Abstract
Background: Occupational exposure to trichloroethylene (TCE) has been linked to adverse health outcomes including non-Hodgkin’s lymphoma and kidney and liver cancer; however, TCE’s mode of action for development of these diseases in humans is not well understood.
Methods: Non-targeted metabolomics analysis of plasma obtained from 80 TCE-exposed workers [full shift exposure range of 0.4 to 230 parts-per-million of air (ppma)] and 95 matched controls were completed by ultra-high resolution mass spectrometry. Biological response to TCE exposure was determined using a metabolome-wide association study (MWAS) framework, with metabolic changes and plasma TCE metabolites evaluated by dose-response and pathway enrichment. Biological perturbations were then linked to immunological, renal and exposure molecular markers measured in the same population.
Results: Metabolic features associated with TCE exposure included known TCE metabolites, unidentifiable chlorinated compounds and endogenous metabolites. Exposure resulted in a systemic response in endogenous metabolism, including disruption in purine catabolism and decreases in sulphur amino acid and bile acid biosynthesis pathways. Metabolite associations with TCE exposure included uric acid (β = 0.13, P-value = 3.6 × 10−5), glutamine (β = 0.08, P-value = 0.0013), cystine (β = 0.75, P-value = 0.0022), methylthioadenosine (β = −1.6, P-value = 0.0043), taurine (β = −2.4, P-value = 0.0011) and chenodeoxycholic acid (β = −1.3, P-value = 0.0039), which are consistent with known toxic effects of TCE, including immunosuppression, hepatotoxicity and nephrotoxicity. Correlation with additional exposure markers and physiological endpoints supported known disease associations.
Conclusions: High-resolution metabolomics correlates measured occupational exposure to internal dose and metabolic response, providing insight into molecular mechanisms of exposure-related disease aetiology.
Keywords: Exposome, Metabolic phenotype, VOC exposure, Bioeffect monitoring, Population screening, High-resolution metabolomics, Trichlororoethylene
Key Messages
Using non-targeted, high-resolution metabolomics analysis of blood obtained from trichloroethylene-exposed workers, we linked exposure to internal dose and perturbations in endogenous metabolism.
A systemic metabolic response to trichloroethylene was observed in exposed workers, which included a large number of unidentified chlorinated chemicals and alterations in endogenous metabolism consistent with known toxic targets, including renal, liver and immune systems.
Metabolic changes were consistent with risk factors for disease associated with trichloroethylene exposure, even though the majority of exposure levels were below the U.S. Occupational Safety and Health Administration limit.
Findings from this study provide insight into the underlying toxic mechanisms of trichloroethylene and exposure-related changes to metabolic phenotype in an otherwise healthy population.
Introduction
Trichloroethylene (TCE) is a widely used industrial solvent and common organic contaminant in groundwater.1 TCE is classified as carcinogenic to humans by the IARC2 [http://monographs.iarc.fr/ENG/Monographs/vol106/index.php] and as both a carcinogenic and non-carcinogenic health hazard by the US Environmental Protection Agency (EPA).3 TCE exposure has been linked to increased risk for kidney cancer,4,5 non-Hodgkin’s lymphoma (NHL)6,7 and liver cancer.8,9 Human and animal studies have identified numerous non-cancer adverse effects of TCE, including immune dysfunction10,11 and nervous system,12 renal13,14 and liver15 toxicity.
The biological plausibility of TCE toxicity has been verified in animal and in vitro studies,3 but limited data exist establishing biochemical changes in humans. Recently, cross-sectional molecular epidemiology studies evaluating TCE exposure on immune function16–18 and nephrotoxicity19 have observed effects that occurred at levels of exposure below the current US Occupational Safety and Health Administration (OSHA) permissible limits of 100 parts-per-million of air (ppma). Participants were selected using extensive screening procedures to avoid confounding exposures, past occupational use of volatile organic compounds (VOC) and previous history of cancer. The results indicated immunosuppression consistent with increased NHL risk,17 elevated urinary nephrotoxicity markers19 and immunotoxic effects including decreased serum immunoglobulin G (IgG), immunoglobulin M (IgM) and interleukin 10 (IL-10).16,18 These findings suggest a complex biochemical response to TCE exposure, with multiple targets of toxicity.
Comprehensive profiling of the metabolic phenotype associated with TCE exposure can provide insight into the biochemical changes occurring within exposed workers. High-resolution metabolomics (HRM) using ultra-high resolution mass spectrometry20,21 with data extraction algorithms22–25 now enables measurement of greater than 10 000 chemicals in biological samples with quantitative reproducibility.26 With mass spectrometry-based HRM platforms, chemicals are detected as ions in a gas phase and measured as a mass-to-charge ratio (m/z). The m/z signals arise from interaction of a neutral molecule with cations present in solution, often resulting in multiple signals corresponding to a single chemical species. Thus, initial measures are most accurately described as an m/z feature, which is defined by the accurate m/z, retention time and associated intensity, and a non-targeted analytical structure is used with chemometric approaches for selection and identification of metabolic species present in a sample. The resulting chemical profile provides in-depth coverage of the metabolic phenotype, including detection of metabolic intermediates, dietary chemicals, xenobiotics and microbiome-related metabolites.27–34 A previous metabolomic study of TCE exposure in mice linked liver-related effects to increased expression of peroxisome proliferator-activated receptor α (PPRα) target genes, which influences inflammatory response, cell proliferation, lipid metabolism and glucose metabolism.35
In this study, we used a metabolome-wide association study (MWAS)30,36,37 to identify dose-dependent metabolic changes in TCE-exposed workers and unexposed controls, using a well-characterized, worker population.17,19 Direct TCE and endogenous metabolites were characterized, and pathway analysis was used to identify biological response to TCE exposure. We further examined metabolite association with separately measured TCE exposure and immunological and nephrotoxicity markers.
Methods
Study design and exposure measurement
Samples were collected in 2006 as part of a cross-sectional study conducted in Guangdong, China, to assess the early biological effects of occupational exposure to TCE; a full description of factory and subject selection is described in Lan and Zhang17 and is summarized in Supplementary Text 1 (available as Supplementary data at IJE online). Replicate full-shift personal air exposure measurements were taken for 80 exposed workers and 95 controls, using 3M organic vapour-monitoring badges. All samples were analysed for TCE, with a subset (48 from TCE-exposed workers) selected for additional VOCs including benzene, methylene chloride, perchloroethylene and epichlorohydrin. Each study subject was asked to provide a 29-ml peripheral blood sample, post-shift urine sample and undergo a brief physical examination. The study was approved by Institutional Review Boards at the US National Cancer Institute and the Guangdong National Poison Control Center, China.
High-resolution metabolomics
Plasma was prepared and analysed daily in batches of 20 by HRM with C18 liquid chromatography using the methodology of G and, Walker.26 Data were extracted with apLCMS38 and xMSanalyser.22 Full details and quality control are included in Supplementary Text 1. From the 10 017 ions detected, we removed all features that were not detected in greater than 50% of the individuals from at least one group. The remaining 7830 m/z features were log2 transformed and used for defining a TCE exposure metabolic phenotype.
Data analysis
Statistical analysis was carried out using R version 3.1.239. A flow diagram detailing all data analysis steps is provided in Supplementary Text 3 (available as Supplementary data at IJE online).
Step 1: TCE exposure MWAS
The TCE exposure MWAS was completed using a linear regression framework. For each m/z feature, the log2 transformed intensity was used to test for dose-response across the categories of exposure, which included controls, low-exposed (< 12 ppma) and high-exposed (≥ 12 ppma), defined as a continuous variable (control = 0; < 12 ppma = 1; ≥ 12 ppma = 2). Equal spacing between the groups was used to bias feature selection towards those showing dose-response while providing representative metabolic associations with TCE exposure across the population and reducing false-positives. The statistical model included adjustments for age (continuous), sex (factor) and body mass index (BMI; continuous), which are known to influence chemical disposition and toxicokinetics.40,41 To evaluate if worker smoking and alcohol use should be considered, the data were re-analysed separately with smoking and alcohol use status included as a covariate. The resulting analysis increased the number of m/z features with false discovery rate (FDR) ≤ 20% by one and eight when accounting for smoking and alcohol use, respectively. Thus, neither was included as covariate in the regression model. To account for multiple comparisons, we applied a Benjamini and Hochberg42 (FDR) threshold of 20% (raw P-value = 4.4 × 10−3), which controls the rate of false findings rather than falsely rejected null hypotheses.43,44
Step 2: Identification of TCE exposure products
We first characterized the m/z features associated with TCE exposure, to identify direct exposure products by matching the mass for common positive electrospray ionization adducts of TCE additives and metabolites,45 at match accuracy of ± 10 ppm (± 10e-5 × theoretical m/z mass), to TCE-associated m/z features. Unidentified masses were tested for the presence of 37Cl and 37Cl2 isotopes with the pattern.search() function in the R package nontarget46 with retention time tolerance of ± 10 s and mass accuracy of ± 5 ppm.
Step 3: Biological response to TCE exposure
The remaining m/z features were matched to a reference database of 75 metabolites previously confirmed with MS2 and co-elution studies.26 Additional features not matching these metabolites were annotated with the KEGG database,47,48 which provides information on 487 pathways containing 17 620 unique metabolites and is a common metabolic reference for metabolomic studies. Identities were assigned using evidence-scoring provided in Mummichog49 for matching to +H, +Na, +K, -H2O+H, -2H2O+H, +ACN+H, +ACN+Na, +2ACN+H, +2Na-H and +2 H adducts at ± 10 ppm mass tolerance. Enriched metabolic pathways were selected using Mummichog scoring threshold of 0.05.
Step 4: Integration with TCE exposure and physiological markers
Correlation for each of the 188 m/z features with biomarkers of TCE exposure, immune function and renal damage were calculated using MetabNet50 in exposed workers only. These measures have been described previously17–19,51 and include white blood cell count (WBC), lymphocytes (LY), CD4 + T cells (CD4), soluble CD27, soluble CD30, mitochondrial DNA copy number (mtDNA), IgG, IgM, urinary isoenzymes of glutathione-s-transferase (αGST, piGST), kidney injury molecule 1 (KIM-1), N-acetyl-beta-glucosaminidase (NAG), trichloroacetic acid (TCA) and total, free and conjugated trichloroethanol (TCE-EtOH). All urinary measurements were normalized by creatinine to account for differences in urinary output.52 Marker-metabolite correlations were selected for consideration and plotting in Cytoscape,53 based upon effect size (Spearman |r| ≥ 0.3) and P-value threshold ≤ 0.05. Due to the use of effect size for prioritizing network connectivity, correction for multiple hypothesis testing was not applied.
Results
Study population
Demographics, including age, sex, BMI, current smoking and alcohol use status were comparable among the exposed and non-exposed workers (Table 1). The median exposure level of 12.0 ppma was used to define low- and high-exposure thresholds. Measured TCE levels for exposed factory worker levels ranged from 0.4 ppma to 229 ppma (Table 1); 96% of the workers exposed to TCE levels were below the OSHA 8-h limit.
Table 1.
Demographic characteristics and TCE exposure level
| Subjects | Controls | Exposed | ||
|---|---|---|---|---|
| controls (n = 95) | Total (n = 80) | <12 ppm (n = 39) | ≥ 12 ppm (n = 41) | |
| Demographic characteristics | ||||
| Age, mean (SD)a | 27 (7) | 25 (7) | 24 (5) | 27 (8) |
| BMIb, mean (SD)a | 22 (3) | 21 (3) | 21 (2) | 22 (3) |
| Current smoking, n (%)c | 37 (39%) | 34 (43%) | 17 (44%) | 17 (41%) |
| Alcohol use, n (%)c | 40 (42%) | 26 (33%) | 13 (33%) | 13 (32%) |
| Sex, n (%)c | ||||
| Female | 23 (24%) | 23 (29%) | 15 (38%) | 8 (20%) |
| Male | 72 (76%) | 57 (71%) | 24 (62%) | 33 (80%) |
| TCE exposure | ||||
| TCE air level (ppma), mean (SD)d | < 0.03 | 22.19 (35.9) | 5.19 (3.5) | 38.36 (44.6) |
| Minimum exposure | NA | 0.4 | 0.4 | 12.0 |
| Maximum exposure | NA | 229 | 11.7 | 229 |
aMean ± standard deviation (SD).
bBMI, body mass index.
cNumber.
dMean ± standard deviation of TCE exposure levels, expressed as parts-per-million (ppm) of air. NA, not available.
At FDR threshold ≤ 20%, MWAS identified 188 m/z features associated with TCE exposure (Figure 1 ). These features were selected for characterization and identification of the metabolic response to TCE exposure: 61 exhibited P-values ≤ 10−4 with adjusted r2 ranging from 0.08 to 0.76. Regression coefficient for exposure categorization ranged from -5.4 to 9.9. The list of m/z features and regression results are provided in Supplementary Text 2 (available as Supplementary data at IJE online).
Figure 1.
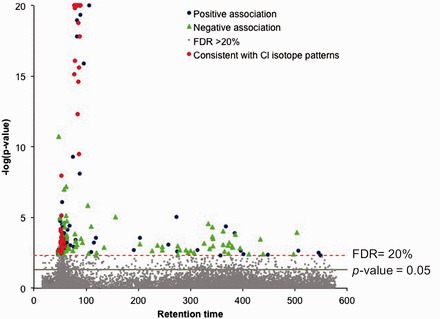
Manhattan plot for TCE MWAS in exposed and non-exposed workers. Linear regression identified m/z features that were associated with TCE exposure at FDR ≤ 20% after correcting for age, sex and BMI. Each m/z features is coloured by association (0 ≤ β ≥ 0). Those with isotopic patterns matching halogenated compounds (based upon the presence of 37Cl and 37Cl2 masses within a retention time window of ± 10 s) are indicated by a red triangle.
Identification of TCE exposure products
We tested for the presence of exposure products by searching the 188 m/z features for TCE metabolites from cytochrome P450, glutathione (GSH) conjugation by GSH S-transferase14,45,54 and nine common additives used in commercial formulations of TCE.55 Three m/z features were consistent with TCE metabolites. A match to the +2Na-H adduct of TCA was detected, and exhibited the highest association with exposure (Table 2). Additional adduct masses of TCA included the +Na+K form. The +2 K-H adduct of TCE-EtOH was also present, as was the stabilizer triethanolamine.
Table 2.
Top m/z features matching chlorinated isotope patterns
| m/za | Retention time (s) | Identity | Isotopesb | Adjusted r2 | β coefficient | P-value |
|---|---|---|---|---|---|---|
| 206.8750 | 81.3 | Trichloroacetic acidc | M + 2, M + 4 | 0.76 | 9.87 | 3.1E-55 |
| 224.8456 | 88.4 | Trichloroethanold | M + 2 | 0.68 | 8.37 | 1.0E-43 |
| 264.8338 | 78.7 | Unknown | M + 2, M + 4 | 0.55 | 7.16 | 9.8E-32 |
| 332.8201 | 83.3 | Unknown | M + 2, M + 4 | 0.43 | 5.69 | 3.9E-23 |
| 280.8067 | 88.5 | Unknown | M + 2, M + 4 | 0.41 | 5.08 | 1.2E-21 |
| 324.7895 | 77.2 | Unknown | M + 2 | 0.39 | 5.55 | 7.2E-21 |
| 340.7621 | 77.1 | Unknown | M + 2, M + 4 | 0.32 | 4.11 | 7.1E-16 |
| 274.8629 | 84.8 | Unknown | M + 2 | 0.29 | 4.01 | 2.4E-15 |
| 392.7587 | 86.1 | Unknown | M + 2 | 0.20 | 4.14 | 3.2E-10 |
| 380.7691 | 52.6 | Unknown | M + 2, M + 4 | 0.16 | 0.08 | 2.6E-04 |
Regression parameters for the monoisotopic mass only are listed. Regression results for all features with FDR ≤ 20% are provided in Supplementary Text 2, available as Supplementary data at IJE online.
aMass-to-charge ratio.
bM + 2 = 37Cl (+1.9971); M + 4 = 37Cl2 (+3.9941).
cDetected as +2Na-H adduct form.
dDetected as +2K-H adduct form.
TCA and TCE-EtOH alone did not explain m/z features associated with exposure, and the presence of features with a negative mass defect suggests additional chlorinated chemicals.56,57 To test for additional chlorinated metabolites, we applied isotopic pattern-searching for m/z ions exhibiting M + 2 and M + 4 mass spacing, which corresponds to the presence 37Cl (+1.9971) and 37Cl2 (+3.9941). We identified 45 m/z features corresponding to 19 unique isotopic pairs (Figure 1). The top 10 monoisotopic m/z masses ranked according to P-values are provided in Table 2. Two included the C2HCl2O237Cl and C2HClO237Cl2 forms of TCA and the C2H3Cl2O37Cl isotope of TCE-EtOH; however, the remaining showed no matches to known TCE-related metabolites.
Biological response to TCE exposure
After removal of the 49 m/z features identified as probable chlorinated chemicals, we annotated the remaining 135 m/z features; 19 m/z features corresponded to 12 confirmed metabolites (Table 3). The remaining had identities assigned based on systematic evidence-scoring by isotopic and adduct pairing, which identified an additional 35 m/z features (Supplementary Text 2, available as Supplementary data at IJE online). In total, 54 m/z features matching 46 unique metabolites were identified. The remaining had no probable matches.
Table 3.
Identified endogenous metabolites associated with TCE exposure and relevant to biological response to exposure
| m/za | Retention time (s) | Identity | ID levelb | Adjusted r2 | β coefficient | P-value |
|---|---|---|---|---|---|---|
| 439.3008 | 503.3 | 7α-Hydroxycholest-4-en-3-one | 3 | 0.12 | −0.15 | 1.1E-04 |
| 394.3031 | 497.2 | Chenodeoxycholic acid | 4 | 0.03 | −1.31 | 3.9E-03 |
| 132.0764 | 59.2 | Creatine | 1 | 0.21 | −0.22 | 4.9E-05 |
| 241.0310 | 75.3 | Cystine | 1 | 0.05 | −0.75 | 2.2E-03 |
| 191.0397 | 49.4 | Glutamine | 1 | 0.14 | 0.07 | 1.3E-03 |
| 307.0197 | 53.8 | Homocysteine | 3 | 0.07 | −0.06 | 2.4E-03 |
| 188.0705 | 63.1 | Indolelactic acid | 1 | 0.07 | −0.16 | 2.9E-04 |
| 336.0527 | 276.1 | Methylthioadenosine | 3 | 0.09 | −1.00 | 4.4E-03 |
| 400.3405 | 363.0 | Palmitoylcarnitine | 1 | 0.20 | −0.18 | 9.3E-05 |
| 119.0488 | 257.4 | Phenylacetic acid | 1 | 0.05 | 0.11 | 8.0E-04 |
| 163.9776 | 63.8 | Taurine | 1 | 0.08 | −2.39 | 1.1E-03 |
| 249.0609 | 61.7 | Tryptophan | 1 | 0.15 | −0.15 | 6.5E-08 |
| 146.0596 | 66.5 | Tyrosine | 1 | 0.05 | −0.09 | 1.5E-03 |
| 212.9993 | 67.7 | Uric acid | 1 | 0.34 | 0.13 | 3.6E-05 |
| 279.2310 | 411.3 | α-Linolenic acid | 1 | 0.03 | −0.09 | 3.9E-03 |
aWhen multiple adducts or isotopes were detected, the highest ranked m/z was used for regression parameters. All TCE exposure association m/z features are listed in Supplementary Text 2, available as Supplementary data at IJE online.
bID level indicates degree of annotation with 1: m/z and retention time matched to authentic standards confirmed with MS2, 2: Multiple/isotopes present; 3: m/z matched single adduct mass within 10 ppm mass error, 4: m/z matched adduct mass of multiple isobaric species, most probable identification used.
We used Mummichog49 to evaluate metabolic pathway enrichment, which identified 12 pathways at P-value threshold ≤ 0.05 (Figure 2 ). Detected metabolites in each pathway are provided in Supplementary Text 2. Metabolites from sulphur amino acid metabolism were decreased with exposure, and included cystine and homocystine (Table 3). Disruption in bile acid biosynthesis was also detected, with decreased levels of taurine, chenodeoxycholic acid and 7α-hydroxy-cholestene-3-one in exposed workers (Table 3). Changes in fatty acid metabolism were observed, including reduced levels of palmitoylcarnitine. Metabolites from purine metabolism, including uric acid and glutamine, were increased in a dose-dependent manner in association with TCE exposure, whereas methylthioadenosine (MTA), a metabolite of methionine and purine salvage, was decreased in TCE-exposed workers (Figure 3 ). Metabolic changes consistent with other physiological effects of TCE were also detected, including tryptophan and tyrosine (Table 3).
Figure 2.
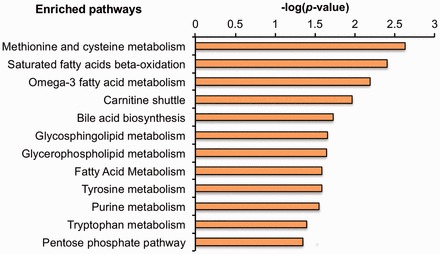
Mummichog enriched pathways for metabolites associated with TCE exposure.
Figure 3.
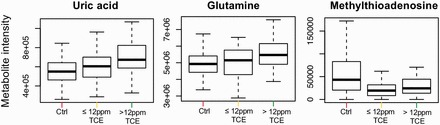
Distribution of uric acid, glutamine and MTA in control, low-exposed and high-exposed workers. Uric acid and glutamine increased in an exposure-dependent manner, whereas MTA was decreased in all exposed workers relative to controls.
Integration with immunological, renal and TCE markers
To visualize systemic effects, we tested for metabolic associations with molecular markers of immune function, nephrotoxicity and TCE exposure. For all 188 m/z features selected through MWAS, 88 were correlated with at least one marker and included 48 identified as metabolites or halogenated chemical species (Figure 4 ). Excluding sCD27 and mtDNA, a high degree of connectivity was present. Chlorinated metabolites were negatively correlated with IgG, IgM and CD4 + T cells, none of which matched known TCE biotransformation products. KIM-1 and additional nephrotoxicity markers were positively associated with chlorinated isotopes. A negative association of KIM-1 with uric acid was present, whereas MTA was negatively associated with the urinary isoenzymes. Correlation with urinary markers of TCE exposure showed positive associations with unidentified and identified chlorinated metabolites, including TCA, TCE-EtOH and unidentified chlorinated chemicals.
Figure 4.
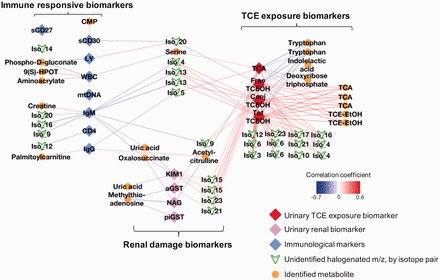
Network correlation analysis of molecular markers previously tested for association with TCE exposure and metabolic features detected in this study. Only correlations with Spearman |r| ≥ 0.3 and P-value ≤ 0.05 corresponding to identified metabolites or probable halogens were included. Immune markers: CD4 = CD4+ T cell count; IgG, immunoglobulin G; IgM, immunoglobulin M; LY, lymphocyte cell count; mtDNA, mitochondrial DNA turnover rate; sCD27, soluble CD27; sCD30, soluble CD30; WBC, white blood cell count. Nephrotoxicity markers: aGST, urinary α glutathione-s-transferase; piGST, urinary pi glutathione-s-transferase; KIM1, urinary kidney injury marker 1; NAG, urinary n-acetyl-beta-glucosaminidase. TCE exposure markers: Conj. TCEOH: urinary trichloroethanol glucuronide; Free TCEOH: urinary trichloroethanol; Tot TCEOH: sum of urinary trichloroethanol and trichloroethanol glucuronide; TCA: urinary trichloroacetic acid.
Discussion
Metabolic phenotyping with HRM enables an unbiased approach to investigate biological changes and mode of action for environmental and occupational exposures. The present study provides an application of HRM to occupational exposure effects in humans and demonstrates the utility of HRM to link VOC exposure to metabolic pathways associated with relevant disease. MWAS of TCE exposure showed alterations to metabolic features characterized as exposure by-products and endogenous metabolites. In addition to detecting known metabolites of TCE, isotopic spacing analysis and strong positive association with exposure categorization suggests the presence of additional chlorinated metabolites, likely derived from TCE. The chemical identity of these metabolites was not determined, but it suggests the existence of additional, uncharacterized TCE-related metabolic products.
Pathway enrichment of metabolites associated with exposure was consistent with TCE detoxification. The presence of cysteine and methionine metabolites is in agreement with metabolism of TCE by glutathione (GSH) conjugation. Association with bile acid biosynthesis is likely related to detoxification of TCE by glucuronidation, with TCE rapidly metabolized to TCE-EtOH in the liver, glucuronidated by glucuronyltransferase and secreted in bile. TCE-EtOH glucuronide undergoes enterohepatic recirculation3 followed by metabolism to TCA.58 Blood bile acid alterations have been identified as a marker of liver damage and hepatotoxicity.59,60 Exposure-related alterations in fatty acid metabolism and reduced palmitoylcarnitine suggest alteration in peroxisome proliferator-activated receptor α, which was also observed in animal TCE exposure models.35
Enriched pathways provided additional evidence of toxicity, with the identified endogenous metabolites relevant to diseases associated with TCE exposure. Elevated levels of blood uric acid have been identified as both an independent risk factor and a marker of kidney disease.61,62 TCE-related alterations in purine catabolism resulting from changes in expression of adenosine deaminase or purine nucleoside phosphorylase could account for the immunosuppressive effects observed by Lan and Zhang.17 Both enzymes are essential for developing and maintaining immune function,63,64 with abnormalities reported in immunodeficiency, leukemia diseases and neoplastic transformations.65–68 The detected TCE-associated changes in plasma glutamine further suggest alterations in immune-related purine catabolism rather than hepatic cells, since glutamine is the primary transporter of ammonia to the liver following deamination of guanine and adenine.69 MTA can act as a secondary precursor to adenine biosynthesis through 5’-methylthioadenosine phosphorylase (MAP).70 Disruption to MAP has been observed in tumour cells, which rely on de novo purine biosynthesis for adenine and DNA synthesis.71 MTA has been observed to be hepatoprotective following treatment with carbon-tetrachloride (CCl4).72 Interestingly, MTA enhances production of IL-1070, and MTA alterations in exposed workers were consistent with TCE-related decreases in IL-1016. Finally, TCE is known to have central nervous system (CNS) depressant effects, with in vitro and long-term exposure associated with dopaminergic neurodegeneration.73–75 Tryptophan and tyrosine both act as precursors for the synthesis of neurotransmitters, and were decreased with exposure
TCE metabolic products are transported through multiple tissues and organ systems.3 The presence of metabolic features consistent with unidentified chlorinated chemicals indicates a complex detoxification response to TCE exposure. A different correlation network for each class of markers suggests the possibility of uncharacterized TCE metabolites being toxic to specific organ systems. Association with immune function markers was consistent with the immunosuppressive effects observed by Lan and Zhang17 and Zhang and Bassig.18 Chlorinated metabolites were negatively correlated with IgG, IgM and CD4+ T cells, suggesting that unidentified metabolites could be directly involved in mediating immunotoxicity. KIM-1 and additional nephrotoxicity markers were positively associated with chlorinated isotopes, consistent with the findings of Vermeulen and Zhang,19 whereas plasma uric acid and MTA showed negative correlation. The negative association of uric acid with KIM-1 was unexpected, since urinary expression of KIM-1 is increased with tubular necrosis and various renal diseases, suggesting perturbed purine metabolism as a potential causal mechanism. A similar trend was observed for MTA and the urinary isoenzyemes: αGST and piGST are shed from the proximal and distal tubular cells, respectively, and are upregulated following kidney damage.76 A large number of suspected chlorinated chemicals endogenous metabolites were correlated with urinary TCA and TCE-EtOH measures. Although a correlation for identified TCE metabolites (TCA and TCE-EtOH) with immune and renal markers was not detected, many unidentified chlorinated chemicals were, suggesting greater toxicity to immune and renal systems.
We acknowledge some limitations in this work. First, there is a predominance of males in our study population; thus, though sex was included as a covariate in the MWAS model, sex-based differences were not evaluated. Second, a limited sample population of 175 individuals from a specific geographical location was used, and an independent cohort was not available to replicate the findings. Third, this study was focused on known TCE and endogenous metabolites because the limited sample availability precluded in-depth structural characterization of unidentified metabolites. Lack of reference standards and low abundance will make structural elucidation of these features challenging. Finally, the results of this study are correlative in nature. We could not account for unknown confounders, nor identify the exact mechanism through which these metabolic associations occurred. Despite these limitations, MWAS identification of pathways known to participate in detoxification of TCE and pathways consistent with toxicological targets of TCE suggests biological relevance. In addition, molecular markers previously shown to be associated with TCE exposure were correlated with TCE metabolites and exposure-associated endogenous metabolites. The results therefore show that HRM provides a useful approach to link occupational exposures to metabolic perturbations and obtain leads to underlying mechanisms of exposure-related disease.
Conclusions
Environmental and workplace exposures are a significant contribution to chronic disease burden. This study of a population with rigorous selection criteria and TCE exposure assessment shows that HRM is able to link internal TCE metabolites and perturbations of endogenous metabolism with isolated occupational TCE exposure. The results show associations with unidentified chlorinated products as well as a multiple mechanistic and disease markers of hepatic function, kidney damage and immune dysfunction. Whereas our study was limited to a high occupational exposure, the metabolic associations represent an important first step in identifying how TCE alters metabolism and leads to disease risk. More generally, the results establish the feasibility of using HRM as an occupational surveillance tool to assist in epidemiological studies of specific exposure risks, underlying toxic mechanisms and exposure-related disease susceptibility.
Funding
This work was supported by intramural funds received from the National Institutes of Health and National Cancer Society; National Institute of Environmental Health Science to D.P.J. [award numbers P30 ES019776, ES023485, RO1 ES009047]; National Heart, Blood and Lung Institute [award number IHL113451]; and Department of Science and Technology of Guangdong Province, China [award number 2007A050100004].
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical expertise of ViLinh Tran for mass spectrometry analyses.
Conflict of interest: None declared.
References
- 1. Moran MJ, Zogorski JS, Squillace PJ. Chlorinated solvents in groundwater of the United States. Environ Sci Technol 2007;41:74–81. [DOI] [PubMed] [Google Scholar]
- 2. Guha N, Loomis D, Grosse Y, et al. Carcinogenicity of trichloroethylene, tetrachloroethylene, some other chlorinated solvents, and their metabolites. Lancet Oncol 2012;13:1192–93. [DOI] [PubMed] [Google Scholar]
- 3. Chiu WA, Jinot J, Scott CS, et al. Human health effects of trichloroethylene: key findings and scientific issues. Environ Health Perspect 2013;121:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moore LE, Boffetta P, Karami S, et al. Occupational trichloroethylene exposure and renal carcinoma risk: evidence of genetic susceptibility by reductive metabolism gene variants. Cancer Res 2010;70:6527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charbotel B, Fevotte J, Hours M, Martin JL, Bergeret A. Case-control study on renal cell cancer and occupational exposure to trichloroethylene. Part II:Epidemiological aspects. Ann Occup Hyg 2006;50:777–87. [DOI] [PubMed] [Google Scholar]
- 6. Raaschou-Nielsen O, Hansen J, McLaughlin JK, et al. Cancer risk among workers at Danish companies using trichloroethylene: a cohort study. Am J Epidemiol 2003;158:1182–92. [DOI] [PubMed] [Google Scholar]
- 7. Purdue MP, Bakke B, Stewart P, et al. A case-control study of occupational exposure to trichloroethylene and non-Hodgkin lymphoma. EnviroHealth Perspect 2011;119:232–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen J, Sallmen M, Selden AI, et al. Risk of cancer among workers exposed to trichloroethylene: analysis of three Nordic cohort studies. J Natl Cancer Inst 2013;105:869–77. [DOI] [PubMed] [Google Scholar]
- 9. Scott CS, Jinot J. Trichloroethylene and cancer: systematic and quantitative review of epidemiologic evidence for identifying hazards. Int J Environ Res Public Health 2011;8:4238–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooper GS, Makris SL, Nietert PJ, Jinot J. Evidence of autoimmune-related effects of trichloroethylene exposure from studies in mice and humans. Environ Health Perspect 2009;117:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iavicoli I, Marinaccio A, Carelli G. Effects of occupational trichloroethylene exposure on cytokine levels in workers. J Occup Environ Med 2005;47:453–57. [DOI] [PubMed] [Google Scholar]
- 12. Kilburn KH. Is neurotoxicity associated with environmental trichloroethylene (TCE)?. Arch Environ Health 2002;57:113–20. [DOI] [PubMed] [Google Scholar]
- 13. Bruning T, Bolt HM. Renal toxicity and carcinogenicity of trichloroethylene: key results, mechanisms, and controversies. Crit Rev Toxicol 2000;30:253–85. [DOI] [PubMed] [Google Scholar]
- 14. Lash LH, Xu Y, Elfarra AA, Duescher RJ, Parker JC. Glutathione-dependent metabolism of trichloroethylene in isolated liver and kidney cells of rats and its role in mitochondrial and cellular toxicity. Drug Metab Dispos 1995;23:846–53. [PubMed] [Google Scholar]
- 15. Brautbar N, Williams J II. Industrial solvents and liver toxicity: risk assessment, risk factors and mechanisms. Int J Hyg Environ Health 2002;205:479–91. [DOI] [PubMed] [Google Scholar]
- 16. Bassig BA, Zhang L, Tang X, et al. Occupational exposure to trichloroethylene and serum concentrations of IL-6, IL-10, and TNF-alpha. Environ Mol Mutagen 2013;54:450–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lan Q, Zhang L, Tang X, et al. Occupational exposure to trichloroethylene is associated with a decline in lymphocyte subsets and soluble CD27 and CD30 markers. Carcinogenesis 2010;31:1592–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang L, Bassig BA, Mora JL, et al. Alterations in serum immunoglobulin levels in workers occupationally exposed to trichloroethylene. Carcinogenesis 2013;34:799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vermeulen R, Zhang L, Spierenburg A, et al. Elevated urinary levels of kidney injury molecule-1 among Chinese factory workers exposed to trichloroethylene. Carcinogenesis 2012;33:1538–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier transform mass spectrometry. Mol Cell Proteomics 2011;10:M111 009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarov A, Denisov E, Kholomeev A, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 2006;78:2113–20. [DOI] [PubMed] [Google Scholar]
- 22. Uppal K, Soltow QA, Strobel FH, et al. xMSanalyser:automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu T, Park Y, Li S, Jones DP. Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J Proteome Res 2013;12:1419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Libiseller G, Dvorzak M, Kleb U, et al. IPO:a tool for automated optimization of XCMS parameters. BMC Bioinformatics 2015;16:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu T, Jones DP. Improving peak detection in high-resolution LC/MS metabolomics data using preexisting knowledge and machine learning approach. Bioinformatics 2014;30:2941–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Go YM, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol Sci 2015;148:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Athersuch TJ, Keun HC. Metabolic profiling in human exposome studies. Mutagenesis 2015;30:755–62 [DOI] [PubMed] [Google Scholar]
- 28. Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008;453:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Ann Rev Nutr 2012;32:183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicholson JK, Holmes E, Elliott P. The metabolome-wide association study:ba new look at human disease risk factors. J Proteome Resh 2008;7:3637–38. [DOI] [PubMed] [Google Scholar]
- 31. Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012;295:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scalbert A, Brennan L, Manach C, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr 2014;99:1286–308. [DOI] [PubMed] [Google Scholar]
- 33. Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell 2008;134:714–17. [DOI] [PubMed] [Google Scholar]
- 34. Psychogios N, Hau DD, Peng J, et al. The human serum metabolome. PloS One 2011;6:e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang ZZ, Krausz KW, Tanaka N, et al. Metabolomics reveals trichloroacetate as a major contributor to trichloroethylene-induced metabolic alterations in mouse urine and serum. Arch Toxicol 2013;87:1975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burgess LG, Uppal K, Walker DI, et al. Metabolome-wide association study of primary open angle glaucoma. Invest Ophthalmol Vis Sci 2015;56:5020–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Go YM, Walker DI, Soltow QA, et al. Metabolome-wide association study of phenylalanine in plasma of common marmosets. Amino Acids 2015;47:589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu T, Park Y, Johnson JM, Jones DP. apLCMS – adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25:1930–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R Core Team. R: A language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2014. [Google Scholar]
- 40. Ernstgard L, Sjogren B, Warholm M, Johanson G. Sex differences in the toxicokinetics of inhaled solvent vapors in humans 2. 2-propanol. Toxicol Appl Pharmacol 2003;193:158–67. [DOI] [PubMed] [Google Scholar]
- 41. Gochfeld M. Framework for gender differences in human and animal toxicology. Environ Res 2007;104:4–21. [DOI] [PubMed] [Google Scholar]
- 42. Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc B Met 1995;57:289–300. [Google Scholar]
- 43. Patel CJ. Analytical complexity in detection of gene variant-by-environment exposure interactions in high-throughput genomic and exposomic research. Curr Environ Health Rep 2016;3:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vinaixa M, Samino S, Saez I, Duran J, Guinovart JJ, Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites 2012;2:775–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lash LH, Fisher JW, Lipscomb JC, Parker JC. Metabolism of trichloroethylene. Environ Health Perspect 2000;108(Suppl 2):177–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loos M. nontarget: Detecting, Combining and Filtering Isotope, Adduct and Homologue Series Relations in High-resolution Mass Spectrometry (HRMS) Data. R package version 1.7 ed; 2015, https://cran.r-project.org/web/packages/nontarget/index.html (7 July 2015, date last accessed). [Google Scholar]
- 47. Tanabe M, Kanehisa M. Using the KEGG database resource. Curr Protoc Bioinformatics 2012;1:Unit1 12. [DOI] [PubMed] [Google Scholar]
- 48. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, Jones DP. MetabNet: An R package for metabolic association analysis of high-resolution metabolomics data. Front Bioeng Biotechnol 2015;3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim S, Collins LB, Boysen G, et al. Liquid chromatography electrospray ionization tandem mass spectrometry analysis method for simultaneous detection of trichloroacetic acid, dichloroacetic acid, S-(1,2-dichlorovinyl)glutathione and S-(1,2-dichlorovinyl)-L-cysteine. Toxicology 2009;262:230–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cone EJ, Caplan YH, Moser F, Robert T, Shelby MK, Black DL. Normalization of urinary drug concentrations with specific gravity and creatinine. J Anal Toxicol 2009;33:1–7. [DOI] [PubMed] [Google Scholar]
- 53. Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. Curr Protoc Bioinformatics 2014;47:8 13 1–8 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chiu WA, Micallef S, Monster AC, Bois FY. Toxicokinetics of inhaled trichloroethylene and tetrachloroethylene in humans at 1 ppm:empirical results and comparisons with previous studies. Toxicol Sci 2007;95:23–36. [DOI] [PubMed] [Google Scholar]
- 55. IARC. Trichloroethylene. IARC monographs on the evaluation of carcinogenic risks to humans;Vol 106. Lyon, France: International Agency for Research on Cancer, 2014. [Google Scholar]
- 56. Jobst KJ, Shen L, Reiner EJ, et al. The use of mass defect plots for the identification of (novel) halogenated contaminants in the environment. Anal Bioanal Chem 2013;405:3289–97. [DOI] [PubMed] [Google Scholar]
- 57. Zhu M, Ma L, Zhang D, et al. Detection and characterization of metabolites in biological matrices using mass defect filtering of liquid chromatography/high resolution mass spectrometry data. Drug metab Dispos 2006;34:1722–33. [DOI] [PubMed] [Google Scholar]
- 58. Stenner RD, Merdink JL, Stevens DK, Springer DL, Bull RJ. Enterohepatic recirculation of trichloroethanol glucuronide as a significant source of trichloroacetic acid. Metabolites of trichloroethylene. Drug Metab Dispos 1997;25:529–35. [PubMed] [Google Scholar]
- 59. Li T, Chiang JY. Bile acid signaling in liver metabolism and diseases. J Lipids 2012;2012:754067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lake AD, Novak P, Shipkova P, et al. Decreased hepatotoxic bile acid composition and altered synthesis in progressive human nonalcoholic fatty liver disease. Toxicol Appl Pharmacol 2013;268:132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sonoda H, Takase H, Dohi Y, Kimura G. Uric acid levels predict future development of chronic kidney disease. Am J Nephrol 2011;33:352–57. [DOI] [PubMed] [Google Scholar]
- 62. Mohandas R, Johnson RJ. Uric acid levels increase risk for new-onset kidney disease. J Am Soc Nephrol 2008;19:2251–53. [DOI] [PubMed] [Google Scholar]
- 63. Moriwaki Y, Yamamoto T, Higashino K. Enzymes involved in purine metabolism– a review of histochemical localization and functional implications. Histol Histopathol 1999;14:1321–40. [DOI] [PubMed] [Google Scholar]
- 64. Cristalli G, Costanzi S, Lambertucci C, et al. Adenosine deaminase: functional implications and different classes of inhibitors. Med Res Rev 2001;21:105–28. [DOI] [PubMed] [Google Scholar]
- 65. Kopff M, Zakrzewska I, Czernicki J, Klem J, Strzelczyk M. Red blood cell adenosine deaminase activity in multiple sclerosis. Clin Chim Acta 1993;214:97–101. [DOI] [PubMed] [Google Scholar]
- 66. Koizumi H, Ohkawara A. Adenosine deaminase activity in sera of patients with psoriasis, mycosis fungoides and adult T cell leukemia. Acta Derm Venereol 1992;72:410–12. [PubMed] [Google Scholar]
- 67. Smyth JF, Poplack DG, Holiman BJ, Leventhal BG, Yarbro G. Correlation of adenosine deaminase activity with cell surface markers in acute lymphoblastic leukemia. J Clin Invest 1978;62:710–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Segura RM, Pascual C, Ocana I, et al. Adenosine deaminase in body fluids: a useful diagnostic tool in tuberculosis. Clin Biochem 1989;22:141–48. [DOI] [PubMed] [Google Scholar]
- 69. Mathews CK, van Holde KE, Appling DR, Anthony-Cahill SJ. Biochemistry. 4th edm Toronto, ON: Pearson, 2012. [Google Scholar]
- 70. Avila MA, Garcia-Trevijano ER, Lu SC, Corrales FJ, Mato JM. Methylthioadenosine. Int J Biochem Cell Biol 2004;36:2125–30. [DOI] [PubMed] [Google Scholar]
- 71. Nobori T, Takabayashi K, Tran P, et al. Genomic cloning of methylthioadenosine phosphorylase: a purine metabolic enzyme deficient in multiple different cancers. Proc Natl Acad Sci U S A 1996;93:6203–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simile MM, Banni S, Angioni E, et al. 5’-Methylthioadenosine administration prevents lipid peroxidation and fibrogenesis induced in rat liver by carbon-tetrachloride intoxication. J Hepatol 2001;34:386–94. [DOI] [PubMed] [Google Scholar]
- 73. Caudle WM, Guillot TS, Lazo CR, Miller GW. Industrial toxicants and Parkinson’s disease. Neurotoxicology 2012;33:178–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zaheer F, Slevin JT. Trichloroethylene and Parkinson disease. Neurol Clin 2011;29:657–65. [DOI] [PubMed] [Google Scholar]
- 75. Liu M, Choi DY, Hunter RL, et al. Trichloroethylene induces dopaminergic neurodegeneration in Fisher 344 rats. J Neurochem 2010;112:773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Branten AJ, Mulder TP, Peters WH, Assmann KJ, Wetzels JF. Urinary excretion of glutathione S transferases alpha and pi in patients with proteinuria: reflection of the site of tubular injury. Nephron 2000;85:120–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


