Abstract
From the initial discovery of the neural crest over 150 years ago to the seminal studies of Le Douarin and colleagues in the latter part of the 20th century, understanding of the neural crest has moved from the descriptive to the experimental. Now, in the 21st century, neural crest research has migrated into the genomic age. Here we reflect upon the major advances in neural crest biology and the open questions that will continue to make research on this incredible vertebrate cell type an important subject in developmental biology for the century to come.
Keywords: neural crest, craniofacial skeleton, peripheral nervous system, embryo, vertebrates
Introduction
The neural crest is a remarkable cell type, unique to vertebrate embryos. Induction of the neural crest occurs at the neural plate border via a signaling interaction between neural and non-neural ectoderm (Selleck and Bronner-Fraser, 1995). After their specification, neural crest precursors reside within the elevating neural folds and dorsal neural tube after its closure. Neural crest cells then undergo an epithelial to mesenchymal transition (EMT) to emigrate from the neuroepithelium and become a motile cell population (Figure 1). These cells migrate collectively or individually, navigating along stereotypic pathways to numerous and often distant sites throughout the embryo, where they eventually differentiate into an incredibly diverse array of cell types. Thus, the neural crest is a transient cell type that rapidly transitions from multipotent precursors to cell types ranging from neurons and glia of the peripheral nervous system to pigment cells of the skin and cartilage and bone cells of the craniofacial skeleton.
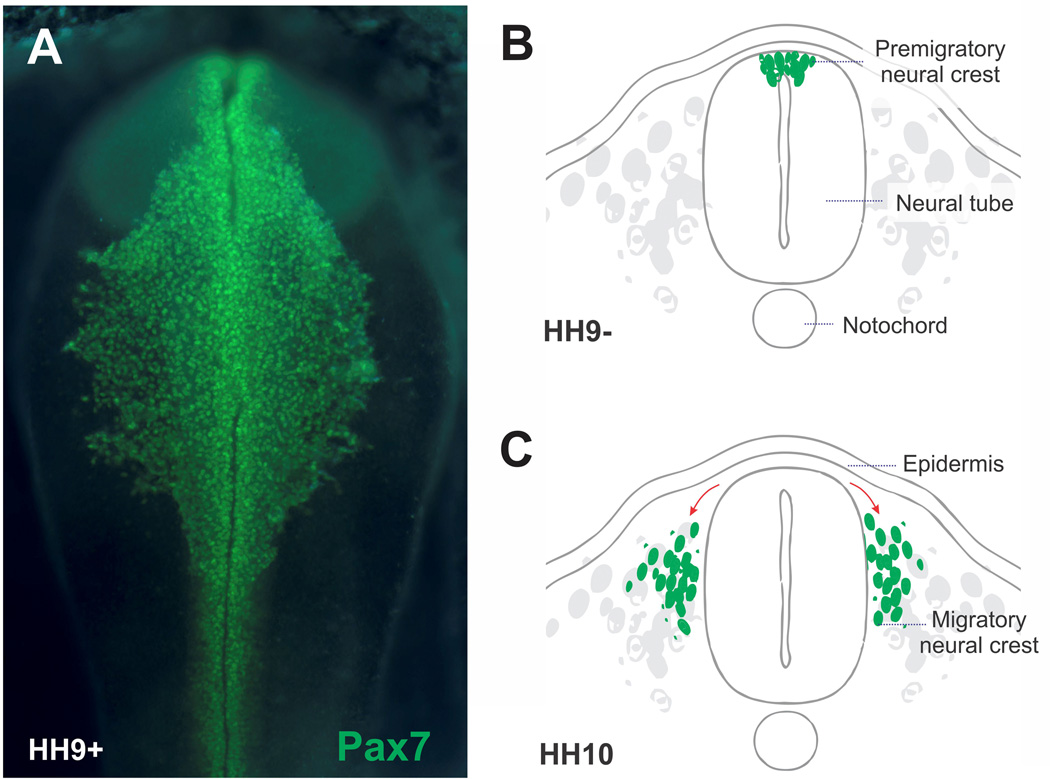
Figure 1. Neural crest cells delaminate from the neural tube and undergo extensive migration.
(A) Immunohistochemistry with an antibody for neural crest marker Pax7 highlights neural crest cells delaminating from the neural tube of a stage HH9 chicken embryo. (B) Diagram representing a cross section of a neurulating chicken embryo. At the stage represented, the neural cells occupy the dorsal portion of the neural tube. (C) Neural crest cells eventually undergo epithelial to mesenchymal transition to delaminate from the neural tube and undergo extensive migration.
Understanding neural crest development has important medical implications because of the variety of birth defects and cancers that arise from mistakes in neural crest development or mutations that effect neural crest derivatives post-natally. In humans, these ‘neurocristopathies’ or defects in neural crest development are amongst the most common birth defects in liveborn infants, including craniofacial malformations, defects in the cardiac outflow tract, familial dysautonomia and a variety of syndromes including Treacher-Collins, Waardenberg, DiGeorge and Charge syndromes (see chapter by Paul Trainor). In addition, neural crest-derived cell types seem to be particularly prone to metastasis, giving rise to melanoma, neuroblastoma, pheochromocytoma, among others.
How these cells arise, migrate and differentiate has fascinated developmental biologists for over 150 years. In this review, we reflect upon the history of neural crest research, focusing on some major discoveries over the past century and speculating upon the fascinating mysteries that are likely to occupy research on this cell type going forward.
A Brief History
In the latter half of the 19th century, the invention of the microtome by Wilhelm His together with improved histological techniques enabled visualization of embryonic tissues with previously unprecedented resolution. In 1868, His first described the neural crest as a "Zwischenstrang“ or cord between the neural and non-neural ectoderm (Figure 2A). He also recognized that neural crest cells formed spinal ganglia and for this reason named it “ganglionic crest.”
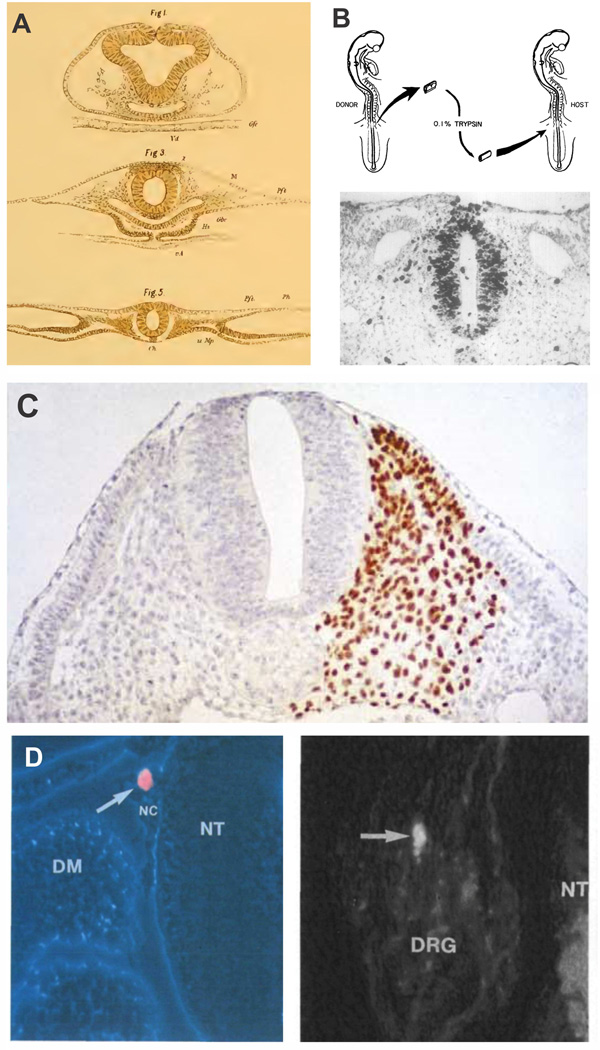
Figure 2. Classical experiments in neural crest research utilizing avian embryos.
(A) Reproduction of the original drawings of Wilhelm His, including the first depiction of neural crest cells (His, 1898). The neural crest is identified with a “Z” for "Zwischenstrang” or cord between the neural and non-neural ectoderm. (B) Transplant experiments by Weston (1963), where neural tubes from radioactively labeled embryos were transplanted to unlabeled hosts. This allowed for the tracking of neural crest cells after delamination and identification of distinct migratory pathways. (C) The chick-quail chimeras of Nicole Le Douarin. Grafting of quail tissue in chicken embryos permitted extensive fate mapping of progenitor cells (Le Douarin, 2004). This approach was used to map migratory pathways and derivatives of the neural crest at distinct axial levels of the avian embryo (Le Douarin, 1982). (D) Clonal analysis of neural crest cells in the developing embryo. Labeling of single neural crest cells with vital dyes showed that many individual progenitor cells are multipotent and differentiate into diverse cell types in vivo (Fraser and Bronner-Fraser, 1991).
In the early to mid-20th century, experimental embryology came to the forefront of research of developmental biology. During this time, “cut and paste” experiments examined the contributions of neural crest cells to various structures, largely by removing the neural crest via surgical ablation in amphibians (Horstadius, 1950), but also in birds (Yntema and Hammond, 1945) as well as by transplantation experiments to examine patterns of pigment cell migration between different species (Twitty, 1936). These experiments revealed many intriguing properties of the neural crest from their extensive migratory ability to their contributions to melanocytes and ganglia of the peripheral nervous system. For a detailed overview of these early experiments, there is a wonderful monograph written by Sven Horstadius (1950).
The 1960s ushered in a new era of experimentation with the application of novel cell marking techniques to neural crest biology. First, Weston (1963) and Chibon (1967) in avian and amphibian embryos, respectively, radioactively labeled embryos with 3H-thymidine and then transplanted neural tubes from labeled donors to unlabeled host embryos (Figure 2B). This made it possible to follow the migration of the labeled cells deep into the embryo. In amniotes, Weston found that neural crest cells in the trunk followed different pathways to contribute to ventral (dorsal root and sympathetic ganglia) or dorsolateral (melanocytes) derivatives. In addition, it was possible to perform these transplantation at different times, revealing that trunk neural crest cells contribute to their derivatives in a ventral to dorsal order, with the first cells to leave the neural tube migrating to the most ventral sites (Weston and Butler, 1966). Later, this was confirmed by experiments using the lipophilic dye, DiI, which made it possible to label the neural tube and neural crest cells without the need for grafting (Serbedzija et al., 1989). However, both radioactive and DiI labeling methods suffered from the issue of dilution of the label in the rapidly dividing neural crest population and thus were only amenable to relatively short term studies.
A major technological advance was made by Le Douarin (1973) with her discovery that quail and chick embryos developed at relatively similar rates, but could be easily and indelibly distinguished since quail cells have a condensed mass of heterochromatin in their nucleolus whereas chick cells are euchromatic. By applying the histological Feulgen stain (Le Douarin, 1974) and later using the quail-specific QCPN antibody to these quail-chick chimeras, it was possible to follow the long-term fate of grafted quail neural crest cells (Figure 2C). Thus, by transplanting neural folds or whole neural tubes from stage matched donor quails into chick hosts, Le Douarin and colleagues (Le Douarin and Teillet, 1973; 1974) could follow quail cells that originated from within the neural tube but subsequently migrated and invaded distant sites in the periphery. In this way, they were able to map the migration pathways and derivatives formed by neural crest cells at all axial levels of the bird embryo. These seminal experiments (see Chapter by Le Douarin) lead to the discovery that neural crest cells give rise to a stunning array of derivatives.
Importantly, Le Douarin and colleagues showed that the cell types formed by neural crest cells vary with the axial level from which they originate (summarized in Le Douarin, 1982). At cranial levels, neural crest cells populated cranial ganglia but also contributed to mesenchymal derivatives like connective tissue, cartilage and bones of the face (Le Lievre and Le Douarin, 1975; Couly and Le Douarin, 1987). In the neck region, the vagal neural crest contributed to parts of the cardiac outflow tract, but also invaded the foregut, and migrated along the entire length of the gut to form enteric ganglia (Le Douarin and Teillet, 1973; Burns and Le Douarin, 2001). A small contribution to the enteric nervous system was also noted for the most caudal (lumbosacral) crest population. At trunk levels, neural crest cells migrated dorsolaterally to form pigment cells and ventrally to form dorsal root, sympathetic and adrenomedullary cells in agreement with the radioactively labeling experiments of Weston (1963).
The diverse nature of derivatives along the body axis led to the question of whether there were intrinsic differences between neural crest populations at different axial levels or whether the choice of derivative was dictated by the environment. To address this, Le Douarin and colleagues performed “heterotopic” grafts in which the axial level was switched. For example, cranial neural crest cells were grafted to truncal levels and vice versa. The results revealed that there indeed were intrinsic differences between cranial and trunk neural crest cells. While cranial crest grafted to the trunk formed all normal trunk derivatives, they also differentiated into ectopic cartilage nodules. In the reciprocal experiment, trunk neural crest cells grafted to the head formed neuronal and glia derivatives in cranial sensory ganglia, but failed to form cartilage (Le Douarin, 1982). These results showed that, although there is flexibility in the neural crest program, there are also intrinsic, axial level specific differences.
While the grafting experiments were useful for examining the flexibility of neural crest cell fate at a population level, only clonal analysis of the progeny of single cells can inform on the developmental potential of individual neural crest cells. Clonal analysis in vitro was first applied to the neural crest by Alan Cohen and colleagues, who showed that clones derived from individual trunk neural crest cells in culture could indeed form multiple neural crest derivatives like neurons, pigment cells and support cells (Cohen and Konigsberg, 1975). David Anderson and colleagues went on to show that not only were migratory neural crest cells multipotent but that they also had the ability to self-renew in culture (Stemple and Anderson, 1992).
The most extensive clonal analyses of neural crest cells in culture were performed by Dupin, Le Douarin and colleagues (Baroffio et al., 1988; Dupin et al., 2010), who showed that some clones of both cranial and trunk neural crest cells can give rise to a very broad array of derivatives, showing a large degree of multipotency, whereas others formed fewer derivatives. Interestingly, they found that both cranial and trunk clones formed some mesenchymal derivatives (i.e. express markers for cartilage and bone) though environmental factors like Shh greatly increased the percentage of trunk neural crest clones with chondrogenic ability (Calloni et al., 2007). These results may suggest that the developmental potential of neural crest cells is broader after exposure to culture conditions in vitro than those observed under normal developmental conditions in vivo.
An important adjunct to in vitro experiments above is clonal analysis in the developing embryo. This was first applied to premigratory (Bronner-Fraser and Fraser, 1988) and migratory (Bronner-Fraser and Fraser, 1989) neural crest cells in avian embryos using intracellular injection of vital dye into single neural crest precursors (Figure 2D). In agreement with cell culture experiments described above, the results showed that many individual progenitor cells are multipotent, contributing to progeny in distinct anatomical structures and differentiating into diverse cell types in vivo. Recently, Sommer and colleagues (Baggiolini et al., 2015) found similar results by driving elegant “confetti” labels in conditional transgenic mouse lines that specifically label either premigratory or early migrating trunk neural crest cells. Given such similar findings in birds and mammals using different but complementary approaches, it is clear that the vast majority of both premigratory and early migrating neural crest cells in vivo are multipotent in amniotes and able to contribute to a broad range of neural crest derivatives. This is a lovely illustration of how important questions in neural crest biology (e.g. what is the developmental potential of individual neural crest cells?) remain highly relevant over decades even as the technology for lineage analysis has evolved from using dye labeling to elegant modern confetti technology. The interest of the question remains pertinent and can be examined at ever better resolution as technology advances.
Neural crest migration
Quail-chick chimeras gave a general sense of the pathways of neural crest migration by identifying derivatives and showing that trunk neural crest cells migrated ventrally to form dorsal root and sympathetic ganglia (ventral pathway) or between the ectoderm and somites (dorsolateral pathway) to form melanocytes. However, the trajectories of individual cells could not be followed with the nuclear quail marker. Therefore, there was poor understanding of the mechanisms underlying migration prior to the discovery of neural crest markers that enabled visualization of cell membranes and/or live cell imaging using vital dyes.
The first insight into the pathways followed by ventrally migrating trunk neural crest cells came from analyses using the HNK-1 antibody, which labels most migratory crest cells (Rickmann et al., 1986; Bronner-Fraser, 1987). Although it had been assumed previously that trunk neural crest cells migrated around the somites, careful analysis showed that they actually migrate in a segmental pattern through the rostral half of each somitic sclerotome, and were restricted from the caudal half. This launched a search for possible rostrally localized attractants and/or caudally localized repellants. Although it was initially thought that ephrins in the caudal sclerotome were primarily responsible for repelling trunk neural crest cells because of their ability to do so in vitro (Wang and Anderson, 1997; Krull et al., 1998), the fact that ephrin null mice lacked an obvious neural crest phenotype made this less clear (Wang and Anderson, 1998). The discovery that neural crest cells expressed the neuropilin2 receptor and that Semaphorin3F, its inhibitory ligand, was expressed in the caudal sclerotome solved the puzzle; importantly, mice null for either the receptor or the ligand lost the segmental pattern of neural crest migration in the trunk (Gammill et al., 2006). In addition to inhibitory cues, trunk neural crest cells also display chemotactic behavior in response to SDF, which is expressed in a graded manner along the ventral to dorsal axis of the trunk region (Kasemeier-Kulesa et al., 2010), suggesting that a combination of attractive and repulsive cues guide their trunk neural crest migration.
In contrast to the relatively simple patterns of neural crest migration in the trunk, cranial neural crest cells migrate along many different pathways, ultimately filling most of the frontonasal process and branchial arches. With the advent of live-imaging in chick, frog and zebrafish embryos, much of what we know about the molecular mechanisms governing neural crest cell migratory behavior has come from analyses of the cranial neural crest population.
The collective migration of the cranial neural crest cells involves a combination of mechanisms, from contact inhibition to chemoattraction and co-attraction. As neural crest cells undergo EMT, they switch the nature of their cadherins (Rogers et al., 2013; Scarpa et al., 2015) and transition from non-motile epithelial cells to migratory mesenchymal cells. This cadherin switch is essential for acquisition of contact inhibition of locomotion and cell repolarization (Scarpa et al., 2015). Contact inhibition of locomotion, originally described for fibroblast cells (Abercrombie and Heaysman, 1953), occurs when two neural crest cells collide; they stop moving at their site of contact and instead move away in the opposite direction. Using Xenopus neural crest cells, Mayor and colleagues have established an important role for contact inhibition of locomotion for cranial neural crest cells both in vitro and in vivo (Carmona-Fontaine et al., 2008). In addition, they have found that neural crest cells are attracted to each other via a ligand receptor interaction mediated by Complement3a (C3a) and the C3a receptor (Carmona-Fontaine et al., 2011) as well as by chemoattraction to the secreted factor SDF (Theveneau et al., 2010). These three systems of contact inhibition of locomotion, co-attraction and chemotaxis function cooperatively to pattern the cranial neural crest into forward moving streams in the frog embryo.
Other types of chemoattractants for neural crest cells include VEGF, which is important for cranial neural crest migration in the chick (McLennan et al., 2015) and GDNF and endothelin, which are critical for migration of vagal neural crest cells in populating the enteric nervous system (Sasseli et al., 2012). The observation that these chemoattractants sometimes uniformly surround neural crest cells, has lead to the idea that there may be a cell-induced gradient in which the ‘leader’ neural crest cells acquire directional information that in turn instructs ‘trailer’ cells to follow (McLellan et al., 2015). This is supported by the fact that leader cells have a different molecular profile that trailers (McLellan et al., 2015).
Studies that combine molecular perturbation and quantitative analysis to examine cell speed, trajectories, and perdurance of migration have provided important insights into neural crest migratory behavior (Scarpa et al., 2015; McLellan et al., 2015). In particular, the blend of imaging, molecular, and mathematical modeling approaches has greatly increased understanding of the molecular mechanisms underlying neural crest motility.
Gene regulatory network controlling neural crest formation
With the development of perturbation approaches using antisense morpholinos, shRNAs and dominant negative constructs, many individual genes have been implicated in the process of neural crest induction and migration in several vertebrates, most notably Xenopus, zebrafish and chick. Taken together, these studies have lead to the assembly of a gene regulatory network (GRN) (Meulemans and Bronner-Fraser, 2004; Simoes-Costa and Bronner, 2015) that helps explain the progression of neural crest development (Figure 3).
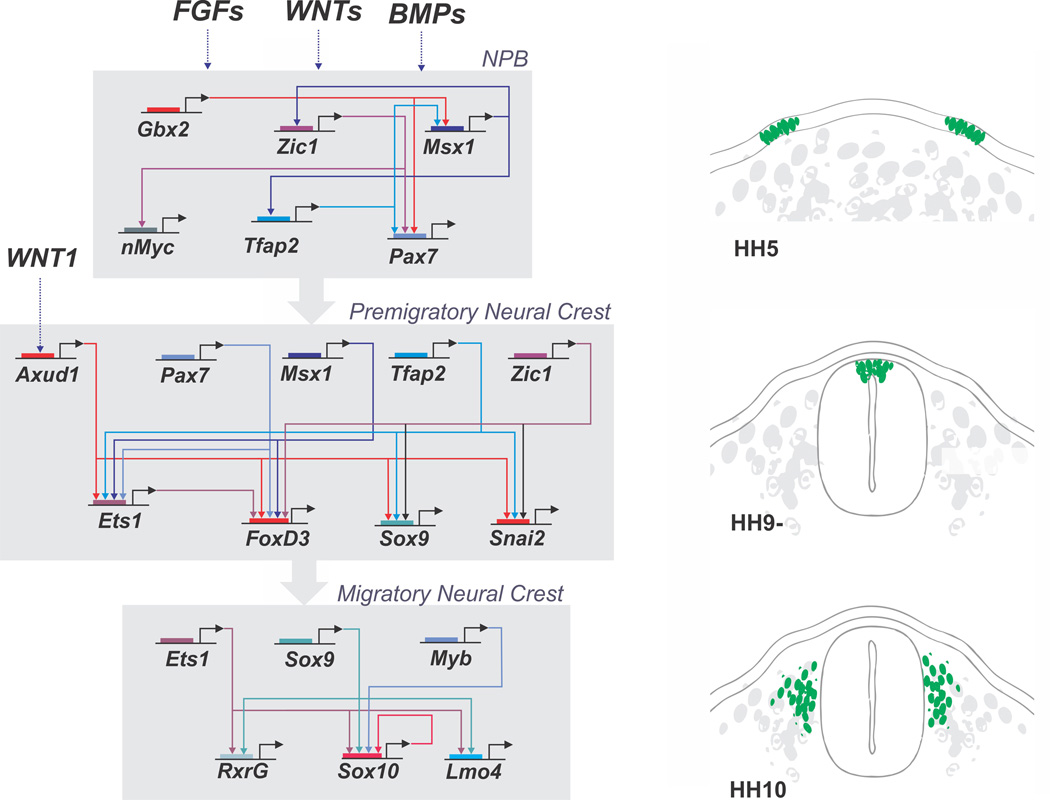
Figure 3. A gene regulatory network (GRN) controls neural crest formation.
A complex genetic program comprised of numerous transcription factors underlies neural crest development. These factors are arranged in distinct hierarchical modules corresponding to the different stages of neural crest formation. First, in the neural plate border, the interplay of signaling systems such as Wnts, Bmps and Fgfs activates the neural plate border specifier genes, which include Gbx2, Msx1, Zic1, Tfap2 and Pax7. These genes drive the transition of the GRN to the neural crest specification module. Signaling systems such as Wnts also feed into the system by activating transcription factor Axud1, which interacts with PAX7 and MSX1 to activate expression of neural crest specifier genes (FoxD3, Ets1, Sox9, etc.) in the dorsal neural folds. The neural crest specifier genes endow the neural crest with its defining features and activate the process of epithelial to mesenchymal transition. In the migratory neural crest, there is activation of additional factors that will affect migration and drive differentiation in multiple cell types.
In its current formulation, the neural crest GRN posits that inductive signals mediated by Wnts, BMPs and FGFs establish the neural plate border between neural and non-neural ectoderm. These signals are thought to activate a suite of transcription factors, including Pax3/7, Msx1/2 and Zic1, whose expression overlaps at the neural plate border region. For example, in chick Pax7 is expressed in the neural plate border at gastrula stages and required for subsequent neural crest formation (Basch et al., 2006). These ‘neural plate border specifier genes’ in turn cooperate with signaling molecules to regulate the next tier of transcription factors, termed ‘neural crest specifier genes’ whose expression initiates in neural crest precursors in the closing neural tube and prior to their emigration. These transcription factors are markers of bona fide neural crest precursors and include genes like FoxD3, Sox9, Sox10, Myc, tfAP2, Id2, and Ets1 among others. The neural crest specifier genes regulate downstream factors necessary for neural crest EMT and lineage specification. For example, Sox10 is critical for differentiation of neuronal, glial and melanocytic lineages whereas Sox9 is involved in cartilage differentiation. The neural crest GRN has been best described at cranial levels and there are some apparent differences between axial levels; for example, Ets1 is only expressed in cranial but not trunk neural crest cells.
With improved genomic analysis in multiple species and the application of transcriptomics to developmental systems, the neural crest field has migrated into the genomic age. Comparative genome analysis made it possible to identify enhancers that mediate expression of key neural crest genes like FoxD3 (Simoes-Costa et al., 2012) and Sox10 (Betancur et al., 2010). Interestingly, these studies have shown that distinct enhancers drive expression of these neural crest specifier genes at the cranial and trunk levels. For example, in the case of FoxD3, both cranial and trunk enhancers require common inputs from neural plate border genes PAX7 and MSX1/2. Whereas the cranial enhancer also requires ETS1 input, the trunk enhancer instead requires ZIC1 (Simoes-Costa et al., 2012). Thus, there are both common and differential inputs mediating FoxD3 expression in the head versus the trunk.
The availability of neural crest enhancers has facilitated isolation of pure populations of neural crest cells for transcriptome analysis. Importantly, this has greatly expanded the number of known neural crest transcriptional regulators (Simoes-Costa, 2014). As the function of individual factors is examined in detail, the number of known inputs into the neural crest GRN is expanded. As case in point, transcriptome analysis identified Axud1, a transcriptional regulator thought to function as a Wnt effector, as highly upregulated in the cranial neural crest (Simoes-Costa et al., 2015). Detailed functional analysis revealed the AXUD1 is indeed downstream of Wnt signaling and a direct input required for expression of neural crest specifier genes like FoxD3 (Figure 3). Importantly, the results showed that AXUD1 directly interacts with PAX7 and MSX1 on the FoxD3 enhancer, thus functioning as an integrator of Wnt signaling and transcriptional input from neural plate border genes. This is an important example of how it is possible to go from a broad genomic database, zoom in and perform detailed analysis of potentially interesting components and gain novel insights that inform upon the structure of the neural crest GRN.
Amazing advances in technology that have rendered sequencing better, faster and cheaper have enabled analysis of neural crest development at multiple stages, axial levels, and in numerous species by RNA-seq. Moreover, the use of ChIP-seq on human embryonic stem cells differentiated in neural crest cells has identified numerous neural crest enhancers (Rada-Iglesias et al., 2012). Going forward, the combination of transcriptomics and novel methods of enhancer analysis, including ChIP-seq with epigenetic markers and ATAC-seq, will greatly aid in establishing direct connections within the neural crest GRN.
Differentiation of neural crest cells into multiple lineages
After completion of migration, neural crest cells settle in discrete sites throughout the embryo where they differentiate into many and diverse cell types that differe depending upon their axial level of origin. The cranial neural crest gives rise to the craniofacial skeleton, including bone, cartilage and connective tissue as well as most of the dental tissues. In addition to enteric nervous system, the vagal neural crest contributes to the cardiac mesenchyme and smooth muscle of the great vessels. The trunk neural crest also differentiates into a number of secretory cell types such as chromaffin and parafollicular cells of the adrenal and the thyroid glands, respectively. Neural crest cells from all axial levels differentiate into melanocytes and contribute to the peripheral nervous system, giving rise to sensory, sympathetic and parasympathetic neurons and multiple types of glial cells (Le Douarin, 1982).
The process of cell differentiation requires the deployment of differentiation gene batteries, which are sets of genes that endow each cell with its terminal identity (Erwin and Davidson, 2009). For instance, the differentiation gene battery of a muscle cell includes a large number of contractile proteins, which allow this cell to exert its function. Gene batteries are controlled by differentiation drivers, which are regulators that orchestrate the transition between the progenitor and differentiated states (Peter and Davidson, 2015). In the neural crest, these differentiation drivers are often genes from the neural crest specifier module that are retained during cell migration. In particular, SoxE transcription factors seem to play a central role in activating differentiation gene batteries during neural crest differentiation. Sox10 is involved in the differentiation of the neural crest into neurons, glia and melanocytes (reviewed by Kelsh, 2006), while Sox9 drives neural crest progenitors toward a chondrocytic fate (Mori-Akiyama et al., 2003).
As expected, the activation of gene batteries also requires inputs from signaling systems, which cooperate with differentiation drivers to activate multiple downstream targets. Neural crest differentiation is contingent upon the interactions between the regulatory machinery present in each cell and extrinsic signals that they encounter during migration (Simões-Costa and Bronner, 2015). Since migratory neural crest cells are multipotent, environmental cues play a crucial part in determining the fate of their progeny. These cues are conveyed through signaling systems that are active along the neural crest migratory pathways. For instance, Kit signaling plays a central role in differentiation of the neural crest into melanocytes (Parichy et al., 1999), and BMP signaling plays a central role in the differentiation during autonomic nervous system formation (Saito et al., 2012; Shah et al., 1996). To interpret such signals, neural crest cells are endowed with a rich repertoire of receptor molecules (Simões-Costa et al., 2014). Activation of signal transduction pathways impacts gene expression in the migrating cells and influences the fate they will subsequently adopt.
As is often the case in embryonic development, the same signaling systems and transcription factors are used reiteratively during neural crest formation and differentiation. For instance, Sox9 has an early role in neural crest specification (Cheung and Briscoe, 2003), and is later a driver of differentiation to cartilage and bone. Similarly, Wnt-signaling plays a critical role first in the determination of the neural plate border territory (reviewed by Prasad et al., 2012), second during neural crest specification (Garcia-Castro et al., 2002; Simões-Costa et al., 2015) and finally in the differentiation of neural crest cells into sensory neurons (Lee et al., 2004). At later stages, Wnts also impact differentiation of neural crest cells into melanocytes, through the activation of the melanocytic driver MITF (Dorsky et al., 2000). Thus, neural crest cells can interpret similar signals differently in distinct moments of their ontogeny, reflecting the profound shifts in GRN architecture that take place during development.
Evolution of the neural crest
All vertebrates including basal jawless vertebrates (agnathans) have neural crest cells that arise from the dorsal tube, undergo an epithelial to mesenchymal transition and migrate extensively. In fact, Gans and Northcutt (1983) proposed that the “New Head” of vertebrates was facilitated by the invention of the neural crest which enabled formation of specific vertebrate features like peripheral ganglia and the craniofacial skeleton. In all vertebrates, there is also a common molecular signature in neural crest cells, such that the premigratory neural crest expresses a suite of transcription factors, most notably FoxD3 and SoxE (Sauka-Spengler et al., 2007; Green et al., 2015). However gnathostomes (jawed vertebrates) have some neural crest derivatives that are absent from jawless agnathans like lamprey, including sympathetic ganglia and jaws. Thus, there seems to have been an expansion of cell types formed by the neural crest during the course of vertebrate evolution.
In contrast to vertebrates, basal chordates like amphioxus completely lack neural crest, even though their body plan shares morphological characteristics similar to those of vertebrates, including a dorsal hollow nerve cord, notochord, and segmented mesoderm. From a GRN point of view, amphioxus has neural plate border genes that exhibit similar expression to those of vertebrates. However, neural crest specifier genes, although present in the genome appear to be absent from the neural folds/dorsal neural tube of amphioxus with the exception AmphiSnail, which is transiently expressed in the lateral portion of the neural tube (Yu et al., 2008).
Although cephalochordates like amphioxus resemble vertebrates morphologically, comparative genomic analysis has shown that urochordates, rather than cephalochordates, are the sister group of vertebrates (Holland et al., 2008). Several studies of ascidian embryos have led to the speculation that tunicates may possess cell types with some but not all characteristics present in vertebrate neural crest cells. Most notably, they possess cells that arise at the neural plate border and can differentiate into pigment cells and/or neurons. For example, Abitua and colleagues (2012) have found that the a9.49 lineage of Ciona intestinalis gives rise to non-motile pigmented sensory cells. Precursor cells of this lineage express several transcription factors like Snail, ETS and FoxD genes that are part of the vertebrate neural crest specifier module. Moreover, ectopic expression of Twist in these cells causes them to become migratory. Thus, these cells may represent some type of intermediate that has some but not all of the neural crest GRN. An intriguing possibility is that acquisition of other neural crest specifier genes like SoxE genes to the neural plate border and neural folds may have conferred multipotency and the ability to undergo EMT to these border cells in order to generate a bona fide neural crest.
Recent insights into neural crest stem cells in post-natal tissue: ‘Schwann cell precursors’
One of the most exciting recent advances in the neural crest field has been the discovery of neural crest stem cells in post-natal tissues. Although it has long been speculated that neural crest cells are “stem” cells, the reality is that the embryonic neural crest is a transient population that quickly transitions from multipotent to restricted progenitors, and their capacity to self-renew is rather limited. This has raised a conundrum in the field: with a cell population like neural crest cells, that have a feed-forward developmental program going from multipotency to relatively rapid restriction in cell fate and differentiation, how do vertebrates continue to add differentiated cells to neural crest derivatives with post-embryonic growth?
The answer may lie in the fact that a subset of the neural crest population is able to maintain multipotency after embryonic development has concluded. Some evidence suggests that many neural crest derivatives, including skin and peripheral nerves, contain stem cells with the ability to give rise to multiple derivatives (Morrison et al., 1999; Jinno et al., 2010). Recently it has been found that ‘Schwann cell precursors’ along nerve processes are able to differentiate into numerous derivatives post-natally. These include pigment cells of the skin (Adameyko et al., 2009), parasympathetic ganglia of the head (Dyachuk et al., 2014; Espinosa-Medina et al., 2014), as well as enteric neurons of the gut (Uesaka et al., 2015). These studies definitively demonstrate the existence of neural crest-derived stem cells with the ability to contribute to melanocytes and neurons at much later times in development than normally associated with neural crest differentiation.
An intriguing possibility is that these may represent the true neural crest stem cells. The degree to which these crest-derived stem cells contribute to derivatives of the adult is not yet known. We speculate that this cell population may contribute to many derivatives classically attributed to the embryonic neural crest. This will be a research area ripe for future discovery.
Conclusion
This is an incredibly exciting time in biology due to the amazing advances in technology that have emerged in the past decade and continue to develop. We live in an era of high throughput sequencing, ATAC- and ChIP-seq, CRISPR-Cas9 genome editing, super high resolution microscopy, and Mass-spec. From improvements in genomic analysis, genome editing, imaging and data acquisition, scientists can examine important question at ever higher resolution. Because of these technologies, the possibility of unraveling mysteries of neural crest formation, migration, differentiation and evolution is in closer reach than was previously imaginable. And yet the important questions remain unresolved: How do cells at the neural plate border become a neural crest cell rather than part of the CNS, ectoderm or placodes? Why do some cells undergo EMT and what are the accompanying molecular changes? What determines their pathways of migration at a single cell versus population level? How does a cell decide to differentiate into a neuron versus a glial or pigment cell, etc.? How are neural crest derivatives replaced upon loss in the adult? New tools and innovations hold the promise of answering these long standing questions that have intrigued neural crest investigators for over one hundred years.
References
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture: I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res. 1954;5:111–131. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Müller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Baggiolini A, Varum S, Mateos JM, Bettosini D, Nessy J, Ziegler U, Dimou L, Clevers H, Furrer R, Sommer L. Genetic Lineage Tracing Demonstrates Multipotency of Premigratory and Migratory Neural Crest Cells in Vivo. Cell Stem Cell. 2015;6(3):314–322. doi: 10.1016/j.stem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Baroffio A, Dupin E, Le Douarin NM. Clone-forming ability and differentiation potential of migratory neural crest cells. Proc Natl Acad Sci USA. 1988;85:5325–5329. doi: 10.1073/pnas.85.14.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch M, Bronner-Fraser M, Garcia-Castro M. Specification of neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10, a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. USA. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. Analysis of the early stages of trunk neural crest migration in avian embryos using the monoclonal antibody HNK-1. Devl. Biol. 1986;115:44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser S. Cell lineage analysis shows multipotentiality of some avian neural crest cells. Nature. l988;335(8):161–164. doi: 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser S. Developmental potential of avian trunk neural crest cells in situ. Neuron. 1989;3(6):755–766. doi: 10.1016/0896-6273(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Burns AJ, Le Douarin NM. Enteric nervous system development: analysis of the selective developmental potentialities of vagal and sacral neural crest cells using quail-chick chimeras. Anat Rec. 2001;262(1):16–28. doi: 10.1002/1097-0185(20010101)262:1<16::AID-AR1007>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell. 2011;21:1026–1037. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell. 2011;21(6):1026–1037. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni GW, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Sonic Hedgehog promotes the development of multipotent neural crest progenitors endowed with both mesenchymal and neural potentials. Proc Natl Acad Sci U S A. 2007;104(50):19879–19884. doi: 10.1073/pnas.0708806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- Chibon P. Marquage nucleaire par la thymidine tritiee des derives de la crete neurale chez FAmphibien Urodele Pleurodeles waltlii Michah. J Embryol Exp Morphol. 1967;18(3):343–358. [PubMed] [Google Scholar]
- Cohen AM, Konigsberg IR. A clonal approach to the problem of neural crest determination. Dev Biol. 1975;46(2):262–280. doi: 10.1016/0012-1606(75)90104-9. [DOI] [PubMed] [Google Scholar]
- Couly GF, Le Douarin NM. Mapping of the early neural primordium in quail-chick chimeras. II. The prosencephalic neural plate and neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev Biol. 1987;120(1):198–214. doi: 10.1016/0012-1606(87)90118-7. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Le Douarin NM. The regeneration of the cephalic neural crest, a problem revisited: the regenerating cells originate from the contralateral or from the anterior and posterior neural fold. Development. 1996;122(11):3393–3407. doi: 10.1242/dev.122.11.3393. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Raible DW, Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes & development. 2000;14:158–162. [PMC free article] [PubMed] [Google Scholar]
- Dupin E, Calloni GW, Le Douarin NM. The cephalic neural crest of amniote vertebrates is composed of a large majority of precursors endowed with neural, melanocytic, chondrogenic and osteogenic potentialities. Cell Cycle. 2010;9(2):238–249. doi: 10.4161/cc.9.2.10491. [DOI] [PubMed] [Google Scholar]
- Dyachuk V, Furlan A, Shahidi MK, Giovenco M, Kaukua N, Konstantinidou C, Pachnis V, Memic F, Marklund U, Müller T, Birchmeier C, Fried K, Ernfors P, Adameyko I. Neurodevelopment. Parasympathetic neurons originate from nerve-associated peripheral glial progenitors. Science. 2014;345:82–87. doi: 10.1126/science.1253281. [DOI] [PubMed] [Google Scholar]
- Erwin EH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nature Reviews Genetics. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Espinosa-Medina I, Outin E, Picard CA, Chettouh Z, Dymecki S, Consalez GG, Coppola E, Brunet JF. Neurodevelopment. Parasympathetic ganglia derive from Schwann cell precursors. Science. 2014;345:87–90. doi: 10.1126/science.1253286. [DOI] [PubMed] [Google Scholar]
- Fraser SE, Bronner-Fraser M. Migrating neural crest cells in the trunk of the avian embryo are multipotent. Development. 1991;112(4):913–920. doi: 10.1242/dev.112.4.913. [DOI] [PubMed] [Google Scholar]
- Gammill L, Gonzalez C, Gu C, Bronner-Fraser M. Guidance of trunk neural crest migration requires Neuropilin-2/Semaphorin3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural crest and the origin of vertebrates: A new head. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Green S, Simões-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörstadius S. The Neural Crest: Its Properties and Derivatives in the Light of Experimental Research. Oxford University Press; 1950. [Google Scholar]
- Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, Rudnicki MA, Chai Y, Rossi F, Marra MA, Miller FD. Convergent genesis of an adult neural crest-like dermal stem cell from distinct developmental origins. Stem Cells. 2010;28:2027–2040. doi: 10.1002/stem.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, McLennan R, Romine MH, Kulesa PM, Lefcort F. CXCR4 controls ventral migration of sympathetic precursor cells. J Neurosci. 2010;30:13078–13088. doi: 10.1523/JNEUROSCI.0892-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN. Sorting out Sox10 functions in neural crest development. BioEssays : news and reviews in molecular, cellular and developmental biology. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Marcelle C, Collazo A, Yancopoulos G, Fraser SE, Bronner-Fraser M. Interactions between Eph-related receptors and ligands confer rostrocaudal polarity to trunk neural crest migration. Current Biology. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. A biological cell labeling technique and its use in experimental embryology. Dev Biol. 1973;30:217–222. doi: 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. A Feulgen-positive nucleolus. Exp Cell Res. 1973;77(1):459–468. doi: 10.1016/0014-4827(73)90600-9. [DOI] [PubMed] [Google Scholar]
- Le Douarin N. The Neural Crest. Cambridge: Cambridge University Press; 1982. [Google Scholar]
- Le Douarin N. The avian embryo as a model to study the development of the neural crest: a long and still ongoing story. Mechanisms of Development. 2004;121(9):1089–1102. doi: 10.1016/j.mod.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA. Experimental analysis of the migration and differentiation of neuroblasts of the autonomic nervous system and of neurectodermal mesenchymal derivatives, using a biological cell marking technique. Dev Biol. 1974;41(1):162–184. doi: 10.1016/0012-1606(74)90291-7. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA. The migration of neural crest cells to the wall of the digestive tract in avian embryo. J Embryol Exp Morphol. 1973;1:31–48. [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: Analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lee HY, Kleber M, Hari L, Brault V, Suter U, Taketo MM, Kemler R, Sommer L. Instructive role of Wnt/beta-catenin in sensory fate specification in neural crest stem cells. Science. 2004;303:1020–1023. doi: 10.1126/science.1091611. [DOI] [PubMed] [Google Scholar]
- McLennan R, Teddy JM, Kasemeier-Kulesa JC, Romine MH, Kulesa PM. Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Dev Biol. 2010;339:114–125. doi: 10.1016/j.ydbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R, Schumacher LJ, Morrison JA, Teddy JM, Ridenour DA, Box AC, Semerad CL, Li H, McDowell W, Kay D, Maini PK, Baker RE, Kulesa PM. Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development. 2015;142(11):2014–2025. doi: 10.1242/dev.117507. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-Regulatory Interactions in Neural Crest Development and Evolution. Developmental Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96:737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Peter I, Davidson EH. Genomic Control Process: Development and Evolution. Academic Press; 2015. [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Developmental biology. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11(5):633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmann M, Fawcett JW, Keynes RJ. The migration of neural crest cells and the growth of motor axons through the rostral half of the chick somite. J Embryol Exp Morphol. 1985;90:437–455. [PubMed] [Google Scholar]
- Rogers C, Saxena A, Bronner ME. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J Cell Biol. 2013;203:835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito D, Takase Y, Murai H, Takahashi Y. The Dorsal Aorta Initiates a Molecular Cascade That Instructs Sympatho-Adrenal Specification. Science. 2012;336:1578–1581. doi: 10.1126/science.1222369. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366(1):64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Scarpa E, Szabó A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev Cell 24. 2015;34(4):421–434. doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck MAJ, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development. 1995;121:526–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Serbedzija G, Bronner-Fraser M, Fraser SE. Vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development. 1989;106:806–816. doi: 10.1242/dev.106.4.809. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M, McKeown S, Tan-Cabugoa J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genetics. 2012:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Stone M, Bronner ME. Axud1 integrates Wnt signaling and transcriptional inputs to drive neural crest formation. Dev. Cell. 2015 doi: 10.1016/j.devcel.2015.06.024. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Tan-Cabugao J, Antoshechkin I, Sauka-Spengler T, Bronner ME. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19(1):39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twitty VC. Correlated genetic and embryological experiments in Triturus: I and II. J. Exp. Zool. 1936;74:239–302. [Google Scholar]
- Uesaka T, Nagashimada M, Enomoto H. Neuronal Differentiation in Schwann Cell Lineage Underlies Postnatal Neurogenesis in the Enteric Nervous System. J Neurosci. 2015;35(27):9879–9888. doi: 10.1523/JNEUROSCI.1239-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JK, Meulemans D, McKeown S, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008;18:1127–1132. doi: 10.1101/gr.076208.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yntema CL, Hammond WS. Depletions and abnormalities in the cervical sympathetic system of the chick following extirpation of neural crest. J Exp Zool. 1945;100:237–263. doi: 10.1002/jez.1401000207. [DOI] [PubMed] [Google Scholar]
- Wang HU, Anderson DJ. Eph family transmembrane ligands can mediate repulsive guidance of trunk neural crest migration and motor axon outgrowth. Neuron. 1997;18(3):383–396. doi: 10.1016/s0896-6273(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93(5):741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Weston JA. A radioautographic analysis of the migration and localization of trunk neural crest cells in the chick. Dev Biol. 1963;6:279–310. doi: 10.1016/0012-1606(63)90016-2. [DOI] [PubMed] [Google Scholar]
- Weston JA, Butler SL. Temporal factors affecting localization of neural crest cells in tbe chicken embryo. Dev Biol. 1966;14(2):246–266. doi: 10.1016/0012-1606(66)90015-7. [DOI] [PubMed] [Google Scholar]