The NAC transcription factor of Poncirus trifoliata, PtrNAC72, interacts in arginine decarboxylase gene expression.
Abstract
Arginine decarboxylase (ADC)-mediated putrescine biosynthesis plays an important role in plant stress responses, but the transcriptional regulation of ADC in response to abiotic stress is not well understood. We isolated a NAM, ATAF1/2, and CUC (NAC) domain-containing transcription factor, PtrNAC72, from trifoliate orange (Poncirus trifoliata) by yeast one-hybrid screening. PtrNAC72, localized to the nucleus, binds specifically to the promoter of PtADC and acts as a transcriptional repressor. PtrNAC72 expression was induced by cold, drought, and abscisic acid. ADC messenger RNA abundance and putrescine levels were decreased in transgenic tobacco (Nicotiana nudicaulis) plants overexpressing PtrNAC72 but increased, compared with the wild type, in an Arabidopsis (Arabidopsis thaliana) transfer DNA insertion mutant, nac72. While transgenic tobacco lines overexpressing PtrNAC72 were more sensitive to drought, plants of the Arabidopsis nac72 mutant exhibited enhanced drought tolerance, consistent with the accumulation of reactive oxygen species in the tested genotypes. In addition, exogenous application of putrescine to the overexpression lines restored drought tolerance, while treatment with d-arginine, an ADC inhibitor, compromised the drought tolerance of nac72. Taken together, these results demonstrate that PtrNAC72 is a repressor of putrescine biosynthesis and may negatively regulate the drought stress response, at least in part, via the modulation of putrescine-associated reactive oxygen species homeostasis.
Abiotic stresses, including drought, extreme temperatures, salinity, and nutrient imbalance, can severely perturb plant growth and development and limit crop productivity. To adapt to such stresses, plants have evolved a broad range of adaptive responses, involving stress perception coupled with signal transduction and amplification (Xiong et al., 2002). Numerous studies have shown that, following exposure to abiotic stresses, the transcript levels of many genes undergo significant increases or decreases, indicating that transcriptional reprogramming plays an important role in stress adaption (Buscaill and Rivas, 2014).
Stress-responsive genes are generally categorized as regulatory or functional. Genes in the former group encode regulatory proteins, including protein kinases, phosphatases, and transcription factors (TFs), which orchestrate stress signaling pathways. Genes in the latter group encode proteins with a direct role in stress tolerance, such as enzymes involved in the synthesis of various metabolites, phytohormones, and antioxidant enzymes. There is substantial evidence that abiotic stresses can induce profound changes in the accumulation of various metabolites that provide protection against stress-associated damage (Vigeolas et al., 2008). However, the transcriptional cascades pertinent to the accumulation of these metabolites have yet to be identified. The elucidation of the TF networks that regulate key enzyme(s) responsible for the biosynthesis of such metabolites may help in developing strategies for stress tolerance enhancement in crops.
Polyamines (PAs), low-molecular-weight compounds, are some of the metabolites that are ubiquitously present in plants and play important roles in the plant response to various environmental stresses (Sharma et al., 2013; Kotakis et al., 2014; Kubiś et al., 2014). PAs are proposed to function in stress tolerance by maintaining membrane stability, adjusting osmotic potential, or promoting the scavenging of reactive oxygen species (ROS; Ha et al., 1998; Shi et al., 2013; Liu et al., 2015). Plants have three major PAs, putrescine, spermidine, and spermine. In higher plants, putrescine can be produced directly from Orn via ornithine decarboxylase (ODC; EC 4.1.1.17) or indirectly from Arg via arginine decarboxylase (ADC; EC 4.1.1.19). It has been proposed that the ADC pathway is associated with putrescine accumulation in response to abiotic stresses while the ODC pathway may be involved in cell differentiation (Paschalidis and Roubelakis-Angelakis, 2005; Liu et al., 2006).
The importance of ADC and putrescine in abiotic stress tolerance is suggested by several lines of evidence. First, ADC gene transcripts and putrescine levels have been shown to be increased substantially by abiotic stresses in a range of plant species (Urano et al., 2004; Zhang et al., 2014). Second, elevated putrescine levels in plants via genetic engineering of the ADC gene were shown to improve stress tolerance. For example, overexpression of an oat (Avena sativa) adc gene in rice (Oryza sativa) increased the biomass of the transgenic plants grown under salt stress conditions compared with the wild type (Roy and Wu, 2001). In another study, transgenic rice plants overexpressing an oat adc gene showed a 10-fold increase in putrescine levels and exhibited marked drought tolerance (Capell et al., 2004). Similarly, overexpression of the trifoliate orange (Poncirus trifoliata) PtADC gene in Arabidopsis (Arabidopsis thaliana) was reported to confer enhanced tolerance to multiple abiotic stresses, including high levels of osmoticum, dehydration, drought, and low temperatures (Wang et al., 2011). In contrast, application of an ADC inhibitor, or mutation of an ADC gene, can repress putrescine synthesis and compromise stress tolerance (Tiburcio et al., 1986; Soyka and Heyer, 1999; Zhang et al., 2015). Thus, there is a strong association between increased putrescine levels and tolerance of several abiotic stresses.
TFs operate as key regulators of signaling networks and function by recognizing specific cis-acting elements within the promoters of their target genes. Numerous TFs involved in abiotic stress responses have been identified in a range of plant species, some of which regulate biosynthetic genes involved in the accumulation of various metabolites, such as vitamin C (Hu et al., 2016) and anthocyanin (Li et al., 2016a). Several studies also have characterized TFs that regulate PA biosynthetic genes. For example, a stress-responsive trifoliate orange MYB family TF, PtsrMYB, was reported to be a possible regulator of PtADC via its interaction with sequences in the PtADC promoter (Sun et al., 2014). In another study, PtADC was reported to be a potential target of PtrABF, which can specifically recognize the abscisic acid (ABA) response element within the promoter of PtADC (Zhang et al., 2015). Recently, WRKY70 was demonstrated to interact with W-box elements within the promoter of a Fortunella crassifolia ADC gene (Gong et al., 2015).
TFs can function to either activate or repress their target genes. The above-mentioned TFs that interact with ADC promoters are transcriptional activators that enhance ADC gene expression and putrescine synthesis. However, whether ADC expression and/or putrescine accumulation are subject to negative regulation at the transcriptional level has not been reported. Here, we describe the identification and characterization of a TF, PtrNAC72, which binds to the PtADC promoter and acts a transcriptional repressor. We demonstrate that PtrNAC72 is a negative regulator of ADC expression and putrescine biosynthesis. Furthermore, we present data to support its suppressive effects on drought tolerance, which may be due, at least in part, to the modulation of putrescine-mediated ROS scavenging. Our findings offer a better understanding of the molecular mechanism underlying the transcriptional regulation of putrescine biosynthesis in response to abiotic stresses.
RESULTS
Yeast One-Hybrid-Based Screening and Sequence Analysis of PtrNAC72
We reported previously that PtADC contributes to abiotic stress tolerance (Wang et al., 2011). To identify TF(s) involved in the regulation of PtADC and putrescine biosynthesis under stress, we used yeast (Saccharomyces cerevisiae) one-hybrid (Y1H) analysis to screen a trifoliate orange cDNA library derived from leaves exposed to dehydration. Of the genes identified through the screen, two were annotated as encoding TFs of the ABF (C8) and NAC (E8) families. Since we have already demonstrated that members of the ABF family of TFs may serve as regulators of PtADC (Zhang et al., 2015), we focused on the E8 protein in this study. A BLAST search against the Arabidopsis genome database using the E8 nucleotide sequence as a query indicated that it corresponded to a full-length cDNA with an open reading frame (ORF) of 1,035 nucleotides, encoding a protein of 344 amino acid residues. A phylogenetic tree was constructed using the amino acid sequences of E8 and 109 NAC members from Arabidopsis, in which E8 was classified into an identical phylogenetic clade with ANAC072 (Supplemental Fig. S1). They share about 78% sequence identity (BLASTN, E = 3e-28), so the gene encoding this TF was named PtrNAC72.
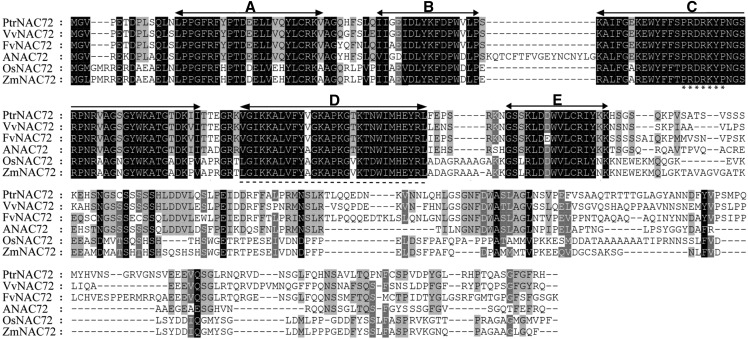
PtrNAC72 had a deduced molecular mass of 38.49 kD and a predicted pI of 8.99. A multiple sequence alignment showed that it shared high sequence similarity at the N terminus with related proteins from grape (Vitis vinifera), strawberry (Fragaria vesca ssp. vesca), Arabidopsis, rice, and maize (Zea mays), whereas the C-terminal regions were generally less conserved among the proteins (Fig. 1). The well-conserved N-terminal region of PtrNAC72, composed of 125 amino acids, was shown to contain five highly conserved subdomains (A–E) that were reported previously by Ooka et al. (2003). Using PSORTII software, a nuclear localization signal (PRDRKYP) was detected in the C subdomain and a NAC repression domain was identified in the D subdomain (Wang et al., 2016c).
Figure 1.
Multiple sequence alignment of PtrNAC72. Multiple sequence alignment is shown for the deduced PtrNAC72 amino acids and its counterparts from other plant species, including rice (Os), Arabidopsis (At), maize (Zm), grape (Vv), and strawberry (Fv). Letters A to E with arrows indicate the five conserved subdomains. Identical amino acids are shown with a black background, and analogous amino acids are shaded in gray. The nuclear localization signal peptides are indicated with asterisks, and the NAC repression domain is represented by the dotted line.
Expression of PtrNAC72 in Response to Different Stresses and ABA
We examined the expression patterns of PtrNAC72 in 1-month-old in vitro-grown trifoliate orange seedlings that had been subjected to various abiotic stresses or treated with exogenous ABA using quantitative real-time (qRT) PCR analysis. PtrNAC72 transcript levels were slightly induced by low temperature within 1 d, followed by a sharp increase, peaking at 5 d. However, PtrNAC72 expression quickly returned to the basal level when the plants were placed in ambient temperature (Supplemental Fig. S2A). Dehydration led to progressive up-regulation of PtrNAC72 expression during the 7-h treatment (Supplemental Fig. S2B). In addition, PtrNAC72 expression was induced within 2 h by treatment with ABA, declined at 4 h, then rose to the highest level at 6 h, followed by a decrease at the last two time points (Supplemental Fig. S2C). These results indicated that PtrNAC72 is indeed a stress-responsive gene.
PtrNAC72 Functions as a TF
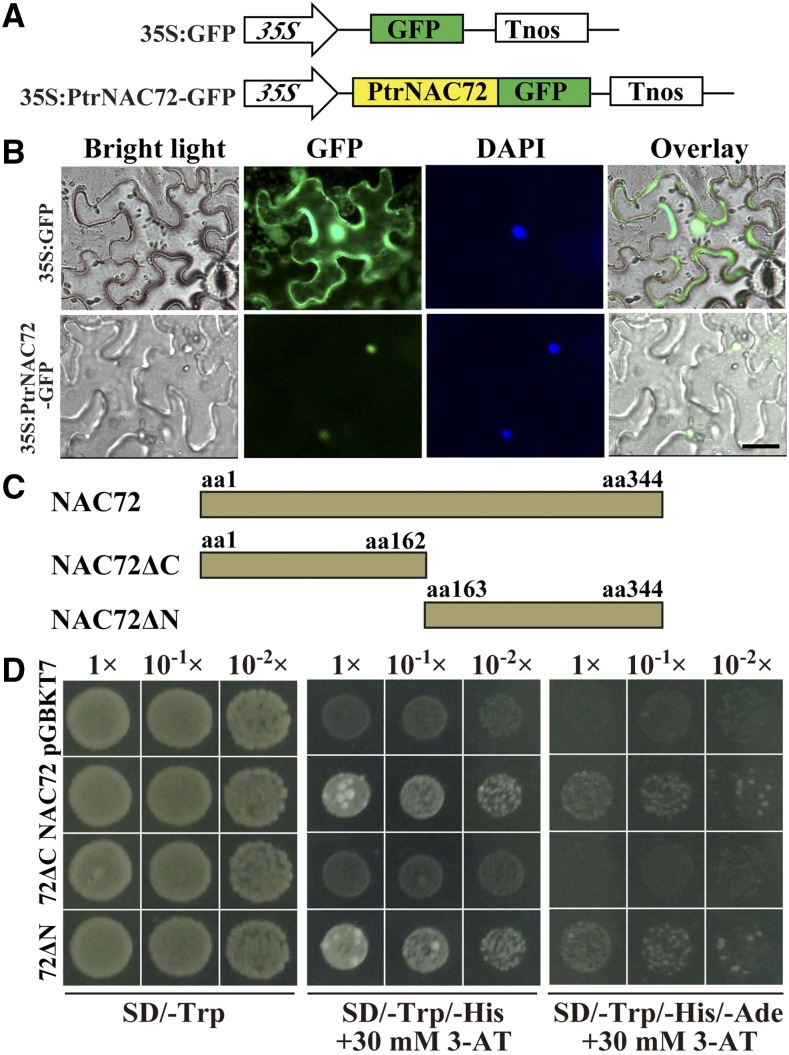
Bioinformatic analysis indicated that the C subdomain of PtrNAC72 contains a nuclear localization signal, PRDRKYP, suggesting that the protein might be targeted to the nucleus. To verify this, we generated a construct (35S:PtrNAC72-GFP) by fusing GFP to the C terminus of PtrNAC72, under the control of the cauliflower mosaic virus 35S promoter (CaMV 35S), while the vector 35S:GFP was used as a control (Fig. 2A). The two constructs were transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells using Agrobacterium tumefaciens-mediated transformation. We observed that GFP signal was distributed evenly in the cytoplasm and the nucleus of epidermal cells when the control vector was used. However, the fluorescence signal was detected only in the nucleus of cells expressing the fusion protein PtrNAC72-GFP (Fig. 2B), indicating that PtrNAC72 was localized to the nucleus. Nuclear localization was confirmed by staining with DAPI.
Figure 2.
Subcellular localization and transcriptional activity of PtrNAC72. A, Schematic diagrams of the constructs used for the subcellular localization assay. B, Subcellular localization of PtrNAC72. The fusion construct (35S:PtrNAC72-GFP) and an empty vector (35S:GFP) were transformed into tobacco (Nicotiana benthamiana) epidermal cells via A. tumefaciens-mediated transformation. Images under bright light and GFP fluorescence were taken. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the nuclei. The overlaid images are shown on the right. Bar = 20 μm. C, Schematic diagrams of the full-length (NAC72) and truncated (N-terminal part, 72ΔC; C-terminal part, 72ΔN) PtrNAC72, which were fused to the GAL4 DNA-binding domain. aa, Amino acids. The numbers following aa indicate the positions of the amino acids. D, Growth of yeast cells, diluted or undiluted, transformed with different constructs on selective medium, using pGBKT7 as a control.
NAC TFs have been shown to have transcriptional activity (Nakashima et al., 2012). To investigate whether this was also true for PtrNAC72, we fused the full-length PtrNAC72 coding sequence (CDS) or various truncated fragments in frame with a GAL4 DNA-binding domain (GDBD) in the yeast expression vector pGBKT7 (Fig. 2C). A construct used to express only the GDBD was employed as a negative control. The constructs were transformed separately into the yeast strain AH109, which carries two reporter genes, ADE2 and HIS3, under the control of a GAL4-responsive upstream activating sequence (UAS) and promoter elements. The different transformants were assayed visually using dilution growth tests. Yeast cells transformed with the pGBKT7 control vector or the different pGBKT7-PtrNAC72 derivatives (full-length or truncated PtrNAC72) all grew well on synthetic dextrose synthetic dropout (SD)/-Trp medium. However, the yeast cells transformed with the control vector or the N-terminal region of PtrNAC72 did not survive on selective SD/-Trp/-His and SD/-Trp/-His/-Ade media supplemented with 30 mm 3-aminotriazole 3-amino-1,2,4-triazole (3-AT). In contrast, yeast cells transformed with the full-length or C-terminal region of PtrNAC72 showed strong growth on the same medium when 10−1 and 10−2 dilutions were used (Fig. 2D). We concluded that PtrNAC72 exhibited transcriptional activity in yeast cells and that the C terminus of PtrNAC72 is necessary for this process.
PtrNAC72 Acts as a Transcriptional Repressor of PtADC
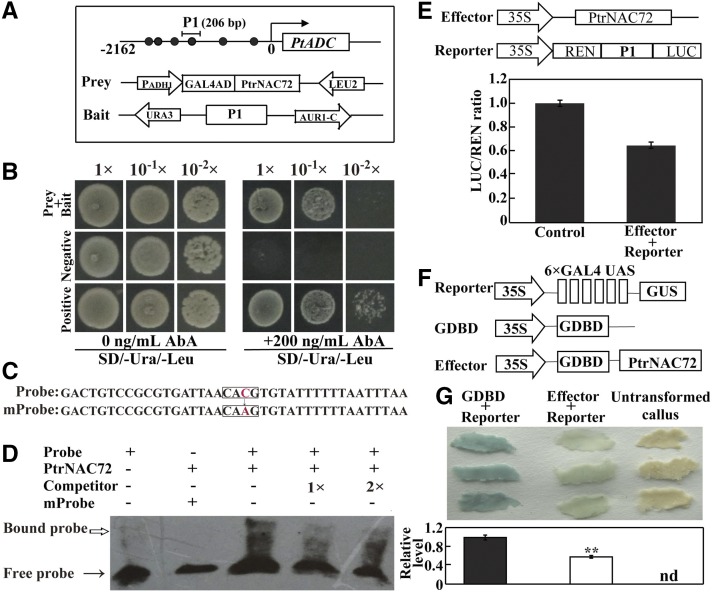
NAC proteins have been reported previously to recognize and bind to a DNA sequence with a core 4-mer motif, CACG, that allows them to regulate downstream genes (Simpson et al., 2003; Tran et al., 2004). Six CACG motifs were identified in the promoter of PtADC. Y1H analysis was first used to verify an interaction between PtrNAC72 and a promoter fragment (P1) containing one CACG motif (−1,496 to −1,493 bp; Fig. 3A). P1 was tested in this assay because PtrNAC72 was fished out using the promoter fragment as a bait for library screening. All of the yeast cells grew normally on SD/-Ura/-Leu medium; however, when antibiotic (200 ng mL−1) was added to the medium, only the positive control yeast cells or cells cotransformed with the effector (PtrNAC72) and the reporter containing P1 survived, in a concentration-dependent manner (Fig. 3B).
Figure 3.
PtrNAC72 binds to the PtADC promoter and acts as a transcriptional repressor. A, Schematic diagrams of the PtADC promoter and the effector and reporter constructs used for the Y1H assay. The circles indicate the cis-acting element, CACG, within the promoter. P1 indicates the partial promoter fragment used to construct the bait plasmid. B, Growth of yeast cells, with or without dilutions, cotransformed with prey and bait, the negative control (bait/pGADT7), or the positive control (p53-AbAi/pGAD-p53) on selective medium without (left) or with (right) 200 ng mL−1 antibiotic (AbA). C, Probes used for EMSA. The top one is a probe synthesized based on the promoter sequence, and the bottom one has CACG replaced with CAAG. D, Binding of PtrNAC72 to the promoter in EMSA. The His-6-PtrNAC72 protein was incubated with the biotin-labeled promoter fragment containing the wild-type CACG or the mutated CAAG form; the nonlabeled fragment was used as a competitor. −, Absence; +, presence. The arrows point to the protein-DNA complex (white arrow) or free probe (black arrow). E, Transient expression assay in tobacco (N. benthamiana) to examine the interaction between PtrNAC72 and the PtADC promoter. Top, Schematic diagrams of the effector and reporter constructs used for the dual LUC assay. The promoter fragment P1 was inserted into the reporter vector pGreen II 0800-LUC, and Renilla luciferase (REN) was used as a control for activity normalization. Bottom, Promoter activities, shown as a ratio of LUC to REN, of tobacco (N. benthamiana) protoplasts cotransformed with the effector and the reporter. The LUC-REN ratio of protoplasts transformed with the empty vector (pGreen II 62-SK/pGreen II 0800-LUC) was set to 1. Data are means ± se (n = 3). F, Schematic diagrams of the three vectors used for the transient expression assay of the transcriptional activity of PtrNAC72 in sweet orange callus using a GAL4/UAS-based system. 6×GAL4 UAS, Six copies of the GAL4-binding site; Effector, PtrNAC72 was inserted downstream of GDBD. G, GUS staining (top) and relative expression level (bottom) of the callus cotransformed with the indicated plasmids. Untransformed callus was used to show the original color. The asterisks indicate a value that is significantly different (**, P < 0.01). nd, Not detected.
Next, we used an electrophoretic mobility shift assay (EMSA) to investigate whether PtrNAC72 specifically binds to the core binding site, CACG. A 39-bp oligonucleotide containing the genuine cis-acting element was synthesized based on the promoter sequence and labeled as a probe (defined as genuine probe), and the same oligonucleotide that was unlabeled was used as a competitor (Fig. 3C). We expressed PtrNAC72 protein in Escherichia coli cells and tested the capacity of this recombinant protein to bind to the oligonucleotides. When the purified PtrNAC72 protein was incubated with the labeled genuine probe, a protein-DNA complex with reduced migration was detected, indicating the binding of PtrNAC72 to the labeled probe. This binding was abolished when unlabeled competitor probe was added. In addition, when the cis-acting element was mutated from CACG to CAAG, a protein-DNA complex was not detected in the presence of the PtrNAC72 protein, indicating that the binding was specific for the CACG sequence (Fig. 3D). We concluded that PtrNAC72 recognized and bound specifically to the CACG motif within the PtADC promoter.
The data above show that PtrNAC72 has transcriptional activity, but it remains to be determined whether it is an activator or a repressor. To address this, we then used a dual luciferase (LUC) assay to investigate how PtrNAC72 interacts with the PtADC promoter. The P1 fragment was introduced into the pGreen II 0800-LUC vector to generate the reporter construct, pADC:LUC. Tobacco (Nicotiana benthamiana) mesophyll protoplasts were cotransformed with both effector (PtrNAC72) and reporter constructs, and the relative LUC activity was determined. Surprisingly, LUC activity was lower in the presence of both the effector and reporter constructs than in the negative control (Fig. 3E), implying that PtrNAC72 may function as a transcriptional repressor. To further confirm this, a GAL4/UAS-based assay was performed, where GDBD binds to six copies of the UAS to activate GUS expression (Tao et al., 2013; Wang et al., 2014). PtrNAC72 CDS was fused to GDBD to generate the fusion protein, GDBD-PtrNAC72, which was coexpressed with 35S-UAS-GUS in sweet orange (Citrus sinensis) embryogenic callus. Histochemical staining showed that GUS expression was prominently repressed in the cotransformed calli compared with the calli transformed with the empty vector control (Fig. 3F). These results suggest that PtrNAC72 might act as a transcriptional repressor of PtADC.
PtrNAC72 Functions to Suppress Putrescine Biosynthesis
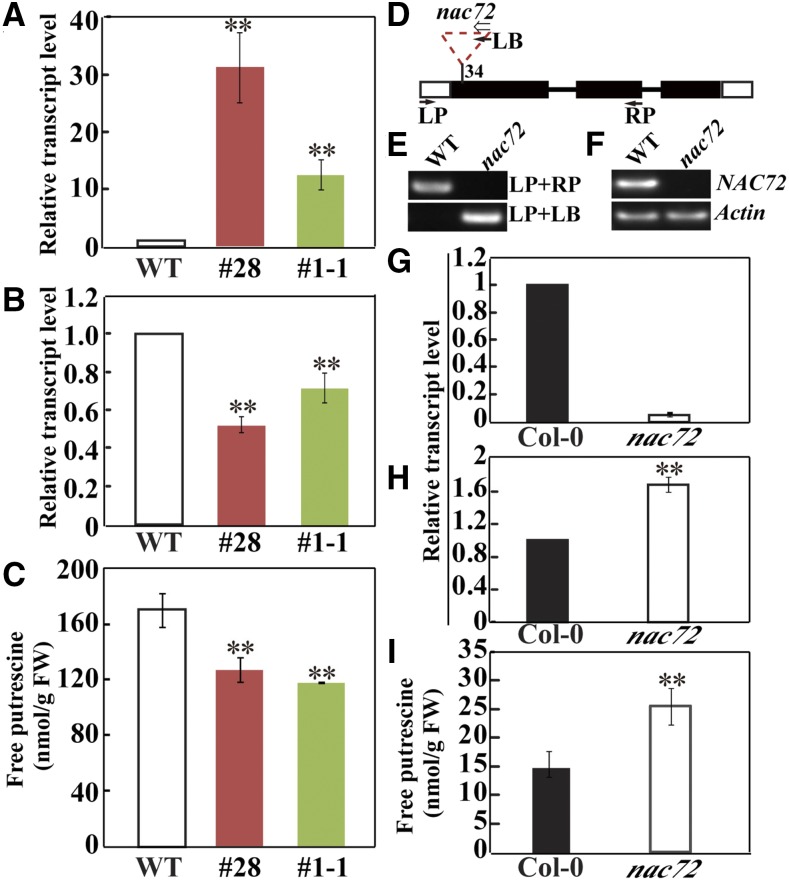
To better understand the function of PtrNAC72, we introduced the 35S:PtrNAC72 binary vector into transgenic tobacco (Nicotiana nudicaulis) by A. tumefaciens-mediated leaf disc transformation. The characterization of these transgenic tobacco plants was considered to be an effective approach, as it is extraordinarily time consuming to obtain progeny of trifoliate orange. Two T3 generation overexpressing lines (designated #1-1 and #28) were selected for further analysis. The transgenic tobacco lines were morphologically indistinguishable from wild-type plants under normal growing conditions. The transcript level of PtrNAC72 was increased prominently in the two overexpression lines (Fig. 4A). Since PtrNAC72 was screened with the PtADC promoter as a bait, we examined how overexpression of this gene may affect the expression of the transgenic tobacco ADC gene and found that the ADC transcript levels were lower in the two transgenic lines compared to that in the wild type (Fig. 4B). Consistent with the lower ADC mRNA abundance, the level of free putrescine, a product of ADC, was lower in the transgenic lines than in the wild type (Fig. 4C).
Figure 4.
Analysis of ADC gene expression and putrescine levels in tobacco and Arabidopsis. A to C, Levels of PtrNAC72 mRNA (A), ADC mRNA (B), and free putrescine (C) in wild-type (WT) tobacco and two PtrNAC72-overexpressing lines, #28 and #1-1. D, Schematic diagram of the Arabidopsis NAC72 gene and the positions of the T-DNA insertion in nac72. The black blocks show exons, lines denote introns, and the white blocks indicate the untranslated regions. The inverted triangle shows the T-DNA, and the direction of the T-DNA is indicated with the white arrow. Primers (LP, RP, and LB) used for genotyping are shown with black arrows. E and F, Genotyping nac72 plants by genomic PCR (E) using gene-specific and T-DNA-specific primers, indicated in D, and semiquantitative RT-PCR (F). G to I, Levels of NAC72 mRNA (G), ADC mRNA (H), and free putrescine (I) in wild-type Arabidopsis (Col-0) and its T-DNA insertion mutant, nac72. For expression analysis, ACTIN and UBIQUITIN were internal controls for Arabidopsis and tobacco, respectively. Data are means ± se (n = 4). Asterisks show values that are significantly different from that of the wild type (**, P < 0.01). FW, Fresh weight.
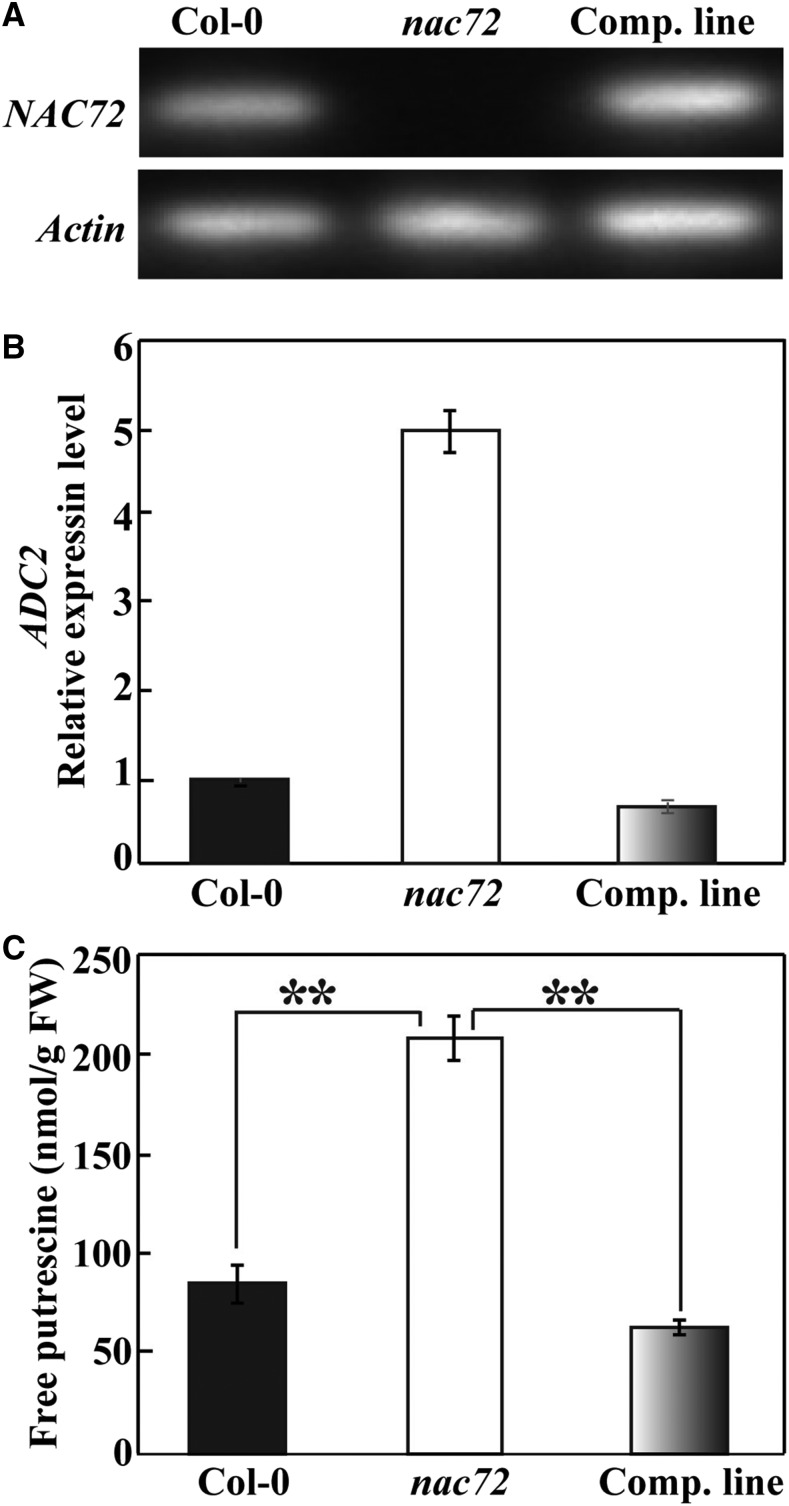
Generating transgenic trifoliate orange lines is technically very challenging, making it difficult to further elucidate the function of PtrNAC72 through the production of RNA interference lines with reduced PtrNAC72 expression. As PtrNAC72 is most closely related to NAC72 of Arabidopsis, efforts were made to investigate the putative function of NAC72 in regulating putrescine synthesis. To this end, an Arabidopsis mutant line was obtained from the publicly available SALK collections (Alonso et al., 2003). The T-DNA homozygous line (SALK_063576) was named nac72 in this study (previously named anac072; Li et al., 2016b), corresponding to NAC72 (At4g27410) in the Columbia-0 (Col-0) accession. Insertion of the T-DNA in this mutant was confirmed by genomic PCR genotyping with two sets of primers. Sequencing of the PCR amplicons and analysis of the flanking sequence demonstrated that the T-DNA is inserted in the first exon of the NAC72 gene, 34 bp downstream of the start codon (Fig. 4, D and E). Semiquantitative real-time (RT) PCR analysis showed that the NAC72 transcript level was barely detectable in nac72, suggesting that nac72 was a true knockout mutant (Fig. 4F), which was further confirmed by qRT-PCR assay (Fig. 4G). In contrast, ADC mRNA abundance and free putrescine levels were higher in the mutant compared with Col-0 (Fig. 4, H and I). Recently, Li et al. (2016b) obtained nac72 complementation plants that had been transformed with a 3.5-kb genomic fragment containing the full-length NAC72 coding region together with its 1.4-kb promoter fragment and 1 kb of the 3′ untranslated region. We used one of these complemented lines (provided by Benke Kuai) to confirm that the increase in the ADC mRNA abundance and putrescine levels could be ascribed to the NAC72 mutation caused by the T-DNA insertion. In contrast to the absence of NAC72 mRNA in the mutant, NAC72 expression in the complemented line was recovered to the level of that in Col-0 (Fig. 5A). We also observed that the ADC transcript (Fig. 5B) and putrescine levels (Fig. 5C) of the complemented line were greatly decreased relative to those of nac72 and were similar to the levels in Col-0 plants. The absolute putrescine levels were different from those of Figure 4I, which may be due to the differences in growth conditions and developmental stages of the tissues sampled for analysis. These data indicated that the T-DNA insertion in the exon of NAC72 accounted for the observed changes in ADC expression and putrescine accumulation. We concluded that NAC72 acts as a negative regulator of ADC expression and putrescine biosynthesis.
Figure 5.
ADC transcripts and putrescine levels in wild-type Arabidopsis (Col-0), the nac72 mutant, and the complemented (Comp.) line. A, NAC72 expression in Col-0, nac72, and the complemented line, as revealed by RT-PCR. B, ADC transcript levels in Col-0, nac72, and the complemented line, as revealed by qRT-PCR. C, Free putrescine levels in Col-0, nac72, and the complemented line. Asterisks indicate significant differences between the genotypes (**, P < 0.01). FW, Fresh weight.
Drought Tolerance Assay Using PtrNAC72-Overexpressing Plants and the nac72 Mutant
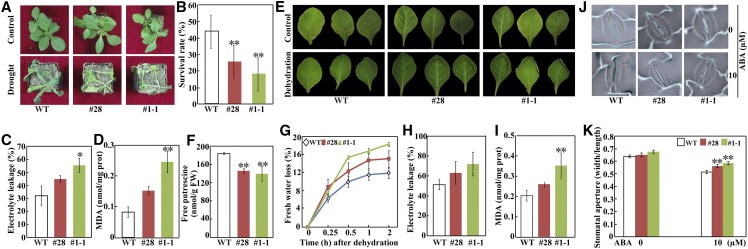
Given that PtADC is involved in drought tolerance (Wang et al., 2011), we hypothesized that increased or reduced NAC72 abundance influences drought responses. To test this hypothesis, we examined the drought tolerance of tobacco transgenic lines overexpressing PtrNAC72. We exposed 30-d-old wild-type and transgenic tobacco plants to drought stress by withholding water for 20 d. The two transgenic lines showed greater sensitivity to the drought stress, as manifested by stronger leaf-wilting symptoms and a lower survival rate, compared with wild-type plants (Fig. 6, A and B). We then measured electrolyte leakage (EL) and the level of malondialdehyde (MDA), two widely used indicators of damage caused by abiotic stresses (Huang et al., 2013). In agreement with the enhanced drought susceptibility, the transgenic lines had greater values of EL and MDA following exposure to drought conditions than did the wild type (Fig. 6, C and D). Since water loss through transpiration is one of the major factors affecting drought tolerance, we assessed the rate of water loss in leaves detached from 30-d-old plants. After dehydration, wilting was more severe in leaves of the transgenic lines than in wild-type leaves (Fig. 6E). The transgenic lines contained lower levels of free putrescine in comparison with the wild type (Fig. 6F). The fresh weight of the detached leaves was measured over 120 min, and water loss was found to be faster for the transgenic lines than for the wild type. At the end of dehydration, the rates of water loss for the two transgenic lines were calculated as being 15.1% (#28) and 18.4% (#1-1) and 11.93% for the wild type (Fig. 6G). Moreover, EL and MDA levels were higher in the transgenic lines than in the wild type after the dehydration treatment (Fig. 6, H and I). Thus, overexpression of PtrNAC72 led to a drought-hypersensitive phenotype in the transgenic lines. In order to investigate whether there was a difference in stomatal movement between the transgenic lines and the wild type, we checked the leaf stomatal apertures in the presence or absence of exogenous ABA, which is an important plant hormone associated with drought stress (Ding et al., 2015). In the absence of ABA, stomatal apertures of the transgenic lines were similar to those of the wild type. However, a noticeable difference was observed with the use of 10 μm ABA. Although ABA treatment led to stomatal closures, stomatal apertures of the transgenic lines were significantly larger than those of the wild type in the presence of ABA (Fig. 6, J and K).
Figure 6.
Drought and dehydration tolerance assay of transgenic tobacco plants overexpressing PtrNAC72. A, Phenotypes of 30-d-old wild-type (WT) tobacco and transgenic lines (#28 and #1-1) before (top) and after (bottom) drought treatment for 20 d. B, Survival rates of the wild type and the transgenic lines after rewatering for 3 d. C and D, EL (C) and MDA levels (D) of the wild type and the transgenic lines after drought treatment. E, Phenotypes of leaves detached from 30-d-old wild-type and transgenic plants before (top) and after (bottom) dehydration for 2 h under ambient conditions. F, Free putrescine levels of the wild type and the transgenic lines at the end of dehydration treatment. FW, Fresh weight. G, Rates of fresh water loss from detached leaves exposed to dehydration treatment at the designated time points. Data are means ± se (n = 3). H and I, EL (H) and MDA content (I) of leaves sampled at the end of dehydration. J and K, Representative images of stomata (J) and quantitative measurement of stomatal apertures (K) in leaves of the wild type and the transgenic lines in the presence of 0 or 10 µm ABA. For stomatal aperture measurement, at least 200 stomata were counted. Asterisks indicate significant differences between the transgenic lines and the wild type (*, P < 0.05 and **, P < 0.01).
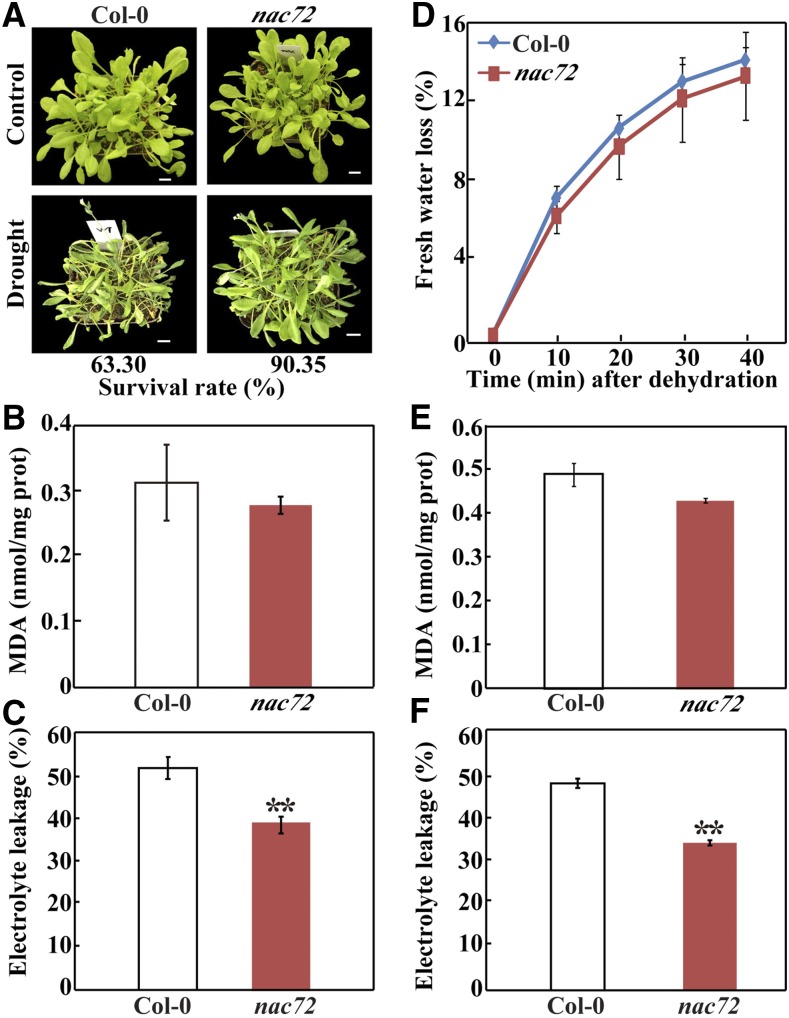
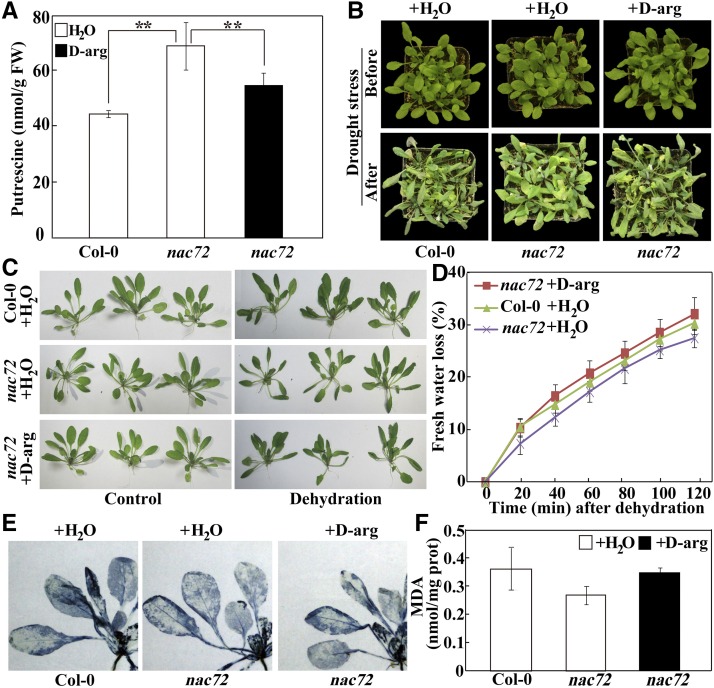
Next, we examined the drought tolerance of the nac72 mutant. Under normal watering conditions, the mutant was phenotypically similar to Col-0. However, the nac72 plants remained relatively healthy after 20 d of water deprivation, while most of the wild-type plants wilted (Fig. 7A). In addition, after rewatering for 3 d, 90% of the mutant plants survived but only 63% of the wild-type plants did. MDA content and EL levels in the mutant were lower than in Col-0 under drought stress (Fig. 7, B and C). In addition, a water-loss assay using rosette leaves indicated that water loss was slightly lower in nac72 than in Col-0 (Fig. 7D), which is consistent with the drought phenotype. Moreover, the mutant had reduced MDA content and lower EL levels compared with Col-0 under dehydration conditions (Fig. 7, E and F). No statistically significant differences in MDA levels were observed between Col-0 and the nac72 mutant after the stress treatments. Following the dehydration treatment, free putrescine levels in nac72 were higher than in Col-0 (174.9 versus 152.4 nmol g−1 fresh weight); however, the difference was not statistically significant (P = 0.341). Thus, the loss of NAC72 function in Arabidopsis conferred enhanced drought tolerance. Taken together, these data suggest that NAC72 functions as a negative regulator in plant responses to drought stress.
Figure 7.
Drought and dehydration tolerance assay of Arabidopsis plants. A, Phenotypes of 30-d-old wild-type (Col-0) and mutant (nac72) plants before (top) and after (bottom) water deprivation for 20 d, followed by rewatering for 3 d. Survival rates of the tested lines are shown below the bottom row. B and C, MDA content (B) and EL (C) of Col-0 and nac72 after drought treatment. D, Water loss rate of Col-0 and nac72 during 40 min of dehydration. For dehydration, the aerial parts of Col-0 and nac72 plants were excised and placed on filter papers in an ambient environment. To quantify water loss, fresh weight was measured every 10 min. E and F, MDA content (E) and EL (F) of Col-0 and nac72 after dehydration treatment. Data are means ± se (n = 3). Asterisks indicate significant differences between Col-0 and nac72 (**, P < 0.05).
ROS Accumulation and Antioxidant Enzyme Activities under Drought Stress Conditions
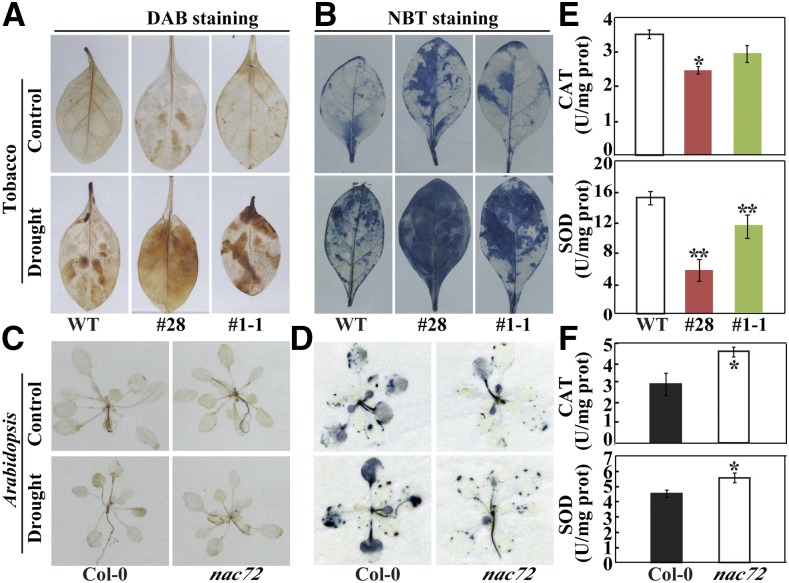
ROS are chemically reactive molecules containing oxygen that are detrimental to plant cells due to the induction of oxidative stress (Andronis et al., 2014; Gong et al., 2014). ROS accumulation can be used as an important variable for measuring the degree of stress tolerance. We examined the in situ accumulation of two major ROS species, hydrogen peroxide (H2O2) and superoxide anion (O2·−), in overexpression tobacco lines and the nac72 mutant before and after dehydration by histochemical staining with 3,3′-diaminobenzidine (DAB; for H2O2) and nitroblue tetrazolium (NBT; for O2·−). DAB staining of the transgenic tobacco lines showed no major difference in H2O2 content between the wild type and PtrNAC72-overexpressing lines prior to dehydration. Dehydration treatment led to an increase in H2O2 levels in both genotypes, but with an accelerated accumulation in the transgenic leaves compared with the wild type (Fig. 8A). Blue coloration, reflecting staining with NBT, was greater in the transgenic than in wild-type leaves both before and after the dehydration treatment, but this difference was particularly notable under stress conditions, indicating greater O2·− accumulation in the transgenic plants (Fig. 8B). Under normal conditions, nac72 and Col-0 leaves were similarly stained by DAB, but the staining was slightly less intense in nac72 than in wild-type leaves after dehydration (Fig. 8C). In addition, NBT staining was evidently less intense in nac72 than in Col-0 leaves before and after the dehydration stress (Fig. 8D). Thus, ROS accumulation was promoted when NAC72 was overexpressed but was reduced when NAC72 expression was knocked out.
Figure 8.
In situ accumulation of ROS and activity of antioxidant enzymes in tobacco and Arabidopsis after dehydration treatment. A to D, Accumulation of H2O2 (A and C) and O2·− (B and D) in tobacco (A and B) and Arabidopsis (C and D) as revealed by histochemical staining with DAB (A and C) and NBT (B and D). E and F, Activities of CAT and SOD in tobacco (E) and Arabidopsis (F) after the dehydration treatment. Asterisks indicate significant differences between the transgenic lines or the mutant and the corresponding wild types (WT; *, P < 0.05 and **, P < 0.01).
Catalase (CAT) and superoxide dismutase (SOD) are two important antioxidant enzymes responsible for scavenging H2O2 and O2·−, respectively (Jaleel et al., 2009). When the activities of these two enzymes were measured in tobacco and Arabidopsis plants after the dehydration treatment, it was found that the tobacco overexpression lines contained lower CAT and SOD activities than the wild type (Fig. 8E), while the activities in the nac72 mutant were significantly higher than in Col-0 (Fig. 8F).
Putrescine Levels Correlate with Drought Tolerance in PtrNAC72-Overexpressing Lines and nac72
Since putrescine levels were higher in the nac72 mutant than in the wild type, and because putrescine is involved in stress tolerance (Cuevas et al., 2009; Alet et al., 2011), we investigated whether the enhanced drought tolerance of nac72 is due to greater accumulation of putrescine. To address this, we treated nac72 with d-Arg, an inhibitor of ADC (Roberts et al., 1984; Liu et al., 2006), before exposing it to drought and dehydration stress. Pretreatment of nac72 with d-Arg resulted in reduced endogenous putrescine levels compared with the levels in water-treated mutant plants, and the levels were similar to those in water-treated wild-type plants (Fig. 9A). After the drought treatment, the d-Arg-treated nac72 plants displayed more severe wilting than did water-treated nac72 plants, and the phenotype was similar to that of the water-treated wild type (Fig. 9B). In addition, d-Arg-treated nac72 showed a reduction in leaf turgor to a greater extent than did the water-treated counterpart under dehydration, which is congruent with the water loss rates (Fig. 9, C and D). In addition, ROS and MDA levels in nac72 plants pretreated with d-Arg were higher than in the water-treated mutant, and the levels in the former were similar to those in the water-treated wild type (Fig. 9, E and F). These results supported the idea that putrescine accumulation accounted for the improved drought tolerance of nac72.
Figure 9.
Drought and dehydration tolerance assay of the nac72 mutant plants pretreated with d-Arg. A, Endogenous putrescine content of nac72 pretreated with water or 1 mm d-Arg compared with the water-treated wild type (Col-0). Asterisks show significant differences between the genotypes (**, P < 0.01). FW, Fresh weight. B, Phenotypes of water- or d-Arg-pretreated nac72, along with water-treated Col-0, before (top) and after (bottom) drought stress for 3 d. C, Phenotypes of water- or d-Arg-pretreated nac72 and water-treated Col-0 before (left) and after (right) dehydration for 2 h in an ambient environment. D, Water loss rates of water- or d-Arg-pretreated nac72 and water-treated Col-0 during a 120-min dehydration treatment. E and F, NBT staining (E) and MDA content (F) of water- or d-Arg-pretreated nac72 leaves after dehydration compared with water-treated Col-0.
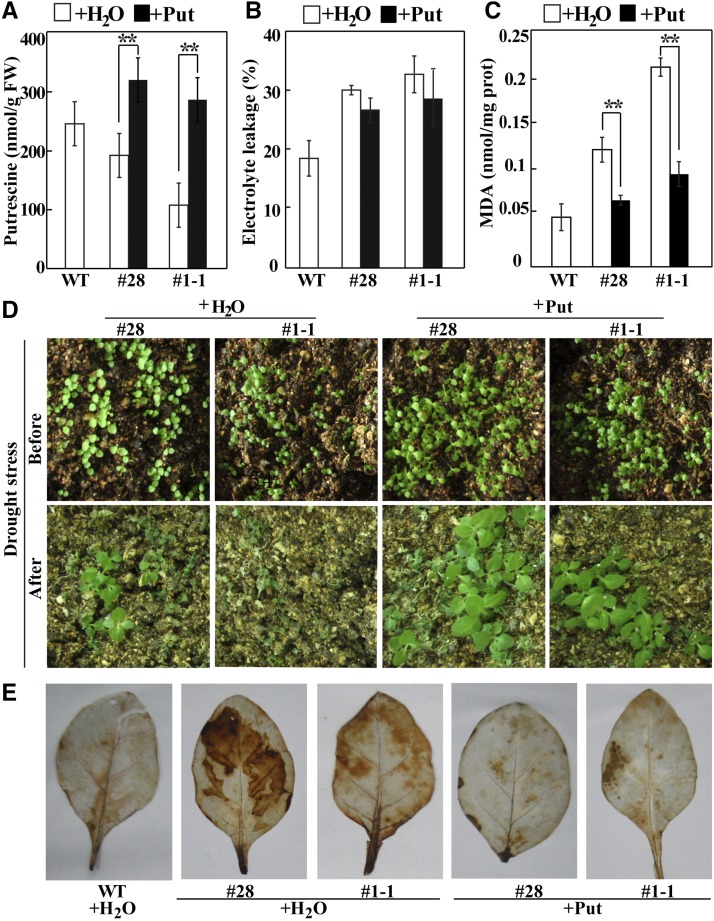
Since the tobacco overexpression lines contained less putrescine and showed impaired drought tolerance, we investigated whether exogenous supply of putrescine could restore, to some extent, the drought and dehydration tolerance of the overexpression lines. To this end, we pretreated the transgenic lines with 10 mm putrescine for 3 d before exposure to drought conditions. Compared with the water treatment, exogenous putrescine supplementation led to a significant increase in the endogenous putrescine levels of the two overexpression lines (Fig. 10A). EL and MDA levels, particularly the latter, were reduced in the transgenic lines that had been pretreated with putrescine compared with the water-treated plants, and the levels were close to that of water-treated wild-type plants (Fig. 10, B and C). In addition, the seedlings of overexpressing lines pretreated with putrescine displayed healthier growth when compared with the water-pretreated seedlings after a 22-d water deprivation (Fig. 10D). Survival rates of water-treated #28 and #1-1 plants were 9.7% and 0%, respectively, which were lower than those of plants of the same lines pretreated with putrescine (18% for #28 and 11.7% for #1-1). DAB staining of the leaves of transgenic lines treated with putrescine, similar to that of the water-treated wild type, was far less than that of the transgenic lines treated with water (Fig. 10E). Thus, exogenous treatment of the transgenic lines with putrescine restored, to some extent, their tolerance of drought stress.
Figure 10.
Drought and dehydration tolerance assay of transgenic tobacco plants pretreated or not with putrescine. A, Endogenous putrescine levels of transgenic lines #28 and #1-1 treated with 10 mm putrescine (Put; black columns) or water (white columns) and the wild type (WT) treated with water. FW, Fresh weight. B and C, EL level (B) and MDA content (C) in the transgenic plants pretreated with putrescine or water, in comparison with the water-treated wild type, measured after the dehydration treatment. Asterisks indicate significant differences between water and putrescine treatment of the same line (**, P < 0.01). D, Phenotypes of water- or putrescine-pretreated overexpression lines before (top) and after (bottom) drought stress for 22 d. E, DAB staining of leaves from transgenic tobacco plants pretreated with putrescine or water and from the water-treated wild type.
DISCUSSION
ADC-mediated putrescine biosynthesis is thought to play an important role in stress responses. However, compared with the well-documented changes in ADC mRNA abundance and putrescine levels in response to different abiotic stresses, relatively little is known about the TFs involved in regulating putrescine biosynthesis via the ADC pathway. In this study, we used Y1H screening to identify a TF, PtrNAC72, with a potential role in regulating ADC expression and putrescine accumulation. We demonstrated that PtrNAC72 recognizes and specifically binds to the promoter of PtADC, thereby acting as a transcriptional repressor. In addition, ADC transcript levels were decreased in PtrNAC72-overexpressing tobacco plants but increased in the nac72 mutant compared with wild-type plants. These results indicate that PtrNAC72 acts as a negative regulator of ADC expression. Prior to this study, several TFs, such as PtrABF (Zhang et al., 2015), FcWRKY70 (Gong et al., 2015), and PtsrMYB (Sun et al., 2014), were shown to activate ADC expression and putrescine synthesis. Identification of these suppressors or activators suggests that ADC is regulated by different TFs, which may in part account for the up- or down-regulation of ADC in response to various abiotic stresses (Liu et al., 2009; Wang et al., 2011). Stress-related signaling pathways are highly complex but are individually regulated by different phytohormones, and ABA has been shown to play a central role in mediating plant responses to abiotic stresses (Urano et al., 2009). It has been suggested that stress-responsive TFs work in an ABA-dependent or -independent manner (Lata and Prasad, 2011). Of the identified TFs, ABFs and MYBs are included in the ABA-dependent signaling system, whereas NAC TFs belong to the ABA-independent group. Thus, complex cellular signaling networks appear to be associated with the regulation of ADC transcription under different environmental conditions. However, whether these TFs work synergistically or independently to regulate the ADC pathway remains to be determined.
NAC proteins are one of the largest TF families and are thought to be unique to plants (Puranik et al., 2012). On the basis of their structural features, they can be divided into two major categories, typical and atypical. Comparison of amino acids between PtrNAC72 and NAC proteins from other plant species suggests that PtrNAC72 may be clustered into the typical NAC group because it has a highly conserved NAC domain, consisting of 125 amino acids at the N terminus. Sequence characteristics in this region have been widely observed in typical NAC proteins from other plant species, suggesting that they may share common biological features in terms of DNA binding. However, it is noted that the NAC proteins vary largely at the C terminus, which may account for the distinct functions of different proteins derived from various clades.
TFs function to regulate the expression of downstream genes by interacting with cis-acting elements present within the target gene promoters. The cis-acting elements contain consensus sequences that can be specifically recognized and bound by corresponding TFs. The NAC cis-acting element contains a NAC recognition sequence with the CATGTG sequence and a consensus CACG sequence as the DNA-binding site (Tran et al., 2004). Most NAC proteins bind to the CACG consensus sequence to regulate the expression of their target genes. This binding was observed in tomato (Solanum lycopersicum) JA2, Arabidopsis NAC019, and rice OsNAC5 (Takasaki et al., 2010; Du et al., 2014; Guan et al., 2014). Consistent with these results, the Y1H and EMSA assays described here revealed that PtrNAC72 can recognize and specifically bind to the CACG core sequence within the PtADC promoter. In addition, potential CACG core sequences also are found in the ADC gene promoter of Arabidopsis based on genome sequence analysis. This may explain the regulatory effects of NAC72 on ADC expression in the nac72 mutant. In addition, it also implies that transcriptional regulation of ADC by NAC72 may be conserved among different plants. However, some NAC proteins can bind a sequence other than CACG. For example, Kim et al. (2007) and Hao et al. (2011) found that two NAC proteins, an Arabidopsis calmodulin-binding NAC and cotton (Gossypium hirsutum) GmNAC11, bound to a GCTT core sequence but not CACG within the target gene promoter. In another study, an NAC016-specific binding motif (NAC16BM), GATTGGAT[AT]CA, was identified in the promoters of NAC016 target genes (Sakuraba et al., 2015). Recently, Chen et al. (2016) reported that sweet potato (Ipomoea batatas) IbNAC1 bound to both CACG and another specific motif, 5′-TACAATATC-3′, in the SWRE (for sporamin wound response cis-element) region in the promoter of sporamin. Neither NAC16BM nor SWRE contained the previously identified CACG core motif or its reverse complement CGTG. Therefore, NAC proteins may vary in their recognition specificity to accomplish their regulatory functions, which implies that the interactions between NAC proteins and the relevant cis-acting elements of downstream target genes are disparate in different plant species or under various environmental conditions. The presence of various binding sequences may be ascribed to difference in the NAC domain, which has been shown to be responsible for DNA binding (Kim et al., 2007). Identification of the DNA-binding sequences may provide insight into the regulation mechanisms of the NAC family members.
NAC proteins play a role in a spectrum of biological processes, including growth and development, morphogenesis, and senescence (Olsen et al., 2005). Increasing evidence indicates that NAC proteins are associated with responses to abiotic stresses (Liu et al., 2014). For example, the transcript levels of many NAC genes show substantial changes in response to various abiotic stresses and that overexpression or mutation of a single NAC gene leads to noticeable changes in abiotic stress tolerance. A number of NAC genes associated with drought tolerance have been identified and characterized from diverse plant species such as rice, Arabidopsis, and soybean (Glycine max). Some of the NACs play a positive role in drought tolerance. For example, overexpression of OsNAC10 and SNAC1 in rice conferred improved drought tolerance and grain yield under drought conditions (Hu et al., 2006). Similarly, overexpression of Arabidopsis ATAF1 and NAC61 and tomato SlNAC35 markedly enhanced drought tolerance in transgenic plants (Wu et al., 2009; Wang et al., 2016a). In contrast, a few NACs have been shown to suppress drought responses, including GmNAC2 (Jin et al., 2013), OMTN2/3/4/6 (Fang et al., 2014), and NAC016 (Sakuraba et al., 2015). Overexpression of these NACs confers sensitivity to drought stress, whereas their mutation enhances drought tolerance. We found that drought tolerance was impaired when PtrNAC72 was overexpressed in tobacco and was promoted in the nac72 mutant of Arabidopsis, suggesting that PtrNAC72 acts as a suppressor of drought tolerance. Taken together, these results indicate that NACs participate in regulating drought responses, either as suppressors or activators, depending on the family member and the nature of the stress (Wu et al., 2009).
TFs are crucial components of the signaling pathways implicated in abiotic stress response cascades. Therefore, the interactions between TFs and downstream target genes play a pivotal role in the defense against stresses. Much progress has been made in the functional characterization of a large spectrum of NAC genes involved in various kinds of abiotic stresses. Concurrent with this functional analysis, a number of studies have identified NAC target genes, some of which encode regulatory proteins. For example, rice OsNAC10 regulates seven protein kinases and five TFs that may function in stress tolerance pathways (Jeong et al., 2010). NAC016 regulates ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN1, a central TF in the stress-responsive ABA signaling pathway (Sakuraba et al., 2015). NAC proteins also regulate genes encoding enzymes that play a direct role in protecting against external stresses. For instance, OsNAC10 targets include several genes that are known to function directly in stress responses: cytochrome P450, NCED, and the potassium transporter HAK5. In addition, Jeong et al. (2010) and Mendes et al. (2013) reported that soybean GmNAC81 and GmNAC30 could regulate a caspase1-like vacuolar processing enzyme to activate plant cell death. Here, we demonstrate that ADC is a target gene of PtrNAC72, based on Y1H, EMSA, and transient expression assays. ADC is a key enzyme involved in PA biosynthesis, and previous studies have shown that ADC expression is induced in different plant species by different abiotic stresses (Urano et al., 2009). In parallel with up-regulation of the ADC gene, endogenous putrescine levels have been reported to increase in plants exposed to abiotic stresses such as drought (Alcázar et al., 2010), low temperatures (Cuevas et al., 2008; Alet et al., 2011), osmotic stress (Kotakis et al., 2014), and salinity (Liu et al., 2006), indicating a close association between the ADC and abiotic stress responses. In this study, we found that transgenic tobacco plants overexpressing PtrNAC72 contained less putrescine than wild-type plants, whereas putrescine levels were elevated in the nac72 mutant. Notably, the drought tolerance capacity of the transgenic overexpressing plants or the nac72 mutant was inversely associated with putrescine levels, suggesting that the magnitude of the drought tolerance in these genotypes depended, at least in part, on the putrescine synthesis/accumulation. This idea is supported by the finding that decreased putrescine levels in the mutant, caused by the application of an ADC inhibitor, compromised drought tolerance, whereas the elevation of putrescine levels in transgenic plants with exogenous putrescine conferred enhanced stress tolerance. Taken together, these data are congruent with a model where PtrNAC72 targets and regulates ADC to modulate the drought response, thereby establishing a newly identified NAC regulon and providing a clue to understanding the physiological relevance of the NACs in stress tolerance. However, it has to be pointed out that NAC72 also may regulate other stress-responsive genes. In this regard, products of these genes, which are not identified here, may directly or indirectly participate in the drought stress response to compensate or offset the influence of NAC72 on the ADC-mediated pathway. This conjecture may be reasonable, as MDA was influenced only slightly in the nac72 mutant after the stress treatment. Our data, together with those of other studies, show that the target genes may vary among different NAC family members. In addition, NAC proteins may activate different sets of target genes in different tissues, as has been reported by Jeong et al. (2010), who showed that the number of genes up-regulated by OsNAC10 in rice roots differed from that in leaves, and only four genes were commonly activated in the two organs.
A well-documented physiological perturbation under drought conditions is the accumulation of ROS (Miller et al., 2010). Overproduction of ROS leads to oxidative stress and consequent damage to cellular components such as membranes and protein synthetic machinery, or even cell death. Plants have evolved scavenging systems composed of both enzymatic and nonenzymatic antioxidants to detoxify ROS that are produced under abiotic stress conditions (Jaleel et al., 2009; Miller et al., 2010). PAs may act as nonenzymatic antioxidants to protect cells against ROS-mediated oxidative stress, although their exact mode of action remains unclear (Gupta et al., 2013; Shi and Chan, 2014; Liu et al., 2015). In this work, we found that ROS accumulation was obviously stimulated in PtrNAC72-overexpressing tobacco lines under drought stress but greatly reduced in the nac72 mutant compared with wild-type plants. Concomitant with the ROS accumulation, EL and MDA levels were increased in the overexpressing lines but decreased in the mutant. Thus, overexpression of PtrNAC72 is accompanied by greater oxidative stress and membrane damage, which were otherwise ameliorated when NAC72 was knocked out. Moreover, the ROS profiles of the tested genotypes were negatively associated with the endogenous putrescine levels. We noted that reduced endogenous putrescine levels in the mutant caused by treatment with an ADC inhibitor promoted ROS accumulation, whereas exogenous putrescine decreased ROS accumulation in the overexpressing lines. These data indicate that putrescine-mediated ROS scavenging plays a major role in modulating the drought tolerance of these plants. This finding is in agreement with earlier studies showing that elevated endogenous putrescine levels mitigated ROS accumulation and enhanced stress tolerance (Liu et al., 2006; Mohapatra et al., 2009; Yiu et al., 2009; Kamiab et al., 2014; Zhang et al., 2015). However, the involvement of the two antioxidant enzymes, CAT and SOD, in ROS detoxification cannot be ruled out, as their activities also were altered in the examined plants. In this regard, the relationship between putrescine and the antioxidant enzymes remains to be clarified.
CONCLUSION
In conclusion, we identified an NAC TF, PtrNAC72, that is involved in regulating PA biosynthesis. PtrNAC72 acts as a repressor of ADC expression and putrescine synthesis. Moreover, PtrNAC72 plays a negative role in modulating drought tolerance, at least in part, due to putrescine-mediated ROS scavenging. Taken with the results of previous studies on the identification of several ADC activators, our study indicates that ADC-mediated putrescine biosynthesis is controlled by a complex regulatory module involving various TFs. Defining the cross talk or interaction between these TFs will help elucidate the molecular mechanisms underlying PA metabolism in response to drought and other abiotic stresses.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatments
Trifoliate orange (Poncirus trifoliata) leaves from 60-d-old seedlings grown in a plant growth chamber were used for cDNA library construction. To examine PtrNAC72 expression patterns, 1-month-old in vitro-grown trifoliate orange seedlings were subjected to various abiotic stresses or exogenous ABA. For the cold treatment, plants were placed in an incubator at 4°C. For dehydration treatment, leaves were detached from the seedlings and desiccated on filter papers at ambient laboratory temperature. ABA treatment was applied by immersing the seedlings in 0.25 mm ABA. Leaves were then sampled at the designated time points and immediately frozen in liquid nitrogen until further analysis.
Seeds of the Arabidopsis (Arabidopsis thaliana) wild-type Col-0 ecotype and the T-DNA insertion mutant nac72 were obtained from the Arabidopsis Biological Resource Center. For PCR-based genotyping, genomic DNA was prepared from Col-0 and mutant leaves using a modified cetyl-trimethyl-ammonium bromide method. PCR amplification was carried out with two sets of primers (Supplemental Table S1) based on the procedures described by Wang et al. (2016b). The PCR-amplified products derived from Col-0 and nac72 were sequenced and aligned to determine the T-DNA insertion position. The expression levels of NAC72 in Col-0 and nac72 were examined by RT-PCR. Complemented plants of nac72 (Li et al., 2016b) were provided by Benke Kuai. Transgenic tobacco (Nicotiana nudicaulis) plants were generated by Agrobacterium tumefaciens-mediated transformation of leaf discs (see below). Arabidopsis and tobacco seeds were surface sterilized with 75% ethanol and 2% NaClO, washed three times with sterile water for 5 min, and then plated on a solid medium comprising one-half-strength Murashige and Skoog (1962) salts supplemented with 3% (w/v) Suc and 0.75% (w/v) agar (pH 5.8). Tobacco and Arabidopsis seedlings were grown in soil pots at 22°C to 24°C with 70% humidity under long-day conditions (16 h of light/8 h of dark) in a growth chamber with a light intensity of 100 µmol m−2 s−1 provided by cool-white fluorescent lights.
Y1H Screening of the cDNA Library
Three promoter fragments of PtADC were amplified from trifoliate orange genomic DNA with specific primers (PY1H-1/2/3; Supplemental Table S1) and fused separately to the pAbAi vector harboring the AUR-1C gene to obtain three bait constructs (P1/2/3). The three bait plasmids were integrated into the Y1HGold yeast (Saccharomyces cerevisiae) genome to create bait-reporter strains. A cDNA library was constructed from RNA extracted from trifoliate orange leaves exposed to dehydration for 1 and 3 h. The cDNA library was screened with the bait-reporter strains using the Matchmaker Gold Yeast One-Hybrid Library Screening System (TaKaRa). Approximately 5 × 105 transformants were initially screened on selective medium SD/-Leu containing 50 ng mL−1 antibiotic (Zhu et al., 2015). The prey fragments from the positive colonies were identified by DNA sequencing.
A retransformation assay was carried out according to Zhu et al. (2015) with minor modifications. The full-length PtrNAC72 ORF was amplified with gene-specific primers (Supplemental Table S1) and fused to the GAL4 activation domain of the pGADT7 vector to generate the prey vector GAL4AD-PtrNAC72. The prey vector was transformed into the bait-reporter strain, and the yeast cells in two dilutions were grown for 3 d on SD/-Ura/-Leu medium with or without antibiotic (200 ng mL−1) at 30°C. Both positive (pGAD-p53+p53-AbAi) and negative (pGADT7+P1-AbAi) controls were included as described by the Y1H system protocol.
Sequence Analysis of PtrNAC72
Bioinformatic analyses of the PtrNAC72 sequence included a homology search of the National Center for Biotechnology Information database, multiple alignments using GeneDoc 2.7, analysis of typical motifs, and prediction of the pI and molecular mass (Liu et al., 2009). In addition, a phylogenetic tree was generated between PtrNAC72 and 109 Arabidopsis NAC proteins using MEGA 6.06 with the neighbor-joining algorithm (Tamura et al., 2007). The putative nuclear localization signal was detected using the protein subcellular localization prediction tool PSORT. Bootstrap analysis was performed using 1,000 replicates in MEGA to evaluate the reliability of different phylogenetic groups (Ying et al., 2011).
RNA Isolation, RT-PCR, and qRT-PCR
Total RNA was extracted using Trizol reagent (TaKaRa Bio Group). First-strand cDNAs were synthesized using the PrimeScript RT Reagent Kit with the gDNA Eraser (Toyobo). RT-PCR was carried out using procedures described by Huang et al. (2010). Quantitative real-time PCR (qRT-PCR) was performed on the LightCyclerW480 Detection System (Roche) with TransStart Top Green qRT-PCR SuperMix (Transgen Biotech). The reaction solution, in a total volume of 10 μL, contained 5 μL of 2× RT-PCR Mix (containing SYBR GREEN I), 0.5 μL of each primer, 3 μL of water, and 0.2 μm cDNA. The thermal profiles were 95°C for 5 min followed by 45 cycles of 95°C for 10 s, 58°C for 30 s, and 72°C for 10 s. Expression of the UBIQUITIN gene was used as an internal reference for tobacco, while ACTIN genes were used for Arabidopsis and trifoliate orange. Gene expression was determined relative to that of the reference gene (Livak and Schmittgen, 2001). The primers for RT-PCR and qRT-PCR are listed in Supplemental Table S1.
Analysis of Subcellular Localization
The PtrNAC72 CDS without the stop codon was amplified by PCR and fused to the 5′ terminus of GFP in the pCAMBIA 1302 vector under the control of CaMV 35S to generate 35S:PtrNAC72-GFP vector. The fusion construct was transformed into A. tumefaciens strain GV3101. Transient transformation of tobacco (Nicotiana benthamiana) epidermis with GV3101 carrying either the fusion construct or the control (35S:GFP) was performed as described previously (Selote et al., 2015). DAPI was used for staining of nuclei, and GFP fluorescence was observed with a confocal laser scanning microscope (Nikon Eclipse 90i).
Transcriptional Activation Assay
The full-length or truncated (ΔC, amino acids 1–162; ΔN, amino acids 163–344) PtrNAC72 ORFs were amplified by PCR with specific primers (Supplemental Table S1) and cloned into the pGBKT7 vector (Clontech) containing GDBD. The recombinant vectors (GDBD-NAC72, GDBD-NAC72ΔC, and GDBD-NAC72ΔN) were transformed into the yeast (Saccharomyces cerevisiae) strain AH109. The transformants with different dilutions were spotted on plates containing three types of medium: SD/-Trp, SD/-Trp-His + 3-AT (30 mm), and SD/-Trp-His-Ade + 3-AT (30 mm). The transactivation activity was evaluated by observing the growth of the transformed cells.
Transient Expression of PtrNAC72 in Tobacco Leaves and Citrus spp. Callus
The full-length PtrNAC72 ORF was amplified from trifoliate orange cDNA with gene-specific primers (Supplemental Table S1) and cloned into the effector vector, pGreenII 62-SK (Hellens et al., 2005), under the control of CaMV 35S. A 206-bp PtADC promoter fragment was amplified with specific primers and ligated into the reporter vector, pGreen II 0800-LUC (Hellens et al., 2005). The effector and reporter constructs were transformed into A. tumefaciens GV3101 cells. Tobacco (N. benthamiana) protoplast isolation and polyethylene glycol-mediated cotransformation of the effector and reporter constructs were performed as described previously (Yoo et al., 2007) with minor modifications. The transformed protoplasts were incubated for 16 h at 22°C before activity assay of firefly luciferase (LUC) and Renilla luciferase (REN) via the Dual-Luciferase Reporter Assay System (Promega) with an Infinite200 Pro microplate reader (Tecan). The promoter activity was expressed as a ratio of LUC to REN.
In addition, a 35S:UAS-GUS reporter system was used to assess the transcriptional activity of PtrNAC72 as described by Wang et al. (2014). For this purpose, the CDS of PtrNAC72 was amplified and ligated to the pYF503 vector, provided by Xia Li, generating an effector GDBD-PtrNAC72 construct. The effector construct, empty control (pYF503; designated as GDBD), and reporter plasmid 35S-UAS-GUS were transformed into A. tumefaciens strain EHA105 cells. Sweet orange (Citrus sinensis) embryogenic callus was cotransformed with the reporter and either of the two effectors and then incubated in the dark for 24 h, followed by histochemical GUS staining. For GUS staining, the calluses were vacuum infiltrated for 10 min with a staining buffer composed of 100 mm sodium phosphate (pH 7), 0.1% Triton X-100, 0.1% N-laurylsarcosine, 10 mm Na2EDTA, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid and then incubated at 37°C for 24 h, followed by a 70% (v/v) ethanol wash.
EMSA
The CDS of PtrNAC72 was amplified and inserted into the pET30a vector to generate the recombinant His-6-PtrNAC72 protein. The plasmid was introduced into Escherichia coli (BL21) cells as described previously (Stone et al., 2005), induction of expression was performed by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm, and then the culture was incubated at 37°C for 3 to 5 h. The recombinant protein was purified using nickel-nitrilotriacetic acid magnetic agarose (Qiagen) according to the manufacturer’s instructions. EMSA involved use of the Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). A 39-bp single-strand fragment containing CACG was synthesized (TsingKe) based on the PtADC promoter sequence and labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific). The same fragment without biotin labeling was used as a competitor. In addition, a mutated fragment of the probe was synthesized and labeled, in which CACG was replaced by CAAG. The probes were incubated with the fusion protein in a 20-μL reaction solution with or without the competitor (1:1 and 1:2 labeled:unlabeled) for 30 min at room temperature. The reaction products were then separated by electrophoresis on a 6% native polyacrylamide gel and electrophoretically transferred to nylon membranes (Beyotime Biotechnology). After UV cross-linking, migration of the biotin-labeled probe on the membrane was visualized by chemiluminescence (Chemiluminescent Nucleic Acid Detection Module; Thermo Scientific).
Binary Vector Construction and Plant Transformation
The full-length PtrNAC72 ORF was PCR amplified from trifoliate orange cDNA using primers containing the restriction sites BamHI and SacI and inserted into the binary vector pBI121 under the control of CaMV 35S. The plasmid was sequenced and introduced into A. tumefaciens strain GV3101 cells by heat shock, which were then used for transgenic tobacco transformation. A. tumefaciens-mediated tobacco leaf disc transformation, regeneration, and selection were performed as described previously (Horsh et al., 1985; Huang et al., 2010). The presence of the transgene in kanamycin-resistant T0 generation overexpression plants was confirmed by genomic PCR analysis. PtrNAC72 transcript levels in the positive transgenic plants were examined by qRT-PCR, performed as described above. T3 homozygous plants were used for subsequent experiments.
Stress Tolerance Assay
For the drought tolerance assay, 1-month-old soil-grown tobacco (wild-type and transgenic plants) and Arabidopsis (Col-0 and nac72 mutant) were deprived of water for 20 d and then returned to regular irrigation for 3 d. Survival rates were then scored. To estimate the water loss under dehydration conditions, leaves were detached from 30-d-old tobacco or 35-d-old Arabidopsis plants and placed on filter paper in the ambient environment for 2 h (tobacco) or 40 min (Arabidopsis). Fresh weights of the leaves were measured at the designated times, and the water loss rate was calculated by comparison with the initial weight. After the drought or dehydration treatments, leaves were sampled for analysis of EL, MDA, and ROS level as well as for the measurement of antioxidant enzyme activities. Free putrescine levels were measured at the end of the dehydration treatment. To investigate the ABA response, leaves of tobacco wild-type and transgenic lines were incubated for 3 h in a stomatal opening solution (10 mm MES-Tris, 50 μm CaCl2, and 10 mm KCl, pH 6.15) under light conditions. Subsequently, ABA was added to the solution to a final concentration of 10 μm (Jeon et al., 2008). The stomata were imaged before and 1 h after ABA treatment using an upright light microscope (Nikon 80i), and stomatal apertures were determined by measuring the inner width and length of stomata using ImageJ software (https://imagej.nih.gov/ij/). At least 200 stomata measurements were performed for each genotype before and after ABA treatment.
In another experiment, the Arabidopsis mutant plants were treated with 1 mm d-Arg or water for 3 d, using water-treated Col-0 as a control, before exposure to dehydration for 2 h or drought for 3 d. For dehydration, the fresh weight of the leaves was measured every 20 min. As for the tobacco plants, the transgenic lines were pretreated with water or 10 mm putrescine for 3 d, using the water-treated wild type as a control, before being subjected to a dehydration treatment for 3 h. For phenotypic observation, 10-d-old seedlings pretreated with putrescine or water were deprived of water for 22 d, followed by scoring of survival rates. EL, MDA, and ROS accumulation were examined after the stress treatments.
Physiological Measurements and Histochemical Staining
EL was measured as described (Dahro et al., 2016). MDA content, and CAT and SOD activities, expressed as units mg−1 protein, were measured using analytical kits (Nanjing Jiancheng Bioengineering Institute). Total protein was colorimetrically determined using Coomassie Brilliant Blue G-250 staining according to Bradford (1976). In situ generation of H2O2 and O2·− was detected by histochemical staining with DAB and NBT, respectively (Zhang et al., 2015). The stained leaves were transferred to a destaining solution consisting of acetic acid:glycerol:ethanol (1:1:3, v/v/v) in a water bath at 95°C for 10 min and kept in 70% ethanol until imaging.
Quantification of Free Putrescine Levels
Putrescine levels were measured as described previously (Liu et al., 2009; Fu et al., 2011; Gong et al., 2015; Zhang et al., 2015). Briefly, approximately 0.3 g of tissue powder was extracted in 5% cold perchloric acid containing dithiothreitol (0.5 g L−1) to extract free PAs. The extracted PAs were derivatized with benzoyl chloride, with 1,6-hexanediamine used as an internal standard, and then separated and quantified at room temperature on an Agilent HPLC system equipped with a C18 reverse-phase column (4.6 mm × 150 mm, particle size of 5 μm) and a UV light detector (230 nm). The mobile phase consisted of HPLC-grade methanol (eluent A) and water (eluent B), changing from 50%:50% (v/v, A:B) to 95%:5% in 10 min at a flow rate of 0.7 mL min–1. The PA level, expressed as nmol g−1 fresh weight, was the average of three replicates for each independent sample.
Statistical Analysis
Stress treatments were repeated at least three times with consistent results. Data are presented as means ± se of at least three independent replicates from one representative experiment. Data were analyzed by Fisher’s lsd test in the ANOVA program of SPSS (IBM SPSS 22). P < 0.05 was considered statistically significant.
Accession Numbers
Sequence data from this article can be found at the National Center for Biotechnology Information database with the following accession numbers: grape VvNAC72, XM_002284632; strawberry FvNAC72, XM_004291619; Arabidopsis ANAC072, NM_118875; rice OsNAC72, AK107746; and maize ZmNAC72, KJ727003. The 109 Arabidopsis NAC family member sequences were obtained based on data from the Plant Transcription Factor Database (Olsen et al., 2005).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of PtrNAC72.
Supplemental Figure S2. Expression of PtrNAC72 in response to abiotic stress and ABA treatment.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Xia Li (Huazhong Agricultural University) for providing the vectors used for the 35S-UAS-GUS reporter system and Benke Kuai (Fudan University) for providing complemented nac72 lines.
Glossary
- TF
transcription factor
- PA
polyamine
- ROS
reactive oxygen species
- ABA
abscisic acid
- ORF
open reading frame
- Y1H
yeast one-hybrid
- qRT
quantitative real-time
- CaMV 35S
cauliflower mosaic virus 35S promoter
- DAPI
4′,6-diamidino-2-phenylindole
- CDS
coding sequence
- 3-AT
3-aminotriazole
- SD
synthetic dextrose
- EMSA
electrophoretic mobility shift assay
- Col-0
Columbia-0
- RT
real-time
- EL
electrolyte leakage
- MDA
malondialdehyde
- DAB
3,3′-diaminobenzidine
- NBT
nitroblue tetrazolium
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31320103908 and 31521092).
References
- Alcázar R, Planas J, Saxena T, Zarza X, Bortolotti C, Cuevas J, Bitrián M, Tiburcio AF, Altabella T (2010) Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol Biochem 48: 547–552 [DOI] [PubMed] [Google Scholar]
- Alet AI, Sanchez DH, Cuevas JC, Del Valle S, Altabella T, Tiburcio AF, Marco F, Ferrando A, Espasandín FD, González ME, et al. (2011) Putrescine accumulation in Arabidopsis thaliana transgenic lines enhances tolerance to dehydration and freezing stress. Plant Signal Behav 6: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andronis EA, Moschou PN, Toumi I, Roubelakis-Angelakis KA (2014) Peroxisomal polyamine oxidase and NADPH-oxidase cross-talk for ROS homeostasis which affects respiration rate in Arabidopsis thaliana. Front Plant Sci 5: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Buscaill P, Rivas S (2014) Transcriptional control of plant defence responses. Curr Opin Plant Biol 20: 35–46 [DOI] [PubMed] [Google Scholar]
- Capell T, Bassie L, Christou P (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SP, Lin IW, Chen X, Huang YH, Chang SC, Lo HS, Lu HH, Yeh KW (2016) Sweet potato NAC transcription factor, IbNAC1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant J 86: 234–248 [DOI] [PubMed] [Google Scholar]
- Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A (2008) Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol 148: 1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A (2009) Putrescine as a signal to modulate the indispensable ABA increase under cold stress. Plant Signal Behav 4: 219–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahro B, Wang F, Peng T, Liu JH (2016) PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol 16: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27: 3228–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Zhai Q, Deng L, Li S, Li H, Yan L, Huang Z, Wang B, Jiang H, Huang T, et al. (2014) Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26: 3167–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65: 2119–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XZ, Chen CW, Wang Y, Liu JH, Moriguchi T (2011) Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H2O2 production and transcriptional alteration. BMC Plant Biol 11: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Zhang J, Hu J, Wang W, Wu H, Zhang Q, Liu JH (2015) FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ 38: 2248–2262 [DOI] [PubMed] [Google Scholar]
- Gong XQ, Hu JB, Liu JH (2014) Cloning and characterization of FcWRKY40, a WRKY transcription factor from Fortunella crassifolia linked to oxidative stress tolerance. Plant Cell Tissue Organ Cult 119: 197–210 [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26: 438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Dey A, Gupta B (2013) Plant polyamines in abiotic stress responses. Acta Physiol Plant 35: 2015–2036 [Google Scholar]
- Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA Jr (1998) The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci USA 95: 11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, et al. (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68: 302–313 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsh R, Fry J, Hoffman N, Eicholts D, Rogers S, Fraley R (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Ye J, Tao P, Li H, Zhang J, Zhang Y, Ye Z (2016) The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J 85: 16–29 [DOI] [PubMed] [Google Scholar]
- Huang XS, Liu JH, Chen XJ (2010) Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol 10: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XS, Wang W, Zhang Q, Liu JH (2013) A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162: 1178–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel CA, Riadh K, Gopi R, Manivannan P, Ines J, Al-Juburi H, Chang-Xing Z, Hong-Bo S, Panneerselvam R (2009) Antioxidant defense responses: physiological plasticity in higher plants under abiotic constraints. Acta Physiol Plant 31: 427–436 [Google Scholar]
- Jeon BW, Hwang JU, Hwang Y, Song WY, Fu Y, Gu Y, Bao F, Cho D, Kwak JM, Yang Z, et al. (2008) The Arabidopsis small G protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 20: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Huang F, Cheng H, Song H, Yu D (2013) Overexpression of the GmNAC2 gene, an NAC transcription factor, reduces abiotic stress tolerance in tobacco. Plant Mol Biol Rep 31: 435–442 [Google Scholar]
- Kamiab F, Talaie A, Khezri M, Javanshah A (2014) Exogenous application of free polyamines enhance salt tolerance of pistachio (Pistacia vera L.) seedlings. Plant Growth Regul 72: 257–268 [Google Scholar]
- Kim SG, Kim SY, Park CM (2007) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226: 647–654 [DOI] [PubMed] [Google Scholar]
- Kotakis C, Theodoropoulou E, Tassis K, Oustamanolakis C, Ioannidis NE, Kotzabasis K (2014) Putrescine, a fast-acting switch for tolerance against osmotic stress. J Plant Physiol 171: 48–51 [DOI] [PubMed] [Google Scholar]
- Kubiś J, Floryszak-Wieczorek J, Arasimowicz-Jelonek M (2014) Polyamines induce adaptive responses in water deficit stressed cucumber roots. J Plant Res 127: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–4748 [DOI] [PubMed] [Google Scholar]
- Li P, Chen B, Zhang G, Chen L, Dong Q, Wen J, Mysore KS, Zhao J (2016a) Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol 210: 905–921 [DOI] [PubMed] [Google Scholar]
- Li S, Gao J, Yao L, Ren G, Zhu X, Gao S, Qiu K, Zhou X, Kuai B (2016b) The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep 35: 1729–1741 [DOI] [PubMed] [Google Scholar]
- Liu J, Peng T, Dai W (2014) Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol Biol Rep 32: 303–317 [Google Scholar]
- Liu JH, Ban Y, Wen XP, Nakajima I, Moriguchi T (2009) Molecular cloning and expression analysis of an arginine decarboxylase gene from peach (Prunus persica). Gene 429: 10–17 [DOI] [PubMed] [Google Scholar]
- Liu JH, Nada K, Honda C, Kitashiba H, Wen XP, Pang XM, Moriguchi T (2006) Polyamine biosynthesis of apple callus under salt stress: importance of the arginine decarboxylase pathway in stress response. J Exp Bot 57: 2589–2599 [DOI] [PubMed] [Google Scholar]
- Liu JH, Wang W, Wu H, Gong X, Moriguchi T (2015) Polyamines function in stress tolerance: from synthesis to regulation. Front Plant Sci 6: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragão FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110: 19627–19632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mohapatra S, Minocha R, Long S, Minocha SC (2009) Putrescine overproduction negatively impacts the oxidative state of poplar cells in culture. Plant Physiol Biochem 47: 262–271 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Paschalidis KA, Roubelakis-Angelakis KA (2005) Spatial and temporal distribution of polyamine levels and polyamine anabolism in different organs/tissues of the tobacco plant: correlations with age, cell division/expansion, and differentiation. Plant Physiol 138: 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Roberts DR, Walker MA, Thompson JE, Dumbroff EB (1984) The effects of inhibitors of polyamine and ethylene biosynthesis on senescence, ethylene production and polyamine levels in cut carnation flowers. Plant Cell Physiol 25: 315–322 [Google Scholar]
- Roy M, Wu R (2001) Arginine decarboxylase transgene expression and analysis of environmental stress tolerance in transgenic rice. Plant Sci 160: 869–875 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC (2015) The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27: 1771–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma HSS, Fleming C, Selby C, Rao JR, Martin T (2013) Plant biostimulants: a review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. J Appl Phycol 26: 465–490 [Google Scholar]
- Shi H, Chan Z (2014) Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J Integr Plant Biol 56: 114–121 [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, Zhang Y, Chan Z (2013) Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J Exp Bot 64: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J 33: 259–270 [DOI] [PubMed] [Google Scholar]
- Soyka S, Heyer AG (1999) Arabidopsis knockout mutation of ADC2 gene reveals inducibility by osmotic stress. FEBS Lett 458: 219–223 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Zhu X, Huang X, Liu JH (2014) Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol Biochem 78: 71–79 [DOI] [PubMed] [Google Scholar]
- Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284: 173–183 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio AF, Kaur-Sawhney R, Galston AW (1986) Polyamine metabolism and osmotic stress. II. Improvement of oat protoplasts by an inhibitor of arginine decarboxylase. Plant Physiol 82: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Urano K, Yoshiba Y, Nanjo T, Ito T, Yamaguchi-Shinozaki K, Shinozaki K (2004) Arabidopsis stress-inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem Biophys Res Commun 313: 369–375 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Chinoy C, Zuther E, Blessington B, Geigenberger P, Domoney C (2008) Combined metabolomic and genetic approaches reveal a link between the polyamine pathway and albumin 2 in developing pea seeds. Plant Physiol 146: 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang S, Ma X, Wang Y, Kong F, Meng Q (2016a) A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol Plant 158: 45–64 [DOI] [PubMed] [Google Scholar]
- Wang H, Jiao X, Kong X, Hamera S, Wu Y, Chen X, Fang R, Yan Y (2016b) A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol 170: 2365–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun PP, Chen CL, Wang Y, Fu XZ, Liu JH (2011) An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J Exp Bot 62: 2899–2914 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao H, Chen D, Li L, Sun H, Lou Y, Gao Z (2016c) Characterization and primary functional analysis of a bamboo NAC gene targeted by miR164b. Plant Cell Rep 35: 1371–1383 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang L, Zou Y, Chen L, Cai Z, Zhang S, Zhao F, Tian Y, Jiang Q, Ferguson BJ, et al. (2014) Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26: 4782–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, et al. (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Zhang DF, Li HY, Liu YH, Shi YS, Song YC, Wang TY, Li Y (2011) Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Rep 30: 1683–1699 [DOI] [PubMed] [Google Scholar]
- Yiu JC, Juang LD, Fang DYT, Liu CW, Wu SJ (2009) Exogenous putrescine reduces flooding-induced oxidative damage by increasing the antioxidant properties of Welsh onion. Sci Hortic (Amsterdam) 120: 306–314 [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang GW, Xu SC, Hu QZ, Mao WH, Gong YM (2014) Putrescine plays a positive role in salt-tolerance mechanisms by reducing oxidative damage in roots of vegetable soybean. J Integr Agric 13: 349–357 [Google Scholar]
- Zhang Q, Wang M, Hu J, Wang W, Fu X, Liu JH (2015) PtrABF of Poncirus trifoliata functions in dehydration tolerance by reducing stomatal density and maintaining reactive oxygen species homeostasis. J Exp Bot 66: 5911–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B (2015) Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84: 597–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.