Abstract
Weak interactions mediated by dynamic linkers are key determinants of allosteric regulation in multidomain signalling proteins. However, the mechanisms of linker-dependent control have remained largely elusive. In the present article, we review an allosteric model introduced recently to explain how signalling proteins effectively sense and respond to weak interactions, such as those elicited by flexible linkers flanking globular domains. Central to this model is the idea that near degeneracy within the free energy landscape of conformational selection maximally amplifies the response to weak (~2RT), but conformation-selective interactions. The model was tested as proof of principle using the prototypical regulatory subunit (R) of protein kinase A and led to the unanticipated finding that dynamic linkers control kinase activation and inhibition by tuning the inhibitory pre-equilibrium of a minimally populated intermediate (apo R). A practical implication of the proposed model is a new strategy to design kinase inhibitors with enhanced potency through frustration-relieving mutations.
Keywords: allostery, cAMP, cGMP, covariance, NMR, protein dynamics, singular value decomposition (SVD)
Introduction
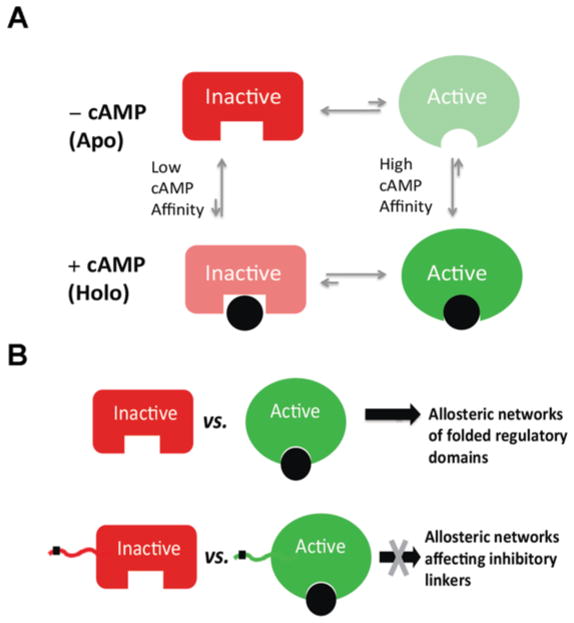
Allostery is a central mechanism of regulation in biological systems. Although initially discovered for multimeric proteins, allostery is now also accepted as a general regulatory mechanism for monomeric proteins [1–11]. Hence one of the simplest allosteric systems is a single domain that binds an effector ligand with a 1:1 stoichiometric ratio. For such as system, allostery is modelled in terms of a four-state thermodynamic cycle that results from the coupling of two equilibria, the ligand-binding equilibrium and the equilibrium between two states that are functionally distinct (i.e. active and inactive states; Figure 1A). These states typically differ in terms of structure and/or dynamics. In the absence of ligand (‘apo’ form) the inactive state is often the most populated (i.e. Lapo = [active]apo/[inactive]apo<1). However, if the effector ligand exhibits active versus inactive state selectivity, the population of the active state is increased upon binding (i.e. Lapparent = [active]total/[inactive]total>Lapo) and at saturation the relative active versus inactive fractions depend on both Lapo and the ratio between the state-specific dissociation constants (i.e. Lapparent,saturation = Lapo×Kd,inactive/Kd,active). Hence it is the active versus inactive state selectivity of the effector ligand that introduces the coupling between the binding and the inhibitory equilibria.
Figure 1. Thermodynamics of allostery and allosteric networks.
(A) Thermodynamic cycle for the allosteric coupling between activation and binding equilibria of a single regulatory domain. The inactive states are indicated by red rectangles, whereas the active states are shown as green ovals. The allosteric effector (e.g. cAMP) is shown as a black circle. The most populated states in either the apo or the holo forms are displayed with opaque colours. (B) Scheme of the relationship between the thermodynamics of allostery and allosteric networks. In the case of a well-folded regulatory domain, the comparative inactive versus active structural analysis is likely to reveal the residues that defined the allosteric networks within the structural module under investigation. However, key inhibitory sites are often located within partially unfolded linkers adjacent to globular domains (e.g. black square on linker). In this case, the comparative structural approach typically utilized for globular domains may not reveal the allosteric networks controlling the inhibitory linker.
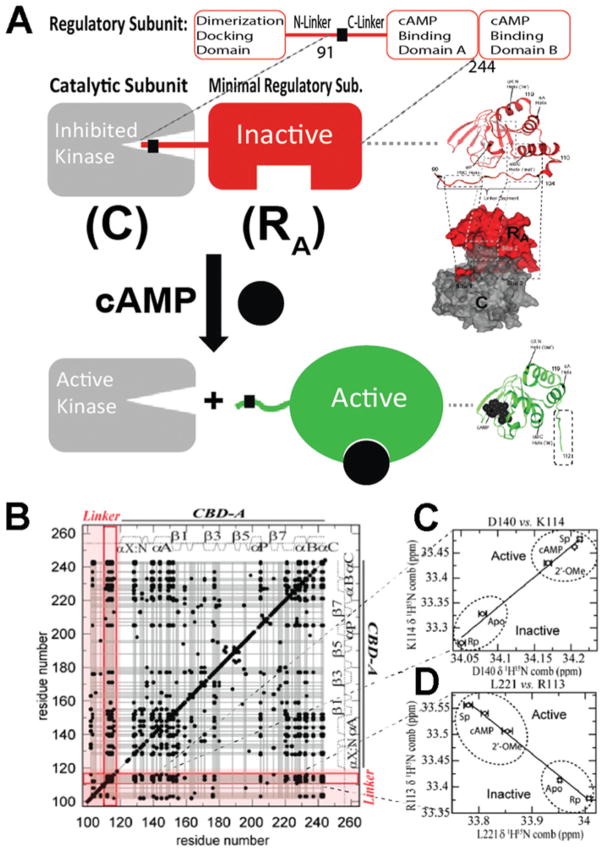
As a first step towards understanding the molecular basis for the state selectivity of the allosteric effector, structures of allosteric domains are typically solved for the apo form and in the presence of excess ligand [7,9–11]. Traditional structure-determination methods provide a valuable picture of the most populated states for either the apo domain (i.e. inactive state) or the ligand-saturated form (i.e. active state). When allostery arises from structural as opposed to purely dynamic changes, the comparison of active and inactive structures has the potential to reveal the network of residues mediating the allosteric propagation of the signal carried by the allosteric effector. Although this approach is highly informative for well-folded domains, limitations are encountered when allostery involves protein regions that are flexible and only partially structured (Figure 1B). This is often the case for inter-domain linkers, where critical functional sites are frequently located [12–15] (Figure 1B). For example, in PKA (protein kinase A), the key inhibitory element (RA) is composed of an N-terminal linker [RIα-(91–118)] followed by a CBD (cAMP-binding domain) [RIα-(119–244)] (Figure 2A). Under resting conditions, the RA linker docks into the active site of the catalytic subunit (C), is well-structured and clearly resolved in the crystal structure of the RA–C complex [16] (Figure 2A). However, when RA binds cAMP, the kinase is released and the RA linker becomes more solvent-exposed and partially flexible (Figure 2A). Hence, in the crystal structure of the RA–cAMP complex, only partial electron density is observed for the linker and several linker residues that are resolved are also affected by crystal packing [17] (Figure 2A). As a result, only partial information about the linker is obtained from traditional structure determination methods. However, linkers, even when not fully structured, have been shown to be functionally relevant [12–15]. Hence a full map of the structural dynamics of linkers is necessary to understand how function is allosterically regulated [17–22]. In the present article, we review recent advances [22] on how signalling can occur through linkers in the prototypical PKA (Figure 2A).
Figure 2. Role of the RA linker in PKA inhibition and allostery.
(A) cAMP-dependent activation of PKA. cAMP binding releases the inhibition of the catalytic subunit of PKA (C) by stabilizing a conformation of the regulatory subunit (R) that binds C only weakly. RA is defined as the minimal fragment of R that inhibits C in a cAMP-dependent manner. RA includes the linker and the adjacent CBD [i.e. RIα-(91–244)]. In the R–C complex, the inhibitory linker of R docks in the active site of C. Hence it is well-structured and clearly visible with conventional structure determination methods. In the R–cAMP complex, the linker of R is more solvent-exposed and flexible. Hence it is poorly resolved and not always visible through X-ray crystallography. However, the interactions between the flexible linker and the adjacent domain are effectively probed through NMR CHESCA, which establishes a residue–residue correlation matrix (B) based on pairwise inter-residue linear chemical shift relationships (C and D). The linker–domain CHESCA correlations are highlighted in red. Each black dot corresponds to a residue–residue combined chemical shift correlation with a Pearson’s correlation coefficient >0.98 in absolute value. (B–D) Adapted from [22]: Akimoto, M., Selvaratnam, R., McNicholl, E.T., Verma, G., Taylor, S.S. and Melacini, G. (2013) Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. U.S.A. 110, 14231–14236 with permission.
The linker adjacent to the N-terminal cAMP-binding domain (CBD-A) of PKA is flexible, but controls the CBD-A through state-selective interactions
The linker of RA [RIα-(91–118)] does not adopt a well-defined secondary structure, as indicated by the secondary chemical shifts, and is flexible in the picosecond–nanosecond time scale, as shown by overall low HN-NOE (nuclear Overhauser effect) values [22]. However, removal of the 91–118 linker results in pervasive and major chemical shift changes in the adjacent CBD (residues 119–244), suggesting that the 91–118 linker, although flexible, interacts with the latter [22]. The linker–CBD coupling is effectively probed through NMR using CHESCA (chemical shift covariance analysis). CHESCA relies on linear inter-residue chemical shift correlations (Figures 2B–2D) to unveil concerted responses to a common set of perturbations [19]. When such a perturbation set is a small library of cAMP analogues that modulate the activation equilibrium (Figures 2C and 2D), CHESCA provides a new way to dissect residue-specific contributions to binding and allostery, identifying otherwise elusive allosteric networks. The pairwise linear inter-residue correlations are best summarized through a correlation matrix, as illustrated in Figure 2(B). A striking feature of the CHESCA correlation matrix measured for RA is that it shows not only a large number of intradomain correlations, but also multiple linker–CBD correlations (Figure 2B). The point distribution for these correlations (Figures 2C and 2D) reflects the extent of PKA activation elicited by the cAMP analogues utilized for the CHESCA [i.e. Rp-cAMPS (Rp isomer of adenosine 3′,5′-monophosphothioate) is an inverse agonist, 2′-OMe-cAMP (2′-O-methyl-cAMP) is a partial agonist and Sp-cAMPS (Sp isomer of adenosine 3′,5′-monophosphothioate) is an agonist of cAMP] [20,21]. This observation suggests that the allosteric networks identified by CHESCA and spanning the flexible linker are functionally relevant.
The linker residues identified by CHESCA as belonging to the functional allosteric network of RA are affected by the adjacent domain in a state-selective manner, and it is the state-selectivity of these interactions that causes the linker–CBD coupling. This conclusion was confirmed independently by differential unfolding experiments in the presence and absence of the linker for apo, cAMP- and Rp-cAMPS-bound RA, revealing that the linker, although flexible, interacts selectively with the active state of the adjacent CBD. Even though weak (i.e. ~2RT) this state-selective interaction significantly affects the active–inactive equilibrium of the CBD due to its near-degeneracy [22]. The state-selectivity of the linker–CBD interactions was also corroborated by direct measurements of linker–CBD distances through PREs (paramagnetic relaxation enhancements). The measurement of PREs requires the covalent linkage of small molecules with an unpaired electron (‘spin label’). Spin labels should be judiciously located, as they must be close to the site to be probed through PREs, but not so close as to perturb or disrupt the native interactions [23]. In this respect, the CHESCA networks are useful, since they can be used to guide the choice of the spin label location. For instance, the CHESCA correlation matrix indicates the presence of multiple clusters between the CBD and Arg113 in the linker, but Ala109, although in the linker [16], does not appear to correlate significantly with the CBD. Hence Ala109 was considered a non-invasive site ideally suited for an A109C mutation and to link a spin label designed to probe the Arg113–CBD interactions. The PREs resulting from MTSL (methanethiosulfonate spin label) coupled to A109C were then measured for both cAMP- and Rp-cAMPS-bound RA, which stabilize the active and inactive states of the CBD respectively. As expected on the basis of the linker selectivity for the active state of the CBD, the PREs measured in the presence of cAMP were significantly larger than those observed for the Rp-cAMPS-bound form [22].
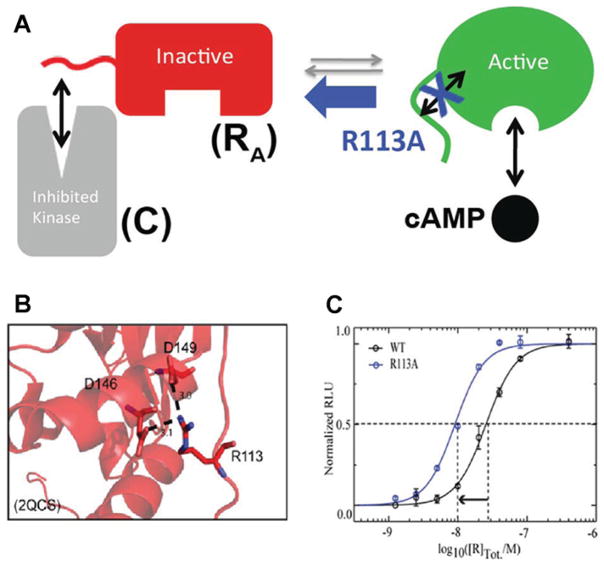
Using CHESCA to design a kinase inhibitor with enhanced affinity without compromising specificity
CHESCA suggests that Arg113 in the linker interacts selectively with the active rather than the inactive state of the adjacent CBD (Figure 3A). This result was exploited to design a mutant of RA with enhanced affinity and uncompromised specificity for C. The R113A mutations weakens the interactions between the linker and the active state of the CBD (Figure 3A), leading to increased populations of the inactive state, which are expected to result in higher affinity for C. This prediction was proven by measuring through a luminescent kinase assay the IC50 for the inhibition of C by RA. As shown in Figure 3(C), R113A RA inhibits C with an IC50 value significantly reduced relative to that measured for wild-type RA. Since Arg113 is not at the R–C interface [16], the R113A mutation is not expected to alter the specificity of R for C. In addition, it is notable that in the RA–C complex Arg113 forms salt bridges with the CBD [24] (Figure 3B), although C selects for the inactive state of the CBD (Figure 3A) and Arg113 preferentially interacts with the active state of the CBD. This observation has two remarkable implications. First, it means that the effect of the R113A mutation would have been challenging to anticipate solely on the basis of structures. To predict the functional outcome of a mutation, it is essential to consider how it affects not only the bound states of R, but also the inhibitory equilibria of apo R, even when the latter is only minimally populated. This is the case for the PKA R subunit, which in vivo typically exists in either the C- or the cAMP-bound forms. Secondly, the RA–C complex is ‘frustrated’, as the formation of stable R–C intermolecular interactions occurs at the price of weakening intramolecular interactions, such as those between the linker and the CBD of RA. We hypothesize that this type of molecular frustration has evolved to ensure that signalling complexes, e.g. RA–C, can be disrupted by low-molecular-mass allosteric effectors, e.g. cAMP. Hence the frustration of RA–C helps to make the inhibition of C by RA reversible and controllable by cAMP.
Figure 3. Role of the RA linker in conformational selection.
Apo RA samples both C-binding-competent conformations (‘inactive’) and cAMP-binding-competent conformations (‘active’). (A) Apo RA samples both C-binding competent conformations (‘inactive’) and cAMP-binding competent conformations (‘active’). The linker is one of the critical elements of RA that controls the position of this inactive versus active conformational equilibrium. When the interactions between the linker and the adjacent domain in the active conformation are weakened by a mutation (e.g. R113A) selected on the basis of CHESCA, the equilibrium shifts towards the C-binding-competent state, resulting in enhanced RA –C affinity (C), despite the fact that Arg113 is surprisingly forming salt bridges in the RA –C complex (B). (B and C) Adapted from [22]: Akimoto, M., Selvaratnam, R., McNicholl, E.T., Verma, G., Taylor, S.S. and Melacini, G. (2013) Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. U.S.A. 110, 14231–14236 with permission.
Conclusions
We have shown that, even when flexible, it is possible for linkers to function not only as covalent threads between domains, but also as active elements of allosteric signalling networks. In the case of PKA, the signalling function of the linker containing the inhibitory site arises from state-selective interactions of the linker with the adjacent CBD. The state-selectivity of the linker–CBD interactions was confirmed through four independent methods, i.e. CHESCA, PREs, unfolding experiments and mutations monitored by kinase assays. Although weak (i.e. ~2RT), the interactions elicited by the flexible linker of PKA are amplified by the near-degeneracy of the free energy landscape of the CBD in apo R, which samples both active and inactive conformers in full agreement with a conformational selection model. Hence apo R, even though only minimally populated, is still a key determinant of PKA allostery and signalling. In addition, it should be noted that the studies described in the present paper are limited to a monomeric fragment of the R subunit of PKA. However, full-length R is dimeric and, in the context of the R dimer, there might be other ways through which the R linker investigated in the studies described could influence the overall function and co-operativity of cAMP binding to R.
Acknowledgments
Funding
This work was supported by the Canadian Institute of Health Research (CIHR) and the National Sciences and Engineering Research Council (NSERC).
Abbreviations
- CBD
cAMP-binding domain
- CHESCA
chemical shift covariance analysis
- PKA
protein kinase A
- PRE
paramagnetic relaxation enhancement
- Rp-cAMPS
Rp isomer of adenosine 3′,5′-monophosphothioate
References
- 1.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 2.Das R, Esposito V, Abu-Abed M, Anand GS, Taylor SS, Melacini G. cAMP activation of PKA defines an ancient signalling mechanism. Proc Natl Acad Sci USA. 2007;104:93–98. doi: 10.1073/pnas.0609033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vendruscolo M. Protein regulation: the statistical theory of allostery. Nat Chem Biol. 2011;7:411–412. doi: 10.1038/nchembio.603. [DOI] [PubMed] [Google Scholar]
- 4.Das R, Chowdhury S, Mazhab-Jafari MT, Sildas S, Selvaratnam R, Melacini G. Dynamically driven ligand selectivity in cyclic nucleotide binding. J Biol Chem. 2009;284:23682–23696. doi: 10.1074/jbc.M109.011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvaratnam R, Mazhab-Jafari MT, Das R, Melacini G. The auto-inhibitory role of the EPAC hinge helix as mapped by NMR. PLoS ONE. 2012;7:e487076. doi: 10.1371/journal.pone.0048707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNicholl ET, Das R, SilDas S, Taylor SS, Melacini G. Communication between tandem cAMP binding domains in the regulatory subunit of protein kinase A-Ia as revealed by domain-silencing mutations. J Biol Chem. 2010;285:15523–15537. doi: 10.1074/jbc.M110.105783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 8.VanSchouwen B, Selavaratnam R, Fogolari F, Melacini G. Role of dynamics in the autoinhibition and activation of the exchange protein directly activated by cyclic AMP (EPAC) J Biol Chem. 2011;286:42655–42669. doi: 10.1074/jbc.M111.277723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selvaratnam R, VanSchouwen B, Fogolari F, Mazhab-Jafari MT, Das R, Melacini G. The projection analysis of NMR chemical shifts reveals extended EPAC autoinhibition determinants. Biophys J. 2012;102:630–639. doi: 10.1016/j.bpj.2011.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ER. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci USA. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B, Tsai CJ, Haliloğlu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci USA. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Jones JM, Nguyen-Huu X, Ten Eyck LF, Taylor SS. Crystal structures of RIαsubunit of cyclic adenosine 5′-monophosphate (cAMP)-dependent protein kinase complexed with (Rp)-adenosine 3′,5′-cyclic monophosphothioate and (Sp)-adenosine 3′,5′-cyclic monophosphothioate, the phosphothioate analogues of cAMP. Biochemistry. 2004;43:6620–6629. doi: 10.1021/bi0302503. [DOI] [PubMed] [Google Scholar]
- 18.Das R, Mazhab-Jafari MT, Chowdhury S, SilDas S, Selvaratnam R, Melacini G. Entropy-driven cAMP-dependent allosteric control of inhibitory interactions in exchange proteins directly activated by cAMP. J Biol Chem. 2008;283:19691–19703. doi: 10.1074/jbc.M802164200. [DOI] [PubMed] [Google Scholar]
- 19.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand GS, Krishnamurthy S, Bishnoi T, Kornev A, Taylor SS, Johnson DA. Cyclic AMP- and (Rp)-cAMPS-induced conformational changes in a complex of the catalytic and regulatory (RIα) subunits of cyclic AMP-dependent protein kinase. Mol Cell Proteomics. 2010;9:2225–2237. doi: 10.1074/mcp.M900388-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen AE, Selheim F, de Rooij J, Dremier S, Schwede F, Dao KK, Martinez A, Maenhaut C, Bos JL, Genieser HG, Døskeland SO. cAMP analog mapping of Epac1 and cAMP kinase: discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J Biol Chem. 2003;278:35394–35402. doi: 10.1074/jbc.M302179200. [DOI] [PubMed] [Google Scholar]
- 22.Akimoto M, Selvaratnam R, McNicholl ET, Verma G, Taylor SS, Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci USA. 2013;110:14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]