Abstract
Several complementary mass spectrometric imaging techniques allow mapping of various analytes within biological tissue sections. Laser ablation inductively coupled plasma mass spectrometry (LA-ICPMS) quantitatively detects elements and isotopes with very high sensitivity and a particularly high dynamical range. Matrix-assisted laser desorption/ionization ion mobility mass spectrometry (MALDI-IM-MS) allows a pixel-by-pixel classification and identification of biomolecules.In order to dispose of the healthy hemisphere as an internal calibrant in addition to routinely used external standards, adjacent brain sections of mice with a unilateral 6-OHDA lesion of the medial forebrain bundle were chosen as exemplary samples. We demonstrate a comprehensive way of data acquisition and analysis by coregistering mass spectrometric data on photomicrographs as common reference space and thus providing trimodal spatial information. Registering subsequent planar element maps yielded continuous 3-dimensional data sets. Furthermore, we introduce a correction of MSI data for variable slice thickness applicable to all MSI techniques. In the present case, we observed increased concentrations of iron, manganese, and copper in the lesioned substantia nigra while monounsaturated lipid levels were decreased in the identical region of interest. Our techniques provide new insights into the intricate spatial relationship of morphology and chemistry within tissue.
Graphical abstract
Mass spectrometry imaging (MSI) is a rapidly growing analytical technology allowing the creation of spatially resolved maps for a multitude of molecular species or elements in a tissue section.1–3 Most of the recent achievements in mapping the molecular inventory of tissues were made on a mesoscopic scale of 5–200 μm spatial resolution. Different MSI-techniques are mutually complementary with respect to the range of mass-to-charge ratios (m/z) and type of molecules that are accessible.
Matrix-assisted laser desorption ionization (MALDI)-MSI is the most widespread technique for analyzing biomolecules on a large scale and in a wide mass range using different types of mass analyzers such as time-of-flight (TOF) analyzers, quadrupole analyzers, linear ion traps, and Orbitrap or Fourier transform ion cyclotron resonance (FTICR) instruments.1–6 Since its introduction, considerable achievements have been made in MALDIMSI while improving the spatial resolution down to 5 μm,6 multiplyingthe numberofaccessible analytes by introducingnew matrixes and matrix application procedures,1 using label and tracer techniques, collecting reference data, and establishing software for image reconstruction, processing, and analysis. Especially, coupling of MALDI sources to high mass resolution instruments multiplied the number of identifiable compounds and changed confidence in peak assignments from some probability to near certainty. Additionally, soft ionization in Orbitrap instruments permitted analysis of samples not specially mounted onto steel plates but from conventional glass slides.6 Applications in MALDI-MSI have mainly focused on the identification of specific protein markers and molecular fingerprints of tumors, on tracking neurodegenerative processes, and on the characterization of transgenic animal models and studies of the in situ distribution of xenobiotics.1–9 Interposition of an ion mobility (IM) separator adds an additional postionization dimension into MALDI-MS.10,11 MALDI-IM-MSI provides the ability to rapidly separate and classify various types of biomolecules such as nucleotides, carbohydrates, peptides, and lipids including isobaric species.11–13
IM separations utilize nonfragmenting low energy gas-phase collisions to differentiate ions based on their apparent surface area or collision cross section. In the present instrumentation, ions migrate through a drift tube filled with a neutral drift gas under the influence of an electric traveling wave that is generated by individually changing the potential of the stacked rings of the ion guide. Compact ions cross the drift tube in shorter time than larger ions (typically 0.1–100 ms).11 Like traditional MS/MS, the coupled arrangement of IM and MS yields the ability to obtain additional structural information in the MS dimension by performing IM-MS/MS. By placing an ion activation region between the drift tube and mass analyzer, ions may be selected for fragmentation according to drift time. In imaging IM-MS/MS experiments, multiple parent ions can be fragmented whereby fragment ions are correlated to their respective parent ions by drift time. This application is a highly promising yet virtually untapped resource for biomolecular imaging MS, where limited sample exists at each pixel location. MALDI-IM-MS has been applied to lipid imaging in rat brain, low abundance protein detection and profiling in tumors, and also for drug imaging.1,2,12,13
Laser ablation inductively coupled plasma (LA-ICP)MSI was established for quantitative distribution analysis of elements and isotopes.14 It utilizes a focused laser beam to ablate sample material which is transported by a carrier gas (Ar) into the ICP source. In the ICP operated at 7000–8000 K, the sample material is atomized and ionized, preferentially to singly charged ions. In contrast to other contemporary MS techniques, ICPMS provides by far the highest concentration dynamic range which spans over nine decades for quadrupole based instruments, as used in this study, and thirteen decades for double focusing sector field instruments, respectively. ICPMS is the technique of choice for sensitive and efficient trace element and isotopic analysis.
Scanning a native tissue section placed onto a conventional glass slide without any pretreatment, such as matrix application, provides a 2D image of its elemental and isotopic composition at a spatial resolution of 100 to 5 μm and limits of detection in the range of 0.001–1 μg g−1.14,15 In this study, a quadrupole based ICPMS was used, which is very robust and stable over many hours. This is an important precondition for imaging studies that need, as here, ca. 4 h to analyze 1 cm2 of tissue. In ICPMS, special attention is paid to the choice of the measuring isotopes for those elements of which several stable isotopes exist in order to avoid isobaric interferences of analyte ions with isobaric atomic ions (e.g., 40Ca+ and 40Ar+; 58Ni+ and 58Fe+) or with polyatomic ions (e.g., 41K+ and 40Ar1H+; 52Cr+ and 40Ar12C+) at the same nominal m/z. However, Fe can be measured in tissue sections by LA-ICPMS at m/z 56 as the background formed by 40Ar16O+ ions is sufficiently low and remains constant. In the absence of commercial software, inhouse written algorithms (IMAGENA)16 have been utilized for the reconstruction of images out of a continuous sequence of raw-data points.
Data are calibrated using matrix matched standards.14,15 In contrast to histochemical techniques or fluorescent dye staining techniques that measure only free metal cations or various fractions of bound metals, LA-ICPMS measures the total content of the selected elements. For images obtained using LA-ICPMS, it is not necessary to normalize element signals to a reference element on a pixel-by-pixel basis as images are of high continuity, symmetry, exhibit a homogeneous background, are free of irregular drift effects, and are highly reproducible. The pixel-wise use of a reference element would introduce additional bias because elemental concentration varies as a function of the water content in tissue which has been determined at 80% in cerebral gray matter and at 70% in white matter.17 Notwithstanding, the correction for variable slice thickness proposed here uses net 13C+ signals averaged across the entire sample section and the entire section of standard homogenate. The gray matter/white matter ratio and the water content of different sections within one study can rather be assumed as constant. In the same way, a representative signal such as the total ion count may be used in other MSI techniques. Alternatively, the average slice thickness is directly accessible by white light interferometry.
LA-ICPMSI has been used to study healthy and diseased brain and tumors.14,15 The distribution of Pt, Se,14 Gd, and Lu18 has also been mapped following in vivo administration of respective metal containing compounds.
The methodological focus of this work was to integrate several spatially resolved analytical techniques into a common reference space. Here, we first combined quantitative element mapping by LA-ICPMS and lipid imaging by MALDI-IM-MS as proposed by Becker and Jakubowski,19 which allows one to correlate different sets of analytes from the same sample, to develop a comprehensive data analysis, and to finally draw functional patho-biochemical conclusions. Precise coregistration of all imaging modalities to the optical image obtained before MSI allows one to assign MSI signals to defined anatomical structures, image merging, data correlation, and inclusion into crossmodality calculations. In a multimodal approach, as presented here, the advantages outweigh the increased expenditure especially when the sample material is scarce. This is the case in mouse brain where only a few cryo-sections are available across a given nucleus of high functional differentiation.
We chose a unilateral lesion model in order to dispose of the healthy hemisphere as an internal calibration in addition to external standards. Injection of the neurotoxin 6-hydroxydopamine (6-OHDA) into the striatum, the substantia nigra (SN), or the medial forebrain bundle, constituting the destination, the origin, and the junction of nigrostriatal dopaminergic neurons, respectively, is a well established and the most common animal model of Parkinson’s disease (PD). 6-OHDA is selectively taken up into catecholaminergic neurons by the dopamine and noradrenaline transporters DAT and NAT. Blockade of NAT by desipramine further restricts the uptake of 6-OHDA to dopaminergic neurons and processes that succumb to retrograde degeneration. The mechanism of toxicity is assumed to be inhibition of complexes I and IV of the mitochondrial respiratory chain.20 Complex III is known to produce a small rate of •O2−. Upon blockade of complex IV, a higher fraction of electrons may prematurely leak to oxygen, resulting in the formation of the toxic free radical superoxide localized at the outer leaflet of the inner mitochondrial membrane. Other sources of radicals are NADPH dependent oxidases driven by excess NADPH and NADH. An increase of iron in the substantia nigra (SN) and other basal ganglia of human PD patients and 6-OHDA-lesioned rodents has been observed using other techniques such as analysis of homogenates of dissected material,21 synchrotron X-ray fluorescence,22 or Prussian blue staining23,24 in situ. This increase of Fe was not paralleled by an increase of ferritin.23 For a long time, this was poorly understood, but recently, this rise in Fe has been attributed to an up-regulation of the divalent metal transporter 1 isoform containing iron responsive elements (DMT1 + IRE) in humans and in rodents treated with MPTP or 6-OHDA.25 Due to these advantages and its popularity, the 6-OHDA rodent model has already been subject of one proteome MALDI-MS imaging study,26 one in situ MALDI-MS profiling study,27 and several classical proteomics studies.28,29 The first mapped a section crossing the SN and included some proteome speciation conducted on the striatum but not the SN. The other studies did not use imaging and only assessed the striatum. These studies found a decrease of cytochrome c and cytochrome c oxidase as could be expected as a consequence of respiratory chain inhibition by 6-OHDA. Interestingly, other proteins not directly involved into the respiratory chain and expected to be unchanged in 6-OHDA lesions before were found to be differentially expressed such as calmodulin (up-regulated), α-enolase, F1-ATPase, CRMP2B, and calmodulin (down-regulated).27,28
EXPERIMENTAL SECTION
Mouse Model
All animal experiments were institutionally approved (Regierungspräsidium Gießen) and were performed in accordance with current guidelines. Wildtype C57Bl6 male mice (Charles River), 12–14 weeks of age, were kept at 23 ± 1 °C under 12 h dark/light conditions with ad libitum access to food and water. Each mouse was intraperitoneally (i.p.) treated with 25 mg kg1− desipramine 40 min before surgery. After i.p. anesthesia with 10 mL kg1− of 1% ketamine and 0.2% xylazine, the mice received an injection of 6-OHDA unilaterally into the right medial forebrain bundle at the stereotactic coordinates 0.35 mm anterior and 1.15 mm lateral from bregma and 3.75 mm ventral from dura (corresponding to Paxinos and Watson30). 6-OHDA was dissolved at a concentration of 1 μg μL−1 in saline containing 0.2% ascorbic acid. A volume of 2 μL was injected at a rate of 0.5 μL min−1. The needle was left in situ for 5 min after the injection before it was slowly retracted. The successful induction of hemiparkinsonism was confirmed by neuropathological assessment of the loss of nigral neurons with positive staining for tyrosine hydroxylase (TH; the rate limiting enzyme in the production of the neurotransmitter dopamine). Twenty-one days after surgery, mice were euthanized i.p. with 100 mg kg−1 pentobarbital (Sigma-Aldrich) and transcardially perfused with 50 mL of saline. Brains were immediately removed, snap frozen in –70 °C isopentane, and wrapped after evaporation of isopentane. From the entire SN, cryo-sections were prepared, thaw mounted onto glass slides, and stored at −80 °C. In situ hybridization (ISH) with 35S-labeled riboprobes against mouse TH mRNA was performed on 14 μm tick cryo-sections of SN to determine the 6-OHDA-induced dopaminergic lesion severity compared to the control hemisphere.31 20 μm thick cryo-sections adjacent to the cryo-sections used for ISH were used for MSI.
LA-ICPMS
A commercial laser ablation (LA) system (New Wave UP 266, Fremont, CA, USA) operated with a frequencyquadrupled Nd:YAG laser (wavelength of 266 nm, repetition frequency of 20 Hz, spot diameter of 110 μm, distance between lines of 50 μm, scan speed of 30 μm/s) was coupled to a quadrupole ICPMS (XSeries2, Thermo Fisher Scientific, Germany). The experimental parameters of LA-ICPMS were optimized with respect to the maximum ion intensity of 63Cu+ using a laboratory standard. Maximum ion intensity was observed at an rf ICP power of 1500W and a carrier gas flow rate of 1.2 L min−1 for the transport of ablated material to the ICPMS. Twenty-seven different mass-to-charge ratios (m/z), corresponding to selected isotopes of 16 elements, in the range from m/z 6 to m/z 300 were preset and determined within a cycle time of 2.7 s that constitute one pixel. Thus, the resolution was 30 μm/s × 2.7 s = 83 μm in ×- and 110 μm + 50 μm = 160 μm in y-direction. As the entire thickness (<100 μm) of a tissue section is ablated, the data represent an average across the thickness of the tissue section. Thus, the spatial precision of the distribution of analytes is maximized, and challenges associated with the vertical transport of solutes during drying, by surface contaminations or by analyte fractionation effects during ablation, are minimized. The spatial resolution in x-direction is given by the product of the x-speed of the piezo-driven xyz-stage and the cycle time; the spatial resolution in y-direction by the preset distance between the centers of lines. The spot size of the laser does not directly influence the spatial resolution. We obtained best results with xand y-step width smaller than the laser spot diameter in the sense of oversampling, leaving about 5 μm wide bars of residual tissue between ablated lines.
Matrix matched standards were prepared from homogenates of mouse brains spiked with 1/9 volume of the dilutions 0, 0.2, 0.4, 0.6, 0.8, and 1 of a multielement standard solution that contained the 10-fold of the expected maximal physiological concentrations, i.e., of 30.0, 20.0, 19.8, and 0.37 μg g−1 Zn, Cu, Fe, and Mn, respectively. Every step was weight monitored, and aliquots of the spiked homogenates further characterized by ICPMS of microwave induced acidic digests. Starting with added concentration 0 (blank), homogenates were subsequently palmed off and frozen as 2 mm thick layers in a cylindric mold resulting in a stack that was chucked in the cryo-microtome with the cylinder axis almost parallel to the blade. Hundreds of 30 μm thick cryosections of these standards were obtained and placed onto glass slides.
In order to correct for the variation of slice thickness of samples and standards, the net ion intensity of 13C+ averaged over the section or the rectangle ablated from the standard stripes proportional to the content of dry organic matter was considered as a substitute of the slice thickness. Net count rates from the standard and the section were normalized to the average net 13C+ signal: CX, corr = ([13C+]Std/[13C+]Sec) × [X]/mx, with CX, corr, corrected concentration of element X; [13C+]Std, [13C+]Sec, averaged net 13C+ ion intensities of the standard and the section; [X], ion intensity of the m/z corresponding to element X within the pixel or within the region of interest; mx, slope of the calibration line for element X.
MALDI-IM-MS
Adjacent 20 μm thick cryo-sections were thaw mounted onto a stainless steel conductive MALDI plate (Applied Biosystems). Matrix was applied using a dry-coating methodology developed by Puolitaival and co-workers.32 Briefly, 2,5-dihydroxybenzoic acid (DHB, Sigma, St. Louis, MO) was milled for approximately 20 min using a mortar and pestle. The finely ground powder was then distributed onto the tissue with a small brush. Excess matrix was removed by directing a soft stream of air over the target. The depth of surface ablation during MALDI is estimated at typically 1 μm.1 Therefore, several subsequent matrix applications and MALDI experiments can be performed on the same tissue section of 20 μm.
IM-MS biomolecular imaging experiments were performed on a Synapt HDMS (Waters Corp., Manchester, UK) equipped with an interchangeable MALDI or ESI source. IM was performed using traveling wave separation through nitrogen gas. MALDI Imaging Pattern Creator (Waters Corp.) was used to specify the areas of interest on the plate where the tissue was located. The pattern was then uploaded into MassLynx (Waters Corp.) where the experimental parameters for the images were designated. Mass calibration was performed externally with fullerenes, C60 and C70. MALDI ionization was performed at the pressure of the drift cell (3.8 Torr). The image was acquired in positive ion V-mode with a mass range from m/z 500 to 3000. The spatial resolution was set to 150 μm, and the Nd:YAG laser (frequency-tripled, 355 nm) was operated at 200 Hz. The ion guide T-wave was operated at 300 m/s and linearly ramped in amplitude from 3 to 18 V over each experiment. The transfer guide T-wave was operated at 248 m s−1 and with a constant 3 V amplitude. Ion injection voltages in the trap and transfer were set at 6 and 4 V, respectively. Peak identification first used the database of m/z × drift time data of pure compounds accumulated on this instrument in the recent years.33 Selected areas in the sample were studied after a second matrix application in the profiling and MS2 mode. Parent ions of interest out of the set of candidate lipids with ambiguous assignment were fragmented while operating the transfer traveling wave drift chamber (the last of a series of 3) as a collision cell. The specific conditions used for lipid fragmentation are described elsewhere.34 Spectra of standard reference compounds were obtained under identical conditions to those that were used from on tissue. MassLynx was also used for instrument control and data analysis.
Data Treatment and Statistics
Images of LA-ICPMS tabular raw data were reconstructed using IMAGENA.16 Concentrations and ion intensities were read out from parametric grayscale TIFF files using Pmod3.1 (Pmod, Zurich, Switzerland) from regions of interest. These data were analyzed using MS-Excel. Data from healthy and lesioned hemispheres were compared using heteroscedastic paired two sample t tests.
RESULTS
Essential steps within the workflow while analyzing the exemplary samples of this work are depicted within Figure 1 paralleling elemental and bio-organic MS including the resulting element and molecular maps, respectively.
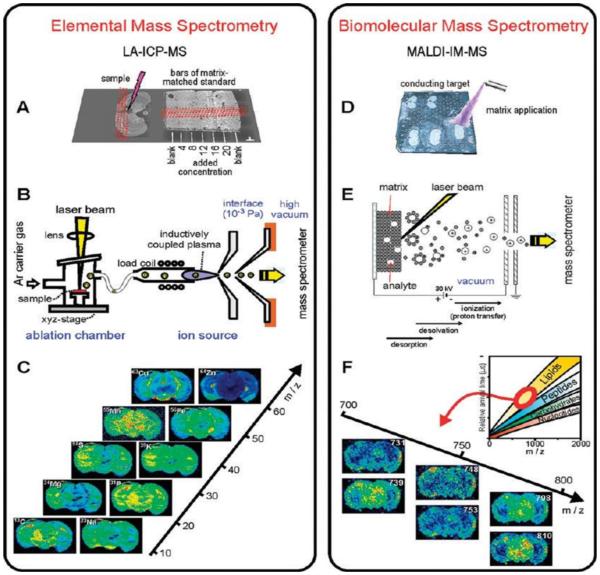
Figure 1.
Workflow of LA-ICPMS imaging (A–C) and MALDI-IMMS imaging (D–F): Cryosections were placed together with a ladder of matrix matched standards on a glass slide (A) and on a conductive MALDI plate and DHB was applied as matrix onto the samples (D). B and E show the processes preceding ion separation which is laser ablation followed by inductively coupled plasma ionization (B) or the sequence of laser-induced desorption, desolvation, and ionization within the MALDI source. Exemplary resulting ion maps on a m/z scale are given in (C) and (F). The inset in (F) shows schematically the position of molecule classes in IM-MS conformational space constituted by m/z and the drift time reflecting the collision cross section. Ion images selectively corresponding to lipids are displayed.
Elemental Imaging Using LA-ICPMS
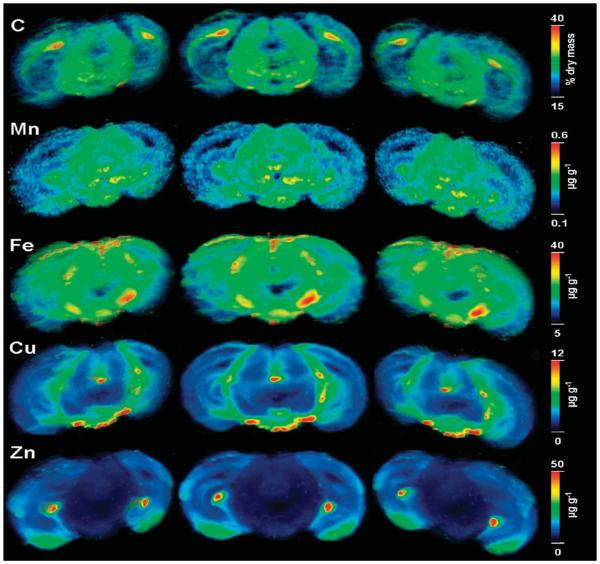
In this work, 11 adjacent sections through the entire SN were analyzed using LA-ICPMSI. Average metal concentrations in free drawn regions of interest (ROIs) including cortex, SN, and characteristic structures with particular high concentrations of the respective metal are given in Table 1. Statistically significant increases of Fe, Cu, and Mn in the lesioned SN of 21%, 17%, and 20%, respectively, were observed while Zn was constant (−1%). Side differences and element concentrations within the limits of one standard deviation could be replicated in a second animal. The correction for variable slice thickness allowed the relative standard deviation (RSD) of regional metal concentrations as a measure of precision to be reduced to about 10–13% for Fe, Cu, and Zn and 14–18% for Mn in the 11 serial sections (Table 1) compared to 25–30% without correction. The 11 respective concentration maps underwent an affine registration to z -stacks using in-house written software. Conversion into 3D-data sets present in 3D file formats such as ANALYZE, INTERFILE, and visualization used Pmod3.1. Semitransparent frontal and tilted maximum intensity projections are presented in Figure 2. In comparison to single image planes (see Figure 1), these 3D views contain less noise, the fine layered pattern of the hippocampus becomes discernible, and small curved tube shaped structures like the Cu rich nucleus supramammillarius can be traced in space and clearly differentiated from artifacts. The present data confirm and build upon previous data on the normal metallo-architecture of the mouse brain.17 The distribution of carbon was congruent to that of white matter reciprocally correlated with the water content.
Table 1.
Element Concentrations Determined by LA-ICPMS in Selected Regions of a 6-OHDA-Lesioned Mouse Brain Averaged Throughout 11 Sectionsa
| hemispere | ||||
|---|---|---|---|---|
| concertration, μg g−1 | ||||
| region | lesioned | healthy | RSD (%)c | |
| Fe | subcallosal blood vessels | 23 | 27 | 14 |
| substantia nigra | 16.5b | 13.6 | 11 | |
| ncl. Interpedumcularis | 1 | 4 | 9 | |
| colliculus superior | 11 | 11 | 11 | |
| cortex | 8 | 8 | 12 | |
| Cu | aqueduct | 1 | 5 | 23 |
| ventral fascia dentata | 9 | 9 | 14 | |
| dorsal fascia dentata | 5.4b | 4.3 | 12 | |
| ncl. supramillarius | 5. | 5 | 30 | |
| substantia nigra | 2.3b | 1.9 | 12 | |
| cortex | 2.2b | 1.9 | 13 | |
| ventral tegmental area | 3. | 0 | 20 | |
| Zn | polymorph layer ca3 | 34 | 36 | 15 |
| amygdala | 18 | 19 | 12 | |
| cortex | 10 | 10 | 12 | |
| corpus callosum | 7 | 7 | 13 | |
| Mn | ncl. oculomotorius access. | 0.34 | 0.34 | 16 |
| nucleus ruber | 0.33 | 0.33 | 16 | |
| colliculos superior | 0.28 | 0.28 | 19 | |
| cortex | 0.17 | 0.15 | 17 | |
| substantia nigra | 0.25b | 0.21 | 14 | |
| corpus callosum | 0.13 | 0.12 | 18 | |
Acces., accessorius; ncl., nucleus.; RSD, relative standard deviation.
Significant side difference p < 0.00005, t test for paired samples.
Average of the at left and right side.
Figure 2.
Three-dimensional reconstructions obtained by affine registration of 11 LA-ICPMS elemental concentration maps of adjacent coronal sections through the substantia nigra (SN). Three semitransparent maximum intensity projections from different angels of five elements are shown. The SN at the lesioned (right) side clearly shows higher iron than the contralateral healthy SN. The width of one section is 9 mm.
Biomolecular Imaging Using MALDI-IM-MS
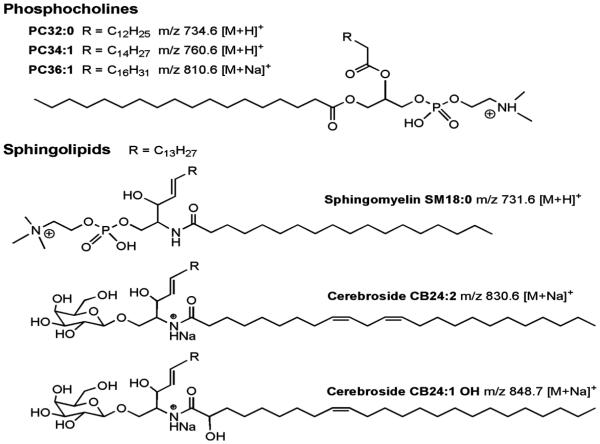
Figure 1 shows the MALDI plate analyzed in this study where 5 brain tissue sections were mounted. MALDI-IM-MS imaging of these sections, intercalated to those subdued to LA-ICPMSI, yielded a plethora of exploitable m/z signals. For example, a series of 18 m/z signals out of the lipid conformational space (Figure 1) gave rise to anatomically meaningful image patterns and was chosen for further analysis. Of these signals, 16 could be assigned with sufficient probability by comparison with the mass × drift time spectra of pure standard substances and fragmentation experiments. Structures of several of the identified compounds are depicted in Figure 4. The number of possible assignments was further reduced when identical anatomical patterns were observed for m/z species supposed to emerge from one compound such as [M + H]+, [M + Na]+, or [M+ K]+. MALDI-IM-MS images of lipids showed essentially three types of distribution patterns. First, with a cortical distribution were a sphingomyeline (SM18:0 at m/z 731.6, 753.5, and 769.6; [M + H]+, [M + Na]+, and [M + K]+, respectively), a saturated phosphatidylcholine (PC32:0 at m/z 734.6, 756.6, and 772.5; [M + H]+, [M + Na]+, and [M + K]+, respectively) and a saturated phosphatidyl-ethanolamine (PE36:0 at m/z 748.6; [M + H]+). Second, with a preferentially cortical and mesencephalic pattern sparing the hippocampus was a monounsaturated phosphatidylcholine (PC34:1 at m/z 739.5, 760.6, 782.6, and 798.5; [M + K – N(CH3)3]+, [M + H]+, [M + Na]+, and [M + K]+ respectively), a sphingomyeline (SM16:0 at m/z 741.6; [M + K]+), and the signals at m/z 814.6 and 863.7 that have been unassigned so far, but must correspond to di- and triunsaturated compounds as only comparatively abundant lipids are likely candidates. Third, preferentially enriched in white matter were a monounsaturated phosphatidylcholine (PC36:1 at m/z 810.6 and 826.6; [M+Na]+ and [M+K]+, respectively) and polyunsaturated cerebrosides (CB24:2 at m/z 830.7; [M + Na]+ and CB24:1 OH at 848.6; [M + K]+). Congruent distribution patterns of SM16:0, SM18:0, PC32:0, PC34:1, and PC36:1 were previously observed using MALDI-MS in dry matrix coated mouse brain.32 Using MALDI-IM-MS comparable patterns of PC32:012 and of PC34:1, PC36:1 and CB24:1-OH13 have been detected at a more anterior level of the brain. A clearly different distribution of PC32:0, PC34:1, and PC36:1 was obtained from α-cyano-4-hydroxy cinnamic acid wet coated rat brain with higher levels in the hippocampus and lower levels in the midbrain.35 One example of each distribution type is depicted in the right column of Figure 3; further images are shown in Figure 1. A ROI based analysis of MALDI-IM-MS images from 5 sections of 2 animals coregistered to microphotographs of adjacent sections showed no significant side difference of any compound or compound class (saturated versus mono- and polyunsaturated) within any region (p > 0.3 in cortex, hippocampus, colliculus superior, amygdala, mesencephalon, corpus callosum) except the SN. Within the lesioned SN, the saturated PC32:0 was unchanged (±0%) while the monounsaturated compounds PC36:1 and PC34:1 were reduced significantly by 11% (p = 0.01; n = 6) and by 7% (p = 0.06; n = 12), respectively, compared to the contra-lateral healthy SN. Notwithstanding, analyzing the signals at m/z 814.6 and 863.7 suggested an increase of the underlying, presumably polyunsaturated, lipids by 10% and 25% (n = 1 each). The ceramide compounds SM16:0, SM18:0, CB24:2, and CB24:1 OH showed insignificant decreases by −2%, −3%, −3%, and −8%.
Figure 4.
Structures of selected lipids and ion species detected by MALDI-IM-MS.
Figure 3.
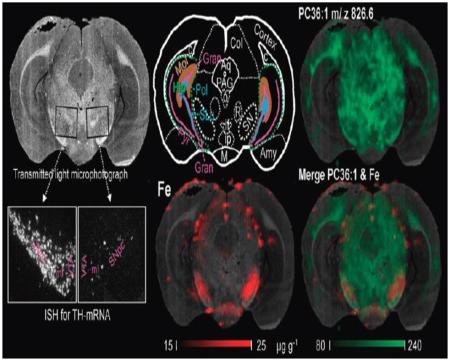
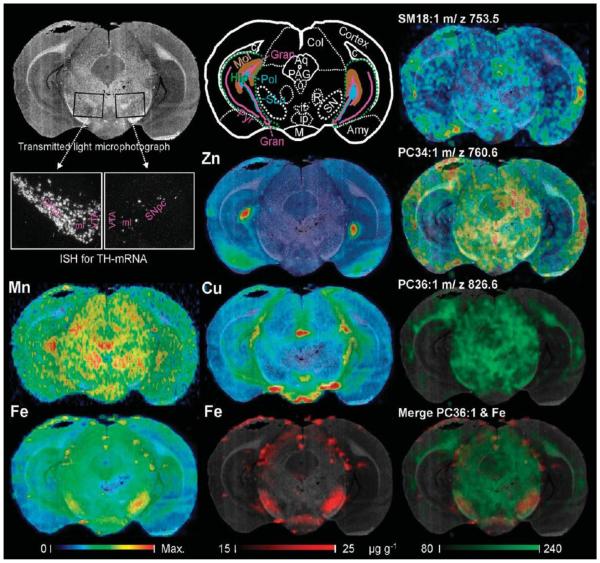
Multimodal imaging approach integrating light microscopy underlain as background of each image facilitating morphological orientation, elemental concentration maps obtained by LA-ICPMS (left and middle column) and lipid maps obtained by MALDI-IM-MS imaging. The respective maximum concentrations of the color scale are Zn, 50; Mn, 0.6 Cu, 10 and Fe, 35 μg g−1. The lipid maps shown represent the tree major distribution types observed, cortical (m/z 753.5 assigned to sphingomyelin SM18:1), cortico-mesencephalic (m/z 760.6 assigned to phosphatidylcholine PC34:1), and of white matter predominance (m/z 826.6 assigned to phosphatidylcholine PC36:1). The picture at the bottom right shows an overlay of light microscopy, LA-ICPMS image (Fe) and MALDI-IM-MS image. There is an increase of Fe and a decrease of the monounsaturated lipid within the right substantia nigra (SN). Darkfield images across the entire SN of sections subjacent to sections used for MS demonstrate ISH signals of TH-positive dopaminergic neurons. Note the nearly complete loss of dopaminergic neurons in the SN pars compacta and ventral tegmental area in the lesioned right side compared to control left side. The schema in the middle of the upper row explains the corresponding anatomy. Abbreviations: Amy, amygdala; Aq, aqueduct; c, corpus callosum (composed of white matter); Col, colliculus superior; Gran, granular layer of the fascia dentata (cut at two positions, dorsal and a ventral); Hip, hippocampus (green dashed conture); if, interfascicular nucleus; ip, interpeduncular region; Mol, stratum moleculare of the fascia dentate; o, accessory oculomotor nuclei; PAG, periaqueductal gray; R, nucleus ruber; Pol, polymorph layer (= stratum multiforme) of the fascia dentata; SLu, stratum lucidum of cornu ammonis portion 3 (of CA3); SN, substantia nigra.
Integration and Coregistration of Elemental, Biomolecular and Optical Images
MS images were coregistered to a photomicrograph of a native section presenting a high contrast between white and gray matter, using Pmod3.1 (Pmod, Zurich, Switzerland) as presented in Figure 3. Signals now could be clearly assigned to anatomical structures such as layers, bundles, and nuclei. Also, the set of ROIs used is delineated in Figure 3. High Fe concentrations were detected in the SN, the interpeduncular region, subcallosal and dorsal blood vessels, the stratum oriens of CA1, the dorsal layers of the colliculus superior, and the outer cortical layers. Particularly high Cu concentration was present in the periventricular, tuberomammillar, and inferior mammillar regions followed by the molecular layer of the fascia dentata, the praesubiculum, and the supramammillar nucleus. Zn was very high in the polymorph layer of the facia dentata, followed by the stratum lucidum of CA3 and the amygdala. Mn preferentially occurred in the nucleus ruber, the complex of accessory oculomotor nuclei, the colliculus superior, and polymorph layer of the facia dentata.
Co-registration to the optical image was particularly informative for the MALDI-IM-MS images, less rich in contrast compared to LA-ICPMS images, wherein for example, signals at m/z 826 could be identified to originate fromwhite matter rich structures such as the corpus callosum or the ventral tegmental decussation. In the trimodal fusion in the lesioned SN, a higher signal for m/z 864 overlapping and surmounting the increase of Fe became obvious while the signal at m/z 826 was higher in the healthy left side merging to yellow with the iron signal coded in red as depicted in Figure 3, right bottom.
DISCUSSION
In the present study, inorganic MSI using LA-ICPMS and biomolecular MSI using MALDI-IM-MS were conducted on adjacent sections of the same brain bearing a unilateral lesion and analyzed within a common reference space defined by the microphotograph.
As an advantage of such an integrated approach, quantitative results are provided from the same material prepared under identical experimental conditions that can be directly compared and correlated. In the given animal model, the 6-OHDA lesion, there is a high variability among animals with respect to severity, time, and location of the lesion. As a function of the part of the medial forebrain bundle hit by the stereotactic injection, various parts or the totality of the SN can succumb to retrograde damage. Moreover, insights in the chemical inventory at the microlocal scale may allow discoveries of novel interrelationships and interactions. In spatially resolved MS techniques, especially in MALDIIM-MS, large quantities of comprehensive data are gathered nearly simultaneously at each pixel position. Thus, data are conserved for hypothesis driven assessments at later time points, for example, when new compounds have been assigned to m/z signals or different analytes become the focus of future research.
Using quantitative LA-ICPMS imaging developed at Research Centre Jülich, we could obtain 3D-data Z-stacks rich in detail and with high signal-to-noise ratio. The normal metalloarchitecture was preserved, and the SN regional element concentrations were very similar to those measured previously by us and others in rodent brain. Our findings were in congruence to changes of +20% s for Fe, −3% ns for Cu, −1% ns for Zn, and 7% s for Mn (s significant, ns not significant) in a preliminary LA-ICPMS study on 6-OHDA mice from another group36 and similar to an increase of 35% s of Fe37 and of 17% s of Cu21 observed in studies on homogenates of dissected tissue of 6-OHDA-rats. Influences of lower availability of metals by food and drinking water and species differences (rat/mouse) may account for diverging concentrations in healthy SN; e.g., we measured 14 μg g−1 of Fe, and others had 14 μg g−1 37 and 34 μg g−1 36 In human fresh frozen tissue (SN) from cohorts of Parkinson’s disease patients compared to controls, changes of +35%, −33%, +49%, and +3% were observed of Fe, Cu, Zn, and Mn, respectively.38 Given the comparatively low changes of metal concentrations, it is noteworthy that in the present study significant changes in the lipid pattern could be detected. MALDI-IM-MS evidenced a constant amount of lipids containing saturated fatty acids and a decrease of monounsaturated lipids. These lipid assignments are tentatively based both on the resolution of the instrument and drift time data consistent with standard reference materials. Note that contemporary lipid analysis is particularly challenging to unambiguously assign total structure and identity, but using all available data about these species underscores our assignments.
The signals at m/z 749.6, 814.6, and 863.7 that were increased in the lesion are likely attributed to polyunsaturated lipids. Hydroperoxides of lipids may already decompose in situ or during laser desorption, so it is likely that these signals may reflect the sum of the respective polyunsaturated lipids and their peroxides. It is unlikely that isobaric endoperoxide species contributed to polyunsaturated lipid signals such as phosphocholine peroxide (PC34:0-O2, [M + Na]+) at m/z 814.6 as higher ion mobilities might be expected for endoperoxides. These findings including constant saturated and decreased monounsaturated lipids are in line with reports on increased peroxidation of lipids using bioassays.24 For the most part, these bioassays detect only polyunsaturated fatty acids and data is lacking on monounsaturated fatty acids in PD and related models. Furthermore, it should be noted that saturated lipids lack oxidation sites; an isolated double bond is easily accessible whereas doubly and triply unsaturated fatty acids are sterically less accessible and may arise from peroxidation of higher unsaturated fatty acid residues. The combination of increased Fe, with a decrease of monounsaturated fatty acids and higher lipid peroxidation, is compatible with a more reductive metabolic state and higher availability of oxygen radicals resulting from respiratory chain inhibition as hypothesized within the sequelae of 6-OHDA lesion and Parkinson’s disease. Consequently, due to the position of the electron transfer site within respiratory chain complex III the highest rate of lipid peroxidation should be expected at the outer surface of the inner mitochondrial membrane. Once initiated by a hydroxyl radical (•OH), lipid peroxidation is a selfpropagating process. Moreover, Prussian blue staining of human postmortem material confirmed an overproportional increase of Fe2+ in the SN of PD, which would further enhance the formation of hydroxyl radicals (•OH) via the Fenton reaction.23
The integration of the methodologies described here creates a powerful tool especially when used to differentially assess disease versus nondiseased states with respect to element and biomolecular distributions. Our techniques provide new insights into the intricate spatial relationship of morphology and chemistry within tissue.
ACKNOWLEDGMENTS
The last author (J.S.B.) would like to acknowledge Hans-Joachim Dietze for fruitful discussion and useful input. We are grateful to Thermo Scientific, Bremen, Germany, and to Deutsche Forschungsgemeinschaft (DFG grant No. BE 2649/5-1) for instrumental support of the BrainMet Laboratory. We thank Astrid Zimmermann for assistance with LA-ICPMS measurements. Martin KH. Schäfer and Eberhard Weihe from the Department of Anatomy; Günter U. Höglinger, Frauke Neff, and Wolfgang H. Oertel from the Department of Neurology of the Philipps University Marburg, Germany, are thankfully acknowledged. L.S.F., M.K., and J.A.M would like to acknowledge funding from the National Institutes of Health (RC2DA028981 and 1R01GM092218-01), the US Defense Threat Reduction Agency (HDTRA1-09-1-0013), the Vanderbilt University College of Arts and Sciences, the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Institute for Integrative Biosystems Research and Education. L.S.F was further supported by a VU College of Arts and Sciences Disseration Enhancement Grant. We also thank R. M. Caprioli (VU Biochemistry) and the VUMC Mass Spectrometry Research Core for utilization of the Synapt HDMS.
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Chughtai K, Heeren RMA. Chem. Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rubakhin SS, Sweedler J. V. e. Mass Spectrometric Imaging: Principles and Protocols. Vol. 656. Springer; Heidelberg: 2010. [Google Scholar]
- (3).Altelaar AFM, Luxembourg SL, McDonnell LA, Piersma SR, Heeren RMA. Nat. Protoc. 2007;2:1185–1196. doi: 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- (4).Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Nat. Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- (5).Caprioli RM, Farmer TB, Gile J. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- (6).Rompp A, Guenther S, Schober Y, Schulz O, Takats Z, Kummer W, Spengler B. Angew. Chem., Int. Ed. 2010;49:3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- (7).Seeley EH, Caprioli RM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chaurand P, Cornett DS, Angel PM, Caprioli RM. Mol. Cell. Proteomics. 2011;10(2) doi: 10.1074/mcp.O110.004259. O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Rapid Commun. Mass Spectrom. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- (10).Vonhelden G, Wyttenbach T, Bowers MT. Science. 1995;267:1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- (11).Fenn LS, McLean JA. Anal. Bioanal. Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- (12).McLean JA, Ridenour WB, Caprioli RMJ. Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- (13).Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods ASJ. Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Becker JS. Inorganic Mass Spectrometry: Principles and Applications. John Wiley and Sons; Chichester: 2007. [Google Scholar]
- (15).Becker JS, Zoriy M, Matusch A, Wu B, Salber D, Palm C, Becker J. Su. Mass Spectrom. Rev. 2010;29:156–175. doi: 10.1002/mas.20239. [DOI] [PubMed] [Google Scholar]
- (16).Osterholt T, Salber D, Matusch A, Becker JS, Palm C. Int. J. Mass Spectrom. 2011;307:232–239. [Google Scholar]
- (17).Matusch A, Depboylu C, Palm C, Wu B, Hoglinger GU, Schafer MK, Becker JS. J. Am. Soc. Mass Spectrom. 2010;21:161–171. doi: 10.1016/j.jasms.2009.09.022. [DOI] [PubMed] [Google Scholar]
- (18).Kalber TL, Kamaly N, So PW, Pugh JA, Bunch J, McLeod CW, Jorgensen MR, Miller AD, Bell J. D. Mol. Imaging Biol. 2011;13:653–662. doi: 10.1007/s11307-010-0400-3. [DOI] [PubMed] [Google Scholar]
- (19).Becker JS, Jakubowski N. Chem. Soc. Rev. 2009;38:1969–1983. doi: 10.1039/b618635c. [DOI] [PubMed] [Google Scholar]
- (20).Glinka YY, Youdim MB. Eur. J. Pharmacol. 1995;292:329–332. doi: 10.1016/0926-6917(95)90040-3. [DOI] [PubMed] [Google Scholar]
- (21).Tarohda T, Ishida Y, Kawai K, Yamamoto M, Amano R. Anal. Bioanal. Chem. 2005;383:224–234. doi: 10.1007/s00216-005-3423-x. [DOI] [PubMed] [Google Scholar]
- (22).Popescu B, George M, Bergmann U, Garachtchenko A, Kelly M, McCrea R, Lüning K, Devon R, George G, Hanson A, Harder S, Chapman L, Pickering I, Nichol H. Phys. Med. Biol. 2009;54:651–663. doi: 10.1088/0031-9155/54/3/012. [DOI] [PubMed] [Google Scholar]
- (23).Faucheux BA, Martin ME, Beaumont C, Hauw JJ, Agid Y, Hirsch EC. J Neurochem. 2003;86:1142–1148. doi: 10.1046/j.1471-4159.2003.01923.x. [DOI] [PubMed] [Google Scholar]
- (24).Dexter DT, Carter CJ, Wells FR, Javoy-Agid F, Agid Y, Lees A, Jenner P, Marsden CD. J. Neurochem. 1989;52:381–389. doi: 10.1111/j.1471-4159.1989.tb09133.x. [DOI] [PubMed] [Google Scholar]
- (25).Salazar J, Mena N, Hunot S, Prigent A, Alvarez-Fischer D, Arredondo M, Duyckaerts C, Sazdovitch V, Zhao L, Garrick LM, Nunez MT, Garrick MD, Raisman-Vozari R, Hirsch EC. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18578–18583. doi: 10.1073/pnas.0804373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. J. Proteome Res. 2008;7:969–978. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- (27).Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. J. Proteome Res. 2004;3:289–295. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- (28).Lessner G, Schmitt O, Haas SJ, Mikkat S, Kreutzer M, Wree A, Glocker MO. J. Proteome Res. 2010;9:4671–4687. doi: 10.1021/pr100389u. [DOI] [PubMed] [Google Scholar]
- (29).Park B, Yang J, Yun N, Choe KM, Jin BK, Oh Y. J. Neurochem. Int. 2010;57:16–32. doi: 10.1016/j.neuint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- (30).Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates; Academic Press; San Diego: 2001. [Google Scholar]
- (31).Schäfer MK-H, Herman JP, Watson SJ. In Imaging Drug Action in the Brain. CRC Press; London: Boca Raton: 1992. p. 337. D. [Google Scholar]
- (32).Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. J. Am. Soc. Mass Spectrom. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Anal. Bioanal. Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kliman M, Vijayakrishnan N, Wang L, Tapp JT, Broadie K, McLean JA. Mol. Biosyst. 2010;6:958–966. doi: 10.1039/b927494d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mikawa S, Suzuki M, Fujimoto C, Sato K. Neurosci. Lett. 2009;451:45–49. doi: 10.1016/j.neulet.2008.12.035. [DOI] [PubMed] [Google Scholar]
- (36).Hare DJ, George JL, Grimm R, Wilkins S, Adlard PA, Cherny RA, Bush AI, Finkelstein DI, Doble P. Metallomics. 2010;2:745–753. doi: 10.1039/c0mt00039f. [DOI] [PubMed] [Google Scholar]
- (37).Oestreicher E, Sengstock GJ, Riederer P, Olanow CW, Dunn AJ, Arendash GW. Brain Res. 1994;660:8–18. doi: 10.1016/0006-8993(94)90833-8. [DOI] [PubMed] [Google Scholar]
- (38).Dexter DT, Wells FR, Lees AJ, Agid F, Agid Y, Jenner P, Marsden CD. J. Neurochem. 1989;52:1830–1836. doi: 10.1111/j.1471-4159.1989.tb07264.x. [DOI] [PubMed] [Google Scholar]