Abstract
Oncolytic measles virus strains are currently being evaluated in several clinical trials, as a promising novel oncolytic platform. Poliovirus receptor‐related 4 (PVRL4) was recently identified as a potent measles virus (MV) receptor; however, its regulation is not yet understood. Increased levels of PVRL4 protein were observed in cell membrane, cytoplasm and nuclei of glioblastoma, breast and ovarian tumor clinical samples with no significant change in PVRL4 mRNA levels in glioblastoma and breast cancer compared with their corresponding control samples, suggesting that PVRL4 is likely post‐transcriptionally regulated. Therefore, we sought to investigate the potential role of miRNAs in PVRL4 regulation and thus MV infectivity. We demonstrated that miR‐31 and miR‐128 can bind to the 3′UTR of PVRL4 and decrease PVRL4 levels while anti‐miR‐31/128 increase PVRL4 levels suggesting that PVRL4 is miRNA targeted. Furthermore, miR‐31/128 expression levels were down‐regulated in glioblastoma and breast tumor samples and showed significant negative correlations with PVRL4 levels. Infection with an MV strain that exclusively utilizes PVRL4 as its receptor showed that over‐expression of miR‐31/128 decreases MV infectivity while inhibition of the respective miRNAs via anti‐miRs increase MV infectivity and reduce tumor size in mouse xenograft models of glioblastoma, breast and ovarian cancer. Additionally, miR‐128 levels showed significant correlations with MV infection and in vivo anti‐tumor effect, while MV infection increased miR‐31 expression and thereby contributed to the observed decrease in PVRL4 levels. This study suggests that PVRL4 is post‐transcriptionally regulated by miR‐128 and miR‐31 and harbors possible miRNA targets that could modulate MV infectivity and in turn enhance MV based oncolytic therapeutic strategies.
Keywords: PVRL4, Measles virus, miRNA, Glioblastoma, Breast cancer, Ovarian cancer
Highlights
PVRL4 is post‐transcriptionally regulated by miR‐31 and miR‐128.
MiR‐31 and miR‐128 regulate measles virus infectivity and oncolysis through PVRL4.
Measles virus infection down‐regulates PVRL4 by up‐regulating miR‐31.
Abbreviation
- MV
Measles virus
1. Introduction
Measles virus (MV) is a primate specific virus that naturally infects humans, rhesus, marmosets, and squirrel monkeys (Albrecht et al., 1980; de Swart, 2009; Kobune et al., 1996). Despite the availability of a safe and cost‐effective vaccine, the increased measles related deaths worldwide in 2013 (Perry et al., 2014), and the recent outbreaks in the USA (Clemmons et al., 2015; Zipprich et al., 2015) signifies the importance of understanding the factors determining MV infectivity. MV is among the first viruses to be documented as an oncolytic virus (Bluming and Ziegler, 1971; Zygiert, 1971) and currently recombinant strains are being tested in clinical trials of glioblastoma (Institute) and ovarian cancer (Institute; Institute) as oncolytic therapy, while clinical applications for breast cancer are in development. MV attenuated vaccine strains have proceeded as promising oncolytic strategies due to their genetic stability, safety records (Nakamura and Russell, 2004) and early evidence of biologic activity (Delpeut et al., 2014; Msaouel et al., 2013). MV, like most morbilliviruses, has a well‐established receptor‐dependent lymphotropism and epitheliotropism (Noyce et al., 2011; Pratakpiriya et al., 2012; Tatsuo et al., 2000). MV receptor tropism to CD46 (Dorig et al., 1993; Naniche et al., 1993) (attenuated vaccine strains), SLAM/CD150 (Erlenhoefer et al., 2001; Tatsuo et al., 2000) and PVRL4/nectin‐4 (Delpeut et al., 2014; Muhlebach et al., 2011; Noyce et al., 2011; Noyce and Richardson, 2012) (both the vaccine and wild‐type viruses), has been well documented.
Since its discovery as an MV receptor in 2011 (Muhlebach et al., 2011; Noyce et al., 2011), PVRL4 has garnered increasing interest. Reymond et al. (2001) and the Human Protein Atlas Project (www.proteinatlas.org) have shown that PVRL4 is expressed not only in the placental trophoblasts, brain, lung, testis glandular cells of the stomach, but also adenocarcinomas of the lung, breast, ovary and glioblastoma. Elevated levels of PVRL4 have been observed in breast cancer cell lines (Fabre‐Lafay et al., 2005), and ovarian cancer tissues and sera (DeRycke et al., 2010). Furthermore, a recombinant SLAM blind wild‐type MV strain that singularly used PVRL4 as its receptor demonstrated greater oncolytic activity than the Edmonston vaccine strain using CD46 in subcutaneous xenograft models of breast cancer (Sugiyama et al., 2013). Moreover, PVRL4 has the strongest biochemical binding affinity to MV (K d) compared with both SLAM and CD46 (Muhlebach et al., 2011). Due to its selective distribution in comparison with the ubiquitously expressed CD46 and the lymphotropic SLAM, PVRL4 represents a promising tumor‐associated receptor for MV oncolytic therapy (Delpeut et al., 2014; Muhlebach et al., 2011; Noyce et al., 2011; Noyce and Richardson, 2012; Sugiyama et al., 2013). Noyce et al. (2011) has observed that most adenocarcinomas are susceptible to MV infection independent of CD46 and SLAM receptor use. In order to exploit PVRL4 to its full potential and thus selectively target, infect and eradicate cancer cells, it is important to understand the molecular mechanisms governing its regulation. However, the regulation of PVRL4 is not well understood.
MiRNAs have emerged as key regulators of gene expression in the past decade (Ambros, 2008; Berezikov, 2011). Most miRNAs regulate gene expression post‐transcriptionally by binding to the 3′UTR of the target mRNA and causing translational repression or degradation of the mRNAs (He and Hannon, 2004). Notably, miRNAs target ∼30% human genes (Lewis et al., 2005), regulate a remarkable number of cellular functions including differentiation, metabolism, aging, apoptosis, and tumorigenesis (Ameres and Zamore, 2013; Amiel et al., 2012; He and Hannon, 2004) and are differentially expressed in cancers (Croce, 2009; Lu et al., 2005; Volinia et al., 2006) including glioblastoma (Ciafre et al., 2005; Rao et al., 2010), breast (Blenkiron et al., 2007; Iorio et al., 2005) and ovarian (Yang et al., 2013; Zhang et al., 2006) cancers. Similarly, a growing number of reports suggest that oncolytic viruses (Barnes et al., 2008; Fu et al., 2012; Lee et al., 2009; Mazzacurati et al., 2015; Yao et al., 2014; Zhang et al., 2012) including MV (Baertsch et al., 2014; Leber et al., 2011), show preferential tropism to differential miRNA expression patterns where (artificial) insertion of miRNA target sequences (miRTs) is explored in this context. Therefore, we sought to investigate whether post‐transcriptional regulation of PVRL4 and thus MV infectivity is mediated by miRNAs.
2. Material and methods
2.1. Patient sample information
Frozen glioblastoma tumor samples (n = 18) and gliosis (control) samples (n = 12), breast cancer tumor samples (n = 15) and their corresponding control (non‐tumor) samples (n = 15), ovarian malignant tumor samples (n = 13 + 5) and corresponding benign tumor samples (control) (n = 13) from the same patient, were obtained from the Mayo clinic Glioma/Breast/Ovarian Specialized Program of Research Excellence (SPORE) programs. Patient information is included in Tables S1–S4. Out of the 12 gliosis samples 1 sample showed late CT (threshold cycle) values for the normalizing gene, RNU6, and thus had higher uncertainty in qRT‐PCR measurements. Subsequently, this sample was eliminated from miRNA analysis.
2.2. Lentivirus‐vector particle production
The miRs, anti‐miRs and PVRL4 lentiviral‐vector plasmids (Genecopoeia), Gag‐pol (packaging protein) and VSV‐G (envelop protein) (gifts from Eric Poeschla, MD) were produced and purified using TOP 10 competent cells (Invitrogen) and Hispeed plasmid maxi kit (Qiagen), and then transfected into 293T cells using Lentifectin (Applied biological materials Inc.) according to manufacturer's instructions. The constructs for miRs contained GFP reporter genes and puromycin selection markers while the constructs for anti‐miRs contained m‐cherry reporter genes and hygromycin selection markers. The vector particles were tittered via flow cytometry using the reporter genes.
2.3. Wild‐type MV production
MV vaccine strains utilize CD46, SLAM and PVRL4 as their receptors while wild‐type MV uses SLAM and PVRL4 as its receptor. This study addresses the regulation of an MV receptor, PVRL4. In order to effectively quantify the regulatory role of the respective miRNAs on PVRL4 and thus on MV infectivity it was important that the other receptors, CD46 and SLAM, did not interfere on MV infectivity. Thus, a wild‐type eGFP expressing MV strain that is SLAM blind and thus exclusively uses PVRL4 as its receptor (Leonard et al., 2010), a gift from Roberto Cattaneo, PhD, was used in this study. The virus was grown in MCF7 cells and tittered according to Reed and Muench, 1938 using the GFP reporter gene. The tittered virus was used in subsequent cell culture and mouse experiments. All MV and lenti‐viral vector studies were conducted according to BSL2+ guidelines.
2.4. Mice
2.4.1. U87 cell xenografts
Six week old athymic nude mice were injected subcutaneously with 1 × 106 stably transduced U87 cells containing lentiviral‐vectors expressing scramble miRNA (both scramble miR construct and anti‐miR construct) (5 mice), miR‐31/‐128 (5 mice), miR‐31/‐128 + PVRL4 (5 mice) and anti‐miRs‐31/128 (5 mice). Out of the 20 mice in the 4 groups, 1 mouse from the miR‐31/128 and 2 mice from the miR‐31/‐128 + PVRL4 group did not develop and thus, were eliminated from the study. At 6 weeks after tumor cell injection, tumor sizes were measured using Vernier caliper measurements and tumor volumes were calculated according to the formula, (greater diameter × greater diameter × smaller diameter)/2. Following tumor measurement all mice were treated intratumorally with SLAM blind WT‐measles virus strain at 2.5 × 103 infectious units (IU) per 62.5 mm2 tumor volume (Poisson distribution applied on MV batch titer). The mice received three consecutive viral doses at 3 and 2 day intervals respectively. Tumors were collected for RNA analysis and histopathology studies following tumor volume calculations and animal euthanasia. Tumor sizes were measured before every treatment. The tumor sizes calculated prior to the first MV dose and prior to euthanasia (48 h following the final MV dose in order to allow oncolytic activity from the final dose) were used for tumor size difference calculations. The tumor size difference was calculated by subtracting tumor volume prior to euthanasia from tumor volume prior to the first MV dose. Size differences were normalized to the mean scrambled miRNA size difference and calculated as percentages. At the end of the experiment, 1 mouse from the scramble miRNA group had no visible tumor and thus tissue from this mouse was not available for RNA analysis and histopathology analysis. Similarly, another mouse (1 mouse) from the same group, scramble miRNA, had a CT values that were too late and thus had higher uncertainty in qRT‐PCR measurements. Therefore, this mouse was eliminated from RNA analysis.
In a second experiment, 4 week old athymic nude mice were injected subcutaneously with stably transduced U87 containing lentiviral‐vectors expressing scramble miRNA (both scramble miR construct and anti‐miR construct) (5 mice), miR‐31/‐128 (5 mice), miR‐31/‐128 + PVRL4 (5 mice) and anti‐miRs‐31/128 (5 mice). For this experiment 1 × 108 cells were injected with 150 μL of PBS and equal volumes of matrigel (Corning). Two weeks after tumor cell injection, tumor sizes were measured using Vernier caliper measurements and tumor volumes were calculated according to the formula, (greater diameter × greater diameter × smaller diameter)/2. Following tumor measurement all mice were treated intratumorally with SLAM blind WT‐measles virus strain. The mice received four consecutive viral doses at 5 × 104 IU, 1.5 × 103 IU, 3 × 104 IU and 6 × 104 IU per 62.5 mm2 tumor volume respectively at 4, 3 and 2 day intervals. Tumors were collected for RNA analysis and histopathology studies following tumor volume calculations and animal euthanasia. Tumor sizes were measured before every treatment. The tumor sizes calculated prior to the first MV dose and prior to euthanasia (48 h following the final MV dose in order to allow oncolytic activity from the final dose) were used for tumor size difference calculations. The tumor size difference was calculated by subtracting tumor volume prior to euthanasia from tumor volume prior to the first MV dose. Size differences were normalized to the mean scrambled miRNA size difference and calculated as percentages.
In a third experiment, 4 week old athymic nude mice were injected subcutaneously with stably transduced U87 containing lentiviral‐vectors expressing scramble miRNA (both scramble miR construct and anti‐miR construct) (10 mice), miR‐31 (9 mice), anti‐miR‐31 (10 mice), miR‐128 (10 mice) and anti‐miR‐128 (10 mice). For this experiment 2 × 107 cells were injected with equal volumes of matrigel (Corning). 2.5 weeks after tumor cell injection, tumor sizes were measured using Vernier caliper measurements and tumor volumes were calculated according to the formula, (greater diameter × greater diameter × smaller diameter)/2. Following tumor measurement all mice were treated intratumorally with SLAM blind WT‐measles virus strain. The mice received four consecutive viral doses at 2.5 × 106 IU, 1.5 × 106 IU, 2 × 106 IU and 2.5 × 106 IU per 62.5 mm2 tumor volume respectively at 3, 4 and 2 day intervals. Tumors were collected for RNA analysis following tumor volume calculations and animal euthanasia. Tumor sizes were measured before every treatment. The tumor sizes calculated prior to the first MV dose and prior to euthanasia (48 h following the final MV dose in order to allow oncolytic activity from the final dose) were used for tumor size difference calculations. The tumor size difference was calculated by subtracting tumor volume prior to euthanasia from tumor volume prior to the first MV dose. Size differences were normalized to the mean scrambled miRNA size difference and calculated as percentages.
2.4.2. MCF7 cell xenografts
Four week old athymic nude mice were injected subcutaneously with 1 × 108 stably transduced MCF7 containing lentiviral‐vectors expressing scramble miRNA (both scramble miR construct and anti‐miR construct) (5 mice), miR‐31/‐128 (5 mice), miR‐31/‐128 + PVRL4 (5 mice) and anti‐miRs‐31/128 (5 mice) with 150 μL of PBS and equal volumes of matrigel. Two weeks after tumor cell injection, tumor sizes were measured using Vernier caliper measurements and tumor volumes were calculated according to the formula, (greater diameter × greater diameter × smaller diameter)/2. Following tumor measurement all mice were treated intratumorally with SLAM blind WT‐measles virus strain. The mice received four consecutive viral doses at 5 × 104 IU, 2 × 104 IU, 4 × 104 IU and 4 × 104 IU per 62.5 mm2 tumor volume respectively 4, 3 and 2 day intervals. Tumors were collected for RNA analysis and histopathology studies following tumor volume calculations and animal euthanasia. Tumor sizes were measured before every treatment. The tumor sizes calculated prior to the first MV dose and prior to euthanasia (48 h following the final MV dose in order to allow oncolytic activity from the final dose) were used for tumor size difference calculations. The tumor size difference was calculated by subtracting tumor volume prior to euthanasia from tumor volume prior to the first MV dose. Size differences were normalized to the mean scrambled miRNA size difference and calculated as percentages.
2.4.3. A2780 cell xenografts
Nine week old athymic nude mice were injected subcutaneously with 1 × 106 stably transduced A2780 containing lentiviral‐vectors expressing scramble miRNA (both scramble miR construct and anti‐miR construct) (5 mice), miR‐31/‐128 (5 mice), miR‐31/‐128 + PVRL4 (5 mice) and anti‐miRs‐31/128 (5 mice). Two weeks after tumor cell injection, tumor sizes were measured using Vernier caliper measurements and tumor volumes were calculated according to the formula, (greater diameter × greater diameter × smaller diameter)/2. Following tumor measurement all mice were treated intratumorally with SLAM blind WT‐measles virus strain. The mice received a single dose at 5 × 104 IU, per 62.5 mm2 tumor volume. A2780 cell xenografts are rapid growing tumors and thus reached IACUC approved tumor volumes before the opportunity to administer additional MV doses. Tumors were collected for RNA analysis and histopathology studies following tumor volume calculations and animal euthanasia. Tumor sizes were measured before every treatment. The tumor sizes calculated prior to the first MV dose and prior to euthanasia (96 h following the final MV dose in order to allow oncolytic activity from the final dose) were used for tumor size difference calculations. The tumor size difference was calculated by subtracting tumor volume prior to euthanasia from tumor volume prior to the first MV dose. Size differences were normalized to the mean scrambled miRNA size difference and calculated as percentages.
All animal procedures conducted were approved by the Institutional Animal Care and Use Committee at Mayo Clinic.
2.5. Luciferase assay
PVRL4 luciferase wild‐type 3′UTR expression clone and the mutant clones for miR‐31 and miR‐128 seed regions, containing firefly luciferase reporter gene and Renilla tracking gene, driven by the SV40 promoter (Genecopoeia) were transfected at 0.8 μg in to confluent 293T cells in 96 well plates using Lipofectamine 2000 (Invitrogen), following miRNA lenti‐vectors transduction at MOI = 2. The luciferase assay was conducted with a dual luciferase assay kit (Luc‐Pair miR Luciferase Assay Kit) (Genecopoeia) where the luciferase expression levels were normalized to Renilla expression levels.
2.6. Cell culture
U87 and MCF7 cells were from American Type Culture Collection and cultured according to Iankov et al. (2012, 2003). A2780 cells from Sigma were cultured in RPMI‐1640 with Glutamine + 10% fetal bovine serum.
2.7. Protein extraction and western blot analysis
Cells were lysed and human tissue were homogenized (using Cole‐Palmer, EW‐04727‐07 Labgen 7 Series Homogenizer) in lysis buffer (cell signaling) (Geekiyanage and Chan, 2011) using protease inhibitor cocktail (Sigma), NuPAGE 4–12% Bis‐Tris gels (Life technologies) and iblot gel transfer device (Life technologies). Blots were imaged using ChemiDoc (BioRad) and protein quantifications (intensity of bands) were conducted using Quantity One (BioRad) version 4.5, by normalizing to the respective loading controls. Primary antibodies included PVRL4 antibodies (Sigma, HPA010775, 1:1000 dilution), (Abcam, 155692, 1:1000 dilution), (Proteintech, 21903‐I‐AP, 1:1000 dilution), Anti‐Measles nucleoprotein antibody (Abcam, 23974, 1:1000 dilution), GAPDH antibody (Cell signaling, 14C10, 2118L, 1:2000 dilution), Histone H3 antibody (Cell signaling, 9715S, 1:1000 dilution), Cyclophilin A antibody (Cell signaling, 2175S, 1:1000 dilution) and α‐tubulin antibody (Sigma, T5168, 1:2000 dilution). Primary antibodies were incubated overnight at 4 °C. Secondary antibodies included HRP Goat Anti‐Mouse Ig (BD Pharmingen, 554002, 1:1000 dilution) and Anti‐rabbit IgG, HRP‐linked Antibody (Cell signaling, 7074S, 1:1000 dilution). Secondary antibodies were incubated for 1 h at room temperature.
PVRL4 is a 510‐amino acid transmembrane protein that has a predicted/calculated molecular weight of ∼55 kDa that has observed bands at ∼44, 55 and 66 kDa, depending on the antibody, cell/tissue type and N‐glycosylation in SDS polyacrylamide gels. The bands between molecular weights 50–75 kDa was used for quantification. We have tried >5 different commercially available antibodies against PVRL4. While most antibodies detect the recombinant PVRL4 protein they fail to detect the endogenous protein in cell lysates. Out of the antibodies we tried Abcam 155692, Proteintech 21903‐I‐AP and Sigma prestige HPA010775, were able to detect endogenous PVRL4 in cell lysates. After we observed that the Protein Atlas Project has validated Sigma prestige HPA010775 antibody, we conducted our studies using Sigma prestige HPA010775 antibody (all data in the main manuscript and most in supplemental data). Endogenous and recombinant PVRL4 protein migration profiles in breast cancer cells (MCF7), glioblastoma cells (U87) and ovarian cancer cells (A2780) are represented in Figure S1. It should be noted that when a protein is over‐expressed, at times cells may not be able to complete the necessary translational modifications and thus, may not show the higher molecular weight band but only show the lower molecular weight band. (The migration distance between molecular markers 50 kDa and 75 kDa may differ depending on the running durations of the SDS page).
2.8. Immunohistochemistry
Frozen human tissues and formalin fixed xenograft tissues were sectioned at 5 μm. Antigen retrieval and staining was conducted using Bond III biosystem, (PVRL4‐citrate, PH 6; MV‐F‐EDTA, pH 9) and polymer refine detection kit (Leica) according to manufacturer's recommendation. Primary antibodies were diluted in Dako background reducing diluent and incubated for 30 min. The frozen tissue samples were incubated in Dako‐X0909, while xenograft tissues were incubated in Rodent Block‐M (Biocare). PVRL4 primary antibody (Sigma, HPA010775) was used on brain (1:50), breast (1:100) and ovarian (1:200) tissue samples. MV fusion protein primary antibody (Abbiotec, 251363) was used on xenograft tissues at 1:100 dilutions. An Axioplan2 microscope was used with AxioCam HR camera and Axiovision software (version 4.7) to generate images under bright field, configured with 20x PlanNEOFLUAR (NA 0.50) and 40x N‐ACHROPLAN (NA 0.65).
2.9. Quantitative RT‐PCR
Total mRNA and miRNA analysis were performed according to Geekiyanage and Chan (2011) using LightCycler 480‐II (Roche) and software (version 1.5.0). MRNA samples with RIN values ≥ 7 were used in this study. Primers included human GAPDH: 5′‐GAGTCAACGGATTTGGTCGT‐3′ and 5′‐TTGATTTTGGAGGGATCTCG‐3′; PVRL4: 5′‐AGCCACTGACTTGTGTGGTG‐3′ and 5′‐CAGCCGTGTCCAGTTGTATG‐3′; MV‐nucleocapsid protein (N‐protein) RNA: 5′‐AGTGAGAATGAGCTACCG‐3′ and 5′‐TGTCTAGGGGTGTGCC‐3′ (Plumet and Gerlier, 2005). All miRNA primers (miScript primers, miR‐31, miR‐128, RNU6) were purchased from Qiagen and their efficiencies have been tested by the manufacturer. PVRL4 and GAPDH primers were designed using Primer3 (version 4.0) (Koressaar and Remm, 2007; Untergasser et al., 2012). The primer sequences for MV‐N were according to the original article (Plumet and Gerlier, 2005) where their efficiencies have been tested. We tested PVRL4 and GAPDH primer efficiencies and they had similar efficiencies (slope = ∼ −3.32). Relative expressions were calculated using the comparative CT method (2−ΔΔCT) (Livak and Schmittgen, 2001) normalizing to their corresponding GAPDH (for mRNA analysis) or RNU6B (for miRNA analysis) expressions.
3. Results
3.1. Elevated PVRL4 protein in malignant tumors
PVRL4 protein levels were detected in malignant tumor samples including, glioblastoma (n = 18) and their control gliosis (n = 15) samples, breast tumor (n = 15) and their corresponding normal breast tissue (control) (n = 15), and malignant ovarian tumor (matched (n = 13) or un‐matched (n = 5) to control) and their respective control benign tumor samples (n = 13). PVRL4 protein levels were significantly (Mann–Whitney U test) elevated in glioblastoma (1, 2A–H and Figure S2A–D), breast tumors (1, 2,J and Figure S8A–L) and malignant ovarian tumors (1, 2M,N and Figure S9A–D) compared with their respective control samples. IHC studies showed PVRL4 protein expression in cell membrane, cytoplasm and nuclei with punctated distribution, in glioblastoma (Figure 2Q–T, Figures S2F,H, S3–S7), gliosis (Figure S2E) breast tumors (Figure 2K,L and Figure S8M–P) and ovarian tumors (Figure 2O,P and Figure S9E–H). The presence of PVRL4 in glioblastoma, although at strongly varying degrees, was further validated in patient derived primary glioblastoma cells via western blotting (Figure S10A) and flow cytometry analysis (Figure S10B–O). Additionally, the expression of PVRL4 in both the cytoplasm and nuclei were further observed through western blot by separating the cellular cytoplasmic and the nuclear fractions (Figure S10P). Only ovarian malignant tumors (Figure 1I) expressed increased PVRL4 mRNA levels corresponding to the elevated PVRL4 protein levels, while glioblastoma (Figure 1C) and breast tumor (Figure 1F) samples showed predominantly similar PVRL4 mRNA expression levels compared with their control samples. Thus, these results suggest that PVRL4 is likely post‐transcriptionally regulated at least in glioblastoma and possibly in breast tumors. Therefore, we further investigated the potential post‐transcriptional regulation of PVRL4 by miRNAs.
Figure 1.
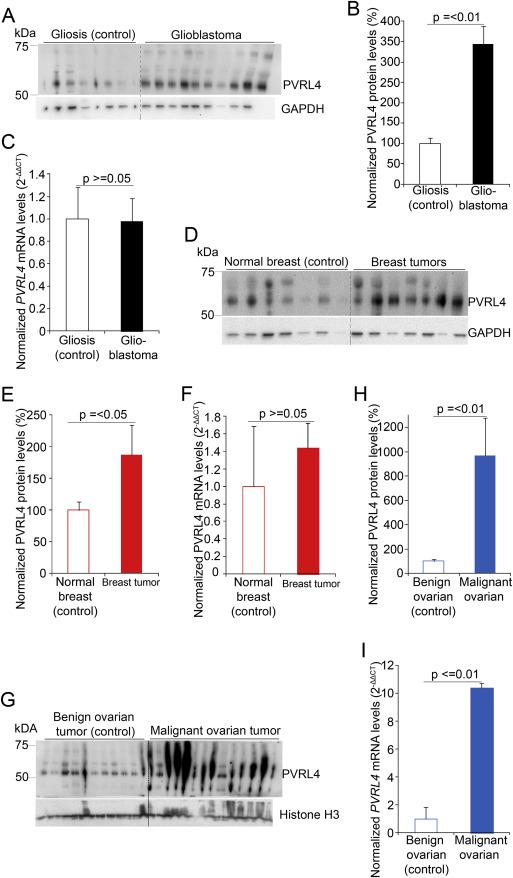
PVRL4 protein levels are increased with unchanged PVRL4 mRNA levels in malignant tumors. Representative western blots and quantification of blots in glioblastoma (A and B) (n = 18), breast cancer (D and E) (n = 15) and ovarian cancer (G and H) (n = 18) patient samples showing increased PVRL4 protein levels in malignant tumor samples compared with control tissues, gliosis (n = 12), normal breast (n = 15) or benign ovarian tumors (n = 13). PVRL4 protein levels were quantified by normalizing to GAPDH or Histone H3 and represented as a percentage of the mean values of the control tissues. PVRL4 mRNA expressions were quantified by qRT‐PCR for glioblastoma (C), breast tumor (F) and ovarian tumor (I) samples. Relative expressions shown are normalized to GAPDH and mean expression levels of control samples. Error bars represent standard error of mean (SEM). Statistical significances were determined by Mann–Whitney U tests.
Figure 2.
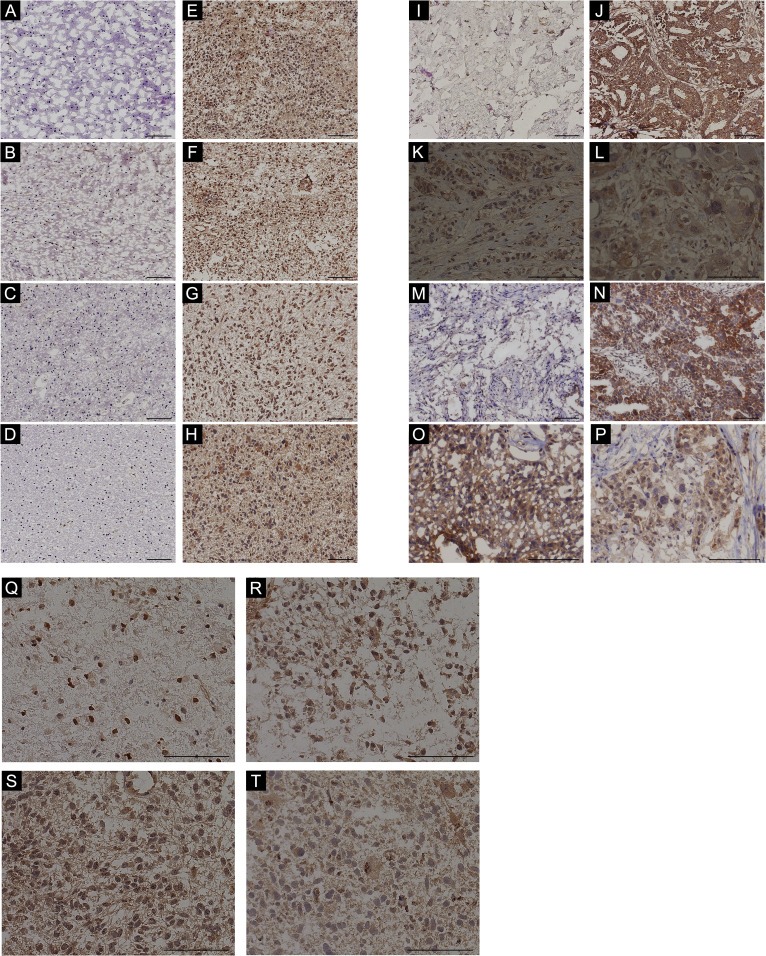
Increased PVRL4 IHC staining in malignant tumors. Gliosis (control) (A–D) glioblastoma (E–F), normal breast (G), breast tumor (H), benign ovarian (K) and malignant ovarian tumor (L) samples stained with PVRL4 antibody (X20). Representation of PVRL4 staining (X40) in cytoplasm and nuclei with punctated distribution in glioblastoma (O–R), breast (I and J) and ovarian malignant (M and N) tumor tissues. Scale bars indicate 100 μm.
3.2. PVRL4 is a miRNA targeted gene
Prediction algorithms, miRNA.org (Betel et al., 2010) and RegRNA (Chang et al., 2013), were incorporated to select potential miRNAs that target PVRL4 mRNA. Common miRNA predictions between miRNA.org and RegRNA were selected and these miRNAs were further filtered according to literature reports of miRNA down‐regulated in glioblastoma, breast cancer and ovarian cancer. Of the miRNAs predicted by the two algorithms, miR‐31 and miR‐128 are reportedly down‐regulated in glioma (Karsy et al., 2012; Mizoguchi et al., 2012; Visani et al., 2014), breast (Qian et al., 2012; Yan et al., 2008; Zhang et al., 2006) and ovarian cancers (Banno et al., 2014; Dahiya and Morin, 2010; Wyman et al., 2009). Luciferase reporter constructs were generated containing the human 3′UTR of PVRL4. Plasmids expressing the respective miRNAs were co‐transfected with the constructs and the luciferase expression was detected in 293T cells. MiR‐31 and miR‐128 significantly decreased the luciferase expression (Figure 3A). These results were confirmed by transducing cells (glioblastoma, breast and ovarian cancer) with lentiviral‐vectors expressing miRs or anti‐miRs (antisense) for miR‐31 and miR‐128. MiR‐31 and miR‐128 suppressed endogenous PVRL4 expression and anti‐miRs‐31 and ‐128 rescued the endogenous PVRL4 levels upon transduction in glioblastoma (U87) (Figure 3B), breast tumors (MCF7) (Figure 3C) and ovarian tumors (A2780) (Figure 3D). It should be noted that many factors could affect the demonstrated in vitro miRNA‐mRNA interaction. First, the cellular machinery may not be able to support the biogenesis of miRNA at the concentration of transduction (lentiviral‐vectors expressing miRs or anti‐miRs) and therefore, may only produce a fraction of the miRNAs or anti‐miRS transduced. Second, miRNA‐mRNA interactions are both spatial and temporal. Thus, the full effect of miR‐31 and miR‐128 may only be detected at a particular time point. The half‐life of PVRL4 mRNA/protein also affects this time point. Third, miRNAs are moderate regulators and the naturally occurring noise in biological samples could interfere with the accurate measurement of relative changes. Fourth, the abundance of endogenous PVRL4 mRNA/protein can influence the prominence of the changes (van Rooij, 2011). Taking these points to account, miR‐31 and miR‐128 could possibly assert more prominent changes than what we observe. Furthermore, due to the high affinity between PVRL4 and MV, subtle changes in PVRL4 protein levels could have significant impact on infection outcome. Confocal microscopy studies further validated the suppressing effect of miRs‐31 and ‐128 and the enhancing effect of anti‐miRs‐31 and ‐128 on PVRL4 levels in glioblastoma cells (U87) (Figure S5).
Figure 3.
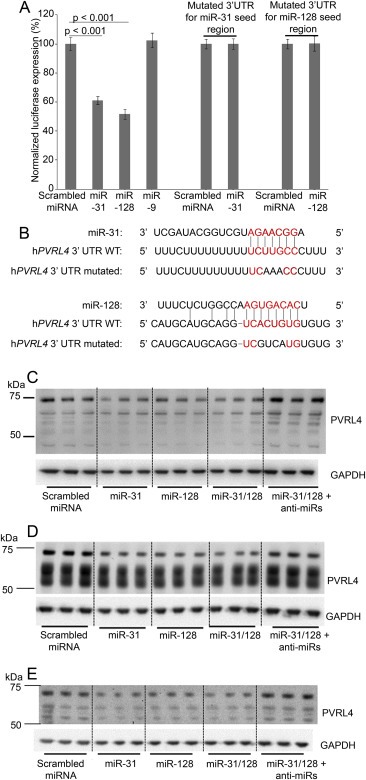
MiR‐31 and miR‐128 regulates PVRL4. Human PVRL4 3′UTR firefly luciferase (wild‐type or mutant (see Figure 3B)) and Renilla luciferase plasmid constructs were transfected (0.8 μg in 96 well plates) into 293T cells with scrambled miRNA, miR‐31, miR‐128 and miR‐9 transduced at MOI = 2. Normalized (to Renilla) sensor luciferase activity is shown as a percentage of luciferase expression in scrambled miRNA transfection (A). Error bars represent standard error of mean (SEM) derived from three or more experiments. Statistical significance between scrambled miRNA and candidate miRs treated cells were determined by 2‐tailed student t tests. The putative binding sites of PVRL4 to miR‐31 and miR‐128 and miRNA seed regions are shown in red font. The binding sites of hPVRL4 3′UTR mutant to miR‐31 and miR‐128 seed regions are indicated in black font within the red font binding sites (B). Western blot analysis of endogenous PVRL4 in U87 (C), MCF7 (D) and A2780 (E) cells, following transduction with lentivirus vectors containing miRs or anti‐miRs (inhibitor) using scrambled miRs/anti‐miRs as controls. Three or more independent experiments were conducted with cells collected at 4 or 7 days post transduction (MOI = 1) for the respective miRs and anti‐miRs.
3.3. Reduced expressions of miR‐31 and ‐128 correlate with PVRL4 levels in glioblastoma
The expression levels of miR‐31 and miR‐128 were significantly down‐regulated in glioblastoma (n = 18) (Figure 4A and B) and breast tumors (Figure 4C and D) compared with their respective controls, gliosis (n = 11) and normal breast tissue (n = 15). Although, other reports (Banno et al., 2014; Dahiya and Morin, 2010; Wyman et al., 2009) have shown differential expression levels of miR‐31 and miR‐128 in ovarian tumors, the malignant ovarian tumors tested in our study did not show a significant change in miR‐31 and ‐128 compared with their benign tumor controls. Differences in cellularity observed between ovarian tumors may have contributed to the differences in observations. Alternatively, benign ovarian tumors may not represent the ideal controls for malignant ovarian tumors. Statistically significant negative correlations were observed between miR‐31 (Figure 4E), miR‐128 (Figure 4F) and PVRL4 protein in gliosis and glioblastoma tissues. A statistically significant negative correlation was observed between miR‐31 (Figure 4G) and PVRL4 protein in normal breast and breast tumor tissues, while there was no significant correlation between miR‐128 and PVRL4.
Figure 4.
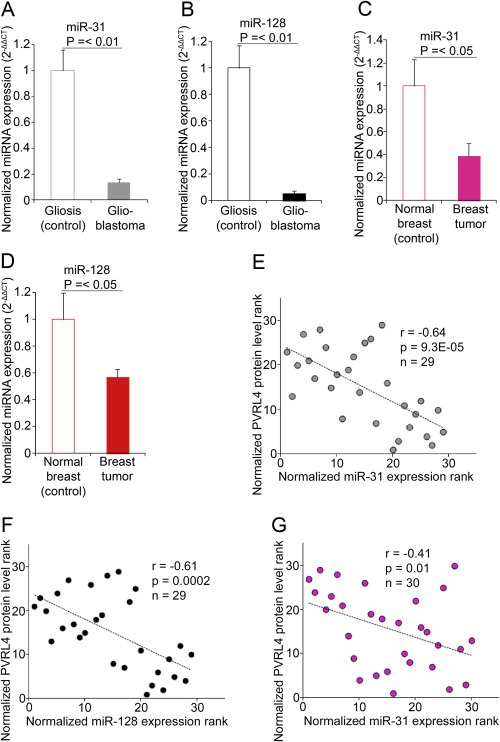
MiR‐31 and miR‐128 expressions correlate with PVRL4 levels. QRT‐PCR quantification of miR‐31(A) and miR‐128 (B) expression levels in gliosis (n = 11) and glioblastoma (n = 18) tissues. QRT‐PCR quantification of miR‐31 (C) and miR‐128 (D) expression levels in normal breast (n = 15) and breast tumor (n = 15) tissue. Relative expressions shown are normalized to RNU6B and mean control expressions. Statistical comparisons were performed using Mann–Whitney U tests. Spearman's correlation analysis demonstrated significant negative correlations between miR‐31 (E),‐miR‐128 (F) expressions and PVRL4 protein levels in gliosis and glioblastoma samples. Spearman's correlation analysis demonstrated significant negative correlations between miR‐31 expressions (G) and PVRL4 protein levels in breast tumor and their respective control samples. The significance of the correlation was determined by two‐tailed T distribution tests.
The tumor microenvironments vary between tumors type, tumor grade and between patients, and therefore, prone to variations in biomarker expression levels. Although a causal relationship has not been established, the PVRL4 promoter has transcription factor binding sites for GATA‐3 and P53 (Kent et al., 2002) and show correlation with their expressions (Fabre‐Lafay et al., 2007), suggesting possibility of transcriptional regulation. This transcriptional regulation can contribute to less than perfect co‐efficient. In the ever changing tumor microenvironment biomarker levels can vary in response to cellular functions and contribute to variations and co‐efficient.
3.4. MiR‐31 and mir‐128 modulates MV infectivity
High copy numbers levels have been observed for miR‐31 in normal astrocytes (340 copies/pg small RNA) (Wu et al., 2009) and miR‐128 in normal brain tissue (Nuovo et al., 2009) (Most of our experiments, cell culture and mouse studies, were conducted by transducing lenti‐vectors expressing miRNAs at MOI of 1–2 (1–2 copies per cell)). In respect to glioblastoma (U87), consistent with other research (Skalsky and Cullen, 2011; Ciafre et al., 2005) we demonstrate very low miR‐128 expression levels in comparison with breast cancer cells (MCF7) (Figure S11B). Comparable miR‐31 expression levels were observed in glioblastoma (U87) in comparison with breast cancer cells (MCF7) (Figure S11A). MiR‐31 and miR‐128 expression levels although low in glioblastoma U87 cells, are detectable. In our studies, miR‐31 and miR‐128 levels in cells are manipulated via lentiviral‐vectors in respect to miR‐31 and miR‐128 base levels of expressions (cells expressing scrambled miRNA) (Figure S11C and D).
Gain‐ and loss‐of‐function experiments were performed to identify the effect of miR‐31 and miR‐128 on MV infectivity. A wild‐type MV that exclusively utilizes PVRL4 as its receptor was employed (Leonard et al., 2010). This is the MV strain used throughout this study. Cells transduced with miRs alone or in combination with PVRL4 and anti‐miRs were infected with MV. MV N‐protein levels were evaluated to assess virus infectivity. Over‐expression of miR‐31 and ‐128 via lentiviral‐vectors down‐regulated the endogenous PVRL4 and MV N‐protein levels in U87 (Figure 5A,B and D, Figure S12), MCF7 (Figure 5C,E and Figure S13A) and A2780 (Figure S13B–D) cells. Over‐expression of PVRL4 via transduction with lentiviral‐vectors restored/increased MV infectivity in U87 glioma cells (Figure 5A,B and C), MCF7 breast cancer cells (Figure 5C,E and Figure S13A) and A2780 ovarian cancer cells (Figure S13B–D) cells as shown by increased MV N‐protein levels. Inhibition of miR‐31 and ‐128, through transduction of anti‐miRs‐31 and ‐128, increase endogenous PVRL4 and in turn, up‐regulate MV infectivity in U87 (Figure 5A and C), MCF7 (Figure 5B,D and Figure S13A) and A2780 (Figure S13B–D) cells. It should be noted that MV infection down‐regulates PVRL4 levels in cells (Figure 6L and Figure S18) and therefore, PVRL4 levels post MV infection may not indicate the PVRL4 levels prior to MV infection.
Figure 5.
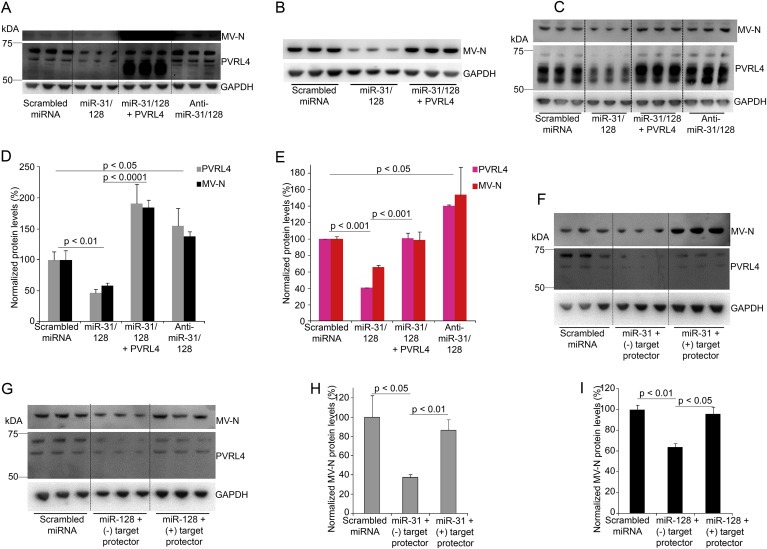
MiR‐31 and miR‐128 modulates MV infectivity. Western blot analysis and blot quantification of PVRL4 and MV N‐protein levels following transduction with lentiviral‐vectors (MOI = 1) expressing miRNA and anti‐miRs alone or in combination with lentiviral‐vectors expressing human PVRL4, and 2, 3, 5 or 7 days post MV (exclusively utilizes PVRL4 as receptor) infection (MOI = 0.1, 2 and 1) in U87 (A, B and D) and MCF7 (C and E) cell lines. Western blot quantification of virus N‐protein demonstrating MV infection (2–3 days post infection (7000–15,000 IU/well)) following transfection of miR‐31 and miR‐128 mimics at a final concentration of 10 nM along with negative or positive “target protectors” for miR‐31 (F and H) or miR‐128 (G and I) seed regions at a final concentration of 500 nM in U87 cells. Error bars represent standard error of mean (SEM) derived from three or more experiments. Statistical analyses were conducted by student t tests.
Figure 6.
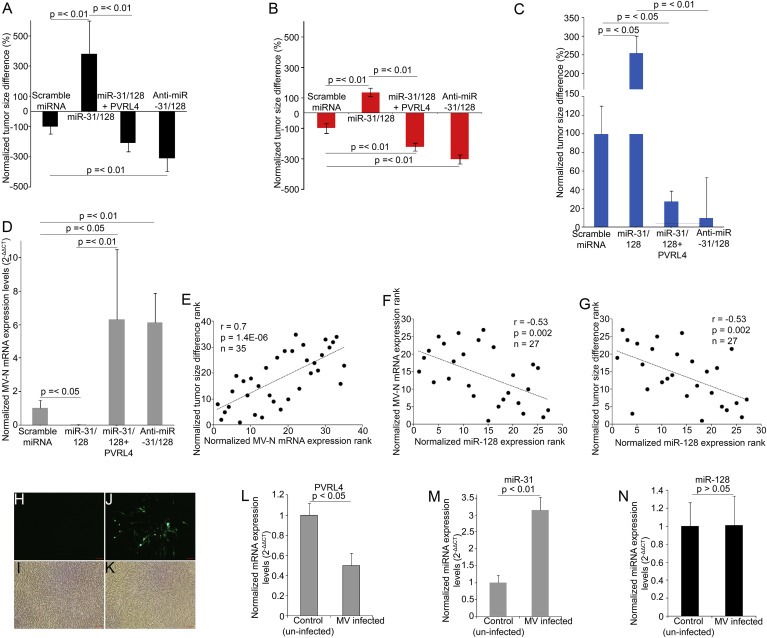
Anti‐miR‐31/‐128 increases MV infectivity and reduces tumor size while miR‐31 regulates MV mediated PVRL4 decrease. Tumor size differences post subcutaneous intratumoral MV administration in U87 (A), MCF7 (B) and A2780 (C) xenografts that were stably transduced with lentiviral‐vectors containing miRs alone or in combination with PVRL4 and anti‐miRs. Tumor sizes were measured before the administration of the first dose of the virus and two days after the last dose immediately prior to euthanasia in U87 groups, scrambled miRNA (n = 10), miR‐31/‐128 (n = 9), miR‐31/‐128 + PVRL4 (n = 8) and anti‐miRs‐31/‐128 (n = 10), MCF7 and A2780 groups, scrambled miRNA (n = 5), miR‐31/‐128 (n = 5), miR‐31/‐128 + PVRL4 (n = 5) and anti‐miRs‐31/‐128 (n = 5). RT‐PCR quantification of tumor MV N‐protein mRNA levels in stably transduced U87 mice at euthanasia (D). Statistical analysis was performed by Mann–Whitney U tests. Spearman's correlation test demonstrates significant positive correlation between tumor MV N‐protein mRNA levels and tumor size differences (E). Spearman's correlation test demonstrates significant negative correlation between tumor miR‐128 expression levels and tumor MV N‐protein mRNA levels (F). Spearman's correlation test demonstrates significant negative correlation between tumor miR‐128 expression levels and tumor size differences (G). The significances of the correlations were determined by two‐tailed T distribution tests. Confluent U87 cells (6 well plates) were infected with MV (150 μL of 106 TCID50). Cells were imaged and collected 7 days post infection. Representative fluorescence (H) and phase contrast of un‐infected (I) cells. Representative fluorescence (J) and phase contrast (K) of GFP expressing MV infected cells. PVRL4 (L) miR‐31 (M) and miR‐128 (N) expression levels were quantified by qRT‐PCR in MV infected and un‐infected cells. Relative expression levels shown are normalized to GAPDH or RNU6B and mean control (un‐infected cells) expressions. Error bars represent standard error of mean (SEM) derived from three or more experiments. Statistical significances were determined by student t tests.
To evaluate the direct role of miR‐31, miR‐128, and thus PVRL4 on MV infectivity, target protectors that prevent the binding of the respective miRNA seed regions to the target mRNA, were designed against the targeted sites on PVRL4 for miR‐31 and miR‐128. U87 cells were transiently transfected with miR‐31 or miR‐128 mimics along with their respective target protectors. MV N‐protein levels were assessed, to identify virus infectivity levels in miRs and target protector transfected cells. MV N‐protein levels decreased significantly upon transfection with miR‐31 (Figure 5F and H) or miR‐128 (Figure 5G and I) along with a negative target protector. MV N‐protein levels remained unchanged upon transfection with miR‐31 (Figure 5F and H) or miR‐128 (Figure 5G and I) along with their respective target protectors. The MV strain used in this study expresses GFP, allowing fluorescence imaging to detect MV infection. By preventing the binding of miR‐31 (Figure S14A–F) and miR‐128 (Figure S14G–L) to PVRL4, target protectors for miR‐31 and miR‐128 allowed MV infection comparable to scrambled miRNA transfected cells, while transfection of miR‐31 or miR‐128 with negative target protectors that do not bind to any specific mRNA and therefore allow miR‐31 and ‐128 to reduce PVRL4 levels, showed decreased MV infectivity. Similar results were observed with co‐transfection with miRNA and target protectors for miR‐31 and miR‐128 in MCF7 (Figure S15A,B and E–P) and A2780 (Figure S15C and D) cells respectively. These results together suggest that miR‐31 and miR‐128 modulates MV infectivity by directly binding and regulating PVRL4.
3.5. MiR‐31 and ‐128 modulates MV infectivity and tumor size
In order to investigate the regulatory effect of miR‐31 and ‐128 on MV infectivity in tumors, U87, MCF7 and A2780 cells were stably co‐transduced with lentiviral‐vectors containing miRs alone or in combination with PVRL4 and anti‐miRs, and were subcutaneously xenografted into athymic nude mice. Implanted U87 and MCF7 tumors were subjected to 3 or 4 consecutive MV injections at 1 × 106 TCID50, while A270 groups received a single dose of virus prior to euthanasia (see full description in Material and methods). Tumor size measurements obtained prior to administration of the first dose of virus and immediately prior to euthanasia were used for tumor size difference calculations.
Tumor sizes were decreased in U87 and MCF7 tumors containing scrambled miRNA, miR‐31/‐128 + PVRL4 and anti‐miRs‐31/‐128, while tumors containing miR‐31/‐128 showed increase in size after MV administration. The size differences between miR‐31/‐128, anti‐miRs‐31/‐128 and scrambled miRNA (control) groups and size differences between miR‐31/‐128 and miR‐31/‐128 + PVRL4 groups are significantly different in U87 (Figure 6A) and MCF7 (Figure 6B) xenografted tumor groups, where the reduction in tumor size in anti‐miRs‐31/‐128 groups are significantly greater than the reduction of tumor size in scrambled miRNA (control) groups. In A2780 (Figure 6C) xenografts, the miR‐31/‐128 group showed increase in tumor size compared with the scrambled miRNA (control) group while the miR‐31/‐128 + PVRL4 group showed less tumor growth compared with the scrambled miRNA (control) and miR‐31/‐128 groups. However, in this A2780 model, the least tumor growth observed in the anti‐miRs‐31/‐128 group did not reach statistical significance (p = 0.09, Mann–Whitney U test) compared with the scrambled miRNA (control) group. This may possibly be due to the small animal number, the fast growth of the tumors compared with U87 and MCF7 models and the single dose of MV administered compared with the multiple doses in the other two models.
Statistically significant increase in the MV N‐protein mRNA levels were observed in miR‐31/‐128 + PVRL4 and anti‐miRs‐31/‐128 groups compared with scrambled miRNA (control) group in U87 (Figure 6D) tumor xenografts, while a statistically significant reduction in MV N‐protein mRNA levels was observed in the miR‐31/‐128 group compared with the scrambled miRNA (control) group, explaining the tumor size differences. MV infection was visualized in U87 tumor xenograft by IHC staining for MV fusion protein (Figure S16). A statistically significant positive correlation was observed between tumor size differences and MV N‐protein mRNA levels in U87 xenografts (Figure 6E) validating the impact of miRNAs on MV infectivity and thus virus oncolysis. Furthermore, statistically significant negative correlations were observed between tumor miR‐128 expression levels and MV N‐protein mRNA levels (Figure 6F) and tumor size differences (Figure 6G), in scrambled miRNA, miR‐31/‐128 and anti‐miRs‐31/‐128 groups, further confirming that miR‐128 modulates MV infectivity in glioblastoma. Statistically significant correlations were not observed between miR‐31 expression levels and MV levels and tumor size differences.
Similarly, miR‐31 and miR‐128 were over‐expressed and inhibited individually in U87 cells via lentiviral‐vectors. 2 × 107 cells were subcutaneously xenografted on to athymic nude mice and MV was administered 2.5 weeks after tumor implantation at 1 × 108 TCID50. Tumor size difference showed (Figure S17A) similar trend to Figure 6A, where over‐expression of the respective miRNAs led to tumor growth, possibly due to lack of MV infection. Inhibition of the miRNAs resulted in reduction in tumor growth, a possible outcome of increased MV infection and anti‐tumor activity. Statistically significant positive correlation was observed between tumor MV infection (MV‐N mRNA expression levels) and tumor size difference indicating the relationship between MV infection and anti‐tumor activity (Figure S17B). While tumor miR‐128 levels showed statistically significant negative correlations with tumor size differences (Figure S17C) and MV infection levels (Figure S17D), miR‐31 did not.
Consistent with prior research (Noyce et al., 2011), PVRL4 protein levels are decreased following MV infection (Figure 6L and Figure S18). In order to investigate whether this down‐regulation of PVRL4 is mediated by a change in miR‐31 expression induced by MV infection, we measured the miR‐31 expression levels in U87 cells following MV infection (Figure 6H–K). MV infection significantly up‐regulated miR‐31 expression levels (Figure 6M) while miR‐128 expression levels (Figure 6N) remained unchanged between infected and un‐infected cells.
4. Discussion
We found that PVRL4 protein levels were increased in glioblastoma patient samples with PVRL4 present in cell membrane, cytoplasm and the nuclei. Also confirmed by our study, other studies have demonstrated increased levels of PVRL4 in breast cancer and ovarian cancer suggesting it as a potential biomarker for cancer diagnosis. Similarly, our findings suggest that PVRL4 may potentially serve as a diagnostic marker for glioblastoma. Prospective independent validations with larger cohorts are warranted. The increase in PVRL4 protein was observed with no change in PVRL4 mRNA levels suggesting that PVRL4 is post‐transcriptionally regulated. We and others (Karsy et al., 2012; Mizoguchi et al., 2012; Visani et al., 2014) have shown that miR‐31 and miR‐128 expression levels are down‐regulated in glioblastoma. We further observed negative correlations between the expression levels of miR‐31/‐128 and PVRL4 protein levels in glioblastoma patient samples. This in combination with our luciferase UTR assays, gain‐ and loss‐of‐function cancer cell culture data and target protector studies suggests that PVRL4 is directly regulated through miR‐31 and miR‐128.
PVRL4 can enhance tumor growth through enabling cell‐to‐cell attachment and matrix‐independent integrin β4/SHP‐2/c‐Src activation (Pavlova et al., 2013) while blocking of PVRL4 can inhibit tumor growth suggesting PVRL4 as a potential therapeutic strategy against breast cancer (Pavlova et al., 2013). MiR‐128 has been shown to inhibit proliferation, tumor growth (Dong et al., 2014; Peruzzi et al., 2013; Zhang et al., 2009), differentiation (Guidi et al., 2010) and angiogenesis (Shi et al., 2012) in glioma. Similarly, miR‐31 has been identified to inhibit migration and proliferation while the loss of miR‐31 increases tumor growth of glioblastoma cells (Rajbhandari et al., 2015). Correspondingly, miR‐31 and miR‐128 inhibit metastasis of breast (Korner et al., 2013; Liu et al., 2014; Masri et al., 2010) and ovarian tumors (Creighton et al., 2010; Woo et al., 2012). These reports further support our findings that PVRL4 is post‐transcriptionally regulated by miR‐31/‐128.
In addition to the tumor types investigated in this study, miR‐128 is down‐regulated in prostate (Schaefer et al., 2010), lung (Weiss et al., 2008), colorectal (Takahashi et al., 2014) and acute lymphocytic leukemia (Kotani et al., 2010). MiR‐128 can be down‐regulated due to loss of heterozygosity in chromosome 3p, the loci for miR‐128‐2, as observed in several cancers including neuroblastoma/glioblastoma (Ciafre et al., 2005) and breast cancer (Qian et al., 2012). Similarly, the miR‐128 aberrations observed could be a result of DNA methylations of the miR‐128 promoter region (Takahashi et al., 2014) and point mutations (Kotani et al., 2010). The down‐regulation of miR‐128 has also been attributed to direct binding of the zinc finger regulatory transcription factor SNAIL, which is expressed in elevated levels in tumors, to the miR‐128 promoter (Qian et al., 2012). Implicated in neurodegenerative diseases (Lee et al., 2011; Lukiw, 2007), miR‐128 is one of the most abundant and highly enriched miRNA in the human brain (Skalsky and Cullen, 2011; tenOever, 2013).
MIR31HG, which encodes miR‐31, flanks the most frequently deleted loci, 9p21.3, CDKN2A/B (cyclin‐dependent kinase inhibitor 2A) in glioblastoma (Network, 2008; Parsons et al., 2008) and this deletion is associated with poor prognosis (Rajbhandari et al., 2015). Other than glioblastoma (Visani et al., 2014), breast (Yan et al., 2008; Zhang et al., 2006) and ovarian (Creighton et al., 2010) cancers, miR‐31 expression is down‐regulated in prostate (Schaefer et al., 2010), stomach (Zhang et al., 2010), urothelial (Veerla et al., 2009), acute lymphoblastic leukemia (Usvasalo et al., 2010), melanoma (Zhang et al., 2006) and bladder cancers (Wszolek et al., 2011). Altered miR‐31 expressions could be due to regulation by TNF and BMP‐2, epigenetic silencing due to promoter hypermethylation (Augoff et al., 2012) or demethylation of H3K4me3 by EMSY (Vire et al., 2014) or post‐transcriptional regulation (Lee et al., 2008).
The wild‐type MV used in this study exclusively utilizes PVRL4 as its receptor and is blind to other receptors, CD46 and SLAM, thus providing a platform to effectively investigate the miRNA mediated PVRL4 regulation of MV infectivity. Cell culture and mouse xenograft data indicate that miR‐31/‐128 regulate MV infectivity and thus can have a significant impact on MV mediated tumor size. The similar negative correlations (r = 0.53) between both miR‐128 and virus infectivity and miR‐128 and tumor size further support that PVRL4 and MV infectivity are regulated by miR‐128. The strong correlation (r = −0.86) between miR‐128 (individual transduction) and tumor size difference in comparison with the moderate correlations between, miR‐128 (individual transduction) and MV infection (r = −0.86), and MV infection and tumor size difference, may suggest interplays between direct impact of miRNA levels on tumorigenesis, varying miRNA and PVRL4 levels in response to MV infection and other off‐target mechanisms. We demonstrated that miR‐31 and miR‐128 mediate MV infectivity through direct regulation of PVRL4 using site‐directed mutagenesis and target protector studies. However, we cannot rule out the effects of off‐target activities in a relatively uncontrolled (compared to in vitro) in vivo model.
Cellular miRNAs could regulate viral infectivity by binding to viral RNAs and thereby directly promoting or limiting viral replication as in the case of miR‐122, where it promotes HCV RNA replication (Jopling et al., 2005; Randall et al., 2007), or by modulating endogenous mRNA targets and indirectly promoting or limiting virus replication (Gottwein and Cullen, 2008). Oncolytic viruses are not only used to deliver miRNAs to tissues (Lou et al., 2013) but they can be engineered to utilize host miRNAs to control viral tropism (Baertsch et al., 2014; Barnes et al., 2008; Leber et al., 2011; tenOever, 2013). PVRL4 levels are known to decrease with MV infection (Noyce et al., 2011) similar to other MV receptors, CD46 (Schneider‐Schaulies et al., 1995) and SLAM (Welstead et al., 2004). Furthermore, several viruses including cytomegalovirus (Buck et al., 2010), Herpesvirus saimiri (Cazalla et al., 2010) and hepatitis C virus (Luna et al., 2015) regulate host miRNAs. Our data demonstrate that miR‐31 expression levels were up‐regulated post MV infection in U87 glioblastoma cells. We also show that miR‐31 post‐transcriptionally regulates PVRL4. Therefore, we hypothesize that this up‐regulation of miR‐31 following MV infection could contribute to the down‐regulation of PVRL4. Additional ongoing work will help us to understand whether MV up‐regulates host miR‐31 (down‐regulate PVRL4) as a survival mechanism (low receptor levels lead to slow infectivity and thus long‐term survival in the host by keeping the host alive) or whether the host up‐regulates its miR‐31 (down‐regulate PVRL4) as a defense mechanism to MV infection.
Although patient breast and ovarian tumor PVRL4 levels did not show strong correlations with miR‐31 and miR‐128, breast and ovarian cell culture results suggest that miR‐31/‐128 may contribute at least in part to the regulation of PVRL4 in breast and ovarian tumors. The concurrent increase in PVRL4 mRNA with PVRL4 protein in ovarian tumors also suggests potential transcriptional regulation. The strong statistically significant negative correlations between miR‐128 and PVRL4 levels in glioblastoma patient samples, and miR‐128 levels and MV infectivity levels in glioblastoma tumor xenografts, in combination with tumor size differences and gain‐ and loss‐of‐function tissue culture study results suggest that loss‐of‐function of miR‐128 results in reduced suppression of PVRL4 and thereby increased MV infectivity. This relationship between miR‐128 and MV infectivity can possibly explain selective MV infection in tumor with down‐regulated miR‐128 expression levels. The in vivo mouse xenograft studies suggest that the impact of miR‐128 and possibly miR‐31 on oncolytic infectivity is far greater than any possible proliferative effect caused by PVRL4 and the anti‐miRs. Although, it may not be practical to further inhibit miRNAs that are already down‐regulated in malignant tumors in efforts to increase MV infectivity and thus oncolysis, depending on the tumor type, stage, grade and miRNA expression profiles, the respective miRNAs can potentially be utilized at least in part to regulate MV infectivity and MV based oncolytic strategies.
PVRL4 was identified as a MV receptor in 2011, while CD46 and SLAM was identified as receptors in 1993 and 2000 respective. Much research on MV infection and anti‐tumor activity has been in respect to CD46 and SLAM in comparison to the more recently identified PVRL4 and therefore, we may not fully understand the contribution of PVRL4 as a MV receptor. Further, studies are warranted to understand the contribution of PVRL4 in MV oncolytic virotherapy in comparison with CD46. The scope of this study was to understand the regulation of PVRL4 in the understood capacity of a MV receptor. The anti‐tumor activity of MV is used to further elucidate the role of the regulatory miRNA on PVRL4 and in turn MV infectivity.
In conclusion, this study demonstrates that host miR‐31 and miR‐128 contribute at least in part to regulate MV infectivity, through post‐transcriptional regulation of the receptor, PVRL4. In cell culture, we have also observed that MV infection can similarly regulate host miRNA, i.e. miR‐31, expression levels.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
This research was supported by R01 CA154348, Mayo Clinic Glioma, Breast and Ovarian SPORE grants CA108961, CA116201 and CA136393, respectively. We thank Mark Jenotoft, MD for assisting with pathology assessment. We acknowledge the support of the Mayo Clinic Pathology Research Core in the Center for Individualized Medicine under the direction of Thomas Flotte, MD, confocal and light microscope facility and the flow cytometry research facility under the Mayo Clinic Microscopy and Cell Analysis Core (MN) supported by the Mayo Clinic Cancer Center grant P30 CA15083. We thank Roberto Cattaneo, PhD and Eric Poeschla, MD for providing with the SLAM blind wild‐type measles virus, and Gag‐pol and VSV‐G plasmids respectively.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.07.007.
Geekiyanage Hirosha, Galanis Evanthia, (2016), MiR‐31 and miR‐128 regulates poliovirus receptor‐related 4 mediated measles virus infectivity in tumors, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.07.007.
Contributor Information
Hirosha Geekiyanage, Email: geekiyanage.donahirosha@mayo.edu.
Evanthia Galanis, Email: galanis.evanthia@mayo.edu.
References
- Albrecht, P. , Lorenz, D. , Klutch, M.J. , Vickers, J.H. , Ennis, F.A. , 1980. Fatal measles infection in marmosets pathogenesis and prophylaxis. Infect. Immun. 27, 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. , 2008. The evolution of our thinking about microRNAs. Nat. Med. 14, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Ameres, S.L. , Zamore, P.D. , 2013. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. [DOI] [PubMed] [Google Scholar]
- Amiel, J. , de Pontual, L. , Henrion-Caude, A. , 2012. miRNA, development and disease. Adv. Genet. 80, 1–36. [DOI] [PubMed] [Google Scholar]
- Augoff, K. , McCue, B. , Plow, E.F. , Sossey-Alaoui, K. , 2012. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer. 11, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch, M.A. , Leber, M.F. , Bossow, S. , Singh, M. , Engeland, C.E. , Albert, J. , Grossardt, C. , Jager, D. , von Kalle, C. , Ungerechts, G. , 2014. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 21, 373–380. [DOI] [PubMed] [Google Scholar]
- Banno, K. , Yanokura, M. , Iida, M. , Adachi, M. , Nakamura, K. , Nogami, Y. , Umene, K. , Masuda, K. , Kisu, I. , Nomura, H. , Kataoka, F. , Tominaga, E. , Aoki, D. , 2014. Application of microRNA in diagnosis and treatment of ovarian cancer. Biomed. Res. Int. 2014, 232817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, D. , Kunitomi, M. , Vignuzzi, M. , Saksela, K. , Andino, R. , 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 4, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov, E. , 2011. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. [DOI] [PubMed] [Google Scholar]
- Betel, D. , Koppal, A. , Agius, P. , Sander, C. , Leslie, C. , 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron, C. , Goldstein, L.D. , Thorne, N.P. , Spiteri, I. , Chin, S.F. , Dunning, M.J. , Barbosa-Morais, N.L. , Teschendorff, A.E. , Green, A.R. , Ellis, I.O. , Tavare, S. , Caldas, C. , Miska, E.A. , 2007. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 8, R214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluming, A.Z. , Ziegler, J.L. , 1971. Regression of Burkitt's lymphoma in association with measles infection. Lancet. 2, 105–106. [DOI] [PubMed] [Google Scholar]
- Buck, A.H. , Perot, J. , Chisholm, M.A. , Kumar, D.S. , Tuddenham, L. , Cognat, V. , Marcinowski, L. , Dolken, L. , Pfeffer, S. , 2010. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 16, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla, D. , Yario, T. , Steitz, J.A. , 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 328, 1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, T.H. , Huang, H.Y. , Hsu, J.B. , Weng, S.L. , Horng, J.T. , Huang, H.D. , 2013. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform. 14, (Suppl. 2) S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre, S.A. , Galardi, S. , Mangiola, A. , Ferracin, M. , Liu, C.G. , Sabatino, G. , Negrini, M. , Maira, G. , Croce, C.M. , Farace, M.G. , 2005. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 334, 1351–1358. [DOI] [PubMed] [Google Scholar]
- Clemmons, N.S. , Gastanaduy, P.A. , Fiebelkorn, A.P. , Redd, S.B. , Wallace, G.S. , 2015. Measles – United States, January 4-April 2, 2015. MMWR Morb. Mortal. Wkly. Rep. 64, 373–376. [PMC free article] [PubMed] [Google Scholar]
- Creighton, C.J. , Fountain, M.D. , Yu, Z. , Nagaraja, A.K. , Zhu, H. , Khan, M. , Olokpa, E. , Zariff, A. , Gunaratne, P.H. , Matzuk, M.M. , Anderson, M.L. , 2010. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 70, 1906–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, C.M. , 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya, N. , Morin, P.J. , 2010. MicroRNAs in ovarian carcinomas. Endocr. Relat. Cancer. 17, F77–F89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Swart, R.L. , 2009. Measles studies in the macaque model. Curr. Top. Microbiol. Immunol. 330, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpeut, S. , Noyce, R.S. , Richardson, C.D. , 2014. The tumor-associated marker, PVRL4 (Nectin-4), is the epithelial receptor for morbilliviruses. Viruses-Basel. 6, 2268–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRycke, M.S. , Pambuccian, S.E. , Gilks, C.B. , Kalloger, S.E. , Ghidouche, A. , Lopez, M. , Bliss, R.L. , Geller, M.A. , Argenta, P.A. , Harrington, K.M. , Skubitz, A.P.N. , 2010. Nectin 4 overexpression in ovarian cancer tissues and serum. Am. J. Clin. Pathol. 134, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Q. , Cai, N. , Tao, T. , Zhang, R. , Yan, W. , Li, R. , Zhang, J. , Luo, H. , Shi, Y. , Luan, W. , Zhang, Y. , You, Y. , Wang, Y. , Liu, N. , 2014. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS One. 9, e98651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorig, R.E. , Marcil, A. , Chopra, A. , Richardson, C.D. , 1993. The human Cd46 molecule is a receptor for measles-virus (Edmonston strain). Cell. 75, 295–305. [DOI] [PubMed] [Google Scholar]
- Erlenhoefer, C. , Wurzer, W.J. , Loffler, S. , Schneider-Schaulies, S. , Ter Meulen, V. , Schneider-Schaulies, J. , 2001. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J. Virol. 75, 4499–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre-Lafay, S. , Garrido-Urbani, S. , Reymond, N. , Goncalves, A. , Dubreuil, P. , Lopez, M. , 2005. Nectin-4, a new serological breast cancer marker, is a substrate for tumor necrosis factor-alpha-converting enzyme (TACE)/ADAM-17. J. Biol. Chem. 280, 19543–19550. [DOI] [PubMed] [Google Scholar]
- Fabre-Lafay, S. , Monville, F. , Garrido-Urbani, S. , Berruyer-Pouyet, C. , Ginestier, C. , Reymond, N. , Finetti, P. , Sauvan, R. , Adelaide, J. , Geneix, J. , Lecocq, E. , Popovici, C. , Dubreuil, P. , Viens, P. , Goncalves, A. , Charafe-Jauffret, E. , Jacquemier, J. , Birnbaum, D. , Lopez, M. , 2007. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer. 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.P. , Rivera, A. , Tao, L.H. , De Geest, B. , Zhang, X.L. , 2012. Construction of an oncolytic herpes simplex virus that precisely targets hepatocellular carcinoma cells. Mol. Ther. 20, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage, H. , Chan, C. , 2011. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer's disease. J. Neurosci. 31, 14820–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein, E. , Cullen, B.R. , 2008. Viral and cellular MicroRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 3, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi, M. , Muinos-Gimeno, M. , Kagerbauer, B. , Marti, E. , Estivill, X. , Espinosa-Parrilla, Y. , 2010. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol. Biol. 11, 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Hannon, G.J. , 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522 [DOI] [PubMed] [Google Scholar]
- Iankov, I.D. , Allen, C. , Federspiel, M.J. , Myers, R.M. , Peng, K.W. , Ingle, J.N. , Russell, S.J. , Galanis, E. , 2012. Expression of immunomodulatory neutrophil-activating protein of Helicobacter pylori enhances the antitumor activity of oncolytic measles virus. Mol. Ther. 20, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute, M.C.A.N.C., MV-NIS infected mesenchymal stem cells in treating patients with recurrent ovarian Cancer. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2000-[2014 May 13]. Available from: https://clinicaltrials.gov/ct2/show/NCT02068794 NLM Identifier: NCT02068794.
- Institute, M.C.A.N.C., Recombinant measles virus vaccine therapy and oncolytic virus therapy in treating patients with progressive, recurrent, or refractory ovarian epithelial cancer or primary peritoneal cancer. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2000-[2014 May 13]. Available from: https://clinicaltrials.gov/ct2/show/NCT00408590 NLM Identifier: NCT00390299.
- Institute, M.C.A.N.C., Viral therapy in treating patients with recurrent glioblastoma multiforme. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2000-[2014 May 13]. Available from: https://clinicaltrials.gov/ct2/show/NCT00390299 NLM Identifier: NCT00390299.
- Iorio, M.V. , Ferracin, M. , Liu, C.G. , Veronese, A. , Spizzo, R. , Sabbioni, S. , Magri, E. , Pedriali, M. , Fabbri, M. , Campiglio, M. , Menard, S. , Palazzo, J.P. , Rosenberg, A. , Musiani, P. , Volinia, S. , Nenci, I. , Calin, G.A. , Querzoli, P. , Negrini, M. , Croce, C.M. , 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L. , Yi, M. , Lancaster, A.M. , Lemon, S.M. , Sarnow, P. , 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 309, 1577–1581. [DOI] [PubMed] [Google Scholar]
- Karsy, M. , Arslan, E. , Moy, F. , 2012. Current progress on understanding microRNAs in glioblastoma multiforme. Genes Cancer. 3, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. , Sugnet, C.W. , Furey, T.S. , Roskin, K.M. , Pringle, T.H. , Zahler, A.M. , Haussler, D. , 2002. The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobune, F. , Takahashi, H. , Terao, K. , Ohkawa, T. , Ami, Y. , Suzaki, Y. , Nagata, N. , Sakata, H. , Yamanouchi, K. , Kai, C. , 1996. Nonhuman primate models of measles. Lab. Anim. Sci. 46, 315–320. [PubMed] [Google Scholar]
- Koressaar, T. , Remm, M. , 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics. 23, 1289–1291. [DOI] [PubMed] [Google Scholar]
- Korner, C. , Keklikoglou, I. , Bender, C. , Worner, A. , Munstermann, E. , Wiemann, S. , 2013. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 288, 8750–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, A. , Ha, D. , Schotte, D. , den Boer, M.L. , Armstrong, S.A. , Lodish, H.F. , 2010. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle. 9, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber, M.F. , Bossow, S. , Leonard, V.H. , Zaoui, K. , Grossardt, C. , Frenzke, M. , Miest, T. , Sawall, S. , Cattaneo, R. , von Kalle, C. , Ungerechts, G. , 2011. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol. Ther. 19, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.Y.F. , Rennie, P.S. , Jia, W.W.G. , 2009. MicroRNA regulation of oncolytic herpes simplex virus-1 for selective killing of prostate cancer cells. Clin. Cancer Res. 15, 5126–5135. [DOI] [PubMed] [Google Scholar]
- Lee, E.J. , Baek, M. , Gusev, Y. , Brackett, D.J. , Nuovo, G.J. , Schmittgen, T.D. , 2008. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 14, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.T. , Chu, K. , Im, W.S. , Yoon, H.J. , Im, J.Y. , Park, J.E. , Park, K.H. , Jung, K.H. , Lee, S.K. , Kim, M. , Roh, J.K. , 2011. Altered microRNA regulation in Huntington's disease models. Exp. Neurol. 227, 172–179. [DOI] [PubMed] [Google Scholar]
- Leonard, V.H. , Hodge, G. , Reyes-Del Valle, J. , McChesney, M.B. , Cattaneo, R. , 2010. Measles virus selectively blind to signaling lymphocytic activation molecule (SLAM; CD150) is attenuated and induces strong adaptive immune responses in rhesus monkeys. J. Virol. 84, 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P. , Burge, C.B. , Bartel, D.P. , 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Gu, X. , Sun, L. , Flowers, A.B. , Rademaker, A.W. , Zhou, Y. , Kiyokawa, H. , 2014. Downregulation of Smurf2, a tumor-suppressive ubiquitin ligase, in triple-negative breast cancers: involvement of the RB-microRNA axis. BMC Cancer. 14, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. , Schmittgen, T.D. , 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lou, W. , Chen, Q. , Ma, L. , Liu, J. , Yang, Z. , Shen, J. , Cui, Y. , Bian, X.W. , Qian, C. , 2013. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J. Mol. Med. (Berl). 91, 715–725. [DOI] [PubMed] [Google Scholar]
- Lu, J. , Getz, G. , Miska, E.A. , Alvarez-Saavedra, E. , Lamb, J. , Peck, D. , Sweet-Cordero, A. , Ebert, B.L. , Mak, R.H. , Ferrando, A.A. , Downing, J.R. , Jacks, T. , Horvitz, H.R. , Golub, T.R. , 2005. MicroRNA expression profiles classify human cancers. Nature. 435, 834–838. [DOI] [PubMed] [Google Scholar]
- Lukiw, W.J. , 2007. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 18, 297–300. [DOI] [PubMed] [Google Scholar]
- Luna, J.M. , Scheel, T.K. , Danino, T. , Shaw, K.S. , Mele, A. , Fak, J.J. , Nishiuchi, E. , Takacs, C.N. , Catanese, M.T. , de Jong, Y.P. , Jacobson, I.M. , Rice, C.M. , Darnell, R.B. , 2015. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 160, 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri, S. , Liu, Z. , Phung, S. , Wang, E. , Yuan, Y.C. , Chen, S. , 2010. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res. Treat. 124, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzacurati, L. , Marzulli, M. , Reinhart, B. , Miyagawa, Y. , Uchida, H. , Goins, W.F. , Lil, A. , Kaur, B. , Caligiuri, M. , Cripe, T. , Chiocca, E.A. , Amankulor, N. , Cohen, J.B. , Glorioso, J.C. , Grandi, P. , 2015. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol. Ther. 23, 99 215–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, M. , Guan, Y. , Yoshimoto, K. , Hata, N. , Amano, T. , Nakamizo, A. , Sasaki, T. , 2012. MicroRNAs in human malignant gliomas. J. Oncol. 2012, 732874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msaouel, P. , Opyrchal, M. , Musibay, E.D. , Galanis, E. , 2013. Oncolytic measles virus strains as novel anticancer agents. Expert Opin. Biol. Ther. 13, 483–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlebach, M.D. , Mateo, M. , Sinn, P.L. , Prufer, S. , Uhlig, K.M. , Leonard, V.H. , Navaratnarajah, C.K. , Frenzke, M. , Wong, X.X. , Sawatsky, B. , Ramachandran, S. , McCray, P.B. , Cichutek, K. , von Messling, V. , Lopez, M. , Cattaneo, R. , 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 480, 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T. , Russell, S.J. , 2004. Oncolytic measles viruses for cancer therapy. Expert Opin. Biol. Ther. 4, 1685–1692. [DOI] [PubMed] [Google Scholar]
- Naniche, D. , Variorkrishnan, G. , Cervoni, F. , Wild, T.F. , Rossi, B. , Rabourdincombe, C. , Gerlier, D. , 1993. Human membrane cofactor protein (Cd46) acts as a cellular receptor for measles-virus. J. Virol. 67, 6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network, C.G.A.R. , 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce, R.S. , Bondre, D.G. , Ha, M.N. , Lin, L.T. , Sisson, G. , Tsao, M.S. , Richardson, C.D. , 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7, e1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce, R.S. , Richardson, C.D. , 2012. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 20, 429–439. [DOI] [PubMed] [Google Scholar]
- Nuovo, G. , Lee, E.J. , Lawler, S. , Godlewski, J. , Schmittgen, T. , 2009. In situ detection of mature microRNAs by labeled extension on ultramer templates. Biotechniques. 46, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, D.W. , Jones, S. , Zhang, X. , Lin, J.C. , Leary, R.J. , Angenendt, P. , Mankoo, P. , Carter, H. , Siu, I.M. , Gallia, G.L. , Olivi, A. , McLendon, R. , Rasheed, B.A. , Keir, S. , Nikolskaya, T. , Nikolsky, Y. , Busam, D.A. , Tekleab, H. , Diaz, L.A. , Hartigan, J. , Smith, D.R. , Strausberg, R.L. , Marie, S.K. , Shinjo, S.M. , Yan, H. , Riggins, G.J. , Bigner, D.D. , Karchin, R. , Papadopoulos, N. , Parmigiani, G. , Vogelstein, B. , Velculescu, V.E. , Kinzler, K.W. , 2008. An integrated genomic analysis of human glioblastoma multiforme. Science. 321, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova, N.N. , Pallasch, C. , Elia, A.E. , Braun, C.J. , Westbrook, T.F. , Hemann, M. , Elledge, S.J. , 2013. A role for PVRL4-driven cell-cell interactions in tumorigenesis. Elife. 2, e00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R.T. , Gacic-Dobo, M. , Dabbagh, A. , Mulders, M.N. , Strebel, P.M. , Okwo-Bele, J.M. , Rota, P.A. , Goodson, J.L. , 2014. Progress toward regional measles elimination–worldwide, 2000–2013. MMWR Morb. Mortal. Wkly. Rep. 63, 1034–1038. [PMC free article] [PubMed] [Google Scholar]
- Peruzzi, P. , Bronisz, A. , Nowicki, M.O. , Wang, Y. , Ogawa, D. , Price, R. , Nakano, I. , Kwon, C.H. , Hayes, J. , Lawler, S.E. , Ostrowski, M.C. , Chiocca, E.A. , Godlewski, J. , 2013. MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol. 15, 1212–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuong, L.K. , Allen, C. , Peng, K.W. , Giannini, C. , Greiner, S. , TenEyck, C.J. , Mishra, P.K. , Macura, S.I. , Russell, S.J. , Galanis, E.C. , 2003. Use of a vaccine strain of measles virus genetically engineered to produce carcinoembryonic antigen as a novel therapeutic agent against glioblastoma multiforme. Cancer Res. 63, 2462–2469. [PubMed] [Google Scholar]
- Plumet, S. , Gerlier, D. , 2005. Optimized SYBR green real-time PCR assay to quantify the absolute copy number of measles virus RNAs using gene specific primers. J. Virol. Methods. 128, 79–87. [DOI] [PubMed] [Google Scholar]
- Pratakpiriya, W. , Seki, F. , Otsuki, N. , Sakai, K. , Fukuhara, H. , Katamoto, H. , Hirai, T. , Maenaka, K. , Techangamsuwan, S. , Lan, N.T. , Takeda, M. , Yamaguchi, R. , 2012. Nectin4 is an epithelial cell receptor for canine distemper virus and involved in neurovirulence. J. Virol. 86, 10207–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, P. , Banerjee, A. , Wu, Z.S. , Zhang, X. , Wang, H. , Pandey, V. , Zhang, W.J. , Lv, X.F. , Tan, S. , Lobie, P.E. , Zhu, T. , 2012. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 72, 6036–6050. [DOI] [PubMed] [Google Scholar]
- Rajbhandari, R. , McFarland, B.C. , Patel, A. , Gerigk, M. , Gray, G.K. , Fehling, S.C. , Bredel, M. , Berbari, N.F. , Kim, H. , Marks, M.P. , Meares, G.P. , Sinha, T. , Chuang, J. , Benveniste, E.N. , Nozell, S.E. , 2015. Loss of tumor suppressive microRNA-31 enhances TRADD/NF-kappaB signaling in glioblastoma. Oncotarget. 6, 17805–17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, G. , Panis, M. , Cooper, J.D. , Tellinghuisen, T.L. , Sukhodolets, K.E. , Pfeffer, S. , Landthaler, M. , Landgraf, P. , Kan, S. , Lindenbach, B.D. , Chien, M. , Weir, D.B. , Russo, J.J. , Ju, J. , Brownstein, M.J. , Sheridan, R. , Sander, C. , Zavolan, M. , Tuschl, T. , Rice, C.M. , 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104, 12884–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S.A. , Santosh, V. , Somasundaram, K. , 2010. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod. Pathol. 23, 1404–1417. [DOI] [PubMed] [Google Scholar]
- Reed, L.J. , Muench, H. , 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27, 493–497. [Google Scholar]
- Reymond, N. , Fabre, S. , Lecocq, E. , Adelaide, J. , Dubreuil, P. , Lopez, M. , 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with Nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276, 43205–43215. [DOI] [PubMed] [Google Scholar]
- Schaefer, A. , Jung, M. , Mollenkopf, H.J. , Wagner, I. , Stephan, C. , Jentzmik, F. , Miller, K. , Lein, M. , Kristiansen, G. , Jung, K. , 2010. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer. 126, 1166–1176. [DOI] [PubMed] [Google Scholar]
- Schneider-Schaulies, J. , Schnorr, J.J. , Brinckmann, U. , Dunster, L.M. , Baczko, K. , Liebert, U.G. , Schneider-Schaulies, S. , Ter Meulen, V. , 1995. Receptor usage and differential downregulation of CD46 by measles virus wild-type and vaccine strains. Proc. Natl. Acad. Sci. U. S. A. 92, 3943–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Z.M. , Wang, J. , Yan, Z. , You, Y.P. , Li, C.Y. , Qian, X. , Yin, Y. , Zhao, P. , Wang, Y.Y. , Wang, X.F. , Li, M.N. , Liu, L.Z. , Liu, N. , Jiang, B.H. , 2012. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 7, e32709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky, R.L. , Cullen, B.R. , 2011. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 6, e24248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, T. , Yoneda, M. , Kuraishi, T. , Hattori, S. , Inoue, Y. , Sato, H. , Kai, C. , 2013. Measles virus selectively blind to signaling lymphocyte activation molecule as a novel oncolytic virus for breast cancer treatment. Gene Ther. 20, 338–347. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y. , Iwaya, T. , Sawada, G. , Kurashige, J. , Matsumura, T. , Uchi, R. , Ueo, H. , Takano, Y. , Eguchi, H. , Sudo, T. , Sugimachi, K. , Yamamoto, H. , Doki, Y. , Mori, M. , Mimori, K. , 2014. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann. Surg. Oncol. 21, 205–212. [DOI] [PubMed] [Google Scholar]
- Tatsuo, H. , Ono, N. , Tanaka, K. , Yanagi, Y. , 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 406, 893–897. [DOI] [PubMed] [Google Scholar]
- tenOever, B.R. , 2013. RNA viruses and the host microRNA machinery. Nat. Rev. Microbiol. 11, 169–180. [DOI] [PubMed] [Google Scholar]
- Untergasser, A. , Cutcutache, I. , Koressaar, T. , Ye, J. , Faircloth, B.C. , Remm, M. , Rozen, S.G. , 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usvasalo, A. , Ninomiya, S. , Raty, R. , Hollmen, J. , Saarinen-Pihkala, U.M. , Elonen, E. , Knuutila, S. , 2010. Focal 9p instability in hematologic neoplasias revealed by comparative genomic hybridization and single-nucleotide polymorphism microarray analyses. Genes Chromosomes Cancer. 49, 309–318. [DOI] [PubMed] [Google Scholar]
- van Rooij, E. , 2011. The art of microRNA research. Circ. Res. 108, 219–234. [DOI] [PubMed] [Google Scholar]
- Veerla, S. , Lindgren, D. , Kvist, A. , Frigyesi, A. , Staaf, J. , Persson, H. , Liedberg, F. , Chebil, G. , Gudjonsson, S. , Borg, A. , Mansson, W. , Rovira, C. , Hoglund, M. , 2009. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int. J. Cancer. 124, 2236–2242. [DOI] [PubMed] [Google Scholar]
- Vire, E. , Curtis, C. , Davalos, V. , Git, A. , Robson, S. , Villanueva, A. , Vidal, A. , Barbieri, I. , Aparicio, S. , Esteller, M. , Caldas, C. , Kouzarides, T. , 2014. The breast cancer oncogene EMSY represses transcription of antimetastatic microRNA miR-31. Mol. Cell. 53, 806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visani, M. , de Biase, D. , Marucci, G. , Cerasoli, S. , Nigrisoli, E. , Bacchi Reggiani, M.L. , Albani, F. , Baruzzi, A. , Pession, A. , 2014. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I–III. Mol. Oncol. 8, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia, S. , Calin, G.A. , Liu, C.G. , Ambs, S. , Cimmino, A. , Petrocca, F. , Visone, R. , Iorio, M. , Roldo, C. , Ferracin, M. , Prueitt, R.L. , Yanaihara, N. , Lanza, G. , Scarpa, A. , Vecchione, A. , Negrini, M. , Harris, C.C. , Croce, C.M. , 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, G.J. , Bemis, L.T. , Nakajima, E. , Sugita, M. , Birks, D.K. , Robinson, W.A. , Varella-Garcia, M. , Bunn, P.A. , Haney, J. , Helfrich, B.A. , Kato, H. , Hirsch, F.R. , Franklin, W.A. , 2008. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 19, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Welstead, G.G. , Hsu, E.C. , Iorio, C. , Bolotin, S. , Richardson, C.D. , 2004. Mechanism of CD150 (SLAM) down regulation from the host cell surface by measles virus hemagglutinin protein. J. Virol. 78, 9666–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.H. , Laszlo, C.F. , Greco, S. , Chambers, S.K. , 2012. Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol. Cancer. 11, 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wszolek, M.F. , Rieger-Christ, K.M. , Kenney, P.A. , Gould, J.J. , Silva Neto, B. , Lavoie, A.K. , Logvinenko, T. , Libertino, J.A. , Summerhayes, I.C. , 2011. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol. Oncol. 29, 794–801 e791. [DOI] [PubMed] [Google Scholar]
- Wu, C. , Lin, J. , Hong, M. , Choudhury, Y. , Balani, P. , Leung, D. , Dang, L.H. , Zhao, Y. , Zeng, J. , Wang, S. , 2009. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol. Ther. 17, 2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, S.K. , Parkin, R.K. , Mitchell, P.S. , Fritz, B.R. , O'Briant, K. , Godwin, A.K. , Urban, N. , Drescher, C.W. , Knudsen, B.S. , Tewari, M. , 2009. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 4, e5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L.X. , Huang, X.F. , Shao, Q. , Huang, M.Y. , Deng, L. , Wu, Q.L. , Zeng, Y.X. , Shao, J.Y. , 2008. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 14, 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D. , Sun, Y. , Hu, L. , Zheng, H. , Ji, P. , Pecot, C.V. , Zhao, Y. , Reynolds, S. , Cheng, H. , Rupaimoole, R. , Cogdell, D. , Nykter, M. , Broaddus, R. , Rodriguez-Aguayo, C. , Lopez-Berestein, G. , Liu, J. , Shmulevich, I. , Sood, A.K. , Chen, K. , Zhang, W. , 2013. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 23, 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, W.C. , Guo, G.C. , Zhang, Q.Q. , Fan, L.Y. , Wu, N. , Bo, Y.L. , 2014. The application of multiple miRNA response elements enables oncolytic adenoviruses to possess specificity to glioma cells. Virology. 458, 69–82. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Huang, J. , Yang, N. , Greshock, J. , Megraw, M.S. , Giannakakis, A. , Liang, S. , Naylor, T.L. , Barchetti, A. , Ward, M.R. , Yao, G. , Medina, A. , O'Brien-Jenkins, A. , Katsaros, D. , Hatzigeorgiou, A. , Gimotty, P.A. , Weber, B.L. , Coukos, G. , 2006. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. U. S. A. 103, 9136–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Chao, T. , Li, R. , Liu, W. , Chen, Y. , Yan, X. , Gong, Y. , Yin, B. , Qiang, B. , Zhao, J. , Yuan, J. , Peng, X. , 2009. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. (Berl). 87, 43–51. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Guo, J. , Li, D. , Xiao, B. , Miao, Y. , Jiang, Z. , Zhuo, H. , 2010. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med. Oncol. 27, 685–689. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.W. , Zhang, X.M. , Newman, K. , Liu, X.Y. , 2012. MicroRNA regulation of oncolytic adenovirus 6 for selective treatment of castration-resistant prostate cancer. Mol. Cancer Ther. 11, 2410–2418. [DOI] [PubMed] [Google Scholar]
- Zipprich, J. , Winter, K. , Hacker, J. , Xia, D. , Watt, J. , Harriman, K. , 2015. Measles outbreak–California, December 2014-February 2015. MMWR Morb. Mortal. Wkly. Rep. 64, 153–154. [PMC free article] [PubMed] [Google Scholar]
- Zygiert, Z. , 1971. Hodgkin's disease: remissions after measles. Lancet. 1, 593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


