Genetic analysis of sequenced Arabidopsis accessions highlights the mutational landscape of an essential metabolic enzyme, with implications for chloroplast transformation and plastid protein import.
Abstract
Natural accessions of Arabidopsis (Arabidopsis thaliana) differ in their ability to tolerate a loss of chloroplast translation. These differences can be attributed in part to variation in a duplicated nuclear gene (ACC2) that targets homomeric acetyl-coenzyme A carboxylase (ACCase) to plastids. This functional redundancy allows limited fatty acid biosynthesis to occur in the absence of heteromeric ACCase, which is encoded in part by the plastid genome. In the presence of functional ACC2, tolerant alleles of several nuclear genes, not yet identified, enhance the growth of seedlings and embryos disrupted in chloroplast translation. ACC2 knockout mutants, by contrast, are hypersensitive. Here we describe an expanded search for hypersensitive accessions of Arabidopsis, evaluate whether all of these accessions are defective in ACC2, and characterize genotype-to-phenotype relationships for homomeric ACCase variants identified among 855 accessions with sequenced genomes. Null alleles with ACC2 nonsense mutations, frameshift mutations, small deletions, genomic rearrangements, and defects in RNA splicing are included among the most sensitive accessions examined. By contrast, most missense mutations affecting highly conserved residues failed to eliminate ACC2 function. Several accessions were identified where sensitivity could not be attributed to a defect in either ACC2 or Tic20-IV, the chloroplast membrane channel required for ACC2 uptake. Overall, these results underscore the central role of ACC2 in mediating Arabidopsis response to a loss of chloroplast translation, highlight future applications of this system to analyzing chloroplast protein import, and provide valuable insights into the mutational landscape of an important metabolic enzyme that is highly conserved throughout eukaryotes.
Understanding the relationship between genotype and phenotype is a central question in biology. With recent advances in genome sequencing, this relationship has received increased attention on a broad scale, from humans to model organisms. In the area of human genetics, emphasis has been placed on interpreting variants in Mendelian disease genes (Stenson et al., 2014; Landrum et al., 2016) and intergenic regions associated with disease phenotypes (Zhang and Lupski, 2015). With Arabidopsis (Arabidopsis thaliana), genome sequence information for more than 1,000 natural accessions (The 1001 Genomes Consortium, 2016) has provided a valuable resource for evaluating genetic contributions to plant growth and development, and for probing the nature of plant interactions with the environment (Alonso-Blanco et al., 2009; Weigel, 2012). Natural variation in essential genes, however, has been difficult to study in any system because deleterious mutations are eliminated through natural selection.
Acetyl-coenzyme A carboxylase (ACCase) is an essential enzyme that functions in fatty acid metabolism. Typically, plants have two different versions of this enzyme: a heteromeric protein encoded in part by the chloroplast genome, which resembles the bacterial enzyme and catalyzes an early, rate-limiting reaction in fatty acid biosynthesis within the plastid (Salie and Thelen, 2016); and a homomeric, nuclear-encoded, cytosolic version required for the production of very-long-chain fatty acids (Shang et al., 2016). Disruption of the cytosolic enzyme (ACC1) in Arabidopsis causes a distinctive embryo-defective (emb) phenotype and results in seedling lethality (Meinke, 1985; Baud et al., 2004). Weak alleles exhibit altered cuticular wax composition, cold sensitivity, and glossy inflorescence stems (Lü et al., 2011; Amid et al., 2012). Loss of the heteromeric, chloroplast-localized enzyme, by contrast, results in early embryo lethality (Li et al., 2011), with the stage of arrest dependent on genetic background (Parker et al., 2014). In some members of the Brassicaceae, including Arabidopsis, a tandem ACC1 gene duplication enables the production of a chloroplast-targeted homomeric enzyme (ACC2) that can partially suppress the early lethality associated with a loss of chloroplast translation and the absence of heteromeric ACCase (Yanai et al., 1995; Babiychuk et al., 2011; Bryant et al., 2011). Most grasses lack the heteromeric enzyme altogether and instead utilize a plastid-localized homomeric enzyme produced from a duplicated nuclear gene (Konishi and Sasaki, 1994; Chalupska et al., 2008). Previously, we showed that natural variation in the response of Arabidopsis embryos and seedlings to a loss of chloroplast translation is mediated by ACC2, and that tolerant accessions likely contain functional ACC2 along with an unlinked enhancer of ACC2 function and modifiers that extend the growth response (Parker et al., 2014). Overall, the focus of that initial study was on identifying factors that improved survival in the absence of chloroplast translation.
In this report, we explore the diversity of genetic defects in natural populations of Arabidopsis that determine hypersensitivity to a loss of chloroplast translation. One question we sought to address was whether sensitivity is consistently associated with defects in ACC2 function alone or whether variation in other genes can lead to a hypersensitive phenotype. To address that question, we characterized the spectinomycin responses of seedlings from 100 sequenced accessions not examined in our initial report, identified hypersensitive accessions similar to the most sensitive accessions from our original screen, and analyzed progeny from crosses with the acc2 knockout to determine whether a defect in ACC2 was responsible. A second objective was to characterize the full range of deleterious ACC2 mutations found in natural populations of Arabidopsis. For that approach, we identified all of the ACC2 variants included among 855 accessions with sequenced genomes, focused on variants most likely to cause hypersensitivity based on sequence conservation among 667 eukaryotic ACCases, analyzed the spectinomycin responses of accessions with variants of interest, and evaluated progeny from crosses with acc2 knockouts.
Because the elimination of ACC2 function in Arabidopsis has no apparent impact on plant growth and development in natural populations, but can be detected in the laboratory as increased sensitivity to spectinomycin, an inhibitor of chloroplast translation, we have in essence created an experimental system to study natural variation in the loss of an essential protein function. This work provides valuable insights into essential domains within a large, homodimeric enzyme that represents a target for herbicide action (Kaundun, 2014) and pharmaceuticals for human diseases (Harriman et al., 2016). Identification of Arabidopsis accessions hypersensitive to spectinomycin should also advance research on chloroplast transformation, where spectinomycin is the selection agent of choice (Svab and Maliga, 1993), and may help to uncover a novel function for ACC2 in some Arabidopsis populations. In addition, the experimental system can be used to evaluate plastid protein import complexes from a unique genetic perspective, as ACC2 uptake appears to occur exclusively through the Tic20-IV complex thought to be associated with housekeeping proteins (Inoue et al., 2010; Hirabayashi et al., 2011). The broad range of ACC2 defects reported here document the degeneration of a duplicated nuclear gene and extend information about eukaryotic ACCase function obtained primarily from protein structure and ACC1 mutagenesis studies.
RESULTS
Several Classes of ACC2 Null Mutations Are Present in Sequenced Accessions
The first example of a naturally occurring null mutation in ACC2 was uncovered by cDNA sequencing the “Nossen” accession (Parker et al., 2014). Our initial objective was to determine why “Nossen” was more sensitive to a loss of chloroplast translation than the Tsu-0 accession. The nonsense mutation identified (R865X) results in a truncated protein missing the C-terminal 1,490 amino acids. After evaluating sequences from the 1001 Genomes Project (http://signal.salk.edu/atg1001), we found three more examples of nonsense mutations: Y753X (Kb-0 and Kl-5), K1225X (Blh1-1), and Q2325X (Hod). All of these variants were confirmed by Sanger sequencing. Next, we searched for evidence of small deletions predicted to cause a frameshift mutation. Two examples of single nucleotide deletions that altered the reading frame were confirmed: 1171fs1190X (Ip-Alo-0 and Ip-Vin-0) and 2020fs2021X (Lu3-30 and Lu4-2). Large deletions affecting the C-terminal region of ACC2 are predicted for Ob-0 and Old-1. We confirmed that ACC2 sequences are missing from exon 31 but were unable to amplify downstream sequences to determine the precise nature of the defect. We suspect that both accessions contain a deletion followed by some type of rearrangement.
Several defects in RNA splicing were uncovered by searching for altered ACC2 splice donor and acceptor sites. In one case (Gn-1), an AG→TG substitution in the acceptor site for intron 10 results in a 10 nucleotide deletion (distance to the first AG in exon 11) and a frameshift mutation. In the Wl-0 accession, an AG→GG substitution in the acceptor site for intron 19 also results in a 10 nucleotide deletion and frameshift. Four accessions (Spro-2, Ste-2, Ste-3, and Vimmerby) harbor the same confirmed GT→TT mutation affecting the intron 29 donor site. The Spro-2 accession, chosen to represent the group, produced a mixture of defective ACC2 transcripts, with mRNA sequences that either included intron 29 or matched those predicted by locations of alternative donor sites (Supplemental Fig. S1). Insertion of five nucleotides just upstream of the intron 4 acceptor site in Hof-1, by contrast, had no effect on splicing.
Seedlings from most of the accessions listed above are highly sensitive to spectinomycin, consistent with the elimination of ACC2 function (Table I). Accessions that typically produced seedlings with cotyledons but no shoot meristem outgrowths on spectinomycin were labeled hypersensitive. The exceptional accession with an intermediate response (Hod) was not unexpected, because the truncated ACC2 protein lacks only the final 30 residues. Surprisingly, the “Nossen” accession was slightly less sensitive than other null alleles, despite the presence of a nonsense mutation predicted to remove much of the protein, possibly reflecting some level of nonsense suppression. Overall, we conclude that a remarkable diversity of ACC2 null alleles is maintained among Arabidopsis accessions.
Table I. ACC2 null mutations identified in sequenced accessions of Arabidopsis.
| Mutation Class | Accession | Reported Country of Origin | Mutationa | Mutation Location | Spectinomycin Response |
||
|---|---|---|---|---|---|---|---|
| Category | Scoreb | Seedlings | |||||
| Nonsense | Kb-0 | Germany | Y753X | Exon 17 | Sensitive | 1.4 | 73 |
| Kl-5 | Germany | Y753X | Exon 17 | Hypersensitive | 1.1 | 76 | |
| “Nossen” | Uncertain | R865X | Exon 19 | Sensitive | 2.3 | 571 | |
| Blh-1 | Czech Republic | K1225X | Exon 26 | Sensitive | 1.3 | 71 | |
| Frameshift | Ip-Alo-0 | Portugal | 1171fs | Exon 25 | Hypersensitive | 1.1 | 51 |
| Ip-Vin-0 | Spain | 1171fs | Exon 25 | Hypersensitive | 1.2 | 33 | |
| Lu3-30 | Germany | 2020fs | Exon 31 | Hypersensitive | 1.1 | 54 | |
| Lu4-2 | Germany | 2020fs | Exon 31 | Hypersensitive | 1.3 | 55 | |
| Splicing | Gn-1 | Germany | GT…TG | Intron 10 | Hypersensitive | 1.1 | 83 |
| Gn2-3 | Germany | GT…TG | Intron 10 | Hypersensitive | 1.1 | 191 | |
| Wl-0 | Germany | GT…GG | Intron 19 | Sensitive | 1.4 | 79 | |
| Spro-2 | Sweden | TT…AG | Intron 29 | Sensitive | 1.3 | 80 | |
| Ste-2 | Sweden | TT…AG | Intron 29 | Hypersensitive | 1.1 | 83 | |
| Ste-3 | Sweden | TT…AG | Intron 29 | Hypersensitive | 1.0 | 82 | |
| Vimmerby | Sweden | TT…AG | Intron 29 | Hypersensitive | 1.0 | 67 | |
| Rearrangement | Ob-0 | Germany | Unresolved | Exon 32 | Hypersensitive | 1.2 | 74 |
| Old-1 | Germany | Unresolved | Exon 32 | Hypersensitive | 1.2 | 75 | |
| Small deletion | Ip-Ber-0 | Spain | Deletion (23 bp) | Intron 17, exon 18 | Sensitive | 1.3 | 49 |
All variants except details of Ob-0 and Old-1 rearrangements were confirmed by Sanger sequencing. bHigher scores reflect increasing levels of tolerance; these scores were among the lowest of all accessions evaluated.
To evaluate other candidate null alleles, we confirmed two small deletions, three nucleotides each, which alter the variable central domain of the protein. In the Qar-8a accession, the deletion (K1376R; ∆1377) results in an amino acid substitution followed by a single amino acid deletion. Because this accession is sensitive to spectinomycin but was not among the most sensitive accessions examined, we suspect that ACC2 function is severely reduced but not eliminated. Another deletion (∆1479) affecting the central domain was found in Ip-Ren-6 and Ip-Voz-0. The intermediate response of Ip-Voz-0 seedlings indicates that this deletion has a less severe effect on ACC2 function. A 23 nucleotide deletion that removes the end of intron 17 and part of the adjacent exon was found in Ip-Ber-0. Sequencing cloned reverse transcription-PCR products confirmed that a complex mixture of transcripts encoding a variety of defective and truncated proteins is produced (Supplemental Fig. S2). The sensitive response of this accession on spectinomycin is consistent with a dramatic loss of ACC2 function.
The Sav-0 Accession Exhibits a Hypersensitive Response on Spectinomycin
In our initial screen of 50 Arabidopsis accessions for seedling growth on spectinomycin, one accession (Sav-0) stood out as consistently hypersensitive, with a seedling response similar to the acc2 knockout (Parker et al., 2014). Because of the strong phenotype, we reasoned that a null mutation or promoter defect was likely present. However, ACC2 transcript levels measured with reverse transcription-quantitative PCR were indistinguishable from tolerant accessions, and cDNA sequencing revealed no frameshift or nonsense mutations or defects in RNA splicing. Instead, we uncovered 15 missense mutations that were initially viewed as variants of unknown significance relative to the Col-0 reference sequence.
To determine whether hypersensitivity was caused by a defect in ACC2, we crossed Sav-0 with the tolerant Tsu-0 accession and genotyped sensitive and tolerant seedlings in the F2 generation. The same approach was used before to evaluate spectinomycin responses of the “Nossen” and Tsu-0 accessions (Parker et al., 2014). Tolerant seedlings in this case were either heterozygous or homozygous Tsu-0 for ACC2, consistent with a requirement of functional ACC2 for tolerance and with the model that a defect in ACC2 is responsible for hypersensitivity in Sav-0. Unexpectedly, several of the sensitive F2 seedlings were not homozygous for the Sav-0 allele of ACC2. To determine whether these exceptional seedlings represented recombinants between ACC2 and a linked locus responsible for sensitivity, additional polymorphisms along chromosome 1 were evaluated. Because perfect linkage was not detected, the significance of those exceptional seedlings remains obscure.
A different result was obtained with F2 seedlings from crosses between Tsu-0 and Nie1-2, another accession initially labeled as sensitive. In this case, the locus that distinguished sensitive from tolerant F2 seedlings was linked to the enhancer on chromosome 5 (Parker et al., 2014). Some of the most tolerant seedlings were homozygous for the Nie1-2 allele of ACC2, which suggests that ACC2 in Nie1-2 is functional and that this accession differed from other sensitive accessions. This was later confirmed when additional seedlings revealed an intermediate phenotype. Residual ACC2 function also was indicated for the sensitive Oy-0 accession. Collectively, these results underscore the fact that we initially focused on tolerance and that a larger collection of sensitive accessions was needed to understand sensitivity.
A Forward Genetic Screen Expands the List of Hypersensitive Accessions
One hundred additional accessions with sequenced genomes and diverse geographical locations were tested for seedling response after 5 weeks on 50 mg L−1 spectinomycin. To facilitate comparison among accessions, we further defined the difference between sensitive, intermediate, and tolerant seedlings, and their corresponding accessions, and included a response score (1–11) to reflect increased levels of tolerance (Table II; Fig. 1). More than 8,000 seedlings from 152 accessions were evaluated in the combined screens. Three categories of sensitive seedlings were recognized: expanded cotyledons (score 1); slight growth of first leaf initials (2); and meristem with slight callus or multiple leaf initials (3). The first category of intermediate seedlings (score 5) had further leaf growth. Hypersensitive accessions had more than 50% of seedlings in the first category and 95% or more in the first or second category. Sensitive accessions, by contrast, had more than 70% of seedlings in a sensitive category but also more intermediate seedlings. Overall, three hypersensitive, 22 sensitive, 107 intermediate, including low and high intermediate, and 20 tolerant accessions were identified (Table II). Differences between categories were subtle but informative and were generally reproducible, with most problematic accessions exhibiting poor seed germination. Considerable variation in seedling response was noted with some intermediate accessions, for unknown reasons. Most accessions with ACC2 null alleles (Table I) were hypersensitive, with response scores similar to the acc2 knockout. Ten accessions with the most sensitive initial scores and without obvious null mutations were chosen for further analysis: Knox-18, Gn2-3, Sav-0, RRS-10, Gifu-2, Pna-10, Aitba-1, Tul-0, La-0, and Tol-0.
Table II. Seedling responses of 152 natural accessions of Arabidopsis germinated on spectinomycin.
| Accession Response Category | Total Accessions Classified | Total Seedlings Classified | Accession Phenotype Scores | Percentage Distribution of Seedling Phenotypes on Spectinomycina |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive |
Intermediate |
Tolerant |
||||||||||
| 1 | 2 | 3 | 5 | 6 | 7 | 9 | 10 | 11 | ||||
| Tolerant | 20 | 1,861 | 8.1–9.7 | 0.5 | 0.8 | 0.5 | 1.2 | 0.6 | 9.3 | 52.0 | 30.0 | 5.1 |
| High intermediate | 11 | 477 | 6.4–8.3 | 2.5 | 1.0 | 0.4 | 4.6 | 1.7 | 32.5 | 49.1 | 8.0 | 0.2 |
| Intermediate | 83 | 2,824 | 3.9–7.8 | 5.2 | 8.0 | 6.4 | 23.7 | 9.8 | 35.2 | 10.4 | 1.3 | |
| Low intermediate | 13 | 427 | 3.2–4.5 | 12.9 | 33.7 | 12.2 | 15.2 | 15.9 | 6.6 | 3.5 | ||
| Sensitive | 22 | 1,872 | 1.3–3.2 | 34.1 | 39.6 | 19.0 | 3.5 | 2.4 | 1.0 | 0.4 | ||
| Hypersensitive | 3 | 546 | 1.1–1.2 | 86.8 | 10.6 | 2.2 | 0.4 | |||||
For examples of sensitive, intermediate, and tolerant seedling phenotypes, see Figure 1. Boldface font indicates the most common phenotypes.
Figure 1.
Classification of Arabidopsis seedling responses on spectinomycin. Higher response scores reflect increased tolerance. A to F, Sensitive seedlings, categories 1 (A and B), 2 (C and D), and 3 (E and F). G to L, Intermediate seedlings, categories 5 (G and H), 6 (I and J), and 7 (K and L). M to P, Tolerant seedlings, categories 9 (M and N), 10 (O), and 11 (P). For additional details on classification criteria, see “Materials and Methods.” Bar = 1 mm.
An Alignment of Homomeric ACCase Sequences Highlights Conserved Residues Likely To Be Essential
Previously, we used an alignment of 20 homomeric ACCase sequences from model eukaryotes to compare levels of variation in ACC1 and ACC2 from sequenced accessions of Arabidopsis (Parker et al., 2014). To highlight conserved residues likely to be essential for protein function, we expanded that alignment to include 667 homomeric ACCase sequences from a wide range of eukaryotes. The Arabidopsis ACC2 protein contains 2,355 amino acid residues encoded by 32 exons. Among the aligned sequences, 526 residues are at least 90% conserved, including 222 that are at least 99% conserved. Four conserved protein domains are recognized: biotin carboxylase, biotin carboxyl carrier protein, and two subunits (α and β) of carboxyltransferase. There also is a variable central domain. After adding our Sav-0 sequence to predicted ACC2 sequences for the 10 sensitive accessions listed above, we identified every substitution that altered a conserved residue and evaluated spectinomycin responses of other accessions with the same variants. Sensitive accessions chosen for analysis were initially placed into four groups: Knox-18, RRS-10, Tul-0, and Tol-0 were predicted to contain the same variants of interest (I404K and T1902K); Gifu-2 had one of those variants but not the other; Sav-0, Aitba-1, and Pna-10 had variants affecting other conserved residues; and both La-0 and Gn2-3 lacked promising ACC2 variants.
These groups changed once we realized that some of our ACC2 sequences differed from those reflected in 1001 Genomes Project data. Sanger sequencing revealed that Gifu-2 shared both variants found in the Knox-18 group, whereas Pna-10 lacked the expected S1883T variant and contained instead the Knox-18 variants. Sixteen accessions predicted to have these two variants were then evaluated on spectinomycin. Two accessions (SLSP-35 and UKSW06-333) exhibited an intermediate response, lacked the expected variants, and were eliminated. The other 20 accessions, including six identified through our forward screen, were all highly sensitive to spectinomycin (Supplemental Table S1). A third variant affecting a conserved residue (E1355G) in the central domain remains of unknown significance because it is found in many accessions that are not sensitive. One accession (Mdn-1) predicted to contain I404K and T1902K but not another common variant (E1567K) was later found to contain all three variants, consistent with the Knox-18 group. Interestingly, all of these accessions, with the exception of Gifu-2, are reported to be from the United States. Collectively, these results implicate I404K, T1902K, and possibly E1355G in the loss of ACC2 function. However, they also underscore the need to confirm variants of interest to be certain that accessions evaluated correspond to those reported by the 1001 Genomes Project. To further illustrate this point, the cDNA sequence of ACC2 that we determined for the Gn2-3 accession (CS76881) differed from that reflected in 1001 Genomes Project data but was identical to Gn-1, another accession (CS76880) from the same geographical region. We conclude that hypersensitivity in Gn2-3 is caused by the same splicing defect already described for Gn-1.
The Sav-0 accession contains one missense mutation of interest (G135E) that alters a conserved residue immediately preceding the biotin carboxylase domain. We postulate that this variant is responsible for the hypersensitivity observed. The Aitba-1 accession, which is less sensitive than Sav-0, contains another variant of interest (F1206L) that affects a conserved residue in the central domain. We sequenced the ACC2 cDNA from the La-0 accession because of the apparent lack of deleterious variants. Our sequence was identical to that from the 1001 Genomes Project, which suggests that a single missense mutation in ACC2 is not responsible for the sensitivity observed. Overall, the 10 most sensitive accessions identified through our forward screen contained at most four missense mutations likely to reduce ACC2 function (Table III).
Table III. Accessions with strong missense mutations affecting conserved ACC2 residues.
| Genetic Screen | Accessions Analyzed | Variant Analyzeda | Protein Domainb | Percentage Conservationc | Sequenced Accessions with Predicted Variant | Tolerant or Intermediate Accessions | Variant Impactd |
|---|---|---|---|---|---|---|---|
| Forward | Sav-0 | G135E | (BC) | 95.7 | 0e | 0 | LD |
| V472I | BC | 83.7 | 51 | 13 | VUS | ||
| Forward | Knox-18 | I404K | BC | 94.8 | 18 | 0 | LD |
| RRS-10 | T1902K | CT-α | 87.6 | 18 | 0 | LD | |
| Gifu-2 | E1355G | Central | 98.7 | 116 | 18 | VUS | |
| Tul-0 | |||||||
| Tol-0 | |||||||
| Pna-10 | |||||||
| Forward | Aitba-1f | F1206L | Central | 96.3 | 1 | 0 | LD |
| Reverse | Etna-2 | Y443C | BC | 94.0 | 1 | 0 | PD |
| Reverse | Ts-1 | E1689G | CT-β | 97.0 | 1 | 0 | D |
The first letter denotes the most common residue found among 855 sequenced accessions. bBC, Biotin carboxylase; (BC), immediately preceding the BC domain; CT, carboxyltransferase. cIn the alignment of 667 homomeric (eukaryotic) ACCases. dD, Deleterious to protein function; LD, likely deleterious; PD, possibly deleterious; VUS, variant of unknown significance. eSav-0 was not included in the 1001 Genomes Project dataset. fThe phenotype of Aitba-1 seedlings on spectinomycin is less severe than in other accessions listed here.
A Reverse Genetic Approach Evaluates ACC2 Missense Mutations in Sequenced Accessions
To expand our search for deleterious missense mutations, we identified all of the variants affecting residues that were 90% or greater conserved in our alignment and evaluated spectinomycin responses of the corresponding accessions. Of the 44 variable residues identified, five were eliminated because of problems with seed availability or a known null mutation in the same accession. Predicted variants in four other accessions were not confirmed by Sanger sequencing. In another 27 cases, the variant was confirmed but the accession was not sensitive, which indicates that ACC2 function was not eliminated. One variant with an intermediate phenotype was assumed to be correct because it was found in multiple accessions, and another variant (Y1594H) was not evaluated. The remaining six variants were consistently associated with sensitivity. Three of these involved accessions (Sav-0, Knox-18, and Aitba-1) already identified through our forward screen, and one (Ts-1) altered the same residue found in the strong pasticcino (pas3-1) mutant allele of ACC1. Our reverse genetic approach, therefore, yielded two sensitive accessions (Etna-2 and Grivo-1) with novel missense mutations (Y443C and A2059V) that altered a highly conserved residue. Analysis of eight ACC2 transit peptide variants revealed another candidate (R4T) in a sensitive accession (Ip-Cum-1), but that variant was also found in an intermediate accession (Ip-Gua-1). Overall, ACC2 missense mutations with a strong phenotype were much less common than expected relative to other types of null mutations (Table III; Supplemental Table S2). Spectinomycin responses and stock numbers for all 245 accessions evaluated are listed in Supplemental Table S3.
Surprisingly, altering some of the most conserved ACC2 residues did not visibly reduce protein function. These variants (P475L, Q478K, N725S, R762C, E1355G, and G1766D) were found in tolerant or high intermediate accessions, which likely contain functional ACC2 (Table IV). Other missense mutations that altered conserved residues were associated with an intermediate response, indicating either a partial loss of ACC2 function or the presence of sensitive alleles of other modifiers required for tolerance. We then confirmed four variants affecting ACC1 residues that are at least 90% conserved (A193V, I493L, P528S, and G1794A). These changes must not reduce protein function because ACC1 is an essential gene. Because ACC1 and ACC2 are close paralogs, the number of conserved residues with variants that do not reduce protein function should be similar. This was true when comparing ACC1 variants affecting conserved residues (four) with ACC2 variants among tolerant accessions (seven), but not with tolerant and intermediate accessions (25) combined (z test P < 0.001). Some ACC2 missense mutations that alter conserved residues in accessions with an intermediate response are therefore likely to cause a partial loss of protein function.
Table IV. Accessions with evidence of residual ACC2 function despite substitutions in highly conserved residues.
| Variant Analyzeda | Protein Domainb | Percentage Conservationc | Sequenced Accessions with Predicted Variant | Intermediate; Low-Intermediate Accessionsd | Tolerant; High-Intermediate Accessionse |
|---|---|---|---|---|---|
| F363L | BC | 99.3 | 5 | Sei-0 | |
| V376A | BC | 100.0 | 12 | Col-0 | |
| L474F | BC | 94.5 | 1 | Chi-0 | |
| P475L | BC | 99.7 | 1 | Lm-2 | |
| Q478K | BC | 97.6 | 28 | Multiple | Uod-1 |
| R494G | BC | 99.9 | 1 | Ip-Pal-0 | |
| T538A | BC | 99.9 | 1 | IP-Tor-1 | |
| N725S | 96.9 | 44 | Multiple | Pog-0 | |
| G739E | 95.2 | 1 | Wa-1 | ||
| R762C | 96.6 | 6 | Mh-0 | Tsu-0; Tu-0 | |
| D777N | (BCCP) | 97.2 | 4 | Leska-1-44; Smolj-1 | |
| G833R | BCCP | 99.3 | 3 | Dja-1 | |
| L847P | 96.0 | 1 | WAR | ||
| E1355G | Central | 98.7 | 116 | Multiple | Si-0; Ema-1 |
| R1405Q | 96.1 | 1 | Db-1 | ||
| G1766D | CT-β | 97.6 | 39 | Multiple | Pog-0 |
| I1821V | CT-β | 98.2 | 1 | MNF-Che-2 | |
| T1834S | CT-β | 99.4 | 2 | Nemrut-1 | |
| S1883T | CT-β | 97.0 | 8 | Multiple | |
| G1897S | CT-α | 99.4 | 2 | Sch1-7; WalHaesB4 | |
| P2013L | CT-α | 98.5 | 3 | Balan-1 | |
| A2014E | CT-α | 99.0 | 1 | App1-16 | |
| I2115R | CT-α | 98.2 | 1 | Iasi-1 | |
| H2207Q | CT-α | 98.1 | 1 | Ip-Lso-0 |
All variants in accessions listed were confirmed by Sanger sequencing. bBC, Biotin carboxylase; BCCP, biotin carboxyl carrier protein; (BCCP), immediately preceding the BCCP domain; CT, carboxyltransferase. cIn the alignment of 667 homomeric (eukaryotic) ACCases. dMay contain partial loss-of-function alleles of ACC2. eLikely contains fully functional alleles of ACC2.
To determine whether a single missense mutation can eliminate homomeric ACCase function in Arabidopsis, and thus provide indirect evidence that some ACC2 missense mutations might result in a hypersensitive phenotype, we compared mutant phenotypes of pas3-1 and pas3-2 missense alleles of ACC1 with a known null allele (emb22) that contains a nonsense mutation. Although the terminal phenotypes of emb22 and pas3-1 embryos were similar, and slightly more severe than that of pas3-2, pas3-1 seedlings exhibited more extensive growth than emb22 after germination (Fig. 2; Supplemental Table S4). We conclude that pas3-1 is not a null allele, and that existing collections of acc1 missense mutations of Arabidopsis are insufficient to demonstrate that a single amino acid substitution can eliminate ACCase function.
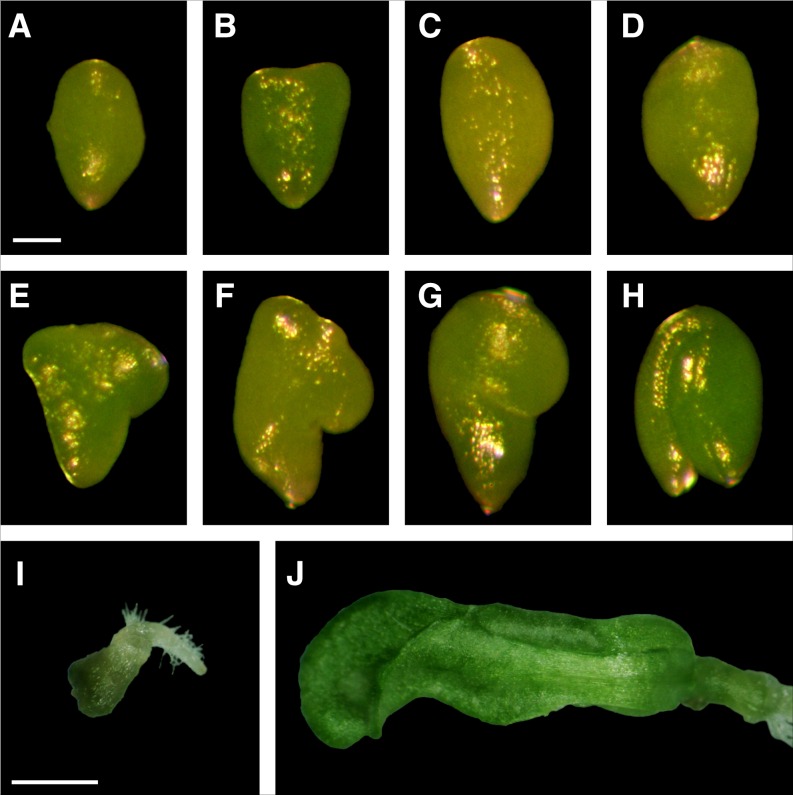
Figure 2.
Phenotypes of acc1 mutant alleles reveal different levels of residual gene function (emb22 < pas3-1 < pas3-2). A to H, Terminal embryo phenotypes for pas3-1 (A–D), which resemble emb22; pas3-2 (E–G); and the wild type (H). I and J, Seedling phenotypes for emb22 (I) and pas3-1 (J) after 5 weeks of growth on basal medium. See also Supplemental Table S4. Bars = 100 μm (A–H) and 1 mm (I and J).
Crossing Sensitive Accessions with Informative Knockout Mutants Assesses the Impact of ACC2 Variants on Protein Function
To obtain further evidence that missense mutations in some accessions reduce ACC2 function and lead to a sensitive phenotype, we performed crosses with the hypersensitive acc2 and tic20-iv knockout mutants. For accessions with acc2 null alleles, we expected to find 100% sensitive F1 seedlings in the acc2 cross, because both acc2 alleles are defective, and 100% intermediate F1 seedlings in crosses with tic20-iv, consistent with the Columbia background, which should provide functional ACC2 in heterozygotes. All F1 seedlings should be intermediate (less sensitive than either parent) in both crosses if disruption of a different gene is involved. In the F2 generation for the acc2 cross, we expected all sensitive seedlings, corresponding to parental knockouts and compound heterozygotes, if the accession is defective in ACC2, and a mixture (9:7 ratio) of intermediate and sensitive seedlings with the tic20-iv cross. The same ratio of F2 phenotypes is expected for both crosses if the accession is defective in an unlinked gene.
Initially, we focused on F2 seedlings because the genotypes of F1 parent plants could be confirmed (Supplemental Table S5). With the Gn2-3 accession, which contains a known acc2 null mutation, the hypersensitive response obtained with acc2 crosses differed from the more varied responses found with tic20-iv crosses, consistent with expectations. About 56% of the seedlings reached a later stage of development than expected for either parent with the tic20-iv crosses, which closely matches the predicted (9:7) ratio for a loss of ACC2 function. A similar pattern was observed with the Sav-0 and Knox-18 group of crosses, providing further evidence that the confirmed acc2 missense mutations in these accessions severely reduce ACC2 function and result in a hypersensitive phenotype. The G135E variant unique to Sav-0 is likely responsible for the hypersensitivity observed; all other Sav-0 variants are also found in more tolerant accessions (Supplemental Table S6). With Knox-18, we sequenced the ACC2 cDNA to confirm that the expected transcript (devoid of additional null mutations) was present. By contrast, results of crosses with the La-0 accession, which lacks an obvious defect in ACC2, clearly differed from those for ACC2 null alleles, but also did not appear to match expectations for a single unlinked locus responsible for sensitivity. Results with the Aitba-1 accession were also not definitive.
We then examined F1 progeny on spectinomycin (Table V) in order to evaluate additional accessions and resolve some of the uncertainties noted above. Progeny from crosses with accessions thought to contain acc2 null alleles were hypersensitive in acc2 crosses and more intermediate in tic20-iv crosses, consistent with expectations. Differences between F1 progeny from Aitba-1 crosses with acc2 and tic20-iv provided further evidence that the F1206L variant within the central domain reduces ACC2 function in this accession. By contrast, results of the La-0 and Grivo-1 crosses differed from those expected for an accession with compromised ACC2 function. Overall, we conclude that sensitivity to a loss of chloroplast translation is caused by deleterious acc2 missense mutations in Sav-0, the Knox-18 group, Ts-1, Aitba-1, and possibly Etna-2, where crosses remain to be analyzed. Our initial question can therefore be answered as follows: accessions with no residual ACC2 function are consistently sensitive to spectinomycin, regardless of genetic background; hypersensitive and most of the highly sensitive accessions have defects in ACC2; and sensitivity in some exceptional accessions may be caused by disruption of other genes that remain to be identified.
Table V. Spectinomycin responses of F1 seedlings from crosses between sensitive accessions and informative knockout mutants.
| Accession Parent | F1 Progeny from acc2 Cross |
F1 Progeny from tic20-iv Cross |
||||
|---|---|---|---|---|---|---|
| Category | Score | Seedlings | Category | Score | Seedlings | |
| Gn2-3 | Hypersensitive | 1.1 | 106 | Intermediate | 4.9 | 98 |
| Sav-0 | Hypersensitive | 1.0 | 71 | Intermediate | 4.4 | 45 |
| Knox-18a | Sensitive | 1.3 | 78 | Intermediate | 7.2 | 83 |
| Tul-0a | Hypersensitive | 1.1 | 83 | Intermediate | 5.8 | 81 |
| RRS-10a | Sensitive | 1.2 | 80 | Intermediate | 5.8 | 81 |
| Gifu-2a | Sensitive | 1.2 | 82 | Intermediate | 5.9 | 72 |
| Pna-10a | Hypersensitive | 1.0 | 74 | Intermediate | 5.0 | 64 |
| Aitba-1 | Sensitive | 2.1 | 149 | Intermediate | 5.4 | 148 |
| La-0 | Intermediate | 4.6 | 61 | Intermediate | 4.6 | 67 |
| Grivo-1 | Intermediate | 7.3 | 74 | Intermediate | 6.6 | 52 |
Part of the Knox-18 group of sensitive accessions with shared variants of interest.
Establishing the Mutational Landscape of Homomeric ACCase in Arabidopsis
To document the extent of natural and induced variation in ACCase function in Arabidopsis, we combined the ACC1 and ACC2 accession data described here with genotype and phenotype information for induced acc1 mutations published elsewhere. The results are presented in Figure 3 and Supplemental Figure S3. Informative variants and residues of interest, including those from other model organisms, are listed in Supplemental Table S7. The Arabidopsis dataset includes five nonsense mutations, two frameshift mutations, four examples of aberrant splicing, three small deletions, four T-DNA insertion mutants analyzed in detail, two deletions associated with a chromosomal aberration, seven strong and three weak missense mutations, nine missense mutations that alter highly conserved residues without causing an obvious loss of protein function, and 24 other missense mutations that change a conserved residue. Based on predicted variants in the 1001 Genomes Project data, 7.6% of the 526 conserved (90% level) ACC2 residues exhibit variation among the 855 sequenced accessions. The number of variable residues is substantial, due to the size of the protein, but the level of saturation is low. Overall, 267 variable ACC2 residues (11.3% of 2,355 total) and 110 variable ACC1 residues (4.9% of 2,254) are included in the 1001 Genomes Project data. Another 178 ACC1 consensus residues differ from those found in ACC2, including two conserved at the 90% level (Y383H and K1603Q). Fourteen of the ACC2 variants affect residues conserved at the 99% level, whereas none of the ACC1 variants in sequenced accessions affect such residues, consistent with an essential cellular function. Among ACC2 residues conserved at the 90% level, variants are limited to a single accession for 21 residues (52.5%), two or three accessions for six residues (15%), and four or more accessions for 13 residues (32.5%). Isolated variants that failed to confirm were excluded from the totals above, although some may have been present in the sequenced material.
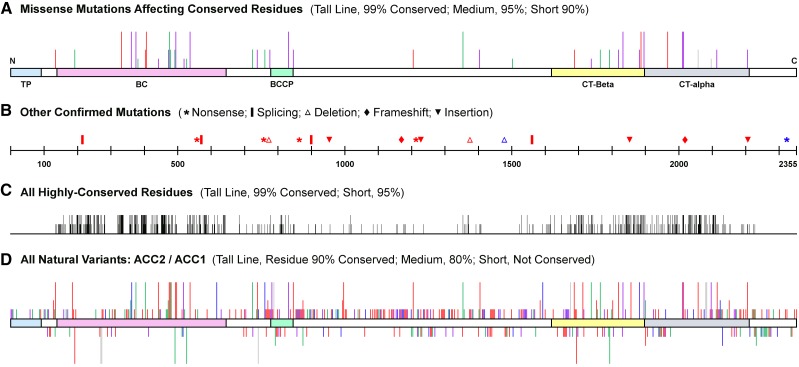
Figure 3.
Mutational landscape of homomeric ACCase in Arabidopsis. Conservation percentages are based on the alignment of 667 eukaryotic protein sequences. A, Induced and natural variants in ACC1 and ACC2 combined. Red lines, Deleterious or likely deleterious variants; purple lines, possibly deleterious (most are likely weak alleles); green lines, not deleterious or likely not deleterious; gray lines, not analyzed or variant of unknown significance. TP, Transit peptide; BC, biotin carboxylase; BCCP, biotin carboxyl carrier protein; CT, carboxyltransferase domains. B, Induced and natural variants combined. Red symbols, Strong alleles; blue symbols, weak or intermediate alleles. C, Highly conserved residues based on the alignment of 667 eukaryotic protein sequences. D, ACC2 variants are shown above the horizontal bar, and ACC1 variants are shown below. Numbers of accessions with predicted variants are as follows: red lines, one; purple lines, two to three; blue lines, four to 10; green lines, more than 10; gray lines, variant not confirmed in the only accession where it was predicted. Shaded areas of the horizontal bar are labeled in A. See also Supplemental Figure S3.
Sequenced Accessions Lack Apparent Tic20-IV Null Mutations
Because tic20-iv knockout mutants resemble acc2 knockouts on spectinomycin and lack a discernible phenotype on soil, we wondered if elimination of Tic20-IV function might contribute to the hypersensitivity of an accession not evaluated on spectinomycin. The Tic20-IV gene is small compared with ACC2 (1.7 versus 12.5 kb), contains only two introns, and encodes a short protein (284 residues). These features reduce the likelihood that variants of interest will be found. Missense mutations affecting 24 residues were still uncovered, but nonsense and frameshift mutations, altered splice sites, and large deletions were all absent from sequenced accessions. Several accessions were predicted to contain a small deletion that eliminated one residue from a variable region of the transit peptide. We confirmed this deletion in one accession (WAR) with a low intermediate phenotype on spectinomycin but did not evaluate the potential impact on protein targeting. Whether any of the missense mutations reduce protein function or limit the uptake of ACC2 into chloroplasts remains an open question. However, we found no evidence that a complete loss of Tic20-IV function can be tolerated in natural environments.
DISCUSSION
In this report, we describe an experimental system that permits the analysis of natural variation in an essential gene, highly conserved among eukaryotes, that performs a critical function in basic metabolism. This work builds upon our longstanding efforts to isolate and characterize embryo-defective mutants of Arabidopsis (Meinke and Sussex, 1979; Meinke, 2013), identify mutants disrupted in chloroplast-localized proteins (Bryant et al., 2011), and establish a comprehensive dataset of Arabidopsis genes with essential functions (Meinke et al., 2008; Muralla et al., 2011; Lloyd and Meinke, 2012). It also extends our recent efforts to understand why plant species in general, and Arabidopsis accessions in particular, differ in their response to a loss of chloroplast translation (Parker et al., 2014). Homomeric ACCase offers several advantages for characterizing genotype-to-phenotype relationships of an essential gene: (1) ACC2 is a duplicated gene whose function can be studied independent of its essential paralog; (2) ACC2 is among the largest proteins targeted to plastids, with multiple conserved domains, which increases the likelihood of finding deleterious mutations in sequenced accessions; (3) ACC2 null mutations can be readily identified by their characteristic hypersensitive response on spectinomycin; and (4) strong and weak alleles of ACC1 can be distinguished by their distinctive embryo, seedling, and vegetative phenotypes in the absence of spectinomycin.
Homomeric ACCases
Homomeric ACCases have been studied most extensively in humans, yeast, and plants (Baud et al., 2004; Hoja et al., 2004; Huerlimann and Heimann, 2013; Tong, 2013). Crystal structures of the yeast (Saccharomyces cerevisiae) holoenzyme dimer and conserved domains have identified residues required for dimerization and catalysis (Wei and Tong, 2015; Hunkeler et al., 2016). Although the enzyme is typically found in the cytosol, gene duplications have on occasion generated a second isoform that functions in mitochondria or chloroplasts. The mitochondrial ACCase in S. cerevisiae (Hfa1p) is translated from a noncanonical AUU codon located upstream of the first AUG codon, consistent with a model in which both forms were originally encoded by a single gene that targeted different cell compartments (Suomi et al., 2014). Drosophila melanogaster and zebrafish also contain ACCases with N-terminal extensions consistent with mitochondrial localization. In humans, the cytosolic version (ACC1) is found in adipose and liver tissues and functions in fatty acid biosynthesis, whereas the mitochondrial version (ACC2) is present in oxidative tissues, including heart and skeletal muscle, and modulates fatty acid oxidation (Kim, 1997; Abu-Elheiga et al., 2000). Human ACCase is controlled by allosteric regulation, protein-protein interactions, reversible phosphorylation, and oligomerization (Brownsey et al., 2006; Tong, 2013). Chemical inhibitors have been developed that interfere with dimerization of one or both isoforms, modify fatty acid metabolism, and represent promising treatments for diabetes, obesity, and other human diseases (Marjanovic et al., 2010; Harriman et al., 2016).
Chloroplasts with homomeric ACCase are found in grasses, which uniquely lack the heteromeric enzyme (Jansen et al., 2007; Chalupska et al., 2008), some members of the Brassicaceae, including Arabidopsis (Yanai et al., 1995; Babiychuk et al., 2011; Bryant et al., 2011; Parker et al., 2014), and several algal lineages derived from additional symbiotic events (Huerlimann et al., 2015). The apicoplast of Toxoplasma gondii, the causative agent for toxoplasmosis, also contains a duplicated ACCase (Jelenska et al., 2001). Several common herbicides target the plastid-localized homomeric enzyme in grasses while not interfering with the cytosolic or heteromeric enzymes found elsewhere. Resistant plants have alterations in one of seven residues located within the carboxyltransferase-binding domain (Zhang et al., 2004; Liu et al., 2007; Kaundun, 2014). The cytosolic ACC1 protein in Arabidopsis enables the biosynthesis of very-long-chain fatty acids (Baud et al., 2004; Lü et al., 2011; Amid et al., 2012), which function in plant protection and signaling pathways and limit the regeneration capacity of roots (Shang et al., 2016). Mitochondria in Arabidopsis generate malonyl-CoA, the product of ACCase action, using an alternative pathway catalyzed by malonyl-CoA synthase (Guan and Nikolau, 2016).
Despite the extensive literature on homomeric ACCases and recent attention focused on herbicide resistance and human health, few mutants altered in this important metabolic enzyme have been described, largely because of problems with lethality. Published examples from S. cerevisiae, D. melanogaster, and Caenorhabditis elegans are limited to a handful of mutant alleles (Schneiter et al., 1996, 2000; Rappleye et al., 2003; Parvy et al., 2012; Sasamura et al., 2013), including several with conditional phenotypes. Because recessive lethals can be readily maintained as heterozygotes in Arabidopsis (Meinke et al., 2008), a variety of acc1 mutants have been isolated and characterized over the past 30 years, including both strong alleles (Meinke, 1985; Torres-Ruiz et al., 1996; Baud et al., 2003, 2004; Kajiwara et al., 2004) and weak alleles (Lü et al., 2011; Amid et al., 2012). The number of informative missense mutations affecting conserved domains, however, remains limited.
Utilizing 1001 Genomes Project Data to Explore Genotype-to-Phenotype Relationships
Genome-wide association studies involving natural variation in Arabidopsis have identified a wide range of novel alleles that underlie phenotypes of interest (Weigel, 2012), including altered plant metabolism (Jasinski et al., 2012; Strauch et al., 2015; Branham et al., 2016). We chose a different approach: evaluating sequence variation in an essential metabolic protein to expand the list of informative variants available for further analysis. With ACC1 and ACC2 combined, we identified a total of 365 residues with predicted missense substitutions among 855 sequenced accessions. The 111 ACC1 variant residues, and six ACC2 variants found in accessions with tolerant responses on spectinomycin, highlight structural changes in homomeric ACCases that can be accommodated without an obvious loss of protein function.
One problem encountered throughout this project was the occasional failure to confirm, through Sanger sequencing, variants predicted in the 1001 Genomes Project dataset. Remarkably, this was true for almost 20% of the 82 accessions genotyped (Supplemental Table S8), even though three full-length cDNAs (La-0, Tsu-0, and Knox-18) sequenced in our laboratory perfectly matched the 7.1-kb predicted sequence. We suspect these inconsistencies represent a mixture of rare sequencing errors; labeling mistakes during stock collection, distribution, or utilization; accidental introgression from unrelated plants (Shao et al., 2016); and genetic heterogeneity in the source population. Variants limited to a single accession are most prone to misinterpretation in the absence of sequence confirmation. We also found some accessions with ACC2 sequences that closely matched accessions from a distant location, a problem analyzed in detail elsewhere (Anastasio et al., 2011). Collectively, these results underscore the need to confirm variants of interest whenever analyzing diverse accessions, consistent with a recent commentary on validating genetic stocks in plant research (Bergelson et al., 2016).
The low level of saturation for variant ACCase residues (15%) and the small number of substitutions available for each affected residue (usually one) point to further limitations of this experimental system. For example, sequenced accessions lacked variants in the seven residues associated with resistance to ACCase herbicides (Kaundun, 2014), three conserved residues required for dimerization of the yeast biotin carboxylase domain (Wei and Tong, 2015), and all but one of the 17 conserved residues located within the CoA binding site (Zhang et al., 2004). Nevertheless, we identified 11 conserved residues that can be altered without an apparent loss of protein function in Arabidopsis. Recent studies of human disease genes have examined deleterious variants that are fixed in the genomes of other vertebrates, often with a single epistatic substitution elsewhere in the protein (Jordan et al., 2015). Widespread epistasis has also been documented in large-scale gene replacement studies in microorganisms (Podgornaia and Laub, 2015; Li et al., 2016). Whether the unexpected tolerance of some ACCase variants is also context dependent, and would be reversed in a different sequence background, remains to be determined.
Most Homomeric ACCase Missense Mutations Do Not Eliminate Protein Function
Using a combination of forward and reverse genetic approaches, we identified only six substitutions in conserved residues that appear to severely reduce ACC2 function in Arabidopsis. This represents about 2% of the variable ACC2 residues uncovered. Three of these variants are located in the biotin carboxylase domain and two in the carboxyltransferase domain. Eighteen variants that alter conserved residues are associated with an intermediate spectinomycin response, which likely reflects either a partial loss of ACC2 function or the presence of sensitive alleles of other genetic modifiers that enhance growth when ACC2 is functional. The most intriguing example is V376A, which alters a residue perfectly conserved among the 667 homomeric ACCases in our alignment. The sole exception is the Col-0 sequence. This suggests that ACC2 function may be reduced in the reference accession, which could help to explain why Columbia embryos defective in chloroplast translation fail to progress beyond a globular stage of development (Parker et al., 2014).
In the Human Gene Mutation Database of disease-causing mutations (www.hgmd.cf.ac.uk), 44% of the entries are missense mutations, 23% are small insertions or deletions, 11% are nonsense mutations, and 9% are changes in splice sites (Stenson et al., 2014). We found a similar variety of mutations associated with a loss of ACC2 function. One major difference is that our screen was most effective at identifying null alleles and was less able to detect weak alleles harboring missense mutations. With continued advances in gene editing technologies, this limitation could be addressed by introducing candidate mutations of interest, based on the responses of ACC2 variants on spectinomycin, into the ACC1 gene of Arabidopsis, where subtle differences in phenotype can be more readily evaluated.
In human genetics, variants are often placed into categories to reflect the likelihood that they cause the observed phenotype. One common system uses five different classes, with pathogenic and likely pathogenic at one end, benign and likely benign at the other, and variants of unknown significance in between. Further guidelines have been proposed to standardize confidence levels using additional criteria (MacArthur et al., 2014). In our study, we placed missense variants into six categories: deleterious to protein function; likely deleterious; potentially deleterious; variant of unknown significance; likely not deleterious; and not deleterious. E1689G is considered deleterious because it was found in both ACC1 (pas3-1 mutant) and ACC2 (Ts-1 accession) alleles with reduced function. Other variants (G135E, I404K, F1206L, and T1902K) are classified as likely deleterious because the altered residue is highly conserved, consistently associated with sensitivity to spectinomycin, and the results of crosses with acc2 knockouts indicated a loss of ACC2 function. Potentially deleterious variants also change conserved residues but are either associated with a less severe phenotype on spectinomycin or remain to be crossed with informative knockouts. Variants of unknown significance have not been evaluated, represent promising candidates based on some criteria but not others, or are present in combination with another variant that reduced ACC2 function. None of the ACC1 variants in sequenced accessions are considered deleterious because they are all maintained in natural populations, where ACC1 performs an essential function. Collectively, these results present a detailed, initial view of the mutational landscape of an essential gene in basic metabolism.
Loss of Tic20-IV-Mediated Chloroplast Protein Import Results in Hypersensitivity to Spectinomycin
Although the elimination of ACC2 function consistently results in sensitivity to spectinomycin, regardless of genetic background, the tic20-iv knockout mutant of Arabidopsis also exhibits a striking, hypersensitive response (Parker et al., 2014). The Tic20 gene family encodes an inner chloroplast membrane protein that likely functions as a channel for protein import (Kasmati et al., 2011; Kikuchi et al., 2013; Campbell et al., 2014). The severe tic20-iv phenotype on spectinomycin indicates that ACC2 is unable to enter the chloroplast and contribute to fatty acid biosynthesis when Tic20-IV is eliminated. The ACC2 protein itself must remain functional in tic20-iv knockouts, because crosses with accessions harboring acc2 null alleles produce F1 seedlings with an intermediate (not hypersensitive) phenotype on spectinomycin. Based on these results, we propose that (1) ACC2 uptake into chloroplasts occurs exclusively via the Tic20-IV complex; (2) the Tic20-I paralog, thought to be required for the import of photosynthetic proteins (Inoue et al., 2010; Hirabayashi et al., 2011), fails to compensate for the absence of Tic20-IV in knockouts grown on spectinomycin, despite the normal appearance of these plants on soil; and (3) hypersensitivity to a loss of chloroplast translation can be used to identify proteins with a non-redundant role in ACC2 production, stabilization, and import into chloroplasts. Hypersensitivity to spectinomycin, therefore, is a key indicator of the failure of functional ACC2 to be localized to plastids, and represents a novel genetic tool for studying the composition and specificity of chloroplast targeting mechanisms and protein import complexes.
Tolerant accessions also represent a valuable tool for studying chloroplast protein import, but for a different reason. With these accessions, a significant amount of tissue can be obtained that lacks Ycf1, Ycf2, and ClpP1, three essential proteins encoded by the chloroplast genome. These accessions can therefore be utilized as conditional triple knockouts to study the functions of three intriguing chloroplast proteins and their associated complexes. Based on the known lethality of Arabidopsis mutants defective in ClpP subunits (Kim et al., 2013), the presumed lethality of tobacco (Nicotiana tabacum) cells harboring plastid clpP1 gene disruptions (Kuroda and Maliga, 2003), and the viability of white leaf sectors in the maize (Zea mays) iojap mutant, which lack ClpP1 because they are devoid of chloroplast ribosomes (Walbot and Coe, 1979), we postulate that ClpP1 functions primarily to modulate chloroplast translation. This model predicts that in our system, where chloroplast translation is blocked through exposure to spectinomycin or mutations that disrupt chloroplast ribosomes, loss of ClpP1 activity should not further limit the growth of cells already defective in chloroplast translation. Thus, given that Ycf1 and Ycf2 likely both function in chloroplast protein import (Kikuchi et al., 2013; Masato Nakai, personal communication), seedlings from tolerant Arabidopsis accessions grown on spectinomycin offer a unique opportunity to evaluate the structural and physiological effects of the simultaneous loss of two major protein import complexes on chloroplast function.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Natural accessions of Arabidopsis (Arabidopsis thaliana) were chosen to represent a wide geographical distribution. Seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. Stock numbers for each accession are listed in Supplemental Table S3. For the forward genetic screen, which involved 100 accessions beyond the initial collection of 52 accessions described by Parker et al. (2014), we focused on seed stocks that were reportedly derived from siblings of sequenced plants. The same was true for accessions chosen for reverse genetics based on predicted variants. Some lines required extended cold treatment for germination or flowering. These requirements are noted in Supplemental Table S3 and in the ABRC stock description. Mature seeds were germinated on agar plates (Meinke et al., 2009), stored in a refrigerator at 4°C for 2 d (standard) to 1 week (extended treatment), and placed beneath fluorescent lights for about 14 d. Seedlings were then transplanted to pots containing a mixture of soil, sand, and vermiculite. Plants were grown under fluorescent lights (16-h-light/8-h-dark cycle) in a growth room maintained at 23°C ± 1°C and watered daily with a nutrient solution as described by Berg et al. (2005). Plants requiring vernalization were transferred at the rosette stage to a cold room for 5 to 6 weeks at 5°C (8-h-light/16-h-dark cycle) and then returned to the growth room for flowering. Seeds for pas3-1 and pas3-2 (Baud et al., 2004) were provided by Jean-Denis Faure. Seeds for gk-101 and acc1-3 (Kajiwara et al., 2004) were obtained from Masahiko Furutani (Nara Institute of Science and Technology) but unfortunately were not viable. Seeds for emb22, initially known as mutant 115D-4A (Meinke, 1985), were derived from internal laboratory stocks and are available through the ABRC.
Seedling Responses on Spectinomycin
To determine seedling growth responses in the absence of chloroplast translation, mature seeds were germinated on a nutrient medium containing Murashige and Skoog salts, 3% (w/v) Glc, 50 mg L−1 spectinomycin, and 0.8% (w/v) agar. Spectinomycin was added by sterile filtration to autoclaved medium before the plates were poured and was included in the final wash solution during seed sterilization. Plates were stored at 4°C for several days as described above, then returned to room temperature and placed under fluorescent lights (16-h-light/8-h-dark cycle). Seedling responses were evaluated 5 weeks after plating, with accommodation for plates refrigerated longer, using a Wild dissecting microscope equipped with an ocular micrometer. Occasional seedlings with evidence of slight greening, often caused by limited root contact with the medium, were excluded from evaluation. Images of embryos and seedlings were taken with a Nikon DXM1200 digital camera attached to a Wild M-8 dissecting microscope. For printed images, the surrounding background was uniformly darkened to highlight the subject.
The seedling classification system initially described by Parker et al. (2014) was updated to enable the rapid calculation of a phenotype score for each accession. Three levels of sensitive seedlings were recognized: (1) cotyledons only (no visible leaf initials; at most a little smooth callus from the shoot apical meristem); (2) first pair of leaf initials (≤ 1.5 mm combined leaf span from tip to base); and (3) more extensive callus or multiple leaf initials (≤ 1.5 mm combined for the two largest initials, including callus). Intermediate seedling categories were as follows: (5) multiple leaves (> 1.5 and ≤ 2.5 mm combined for the two largest); (6) one pair of leaves (> 1.5 mm combined); and (7) multiple leaves (> 2.5 and ≤ 4 mm combined for the two largest). Tolerant seedlings with multiple leaves were assigned to the following categories: (9) > 4 and ≤ 6 mm; (10) > 6 and ≤ 9 mm; and (11) > 9 mm combined for the two largest leaves. For curled leaves, extended lengths were estimated. Overall scores represent the mean score of individual seedling scores combined. The following criteria were used to classify each accession: hypersensitive, 50% or more of seedlings in category 1 and 95% or more of seedlings in categories 1 or 2; sensitive, 70% or more of seedlings in a sensitive category; low intermediate, 50% or more of seedlings in a sensitive category; tolerant, 70% or more of seedlings in a tolerant category; high intermediate, 50% or more of seedlings in a tolerant category; intermediate, everything else that fails to meet the criteria above.
Homomeric ACCase Alignments
Eukaryotic homomeric ACCases were selected based on the presence of a large central domain as defined in the Pfam database (http://pfam.xfam.org/family/PF08326). Of the 744 protein sequences that initially matched the ACCase domain architecture in Arabidopsis, four sequences from C. elegans were removed because they lacked the essential Lys residue required for biotin binding, five fragmented plant sequences were eliminated, and 104 bacterial sequences were removed because the central domain of ACCase should be unique to eukaryotes. Thirty-six other plant sequences identified through BLAST searches with Arabidopsis ACC1 and ACC2 were then added, to bring the final collection to 667 total sequences (198 animal, 139 plant, 276 fungal, and 54 other). Because some of these sequences still contained small gaps indicative of annotation errors, the actual percentage conservation for some residues may be slightly higher than what was calculated. All homomeric ACCase sequences were aligned using the MUSCLE program (Edgar, 2004) through the Jalview 2.8.2 analysis workbench (Waterhouse et al., 2009). ACC2 protein sequences from 855 Arabidopsis accessions with sequenced genomes were downloaded from the Salk Institute 1001 Genomes Web site (http://signal.salk.edu/atg1001/index.php) and entered into an Excel spreadsheet to track individual variants.
Confirmation of Expected Variants
Genomic DNA was isolated from seedlings and inflorescences derived from ABRC seed stocks. A modified cetyltrimethylammonium bromide protocol was followed (Lukowitz et al., 2000). PCR primers (Supplemental Table S9) were designed to amplify genomic regions containing predicted variants of interest while avoiding amplification of the adjacent ACC1 locus. Variants confirmed and not confirmed are listed in Supplemental Table S8. All primers were purchased from Integrated DNA Technologies. PCRs utilized the Qiagen PCR Master Mix and a Biometra Uno II thermocycler. PCR products were separated in 1% agarose gels containing GelRed Nucleic Acid Stain (Phenix). The AlphaImager HP system (Proteinsimple) was used to visualize the bands. For DNA sequencing, all PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced at the Oklahoma State University Recombinant DNA/Protein Resource Facility.
Genomic DNA and cDNA Sequencing
Genomic DNA spanning the ACC2 locus was sequenced from Sav-0 plants grown in our laboratory. PCR primers were located in the 5′ untranslated region (UTR; ACC2-109) and 3′ UTR (ACC2-65) to avoid amplification of the adjacent ACC1 locus. Takara (Clontech) PrimeSTAR GXL DNA Polymerase was used to amplify large fragments of ACC2. PCR products were purified and sequenced with primers located within the ACC2 gene (Supplemental Table S9). For ACC2 cDNA sequencing from the Sav-0, Gn2-3, La-0, Tsu-0, and Knox-18 accessions, RNA was prepared from seedlings grown on basal medium using the RNeasy Plant Mini Kit (Qiagen). cDNAs were synthesized using the Takara PrimeScript RT Reagent Kit. For PCR, the same primers and PrimeSTAR DNA polymerase used for genomic ACC2 sequencing were employed. PCR products were purified and sequenced with primers located within ACC2 exons and UTRs (Supplemental Table S9). To characterize accessions with predicted splicing defects, the same RNA preparation, cDNA synthesis, and PCR procedures were carried out as described above. When cloning PCR products into the T-Easy vector (Promega), nucleotide A was added using Taq DNA polymerase (Invitrogen). The GeneJET Plasmid Miniprep Kit (Thermo Scientific) was used to prepare plasmids. Eight to 10 clones were sequenced for each accession to characterize the diversity of transcripts produced.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Variant ACC2 splice site and altered transcripts in the Spro-2 accession.
Supplemental Figure S2. Short ACC2 deletion and altered transcripts in the Ip-Ber-0 accession.
Supplemental Figure S3. ACC2 consensus protein sequence with conserved residues and variants highlighted.
Supplemental Table S1. Seedling responses of 20 accessions with the ACC2 variants I404K and T1902K.
Supplemental Table S2. ACC2 variants that alter conserved residues in sequenced Arabidopsis accessions.
Supplemental Table S3. Natural accessions analyzed and spectinomycin responses obtained.
Supplemental Table S4. Terminal phenotypes and allele strengths of ACC1 nonsense and missense mutants.
Supplemental Table S5. Spectinomycin responses of F2 seedlings from crosses between sensitive accessions and informative knockout mutants.
Supplemental Table S6. ACC2 variants in the Sav-0 accession that differ from the consensus among sequenced accessions.
Supplemental Table S7. Informative variants that alter conserved residues or disrupt protein structure in eukaryotic, homomeric ACCases.
Supplemental Table S8. Confirmation status of predicted variants among natural accessions analyzed.
Supplemental Table S9. PCR primers used for genomic DNA and cDNA sequencing and variant confirmation.
Supplementary Material
Acknowledgments
We thank Jean-Denis Faure (Institut National de la Recherche Agronomique) for pas3 seeds, Kayla Cook (Oklahoma State University) for assistance with pas3 seedling screens, Paul Jarvis (University of Oxford) for tic20-iv seeds, Masato Nakai (Osaka University) for information on chloroplast ycf2 function, and the ABRC for multiple seed stocks and information on Arabidopsis natural accessions.
Glossary
- ABRC
Arabidopsis Biological Resource Center
- UTR
untranslated region
Footnotes
This work was supported by the Developmental Systems Cluster, Biological Sciences Directorate, National Science Foundation.
Articles can be viewed without a subscription.
References
- Abu-Elheiga L, Brinkley WR, Zhong L, Chirala SS, Woldegiorgis G, Wakil SJ (2000) The subcellular localization of acetyl-CoA carboxylase 2. Proc Natl Acad Sci USA 97: 1444–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amid A, Lytovchenko A, Fernie AR, Warren G, Thorlby GJ (2012) The sensitive to freezing3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies. J Exp Bot 63: 5289–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasio AE, Platt A, Horton M, Grotewold E, Scholl R, Borevitz JO, Nordborg M, Bergelson J (2011) Source verification of mis-identified Arabidopsis thaliana accessions. Plant J 67: 554–566 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep 5: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Bergelson J, Buckler ES, Ecker JR, Nordborg M, Weigel D (2016) A proposal regarding best practices for validating the identity of genetic stocks and the effects of genetic variants. Plant Cell 28: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham SE, Wright SJ, Reba A, Linder CR (2016) Genome-wide association study of Arabidopsis thaliana identifies determinants of natural variation in seed oil composition. J Hered 107: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM (2006) Regulation of acetyl-CoA carboxylase. Biochem Soc Trans 34: 223–227 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Hoang T, Jelokhani-Niaraki M, Smith MD (2014) Folding and self-association of atTic20 in lipid membranes: implications for understanding protein transport across the inner envelope membrane of chloroplasts. BMC Biochem 15: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P (2008) Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA 105: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Nikolau BJ (2016) AAE13 encodes a dual-localized malonyl-CoA synthetase that is crucial for mitochondrial fatty acid biosynthesis. Plant J 85: 581–593 [DOI] [PubMed] [Google Scholar]
- Harriman G, Greenwood J, Bhat S, Huang X, Wang R, Paul D, Tong L, Saha AK, Westlin WF, Kapeller R, et al. (2016) Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats. Proc Natl Acad Sci USA 113: E1796–E1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Kikuchi S, Oishi M, Nakai M (2011) In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant Cell Physiol 52: 469–478 [DOI] [PubMed] [Google Scholar]
- Hoja U, Marthol S, Hofmann J, Stegner S, Schulz R, Meier S, Greiner E, Schweizer E (2004) HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J Biol Chem 279: 21779–21786 [DOI] [PubMed] [Google Scholar]
- Huerlimann R, Heimann K (2013) Comprehensive guide to acetyl-carboxylases in algae. Crit Rev Biotechnol 33: 49–65 [DOI] [PubMed] [Google Scholar]
- Huerlimann R, Zenger KR, Jerry DR, Heimann K (2015) Phylogenetic analysis of nucleus-encoded acetyl-CoA carboxylases targeted at the cytosol and plastid of algae. PLoS ONE 10: e0131099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkeler M, Stuttfeld E, Hagmann A, Imseng S, Maier T (2016) The dynamic organization of fungal acetyl-CoA carboxylase. Nat Commun 7: 11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Rounds C, Schnell DJ (2010) The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Lécureuil A, Miquel M, Loudet O, Raffaele S, Froissard M, Guerche P (2012) Natural variation in seed very long chain fatty acid content is controlled by a new isoform of KCS18 in Arabidopsis thaliana. PLoS ONE 7: e49261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Crawford MJ, Harb OS, Zuther E, Haselkorn R, Roos DS, Gornicki P (2001) Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc Natl Acad Sci USA 98: 2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DM, Frangakis SG, Golzio C, Cassa CA, Kurtzberg J, Task Force for Neonatal Genomics, Davis EE, Sunyaev SR, Katsanis N (2015) Identification of cis-suppression of human disease mutations by comparative genomics. Nature 524: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara T, Furutani M, Hibara K, Tasaka M (2004) The GURKE gene encoding an acetyl-CoA carboxylase is required for partitioning the embryo apex into three subregions in Arabidopsis. Plant Cell Physiol 45: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P (2011) Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J 66: 877–889 [DOI] [PubMed] [Google Scholar]
- Kaundun SS. (2014) Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag Sci 70: 1405–1417 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kim J, Olinares PD, Oh SH, Ghisaura S, Poliakov A, Ponnala L, van Wijk KJ (2013) Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol 162: 157–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH. (1997) Regulation of mammalian acetyl-coenzyme A carboxylase. Annu Rev Nutr 17: 77–99 [DOI] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y (1994) Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA 91: 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44: D862–D868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qian W, Maclean CJ, Zhang J (2016) The fitness landscape of a tRNA gene. Science 352: 837–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ (2011) Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol 155: 293–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Harrison DK, Chalupska D, Gornicki P, O’Donnell CC, Adkins SW, Haselkorn R, Williams RR (2007) Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA 104: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Meinke D (2012) A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol 158: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Zhao H, Parsons EP, Xu C, Kosma DK, Xu X, Chao D, Lohrey G, Bangarusamy DK, Wang G, et al. (2011) The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol 157: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. (2014) Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic J, Chalupska D, Patenode C, Coster A, Arnold E, Ye A, Anesi G, Lu Y, Okun I, Tkachenko S, et al. (2010) Recombinant yeast screen for new inhibitors of human acetyl-CoA carboxylase 2 identifies potential drugs to treat obesity. Proc Natl Acad Sci USA 107: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D. (2013) Large-scale mutant analysis of seed development in Arabidopsis. In Becraft PW, ed, Seed Genomics. Wiley-Blackwell, Ames, IA, pp 5–20 [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Meinke D, Sweeney C, Muralla R (2009) Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE 4: e7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. (1985) Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor Appl Genet 69: 543–552 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Sussex IM (1979) Embryo-lethal mutants of Arabidopsis thaliana: a model system for genetic analysis of plant embryo development. Dev Biol 72: 50–61 [DOI] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N, Wang Y, Meinke D (2014) Natural variation in sensitivity to a loss of chloroplast translation in Arabidopsis. Plant Physiol 166: 2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvy JP, Napal L, Rubin T, Poidevin M, Perrin L, Wicker-Thomas C, Montagne J (2012) Drosophila melanogaster acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genet 8: e1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgornaia AI, Laub MT (2015) Pervasive degeneracy and epistasis in a protein-protein interface. Science 347: 673–677 [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Le Bot N, Ahringer J, Aroian RV (2003) Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev Biol 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salie MJ, Thelen JJ (2016) Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim Biophys Acta 1861: 1207–1213 [DOI] [PubMed] [Google Scholar]
- Sasamura T, Matsuno K, Fortini ME (2013) Disruption of Drosophila melanogaster lipid metabolism genes causes tissue overgrowth associated with altered developmental signaling. PLoS Genet 9: e1003917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Guerra CE, Lampl M, Tatzer V, Zellnig G, Klein HL, Kohlwein SD (2000) A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol Cell Biol 20: 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM (1996) A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16: 7161–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B, Xu C, Zhang X, Cao H, Xin W, Hu Y (2016) Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc Natl Acad Sci USA 113: 5101–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao MR, Shedge V, Kundariya H, Lehle FR, Mackenzie SA (2016) Ws-2 introgression in a proportion of Arabidopsis thaliana Col-0 stock seed produces specific phenotypes and highlights the importance of routine genetic verification. Plant Cell 28: 603–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch RC, Svedin E, Dilkes B, Chapple C, Li X (2015) Discovery of a novel amino acid racemase through exploration of natural variation in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 11726–11731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomi F, Menger KE, Monteuuis G, Naumann U, Kursu VA, Shvetsova A, Kastaniotis AJ (2014) Expression and evolution of the non-canonically translated yeast mitochondrial acetyl-CoA carboxylase Hfa1p. PLoS ONE 9: e114738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L. (2013) Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70: 863–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Lohner A, Jürgens G (1996) The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. Plant J 10: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Walbot V, Coe EH (1979) Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc Natl Acad Sci USA 76: 2760–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Tong L (2015) Crystal structure of the 500-kDa yeast acetyl-CoA carboxylase holoenzyme dimer. Nature 526: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Zhang F, Lupski JR (2015) Non-coding genetic variants in human disease. Hum Mol Genet 24: R102–R110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tweel B, Tong L (2004) Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA 101: 5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.