Abstract
Proton nuclear magnetic resonance spectroscopy of biofluids has become one of the key techniques for metabolic profiling and phenotyping. This technique has been widely used in a number of epidemiological studies and in a variety of health disorders. However, its utilization in brain disorders is limited due to the blood–brain barrier, which not only protects the brain from unwanted substances in the blood, but also substantially limits the potential of finding biomarkers for neurological disorders in serum. This review article focuses on the potential of localized in vivo proton magnetic resonance spectroscopy (1H-MRS) for non-invasive neurochemical profiling in the human brain. First, methodological aspects of 1H-MRS (data acquisition, processing and metabolite quantification) that are essential for reliable non-invasive neurochemical profiling are described. Second, the power of 1H-MRS-based neurochemical profiling is demonstrated using some examples of its application in neuroscience and neurology. Finally, the authors present their vision and propose necessary steps to establish 1H-MRS as a method suitable for large-scale neurochemical profiling in epidemiological research.
Keywords: 1H-MRS, methodology, spectral quality, neurochemical profiling, CNS disorders
Key Messages
In vivo neurochemical profiling in the human brain is feasible, but the widespread use requires the implementation of the most advanced methodologies of 1H-MRS.
There is a widening gap between what is feasible at specialized MR research centres and what is currently available for routine clinical practice.
The software upgrade (B0 shimming, localization pulses sequence) on clinical 3T MR scanners is sufficient for a significant improvement of the 1H-MRS data quality and for an increase in the range of reliably quantified metabolites.
Introduction
Applications of comprehensive metabolic profiling are increasingly common in epidemiology.1,2 In general, ‘metabolic profiling’ refers to the simultaneous quantification of a wide range of low-molecular-weight metabolites in a variety of tissues or biofluids. As the fundamental principles of epidemiology require large-scale cohort studies, only minimally invasive, high-throughput screening methods are acceptable for these types of studies. Consequently, metabolic profiling has been almost exclusively applied to biofluids, such as plasma (serum) or urine, using high-resolution liquid-state 1H nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS) combined with chromatographic separation.1 These metabolic profiles provide a top-down ‘systems-level’ readout of the biochemistry, physiological status and environmental exposure of individuals and populations that can be exploited in personalized medicine and public healthcare.3 Wide-range metabolic phenotyping has been successfully applied to different health issues, such as nutritional problems,4 effects of long-term physical activity,5 type 1 and 2 diabetes6,7 and cardiovascular diseases.2 Other studies indicated the potential of blood plasma metabolic phenotyping as a tool to discriminate between cancer types.8 However, these types of preliminary results must be taken with caution due to the tendency of multivariate classification to overestimate the predictive power of the method when the number of subjects is small.9 Metabolic phenotyping is becoming important in disease prevention due to its potential to identify individuals or sub-populations at risk, allowing for interventions (change in lifestyle, diet or appropriate medication) before the onset of the disease.3 Metabolic phenotyping can be further utilized in personalized medicine for patient stratification by analysing pre-dose biofluid metabolite profiles to predict drug effects,10 which would increase the efficacy of treatment and decrease unwanted side effects.11,12
Metabolic phenotyping and neurological disorders
Applications of metabolite phenotyping to neurological disorders are rather sparse.13,14 The brain metabolism and its homeostasis are well protected from the rest of the body and circulating blood by the blood–brain barrier (BBB), which substantially limits the potential for finding biomarkers for neurological disorders in serum15 and even more in urine.16 For that reason, metabolic profiling of cerebrospinal fluid (CSF) should be able to characterize the neurochemical status of the central nervous system (CNS) more directly than the profiling of plasma or urine.17–19 However, the CSF sample collection by lumbar puncture is invasive and is justified only for diagnostic purposes. Such an invasive procedure is not ideal for repeated follow-up visits to assess the efficacy of treatment and cannot be applied for screening in the general population.19 Moreover, metabolic profiling of CSF has a limited capability to map regional metabolic deviations from brain homeostasis due to the high complexity of the CNS and its regional heterogeneity (e.g. cortical and subcortical structures, grey and white matter), diversity of neural cell types (neurons, glia), neuronal morphology (soma, dendrites, axons) and neuronal diversity (e.g. glutamatergic and GABAergic neurons). For these reasons, profiling techniques directly targeting the brain tissue would be preferential. Although ex vivo metabolic profiling of intact brain tissue (magic-angle-spinning NMR) or tissue extracts is a powerful tool for animal model studies,20 sample collection using brain biopsy is totally unsuitable for any type of epidemiological screening in the general population. Considering the pros and cons of previously mentioned methodologies, localized in vivo 1H magnetic resonance spectroscopy (MRS) might be the method of choice for metabolic profiling and phenotyping of CNS diseases.
Does 1H-MRS meet the criteria for epidemiology screening?
In epidemiology, prerequisites for metabolomics methodologies are high cost-effectiveness via high-throughput and absolute quantification of molecular measures.2 Does the current status of neurochemical profiling by in vivo 1H-MRS meet the criteria for molecular epidemiological studies? The answer is NO. However, 1H-MRS may have the potential to serve as a reliable method for neurochemical profiling in clinical diagnostics as well as for metabolic phenotyping in epidemiologic studies. Our optimism is based on our own experience with 1H-MRS and on literature that has demonstrated the feasibility of advanced, non-invasive neurochemical profiling in animals and humans.21–27 In this review, we will first discuss the methodological requirements for MRS data acquisition, MRS data processing and metabolite quantification that are essential for reliable non-invasive neurochemical profiling. Then we will demonstrate the power of 1H-MRS-based neurochemical profiling by showing some examples of its application in neuroscience. Finally, we will present our vision and propose the necessary steps to establish 1H-MRS as a method suitable for large-scale neurochemical profiling in epidemiological research.
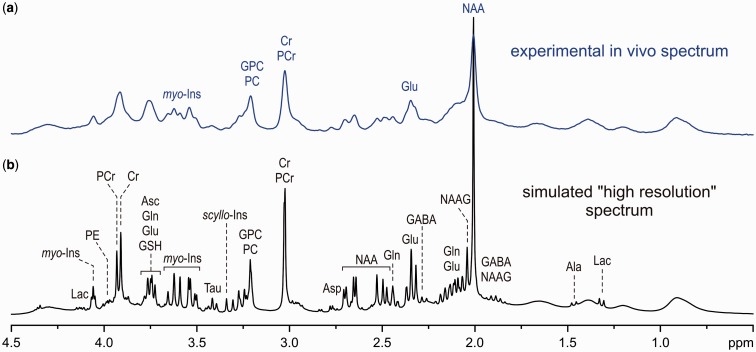
NMR-based analysis of biofluids clearly demonstrates that the entire metabolic profile, rather than individual metabolites, is more powerful for capturing (patho)physiological processes.2 Thus, meaningful metabolic phenotyping requires a comprehensive quantification of metabolites. Unfortunately, in vivo 1H-MRS faces several challenges that limit the range of detectable metabolites. One of the key limiting factors is the spectral resolution, which is substantially reduced under in vivo conditions relative to liquid 1H NMR spectroscopy. This can be seen in Figure 1, where a theoretical human brain spectrum, simulated with a resolution typical for 1H NMR spectroscopy of biofluids, is compared with the spectrum experimentally achievable at 7 T. At this reduced spectral resolution, accurate and precise quantification of metabolites is complicated by the strong overlap of their individual spectra. The spectral resolution is not the only challenge for non-invasive metabolite quantification by 1H-MRS. In addition, the detection sensitivity of 1H-MRS is much lower than the sensitivity typical for 1H NMR spectroscopy of biofluids. Furthermore, the volume selection, which is essential for localized 1H-MRS, may affect the overall spectral pattern and degrade the spectral quality. Last but not least, metabolite spectra overlap with broad signals of fast-relaxing macromolecules (proteins), which are not possible to eliminate by techniques commonly used for plasma 1H NMR spectroscopy28 due to the short T2 relaxation times of metabolites and limited RF radio frequency (RF) power for a multi-pulse refocusing train. Consequently, any deterioration in spectral quality, such as increased line width, decreased signal-to-noise ratio (SNR), baseline distortions or contamination by unwanted coherences, has a major impact on the reliability of metabolite quantification. Because of these challenges, the number of metabolites that can currently be non-invasively quantified in the human brain using 1H-MRS routinely available on clinical MRI scanners is limited to three or four: N-acetylaspartate (NAA), total creatine (tCr), total choline (tCho) and occasionally lactate (Lac). It would be rather naive to expect that concentration changes in just these few brain metabolites would be capable of probing the complex network of neurochemical processes. Therefore, improvements in 1H-MRS methodology that expand the range of reliably quantifiable metabolites are critical for making this method suitable for metabolic profiling. In the following sections, we will describe basic concepts of advanced 1H-MRS methodology (data acquisition, data preprocessing and metabolite quantification) that are critical for extending the range of detectable metabolites and for improving the precision and accuracy of their quantification. For those interested in more technical details of 1H-MRS data acquisition and processing, additional text and figures can be found in the Supplementary data, available at IJE online.
Figure 1.
Comparison of an experimental in vivo 1H MR spectrum of the brain achievable at 7 T (a) to a simulated ‘high-resolution’ 1H NMR spectrum (b).
Methodology of advanced 1H-MRS
Similarly to 1H NMR spectroscopy of biofluids, high magnetic field strength is also advantageous for in vivo 1H-MRS because of increased detection sensitivity and chemical shift dispersion at high fields. However, moving to high magnetic fields is not as straightforward for in vivo applications because wide-bore magnets for human body MRI/MRS are much more complex and costly than the narrow-bore magnets used for 1H NMR spectroscopy of biofluids. In addition, adjustment of the B0 field homogeneity (B0 shimming) and the localization performance of MRS techniques are much more challenging at high fields. Accordingly, localized in vivo 1H MRS of the brain can only benefit from high magnetic field when these challenges are successfully resolved. Consequently, in vivo 1H-MRS of the human brain is typically limited to 7 T for research applications and only to 3 T in clinical practice. It should be emphasized that the term ‘ultra-high field’ (UHF) is used differently in analytical chemistry and MRI. This term is typically reserved for magnetic fields B0 > 20 T in NMR spectroscopy of liquids, whereas it is commonly used for human body MRI/MRS at 7 T. Even 3T MRI scanners are considered high-field systems in clinical practice.
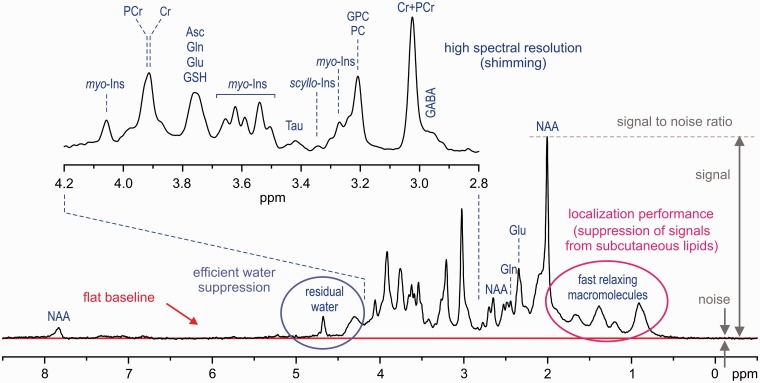
The first and utmost requirement for in vivo neurochemical profiling by 1H-MRS is high quality of the acquired spectra (Figure 2). Spectral resolution is the key factor of the spectral quality and directly affects the reliability of metabolite quantification. A resolution typical for 1H NMR spectroscopy of biofluids is not achievable by 1H-MRS due to the line broadening caused by microscopic B0 field heterogeneity on a cellular and microvascular level. Therefore, additional line broadening caused by macroscopic B0 inhomogeneity within the selected volume of interest (VOI) is not acceptable and has to be eliminated by B0 shimming. Successful B0 shimming requires an efficient B0 mapping technique, such as FASTMAP29,30 and a powerful B0 shimming system (shim coils and drivers).
Figure 2.
Characteristic factors for in vivo 1H-MR spectra quality assessment (STEAM, B0 = 7 T, TE = 6 ms, TR = 5 s, VOI = 8 ml, number of transients (NT) = 160, grey-matter-rich occipital cortex).
Volume selection is a fundamental feature of in vivo 1H-MRS data-acquisition techniques, which makes it significantly different from high-resolution 1H NMR spectroscopy of liquids. In theory, the 1H-MR spectrum exclusively originates from the selected VOI without any contamination from the surrounding tissue. These unwanted coherences, especially from subcutaneous lipids (1–2 ppm chemical shift range), can significantly degrade the spectral quality (Figure 2). In addition, volume selection at high fields is challenged by the chemical shift displacement error (CSDE), i.e. spatial displacement of volumes for off-resonance signals from the prescribed VOI (see more detailed explanation in the Supplementary data, available at IJE online). Last, but not least, high spectral quality requires highly efficient water suppression (Figure 2) because the residual water signal may cause major distortions of spectrum baseline and overlap with surrounding metabolite signals. Different localization strategies have been successfully utilized in pulse sequences developed for ultra-high magnetic fields23,25,31–34 in order to improve the localization performance and, consequently, to provide spectra of the best quality. Each of these techniques has its pros and cons, but it appears that the semi-LASER localization sequence33 combined with VAPOR water suppression25,34 is currently the best compromise for operational feasibility and performance. Despite excellent B0 shimming and high-performance localization, the final spectral quality can be severely compromised by frequency and phase fluctuations induced by physiological motion (respiratory and cardiac cycles) during data collection. For these reasons, a single scan averaging mode (where each individual scan is stored separately) is highly advantageous for in vivo applications because it allows for frequency and phase correction of individual scans and elimination of uncorrectable scans before summation. This approach helps to maintain the best achievable spectral quality (Figure 2).
Metabolite quantification
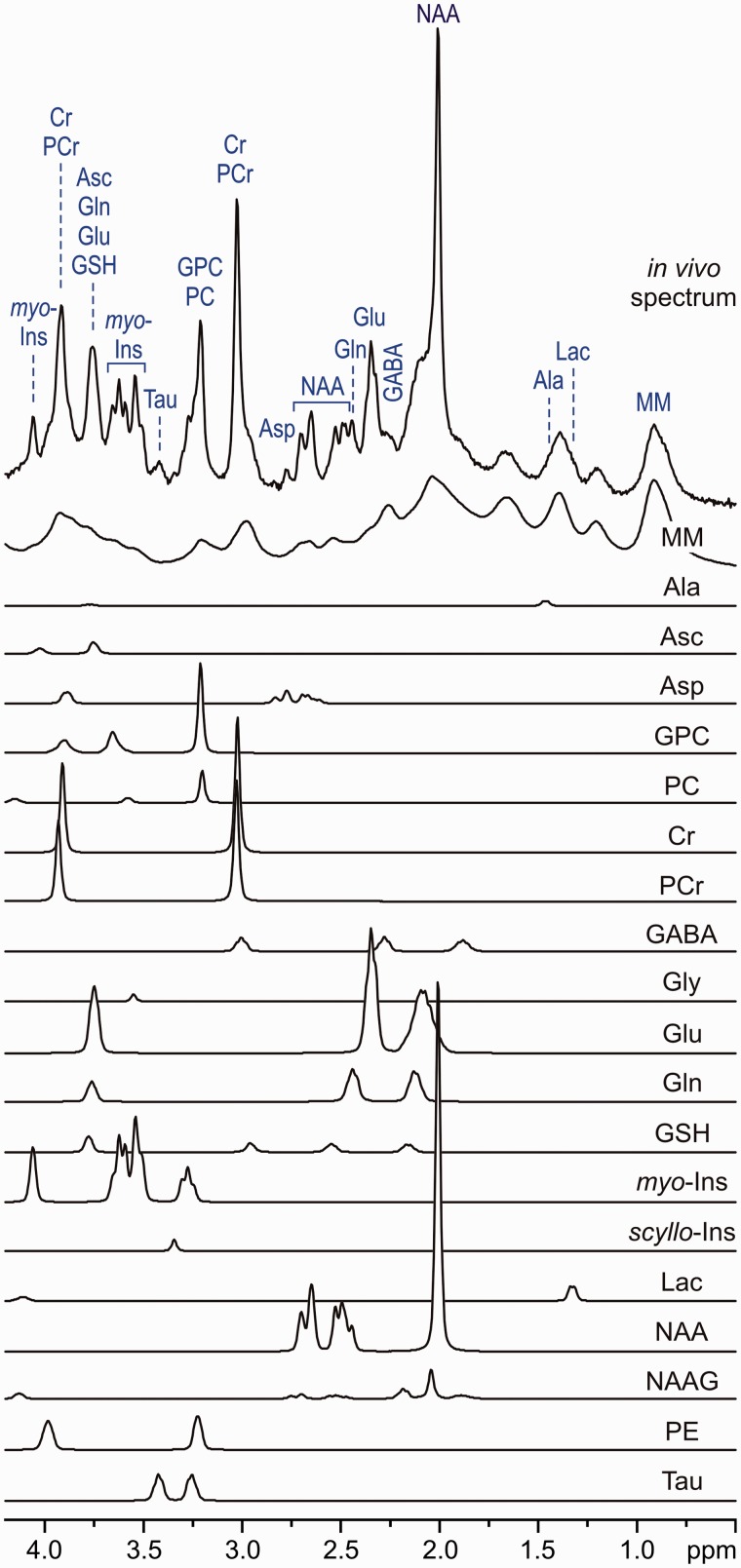
The natural (intrinsic) linewidth of in vivo 1H-MR spectra increases with the magnetic field strength due to shortened metabolite T2 relaxation times and microscopic B0 field inhomogeneity.35,36 Consequently, the overlap of metabolite spectra cannot be eliminated even at UHFs, despite increased chemical shift dispersion. Moreover, metabolite spectra overlap with underlying broad signals of fast-relaxing macromolecules, which makes the quantification even more challenging. Fortunately, the range of metabolites that contribute dominantly to brain spectra is typically known. Except in some rare metabolic diseases, the concentrations of only about 20–25 metabolites reach the current 1H-MRS detection threshold of ∼0.4 µmol/g. The primary goal of 1H-MRS is precise and accurate quantification of this well-defined set of metabolites detectable in the brain. The 1H MR spectra of these metabolites, which are used as a prior knowledge, are essential for a meaningful quantification of in vivo 1H-MRS data. Although metabolite 1H MR spectra can be measured experimentally (T = 37 °C, pH = 7.2), the simulation of these spectra using published chemical shifts and J-couplings of brain metabolites37 is recently preferred. The commonly used fitting programmes for 1H-MRS data analysis work in either the time domain (jMRUI38,39) or the frequency domain (LCModel40). Figure 3 shows the basic principle of the LCModel analysis of a 1H MR spectrum acquired from the human brain at 7 T. The experimental in vivo spectrum is modelled as a linear combination of metabolite spectra from the LCModel basis set. This basis set also includes the spectrum of fast-relaxing macromolecules, which was measured by the metabolite nulling technique using an inversion-recovery experiment.24–26 The inclusion of the macromolecule spectrum into the basis set significantly improves the robustness of metabolite quantification, especially for metabolites that are present at low concentrations and whose spectra overlap with other strong signals, such as ascorbate (Asc), γ-aminobutyric acid (GABA) or glutathione (GSH). Quantification errors are estimated by Cramér-Rao lower bounds (CRLB).
Figure 3.
LCModel analysis of an in vivo 1H MR spectrum acquired from the human brain at 7 T (STEAM, TE = 6 ms, TR = 5 s, NT = 160, grey-matter-rich occipital cortex). In vivo spectrum can be modelled as a linear combination of brain metabolite spectra from the LCModel basis set: macromolecules (MM), alanine (Ala), ascorbate (Asc), glycerophosphocholine (GPC), phosphocholine (PC), creatine (Cr), phosphocreatine (PCr), γ-aminobutyric acid (GABA), glycine (Gly), glutamate (Glu), glutamine (Gln), glutathione (GSH), myo-inositol (myo-Ins), scyllo-inositol (scyllo-Ins), lactate (Lac), N-acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphoethanolamine (PE), taurine (Tau). Modified with permission from Tkac et al.25 © 2005 Springer.
In vivo metabolite quantification requires appropriate referencing. The signal of total creatine has been widely used as an internal reference. However, this approach does not fulfil the criteria for ‘absolute’ quantification, because the tCr content differs between brain regions22 and between grey and white matter41, and may change due to neurodegenerative processes.42 Using unsuppressed water signal as a reference appears to be the best available approximation for metabolite quantification because it is robust and only requires the knowledge of the brain tissue water content.
Finally, T1 and T2 relaxation effects have to be taken into account for ‘absolute’ metabolite quantification. Correcting for relaxation effects requires knowledge of T1 and T2, but it is rather difficult to measure them precisely.43,44 In addition, these values vary across different brain regions and with age.43,44 Contrary to metabolite T1 relaxation times, which are nearly independent of the field strength, T2 relaxation times become significantly shorter with increasing B0 field.35 This shortening of T2 values leads to a major loss in signal intensity if long echo-times (TEs) are used at high fields. Moreover, signals of metabolites with spin–spin coupled protons (e.g. glutamate, glutamine, GABA) attenuate at longer TEs because of destructive interferences caused by the J-evolution. Therefore, neurochemical profiling strictly requires localization sequences with short TEs because the substantial part of the neurochemical information available at short TEs is irreversibly lost at long TEs. The ‘absolute’ metabolite quantification can be substantially simplified by acquiring data using an ultra-short TE localization sequence with a long repetition time (TR), when relaxation effects become negligible and can be ignored. The STEAM sequence is the method of choice for ultra-short TE localization, as it allows a reduction of the TE to 6 ms.25,26,36
Advanced 1H-MRS neurochemical profiling at 7 T
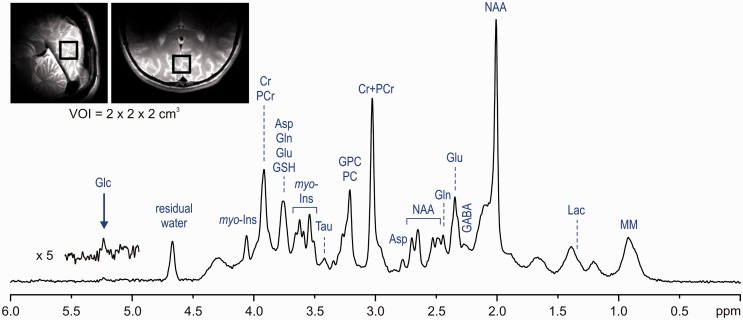
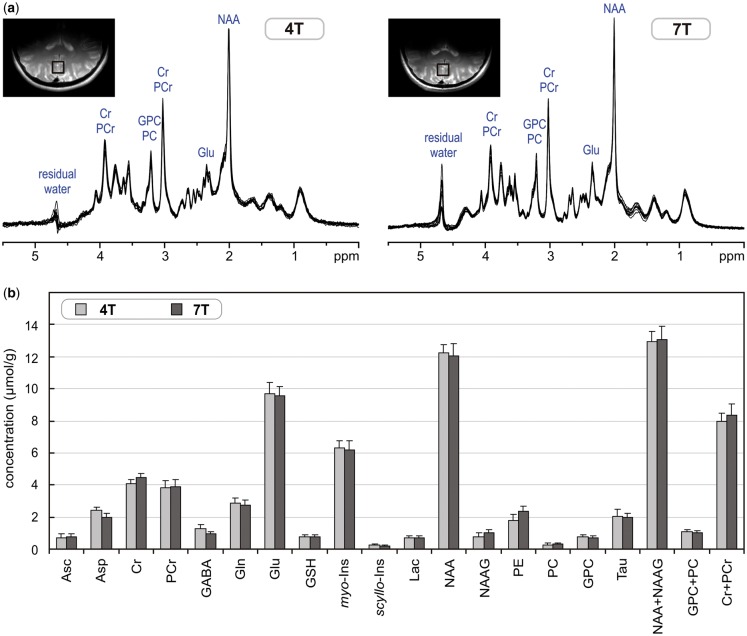
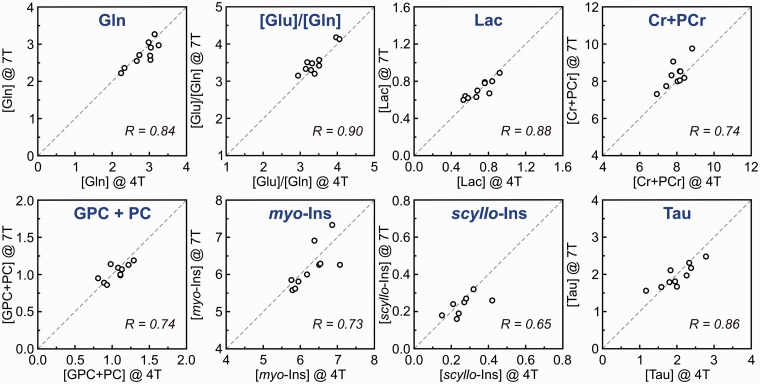
The following paragraph demonstrates the potential and limitations of non-invasive neurochemical profiling using the most advanced methodologies currently available for 1H-MRS at ultra-high magnetic fields. An ultra-short echo-time STEAM sequence with interleaved OVS and VAPOR water suppression25,36 has an excellent localization performance and, combined with FASTMAP B0 shimming, can provide high-quality in vivo 1H MR spectra from the human brain at 7 T (Figure 4). This type of artefact-free spectra with the best available spectral resolution is the critical requirement for reliable neurochemical profiling. This method was used in a study comparing neurochemical profiling at 4 T and 7 T.26 The results of this study clearly demonstrate that spectra of high quality can be routinely achieved (Figure 5a). The neurochemical profiles from both 4 T and 7 T, quantified from the grey-matter-rich occipital lobe of the same group of subjects, were in very good agreement (Figure 5b). Surprisingly, standard deviations (SDs) of quantified metabolite concentrations were not lower at 7 T than at 4 T. The similarity of SDs indicated that the precision in metabolite quantification achieved at both field strengths was already better than the variation in metabolite levels between subjects. This ability of 1H-MRS to map the actual inter-subject variability in metabolite concentration in a specific brain region was demonstrated by a strong correlation between metabolite levels of individual subjects determined at 4 T and 7 T (Figure 6).
Figure 4.
In vivo 1H-MR spectrum acquired from the human brain at 7 T (STEAM, TE = 6 ms, TR = 5 s, VOI = 8 ml, NT = 160). Modified with permission from Tkac et al.25 © 2005 Springer.
Figure 5.
(a) Superposition of in vivo 1H-MR spectra acquired from the grey-matter-rich occipital cortex of 10 healthy subjects at 4 T and 7 T (STEAM, VOI = 8 ml, NT = 160, TE = 4 ms at 4 T and TE = 6 ms at 7 T). (b) Comparison of neurochemical profiles determined from these 4T and 7T spectra. Error bars indicate SD. Only concentrations of Asp, GABA, NAAG and PE were statistically different between 4 T and 7 T. Modified with permission from Tkac et al.26 © 2009 John Wiley and Sons.
Figure 6.
Correlation between metabolite concentrations and concentration ratios of individual subjects determined from 4T and 7T 1H-MR spectra. Concentrations are expressed in µmol/g. Modified with permission from Tkac et al.26 © 2009 John Wiley and Sons.
Similar results were found in a recent multi-centre study that focused on the accuracy and reproducibility of neurochemical profiles in the human brain at 7 T.27 This study was performed at four different institutions (University Duisburg-Essen, University of Minnesota, Leiden University Medical Center and University Medical Center Utrecht) using 7T MRI scanners from two different vendors (Philips or Siemens). The same seven subjects were each examined twice at each site using the same protocol: FASTMAP B0 shimming, semi-LASER localization sequence33 combined with VAPOR water suppression,25,34 LCModel metabolite quantification. 1H-MRS data were acquired from the grey-matter-rich posterior cingulate cortex and the white-matter-rich corona radiata. Concentrations of 10 metabolites were consistently quantified with CRLB < 20%. Neurochemical profiles measured at different sites equipped with MRI scanners from two different vendors were in reasonably good agreement. The test–retest measurements showed a high level of reproducibility and, for several metabolites, such as glutamate (Glu), tCr, tCho and myo-inositol, the primary source of variance was found between subjects. This study clearly demonstrates that harmonization of data acquisition and post-processing of 1H-MRS can produce very similar results at four different 7T MRI sites.
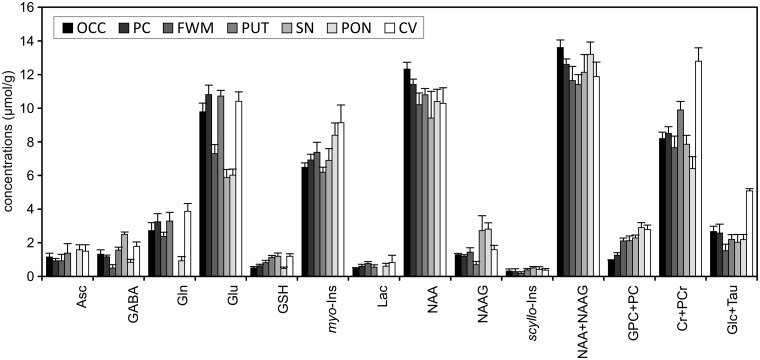
1H-MRS can measure regional differences between neurochemical profiles in the human brain.22 The neurochemical profiles measured from the brain regions that are of interest for various neurological disorders (frontal white matter, posterior cingulate, putamen, substantia nigra, pons and cerebellar vermis) show noticeable differences (Figure 7). Because of these regional differences in neurochemical profiles, highly consistent and reproducible VOI placement is extremely important in order to minimize variations in quantified metabolite levels. In addition, Figure 7 clearly demonstrates that using tCr as a universal internal reference for ‘absolute’ metabolite quantification is not appropriate.
Figure 7.
Comparison of neurochemical profiles quantified from different regions in the human brain at 7 T. STEAM, TE = 8, TR = 5 s, N = 5. Error bars indicate SD. Significant regional differences were found for all metabolites except Lac and scyllo-Ins (one‐way analysis of variance with Tukey post hoc, p < 0.005). Brain regions: occipital cortex (OCC), posterior cingulate (PC), frontal white matter (FWM), putamen (PUT), substantia nigra (SN), pons (PON), cerebellar vermis (CV). Modified with permission from Emir et al.22 © 2012 John Wiley and Sons.
The current capabilities of advanced 1H-MRS at 7 T can be nicely demonstrated by studies that aimed to measure neurochemical changes in the activated human brain.45–49 These studies clearly illustrate that the advanced technology and methodology of 1H-MRS currently available has the power to detect metabolite concentration changes as low as 0.2 µmol/g. Visual stimulation studies45–48 show very consistent results of small, but significant, changes in four metabolites (Lac, Glu, glucose, aspartate) that are directly linked to energy metabolism. Techniques used in these studies provide neurochemical profiles with up to 19 brain metabolites and enable quantification of their changes during stimulation paradigm with precision better than 0.1 µmol/g.45,46,48
Feasibility of 1H-MRS neurochemical profiling at 3 T
Although 1.5 T is the most common field strength for clinical MRI (approximately 15 000 units already installed and more than 50% of 2500 new units sold worldwide), the sensitivity and spectral resolution at this field are not sufficient for a meaningful neurochemical profiling (i.e. reliable quantification of metabolites beyond NAA, tCr and tCh). An ultra-high magnetic field is better-suited for non-invasive neurochemical profiling, but widespread availability of 7T MRI scanners for large-scale cohort studies is not realistic in the near future. For that reason, 3T MRI scanners, which are increasingly available on clinical sites, seem to be a feasible platform for neurochemical profiling. Although slow implementation of newly developed 1H-MRS techniques on clinical MRI scanners has created a widening gap between what is feasible at specialized MR research centres and what is currently available for routine clinical practice, advanced 1H-MRS methodology developed for 7 T can be implemented on 3T MRI scanners without any requirement on hardware upgrades. It was already successfully demonstrated that simply upgrading the pulse sequence (software), which consisted of FASTMAP shimming29,30 and semi-LASER localization33 with VAPOR water suppression,25,34 significantly improved the 1H-MRS data quality and increased the range of reliably quantified metabolites from the original 3–5 (using vendor-provided sequences) up to 13.21 In addition, the same methodology was implemented at two different institutions (the University of Minnesota, in Minneapolis, and the Institut du Cerveau et de la Moelle, in Paris) and the measured neurochemical profiles (vermis, ponds) were nearly identical between these sites. Moreover, this approach showed the feasibility to detect inter-individual differences in the healthy brain.21
A similar approach was used to investigate the feasibility and reproducibility of neurochemical profiling in the human hippocampus at 3 T.43,50 The hippocampus is a brain region of high clinical interest because its dysfunction is associated with several neurological and neuropsychiatric disorders such as Alzheimer’s disease, epilepsy, schizophrenia and depression. However, 1H-MRS of the hippocampus is challenging due to its size and shape and the severe B0 inhomogeneity in that brain region. Using a semi-LASER localization sequence and FASTMAP shimming routinely provided 1H-MR spectra of high quality from a relatively small VOI of ∼4 ml.50 Achieved spectral quality allowed reliable quantification of up to 12 brain metabolites. This study clearly demonstrates that neurochemical profiling of the hippocampus is feasible at 3 T despite technical challenges. For most metabolites, the between-session variation was lower than between-subject variation, indicating that the method has the sensitivity to detect inter-individual differences in the healthy brain. Our preliminary 1H-MRS results from the occipital cortex demonstrate that it is even possible to increase the number of quantifiable metabolites at 3 T close to the range achievable at 7 T, but it requires exceptional spectral quality (see Supplementary Figures 1 and 2, available as Supplementary data at IJE online).
Applications of 1H-MRS for CNS disorders
In vivo 1H-MRS has been widely applied to investigate CNS disorders. A PubMed search for keywords ‘MR spectroscopy human CNS disorders’ retrieved over 6500 original papers and 1500 reviews. These MRS studies report valuable observations about a variety of CNS disorders, but the range of detectable metabolites is almost completely limited only to NAA, tCr and tCho. Some recent publications of interest include reviews on Alzheimer disease and mild cognitive impairments,51,52 common dementias,53 psychiatric disorders,54 autism spectrum disorders55 and alcohol abuse disorders.56 Recently, the MRS Consensus Group published useful recommendations for clinical 1H-MRS in CNS disorders.57 However, there are few published studies utilizing the true potential of 1H-MRS-based neurochemical profiling to investigate metabolic changes associated with the brain disorders. For example, in vivo neurochemical profiling was used to study biochemical changes in patients with spinocerebellar ataxias,58–61 type 1 diabetes62 and adrenoleukodystrophy.63
Potential of in vivo neurochemical profiling in epidemiology
As stated in the previous section of this paper, non-invasive neurochemical profiling is, in principle, feasible on 3T MRI scanners that are commonly available in hospitals. However, successful neurochemical profiling by 1H-MRS requires the implementation of efficient B0 shimming methods (e.g. FASTMAP) and high-performance localization techniques (e.g. semi-LASER, VAPOR water suppression). In addition, preprocessing software and appropriate metabolite quantification technique (e.g. LCModel), with a database of well-constructed prior knowledge, are also required. The main problem for wider use of 1H-MRS for screening purposes is the duration of the scanning session and consequently the cost of this examination. The duration of a typical 1H-MRS scanning session, including MRI for positioning of the VOI, might be as long as 60 min, which is far too long for screening. Using a fully automated approach, it should be possible to reduce the time period necessary for parameter optimization (B0 shimming, power adjustment, water suppression) from about 10 min to 2 min or less. With 5 min of 1H-MRS data acquisition and an additional 3 min for structural MRI, reduction of the scanning time below 10 min seems to be feasible. Such an approach would significantly increase the throughput of the method and thereby reduce the cost of examination. Although higher throughput for 1H-MRS-based screening is desirable, any improvements in the throughput cannot come at the expense of the overall spectral quality.
As neurochemical profiles show significant regional differences (Figure 7), placement of the VOI plays an important role in data reproducibility. For this reason, automatic VOI positioning methods have been developed for 1H-MRS.64 These regional differences in metabolite concentrations further complicate 1H-MRS by making it challenging to choose the most appropriate brain region for neurochemical profiling. From this viewpoint, comprehensive neurochemical profiling of multiple brain regions would be preferential but would substantially decrease the throughput. Thus, it appears that MR spectroscopic imaging (MRSI), a non-invasive technique that provides information about the spatial distribution of metabolites, might be the method of choice for a potential non-invasive neurochemical profiling. However, even for the most advanced MRSI techniques currently available,65,66 the range of quantifiable metabolites and the precision of their quantification are behind the capabilities of single voxel MRS techniques.
Finally, recently published 1H-MRS trials clearly demonstrated that it is feasible to obtain highly reproducible data at different sites (in the USA and Europe) using MRI scanners from different vendors.27 This was possible by implementing the same MRS methodology including data acquisition and processing (FASTMAP shimming, semi-LASER localization sequence, VAPOR water suppression, LCModel processing). There is an ongoing multi-centre effort (NIH grant: Partnership for MRS biomarker development, PI: Gülin Öz, University of Minnesota) that seeks to simplify the operator input necessary to acquire MRS data, to make it robust and user-friendly and to make it available for all major clinical MR platforms. This effort will enhance the possibility to make high-quality neurochemical profiling widely available on 3T clinical MRI scanners, which will increase the probability for successful in vivo large-scale cohort screening in the near future.
In conclusion, in vivo neurochemical profiling in the human brain is feasible, but the widespread use requires the implementation of the most advanced methodology of 1H-MRS, including B0 shimming, localization pulse sequences, data processing and metabolite quantification. In addition, these techniques have to be broadly available on 3T MRI scanners from major MR systems vendors (Siemens, Philips and GE). Easy access to in vivo neurochemical profiling for the general population is probably not feasible in the near future, but this screening technique should be made available as soon as possible for disease prevention, at least for at-risk sub-populations.
Supplementary Data
Supplementary data are available at IJE online.
Acknowledgements
This work was supported by grants of the National Institutes of Health to the Center for Magnetic Resonance Research (P41 EB015894 and P30 NS076408).
Conflict of interest: None declared.
References
- 1.Bictash M, Ebbels TM, Chan Q. et al. Opening up the ‘Black Box’: metabolic phenotyping and metabolome-wide association studies in epidemiology. J Clin Epidemiol 2010;63:970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soininen P, Kangas AJ, Wurtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circulation Cardiovascular Genetics 2015;8:192–206. [DOI] [PubMed] [Google Scholar]
- 3.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell 2008;134:714–17. [DOI] [PubMed] [Google Scholar]
- 4.Kinross J, Li JV, Muirhead LJ, Nicholson J. Nutritional modulation of the metabonome: applications of metabolic phenotyping in translational nutritional research. Current Opinion in Gastroenterology 2014;30:196–207. [DOI] [PubMed] [Google Scholar]
- 5.Kujala UM, Makinen VP, Heinonen I. et al. Long-term leisure-time physical activity and serum metabolome. Circulation 2013;127:340–8. [DOI] [PubMed] [Google Scholar]
- 6.Faber JH, Malmodin D, Toft H. et al. Metabonomics in diabetes research. Journal of Diabetes Science and Technology 2007;1:549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makinen VP, Kangas AJ, Soininen P, Wurtz P, Groop PH, Ala-Korpela M. Metabolic phenotyping of diabetic nephropathy. Clin Pharmacol Ther 2013;94:566–9. [DOI] [PubMed] [Google Scholar]
- 8.Louis E, Adriaensens P, Guedens W. et al. Metabolic phenotyping of human blood plasma: a powerful tool to discriminate between cancer types? Ann Oncol 2016;27:178–84. [DOI] [PubMed] [Google Scholar]
- 9.Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics: Official Journal of the Metabolomic Society 2013;9:280–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett JR. Pharmacometabonomics in humans: a new tool for personalized medicine. Pharmacogenomics 2015;16:737–54. [DOI] [PubMed] [Google Scholar]
- 11.Lindon JC, Nicholson JK. The emergent role of metabolic phenotyping in dynamic patient stratification. Expert Opinion on Drug Metabolism & Toxicology 2014;10:915–19. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Molecular Systems Biology 2006;2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumas ME, Davidovic L. Metabolic profiling and phenotyping of central nervous system diseases: metabolites bring insights into brain dysfunctions. Journal of Neuroimmune Pharmacology: The Official Journal of the Society on NeuroImmune Pharmacology 2015;10:402–24. [DOI] [PubMed] [Google Scholar]
- 14.Jove M, Portero-Otin M, Naudi A, Ferrer I, Pamplona R. Metabolomics of human brain aging and age-related neurodegenerative diseases. Journal of Neuropathology and Experimental Neurology 2014;73:640–57. [DOI] [PubMed] [Google Scholar]
- 15.Tukiainen T, Tynkkynen T, Makinen VP. et al. A multi-metabolite analysis of serum by 1H NMR spectroscopy: early systemic signs of Alzheimer’s disease. Biochem Biophys Res Commun 2008;375:356–61. [DOI] [PubMed] [Google Scholar]
- 16.An M, Gao Y. Urinary biomarkers of brain diseases. Genomics, Proteomics & Bioinformatics 2015;13:345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czech C, Berndt P, Busch K. et al. Metabolite profiling of Alzheimer’s disease cerebrospinal fluid. PloS One 2012;7:e31501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohman A, Forsgren L. NMR metabonomics of cerebrospinal fluid distinguishes between Parkinson’s disease and controls. Neurosci Lett 2015;594:36–9. [DOI] [PubMed] [Google Scholar]
- 19.Trushina E, Mielke MM. Recent advances in the application of metabolomics to Alzheimer’s Disease. Biochim Biophys Acta 2014;1842:1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang TM, Woodman B, McLoughlin GA. et al. Metabolic characterization of the R6/2 transgenic mouse model of Huntington’s disease by high-resolution MAS 1H NMR spectroscopy. J Proteome Res 2006;5:483–92. [DOI] [PubMed] [Google Scholar]
- 21.Deelchand DK, Adanyeguh IM, Emir UE. et al. Two-site reproducibility of cerebellar and brainstem neurochemical profiles with short-echo, single-voxel MRS at 3T. Magn Reson Med 2015;73:1718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emir UE, Auerbach EJ, Van De Moortele PF. et al. Regional neurochemical profiles in the human brain measured by (1)H MRS at 7 T using local B(1) shimming. NMR Biomed 2012;25:152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med 2009;61:1279–85. [DOI] [PubMed] [Google Scholar]
- 24.Pfeuffer J, Tkac I, Provencher SW, Gruetter R. Toward an in vivo neurochemical profile: quantification of 18 metabolites in short-echo-time (1)H NMR spectra of the rat brain. J Magn Reson 1999;141:104–20. [DOI] [PubMed] [Google Scholar]
- 25.Tkac I, Gruetter R. Methodology of H NMR spectroscopy of the human brain at very high magnetic fields. Appl Magn Reson 2005;29:139–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med 2009;62:868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Bank BL, Emir UE, Boer VO. et al. Multi-center reproducibility of neurochemical profiles in the human brain at 7 T. NMR Biomed 2015;28:306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckonert O, Keun HC, Ebbels TM. et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols 2007;2:2692–703. [DOI] [PubMed] [Google Scholar]
- 29.Gruetter R. Automatic, localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med 1993;29:804–11. [DOI] [PubMed] [Google Scholar]
- 30.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med 2000;43:319–23. [DOI] [PubMed] [Google Scholar]
- 31.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson 2001;153:155–77. [DOI] [PubMed] [Google Scholar]
- 32.Mlynarik V, Gambarota G, Frenkel H, Gruetter R. Localized short-echo-time proton MR spectroscopy with full signal-intensity acquisition. Magn Reson Med 2006;56:965–70. [DOI] [PubMed] [Google Scholar]
- 33.Oz G, Tkac I. Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem. Magn Reson Med 2011;65:901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med 1999;41:649–56. [DOI] [PubMed] [Google Scholar]
- 35.Deelchand DK, Van de Moortele PF, Adriany G. et al. In vivo 1H NMR spectroscopy of the human brain at 9.4 T: initial results. J Magn Reson 2010;206:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at 7 T. Magn Reson Med 2001;46:451–6. [DOI] [PubMed] [Google Scholar]
- 37.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed 2000;13:129–53. [DOI] [PubMed] [Google Scholar]
- 38.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med 2001;31:269–86. [DOI] [PubMed] [Google Scholar]
- 39.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed 2005;18:1–13. [DOI] [PubMed] [Google Scholar]
- 40.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med 1993;30:672–9. [DOI] [PubMed] [Google Scholar]
- 41.Adany P, Choi IY, Lee P. B0-adjusted and sensitivity-encoded spectral localization by imaging (BASE-SLIM) in the human brain in vivo. NeuroImage 2016;134:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacharoff L, Tkac I, Song Q. et al. Cortical metabolites as biomarkers in the R6/2 model of Huntington’s disease. J Cereb Blood Flow Metab 2012;32:502–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marjanska M, Auerbach EJ, Valabregue R, Van de Moortele PF, Adriany G, Garwood M. Localized 1H NMR spectroscopy in different regions of human brain in vivo at 7 T: T2 relaxation times and concentrations of cerebral metabolites. NMR Biomed 2012;25:332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xin L, Schaller B, Mlynarik V, Lu H, Gruetter R. Proton T1 relaxation times of metabolites in human occipital white and gray matter at 7 T. Magn Reson Med 2013;69:931–6. [DOI] [PubMed] [Google Scholar]
- 45.Bednarik P, Tkac I, Giove F. et al. Neurochemical and BOLD responses during neuronal activation measured in the human visual cortex at 7 Tesla. J Cereb Blood Flow Metab 2015;35:601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y, Stephenson MC, Xin L, Napolitano A, Morris PG. Investigating the metabolic changes due to visual stimulation using functional proton magnetic resonance spectroscopy at 7 T. J Cereb Blood Flow Metab 2012;32:1484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangia S, Tkac I, Gruetter R, Van de Moortele PF, Maraviglia B, Ugurbil K. Sustained neuronal activation raises oxidative metabolism to a new steady-state level: evidence from 1H NMR spectroscopy in the human visual cortex. J Cereb Blood Flow Metab 2007;27:1055–63. [DOI] [PubMed] [Google Scholar]
- 48.Schaller B, Mekle R, Xin L, Kunz N, Gruetter R. Net increase of lactate and glutamate concentration in activated human visual cortex detected with magnetic resonance spectroscopy at 7 tesla. J Neurosci Res 2013;91:1076–83. [DOI] [PubMed] [Google Scholar]
- 49.Schaller B, Xin L, O’Brien K, Magill AW, Gruetter R. Are glutamate and lactate increases ubiquitous to physiological activation? A (1)H functional MR spectroscopy study during motor activation in human brain at 7Tesla. NeuroImage 2014;93:138–45. [DOI] [PubMed] [Google Scholar]
- 50.Bednarik P, Moheet A, Deelchand DK. et al. Feasibility and reproducibility of neurochemical profile quantification in the human hippocampus at 3 T. NMR Biomed 2015;28:685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao F, Barker PB. Various MRS application tools for Alzheimer disease and mild cognitive impairment. AJNR Am J Neuroradiol 2014;35(Suppl 6):S4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang N, Song X, Bartha R. et al. Advances in high-field magnetic resonance spectroscopy in Alzheimer’s disease. Current Alzheimer Research 2014;11:367–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kantarci K. Magnetic resonance spectroscopy in common dementias. Neuroimaging Clin N Am 2013;23:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maddock RJ, Buonocore MH. MR spectroscopic studies of the brain in psychiatric disorders. Current Topics in Behavioral Neurosciences 2012;11:199–251. [DOI] [PubMed] [Google Scholar]
- 55.Baruth JM, Wall CA, Patterson MC, Port JD. Proton magnetic resonance spectroscopy as a probe into the pathophysiology of autism spectrum disorders (ASD): a review. Autism Research: Official Journal of the International Society for Autism Research 2013;6:119–33. [DOI] [PubMed] [Google Scholar]
- 56.Meyerhoff DJ. Brain proton magnetic resonance spectroscopy of alcohol use disorders. Handbook of Clinical Neurology 2014;125:313–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oz G, Alger JR, Barker PB. et al. Clinical proton MR spectroscopy in central nervous system disorders. Radiology 2014;270:658–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adanyeguh IM, Henry PG, Nguyen TM. et al. In vivo neurometabolic profiling in patients with spinocerebellar ataxia types 1, 2, 3, and 7. Mov Disord 2015;30:662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iltis I, Hutter D, Bushara KO. et al. (1)H MR spectroscopy in Friedreich’s ataxia and ataxia with oculomotor apraxia type 2. Brain Res 2010;1358:200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oz G, Hutter D, Tkac I. et al. Neurochemical alterations in spinocerebellar ataxia type 1 and their correlations with clinical status. Mov Disord 2010;25:1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oz G, Iltis I, Hutter D, Thomas W, Bushara KO, Gomez CM. Distinct neurochemical profiles of spinocerebellar ataxias 1, 2, 6, and cerebellar multiple system atrophy. Cerebellum 2011;10:208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangia S, Kumar AF, Moheet AA. et al. Neurochemical profile of patients with type 1 diabetes measured by (1)H-MRS at 4 T. J Cereb Blood Flow Metab 2013;33:754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oz G, Tkac I, Charnas LR. et al. Assessment of adrenoleukodystrophy lesions by high field MRS in non-sedated pediatric patients. Neurology 2005;64:434–41. [DOI] [PubMed] [Google Scholar]
- 64.Dou W, Speck O, Benner T. et al. Automatic voxel positioning for MRS at 7 T. MAGMA 2015;28:259–70. [DOI] [PubMed] [Google Scholar]
- 65.Lecocq A, Le Fur Y, Maudsley AA. et al. Whole-brain quantitative mapping of metabolites using short echo three-dimensional proton MRSI. J Magn Reson Imaging 2015;42:280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Posse S, Otazo R, Dager SR, Alger J. MR spectroscopic imaging: principles and recent advances. J Magn Reson Imaging 2013;37:1301–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.