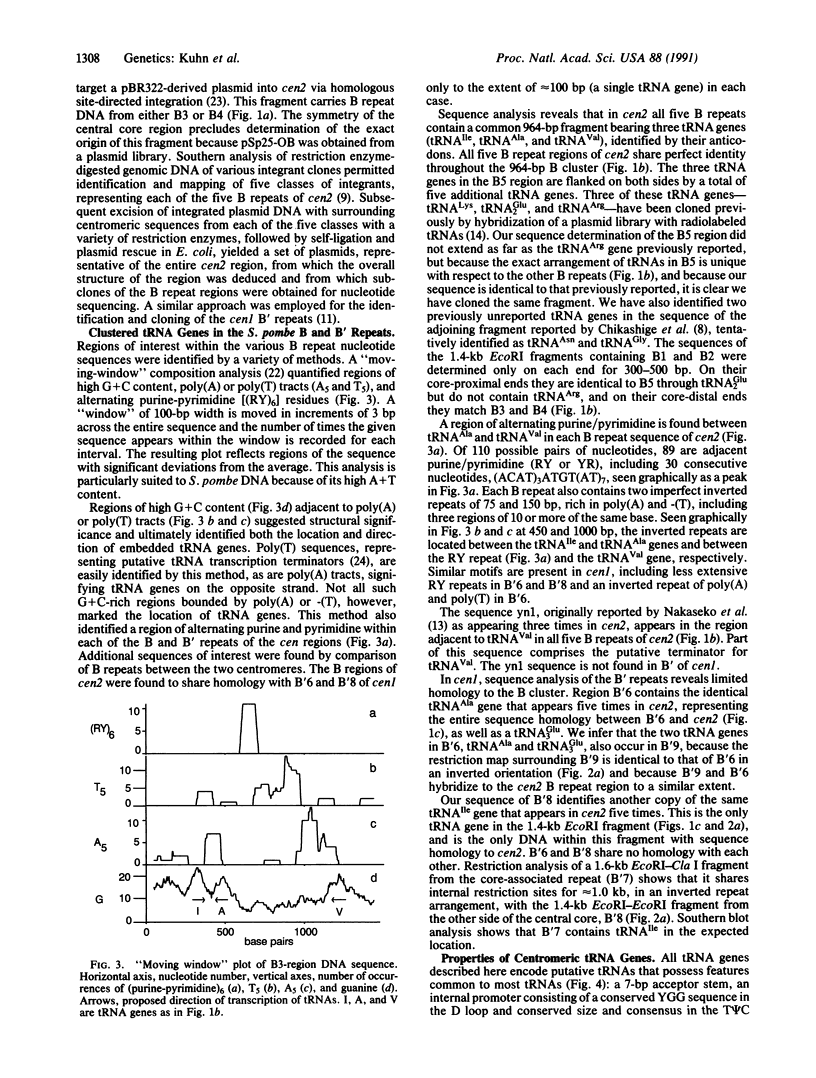
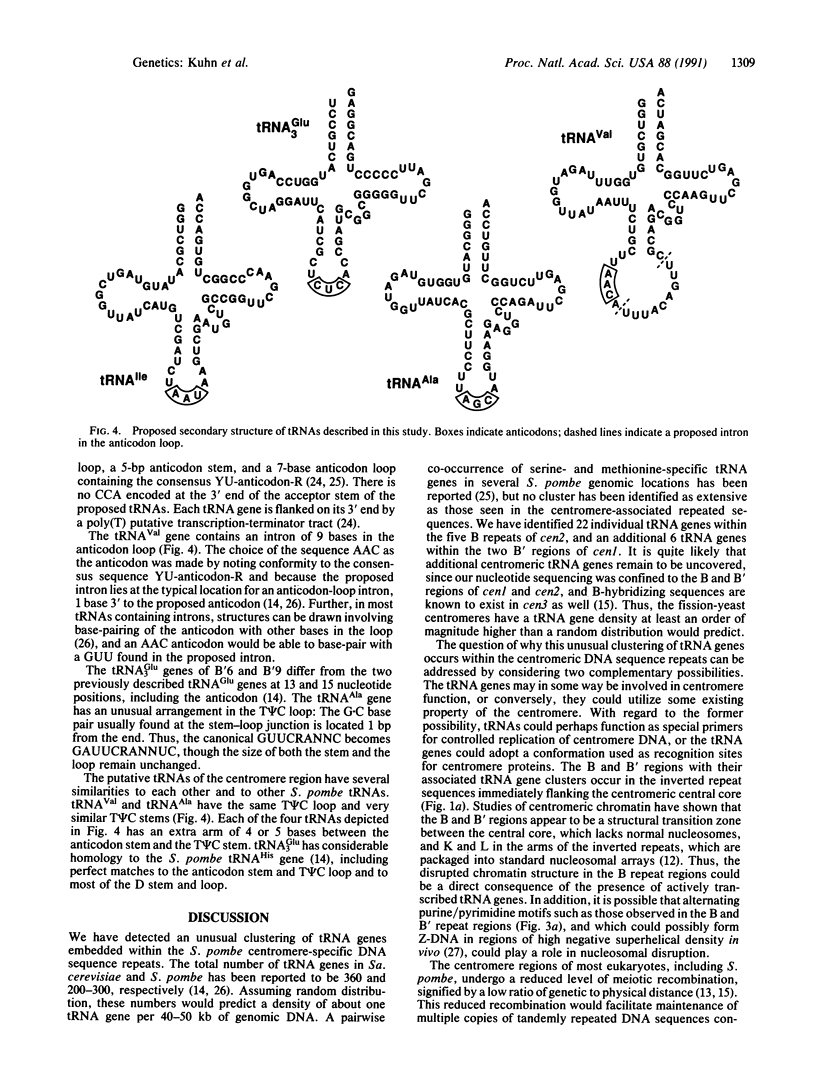
The centromere-associated B' and B DNA sequence repeats of Schizosaccharomyces pombe chromosomes I and II have been found to contain clusters of tRNA genes. The centromere II region (cen2) includes at least 22 tRNA genes distributed among five copies of the B sequence repeat containing genes specifying tRNA(Ile), tRNA(Ala), and tRNA(Val). Individual B repeats are variously associated with other tRNA genes, including those specifying tRNA(Lys), tRNA(Arg), and tRNA(Glu2). The centromere I region (cen1) contains at least six tRNA genes in two copies of the B' repeated element, including genes specifying tRNA(Ile), tRNA(Ala), and tRNA(Glu3). Multiple tandemly arranged clusters of tRNA genes are presumably conserved due to restricted recombination frequencies in the centromere regions.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amstutz H., Munz P., Heyer W. D., Leupoid U., Kohli J. Concerted evolution of tRNA genes: intergenic conversion among three unlinked serine tRNA genes in S. pombe. Cell. 1985 Apr;40(4):879–886. doi: 10.1016/0092-8674(85)90347-2. [DOI] [PubMed] [Google Scholar]
- Carbon J., Clarke L. Centromere structure and function in budding and fission yeasts. New Biol. 1990 Jan;2(1):10–19. [PubMed] [Google Scholar]
- Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989 Jun 2;57(5):739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Clarke L., Amstutz H., Fishel B., Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Baum M. P. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990 May;10(5):1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. The structure and function of yeast centromeres. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres of budding and fission yeasts. Trends Genet. 1990 May;6(5):150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B., Amstutz H., Baum M., Carbon J., Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988 Feb;8(2):754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamulin V., Mao J., Appel B., Sumner-Smith M., Yamao F., Söll D. Six Schizosaccharomyces pombe tRNA genes including a gene for a tRNALys with an intervening sequence which cannot base-pair with the anticodon. Nucleic Acids Res. 1983 Dec 20;11(24):8537–8546. doi: 10.1093/nar/11.24.8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Guo L. H., Wu R. Exonuclease III: use for DNA sequence analysis and in specific deletions of nucleotides. Methods Enzymol. 1983;100:60–96. doi: 10.1016/0076-6879(83)00046-4. [DOI] [PubMed] [Google Scholar]
- Hahnenberger K. M., Baum M. P., Polizzi C. M., Carbon J., Clarke L. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1989 Jan;86(2):577–581. doi: 10.1073/pnas.86.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Mao J., Schmidt O., Söll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980 Sep;21(2):509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P., Amstutz H., Kohli J., Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982 Nov 18;300(5889):225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- Munz P., Leupold U., Agris P., Kohli J. In vivo decoding rules in Schizosaccharomyces pombe are at variance with in vitro data. Nature. 1981 Nov 12;294(5837):187–188. doi: 10.1038/294187a0. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Adachi Y., Funahashi S., Niwa O., Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986 May;5(5):1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R., Kuhn R. M., Newman J. L., Meade J. C. Unequal homologous recombination between tandemly arranged sequences stably incorporated into cultured rat cells. Mol Cell Biol. 1985 Oct;5(10):2613–2622. doi: 10.1128/mcb.5.10.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Carbon J., Bloom K. Tightly centromere-linked gene (SPO15) essential for meiosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):158–167. doi: 10.1128/mcb.6.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]