NAD-mediated defense responses stimulate PAMP-triggered immunity in Arabidopsis by stimulating redox signaling and modulating the hormonal balance.
Abstract
Pyridine nucleotides, such as NAD, are crucial redox carriers and have emerged as important signaling molecules in stress responses. Previously, we have demonstrated in Arabidopsis (Arabidopsis thaliana) that the inducible NAD-overproducing nadC lines are more resistant to an avirulent strain of Pseudomonas syringae pv tomato (Pst-AvrRpm1), which was associated with salicylic acid-dependent defense. Here, we have further characterized the NAD-dependent immune response in Arabidopsis. Quinolinate-induced stimulation of intracellular NAD in transgenic nadC plants enhanced resistance against a diverse range of (a)virulent pathogens, including Pst-AvrRpt2, Dickeya dadantii, and Botrytis cinerea. Characterization of the redox status demonstrated that elevated NAD levels induce reactive oxygen species (ROS) production and the expression of redox marker genes of the cytosol and mitochondrion. Using pharmacological and reverse genetics approaches, we show that NAD-induced ROS production functions independently of NADPH oxidase activity and light metabolism but depends on mitochondrial respiration, which was increased at higher NAD. We further demonstrate that NAD primes pathogen-induced callose deposition and cell death. Mass spectrometry analysis reveals that NAD simultaneously induces different defense hormones and that the NAD-induced metabolic profiles are similar to those of defense-expressing plants after treatment with pathogen-associated molecular patterns. We thus conclude that NAD triggers metabolic profiles rather similar to that of pathogen-associated molecular patterns and discuss how signaling cross talk between defense hormones, ROS, and NAD explains the observed resistance to pathogens.
NAD is an ubiquitous redox carrier that controls virtually all metabolic reactions in the cell (Noctor et al., 2006). In plants, pools of NAD and its phosphate derivative NADP are essentially oxidized (i.e. NAD+ and NADP+, respectively) and tightly maintained at the cellular level (Hunt et al., 2004; Noctor et al., 2006). Beside its redox properties, NAD(P) is consumed during intracellular signaling processes, including calcium (Ca2+) signaling, DNA repair via poly-ADP-ribosylation (i.e. PARP), and histone deacetylation (i.e. sirtuins; Hunt et al., 2004; Noctor et al., 2006; Briggs and Bent, 2011). A growing body of evidence indicates that NAD biosynthesis acts in plant stress responses related to plant immunity (for review, see Pétriacq et al., 2013). Indeed in Arabidopsis (Arabidopsis thaliana), genetic knockdown of aspartate oxidase (AO) activity (EC 1.4.3.16; the first enzyme of de novo NAD synthesis in the chloroplast; Katoh et al., 2006) hampers stomatal immunity against the hemibiotrophic pathogen Pseudomonas syringae pv tomato (Pst; Macho et al., 2012), while AO transcripts consistently build up in response to bacterial infection, presumably to sustain NAD consumption (Pétriacq et al., 2012). Furthermore, we have demonstrated previously a direct role for intracellular NAD+ in defense, using an inducible system that is based on overexpression of the nadC gene from Escherichia coli, which encodes the NAD biosynthesis enzyme quinolinate phosphoribosyltransferase (EC 2.4.2.19; Pétriacq et al., 2012). Transgenic nadC Arabidopsis plants accumulate NAD upon treatment with quinolinic acid (Q), thereby allowing one to determine the biochemical and physiological consequences of increased NAD content in leaves. Transient increase in NAD+ pools induced resistance to the avirulent bacterial strain Pst-AvrRpm1 via stimulation of the defense hormone salicylic acid (SA). Transcriptomic analyses of nadC plants also pointed to NAD-dependent up-regulation of pathogen-inducible genes associated with Ca2+ signaling and various redox targets, including the hypersensitive response (HR; Pétriacq et al., 2012, 2013). In support of these results, Zhang and Mou (2009, 2012) suggested that exogenous NAD+ in the apoplast plays a role in defense-related Ca2+ signaling via both SA-dependent and SA-independent signaling pathways. Manipulating plant NAD catabolism also has proven important for defense responses (Pétriacq et al., 2013). Several studies have found that disruption of ADP-ribose/NADH pyrophosphohydrolase (i.e. NUDIX hydrolase or NDUT in Arabidopsis) metabolism (Ge et al., 2007; Ge and Xia, 2008; Ishikawa et al., 2010; Jambunathan et al., 2010) and poly-ADP-ribosylation (Adams-Phillips et al., 2008, 2010; Briggs and Bent, 2011; Song et al., 2015) impacts the cellular NADH-NAD+ ratio and SA-dependent and SA-independent immunity. In line with this conclusion, the Arabidopsis NUDT7 gene was found to regulate both SA-dependent and SA-independent defense signaling (Bartsch et al., 2006; Ge et al., 2007). Hence, NAD-mediated regulation of plant defense involves SA-dependent and SA-independent signaling mechanisms.
While reactive oxygen species (ROS)-mediated defense responses are well documented (Dietz, 2003; Torres, 2010; Mittler et al., 2011; O’Brien et al., 2012; Frederickson Matika and Loake, 2014; Lehmann et al., 2015; Trapet et al., 2015), the precise role of NAD in ROS-related plant immunity remains poorly understood. ROS bursts contribute to basal defense responses after the perception of pathogen-associated molecular patterns (PAMPs), which are conserved molecules for a whole class of microbes, or via damage-associated molecular patterns (DAMPs), which are signals of cell disintegration (Heil and Land, 2014; Macho and Zipfel, 2014). Although some evidence indicates that exogenous NAD could act as a DAMP by leaking from an extracellular compartment and then stimulating immune responses (Zhang and Mou, 2009), this scenario awaits further investigation to determine how NAD intervenes in DAMP-triggered immunity. We have substantiated the hypothesis that NAD interacts with redox signaling by stimulating ROS-producing oxidase systems (Pétriacq et al., 2012). However, no direct evidence for NAD impacts on ROS production have been reported (Pétriacq et al., 2013). In plants, although many NADPH-consuming oxidases are capable of generating ROS, it is still assumed that the apoplastic NADPH oxidase complexes (also named respiratory burst homologs [RBOHs]) are the main ROS-producing enzymes involved in defense against pathogens (Miller et al., 2009; Torres, 2010; Marino et al., 2012). In Arabidopsis, RBOHD and RBOHF were initially described as key players in HR-associated ROS production against Pst-AvrRpm1 (Torres et al., 2002). Surprisingly, however, rbohD and rbohF mutants still showed induced defense by intercellular NAD (Zhang and Mou, 2009). On the other hand, AO activity (the committed step of NAD biosynthesis) has been shown to be essential for RBOHD-dependent ROS production after treatment with PAMPs, while RBOHD-independent PAMP responses do not require full AO activity (Macho et al., 2012). Together, these data suggest that manipulating endogenous NAD levels might impact ROS production by RBOH, but direct evidence of how NAD and ROS interact is still missing.
As an aid to clarify the mechanisms of NAD-mediated immunity, we used inducible NAD biosynthesis in the transgenic Arabidopsis nadC line and analyzed its response to pathogens. We provide evidence that NAD plays a role in defense against various plant pathogens and demonstrate that ROS production is stimulated directly by NAD. We also report that the effect of NAD encompasses callose deposition and cell death, the latter being responsible for enhanced resistance to Pst-AvrRpm1. Furthermore, metabolomics analyses show that NAD induces a metabolomics pattern similar to PAMP-triggered immunity.
RESULTS
Inducible Elevation of Intracellular Pyridine Nucleotides Correlates with Broad Resistance to Diverse Plant Pathogens
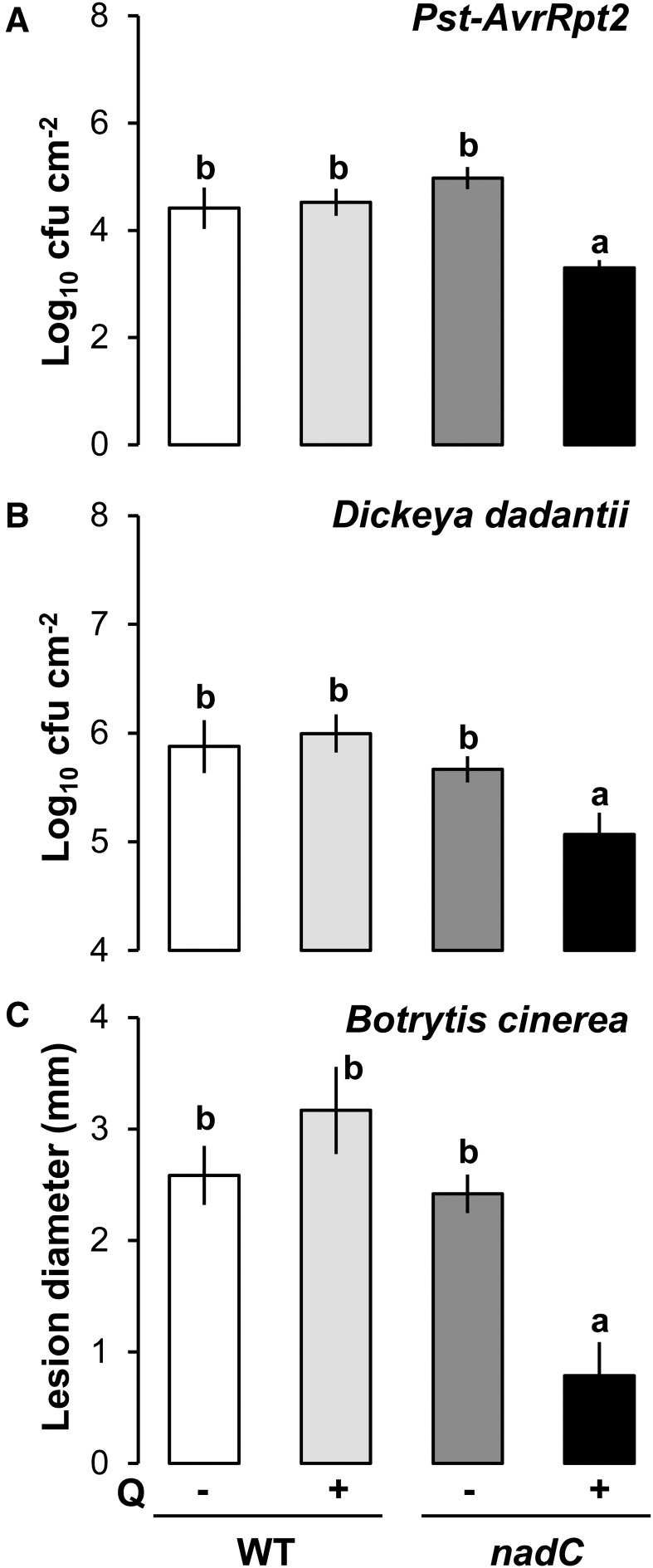
In a previous study, we demonstrated that inducible intracellular NAD stimulates resistance against avirulent Pst-AvrRpm1 in nadC plants after treatment with Q (Pétriacq et al., 2012). This enhanced resistance correlates with an increased accumulation of NAD (Supplemental Fig. S1A), an oxidized redox state (Supplemental Fig. S1B), and increased SA pools (Supplemental Fig. S1C) in Q-treated nadC plants (nadC Q) as compared with the wild type and a Q-treated control. Higher pools of NAD derivatives, such as nicotinic acid (Supplemental Fig. S1D) and nicotinamide (Pétriacq et al., 2012), were observed in Q-treated wild-type and nadC plants. This suggests that NAD catabolism was induced, but it has been shown to be unrelated to the bacterial resistance in Q-treated nadC plants (Pétriacq et al., 2012). To further investigate the role of endogenous NAD in resistance against other pathogens (Fig. 1), we compared NAD-associated resistance against avirulent Pst-AvrRpt2, the hemibiotrophic virulent bacterial pathogen Dickeya dadantii, and the necrotrophic fungus Botrytis cinerea. Prior to pathogen challenge (see scheme in Supplemental Fig. S2), intact leaves of wild-type and nadC plants were syringe infiltrated with Q to induce NAD production (Pétriacq et al., 2012) and 48 h later challenged with Pst-AvrRpt2 (Fig. 1A), D. dadantii (Fig. 1B), or B. cinerea (Fig. 1C). For all three pathosystems, Q-treated nadC plants showed statistically enhanced levels of resistance in comparison with the other conditions (Fig. 1). Hence, NAD-associated defense is effective against pathogenic microbes with different infection strategies.
Figure 1.
Resistance phenotypes to pathogens with different lifestyles in the NAD-overproducing nadC line. A and B, Bacterial growth of Pst-AvrRpt2 and D. dadantii, respectively, at 48 h post infection (hpi) in wild-type (WT) Columbia-0 (Col-0) and nadC plants with (+) or without (−) Q pretreatment. C, Lesion diameters after fungal infection with B. cinerea (24 hpi). Values shown are means of four to six biological replicates ± se. Letters above the bars denote statistically significant differences (P < 0.05, multiple Student’s t tests).
Elevated NAD Is Associated with an Oxidative Burst
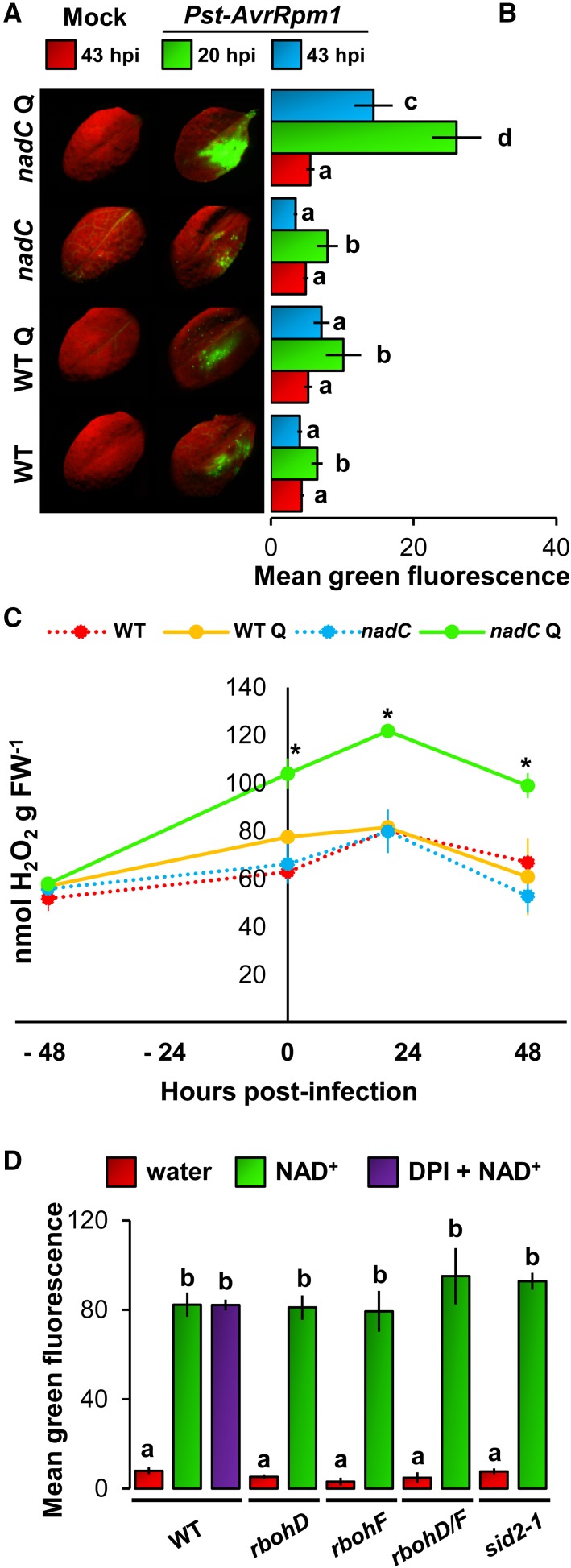
An increased availability of pyridine nucleotides might lead to biochemical stimulation of ROS production through various oxidases that control the immune-related oxidative burst preceding the HR (Hunt et al., 2004; Mahalingam et al., 2007; Pétriacq et al., 2013). To test this hypothesis, in situ detection of ROS was performed using the fluorescence dye 2,7-dichlorofluorescein diacetate (DCFH-DA), which is sensitive to intracellular ROS (Degrave et al., 2008; Queval et al., 2008; Jambunathan, 2010). Illuminated intact leaves of wild-type and nadC plants were infiltrated with Q and 48 h later challenged with Pst-AvrRpm1 (Supplemental Fig. S2). In mock-inoculated plants (68 h after Q treatment), no substantial difference in ROS production was observed between wild-type and nadC plants with or without Q application (Fig. 2, A and B). Conversely, challenge with Pst-AvrRpm1 stimulated ROS production at 20 hpi, which was strongly enhanced in Q-pretreated nadC plants at 20 and 43 hpi compared with all other conditions (Fig. 2, A and B). To confirm the DCFH-DA quantification of ROS production, we assayed ROS production by hydrogen peroxide (H2O2)-dependent chemiluminescence of luminol (Fig. 2C). This technique provides a quantitative measurement of ROS in whole cells after biochemical extraction from ground foliar tissues (Veljovic-Jovanovic et al., 2002; Jambunathan, 2010). In agreement with the DCFH-DA-based quantitation, we detected augmented ROS production after challenge with Pst-AvrRpm1 in Q-treated nadC plants, with maximal ROS production observed at 20 hpi (Fig. 2C). To test whether this ROS production also is effective against virulent bacteria, we quantified ROS by DCFH-DA fluorescence in wild-type and nadC plants after challenge with D. dadantii (Supplemental Fig. S3). As observed for the interaction with Pst-AvrRpm1, the NAD-associated resistance to D. dadantii (Fig. 1B) correlated with augmented ROS production in nadC Q (Supplemental Fig. S3). Taken together, these results provide in situ evidence that increased NAD pools correlate with a stimulated oxidative burst.
Figure 2.
Higher NAD is associated with an increased ROS burst. A, Typical photographs of in situ detection of ROS by DCFH-DA staining at 20 hpi in wild-type (WT) Col-0 and nadC Arabidopsis lines with or without Q and after inoculation with mock solution (left) or Pst-AvrRpm1 (right) in half of each leaf. B, Relative quantification of ROS-related green fluorescence of DCFH-DA at 20 and 43 hpi. Values shown represent means of 12 leaves ± se. Letters indicate statistically significant differences (P < 0.05, Student’s t test). C, H2O2 quantification using luminol-based chemiluminescence (n = 4; ±se). Asterisks indicate statistically significant differences between wild-type Q and nadC Q (P < 0.05, Student’s t test). FW, Fresh weight. D, Disparities in ROS production in NAD+-infiltrated signaling mutants. Leaves of wild-type, rbohD, rbohF, rbohD/F, and sid2-1 genotypes were infiltrated with water or 1 mm NAD+, and DCFH-DA fluorescence (n = 12; ±se) was quantified 20 h later. The purple bar shows coinfiltration with the NADPH oxidases inhibitor diphenylene iodonium (DPI; 25 µm) in Col-0. Letters denote statistically significant differences (P < 0.05, Student’s t test).
The NAD-Related Oxidative Burst Is Not under the Control of SA Biosynthesis and NADPH Oxidase
To check whether ROS and SA might interact in the nadC lines to synergistically amplify the defense loop (Pétriacq et al., 2013), we measured the NAD-associated ROS burst in the SA biosynthesis mutant sid2-1 and the RBOH-dependent ROS production mutants rbohD, rbohF, and rbohD/F. Illuminated intact leaves were infiltrated with water or NAD+ (1 mm), and ROS-dependent DCFH-DA fluorescence was analyzed 20 h later. Wild-type Col-0 plants showed higher levels of NAD-induced ROS production compared with water-infiltrated leaves (Fig. 2D). Thus, this indicates that NAD+ alone is able to increase ROS production in Arabidopsis leaves. Since Q leads to much lower NAD contents than direct NAD addition (Supplemental Fig. S1), in the second case, effects are visible even without inoculation. Remarkably, however, coinfiltration with the NADPH oxidase inhibitor DPI (Li and Trush, 1998; Müller et al., 2009) did not reduce NAD-induced ROS production. In addition, rbohD, rbohF, rbohD/F, and sid2-1 mutants showed similar levels of NAD-associated ROS to wild-type plants, indicating that neither RBOH D/F activities, nor SA synthesis, were required for NAD-associated production of ROS (Fig. 2D). To corroborate these results, we used nadC plants to increase endogenous NAD contents after Q treatment for 48 h. One hour prior to infection with Pst-AvrRpm1 or D. dadantii, we infiltrated leaves with DPI. In both pathosystems, NAD-correlated augmentation of pathogen-inducible ROS was remarkably insensitive to DPI, which was evident for luminol assay and DCFH-DA fluorescence (Supplemental Fig. S4). This confirms that RBOH is not necessary for NAD-associated ROS production, thereby suggesting that NAD-associated ROS originate from a different enzymatic pathway.
NAD-Based ROS Production Depends on Mitochondrial Respiration But Not on Light Metabolism
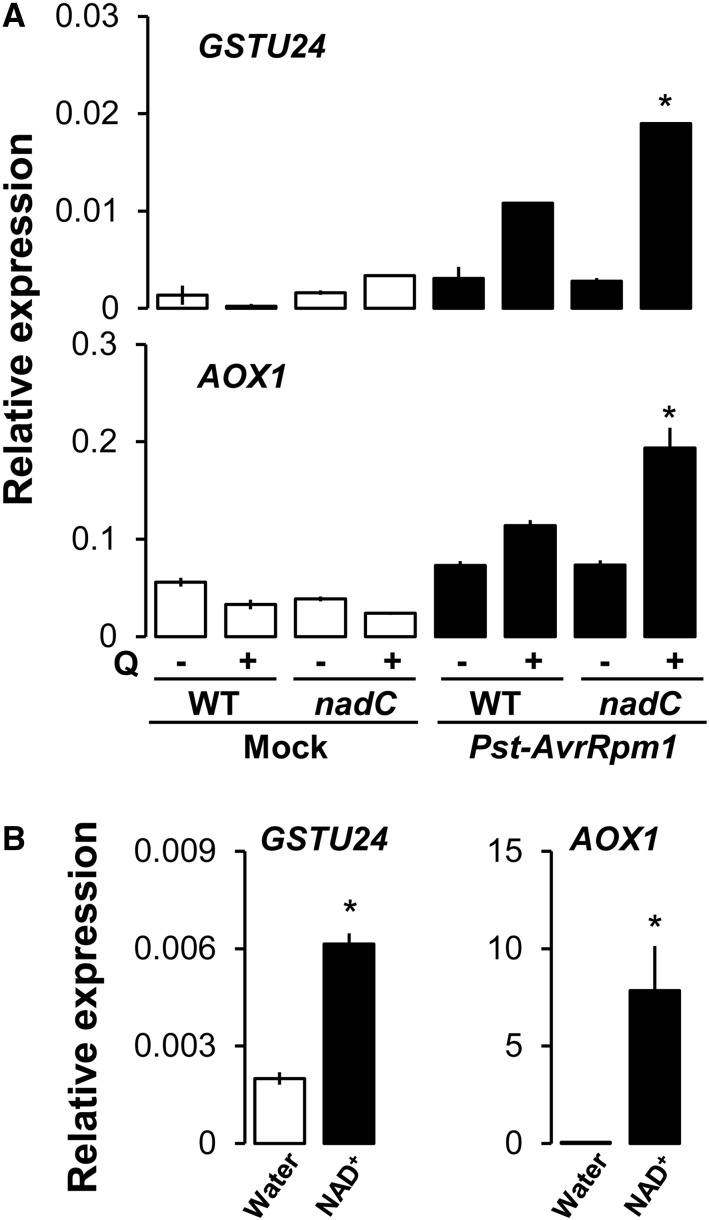
To get further insights on the source of the NAD-based ROS burst, we assessed the expression of GSTU24, a cytosolic oxidative stress marker (Queval et al., 2009), and the mitochondrial redox marker AOX1 (Simons et al., 1999; Polidoros et al., 2009). This allowed us to detect oxidative stress in different cellular compartments known for ROS production (Dixon et al., 2010; Gill and Tuteja, 2010). Illuminated intact wild-type and nadC leaves were infiltrated with Q and challenged with Pst-AvrRpm1 48 h later (Supplemental Fig. S2). Mock-infiltrated and Pst-AvrRpm1-infected leaves were analyzed for wild-type and nadC plants at 20 hpi (Fig. 3A). Both genes showed increased expression in wild-type and nadC plants following Pst-AvrRpm1 (Fig. 3), which was augmented in Q-pretreated nadC plants. Since NAD alone caused an oxidative burst (Fig. 2D), we further checked the expression levels of GSTU24 and AOX1 in response to direct NAD treatment and observed an up-regulation of both redox markers (Fig. 3B). This suggests cytosolic and mitochondrial redox perturbations at higher NAD levels.
Figure 3.
Oxidative stress transcripts are up-regulated at higher NAD. Transcriptional expression is shown relative to the reference gene ACTIN2 of redox marker genes of the cytosol (GSTU24) and the mitochondrion (AOX1) in mock-infiltrated (white bars) and Pst-AvrRpm1-infected (black bars) leaves of wild type (WT) Col-0 and nadC plants at 20 hpi, with (+) or without (−) Q pretreatment (A), and 24 h after infiltration of wild-type Col-0 leaves with water (white bars) or 1 mm NAD+ (black bars; B). Values shown indicate means of three biological replicates ± se (n = 3). Asterisks in A denote statistically significant differences between wild-type Q and nadC Q, and asterisks in B denote statistically significant differences between water and NAD+ treatments (P < 0.05, Student’s t test).
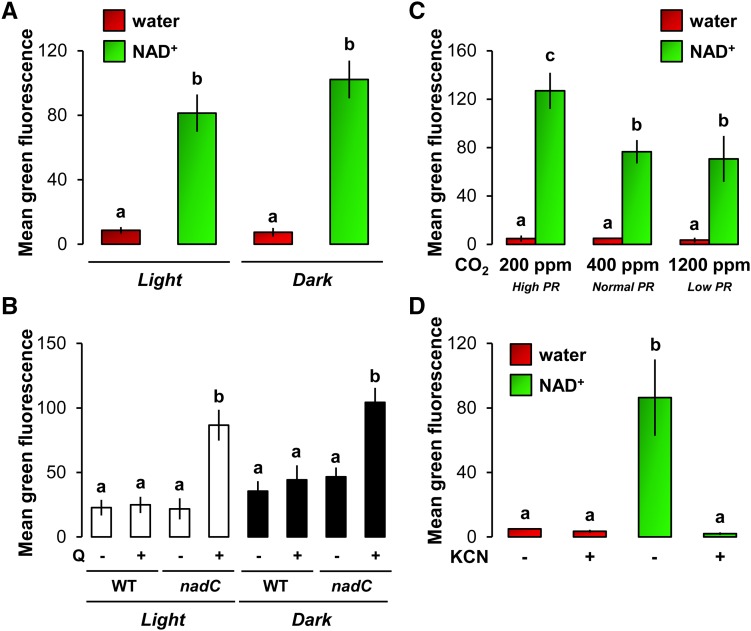
In addition, we detected ROS by DCFH-DA fluorescence in response to exogenous NAD+ as a proxy to test the source of ROS production (Fig. 4). Remarkably, NAD-based ROS accumulation was not reduced in the dark after NAD+ treatment alone (Fig. 4A) and in infected Q-treated nadC leaves compared with normal light conditions (Fig. 4B). ROS detection in NAD+-treated Col-0 plants grown at different photorespiration regimes showed that increased photorespiration rates stimulated the NAD-based ROS burst compared with normal photorespiration conditions (Fig. 4C). However, lower photorespiration in plants grown at elevated CO2 only led to a slightly nonsignificant decrease in NAD-based ROS burst (Fig. 4C). This indicates that NAD-based production of ROS is not likely to come from photosynthetic and photorespiration metabolisms. More importantly, KCN-dependent inhibition of mitochondrial respiration revealed that the NAD-related ROS was totally abolished when respiration was inhibited (Fig. 4D). We then tested cellular respiration after direct NAD+ treatment and upon intracellular NAD buildup in Q-treated nadC (Supplemental Fig. S5). We measured a higher capacity of respiration (Supplemental Fig. S5A), which was correlated in Q-treated nadC with enriched pools of the mitochondrial TCAP metabolites 2-oxoglutarate and succinate (Supplemental Fig. S5B). Taken together, our results demonstrate that respiration capacity is increased when NAD accumulates, which in turns augments mitochondria-derived ROS burst.
Figure 4.
Relationships between NAD-associated ROS production, light metabolism, and (photo)respiration. A, Analysis of ROS production in Col-0 leaves 24 h after infiltration with water (red bars) or 1 mm NAD+ (green bars) under normal light conditions (Light) or in the absence of light (Dark; see “Materials and Methods”). B, Leaves of wild-type (WT) Col-0 and nadC plants were infiltrated with (+) or without (−) 5 mm Q to increase NAD contents in Q-treated nadC, and 48 h later, the same leaves were infected with Pst-AvrRpm1 and analyzed at 24 hpi for ROS production by DCFH-DA fluorescence under normal light conditions (Light; white bars) or in the absence of light (Dark; black bars). C, Analysis of ROS in Col-0 leaves from plants grown continuously at decreasing photorespiration (PR) rates and treated for 24 h with water (red bars) or 1 mm NAD+ (green bars) under normal light conditions. D, Analysis of ROS in Col-0 leaves 24 h after infiltration with water (red bars) or 1 mm NAD+ (green bars) under normal light conditions. To inhibit mitochondrial respiration, leaves were coinfiltrated with or without the respiration inhibitor KCN (1 mm). Values shown represent means of ROS-related DCFH-DA fluorescence ± se (n = 12). Letters above the bars indicate statistically significant differences (P < 0.05, Student’s t test).
Higher NAD Correlates with Increased Callose Deposition and Cell Death in nadC Leaves
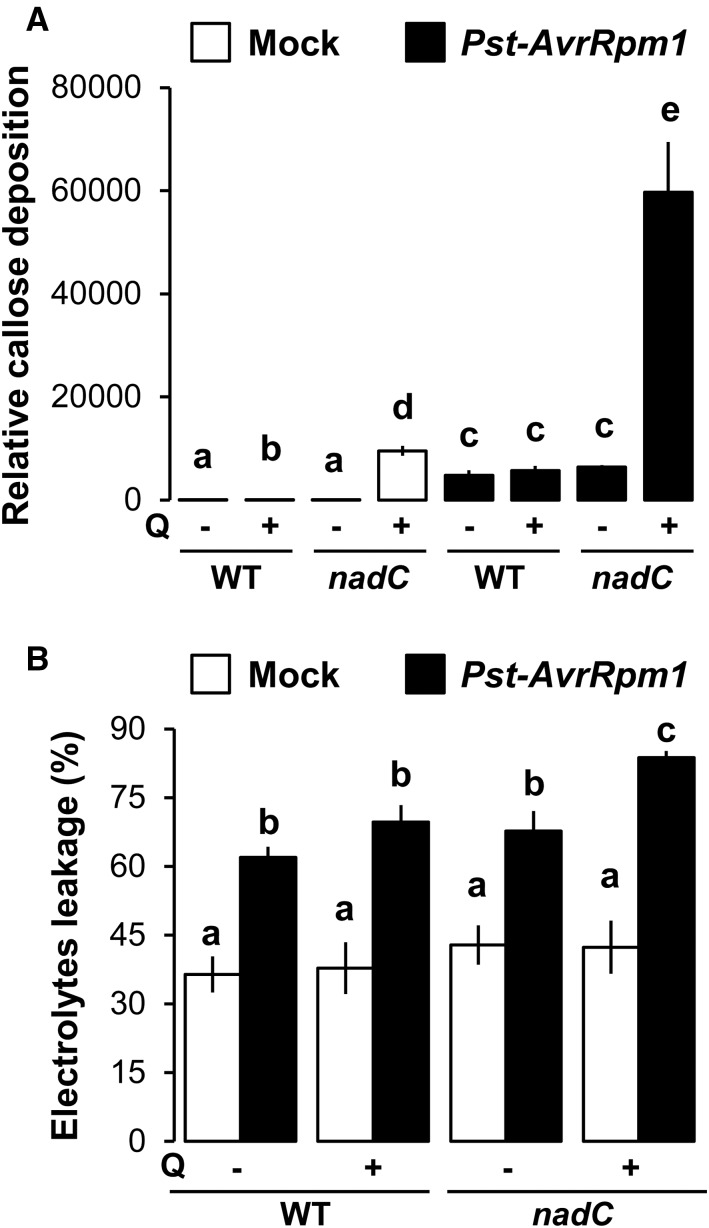
Callose deposition often is used as a marker for PAMP-triggered and effector-triggered immunity in plants (Luna et al., 2011; Pastor et al., 2013). A recent study revealed a link between NAD catabolism (i.e. PARP) and callose deposition (Song et al., 2015). Furthermore, higher NAD contents in nadC have been shown to correlate with increased pools of SA, which can lead to cell death (Supplemental Fig. S1; Yoshimoto, 2010; Pétriacq et al., 2012). To gain further insight in the physiological mechanisms underlying the NAD-mediated resistance to Pst-AvrRpm1, we examined callose deposition by Aniline Blue staining. In addition, we measured cell death by conductivity measurements of electrolyte leakage before and following infection with Pst-AvrRpm1 (48 hpi). Following mock inoculation (white bars in Fig. 5A and Supplemental Fig. S6), Q-treated wild-type plants showed a relatively minor increase in callose compared with the wild type, suggesting that redox perturbations by Q treatment (Fig. 2) mildly induced callose deposition. In Q-treated nadC leaves, levels of callose were higher (92-fold compared with the Q-treated wild type), indicating a direct impact of Q-induced NAD on callose. After challenge inoculation with Pst-AvrRpm1 (black bars in Fig. 5A and Supplemental Fig. S6), pathogen-induced callose deposition was strongly augmented in Q-treated nadC plants, which was 10-fold higher than callose levels in Q-treated wild-type plants after pathogen challenge.
Figure 5.
Callose deposition and cell death are stimulated against Pst-AvrRpm1 when NAD pools are higher. A, Aniline Blue staining of callose at 48 hpi in wild type (WT) Col-0 and nadC leaves with (+) or without (−) Q and treated with mock solution (white bars) or infected with Pst-AvrRpm1 (black bars). Relative callose intensity was analyzed by UV epifluorescence microscopy and quantified from digital photographs. Values shown are mean areas of callose per leaf relative to total leaf area ± se (n = 12). B, Electrolyte leakage was assessed by conductivity measurements at 48 hpi after mock solution (white bars) or Pst-AvrRpm1 (black bars) infiltration in wild-type and nadC plants with or without Q from four biologically replicated samples (n = 4; ±se). Letters above the bars denote statistically significant differences (P < 0.05, Student’s t test).
Despite the increase in SA levels (Supplemental Fig. S1C), Q-induced NAD production in nadC (i.e. mock-inoculated nadC Q; white bars in Fig. 5B) failed to induce cell death, as evidenced by unaltered levels of electrolyte leakage. As expected, Pst-AvrRpm1 stimulated cell death under all conditions (Fig. 5B, black versus white bars). However, Q-treated nadC plants showed reproducibly augmented levels of electrolyte leakage at 48 hpi with Pst-AvrRpm1. This indicates that NAD buildup in Q-treated nadC stimulates pathogen-induced HR. Consistent with a more oxidized medium state (Supplemental Fig. S1B), a stimulation of ROS production (Fig. 2), and an up-regulation of redox markers (Fig. 3), our results suggest that elevated pools of NAD in Q-treated nadC exacerbate effector-triggered immunity that could participate in the increased resistance to Pst-AvrRpt2 (Fig. 1) and Pst-AvrRpm1 (Pétriacq et al., 2012).
NAD Elicits a Defense-Related Metabolic Signature That Is Similar to PAMP-Triggered Immunity
It is not clear how NAD-related defense responses favor the resistance to both biotrophic and necrotrophic pathogens (Fig. 1), possibly by altering the hormonal balance controlling resistance to these pathogens (Pétriacq et al., 2013). Since NAD, like ATP and Glc, is essential for the primary functions of metabolism, we hypothesized that NAD could act as a defense-eliciting DAMP signal during tissue disruption. To compare the effects of NAD with those of other defense elicitors (PAMPs and DAMPs), we carried out untargeted metabolic profiling by liquid chromatography-mass spectrometry (UPLC-qTOF-MSE; see Supplemental Methods S1) from leaf material of two independent experiments. The first experiment compared metabolic profiles from intact illuminated wild-type leaves that had been infiltrated for 48 h with NAD+, two DAMPs (ATP and Glc), two PAMPs (flagellin [Flg22] and chitin), and the defense phytohormones SA and jasmonic acid (JA; which regulate resistance to biotrophic and necrotrophic pathogens, respectively; Pieterse et al., 2012). The second experiment compared profiles from water- or Q-treated leaves of wild-type and nadC plants. Metabolomics data were subjected to supervised multivariate statistics by OPLS-DA for both experiments (elicitors or nadC) in order to reveal the changes in the metabolic profiles that were associated with the resistance response to NAD and elicitors. The corresponding OPLS-DA statistical models displayed high levels of correlation and predictability (detailed in Supplemental Table S1), suggesting that sample grouping and associated metabolomics patterns were statistically significant (Worley and Powers, 2013).
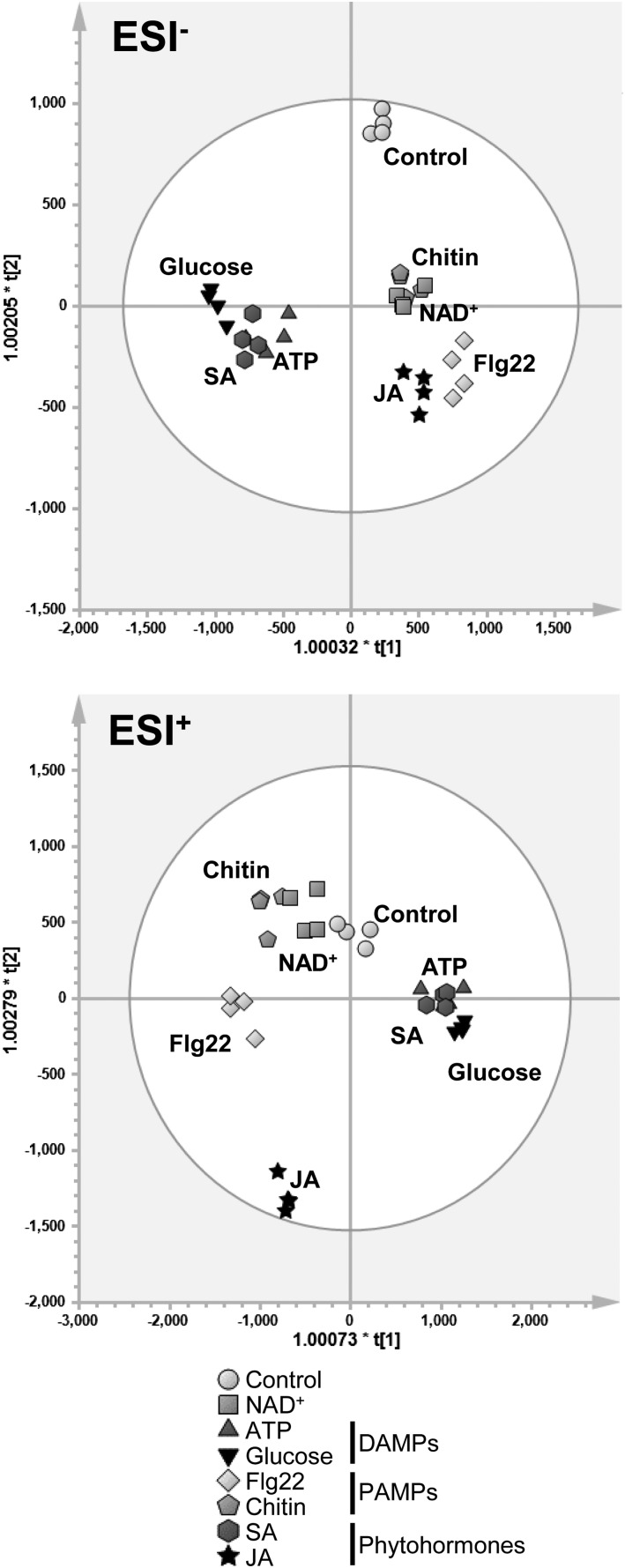
Based on 5,112 and 10,227 metabolic features (mass-to-charge ratio [m/z] ions) detected in negative (ESI−) and positive (ESI+) ionization modes from the first experiment (Fig. 6), the metabolic profile of NAD+-treated plants resembled that of chitin-treated plants. Metabolic similarities also were observed between NAD+ and Flg22 samples in both ESI− and ESI+ (Fig. 6) and between NAD+ and JA samples in ESI− (Fig. 6, top). This suggests that NAD+ elicits metabolic patterns that are similar to those triggered by defense hormones and PAMPs. Furthermore, profiles of ATP- and Glc-treated plants clustered together in both ionization modes, supporting their role as common DAMPs. In addition, hierarchical clustering analysis (HCA) of samples clearly showed levels of similarity between metabolic profiles of NAD+-, chitin-, and flagellin-treated plants (Supplemental Fig. S7). Overall, these results indicate that exogenously applied NAD+ elicits metabolic signatures that are more similar to those of PAMPs than those of DAMPs. Surprisingly, SA and NAD+ treatments did not plot together (Fig. 6, hexagons and squares). This suggests that, despite being positively correlated, SA and NAD+ elicit distinct chemical responses, possibly due to the effect of NAD+ as an enzymatic cofactor in primary metabolism or the induction of SA-independent defense responses by NAD (Bartsch et al., 2006; Ge et al., 2007; Ge and Xia, 2008; Zhang and Mou, 2009, 2012; Ishikawa et al., 2010; Jambunathan et al., 2010).
Figure 6.
OPLS-DA of unfiltered 5,112 anions (ESI−) and 10,273 cations (ESI+) from untargeted UPLC-qTOF-MSE metabolic profiling. Five-week-old illuminated leaves of Col-0 were harvested 48 h after treatment with PAMPs, DAMPs, or phytohormones, and leaf extracts were analyzed by UPLC-qTOF-MSE. Data were Pareto scaled prior to multivariate analyses in SIMCA (version 13.0.3). Each biologically replicated sample is indicated by a single data point in the projection (n = 4). For both ionization modes, validation parameters were highly satisfactory (Supplemental Table S1). ESI, Electrospray ionization.
To examine the impacts of endogenous NAD accumulation in the second experiment, we first conducted supervised OPLS-DA of water- or Q-treated wild-type and nadC plants, using data collected in both negative (5,107 features) and positive (10,207 features) ionization modes (Supplemental Fig. S8A). The resulting OPLS-DA plot differentiated samples according to Q treatment and genotype, demonstrating that each condition is associated with specific metabolic patterns. To obtain a global impression of the impacts of each condition on the plant metabolome, we conducted an unsupervised principal component analysis (Supplemental Fig. S8B) and HCA (Supplemental Fig. S8C). As expected, both unsupervised analyses revealed that samples from Q-treated nadC plants clustered apart from samples of untreated nadC, untreated wild-type, and Q-treated wild-type plants. Hence, intracellular NAD accumulation in Q-treated nadC has a profoundly bigger impact on the plant metabolic profiles than Q treatment of wild-type plants or nadC overexpression alone.
Next, we followed a top-down approach to identify common metabolic markers (m/z ions) that are responsible for the specific metabolic signatures captured in the OPLS-DA plots of both experiments (Fig. 6; Supplemental Fig. S8A). To this end, we retained all metabolic discriminators of each plot with a variable importance for the projection (VIP) score higher than 1 in both experiments, yielding 185 anions (ESI−) and 244 cations (ESI+; Supplemental Table S2). Subsequently, the selected VIP scores from each experiment were plotted against each other (Supplemental Fig. S9) in order to visualize markers with the highest discriminatory power in both experiments. In such a graphical representation, metabolic markers that appear at the extreme ends of the y and x axes are the best discriminators specific to the treatment with elicitors and the nadC + Q effect, respectively. Metabolic markers that have both high x and y coordinates are good discriminators in both experiments. This graphical representation allows for the selection of metabolic markers that explain the highest level of metabolic variation in both the first experiment (which determined metabolic impacts of defense elicitors) and the second experiment (which assessed metabolic impacts of nadC overexpression and Q treatment). By selecting metabolic markers with a sum of each VIP value greater than 10, we identified a set of 32 markers that exerted the highest discriminatory power in both experiments. The statistical significance of these markers was then validated by univariate statistics (ANOVA, P < 0.01 adjusted for multiple testing). This stringent analysis of metabolites, annotated from accurate mass and online databases (Supplemental Methods S1), included two anions of SA-related metabolism, two anions and 12 cations associated with lipids, five anions and four cations of the phenylpropanoid family, and six anions of the glucosinolate pathway (Supplemental Table S3). Based on these putative identifications, a biochemical pathway was considered relevant when two or more predicted markers belonged to the same pathway. These top metabolic markers (32) were further analyzed by clustering analysis (Supplemental Fig. S10) to confirm their differential patterns in both experiments (elicitors and the nadC Q). After exogenous application of NAD+ (Supplemental Fig. S10A) and in Q-treated nadC (Supplemental Fig. S10B), heat maps showed higher intensities of ions that mapped to defense metabolism, such as SA-related compounds (e.g. m/z = 299.077), phenylpropanoids (e.g. m/z = 377.086), and lipids (e.g. m/z = 326.379). Interestingly, these conditions also triggered changes in glucosinolate ion signals (e.g. m/z = 436.040 and 477.064, respectively), suggesting a dynamic regulation of the glucosinolate pathway in response to NAD. Furthermore, exogenous application of NAD+ and Q-treated nadC showed only partial overlap (Supplemental Fig. S10). This indicates that exogenous and endogenous NAD+ trigger partly distinct defense-related metabolites, suggesting differential mechanisms for intracellular and intercellular supplemental NAD in plant defense. The HCA presented in Supplemental Fig. S10B also reveals a separate cluster for samples from untreated nadC-overexpressing plants, which results from the discriminating analysis (OPLS-DA) shown in Supplemental Fig. S8A. This suggests that nadC overexpression alone has an effect on the plant’s metabolic profile that is not related to endogenous NAD.
Higher NAD Correlates with the Accumulation of Defense Regulatory Phytohormones
To analyze the impacts of NAD on defense regulatory hormones, we quantified relative amounts of SA, JA, and abscisic acid (ABA) in samples from both experiments (Supplemental Methods S1). This defense hormone profiling showed highly significant variation between conditions (Table I; P < 9 × 10−5, ANOVA adjusted for multiple testing). While Flg22 increased both SA and ABA, only NAD+-treated wild-type plants and Q-treated nadC plants showed elevated levels of JA, SA, and ABA in comparison with the corresponding controls (Table I; Supplemental Fig. S11). Thus, both exogenously applied NAD+ in wild-type plants and enhanced production of intracellular NAD+ in Q-treated nadC plants induced defense hormones, thereby stimulating resistance to the contrasted pathogens (Fig. 1).
Table I. Relative quantification of defense hormones.
Fold change analysis is shown for defense phytohormones detected by UPLC-qTOF-MSE in wild-type Col-0 and nadC illuminated leaves treated for 48 h with elicitors/chemicals. Data are means ± se (n = 4). For all hormones, P values across the whole data set were very low (P ≤ 9E-05). Asterisks indicate statistically significant differences between treatment and control conditions (Student’s t test, P < 0.05). The charts corresponding to these data are presented in Supplemental Figure S11.
| Samples |
SA |
JA |
ABA |
|||
|---|---|---|---|---|---|---|
| Mean | se | Mean | se | Mean | se | |
| Control | 1.0 | 0.4 | 1.0 | 0.1 | 1.0 | 0.2 |
| NAD | 3.9* | 0.2 | 2.1* | 0.5 | 2.1* | 0.3 |
| ATP | 0.8 | 0.0 | 0.7 | 0.1 | 1.2 | 0.3 |
| Glc | 0.8 | 0.1 | 0.9 | 0.1 | 1.2 | 0.1 |
| Flg22 | 4.8* | 1.2 | 1.0 | 0.2 | 1.8* | 0.4 |
| Chitin | 1.6 | 0.3 | 1.0 | 0.1 | 1.8* | 0.3 |
| SA | 81.2* | 4.1 | 1.0 | 0.1 | 1.9* | 0.1 |
| JA | 1.8 | 0.6 | 35.2* | 1.6 | 2.1* | 0.4 |
| Wild type | 1.0 | 0.3 | 0.9 | 0.2 | 1.0 | 0.4 |
| Wild-type Q | 0.7 | 0.2 | 1.6 | 0.2 | 1.5 | 0.1 |
| nadC | 0.9 | 0.1 | 1.1 | 0.4 | 1.0 | 0.3 |
| nadC Q | 2.7* | 0.1 | 2.5* | 0.1 | 2.7* | 0.1 |
We finally validated the outcome of our metabolic profiling by measuring the expression of defense-related genes after direct NAD+ treatment (Supplemental Fig. S12). Reverse transcription-quantitative PCR (RT-qPCR) analysis revealed the induction of marker genes of SA (PR1, PR5, and ICS1), JA (VSP2 and PDF1.2), chitin (CHIT), flagellin (FRK1), and glucosinolates (PEN2 and CYP81F2), thus confirming a role for exogenous NAD+ in these defense pathways.
DISCUSSION
A growing body of evidence suggests that the redox carrier NAD is a key player in plant immunity, although the specific mechanisms involved remain poorly understood and the lack of loss-of-function genetic resources renders it difficult to test all the hypotheses (Zhang and Mou, 2009, 2012; Macho et al., 2012; Pétriacq et al., 2012, 2013). Here, we have investigated the beneficial effects of supplemental NAD+ to defense efficacy using both exogenous treatment with NAD+ and the Q-inducible nadC line.
NAD-Mediated Defense Is Effective against Pathogens with Diverse Infection Strategies
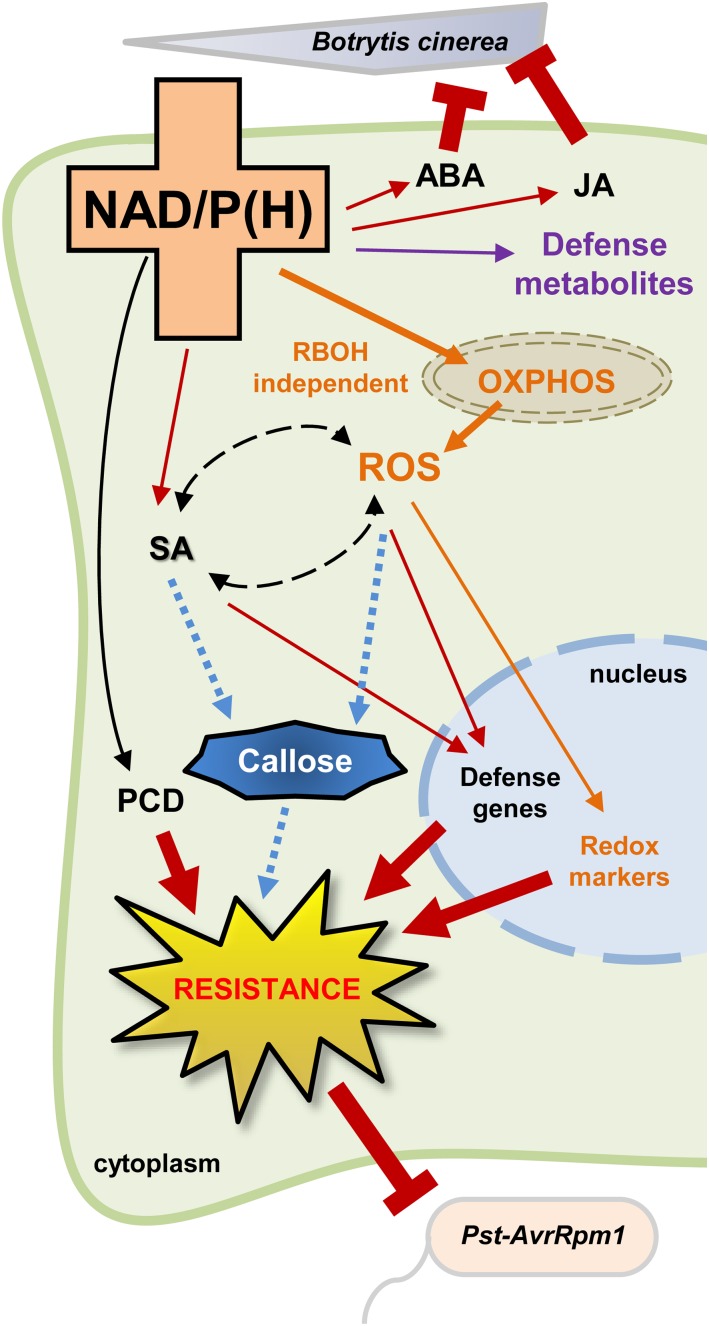
It is commonly believed that responses against (hemi)biotrophic and necrotrophic pathogens are rather distinct (Thomma et al., 1998; Glazebrook, 2005). Typically, the plant immune system activates SA- and ROS-related defenses against (hemi)biotrophs that feed on plant living cells and employ effector molecules that deregulate plant immunity (e.g. Pst-AvrRpm1 and Pst-AvrRpt2). In contrast, necrotrophs (e.g. B. cinerea) thrive on dead or decomposing tissues and are resisted by JA- and ethylene-controlled defense mechanisms (Glazebrook, 2005; Pel and Pieterse, 2013). Surprisingly, our pathogenicity assays clearly showed that NAD-induced defense is effective against a range of pathogens with different lifestyles (Fig. 1), including the avirulent hemibiotrophic pathogen Pst-AvrRpt2 and the virulent hemibiotrophic pathogen D. dadantii (which are resisted by SA- and ROS-related defenses; Katagiri et al., 2002; Fagard et al., 2007; Kraepiel et al., 2011) as well as the necrotrophic pathogen B. cinerea (which is sensitive to JA-dependent defenses; Glazebrook, 2005). Our metabolomics analysis has demonstrated that exogenously applied NAD and intracellular accumulation of NAD simultaneously increased pools of SA, JA, and ABA (Table I; Supplemental Fig. S11), and this was consistent with NAD stimulation of marker genes of SA and JA pathways (Supplemental Fig. S12). Thus, the stimulation of hormonal signaling probably contributed to the enhanced resistance of the NAD-enriched nadC plants (with Q) against different pathogens (Fig. 1). These results support our previous finding that intracellular NAD increases SA accumulation and induces the expression of both SA-dependent (PR1, ICS1, and AIG2) and JA-dependent (JAZ8, GLIP1, and CEJ1) marker genes (Pétriacq et al., 2012). Data mining on these transcriptomic data also showed that NAD-related expression profiles were similar to several plant-pathogen interactions, including B. cinerea (Pétriacq et al., 2012). This also agrees with previous studies on NAD catabolism that have reported that NAD can increase the activity of both SA-dependent and SA-independent pathways (Bartsch et al., 2006; Ge et al., 2007; Ge and Xia, 2008; Zhang and Mou, 2009, 2012; Ishikawa et al., 2010; Jambunathan et al., 2010). Our results provide further compelling evidence that elevated NAD content activates a broad spectrum of SA-dependent and SA-independent immune responses (Fig. 7).
Figure 7.
Higher NAD modulates defense responses by stimulating ROS and hormonal balance. OXPHOS, Mitochondrial oxidative phosphorylation; PCD, programmed cell death.
NAD Stimulates an Oxidative Stress
Until now, the link between NAD signaling and ROS production was somewhat uncertain. Our results demonstrate that increased pools of intracellular NAD+ in Q-treated nadC plants, as well as exogenous leaf infiltration of NAD+ in wild-type plants, considerably stimulate ROS accumulation (Fig. 2; Supplemental Fig. S3). Given the comparable augmentation in ROS bursts between Q-treated nadC plants challenged with different bacterial pathogens (Pst-AvrRpm1 and D. dadantii), our data suggest that NAD-induced ROS accumulation is not involved solely in the incompatible interaction with avirulent pathogens. A plausible hypothesis to link ROS production to higher NAD would be through the redox potential of pyridine nucleotides, which can be used directly by NADPH oxidases to boost pathogen-induced ROS production (Torres, 2010; Marino et al., 2012). Surprisingly, however, our results showed that neither genetic mutation of RBOHD and RBOHF (Fig. 2D) nor pharmacological inhibition of NADPH oxidases by DPI (Supplemental Fig. S4) was effective in blocking NAD-induced ROS production. NAD+ application also failed to induce the expression of the RBOHD gene (Supplemental Fig. S12), and NAD-mediated ROS production was RBOH independent after infection by Pst-AvrRpm1 and D. dadantii (Supplemental Fig. S4).
Besides RBOH, it is believed that stress-related ROS are mostly generated by the electron transport chains of the chloroplast and the mitochondrion and by glycolate oxidase activity involved in the photorespiration cycle (Torres, 2010; Rojas and Mysore, 2012; Rojas et al., 2012; Hodges et al., 2016). Here, analysis of ROS accumulation in the dark showed that the NAD-related oxidative burst was uncoupled from light metabolism (Fig. 4). This was supported by gene expression analysis of the chloroplastic redox markers FER1 and APX2 (Oelze et al., 2012), which did not show significant regulation upon NAD+ treatment (Supplemental Fig. S12). Previously, we reported unchanged photosynthetic metabolites and parameters between wild-type plants and the NAD-overproducing system nadC (Guérard et al., 2011). While higher photorespiration rates led to a more pronounced ROS burst in response to NAD+, decreasing photorespiration did not significantly lower the oxidative stress (Fig. 4C). Hence, a possible role of photorespiration in NAD-mediated defense signaling cannot be excluded (Sørhagen et al., 2013), but it is not strictly required to increase the NAD-associated accumulation of ROS.
The cytosolic GST gene GSTU24 is H2O2 and SA inducible and up-regulated by pathogens (Queval et al., 2009). AOX1 is a mitochondrial alternative oxidase that is induced by avirulent Pst (Simons et al., 1999) and links mitochondrial ROS to cell death (Garmier et al., 2007; Vidal et al., 2007). Remarkably, it has been proposed that mitochondrial H2O2 is quantitatively more important than H2O2 coming from chloroplast- and RBOH-originated ROS in elicitor-induced Arabidopsis and tobacco (Nicotiana tabacum) cells (Garmier et al., 2007). Here, we show that higher NAD might link to mitochondrial metabolism upon biotic stress. First, the NAD-based ROS production is not under the control of RBOH (Fig. 2; Supplemental Fig. S4) and is totally abolished when mitochondrial respiration is inhibited (Fig. 4D). This strongly suggests that the NAD-based ROS burst originates from the mitochondria. Second, analysis of cytosolic and mitochondrial redox markers revealed an oxidative stress in the cytosol and the mitochondrion in response to NAD+ (Fig. 3). Third, respiration capacity was increased in response to exogenous NAD+ and in Q-treated nadC (Supplemental Fig. S5A) together with enriched pools of 2-oxoglutarate and succinate (Supplemental Fig. S5B). These organic acids have been shown to be closely associated in the mitochondria (Araújo et al., 2008), where succinate dehydrogenase is involved in ROS production in plants (Jardim-Messeder et al., 2015). Higher NAD could possibly stimulate respiration fluxes that lead to higher mitochondrial ROS. Taken together, our results exemplify an important role of NAD in mitochondrial metabolism and stress. This is in line with studies on NAD-enriched mitochondrial mutants that were more tolerant to stress, including pathogens (Dutilleul et al., 2003, 2005; Djebbar et al., 2012).
After infection with Pst-AvrRpm1, we detected enhanced cell death levels in Q-treated nadC plants (Fig. 5B). Despite an accumulation of ROS (Fig. 2) and SA in basal conditions (Pétriacq et al., 2012; Supplemental Figs. S1 and S11), cell death was not stimulated in Q-treated nadC plants after mock inoculation. This suggests that, upon NAD addition, cell death was independent of SA and ROS accumulation in the absence of bacterial infection; conversely, it was at least partly SA and ROS dependent during infection by avirulent pathogens. NPR1, an SA receptor and master regulator of SA-related immunity, is redox activated (Mou et al., 2003; Tada et al., 2008; Wu et al., 2012), and Zhou et al. (2015) reported that a treatment with SA under constant light significantly perturbs NADP(H) pools and the NADP-NADPH ratio. Reciprocally, NAD can induce the expression of PR1 via both SA-dependent and SA-independent pathways (Zhang and Mou, 2009; Pétriacq et al., 2012). However, mutation in SA biosynthesis (sid2-1) had no effect on ROS production (Fig. 2), and NAD and SA treatments elicited distinct metabolic responses (Fig. 6; Supplemental Fig. S7). These observations can be reconciled with the assumption that NAD can stimulate ROS production requiring neither SA synthesis nor SA-stimulated RBOH activity. This would agree with the induction of PR genes in the rboh mutant after extracellular NAD treatment (Zhang and Mou, 2009). In summary, NAD increases both SA synthesis (Supplemental Figs. S11 and S12) and ROS production in an SA-independent manner (Fig. 2).
The Effect of NAD on Metabolomics Pattern Matches That of Elicitors
Exogenous NAD has been postulated to act as a DAMP following its release from damaged plant cells into the extracellular compartment (Zhang and Mou, 2009, 2012). Thus far, a plant receptor for NAD has not been identified, and the biological relevance of NAD signaling in DAMP-triggered immunity remains uncertain. Both PAMPs and DAMPs induce the deposition of callose at the cell wall, which contributes to the inhibition of pathogen colonization in the apoplast (Ellinger and Voigt, 2014). Callose deposition is preceded and potentiated by ROS (Luna et al., 2011; Pastor et al., 2013). Furthermore, studies on parp and parg mutants, which are impaired in NAD-consuming ADP-ribosylation, have reported that NAD catabolism is important for callose deposition (Briggs and Bent, 2011; Song et al., 2015). In line with these works, we have shown that elevated NAD levels in Q-treated nadC plants enhanced callose deposition under basal conditions and drastically augmented callose levels after infection with Pst-AvrRpm1 (Fig. 5A; Supplemental Fig. S6). Hence, NAD acts as an elicitor, thereby stimulating callose deposition.
Our metabolic analysis further revealed changes in secondary defense metabolism induced by NAD. These changes were similar to those caused by resistance-inducing PAMPs. Multivariate analysis of metabolic discriminators (OPLS-DA) indicated striking similarities in the metabolic response to chitin and NAD (Fig. 6), which were confirmed by clustering analysis (Supplemental Fig. S7). After cellulose, chitin is the second most ubiquitous polysaccharide in fungal cell walls (Sánchez-Vallet et al., 2015; Shinya et al., 2015). In Arabidopsis, chitin perception by CERK1 leads to a burst of ROS (Miya et al., 2007), and chitin, or its derivative chitosan, increases callose deposition and cell death (Hamel and Beaudoin, 2010; Luna et al., 2011; Hadwiger, 2013). These effects of chitin signaling are supported by our data, since NAD is found to be a potent elicitor of ROS (Fig. 2), callose deposition, and cell death (Fig. 4) and directly induces the expression of CHIT (Supplemental Fig. S12), a chitin-responsive gene (Gouhier-Darimont et al., 2013). Remarkably, an analysis of ROS burst in the cerk1-2 mutant impaired in the chitin response indicated reduced ROS production after NAD application (Supplemental Fig. S13). Since chitin-induced defense contributes to resistance against fungal infection (Tanabe et al., 2006; Povero et al., 2011; Hadwiger, 2013), it supports the view that NAD plays a role in PAMP-triggered immunity against necrotrophic fungi, such as B. cinerea (Fig. 1). This also supports our previous transcriptome study, which reported NAD-specific induction of genes coding for chitinases and cell wall-related metabolism (Pétriacq et al., 2012).
Multivariate analysis of important metabolic discriminators of the NAD-induced metabolome further demonstrated a role for various defense pathways (Supplemental Figs. S9 and S10; Supplemental Table S3). This analysis revealed enhanced accumulation of glucosinolates in particular in response to exogenously applied NAD+ (Supplemental Figs. S9 and S10). In addition to metabolomics data, we also found an up-regulation of the glucosinolate-responsive genes PEN2 and CYP81F2 after direct NAD+ treatment (Supplemental Fig. S12). This conclusion is further supported by previous transcriptomics data indicating NAD-specific regulation in Q-treated nadC of genes involved in glucosinolate metabolism (e.g. CYP81F2, BGLU38, and BGLU37; Pétriacq et al., 2012). Glucosinolates are amino acid-derived sulfur-containing defense metabolites known for their role in defense responses (Brader et al., 2001; Halkier and Gershenzon, 2006), in particular against (hemi)biotrophic pathogens, where they act synergistically with SA (Bednarek and Osbourn, 2009; Schlaeppi et al., 2010; Wang et al., 2013). Hence, NAD participates in adjusting glucosinolate pools, since (1) higher NAD stimulates amino acid metabolism (Pétriacq et al., 2012) from which glucosinolates are synthesized, and (2) NAD induces SA-related responses that might regulate glucosinolate signaling. Besides glucosinolates, untargeted metabolomics also reported the role of phenylpropanoids and lipid-derived molecules in response to NAD (Supplemental Figs. S9 and S10; Supplemental Table S3). These pathways have been shown to play a role in defense, notably SA-related pathways (for review, see Cheynier et al., 2013; Zhang and Xiao, 2015). Here again, the outcome of our metabolome analysis fits with the NAD-regulated expression of genes involved in phenolics and lipid metabolisms (e.g. O-methyltransferases and glucosidases; Pétriacq et al., 2012).
CONCLUSION
We have shown that NAD triggers the production of ROS and defense hormones and reorchestrates the defense metabolome. In addition to a direct induction of callose, NAD also primes pathogen-induced callose and cell death. Furthermore, we have demonstrated that the metabolic response to NAD resembles the metabolic response to resistance-inducing PAMPs. In this regard, NAD can be viewed as an integral regulator of broad-spectrum defense. Nevertheless, we recognize that NAD-dependent pleiotropic effects also might impact defense responses, and further investigations are required to disentangle the highly interconnected metabolic signaling networks in which NAD is involved. Genetic studies coupled to high-resolution metabolomics and transcriptomics analyses would help to address the contributions of different signaling pathways. In particular, loss-of-function strategies, not yet available, would be critical to verify the NAD-stimulated defense components when NAD levels are lower. Our results also raise the question of whether NAD itself can be considered as a DAMP, and particular effort is required to determine which receptor is able to sense NAD and transduce the signal. The fact that NAD is an efficient inducer of resistance against a broad spectrum of pathogens, including B. cinerea (i.e. the second most important fungal pathogen in plants; Dean et al., 2012), justifies further research into the mechanistic basis of NAD-dependent plant responses.
MATERIALS AND METHODS
Statistical Analyses
Data shown are means ± se of three to 16 independent biologically replicated samples from different plants. Statistically significant differences were determined in Microsoft Excel using Student’s t test with a P value of 0.05, 0.01, or 0.001 or Fisher’s exact test for differences in class distribution between treatments (Dickeya dadantii disease severity; P < 0.05). All experiments were repeated at least three times with similar results. For large-scale metabolomics analyses, two independent experiments were conducted.
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild-type control plant along with the transgenic line nadC 15.3 overexpressing the bacterial NAD biosynthetic gene nadC (Pétriacq et al., 2012), the SA biosynthesis mutant sid2-1 (Wildermuth et al., 2001), the chitin-insensitive mutant cerk1-2 (provided by Naoto Shibuya; Miya et al., 2007), and the ROS production mutants rbohD, rbohF, and rbohD/F (Torres et al., 2002). Plants were grown in a cabinet under 150 µmol quanta m−2 s−1 at leaf level using an 8/16-h (light/dark) photoperiod and a 20°C/18°C (day/night) temperature with 65% relative humidity. Seeds were sown on soil and stratified at 4°C for 2 d, then seedlings were grown for 2 weeks before being potted individually into separate pots. The resulting plants were watered twice per week and analyzed at the age of 5 weeks. In all experiments, we considered intact leaves of similar age and physiological stage.
Pathogen Test
In this study, four different pathogen strains were used: the avirulent strains of Pseudomonas syringae pv tomato DC3000 carrying the avirulence gene AvrRpm1 (Pst-AvrRpm1) or AvrRpt2 (Pst-AvrRpt2; a gift from Jane Glazebrook), the virulent bacterial strain 3937 of D. dadantii (a gift from Dominique Expert), and the ascomycete Botrytis cinerea (a gift from Marie Garmier). Microbe resistance tests were carried out as described previously (Pétriacq et al., 2012) and are detailed in Supplemental Figure S2. Briefly, whole leaves of 5-week-old plants were infiltrated with 5 mm Q (pH 6) using a needle-free 1-mL syringe, and 48 h later, the same leaves were infiltrated with mock solution (10 mm MgCl2 or MgSO4) or the corresponding pathogen suspension prepared as documented (Fagard et al., 2007; Adams-Phillips et al., 2008; Pétriacq et al., 2012). Bacterial growth (Pst and D. dadantii) was assessed 48 hpi with an inoculum at 105 colony-forming units mL−1 by recording the number of colonies as described previously (Pétriacq et al., 2012). Growth and harvesting of spores of the necrotrophic ascomycete B. cinerea were done as described previously (Thomma et al., 1998), and the infection was determined by measuring lesion diameters 24 hpi with a fungal inoculum of 106 spores mL−1. To facilitate the inoculation, plants were watered 24 h before inoculation and covered with a lid to stimulate stomatal opening. Pathoassays (n = 4–6) were conducted three times with similar results.
Histochemical Detection of ROS
In situ detection of mainly intracellular ROS was conducted in vivo by DCFH-DA (Sigma-Aldrich) staining as described previously (Fagard et al., 2007; Degrave et al., 2008). Intact leaves treated with mock solution (10 mm MgCl2 or MgSO4), infected with Pst-AvrRpm1/D. dadantii, or infiltrated with water/1 mm NAD+ were harvested, stained at corresponding time points by immersing in a 300 µm DCFH-DA solution, and vacuum infiltrated. Green fluorescence was detected from whole-leaf images using an Olympus SZX12 binocular magnifier (HQ510 1p emission filter) and quantified using ImageJ (http://imagej.nih.gov/ij/). Experiments were conducted at least three times with 12 leaves from different plants in each experiment. Pharmacological inhibition of NADPH oxidases was performed using the flavoenzyme inhibitor DPI at a concentration of 25 μm in 5-week-old Col-0. For bacterial infections, DPI was syringe infiltrated 1 h before infection with pathogens. For chemical treatments, DPI was coinfiltrated with water or NAD+. DPI on its own did not induce ROS production (data not shown). The respiration inhibition assay was carried out on intact leaves by coinfiltration of 1 mm KCN with water or NAD+ (1 mm). ROS detection in the dark was conducted by covering the plant just after treatment (NAD+ or Q) with a tray to block the light until sampling for the DCFH-DA assay. The photorespiration assay was performed on Col-0 plants grown continuously in three different growth cabinets that differ only in CO2 concentrations: high photorespiration (200 µL L−1), normal photorespiration (400 µL L−1), and low photorespiration (1,200 µL L−1).
H2O2 Quantification in Whole-Leaf Tissue
Total soluble foliar H2O2 was determined by the luminol chemiluminescence assay according to a method adapted from Veljovic-Jovanovic et al. (2002). At the specified time point and treatment (Supplemental Fig. S2), fresh leaves (n = 4; 100 mg) were flash frozen in liquid nitrogen and subsequently ground and extracted in 1 mL of 5% TCA. After centrifugation (10 min, 12,000g, and 4°C), 200 μL of supernatant was neutralized to pH 5.6 with 5 m K2CO3 and incubated with 1 unit of ascorbate oxidase to remove ascorbate that may inhibit luminol chemiluminescence (Veljovic-Jovanovic et al., 2002). For each sample, 20 μL was injected into 500 μL of 0.5 mm luminol (prepared in 0.5 mm NH4OH, pH 9.5), and chemiluminescence was initiated by adding 50 μL of 0.5 mm ferricyanide (in 0.2 m NH4OH, pH 9.5). Chemiluminescence was recorded for 2 s using a Berthold luminometer (Lumat LB9507). The results were expressed as H2O2 compared with a calibration curve obtained from fresh H2O2. The luminol assay was conducted three times with comparable outputs.
Transcript Abundance
Analysis of gene expression was performed as described by Pétriacq et al., (2012). Briefly, four leaves from 5-week-old plants were pooled (n = 3), flash frozen in liquid nitrogen 20 h after infection with Pst-AvrRpm1 or 24 h after syringe infiltration with water or NAD+ (1 mm), and then stored at −80°C until RNA extraction and RT-qPCR analyses. mRNA abundances were expressed relative to the ACTIN2 reference gene (At3g18780) using previously described gene-specific primers (Oelze et al., 2012; Pétriacq et al., 2012; Gouhier-Darimont et al., 2013; Gruner et al., 2013; Pétriacq et al., 2016). The sequences of RT-qPCR primers are given in Supplemental Table S4.
Cell Death Measurements
Cell death was assessed by conductivity measurements of electrolyte leakage from four biological replicates (n = 4) as reported (Pétriacq et al., 2016). Briefly, 5-week-old plants were syringe infiltrated with 10 mm MgCl2 or Pst-AvrRpm1 (105 colony-forming units mL−1), and 48 hpi, six discs (total area of 1.2 cm2) from leaves of the same developmental age were punched on different plants using a cork borer and floated onto 2 mL of double-deionized sterile water in 2-mL tubes. Samples were then agitated at room temperature with an orbital shaker (200 rpm) for 2 h, and conductivity (CDM210 MeterLab; Radiometer Analytical) was measured on the balanced bathing solution. To determine the values corresponding to 100% electrolyte leakage from lysed leaf material, all samples were boiled for 15 min and then measured for total conductivity. Cell death was expressed as the percentage of ion leakage between initial and total electrolyte leakage. The experiment was repeated three times with comparable results.
Quantification of Callose Deposition
Callose deposition was measured from 5-week-old leaves stained with Aniline Blue using a UV epifluorescence microscope (Olympus BX51) as described previously (Pétriacq et al., 2016). For each condition, 12 leaves from different plants (n = 12) were analyzed in Photoshop CS4 (Adobe), and callose intensity was expressed as relative callose area as described previously (Pétriacq et al., 2016).
Metabolomics
Extraction and analysis of leaf metabolites by mass spectrometry are described in Supplemental Methods S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pools of pyridine nucleotides, SA, and nicotinic acid.
Supplemental Figure S2. Chemical treatments and pathogen infections.
Supplemental Figure S3. Disease phenotype of D. dadantii and ROS contents.
Supplemental Figure S4. NADPH oxidase inhibitor assay.
Supplemental Figure S5. Higher NAD and respiration.
Supplemental Figure S6. Epifluorescence microscopy of callose deposition.
Supplemental Figure S7. HCA of metabolic markers.
Supplemental Figure S8. Multivariate analyses of nadC metabolomics.
Supplemental Figure S9. Analysis of important metabolic discriminators.
Supplemental Figure S10. Heat maps of selected metabolic markers.
Supplemental Figure S11. NAD-associated hormonal perturbations.
Supplemental Figure S12. Expression of redox and defense marker genes.
Supplemental Figure S13. NAD-induced ROS burst in cerk1-2.
Supplemental Table S1. Validation parameters of OPLS-DA.
Supplemental Table S2. List of best discriminators (VIPs).
Supplemental Table S3. Putative identification of metabolic markers.
Supplemental Table S4. List of RT-qPCR primers.
Supplemental Methods S1. Metabolomics and respiration measurements.
Supplementary Material
Acknowledgments
We thank Ana López Sánchez and Alex Williams for revising the article; Dominique Expert for helpful scientific discussions; Linda de Bont and Victoria Pastor for technical assistance; Graham Noctor for contributing to the original nadC project; and Michael Hodges for support as head of the Regulation, Signaling, and Metabolic Interactions laboratory.
Glossary
- SA
salicylic acid
- HR
hypersensitive response
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- DAMP
damage-associated molecular pattern
- Q
quinolinic acid
- DCFH-DA
2,7-dichlorofluorescein diacetate
- hpi
hours post inoculation
- Col-0
Columbia-0
- DPI
diphenylene iodonium
- JA
jasmonic acid
- HCA
hierarchical clustering analysis
- VIP
variable importance for the projection
- ABA
abscisic acid
- RT-qPCR
reverse transcription-quantitative PC
Footnotes
This work was supported by the Université Paris-Sud and the Leverhulme Trust (Research Leadership Award no. RL–2012–042 to J.T.).
Articles can be viewed without a subscription.
References
- Adams-Phillips L, Briggs AG, Bent AF (2010) Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol 152: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L, Wan J, Tan X, Dunning FM, Meyers BC, Michelmore RW, Bent AF (2008) Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact 21: 646–657 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Trenkamp S, Bunik VI, Fernie AR (2008) Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Osbourn A (2009) Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748 [DOI] [PubMed] [Google Scholar]
- Brader G, Tas E, Palva ET (2001) Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol 126: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AG, Bent AF (2011) Poly(ADP-ribosyl)ation in plants. Trends Plant Sci 16: 372–380 [DOI] [PubMed] [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S (2013) Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol Biochem 72: 1–20 [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave A, Fagard M, Perino C, Brisset MN, Gaubert S, Laroche S, Patrit O, Barny MA (2008) Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Mol Plant Microbe Interact 21: 1076–1086 [DOI] [PubMed] [Google Scholar]
- Dietz KJ. (2003) Redox control, redox signaling, and redox homeostasis in plant cells. Int Rev Cytol 228: 141–193 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Edwards R (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71: 338–350 [DOI] [PubMed] [Google Scholar]
- Djebbar R, Rzigui T, Pétriacq P, Mauve C, Priault P, Fresneau C, De Paepe M, Florez-Sarasa I, Benhassaine-Kesri G, Streb P, et al. (2012) Respiratory complex I deficiency induces drought tolerance by impacting leaf stomatal and hydraulic conductances. Planta 235: 603–614 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, de Paepe R (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, Voigt CA (2014) Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann Bot (Lond) 114: 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Dellagi A, Roux C, Périno C, Rigault M, Boucher V, Shevchik VE, Expert D (2007) Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol Plant Microbe Interact 20: 794–805 [DOI] [PubMed] [Google Scholar]
- Frederickson Matika DE, Loake GJ (2014) Redox regulation in plant immune function. Antioxid Redox Signal 21: 1373–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmier M, Priault P, Vidal G, Driscoll S, Djebbar R, Boccara M, Mathieu C, Foyer CH, De Paepe R (2007) Light and oxygen are not required for harpin-induced cell death. J Biol Chem 282: 37556–37566 [DOI] [PubMed] [Google Scholar]
- Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145: 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Xia Y (2008) The role of AtNUDT7, a Nudix hydrolase, in the plant defense response. Plant Signal Behav 3: 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gouhier-Darimont C, Schmiesing A, Bonnet C, Lassueur S, Reymond P (2013) Signalling of Arabidopsis thaliana response to Pieris brassicae eggs shares similarities with PAMP-triggered immunity. J Exp Bot 64: 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérard F, Pétriacq P, Gakière B, Tcherkez G (2011) Liquid chromatography/time-of-flight mass spectrometry for the analysis of plant samples: a method for simultaneous screening of common cofactors or nucleotides and application to an engineered plant line. Plant Physiol Biochem 49: 1117–1125 [DOI] [PubMed] [Google Scholar]
- Hadwiger LA. (2013) Multiple effects of chitosan on plant systems: solid science or hype. Plant Sci 208: 42–49 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Beaudoin N (2010) Chitooligosaccharide sensing and downstream signaling: contrasted outcomes in pathogenic and beneficial plant-microbe interactions. Planta 232: 787–806 [DOI] [PubMed] [Google Scholar]
- Heil M, Land WG (2014) Danger signals: damaged-self recognition across the tree of life. Front Plant Sci 5: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M, Dellero Y, Keech O, Betti M, Raghavendra AS, Sage R, Zhu XG, Allen DK, Weber APM (2016) Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. J Exp Bot 67: 3015–3026 [DOI] [PubMed] [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45: 703–714 [DOI] [PubMed] [Google Scholar]
- Hunt L, Lerner F, Ziegler M (2004) NAD: new roles in signalling and gene regulation in plants. New Phytol 163: 31–44 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Yoshimura K, Ogawa T, Shigeoka S (2010) Distinct regulation of Arabidopsis ADP-ribose/NADH pyrophosphohydrolases, AtNUDX6 and 7, in biotic and abiotic stress responses. Plant Signal Behav 5: 839–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N. (2010) Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol Biol 639: 292–298 [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Penaganti A, Tang Y, Mahalingam R (2010) Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol 10: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim-Messeder D, Caverzan A, Rauber R, de Souza Ferreira E, Margis-Pinheiro M, Galina A (2015) Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol 208: 776–789 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh A, Uenohara K, Akita M, Hashimoto T (2006) Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel Y, Pédron J, Patrit O, Simond-Côte E, Hermand V, Van Gijsegem F (2011) Analysis of the plant bos1 mutant highlights necrosis as an efficient defence mechanism during D. dadantii/Arabidopsis thaliana interaction. PLoS ONE 6: e18991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S, Serrano M, L’Haridon F, Tjamos SE, Metraux JP (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112: 54–62 [DOI] [PubMed] [Google Scholar]
- Li Y, Trush MA (1998) Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem Biophys Res Commun 253: 295–299 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Macho AP, Boutrot F, Rathjen JP, Zipfel C (2012) Aspartate oxidase plays an important role in Arabidopsis stomatal immunity. Plant Physiol 159: 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Jambunathan N, Penaganti A (2007) Pyridine nucleotide homeostasis in plant development and stress. Int J Plant Dev Biol 1: 194–201 [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9–15 [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G (2009) The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 184: 885–897 [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Gakière B (2006) NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57: 1603–1620 [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Daudi A, Butt VS, Bolwell GP (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236: 765–779 [DOI] [PubMed] [Google Scholar]
- Oelze ML, Vogel MO, Alsharafa K, Kahmann U, Viehhauser A, Maurino VG, Dietz KJ (2012) Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J Exp Bot 63: 1297–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor V, Luna E, Ton J, Cerezo M, García-Agustín P, Flors V (2013) Fine tuning of reactive oxygen species homeostasis regulates primed immune responses in Arabidopsis. Mol Plant Microbe Interact 26: 1334–1344 [DOI] [PubMed] [Google Scholar]
- Pel MJC, Pieterse CMJ (2013) Microbial recognition and evasion of host immunity. J Exp Bot 64: 1237–1248 [DOI] [PubMed] [Google Scholar]
- Pétriacq P, de Bont L, Hager J, Didierlaurent L, Mauve C, Guérard F, Noctor G, Pelletier S, Renou JP, Tcherkez G, et al. (2012) Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J 70: 650–665 [DOI] [PubMed] [Google Scholar]
- Pétriacq P, de Bont L, Tcherkez G, Gakière B (2013) NAD: not just a pawn on the board of plant-pathogen interactions. Plant Signal Behav 8: e22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétriacq P, Stassen JH, Ton J (2016) Spore density determines infection strategy by the plant-pathogenic fungus Plectosphaerella cucumerina. Plant Physiol 170: 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Polidoros AN, Mylona PV, Arnholdt-Schmitt B (2009) AOX gene structure, transcript variation and expression in plants. Physiol Plant 137: 342–353 [DOI] [PubMed] [Google Scholar]
- Povero G, Loreti E, Pucciariello C, Santaniello A, Di Tommaso D, Di Tommaso G, Kapetis D, Zolezzi F, Piaggesi A, Perata P (2011) Transcript profiling of chitosan-treated Arabidopsis seedlings. J Plant Res 124: 619–629 [DOI] [PubMed] [Google Scholar]
- Queval G, Hager J, Gakière B, Noctor G (2008) Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J Exp Bot 59: 135–146 [DOI] [PubMed] [Google Scholar]
- Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakière B, Noctor G (2009) H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant 2: 344–356 [DOI] [PubMed] [Google Scholar]
- Rojas C, Mysore KS (2012) Glycolate oxidase is an alternative source for H2O2 production during plant defense responses and functions independently from NADPH oxidase. Plant Signal Behav 7: 752–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu CM, Kaundal A, Mysore KS (2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A, Mesters JR, Thomma BPHJ (2015) The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol Rev 39: 171–183 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Abou-Mansour E, Buchala A, Mauch F (2010) Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J 62: 840–851 [DOI] [PubMed] [Google Scholar]
- Shinya T, Nakagawa T, Kaku H, Shibuya N (2015) Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr Opin Plant Biol 26: 64–71 [DOI] [PubMed] [Google Scholar]
- Simons BH, Millenaar FF, Mulder L, Van Loon LC, Lambers H (1999) Enhanced expression and activation of the alternative oxidase during infection of Arabidopsis with Pseudomonas syringae pv tomato. Plant Physiol 120: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Keppler BD, Wise RR, Bent AF (2015) PARP2 is the predominant poly(ADP-ribose) polymerase in Arabidopsis DNA damage and immune responses. PLoS Genet 11: e1005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørhagen K, Laxa M, Peterhänsel C, Reumann S (2013) The emerging role of photorespiration and non-photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol (Stuttg) 15: 723–736 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Okada M, Jikumaru Y, Yamane H, Kaku H, Shibuya N, Minami E (2006) Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosaccharide. Biosci Biotechnol Biochem 70: 1599–1605 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapet P, Kulik A, Lamotte O, Jeandroz S, Bourque S, Nicolas-Francès V, Rosnoblet C, Besson-Bard A, Wendehenne D (2015) NO signaling in plant immunity: a tale of messengers. Phytochemistry 112: 72–79 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S, Noctor G, Foyer CH (2002) Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol Biochem 40: 501–507 [Google Scholar]
- Vidal G, Ribas-Carbo M, Garmier M, Dubertret G, Rasmusson AG, Mathieu C, Foyer CH, De Paepe R (2007) Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19: 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, van de Mortel JE, Shan W, Govers F (2013) A novel Arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ 36: 1192–1203 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Worley B, Powers R (2013) Multivariate analysis in metabolomics. Curr Metabolomics 1: 92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1: 639–647 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2010) Plant autophagy puts the brakes on cell death by controlling salicylic acid signaling. Autophagy 6: 192–193 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Xiao S (2015) Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Front Plant Sci 6: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mou Z (2009) Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J 57: 302–312 [DOI] [PubMed] [Google Scholar]
- Zhang X, Mou Z (2012) Expression of the human NAD(P)-metabolizing ectoenzyme CD38 compromises systemic acquired resistance in Arabidopsis. Mol Plant Microbe Interact 25: 1209–1218 [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, Dong X (2015) Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.