BTB-TAZ proteins bridge and scaffold the CRL3 complex to modulate iron homeostasis by ubiquitinating and degrading MdbHLH104 proteins in response to Fe status in apple.
Abstract
Iron (Fe) homeostasis is crucial for plant growth and development. A network of basic helix-loop-helix (bHLH) transcription factors positively regulates Fe uptake during iron deficiency. However, their up-regulation or overexpression leads to Fe overload and reactive oxygen species generation, thereby damaging the plants. Here, we found that two BTB/TAZ proteins, MdBT1 and MdBT2, interact with the MbHLH104 protein in apple. In addition, the function of MdBT2 was characterized as a regulator of MdbHLH104 degradation via ubiquitination and the 26S proteasome pathway, thereby controlling the activity of plasma membrane H+-ATPases and the acquisition of iron. Furthermore, MdBT2 interacted with MdCUL3 proteins, which were required for the MdBT2-mediated ubiquitination modification of MdbHLH104 and its degradation. In sum, our findings demonstrate that MdBT proteins interact with MdCUL3 to bridge the formation of the MdBTsMdCUL3 complex, which negatively modulates the degradation of the MdbHLH104 protein in response to changes in Fe status to maintain iron homeostasis in plants.
Iron (Fe) is an essential micronutrient for plants, animals, and humans. Although Fe is usually quite abundant on Earth, it limits plant growth in approximately 30% of the world’s soils due to unsuitable environmental factors (Kobayashi and Nishizawa, 2012). In calcareous soils, Fe precipitates into insoluble Fe(III)-oxyhydroxide complexes, which limits the availability of Fe to plants. Thus, the molecular mechanisms utilized by all dicot and nongrass monocot plants for Fe acquisition often include a first step that solubilizes ferric Fe followed by a second step in which Fe is uptaken from the soil and transported into the root cells (Ivanov et al., 2012).
A network of transcription factors (TFs), such as the basic helix-loop-helix (bHLH) TFs, plays a central role in positively regulating the expression of the major Fe acquisition and transportation genes in response to internal and external iron availability (Kobayashi and Nishizawa, 2012; Ivanov et al., 2012). In Arabidopsis (Arabidopsis thaliana), the Ib and IVc subgroups of bHLH TFs have been shown to modulate responses to Fe deficiency (Ivanov et al., 2012). Tomato (Solanum lycopersicum) FER was the first bHLH TF of the Ib subgroup characterized to have a function in Fe acquisition by activation of the strategy I pathway (Ling et al., 2002; Brumbarova and Bauer, 2005). Its Arabidopsis ortholog FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) controls iron uptake by directly regulating the expression of the FRO2 and IRT1 genes under Fe-deficient conditions (Colangelo and Guerinot, 2004; Yuan et al., 2005, 2008). Meanwhile, several other bHLH TFs such as IAA-Leu Resistant 3 (ILR3, also called bHLH105), PYE, bHLH104, OsbHLH133, and CmbHLH1 regulate plant growth and development by mediating the activity of Fe-chelate reductase, the acidification of rhizospheres, and the transportation of iron under Fe-deficient conditions in Arabidopsis, rice (Oryza sativa), and chrysanthemum (Chrysanthemum morifolium; Li et al., 2016; Long et al., 2010; Selote et al., 2015; Wang et al., 2013; Zhang et al., 2015; Zhao et al., 2014). In rice, IRON-RELATED TRANSCRIPTION FACTOR 2 (IRO2) is a homolog of bHLH38 and bHLH39. Its overexpression promotes both Fe uptake and its translocation to seeds (Ogo et al., 2011; Zheng et al., 2010).
Although iron is absolutely necessary for plants and animals, its excessive accumulation or inappropriate distribution causes toxicity by generating reactive oxygen species (ROS) via the Fenton reaction, leading to the oxidative destruction of cells (Thomine and Vert, 2013). In animals, the excess iron is not excreted and is deposited in the body, which causes various diseases (Hentze et al., 2004). In plants, iron toxicity is a major physiological toxicity that occurs in anaerobic fields or acidic soils (which contain an excess of Fe2+ in the soil). Fe content and ROS levels are higher in the leaves, and they exhibit the symptom of bronzing under conditions of iron toxicity compared with normal conditions in various plant species (Thongbai and Goodman, 2000). The first mutant that accumulates excessive Fe, the bronze (brz) mutant, was identified in pea (Pisum sativum). It is controlled by a monogenic recessive locus that is located on chromosome 4. However, the causative genes have not been cloned (Grusak et al., 1990). In addition, transgenic plants overexpressing Fe-related genes, such as the bHLH TF genes bHLH104 and bHLH105 and the ferroxidase gene LOW PHOSPHATE ROOT1 (LPR1), take up large amounts of Fe, resulting in a high iron content that is not fully utilized and induces the accumulation of ROS (Müller et al., 2015; Zhang et al., 2015). To prevent damage due to Fe-oversupply-induced ROS, plants have evolved a series of sophisticated regulatory networks at the transcriptional and posttranscriptional levels to maintain Fe homeostasis. There is increasing evidence that the ubiquitin-proteasome system (UPS) is a quick and flexible mechanism for coping with various environmental factors including Fe-sufficient or Fe-deficient conditions (Hua and Vierstra, 2011). In mammals, the F-box and Leu-rich repeat protein 5 (FBXL5), which works with CUL1 and SKP1 to form the E3 ubiquitin ligase complex, promotes the iron-dependent degradation of IRON REGULATORY PROTEIN2 (IRP2) under conditions of sufficient Fe (Salahudeen et al., 2009; Vashisht et al., 2009). In plants, the ubiquitin conjugase UBC13 is involved in the up-regulation of the IRT1 and AHA2 genes and the activation of iron reductase activity under conditions of iron deficiency in cucumber (Li and Schmidt, 2010).
In Arabidopsis, FIT is a short-lived protein. It is degraded by an unknown ubiquitin E3 ligase in response to Fe starvation, which clears the spent FIT proteins and allows new ones to activate the transcription of target genes, such as FRO2 and IRT1 (Sivitz et al., 2011). Most recently, it has been reported that ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) is enhanced by an H2O2 signal triggered by prolonged Fe deficiency. It acts as a negative regulator of Fe uptake in a FIT-dependent manner (Le et al., 2016). In addition, the ubiquitin E3 ligase BTS is induced by Fe deficiency and acts as a negative regulator involved in the 26S proteasome-mediated degradation of ILR3 and bHLH115, but not that of bHLH104 and PYE under prolonged iron deficiency (Selote et al., 2015). As BTS homologs, rice OsHRZ1 and OsHRZ2 are also transcriptionally induced by Fe deficiency. They negatively regulate the expression of Fe deficiency-induced genes, probably in a manner similar to BTS, although their direct target proteins have not been identified (Kobayashi et al., 2013).
In apple (Malus domestica), an IVc subgroup of bHLH TF gene MdbHLH104 has been characterized and found to function in Fe acquisition by directly binding to the promoter of the MdAHA8 gene, thereby modulating the activity of plasma membrane (PM) H+-ATPases and the uptake of Fe under Fe-deficient conditions (Zhao et al., 2016). Here, we examined if its overexpression resulted in Fe overload and ROS generation under conditions of sufficient Fe. Furthermore, two MdbHLH104-interacting BTB-TAZ proteins, MdBT1 and MdBT2, were identified as negative regulators of Fe acquisition by screening with a yeast two-hybrid method. Subsequently, their role in the ubiquitin-associated degradation of the MdbHLH104 protein was verified, especially under conditions of sufficient Fe. Finally, we investigated and discussed how the Cullin-RING ubiquitin ligase 3 (CRL3) complex MdBT2MdCUL3 controls Fe homeostasis by negatively regulating the stability of the MdbHLH104 protein.
RESULTS
Overexpression of MdbHLH104 Leads to Fe Overload and ROS Accumulation in Transgenic Apple Plants
The MdbHLH104 transgenic apple lines were obtained in our previous study (Zhao et al., 2016). After being transferred to and grown in soil with a normal iron supply for 4 months, the apple plantlets of three transgenic lines exhibited bronze spots, and even necrotic lesions, all over their leaves, especially the mature ones near the base of the stem; the wild-type controls grew well (Fig. 1, A and B). Here, Perls staining was carried out to examine the accumulation and distribution of Fe3+ in the leaves of the wild-type and transgenic apple plants. The results showed that the transgenic apple plants accumulated excessive Fe3+ in the mature leaves of three transgenic apple lines, especially within the bronze spots, whereas the wild-type control contained normal levels of Fe3+ (Fig. 1C). Subsequently, Trypan blue staining was conducted to detect cell death. The results demonstrated that cell death spots, as indicated by strong staining, occurred throughout the mature leaves of the MdbHLH104 transgenic apple lines like the bronze spots and necrotic lesions, but not in those of the wild-type control (Fig. 1D).
Figure 1.
Overexpression of MdbHLH104 results in Fe overaccumulation in transgenic apple plantlets. A, Phenotypes of the MdbHLH104 transgenic apple lines and the wild-type controls grown in soil under conditions of sufficient Fe. B, Magnified image of the mature leaves near the basal stem, indicated by arrowheads. C, Perls stain for Fe signals in the mature leaves. D, Trypan blue staining for cell death in the mature leaves. E, Histochemical detection of O2·− and H2O2 in the mature leaves. Leaves were infiltrated with 0.5 mg/mL NBT for 1 h for O2·− staining and 1 mg/mL DAB for 8 h for H2O2 staining in the dark. F, O2·− accumulation and H2O2 contents in the mature leaves.
Histochemical staining with 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) were performed to examine if ROS were generated in either the wild-type or transgenic apple lines. The results showed that the leaves of the transgenic apple plants appeared to be much more deeply colored than the leaves of the wild-type control, which indicated that there were more polymeric oxidation products resulting from ROS accumulation in the transgenic apple plants (Fig. 1E). To further verify the generation of ROS, the O2.− and H2O2 contents were determined. The results showed that the transgenic apple lines generated much more O2.− and H2O2 in their leaves than the wild-type control (Fig. 1F).
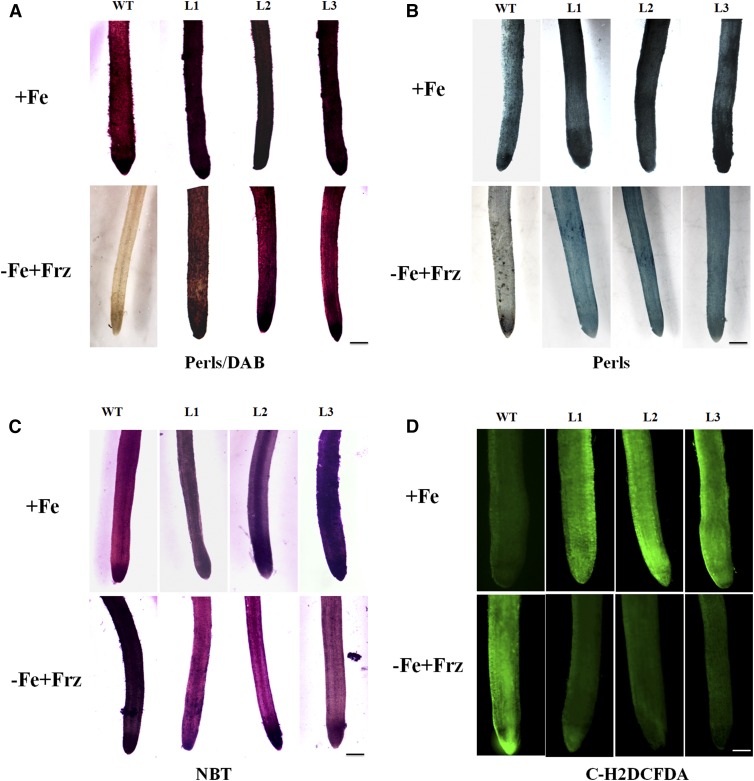
In addition, a sensitized Perls/DAB staining and a less-sensitive Perls method (no DAB enhancement) were conducted to monitor Fe distribution in roots. The results showed that transgenic apple plants deposited much more Fe in their roots than the wild-type control under both Fe-sufficient and -deficient conditions (Fig. 2, A and B). Subsequently, carboxylated 2′,7′-dichlorodihydrofluorescein diacetate [C-H2DCFDA] staining and NBT staining were carried out to visualize ROS accumulation in roots. The results indicated that the transgenic apple plants generated more ROS in roots under Fe-sufficient conditions but less ROS under Fe-deficient conditions than the wild-type controls (Fig. 2, C and D).
Figure 2.
Fe accumulation and ROS generation in the roots of the transgenic apple plants and the wild-type control under iron-sufficient (+Fe) and -deficient (−Fe+Frz) conditions. A, Perls/DAB staining of Fe distribution in the roots. Scale bar, 200 μm. B, Fe staining (Perls only) in the roots. Scale bar, 200 μm. C, C-H2DCFDH staining of ROS in the roots. Scale bar, 200 μm. D, NBT staining of superoxide in the roots. Scale bar, 200 μm.
MdbHLH104 Interacts with the Two BTB Scaffold Proteins MdBT1 and MdBT2
To identify proteins that interact with MdbHLH104, the MdbHLH104 protein was used as bait in a yeast two-hybrid (Y2H) screening of an apple cDNA library. As a result, the MdBT proteins were identified as candidates (Supplemental Fig. S1, A and B). To verify the interaction between MdbHLH104 and each of the MdBTs, Y2H assays were performed. The full-length cDNA of each MdBT was integrated into vector pGBT9 (BD-MdBTs) as bait, whereas that of MdbHLH104 was integrated into pGAD424 (AD-MdbHLH104) as prey. Positive X-α-gal activity was observed in yeast containing either pGBT9-MdBT1 or pGBT9-MdBT2 plus pGAD424-MdbHLH104, but not in those containing pGBT9-MdBT1 or pGBT9-MdBT2 plus the empty pGAD424 vector, indicating that MdbHLH104 interacted with MdBT1 and MdBT2, respectively (Fig. 3A). In contrast, it was found that the MdbHLH104 protein did not interact with MdBT3.1, MdBT3.2, or MdBT4 (Supplemental Fig. S1C).
Figure 3.
MdBT1 and MdBT2 interact with the MdbHLH104 protein. A, MdbHLH104 interacted with MdBT1 and MdBT2 in Y2H assays. B and C, The TAZ domain of MdBT2 (B) and the C terminus of MdbHLH104 (C) are responsible for their interaction. Left, Diagram of full-length and truncated MdBT2 (B) and MdbHLH104 (C) constructs with specific deletions. Right, Interactions were indicated by the ability of yeast cells to grow on SDII and SDIV media. D, Pull-down assays with E. coli-expressed His-MdbHLH104 proteins resulted in the precipitation of GST-MdBT1 and GST-MdBT2; His alone was used as the control. Blots were first probed with the anti-His antibody and then stripped and probed with the anti-GST antibody. E, Co-IP assays of MdBT1 (or MdBT2) and MdbHLH104 in the 35S::MdbHLH104-MYC+35S::MdBT2-GFP transgenic apple calli. The MdBT-GFP proteins were immunoprecipitated with an anti-MYC antibody and immunoblotted with an anti-GFP antibody.
To examine which regions of the MdBT proteins are required for their interaction with the MdbHLH104 protein, MdBT2 was divided into its N-terminal BTB domain and C-terminal TAZ domain. The Y2H assay showed that MdbHLH104 only interacted with the C-terminal TAZ domain (Fig. 3B). Meanwhile, five truncated MdbHLH104 variants were inserted into the Gal4 activation domain of the prey vector. The Y2H assay indicated that deletion of the 150 residues from the N terminus of MdbHLH104 did not affect its interaction with MdBT2. In contrast, the MdbHLH104 protein without its 60 C-terminal residues completely failed to interact with the MdBT2 protein (Fig. 3C). Therefore, the C terminus of MdbHLH104 is essential for its interaction with the MdBT2 protein.
To further verify the interaction between the MdBTs and MdbHLH104, in vitro pull-down analyses and in vivo coimmunoprecipitation (Co-IP) assays were carried out. Pull-down analysis was conducted with GST-MdBT1 or GST-MdBT2 and His-MdbHLH104 proteins that were expressed in and purified from Escherichia coli BL21. The result showed that His-MdbHLH104, but not His alone, interacted with the MdBT1 and MdBT2 proteins, respectively (Fig. 3D).
For the Co-IP assay, 35S::MdbHLH104-MYC transgenic calli were obtained. Subsequently, the expression vectors containing 35S::MdBT1-GFP and 35S::MdBT2-GFP were genetically transformed into the 35S::MdbHLH104-MYC transgenic calli, respectively. Nuclear proteins isolated from 35S::MdBT1-GFP (or 35S::MdBT2-GFP)+35S::MdbHLH104-MYC and 35S::MdbHLH104-MYC calli that had been treated with the proteasome inhibitor MG132 were immunoprecipitated after incubation with an anti-MYC antibody. The eluted purified proteins were blotted and detected with anti-GFP antibody. A clear signal was observed in 35S::MdBT1-GFP (or 35S::MdBT2-GFP)+35S::MdbHLH104-MYC calli, but not in control extracts from the 35S::MdbHLH104-MYC calli (Fig. 3E). These findings indicated that MdbHLH104 interacted with both the MdBT1 and MdBT2 proteins, respectively.
MdBT1 and MdBT2 Are Involved in the Degradation of the MdbHLH104 Protein
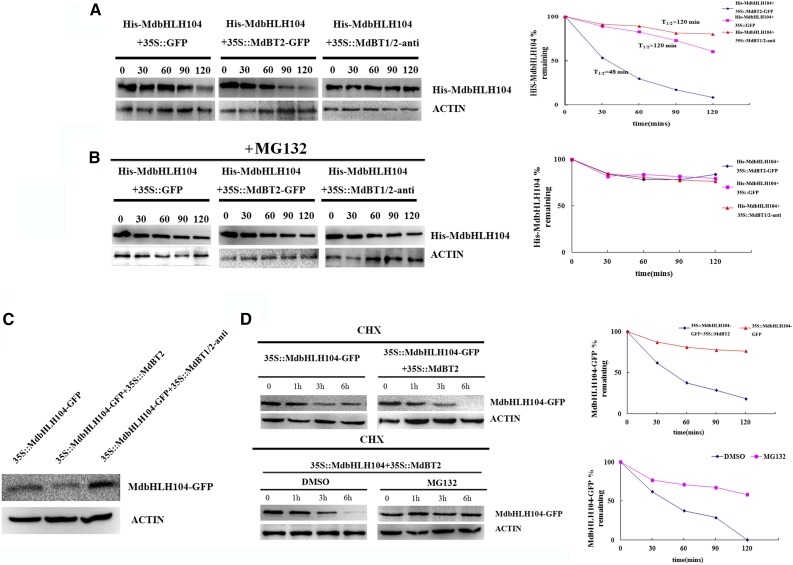
To examine if MdBT2 is involved in the degradation of the MdbHLH104 protein, cell-free degradation assays of the recombinant His-MdbHLH104 protein were conducted using protein samples extracted from the 35S::MdBT2 transgenic calli in the presence or absence of MG132; protein samples extracted from wild-type calli were used as controls. Protein gel blots showed that His-MdbHLH104 degraded much faster in protein extracts of 35S::MdBT2 calli than in wild-type extracts (Fig. 4A). Additionally, MG132 noticeably inhibited the degradation of the recombinant His-MdbHLH104 protein (Fig. 4B).
Figure 4.
The abundance and stability of the MdbHLH104 protein. A and B, Cell-free degradation assays of the purified recombinant His-MdbHLH104 protein in the protein extract of transgenic apple calli as labeled. Samples were incubated in the degradation buffer with or without proteasome inhibitor (50 μm MG132). His-MdbHLH104 levels were visualized by immunoblotting using the anti-His antibody. ACTIN was used as the loading control. Right, The half-life plot for the cell-free degradation assay of His-MdbHLH104 was calculated based on densitometric analyses of the immunoblots of His-MdbHLH104 and MdACTIN. C, The abundance of MdbHLH104 proteins in three transgenic apple calli as labeled. D, MdBT2-mediated destabilization of the MdbHLH104 protein is dependent on the ubiquitin/proteasome. 35S::MdbHLH104-GFP apple calli were transformed with or without the expression of MdBT2. At time 0, 250 μm CHX was added, and total proteins were extracted at the indicated times and then analyzed by immunoblotting using the anti-GFP antibody (top). 35S::MdbHLH104-GFP transgenic apple calli were transformed with or without 35S::MdBT2, and the resultant 35S::MdbHLH104-GFP+35S::MdBT2 transgenic calli were either treated with DMSO or MG132 for the indicated amounts of time. The total proteins were extracted at the indicated times and then analyzed by immunoblotting using the anti-GFP antibody (bottom). Quantification of the protein levels using Quantity One software is shown on the right.
To determine if MdBT1 and MdBT2 are required for MdbHLH104 degradation, an antisense cDNA that is highly conserved in MdBT1 and MdBT2 but not in other MdBTs was used to construct the suppression vector 35S::MdBT1/2-anti, which was then transformed into apple calli. Subsequently, quantitative real-time reverse transcription-PCR (qRT-PCR) showed that 35S::MdBT1/2-anti apple transgenic calli had been obtained (Supplemental Fig. S2A). The resultant transgenic calli were then used in a cell-free degradation assay. The results indicated that suppression of MdBT1 and MdBT2 inhibited the degradation of MdbHLH104 (Fig. 4, A and B). Therefore, MdBT1 and MdBT2 are involved in the 26S proteasome-related degradation of MdbHLH104 proteins in the cell-free system.
To assess the contribution of MdBT1 and MdBT2 to the degradation of MdbHLH104 proteins in vivo, three types of transgenic calli, i.e. 35S::MdbHLH104-GFP, 35S::MdbHLH104-GFP+35S::MdBT2, and 35S::MdbHLH104-GFP+35S::MdBT1/2-anti were obtained (Supplemental Fig. S2B) and used for western-blotting assays with an anti-GFP antibody. The results showed that the 35S::MdbHLH104-GFP+35S::MdBT2 calli accumulated less and the 35S::MdbHLH104-GFP+35S::MdBT1/2-anti calli more MdbHLH104-GFP protein than the 35S::MdbHLH104-GFP calli (Fig. 4C).
To check if MdbHLH104 degradation is associated with the MdBT2-mediated 26S proteasome pathway in vivo, the 35S::MdbHLH104-GFP and 35S::MdbHLH104-GFP+35S::MdBT2 transgenic calli were treated with the translational inhibitor cycloheximide (CHX), followed by a western-blotting assay. The result showed that MdbHLH104-GFP proteins degraded much faster in 35S::MdbHLH104-GFP+35S::MdBT2 calli treated with CHX than in 35S::MdbHLH104-GFP calli treated with CHX, indicating that MdBT2 promotes the degradation of MdbHLH104-GFP proteins in apple calli. However, after 35S::MdbHLH104-GFP+35S::MdBT2 calli were treated with CHX plus the proteasome inhibitor MG132, the MdBT2-mediated degradation of MdbHLH104 was inhibited, similar to the cell-free system (Fig. 4B), suggesting the involvement of the 26S proteasome pathway in the MdBT2-mediated degradation of the MdbHLH104 protein (Fig. 4D).
MdBT2 Ubiquitinates MdbHLH104 by Bridging the Assembly of the E3 Ligase MdCUL3
To determine whether MdbHLH104 is degraded by the 26S proteasome, 35S::MdbHLH104-GFP transgenic apple calli were treated with the translational inhibitor CHX and the proteasome inhibitor MG132 and their protein extracts used for western blotting. The result showed that the accumulation of MdbHLH104 protein was completely inhibited by CHX in the transgenic calli. In contrast, MdbHLH104 protein accumulated in the transgenic calli that had been treated with both CHX and MG132 (Fig. 5A), demonstrating that MdbHLH104 is degraded in a 26S proteasome-dependent manner.
Figure 5.
The MdBT2-mediated ubiquitination and degradation of MdbHLH104 requires MdCUL3s. A, Immunoblotting assays of MdbHLH104-GFP proteins with the anti-GFP antibody in transgenic calli that had been treated for 6 h with CHX alone or CHX plus MG132 (50 μm). B, Detection of ubiquitination of the MdbHLH104 protein in 35S::MdbHLH104-MYC and 35S::MdbHLH104-MYC+35S::MdBT2 transgenic apple calli pretreated with 50 μm MG132. MdbHLH104-MYC protein was IPed with the anti-MYC antibody. Anti-MYC and anti-ubiquitin antibodies were used to detect MdbHLH104-MYC and ubiquitinated MdbHLH104-MYC, respectively. IB, Immunoblot. C, Pull-down assays with E. coli-expressed His-MdCUL3 result in the precipitation of GST-MdBT2. His was used as the control. Blots were first probed with the anti-His antibody and then stripped and probed with the anti-GST antibody. D, IP assays using the anti-FLAG antibody show the Co-IP of FLAG-MdCUL3 or FLAG with MdBT2 from 35S::FLAG-MdbCUL3+35S::MdBT2-GFP or 35S:: FLAG+35S::MdBT2-GFP transgenic apple calli. E and F, Detection of ubiquitination of the MdbHLH104 protein in the different transgenic apple calli as labeled. The MdbHLH104 protein was IPed with the anti-MYC antibody. The anti-ubiquitin antibody was used to detect ubiquitination of the MdbHLH104 protein.
Generally, 26S proteasome-mediated protein degradation is closely associated with the modification of ubiquitination. To check if MdBT2 influences the ubiquitination of the MdbHLH104 protein, two types of transgenic calli, 35S::MdbHLH104-MYC and 35S::MdbHLH104-MYC+35S::MdBT2-GFP, were obtained and used. The anti-MYC antibody was used to immunoprecipitate the MdbHLH104-MYC proteins. The change in molecular mass of the immunoprecipitated proteins was then checked with an anti-MYC antibody. The result indicated that many more high-molecular-mass forms of MdbHLH104 were detected in 35S::MdbHLH104-MYC+35S::MdBT2-GFP calli than in 35S::MdbHLH104-MYC calli (Fig. 5B). The modified forms of the MdbHLH104-MYC proteins were further checked for their level of ubiquitination with an anti-ubiquitin antibody that recognized only ubiquitinated MdbHLH104-MYC proteins. The anti-ubiquitin antibody detected a much higher level of ubiquitinated MdbHLH104-MYC proteins in 35S::MdbHLH104-MYC+35S::MdBT2-GFP calli than in 35S::MdbHLH104-MYC calli (Fig. 5C). Therefore, MdBT2 is involved in the ubiquitination of MdbHLH104 proteins in apple calli.
In Arabidopsis, the BTB protein ubiquitinates its target proteins by bridging the assembly of the E3 ligase CUL3 (Figueroa et al., 2005). There are three MdCUL3 genes in the apple genome, MdCUL3-1, MdCUL3-2, and MdCUL3-3 (Supplemental Fig. S3, A and B). To examine if the MdBT2 function is conserved, the interaction between MdBT2 and MdCUL3-1 was assayed with Y2H, pull-down, and Co-IP assays. The Y2H assay did not detect an interaction between MdBT2 and MdCUL3-1 (Supplemental Fig. S3C). However, when the GST-MdBT2 and His-MdCUL3-1 proteins, which were expressed in and purified from Escherichia coli BL21, were used in a pull-down assay, MdBT2 physically interacted with MdCUL3-1 (Fig. 5D).
For the Co-IP assay, 35S::MdBT2-GFP transgenic calli were obtained. Subsequently, 35S::FLAG and 35S::FLAG-MdCUL3-1 were genetically transformed into the 35S::MdBT2-GFP transgenic calli background. Proteins were isolated from the 35S::MdBT2-GFP, 35S::MdBT2-GFP+35S::FLAG, and 35S::MdBT2-GFP+35S::FLAG-MdCUL3-1 calli, which had been treated with the proteasome inhibitor MG132 and immunoprecipitated after incubation with an anti-GFP antibody. The eluted purified proteins were blotted and detected with the anti-FLAG antibody. The result showed that MdBT2 interacted with MdCUL3-1 (Fig. 5E).
Furthermore, cell-free degradation assays were conducted to examine if MdCUL3s are essential for MdbHLH104 degradation. The result showed that MdCUL3 promoted the degradation of MdbHLH104 protein via the 26S proteasome (Supplemental Fig. S4, A and B). To examine if MdCUL3 is involved in the MdBT2-mediated ubiquitination of MdbHLH104, a viral vector-based method was carried out. The above-mentioned antisense cDNA, which specifically suppresses three MdCUL3 genes, was inserted into a viral vector. Subsequently, the resultant pIR-MdCUL3-anti vector and the empty control pIR vector were transiently transformed into 35S::MdbHLH104-MYC+35S::MdBT2 calli, respectively. The proteins were immunoprecipitated with anti-MYC antibody and then anti-ubiquitin antibody was used to detect the level of MdbHLH104 ubiquitination. The result showed that the suppression of MdCUL3s resulting from pIR-MdCUL3-anti transformation noticeably inhibited the MdBT2-mediated ubiquitination of the MdbHLH104 protein (Fig. 5F), indicating the requirement of MdCUL3s for this process. Meanwhile, a sense full-length cDNA of MdCUL3-1 was also inserted into the viral vector pIR. The vectors pIR-MdCUL3-1 and pIR were transiently transformed into 35S::MdbHLH104-MYC+35S::MdBT1/2-anti calli. As a result, MdCUL3-1 overexpression only had a slight influence on the ubiquitination of the MdbHLH104 protein, suggesting that MdCUL3-associated ubiquitination requires the involvement of MdBT2 (Fig. 5G). Finally, the cell-free degradation assays demonstrated that MdCUL3s and MdBTs are necessary for the degradation of MdbHLH104 proteins (Supplemental Fig. S5, A and B).
MdBTs Are Induced and Required for the Ubiquitination and Degradation of MdbHLH104 in Response to a Surplus of Fe
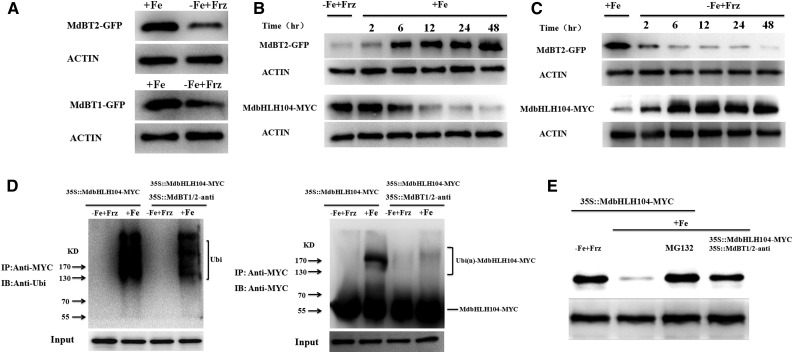
To determine whether the abundance of MdBT1 and MdBT2 proteins is influenced by iron, the 35S::MdBT1-GFP and 35S::MdBT2-GFP constructs were transformed into apple calli. The resultant transgenic calli were treated with or without iron and then used for western blotting with an anti-GFP antibody. The result showed that Fe treatment remarkably increased the abundancies of both the MdBT1 and MdBT2 proteins (Fig. 6A). Furthermore, the construct 35S::MdbHLH104-MYC was transformed into the 35S::MdBT2-GFP transgenic calli. The resultant 35S::MdBT2-GFP+35S::MdbHLH104-MYC transgenic apple calli were treated with iron and used for western blotting with anti-GFP and anti-MYC antibodies, respectively. Time-course western blots demonstrated that the abundance of MdBT2-GFP proteins gradually increased, whereas that of MdbHLH104-MYC decreased correspondingly, with the length of iron treatment (Fig. 6B). In contrast, when the calli grown under conditions of sufficient Fe were exposed to Fe-deficient conditions, the abundance of the MdBT2-GFP proteins gradually decreased, whereas that of MdbHLH104-MYC correspondingly increased, with the length of iron starvation (Fig. 6C). Therefore, the abundance of MdBT2 was negatively regulated by iron at both transcriptional and posttranslational levels, which underlies the Fe-responsive accumulation of MdbHLH104 proteins (Fig. 6, A–C; Supplemental Fig. S5C).
Figure 6.
MdBT protein accumulation is dependent on iron. A, Immunoblot analysis of MdBT1-GFP and MdBT2-GFP proteins with the anti-GFP antibody under iron-deplete or iron-replete conditions. The 35S::MdBT1-GFP and 35S::MdBT2-GFP transgenic apple calli were used. B and C, Time course of changes in the abundance of the MdBT2 and MdbHLH104 proteins in response to iron availability. Transgenic apple calli were incubated with (+Fe) or without (−Fe+Frz) iron for 1 d, and then switched to no (−Fe+Frz) iron or iron (+Fe) conditions, respectively. D, The 35S::MdbHLH104-MYC and 35S::MdbHLH104-MYC+35S::MdBT1/2-anti transgenic apple calli were treated with (+Fe) or without (−Fe+Frz) iron and IPed with the anti-MYC antibody. Ubiquitin conjugates that were immunoprecipitated were detected with anti-ubiquitin and anti-MYC antibodies, respectively. IP, Immunoprecipitated; IB, immunoblotted. E, Fe-oversupply-mediated degradation of the MdbHLH104 protein is dependent on MdBT1 and MdBT2. 35S::MdbHLH104-MYC transgenic apple calli were treated without iron (−Fe+Frz) and then given iron (+Fe) in the presence or absence of MG132 (50 μm). The 35S::MdbHLH104-MYC+35S::MdBT1/2-anti transgenic apple calli were given iron (+Fe). Total protein extracts were used for immunoblotting with the anti-MYC antibody. ACTIN was used as the loading control.
To examine if ubiquitination mediates the Fe-oversupply-induced degradation of MdbHLH104 proteins, 35S::MdbHLH104-MYC transgenic apple calli were either treated with iron or given none and used for Co-IP with the anti-MYC antibody and for western blotting with the anti-MYC and anti-ubiquitin antibodies, respectively. The result showed that Fe treatment induced noticeable ubiquitination of the MdbHLH104-MYC proteins (Fig. 6D). Furthermore, 35S::MdbHLH104-MYC+35S::MdBT1/2-anti transgenic apple calli were used to examine the role of the MdBT proteins in the Fe-oversupply-induced ubiquitination of MdbHLH104 proteins. The result showed that the Fe-oversupply-induced ubiquitination level of MdbHLH104 proteins was much lower in the 35S::MdbHLH104-MYC+35S::MdBT1/2-anti transgenic apple calli than in the 35S::MdbHLH104-MYC ones, indicating that the Fe-oversupply-induced ubiquitination is at least partially attributable to the presence of MdBT proteins (Fig. 6D).
Finally, western blotting was conducted with an anti-MYC antibody to examine the role of MdBT proteins in the Fe-oversupply-induced degradation of MdbHLH104-MYC proteins. The result showed that suppression of the MdBT proteins inhibited the degradation of MdbHLH104-MYC in response to a surplus of Fe (Fig. 6E).
MdBTs Negatively Regulate the Ability of MdbHLH104 to Affect PM H+-ATPase Activity and Iron Homeostasis
It is difficult to obtain transgenic apple plants containing two or more genes. Therefore, Agrobacterium rhizogenes-mediated transformation was conducted to examine how MdBTs influence the function of MdbHLH104 (Supplemental Fig. S6, A–C). The roots containing 35S::MdbHLH104-GFP+35S::GUS, 35S::MdbHLH104-GFP+35S::MdBT1-GUS, and 35S::MdbHLH104-GFP+35S::MdBT2-GUS were used to determine if MdBTs negatively regulate the function of MdbHLH104 in the control of PM H+-ATPase activity, Fe uptake, and ROS generation. The roots containing 35S::MdbHLH104-GFP+35S::GUS exhibited the same phenotype as the 35S::MdbHLH104-GFP background control (Fig. 7), indicating that A. rhizogenes-mediated transformation did not influence the function of MdbHLH104. Because PM H+-ATPase activity results in acidification of the rhizosphere, it was visualized by using the pH indicator Bromocresol Purple. The result showed that overexpression of MdbHLH104-GFP did not influence rhizosphere acidification under conditions of sufficient Fe (Fig. 7A), presumably because the expression of MdbHLH104-activated MdAHAs and the resulting rhizosphere acidification require other bHLH genes that are induced by Fe deficiency. In contrast, 35S::MdbHLH104-GFP transgenic apple plants exhibited increased rhizosphere acidification compared with the wild-type control under Fe-deficient conditions (Fig. 7A). Furthermore, the overexpression of MdBT1 and MdBT2 noticeably inhibited the MdbHLH104-induced rhizosphere acidification under Fe-deficient conditions (Fig. 7A), indicating that MdBT1 and MdBT2 repressed the function of MbHLH104 in rhizosphere acidification.
Figure 7.
Rhizosphere acidification, Fe distribution, and ROS generation in the roots of the 35S::MdbHLH104-GFP transgenic apple plants and the wild-type control under iron-sufficient (+Fe) and -deficient (−Fe+Frz) conditions. A, Rhizosphere acidification of the wild-type control and transgenic apple lines. The yellow color around the roots stained with Bromocresol Purple indicates acidification. B and C, Phenotypic comparison of Fe (Perls/DAB) and ROS (C-H2DCFDH) staining in the roots of the transgenic apple plants as indicated. Scale bar, 200 μm.
In addition, Perls/DAB and Perls stainings were conducted to detect Fe accumulation. The results showed that the overexpression of each of MdBT1 and MdBT2 remarkably inhibited the MdbHLH104-induced Fe accumulation in roots under both Fe-sufficient and -deficient conditions (Fig. 7B; Supplemental Fig. S7A). Meanwhile, C-H2DCFDA and NBT stainings were used to visualize the accumulation of ROS in roots. The results indicated that the overexpression of each MdBT1 and MdBT2 gene noticeably inhibited the MdbHLH104-induced ROS accumulation in roots under conditions of sufficient Fe (Fig. 7C; Supplemental Fig. S7B). Under Fe-deficient conditions, MdbHLH104 overexpression inhibited the accumulation of ROS in roots compared with the wild-type control. The overexpression of either the MdBT1 or MdBT2 gene resulted in partial reversal of the MdbHLH104 inhibition of ROS accumulation in roots (Fig. 7C; Supplemental Fig. S7B).
In addition, the vectors 35S::MdBT2 and 35S::MdBT1/2-anti were transformed into apple calli to explore the physiological functions of the MdBTs in the regulation of cell acidification, PM H+-ATPase activity, and iron uptake. The results showed that 35S::MdBT1/2-anti and 35S::MdBT2 calli exhibited higher and lower levels, respectively, of cell acidification, PM H+-ATPase activity, and iron than the wild-type control (Fig. 8); these results indicate that MdBT2 negatively regulates PM H+-ATPase activity and iron uptake, especially under conditions of iron deficiency.
Figure 8.
MdBT2 regulates H+-ATPase activity and iron uptake. A, Acidification of transgenic apple calli. After allowing the calli to grow in the iron-deficient media for 5 d, the calli were exposed to the pH indicator dye Bromocresol Purple for 24 h. Acidification is indicated as the yellow color around the apple calli. B, PM H+-ATPase activity in vesicles isolated from the wild-type control, 35S::MdBT2 and 35S::MdBT1/2-anti transgenic apple calli that had been treated with (+Fe) or without (−Fe+Frz) iron for 7 d. C, Visualization of ferrous iron in the wild-type control, 35S::MdBT2-GFP and 35S::MdBT1/2-anti transgenic apple calli with FerroZine under iron-sufficient (+Fe) or iron-deficient (−Fe+Frz) conditions. The resultant Fe(II) was trapped by FerroZine to produce a red product. D, Fe content of the wild-type control, 35S::MdBT2-GFP and 35S::MdBT1/2-anti transgenic apple calli treated with (+Fe) or without (−Fe+Frz) Fe-EDTA for 7 d. The data represent the means ± sd of three independent experiments. DW, Dry weight.
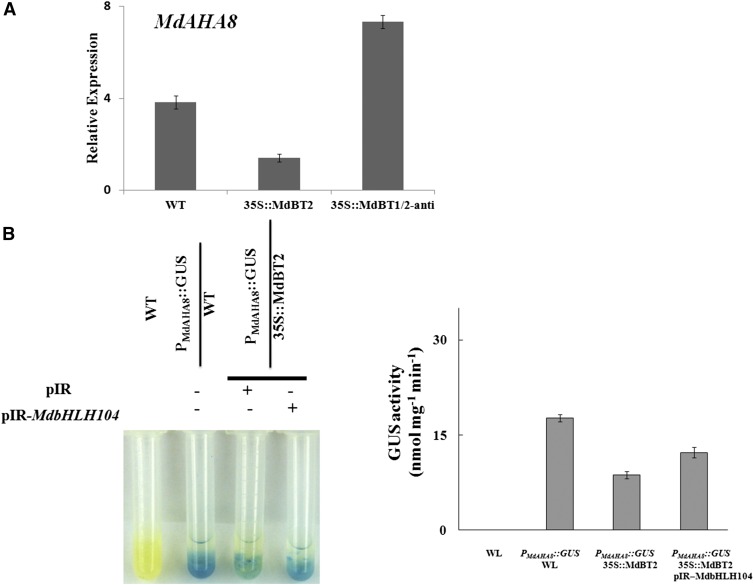
Furthermore, our previous work indicated that MdbHLH104 directly regulates the expression of the MdAHA8 gene (Zhao et al., 2016). Here, it was found that 35S::MdBT2 and 35S::MdBT1/2-anti transgenic calli generated fewer and more MdAHA8 transcripts, respectively, than the wild-type control, indicating that MdBT2 inhibited the expression of the MdAHA8 gene (Fig. 9A). To determine if MdbHLH104 is involved in the suppression of MdAHA8 gene expression by MdBT2, the viral vectors pIR and pIR-MdbHLH104 were used to transiently transform PMdAHA8::GUS and PMdAHA8::GUS+35S::MdBT2 transgenic calli, respectively. The GUS reporter assay demonstrated that PMdAHA8::GUS+35S::MdbBT2 calli exhibited lower GUS activity than PMdAHA8::GUS calli, indicating that MdBT2 inhibited the transcriptional activity of the MdAHA8 promoter. However, transient overexpression of MdbHLH104 counteracted the MdBT2 reduction of GUS activity (Fig. 9B), indicating that MdBT2 represses the effects of ectopic MdbHLH104 on the transcriptional activity of the MdAHA8 promoter.
Figure 9.
The MdBT2-MdbHLH104 pathway negatively regulates the expression of MdAHA8. A, qRT-PCR analysis of the expression of MdAHA8 in the transgenic apple calli and the wild-type control. B, Promoter activity assays using GUS staining and activity assays. The data represent means ± sd of three independent experiments.
Taken together, we concluded that MdBT2 negatively regulates Fe acquisition partially, if not completely, via the Fe-MdBTs-MdbHLH104-MdAHA8-PM H+-ATPase pathway.
DISCUSSION
In plants, both Fe deficiency and Fe overload negatively affect plant development and growth, which is at least partially due to the generation of ROS. Therefore, plants have evolved a series of metal-ion detoxification mechanisms to control Fe uptake and homeostasis to maintain appropriate Fe concentrations (Guerinot and Yi, 1994). Evidence is accumulating to indicate that the proteasome-mediated turnover of bHLH transcriptional activators is required for the uptake and homeostasis of iron (Kobayashi and Nishizawa, 2012). Generally, the Fe-deficiency-induced bHLH TFs are negatively modulated via transcriptional regulation or a UPS under conditions of prolonged Fe deficiency (Long et al., 2010; Selote et al., 2015; Sivitz et al., 2011; Le et al., 2016; Lingam et al., 2011). ZAT12 negatively regulates the function of FIT in response to H2O2 signaling triggered by prolonged Fe deficiency (Le et al., 2016). The ubiquitin E3 ligases that are involved in the proteasome-mediated turnover of the bHLH transcriptional activators FIT, ILR3, and bHLH115, but not the turnover of bHLH104, generally only work under conditions of Fe deficiency (Selote et al., 2015; Sivitz et al., 2011). The IVc subgroup bHLH TF MdbHLH104 binds directly to the promoter of the MdAHA8 gene, thereby positively regulating the activity of PM H+-ATPase and the uptake of Fe in apple (Zhao et al., 2016). It interacts with the BTB/TAZ proteins MdBT1 and MdBT2, respectively (Fig. 3, A–C).
In various organisms, BTB/POZ proteins operate as scaffold proteins, especially in the CRL3 complex, where they interact with the ubiquitin ligase CUL3 through their BTB domains and select target proteins for ubiquitination through a second protein-protein interaction domain (Pintard et al., 2003). For example, the Arabidopsis BTB/POZ protein NPR1 interacts with the TF TGA2.2 to negatively regulate its stability during the systemic acquired response (Boyle et al., 2009). Based on their secondary domains, BTB/POZ proteins are classified into different subfamilies (Chen et al., 2009), including the BTB/TAZ subfamily, which is specific to plants. There are five BTB/TAZ (BT) members, i.e. BT1 to BT5 in Arabidopsis (Du and Poovaiah, 2004), and also the five members MdBT1, MdBT2, MdBT3.1, MdBT3.2, and MdBT4 in apple (Supplemental Fig. S1A). All of these proteins share two similar structures, i.e. BTB domains in their N-terminal parts and TAZ domains in their C-terminal parts (Supplemental Fig. S1B).
The TAZ domain of the BTB-TAZ proteins is a versatile protein-protein binding domain involved in interactions with various TFs (Chan and La Thangue, 2001). In apple, the TAZ domains of MdBT1 and MdBT2 have been shown to interact with the C terminus of the MdbHLH104 protein (Fig. 3, D and E). In addition, the BTB domain of BT proteins are responsible for their interactions with CUL3s (CUL3a and CUL3b) and their associated partners, such as RBX1 and bromodomain proteins (AtBET10 and AtBET9), to form the CRL3 complex (Figueroa et al., 2005). Most recently, it was found that the Arabidopsis BTB protein Light-Response BTB (LRB) interacts with CUL3 through its BTB domain to assemble the CRL3 complex. Meanwhile, LRB binds to PIF3-PHYB and delivers them to the CRL3 complex for polyubiquitination and degradation via the 26S proteasome (Ni et al., 2014). Like LRB in Arabidopsis, MdBT2 interacted with MdCUL3 and mediated the ubiquitination and degradation of the MdbHLH104 protein (Fig. 5).
In mammals and plants, a few proteins such as FBXL5, OsHRZ1, OsHRZ2, and AtBTS are reported to be Fe sensors (Kobayashi et al., 2013; Salahudeen et al., 2009; Selote et al., 2015; Vashisht et al., 2009). Among them, the human FBXL5 protein accumulates under iron-replete conditions, whereas it degrades upon iron depletion. However, its gene is constitutively expressed regardless of Fe status (Salahudeen et al., 2009; Vashisht et al., 2009). In contrast with FBXL5, OsHRZs and AtBTS are transcriptionally induced under conditions of Fe deficiency (Kobayashi et al., 2013; Selote et al., 2015). At the posttranslational level, OsHRZs are degraded under both iron-replete and -deplete conditions (Kobayashi et al., 2013), whereas BTS degrades under iron-replete conditions and accumulates during iron depletion (Selote et al., 2015). Similar to these putative Fe sensors, the abundancy of MdBT1 and MdBT2 proteins was influenced by iron. Their transcript levels and protein abundancy were decreased after Fe treatment (Fig. 6, A–C; Supplemental Fig. S5C), which is different from the OsHRZs and BTS but similar to the FBXL5 responses to Fe status (Salahudeen et al., 2009; Vashisht et al., 2009).
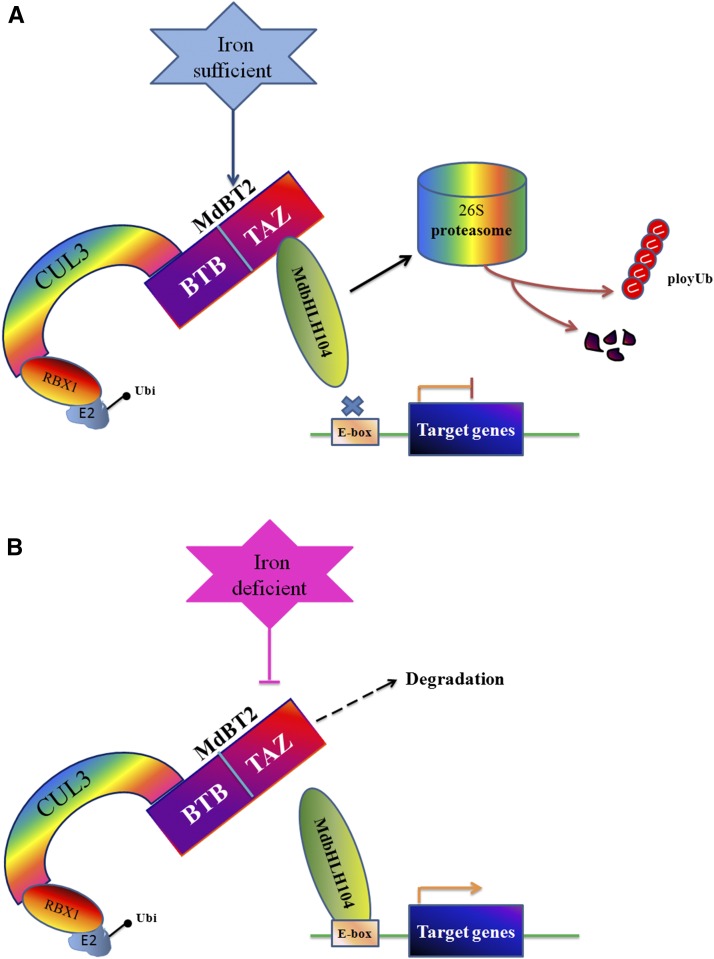
Under prolonged conditions of Fe deficiency, OsHRZs and BTS function as negative modulators of Fe-deficiency-inducible TFs via protein degradation to limit Fe uptake and translocation, thereby preventing cellular Fe toxicity. This regulatory mechanism enables them to mediate the degradation of Fe-deficiency-inducible proteins when iron is once again available under low iron conditions (Long et al., 2010; Selote et al., 2015; Kobayashi et al., 2013). The MdBT proteins are upregulated under conditions of Fe sufficiency. In this case, they accumulate and recruit the MdCUL3-containing CRL3 (MdBT2MdCUL3) complex to degrade the MdbHLH104 protein via the 26S proteasome pathway, thereby repressing the expression of target genes involved in Fe acquisition to inhibit iron acquisition, maintain homeostasis, and prevent the generation of ROS. In contrast, they are downregulated under Fe-deficient conditions. As a result, MdbHLH104 abundance enhances and activates the expression of target genes to promote Fe uptake. A model summarizing our findings regarding this regulatory pathway, i.e. the MdBT2MdCUL3-MdbHLH104-iron response and homeostasis pathway, is presented in Figure 10. Our study provides novel evidence that BTB-TAZ proteins bridge and scaffold the CRL3 complex and that the target protein MdbHLH104 can modulate iron homeostasis in response to Fe status.
Figure 10.
Model for the regulation of Fe uptake and homeostasis by MdBT2 by posttranslational modulation of the MdbHLH104 protein. High levels of iron result in the increased accumulation of the MdBTs. The stabilized MdBT proteins interact with MdCUL3s to form an active E3 ligase complex and catalyze the ubiquitination modification and the subsequent degradation of MdbHLH104 proteins via the 26S proteasome pathway. Finally, MdBTs regulate the expression of downstream target genes, such as MdAHA8, and modulate Fe absorption and homeostasis. Under low iron conditions, MdBTs are destabilized, resulting in the accumulation of MdbHLH104 protein to high levels, which promotes Fe uptake.
As is well known, Fe uptake and transportation in plants are influenced by various internal and external stimuli, such as phytohormones (ethylene, cytokinins and abscisic acid, gibberellins, jasmonic acid, brassinosteroids, and auxins) and signaling molecules (nitric oxide; Brumbarova et al., 2015). For example, ETHYLENE-INSENSITIVE3 (EIN3) and ETHYLENE-INSENSITIVE3-LIKE1 (EIL1) interact with FIT and prevent its degradation via the UPS under conditions of Fe deficiency, indicating that ethylene signaling is involved in the posttranslational turnover of the FIT protein (Sivitz et al., 2011; Lingam et al., 2011).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tissue cultures of Malus × domestica cv Royal Gala apples were used as the wild type in this study. The ‘Gala’ cultures and transgenic apple plantlets were grown on Murashige and Skoog medium containing 0.6 mg L−1 6-benzylaminopurine (6-BA), 0.2 mg L−1 indole-3-acetic acid, and 0.1 mg L−1 gibberellins and rooted in Murashige and Skoog medium containing 0.15 mg L−1 naphthaleneacetic acid at 25°C under long-day conditions (16 h light/8 h dark). For normal soil growth, apple plantlets were transferred to pots containing a mixture of soil/perlite (1:1) and grown in the greenhouse under a 16-h/8-h light/dark and 25°C/20°C day/night cycle. ‘Orin’ apple calli were cultured on Murashige and Skoog medium containing 1.5 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.4 mg L−1 6-BA at 25°C ± 1°C under dark conditions.
For phenotypic analyses of the Fe-deprivation response, the apple calli were transferred to iron-sufficient (+Fe) media (Murashige and Skoog media plus 1.5 mg L−1 2,4-D and 0.4 mg L−1 6-BA) or iron-deficient (−Fe+Frz) media (Murashige and Skoog medium without Fe-EDTA containing 1.5 mg L−1 2,4-D, 0.4 mg L−1 6-BA, and 100 mm ferrozine).
Plasmid Construction and Genetic Transformation
All primers used are presented in Supplemental Table S1. After PCR, the products were cloned into the vector pMD18-T (TaKaRa). All of the cloned genes were double-digested with SalI and BamHI/EcoRI and ligated into the 35S-GFP/MYC vector, so that the genes were under the control of the CaMV 35S promoter.
The viral vector pIR was used (Peretz et al., 2007). To construct the pIR-MdCUL3-anti silencing vector, the 5′-UTRs of either MdCUL3 or MdbHLH104 (up to 300 bp) were inserted into pIR. To construct the pIR-MdCUL3 or pIR-MdbHLH104 overexpression vector, the open reading frames of MdCUL3 or MdbHLH104 were inserted into the pIR vector between the AvrII and EcoRI sites. The IL-60-1 vector was used as a helper plasmid.
Agrobacterium tumefaciens strain LBA4404 containing the binary constructs were used to transform apple calli as reported by Xie et al. (2012).
Analysis of Gene Expression
Total RNA was extracted from apple calli and transgenic apple lines using the TRIzol Reagent (Invitrogen) and RNAplant Plus Reagent (Tiangen) according to the manufacturers’ instructions. First-strand cDNA was synthesized using the PrimeScript First Strand cDNA Synthesis Kit (TaKaRa) by following the manufacturer’s instructions.
The 20 μL qPCR solutions contained 10 μL of SYBR Green PCR Master Mix, 0.25 μm forward and 0.25 μm reverse primers, and 50 ng of cDNA template. qRT-PCR reactions (95°C, 7 min; 95°C, 15 s; 60°C, 1 min; 40 cycles) were performed by using the SYBR Green method on the iCycler iQ5 Detection System (Bio-Rad). Relative gene expression analyses were calculated by the full quantification method with MdACTIN (CN938023) as the internal control gene. At least three biological replicates were performed for each individual experiment. Primers that were used for qRT-PCR are shown in Supplemental Table S2.
Binary Vectors, Transformation, and Induction of Hairy Roots
The binary vectors 35S::GUS, 35S::MdBT1-GUS, and 35S::MdBT2-GUS containing the GUS reporter gene were introduced into the Agrobacterium rhizogenes K599 strain by electroporation (Eppendorf Electroporator model 2510; 1.8 kV), and the resultant A. rhizogenes K599 containing the binary vector was grown in liquid Luria-Bertani medium containing 50 mg L−1 kanamycin in the dark at 28°C. To obtain fresh cells, a single A. rhizogenes K599 cell colony was inoculated in 10 mL of liquid Luria-Bertani medium containing 50 mg L−1 kanamycin, diluted 1:1000, and incubated overnight with 100 μm acetosyringone at 28°C until the OD600 reached 0.5 to 0.6.
For transformation, the 35S::MdbHLH104-GFP transgenic plants were cut into segments of approximately 1.5 cm in length, immersed in the A. rhizogenes solution, and shaken for 15 to 20 min at 25°C. The inoculated explants were dried on sterile filter paper, transferred to a coculture medium (Murashige and Skoog medium containing 3% Suc and 0.6% agar), and cultured in the dark for 2 to 3 d. After cocultivation, the explants were washed in sterile distilled water three times with 250 mg L−1 carbenicillin for 5 min each time. The explants were then transferred to solid half-strength Murashige and Skoog medium containing 20 mg L−1 kanamycin and 250 mg L−1 carbenicillin for root induction under light/dark cycles of 16 h/8 h and day/night temperatures of 25°C/20°C.
Phenotypic Analyses
Rhizosphere acidification was analyzed as previously described (Yi and Guerinot, 1996). Wild-type and transgenic apple calli were grown on iron-sufficient media (+Fe) for 10 d and then transferred to iron-deficient media (−Fe+Frz) for 5 d. They were finally transferred to 1% agar plates containing 0.006% Bromocresol Purple and 0.2 mm CaSO4 (pH adjusted to 6.5 with NaOH) for 24 h. Acidification was indicated by the appearance of a yellow color.
For visualization of ferrous iron, the FerroZine reagent was used; it forms a red-colored complex with ferrous, but not ferric, iron (Stookey, 1970).
For iron content analysis, transgenic apple calli were dried for 2 d at 80°C and then wet ashed with 2 mL of 13.4 m HNO3 and 2 mL of 8.8 m H2O2 for 1 h at 220°C using a muffle furnace. Measurement of the iron concentration was carried out as described by Kobayashi et al. (2013).
Perls Staining for Fe, ROS Accumulation, and Histochemical Staining Analysis in Leaves and Roots
For Fe3+ localization, the leaves of wild-type and transgenic apple lines were first incubated with a fixative solution (methanol/chloroform/acetic acid, 6:3:1) for 1 h at room temperature and then vacuum infiltrated with Perls stain solution (equal volumes of 4% [v/v] HCl and 4% [w/v] K-ferrocyanide) for 60 min.
The distribution of Fe in roots was determined by Perls and Perls/DAB staining. Transgenic and wild-type apple plantlets were maintained in Fe-sufficient or -deficient media for 10 d. For Perls staining, roots were treated with equal volumes of 4% (v/v) HCl and 4% (w/v) K-ferrocyanide for 60 min. Perls/DAB staining was performed as previously described (Meguro et al., 2003; Roschzttardtz et al., 2009). First, the roots were placed into Perls stain solution for 60 min. The roots were then washed with distilled water and incubated in methanol containing 10 mm Na-azide and 0.3% (v/v) H2O2 for 90 min followed by a wash with 0.1 m Na-phosphate buffer (pH 7.4). Lastly, the roots were incubated in 0.1 m Na-phosphate buffer (pH 7.4) containing 0.025% (w/v) DAB and 0.005% (v/v) H2O2 for 50 min. Staining was stopped by washing the roots with distilled water. The roots were analyzed using a stereo microscope (Olympus).
The O2·− productivity rate and H2O2 content were quantified according to the method of Huang et al. (2013). In situ O2·− and H2O2 accumulation and cell death in leaves were examined via histochemical staining with DAB, NBT (Romero-Puertas et al., 2004), and Trypan blue (Huang et al., 2013), respectively.
The presence of O2·− and ROS in roots was determined by staining with 0.5 mm NBT and the fluorescent dye C-H2DCFDA (Sigma-Aldrich) as described (Müller et al., 2015). For C-H2DCFDA staining, roots were treated with 20 µm C-H2DCFDA (Invitrogen) for 30 min and subsequently imaged with an Axio Observer D1 microscope equipped with a CCD camera (Zeiss). For NBT staining, roots were incubated for 45 min in a 0.5 mg mL−1 NBT solution in 100 mm Na-phosphate buffer (pH 7.2).
Y2H Assays
Y2H studies were carried out as described by Xie et al. (2012). The MdbHLH104 cDNA was amplified (see Supplemental Table S3 for primers) and inserted into the EcoRI and SalI sites of pGAD424. The MdBT1and MdBT2 cDNAs were amplified by PCR (see Supplemental Table S3 for primers) and inserted into the BamHI and SalI sites of pGBT9. The pGAD424-MdbHLH104, pGBT9-MdBT1, and pGBT9-MdBT2 plasmids were cotransformed into yeast Y2HGold (Clontech). The yeasts were grown on -Leu/-Trp selection medium for the transformation control and on -Leu/-Trp/-His/-Ade selection medium with or without 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid for the interaction studies.
Pull-Down Analysis
The full-length cDNAs of MdbHLH104 and MdCUL3 were amplified by PCR (see Supplemental Table S3 for primers) with additional restriction enzyme sites and inserted into the EcoRI-SalI sites of pET-32a to produce a His-tagged recombinant protein. The full-length MdBT1 and MdBT2 coding regions were cloned into the EcoRI-SalI sites of pGEX-4T-1 to produce a GST fusion protein (see Supplemental Table S3 for primers). For protein expression, the plasmids were transformed into Escherichia coli BL21 (DE3; Transgene) and induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside in Luria-Bertani broth for 6 h at 16°C. For pull-down analysis with the GST- and His-tagged proteins, GST-MdBT1/2 proteins were eluted from glutathione-agarose beads before incubation with His-MdbHLH104 or His-MdCUL3 that remained attached to the tetradentate-chelated nickel resin. In general, proteins were incubated for at least 4 h at 4°C with shaking before being centrifuged. Precipitates were washed no less than three times to remove nonspecific binding and boiled (10 min, 100°C).
Co-IP Assays
For the IP assays, 1 mg of freshly extracted protein was precleaned with 30 mL of Protein A agarose beads (4 h, 4°C). The beads were centrifuged, and the supernatant was transferred into a fresh tube and incubated with anti-GFP/MYC/FLAG antibody (overnight, 4°C). After brief centrifugation, four washing steps followed, after which loading buffer was added to the precipitates and boiled as described above.
For pull-down and IP studies, precipitates were further analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting using standard procedures.
Protein Extraction and Western Blotting
A total of 2 g of transgenic apple calli or apple plants for each sample were ground in the following buffer: 20 mm Tris (pH 7.4), 100 mm NaCl, 0.5% Nonidet P-40, 0.5 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, and 0.5% Protease Inhibitor Cocktail (Sigma-Aldrich). MdbHLH104 protein levels were determined by protein gel blotting using an anti-GFP or anti-MYC antibody as described previously (Feng et al., 2012). Protein extracts were separated on a 12% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes (Roche) using an electrotransfer apparatus (Bio-Rad). The membranes were incubated with anti-MYC, anti-GFP (Sigma-Aldrich), or anti-ubiquitin (Sigma-Aldrich) primary antibodies and then peroxidase-conjugated secondary antibodies (Abcam) before visualization of immunoreactive proteins using an ECL detection kit (Millipore). ACTIN served as a protein loading control.
Ubiquitination Assay
For in vivo detection of ubiquitinated MdbHLH104, apple calli were cotransfected with 35S::MdbHLH104-MYC or 35S::MdbHLH104-GFP and 35S::MdBT2 with or without 35S::MdCUL3. After incubation for 5 d, 50 µm MG132 was added to the protoplasts, and they were incubated for another 6 h. Total proteins were extracted as mentioned above. The protein complexes were immunoprecipitated using a Pierce Classic Protein A IP Kit (Thermo Fisher Scientific) with anti-GFP or anti-MYC antibody (Beyotime). The resultant proteins were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membrane (Roche). The gel blot was probed with anti-ubiquitin (Sigma-Aldrich) and anti-MYC or anti-GFP antibodies, respectively, and was visualized by chemiluminescence with the ECL Plus detection kit (Millipore) according to the manufacturer’s instructions.
Cell-Free Degradation
Cells (E. coli, BL21) were induced by 0.1 mm isopropyl β-d-1-thiogalactopyranoside and allowed to grow for 4 h at 16°C. His-MdbHLH104 protein was eluted from the tetradentate-chelated nickel resin. The total proteins of the transgenic apple calli were subsequently extracted in degradation buffer containing 25 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm MgCl2, 4 mm phenylmethylsulfonyl fluoride, 5 mm DTT, and 10 mm ATP as previously described (Wang et al., 2009). The supernatant was collected, and the protein concentration was determined by the using the Bradford assay reagent (Bio-Rad). Each reaction mix contained 100 ng of His-MdbHLH104 and 500 µg of total protein from transgenic apple calli. For the proteasome inhibitor experiments, 50 µm MG132 was added 30 min prior to the experiment. The reaction mixes were incubated at 22°C, and the reactions were stopped by the addition of SDS-PAGE sample buffer and boiling (10 min, 100°C). The results were quantified using Quantity One 1-D Analysis Software (Bio-Rad).
Isolation of PM H+-ATPase and Activity Assays
Transgenic and wild type apple calli were grown in Fe-sufficient medium (+Fe) and then transferred to Fe-deficient medium (−Fe+Frz). The PMs were isolated as described (Yang et al., 2010).
For the measurement of PM H+-ATPase activity, a pH gradient (∆pH) was formed in the vesicles (acidic inside) by the activity of the H+-ATPase and measured as a decrease (quench) in the fluorescence of the pH-sensitive fluorescent probe quinacrine as described (Yang et al., 2010). The assay reactions contained 3 mm MgSO4, 100 mm KCl, 25 mm BTP-MES-HEPES (Sigma-Aldrich), pH 6.5, 250 mm mannitol, 10 μm quinacrine, and 50 mg mL−1 of PM protein. The reactions were mixed and placed in the dark for 5 min. The reactions were then initiated by the addition of 3 mm ATP and the formation of ∆pH was measured at the wavelengths Ex = 430 nm and Ex = 500 nm using a fluorescence spectrophotometer (Hitachi F-4010). After 5 min, 10 μm m-chlorophenylhydrazone was added to stop the reaction. The change in fluorescence was expressed per mass of PM protein in the reaction per unit time (∆F/min per mg of protein) were used to calculate.
GUS Analysis
For histochemical staining, transgenic apple calli were immersed in GUS staining buffer (1 mm 5-bromo-4-chloro-3-indolyl-β-D-GlcA solution in 100 mm sodium phosphate pH 7.0, 0.1 mm EDTA, 0.5 mm ferrocyanide, 0.5 mm ferricyanide, and 0.1% Triton X-100) at 37°C overnight.
GUS activity was determined using the method described by Jefferson et al. (1987).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis and alignments of Arabidopsis and apple BTs and Y2H analyses of the interactions between MdbHLH104 and each of MdBT3.1, MdBT3.2, and MdBT4.
Supplemental Figure S2. Expression analysis of the MdBT genes by qRT-PCR in different transgenic apple calli.
Supplemental Figure S3. Amino acid alignment of MdCUL3 proteins and the results of the Y2H assay to test for the interaction between MdBT2 and MdCUL3-1.
Supplemental Figure S4. qRT-PCR analysis of MdCUL3 genes in different transgenic apple calli and a cell-free assay of the MdCUL3-1-mediated degradation of the MdbHLH104 protein.
Supplemental Figure S5. MdCUL3s and MdBTs are necessary for the degradation of MdbHLH104 protein in an in vitro cell-free system.
Supplemental Figure S6. Identification of the transgenic apple hairy roots obtained by an A. rhizogenes-mediated transformation method.
Supplemental Figure S7. Fe accumulation and ROS formation in the roots of the transgenic plants and the wild-type control under iron-sufficient and -deficient conditions.
Supplemental Table S1. Primers used for gene cloning.
Supplemental Table S2. Primers used for qRT-PCR analysis of different genes.
Supplemental Table S3. Primers used for Y2H and pull-down assays.
Acknowledgments
We thank Dr. Ilan Sela for IL-60-BS and pIR binary vectors and Dr. Takaya Moriguchi of the National Institute of Fruit Tree Science, Japan, for ‘Orin’ apple calli.
Glossary
- TF
transcription factor
- bHLH
basic helix-loop-helix
- ROS
reactive oxygen species
- UPS
ubiquitin-proteasome system
- PM
plasma membrane
- CRL3
Cullin-RING ubiquitin ligase 3
Footnotes
This work was supported by grants from National Natural Science Foundation of China (31430074 and 31272142), Ministry of Agriculture (201203075-3), Ministry of Education of China (IRT15R42), and Shandong Province (SDAIT-06-03).
Articles can be viewed without a subscription.
References
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20: 124–133 [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114: 2363–2373 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35: 841–855 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Poovaiah BW (2004) A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol Biol 54: 549–569 [DOI] [PubMed] [Google Scholar]
- Feng XM, Zhao Q, Zhao LL, Qiao Y, Xie XB, Li HF, Yao YX, You CX, Hao YJ (2012) The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, et al. (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, Welch RM, Kochian LV (1990) Physiological characterization of a single-gene mutant of Pisum sativum exhibiting excess iron accumulation I. Root iron reduction and iron uptake. Plant Physiol 93: 976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC (2004) Balancing acts: molecular control of mammalian iron metabolism. Cell 117: 285–297 [DOI] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Huang XS, Wang W, Zhang Q, Liu JH (2013) A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162: 1178–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5: 27–42 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK (2013) Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat Commun 4: 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Le CTT, Brumbarova T, Ivanov R, Stoof C, Weber E, Mohrbacher J, Fink-Straube C, Bauer P (2016) ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol 170: 540–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Schmidt W (2010) A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J 62: 330–343 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol 170: 2478–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro R, Asano Y, Iwatsuki H, Shoumura K (2003) Perfusion-Perls and -Turnbull methods supplemented by DAB intensification for nonheme iron histochemistry: demonstration of the superior sensitivity of the methods in the liver, spleen, and stomach of the rat. Histochem Cell Biol 120: 73–82 [DOI] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33: 216–230 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK (2011) OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol 75: 593–605 [DOI] [PubMed] [Google Scholar]
- Peretz Y, Mozes-Koch R, Akad F, Tanne E, Czosnek H, Sela I (2007) A universal expression/silencing vector in plants. Plant Physiol 145: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JLF, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. (2003) The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Rodríguez-Serrano M, Corpas FJ, Gómez M, Del Río LA, Sandalio LM (2004) Cadmium-induced subcellular accumulation of O2·− and H2O2 in pea leaves. Plant Cell Environ 27: 1122–1134 [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Stookey LL. (1970) Ferrozine-a new spectrophotometric reagent for iron. Anal Chem 42: 779–781 [Google Scholar]
- Thomine S, Vert G (2013) Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol 16: 322–327 [DOI] [PubMed] [Google Scholar]
- Thongbai P, Goodman BA (2000) Free radical generation and post-anoxic injury in rice grown in an iron-toxic soil. J Plant Nutr 23: 1887–1900 [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ying Y, Narsai R, Ye L, Zheng L, Tian J, Whelan J, Shou H (2013) Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ 36: 224–236 [DOI] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, et al. (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Song A, Li P, Chen S, Jiang J, Chen F (2014) A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci Rep 4: 6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ren YR, Wang QJ, Yao YX, You CX, Hao YJ (2016) Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol J 14: 1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Ying Y, Wang L, Wang F, Whelan J, Shou H (2010) Identification of a novel iron regulated basic helix-loophelix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]