Abstract
Docetaxel (Dtxl) is currently the most common therapeutic option for prostate cancer (PC). However, adverse side effects and problems associated with chemo-resistance limit its therapeutic outcome in clinical settings. A targeted nanoparticle system to improve its delivery to and activity at the tumor site could be an attractive strategy for PC therapy. Therefore, the objective of this study was to develop and determine the anti-cancer efficacy of a novel docetaxel loaded, prostate specific membrane antigen (PSMA) targeted superparamagnetic iron oxide nanoparticle (SPION) (J591-SPION-Dtxl) formulation for PC therapy. Our results showed the SPION-Dtxl formulation exhibits an optimal particle size and zeta potential, which can efficiently be internalized in PC cells. SPION-Dtxl exhibited potent anti-cancer efficacy via induction of the expression of apoptosis associated proteins, downregulation of anti-apoptotic proteins, and inhibition of chemo-resistance associated protein in PC cell lines. J591-SPION-Dtxl exhibited a profound uptake in C4-2 (PSMA+) cells compared to PC-3 (PSMA−) cells. A similar targeting potential was observed in ex-vivo studies in C4-2 tumors but not in PC-3 tumors, suggesting its tumor specific targeting. Overall this study suggests that a PSMA antibody functionalized SPION-Dtxl formulation can be highly useful for targeted PC therapy.
Graphical Abstract
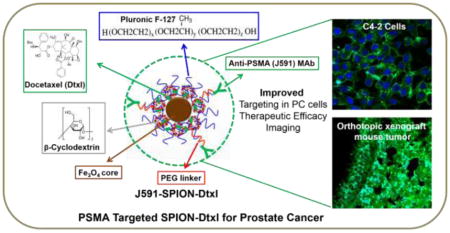
1. Introduction
Prostate cancer (PC) is the most commonly diagnosed cancer and second leading cause of cancer-related deaths in men in the United States [1]. The existing androgen-deprivation therapy is an initial treatment modality ultimately leading to castration-resistant prostate cancer (CRPC). Docetaxel (Dtxl)-based chemotherapy is a standard first-line treatment for CRPC [2–4]. However, Dtxl resistance remains a major concern in clinical oncology [5–7]. Thus, identification of an effective therapeutic approach or reversing Dtxl drug resistance in CRPC is highly desired. Numerous drug nanoformulations have been developed for greater accumulation of loaded therapeutics at the tumor site and to maximize the potent activity [8], thereby offering improved therapeutic applications [9]. In addition, cumulative literature demonstrates that drug nanoformulations can reverse drug resistance. Therefore, we believe that Dtxl-based nanomedicine can provide a novel way to deliver docetaxel in its active form to cancer cells and redefine the drug interaction [10, 11]. About 388 studies report improved Dtxl therapeutic activity using various forms of nanoparticles (PubMed search conducted on Dec. 30, 2015). However, Cellax™ (a covalently conjugated Dtxl onto polyethylene glycol-acetylated carboxymethylcellulose to form self-assembled 120 nm particles) [12–14] and BIND-014 (Dtxl encapsulated in biodegradable polymer based nanoparticles) [15, 16] are emerging as clinically useful nanoformulations. Among various nanoparticle based drug nanoformulations, superparamagnetic iron oxide nanoparticles (SPION) or magnetic nanoparticles (MNPs) have recently received considerable attention as theranostic applications [therapy (drug delivery, hyperthermia) and diagnosis (MR imaging, tumor cell-isolation)] due to their biocompatibility and superparamagnetic properties [17–20]. Therefore, formulation of docetaxel with SPION would not only enhance its biological activity but also add imaging capability to the treatment modality.
Physico-chemical and biological properties are critical components in generating a successful targeted magnetic nanoformulation [21–23]. Therefore, we designed a SPION formulation that is comprised of an iron oxide core with self-assembled layers of β-cyclodextrin and pluronic polymer F127 using the one-pot wet nanoprecipitation method. This formulation possesses novel features that include: i) an iron oxide core of nanoparticles exhibiting super paramagnetic properties that can be used for magnetic resonance imaging (MRI) (diagnosis/imaging); ii) the cyclodextrin and pluronic polymer (F127) double layer coating supports hydrophobic layers to load/encapsulate anti-cancer drugs; and iii) the polyethylene glycol chains of pluronic F127 polymer act as a stealth polymer, which diminishes the nonspecific uptake of formulation and reduces drug resistance. Feasibility of encapsulation and delivery of a model anti-cancer drug molecule (curcumin) in such a formulation has been tested in prostate, breast and pancreatic cancer cells [24–27].
The primary aim of the current study dealt with the use of this novel SPION formulation to deliver docetaxel. Among many cell-surface protein markers, prostate specific membrane antigen (PSMA) is highly overexpressed in prostate cancers [28], and thus it is considered a valid target for PC [29, 30]. In addition, anti-PSMA antibody or PSMA-specific aptamers A10-3.2 aptamer based targeting and imaging have been tested on patients with PC [29, 31, 32]. Therefore, in order to enhance the cancer cell specific targeting, uptake, and retention, we biofunctionalized nanoparticles through the addition of a PEG-linker, notably, the N-hydroxysuccinimide (NHS) group, with J591 monoclonal antibodies, for targeting PSMA-expressing prostate tumors [33–36]. Using a PSMA targeted nanoformulation, we evaluated the therapeutic benefit and targeting using both in vitro and ex vivo models of PSMA+ and PSMA− PC cells and mouse tumors. Our results suggest that encapsulation of Dtxl in SPION significantly enhanced the targeting potential to and efficacy in PC cells, which indicates the applicability of this unique nanotechnology platform for targeted PC therapy.
2. Materials and Methods
All reagents, solvents, chemicals, and cell culture plastics were purchased from Sigma–Aldrich Co. (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) unless otherwise mentioned. All chemicals were used as received without further purification. PC cell lines [C4-2 (PSMA+) and PC-3 (PSMA−)] were received from Dr. Meena Jaggi’s Laboratory, The University of Tennessee Health Science Center, and cultured in RPMI-1640 medium (Gibco, Grand Island, NY) containing 10% (v/v) fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Grand Island, NY), and 1% (w/v) penicillin–streptomycin (Gibco) at 37°C in a humidified 5% CO2–95% air atmosphere (Sanyo Scientific Ltd, Tokyo, Japan). For most of the in vitro cell culture experiments, cells were trypsinized when the confluence level was about 80% and above, seeded in either 6 or 96-well plates, and allowed to attach overnight to the plate before starting any treatments.
2.1. Preparation and characterization
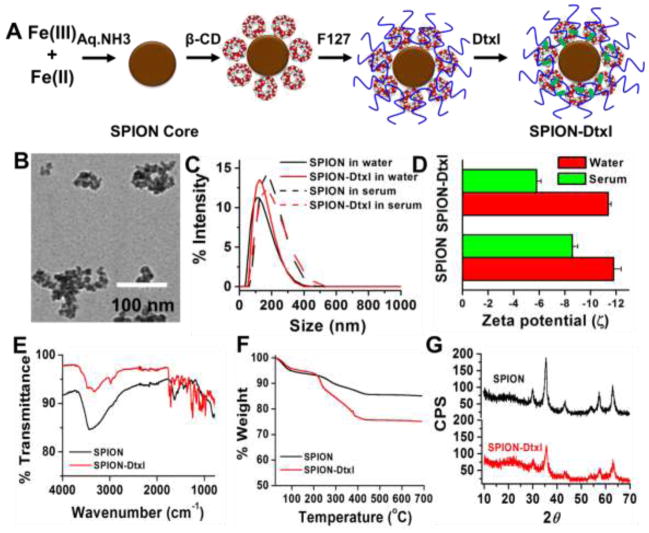
SPION composed of iron oxide cores were prepared from Fe2+ (300 mg) and Fe3+ ions (800 mg) (1:1 molar ratio) using chemical precipitation in the presence of aqueous ammonia environment, followed by cyclodextrin and F127 polymer coating, respectively [25]. These were considered to be SPION or control SPION for all our studies. 100 μL of 10 mg/mL docetaxel in acetone was employed to load docetaxel in SPION at a ratio of 1:10 according to our previous drug loading protocol; these docetaxel loaded SPION are termed SPION-Dtxl throughout the manuscript. A schematic representation of this SPION-Dtxl formulation is presented in Fig. 1A.
Figure 1. Physico-chemical characterization of SPION-Dtxl.
A) Schematic preparative approach and hypothetical structure of SPION-Dtxl formulation. Color and dimensions are not comparable to original formulation. Docetaxel resides in β-cyclodextrin and/or pluronic F127 polymer layers. B) Representative transmission electron microscopic image of SPION-Dtxl. Scale bars on TEM images equal 100 nm. Individual grain of SPION is about 8–10 nm. Size of aggregated SPION-Dtxl ~100 nm. Direct Mag: 100,000X, obtained using AMT camera system. C–D) Particle size and zeta potential of SPION and SPION-Dtxl in water and serum. Note: A slight increase of particle size and zeta potentials was noticed in serum over water. Data represented as mean ± SEM (n = 3). E) FT-IR spectra of Dtxl, SPION and SPION-Dtxl. F) Thermogravimetric analysis of SPION and SPION-Dtxl. Dtxl loading in SPION (SPION-Dtxl) resulted in greater (~9.5–10%) weight compared to SPION. G) X-ray diffraction patterns of SPION and SPION-Dtxl. No significant change in patterns indicated successful entrapment of Dtxl in β-CD and pluronic F127 polymer layers.
2.1.1. Particle size and zeta potential
The average hydrodynamic size and size distribution of SPION and SPION-Dtxl were measured by dynamic light scattering methods using a Zetasizer (Nano ZS, Malvern Instruments, Malvern, UK). Before measurements, the freshly prepared SPION or SPION-Dtxl were diluted to achieve 1 mg/mL nanoparticle suspensions and probe sonicated using a VirSonic Ultrasonic Cell Disrupter 100 (VirTis, Gardiner, NY) for 30 sec. All measurements were carried out at room temperature after equilibration for 10 min. Three runs of each sample suspension were measured for 3 min. The same SPION and SPION-Dtxl suspensions were used to measure zeta potential using the principle of electrophoretic mobility under an electric field. The representation of data was an average of 3 independent measurements for 9 min.
2.1.2. Fourier transform infrared (FTIR), X-ray diffraction, and thermogravimetric analysis
For these studies, SPION and SPION-Dtxl formulations were lyophilized using a Labconco Freeze Dry System (−48 °C, 133 × 10−3 mBar; Labconco, Kansas City, MO) to obtain dry solid nanoparticles.
The composition of SPION and SPION-Dtxl was evaluated using FTIR (Perkin Elmer Series Spectra 100, Waltham, MA) to affirm the drug encapsulation [37]. A Rigaku D/Max-B diffractometer (Rigaku Americas Corporation, The Woodlands, TX) was used to determine the crystalline behavior of SPION/SPION-Dtxl [26]. The thermal behavior of SPION and SPION-Dtxl was assessed using a Perkin Elmer Simultaneous Thermal Analyzer STA6000 [37].
2.1.3. In vitro magnetic resonance imaging
To test their MRI potential, SPION, SPION-Dtxl or SPION/SPION-Dtxl in 2 × 106 C4-2 cells were suspended in 3% agar solution to achieve agar-nanoparticle phantom gels (final concentration of NPs ~ 50 μg iron/mL). A 9.4 T (400 MHz H1) with an 89 mm vertical bore MRI scanner (Agilent Technologies, Santa Clara, CA) was used to measure T1 and T2 relaxation times using a multi-echo multi slice (MEMS) sequence [25].
2.2. Cellular uptake
The uptake of SPION is an indicator of effective delivery of therapeutics in cells. The flow cytometry method was utilized for quantitative evaluation of SPION uptake in C4-2 PC cells. For efficient quantification of SPION-Dtxl in addition to Dtxl, Coumarin 6 dye, which can be detected as green fluorescent in a flow cytometer in the FL1 channel (488 excitation, Blue laser, 530 ± 15 nm, FITC/GFP), was encapsulated. In this study, PC cells (2.5 × 105 cells/well in 2 mL media) in 6-well plates were treated with 2 μg C6-SPION-Dtxl and SPION-Dtxl (no dye) for 2 hrs. After that, cells were washed with PBS, trypsinized, and collected in 2 mL media, centrifuged, washed twice with PBS, and finally collected in 2 mL phenol-red free medium for flow cytometry analysis as mentioned in our previous report [38]. An Accuri C6 Flow Cytometer (BD Biosciences, San Jose, CA) was used to determine the fluorescence levels of C6-SPION-Dtxl. Evaluation of specific endocytosis pathway mechanisms of SPION could provide valuable information. Therefore, uptake mechanisms of SPION were examined in C4-2 and PC-3 (5×106 cells in each well, in 6-well plates) PC cell lines. Cells were treated with various endocytic inhibitors, chlorpromazine (CPZ) (10 μg/ml, clathrin vesicle inhibitor), methyl-β-cyclodextrin (mCD) (1 mM, clathrin independent inhibitor), and nocodazole (NOCO) (10 μg/ml, microtubule-based endocytic inhibitor), for 1 hr at 37°C [39, 40]. Then cells were incubated with 50 μg equivalent SPION-Dtxl for an additional 2 hrs. Subsequently, cells were washed three times with 1X PBS (0.1 M, pH 7.4), prepared for Prussian blue staining [27] and collected in PBS suspension for absorbance measurements. No inhibitor treated cells were considered as actual control. The proportion of nanoparticle uptake was determined by measuring the Prussian blue absorbance at 510 nm using a SpectraMax Plus Plate Reader (Molecular Devices, Sunnyvale, CA).
2.3. Sub-cellular localization
In this study, to track SPION instead of Dtxl, Coumarin 6, which serves as green fluorescent marker, was encapsulated. To determine the cellular localization of Coumarin 6 labeled SPION (C6-SPION), C4-2 and PC-3 cells were seeded at a density of 2×104 on 8-well chamber glass slides (Nalgene Nunc Intl., Rochester, NY). Cells were then treated with 2 μg of Coumarin 6 equivalent C6-SPION for 3 hrs. After 2 hrs of incubation, cells were fixed using 2% paraformaldehyde for 10 min, permeabilized with 0.1% TritonX-100 in 1X PBS for 10 min, and washed twice with an additional 1X PBS, followed by a blocking step with 2% goat serum in PBS for 1 hr. Then cells were incubated with 30 nM Mito Tracker Red or 50 nM Transferrin from Human Serum, Texas Red® Conjugate or 75nM LysoTrackerR Red DND-99 (Life Technologies) to stain as a marker for mitochondria, endosome, and lysosome, respectively. Finally, nuclei were counter stained with DAPI (4′,6-diamidino-2-phenylindole, Life Technologies) and mounted in Vectashield Mounting Medium (Vector Labs, Burlingame, CA). Finally, cells were visualized under a laser confocal microscope (Carl Zeiss LSM 710, Oberkochen, Germany) under 400X magnification using oil immersion objective.
2.4. Proliferation assay
The cytotoxicity of Dtxl or SPION-Dtxl in C4-2 and PC-3 PC cells was measured with the CellTiter 96® AQueous One Solution Cell Proliferation Kit (Promega Corporation, Madison, WI) [41]. In brief, 5 × 103 cells in 100 μL culture medium were seeded into each well of 96-well plates and allowed to attach overnight. Then cells were treated with 1–50 nM of Dtxl or SPION-Dtxl for 48 hrs. After completion of the treatment period, 20 μL MTS reagent + 75 μL of fresh culture medium was replaced in the wells for 2–3 hrs. The intensity of the absorbed color of intracellular formazan was measured at 492 nm using a microplate reader (BioMate 3 UV–Vis Spectrophotometer, Thermo Scientific, Waltham, MA). The percentage of cell growth was calculated as the percentage of absorption of color intensity of treated cells to absorption of color intensity of non-treated cells.
2.5. Clonogenic assay
PC cell lines (C4-2 and PC-3) were treated with Dtxl or SPION-Dtxl to determine the degree of cytotoxic effect. For this assay, PC cells (500 cells/well in 2 mL medium) were suspended uniformly and seeded in 6-well plates. After 3 days, initial growth of colonies was observed, cells were treated with 2.5 and 10 nM Dtxl or Dtxl equivalent SPION-Dtxl for a week under standard cell culture conditions. Equivalent concentrations of DMSO or SPION treated cells were considered as controls for this experiment. Then media was replaced with fresh media without drugs and maintained under standard cultural condition until day 14. At the end of the treatment, cells were washed with 1X PBS, fixed with cold methanol, subsequently stained with hematoxylin for 5 min, and rinsed with double distilled water several times. The colonies were photographed using a BioSpectrum® 500 Imaging System (UVP, Upland, CA) [24].
2.6. Tubulin stabilization assay
The influence of Dtxl or SPION-Dtxl on tubulin stabilization was studied using immunofluorescence assay. For this, PC cells (1×107 cells/mL in each slide) were seeded in chambered slides and grown for 24 hrs. Cells were treated with 10 nM Dtxl or Dtxl equivalent SPION-Dtxl or respective controls (DMSO or SPION) for 8 hrs. After incubation, cells were washed with 1X PBS, fixed with ice-cold methanol for 20 min, blocked with 10% goat serum for 1 hr, incubated overnight with β-tubulin antibody (1:50 cell signaling, #CS2146) at 4°C on a rocker. After washing, cells were then probed with FITC-conjugated goat anti-rabbit secondary antibody (1:200, #111-095-003, Jackson ImmunoResearch Labs, West Grove, PA) for 1 hr, and nuclei were counterstained with DAPI mounting medium and visualized under a laser confocal microscopy (Carl Zeiss LSM 710).
2.7. Immunoblot analysis
For immunoblot analysis, PC cells (1×106 in 10 mL medium) were seeded in a 100 mm culture dish and treated with 5 or 10 nM Dtxl or Dtxl equivalent SPION-Dtxl for 48 hrs. Cells were lysed with 2X SDS lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA). The process of extraction, quantification of proteins, and the Western blotting method were followed as described previously [39]. Primary antibodies [β-actin (#4967), BAD (#9292), Bax (#2772), Bcl-xL (#2762), Bcl-2 (#2876), p53 (#9282), Cleaved PARP (#5625), PSMA (#12815), and MDR1 (#13342)] and secondary antibody [horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody] (Promega) were used in this study. The protein bands were detected and imaged with the Lumi-Light Detection Kit (Roche, Nutley, NJ) using a BioSpectrum® 500 Imaging System. The densitometric studies of the acquired blots were analyzed using GelQuant® Software (DNR Bio-Imaging Systems, Jerusalem, Israel).
2.8. Generation of PSMA targeted SPION-Dtxl
To generate a PSMA targeted SPION-Dtxl formulation, PSMA monoclonal antibody (PSMA MAb, J591, acquired from Dr. Neil Bander, Weill Cornell Medical College, New York by Dr. Chauhan) was used to conjugate on SPION-Dtxl. The conjugation of J591 onto SPION-Dtxl was prepared through two steps as discussed below. In the first step, to obtain an active surface moiety on SPION-Dtxl, 2 mg of SPION-Dtxl in 10 mM sodium bicarbonate reaction buffer, (pH 8.0 with 0.1 PEG) was mixed with 10 mg of the coupling agent Succinate Succinimidyl PEG (NHS-PEG-NHS, MW 5000, NANOCS, Boston, MA). After 1 hr, excess or un-reacted coupling agent was removed by centrifugation (10,000 rpm for 20 min) and the activated NHS coupled SPION-Dtxl was resuspended in reaction buffer. In the second step of reaction, 100 μg of J591 in reaction buffer was slowly added at 4 °C for 18 hrs to this NHS activated SPION-Dtxl to generate PSMA targeted SPION-Dtxl. The unconjugated J591 MAb was removed by centrifugation and thorough washing.
2.9. Cellular targeting
To further verify the quantitative cellular specific targeting of J591-SPION-Dtxl by PC cells, immunofluorescence microscopy was performed. For this study, C4-2 (PSMA+) and PC-3 (PSMA−) PC cells were cultured in 4-well chamber slides at a density of 2×105 cells per well. Cells were blocked with 2% goat serum for 1 hr and then cells were treated with 6 μg SPION-Dtxl or J591-SPION-Dtxl for 2 hrs. Then the culture medium was removed, cells were washed with 1X PBS (twice), fixed in 2% para-formaldehyde, permeabilized, and incubated with Cy3 labeled secondary antibody (Cell Signaling Technologies, Danvers, MA). After further washing, the cover slips were mounted on glass slides using aqueous anti-fade medium (Vector Laboratories) and the slides were visualized under a laser confocal microscope (Carl Zeiss LSM 710). The corrected total cell fluorescence (CTCF) was calculated for 10 cells in each field using ImageJ Software (US NIH, http://imagej.nih.gov/ij/, 1997–2015) using the equation: CTCF = Integrated Density - (Area of selected cell × Mean fluorescence of background readings).
2.10. Proliferation kinetics and clonogenic potential
To examine improved and targeted cytotoxicity of the J591-SPION-Dtxl, a cell proliferation kinetic method was followed instead of an MTS assay. In this study, C4-2 cells (10,000 cells/well in 1 mL medium) were seeded in six different sets of 6-well plates. After overnight cell attachment, cells were treated with 5 nM Dtxl, SPION-Dtxl, or J591-SPION-Dtxl. On each day, cells were trypsinized and viability was counted via a trypan exclusion study employing a Countess™ Automated Cell Counter [26] (Invitrogen, Carlsbad, CA). Cell viability was calculated as a percentage of treated compared to non-treated cells [42]. Cell images were captured under light microscope (200X magnification) (Eclipse Ti, Nikon, Melville, NY). Similarly, J591-SPION-Dtxl targeted efficacy was examined using clonogenic potential assay as mentioned in Section 2.6 but at a low concentration of 0.5 nM instead of 2.5 nM.
2.11. Ex-vivo tumor targeting
The tumor targeting ability of J591-SPION-Dtxl was further evaluated through ex-vivo studies on histological models such as C4-2 orthotopic and subcutaneous and PC-3 orthotopic xenograft mice tumor tissues (tissue slides procured from Dr. Bilal Hafeez). The slides were processed by employing a heat-induced antigen retrieval immunohistochemistry technique using a Biocare Kit (Biocare Medical, Concord, CA) as described previously [43]. Briefly, paraffin-embedded (FFPE) tumor tissues were deparaffinized, rehydrated, treated with 0.3% hydrogen peroxide or peroxidazed solution, and processed for antigen retrieval. Then tissues were blocked with 4% donkey serum (Jackson ImmunoResearch Laboratories) to avoid non-specific binding using background sniper. These tissues were incubated with 30 μg SPION-Dtxl (untargeted) and J591-SPION-Dtxl (PSMA targeted) for 1 hr, then washed with PBS for 30 mins. Finally, tissue slides were mounted using DAPI containing mounting medium. These slides were evaluated for targeting ability using a laser scanning confocal microscope (Carl Zeiss LSM 710).
2.1.2. Statistical analysis
The data values were processed using Microsoft Excel 2007 and graphs were drawn using Graphpad Prism 5 (Graphpad Software, LaJolla, CA) or Origin 6.1 (OriginLab Corp, Northampton, MA) software. All data were presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using an unpaired, two-tailed Student’s t-test. The level of significance was set at *p < 0.05.
3. Results
The main objective of this work was to encapsulate docetaxel (a clinically used anti-cancer drug) in β-cyclodextrin and pluronic F127 polymer inner layers in a self-assembly manner (SPION-Dtxl formulation) to improve its therapeutic efficacy. An additional aim was to achieve a specific targeting capacity of SPION-Dtxl by functionalization of anti-PSMA J591 MAb on the surface of nanoparticles to offer both passive and active targeting of PC cells.
3.1. SPION-Dtxl preparation and characterization
A SPION formulation composed of iron oxide core with β-cyclodextrin and pluronic F127 polymer layers [25] was prepared, which was used to encapsulate docetaxel (SPION-Dtxl). In this method a major portion of Dtxl was entrapped physically in the hydrophobic cavity of β-CD. This formulation was stable in suspension due to the stealth capabilities of surface coating arising from pluronic polymer chains (Fig. 1A).
A representative TEM image of SPION-Dtxl revealed an aggregation size of ~100 nm while individual particle size was 8–10 nm (Fig. 1B). The particle size of SPION-Dtxl was 139.5±2.16 nm, which was a slight increase compared with SPION (127.1±4.97 nm), as determined by DLS (Fig. 1C). This indicates Dtxl was incorporated into SPION. A slight increase in particle size was observed when particles were measured in serum. However, overall particle size distribution was < 0.2. The results indicated that zeta potential of SPION-Dtxl (−10.9 mV) slightly varied compared to SPION (−11.2 mV) (Fig. 1D). FTIR analysis of SPION-Dtxl (Fig. 1E) exhibited additional major peaks at 3,000 cm−1 due to O-H stretch of the carboxyl group, 1,752 cm−1 due to C=O stretch of the ester group, and 1,055/1010 cm−1 due to C-O-C and C-C/C-O vibrations, in addition to the SPION [25] characteristic peaks. FTIR of docetaxel has been provided in supplementary Fig. S1 for a comparison with SPION-Dtxl. Thermogravimetric analysis disclosed greater weight loss in SPION-Dtxl due to Dtxl encapsulation in SPION compared to SPION alone (Fig. 1F). Weight loss of Dtxl as displayed in the second regime may correspond to Dtxl decomposition. The diffraction data of SPION was consistent with JCPDS file no: 01–088-035 which showed XRD patterns at 2θ = 30.1°, 36.2°, 42.4°, 52.5°, 57.5° and 62.2° due to characteristic peaks of face-centered cubic lattice structures 220, 311, 400, 422, 440 and 511 of the Fe3O4 crystal structure, respectively (Fig. 1G). This revealed the nanocrystalline nature of SPION. There were no additional XRD crystalline peaks of Dtxl observed in SPION-Dtxl. This indicates that the Dtxl was well dispersed in the SPION and may exist in its amorphous form at the β-cyclodextrin and pluronic F127 polymer layers. These results following encapsulation of drug(s) in nanoparticles are in agreement with reported literature [44].
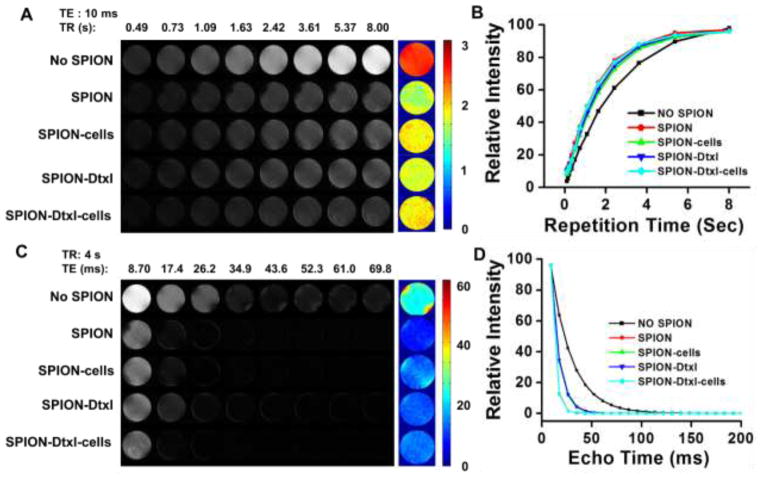
Further, there was no significant change noticed in the MR imaging characteristic of SPION-Dtxl compared to SPION in Phantom gels (Fig. 2). This indicates Dtxl loading in SPION was not affecting its inherent characteristic (Fig. 2A,C). This behavior of SPION-Dtxl persisted even in C4-2 cells (Fig. 2A,C). T1 relaxation times were recorded as 1.81±0.09 s and 1.96±0.12 s for SPION-Dtxl and SPION-Dtxl with C4-2 cells, respectively, which are values close to SPION and SPION with C4-2 cells, 1.68±0.10 s and 1.93±0.11 s, respectively (Fig. 2B). This pattern was true even in T2 relaxation times; i.e., SPION-Dtxl (13.74±1.17 ms) and SPION-Dtxl with C4-2 cells (13.82±1.06 ms) have values that are close to SPION (12.66±0.077 ms) and SPION with C4-2 cells (13.91±1.10 ms) (Fig. 2D).
Figure 2. Magnetic resonance imaging properties of SPION-Dtxl formulation.
A,C) Signal intensity gray scale T1 and T2 relaxation MR images of agarose phantom gels of SPION, SPION in C4-2 cells, SPION-Dtxl, and SPION-Dtxl in C4-2 cells. Representative color T1 and T2 relaxation of phantom gels of SPION, SPION in C4-2 cells, SPION-Dtxl, and SPION-Dtxl in C4-2 cells. B,D) T1 and T2 relaxation curves of phantom gels of SPION, SPION in C4-2 cells, SPION-Dtxl, and SPION-Dtxl in C4-2 cells. Note: No SPION indicates only phantom gel (no sample). A part of the image was reproduced with permission from Elsevier.
3.2. SPION-Dtxl exhibits cellular internalization, accumulation and cytosol delivery in PC cells
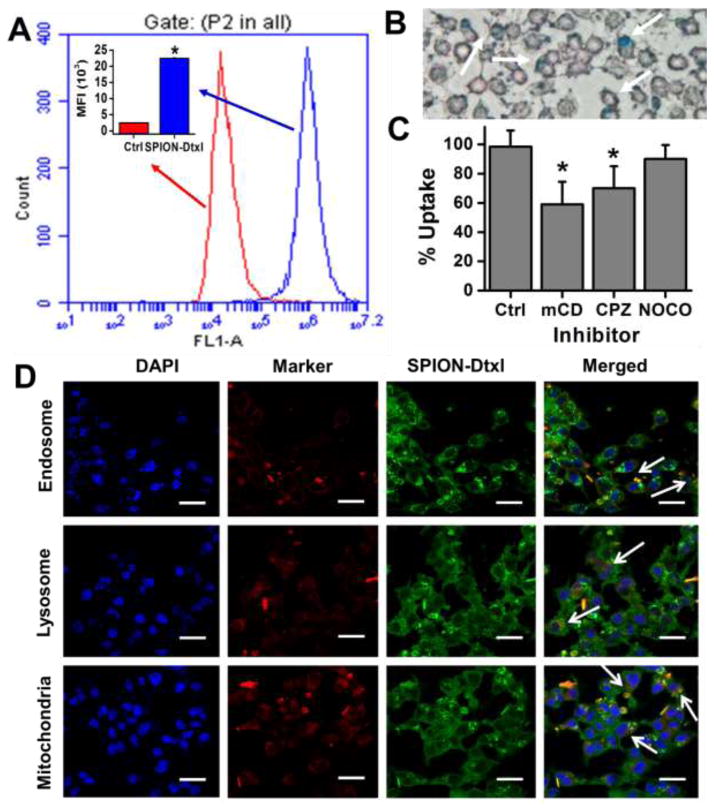
In the current study, the uptake/internalization efficiency of SPION-Dtxl in C4-2 cells at a concentration of 2 μg/ml was measured using mean fluorescence intensity of cells through flow cytometric analysis (Fig. 3A and inset). An intense green fluorescence in cells annotated its internalization ability. A similar observation of internalization was made by microscopy using Prussian blue staining (Fig. 3B). We subsequently focused on assessing the internalization pathways. Upon treatment with endocytosis inhibitors, SPION-Dtxl internalization was prominently reduced. The inhibitory capacity of SPION-Dtxl was: methyl-β-cyclodextrin (mCD) > chlorpromazine (CPZ) > nocodazole (NOCO). Methyl-β-cyclodextrin inhibits up to 40% uptake of SPION-Dtxl, which strongly indicates that uptake of SPION-Dtxl occurred through clathrin independent endocytosis followed by clathrin dependent with minor involvement of tubulin mediation endocytosis (Fig. 3C).
Figure 3. Cellular uptake and sub-cellular fate of SPION-Dtxl.
A–C) Cellular uptake of SPION-Dtxl in C4-2 cells. A) Cells were treated with SPION-Dtxl labeled with Coumarin 6 dye (2 μg) for 3 hrs. Representative cellular uptake chart of SPION-Dtxl in which fluorescence levels in the FL1 channel were analyzed using an Acuri C6 Flow Cytometer. Inset represents mean fluorescence intensity of SPION-Dtxl in C4-2 over C4-2 control cells. Bars represent mean ± SEM; n = 3; *p < 0.05. B) SPION-Dtxl efficiently internalize in C4-2 cancer cells. This was evaluated through Prussian blue staining (blue color staining can be seen visually). Original magnification 200X. White arrows point towards Prussian blue stain. C) Uptake of SPION-Dtxl in the presence of various endocytosis inhibitors. In the presence of endocytosis inhibitors the uptake is significantly controlled. D) Fate and sub-cellular localization of SPION-Dtxl. C4-2 prostate cancer cells (2.5 × 103/well) were exposed to 2 μg Coumarin 6 labeled SPION-Dtxl for 3 hrs, then cells were washed, fixed and permeabilized. For co-localization evaluation, Transferrin from Human Serum, Texas Red® Conjugate LysoTracker Red, and Mito Tracker Red (red fluorescence) were used to stain as a marker for endosome, lysosome, and mitochondria, respectively. DAPI was used to stain nuclei. Yellow color indicates co-localization of Coumarin 6 labeled SPION-Dtxl in cells. Cells were visualized under 400X magnification (Bar = 4 μm). White arrows indicate colocalization of SPION-Dtxl with endo-, lyso-some, and mitochondrial markers.
Since SPION-Dtxl has been shown to participate in the endocytosis internalization process, we sought to examine its sub-cellular fate using confocal microscopic analysis. SPION-Dtxl was efficiently internalized and localized in C4-2 and PC-3 PC cells (3 hrs post-incubation). An intense cytosolic staining and strong co-localization of SPION-Dtxl to the mitochondrial marker, mitotracker, and less co-localization of SPION-Dtxl with endosome(s) and late endosome/lysosome marker(s) were observed in these cells. The degree of co-localization was: mitochondria > endosome > lysosome. This suggests that SPION-Dtxl is capable of escaping from late endosomal and lysosomal compartments and reaching into the cytosol/mitochondria [45, 46], resulting in efficient functioning (Fig. 3D).
3.3. SPION-Dtxl is effective in inducing anti-cancer effect in PC cells
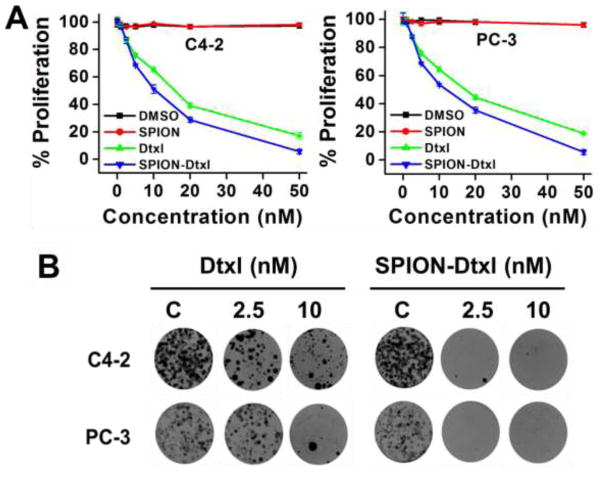
To quantify the effect of Dtxl and SPION-Dtxl, cell proliferation was assessed by an MTS colorimetric method. Both controls, DMSO and SPION, did not affect cell growth; however, Dtxl and SPION-Dtxl exhibited a dose-dependent inhibitory effect in cell proliferation assays (Fig. 4A). The IC50 of SPION-Dtxl was 8.5 and 9.2 nM while Dtxl ranged from 13.5 and 15.6 nM for C4-2 and PC-3 cancer cell lines, respectively. The colony forming potential of cancer cells reflects their growth in vivo. Thus, we examined the effects of the SPION-Dtxl formulation on inhibition of long term colony formation. The results demonstrated that the durable effects of SPION-Dtxl caused significant repression in colony-forming ability of PC cells (Fig. 4B). The number of colonies was decreased upon a 15-day treatment of Dtxl in a dose dependent manner in C4-2 and PC-3 cells (Fig. 3B, left panel). The exposure of cells to SPION-Dtxl was associated with significant repression of cell growth resulting in a decrease in the number of colonies at 2.5 nM (Fig. 4B, right panel), whereas higher concentration of SPION-Dtxl was required for complete inhibition of PC cell growth. Both assays suggest that PC cells are relatively more responsive to SPION-Dtxl treatment over free Dtxl.
Figure 4. SPION-Dtxl formulation inhibits proliferation and clonogenicity of PC cells.
A) PC cells (5×103/well in 96-well plate) were treated with 0–50 nM Dtxl, SPION-Dtxl or respective control for 48 hrs and proliferative ability of cells was assessed by MTS assay. B) PC cells (500) were incubated with 0, 2.5, 5, and 10 nM Dtxl or equivalent SPION-Dtxl or respective controls (DMSO/SPION, represented as C). On Day 14, cells were PBS rinsed and stained with hematoxylin. Photographs of clonogenic pattern represent superior inhibition of clonogenic formulation with SPION-Dtxl.
3.4. SPION-Dtxl induces alteration of cell-regulatory, survival and apoptotic proteins in PC cells
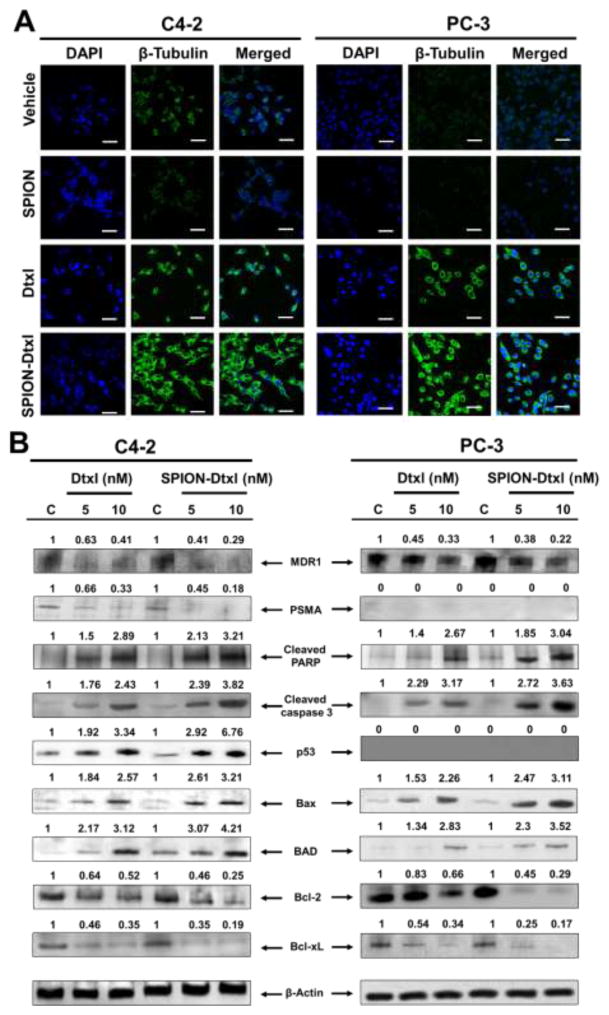
The next goal of this study was to investigate the molecular effects of the SPION-Dtxl formulation, which are expected to be modulated upon efficient cellular uptake [12]. To examine such pronounced effects, we elucidated Dtxl microtubule-stabilizing activity in PC cells. Compared to free Dtxl, SPION-Dtxl treatment demonstrated an increased microtubule polymer mass, thereby inducing microtubule bundles in cells (green fluorescence for microtubulin staining) (Fig. 5A). Significance of this result is related to maintaining Dtxl activity in SPION-Dtxl more effectively.
Figure 5. Tubulin stabilization and apoptotic effects of SPION-Dtxl on PC cells.
A) Comparative effects of SPION-Dtxl and Dtxl through docetaxel-induced microtubule stabilization, where cells have been treated with equivalent concentrations of 10 nM Dtxl or SPION-Dtxl for 8 hrs. Original magnification 400X (Bar = 4 μm). B) SPION-Dtxl inhibits survival and induces pro-apoptotic signaling and activation of caspases. C4-2 and PC-3 PC cells were treated with 5 and 10 nM Dtxl or equivalent concentrations of SPION-Dtxl for 48 hrs, and the cell lysates were collected and immunoblotted. The results were consistent in two independent sets of experiments. Note: PC-3 cells are hemizygous for chromosome 17p and their single copy of the p53 gene has a bp deletion and thus PC-3 cells are p53-null. PC-3 cells are PSMA-null too. Thus we did not observe any changes in these two proteins in western blot analysis.
Immunoblot analysis was performed to investigate the involvement of SPION-Dtxl on mechanistic anti-cancer effects. The effect of Dtxl/SPION-Dtxl on the expression levels of various proteins, such as Bcl2, Bcl-xL, Bax, Bad, cleaved PARP, cleaved caspase 3, p53, PSMA, and MDR1, was assessed in PC cells. Dtxl/SPION-Dtxl exposure to C4-2 cells resulted in an increased expression of p53, a key tumor suppressor protein. Additionally, expression of some key pro-apoptosis associated proteins (Bax, Bad, cleaved PARP and caspase 3) also increased following Dtxl and SPION-Dtxl treatment. Contrary to this, the expression of key pro-survival proteins (Bcl2 and Bcl-xL) was effectively down-regulated by SPION-Dtxl treatment. Additionally, cell regulatory proteins such as PSMA were found to be downregulated in C4-2 cells after treatment (Fig. 5B). The pro-apoptotic Bax protein was upregulated and the antiapoptotic Bcl-2 was downregulated in treated cells; thus, the Bax/Bcl-2 ratio was increased by treatments, suggesting that Dtxl induces apoptosis. Moreover, SPION-Dtxl treatment was found to be more effective in reducing expression of MDR1 protein over Dtxl treatment. All together, co-relation was found for the improved therapeutic activity of SPION-Dtxl, which can be explained by the intrinsic difference in molecular effects.
3.5. J591-SPION-Dtxl promotes specific targeting to PC
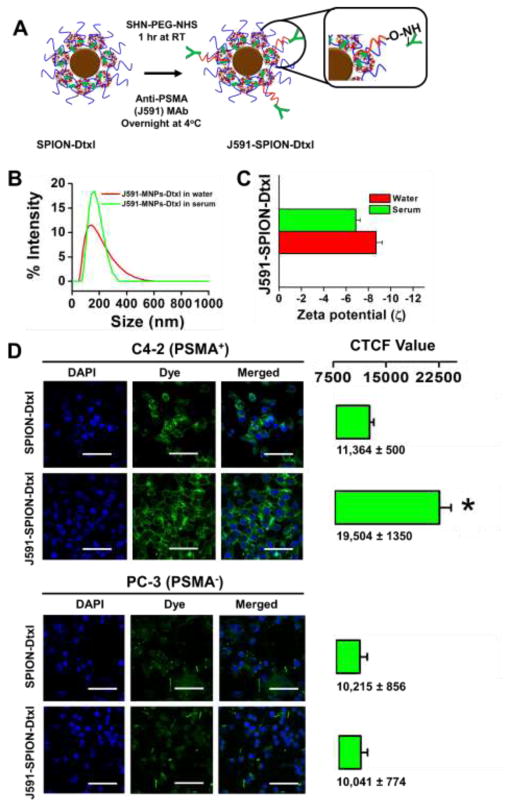
Our studies have confirmed the cytotoxic behavior of SPION-Dtxl against PC cells; however, it is not a specific targeting mechanism, which is a major concern in the drug delivery field. This prompted us to generate a SPION-Dtxl formulation with tumor specific targeting capability. For this, we have chosen PSMA, which overexpresses in almost all prostate tumors with minimal expression in normal tissues. Based on this fact, a human antibody (J591) against PSMA has been approved by FDA as a biomarker and imaging agent against PC [29, 47]. Therefore, we generated a targeted SPION-Dtxl by conjugating J591 through PEG-NHS groups that were coated on the surface of SPION-Dtxl (Fig. 6A). After conjugation with J591, the particle size of J591-SPION-Dtxl (166.9±1.26) varied only slightly from SPION-Dtxl and it still possessed a negative zetapotential (−7.58 mV) (Fig. 6B–C). Specific affinity of J591-SPION-Dtxl was validated using PSMA+ and PSMA− PC cells (Fig. 6D). In this comparative study, both C4-2 and PC-3 cells were treated and processed for confocal microscopy imaging. Our results showed pronounced green fluorescence at peripheral regions of C4-2 cells treated with J591-SPION-Dtxl. However, we did not find a similar observation in PC-3 cells, annotating the targeting and high affinity of J591-SPION-Dtxl with PSMA+ cells (Fig. 6D). We calculated CTCF fluorescence intensity values of 10 different cells in 5 different set of images to quantify the targeting potential, which was significant in the case of C4-2 cells.
Figure 6. Generation of PSMA targeted SPION-Dtxl formulation and its characterization.
A) Schematic illustration of PSMA antibody (J591) conjugation onto SPION-Dtxl. Inset represents conjugation through NHS of PEG groups on SPION-Dtxl. B–C) Characterization studies of J591-SPION-Dtxl for particle size and zeta potential. D) Cellular uptake of PSMA specific targeting using J591-SPION-Dtxl. Cells were treated with 6 μg of Coumarin 6 dye loaded SPION-Dtxl or J591-SPION-Dtxl for 3 hrs. Cells were visualized under confocal microscope with 400X magnification (Bar = 4 μm). Quantitative measurement of cell fluorescence through CTCF intensity using ImageJ Software.
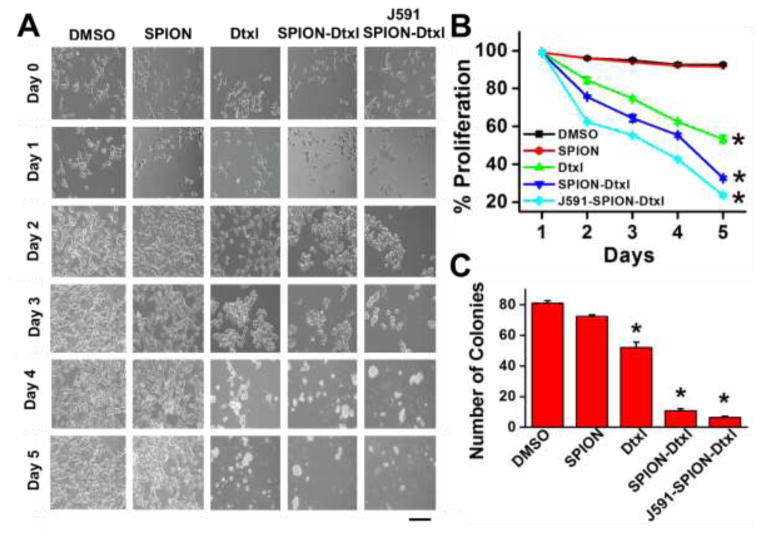
The enhanced efficacy of J591-SPION-Dtxl was reflected in its specificity towards PSMA antigen in the PSMA+ (C4-2) cell line. Treatment with 5 nM of native Dtxl or Dtxl equivalent SPION-Dtxl or J591-SPION-Dtxl for 1–5 days was executed through the kinetic approach and percent viability was calculated each day using trypan blue exclusion studies. We observed a sustained decrease in percent viability on each day. On day 5, cell viability was 5, 17.5 and 26% with J591-SPION-Dtxl, SPION-Dtxl, and Dtxl treatments, respectively (Fig. 7A) and relevant representative images were captured under 200X magnification (Fig. 7B). Cells showed morphological changes such as shrinkage, retraction of filopodia, and spheroids formation after treatment with native Dtxl or Dtxl equivalent to SPION-Dtxl and J591-SPION-Dtxl. These changes in morphology are clear signs of apoptosis with disruption in membrane integrity [48]. These cellular changes were more pronounced with increasing concentrations of SPION-Dtxl and J591-SPION-Dtxl. Also, colonogenic results displayed a progressive decrease in the number of colonies after treatments in the order: J591-SPION-Dtxl > SPION-Dtxl > Dtxl. We observed the lowest number of colonies during J591-SPION-Dtxl treatments in comparison to other groups (Fig. 7C). The colony formation study further substantiated the above results revealing the targeting attribute of J591-SPION-Dtxl.
Figure 7. J591-SPION-Dtxl targeted formulation exhibits superior anticancer effects in PSMA+ PC cells.
A–B) The anti-cancer effects of the J591-SPION-Dtxl formulation were evaluated through a 5 day kinetics study. C4-2 (PSMA+) cells (10,000 cells/well) were treated with 5 nM Dtxl or Dtxl equivalent SPION-Dtxl and J591-SPION-Dtxl for 1, 2, 3, 4, and 5 days. At the end of each day, percent cell viability was counted through trypan blue exclusion using a Countess™ Automated Cell Counter. Data represent mean ± SEM; n = 3; *p < 0.05. B) Cell images were captured on each day under an inverted light microscope. Representative Day 5 cell images were documented. Original magnification 200X (Bar = 8 μm). C) Colonogenicity assay was performed to assess the ability of cells to form colonies following treatment with 0.5 nM Dtxl or equivalent SPION-Dtxl, J591-SPION-Dtxl and respective controls (DMSO, SPION, and J591) for 2 weeks. At the end of treatment, cells were PBS rinsed and stained with hematoxylin. Photographs were taken of the clonogenic pattern in a UVP light cabinet and colonies were analyzed using GelQuant® software.
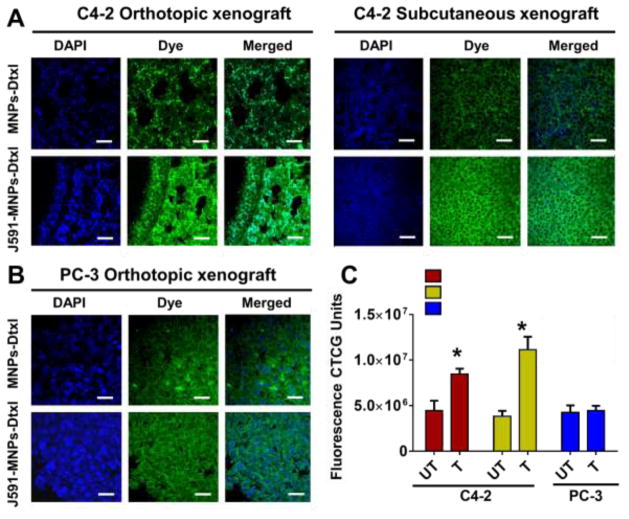
Although J591-SPION-Dtxl showed in vitro specific targeting against PSMA+ PC cells, to further ascertain its active targeting to PC, we selected a panel of C4-2 orthotopic/subcutaneous xenograft tumors (expressing PSMA) and PC-3 tumors (non-expressing PSMA). Interestingly, there was bright and intense fluorescence in C4-2 xenograft and orthotopic tumor cells (Fig. 8A) as compared to PC-3 tumor cells (Fig. 8B). These results indicate that J591-SPION-Dtxl has specific targeting towards PSMA+ compared to PSMA− prostate tumors. Their respective fluorescence CTCF values are represented in Figure 8C. Quantification data showed only a moderate level of SPION-Dtxl in tissues but J591-SPION-Dtxl binds more specifically to the C4-2 tumor cells but not to PC-3 tumor cells. This not only revealed an enhanced targeting phenomenon but also established an approach to target PSMA, which is highly expressed in prostate tumors.
Figure 8. J591-SPION-Dtxl formulation targets PSMA+ PC tumors.
J591-SPION-Dtxl specifically targets A) C4-2 orthotopic and subcutaneous xenograft (PSMA+) tumors tissues, but not B) PC-3 orthotropic xenograft (PSMA−) tumor tissues. Histological tissue sections were treated with Coumarin 6 loaded SPION-Dtxl (untargeted NPs) or J591-SPION-Dtxl (targeted NPs) and processed slides were visualized under confocal microscope. Original magnification 400X (Bar = 4 μm). C) Quantitative measurement of cell fluorescence of SPION-Dtxl (untargeted NPs) or J591-SPION-Dtxl (targeted NPs) in tumor tissues through CTCF intensity using ImageJ Software. Fluorescence data represent mean ± SEM; n = 40 cells from 5 tissue slide images; *p < 0.05.
4. Discussion
Docetaxel is an effective FDA approved drug widely used in the chemotherapy of castration resistant and metastatic PC. However, normal organ toxicity and chemoresistance are major clinical challenges associated with docetaxel chemotherapy. A few strategies (Cellax™ and BIND-014) have been devised and implemented to prevent chemo-resistance, reduce side effects or to further enhance overall anti-cancer efficacy of Dtxl. Cellax™ is a cellulose based-docetaxel nanoparticle formulation while BIND-014 is a PSMA targeted docetaxel loaded biodegradable polymer nanoparticle formulation. Herein, our aim was to use PSMA targeted SPION for docetaxel delivery combined with an MR imaging option for PC treatment. SPION were chosen due to their extensive use in biomedical applications. Additionally, β-CD and pluronic polymers induce Dtxl binding to and stability properties of SPION, as confirmed through physico-chemical properties (particle size, surface charge, functional groups, thermal and crystalline nature) of SPION-Dtxl (Fig. 1). In agreement with previous reports [49], SPION-Dtxl and J591-SPION-Dtxl exhibit tunable particle size in aqueous medium and in serum (< 200 nm) with negative zeta potential. This range of particle size and zeta potentials is suitable for cancer therapeutics. More importantly, MR imaging characteristics of SPION were maintained even after loading with Dtxl and after internalization in PC cells (Fig. 2), which suggests their multi-purpose use as a theranostic modality.
SPIONs are promising as drug delivery carriers for cancer therapy. This is due to their significant cellular uptake/internalization, accumulation and release of loaded therapeutics in cancer cells. The key internalization study demonstrates SPION-Dtxl entry is a combination of caveolae and clathrin associated delivery but not micropinocytosis (Fig. 3). A strong co-localization of SPION-Dtxl with mitochondria with less co-localization of endo- and lyso-somes suggests that the internalized SPION-Dtxl was capable of escaping endo-/lyso-somal degradation (Fig. 3C). Release of SPION-Dtxl from these compartments to the cytosol allows the drug to take effective action (Fig. 3D). Such internalized drug formulation achieves significant improvements to loaded therapeutics.
Earlier studies with SPION loaded with curcumin had demonstrated a lower IC50 value than free curcumin in various cancer cell lines [24–27]. Our current results indeed indicate greater cytotoxic effects of SPION-Dtxl against PC cells over Dtxl in MTS and clonogenic potential assays (Fig. 4). This proves that the loaded therapeutic molecule was actively delivered to cancer cells. This prompted us to further examine the molecular mechanism of how SPION-Dtxl showed improved activity in PC cells (Fig. 5). The primary mechanism of action of Dtxl is binding to tubulin and microtubule stabilization, which resulted in cell cycle arrest and induction of apoptosis. This function of Dtxl was increased with SPION-Dtxl, which can be explained by the demonstrated effective uptake/internalization in PC cells. On the other hand, it may be possible that slow intracellular release contributes to limiting drug efflux and thus induce its greater activity. Disruption to the integrity of DNA induces transcription of pro-apoptotic proteins [50, 51], which counteracts the effects of antiapoptotic members of the Bcl-2 family, including Bcl-2 [52]. Excessive downregulation of Bcl-2 elicits the activation of caspase-dependent apoptosis [53, 54]. The ratio of Bax to Bcl-2 is a crucial determinant of cell survival or its ability to undergo apoptosis [55, 56]. Following Dtxl and SPION-Dtxl treatment, western blot results indicate a decrease in antiapoptotic Bcl-2 and Bcl-xL expression, and an increase in pro-apoptotic Bax and Bad proteins, confirming the involvement of intrinsic pathway to induce apoptosis. Increased expression of P53 (C4-2 only), cleavage in PARP protein, and activation of caspase further support our results of apoptosis. It has been known that increased expression of PSMA is linked to Dtxl resistance. A decrease in the expression of MDR1 and PSMA (C4-2 only) indicates resistance was overcome and PSMA associated growth was reduced in PC cells. However, the effective inhibition/induction of protein levels was observed with SPION-Dtxl, indicating that Dtxl is available through SPION in its active form for anti-cancer activity. These findings are consistent with our results obtained from cytotoxicity and colony formation.
A critical need exists to develop a specific-targeted drug delivery carrier that enhances specificity to PC. Among all PC cell surface proteins (prostate stem cell antigen, prostein, T cell receptor gamma alternate reading frame protein, six-transmembrane epithelial antigen of the prostate 1, and PSMA), PSMA is a cell-surface protein that is highly overexpressed in almost all prostate cancers and within the tumor neovasculature of other solid tumors [28]. Moreover, FDA-approved anti-PSMA antibody is a prostate-specific imaging agent and a target for the delivery of PC therapeutic agents [29, 30]. Therefore, we utilized PSMA as a PC specific target to deliver Dtxl using our SPION based delivery system. Specificity of this targeted formulation (J591-SPION-Dtxl) was confirmed by utilizing PSMA+ and PSMA− PC cells (Fig. 6). In PSMA+ PC cells a greater uptake was found compared to PSMA null PC cells (Fig. 6D). J591 MAb conjugated SPION-Dtxl exhibited superior anti-cancer potential in PSMA+ PC cells indicating its applicability for targeted delivery of therapeutics (Fig. 7). Further, our immunofluorescence data confirmed that J591-SPION-Dtxl specifically targeted PSMA+ PC cell derived xenograft tumors (Fig. 8).
Taken together, our studies reported SPION-Dtxl induced potent anti-cancer effects in PC cells via down-regulation of key prosurvival and multi-drug resistance proteins, and up-regulation of antiapoptotic proteins. In addition, J591 MAb conjugated to SPION-Dtxl increased specific targeting and anticancer potential in PC cells (in vitro), and targeted PC cells (ex-vivo), providing greater confidence for developing a PSMA targeted Dtxl nanoformulation as an adjunct to conventional chemotherapeutics.
5. Conclusions
Docetaxel can be efficiently formulated through binding within β-cyclodextrin and pluronic polymer layers on magnetic nanoparticles. The physico-chemical characterization and enhanced cellular uptake of SPION-Dtxl demonstrates its prospective use in cancer therapeutics. SPION-Dtxl exhibited superior anti-cancer activity (MTS and clonogenic assays), β-tubulin binding efficacy, and induced apoptosis potential. More importantly, SPION-Dtxl exhibited reduced MDR1 protein levels in PC cells that are highly associated with chemo-resistance. Furthermore, specific targeting of SPION-Dtxl to PSMA+ PC cells and tumor tissues advocates the development SPION-Dtxl as a targeted therapy for PC.
Supplementary Material
Highlights.
Docetaxel was successfully loaded in cyclodextrin layers of SPION formulation
SPION-Dtxl exhibits cellular internalization and delivery in prostate cancer cells
SPION-Dtxl induces superior anti-cancer effects in PC cells
PSMA MAb was conjugated via SHN-PEG-PEG linker onto SPION-Dtxl formulation
PSMA targeted J591-SPION-Dtxl promotes specific targeting of prostate cancer cells
Acknowledgments
This work was supported by a K22 award from the NIH (K22CA174841 to MMY). This work was also partially supported by the National Institutes of Health (R01 CA142736 to SCC and U01 CA162106 to SCC and MJ), and the College of Pharmacy Dean’s Seed and Instrument Grant(s) of the University of Tennessee Health Science Center (to SCC, MJ and MMY). The authors wish to thank Cathy Christopherson for editorial assistance.
Footnotes
Conflict of Interest
No conflict of interest exists in this work. However, SCC, MJ and MMY have filed a patent for MNPs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Can J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Figg WD, Woo S, Zhu W, Chen X, Ajiboye AS, Steinberg SM, Price DK, Wright JJ, Parnes HL, Arlen PM, Gulley JL, Dahut WL. A phase I clinical study of high dose ketoconazole plus weekly docetaxel for metastatic castration resistant prostate cancer. J Urol. 2010;183:2219–2226. doi: 10.1016/j.juro.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg JE, Ryan CJ, Weinberg VK, Smith DC, Hussain M, Beer TM, Ryan CW, Mathew P, Pagliaro LC, Harzstark AL, Sharib J, Small EJ. Phase I study of ixabepilone, mitoxantrone, and prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy: a study of the department of defense prostate cancer clinical trials consortium. J Clin Oncol. 2009;27:2772–2778. doi: 10.1200/JCO.2008.19.8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walczak JR, Carducci MA. Phase 3 randomized trial evaluating second-line hormonal therapy versus docetaxel-estramustine combination chemotherapy on progression-free survival in asymptomatic patients with a rising prostate-specific antigen level after hormonal therapy for prostate cancer: an Eastern Cooperative Oncology Group (E1899), Intergroup/Clinical Trials Support Unit study. Urology. 2003;62(Suppl 1):141–146. doi: 10.1016/j.urology.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Corcoran C, Rani S, O’Brien K, O’Neill A, Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, O’Driscoll L. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PloS one. 2012;7:e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang C. Overcoming docetaxel resistance in prostate cancer: a perspective review. Ther Adv Med Oncol. 2012;4:329–340. doi: 10.1177/1758834012449685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cengiz E, Karaca B, Kucukzeybek Y, Gorumlu G, Gul MK, Erten C, Atmaca H, Uzunoglu S, Karabulut B, Sanli UA, Uslu R. Overcoming drug resistance in hormone- and drug-refractory prostate cancer cell line, PC-3 by docetaxel and gossypol combination. Mol Biol Rep. 2010;37:1269–1277. doi: 10.1007/s11033-009-9501-y. [DOI] [PubMed] [Google Scholar]
- 8.Hu CM, Zhang L. Therapeutic nanoparticles to combat cancer drug resistance. Curr Drug Metab. 2009;10:836–841. doi: 10.2174/138920009790274540. [DOI] [PubMed] [Google Scholar]
- 9.Bilensoy E. Cationic nanoparticles for cancer therapy. Expert Opin Drug Deliv. 2010;7:795–809. doi: 10.1517/17425247.2010.485983. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Zhang N. How nanotechnology can enhance docetaxel therapy. Int J Nanomed. 2013;8:2927–2941. doi: 10.2147/IJN.S46921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganju A, Yallapu MM, Khan S, Behrman SW, Chauhan SC, Jaggi M. Nanoways to overcome docetaxel resistance in prostate cancer. Drug Resist Updat. 2014;17:13–23. doi: 10.1016/j.drup.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernsting MJ, Tang WL, MacCallum NW, Li SD. Preclinical pharmacokinetic, biodistribution, and anti-cancer efficacy studies of a docetaxel-carboxymethylcellulose nanoparticle in mouse models. Biomaterials. 2012;33:1445–1454. doi: 10.1016/j.biomaterials.2011.10.061. [DOI] [PubMed] [Google Scholar]
- 13.Ernsting MJ, Foltz WD, Undzys E, Tagami T, Li SD. Tumor-targeted drug delivery using MR-contrasted docetaxel - carboxymethylcellulose nanoparticles. Biomaterials. 2012;33:3931–3941. doi: 10.1016/j.biomaterials.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Hoang B, Ernsting MJ, Murakami M, Undzys E, Li SD. Docetaxel-carboxymethylcellulose nanoparticles display enhanced anti-tumor activity in murine models of castration-resistant prostate cancer. Int J Pharm. 2014;471:224–233. doi: 10.1016/j.ijpharm.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A, Low S, McDonnell K, Peeke E, Retnarajan B, Sabnis A, Schnipper E, Song JJ, Song YH, Summa J, Tompsett D, Troiano G, Van Geen Hoven T, Wright J, LoRusso P, Kantoff PW, Bander NH, Sweeney C, Farokhzad OC, Langer R, Zale S. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4:128ra139. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 16.Von Hoff D, Mita MM, Ramanathan RK, Weiss GJ, Mita AC, LoRusso PM, Burris HA, 3rd, Hart LL, Low SC, Parsons DM, Zale SE, Youssoufian H, Summa J, Sachdev JC. Phase 1 study of PSMA-targeted docetaxel-containing nanoparticle BIND-014 in patients with advanced solid tumors. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-2548. Epub ahead of Print ( http://www.ncbi.nlm.nih.gov/pubmed/26847057) [DOI] [PubMed]
- 17.He H, David A, Chertok B, Cole A, Lee K, Zhang J, Wang J, Huang Y, Yang VC. Magnetic nanoparticles for tumor imaging and therapy: a so-called theranostic system. Pharm Res. 2013;30:2445–2458. doi: 10.1007/s11095-013-0982-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadajkar AS, Menon JU, Kadapure T, Tran RT, Yang J, Nguyen KT. Design and Application of Magnetic-based Theranostic Nanoparticle Systems. Recent Pat Biomed Eng. 2013;6:47–57. doi: 10.2174/1874764711306010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namdeo M, Saxena S, Tankhiwale R, Bajpai M, Mohan YM, Bajpai SK. Magnetic nanoparticles for drug delivery applications. J Nanosci Nanotech. 2008;8:3247–3271. doi: 10.1166/jnn.2008.399. [DOI] [PubMed] [Google Scholar]
- 20.Nandwana V, De M, Chu S, Jaiswal M, Rotz M, Meade TJ, Dravid VP. Theranostic Magnetic Nanostructures (MNS) for Cancer. Cancer Treat Res. 2015;166:51–83. doi: 10.1007/978-3-319-16555-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hola K, Markova Z, Zoppellaro G, Tucek J, Zboril R. Tailored functionalization of iron oxide nanoparticles for MRI, drug delivery, magnetic separation and immobilization of biosubstances. Biotech Adv. 2015;33:1162–1176. doi: 10.1016/j.biotechadv.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Jiang S, Win KY, Liu S, Teng CP, Zheng Y, Han MY. Surface-functionalized nanoparticles for biosensing and imaging-guided therapeutics. Nanoscale. 2013;5:3127–3148. doi: 10.1039/c3nr34005h. [DOI] [PubMed] [Google Scholar]
- 23.Frimpong RA, Hilt JZ. Magnetic nanoparticles in biomedicine: synthesis, functionalization and applications. Nanomedicine. 2010;5:1401–1414. doi: 10.2217/nnm.10.114. [DOI] [PubMed] [Google Scholar]
- 24.Yallapu MM, Othman SF, Curtis ET, Gupta BK, Jaggi M, Chauhan SC. Multi-functional magnetic nanoparticles for magnetic resonance imaging and cancer therapy. Biomaterials. 2011;32:1890–1905. doi: 10.1016/j.biomaterials.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yallapu MM, Othman SF, Curtis ET, Bauer NA, Chauhan N, Kumar D, Jaggi M, Chauhan SC. Curcumin-loaded magnetic nanoparticles for breast cancer therapeutics and imaging applications. Int J Nanomed. 2012;7:1761–1779. doi: 10.2147/IJN.S29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yallapu MM, Chauhan N, Othman SF, Khalilzad-Sharghi V, Ebeling MC, Khan S, Jaggi M, Chauhan SC. Implications of protein corona on physico-chemical and biological properties of magnetic nanoparticles. Biomaterials. 2015;46:1–12. doi: 10.1016/j.biomaterials.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yallapu MM, Ebeling MC, Khan S, Sundram V, Chauhan N, Gupta BK, Puumala SE, Jaggi M, Chauhan SC. Novel curcumin-loaded magnetic nanoparticles for pancreatic cancer treatment. Mol Cancer Ther. 2013;12:1471–1480. doi: 10.1158/1535-7163.MCT-12-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiessling A, Wehner R, Fussel S, Bachmann M, Wirth MP, Schmitz M. Tumor-associated antigens for specific immunotherapy of prostate cancer. Cancers. 2012;4:193–217. doi: 10.3390/cancers4010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouchelouche K, Choyke PL, Capala J. Prostate specific membrane antigen- a target for imaging and therapy with radionuclides. Discovery Med. 2010;9:55–61. [PMC free article] [PubMed] [Google Scholar]
- 30.Damber JE, Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- 31.Olson WC, Heston WD, Rajasekaran AK. Clinical trials of cancer therapies targeting prostate-specific membrane antigen. Rev Recent Clin Trials. 2007;2:182–190. doi: 10.2174/157488707781662724. [DOI] [PubMed] [Google Scholar]
- 32.Haberkorn U, Eder M, Kopka K, Babich JW, Eisenhut M. New Strategies in Prostate Cancer: Prostate-Specific Membrane Antigen (PSMA) Ligands for Diagnosis and Therapy. Clin Cancer Res. 2016;22:9–15. doi: 10.1158/1078-0432.CCR-15-0820. [DOI] [PubMed] [Google Scholar]
- 33.Kasperzyk JL, Finn SP, Flavin R, Fiorentino M, Lis R, Hendrickson WK, Clinton SK, Sesso HD, Giovannucci EL, Stampfer MJ, Loda M, Mucci LA. Prostate-specific membrane antigen protein expression in tumor tissue and risk of lethal prostate cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:2354–2363. doi: 10.1158/1055-9965.EPI-13-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright GL, Jr, Grob BM, Haley C, Grossman K, Newhall K, Petrylak D, Troyer J, Konchuba A, Schellhammer PF, Moriarty R. Upregulation of prostate-specific membrane antigen after androgen-deprivation therapy. Urology. 1996;48:326–334. doi: 10.1016/s0090-4295(96)00184-7. [DOI] [PubMed] [Google Scholar]
- 35.Bostwick DG, Pacelli A, Blute M, Roche P, Murphy GP. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 184 cases. Cancer. 1998;82:2256–2261. doi: 10.1002/(sici)1097-0142(19980601)82:11<2256::aid-cncr22>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 36.Sweat SD, Pacelli A, Murphy GP, Bostwick DG. Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology. 1998;52:637–640. doi: 10.1016/s0090-4295(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 37.Khan S, Chauhan N, Yallapu MM, Ebeling MC, Balakrishna S, Ellis RT, Thompson PA, Balabathula P, Behrman SW, Zafar N, Singh MM, Halaweish FT, Jaggi M, Chauhan SC. Nanoparticle formulation of ormeloxifene for pancreatic cancer. Biomaterials. 2015;53:731–743. doi: 10.1016/j.biomaterials.2015.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yallapu MM, Gupta BK, Jaggi M, Chauhan SC. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J Colloid Interface Sci. 2010;351:19–29. doi: 10.1016/j.jcis.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Yallapu MM, Khan S, Maher DM, Ebeling MC, Sundram V, Chauhan N, Ganju A, Balakrishna S, Gupta BK, Zafar N, Jaggi M, Chauhan SC. Anti-cancer activity of curcumin loaded nanoparticles in prostate cancer. Biomaterials. 2014;35:8635–8648. doi: 10.1016/j.biomaterials.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Effects of transport inhibitors on the cellular uptake of carboxylated polystyrene nanoparticles in different cell lines. PloS one. 2011;6:e24438. doi: 10.1371/journal.pone.0024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manikandan M, Hasan N, Wu HF. Platinum nanoparticles for the photothermal treatment of Neuro 2A cancer cells. Biomaterials. 2013;34:5833–5842. doi: 10.1016/j.biomaterials.2013.03.077. [DOI] [PubMed] [Google Scholar]
- 42.Bernheim JL, Mendelsohn J, Kelley MF, Dorian R. Kinetics of cell death and disintegration in human lymphocyte cultures. Proc Natl Acad Sci U S A. 1977;74:2536–2540. doi: 10.1073/pnas.74.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, Thompson PA, Jaggi M, Chauhan SC. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget. 2014;5:7599–7609. doi: 10.18632/oncotarget.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain S, Spandana G, Agrawal AK, Kushwah V, Thanki K. Enhanced Antitumor Efficacy and Reduced Toxicity of Docetaxel Loaded Estradiol Functionalized Stealth Polymeric Nanoparticles. Mol Pharm. 2015;12:3871–3884. doi: 10.1021/acs.molpharmaceut.5b00281. [DOI] [PubMed] [Google Scholar]
- 45.Weissig V, Boddapati S, Jabr L, D’Souza GG. Mitochondria-specific nanotechnology. Nanomedicine. 2007;2:275–285. doi: 10.2217/17435889.2.3.275. [DOI] [PubMed] [Google Scholar]
- 46.Han M, Kickhoefer VA, Nemerow GR, Rome LH. Targeted vault nanoparticles engineered with an endosomolytic peptide deliver biomolecules to the cytoplasm. ACS Nano. 2011;5:6128–6137. doi: 10.1021/nn2014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne JR, Akhtar NH, Vallabhajosula S, Anand A, Deh K, Tagawa ST. Prostate-specific membrane antigen-based imaging. Urol Oncol. 2013;31:144–154. doi: 10.1016/j.urolonc.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lane JD, Allan VJ, Woodman PG. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J Cell Sci. 2005;118:4059–4071. doi: 10.1242/jcs.02529. [DOI] [PubMed] [Google Scholar]
- 49.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotech. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 51.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006;26:9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13:1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams JM, Cory S. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci. 2001;26:61–66. doi: 10.1016/s0968-0004(00)01740-0. [DOI] [PubMed] [Google Scholar]
- 54.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 55.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 56.Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys J. 2006;90:1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.