Multiple levels of functional divergence contributed to gene retention after whole-genome duplication in the soybean membrane-bound NAC transcription factors gene family.
Abstract
Functional divergence is thought to be an important evolutionary driving force for the retention of duplicate genes. We reconstructed the evolutionary history of soybean (Glycine max) membrane-bound NAC transcription factor (NTL) genes. NTLs are thought to be components of stress signaling and unique in their requirement for proteolytic cleavage to free them from the membrane. Most of the 15 GmNTL genes appear to have evolved under strong purifying selection. By analyzing the phylogenetic tree and gene synteny, we identified seven duplicate gene pairs generated by the latest whole-genome duplication. The members of each pair were shown to have variously diverged at the transcriptional (organ specificity and responsiveness to stress), posttranscriptional (alternative splicing), and protein (proteolysis-mediated membrane release and transactivation activity) levels. The dormant (full-length protein) and active (protein without a transmembrane motif) forms of one pair of duplicated gene products (GmNTL1/GmNLT11) were each separately constitutively expressed in Arabidopsis (Arabidopsis thaliana). The heteroexpression of active but not dormant forms of these proteins caused improved tolerance to abiotic stresses, suggesting that membrane release was required for their functionality. Arabidopsis carrying the dormant form of GmNTL1 was more tolerant to hydrogen peroxide, which induces its membrane release. Tolerance was not increased in the line carrying dormant GmNTL11, which was not released by hydrogen peroxide treatment. Thus, NTL-release pattern changes may cause phenotypic divergence. It was concluded that a variety of functional divergences contributed to the retention of these GmNTL duplicates.
Plant genomes have undergone several rounds of whole-genome duplication (WGD), which have had a significant impact on genome stability, molecular functions, physiology, and adaptation (Adams and Wendel, 2005; De Smet and Van de Peer, 2012; Moghe and Shiu, 2014). Duplicate genes derived from these WGD events as well as tandem duplication, segmental duplication, and transposition are considered the primary source of new genes and novel functions (Ohno, 1970; Zhang, 2003). During evolution, many duplicate genes lost their function and were eventually removed. However, some duplicates were retained and have evolved diverse functions (Blanc and Wolfe, 2004; Roulin et al., 2013; Rensing, 2014; Freeling et al., 2015). To explain how/why duplicated genes are retained after WGD, models have been proposed in which duplicated genes are pseudogenized (loss of regulatory subfunction; Moore and Purugganan, 2005), subfunctionalized (partitioning of the function between daughter copies; Cusack and Wolfe, 2007), and/or neofunctionalized (functional diversification; Ohno, 1970; Force et al., 1999; Zhang, 2003; Blanc and Wolfe, 2004). These provide testable hypotheses suggesting that subfunctionalized gene copies undergo purifying selection, whereas neofunctionalized gene copies are expected to undergo positive selection or relaxed purifying selection. It has been suggested that the functional divergence between duplicated genes can affect either the gene expression pattern or the protein biochemical properties. The prevalence of expression subfunctionalization after polyploidization (variation in the relative expression of homologs among tissues in the polyploids) has been assessed in a few studies (Adams et al., 2003; Flagel et al., 2008; Chaudhary et al., 2009; Buggs et al., 2010; Flagel and Wendel, 2010; Guo et al., 2010). Differential expression between duplicated genes has been shown to contribute to phenotypic variation (Buggs et al., 2010). A genome-wide analysis of soybean (Glycine max) revealed that approximately 50% of paralogs were differentially expressed and, thus, had undergone expression subfunctionalization (Roulin et al., 2013). Recent studies indicated that the changes in gene body DNA methylation or microRNA-binding sites could provide another avenue for duplicate genes to develop differential expression patterns (Keller and Yi, 2014; Wang et al., 2014; Wang and Adams, 2015). Other studies demonstrated or suggested that the divergence in protein characteristics, which includes enzyme activity (Lan et al., 2009; Liu et al., 2015), posttranslational regulatory motifs (Nguyen Ba et al., 2014), DNA-binding specificity (Lehti-Shiu et al., 2015), and subcellular localization (Ren et al., 2014), plays an important role in the subfunctionalization/neofunctionalization of duplicate genes. The experimental evidence of how the functional divergence happened in the expansion of a specific gene family still remains largely unknown.
Transcription factor (TF) genes tend to be retained after duplication (Blanc and Wolfe, 2004; Seoighe and Gehring, 2004; Maere et al., 2005; Shiu et al., 2005). Their expansion often correlates with critical events in the evolution of plants in either development innovation (Liu and Meinke, 1998; Xiao et al., 2008; Blackman et al., 2010; Airoldi and Davies, 2012; Zhang et al., 2013; Rensing, 2014) or stress response diversification (Haake et al., 2002; Mizoi et al., 2012; Kang et al., 2013; Yang et al., 2014). This suggests that the expansion of TF families may provide an adaptive benefit (Lang et al., 2010). Studying the mechanisms underlying the retention of TF duplicate genes can give insights into how plants adapt to abiotic stress.
Membrane-bound transcription factors (MTFs) are recognized by the presence of a distinctive transmembrane domain, which directs their association with the endoplasmic reticulum (ER), nuclear membrane, or plasma membrane. Upon the plant’s exposure to specific developmental or environmental cues, MTFs are proteolytically processed, then transported to the nucleus, where they exercise their regulatory function (Chen et al., 2008). The NTM1 MTF is one of the NAC family proteins (Kim et al., 2006) that feature a highly conserved N-terminal DNA-binding NAC domain, a variable transcription-regulating C-terminal domain, and a C-terminal transmembrane (TM) motif (Ernst et al., 2004). It has been proposed that there are 13 NTM1-like (NTL) sequences in the Arabidopsis (Arabidopsis thaliana) genome (Kim et al., 2007b), 11 in the soybean (Le et al., 2011) and Brachypodium distachyon (Zhu et al., 2015) genomes, seven in the maize (Zea mays) genome (Shen et al., 2009), six in the grapevine (Vitis vinifera) genome (Shen et al., 2009), five in each of the rice (Oryza sativa; Kim et al., 2010), poplar (Populus spp.), switchgrass (Panicum virgatum), and sorghum (Sorghum bicolor) genomes (Shen et al., 2009), 14 in the potato (Solanum tuberosum) genome (Singh et al., 2013), and eight in the Setaria italica genome (Puranik et al., 2013). The transcription of most of the AtNTL genes is inducible by abiotic stress (Kim et al., 2006, 2007b, 2008; Park et al., 2011). The movement of AtNTL proteins into the nucleus follows the proteolytic cleavage of their anchor by an intramembrane protease(s), a process that also is mediated by various stresses (Kim et al., 2008; Yoon et al., 2008; Seo et al., 2010; Park et al., 2011). The biological function of most AtNTLs has been associated with different kinds of stress response (Kim et al., 2007a, 2008; Yoon et al., 2008; Seo et al., 2010; Park et al., 2011; Klein et al., 2012; Lee et al., 2012; De Clercq et al., 2013; Ng et al., 2013). Overall, the NTLs make an important contribution to the interaction with the environment, and their function could be affected by multiple factors, including transcription, membrane release, and transactivation activity. This makes them good models for the study of the variation of functional divergence of duplicate genes in the stress response.
The extant soybean genome represents the outcome of two WGD events, one affecting the progenitor of the papilionoid legumes some 59 million years ago (Mya) and the second, occurring 13 Mya, that was specific to the ancestor of the modern soybean (Schmutz et al., 2010; Cannon et al., 2015). We took advantage of the availability of this genomic resource and the properties of NTLs to study the evolution of duplicated genes. Here, an evolutionary and functional analysis of its NTL gene content is presented. The data have revealed a picture of functional divergence at the transcriptional, posttranscriptional, and protein levels and have demonstrated the contribution of the membrane-release pattern, a new kind of functional divergence, to phenotypic divergence.
RESULTS
Identification of the GmNTL Gene Family
A survey of the soybean genome (https://phytozome.jgi.doe.gov/pz/portal.html) revealed that it has 17 potential NTL sequences, of which 15 (GmNTL1 through GmNTL15) are represented by a full-length open reading frame (Supplemental Table S1). The derived amplicon sequence in each case matched that of the respective genomic sequence. The predicted gene products were all structurally similar to that of AtNTM1, having a NAC domain in their N-terminal region and one or two α-helical TMs in their C-terminal region (Supplemental Fig. S1; Supplemental Table S1). A multiple sequence alignment involving the GmNTLs and a representative sample of Arabidopsis NAC proteins (ANAC013, ANAC055, and ANAC072) showed that the soybean sequences shared a substantial level of sequence similarity in their N-terminal domain and included five conserved subdomains (Supplemental Fig. S1). An in silico analysis placed the genes on 11 different chromosomes, with chromosomes 2, 10, 14, and 20 each having two genes and seven other chromosomes having one gene each (Supplemental Table S1).
Phylogeny of the GmNTL Proteins
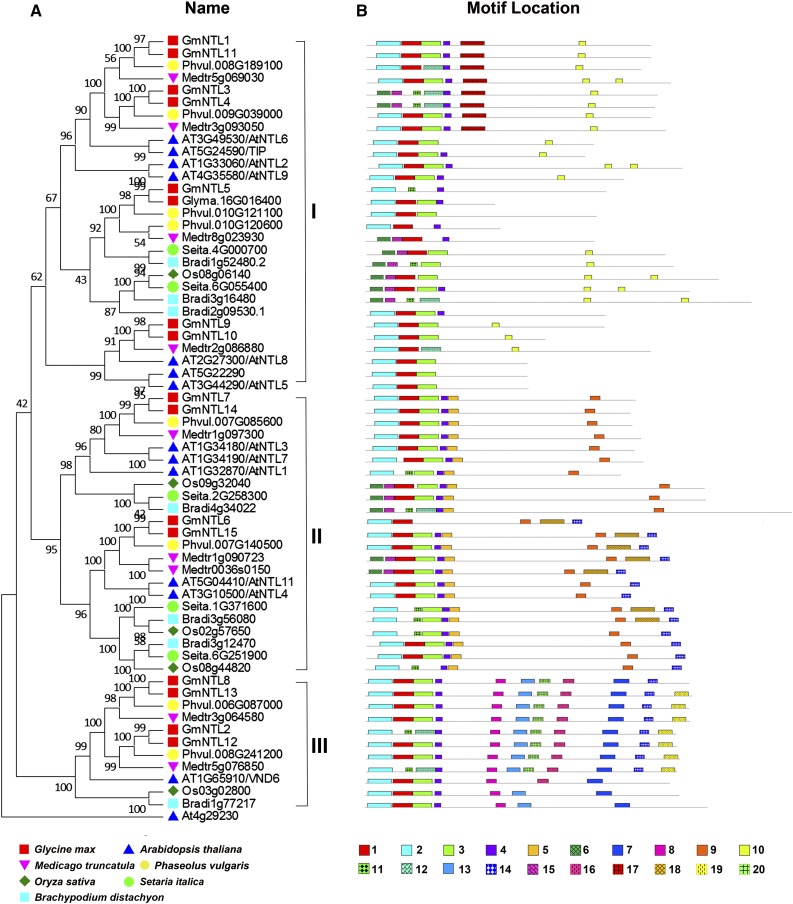
To investigate the phylogenetic relationship of GmNTL genes with those of other dicot and monocot species, a full-length peptide sequence alignment was performed among 15 GmNTLs (Glycine max), 13 AtNTLs (Arabidopsis), five OsNTLs (rice), nine MtNTLs (Medicago truncatula), five SiNTLs (S. italica), seven BdNTLs (B. distachyon), and eight PvNTLs (common bean [Phaseolus vulgaris]), and three clades generated the set of GmNTLs (Fig. 1A). Phylogenetic analysis based on the NTL coding sequences showed a similar tree structure (Supplemental Fig. S2). A total of 29 of the sequences fell into clade I, 23 were clustered into clade II, and 11 were found in clade III. As each of the clades contained sequences derived from all of the diverse species sampled (including both monocotyledonous and dicotyledonous representatives), the inference was that the NTLs within a clade probably shared a common ancestor that predated the divergence of the monocotyledons and dicotyledons. Seven soybean leaf pairs were recognized (GmNTL1/11, GmNTL3/4, GmNTL2/12, GmNTL6/15, GmNTL9/10, GmNTL8/13, and GmNTL7/14; Fig. 1A). A MEME-based analysis showed that GmNTLs clustered within a clade possessing a similar motif content. This was particularly the case for the phylogenetically closely related pairs of proteins (Fig. 1B). Most of the conserved motifs lay within the N-terminal NAC domain, suggesting their probable functional importance. Motifs 2/11/15, 1, 3, and 4 (equivalent to subdomains A/B–E, respectively; Supplemental Table S2) were present in most of the proteins. A number of motifs were present within the more variable C-terminal end of the proteins, notably motif 10 in clade I, motif 9 in clade II, and motifs 7/8/13 in clade III. This result further supports the conclusions.
Figure 1.
GmNTL phylogeny and conserved motifs. A, A maximum likelihood (ML) procedure using the Jones, Taylor, and Thornton model and 500 bootstrap replicates were used for the phylogenic reconstruction; numbers at each node of the ML tree refer to bootstrap values. The sequences used to construct the tree are given in Supplemental Data Set S1. The three clades (I–III) were determined by a phylogenetic analysis based on NTL protein sequences from soybean (red squares), Arabidopsis (blue triangles), M. truncatula (purple triangles), common bean (yellow circles), rice (green diamonds), S. italica (green circles), and B. distachyon (blue squares). The clades were defined on the basis of the similarity of motif distribution and the bootstrap values (greater than 50%). B, Conserved motifs in the NTL proteins as revealed by MEME analysis. Each motif is represented by a numbered box. The proteins are drawn to scale.
A reconstruction of the evolution of the GmNTLs was attempted by reconciling their phylogeny with their pattern of duplication. The WGD event experienced 13 Mya did not occur in the progenitor shared with the genus Phaseolus and M. truncatula (Schmutz et al., 2010; Young et al., 2011). As a result, the NTL loci present as a duplicated pair in soybean are each represented by just a single locus in Phaseolus and M. truncatula (Fig. 1A). The evidence is that, following this WGD event, seven pairs of GmNTLs having a full TM motif content were retained (Fig. 1A); these are hereafter referred to as pairs I through VII. While GmNTL5 does have a full TM motif content, its duplicate Glyma.16g016400 encodes a TM-free NAC protein (pair VIII). A synteny-based study focusing on regions of chromosomes 2, 4, 6, 7, 10 to 14, 16, 18, and 20 (Fig. 2A; Supplemental Fig. S3) showed that synteny has been maintained around GmNTL1/11 (pair I), GmNTL3/4 (pair II), GmNTL2/12 (pair III), and GmNTL8/13 (pair IV), consistent with the outcome of a genome-wide or a segmental duplication event (Fig. 2A; Supplemental Table S3). An evolutionary pathway based on the phylogeny and the pattern of gene duplication is given in Figure 2B. The likelihood is that four ancestral NTL genes were present at the moment of divergence between the monocotyledonous and dicotyledonous species, with two of them being the result of a duplication event, representing the progenitor of each of three NTL clades. One of these genes appears to have undergone at least four rounds of duplication, generating a copy number of seven in the modern soybean genome (GmNTL1/11, GmNTL3/4, GmNTL5, and GmNTL9/10), along with one NAC gene (Glyma.16g016400). The two genes generated by the ancestor of clade II were duplicated once each to give rise to GmNTL6/15 and GmNTL7/14, and the third was duplicated at least twice to produce GmNTL2/12 and GmNTL8/13.
Figure 2.
Synteny in GmNTL-flanking regions and a proposed evolutionary pathway. A, Local synteny based on gene annotation (detailed information is provided in Supplemental Table S3). Flanking genes assumed to form homologous genomic segments are connected by lines. The blocks having GmNTLs are colored red. The numbers shown at left indicate the chromosome number in which each gene resides. Bar = 10 kb. B, The most parsimonious scenario for gene duplication, loss, and rearrangement was deduced by reconciling the phylogeny with the genomic locations of each gene. Black boxes indicate GmNTL genes, and gray boxes indicate GmNAC genes lacking the TM motif. W and S, WGD and segmental duplication, respectively.
Molecular Evolution of GmNTL Sequences
The ratio of nonsynonymous to synonymous substitutions (ω = dN/dS) is an indicator of the history of selection acting on a gene or gene region. A history of purifying selection of a gene is implied when ω lies below unity, whereas an ω value in excess of unity suggests directional selection (Freeling, 2008). To infer the influence of selection on the expansion of GmNTL genes, ω values were computed for each branch of the NTL gene family tree using ML codon models (Supplemental Fig. S5). Four alternative assumptions were tested: the fixed one-ratio model, the free-ratio model, the near-neutral model, and the branch-site model. Based on the free-ratio model, ω was estimated to lie in the range 0.01 to 0.11 for all the soybean genes except GmNTL6 (Supplemental Fig. S5, A–H). Such low ratios suggested that the genes were constrained by strong purifying selection. The dN/dS of the eight pairs of branches of GmNTL were estimated using branch-site models leading to key extant genes. Among the eight branches analyzed, significant positive selection was detected only for branch f (Table I; Supplemental Fig. S5). The overall conclusion was that while most of the GmNTL duplicates have experienced strong purifying selection, the evolution of GmNTL6 was driven by positive selection, which has allowed it to neofunctionalize.
Table I. Likelihood ratio tests of the evolutionary model for all branches after gene duplication.
| Model | Branch | Estimates of Parametersa | ln L (Log-Likelihood) | 2ΔLb | P |
|---|---|---|---|---|---|
| M0 (one ratio) | 0.07432 | −9,248.27 | |||
| M1 (free ratio) | Glyma.10G197600/GmNTL6 | 16.259820 | −9,049.42 | 397.7 | <0.001 |
| M1a (near neutral) | p0 = 0.94552, p1 = 0.05448, ω0 = 0.07298, ω1 = 1 | −9,191.27 | |||
| Ma (branch site) | Glyma.10G197600/GmNTL6 | p0 = 0.84890, p1 = 0.04958, p2a = 0.09592, p2b = 0.00560, ω0 = 0.06996, ω1 = 1, ω2 = 486.62276 | −9,155.89 | 70.76 | <0.01 |
The proportion of sites (p0, p1, etc.) estimated to have ω0, ω1, etc.
2ΔL is twice the log-likelihood difference between Ma and M1a.
Gene and Protein Structure Are Conserved among GmNTL Duplicates
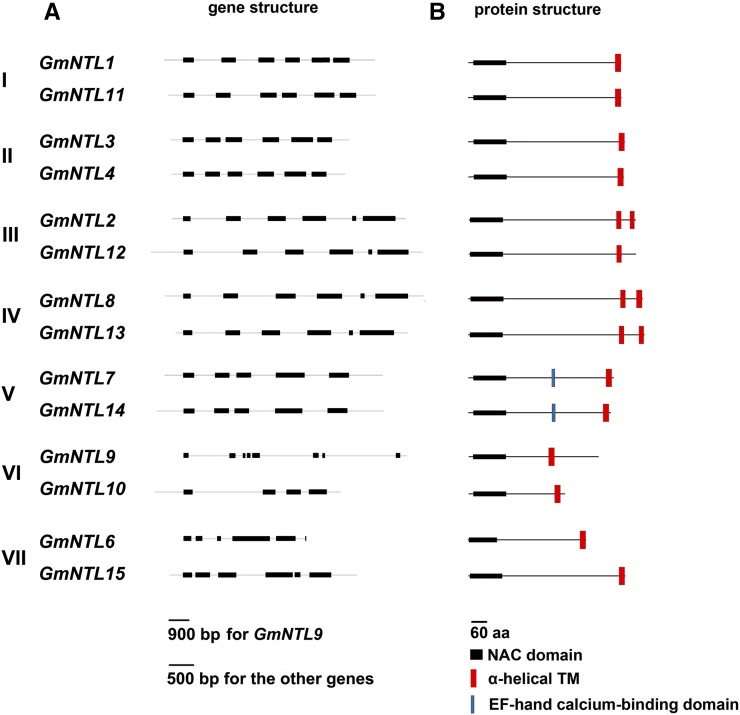
A comparison of the sequences of the various duplicate pairs was based on their gene structure and those of their predicted gene products (Fig. 3). The coding sequence of the two members of a pair was generally split into an identical number of exons: the exceptions were the genes in pairs VI and VII (Fig. 3A; Table II). The gene products were all structurally similar to that of AtNTM1, in that they had a NAC domain in their N-terminal region (Supplemental Fig. S1) along with one or two α-helical TMs in their C-terminal region (Fig. 3B; Table II; Supplemental Table S1). Within a given pair, gene product length was conserved (except for pairs VI and VII; Fig. 1C; Table II). The number and location of TMs were generally conserved within a pair (except in pairs III and VII; Fig. 1C; Table II). The divergence in gene and protein structure (including the MEME motif distribution) between GmNTL duplicates is summarized in Table II.
Figure 3.
GmNTL and GmNTL structures. A, GmNTL exon/intron structure. Exons are represented by black boxes and introns by gray lines; segment lengths are drawn to scale. B, GmNTL structure. The N-terminal region has the highly conserved NAC domain (black boxes). The α-helical TMs (red boxes) lie close to the C terminus. GmNTL7 and GmNTL14 also have an elongation factor (EF)-hand Ca-binding domain (blue lines). Protein sizes are shown to scale. The numerals I to VII refer to the seven duplicated gene pairs (Table I).
Table II. The divergence of duplicated GmNTLs.
The motif number is uniform with that in Figure 1. Dashes, No information provided.
| Pair No. | NTL Pairs | Exon No. | Size | Alter-native Transcripts | No. of TMs | MEME Motifs | Expression Pattern | Transactivation Activity | Subcellular Localization | Membrane-Release Pattern | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ Specificity | Abiotic Stress Response | Proteolysis | Alternative Splicing | |||||||||
| amino acids | ||||||||||||
| I | GmNTL1 | 6 | 589 | – | 1 | – | Root, leaf | Induced | Weak | ER | H2O2, cold | – |
| GmNTL11 | 6 | 590 | Yes | 1 | – | All organs except flower | Induced | Weak | ER | – | – | |
| II | GmNTL3 | 6 | 422 | – | 1 | – | Seed | Induced | Strong | ER | H2O2, cold | – |
| GmNTL4 | 6 | 598 | – | 1 | – | Root, stem, leaf | Induced | Strong | ER | H2O2, cold | – | |
| III | GmNTL2 | 6 | 584 | – | 2 | – | Seed, root | Induced | Weak | ER | – | – |
| GmNTL12 | 6 | 600 | – | 1 | – | Seed, stem, leaf | Induced | Weak | ER | – | – | |
| IV | GmNTL8 | 6 | 671 | – | 2 | Without motif 19 | Stem, flower | Induced | Strong | ER | Normal condition | – |
| GmNTL13 | 6 | 631 | – | 2 | With motif 19 | Root, stem, leaf | Induced | Weak | ER | – | – | |
| V | GmNTL7 | 5 | 560 | – | 1 | – | All organs except flower | Induced | Moderate | ER | – | – |
| GmNTL14 | 5 | 549 | Yes | 1 | – | All organs except flower | Induced | Moderate | ER | – | NaCl, ABA | |
| VI | GmNTL9 | 8 | 493 | – | 1 | – | Flower | – | Weak | ER | – | – |
| GmNTL10 | 4 | 371 | – | 1 | – | Root, stem, leaf | Induced | None | ER | Cold | – | |
| VII | GmNTL6 | 6 | 448 | Yes | 1 | Without motifs 3, 4, 5 | Root, leaf, flower | Repressed | Weak | ER | – | NaCl |
| GmNTL15 | 6 | 604 | – | 1 | With motifs 3, 4, 5 | Root, flower | Induced | Moderate | ER | – | – | |
Variation between Duplicates with Respect to Transcriptional Behavior
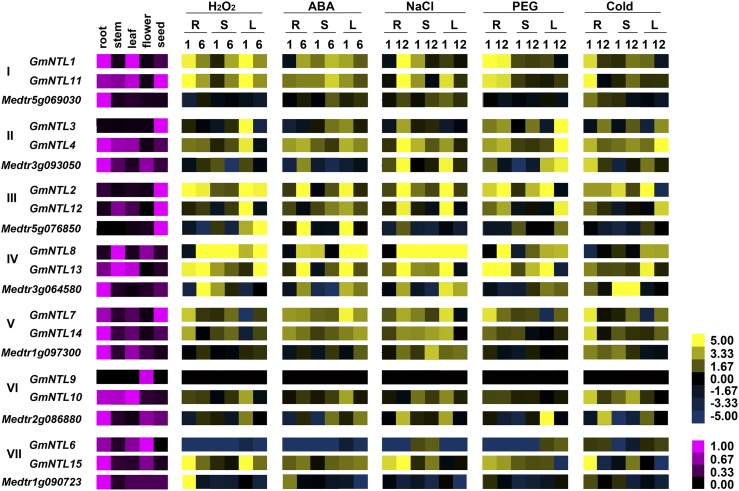
Studies have established that duplicated gene pairs can be differentially transcribed between tissues or cell types (Adams et al., 2003; Flagel et al., 2008; Chaudhary et al., 2009; Buggs et al., 2010; Flagel and Wendel, 2010; Roulin et al., 2013), and all the Arabidopsis NTL genes have been shown to transcriptionally respond to at least one kind of abiotic/biotic stress or stress-related hormone (Kim et al., 2007b; Park et al., 2011). In order to obtain insights into the functional divergence of the GmNTL genes after duplication, their expression patterns were examined in terms of both organ specificity and abiotic stress response. To determine whether differences in the pattern of transcription between duplicated GmNTLs reflect the occurrence of subfunctionalization, the M. truncatula NTLs were used as a reference. A series of quantitative real-time (qRT)-PCR experiments demonstrated that the genes in pairs III (GmNTL2 and GmNTL12) and VII (GmNTL6 and GmNTL15) shared a common pattern of transcription with respect to organ specificity, while those in pairs I (GmNTL1 and GmNTL11), IV (GmNTL8 and GmNTL13), and V (GmNTL7 and GmNTL14) exhibited a degree of divergence, while pairs II (GmNTL3 and GmNTL4) and VI (GmNTL9 and GmNTL10) were strongly divergent (Fig. 4; Table II; Supplemental Fig. S4). On the basis of the transcription patterns of the M. truncatula homologs that were not duplicated, most of the divergence between gene pairs can be interpreted as being due to subfunctionalization. When the plants were subjected to abiotic stress, pairs I, II, and V responded similarly, but for pairs IV and VII, this was not the case. Although the indication from this analysis was that GmNTL6 (pair VII) had experienced neofunctionalization (Table I; Supplemental Fig. S5), it appeared that both GmNTL6 and GmNTL15 were expression subfunctionalized, since they both partially mimic the behavior of Medtr1g090723 (Fig. 4; Table II; Supplemental Fig. S4). Additionally, these data demonstrated that the expression divergence happened not only in organ specificity but also in inducibility by external stimuli.
Figure 4.
Transcription profiling of GmNTL duplicated pairs and their homologs in M. truncatula. Transcript abundances were derived via qRT-PCR. The numerals I to VII refer to the seven duplicated gene pairs (Table I). The column at left details the organ specificity of transcription. Transcript abundances were normalized to the most abundant transcript. The other columns detail the transcriptional responses to salinity (200 mm NaCl), moisture stress (20% [w/v] polyethylene glycol [PEG] 6000), low temperature (4°C), oxidative stress (10 mm H2O2), and 100 µm abscisic acid (ABA) treatment. For the organ specificity assay, the levels were normalized against the transcript abundance in the most strongly transcribed organ. For the stress response assay, fold changes in abundance were log2 transformed. R, Root; S, stem; L, leaf; numbers below these letters indicate hours.
In an attempt to relate their stress response to promoter sequences, the 1,500-bp upstream sequence of each GmNTL coding region was queried with the PlantCARE server (Lescot et al., 2002). Various stress-related cis-acting regulatory elements were detected in the GmNTL promoters, including ABRE, TC-rich repeats, ARE, CGTCA motif, ERE, TGACG motif, SARE, HSE, LTR, MBS, Box W1, and the TCA element (Supplemental Table S4). The assumption is that the duplicated GmNTLs respond transcriptionally to multiple stresses in different ways as a result of the varied composition in their promoters of relevant cis-acting elements.
Transactivation Activity Diverged between GmNTL Genes in Duplicate Pairs
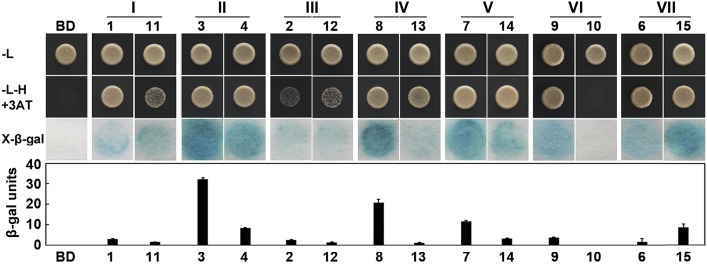
The NAC TFs are classified by the conserved N-terminal DNA-binding domain and the variable C-terminal transcriptional regulation region, which can act as either a repressor or activator in regulatory pathways (Tran et al., 2004). To further characterize their function, the transcriptional activity of the 15 GmNTLs was studied by fusing their coding sequence with the GAL4 DNA-binding domain of the yeast expression vector pDEST32 in yeast MaV203. The NTL fusion protein is expected to bind with GAL4-binding sites present in the promoters of the reporter genes lacZ and HIS3, and if they have acquired transcriptional activation activity, cells expressing them will be able to both grow on medium lacking His and show β-galactosidase activity. All of the yeast transformants, including those containing only the empty vector, grew freely on medium lacking Leu (Fig. 5). However, on medium lacking His and supplemented with 10 mm 3AT, cells having the empty vector pDEST32 could not grow at all, while those carrying GmNTLs grew to a greater or lesser extent, depending on which GmNTL was present. The transactivation activity assay (X-β-gal assay) gave a consistent result (Fig. 5). The products of the members of pairs II, IV, VI, and VII differed with respect to their transactivation activity, but members of the other pairs behaved consistently with one another (Fig. 5; Table II; Supplemental Fig. S5). Although the assay did not indicate the transactivation activity of the GmNTL protein itself (rather, it reported the combined outcomes of membrane release and stability, in addition to transactivation activity), the result indicated some measure of divergence between the products of the GmNTL duplicate pairs.
Figure 5.
GmNTL transcriptional activation activity. The numerals I to VII refer to the seven duplicated gene pairs (Table I). The ability of yeast transformants to grow on medium lacking His (H) and Leu (L) but containing 10 mm 3-aminotriazole (3AT), and the formation of color in the X-β-gal assay, indicate transcriptional activation. BD, Empty vector control. The image shows a representative outcome (detailed results are provided in Supplemental Fig. S6). Measures of β-galactosidase activity represent means of three independent transformants. Error bars indicate se. β-gal units = 1,000 × OD574/(t × V × OD600), where OD = optical density, t = elapsed incubation time (min), and V = the volume of culture (mL). The numbers 1 to 15 represent GmNTL1 to GmNTL15.
All GmNTLs Associated with the ER, But the Pattern of Membrane Release Was Divergent
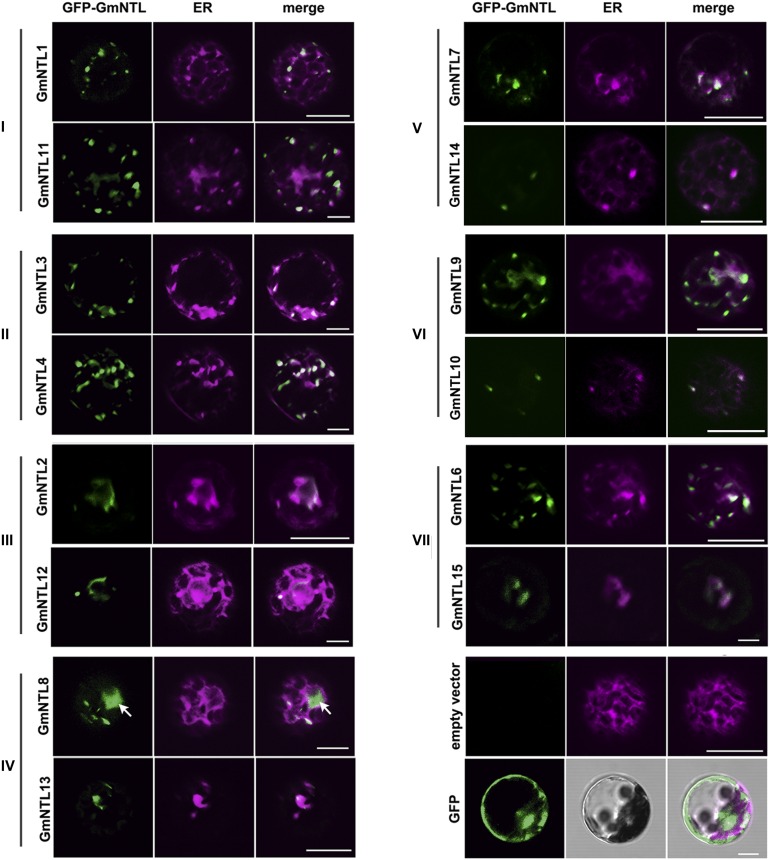
Membrane association is essential for the function of NTLs in signaling transduction. Five NTLs were demonstrated to localize in the ER (Kim et al., 2006; Klein et al., 2012; De Clercq et al., 2013; Ng et al., 2013; Block et al., 2014) and four in the plasma membrane (Kim et al., 2007a; Seo et al., 2010; Park et al., 2011; Puranik et al., 2011) in plants. The product of each of the 15 GFP-GmNTL fusion transgenes transiently expressed in Arabidopsis protoplasts colocalized with the ER marker (Fig. 6; Table II). Exceptionally, GFP-GmNTL8 also was deposited in the nucleus, suggesting its partial release from the ER. Thus, divergence in terms of subcellular localization seemed not to contribute to their functional divergence for the duplicate genes.
Figure 6.
Subcellular localization of the products of GmNTL duplicate pairs. The numerals I to VII refer to the seven duplicated gene pairs (Table I). GFP-GmNTL fusion proteins localized to the ER in transiently transformed Arabidopsis protoplasts. The left images show the GFP signal (green), the middle images show the mCherry signal (magenta; an ER marker), and the right images are merged images. The magenta color in the control image (right column, bottom) is chloroplast autofluorescence. Arrows indicate the nuclei. Bars = 20 μm.
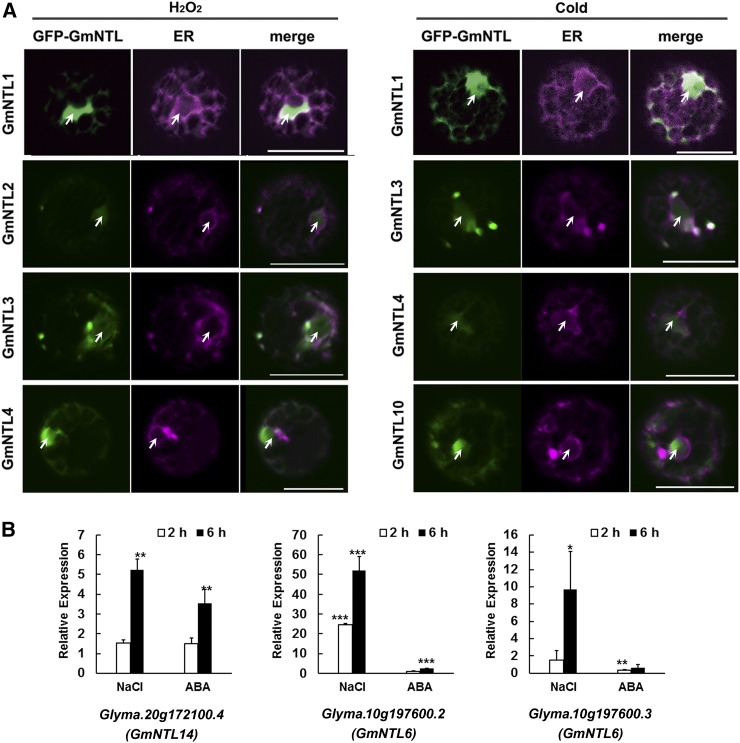
In addition to the transcriptional response, the environmental induction of membrane release is essential for NTL function in stress signaling. To characterize divergence with respect to the environmental signaling of membrane release of the GmNTLs, transiently transformed Arabidopsis protoplasts carrying the various GFP-GmNTL constructs were exposed to various stresses. Both low-temperature and hydrogen peroxide (H2O2) treatments provided evidence for stress-induced processing by some of the GmNTLs (Fig. 7A; Table II). At room temperature, the GFP signal was restricted to the ER, but in plants exposed to either 4°C or 0.1 mm H2O2 for 15 min, the signal was distributed between the ER and the nucleus or totally concentrated in the nucleus. Membrane release was triggered by both H2O2 and low temperature in the case of GmNTL1, GmNTL3, and GmNTL4, only by H2O2 in the case of GmNTL2, and only by low temperature in the case of GmNTL10. Thus, divergence with respect to inducible membrane release was the case for gene pairs I, III, and VI.
Figure 7.
Membrane release of GmNTLs. A, Transiently transformed Arabidopsis protoplasts assayed 5 h after exposure to low temperature or H2O2. The GFP (green) and mCherry (magenta) signals mark the nucleus/cytoplasm and the ER, respectively. Cold, Protoplasts exposed to 4°C for 15 min; H2O2, protoplasts exposed to 10 mm H2O2 for 15 min. Arrows indicate the nuclei. Bars = 20 μm. B, The inducibility of alternative GmNTL transcripts in the roots of plants exposed to either 200 mm NaCl or 100 µm ABA. The data shown are means ± se (n = 3). Statistically significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) in transcript abundance are shown between treated and mock-treated samples.
Divergence in Alternative Splicing-Mediated Membrane Release between GmNTL Duplicates
In addition to the membrane release mediated by protein processing, alternative splicing has been demonstrated to achieve this purpose (Deng et al., 2011; Nagashima et al., 2011; Lu et al., 2012). A detailed analysis of the GmNTL transcripts was carried out to check whether alternative splicing could contribute to diversity in membrane release. GmNTL6 generated three transcripts, one of which (Glyma.10g197600.1) encoded an NTL protein, while the other two (Glyma.10g197600.2 and Glyma.10g197600.3) encoded identical TM-free NAC proteins (which seemed to be the released form of GmNTL6; Supplemental Table S1). Furthermore, GmNTL14 generated four transcripts: two (Glyma.20g172100.1 and Glyma.20g172100.2) encoded the same product as GmNTL14, one (Glyma.20g172100.4) encoded a TM-free NAC protein (which seemed to be the released form of GmNTL14; Supplemental Table S1), and one (Glyma.20g172100.3) encoded a protein lacking a NAC domain (Supplemental Table S1). The Glyma.20g172100.4 (GmNTL14) transcript became highly abundant in the roots in response to either salinity stress or ABA treatment, while the abundance of both Glyma.10g197600.2 and Glyma.10g197600.3 (GmNTL6) was increased by salinity stress but decreased by exposure to ABA (Fig. 7B; Table II). The indication, therefore, was that, since both GmNTL14 and GmNTL6 can be released from the membrane in response to salinity stress via alternative transcription, there has been some divergence in this respect, involving genes in pairs V and VII. When the alternative transcripts of their M. truncatula and common bean homologs were explored, it was shown that Phvul.007G085600 (ancestral homolog of GmNTL7/14) and Phvul.007G140500 (ancestral homolog of GmNTL6/15) produced no alternative transcripts, while Medtr1g097300 (ancestral homolog of GmNTL7/14) and Medtr1g090723 (ancestral homolog of GmNTL6/15) did. However, none of them encodes a TM-free NAC protein. This suggests that the stress-induced membrane release mediated by alternative transcripts in GmNTL14 and GmNTL6 reflects some neofunctionalization.
Membrane Release Is Required for GmNTL Function during an Abiotic Stress Episode, and Its Pattern Contributes to Phenotypic Variation
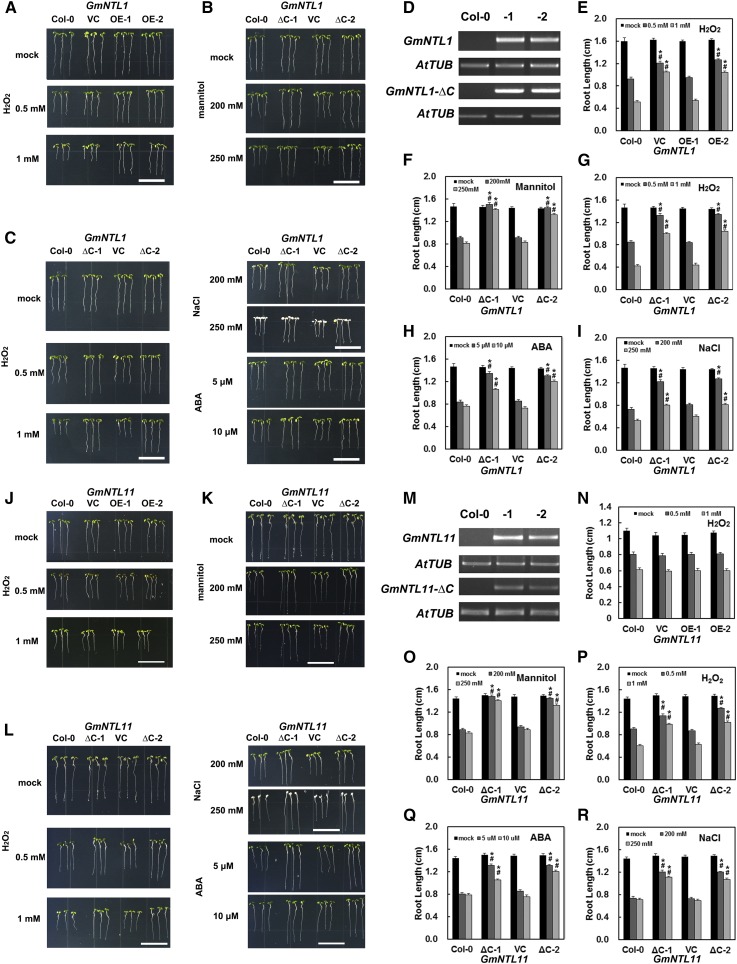
Since all the GmNTL genes were transcriptionally responsive to abiotic stress (Fig. 4; Supplemental Fig. S4) and the membrane release of some of the GmNTLs was induced by abiotic stress (Fig. 7), the assumption was that these proteins have a function within the plant’s response to abiotic stress. The contribution of GmNTL1 was explored by heterologously expressing its encoding gene in Arabidopsis. The transgene was efficiently transcribed in the transgene homozygous lines OE-1 and OE-2 (Fig. 8, A and D). Comparisons of primary root growth were made between these transgenic and appropriate control plants (the wild type and a transgenic line carrying an empty vector) exposed to a variety of abiotic stresses. The performance of the full-length GmNTL1 overexpression (OE) lines under either nonstressed conditions or when exposed to either salinity or mannitol was indistinguishable from that of the controls (Fig. 8, A and E). However, when the stress agent was H2O2, primary root growth of the OE lines was significantly superior to that of the control plants. To examine whether the membrane release of GmNTL1, which was induced by H2O2 treatment (Fig. 7A), was required for its function, Arabidopsis transgenics were constructed having the 35Spro:GmNTL1-ΔC (encoding an active GmNTL1 form that lacks the TM domain and is localized in the nucleus) as represented by the two homozygous transgenic lines (ΔC-1 and ΔC-2), which each produced the GmNTL1-ΔC transcript (Fig. 8D). As was the case for plants having 35Spro:GmNTL1, the 35Spro:GmNTL1-ΔC plants were indistinguishable from the wild type under nonstressed conditions. However, when exposed to various levels of NaCl, mannitol, H2O2, or ABA, their ability to maintain root growth was superior to that of the control plants (Fig. 8, B, C, and F–I). Thus, GmNTL1 has a demonstrable role in abiotic stress tolerance, with the membrane release of GmNTL likely being an important factor.
Figure 8.
The phenotypes of transgenic Arabidopsis plants constitutively expressing GmNTL1 or GmNTL11. A and J, Seedlings carrying 35Spro:GmNTL1 (A) or 35Spro:GmNTL11 (J) exposed to 0 to 1 mm H2O2. B, C, K, and L, Seedlings carrying 35Spro:GmNTL1-ΔC (B and C) or 35Spro:GmNTL11-ΔC (K and L) exposed to 200 to 250 mm mannitol (B and K), 0 to 1 mm H2O2, 200 to 250 mm salinity, or 5 to 10 μm ABA (C and L). The photographs in A to C and J to L were taken 5 d after the stress treatment was initiated. Bars = 1.5 cm. D and M, Semiquantitative real-time PCR profiles of wild-type and transgenic Arabidopsis. AtTUB was the reference gene. E to I and N to R, Primary root length measured 5 d after the imposition of H2O2, mannitol, ABA, or NaCl stress. Error bars indicate se (n = 15). Significant differences (*, between the transgenic line and the wild type; #, between the transgenic line and the empty vector control) were determined by Student’s t test (P < 0.01). Col-0, Columbia-0; ∆C-1 or ∆C-2, transgenic lines having 35Spro:GmNTL1-ΔC or 35Spro:GmNTL11-ΔC; OE-1 or OE-2, transgenic lines having 35Spro:GmNTL1 or 35Spro:GmNTL11; VC, empty vector control.
It remains to be explored whether the divergence of GmNTLs with respect to their protein membrane-release patterns (Fig. 7) contributes to phenotypic variation. GmNTL11 and GmNTL1 constitute one duplicated gene pair. They are very closely related in gene and protein structure (Fig. 3), expression pattern (Fig. 4), transactivation activity (Fig. 5), and subcellular localization (Fig. 6). The only difference between them is the protein membrane-release pattern, where the release of GmNTL1 was induced by H2O2 and cold but the release of GmNTL11 was not (Fig. 7). The 35::GmNTL11 and 35::GmNTL11-ΔC transgenic Arabidopsis homozygous lines also were constructed (Fig. 8M). These plants were exposed to similar stresses to those for 35::GmNTL1 and 35::GmNTL1-ΔC transgenic plants. The same phenotype was found in 35::GmNTL11-ΔC plants as in the 35::GmNTL1-ΔC line (Fig. 8, K, L, and O–R). But 35::GmNTL11 overexpression plants did not display greater tolerance to H2O2 compared with the 35::GmNTL1 OE line (Fig. 8, J and N). This is clearly due to different membrane-release patterns in response to H2O2 of GmNTL1 and GmNTL11. To our knowledge, these results provide the first evidence that divergence in protein membrane-release patterns contributes to plant phenotypic variation.
DISCUSSION
The Subfunctionalization/Neofunctionalization of Duplicated GmNTLs
Functional diversification among gene family members is viewed as an important source of evolutionary innovation in complex organisms, and duplicated gene pairs can experience pseudogenization (Moore and Purugganan, 2005), subfunctionalization (Cusack and Wolfe, 2007), or neofunctionalization (Blanc and Wolfe, 2004). In soybean, the major consequences of polyploidy, especially to TF genes, appear to have been subfunctionalization and/or neofunctionalization (Roulin et al., 2013). Soybean and the common bean diverged from one another approximately 19.2 Mya, soybean and M. truncatula diverged from one another approximately 54 Mya, but their common ancestor experienced a WGD event timed at approximately 56.5 Mya (Lavin et al., 2005; Young et al., 2011). A second WGD event occurred in soybean specifically approximately 13 Mya (Schmutz et al., 2010). Sequence comparisons between the GmNTLs and their bean homologs suggest that seven GmNTL pairs were generated by the latter WGD event (Figs. 1 and 2). The analysis of the duplicated GmNTLs indicated that 14 of the 15 had experienced strong purifying selection and, therefore, have retained their ancestral functions. As suggested by Bush et al. (1999), using a single individual per species to test for positive selection on short terminal branches can generate a high rate of false positives in the calculation of dN/dS. However, the combined evidence points to GmNTL6 having not just evolved under positive selection (Table I; Supplemental Fig. S5); it shows that the different organ specificity with the preduplication reference gene in M. truncatula, and the release of its product from the membrane, can be mediated by alternative splicing (Fig. 7B), but that does not happen in the reference gene in M. truncatula and common bean. This suggests the occurrence of neofunctionalization. Similar neofunctionalization in alternative splicing-mediated membrane release also was found in GmNTL14 (Fig. 7B). On the other hand, in both rice and Arabidopsis, NTLs can be induced by at least one stress agent (Kim et al., 2010). All known NTL functions, with the exception of NTM1, have proven to be involved in abiotic and/or biotic stress signaling (Supplemental Table S5). So we supposed that function in abiotic and/or biotic stress signaling was the ancestor function of NTL genes. The data presented here have added to this picture as follows: the GmNTLs also proved to be inducible by various abiotic stress agents (Fig. 4), some of the GmNTLs were membrane released by the induction of abiotic stresses (Fig. 7), and GmNTL1 appeared to be a positive regulator of abiotic stress tolerance (Fig. 8). Overall, it appears that the GmNTLs have largely retained their function in the sphere of the plant’s response to abiotic stress, and the subfunctionalization or neofunctionalization that has arisen has conferred a diversity of roles within this response.
Functional Divergence in the Evolution of GmNTL Duplicate Genes
The evolutionary mechanisms of the retention of duplicate genes might vary among different gene families. Previous studies have shown that divergence in gene expression or protein function contributes to the retention of duplicate genes (Ganko et al., 2007; Lan et al., 2009; Yang et al., 2013). Protein subcellular relocalization also has been considered another mechanism for gene retention, because proteins could alter their functions when relocalized to a new subcellular structure (Byun-McKay and Geeta, 2007; Marques et al., 2008; Qian and Zhang, 2009; Ren et al., 2014). Alternative splicing involved in the divergence of duplicate genes was shown recently in vertebrate genomes (Lambert et al., 2015), but such studies are limited in plants (Shan et al., 2007). Relatively few studies have empirically analyzed the functional diversification of a gene family at different levels. Whether and how these various functional divergences are involved in the evolution of a gene family remained unknown. Members of the GmNTL pairs showed diversity in their transcription profile, both in space and in response to abiotic stress treatment (Fig. 4). Notably, pair III and VII genes were similar with respect to where the transcription occurred but differed in their stress inducibility, while pair I and II genes behaved in the opposite manner (Fig. 4). Thus, complexity in the regulatory network generated by polyploidization can arise not just as a result of spatial differentiation in patterns of transcription (Roulin et al., 2013) but also through the generation of diversity in the genes’ response to external stimuli. Additionally, Roulin et al. (2013) indicated that the transcriptional divergence and dN/dS were negatively correlated in genes across the sampled tissues, suggesting that expression divergence across tissues has increased evolutionary pressures to maintain gene function. But in this study, this relationship was not observed. For example, dN/dS of genes in the duplicate pair IV and V was extremely low, less than 0.07 (Supplemental Fig. S5), and their expression pattern had little divergence (Fig. 4). One explanation for this result is that expression divergence did not contribute much to the evolutionary pressure of these genes but other kinds of functional divergence did, such as subcellular localization, transactivation activity, and membrane release, as the data showed (Figs. 5–7; Table II). Another explanation is that the expression divergence is the consequence of the change in promoter nucleotide sequences, and the dN/dS from gene-coding sequences may not be consistent with that from the promoter sequence. A supportive result is that the topology of the phylogenetic tree of the promoter sequences of GmNTLs is slightly different from that of protein sequences (Supplemental Fig. S7). Moreover, functional divergence apparently has arisen from the protein character (including transactivation activity, subcellular localization, and proteolytic cleavage-mediated membrane release; Table II, Figs. 5–7A) and alternative splicing (Table II; Fig. 7B; Supplemental Table S1). Therefore, the functional divergence that has evolved reflects variation generated within either the genes’ promoter and/or transcription regions (including coding and intron sequences).
Membrane Release Plays an Important Role in the Functional Divergence of GmNTL Duplicate Genes in Abiotic Stress Response
The MTF’s unique character is to associate with the ER, nuclear, or plasma membrane in a dormant format when the condition is normal, and when the plant is exposed to specific developmental or environmental cues, MTFs are proteolytically processed, then transported to the nucleus and function as TFs (Chen et al., 2008). It has been proposed that the activation of MTFs represents an efficient means of achieving a rapid transcriptional response. The signal that induces the membrane release of MTFs determines when and where they exercise their transcription regulation function. Evidence for the critical role of membrane release in the function of NTLs include the finding that when the full-length NTL protein is overexpressed in plants, there often are no phenotypic differences between transgenic and control seedlings, but transgenic plants overexpressing NTL proteins without the TM motif show a corresponding phenotypic difference from control plants (Kim et al., 2006, 2007b, 2008; Park et al., 2011; Zhang et al., 2011; Lee et al., 2012). Experimental results in this study also demonstrated this conclusion. When the full-length GmNTL1 was heterologously expressed in Arabidopsis, the transgenic lines only showed improved oxidative stress tolerance when the plants were exposed to H2O2, which had been demonstrated to induce its membrane release (Fig. 8). However, heterogenous expression of GmNTL1 proteins without the TM motif conferred numerous abiotic stress tolerances to transgenic Arabidopsis during its early growth (Fig. 8). On the contrary, heterologous expression of GmNTL11 (in the same duplicate gene pair with GmNTL1) full-length proteins conferred no phenotypic difference from control plants under oxidative or other abiotic stresses to transgenic Arabidopsis (Fig. 8), since its membrane release mediated by proteolytic cleavage could not be induced under those conditions. This further demonstrates the critical role of the membrane release in their function in the abiotic stress response and, therefore, provides an efficient evolutionary driving force in subfunctionalization. This kind of subfunctionalization may have contributed to the retention of genes in pairs I, III, and VI (Table II; Fig. 7A). Alternative splicing is confirmed to be another efficient way to generate the active form of bZIP MTFs in Arabidopsis (Nagashima et al., 2011), but it has not been found in any other kind of MTFs. In this study, we surprisingly found that alterative splicing that mediated the activation of GmNTLs may contribute to their subfunctionalization after WGD. The alternative transcript of GmNTL14 codes for a TM-free NTL protein, and its transcription could be induced significantly by NaCl and ABA treatment (Fig. 7B). At the same time, GmNTL7 and GmNTL14 were largely similar, including expression pattern (Fig. 4), transactivation activity (Fig. 5), and subcellular localization (Fig. 6), while the dN/dS of these two genes was as low as 0.01 and 0.05, suggesting strong purifying selection (Supplemental Fig. S2). This caused us to propose that an alternative splicing-mediated activation mechanism mostly contributed to their functional divergence in the evolutionary process.
CONCLUSION
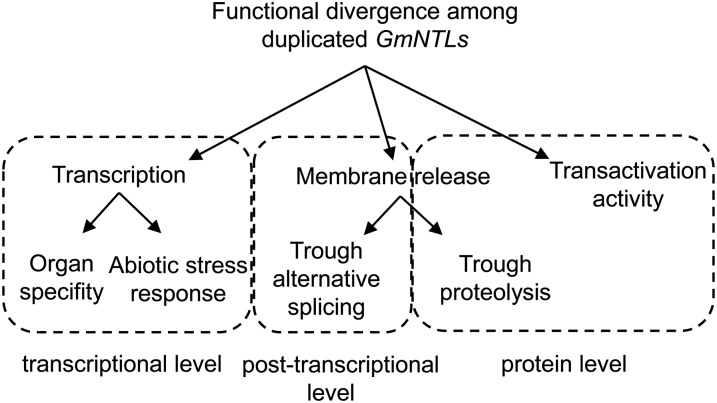
The analysis of complete genome sequences is increasingly revealing the presence of gene duplications, particularly in plants, where polyploidization has played a large part in speciation and evolution. The mechanistic basis of duplicate retention and subsequent functional divergence remains rather obscure. Here, the focus was on a family of seven GmNTL gene pairs after the latest WGD in soybean. A variety of approaches was applied to determine the evolutionary processes likely to have contributed to divergence in the gene family. The findings suggest that multiple levels of functional divergence (Fig. 9), especially a new kind of functional divergence in the membrane-release pattern, and purifying selection have been key to the retention and subsequent functional divergence of these duplicated genes.
Figure 9.
Model of functional divergence among duplicated GmNTLs. The members of each duplicated GmNTL pair variously diverged at the transcriptional (organ specificity and responsiveness to abiotic stress), posttranscriptional (alternative splicing), and protein (proteolysis-mediated membrane release and transactivation activity) levels.
MATERIALS AND METHODS
Identification and Isolation of GmNTL, PvNTL, and MtNTL Genes
Soybean (Glycine max), common bean (Phaseolus vulgaris), and Medicago truncatula protein sequences containing the NAM domain (keyword search by PF02365) were obtained from the Phytozome version 11 database (https://phytozome.jgi.doe.gov/pz/portal.html), and their TM motif content was identified using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/; Sonnhammer et al., 1998; Krogh et al., 2001) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) software. Proteins containing the Pfam domain PF02365 and at least one transmembrane helix predicted by either TMHMM or TMpred were assigned as NTLs. The putative GmNTL sequences having a full open reading frame (as predicted by the Glyma1.1 model; https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Gmax) were subjected to PCR amplification directed at their open reading frames (primer sequences are listed in Supplemental Table S6) using a cDNA template derived from the total RNA extracted from 4-week-old cv Williams 82 seedlings. The amplicons were introduced into the Gateway pDONR221 vector (Invitrogen) for sequencing.
GmNTL Identification, Domain Prediction, and Motif Analysis
The in silico translation of GmNAC was obtained using Phytozome version 11 software (https://phytozome.jgi.doe.gov/pz/portal.html). Protein domain prediction was performed using ScanProsite implemented in the PROSITE database (http://prosite.expasy.org/prosite.html). The hydrophobicity of the various predicted proteins was obtained using the ConPred II program (Arai et al., 2004). Conserved motifs were identified using the MEME version 4.11.1 program (Bailey and Elkan, 1994; Bailey et al., 2009). The analysis was performed by keeping the number of repetitions, any; the maximum number of motifs, 20; and the optimum width of the motif, 6 or greater and 50 or less.
Phylogenetic and Molecular Evolution Analyses
The ClustalW-based alignment of GmNTL polypeptide sequences employed a gap open penalty of 10 and a gap extension penalty of 0.2, as implemented within MEGA6 software (Tamura et al., 2013). The resulting phylogenetic tree was derived by the same software. The cross-species analysis of plant NTLs used the ML method based on the Jones, Taylor, and Thornton amino acid substitution model. The NTL peptide sequences were from soybean, common bean, M. truncatula, Arabidopsis (Arabidopsis thaliana; Kim et al., 2007b), Brachypodium distachyon (Zhu et al., 2015), rice (Oryza sativa; Kim et al., 2010), Setaria italica (Puranik et al., 2013), and Glyma.16G016400, which is a NAC protein without transmembrane helix but is coded by the duplicated gene of GmNTL5. The At4g29230 sequences were adopted as the outgroup in the phylogenetic analysis of the whole NTL subfamily, which encodes a NAC family protein but not a member of NTLs. A total of 500 bootstrap replicates were included to allow for the assigning of confidence levels to each node. To evaluate variation in selective pressure over a phylogeny, the branch models of CODEML and PAML (Yang, 2007) were used to estimate ω under different assumptions. Analyses were conducted using the same alignment as the phylogenetic analysis, along with an ML tree constructed using three a priori assumptions: a one-ratio model in which one ω value was assumed for the entire tree, a free-ratio model that allowed ω to vary throughout the tree, and a two-ratio model in which ω values were allowed to vary between selected branches. To determine whether positive selection had acted at specific sites in the NTL sequences, four models in the PAML package were explored: the one-ratio model (M0), the free-ratio model (M1), the near-neutral model (M1a), and the positive-selection model (M2a).
Plant Materials and Growth Conditions
Soybean ‘Williams 82’ seedlings were grown in a greenhouse in which the day/night temperature was maintained at 28°C/20°C, the photoperiod at 14 h, the level of light intensity at 800 µmol m−2 s−1, and the relative humidity at 60%. At the three fully opened trifoliate leaf stage (approximately 25 d after sowing), roots, leaves, and stems were sampled. Flowers and early R5 stage seeds were harvested at day 55 and day 100, respectively. Groups of 14-d-old seedlings were exposed to various abiotic stresses or exogenous ABA by removing them from the soil, washing their roots, and transferring them to distilled water (mock treatment), 200 mm NaCl, 20% (w/v) PEG 6000, 100 µm ABA, or 10 mm H2O2. A low-temperature stress episode was imposed by holding the seedlings in refrigerated water maintained at 4°C. Following treatment, the seedlings were dissected into their component organs, and the material was snap frozen in liquid nitrogen. M. truncatula ecotype R108 was grown at 22°C/20°C, the photoperiod was 16 h, the level of light intensity was 800 µmol m−2 s−1, and the relative humidity was 70%. All the Arabidopsis material was based on the Columbia ecotype. Seeds were surface sterilized, held at 4°C for 72 h, and then cultured under a 16-h photoperiod at 22°C on one-half-strength Murashige and Skoog solidified medium (0.7% [w/v] agar) supplemented with 2% (w/v) Suc.
Transcriptional Activation Activity Assay
The GmNTL coding sequences in Gateway pDONR221 vector were transferred into the pDEST32 vector (Invitrogen) via an LR reaction, and the resulting constructs were transformed into yeast strain MaV203 containing the HIS3 and lacZ reporter genes. Transformed yeast cells were cultured on yeast nitrogen base plates, either with or without added His, Leu, and 3AT (Sigma), and were assayed for β-galactosidase expression. The quantitative β-galactosidase assays were performed using chlorophenol red-β-d-galactopyranoside as a substrate in liquid cultures. The protocol was as recommended by the supplier of the ProQuest Two-Hybrid System (Invitrogen).
Subcellular Localization of GmNTL Proteins
Individual GmNTL sequences were fused to GFP and inserted into the pB7WGF2 transient expression vector (PSB) in which the fusion protein coding sequence was driven by the 35S promoter. Protoplasts were prepared from the rosette leaves of 4-week-old Arabidopsis seedlings, and the transgene was introduced using PEG-mediated transformation as described by Yoo et al. (2007). The detection of GFP was carried out by laser scanning confocal microscopy (LSM 700; Carl Zeiss) following a 12- to 18-h incubation in the dark at 22°C. The 35S::GFP (Li et al., 2011) and mCherry-tagged HDEL (Nelson et al., 2007) transgenic protoplasts were used as localization controls for expression in the cytoplasm/nucleus and the ER. For the membrane-release experiment, transformed protoplasts were subjected to a 15-min exposure to either 4°C or 10 mm H2O2 and then held at 22°C in the dark for a further 5 h.
cDNA Synthesis and qRT-PCR
The total RNA isolated from plants using the TRIzol reagent (Invitrogen) was treated with Turbo DNA-free DNase I (Ambion), following the manufacturer’s protocol. The first cDNA strand was synthesized from 3 µg of RNA using the Script cDNA Synthesis Kit (Bio-Rad) in a 20-µL reaction, following the manufacturer’s protocol. Gene-specific primers (sequences are given in Supplemental Table S7) were designed using ProbeFinder version 2.44 (https://www.roche-applied-science.com). The 15-µL qRT-PCRs were based on the FS Universal SYBR Green Master (Roche). Each reaction contained 0.4 µm of each primer, and the PCR cycling regime was composed of an incubation at 50°C for 2 min and at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Dissociation curves were obtained after the final cycle by heating the reaction to 95°C for 15 s, followed by a steadily increasing temperature from 60°C to 95°C. GmTUB (GenBank accession no. AY907703) and Medtr7g026230 were used as internal reference genes for profiling across the plant (Song et al., 2014), Gm60S (Glyma.17g05270) and Mt60S (Medtr4g055270) for the PEG 6000, low-temperature, and H2O2 treatments, and GmELF1b (Glyma.02g44460) and MtELF1b (Medtr5g088660) for the salinity and ABA treatments (Le et al., 2012). Three biological replicates were included for each treatment. Relative transcript abundances were estimated via the 2−ΔΔCt method (Livak et al., 2013).
Generation of Transgenic Arabidopsis Plants
The GmNTL1 open reading frame or open reading frame without the C-terminal coding sequence was fused to the cauliflower mosaic virus 35S promoter and introduced into the binary vector pK2GW7 (PSB; primer sequences are given in Supplemental Table S7). The recombinant plasmids were transformed into Agrobacterium tumefaciens GV3101. Arabidopsis was transformed using the floral dipping method (Clough and Bent, 1998). Positive transformants were selected by culturing on solidified one-half-strength Murashige and Skoog medium supplemented with 50 mg L−1 kanamycin. Genomic DNA was extracted from the leaf of putative transformants to confirm the presence of the transgene using PCR, while real-time RT-PCR was used to estimate the GmNTL1 transcript abundance (primer sequences are given in Supplemental Table S8).
Transgenic Phenotyping
To assess primary root elongation, 3-d-old seedlings (at least 20 seedlings per line) were transferred to either fresh one-half-strength Murashige and Skoog medium (control) or to one-half-strength Murashige and Skoog medium containing H2O2, NaCl, mannitol, or ABA of different concentrations and left to grow vertically for 5 d. The primary root length was measured by ImageJ. Student’s t test was applied to test the significance of differences between two data groups, and the se was applied to measure the deviation of the sampling distribution. Both types of experiment were replicated three or four times.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Conserved motifs in the GmNTL N-terminal region.
Supplemental Figure S2. The coding sequence tree of NTL genes.
Supplemental Figure S3. Gene duplication for the GmNTLs.
Supplemental Figure S4. Hierarchical clustering of GmNTL duplicated pairs and their homologs in M. truncatula.
Supplemental Figure S5. Substitution rate analysis using the free-ratio model.
Supplemental Figure S6. The transcriptional activation activity assay of 15 GmNTL proteins.
Supplemental Figure S7. The promoter tree of GmNTLs.
Supplemental Table S1. GmNTLs identified and characterized in this study.
Supplemental Table S2. Conserved motifs in NTLs.
Supplemental Table S3. Flanking sequences of GmNTL within 140 kb.
Supplemental Table S4. Stress-related cis-acting regulatory elements in GmNTL promoters.
Supplemental Table S5. Functions of NTLs in Arabidopsis.
Supplemental Table S6. Sequences of primer pairs used for the isolation of GmNTL genes.
Supplemental Table S7. Sequences of primer pairs used for qRT-PCR analysis.
Supplemental Table S8. Sequences of primer pairs used for the semiquantitative real-time PCR of GmNTL genes.
Supplemental Data Set S1. NTL peptide sequences.
Supplementary Material
Acknowledgments
We thank Laibao Feng (Institute of Botany, Chinese Academy of Sciences) for syntenic analysis of GmNTLs in the soybean genome and Dr. Edward C. Mignot (Shandong University) for linguistic advice.
Glossary
- WGD
whole-genome duplication
- TF
transcription factor
- MTF
membrane-bound transcription factor
- ER
endoplasmic reticulum
- TM
transmembrane
- Mya
million years ago
- dN/dS
ratio of nonsynonymous to synonymous substitutions
- qRT
quantitative real-time
- 3AT
3-aminotriazole
- ABA
abscisic acid
- ML
maximum likelihood
- PEG
polyethylene glycol
Footnotes
This work was supported by the National High-Technology Research and Development Program 863 of China (grant no. 2013AA102602–4 to F.X.), the National Key Research and Development Program of China (grant no. 2016YFD0101902 to S.L.), the National Research and Development Project of Transgenic Crops of the Ministry of Science and Technology of China (grant no. 2013ZX08010002–002 to F.X.), the Key Projects in the National Science and Technology Pillar Program in the 12th Five-Year Plan of China (grant no. 2011BAI06B01 to F.X.), the Chinese National Special Science Research Program (grant no. 2013CB967303 to F.X.), the National Natural Science Foundation of China (grant no. 31201269 to S.L. and grant nos. 30970243 and 31270328 to F.X.), the International Technology Cooperation Project of Shandong Province of China (grant no. 2011176 to F.X.), the Chinese Natural Education Ministry Doctor Station Foundation Fellowship (grant no. 913111006 to F.X. and grant no. 20120131120028 to S.L.), and the Science and Technology Plan of Shandong Province (grant no. 2013GNC11010 to F.X.).
References
- Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Airoldi CA, Davies B (2012) Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genomics 39: 157–165 [DOI] [PubMed] [Google Scholar]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T (2004) ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res 32: W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Toruno TY, Elowsky CG, Zhang C, Steinbrenner J, Beynon J, Alfano JR (2014) The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol 201: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Buggs RJ, Elliott NM, Zhang L, Koh J, Viccini LF, Soltis DE, Soltis PS (2010) Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol 186: 175–183 [DOI] [PubMed] [Google Scholar]
- Bush RM, Fitch WM, Bender CA, Cox NJ (1999) Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol 16: 1457–1465 [DOI] [PubMed] [Google Scholar]
- Byun-McKay SA, Geeta R (2007) Protein subcellular relocalization: a new perspective on the origin of novel genes. Trends Ecol Evol 22: 338–344 [DOI] [PubMed] [Google Scholar]
- Cannon SB, McKain MR, Harkess A, Nelson MN, Dash S, Deyholos MK, Peng Y, Joyce B, Stewart CN Jr, Rolf M, et al. (2015) Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol 32: 193–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, Wendel JF (2009) Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YN, Slabaugh E, Brandizzi F (2008) Membrane-tethered transcription factors in Arabidopsis thaliana: novel regulators in stress response and development. Curr Opin Plant Biol 11: 695–701 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH (2007) When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet 23: 270–272 [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inze A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y (2012) Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15: 168–176 [DOI] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Larsen S, Lo Leggio L (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Udall J, Nettleton D, Wendel J (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF (2010) Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186: 184–193 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2008) The evolutionary position of subfunctionalization, downgraded. Genome Dyn 4: 25–40 [DOI] [PubMed] [Google Scholar]
- Freeling M, Scanlon MJ, Fowler JE (2015) Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr Opin Genet Dev 35: 110–118 [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ (2007) Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol 24: 2298–2309 [DOI] [PubMed] [Google Scholar]
- Guo DM, Ran JH, Wang XQ (2010) Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of bona fide CAD. J Mol Evol 71: 202–218 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Zhang H, Sun T, Shi Y, Wang J, Zhang B, Wang Z, Zhou Y, Gu H (2013) Natural variation of C-repeat-binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol 199: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Keller TE, Yi SV (2014) DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci USA 111: 5932–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Kim SY, Park CM (2007a) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226: 647–654 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee AK, Yoon HK, Park CM (2008) A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 55: 77–88 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, Park CM (2010) Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65 [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM (2007b) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P, Seidel T, Stocker B, Dietz KJ (2012) The membrane-tethered transcription factor ANAC089 serves as redox-dependent suppressor of stromal ascorbate peroxidase gene expression. Front Plant Sci 3: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Lambert MJ, Cochran WO, Wilde BM, Olsen KG, Cooper CD (2015) Evidence for widespread subfunctionalization of splice forms in vertebrate genomes. Genome Res 25: 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, Yang ZL, Yang X, Liu YJ, Wang XR, Zeng QY (2009) Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 21: 3749–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Weiche B, Timmerhaus G, Richardt S, Riano-Pachon DM, Correa LG, Reski R, Mueller-Roeber B, Rensing SA (2010) Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol Evol 2: 488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Ham LH, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2012) Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS ONE 7: e49522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Uygun S, Moghe GD, Panchy N, Fang L, Hufnagel DE, Jasicki HL, Feig M, Shiu SH (2015) Molecular evidence for functional divergence and decay of a transcription factor derived from whole-genome duplication in Arabidopsis thaliana. Plant Physiol 168: 1717–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J (2011) Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Liu HJ, Tang ZX, Han XM, Yang ZL, Zhang FM, Yang HL, Liu YJ, Zeng QY (2015) Divergence in enzymatic activities in the soybean GST supergene family provides new insight into the evolutionary dynamics of whole-genome duplicates. Mol Biol Evol 32: 2844–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Wills QF, Tipping AJ, Datta K, Mittal R, Goldson AJ, Sexton DW, Holmes CC (2013) Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods 59: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SJ, Yang ZT, Sun L, Song ZT, Liu JX (2012) Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol Plant 5: 504–514 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Vinckenbosch N, Brawand D, Kaessmann H (2008) Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol 9: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Moghe GD, Shiu SH (2014) The causes and molecular consequences of polyploidy in flowering plants. Ann N Y Acad Sci 1320: 16–34 [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Strome B, Hua JJ, Desmond J, Gagnon-Arsenault I, Weiss EL, Landry CR, Moses AM (2014) Detecting functional divergence after gene duplication through evolutionary changes in posttranslational regulatory sequences. PLOS Comput Biol 10: e1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer-Verlag, Berlin [Google Scholar]
- Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM (2011) Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol 156: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Bahadur RP, Srivastava PS, Prasad M (2011) Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet Setaria italica (L.) P. Beauv. Mol Biotechnol 49: 138–150 [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Mandal SN, B VS, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8: e64594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhang J (2009) Protein subcellular relocalization in the evolution of yeast singleton and duplicate genes. Genome Biol Evol 1: 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren LL, Liu YJ, Liu HJ, Qian TT, Qi LW, Wang XR, Zeng QY (2014) Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of Populus class III peroxidase family. Plant Cell 26: 2404–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA. (2014) Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol 17: 43–48 [DOI] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73: 143–153 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, Xu G, Zhang J, Chen Z, Kong H (2007) Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phylogenet Evol 44: 26–41 [DOI] [PubMed] [Google Scholar]
- Shen H, Yin Y, Chen F, Xu Y, Dixon R (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. BioEnergy Res 2: 217–232 [Google Scholar]
- Shiu SH, Shih MC, Li WH (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS (2013) Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res 20: 403–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175–182 [PubMed] [Google Scholar]
- Song H, Yin Z, Chao M, Ning L, Zhang D, Yu D (2014) Functional properties and expression quantitative trait loci for phosphate transporter GmPT1 in soybean. Plant Cell Environ 37: 462–472 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Marowsky NC, Fan C (2014) Divergence of gene body DNA methylation and evolution of plant duplicate genes. PLoS ONE 9: e110357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Adams KL (2015) Duplicate gene divergence by changes in microRNA binding sites in Arabidopsis and Brassica. Genome Biol Evol 7: 646–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang Y, Gao Y, Zhou Y, Zhang E, Hu Y, Yuan Y, Liang G, Xu C (2014) Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol Biol 14: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZL, Liu HJ, Wang XR, Zeng QY (2013) Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol 197: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM (2008) Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells 25: 438–445 [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Guo C, Zhang W, Wang P, Li L, Duan X, Du Q, Zhao L, Shan H, Hodges SA, et al. (2013) Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc Natl Acad Sci USA 110: 5074–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Belcram H, Gornicki P, Charles M, Just J, Huneau C, Magdelenat G, Couloux A, Samain S, Gill BS, et al. (2011) Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc Natl Acad Sci USA 108: 18737–18742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ. (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18: 292–298 [Google Scholar]
- Zhu G, Chen G, Zhu J, Zhu Y, Lu X, Li X, Hu Y, Yan Y (2015) Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS ONE 10: e0139794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.