OsHAC1;1 and OsHAC1;2 function as arsenate reductases that play an important role in restricting As accumulation in rice shoots and grain when plants are exposed to arsenate.
Abstract
Rice is a major dietary source of the toxic metalloid arsenic (As). Reducing its accumulation in rice (Oryza sativa) grain is of critical importance to food safety. Rice roots take up arsenate and arsenite depending on the prevailing soil conditions. The first step of arsenate detoxification is its reduction to arsenite, but the enzyme(s) catalyzing this reaction in rice remains unknown. Here, we identify OsHAC1;1 and OsHAC1;2 as arsenate reductases in rice. OsHAC1;1 and OsHAC1;2 are able to complement an Escherichia coli mutant lacking the endogenous arsenate reductase and to reduce arsenate to arsenite. OsHAC1:1 and OsHAC1;2 are predominantly expressed in roots, with OsHAC1;1 being abundant in the epidermis, root hairs, and pericycle cells while OsHAC1;2 is abundant in the epidermis, outer layers of cortex, and endodermis cells. Expression of the two genes was induced by arsenate exposure. Knocking out OsHAC1;1 or OsHAC1;2 decreased the reduction of arsenate to arsenite in roots, reducing arsenite efflux to the external medium. Loss of arsenite efflux was also associated with increased As accumulation in shoots. Greater effects were observed in a double mutant of the two genes. In contrast, overexpression of either OsHAC1;1 or OsHAC1;2 increased arsenite efflux, reduced As accumulation, and enhanced arsenate tolerance. When grown under aerobic soil conditions, overexpression of either OsHAC1;1 or OsHAC1;2 also decreased As accumulation in rice grain, whereas grain As increased in the knockout mutants. We conclude that OsHAC1;1 and OsHAC1;2 are arsenate reductases that play an important role in restricting As accumulation in rice shoots and grain.
Arsenic (As) is a toxic metalloid and is listed as a class-one carcinogen (National Research Council, 2001). Humans are exposed to As mainly through drinking water and food. Rice, the staple food for more than half of the world population, is the most important dietary source of As for populations in south and southeast Asia (Mondal and Polya, 2008; Meharg et al., 2009; Zhao et al., 2010b; Li et al., 2011), and there is evidence linking high As exposure in rice (Oryza sativa) with genotoxic effects in humans (Banerjee et al., 2013). A study on pregnant women in the US also found a significant association between rice consumption and urinary As excretion, a biomarker of As exposure (Gilbert-Diamond et al., 2011). Precooked milled rice is a common ingredient of baby food, and high As levels in some baby rice products present a particular concern (Meharg et al., 2008). It is therefore of critical importance to reduce As accumulation in rice grain.
Rice roots take up arsenate [As(V)] or arsenite [As(III)] depending on the prevailing soil conditions. As(III) is the main As species in anaerobic flooded paddy soil. However, rice grown under upland or water-saving cultivation conditions experience long periods when field soils becomes aerobic. Under such aerobic conditions, As(V) is the main form of As rice roots are exposed to (Xu et al., 2008). In addition, As(III) can also be oxidized in the rhizosphere even during flooded paddy cultivation due to the release of oxygen from rice roots (Liu et al., 2006; Seyfferth et al., 2010). As(III) and As(V) are taken up into roots by the silicic acid and phosphate transporters, respectively (Abedin et al., 2002; Ma et al., 2008; Wu et al., 2011). As(III) is detoxified by complexation with phytochelatins (Ha et al., 1999; Raab et al., 2005; Liu et al., 2010) and transported into the vacuoles via ABCC transporters (Song et al., 2010, 2014). Vacuolar sequestration of As(III)-thiol complexes helps restrict the translocation of As to rice grain (Song et al., 2014; Chen et al., 2015). The first step of As(V) detoxification is the reduction to As(III). Most plant species have an inherently high As(V) reduction capacity, because As(III) is found to be the predominant As species in plants exposed to As(V) (Dhankher et al., 2006; Xu et al., 2007). Reduction of As(V) to As(III) allows the latter to be detoxified via the mechanisms of phytochelatin complexation and vacuolar sequestration. Importantly, As(III) can also be extruded into the external environment following As(V) uptake, with As(III) efflux typically accounting for 60% to 80% of the As(V) uptake by roots of rice and other plants (Xu et al., 2007; Liu et al., 2010; Zhao et al., 2010a). Therefore, As(III) efflux is an efficient way for reducing the cellular As burden without the risk of losing phosphate, a chemical analog of As(V).
Despite the importance of As(V) reduction in plant As metabolism and detoxification, rather little was known about the enzymes catalyzing the reduction reaction until recently. Earlier studies suggested that plant ACR2 proteins, which are homologs of the yeast (Saccharomyces cerevisiae) As(V) reductase and belong to CDC25 phosphatases, may be responsible for As(V) reduction in plant cells (Bleeker et al., 2006; Dhankher et al., 2006; Ellis et al., 2006; Duan et al., 2007). However, these studies are mainly based on heterologous functional assays in Escherichia coli or yeast, which may not reflect the in planta functions of the genes. In the study of Dhankher et al. (2006), RNAi silencing of the Arabidopsis (Arabidopsis thaliana) ACR2 was found to lead to As(V) sensitivity and As hyperaccumulation in shoots. However, these observations could not be reproduced in studies using T-DNA insertion ACR2 null mutants (Liu et al., 2012; Chao et al., 2014). Recently, Chao et al. (2014) and Sánchez-Bermejo et al. (2014) independently identified a new As(V) reductase in Arabidopsis, named HAC1 (for High As Content 1) or ATQ1 (for Arsenate Tolerance QTL 1). The protein is a member of the rhodanase-like family but lacks the HCX5R active site found in the yeast ACR2 (Mukhopadhyay and Rosen, 2001). HAC1/ATQ1 is able to reduce As(V) to As(III) both in vitro and in planta. Weak or null alleles of HAC1/ATQ1 in Arabidopsis accessions are associated with decreased tolerance to As(V) (Chao et al., 2014; Sánchez-Bermejo et al., 2014) and elevated As accumulation in shoots (Chao et al., 2014). Knockout mutants of HAC1 have greatly decreased As(III) efflux to the external medium following As(V) uptake, which causes As hyperaccumulation in shoots (Chao et al., 2014). This study therefore demonstrates a crucial role of HAC1 in mediating As(V) reduction and limiting As accumulation in the above-ground tissues.
There are more than 10 AtHAC1-like genes in the rice genome (Supplemental Fig. S1), but their functions have not been characterized. Here, we show that OsHAC1;1 and OsHAC1;2, close homologs of AtHAC1, function as As(V) reductases and play an important role in regulating As accumulation in rice shoots and grain.
RESULTS
OsHAC1;1 and OsHAC1;2 Function as Arsenate Reductase
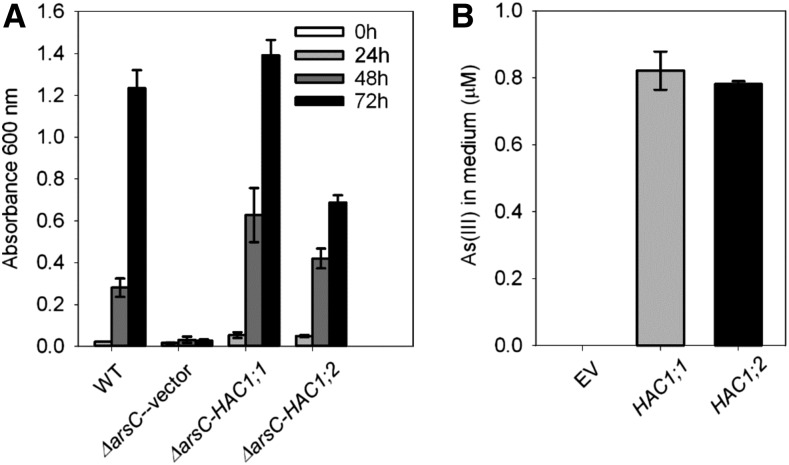
We identify two genes encoding rhodanase-like proteins (Loc_Os02g01220 and Loc_Os04g17660) from rice (cv Nipponbare), which have a high homology (84% and 81% similarity in the amino acid sequence, respectively) with Arabidopsis AtHAC1. These two genes share 90% amino acid sequence similarity and are thereafter named OsHAC1;1 and OsHAC1;2, respectively (Supplemental Fig. S1). To test if OsHAC1;1 and OsHAC1;2 are able to reduce As(V) to As(III), we expressed the two rice genes in a strain of E. coli lacking the endogenous arsenate reductase ArsC. This mutant strain is sensitive to As(V), because it is not able to reduce the absorbed As(V) to As(III) to allow the latter to be extruded from the cell (Oden et al., 1994). Heterologous expression of either OsHAC1;1 or OsHAC1;2 restored the growth of the E. coli strain in the Luria-Bertani (LB) medium in the presence 1 mm As(V) (Fig. 1A). Furthermore, As speciation analysis using high performance liquid chromatography - inductively coupled plasma - mass spectrometry (HPLC-ICP-MS) showed the production of As(III) in the medium in the presence 10 µM As(V) by the E. coli ΔarsC strain expressing OsHAC1;1 or OsHAC1;2, in contrast to the empty vector control that produced no detectable As(III) (Fig. 1B).
Figure 1.
OsHAC1;1 and OsHAC1;2 encode arsenate reductases. A, Expression of OsHAC1;1 or OsHAC1;2 suppresses the As(V) sensitivity of the E. coli mutant lacking the arsC arsenate reductase. Strains were grown at 16°C and cell density measured at OD600 nm after exposure to 1 mm As(V) for 0 to 72 h. Wild type = E. coli wild type (W3110); ΔarsC = arsC mutant in WC3110; Vector = empty pET29a; arsC-HAC1;1 = pET-29a vector containing OsHAC1;1; ΔarsC-HAC1;2 = pET-29a vector containing OsHAC1;2. B, Production of As(III) in LB medium after E. coli expressing the empty vector (EV), OsHAC1;1 or OsHAC1;2 was exposed to 10 µM As(V). n.d., not detected.
The Expression Patterns and Subcellular Localization of OsHAC1;1 and OsHAC1;2 in Rice
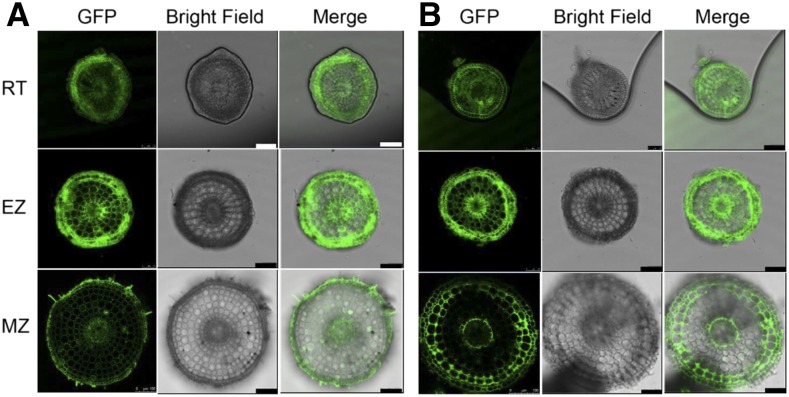
To investigate the expression patterns of the two HAC1 genes in rice, we created stable transgenic rice lines expressing OsHAC1;1-GFP and OsHAC1;2-GFP chimeric protein constructs driven by their native promoters. Based on the GFP signals, we found that both OsHAC1;1 and OsHAC1;2 predominantly accumulate in root, with the epidermis and the pericycle cells, as well as root hairs in the mature zone of roots, showing particularly strong accumulation of OsHAC1;1 (Fig. 2). In contrast, OsHAC1;2 is more abundantly found in the epidermis, the exodermis, the outer layer of cortex, and the endodermis cells (Fig. 2). The expression patterns are similar between the different root zones from the root tip to the mature zone. For both gene constructs, very faint signals of GFP were observed in the shoot tissues (data not shown).
Figure 2.
Expression patterns of OsHAC1;1 and OsHAC1;2 revealed by the accumulation of the OsHAC1;1-GFP (A) or OsHAC1;2-GFP (B) fusion proteins driven by their native promoters. Roots were cut by hand at different zones: EZ, Elongation zone; MZ, mature zone; RT, root tip. Scale bar = 100 μm.
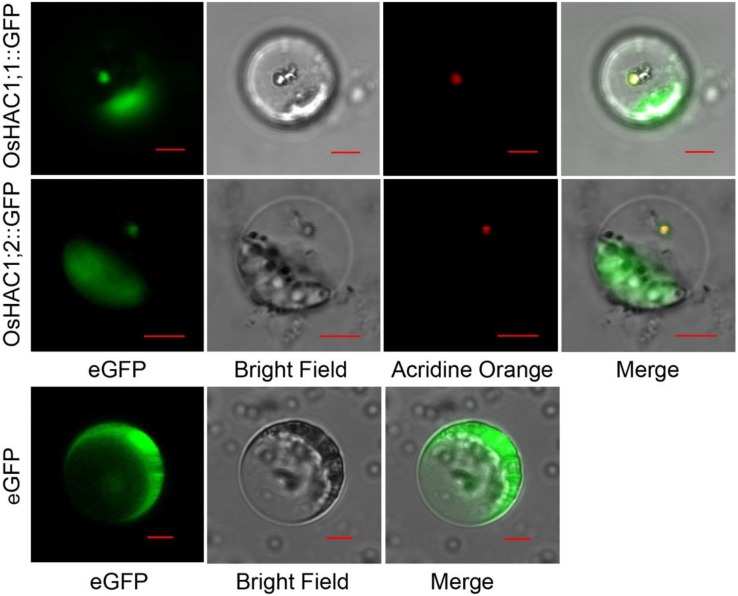
To investigate the subcellular localization of the OsHAC1;1 and OsHAC1;2, we isolated protoplasts from the transgenic rice roots expressing OsHAC1;1-GFP or OsHAC1;2-GFP. For both gene constructs, the GFP signals were localized in the cytoplasm (Supplemental Fig. S2). To further investigate the subcellular localization of the two proteins, we constructed N-terminal OsHAC1;1 or OsHAC1;2 fusions with GFP with expression driven by the cauliflower mosaic virus 35S promoter and transfected the derived expression vector into rice protoplasts. We observed that both the OsHAC1;1::GFP and OsHAC1;2::GFP fusion proteins are localized in the cytoplasm and nucleus (Fig. 3). Because of the relatively small molecular sizes of the two proteins, the possibility of their diffusion from the cytoplasm to the nucleus cannot be ruled out.
Figure 3.
Subcellular localization of OsHAC1;1 and OsHAC1;2. Representative microscopic images of rice protoplasts expressing the OsHAC1;1-GFP (top) or OsHAC1;2-GFP (middle) fusion protein or eGFP (bottom) driven by the cauliflower mosaic virus 35S promoter. Scale bars = 5 μm.
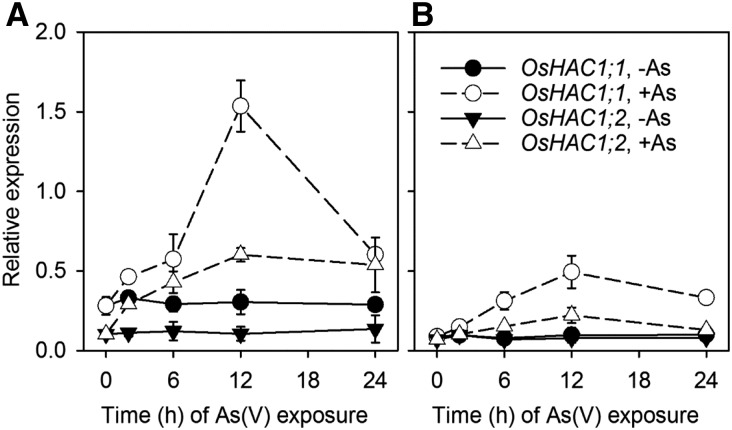
We used quantitative real-time PCR (qRT-PCR) to investigate the expression pattern of OsHAC1;1 and OsHAC1;2 in response to As(V) exposure. Three-week-old rice plants (cv Nipponbare) were exposed to 0 or 10 µM As(V) in a nutrient solution without phosphate for up to 24 h. Phosphate was withdrawn during this short-term experiment to facilitate As(V) uptake. In the control treatment [no As(V)], OsHAC1;1 and OsHAC1;2 were predominantly expressed in roots and moderately in shoots, and there were no temporal changes in their relative expression levels over the 24-h time course (Fig. 4). Exposure to As(V) significantly enhanced the expression of both OsHAC1;1 and OsHAC1;2 during the first 12 h, with OsHAC1;1 showing a greater response than OsHAC1;2 (Fig. 4). In contrast, exposure to 10 µM As(III) decreased the mRNA levels of OsHAC1;1 and OsHAC1;2 in both roots and shoot (Supplemental Fig. S3).
Figure 4.
Induction of OsHAC1;1 and OsHAC1;2 expression in roots (A) and shoots revealed by qRT-PCR. Plants were exposed to 0 or 10 µM As(V) for 24 h. Expression of each gene was calculated as 2−∆CT relative to OsActin. Data are means ± sd (n = 3 biological replicates).
Knocking Out OsHAC1;1 or OsHAC1;2 Affects Arsenate Reduction and Arsenic Accumulation in Rice
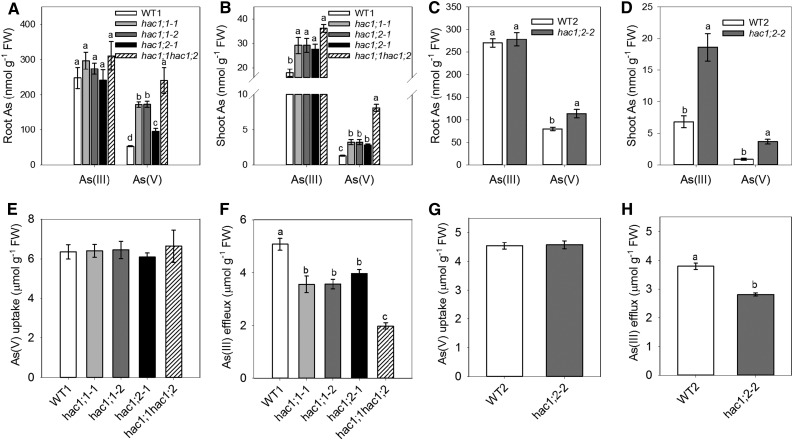
To investigate the in planta function of OsHAC1;1 and OsHAC1;2, we created two independent knockout lines of OsHAC1;1, hac1;1-1 and hac1;1-2, in the cv Zhonghua 11 background using CRISPR-Cas9 technology (Supplemental Fig. S4). We also obtained two independent homozygous T-DNA insertion mutants of OsHAC1;2 (hac1;2-1 and hac1;2-2 in the cv Zhonghua 11 and Dongjin background, respectively; Supplemental Fig. S4). In addition, we obtained a homozygous double mutant hac1;1 hac1;2 in the cv Zhonghua 11 background by crossing hac1;1-1 with hac1;2-1. Analysis using qRT-PCR showed that the expressions of OsHAC1;1 and OsHAC1;2 were abolished in these mutants (Supplemental Fig. S4). If OsHAC1;1 and OsHAC1;2 play a role in As(V) reduction, knocking out of OsHAC1;1 or OsHAC1;2 may impact As speciation in the plants. To test this hypothesis, mutant and wild-type plants were exposed to 10 µM As(V) for 48 h, and As speciation in roots and shoots were determined using HPLC-ICP-MS. hac1;1-1, hac1;1-2, hac1;2-1 and hac1;1 hac1;2 and their common wild type (Zhonghua 11) were compared in the same experiment, and hac1;2-2 and its wild type (Dongjin) in a separate experiment. As(III) and As(V) were the only two As species detected in the plant tissues, with As(III) being the predominant As species. The extraction method does not preserve As(III)-thiol complexes (Raab et al., 2005; Liu et al., 2010), which would be dissociated and determined as As(III) by the method used. Knocking out of OsHAC1;1 resulted in 3.1 times higher As(V) concentration in roots compared with wild type, while the mutation in OsHAC1;2 increased root As(V) concentration by 40% to 80% (Fig. 5, A and C). In the double mutant, root As(V) concentration was 4.7 times higher than wild type. All mutants had significantly higher concentrations of both As(V) and As(III) in shoots than wild-type plants (Fig. 5, B and D). Compared with wild type, hac1;1, hac1;2 and the double mutant had 170%, 150% to 190%, and 230%, respectively, higher total As [sum of As(V) and As(III)] concentration in shoots. The effect of OsHAC1;1 or OsHAC1;2 mutations is also evident from the changes in As(III) as a percentage of the total As in roots, decreasing from 82% in wild type (Zhonghua 11) to 62%, 72%, and 57% in hac1;1, hac1;2-1, and the double mutant, respectively, and from 77% in wild type (Dongjin) to 71% in hac1;2-2. In the shoot tissues, As(III) as a percentage of the total As also decreased from 93% in wild type (Zhonghua 11) to 90% in the three single mutants and 81% in the double mutant. These results support a role for OsHAC1;1 and OsHAC1;2 in As(V) reduction in rice roots, with the double mutant having a greater effect than the single mutants and the single mutants of OsHAC1;1 having a greater effect than those of OsHAC1;2.
Figure 5.
Knocking out OsHAC1;1 or OsHAC1;2 affects As(V) reduction and As accumulation in rice. A to D, As speciation in roots (A and C) and shoots (B and D) after wild-type and knockout single or double mutants were exposed to 10 μm As(V) for 48 h. E to H, Uptake of As(V) (E and G) and efflux of As(III) (F and H) after wild-type and mutant plants were exposed to 10 μm As(V) for 48 h. WT1, cv Zhonghua 11; WT2, cv Dongjin. Data are means ± se (n = 4 biological replicates). Different letters above bars represent significant difference at P < 0.05.
Chao et al. (2014) showed that As(III) efflux from roots to the external medium diminished greatly in mutants with a loss of AtHAC1 function, resulting in a markedly increased As accumulation in Arabidopsis shoots. To test if mutations in OsHAC1;1 or OsHAC1;2 also affect As(III) efflux in rice, plants were exposed to 10 μm As(V) for 48 h. As(V) uptake and As(III) efflux were estimated by measuring the changes in As speciation in the culture solution. There were no differences in As(V) uptake between mutants and wild type (Fig. 5, E and G). In contrast, As(III) efflux from roots was significantly decreased in all single mutants compared with wild-type plants, and a larger decrease was found in the double mutant than the single mutants (Fig. 5, F and H). The As(III) efflux efficiency, calculated as a ratio of As(III) efflux to As(V) uptake, was 0.80 to 0.83 in wild type, 0.56 to 0.66 in the single mutants, and 0.32 in the double mutant. A decreased As(III) efflux to the external medium could explain the enhanced As accumulation in mutant shoots.
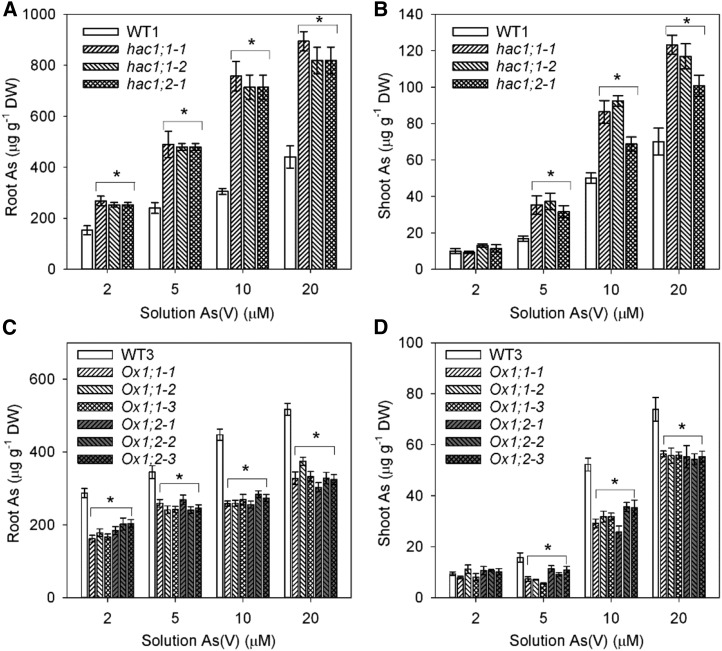
In a further experiment, we tested the effect of OsHAC1;1 or OsHAC1;1 mutations on As accumulation over a range of As(V) concentration from 2 to 20 µM. Knocking out of OsHAC1;1 or OsHAC1;2 resulted in a significant increase in As accumulation in roots at all As(V) concentrations and in shoots at all but the 2-µM As(V) treatment (Fig. 6, A and B).
Figure 6.
Knockout or overexpression OsHAC1;1 or OsHAC1;2 affects As accumulation in rice. Arsenic concentration in roots and shoots of knockout mutant and wild-type (WT1, Zhonghua 11) plants (A and B) and the overexpression lines and wild-type (WT3, Nipponbare) plants (C and D). Plants were exposed to different As(V) concentrations for 48 h. Ox1;1-1, Ox1;1-2, Ox1;1-3 represent independent overexpression lines of OsHAC1;1. Ox1;2-1, Ox1;2-2, Ox1;2-3 represent independent overexpression lines of OsHAC1;2. Data represent means ± se (n = 4 biological replicates). *Significant difference from wild type at P < 0.05. DW, dry weight.
Furthermore, we observed no significant change in the concentrations of As(V) and As(III) in roots or shoots of single or double mutants compared with wild types when plants were exposed to 10 μm As(III) for 48 h (Supplemental Fig. S5), suggesting that the effect of OsHAC1;1 and OsHAC1;1 is specific to As(V).
Overexpression of OsHAC1;1 or OsHAC1;2 Increases Arsenate Reduction and Decreases Arsenic Accumulation
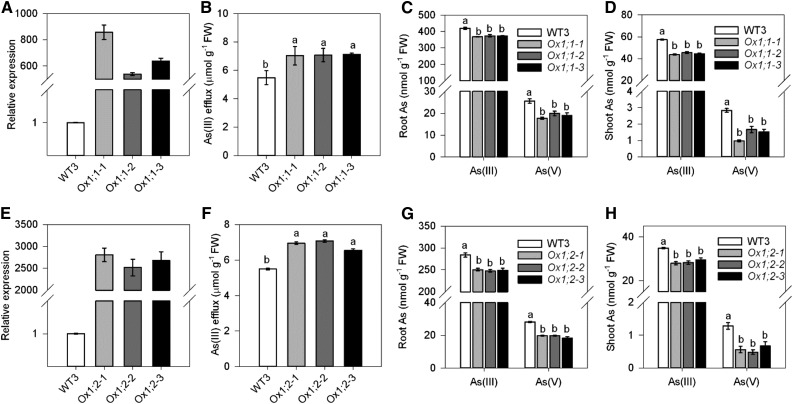
To further investigate the role of OsHAC1;1 and OsHAC1;2 in As metabolism, we overexpressed OsHAC1;1 and OsHAC1;2 in rice (cv Nipponbare) using the Ubiquitin promoter. Three independent lines for each gene were selected for further investigation. qRT-PCR analysis showed that all overexpressing lines had greatly enhanced expression of OsHAC1;1 or OsHAC1;2 in roots compared with wild-type plants (Fig. 7, A and E). After exposure to 10 µM As(V) for 48 h, there was no significant difference in As(V) uptake between different transgenic plants and wild-type plants (Supplemental Fig. S6). In contrast, transgenic plants overexpressing OsHAC1;1 or OsHAC1;2 had 34% to 50% and 20% to 28%, respectively, larger As(III) efflux into the external medium than wild-type plants (Fig. 7, B and F). As(III) efflux as a proportion of As(V) uptake increased from 0.65 in wild type to 0.83 in the OsHAC1;1 overexpression lines, and from 0.76 in wild type to 0.90 in the OsHAC1;2 overexpression lines. As a result, overexpression of OsHAC1;1 or OsHAC1;2 significantly decreased the concentrations of As(III), As(V), and total As in both shoots and roots compared with wild-type plants, with the effect being greater on shoot As concentration than on root As concentration (Fig. 7, C, D, G, and H).
Figure 7.
Overexpression of OsHAC1;1 or OsHAC1;2 increases As(III) efflux and decreases As accumulation in rice shoots. A and E, The expression levels of OsHAC1;1 (A) or OsHAC1;2 (E) in wild-type (WT3, Nipponbare) and transgenic lines by qRT-PC. B and F, Efflux of As(III) after wild-type and overexpression lines of OsHAC1;1 (B) or OsHAC1;2 (F) were exposed to 10 μm As(V) for 48 h. C, D, G, and H, As speciation in roots (C and G) and shoots (D and H) after wild-type and overexpression lines of OsHAC1;1 (C and D) or OsHAC1;2 (G and H) were exposed to 10 μm As(V) for 48 h. Data are means ± se (n = 4 biological replicates). Different letters above bars represent significant difference at P < 0.05.
In a further experiment, plants were exposed to a range of As(V) concentrations varying from 2 to 20 µM for 48 h. Arsenic accumulation in roots and shoots was determined. Overexpression of OsHAC1;1 or OsHAC1;2 resulted in a significant decrease in root As concentration at all four As(V) exposure concentrations, as well as a significant decrease in shoot As concentration in all but the 2 µM As(V) treatment (Fig. 6, C and D).
In contrast, overexpression of OsHAC1;1 or OsHAC1;2 had no significant effect on As(V) and As(III) concentrations in roots and shoots compared with wild type when plants were exposed to 10 µM As(III) (Supplemental Fig. S7).
Knockout or Overexpression of OsHAC1;1 or OsHAC1;2 Affects As Speciation in Xylem Sap
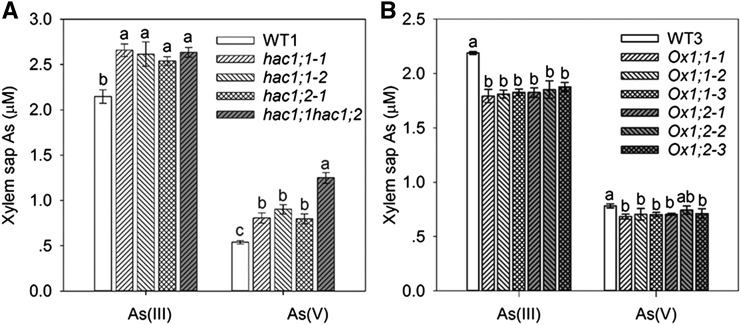
If OsHAC1;1 or OsHAC1;2 plays a role in As(V) reduction in roots, knocking out or overexpression of these genes may affect As speciation in xylem sap. To test this hypothesis, we analyzed As speciation in xylem sap collected from plants exposed to 10 µM As(V) for 24 h. As(III) was found to be the predominant species of As in xylem sap. The concentrations of both As(V) and As(III) in the xylem sap from OsHAC1;1 or OsHAC1;2 single mutants were significantly higher than those from wild type, whereas the double mutant also had a significantly higher As(V) concentration than the single mutants (Fig. 8A). The percentage of As(III) in the xylem sap total As was higher in wild type (80%) than in single mutants (73% to 76%) or double mutant (66%). In contrast, transgenic plants overexpressing either OsHAC1;1 or OsHAC1;2 showed lower As(V) and As(III) concentrations in the xylem sap compared with wild type (Fig. 8B). The differences were significant in all except one of the OsHAC1;2 overexpression lines for As(V) concentration. These results are consistent with a role of OsHAC1;1 and OsHAC1;2 in As(V) reduction in rice roots.
Figure 8.
Knockout or overexpression of OsHAC1;1 or OsHAC1;2 affects As speciation in xylem sap. Concentrations of As(V) and As(III) in xylem sap of knockout single or double mutants and wild-type (WT1, Zhonghua 11) plants (A) and the overexpression lines and wild-type (WT3, Nipponbare) plants (B). Plants were exposed to 10 µM As(V) for 24 h. Data represent means ± se (n = 4 biological replicates). Different letters above bars represent significant difference at P < 0.05.
Overexpression of OsHAC1;1 or OsHAC1;2 Enhances Tolerance To Arsenate
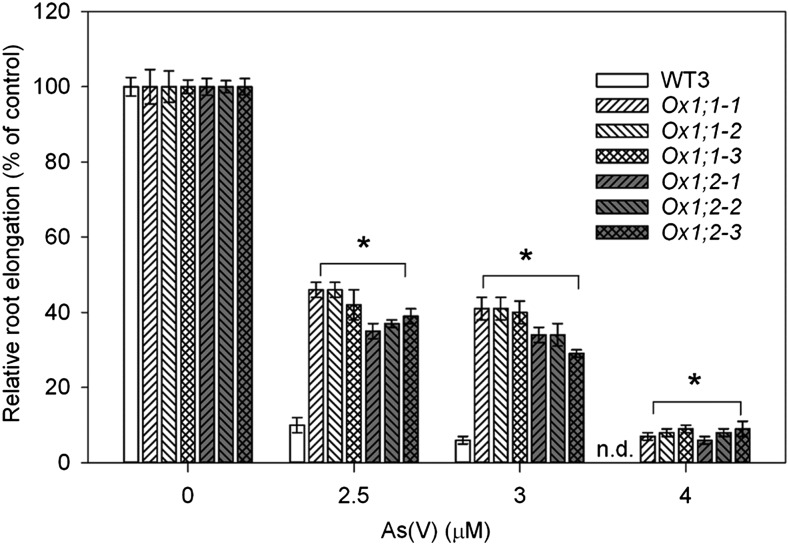
Because overexpression of OsHAC1;1 or OsHAC1;2 increased As(III) efflux to the external medium, we hypothesized that the overexpression lines might be more tolerant to As(V). In a short-term root elongation assay, root elongation of rice seedlings during 24 or 48 h under different As(V) concentrations was measured. The assay was conducted in the absence of phosphate to heighten the toxicity of As(V). Because the response patterns were similar between the 24- and 48-h exposure, only the data of 24-h exposure are shown (Fig. 9; Supplemental Fig. S8). Root growth of wild-type seedlings was inhibited by more than 90% by 2.5 µM As(V) and completely arrested by 4 µM As(V). In contrast, OsHAC1;1 or OsHAC1;2 overexpression lines had significantly larger root elongation than wild type in the presence of 2.5 to 4 µM As(V) (Fig. 9), indicating an increased tolerance to As(V).
Figure 9.
Overexpression of OsHAC1;1 or OsHAC1;2 enhances As(V) tolerance. Root growth of rice seedlings during 24 h under different As(V) concentrations was measured. Ox1;1-1, Ox1;1-2, Ox1;1-3 represent independent overexpression lines of OsHAC1;1. Ox1;2-1, Ox1;2-2, Ox1;2-3 represent independent overexpression lines of OsHAC1;2. Data represent means ± se (n = 10 biological replicates). *Significant difference from wild type (WT3, Nipponbare) at P < 0.05.
OsHAC1;1 and OsHAC1;2 Affect Grain As Accumulation in Soil-grown Rice
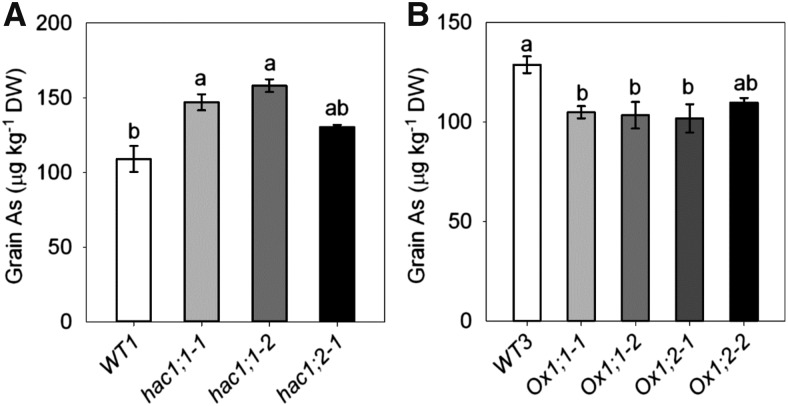
The experiments described above were conducted in hydroponic cultures with young rice plants. To determine if OsHAC1;1 and OsHAC1;2 play a role in regulating As accumulation in rice grain, plants were grown up to maturity in a paddy soil amended with an environmentally relevant dose of As(V) (20 mg kg−1; Zhao et al., 2010b). The soil was irrigated regularly with free drainage, aerobic conditions under which As(V) was likely to be the main species of As in the soil solution (Xu et al., 2008). Under aerobic conditions, hac1;1 and hac1;2 mutants had 36% and 20%, respectively, higher As concentration in the brown rice than wild type (Fig. 10A), whereas the OsHAC1;1 and OsHAC1;2 overexpression lines had approximately 20% lower grain As concentration than wild type (Fig. 10B). Grain yield and straw biomass were not significantly different between the mutants and wild type or between the overexpressing lines and wild type (data not shown).
Figure 10.
Knockout or overexpression of OsHAC1;1 or OsHAC1;2 affect arsenic accumulation in rice grain under aerobic soil conditions. A, As concentration in brown rice of OsHAC1;1 or OsHAC1;2 knockout mutants and wild-type (WT1, Zhonghua 11) plants. B, As concentration in brown rice of OsHAC1;1 or OsHAC1;2 overexpression lines and wild-type (WT3, Nipponbare) plants. Plants were grown in a soil amended with 20 mg As(V) kg−1 under aerobic conditions and rice grain were harvested at maturity. Data are means ± se (n = 4 biological replicates). Different letters above bars represent significant difference at P < 0.05.
DISCUSSION
In the current study, we show that OsHAC1;1 and OsHAC1;2 function as As(V) reductases and are involved in the reduction of As(V) to As(III) in rice plants. OsHAC1;1 and OsHAC1;2 are the closest homologs of the Arabidopsis AtHAC1/ATQ1, which was discovered recently as representing a new type of As(V) reductase in plants (Chao et al., 2014; Sánchez-Bermejo et al., 2014). Similar to AtHAC1, both OsHAC1;1 and OsHAC1;2 were able to complement the As(V)-sensitive E. coli mutant lacking the endogenous As(V) reductase ArsC (Fig. 1). The E. coli mutant expressing either OsHAC1;1 or OsHAC1;2 was able to reduce As(V) and extrude As(III) into the external medium, which is a key mechanism of As(V) detoxification widely employed by microorganisms (Rosen, 2002). Further evidence for a role of OsHAC1;1 and OsHAC1;2 in As(V) reduction in rice plants can be seen in the altered As speciation in the mutants or overexpression lines of the two genes; mutations in either OsHAC1;1 or OsHAC1;2 resulted in an increased proportion of As(V) and a decreased proportion of As(III) in rice roots and xylem saps, whereas overexpression of either gene produced the opposite effect (Figs. 5, 7, and 8). The effect was greater when both genes were knocked out in the double mutant.
We show that OsHAC1;1 protein accumulates predominantly in the root epidermis, root hairs, and the stele, and OsHAC1;2 accumulates mainly in the exodermis, the outer layer of cortex, and the stele (Fig. 2). By mediating As(V) reduction in the root epidermis, root hairs, and possibly also the exodermis, OsHAC1;1 and OsHAC1;2 enable As(III) efflux from the outer layers of root cells to the external medium. In agreement with previous studies (Xu et al., 2007; Zhao et al., 2010a), As(III) efflux was found to represent a large proportion of As(V) influx in wild-type plants (approximately 80%). This proportion decreased to approximately 60% in the single mutants and to approximately 30% in the double mutant (Fig. 5). Efflux of As(III) following As(V) uptake is critical for controlling As accumulation in plant tissues. Decreased As(III) efflux in the mutants leads to more As accumulation in both roots and shoots (Figs. 5 and 6). In contrast, overexpression of either OsHAC1;1 or OsHAC1;2 increased As(V) reduction and As(III) efflux to the external medium, resulting in decreased As accumulation in both roots and shoots (Figs. 6 and 7). Furthermore, the localization of OsHAC1;1 and OsHAC1;2 in the stele implies a role of the two proteins in regulating As translocation from roots to shoots, possibly by blocking xylem loading of As(V) via phosphate transporters such as PHO1 (Poirier et al., 1991; Secco et al., 2010). Increased proportions of As(V) in the xylem sap of the knockout mutants (Fig. 8) are consistent with this interpretation. A previous study showed that As(III) is preferentially stored in the vacuoles of the pericycle and endodermal cells of rice roots (Moore et al., 2011), supporting the notion that these cells are important in regulating the root to shoot translocation of As. It is possible that OsHAC1;1 and OsHAC1;2 also contribute to As(V) reduction in the shoots. However, the altered phenotypes of As accumulation in the knockout mutants and the overexpression lines can be attributed primarily to the function of the two enzymes in the roots.
The presence of substantial amounts of As(III) in the roots and shoots of the oshac1;1 oshac1;2 double mutant indicates the presence of other As(V) reduction mechanisms in plants. There are more than 10 HAC1-like genes in the rice genome (Supplemental Fig. S1), some of which may also play a role in As(V) reduction. Another possibility is that As(V) could be reduced nonenzymatically by glutathione (Delnomdedieu et al., 1994), although the reaction may be slow. However, As(V) can participate in phosphorylation reactions (Byers et al., 1979), forming arsenate esters that are more easily reduced by thiols such as glutathione (Gregus et al., 2009). Unlike AtHAC1, OsHAC1;1, and OsHAC1;2, some of these additional As(V) reduction processes may not be linked to As(III) efflux, either because they do not interact with efflux transporters or are localized in cells not suited for As(III) efflux to the external medium (Chao et al., 2014). The presence of multiple As(V) reduction mechanisms explains why not only As(V), but also As(III), is elevated in the xylem sap and shoots of the mutants (Figs. 5 and 8).
OsHAC1;1- and OsHAC1;2-mediated As(V) reduction is also required for tolerance to As(V), as it likely lessens the cellular burden of As through efficient As(III) efflux. Furthermore, As(V) reduction allows the product As(III) to be complexed with phytochelatins and subsequently sequestered in the vacuoles (Zhao et al., 2009; Liu et al., 2010; Song et al., 2010). The expressions of OsHAC1;1 and OsHAC1;2 were strongly induced by As(V) exposure (Fig. 4), which is consistent with a role of the two genes in As(V) detoxification.
OsHAC1;1 appears to play a greater role in controlling As accumulation than OsHAC1;2. This difference could be attributed to a higher expression of OsHAC1;1 (Fig. 4). The strong localization of OsHAC1;1 in the epidermis and root hairs would also make it more efficient in enabling As(III) efflux, as has been observed for AtHAC1 in Arabidopsis (Chao et al., 2014). Although OsHAC1;1 and OsHAC1;2 are similar to AtHAC1 in a number of aspects discussed above, the impact of OsHAC1;1 or OsHAC1;2 single mutation is not as large as that of AtHAC1 knockout reported by Chao et al. (2014). This difference can be explained by a degree of functional redundancy between OsHAC1;1 and OsHAC1;2, which is clearly demonstrated by larger effects in the double mutant (Figs. 5 and 8). In addition, there are other HAC1-like genes in rice (Supplemental Fig. S1), whose functions remain to be investigated. Although OsHAC1;1 and OsHAC1;2, as well as AtHAC1, function as As(V) reductases, their primary metabolic functions, if any, remain unknown. It is also intriguing that the rice genome contains a considerable number of HAC1-like genes. We observed no growth or developmental phenotypes in oshac1;1, oshac1;2 or athac1 mutants under non-As stressed conditions.
Previously, OsACR2, a CDC-25 protein and a homolog of the yeast As(V) reductase ScACR2, has been suggested to be involved in As(V) reduction in rice (Duan et al., 2007). However, this study was based on heterologous expression of OsACR2 in yeast and in vitro characterization of the OsACR2 enzyme. No knockout or knockdown lines of OsACR2 were included in the study of Duan et al. (2007). Whether OsACR2 plays a role in As(V) reduction in rice plants remains unclear. In the case of Arabidopsis, the report by Dhankher et al. (2006) that silencing AtACR2 by RNA interference leads to As hyperaccumulation in the shoots could not be confirmed by recent studies using two independent T-DNA insertional knockout mutants of the gene (Liu et al., 2012; Chao et al., 2014). Because AtACR2 and AtHAC1 share sequence identity within the region used by Dhankher et al. (2006) to knock down expression of AtACR2 by RNA interference, this sequence may also have suppressed AtHAC1 expression in their RNAi lines, thus resulting in decreased As(V) tolerance and As hyperaccumulation in the shoots (Chao et al., 2014). Nahar et al. (2012) reported increased As(V) sensitivity and As accumulation in the shoots of a single T-DNA line (SALK_005882C) with a T-DNA insertion to the neighboring gene (At5g03452) of AtACR2 (At5g03455), which appeared to knockdown the expression of AtACR2.
Although lowland rice is typically grown under flooded conditions, paddy water is usually drained periodically during the rice growing season. Upland rice often experiences dry periods due to water shortage. There is also an increasing trend of using aerobic to save water usage and to reduce greenhouse gas emissions from paddy fields (Bouman et al., 2005; Linquist et al., 2015). All these agronomic factors lead to aerobic soil conditions under which As(V) is expected to be the dominant As species present in the soil solution and taken up by rice roots. Consequently, As(V) reductases may play an important role in As accumulation in rice grain when plants are exposed to the aerobic soil conditions that can occur when rice is grown under normal field conditions. We tested this hypothesis by growing mutants and overexpression lines of OsHAC1;1 and OsHAC1;2 under aerobic soil conditions to maturity. Under the experimental conditions, loss-of-function mutants of OsHAC1;1 or OsHAC1;2 had a significantly higher concentration of As in rice grain than wild type, whereas overexpression lines contained significantly lower levels of As than wild type (Fig. 10). We therefore conclude that in the field, when rice roots are exposed to irregular oxidizing and reducing cycles, the ability to specifically reduce As(V) to As(III) through the action of the OsHAC1 arsenate reductases is important to restrict As accumulation in rice grain.
Our study has shed light on the mechanism of As(V) reduction in rice, a staple food crop with an unusually high contribution to dietary As intake by humans. Our results and those of Chao et al. (2014) and Sánchez-Bermejo et al. (2014) on Arabidopsis show that As(V) reduction is a key step in As metabolism that controls the accumulation of As in the above-ground tissues of plants. Our results point to a possible strategy for limiting grain As accumulation in rice cultivated under conditions in which the soil is aerobic for extended periods of time. Such a strategy would involve enhancing As(V) reductase activities in rice roots to both enhance As(III) efflux and limit its xylem loading and transport.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa ssp. japonica) cv Nipponbare, Zhonghua 11, or Dongjin were used as wild types in this study and for rice transformation. A T-DNA insertion mutant line oshac1;2-1 (RMD_03Z11FF65) in the Zhonghua11 background was obtained from Huazhong Agricultural University, China. We obtained another T-DNA insertion mutant line oshac1;2-2 (PFG_3A-02094) in the Dongjin background from Zhejiang University. The location of the T-DNA insertion in the mutant was determined by DNA sequence analysis using PCR. A homozygous T-DNA insertion line was identified by PCR using gene-specific primers in conjunction with T-DNA border primers (Supplemental Table S1). Two independent mutants of OsHAC1;1, oshac1;1-1 and oshac1;1-2, were generated using the CRISPR/Cas9 technology (see below). Overexpression lines of OsHAC1;1 and OsHAC1;2 were generated in the cv Nipponbare background (see below). hac1;1-1 and hac1;2-1 (both in the Zhonghua 11 background) were crossed to generate a double mutant. A homozygous double mutant line was identified by PCR using gene-specific primers and sequencing.
Plant Growth Conditions
Rice seeds were surface sterilized in a 30% (v/v) hydrogen peroxide solution for 30 min, washed, and germinated for 3 d at 37°C in the dark. Ten-day-old seedlings were transferred to a one-half-strength Kimura nutrient solution. The composition of the nutrient solution was as follows (in mM): 0.091 KH2PO4, 0.273 MgSO4, 0.182 (NH4)2SO4, 0.091 KNO3, 0.183 Ca(NO3)2, 0.003 H3BO3, 0.0005 MnCl2, 0.001 (NH4)6Mo7O24, 0.0004 ZnSO4, 0.0002 CuSO4, 0.02 Fe(III)-EDTA. The pH of the solution was adjusted to 5.5. The nutrient solution was renewed every 2 d. Hydroponic experiments were conducted inside a growth room with a 14-h-light/10-h-dark period, 250 µmol m−2 s−1 light intensity, 25/20°C day/night temperatures, and a relative humidity at approximately 70%. Arsenic treatments were started by adding As(V) (Na3AsO4) or As(III) (NaAsO2) to the nutrient solution at target concentrations.
A soil pot experiment was conducted with mutants, overexpression lines, and their wild type. A paddy soil was collected from an experimental farm of Nanjing Agricultural University. The soil contains 12 mg As kg−1 and has a pH of 6.6. Basal fertilizers (120 mg N kg−1 as NH4NO3, 25 mg S kg−1 soil as MgSO4, 30 mg P kg−1 soil, and 75.5 mg K kg−1 soil as K2HPO4) were added to the soil and mixed thoroughly. The soil was amended with 20 mg As(V) kg−1. Twelve kg soil was placed in a 15-L plastic pot. The water management regimes with the soil were maintained under aerobic conditions. Each pot contained one seedling each of oshac1;1-1, oshac1;1-2, oshac1;2-1 mutants and their wild type (Zhonghua 11) or two overexpression lines each of OsHAC1;1 and OsHAC1;2 and their wild-type (Nipponbare). There were four replicated pots for each treatment. Plants were harvested at grain maturity.
RNA Extraction and Transcriptional Analysis by qRT-PCR
Total RNA were extracted from shoots and roots using the RNeasy Plant Mini Kit (Biotech). Reverse transcription was carried out using the R233-01 kit (Vazyme). qRT-PCR analysis was performed with a Real-Time PCR Detection System (Bio-Rad CFX96) in a reaction mixture of 20 µL of SYBR Green Master Mix (Vazyme, http://www.vazyme.com). OsActin (accession no. AB047313) was used as the reference gene. Expression of each gene was calculated as 2−∆CT relative to OsActin. The qRT-PCR program was set as follows: 95°C, 3 min; (95°C, 15 s; 58°C, 30 s; 72°C, 15 s) × 39; 60°C to 90°C for melting curve detection. Accession numbers of the rice genes investigated in this study and primer sequences are given in Supplemental Table S1.
Construction of pOsHAC1;1:OsHAC1;1-GFP and pOsHAC1;2:OsHAC1;2-GFP Fusion Proteins and Microscopy Observation
We modified the binary vector pHB (Mao et al., 2005) to construct the expression vectors for expressing the fusion proteins of OsHAC1;1-GFP and OsHAC1;2-GFP driven by the OsHAC1;1 and OsHAC1;2 promoter, respectively. Firstly, we replaced the 35S promoter of pHB with a small fragment containing three restriction sites, EcoRI, SalI, and HindIII, by the two enzymes EcoRI and HindIII, to form the vector pHMS. A fragment fused with a GFP coding sequence and a linker (ggaggaggaggaggagga) coding a 6× Gly peptide was inserted into the PstI and XbaI site of pHMS to form the vector pHMS-GFP. The OsHAC1;1 genomic fragment including 2.3-kb promoter region and gene body with the stop codon replaced with TTA and the OsHAC1;2 genomic fragment including 1.5-kb promoter region and gene body with the stop codon replaced with TTG were amplified from the genomic DNA of rice variety Zhonghua 11 by using primers listed in the Supplemental Table S1. The OsHAC1;1 genomic fragment was inserted in frame into the pHMS-GFP vector by the HindIII and PstI restriction enzymes, while the OsHAC1;2 genomic fragment was homologously recombined in frame into the pHMS-GFP vector with One Step Pcr Cloning Kit (Shawnxin Biotech. Co. Ltd, Shanghai). The expression vectors were transformed into rice variety Zhonghua 11 mediated by Agrobacterium tumeraciens strain EHA105. The positive transgenic lines were observed for GFP signal under stereo fluorescence microscope (LEICA M165 FC, Leica Co. Ltd) and confocal microscope (LEICA TCS SP8, Leica Co. Ltd) in the Core Facility Center of Shanghai Institute of Plant Physiology and Ecology. To check the GFP signal inside of the root, the roots of the transgenic lines were cross sectioned by free hand, and the hand sections were screened under confocal microscope (LEICA TCS SP8, Leica Co. Ltd). To observe the subcellular localization, protoplasts were isolated from the roots of transgenic rice expressing OsHAC1;1-GFP or OsHAC1;2-GFP. Roots were cut into segments and placed in an enzyme digestion solution (MES, pH 5.7, 10 mm mannitol, 0.5 m cellulose, 1.5% RS cellulase, 0.75% macerozyme R-10, 10 mm CaCl2, 0.1% bovine serum albumin) for 4 h in the dark at 28°C with gentle shaking (80 rpm). Thereafter, an equal volume of W5 solution (2 mm MES, pH 5.7, 5 mm KCl, 154 mm NaCl, 125 mm CaCl2) was added, followed by vigorous shaking by hand for 10 s. Protoplasts were released by filtering through 40-μm nylon meshes into a round bottom tube with three to five washes of W5 solution. The pellet was collected by centrifugation at 140 g for 7 min and resuspended with 1 mL W5 solution. The GPF signals in the isolated protoplasts were examined using a confocal microscope.
Construction of OsHAC1;1-GFP and OsHAC1;2-GFP Fusion Proteins, Transient Expression in Rice Protoplasts, and Subcellular Localization of OsHAC1;1-GFP and OsHAC1;2-GFP
The full-length cDNAs of OsHAC1;1 and OsHAC1;2 without the stop codon were amplified and sequenced. The fragments were cloned into the pS1GFP-8 vector driven by the cauliflower mosaic virus 35S promoter. Then, 0.2 mL of protoplast suspension (approximately 2105 cells) was transfected with DNA for various constructs (10 mg each). After transfection, cells were cultured in a protoplast medium (0.4 m mannitol, 4 mm MES [pH 5.7], 4 mm KC1, sterilized) overnight (approximately 12 h). The fluorescence of Acridine orange (a nucleus-selective dye) and GFP in the cells were analyzed with a 543-nm helium-neon laser and a 488-nm argon laser, respectively, using a confocal laser scanning microscope (LSM410; Carl Zeiss).
Generation of OsHAC1;1 Knockout Mutants
We used the CRISPR/cas9 technology to generate oshacl;1 knockout lines in the cv Zhonghua11 background using the protocol described previously (Feng et al., 2013). Firstly, we chose the sequence 5′-TGGCGCCTCCCTATGAAACC-3′ in the first exon of OsHAC1;1 as the target region and designed two oligos: CAS9-OsHAC1;1F and CAS9-OsHAC1;1R (Supplemental Table S1). The two oligos were annealed and ligated with vector SK-OsU6-2-85-sgRNA restricted by enzyme BbsI to form the transition vector SK-OsU6-2-85-OsHAC1;1-sgRNA. The transition vector was then restricted with KpnI and HindIII to harvest a 476-bp fragment containing OsU6 promoter and guide RNA. Meanwhile, the vector SK-35S-CAS9-NOS was restricted with HindIII and EcoRI to harvest a 5.5-kb fragment containing 35S promoter, CAS9 coding gene, and a NOS terminator. The two fragments were subsequently ligated with linearized pCambia1300 with restriction enzymes KpnI and EcoRI to form the final expression binary vector. The final vector was transformed into rice variety Zhonghua 11 mediated by A. tumeraciens strain EHA105. At the T0 generation, all positive transgenic lines were genotyped with the primers HAC1;1-CAS9SF and HAC1;1-CAS9SF (Supplemental Table S1). Heterozygous knockout mutants were picked, and their T1 progenies were further genotyped for homozygous knockout mutants.
Generation of OsHAC1;1 and OsHAC1;2 Overexpression Lines
To generateOsHAC1 and OsHAC1;2 overexpression lines, the full-length coding sequence of OsHAC1;1 and OsHAC1;2 were amplified and sequenced using the specific primers listed in Supplemental Table S1. The fragments were digested with BamHI and SpeI and ligated to the pTCK303 vector (Wang et al., 2004). The verified vectors were used for generating transgenic plants of OsHAC1;1 and OsHAC1;2 in the cv Nipponbare background. We obtained 25 transgenic lines for each gene. Three lines each were selected randomly for hydroponic and soil pot experiments.
Functional Complementation of OsHAC1;1 and OsHAC1;2 in Escherichia coli
For prokaryotic expression of OsHAC1;1 and OsHAC1;2, the full-length coding sequences of OsHAC1;1 and OsHAC1;2 were amplified using gene-specific primers (Supplemental Table S1). The fragments were cloned into the prokaryotic expression vector pET-29a and verified by sequencing. The vector was transformed into E. coli ΔarsC mutant WC3110 (a strain lacking the endogenous arsenate reductase) and its wild type W3110 for complementation. The ΔarsC mutant (WC3110) and its wild type (W3110) with pET-29a empty vector, pET-29a-OsHAC1;1 or pET-29a-OsHAC1;2 were cultured at 37°C overnight. All cultured strains were diluted to OD600 nm = 0.5 and 1 mL was inoculated into 100 mL of LB liquid media containing 1 mm isopropylthio-β-galactoside and different concentrations of As(V). Cells were cultured at 16°C. The cell density was measured at OD600 nm using a spectrophotometer at different time points. The LB medium containing 10 µM As(V) was collected at 72 h and filtered through a 0.22-µm membrane filter before As speciation analysis using HPLC-ICP-MS.
Analysis of Total As Content and As Speciation
For the determination of total As concentration in plant samples, plant tissues were washed with deionized water three times and dried at 70°C for 3 d. Dried plant samples were digested with 5 mL mix acids of HNO3/HClO4 (85:15) in a digestion block. The digests were diluted with 2% HNO3 and As concentrations were determined using ICP-MS (Perkin Elmer NexION 300x, Waltham, MA). As speciation in nutrient solutions, xylem saps and plant extracts was determined using HPLC-ICP-MS (Liu et al., 2010). Plant roots were rinsed briefly in an ice-cold desorption solution containing 1 mm K2HPO4, 0.5 mm Ca(NO3)2, and 5 mm MES (pH 6.0) and immersed in 1 L of the same solution for 10 min to remove apoplastic As. Roots were blotted dry, weighed, and frozen in liquid nitrogen. Plant shoots were rinsed with deionized water, blotted dry, weighed, and frozen in liquid nitrogen. Shoots and roots were ground in liquid nitrogen to fine powder with a mortar and pestle. The finely ground materials were extracted with 10 mL phosphate-buffer solution containing 2 mm NaH2PO4 and 0.2 mm Na2-EDTA (pH 6.0) for 1 h under sonication in a 4°C room (Xu et al., 2007). The extract was filtered through a 0.22 μm membrane filter before analysis. Arsenic species were separated using an anion-exchange column (Hamilton PRP X-100, fitted with a guard column; Reno, NV) with a mobile phase of 6.0 mm NH4H2PO4, 6.0 mm NH4NO3, and 0.2 mm Na2EDTA (pH 6.0), run isocratically at 1 mL min−1. The solution from the separation column was mixed continuously with an internal standard solution (Indium) before being introduced into the ICP-MS. The instrument was set up in the kinetic energy discrimination mode with helium as the collision gas to reduce polyatomic interferences. Signals at m/z75As and 115In were collected with a dwell time of 300 ms; the In counts were used to normalize the As counts. Arsenic species in the sample were quantified by external calibration curves using peak areas.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_001045596 and NP_001052130 for OsHAC1;1 and OsHAC1;2, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence analysis of HAC genes in rice and Arabidopsis.
Supplemental Figure S2. Subcellular localization of OsHAC1;1 and OsHAC1;2 in protoplasts isolated from transgenic rice plants expressing pHAC1;1:OsHAC1;1-GFP or pHAC1;2:OsHAC1;2-GFP.
Supplemental Figure S3. Exposure to As(III) decreases the expression of OsHAC1;1 and OsHAC1;2.
Supplemental Figure S4. Knockout mutants of OsHAC1;1 (hac1;1-1, hac1;1-2) and OsHAC1;2 (hac1;2-1, hac1;2-2).
Supplemental Figure S5. Knocking out OsHAC1;1 or OsHAC1;2 has no significant effect on As(III) uptake and As accumulation in rice.
Supplemental Figure S6. Overexpression of OsHAC1;1 or OsHAC1;2 has no significant effect on As(V) uptake.
Supplemental Figure S7. Overexpression of OsHAC1;1 or OsHAC1;2 has no significant effect on As(III) uptake and As accumulation in rice.
Supplemental Figure S8. Root elongation of OsHAC1;1 and OsHAC1;2 overexpression lines and wild-type plants exposed to different concentrations of As(V).
Supplemental Table S1. The primers used in this study.
Supplementary Material
Glossary
- LB
Luria-Bertani
- qRT-PCR
quantitative real-time PCR
References
- Abedin MJ, Feldmann J, Meharg AA (2002) Uptake kinetics of arsenic species in rice plants. Plant Physiol 128: 1120–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Banerjee N, Bhattacharjee P, Mondal D, Lythgoe PR, Martínez M, Pan J, Polya DA, Giri AK (2013) High arsenic in rice is associated with elevated genotoxic effects in humans. Sci Rep 3: 2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Hakvoort HWJ, Bliek M, Souer E, Schat H (2006) Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. Plant J 45: 917–929 [DOI] [PubMed] [Google Scholar]
- Bouman BAM, Peng S, Castaneda AR, Visperas RM (2005) Yield and water use of irrigated tropical aerobic rice systems. Agric Water Manage 74: 87–105 [Google Scholar]
- Byers LD, She HS, Alayoff A (1979) Interaction of phosphate analogues with glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 18: 2471–2480 [DOI] [PubMed] [Google Scholar]
- Chao DY, Chen Y, Chen J, Shi S, Chen Z, Wang C, Danku JM, Zhao FJ, Salt DE (2014) Genome-wide association mapping identifies a new arsenate reductase enzyme critical for limiting arsenic accumulation in plants. PLoS Biol 12: e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Moore KL, Miller AJ, McGrath SP, Ma JF, Zhao F-J (2015) The role of nodes in arsenic storage and distribution in rice. J Exp Bot 66: 3717–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ (1994) Reduction and binding of arsenate and dimethylarsinate by glutathione: a magnetic resonance study. Chem Biol Interact 90: 139–155 [DOI] [PubMed] [Google Scholar]
- Dhankher OP, Rosen BP, McKinney EC, Meagher RB (2006) Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2). Proc Natl Acad Sci USA 103: 5413–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan GL, Zhou Y, Tong YP, Mukhopadhyay R, Rosen BP, Zhu YG (2007) A CDC25 homologue from rice functions as an arsenate reductase. New Phytol 174: 311–321 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Gumaelius L, Indriolo E, Pickering IJ, Banks JA, Salt DE (2006) A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol 141: 1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang D-L, Wei P, Cao F, Zhu S, Zhang F, Mao Y, et al. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23: 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert-Diamond D, Cottingham KL, Gruber JF, Punshon T, Sayarath V, Gandolfi AJ, Baker ER, Jackson BP, Folt CL, Karagas MR (2011) Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci USA 108: 20656–20660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregus Z, Roos G, Geerlings P, Németi B (2009) Mechanism of thiol-supported arsenate reduction mediated by phosphorolytic-arsenolytic enzymes. II. Enzymatic formation of arsenylated products susceptible for reduction to arsenite by thiols. Toxicol Sci 110: 282–292 [DOI] [PubMed] [Google Scholar]
- Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, O’Connell MJ, Goldsbrough PB, Cobbett CS (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Sun GX, Williams PN, Nunes L, Zhu YG (2011) Inorganic arsenic in Chinese food and its cancer risk. Environ Int 37: 1219–1225 [DOI] [PubMed] [Google Scholar]
- Linquist BA, Anders MM, Adviento-Borbe MAA, Chaney RL, Nalley LL, da Rosa EFF, van Kessel C (2015) Reducing greenhouse gas emissions, water use, and grain arsenic levels in rice systems. Glob Change Biol 21: 407–417 [DOI] [PubMed] [Google Scholar]
- Liu W, Schat H, Bliek M, Chen Y, McGrath SP, George G, Salt DE, Zhao FJ (2012) Knocking out ACR2 does not affect arsenic redox status in Arabidopsis thaliana: implications for as detoxification and accumulation in plants. PLoS One 7: e42408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wood BA, Raab A, McGrath SP, Zhao FJ, Feldmann J (2010) Complexation of arsenite with phytochelatins reduces arsenite efflux and translocation from roots to shoots in Arabidopsis. Plant Physiol 152: 2211–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Zhu YG, Hu Y, Williams PN, Gault AG, Meharg AA, Charnock JM, Smith FA (2006) Arsenic sequestration in iron plaque, its accumulation and speciation in mature rice plants (Oryza sativa L.). Environ Sci Technol 40: 5730–5736 [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ (2008) Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA 105: 9931–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg AA, Sun G, Williams PN, Adomako E, Deacon C, Zhu YG, Feldmann J, Raab A (2008) Inorganic arsenic levels in baby rice are of concern. Environ Pollut 152: 746–749 [DOI] [PubMed] [Google Scholar]
- Meharg AA, Williams PN, Adomako E, Lawgali YY, Deacon C, Villada A, Cambell RCJ, Sun G, Zhu YG, Feldmann J, et al. (2009) Geographical variation in total and inorganic arsenic content of polished (white) rice. Environ Sci Technol 43: 1612–1617 [DOI] [PubMed] [Google Scholar]
- Mondal D, Polya DA (2008) Rice is a major exposure route for arsenic in Chakdaha block, Nadia district, West Bengal, India: a probabilistic risk assessment. Appl Geochem 23: 2987–2998 [Google Scholar]
- Moore KL, Schröder M, Wu ZC, Martin BGH, Hawes CR, McGrath SP, Hawkesford MJ, Ma JF, Zhao FJ, Grovenor CRM (2011) High-resolution secondary ion mass spectrometry NanoSIMS reveals contrasting patterns of arsenic and silicon localization in rice roots. Plant Physiol 156: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP (2001) The phosphatase C(X)5R motif is required for catalytic activity of the Saccharomyces cerevisiae Acr2p arsenate reductase. J Biol Chem 276: 34738–34742 [DOI] [PubMed] [Google Scholar]
- Nahar N, Rahman A, Moś M, Warzecha T, Algerin M, Ghosh S, Johnson-Brousseau S, Mandal A (2012) In silico and in vivo studies of an Arabidopsis thaliana gene, ACR2, putatively involved in arsenic accumulation in plants. J Mol Model 18: 4249–4262 [DOI] [PubMed] [Google Scholar]
- National Research Council (2001) Arsenic in Drinking Water 2001 Update. National Academies Press, Washington, DC [Google Scholar]
- Oden KL, Gladysheva TB, Rosen BP (1994) Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol 12: 301–306 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab A, Schat H, Meharg AA, Feldmann J (2005) Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): formation of arsenic-phytochelatin complexes during exposure to high arsenic concentrations. New Phytol 168: 551–558 [DOI] [PubMed] [Google Scholar]
- Rosen BP. (2002) Biochemistry of arsenic detoxification. FEBS Lett 529: 86–92 [DOI] [PubMed] [Google Scholar]
- Sánchez-Bermejo E, Castrillo G, del Llano B, Navarro C, Zarco-Fernández S, Martinez-Herrera DJ, Leo-del Puerto Y, Muñoz R, Cámara C, Paz-Ares J, et al. (2014) Natural variation in arsenate tolerance identifies an arsenate reductase in Arabidopsis thaliana. Nat Commun 5: 4617. [DOI] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y (2010) Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth AL, Webb SM, Andrews JC, Fendorf S (2010) Arsenic localization, speciation, and co-occurrence with iron on rice (Oryza sativa L.) roots having variable Fe coatings. Environ Sci Technol 44: 8108–8113 [DOI] [PubMed] [Google Scholar]
- Song WY, Park J, Mendoza-Cózatl DG, Suter-Grotemeyer M, Shim D, Hörtensteiner S, Geisler M, Weder B, Rea PA, Rentsch D, et al. (2010) Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc Natl Acad Sci USA 107: 21187–21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Y, Yamaki T, Yamaji N, Ko D, Jung K-H, Fujii-Kashino M, An G, Martinoia E, Lee Y, Ma JF (2014) A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc Natl Acad Sci USA 111: 15699–15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Report 22: 409–417 [Google Scholar]
- Wu Z, Ren H, McGrath SP, Wu P, Zhao FJ (2011) Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol 157: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XY, McGrath SP, Meharg AA, Zhao FJ (2008) Growing rice aerobically markedly decreases arsenic accumulation. Environ Sci Technol 42: 5574–5579 [DOI] [PubMed] [Google Scholar]
- Xu XY, McGrath SP, Zhao FJ (2007) Rapid reduction of arsenate in the medium mediated by plant roots. New Phytol 176: 590–599 [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Ago Y, Mitani N, Li RY, Su YH, Yamaji N, McGrath SP, Ma JF (2010a) The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol 186: 392–399 [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Ma JF, Meharg AA, McGrath SP (2009) Arsenic uptake and metabolism in plants. New Phytol 181: 777–794 [DOI] [PubMed] [Google Scholar]
- Zhao FJ, McGrath SP, Meharg AA (2010b) Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu Rev Plant Biol 61: 535–559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.