Abstract
The epigenetic control of gene expression is central to the development of the hematopoietic system and the execution of lineage-specific transcriptional programs. During the last ten years, mounting evidence implicates the family of lysine-specific histone demethylases as critical regulators of normal hematopoiesis, whereas their deregulation is found in a broad spectrum of hematopoietic malignancies. Here, we review recent findings on the role of these enzymes in normal and malignant hematopoiesis to highlight how aberrant epigenetic regulation facilitates hematopoietic cell transformation through subversion of cell fate and lineage commitment programs.
Keywords: KDM1A, KDM2B, KDM6A, Histone Demethylase, Hematopoiesis, Leukemias, Epigenetics, Histone methylation
Introduction
Although histone methylation was originally thought to be an irreversible chromatin modification, biochemical characterization of KDM1A/LSD1, a flavin-dependent amine oxidase, led to the discovery of the first known histone demethylase. KDM1A is component of the CoREST transcriptional repressor complex and exhibits H3K4me2/me1 demethylase activity (1, 2). Subsequent studies showed that KDM1A also associates with nuclear receptors where it functions as co-activator through demethylation of repressive H3K9me2 (3, 4). The KDM1A homolog KDM1B/LSD2 is also an H3K4me2 demethylase (5, 6). However, KDM1A/B cannot remove tri-methyl groups from modified lysines which suggested that alternative enzymatic activities exist. Indeed, work by several groups discovered that the Jumonji C (JmjC) domain family of Fe(II)- and α-ketoglutarate-dependent dioxygeneases function as histone demethylases in a mechanism that involves oxidative hydroxylation by radical attack of the methyl group (reviewed in (7, 8)). The human genome encodes approximately 30 JmjC domain containing histone demethylases that—based on structural homology of the JmjC domain—can be further divided into groups with activities towards H3K4, H3K9, H3K27, and H3K36 modifications. Several of these demethylases have also been shown to act on non-histone substrates (7, 8). Considering the fast pace advancements in the field of epigenetics, it is likely that additional enzymatic systems capable of demethylating histones will be described in the future. Mass-spectrometry based studies revealed that lysine-specific histone demethylases participate in multi-protein complexes to regulate gene expression, heterochromatin formation, and genome organization (7, 8). In recent years, several of these enzymes have been implicated in the epigenetic regulation of normal hematopoiesis while their deregulation has been linked to the development of leukemias. Given the reversibility of histone methylation, understanding the context dependent roles of histone demethylases will be critical for the development of novel therapies.
Histone demethylases in hematopoiesis
In murine embryogenesis, definitive hematopoiesis commences around E10.5 from the hemogenic endothelium (HE) of the aorta-gonad-mesonephros (AGM) region. The hematopoietic stem and progenitor cells (HSPCs) that arise from the AGM are the first to harbor multi-lineage reconstitution capacity (9, 10). These HSPCs subsequently migrate to the fetal liver where they expand prior to homing the bone marrow, the permanent site for hematopoiesis throughout adulthood (9, 10). In adult mice, HSPCs reside in a compartment defined by lack of lineage specific markers (Linneg) and positive for both c-Kit and Sca-1 (LinnegKS+), while long-term hematopoietic stem cells (HSCs) are further identified by the presence of CD150 and absence of CD48 and can rescue myelo-ablated mice from hematopoietic failure and establish long-term multi lineage reconstitution (11).
The first insight into the role of histone demethylation in hematopoietic development began with the finding that deletion of Kdm1a perturbs terminal differentiation of erythroid, megakaryocytic, and granulocytic cells by derepressing Gfi-1/1b lineage-specific transcriptional programs (12). Further studies revealed an indispensable role for Kdm1a, in the context of Gfi-1/1b, for the emergence of HSCs from the AGM through silencing the endothelial program within the HE both in mice and zebrafish (13, 14), suggesting an evolutionary conserved role in definitive hematopoiesis. Conditional deletion of Kdm1a in fetal (Vav1Cre) and adult (Mx1Cre) HSPCs resulted in severe pancytopenia and compromised terminal differentiation of granulocytic and erythroid lineages (15, 16). At the molecular level, Kdm1a binds enhancers of genes that regulate self-renewal and lineage commitment pathways and demethylates H3K4me1/2 as part of the CoREST repressive complex (12, 13, 15).
More recently, the JmjC domain histone H3K36 di-demethylase KDM2B/FBXL10 has been also shown to play an important role in definitive hematopoiesis (17). Kdm2b is highly expressed in the HE, and its deletion (Tie2Cre) caused embryonic lethality due to a precipitous drop in the number of hemogenic endothelial cells within the AGM. On the other hand, Vav1Cre;Kdm2bfl/fl mice were viable, but displayed a dramatic reduction in the number of long-term HSCs as well as defective lymphopoiesis which was accompanied by a concomitant upregulation of myeloid differentiation (17). A similar phenotype was also observed in Mx1Cre;Kdm2bfl/fl mice upon pIpC administration suggesting an important role for Kdm2b in the maintenance of HSPCs and regulation of lymphopoiesis, the latter in a JmjC domain dependent manner (17). Gene expression and chromatin immunoprecipitation studies coupled to next generation sequencing showed that KDM2B associates, in a mutually exclusive manner, with Trithorax (TrxG)-active and Polycomb (PcG)-repressed chromatin to regulate quiescence, cell fate, and lineage commitment. In mammalian cells, the TrxG proteins MLL1-4 reside in a complex that includes WDR5, RBBP5, ASH2L, and DPY30 which methylates histone H3K4 to activate transcription. Although it does not physically interact, KDM2B co-binds with TrxG proteins on gene promoters to enhance NOTCH1 signaling and promote T-cell commitment. On the other hand, PcG proteins reside in two complexes: the Polycomb Repressive Complex 1 (PRC1) and 2 (PRC2) that function as global transcriptional repressors through H2AK119 ubiquitination and H3K27 tri-methylation, respectively. KDM2B is an integral component of a non-canonical PRC1 and cross-talks with PRC2 to epigenetically repress transcriptional programs of myeloid differentiation. Thus, these distinct functions of KDM2B ensure the faithful execution of transcriptional programs for the initiation of lymphoid and repression of myeloid commitment (17).
Hematopoietic cell migration is critical for normal hematopoiesis (9, 10). KDM6A/UTX, a H3K27me3 demethylase encoded by the X chromosome, has been shown to play an important role in the migration of HSCs in response to SDF-1/CXCR4 signaling, although the exact molecular mechanism(s) remain elusive (18). Female Kdm6a null mice displayed hematopoietic phenotypes, such as myelodysplasia and suppressed erythro-megakaryocytopoiesis, whereas the counterpart male mice showed no phenotype. Given that UTY, encoded by the Y chromosome and exhibiting >80% similarity to KDM6A, does not demethylate H3K27me3 due to mutations in the JmjC domain (19, 20), this suggests that KDM6A regulates those responses in a demethylase–independent manner. KDM6A is an integral component of MLL2/3 H3K4 methyltransferase complexes (21, 22), and likely functions as a scaffold to facilitate the recruitment or the activity of this complex towards its substrate. Interestingly, although KDM6A demethylase activity is dispensable in definitive hematopoiesis, H3K27me3 demethylation by either KDM6A or KDM6B/JMJD3 (encoded by an autosomal gene) is crucial for the terminal steps of T-cell differentiation (23). Consistently, another study showed that T-cell specific ablation of KDM6B promoted Th2 and Th17 and inhibited Th1 and Treg differentiation under different cytokine-induced conditions (24). The latter was also dependent on active demethylation and changes in the expression of key genes, such as Tbx21,Gata3, and Foxp3. The preferential impact of KDM6A/B on late T-cell maturation may be linked to a role for these enzymes in integrating extracellular cues and cytokine signals to regulate differentiation (23, 24). Further studies by using Cre strains at different stages of hematopoietic development and generation of demethylation deficient mutants of KDM6A and KDM6B through CRISPR-mediated genome editing will be useful in determining the demethylase dependent- and independent- functions of those enzymes in hematopoiesis.
Histone demethylases in hematopoietic malignancies
Although epigenetic mechanisms are implicated in the pathogenesis of hematopoietic malignancies, little is known about the role of lysine-specific histone demethylases, and whether manipulation of these enzymes can be translated into targeted therapies. KDM6A is the most frequently mutated histone demethylase in hematopoietic malignancies as well as in a broad spectrum of solid tumors (25, 26). Loss-of-function mutations or deletion of KDM6A are prominent in NOTCH1 driven T-ALL and predominantly occur in male patients (26). Consistently, in the context of NOTCH1, leukemia kinetics ware faster in Kdm6a−/Y mice with shorter latency compared to the wild-type animals (25). Gene expression and ChIP-seq studies showed that KDM6A positively controls tumor suppressor genes, including the regulator of NOTCH1 stability FBXW7, and counteracts the activity of PRC2. The latter rendered T-ALL driven by KDM6A inactivation sensitive to pharmacologic inhibition of H3K27 methylation (26). Intriguingly, a pro-oncogenic role for KDM6A has been recently reported in TAL1-positive leukemias (27), suggesting that the impact of KDM6A inactivation in T-ALL may be context dependent. Furthermore, KDM6B, the other known H3K27 demethylase, was found to be essential for the initiation and maintenance of T-ALL (25). KDM6B is induced by the NF-kB pathway and regulates NOTCH1 targets HES1 and HEY1. Treatment of T-ALL cell lines with the KDM6B small molecule inhibitor GSKJ4 led to cell cycle arrest and increased apoptosis. Overall, these findings provide a basis for the development of personalized epigenetic therapies for T-ALL, with respect to the status of KDM6A and KDM6B, and highlight a context dependent role of those enzymes in T-cell transformation (25-27).
Another example of a histone demethylase with a context dependent role in hematopoietic transformation is KDM2B, originally cloned as a putative oncogene from an insertional mutagenesis screen in Moloney Murine Leukemia virus induced T-cell lymphomas (28, 29). KDM2B is an integral component of a non-canonical PRC1 complex and cooperates with PRC2 to regulate senescence, differentiation, and oncogenesis (17, 28, 30-33). KDM2B is upregulated in and is required for the oncogenicity of human ALL cell lines, whereas its overexpression in Sca-1+ cells suffices to induce mixed lineage leukemias with complete penetrance in mice (17, 34). On the other hand, deletion KDM2B expanded myeloid progenitors and accelerated Kras-driven AML through subversion of lineage specification pathways (17). Although KDM2B is neither mutated nor exhibits copy number changes in leukemias, its expression is downregulated in AML; similarly its interacting partners, such as BCOR and components of PRC2, are frequently inactivated and confer poor prognosis (17, 35, 36). Thus, modulation of KDM2B activity may be directed to restore differentiation in acute leukemias and improve the efficacy of current therapies.
Overexpression of KDM1A is frequently observed in leukemias and confers a poor prognosis by blocking differentiation and maintaining a “stem-like” phenotype of leukemia initiating cells (37, 38). Overexpression of the shortest isoform of Kdm1a, which preferentially demethylates histone H3K9 and is repressed in quiescent HSCs, sufficed to induce T-cell acute lymphoblastic leukemia/lymphoma in mice (39). Inhibition of the enzyme with small molecular inhibitors, as well as knockdown of the endogenous protein, resulted in increased apoptosis and impaired leukemogenicity of MLL-AF9 driven AML (37). Similar results were obtained in Acute Promyelocytic Leukemia (APL), a cytogenetically distinct subtype of AML characterized by the t(15;17)-associated PML-RARA fusion, in which all-trans-retinoic acid (ATRA) is used to differentiate leukemic blasts. Combination of KDM1A inhibition and ATRA administration showed a synergistic therapeutic effect, even in non-APL AML (38). In that context, ChIP-seq studies revealed that ablation of KDM1A increased H3K4me2 and expression of myeloid lineage specific genes, suggesting that differentiation of leukemic blasts with “epi-drugs” holds promise for the treatment of AML (38).
JMJD1C is another JmjC domain protein discovered as putative oncogene in shRNA screens in MLL-AF9, HOXA9, and AML1-ETO driven AML (40-42). Depletion of JMJD1C decreased the frequency of leukemia initiating cells by inducing their differentiation and impaired the growth and establishment of leukemia in serial transplantation experiments. Notably, although JMJD1C is important for the maintenance of the malignant phenotype, it is dispensable for leukemia initiation (41). Although originally reported to be an H3K9me2/me1 demethylase leading to transcriptional activation, subsequent studies failed to detect demethylase activity for JMJD1C (40-42). Thus, the question whether the role of JMJD1C in leukemia maintenance relies on its demethylation activity remains open. Considering that loss of JMJD1C caused minor defects in normal hematopoiesis, its inhibition with small molecule inhibitors may be beneficial in the aforementioned types of AML with minimal adverse effects.
Conclusions and future directions
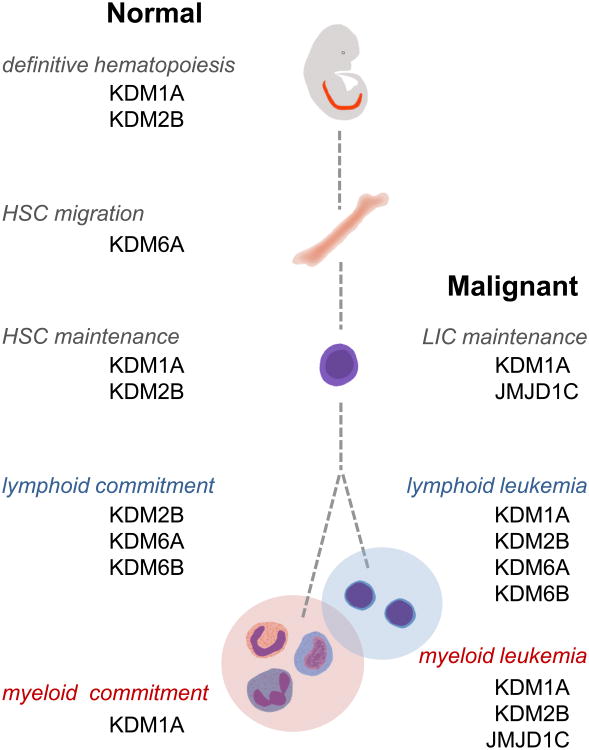
A hallmark of hematopoietic malignancies is a block in cellular differentiation which causes accumulation of progenitor cells and facilitates cell transformation in the context of additional genetic and epigenetic changes. Thus, restoring differentiation may represent an Achilles heel for successful remission and treatment of leukemias by forcing “leukemia initiating cells” to differentiate to more mature cells that can be eradicated by current treatments. As reviewed here, strong evidence indicates that lysine-specific histone demethylases are major regulators of cell fate and lineage commitment decisions and their deregulation facilitates hematopoietic transformation, primarily through maintaining leukemic cells in an undifferentiated and drug-resistant state (Figure 1). Thus, combining histone demethylase specific small molecule inhibitors with current treatments is a promising avenue to promote differentiation and eradication of blast cells. Further studies using animal models are needed to delineate the complexity of those circuitries, and to help determine the appropriate therapeutic window to alleviate adverse effect in normal hematopoiesis.
Figure 1.
Schematic of important stages of normal (left) and malignant (right) hematopoiesis, and the corresponding lysine-specific histone demethylases implicated in those processes. HSC; hematopoietic stem cell, LIC; leukemia initiating cell.
Acknowledgments
This work was supported by an Alex's Lemonade Stand Foundation Young Investigator Award, and National Cancer Institute grants (R00CA158582 and R21CA182662 to A. Tzatsos).
Footnotes
Conflict of interest disclosure: The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 5.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kampranis SC, Tsichlis PN. Histone demethylases and cancer. Adv Cancer Res. 2009;102:103–169. doi: 10.1016/S0065-230X(09)02004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 9.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 12.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 13.Thambyrajah R, Mazan M, Patel R, Moignard V, Stefanska M, Marinopoulou E, Li Y, Lancrin C, Clapes T, Moroy T, et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. 2016;18:21–32. doi: 10.1038/ncb3276. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi M, Fuse Y, Watanabe M, Andrea CS, Nakajima H, Ohashi K, Kaneko H, Kobayashi-Osaki M, Yamamoto M, Kobayashi M. LSD1/KDM1A promotes hematopoietic commitment of hemangioblasts through downregulation of Etv2. Proc Natl Acad Sci U S A. 2015;112:13922–13927. doi: 10.1073/pnas.1517326112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O'Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A, Konig K, Diehl L, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 17.Andricovich J, Kai Y, Peng W, Foudi A, Tzatsos A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J Clin Invest. 2016;126:905–920. doi: 10.1172/JCI84014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thieme S, Gyarfas T, Richter C, Ozhan G, Fu J, Alexopoulou D, Muders MH, Michalk I, Jakob C, Dahl A, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121:2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 19.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 20.Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, Schofield CJ, Johansson C. Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J Biol Chem. 2014;289:18302–18313. doi: 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 22.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manna S, Kim JK, Bauge C, Cam M, Zhao Y, Shetty J, Vacchio MS, Castro E, Tran B, Tessarollo L, et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Zou J, Wang M, Ding X, Chepelev I, Zhou X, Zhao W, Wei G, Cui J, Zhao K, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, Tremblay V, Rakopoulos P, Singh K, Huang S, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci U S A. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci U S A. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park PJ, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 32.He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, Wong KK, Bardeesy N. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda T, Nagamachi A, Takubo K, Yamasaki N, Matsui H, Kanai A, Nakata Y, Ikeda K, Konuma T, Oda H, et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood. 2015;125:3437–3446. doi: 10.1182/blood-2014-03-562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, SpanholRosseto A, Klein HU, Dugas M, Schindela S, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 36.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 37.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada T, Koyama D, Kikuchi J, Honda H, Furukawa Y. Overexpression of the shortest isoform of histone demethylase LSD1 primes hematopoietic stem cells for malignant transformation. Blood. 2015;125:3731–3746. doi: 10.1182/blood-2014-11-610907. [DOI] [PubMed] [Google Scholar]
- 40.Sroczynska P, Cruickshank VA, Bukowski JP, Miyagi S, Bagger FO, Walfridsson J, Schuster MB, Porse B, Helin K. shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood. 2014;123:1870–1882. doi: 10.1182/blood-2013-08-522094. [DOI] [PubMed] [Google Scholar]
- 41.Zhu N, Chen M, Eng R, DeJong J, Sinha AU, Rahnamay NF, Koche R, Al-Shahrour F, Minehart JC, Chen CW, et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J Clin Invest. 2016;126:997–1011. doi: 10.1172/JCI82978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M, Zhu N, Liu X, Laurent B, Tang Z, Eng R, Shi Y, Armstrong SA, Roeder RG. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015;29:2123–2139. doi: 10.1101/gad.267278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]