Class II cytochrome P450 reductase in Madagascar periwinkle displays a prominent contribution toward specialized metabolism by acting as the main partner of P450s dedicated to alkaloid biosynthesis.
Abstract
Expansion of the biosynthesis of plant specialized metabolites notably results from the massive recruitment of cytochrome P450s that catalyze multiple types of conversion of biosynthetic intermediates. For catalysis, P450s require a two-electron transfer catalyzed by shared cytochrome P450 oxidoreductases (CPRs), making these auxiliary proteins an essential component of specialized metabolism. CPR isoforms usually group into two distinct classes with different proposed roles, namely involvement in primary and basal specialized metabolisms for class I and inducible specialized metabolism for class II. By studying the role of CPRs in the biosynthesis of monoterpene indole alkaloids, we provide compelling evidence of an operational specialization of CPR isoforms in Catharanthus roseus (Madagascar periwinkle). Global analyses of gene expression correlation combined with transcript localization in specific leaf tissues and gene-silencing experiments of both classes of CPR all point to the strict requirement of class II CPRs for monoterpene indole alkaloid biosynthesis with a minimal or null role of class I. Direct assays of interaction and reduction of P450s in vitro, however, showed that both classes of CPR performed equally well. Such high specialization of class II CPRs in planta highlights the evolutionary strategy that ensures an efficient reduction of P450s in specialized metabolism.
With more than 200,000 distinct molecules, the specialized metabolism of plants constitutes one of the main sources of bioactive natural compounds, with this large number reflecting the capacity of these sessile organisms to adapt to and interact with the environment. This singular complexity of compounds results from an evolutionary process that involves a dramatic diversification of plant metabolic pathways and the genes encoding the associated metabolic enzymes. From this perspective, cytochromes P450 (P450s) are a prototypical example of ubiquitous enzymes encoded by a gene superfamily that can carry out multiple types of reactions, among them hydroxylation, epoxidation, oxygenation, dealkylation, decarboxylation, C-C cleavage, and ring opening (Bak et al., 2011; Guengerich and Munro, 2013). P450s catalyze a considerable array of chemically challenging reactions in the biosynthesis of plant specialized metabolites, including phenylpropanoids, terpenoids, cyanogenic glycosides, and alkaloids (Mizutani and Ohta, 2010; Mizutani and Sato, 2011), in addition to various roles in plant primary metabolism, such as the biosynthesis of hormones (Takei et al., 2004; Bak et al., 2011). Most characterized P450s perform single oxidation reactions, but growing evidence now also points to the existence of multistep oxidations catalyzed by a single P450 (Guengerich et al., 2011). However, in all P450-catalyzed reactions, the catalytic cycle always requires two one-electron transfer steps from NADPH into the prosthetic heme. This transfer occurs through the FAD and FMN domains of NADPH-cytochrome P450 reductases (CPRs), although, in some cases, the second electron transfer also can arise from cytochrome b5 (Munro et al., 2013). The absolute requirement of CPR thus makes this flavoprotein a cornerstone in P450 activities and, consequently, in specialized and primary metabolic pathways. As such, the physical interactions between P450s and CPRs that guide electron shuffling directly influence P450 activities and are facilitated by membrane anchoring of both types of proteins, which are typically localized to the endoplasmic reticulum (ER; Hasemann et al., 1995; Ro et al., 2002; Denisov et al., 2007). Elucidation of the three-dimensional structure of the rat CPR suggested that P450-CPR interactions occur through both ionic interactions involving the FMN domain of CPRs and hydrophobic interactions between the membrane domains of CPRs and P450s (Wang et al., 1997). The low CPR:P450 ratio, measured at approximately 1:15 in liver, also implies a potential competition between P450s for these accessory proteins in plants; as a consequence, some kind of logistic control must be employed to ensure the coordinated operation of all P450s belonging to similar biosynthetic pathways (Shephard et al., 1983).
In contrast to yeasts and mammals that harbor only a single CPR, vascular plants have evolved two or three CPR isoforms, as reported for instance in Jerusalem artichoke (Helianthus tuberosus; Benveniste et al., 1991), poplar (Populus spp.; Ro et al., 2002), parsley (Petroselinum crispum; Koopmann and Hahlbrock, 1997), cotton (Gossypium hirsutum; Yang et al., 2010), and winter cherry (Withania somnifera; Rana et al., 2013). The Arabidopsis (Arabidopsis thaliana) genome contains two genes encoding functionally active CPR genes (ARABIDOPSIS THALIANA P450 REDUCTASE), named ATR1 and ATR2, and a third more distant gene (ATR3) whose expression product was not able to reduce P450s in vitro (Urban et al., 1997; Mizutani and Ohta, 2010; Varadarajan et al., 2010). By contrast, poplar and Nothapodytes foetida possess three genes encoding genuine CPRs that are similar to ATR1 and ATR2 and an additional predicted ATR3-like isoform (Ro et al., 2002; Huang et al., 2012). Homologs of ATR1 and ATR2 in flowering plants are highly conserved (65%–80%) and are clustered into two distinct phylogenic groups on the basis of their N-terminal sequences. The first cluster (ATR1 homologs, class I) contains sequences from eudicotyledons, while the second (class II) are found in both monocotyledons and eudicotyledons (Ro et al., 2002). Homologs of ATR3 fall into a distinct third cluster (Varadarajan et al., 2010). While CPRs from both class I and class II can reduce P450 with an apparent similar efficiency, their expression profiles are different (Urban et al., 1997; Ro et al., 2002; Mizutani and Ohta, 2010). CPR1s belonging to class I, such as ATR1, are constitutively expressed, while CPR2s from class II (e.g. ATR2) are inducible by environmental stimuli such as wounding, pathogen infection, or light exposure (Koopmann and Hahlbrock, 1997; Ro et al., 2002; Schwarz et al., 2009; Mizutani and Ohta, 2010; Yang et al., 2010, Rana et al., 2013). These observations suggest that each CPR isoform has dedicated physiological roles, ensuring that plants can meet the reductive demand of P450-mediated reactions. It is now believed that constitutively expressed CPRs (CPR1s, class I) ensure basal P450 activities in primary metabolism or in the constitutive synthesis of specialized metabolism, while inducible CPRs (CPR2s, class II) serve in adaptation mechanisms or in defense reactions, including the elicited biosynthesis of specialized metabolites. This hypothesis has been partially confirmed by establishing a correlation between ATR2 activity and lignin biosynthesis in Arabidopsis as well as between CPR1 expression and basal pungent alkaloid synthesis in Capsicum spp. (Mazourek et al., 2009; Sundin et al., 2014). In contrast, CPR-like (ATR3) homologs are poorly characterized but appear to be essential for embryo development (Varadarajan et al., 2010). However, more investigations are required to firmly establish the functional specificity of all CPRs in plants.
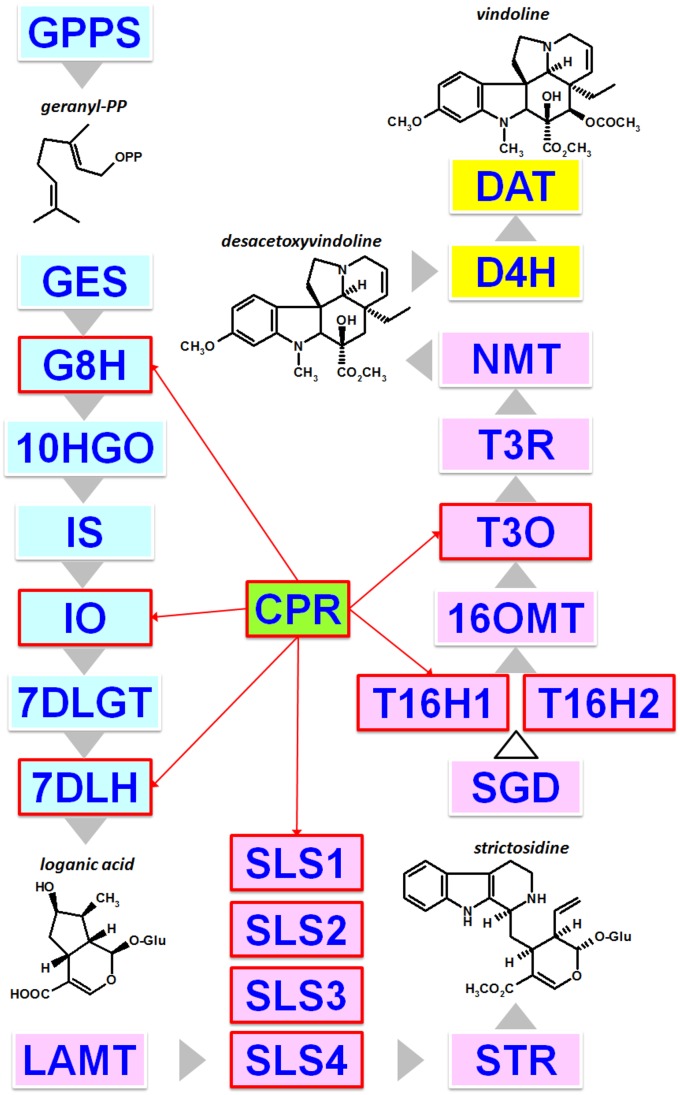
For more than 40 years, specialized metabolism has been investigated using the Madagascar periwinkle (Catharanthus roseus), which serves as an excellent case study for the analysis of plant specialized metabolism. This plant most notably synthesizes alkaloids from the monoterpene indole alkaloid (MIA) family that includes valuable compounds such as the antineoplastic vinblastine and vincristine. These MIAs result from a long and complex biosynthetic pathway whose characterization has made great progress over the last 5 years due to the development of large sets of transcriptomic data and a draft genome sequence (Góngora-Castillo et al., 2012; Van Moerkercke et al., 2013; Xiao et al., 2013; Dugé de Bernonville et al., 2015a; Kellner et al., 2015b). Within the 30 to 50 enzymatic steps predicted to form the network of reactions of the MIA biosynthetic pathway, no less than 11 P450s have been identified to date (Fig. 1): geraniol 8-hydroxylase (G8H [CYP76B6]; Collu et al., 2001), iridoid oxidase (IO [CYP76A26]; Miettinen et al., 2014; Salim et al., 2014), 7-deoxyloganic acid 7-hydroxylase (7DLH [CYP72A224]; Salim et al., 2013; Miettinen et al., 2014), four isoforms of secologanin synthase (SLS1–SLS4 [CYP72A1]; Irmler et al., 2000; Brown et al., 2015; Dugé de Bernonville et al., 2015b), two isoforms of tabersonine 16-hydroxylase (T16H1 and T16H2 [CYP71D12 and CYP71D351]; Schröder et al., 1999; Guirimand et al., 2011b; Besseau et al., 2013), tabersonine 19-hydroxylase (T19H [CYP71BJ1]; Giddings et al., 2011), and 16-methoxytabersonine 3-oxygenase (T3O [CYP71D1]; Kellner et al., 2015a; Qu et al., 2015b). While some of these enzymes catalyze a single oxygenation reaction, including hydroxylation (7DLH, T16H1, T16H2, and T19H) or epoxidation (T3O), unusual reactions also have been reported, such as the ring opening of loganin to yield secologanin (SLS) or the three-step oxidation of nepetalactol to form 7-deoxyloganetic acid (IO). In addition to this diversity of reactions, a complex spatiotemporal organization of these P450-mediated enzymatic steps also has been reported in periwinkle leaves, with the first MIA biosynthetic conversions (G8H to 7DLH) occurring in internal phloem-associated parenchyma (IPAP) and the remaining reactions up to T3O and the next two enzymatic steps in epidermis (Fig. 1; Courdavault et al., 2014; Miettinen et al., 2014; Salim et al., 2014; Qu et al., 2015b). As a matter of fact, this multicellular organization of the MIA biosynthetic pathway constitutes the first layer of the physiological processes regulating MIA formation. Interestingly, the periwinkle CPR was one of the first plant CPRs to be purified and cloned (Madyastha and Coscia, 1979; Meijer et al., 1993). Due to the presence of a hydrophobic residue region at its N-terminal end, this protein groups with class II CPRs, in agreement with its transcriptional regulation in response to fungal elicitor preparation and to jasmonate (Meijer et al., 1993; Collu et al., 2001; Ro et al., 2002). Although only one CPR had been cloned in C. roseus at the start of this study, the occurrence of multiple isoforms was expected and is now supported by transcriptomic and genomic data (Canto-Canché and Loyola-Vargas, 2001). However, no formal relationship had been established between the periwinkle CPRs and the biosynthesis of MIA. This prompted us to take advantage of the richness of the C. roseus MIA metabolism to accurately explore the role of each class of CPR in specialized metabolism. By combining biochemical characterization, protein interaction analyses, mapping of cellular gene expression profiles, and gene-silencing approaches, our data provide new evidence for the predominant role of class II CPRs in MIA/specialized metabolism.
Figure 1.
Main characterized steps of MIA biosynthesis in C. roseus aerial organs highlighting several steps involving P450s. The color code indicates the cellular organization of the pathway as follows: blue, pink, and yellow rectangles for compartmentation in internal phloem-associated parenchyma (IPAP), epidermis, and laticifers/idioblasts, respectively. Noteworthy metabolites are depicted, such as geranyl diphosphate, which constitutes the entry point of MIA biosynthesis; loganic acid and desacetoxyvindoline, which are transported from IPAP to epidermis and from epidermis to laticifers/idioblasts, respectively; strictosidine, the first MIA; and vindoline, which is the main MIA accumulated in leaves. P450s are highlighted by red rectangles, and reducing power by CPR is indicated by red arrow lines. The white arrowhead indicates multiple not yet discovered enzymes. GPPS, Geranyl diphosphate synthase; GES, geraniol synthase; G10H (CYP76B6), geraniol 10-hydroxylase; 10HGO, 10-hydroxygeraniol oxidoreductase; IS, iridoid synthase; 7DLGT, 7-deoxyloganetic acid glucosyltransferase; 7DLH (CYP72A224), 7-deoxyloganic acid hydroxylase; LAMT, loganic acid O-methyltransferase; STR, strictosidine synthase; SGD, strictosidine β-glucosidase; 16OMT, 16-hydroxytabersonine O-methyltransferase; NMT, 3-hydroxy-16-methoxy-2,3-dihydrotabersonine N-methyltransferase; T3R, tabersonine 3-reductase; D4H, desacetoxyvindoline 4-hydroxylase; DAT, deacetylvindoline 4-acetyltransferase.
RESULTS
Identification of C. roseus CPR Homologs
In addition to a contig corresponding to the CPR cDNA (X69791), which was cloned previously by Meijer et al. (1993), interrogation of C. roseus transcriptomic resources led to the identification of two additional contigs homologous to CPR (Supplemental Table S1). Proteins deduced from contig CPR candidate 1 and contig CPR candidate 2 displayed 67% and 25% identity with the originally identified CPR, respectively (Supplemental Fig. S1). According to previously published classifications of CPRs (Ro et al., 2002; Jensen and Moller, 2010; Varadarajan et al., 2010), phylogenetic analyses demonstrated that each deduced protein clusters in distinct phylogenetic subgroups (Supplemental Fig. S2). While the original CPR is categorized as a class II CPR, contig CPR candidate 1 falls into the class I CPRs, which is hypothesized to be mostly dedicated to basal metabolism. Contig CPR candidate 2 clusters in class III CPRs, whose prominent member in Arabidopsis corresponds to ATR3. Based on this result and on the Arabidopsis nomenclature, contig CPR candidate 1, the original CPR, and contig CPR candidate 2 were renamed CPR1, CPR2, and CPR3/diflavin reductase (DFR), respectively. C. roseus genome analysis also revealed that the three corresponding genes were present at one copy per haploid genome and spanned over 18 exons for CPR1 (CRO_T001672) and CPR2 (CRO_T031702) and 12 exons for CRP3 (CRO_T033752; Supplemental Fig. S3). The genomic organization of CPR1 and CPR2 is similar regarding intron positions and intron/exon sizes, except for the first intron of CPR1, which is roughly 7-fold longer than the first intron of CPR2. Such similarity may reflect the gene duplication event that led to both CPR appearances. Therefore, these results suggest that C. roseus contains two CPRs, CPR1 (KJ701028) and CPR2 (X69791), potentially associated with basal and inducible/specialized metabolism, respectively, and one more distant copy, CPR3/DFR (KM111538), a likely ortholog of ATR3, the function of which remains unclear.
Sequence Analysis and Subcellular Localization of C. roseus CPRs
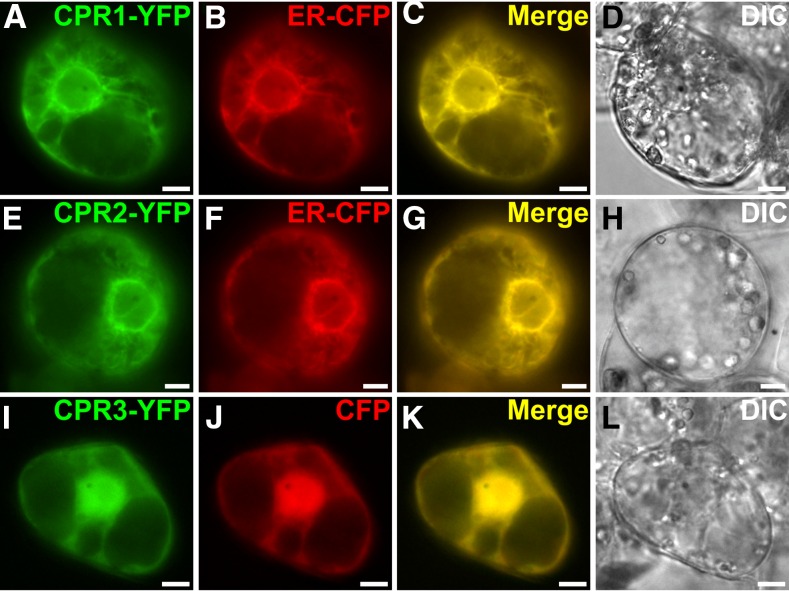
Analysis of the deduced protein sequences of C. roseus CPRs revealed that both CPR1 and CPR2 are characterized by (1) the presence of conserved FMN-, FAD-, and NADPH-binding domains that have been implicated in electron transfer and (2) identical residues predicted to be involved in interactions with P450s (Jensen and Moller, 2010; Supplemental Figs. S4 and S5). Both proteins also bear a predicted membrane-spanning domain at their N-terminal ends (Supplemental Fig. S6) that has been shown to mediate ER anchoring in poplar (Ro et al., 2002). To investigate whether this sequence does in fact anchor the protein to the ER membrane, the subcellular localizations of C. roseus CPR1 and CPR2 were expressed as C-terminal yellow fluorescent protein (YFP) fusion proteins (CPR1-YFP and CPR2-YFP) to avoid interference with the membrane-spanning domain. In C. roseus transiently transformed cells, the fusion proteins exhibited a network-shaped fluorescent signal that perfectly merged with the signal of the ER-cyan fluorescent protein (CFP) marker (Fig. 2, A–H), suggesting that both CPR1 and CPR2 are anchored to the ER, in agreement with the classical localization pattern of P450s. When mutants of CPR1 and CPR2 that lacked this predicted transmembrane domain were expressed as YFP-labeled fusions, the previously observed localization pattern was disrupted, while the fusion of these membrane domains to YFP enabled ER anchoring. These experiments clearly demonstrate that this predicted membrane-spanning domain is necessary and sufficient to ensure ER localization/anchoring (Supplemental Fig. S7). In addition, in agreement with the assigned CPR classification, we noted that CPR1 and CPR2 differ in the length of the protein sequence preceding the membrane-spanning domain: CPR1 has only a short stretch of residues, while CPR2 exhibits an extended amino acid sequence, enriched in Ser residues, that was initially, but wrongly, considered to be a plastid-targeting sequence (Supplemental Fig. S1; Ro et al., 2002). CPR3/DFR also displays conserved FMN-, FAD-, and NADPH-binding domains but shows substantial differences in the P450-interacting region compared with CPR1 and CPR2 (Supplemental Fig. S5). Moreover, CPR3/DFR lacks the N-terminal membrane-spanning domain, explaining the nucleocytosolic localization observed in C. roseus cells transformed with the CPR3-YFP fusion protein (Fig. 2, I–L). This nucleocytosolic localization was observed previously with ATR3 (Varadarajan et al., 2010). Therefore, the distinct subcellular localization patterns, along with the sequence differences in the P450-interacting domain in CPR1 and CPR2 compared with CPR3/DFR, suggest the existence of distinct sets of interacting partners for these proteins.
Figure 2.
CPR1 and CPR2 are located to the ER, while CPR3 is a nucleocytosolic protein. C. roseus cells were transiently transformed with the CPR1-YFP (A), CPR2-YFP (E), or CPR3-YFP (I) expressing vectors in combination with plasmids expressing either an ER-CFP marker (B and F) or a nucleocytosolic marker (CFP; J). Colocalization of the fluorescent signals appears on the merged images (C, G, and K). Cell morphologies (D, H, and L) were observed with differential interference contrast (DIC) imaging. Bars = 10 μm.
CPR1 and CPR2, But Not CPR3/DFR, Interact and Reduce C. roseus P450s Associated with Specialized Metabolism
To gain insight into possible selective interactions among the P450s and CPRs of C. roseus, bimolecular fluorescence complementation (BiFC) analyses were conducted by coexpressing each of the three CPRs with several P450s from the MIA biosynthetic pathway, including G8H, SLS2, T16H1, and T16H2 (Figs. 1 and 3). An additional test also was conducted with cinnamate 4-hydroxylase (C4H; CYP73A5) from C. roseus, since this P450 corresponds to the first P450 of the phenylpropanoid biosynthetic pathway, thus acting in a distinct specialized metabolic pathway. Moreover, the Arabidopsis C4H ortholog has been shown to be reduced with a similar efficiency by both ATR1 and ATR2, thereby providing a point of reference (Hotze et al., 1995; Mizutani and Ohta, 2010).
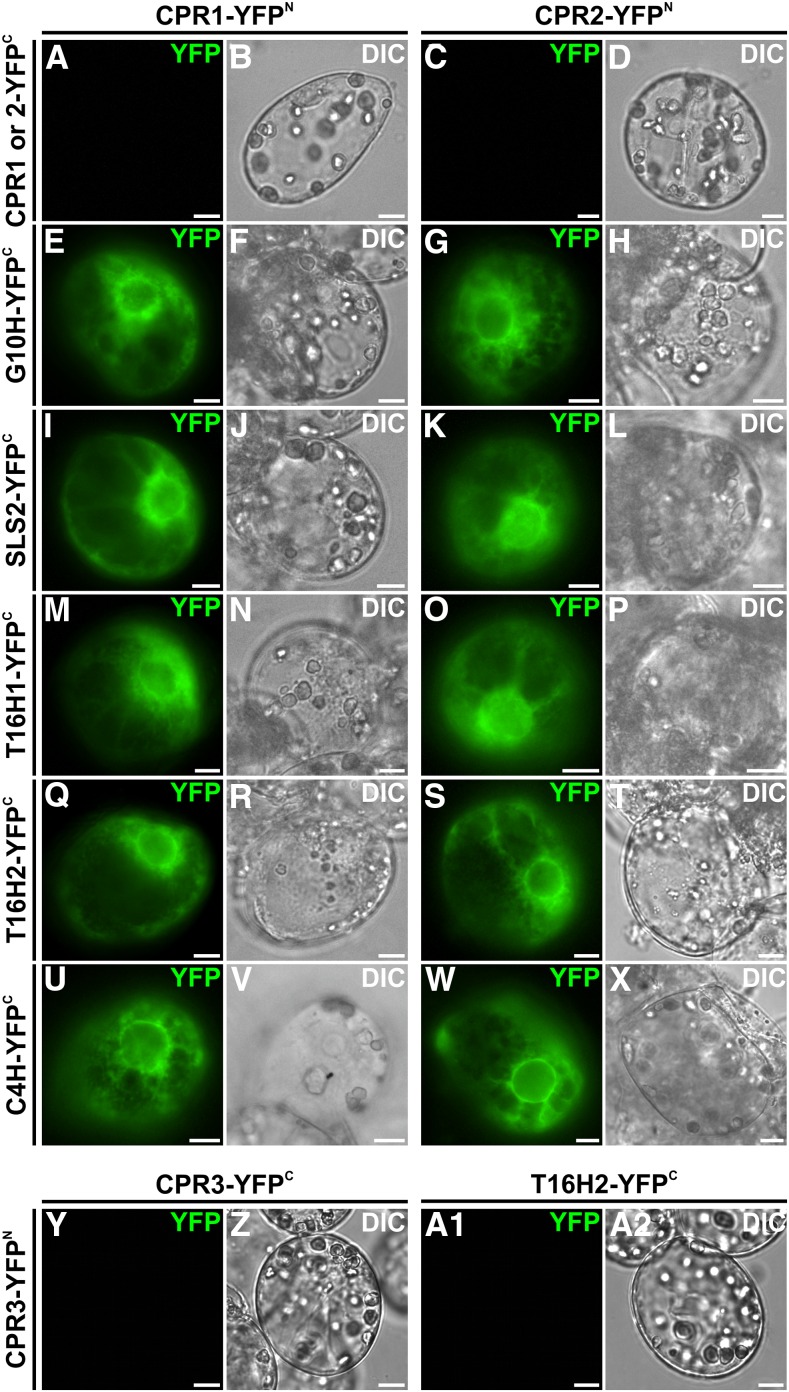
Figure 3.
CPR1 and CPR2 but not CPR3 interact with G10H, SLS2, T16H1, T16H2, and C4H. CPR and P450 interactions were analyzed by BiFC in C. roseus cells transiently transformed by plasmids encoding fusions indicated on the top (CPR1 or CPR2 fused to the split YFPN fragment) and on the left (CPR1, CPR2, G10H SLS2, T16H1, T16H2, and C4H fused to the split YFPC fragment). The YFP signal emanating from BiFC complex reformation is show in green false color (A, C, E, G, I, K, M, O, Q, S, U, W, Y, and A1), and cell morphology is observed with differential interference contrast (DIC) imaging (B, D, F, H, J, L, N, P, R, T, V, X, Z, and A2). Bars = 10 µm.
As described above, CPRs and these five P450s were fused to the N-terminal end of the split YFP fragments (N-terminal split YFP [YFPN] for CPRs and C-terminal split YFP [YFPC] for P450s) to preserve ER-anchoring capacities following transient expression in C. roseus cells. Interestingly, while no self-interactions were observed for either CPR1 or CPR2 (Fig. 3, A–D), the appearance of a strong fluorescent signal in the combinations of each CPR with all of the P450s suggested that CPR1 and CPR2 were capable of interaction with each of the five tested P450s (Fig. 3, E–X). By contrast, no interactions of CPR3/DFR, either with itself or with any of the tested P450s, were observed, as exemplified with T16H2 (Fig. 3, Y–A2). This may be linked to the neofunctionalization of class III CPRs and/or to the lack of the N-terminal membrane-spanning domain in CPR3/DFR.
Additionally, the capacity of the periwinkle CPR1 and CPR2 to reduce G8H, SLS2, T16H1, T16H2, and C4H was analyzed by conducting functional assays with proteins heterologously expressed in Saccharomyces cerevisiae. The resulting activities were compared with those measured in the WAT11 strain expressing an Arabidopsis CPR and with the activity engendered by the endogenous yeast (S. cerevisiae) CPR (Table I; Supplemental Table S2). Since partial losses of activity were observed when C. roseus CPRs were expressed with tags, as reported for CPR from Camptotheca acuminata (Qu et al., 2015a), assays were performed with untagged proteins using unpurified microsomes. Interestingly, in agreement with the BiFC results, we noted that both CPR1 and CPR2 were able to reduce the five tested P450s, as revealed by the conversion of the specific substrates of each of the five P450s. Moreover, although absolute quantification of CPR/P450 activity, which requires tagged protein, was not achievable, the relative activity of each of the P450s was measured to be within the same order of magnitude, suggesting that both CPR1 and CPR2 may reduce P450s in vitro with a similar efficiency, as reported previously for C4H in Arabidopsis (Mizutani and Ohta, 2010). Furthermore, we also noticed that CPR3/DFR was unable to reduce T16H, which is consistent with the lack of interaction observed in BiFC assays. To determine whether the inactivity resulted from a lack of ER membrane anchoring, the first 74 residues of CPR2 encompassing the membrane-spanning domain were fused to the N terminus of CPR3/DFR, which resulted in ER localization, as revealed by YFP imaging in C. roseus cells (Supplemental Fig. S8). However, this modification was unable to restore P450 activities (at least with SLS2, T16H1, and T16H2) in yeast (S. cerevisiae), strongly suggesting that CPR3/DFR reduces different enzymes and acts in different metabolic pathways. As a consequence, CPR3/DFR was renamed DFR in agreement with the presence of two flavin reductase domains and the apparent lack of P450 reduction capacity.
Table I. CPR1 and CPR2 reduce C4H, G8H, SLS2, T16H1, and T16H2 with an apparent similar efficiency.
Substrate conversion rates (%) were determined using crude protein extract of the WT303 yeast strain expressing each CPR/P450 pair (C4H, SLS2, T16H1, and T16H2 assays) or microsomes (G8H assay) by the addition of NADPH as an electron donor. Yeast endogenous CPR activity was estimated by measuring P450 activity in similar conditions without expression of the periwinkle CPRs and was subtracted from activities measured with CPR1 and CPR2. All assays were conducted independently three times with at least three technical replicates.
| Assay | Substrate | CPR1 | CPR2 |
|---|---|---|---|
| C4H (CYP73) | Cinnamic acid | 31.2 ± 0.8 | 26.2 ± 1.61 |
| G10H (CYP76B6) | Geraniol | 65.2 ± 0.95 | 70.3 ± 1.12 |
| SLS2 (CYP71A2) | Loganin | 25.6 ± 1.12 | 39.1 ± 3.7 |
| T16H1 (CYP71D12) | Tabersonine | 46 ± 0.83 | 47.3 ± 0.7 |
| T16H2 (CYP71D351) | Tabersonine | 60.9 ± 1.07 | 60.2 ± 0.25 |
CPR1 and CPR2 Display Ubiquitous Expression Patterns Associated with Specific Sets of Genes
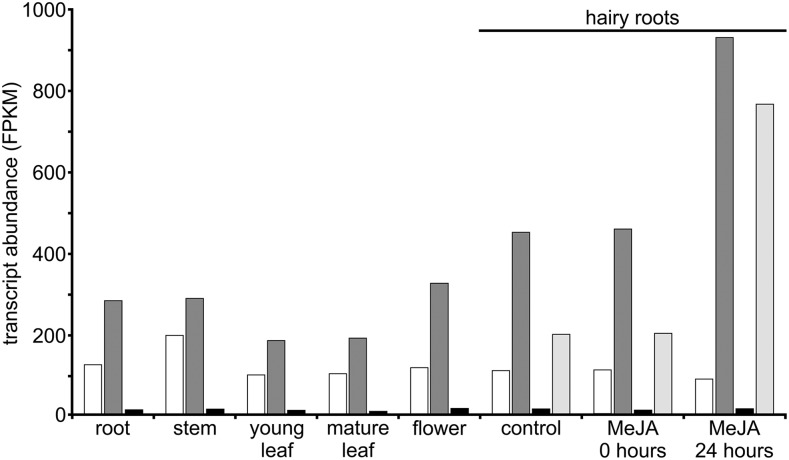
The putative specialization of CPRs with respect to MIA biosynthesis was investigated subsequently at the whole-plant level through gene expression analyses. By studying transcript abundance in C. roseus transcriptomic data sets (Góngora-Castillo et al., 2012; Dugé de Bernonville et al., 2015b), we observed that CPR1, CPR2, and DFR genes were expressed in all organs associated with MIA biosynthesis, including roots, stems, young/mature leaves, and flowers, although DFR expression was very low, as reported previously for ATR3 (Fig. 4; Varadarajan et al., 2010). CPR1 and CPR2 exhibited a similar pattern of expression in organs, but CPR2 expression was always almost 2-fold higher than CPR1 expression. Furthermore, while CPR1 levels were not affected by methyl jasmonate treatment, CPR2 was induced by this hormone, which is consistent with the role of class II CPR in induced pathways. This ubiquitous distribution of CPR1 and CPR2 transcripts in periwinkle organs was confirmed by quantitative PCR (qPCR) and also was compared with the expression of G10H, SLS2, T16H1, T16H2, and C4H (Supplemental Fig. S9). Although we noted that CPR- and P450-encoding genes were coexpressed in the different MIA-producing organs with some profile specificities inherent to each P450, this cooccurrence of gene expression led us to speculate that CPR1 as well as CPR2 both can potentially reduce P450s associated with MIA biosynthesis.
Figure 4.
CPR1, CPR2, and DFR transcript abundance in C. roseus organs and response to methyl jasmonate (MeJA) treatment. The abundance of transcripts was determined by comparing fragments per kilobase of transcript per million mapped reads (FPKM) values of CPR1, CPR2, and DFR loci from the main C. roseus transcriptomic data set. In hairy roots, estimation of the abundance of the transcripts encoding strictosidine synthase (STR), a well-known MIA biosynthetic gene inducible by methyl jasmonate, was included as a control. CPR1, White bars; CPR2, dark gray bars; DFR, black bars; STR, light gray bars.
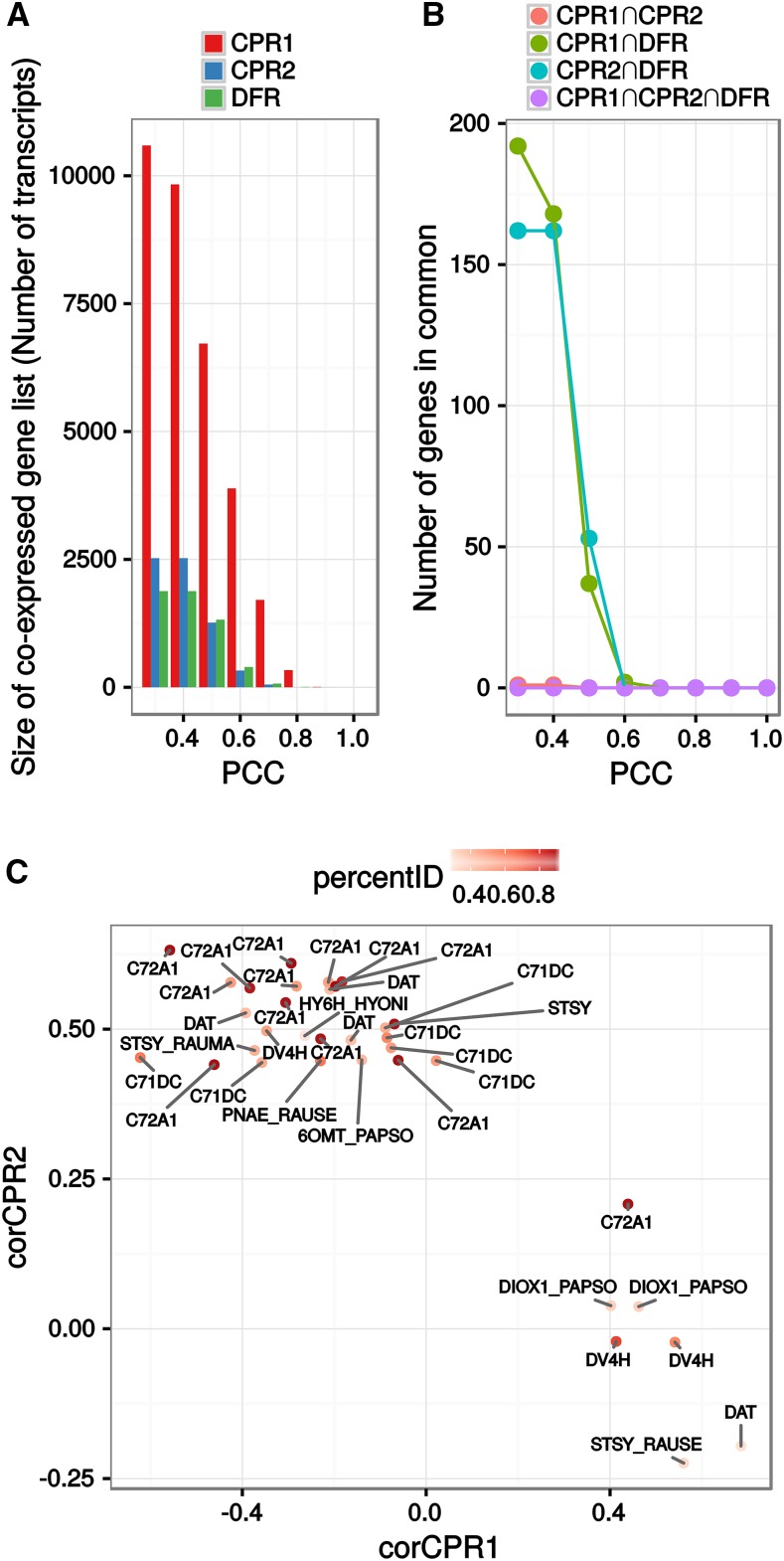
As a consequence, by taking advantage of the optimized transcriptomic C. roseus data set (Dugé de Bernonville et al., 2015b), a global analysis of the periwinkle genes coexpressed with each CPR was undertaken by calculating the Pearson correlation coefficient (PCC) of CPR1, CPR2, or DFR with each of the 58,338 other transcripts found in this assembly. This data set was obtained previously by combining individual transcriptomes assembled from sequencing runs available on the Sequence Read Archive. Expression levels within this transcriptome were computed with Salmon (Patro et al., 2015) on 39 samples (including 16 with paired-end sequencing design). This consensus assembly has the advantage of integrating expression measurements over a wider range of samples than was reported previously while integrating sequence polymorphisms by the use of sequences that were clustered from individual assemblies (Dugé de Bernonville et al., 2015b). PCC were calculated on log2-transformed TPM to switch the expected Poisson distribution of RNA sequencing expression data to a Gaussian shaped distribution (Supplemental Fig. S10). Different lists of coexpressed genes for each CPR/DFR were then established at different PCC values (Fig. 5A), and we subsequently examined their similarities (Fig. 5B). Interestingly, the number of correlated genes differed strongly between the two CPRs and DFR. For instance, at PCC > 0.4 (FDR-corrected P < 0.05), we counted 9,831 transcripts for CPR1, 2,525 for CPR2, and 1,881 for DFR (Fig. 5A). In fact, CPR1 had the highest number of strongly coexpressed genes (for PCC > 0.8, 336 transcripts for CPR1, one for CPR2, and seven for CPR3). In addition, intersection sizes indicated that these coexpressed gene lists did not overlap unless considering genes with lower PCC values (e.g. PCC < 0.3), thus arguing for a specialization of CPR expression. This revealed once again that periwinkle CPRs and DFR are transcriptionally unrelated. In this analysis, coexpressed gene lists for each CPR were set with a PCC threshold of 0.4 and an FDR-corrected P of 0.05. While this PCC might keep weak correlations, our P threshold was expected to conserve only significant positive correlations. In addition, this PCC cutoff value was used to retrieve enough information (in particular for CPR2 and DFR) in each list to perform Gene Ontology (GO) term and UniProt keyword enrichment as well as promoter motif analyses. Indeed, the transcriptional unrelatedness was further confirmed by an analysis of overexpressed GO terms represented in coexpressed genes with a chosen low PCC value and of keywords from UniProt (Supplemental Fig. S11, A and B).
Figure 5.
Analysis of CPR1, CPR2, or DFR gene coexpression correlation. Lists of coexpressed genes were obtained after calculating the PCC of C. roseus CPR expression levels with each other transcript found in the CDF97 assembly and setting different PCC threshold values. For each threshold, only associations with a false discovery rate (FDR)-corrected P < 0.05 were kept. A, Sizes of coexpressed gene lists for each CPR at different PCC thresholds. B, Sizes of intersections between coexpressed gene lists. C, Comparison of PCCs of transcripts related to alkaloid metabolism (according to the UniProt annotation) with CPR1 and CPR2. The corresponding protein name is followed by the name of the initial plant (CATRO, C. roseus; HYONI, Hyoscyamus niger; PAPSO, Papaver somniferum; RAUMA, Rauwolfia mannii; and RAUSE, Rauwolfia serpentina). For full gene information, see Supplemental Table S3B.
While more genes coexpressed with CPR1, we noted that a higher proportion of P450s were associated with CPR2 (54 out of 2,525 coexpressed genes) compared with CPR1 (49 out of 9,831) or with DFR (six out of 1,881; Supplemental Table S3, A and B), based on a total of around 400 predicted P450 coding sequences (that may exhibit redundancy). A basic analysis of the predicted P450 functions revealed that more than 20% of the CPR1-associated P450s (10 within 49 identified P450s but 10 of 35 P450s displaying predicted functions) acted in primary metabolism such as hormone synthesis, whereas the remaining P450s were potentially involved in a specialized metabolism that appears to correspond to phenylpropanoid biosynthesis. By contrast, only three occurrences potentially associated with primary metabolism were found in the CPR2 list, whereas all the others (40 within 54 identified P450s but 40 of 43 with predicted functions) were associated with specialized metabolism, including MIA biosynthesis. Furthermore, the fact that only six P450s were associated with DFR strengthens the proposition that this protein is not involved in the reduction of P450s, in agreement with biochemical activity assays.
Such discrepancies also were confirmed by the analysis of GO terms of the whole gene list of each CPR/DFR. Indeed, GO terms found in the coexpressed gene list of CPR1 correspond to large cellular functions (nucleotide binding and protein binding), supporting its involvement in a basal/primary metabolism (Supplemental Fig. S11; Supplemental Table S3, A and B). By contrast, many genes coexpressed with CPR2 were associated with oxidoreduction and metabolic processes, reflecting its transcriptional association with metabolic processes requiring many P450s, such as specialized metabolisms. In fact, more than 25 genes from the MIA biosynthetic pathway (including P450s) were identified in the coexpressed gene list of CPR2, whereas only a single MIA gene was found in CPR1 and DFR lists (Fig. 5C; Supplemental Table S3, A and B). Accordingly, a motif enrichment analysis in the promoter regions (500 bp upstream the ATG codon, when available) of genes coexpressed with each CPR revealed associations with specific metabolisms (Supplemental Table S3C). Upstream regions of genes coexpressed with CPR1 were enriched with the CACRTR and ATRYAC motifs and were found to be associated with primary metabolism and hormone response. The latter motif also was present in the promoter of CPR1, although only 300 bp was available in the 5′ sequence. Additionally, promoter regions of genes coexpressed with CPR2 were enriched significantly with the CACGHG motif. CPR2 also contained this motif, which was shown previously in the promoter of strictosidine synthase and is able to bind MYC transcription factors, probably involved during jasmonic acid signaling (Chatel et al., 2003). Altogether, these results emphasize the occurrence of specific metabolic associations for each CPR (e.g. CPR1 with primary and basal specialized metabolisms and CPR2 with specialized metabolisms and, notably, the biosynthesis of MIA). Due to the absence of P450 gene expression association and reduction capacity, DFR was not investigated further in this study.
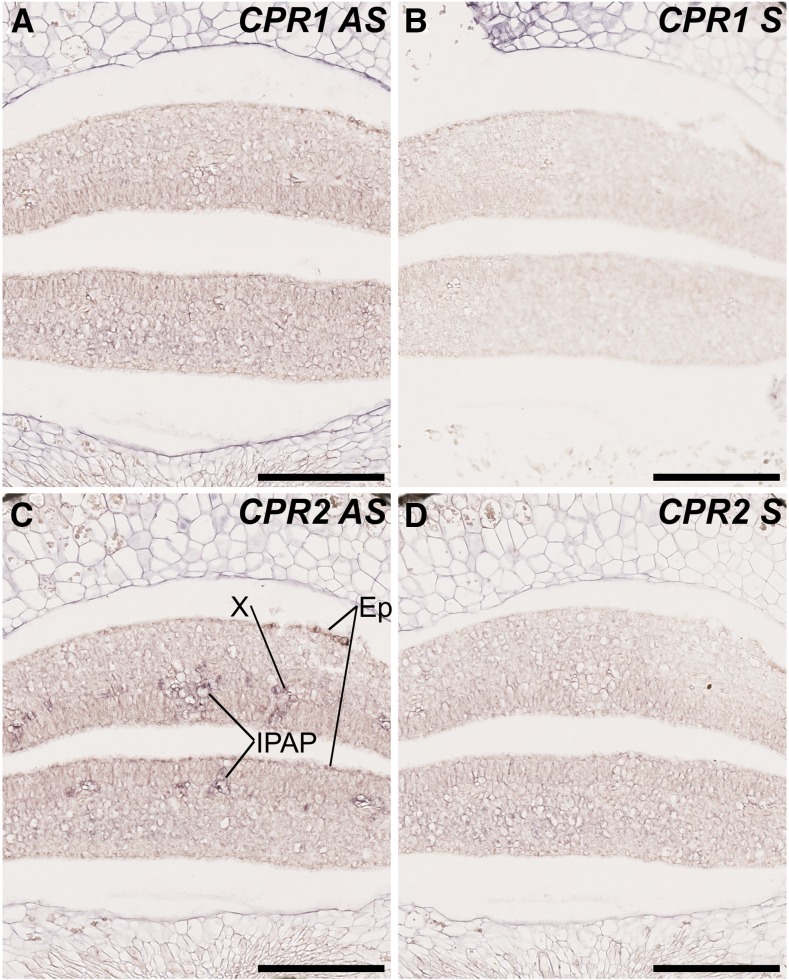
CPR2 Is Expressed in Leaf and Cotyledon Tissues Hosting MIA Biosynthetic Steps Catalyzed by P450s
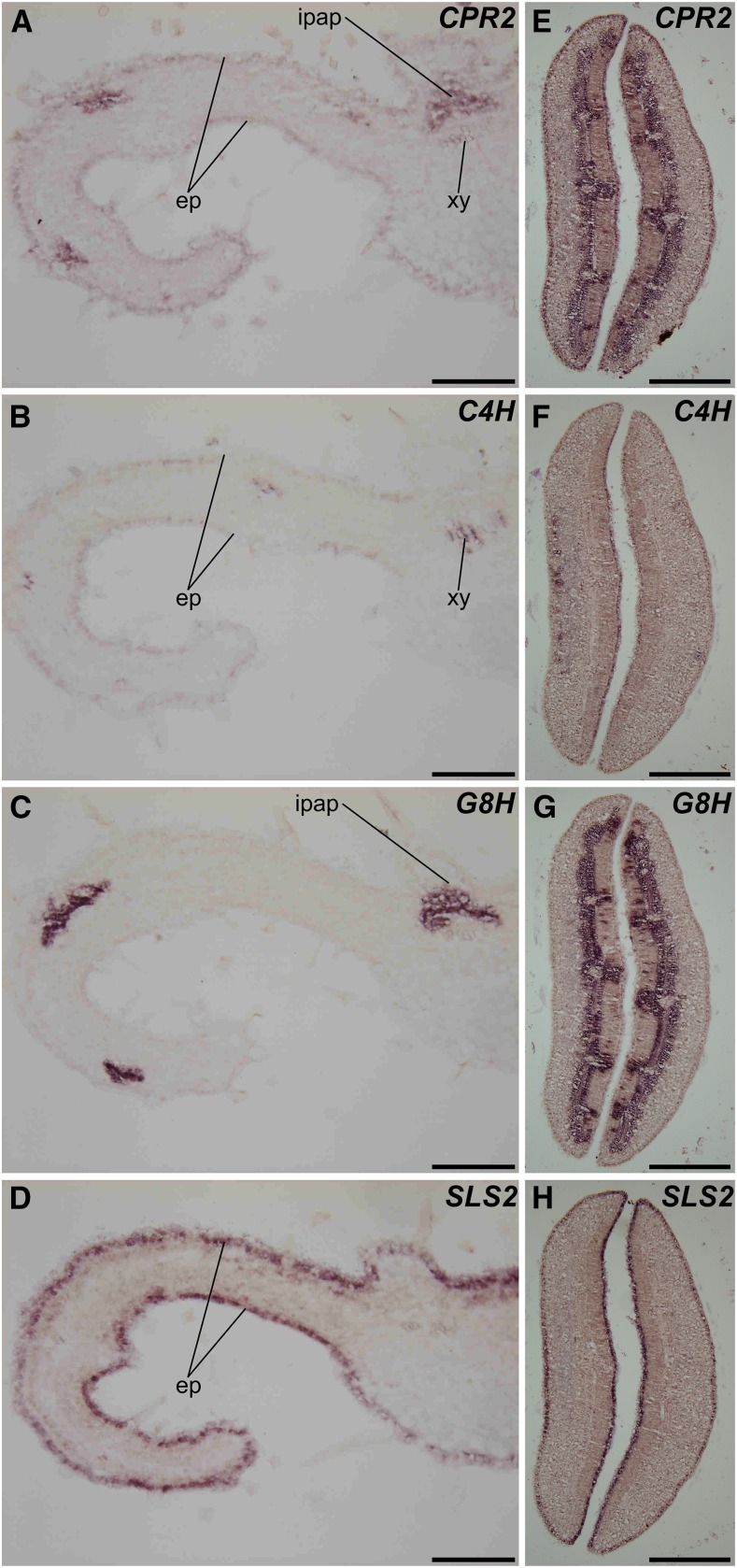
Although our global analysis of gene coexpression provides valuable information regarding CPRs/metabolic pathway associations, these analyses cannot actually reflect the intricate gene coexpression networks occurring at the tissue and/or cellular levels and are required to ensure efficient P450 reduction. Therefore, since the MIA and phenylpropanoid biosynthetic pathways display complex spatial organizations in C. roseus (St-Pierre et al., 1999; Burlat et al., 2004; Mahroug et al., 2006; for review, see Courdavault et al., 2014), we next studied the cellular distribution of CPR1 and CPR2 transcripts in cotyledons of germinating seedlings. Given the high expressions of CPR1/CPR2 in this tissue, RNA in situ hybridization was used to monitor cellular distribution. No specific zones of high accumulation of CPR1 mRNAs were detected, but a diffuse barely detectable signal was detected in the whole cotyledon tissues (mostly visible in the spongy parenchyma) with the antisense probe (Fig. 6A), while no apparent signal was revealed with the sense probe (Fig. 6B). By contrast, intense signals were observed with the CPR2 antisense probe in distinct cotyledon cell types, including IPAP, xylem, and epidermis (Fig. 6C), which were not revealed with the sense probe (Fig. 6D). This complex organization of CPR2 transcript distribution was further compared with the expression of representative P450s from both MIA and phenylpropanoid pathways that also exhibited compartmented expression, such as C4H, G8H, and SLS2. In young leaves, CPR2 mRNAs were detected in IPAP, epidermis, and xylem once again (Fig. 7A). Remarkably, this profile corresponds to a superimposition of the expression patterns of each P450 tested, including C4H, whose transcripts were detected in epidermis and xylem (Fig. 7B), G8H expressed in IPAP, and SLS2 displaying mRNA accumulation in epidermis (Fig. 7D). Such observations also were confirmed in cotyledons of germinating seedlings (Fig. 7, E–H) and support a preferential involvement of CPR2 in the reduction of P450s expressed in tissues hosting high levels of MIA and phenylpropanoid biosynthesis. In contrast, the low ubiquitous expression of CPR1 argues again for a preferential involvement in basal specialized and primary metabolism. These results also suggest that the 79 predicted P450s coexpressed with CPR2 may be involved in metabolic pathways located to one of these three tissues (i.e. IPAP, xylem, and/or epidermis), whereas no such conclusion may be drawn for the P450s coexpressed with CPR1.
Figure 6.
Localization of CPR1 and CPR2 transcripts in cotyledons of C. roseus germinating seedlings. The analysis of CPR1 and CPR2 transcript distribution was performed by in situ RNA hybridization. Serial sections of germinating seedlings were hybridized with CPR1 antisense (AS) probes (A), CPR2 antisense probes (C), CPR1 sense (S) probes (B), or CPR2 sense probes (D) used as negative controls. Ep, Epidermis; X, xylem. Bars = 100 µm.
Figure 7.
CPR2 is expressed in leaf and cotyledon tissues hosting transcripts of P450s involved in MIA and phenylpropanoid metabolism. CPR2, C4H, G8H, and SLS2 transcript localizations were carried out by RNA in situ hybridization performed on young leaves (A–D) and cotyledons (E–H). Serial sections were hybridized with CPR2 antisense probes (A and E), C4H antisense probes (B and F), G8H antisense probes (C and G), or SLS1 antisense probes (D and H). ep, Epidermis; xy, xylem. Bars = 100 µm.
Silencing of CPR1 Does Not Impact the Biosynthesis of MIA
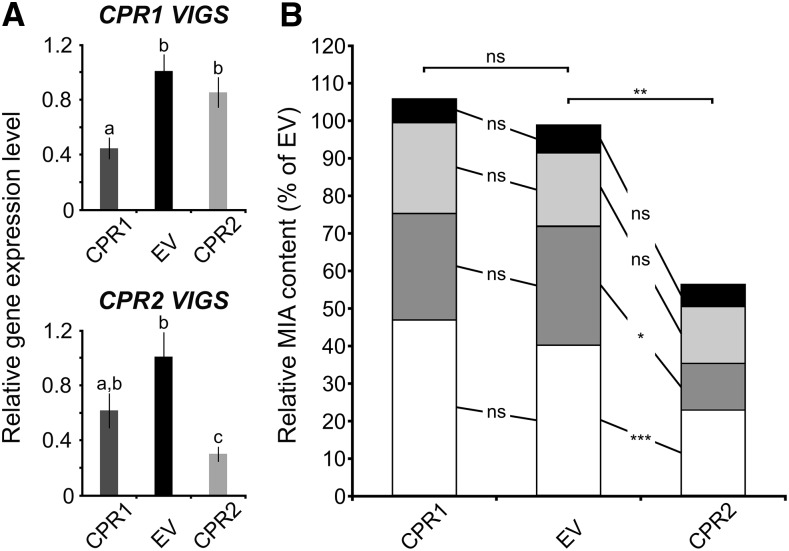
To determine the role of CPR1 and CPR2 in the reduction of P450s associated with specialized metabolism in planta, we used virus-induced gene silencing (VIGS) to target each protein. Given the relatively high sequence similarity between CPR1 and CPR2 and the presence of highly conserved domains, the identification of silencing sequences that did not result in cross silencing remained tricky. For instance, these silencing sequences were first designed in the 3′ untranslated region of each CPR but did not lead to substantial transcript down-regulation (data not shown). Therefore, we subsequently used partial coding sequences of CPR1 or CPR2, but it was difficult to avoid some degree of cross silencing, although the most divergent sequence regions were selected. qPCR of mRNA isolated from silenced plants indicated that CPR1 could be silenced to an expression level of approximately 40% of the empty vector control levels with minimal cross silencing of CPR2 (Fig. 8A). However, when CPR2 was silenced (an expression level down to 30% of empty vector control levels), we also observed a decrease in the levels of CPR1 transcripts (an expression level down to 60% of empty vector control levels; Fig. 8A). Nevertheless, we used these plants to monitor the levels of four monoterpene indole alkaloids that are abundant in leaf, namely catharanthine, vindoline, vindorosine, and serpentine (Fig. 8B). First, we noted that CPR1 silencing did not result in any quantitative modifications of the MIA content of the silenced plants, strongly suggesting that CPR1 is not required for MIA biosynthesis and/or that CPR2 can fully compensate the lack of CPR1 to reduce P450s associated with the production of MIA. By contrast, in CPR2 silenced plants, a 45% decrease of the total analyzed MIA content was observed, which mainly resulted from reduction in the levels of catharanthine and vindorosine. This result also was reinforced by the untargeted metabolite analysis, showing that silencing CPR2 has a much stronger effect on metabolites. In CPR2 silenced tissue, 37 peaks were altered significantly (13 up-regulated and 24 down-regulated; P value threshold of 0.005), while in CPR1, only four were altered (one up-regulated and three down-regulated; Supplemental Table S4). Although the precise identity of these compounds cannot be established from exact mass data, it is possible that some up-regulated metabolites in CPR2 silenced plants correspond to MIA biosynthetic intermediates. Finally, although we cannot exclude that this MIA decrease is a consequence of both CPR1 and CPR2 transcript down-regulation, the more pronounced CPR2 silencing suggests that CPR2 impacts MIA biosynthesis to a greater extent than CPR1.
Figure 8.
Silencing of CPR1 does not impact MIA biosynthesis. A, Down-regulation of CPR1 and CPR2 transcript by VIGS. The relative expression of each gene was determined by real-time quantitative reverse transcription-PCR analyses performed on total RNA extracted from C. roseus leaves of CPR1 or CPR2 silenced plants (dark gray or light gray bars, respectively) or plants transformed with an empty virus control (EV; black bars). CrRPS9 (for RIBOSOMAL PROTEIN S9) was used as a reference gene. Data were normalized to CrRPS9 expression and correspond to average values (n = 6) ± sd of independently transformed plants. Letters indicate statistical classes (Wilcoxon rank-sum test, FDR-adjusted P < 0.05). B, Relative MIA contents of CPR1- and CPR2-VIGS plants expressed relative to that of EV plants. The relative MIA content was determined by measuring peak areas (normalized by sample fresh weight) of catharanthine (white bars), vindorosine (dark gray bars), vindoline (light gray bars), and serpentine (black bars) performed by liquid chromatography-mass spectrometry. The amount of each MIA in silenced plants (eight plants per silencing construct) was expressed relative to that measured in EV plants (eight plants; normalized to 100%). Asterisks denote statistical significance (*, P < 0.005; **, P < 0.0005; and ***, P < 0.00005). ns, Not significant.
DISCUSSION
While the specific functions of class I and class II CPRs in plants have not been clearly deciphered to date, the Madagascar periwinkle and its complex MIA biosynthesis constitute an attractive model in which to study the relationship between CPRs and specialized metabolism. In contrast to fungi and animals that contain a single CPR gene, most vascular plants possess two functional CPRs, originating from an ancestral gene duplication, along with a more distant CPR without a transmembrane domain (DFR) that does not reduce the P450s tested in this study. Based on their distinct N-terminal sequences, the two functional periwinkle CPRs are assigned to class I and class II CPRs. Their responses to jasmonate suggest that class I and class II play active roles in basal primary metabolism/constitutive specialized metabolism and inducible specialized metabolism, respectively (Jensen and Moller, 2010). In support of this hypothesis, functional biochemical assays and tests of interaction between CPR1 or CPR2 and the P450s G8H, SLS2, T16H1, T16H2, and C4H clearly demonstrated that both CPRs interact with and reduce each P450 with an apparent similar efficiency, suggesting that both types of CPR do not display any structural specificity toward individual P450s (Fig. 3; Table I). However, analysis of gene coexpression patterns with CPR1 and CPR2 sheds light on how CPR2 plays a dedicated role in specialized metabolism, in this case MIA biosynthesis. First, CPR1 expression is correlated with a higher number of genes than CPR2 (9,831 versus 2,525 transcripts), suggesting a broader role of CPR1 in general plant physiology (Fig. 5B). Moreover, the genes coexpressed with CPR1 and CPR2 differed strongly, even when considering lower correlations (PCC < 0.3), further supporting the distinct specialization of CPR1 and CPR2 (Fig. 5A). Such a hypothesis is reinforced by the fact that the UniProt keywords associated with genes correlated to CPR1 primarily include basal cellular functions (Supplemental Fig. S10). For example, among the P450s with expression profiles related to CPR1, almost 20% act in primary metabolic pathways such as hormone biosynthesis, while the remaining P450s are mostly associated with phenylpropanoid metabolism (Supplemental Table S3, A and B). In contrast, almost all of the P450s with expression profiles correlating with CPR2 are known to be associated with, or have UnitProt keywords associated with, specialized metabolism, especially the MIA metabolism that predominates in C. roseus (Supplemental Fig. S10; Supplemental Table S3, A and B). The strong correlation between the expression profiles of CPR2 and MIA biosynthetic genes is reinforced by the identification of nearly all known MIA biosynthetic genes in the gene list associated with CPR2 (Fig. 5C; Supplemental Table S3B). Interestingly, although numerous P450s associated with other specialized metabolic pathways such as sesquiterpene metabolism were correlated to CPR2, only a few involved in phenylpropanoid metabolism were identified. The functional specialization of CPRs in other plant species can be less stringent. For example, this is slightly different from the situation described in Arabidopsis, in which previous transcriptomic data analyses showed that both ATR1 and ATR2 were coexpressed with lignin biosynthetic genes during inflorescence stem development, even if, for ATR2, the data set was generated from Arabidopsis leaves subjected to cold treatment under high-light conditions or from leaves of lignin biosynthetic gene mutants (Soitamo et al., 2008; Sundin et al., 2014). Taken altogether, specific gene coexpression with each class of CPR adds another layer of complexity to CPR specialization and the control of specialized metabolism. In periwinkle, the proposed partition of class I and class II CPRs between basal and inducible specialized metabolism cannot be ruled out for phenylpropanoid biosynthesis, but the exclusive association of class II CPRs with subclasses of specialized metabolism such as MIA biosynthesis seems to have been acquired.
The localization pattern of these CPRs also supports a specialized role for CPR2 expression in MIA biosynthesis. Although qPCR demonstrated that CPR1 and CPR2 were both expressed in the same organs, RNA in situ hybridization established cell-specific colocalization of CPR2 transcripts with the tested P450s involved in specialized metabolism, while CPR1 did not show such a correlation (Figs. 4, 6, and 7). The biosynthesis of MIA in C. roseus leaves is a highly compartmentalized process involving at least four distinct cell types: IPAP, hosting all the early steps of the monoterpene precursor synthesis up to loganic acid; epidermis that carries out the incorporation of loganic acid into MIA including desacetoxyvindoline; and the specialized cells laticifers and idioblasts, where the last two steps of vindoline formation from desacetoxyvindoline take place. The expression of P450s known to be associated with MIA biosynthesis has been shown to be restricted to IPAP (G8H, IO, and 7DLH) or to epidermis (SLS1–SLS4, T16H1, T16H2, and 16T3O) cell types. In our localization studies, while the CPR1 transcripts were barely detectable in cotyledons and leaves with an apparent general distribution, we observed that CPR2 mRNAs were highly enriched, together with G8H, in IPAP; in epidermis, similar to SLS2 and T16H2; and to a lower extent in xylem, where C4H transcripts from phenylpropanoid biosynthesis also were detected. Thus, the CPR2 expression profile appears to be highly compartmentalized, being superimposed with selected MIA and phenylpropanoid biosynthetic genes. This high CPR2 expression in IPAP suggests that class II CPR is required for G8H activity in this tissue, and by extension to IO and 7DLH, while class I CPR seems to play a housekeeping role, given its uniform distribution and low transcript level. The same rationale could be applied to leaf epidermis, in which CPR2, SLS2, and T16H2 are highly transcribed, reinforcing the specialization of CPR2 in MIA biosynthesis. Data concerning the tissue-specific expression of CPRs in plants are scarce. Therefore, the demonstration of the specific cooccurrence of CPR and P450 transcripts in similar restricted tissues provides a plausible explanation of how CPR specialization is established in planta, based on similar transcriptional regulatory processes. This also may shed light on a predictive tissue-specific expression for the list of P450s coexpressed with CPR2 and on the colocalization of various specialized metabolic pathways in given specialized cell types, as exemplified for epidermis, which harbors MIA, indole, monoterpene-secoiridoid, and phenylpropanoid pathways (Mahroug et al., 2006).
CPR specialization also was supported by VIGS assays conduced in C. roseus. While the characterization of CPR mutants was already reported in Arabidopsis (Sundin et al., 2014), gene-silencing approaches have only been applied to CPRs in animal systems (Mackenzie et al., 2010; Tang et al., 2012; Liu et al., 2015). Here, we established that silencing CPR1 in C. roseus has no substantial effect on MIA biosynthesis (Fig. 8). This result again suggests that CPR1 does not play a significant role in the reduction of P450s associated with MIA production. Although we cannot exclude a possible compensation by CPR2 in CPR1 silenced plants, this seems unlikely, given their highly specific gene expression profiles (Figs. 6 and 7); moreover, such a complete compensation between ATR1 and ATR2 has not been reported in Arabidopsis systems (Sundin et al., 2014). Due to the high sequence identity, we were not able to specifically down-regulate CPR2; while CPR2 silenced plants had a huge decrease of CPR2 transcripts, this was accompanied by a less marked but substantial decrease (40%) of CPR1 (Fig. 8A). Such transcript down-regulation engendered a significant decrease of the total leaf MIA content (around 45%) in which the most abundant leaf MIAs were most affected (Fig. 8B). Given the absence of such a decrease in CPR1 VIGS lines, it seems likely that this decrease in MIAs is largely the result of CPR2 down-regulation. Interestingly, the lignin content measured in Arabidopsis CPR2 null is altered only slightly (Sundin et al., 2014), suggesting that CPR1 can compensate by providing electrons for the basal lignin metabolism level in Arabidopsis. In contrast, there does not appear to be a compensatory effect for MIA metabolism in C. roseus. Notably, in this plant, the biosynthesis of MIA is a constitutive process that also is stimulated by external stimuli or defense-stimulated phytohormones (El-Sayed and Verpoorte, 2007). However, although CPR1 has been proposed to control basal specialized metabolism, it does not have a major impact on MIA biosynthesis in C. roseus. Thus, this distinction between the roles of class I and class II CPRs with respect to basal/inducible specialized metabolism may not always hold. Overall, our results suggest that, in C. roseus, CPR2 has a distinct function in the specialized metabolism at both the basal and developmentally regulated levels as well as in the response to environmental signals. By contrast, gene expression correlations reinforce the potential involvement of class I CPR with a different set of P450s involved in other metabolic pathways.
CONCLUSION
In summary, our study gives compelling evidence that the acquisition of a second isoform of CPR (class II CPR) in the plant lineage provides an additional level of control of redox power homeostasis. With respect to the hazard of CPR overexpression (causing reactive oxygen species production; Bassard et al., 2012; Paddon et al., 2013), strict adjustment of cytochrome reducing power to metabolic demand may take advantage of the transcriptional regulation of two independent genes, each with their specific regulatory control. The current prevailing hypothesis is that class I CPR provides reducing power required for low-level, basal, primary, and secondary metabolism, while class II CPR provides electrons for highly expressed P450s involved in tissue-specific and intensive/induced specialized metabolism. However, as more work is conducted in this area, this hypothesis will continue to be refined. For example, it was demonstrated recently that, in the Apiales lineage, class I CPR genes have been lost and compensated by the acquisition of additional isoforms of class II CPR (Andersen et al., 2016). In this study, we capitalize on the well-studied, complex MIA pathway of C. roseus to address the role of CPRs in specialized metabolism. With the indubitable requirement of class II CPR in the biosynthesis of MIA, our work provides new insight into the so-far poorly understood specialization of the CPR classes, suggesting another possible strategy of evolution for redox control.
MATERIALS AND METHODS
Chemicals
Secologanin and tabersonine were purchased from Phytoconsult and ChromaDex, respectively. Cinnamic acid, geraniol, and 10-hydroxygeraniol were purchased from Sigma-Aldrich.
Plant and Cell Culture Growth
Fully expanded Catharanthus roseus plants (‘Pacifica Pink’ and ‘Sunstorm Apricot’) were used for microscopy fixation (RNA in situ hybridization experiments) and RNA extraction (cloning experiments), respectively. VIGS assays were performed on cv Sunstorm Apricot. The C. roseus C20A cell suspension culture used for subcellular localization studies was propagated in Gamborg’s B5 medium (Duchefa) at 24°C under continuous shaking (100 rpm) for 7 d as described previously (Guirimand et al., 2009).
Transcriptomic Resources
Identification of CPR homologous sequences was achieved by interrogating C. roseus transcriptomic databases, including the consensus C. roseus transcriptome CDF97 (Dugé de Bernonville et al., 2015b; http://bbv-ea2106.sciences.univ-tours.fr/images/files_to_download/BAFC94.fasta), the Medicinal Plant Genomic Resource database (Góngora-Castillo et al., 2012; http://medicinalplantgenomics.msu.edu), and the Phytometasyn database (Xiao et al., 2013; http://www.phytometasyn.ca).
RNA Extraction and Reverse Transcription
Total RNA was extracted from young leaves using the NucleoSpin RNA Plant Kit (Macherey-Nagel). First-strand cDNA was synthesized from 0.5 µg of total RNA using oligo(dT)18 primers (0.5 µm) and 15 units of Thermoscript reverse transcriptase (Invitrogen). Following reverse transcription, complementary RNA was removed by treatment with Escherichia coli RNase H (Invitrogen) for 20 min at 37°C.
Subcellular Localization, Fusion/Deletion Experiments, and Protein Interaction Studies
The subcellular localizations of CPR1 (KJ701028), CPR2 (X69791), and CPR3/DFR (KM111538) were determined by expressing YFP fusion proteins. The full-length open reading frame of each CPR was amplified using specific pairs of primers (Supplemental Table S5) and cloned into the SpeI restriction site of the pSCA-cassette YFPi plasmid in frame with the 5′ extremity of the YFP coding sequence, to generate the CPR1-YFP, CPR2-YFP, and CPR3-YFP fusion proteins (Guirimand et al., 2009). The functionalities of the membrane-anchoring domains of CPR1 and CPR2 were studied by fusion/deletion experiments as follows. The coding sequences of the first 53 and 74 residues of CPR1 and CPR2 were amplified using primers helixCPR1-for/helixCPR1-rev and helixCPR2-for/helixCPR2-rev (Supplemental Table S5) and cloned into the SpeI restriction site of the pSCA-cassette YFPi plasmid to express the hxCPR1-YFP and hxCPR2-YFP fusion proteins, respectively. To express the CPR1 and CPR2 proteins deprived of their membrane-anchoring domain, the coding sequence of the remaining part of each protein (residues 54–691 for CPR1 and 75–714 for CPR2) was amplified using primers delCPR1/CPRnewrev and delCPR2/CPRoldrev (Supplemental Table S5) and cloned into the SpeI restriction site of pSCA-cassette YFPi. The addition of the CPR2 membrane-anchoring domain to CPR3 was achieved through amplification of the coding sequence of the first 74 residues of CPR2 using primers helixCPR-for and helixCPR-rev, harboring XbaI and SpeI restriction sites at their extremity, respectively. The resulting PCR product was cloned into the SpeI site of pSCA-YFP to generate pSCA-helixYFP. This plasmid was linearized subsequently by SpeI to allow the introduction of the CPR3 coding sequence, yielding the pSCA-hxCPR3-YFP plasmid. For protein interaction analyses, the same CPR amplification products were cloned into the SpeI restriction site of the pSCA-SPYNE 173 plasmid in frame with the 5′ end of the sequence encoding YFPN to express the CPR1-YFPN, CPR2-YFPN, and CPR3-YFPN proteins (Guirimand et al., 2010). The coding sequences of the five tested P450s (G10H, SLS2, T16H1, T16H2, and C4H) were amplified using appropriate primers (Supplemental Table S5) and cloned into the SpeI and/or BglII restriction sites of the pSCA-SPYCE(M) plasmid in frame with the 5′ end of the sequence encoding YFPC to express G10H-YFPC, SLS2-YFPC, T16H1-YFPC, T16H2-YFPC, and C4H-YFPC. These recombinant plasmids were used for the transient transformation of C. roseus cells by particle bombardment and YFP imaging according to Guirimand et al. (2009, 2010) and Foureau et al. (2016). Briefly, plated C. roseus cells were bombarded with DNA-coated gold particles (1 µm) and a 1,100-p.s.i. rupture disc at a stopping-screen-to-target distance of 6 cm, using the Bio-Rad PDS1000/He system. Cells were cultivated for 16 to 38 h before being harvested and observed. The subcellular localization was determined using an Olympus BX-51 epifluorescence microscope equipped with an Olympus DP-71 digital camera and a combination of YFP and CFP filters. The pattern of localization presented in this work is representative of approximately 50 observed cells. The ER or nucleocytosolic localization of the different fusion proteins was confirmed by cotransformation experiments using an ER-CFP marker (CD3-954; Nelson et al., 2007) and a nucleocytosolic CFP marker (Guirimand et al., 2011a). Such plasmid cotransformations were performed using 400 ng of each plasmid or 100 ng for BiFC assays.
Tissue Fixation, Embedding in Paraffin, and Sectioning
RNase-free conditions were strictly observed for all steps. All glassware was baked for 8 h at 180°C, and nondisposable plastic wares were incubated for 10 min in an aqueous 3% hydrogen peroxide solution before washing in diethyl pyrocarbonate-treated water. Leaves from mature C. roseus plants grown in a greenhouse and young germinating seedlings were rapidly fixed in formalin (10%), acetic acid (5%), and ethanol (50%) and embedded in Paraplast (Dominique Dutscher) as described previously (Mahroug et al., 2006; Guirimand et al., 2011b). Serial sections (10 µm) were spread on aminopropyltriethoxysilane-coated slides overnight at 40°C, and paraffin was removed using xylene (twice for 15 min) before rehydration in an ethanol gradient series up to diethyl pyrocarbonate-treated water.
In Situ RNA Hybridization of C. roseus Leaves and Cotyledons
Full-length CPR1 and CPR2 cDNAs cloned into pGEMT-easy vector (Promega) were used for the synthesis of sense and antisense RNA probes as described previously (Mahroug et al., 2006). For G8H, SLS, and C4H, previously described plasmids were used for the riboprobe in vitro transcription (Irmler et al., 2000; Burlat et al., 2004; Mahroug et al., 2006). Paraffin-embedded serial longitudinal sections of young leaves and cross sections of emerging cotyledons were hybridized with digoxigenin-labeled transcripts and localized with anti-digoxigenin-alkaline phosphatase conjugates according to Mahroug et al. (2006).
Heterologous Expression of C. roseus CPRs and P450s in Saccharomyces cerevisiae
Full-length CPR1, CPR2, and CPR3 cDNAs were amplified using the specific yeast expression primers described in Supplemental Table S5. Each pair of primers includes appropriate restriction sites at their extremities to allow cloning of the resulting PCR products into the SpeI site of pESC-Leu yeast expression plasmid harboring the LEU2 nutritional marker (Agilent Technologies). Addition of the CPR2 membrane-anchoring domain to CPR3 was achieved as described in the previous section, except that cloning was performed into the pESC-Leu plasmid. G8H, SLS2, T16H1, T16H2, and C4H coding sequences were amplified using primers including BglII restriction at their extremities to allow cloning into the BamHI site of the pYeDP60 plasmid harboring URA3 and ADE2 nutritional markers (Pompon et al., 1996). pESC-Leu CPRs, pYeDP60 P450 recombinant vectors, and/or empty plasmids were associated by pair and used to transform either the Saccharomyces cerevisiae strain WT303 (containing the endogenous yeast CPR) or the WAT11-expressing ATR1 (Pompon et al., 1996). Leu+, Ura+/Ade+, or Ura+/Ade+/Leu+ transformants were selected onto solid complete synthetic medium plates containing 0.67% yeast nitrogen base, 2% agar, 2% dextrose, and 0.05% DOB-Leu, DOB-Ura-Ade, or DOB-Ura-Ade-Leu as required. Yeast transformants were grown in 10 mL of appropriate liquid complete synthetic medium until reaching the stationary phase of culture and then harvested by centrifugation. Protein expression was induced by cultivating the harvested yeast in 50 mL of liquid YPGal medium (1% bactopeptone, 1% yeast extract, and 2% Gal) for 6 h as described by Besseau et al. (2013).
Enzyme Assays
Following the induction of protein expression, 50 mL of yeast culture was harvested by centrifugation and resuspended in 2 mL of buffer R (50 mm Tris-HCl, pH 7.5, and 1 mm EDTA) in a 50-mL centrifugation tube. An equal volume of glass beads was added (425–600 μm; Sigma-Aldrich), and cells were broken by vigorous shaking. For this purpose, tubes were shaken by hand during 30 s in a cold room (4°C) before being put on ice for an additional 30 s. This operation was repeated 10 times before the addition of 2 volumes of buffer R, allowing the recovery of the yeast crude extracts prior to protein quantification using the Bio-Rad protein microassay. P450 activities were analyzed in a final volume of 100 μL containing 300 μg of protein, 100 μm NADPH(H+), and 20 μm of loganin for SLS2, tabersonine for T16H1 and T16H2, or cinnamate for C4H. Reactions were initiated by the addition of NADPH(H+), incubated at 30°C for 0, 5, 15, and 30 min (T16H1, T16H2, and C4H) or for 10, 30, 60, or 120 min (SLS2), and quenched by the addition of 100 μL of methanol prior to ultra-performance liquid chromatography-mass spectrometry analysis. G10H activity was tested using microsomal membranes purified according to Heitz et al. (2012). Enzymatic assays were conducted according to Höfer et al. (2013) in a final volume of 100 µL of 20 mm citrate-phosphate buffer, pH 7.4, containing 400 µm NADPH, 200 µm geraniol, and normalized amounts of microsomes. After 60 min of reaction, products were extracted with 500 µL of ethyl acetate. The ethyl acetate phase was collected with a glass pipette and dried under a nitrogen gas flow, and products were analyzed by gas chromatography-flame ionization detector. For all P450 tests performed in the WT303 yeast strain including CPR1, CPR2, or CPR3, activities resulting from the yeast endogenous CPR were subtracted from total activity to estimate the activity resulting from each periwinkle CPR. All results were expressed as substrate conversion rate for 1 h.
Ultra-Performance Liquid Chromatography-Mass Spectrometry Analyses
All samples were centrifuged, and the supernatants were stored at 4°C prior to injection. The ultra-performance liquid chromatography system consisted of an ACQUITY UPLC device (Waters). Separation was performed using a Waters Acquity HSS T3 C18 column (150 mm × 2.1 mm, 1.8 μm) with a flow rate of 0.4 mL min−1 at 55°C. The injection volume was 5 μL. The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Chromatographic separation was achieved using an 8-min linear gradient from 10% to 24% solvent B. Mass spectrometry detection was performed using an SQD mass spectrometer equipped with an electrospray ionization source controlled by Masslynx 4.1 software (Waters). The capillary and sample cone voltages were 3,000 and 30 V, respectively. The cone and desolvation gas flow rates were 60 and 800 L h−1. Data collection was carried out in negative mode for secologanin ([M + HCOOH-H]− = 433, retention time = 7.12 min), cinnamate ([M − H]− = 147), and 4-hydroxycinnamate ([M − H]− = 163) and in positive mode for loganin ([M + Na]+ = 413, retention time = 5.61 min), tabersonine ([M + H]+ = 337, retention time = 4.72 min), and 16-hydroxytabersonine ([M + H]+ = 353, retention time = 3.87 min).
Gas Chromatography Analyses
Monoterpenes were analyzed by gas chromatography-flame ionization detection (Alpha-MOS). Samples were injected in the split mode (50 mL min−1), and compounds were separated on a BPX5 capillary column. The injector was heated at 250°C, and the oven was set at 45°C for 60 s. The temperature was next increased to 250°C at a rate of 8°C min−1, followed by an increase to 320°C at a rate of 30°C min−1. The oven was maintained at 320°C for 1 min before the end of analysis. The flame ionization detector was set at 280°C. Peaks were identified by comparing the retention times of authentic standards.
Gene Expression Correlation Analyses
The abundance of transcripts in the CDF97 assembly was estimated by pseudoaligning reads with Salmon (Patro et al., 2015) in the variational bayesian optimized (–vbo) quasi-mapping mode with bias correction (–biasCorrect). Expression was log2 transformed to distribute TPM normally (Supplemental Fig. S10). A total of 16 paired-end samples (SRR1144633, SRR1144634, SRR1271857, SRR1271858, SRR1271859, SRR342017, SRR342019, SRR342022, SRR342023, SRR646572, SRR646596, SRR646604, SRR648705, SRR648707, SRR924147, and SRR924148) and 23 SE samples (SRR122239, SRR122240, SRR122241, SRR122242, SRR122243, SRR122244, SRR122245, SRR122246, SRR122247, SRR122248, SRR122249, SRR122250, SRR122251, SRR122252, SRR122253, SRR122254, SRR122255, SRR122256, SRR122257, SRR122258, SRR122259, SRR122260, and SRR122261) were used as described by Dugé de Bernonville et al. (2015b). PCCs and their significance were calculated with R (R Development Core Team, 2015) for each CPR with each transcript predicted in the CDF97 consensus assembly. Annotation of transcripts was performed by following the Trinotate pipeline (https://trinotate.github.io/). GO terms and UniProt keywords were attributed according to the UniProt database. Gene set enrichment analyses were made by comparing the observed representation of GO terms with a theoretical hypergeometric distribution (phyper function in R). A promoter analysis was performed with the MEME suite (version 4.11; Bailey et al., 2009). R scripts were designed to retrieve promoter sequences of each predicted transcript in the C. roseus genome sequence (Kellner et al., 2015b). Promoter sequences of genes putatively associated with each CPR (PCC > 0.4 and FDR-adjusted PCC P < 0.05) were analyzed with DREME, and the resulting motifs were analyzed for GO term association with GOMO. The most promising motifs were next inspected manually in promoter sequences of each CPR.
Gene Expression Analyses
CPR1, CPR2, and CPR3 expression was first determined by comparing transcript abundance in C. roseus transcriptomic data sets (Góngora-Castillo et al., 2012; Dugé de Bernonville et al., 2015b) and subsequently measured by real-time reverse transcription-qPCR using the primers listed in Supplemental Table S6. The corresponding PCR products were cloned in pGEMT-easy (Promega) and sequenced to ensure the specificity of amplification. Primer efficacy was evaluated on plasmids containing the appropriate cDNA. Distinct C. roseus organs (such as roots, stems, young and mature leaves, and petals; cv Apricot Sunstorm) were immediately frozen in liquid nitrogen after sampling. Samples (50 mg) were ground with a mortar and pestle in liquid nitrogen, and total RNA was extracted with Nucleospin RNA (Macherey-Nagel), quantified with a Nanodrop spectrophotometer (Thermo Fisher), and treated (1.5 μg) with RQ1 RNase-free DNase (Promega) before being used for first-strand cDNA synthesis by priming with 0.5 μm oligo(dT)18. Retrotranscription of 1.5 μg of total RNA was carried out using the SuperScript III reverse transcriptase kit (Invitrogen) at 50°C for 1 h according to the manufacturer’s instructions. Real-time PCR was run on the CFX96 Touch Real-Time PCR System (Bio-Rad) using SYBR Green I technology. Each reaction was performed in a total reaction volume of 25 μL containing an equal amount of cDNAs (1:3 dilution), 0.05 μm forward and reverse primers, and the 1× DyNAmo ColorFlash Probe qPCR Kit (Thermo Fisher Scientific). The amplification program was 95°C for 7 min (polymerase heat activation) followed by 40 cycles containing two steps, 95°C for 10 s and 60°C for 40 s. At the end of the amplification, a melt curve was performed to check amplification specificity. Relative quantification of transcripts was performed with calibration curves and normalization with the C. roseus 40S RPS9 (AJ749993.1) reference gene. All amplifications were performed in triplicate with at least two independent biological repeats.
VIGS
CPR1 and CPR2 silencing fragments were amplified using the primers described in Supplemental Table S5 and cloned into the pTRV2u vector described by Geu-Flores et al. (2012). The resulting plasmids and the empty vector were used to perform the VIGS assays on C. roseus seedlings as described by Liscombe and O’Connor (2011) or Carqueijeiro et al. (2015). Leaves from the first two leaf pairs to emerge following inoculation were harvested from eight plants transformed with each construct and subjected to gene expression analysis by real-time reverse transcription-PCR (primers are given in Supplemental Table S6). The alkaloid content of silenced leaves was determined by liquid chromatography-mass spectrometry as described previously (Liscombe and O’Connor, 2011; Geu-Flores et al., 2012). A detailed version of the silencing procedure is given in Supplemental Methods S1.
Accession Numbers
Sequence data of CPR1, CPR2 and CPR3/DRF from this article can be found in the GenBank/EMBL data libraries under accession numbers KJ701028, X69791, and KM111538, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of periwinkle CPR1, CPR2, and CPR3 deduced amino acid sequences.
Supplemental Figure S2. CPR phylogenic tree.
Supplemental Figure S3. Gene organization of CPR1, CPR2, and CPR3/DFR.
Supplemental Figure S4. Alignment of CPR1 and CPR2 amino acid sequences, highlighting characteristic membrane anchor, FMN, FAD, NADPH, and P450-binding domains.
Supplemental Figure S5. Alignment of periwinkle CPR1, CPR2, and CPR3/DFR deduced amino acid sequences and determination of the putative functional domains of CPR3 according to Varadarajan et al. (2010).
Supplemental Figure S6. Detection of a putative transmembrane helix at the N-terminal end of CPR1, CPR2, and CPR3/DFR.
Supplemental Figure S7. Characterization of the membrane-anchoring domain of CPR1 and CPR2.
Supplemental Figure S8. Addition of the CPR2 membrane-anchoring domain to CPR3 enables ER anchoring.
Supplemental Figure S9. Transcript distribution of CPR1, CPR2, C4H, G8H, SLS2, T16H1, and T16H2 in various C. roseus organs.
Supplemental Figure S10. Distribution of TPM.
Supplemental Figure S11. Functional classification of the nearest coexpressed genes with C. roseus CPR.
Supplemental Table S1. Identification of contigs potentially encoding CPR candidates in the Medicinal Plant Genomic Resource database, the Phytometasyn database, and the C. roseus consensus transcriptome.
Supplemental Table S2. Evaluation of the efficiency of P450 reduction by CPR1, CPR2, CPR3/DFR, hxCPR3/DFR, the yeast endogenous CPR (WT303 yeast strain), and the codon-optimized ATR1 of the WAT11 yeast strain.
Supplemental Table S3A. P450 genes coexpressed with CPR1, CPR2, or DFR.
Supplemental Table S3B. Whole list of genes coexpressed with each CPR.
Supplemental Table S3C. Promoter analysis of coexpressed genes with each CPR.
Supplemental Table S4. Untargeted metabolite analysis in silenced plants.
Supplemental Table S5. Primers used for cDNA cloning.
Supplemental Table S6. Primers used for qPCR analyses.
Supplemental Methods S1. Detailed VIGS procedure.
Supplementary Material
Acknowledgments
We thank Marie-Antoinette Marquet, Marie-Françoise Aury, Evelyne Danos, and Cédric Labarre (EA2106 Biomolécules et Biotechnologies Végétales) for help in maintaining cell cultures and plants; Emelyne Marais and Céline Melin for valuable technical assistance; and the Fédération CaSciModOT for access to the Région Centre computing grid.
Glossary
- MIA
monoterpene indole alkaloid
- IPAP
internal phloem-associated parenchyma
- ER
endoplasmic reticulum
- BiFC
bimolecular fluorescence complementation
- qPCR
quantitative PCR
- PCC
Pearson correlation coefficient
- FDR
false discovery rate
- GO
Gene Ontology
- VIGS
virus-induced gene silencing
Footnotes
This work was supported by the Région Centre of France (ABISAL Project and fellowships to C.P. and I.C.), the Ministère de l’Education Nationale, de l’Enseignement Supérieur, et de la Recherche, France (fellowship to E.F.), the European Research Council (grant no. 311363), the Biotechnology and Biological Sciences Research Council Institute Strategic Programme (grant no. BB/J004561/1 to S.E.O.), and the University of East Anglia (Ph.D. studentship to F.K.).
References
- Andersen TB, Hansen NB, Laursen T, Weitzel C, Simonsen HT (2016) Evolution of NADPH-cytochrome P450 oxidoreductases (POR) in Apiales: POR 1 is missing. Mol Phylogenet Evol 98: 21–28 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D (2011) Cytochromes p450. The Arabidopsis Book 9: e0144 doi: 10.1199/tab.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Mutterer J, Duval F, Werck-Reichhart D (2012) A novel method for monitoring the localization of cytochromes P450 and other endoplasmic reticulum membrane associated proteins: a tool for investigating the formation of metabolons. FEBS J 279: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Benveniste I, Lesot A, Hasenfratz MP, Kochs G, Durst F (1991) Multiple forms of NADPH-cytochrome P450 reductase in higher plants. Biochem Biophys Res Commun 177: 105–112 [DOI] [PubMed] [Google Scholar]
- Besseau S, Kellner F, Lanoue A, Thamm AM, Salim V, Schneider B, Geu-Flores F, Höfer R, Guirimand G, Guihur A, et al. (2013) A pair of tabersonine 16-hydroxylases initiates the synthesis of vindoline in an organ-dependent manner in Catharanthus roseus. Plant Physiol 163: 1792–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Clastre M, Courdavault V, O’Connor SE (2015) De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA 112: 3205–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlat V, Oudin A, Courtois M, Rideau M, St-Pierre B (2004) Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J 38: 131–141 [DOI] [PubMed] [Google Scholar]
- Canto-Canché BB, Loyola-Vargas VM (2001) Multiple forms of NADPH-cytochrome P450 oxidoreductases in the Madagascar periwinkle Catharanthus roseus. In Vitro Cell Dev Biol Plant 37: 622–628 [Google Scholar]
- Carqueijeiro I, Masini E, Foureau E, Sepúlveda LJ, Marais E, Lanoue A, Besseau S, Papon N, Clastre M, Dugé de Bernonville T, et al. (2015) Virus-induced gene silencing in Catharanthus roseus by biolistic inoculation of tobacco rattle virus vectors. Plant Biol (Stuttg) 17: 1242–1246 [DOI] [PubMed] [Google Scholar]
- Chatel G, Montiel G, Pré M, Memelink J, Thiersault M, Saint-Pierre B, Doireau P, Gantet P (2003) CrMYC1, a Catharanthus roseus elicitor- and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J Exp Bot 54: 2587–2588 [DOI] [PubMed] [Google Scholar]
- Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220 [DOI] [PubMed] [Google Scholar]
- Courdavault V, Papon N, Clastre M, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2014) A look inside an alkaloid multisite plant: the Catharanthus logistics. Curr Opin Plant Biol 19: 43–50 [DOI] [PubMed] [Google Scholar]
- Denisov IG, Baas BJ, Grinkova YV, Sligar SG (2007) Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem 282: 7066–7076 [DOI] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Clastre M, Besseau S, Oudin A, Burlat V, Glévarec G, Lanoue A, Papon N, Giglioli-Guivarc’h N, St-Pierre B, et al. (2015a) Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine. Phytochemistry 113: 9–23 [DOI] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Foureau E, Parage C, Lanoue A, Clastre M, Londono MA, Oudin A, Houillé B, Papon N, Besseau S, et al. (2015b) Characterization of a second secologanin synthase isoform producing both secologanin and secoxyloganin allows enhanced de novo assembly of a Catharanthus roseus transcriptome. BMC Genomics 16: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed M, Verpoorte R (2007) Catharanthus terpenoid indole alkaloids: biosynthesis and regulation. Phytochem Rev 6: 277–305 [Google Scholar]
- Foureau E, Carqueijeiro I, Dugé de Bernonville TD, Melin C, Lafontaine F, Besseau S, Lanoue A, Papon N, Oudin A, Glévarec G, et al. (2016) Prequels to synthetic biology: from candidate gene identification and validation to enzyme subcellular localization in plant and yeast cells. Methods Enzymol 576: 167–206 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138–142 [DOI] [PubMed] [Google Scholar]
- Giddings LA, Liscombe DK, Hamilton JP, Childs KL, DellaPenna D, Buell CR, O’Connor SE (2011) A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J Biol Chem 286: 16751–16757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Góngora-Castillo E, Childs KL, Fedewa G, Hamilton JP, Liscombe DK, Magallanes-Lundback M, Mandadi KK, Nims E, Runguphan W, Vaillancourt B, et al. (2012) Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS ONE 7: e52506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Munro AW (2013) Unusual cytochrome p450 enzymes and reactions. J Biol Chem 288: 17065–17073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Sohl CD, Chowdhury G (2011) Multi-step oxidations catalyzed by cytochrome P450 enzymes: processive vs. distributive kinetics and the issue of carbonyl oxidation in chemical mechanisms. Arch Biochem Biophys 507: 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Burlat V, Oudin A, Lanoue A, St-Pierre B, Courdavault V (2009) Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep 28: 1215–1234 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2010) Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Ginis O, Poutrain P, Héricourt F, Oudin A, Lanoue A, St-Pierre B, Burlat V, Courdavault V (2011a) The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J 278: 749–763 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Poutrain P, Héricourt F, Mahroug S, St-Pierre B, Burlat V, Courdavault V (2011b) Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J Plant Physiol 168: 549–557 [DOI] [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 15: 41–62 [DOI] [PubMed] [Google Scholar]
- Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R, Dong L, André F, Ginglinger JF, Lugan R, Gavira C, Grec S, Lang G, Memelink J, Van der Krol S, et al. (2013) Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab Eng 20: 221–232 [DOI] [PubMed] [Google Scholar]
- Hotze M, Schröder G, Schröder J (1995) Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett 374: 345–350 [DOI] [PubMed] [Google Scholar]
- Huang FC, Sung PH, Do YY (2012) Differential expression and functional characterization of the NADPH cytochrome P450 reductase genes from Nothapodytes foetida. Plant Sci 190: 16–23 [DOI] [PubMed] [Google Scholar]
- Irmler S, Schröder G, St-Pierre B, Crouch NP, Hotze M, Schmidt J, Strack D, Matern U, Schröder J (2000) Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J 24: 797–804 [DOI] [PubMed] [Google Scholar]
- Jensen K, Møller BL (2010) Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry. 71: 132–141 [DOI] [PubMed] [Google Scholar]
- Kellner F, Geu-Flores F, Sherden NH, Brown S, Foureau E, Courdavault V, O’Connor SE (2015a) Discovery of a P450-catalyzed step in vindoline biosynthesis: a link between the aspidosperma and eburnamine alkaloids. Chem Commun (Camb) 51: 7626–7628 [DOI] [PubMed] [Google Scholar]
- Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L, et al. (2015b) Genome-guided investigation of plant natural product biosynthesis. Plant J 82: 680–692 [DOI] [PubMed] [Google Scholar]
- Koopmann E, Hahlbrock K (1997) Differentially regulated NADPH:cytochrome P450 oxidoreductases in parsley. Proc Natl Acad Sci USA 94: 14954–14959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, O’Connor SE (2011) A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liang QM, Zhou WW, Jiang YD, Zhu QZ, Yu H, Zhang CX, Gurr GM, Zhu ZR (2015) RNA interference of NADPH-cytochrome P450 reductase of the rice brown planthopper, Nilaparvata lugens, increases susceptibility to insecticides. Pest Manag Sci 71: 32–39 [DOI] [PubMed] [Google Scholar]
- Mackenzie NC, Lillico SG, Brown K, Wolf CR, Whitelaw CB (2010) Evaluation of RNA-knockdown strategies for modulation of cytochrome P450 reductase activity in mouse hepatocytes. J RNAi Gene Silencing 6: 416–421 [PMC free article] [PubMed] [Google Scholar]
- Madyastha KM, Coscia CJ (1979) Detergent-solubilized NADPH-cytochrome c(P-450) reductase from the higher plant, Catharanthus roseus: purification and characterization. J Biol Chem 254: 2419–2427 [PubMed] [Google Scholar]
- Mahroug S, Courdavault V, Thiersault M, St-Pierre B, Burlat V (2006) Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Mazourek M, Pujar A, Borovsky Y, Paran I, Mueller L, Jahn MM (2009) A dynamic interface for capsaicinoid systems biology. Plant Physiol 150: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Lopes Cardoso MI, Voskuilen JT, de Waal A, Verpoorte R, Hoge JHC (1993) Isolation and characterization of a cDNA clone from Catharanthus roseus encoding NADPH:cytochrome P-450 reductase, an enzyme essential for reactions catalysed by cytochrome P-450 mono-oxygenases in plants. Plant J 4: 47–60 [DOI] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Sato F (2011) Unusual P450 reactions in plant secondary metabolism. Arch Biochem Biophys 507: 194–203 [DOI] [PubMed] [Google Scholar]
- Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ (2013) What makes a P450 tick? Trends Biochem Sci 38: 140–150 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1236 [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Kingsford C (2015) Accurate, fast, and model-aware transcript expression quantification with Salmon. bioRxiv 021592 [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Qu X, Pu X, Chen F, Yang Y, Yang L, Zhang G, Luo Y (2015a) Molecular cloning, heterologous expression, and functional characterization of an NADPH-cytochrome P450 reductase gene from Camptotheca acuminata, a camptothecin-producing plant. PLoS ONE 10: e0135397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Easson ML, Froese J, Simionescu R, Hudlicky T, De Luca V (2015b) Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc Natl Acad Sci USA 112: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
- Rana S, Lattoo SK, Dhar N, Razdan S, Bhat WW, Dhar RS, Vishwakarma R (2013) NADPH-cytochrome P450 reductase: molecular cloning and functional characterization of two paralogs from Withania somnifera (L.) Dunal. PLoS ONE 8: e57068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Ehlting J, Douglas CJ (2002) Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol 130: 1837–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim V, Wiens B, Masada-Atsumi S, Yu F, De Luca V (2014) 7-Deoxyloganetic acid synthase catalyzes a key 3 step oxidation to form 7-deoxyloganetic acid in Catharanthus roseus iridoid biosynthesis. Phytochemistry 101: 23–31 [DOI] [PubMed] [Google Scholar]
- Salim V, Yu F, Altarejos J, De Luca V (2013) Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J 76: 754–765 [DOI] [PubMed] [Google Scholar]
- Schröder G, Unterbusch E, Kaltenbach M, Schmidt J, Strack D, De Luca V, Schröder J (1999) Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett 458: 97–102 [DOI] [PubMed] [Google Scholar]
- Schwarz H, Liu B, Peters S, Barillas W, Beerhues L (2009) Purification, cDNA cloning and functional expression of NADPH-cytochrome P450 reductase from Centaurium erythraea cell cultures. Plant Biol (Stuttg) 11: 300–306 [DOI] [PubMed] [Google Scholar]
- Shephard EA, Phillips IR, Bayney RM, Pike SF, Rabin BR (1983) Quantification of NADPH:cytochrome P-450 reductase in liver microsomes by a specific radioimmunoassay technique. Biochem J 211: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro EM (2008) Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota FA, De Luca V (1999) Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L, Vanholme R, Geerinck J, Goeminne G, Höfer R, Kim H, Ralph J, Boerjan W (2014) Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol 166: 1956–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H (2004) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J Biol Chem 279: 41866–41872 [DOI] [PubMed] [Google Scholar]
- Tang T, Zhao C, Feng X, Liu X, Qiu L (2012) Knockdown of several components of cytochrome P450 enzyme systems by RNA interference enhances the susceptibility of Helicoverpa armigera to fenvalerate. Pest Manag Sci 68: 1501–1511 [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Fabris M, Pollier J, Baart GJ, Rombauts S, Hasnain G, Rischer H, Memelink J, Oksman-Caldentey KM, Goossens A (2013) CathaCyc, a metabolic pathway database built from Catharanthus roseus RNA-Seq data. Plant Cell Physiol 54: 673–685 [DOI] [PubMed] [Google Scholar]
- Varadarajan J, Guilleminot J, Saint-Jore-Dupas C, Piégu B, Chabouté ME, Gomord V, Coolbaugh RC, Devic M, Delorme V (2010) ATR3 encodes a diflavin reductase essential for Arabidopsis embryo development. New Phytol 187: 67–82 [DOI] [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ (1997) Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94: 8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Zhang Y, Chen X, Lee EJ, Barber CJ, Chakrabarty R, Desgagné-Penix I, Haslam TM, Kim YB, Liu E, et al. (2013) Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol 166: 122–134 [DOI] [PubMed] [Google Scholar]
- Yang CQ, Lu S, Mao YB, Wang LJ, Chen XY (2010) Characterization of two NADPH:cytochrome P450 reductases from cotton (Gossypium hirsutum). Phytochemistry 71: 27–35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.