A robot system allows automated handling and phenotyping of individual seeds of different sizes, delivering biometric traits relevant for various aspects in seed biology.
Abstract
The enormous diversity of seed traits is an intriguing feature and critical for the overwhelming success of higher plants. In particular, seed mass is generally regarded to be key for seedling development but is mostly approximated by using scanning methods delivering only two-dimensional data, often termed seed size. However, three-dimensional traits, such as the volume or mass of single seeds, are very rarely determined in routine measurements. Here, we introduce a device named phenoSeeder, which enables the handling and phenotyping of individual seeds of very different sizes. The system consists of a pick-and-place robot and a modular setup of sensors that can be versatilely extended. Basic biometric traits detected for individual seeds are two-dimensional data from projections, three-dimensional data from volumetric measures, and mass, from which seed density is also calculated. Each seed is tracked by an identifier and, after phenotyping, can be planted, sorted, or individually stored for further evaluation or processing (e.g. in routine seed-to-plant tracking pipelines). By investigating seeds of Arabidopsis (Arabidopsis thaliana), rapeseed (Brassica napus), and barley (Hordeum vulgare), we observed that, even for apparently round-shaped seeds of rapeseed, correlations between the projected area and the mass of seeds were much weaker than between volume and mass. This indicates that simple projections may not deliver good proxies for seed mass. Although throughput is limited, we expect that automated seed phenotyping on a single-seed basis can contribute valuable information for applications in a wide range of wild or crop species, including seed classification, seed sorting, and assessment of seed quality.
Seeds play a major role in keeping continuity between successive generations (Esau, 1977) and are key for the distribution and evolution (Moles et al., 2005) of higher plants. Fertile seeds carry an embryo and may contain nutrient storage tissues in cotyledons, endosperm, and/or perisperm, supporting germination and seedling development at early developmental stages. Although this is true for all seed plants, various traits of seeds, such as size, shape, weight, and chemical composition, can be very different between plant species or accessions. For example, the Arabidopsis (Arabidopsis thaliana) accession Cape Verde Islands was reported to yield on average 40% fewer seeds than Landsberg erecta, but they are almost twice as heavy (Alonso-Blanco et al., 1999). Considering today’s plant species, single-seed mass may vary over a range of 11.5 orders of magnitude (Moles et al., 2005). Seed mass is under strong genetic control, whereas the total number of seeds of a plant is largely affected by the environment (Paul-Victor and Turnbull, 2009). It has been demonstrated that the size, mass, and shape of Arabidopsis seeds may be regulated by brassinosteroid (Jiang et al., 2013), and it was shown recently that seed size in rice (Oryza sativa) can be influenced by the epiallele Epi-rav6 (Zhang et al., 2015). The ability of plants to switch between small and larger seeds may be understood as an adaptation to novel environments (Igea et al., 2016). However, it is still not fully understood whether, or to what extent, the variability of seed traits within plant species or genotypes has an impact on the development and further performance of a plant.
When comparing biometric seed data of different dimensions such as length (one-dimensional), projected area (two-dimensional [2D]), or volume and mass (both three-dimensional [3D]), one can argue that mass is the most relevant parameter as a proxy for the amount of reserves a seed provides for the offspring. This might be true even when considering that the type of reserves, such as proteins, carbohydrates, or lipids (Rolletschek et al., 2015), and also different seed tissues, such as seed coat, embryo, or endosperm, may contribute differently to seed mass (Alonso-Blanco et al., 1999). While seed mass and time to germination (radicle protrusion) do not necessarily correlate (Norden et al., 2009), in particular under greenhouse conditions, higher seed mass may be advantageous for seedling establishment under adverse environmental conditions (Moles et al., 2005). For example, shade-tolerant species showed largely higher seed masses than cogeneric species growing in open habitats, indicating that seedlings under low-light conditions need more reserves than under good light (Salisbury, 1974). Seedlings of wild radish (Raphanus raphanistrum) emerged more likely from heavier seeds than from small seeds under field conditions but not in the greenhouse (Stanton, 1984), and for Arabidopsis, seed mass was reported to be higher in populations growing naturally at higher altitudes taken as a proxy for harsher conditions (Montesinos-Navarro et al., 2011).
Seed mass can be measured individually (Stanton, 1984), but it is generally collected as an average value of batches of 50 to 1,000 seeds (Jako et al., 2001; Jofuku et al., 2005; Montesinos-Navarro et al., 2011; Tanabata et al., 2012). Alternatively, 2D scans are analyzed to determine parameters such as seed length, width, area, and perimeter length as a measure for seed size (Tanabata et al., 2012). This approach can be implemented in high-throughput facilities to obtain projected areas of seed grains combined with genome-wide association studies (Yang et al., 2014). Although projected seed area can easily be measured with a common office scanner (Herridge et al., 2011; Tanabata et al., 2012; Moore et al., 2013), it is not necessarily a precise or reliable measure of the true seed size because it may depend on the shape (Alonso-Blanco et al., 1999) and the orientation of a seed at scan (see “Results”). These issues also apply when using 2D projections to calculate length-to-width ratios as a simple shape factor (Tanabata et al., 2012). Projected seed area also has been used to calculate seed mass, assuming a fixed relationship between these parameters (de Jong et al., 2011; Herridge et al., 2011). This may hold with sufficient accuracy when averaging a large number of seeds but might be misleading when considering individual seeds.
From a physical point of view, volume should be a much better proxy for mass than 2D traits. Although it has been stated that for 65 species analyzed seed masses can be compared easily with seed volumes (Moles et al., 2005), it is not clear how these seed volumes were determined. Volumes can be assessed using advanced methods such as x-ray computed tomography (CT) on fruits (Stuppy et al., 2003) or synchrotron radiation x-ray tomographic microscopy applied in paleobiological studies (e.g. on fruits and seed; Friis et al., 2014). Nuclear magnetic resonance (NMR) methods are used to measure water uptake in kidney beans (Phaseolus vulgaris) and adzuki beans (Vigna angularis; Kikuchi et al., 2006) or to estimate seed weight and content (Borisjuk et al., 2011; Rolletschek et al., 2015) rather than volumes. To our best knowledge, affordable methods to measure seed volumes directly are not achievable so far. For that reason, we have set up a volume-carving method for 3D seed shape reconstruction that is described briefly here and in more detail in a recent publication (Roussel et al., 2016).
While traits derived from scanning procedures can easily be assigned to individual seeds (Herridge et al., 2011), further handling and processing of phenotyped single seeds is not as simple, in particular for tiny ones like those of Arabidopsis. The aim of this work was to develop an automated seed-handling system that can analyze single seeds of very different sizes or shapes, from Arabidopsis seeds up to barley (Hordeum vulgare) seeds or even bigger. The phenoSeeder system is designed to pick and place seeds, to achieve basic morphometric traits (one-dimensional and 2D data from projections, 3D reconstruction data, and mass) of each individual seed, and to store all analyzed seed traits in a database. Another goal is to use phenoSeeder for seed-to-plant tracking approaches and to analyze whether, or which, particular seed traits have an impact on plant development and performance under various environmental conditions. We describe the main features of the phenoSeeder technology and present results obtained with seeds of three accessions of Arabidopsis, rapeseed (Brassica napus), and barley, respectively. When analyzing the data, we focused particularly on correlations between projected seed area, seed volume, and seed mass, with the hypothesis that the respective seed volume may better correlate with mass than the projected area.
RESULTS
Here, we describe the general concept of the phenoSeeder system. More technical details are given in Supplemental Materials and Methods S1. The terminology used for the measured seed traits is compiled in Table I. Results of test measurements relevant for the performance of the system are presented (Table II), and traits of more than 1,000 seeds (for exact numbers, see Table III) for three accessions of Arabidopsis, rapeseed, and barley, respectively, were evaluated.
Table I. List of analyzed seed traits and abbreviations.
| Seed Trait | Symbol | Unit |
|---|---|---|
| Projected area (or size)a | A | mm2 |
| Length from projectiona | L2D | mm |
| Width from projectiona | W2D | mm |
| Volumeb | V | mm3 |
| Lengthb | L | mm |
| Widthb | W | mm |
| Heightb | H | mm |
| Mass (or weight) | M | mg |
| Volume Aa (=π/6 A3/2) | VA | mm3 |
| Volume 2Da (=π/6 L2D W2D W2D) | V2D | mm3 |
| Density (=M/V) | ρ | mg mm−3 |
| Density A (=M/VA) | ρA | mg mm−3 |
| Density 2D (=M/V2D) | ρ2D | mg mm−3 |
Table II. Variance of seed parameters when single seeds were measured repeatedly.
Mean values ± relative standard deviation (RSD); RSDs are given in percentages. Lag2-2, Accession Lagodechi; n, number of repetitions; No., seed number.
| Species and Accession | No. | n | A | V | M |
|---|---|---|---|---|---|
| mm2 | mm3 | mg | |||
| Rapeseed, Wotan | 1 | 60 | 2.80 ± 5.4 | 2.73 ± 0.23 | 3.13 ± 0.13 |
| 2 | 60 | 1.70 ± 3.8 | 1.47 ± 0.45 | 1.61 ± 0.29 | |
| 3 | 60 | 2.68 ± 5.5 | 3.08 ± 0.20 | 3.42 ± 0.13 | |
| Barley, Barke | 1 | 63 | 24.3 ± 4.1 | 40.1 ± 1.3 | 54.4 ± 0.03 |
| 2 | 60 | 19.3 ± 6.4 | 26.9 ± 1.3 | 45.6 ± 0.02 | |
| 3 | 60 | 23.3 ± 7.8 | 34.5 ± 1.2 | 44.7 ± 0.02 | |
| Arabidopsis, Lag2-2 | 1 | 60 | 0.123 ± 5.1 | 0.0144 ± 3.7 | 0.0169 ± 14.5 |
| 2 | 60 | 0.103 ± 6.1 | 0.0098 ± 5.4 | 0.0099 ± 29.0 | |
| 3 | 60 | 0.131 ± 4.7 | 0.0155 ± 1.8 | 0.0184 ± 11.8 |
Table III. Selected seed traits of rapeseed, barley, and Arabidopsis.
Bn1, Wotan; Bn2, Expert; Bn3, Pirola; Hv1, Barke; Hv2, HOR13719; Hv3, HOR9707; At1, Col-0; At2, Lag2-2; At3, Aguaron (Agu-1); n, number of repetitions. Mean values ± RSD; RSDs are given in percentages. Abbreviations of traits are as listed in Table I. All pairwise differences between the means of the different accessions of a plant species were statistically significant (P < 0.01) except for L (Hv1 versus Hv2), ρA (Bn1 versus Bn2), ρ2D (Bn1 versus Bn2), and ρ2D (Hv1 versus Hv3), denoted by lowercase a, b, c, and d, respectively.
| Trait | Rapeseed | Barley | Arabidopsis | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
Bn1 |
Bn2 |
Bn3 |
Hv1 |
Hv2 |
Hv3 |
At1 |
At2 |
At3 |
|
| n | 1,077 | 1,009 | 1,019 | 1,047 | 1,112 | 1,058 | 1,006 | 1,009 | 1,008 |
| M (mg) | 1.88 ± 24.5 | 2.31 ± 32.3 | 1.32 ± 40.0 | 46.6 ± 15.1 | 51.3 ± 17.5 | 56.8 ± 19.4 | 0.0210 ± 15.7 | 0.0129 ± 26.4 | 0.0236 ± 22.9 |
| A (mm2) | 2.12 ± 15.4 | 2.42 ± 20.6 | 1.97 ± 22.5 | 21.3 ± 11.7 | 21.7 ± 12.6 | 27.6 ± 13.5 | 0.140 ± 9.36 | 0.109 ± 12.8 | 0.159 ± 12.7 |
| V (mm3) | 1.62 ± 25.0 | 2.04 ± 33.5 | 1.19 ± 40.9 | 37.1 ± 13.9 | 39.6 ± 17.6 | 49.3 ± 17.0 | 0.0200 ± 14.2 | 0.0112 ± 22.2 | 0.0215 ± 20.6 |
| VA (mm3) | 2.35 ± 23.1 | 2.88 ± 30.9 | 2.12 ± 34.0 | 74.3 ± 17.0 | 76.5 ± 18.4 | 109.6 ± 19.9 | 0.0395 ± 14.0 | 0.0274 ± 19.2 | 0.0483 ± 18.8 |
| V2D (mm3) | 2.17 ± 24.1 | 2.71 ± 31.9 | 1.98 ± 36.2 | 52.3 ± 18.1 | 54.5 ± 19.5 | 64.8 ± 21.1 | 0.0325 ± 13.5 | 0.0223 ± 18.6 | 0.0384 ± 19.4 |
| L (mm) | 1.77 ± 8.07 | 1.86 ± 11.1 | 1.72 ± 11.6 | 7.16 ± 6.31 a | 7.20 ± 6.77 a | 9.82 ± 8.01 | 0.488 ± 7.04 | 0.433 ± 9.79 | 0.542 ± 8.19 |
| L2D (mm) | 1.81 ± 7.48 | 1.90 ± 10.5 | 1.74 ± 9.94 | 7.76 ± 6.30 | 7.54 ± 6.30 | 10.62 ± 8.76 | 0.527 ± 6.27 | 0.477 ± 8.31 | 0.583 ± 7.21 |
| W (mm) | 1.45 ± 9.13 | 1.61 ± 11.2 | 1.41 ± 14.2 | 3.66 ± 5.54 | 3.73 ± 6.77 | 3.56 ± 6.97 | 0.311 ± 4.97 | 0.256 ± 7.28 | 0.318 ± 8.05 |
| W2D (mm) | 1.50 ± 8.97 | 1.62 ± 11.8 | 1.44 ± 13.9 | 3.57 ± 8.24 | 3.69 ± 8.63 | 3.39 ± 8.81 | 0.343 ± 5.12 | 0.297 ± 7.00 | 0.353 ± 8.09 |
| H (mm) | 1.19 ± 12.6 | 1.29 ± 15.6 | 0.97 ± 19.6 | 2.75 ± 6.83 | 2.83 ± 8.27 | 2.78 ± 8.84 | 0.264 ± 6.11 | 0.210 ± 8.28 | 0.249 ± 10.2 |
| ρ (mg mm−3) | 1.17 ± 2.49 | 1.14 ± 2.44 | 1.12 ± 7.96 | 1.25 ± 4.80 | 1.29 ± 3.39 | 1.15 ± 6.99 | 1.05 ± 12.5 | 1.16 ± 20.4 | 1.10 ± 13.0 |
| ρA (mg mm−3) | 0.80 ± 13.0 b | 0.81 ± 16.4 b | 0.62 ± 19.0 | 0.64 ± 15.6 | 0.68 ± 12.3 | 0.53 ± 17.7 | 0.53 ± 14.3 | 0.47 ± 22.6 | 0.49 ± 17.2 |
| ρ2D (mg mm−3) | 0.87 ± 13.8 c | 0.86 ± 17.9 c | 0.67 ± 21.0 | 0.91 ± 18.1 d | 0.95 ± 14.9 | 0.89 ± 18.6 d | 0.65 ± 14.5 | 0.58 ± 23.0 | 0.62 ± 17.4 |
| r2 (M versus V) | 0.991 | 0.994 | 0.977 | 0.930 | 0.964 | 0.888 | 0.492 | 0.537 | 0.713 |
| r2 (M versus A) | 0.669 | 0.691 | 0.692 | 0.531 | 0.678 | 0.504 | 0.352 | 0.389 | 0.480 |
| r2 (M versus V2D) | 0.680 | 0.706 | 0.798 | 0.370 | 0.566 | 0.327 | 0.275 | 0.385 | 0.471 |
Design and Modules of the phenoSeeder System
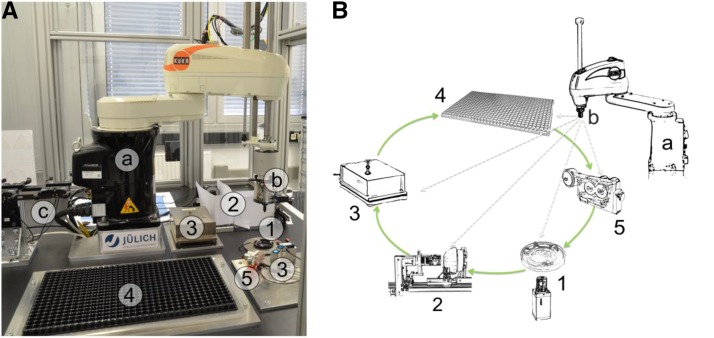
The setting of the system enables routine handling (pick and place) of individual seeds and the measurement of morphometric seed traits. Both hardware and software are modular, allowing easy implementation of new components. Figure 1A shows the main components of phenoSeeder with an industrial robot (details are given in Supplemental Materials and Methods S1), an exchangeable seed-handling tool, a tool magazine, a 2D imaging station, 3D imaging modules, balances, a seed placement station, a nozzle cleaning tool, and a through-beam sensor for tool calibration. Figure 1B shows a schematic of the basic workflow: the cycle starts at the 2D imaging module (station 1), where dispersed seeds are recognized by image processing and a selected seed is picked up; after assigning a unique identifier to the seed, it is then moved to the 3D imaging module (station 2) to obtain volumetric data; thereafter, the seed is placed on a balance (station 3); finally, the seed is either planted or stored (station 4). If necessary, the nozzle gets cleaned (station 5) before a new cycle starts. A more detailed description of the phenoSeeder workflow is presented in Supplemental Materials and Methods S1. The sequence of the different steps is illustrated in Supplemental Movie S1. All information of measured traits and the actual location of the seed are stored in a distributed database system (Schmidt et al., 2013). Seed location can be used in subsequent experiments to reidentify the seed or seedling.
Figure 1.
Overview of phenoSeeder. A, Photograph of the setup located in a security cage. The system is composed of modules that can be placed at various positions in the cruising range of the industrial pick-and-place robot (a), equipped with a seed-handling tool (b) and a tool magazine (c). Default modules are a 2D imaging station (1), 3D imaging stations (2), balances (3), a seed-placement station (4), and a nozzle-cleaning station (5). B, Schematic illustrating the main workflow along the different modules. At the 2D imaging station (1), dispersed seeds are detected, 2D traits are measured, and one seed is picked up by the seed-handling tool; at the 3D imaging stations (2), the volumetric data of the seed are measured; the mass of the seed is taken at a balance (3); the seed is planted or stored (4); and the nozzle gets cleaned if needed (5).
Seed-Handling Tools and Pneumatic System
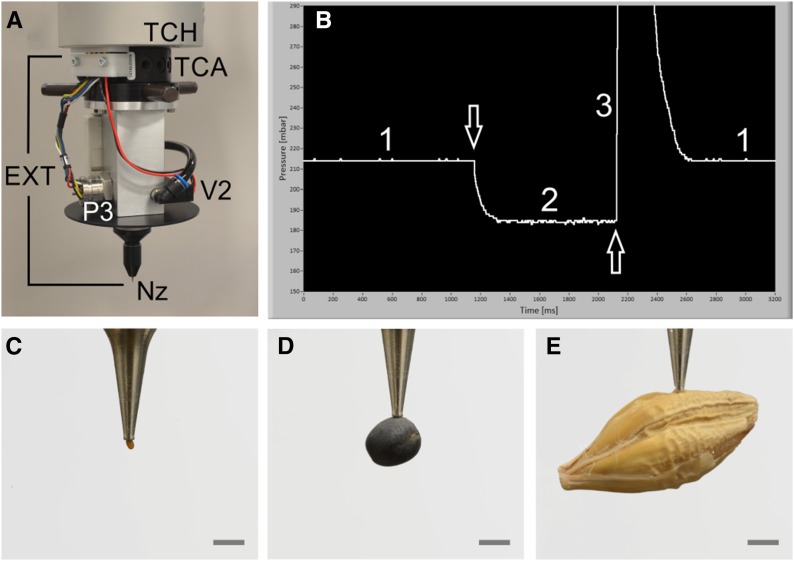
The robot arm is equipped with a tool-changing head to which seed-handling tools or grippers can be plugged or unplugged in an automated fashion (Fig. 2A). A seed-handling tool is equipped with a dedicated nozzle depending on seed size or shape to which either vacuum is provided for sucking or slight overpressure for releasing the seed. The actual air pressure at the nozzle is measured by a pressure sensor, P3 (Fig. 2A), and is fully controlled by a pneumatic system (Supplemental Fig. S1) described in more detail in Supplemental Materials and Methods S1. The air pressure at P3 indicates whether a seed is sucked at the nozzle, whether a seed has been properly released, or whether the nozzle is clogged. The actual pressure values may depend on seed properties and specifications of the seed-handling tool including the nozzle. A typical change in air pressure at P3 is shown in Figure 2B for an Arabidopsis seed handled with a 0.15-mm nozzle (Supplemental Materials and Methods S1): when the nozzle orifice is open with no seed at the nozzle (phase 1), the pressure is about 215 mbar; as soon as a seed is sucked (arrow facing down), the pressure decreases and reaches a new plateau of approximately 180 mbar (phase 2) after about 200 ms; within the first 20 to 30 ms of the pressure drop, it is decided whether a seed has been sucked or not by simple thresholding; the seed is released by providing a slight overpressure to the nozzle (phase 3; arrow facing up) for about 150 ms, and after that short time, air pressure is switched back to vacuum. Evacuation of the system takes a little longer than pressurizing but, after around 320 ms, phase 1 is reestablished and the system is ready for a new cycle. Seeds of Arabidopsis, rapeseed, and barley sucked at a nozzle are shown in Figure 2, C, D, and E, respectively, demonstrating that seeds of very different sizes and shapes can be handled.
Figure 2.
Seed handling and seeds of the three species investigated here. A, Tool change head (TCH) at the robot arm to which, by the aid of a tool change adapter (TCA), an exchangeable seed-handling tool (EXT) is fixed. A pressure sensor (P3), solenoid valve (V2), and dedicated nozzle (Nz) also are shown. B, Screen shot of temporal pressure changes near the nozzle as measured by P3. The denoted phases 1 to 3 and the arrows are described in detail in the text. C to E, Seeds sucked at the nozzle are shown for Arabidopsis (C), rapeseed (D), and barley (E). Bars = 1 mm.
2D Imaging Station, Seed Segmentation, and Selection
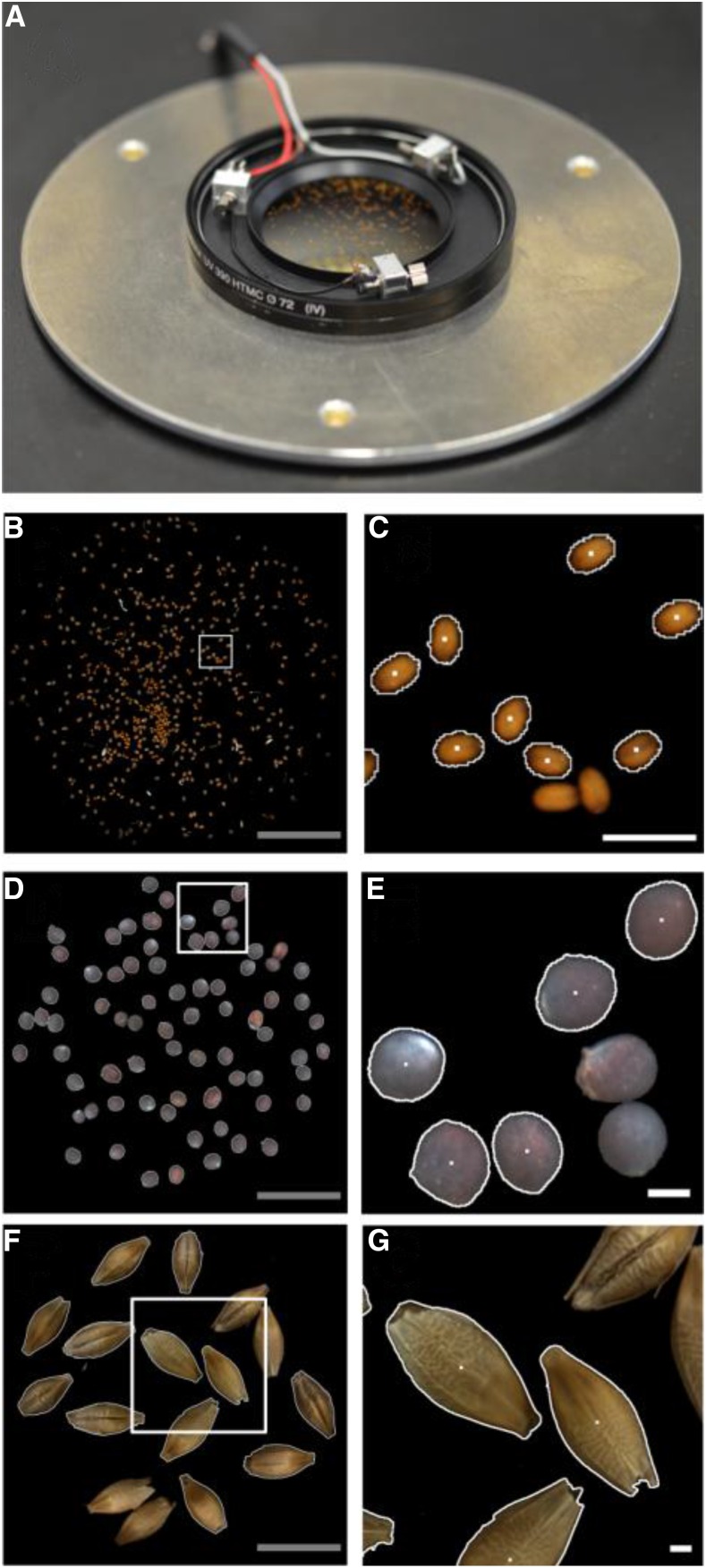
Seeds are dispersed on an optical glass filter equipped with a vibration device to separate seeds touching each other (Fig. 3A). A light-emitting diode ring light for illumination and a calibrated camera with a macro lens are mounted underneath the glass filter (for details, see Supplemental Fig. S2; Supplemental Materials and Methods S1). From the seeds on the glass filter, photographs are taken as demonstrated for Arabidopsis (Fig. 3B), rapeseed (Fig. 3D), and barley (Fig. 3F), for which enlarged areas are shown in Figure 3, C, E, and G, respectively. Such images are acquired repeatedly during each cycle of the workflow to detect and segment individual seeds. The procedure used for seed segmentation is explained in more detail in Supplemental Materials and Methods S1. For each segmented seed, the projected area (A) is determined, both length (L2D) and width (W2D) are calculated from fitting an ellipse to the seed, and red, green, and blue (RGB) color statistics are obtained. The x and y positions of each seed’s center of mass (white dots in Fig. 3, C, E, and G) are used for picking up the seed by the robot. For further processing, a seed is chosen either randomly or by a criterion (e.g. projected area) defined by the user. As soon as a seed is selected and successfully sucked by the robot, the seed gets an identifier and its database record is created.
Figure 3.
2D imaging station. A, Module for 2D imaging with an optical glass filter on which seeds are dispersed and with three vibration motors to separate seeds lying close together. Below the glass filter, a light-emitting diode ring for illumination and a camera are mounted (shown in Supplemental Fig. S2). B to G, Seed images of Arabidopsis (B and C), rapeseed (D and E), and barley (F and G). B, D, and F, Total field of view with a diameter of 41 mm. C, E, and G, Enlarged areas (indicated by white squares in B, D, and F, respectively) where segmented seeds (indicated by white borders) and nonsegmented seeds (without white borders; these are skipped to ensure single-seed handling) are shown. White dots denote the center of gravity of the segmented seeds. Gray bars = 10 mm; white bars = 1 mm.
3D Imaging and Weighing Stations
Two 3D imaging modules are set up differing in their optical layout suited for different seed sizes; the one for small seeds is shown in Figure 4A. When a seed is picked up by the robot, it is positioned in front of the camera lens while hanging at the nozzle (Fig. 4A), stepwise turned by 360°, and at each step of, for example, 10°, an image is acquired. To achieve 3D reconstruction of a seed (Fig. 4B), a volume-carving technique is used to determine the seed volume (V) and to calculate the length (L), width (W), and height (H) of the seed employing an ellipsoid fit. Both the imaging modules and the 3D reconstruction have been described in a previous publication (Roussel et al., 2016), and more information is given in Supplemental Materials and Methods S1. For small seeds (like those of Arabidopsis), a high-resolution balance is used with a custom-made seed receiver (Fig. 4C and inset). For seeds of rapeseed or larger, other weighing cells (Supplemental Fig. S3A) are used. More details are given in Supplemental Materials and Methods S1.
Figure 4.
3D imaging station and high-resolution weighing cell. A, Module for 3D imaging with a camera plus objective lens, light source, and white reflector. In front of the camera, a rapeseed seed is turned by 360° with the aid of the fourth robot axis. B, Images of an Arabidopsis seed from two different angles (left) and the respective reconstructions (right; x and y axes in mm) shown at greater magnification. C, High-resolution weighing cell with a custom-made receiver (inset) dedicated to small seeds such as Arabidopsis.
Seed Placement Stations
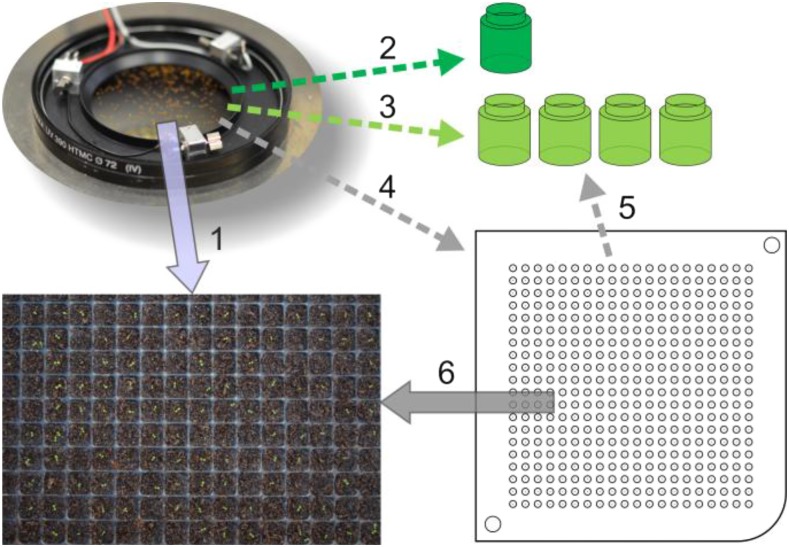
After phenotyping, seeds can be processed in different ways as illustrated in Figure 5. Option 1: seeds are planted directly into substrate. Option 2: seeds can be recollected in just one vessel if only information of a seed batch is needed. Option 3: seeds can be classified by any measurable trait (to be defined a priori) and collected in as many vessels as trait classes were defined. Option 4: alternatively, seeds may be placed individually in a multiwell plate, where each single seed can be identified by its position. Option 5: seeds collected in multiwell plates may be recollected by classification criteria, which can be defined a posteriori (i.e. after all measured traits of all seeds are available). Option 6: seeds stored in multiwell plates are planted into substrate, enabling seed-to-plant tracking. Currently, options 1 to 3 are running while options 4 to 6 are under construction.
Figure 5.
A selection of different options for seed treatment after phenotyping. Options are as follows: direct planting (1); recollection in just one vessel (2); classification and collecting into different vessels according to preselected seed trait(s) and bin size (3); storing in multiwell plates (4); classification of seeds after storage in multiwell plates (5); and planting seeds from multiwell plates into substrate, enabling seed-to-plant tracking (6).
Precision of Seed Placing
The precision of seed placing into a substrate was tested by planting seeds of Arabidopsis accession Columbia-0 (Col-0) on wet blotting paper taken as an example, since it allows easy visual inspection of the seeds (Supplemental Fig. S5). When the blotting paper is well damped, almost all seeds are placed at the position where they were released (Supplemental Fig. S5); however, when the paper (or other substrate) is getting dry, seeds upon release may jump off the position where they first touch the surface.
Cleaning Station
In case the nozzle becomes clogged by a seed, the robot moves the nozzle to a station to get cleaned (Supplemental Fig. S3B). For Arabidopsis seeds, this happens in 1% to 2% of cases; for rapeseed and barley, the percentage is below 1%. Reasons for clogging can be electrostatic charges, stickiness, or glumes adhering to the nozzle.
Data Preprocessing and Reproducibility of Measured Seed Traits
Incomplete or obviously erroneous data sets of single seeds have been excluded from data analysis. Possible causes for exclusion included measurement of two seeds sticking to each other, failed weighing because of seed loss during placement on the balance, or inaccurate 3D reconstruction because of light reflections on the seed or extremely dark or bright seed color. Here, the excluded seed data were well below 1% for all species and accessions studied. These problems are now solved by making adjustments in hardware setup and parametrization of the software. To evaluate the reproducibility of measurements, three individual seeds of each measured species were analyzed repeatedly (n ≥ 60; Table II). To ensure that the positioning of the individual seed at the nozzle varied arbitrarily, a seed was picked up, A, V, and M were measured consecutively, after which the seed was picked up again and a new measuring cycle started. For rapeseed, the RSD of V and M was very small (0.45% or less and 0.29% or less, respectively; Table II); at 3.8% to 5.5%, it was clearly higher for A. Barley also showed high reproducibility with regard to V and M (RSD ≤ 1.3% and RSD ≤ 0.03%, respectively) but not to A (RSD up to 7.8%), as discussed below. For Arabidopsis, determination of V was best (RSD ≤ 5.4%), A was poorer (RSD ≤ 6.1%), and M was worst (RSD > 11.8%), with the highest RSD of 29% for the lightest of the three measured seeds (no. 2; Table II) due to a limited resolution of the balance (see discretization effects in Fig. 9, G–I).
Figure 9.
Seeds of the different investigated plant species and accessions sorted by seed mass. The blue lines represent seed mass (M), red crosses denote measured volume (V), and green circles represent volume calculated from length and width of projection (V2D). For better readability, both V and V2D were scaled by the slopes of linear regressions to meet the range of M. A to C, Rapeseed accessions Wotan (A), Expert (B), and Pirola (C). D to F, Barley accessions Barke (D), HOR13719 (E), and HOR9707 (F). G to I, Arabidopsis accessions Col-0 (G), Lag2-2 (H), and Agu-1 (I).
Distribution of Seed Traits
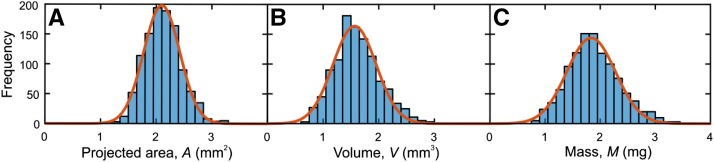
For rapeseed accession Wotan, frequency histograms of A, V, and M are displayed in Figure 6, A, B, and C, respectively, and these traits show a not perfect but fairly good normal distribution. For all species and accessions studied here (see “Materials and Methods”; images of the seeds are presented in Supplemental Fig. S6), this is not always the case, as can be seen from the frequency histograms in Supplemental Figures S7 to S15. The distribution of trait values may be skewed toward higher values, as for V and M of the rapeseed accessions Expert (Supplemental Fig. S8) and Pirola (Supplemental Fig. S9). In other cases, trait values have an asymmetric tail at lower numbers, as for A and M in the barley accessions Barke (Supplemental Fig. S10) and HOR13719 (Supplemental Fig. S11).
Figure 6.
Frequency histograms of seed traits of rapeseed accession Wotan. A, Projected area of seeds. B, Seed volume. C, Seed mass. The bin sizes are 0.15 mm2, 0.15 mm3, and 0.15 mg in A, B, and C, respectively. Red curves represent normal distributions fitted to the histograms.
Correlation Studies between Different Seed Traits
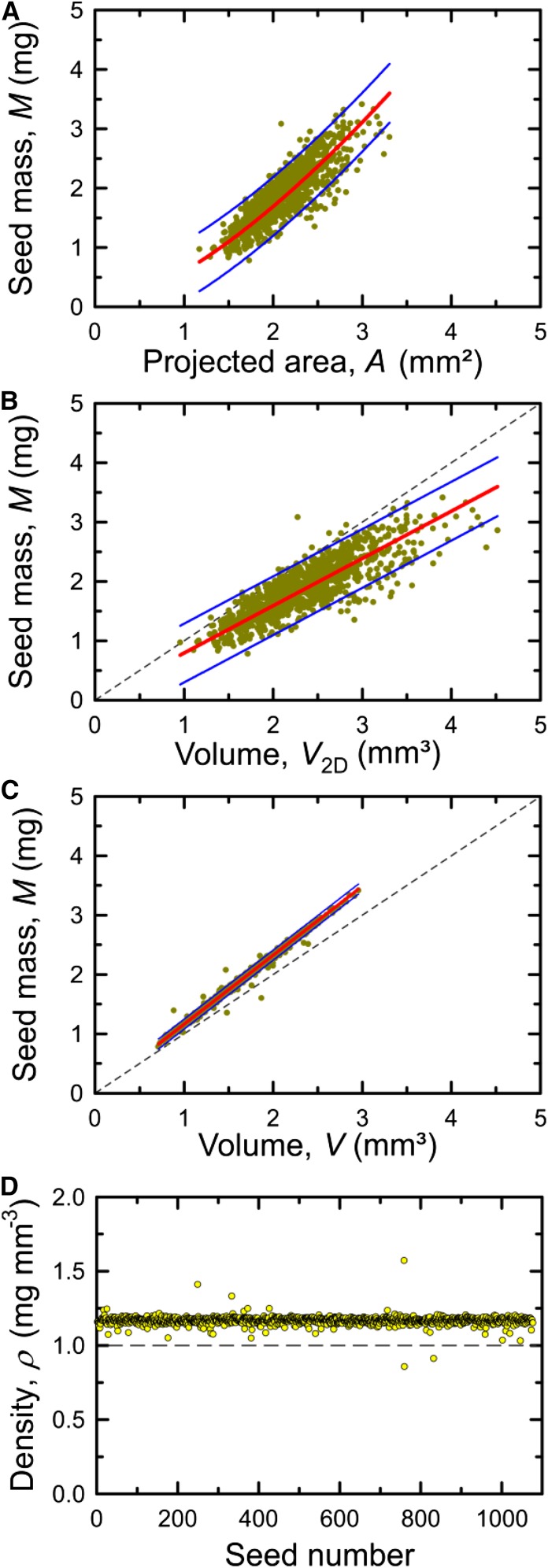
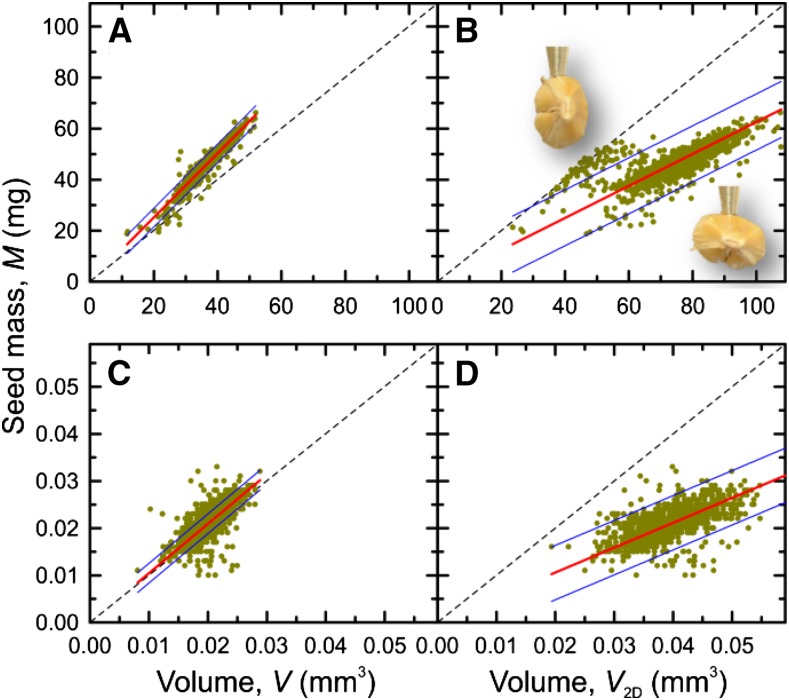
A selection of different correlations is presented for each investigated species and accession in Supplemental Figures S7 to S15, and an overview of calculated numbers is given in Table III. Figure 7 shows examples for seeds of rapeseed accession Wotan when M was compared with A (Fig. 7A), V2D (Fig. 7B), and V (Fig. 7C). When plotting M of individual seeds against the corresponding A, there is a nonlinear relationship (Fig. 7A). Since mass can be regarded as a volume property while area is a 2D entity, the relationship between these traits can be approximately described by the equation y = a A3/2. However, the correlation was not good (r2 = 0.67; Fig. 7A), and the 95% prediction intervals indicate that, when M is calculated from A, the error could be rather high. Almost the same result was obtained for the linear fit of M by V2D (Fig. 7B). However, M versus V showed a very good correlation (r2 = 0.99) and a narrow 95% prediction interval (hardly visible; Fig. 7C). From M and V, seed density (ρ) was obtained for each individual seed (Fig. 7D) and used to calculate mean densities (Table III). For barley and Arabidopsis, correlations between M and V, as well as between M and V2D, are presented in Figure 8. Seeds of barley accession Barke showed a good correlation for M versus V (r2 = 0.93; Fig. 8A) but not for M versus V2D (r2 = 0.37), where two clusters of data were observed (Fig. 8B). The two clusters can be explained by systematic errors in 2D images from which W2D, and thus V2D, are determined, because barley seeds may lie either on the ventral/dorsal side or on the edge, as photographs taken at the 3D imaging station demonstrate (Fig. 8B, insets). For seeds of Arabidopsis accession Col-0 (Fig. 8, C and D), the correlations were even worse (r2 = 0.28) when M versus V2D was plotted (Fig. 8D) but also for M versus V (Fig. 8C; r2 = 0.49). The main reason for the data scattering in Figure 8C was inaccuracy in measuring the seed mass (ranging between 10 and 35 µg) due to limited reproducibility, as mentioned above.
Figure 7.
Correlation between selected seed traits of rapeseed accession Wotan. A, Seed mass versus projected area. B, Seed mass versus volume calculated from length and width of projection. C, Seed mass versus measured volume. D, Seed density plotted for each individual seed. Red lines denote a fit of y = a x3/2 (A) and a linear fit of y = a x (B and C). Short-dashed lines are identity lines (B and C), the long-dashed line denotes y = 1 (D), and blue lines indicate 95% prediction intervals (A–C).
Figure 8.
Correlations between selected seed traits of barley and Arabidopsis. A and B, Barley accession Barke. C and D, Arabidopsis accession Col-0. A and C, Seed mass versus measured volume. B and D, Seed mass versus volume calculated from length and width of projection. Red lines denote linear fits of y = a x, and blue lines indicate 95% prediction intervals. The dashed lines in the panels indicate identity lines. In B, the insets show barley seeds sucked at the nozzle tip, illustrating two different positions of seeds when lying on the optical glass filter of the 2D imaging station; this is responsible for the clustering of data.
Results of all three species with three accessions each are presented in Figure 9, where the seeds were sorted by M and both V and V2D were scaled to meet the range of M. The data of V matched M very well for all measured accessions of rapeseed, whereas V2D matched M rather badly (Fig. 9, A–C). In the case of barley, the data of V matched M rather well for Barke and HOR13719 (Fig. 9, D and E, respectively), while, for HOR9707, deviations between M and V became obvious (Fig. 9F) and V2D matched M rather badly in all three accessions (Fig. 9, D–F). The clustering of V2D values due to the seed orientation can be seen in particular with Barke (Fig. 9D), as already shown in Figure 8B. The data of both V and V2D did not really match M for all three accessions of Arabidopsis (Fig. 9, G–I), and again, V2D matched M rather badly. The stepwise increase in M visible in Figure 9, G to I, reflects the 1-µg resolution steps of the balance, limiting accurate weighing of seeds as light as Arabidopsis. This limitation also might account for the poor congruence of M and V for very small and very large seed sizes in Figure 9, G to I.
Additional correlation results of all measured species and accessions are presented in Table III and Supplemental Figures S7 to S15. Linear fits to M versus VA showed similar correlations (r2 = 0.35–0.7) to M versus V2D (r2 = 0.28–0.8), generally much worse than M versus V (r2 = 0.89–0.99 for rapeseed and barley and r2 = 0.5–0.72 for Arabidopsis). For all investigated species, the analyzed seed traits showed statistically significant differences between the three accessions with very few exceptions (Table III). This shows that the measurement accuracy and sample size possible with phenoSeeder allow a detection of statistically significant differences of trait means between genotypes, even if the absolute differences are rather small. The values of VA and V2D generally were overestimated compared with V (Table III), because the heights used for volume calculation from 2D data (W or W2D; see Table I) were always higher than measured H (Table III; Supplemental Figs. S7–S15, O). Since the volumes calculated from 2D images were too big, the corresponding density values, ρA and ρ2D, turned out too low compared with ρ, calculated from V (Table III; Supplemental Figs. S7–S15, J–L).
DISCUSSION
Seed mass across the plant kingdom is highly defined by plant and genome size rather than by the mode of dispersal or environmental conditions (Linkies et al., 2010). Compared with other morphological traits, seed mass is a most important parameter, indicating the potential to supply both radicle protrusion (germination) and seedling establishment (Salisbury, 1974; Montesinos-Navarro et al., 2011; Igea et al., 2016). Anatomical structures, but also the ratio between carbohydrates and lipids, may affect seed mass (Rolletschek et al., 2015); for example, overexpression of diacylglycerol acyltransferase cDNA in Arabidopsis has been shown to enhance oil deposition and average seed size (Jako et al., 2001). Since weighing individual seeds is laborious, projected area often is used as a proxy for seed mass, which can be highly erroneous, as discussed below. Seed traits are complex, and there is not just one best trait characterizing a seed. Therefore, we propose that measuring 3D properties of individual seeds, such as volume and mass, in addition to generally measured 2D parameters opens new possibilities to study seed biology in greater depth.
For reliable handling of single seeds, a well-coordinated interplay between the phenoSeeder’s robot actions (Fig. 1; Supplemental Fig. S4) and the pneumatics delivering either vacuum or overpressure to the nozzle (Fig. 2; Supplemental Fig. S1) has been implemented. For different seed types, specific parameters (e.g. robot speed, maximum seed height, release pressure, or color thresholds) have to be defined in pretests to ensure proper handling. Once a dedicated parameter set is recorded in the database, automated phenotyping can be started by choosing a given species or accession on the screen. A particular challenge was handling very small seeds of Arabidopsis (Fig. 2C). The main solutions were a very precise positioning robot, dedicated nozzles for seed uptake and release, and fine-tuning of the parameters for seed sucking and release. While seeds of rapeseed (Fig. 2D) are easy to handle, this is not necessarily true for all larger seeds, in particular when the seed surface is rough and irregular (like that of barley; Fig. 2E). After constructing and testing a large number of different nozzles, we predict that three different nozzle tools will be sufficient to handle most seeds ranging in size between Arabidopsis and maize (Zea mays). Particularly when seeds are stored in plastic tubes, electrostatic charging of seeds can be a serious issue and may lead to sucking up several (clustered) seeds at once or prevent the release of a seed from the nozzle. This problem has been minimized by proper grounding of all phenoSeeder parts. Failure rates in seed release are below 1% in most cases, and seed positioning is very precise (Supplemental Fig. S5). Although the primary positioning precision of the robot system is extremely high, the final position of a seed may be very dependent on the properties of the substrate, such as wetness or roughness of the surface, and also properties of the seeds.
Projected area is measured either by microscopic analyses (Alonso-Blanco et al., 1999) or quite often by scanning methods (de Jong et al., 2011; Tanabata et al., 2012; Moore et al., 2013). For 11 natural accessions of Arabidopsis, a quite good correlation (r2 = 0.8879) of A with average seed weight has been reported (Herridge et al., 2011). However, the trend line fitted to the data was linear in A (Herridge et al., 2011) and not proportional to A3/2, as one would expect (Fig. 7A). Statistical correlations are of limited use when single-seed performance is of interest, as observed here, where M was not well presented by A for all investigated genotypes (Supplemental Figs. S7–S15). While for the measured rapeseed genotypes the correlation M versus V2D was little better than M versus A, the opposite was found for barley and Arabidopsis (Table III). In general, a better correlation is not necessarily an unambiguous measure of correctness: when V2D was taken as seed volume, the resulting seed density, ρ2D, was markedly smaller than 1 mg mm−3, which is rather unlikely (see also the slope of the linear fit of Fig. 7B, Supplemental Fig. S7–S15, K, and Table III). Another general problem of scanning methods is the fact that particularly angular seeds may have distinct positions when dispersed on a flat (scanner) surface, as demonstrated by the images of barley grains in Figure 8B. This also would affect other parameters derived from scans, such as L2D and W2D, and indicates that even relatively simple scanning procedures can be prone to systematic errors.
Direct measurement of a single seed volume is not easy to achieve, and published data are based mainly on sophisticated methods such as x-ray CT (Stuppy et al., 2003; Friis et al., 2014). With phenoSeeder, images of seeds sucked at a nozzle are taken from different angles (Fig. 4) and, by 3D reconstruction, single-seed volumes are achieved (Roussel et al., 2016) with good repeatability (Table II). The optical measurement and space-carving procedures used here for 3D reconstruction (Roussel et al., 2016) cannot represent each groove on a seed surface. Therefore, we assume that the measured volumes are slightly overestimated, in particular for seeds with rough surfaces such as barley. Still, the measured volume, compared with A, provided a much better proxy for M, as becomes obvious when comparing the correlations of M versus A and M versus V (Table III). Automated seed mass detection with phenoSeeder was very reliable for both rapeseed and barley (Table III). However, for Arabidopsis seeds having extremely low masses, weighing errors became significant, which also explains the weak correlations of M versus V for the three Arabidopsis accessions and the relatively high RSD values of seed density (Table III).
As reasoned above for estimating M from A, a correlation between M and V may be rather good as long as average values of masses and volumes are compared, but when single seeds are regarded, statistical correlations are of limited use. Different densities (Table III), also represented by different slopes of the M versus V plots (Supplemental Figs. S7–S15), suggest that there is not just one universal equation to calculate M from V (or A) for all seeds. Yet, seed mass calculation from V, or in specific cases even A, may become applicable when calibration parameters for a given species or accession have been assessed. For the tiny seeds of Arabidopsis, the balance readings were randomly scattered when reaching the measurement limits. Estimating M from V might be a valid way to overcome this issue of weighing imprecision. Considering the very good correlation between M and V for larger seeds with rather similar shape, as for rapeseed (Supplemental Figs. S7–S9), we might assume that, for single Arabidopsis seeds, mass can be estimated reliably from V by taking the slopes of a linear fit to M versus V of a particular genotype (Supplemental Figs. S12–S14).
Common methods in seed processing and evaluating size distributions are separating seed batches with sieves of defined meshes, as shown for Arabidopsis (Jofuku et al., 2005), or image analysis methods using 2D seed shape parameters, as shown for rice grains (Tanabata et al., 2012). Since, besides other seed traits, seed size distribution is “significant for the seed companies and processing industry affecting product value” (Shahin and Symons, 2005), both seed sieving and image-analysis approaches were compared with respect to reproducibility and speed (Shahin and Symons, 2005). We are aware of the fact that the throughput of phenoSeeder is not high compared with the standard procedures used by seed companies. Yet, we are convinced that, for example, it is suitable for various research approaches or to assess aliquots of seed batches to evaluate standard methods used for seed classification. A particular feature of phenoSeeder is the potential to classify seeds by all measured traits and to sort seeds by any criterion. The histograms in Figure 6 show that, for rapeseed accession Wotan, frequencies of A, V, and M were almost normally distributed. In Supplemental Figures S7 to S15, the frequencies of VA and V2D are presented for all analyzed species and genotypes. It can be seen that trait distribution does not match the normal distribution in some cases, such as V of rapeseed accession Expert or Pirola (Supplemental Figs. S8D and S9D, respectively) or A of barley accession Barke or HOR13719 (Supplemental Figs. S10A and S11A, respectively). Such deviations from normal distribution can be based on different factors, like impurities, heterogenous genetic background, or hampered seed development caused by disease or abiotic stress, but they also may point to measurement errors, as shown with the 2D scans of barley seeds. Thus, frequency histograms based on single-seed detection considering different traits can be used for quality assessment of seeds.
Seed density is a rather complex trait because different seed parts, such as seed coat (Alonso-Blanco et al., 1999; Young et al., 2007), lipid content in the endosperm (Fuchs et al., 2013), or void space inside a seed (Verboven et al., 2013) may contribute differently. Air space in seeds may mask different oil contents, as reported for rapeseed by measuring the commonly applied seed buoyant density (Young et al., 2006). The seed volumes reconstructed here may be slightly overestimated as discussed, and the true ρ values could be somewhat larger than the ones presented in Table III (1.05–1.29 mg mm−3 throughout all measured genotypes). However, when volumes estimated from 2D scans were used to calculate ρA and ρ2D (Table III), all densities were clearly below 1 mg mm−3, showing that the achieved 2D data were not suited to achieve proper values of seed volumes. The mean density values of the three rapeseed genotypes (1.12–1.14 mg mm−3; Table III) match very well to data of two rapeseed populations, with a main density class at 1.13 mg mm−3 (Young et al., 2006). Phenotyping of individual seeds as presented here can be used to search for seeds with unusual properties, opening possibilities for subsequent in-depth (e.g. anatomical, metabolomic, or genomic) analyses.
At the present stage of development, phenoSeeder is used for routine seed phenotyping as demonstrated, but there are still possibilities for improvement and extension. Due to the modular and flexible structure of the system, future approaches are so manifold that only a few can be mentioned here. Extensions under construction or planned are (1) multiwell plates for intermediate storage of single seeds (Fig. 5, option 4), enabling seed classification a posteriori (option 5), as described above, or fast planting into substrates (option 6); (2) automated seed delivery systems; and (3) color and pattern recognition at the 3D imaging station. We are trying to increase speed, since, so far, a full cycle of phenotyping takes up to 35 s for one seed (Supplemental Table S1) and direct planting of a phenotyped seed may cause a considerable delay between the first and last seed of a large seed batch. Seed planting from multiwell plates (Fig. 5, option 6), however, would need only pick and place, which is much faster. To monitor different traits of interest beyond morphological ones, additional sensors could be implemented: near-infrared spectroscopy to analyze the chemical content of seeds (Agelet and Hurburgh, 2014); chlorophyll fluorescence to score seed maturity and performance (Jalink et al., 1998); spectral imaging to classify common wheat (Triticum aestivum) and durum wheat (Triticum durum; Benoit et al., 2016); low-field NMR to measure both solid and liquid parts of a seed, as demonstrated for growing bean (Phaseolus vulgaris) pods (Windt and Blümler, 2015), or chemical components, such as lipids, carbohydrates, and proteins (Rolletschek et al., 2015); or x-ray CT to image internal seed structures, allowing, for example, the detection of internal defects of seeds (Stuppy et al., 2003; Belin et al., 2011; Yamauchi et al., 2012; Verboven et al., 2013). Further developments of phenoSeeder can be followed at www.phenoseeder.de.
phenoSeeder can be employed for a large number of applications, such as characterizing seeds of unknown collections, evaluating seed quality, or setting up seed-to-plant tracking pipelines. It might even be considered for seed germination studies using blotting paper, allowing one to correlate germination not only with seed traits obtained from projections (Joosen et al., 2010; Demilly et al., 2014) but also with seed volume, mass, or density. Regarding the large diversity of seeds and the high complexity of seed traits, we conclude that the phenoSeeder approach enabling automated identification and monitoring of relevant traits of individual seeds provides a wide range of valuable options for applications in seed biology.
MATERIALS AND METHODS
Plant Material
The term seed is used here in a functional sense including diaspores such as caryopses (see also Supporting Online Material of Moles et al., 2005). The investigated seeds of rapeseed (Brassica napus) with the genotypes Wotan, Expert, and Pirola were from Norddeutsche Pflanzenzucht Hans-Georg Lembke, seeds of barley (Hordeum vulgare) with the genotypes Barke, HOR13719, and HOR9707 were from the gene bank at Leibniz Institute of Plant Genetics and Crop Plant Research, and seeds of Arabidopsis (Arabidopsis thaliana) with the genotypes Col-0, Lag2-2, and Agu-1 were from the 1001 Genomes project (Weigel and Mott, 2009). Images of seeds of the different species and genotypes are presented in Supplemental Figure S6.
System Specifications and Statistical Analyses
More information and detailed specifications of the phenoSeeder components are provided in Supplemental Materials and Methods S1. Statistical analysis was performed using the Matlab (R2015b; MathWorks) Statistics and Machine Learning Toolbox as well as home-built procedures. The Matlab function histfit was used for plotting histograms with included fitted normal distributions in Figure 6 and Supplemental Figures S7 to S15. The Matlab function ttest2 was used for the pairwise comparison of sample means in Table III by a two-sample Student’s t-test, assuming unequal variances of the two samples (also called Welch’s t-test). Linear and nonlinear fits as well as the estimation of 95% prediction intervals shown in Figures 7 and 8 and Supplemental Figures S7 to S15 were performed using procedures described in detail in Supplemental Materials and Methods S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Scheme of the pneumatic system.
Supplemental Figure S2. Photographs of the 2D imaging station.
Supplemental Figure S3. Photographs of stations for weighing, cleaning, and calibration.
Supplemental Figure S4. Activity diagram of the phenoSeeder workflow.
Supplemental Figure S5. Example of seed planting at the seed placement station.
Supplemental Figure S6. Images of seeds of all nine accessions measured.
Supplemental Figure S7. Seed traits of rapeseed accession Wotan.
Supplemental Figure S8. Seed traits of rapeseed accession Expert.
Supplemental Figure S9. Seed traits of rapeseed accession Pirola.
Supplemental Figure S10. Seed traits of barley accession Barke.
Supplemental Figure S11. Seed traits of barley accession HOR13719.
Supplemental Figure S12. Seed traits of barley accession HOR9707.
Supplemental Figure S13. Seed traits of Arabidopsis accession Col-0.
Supplemental Figure S14. Seed traits of Arabidopsis accession Lag2-2.
Supplemental Figure S15. Seed traits of Arabidopsis accession Agu-1.
Supplemental Table S1. Average time needed for seed handling.
Supplemental Movie S1. The phenoSeeder system in action.
Supplemental Materials and Methods S1. Additional information and specifications of the phenoSeeder components and Matlab scripts used for statistical analysis.
Supplementary Material
Acknowledgments
We thank Axel Dahmen, Alexander Putz, Patrick Embgenbroich, Torge Herber, Benjamin Bruns, Marcus Brenscheidt, Felix Geiger, Jonas Bühler, Beate Uhlig, and Simone Gatzke for help with a number of different tasks; Ulrich Schurr for valuable contributions and two anonymous reviewers whose comments helped to improve the article; and the breeding company Norddeutsche Pflanzenzucht Hans-Georg Lembke (Pre-BreedYield project, BMBF Fz. 0315964D) for rapeseed seeds, Genbank Gatersleben of the Leibniz Institute of Plant Genetics and Crop Plant Research for barley seeds, and Detlef Weigel for Arabidopsis seeds.
Glossary
- 2D
two-dimensional
- 3D
three-dimensional
- CT
computed tomography
- Col-0
Columbia-0
- RSD
relative standard deviation
- Lag2-2
Lagodechi
- Agu-1
Aguaron
Footnotes
This work was performed within the German Plant Phenotyping Network funded by the German Federal Ministry of Education and Research (project identification no. 031A053).
Articles can be viewed without a subscription.
References
- Agelet LE, Hurburgh CR Jr (2014) Limitations and current applications of near infrared spectroscopy for single seed analysis. Talanta 121: 288–299 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin E, Rousseau D, Léchappé J, Langlois-Meurinne M, Dürr C (2011) Rate-distortion tradeoff to optimize high-throughput phenotyping systems: application to x-ray images of seeds. Comput Electron Agric 77: 188–194 [Google Scholar]
- Benoit L, Benoit R, Belin E, Vadaine R, Demilly D, Chapeau-Blondeau F, Rousseau D (2016) On the value of the Kullback-Leibler divergence for cost-effective spectral imaging of plants by optimal selection of wavebands. Mach Vis Appl 27: 625–635 [Google Scholar]
- Borisjuk L, Rolletschek H, Fuchs J, Melkus G, Neuberger T (2011) Low and high field magnetic resonance for in vivo analysis of seeds. Materials (Basel) 4: 1426–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong TJ, Hermans CM, van der Veen-van Wijk KC (2011) Paternal effects on seed mass in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 13: 71–77 [DOI] [PubMed] [Google Scholar]
- Demilly D, Ducournau S, Wagner MH, Dürr C (2014) Digital imaging of seed germination. In Dutta Gupta S, Ibaraki Y, eds, Plant Image Analysis. CRC Press, Boca Raton, FL, pp 147–164 [Google Scholar]
- Esau K. (1977) Anatomy of Seed Plants, Ed 2 Wiley, New York [Google Scholar]
- Friis EM, Marone F, Pedersen KR, Crane PR, Stampanoni M (2014) Three-dimensional visualization of fossil flowers, fruits, seeds, and other plant remains using synchrotron radiation x-ray tomographic microscopy (SRXTM): new insights into Cretaceous plant diversity. J Paleontol 88: 684–701 [Google Scholar]
- Fuchs J, Neuberger T, Rolletschek H, Schiebold S, Nguyen TH, Borisjuk N, Börner A, Melkus G, Jakob P, Borisjuk L (2013) A noninvasive platform for imaging and quantifying oil storage in submillimeter tobacco seed. Plant Physiol 161: 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge RP, Day RC, Baldwin S, Macknight RC (2011) Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igea J, Miller EF, Papadopulos AS, Tanentzap AJ (2016) Seed size drives species diversification across angiosperms. bioRxiv doi/org/10.1101/053116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalink H, van der Schoor R, Frandas A, van Pijlen JG, Bino RJ (1998) Chlorophyll fluorescence of Brassica oleracea seeds as a non-destructive marker for seed maturity and seed performance. Seed Sci Res 8: 437–443 [Google Scholar]
- Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH (2013) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162: 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102: 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RVL, Kodde J, Willems LAJ, Ligterink W, van der Plas LHW, Hilhorst HWM (2010) GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 62: 148–159 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Koizumi M, Ishida N, Kano H (2006) Water uptake by dry beans observed by micro-magnetic resonance imaging. Ann Bot (Lond) 98: 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Graeber K, Knight C, Leubner-Metzger G (2010) The evolution of seeds. New Phytol 186: 817–831 [DOI] [PubMed] [Google Scholar]
- Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, Westoby M (2005) A brief history of seed size. Science 307: 576–580 [DOI] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Pico FX, Tonsor SJ (2011) Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol 189: 282–294 [DOI] [PubMed] [Google Scholar]
- Moore CR, Gronwall DS, Miller ND, Spalding EP (2013) Mapping quantitative trait loci affecting Arabidopsis thaliana seed morphology features extracted computationally from images. G3 (Bethesda) 3: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden N, Daws MI, Antoine C, Gonzalez MA, Garwood NC, Chave J (2009) The relationship between seed mass and mean time to germination for 1037 tree species across five tropical forests. Funct Ecol 23: 203–210 [Google Scholar]
- Paul-Victor C, Turnbull LA (2009) The effect of growth conditions on the seed size/number trade-off. PLoS ONE 4: e6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Fuchs J, Friedel S, Börner A, Todt H, Jakob PM, Borisjuk L (2015) A novel noninvasive procedure for high-throughput screening of major seed traits. Plant Biotechnol J 13: 188–199 [DOI] [PubMed] [Google Scholar]
- Roussel J, Geiger F, Fischbach A, Jahnke S, Scharr H (2016) 3D surface reconstruction of plant seeds by volume carving: performance and accuracies. Front Plant Sci 7: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury E. (1974) Seed size and mass in relation to environment. Proc R Soc Lond B Biol Sci 186: 83–88 [Google Scholar]
- Schmidt F, Bruns B, Bode T, Scharr H, Cremers AB (2013) A distributed information system for managing phenotyping data. In Clasen M, Kersebaum KC, Mayer-Aurich A, Theuvsen B, eds, Lecture Notes in Informatics P-211. Gesellschaft für Informatik, Bonn, Germany, pp 303–306 [Google Scholar]
- Shahin MA, Symons SJ (2005) Seed sizing from images of non-singulated grain samples. Can Biosyst Eng 47: 49–55 [Google Scholar]
- Stanton ML. (1984) Seed variation in wild radish: effect of seed size on components of seedling and adult fitness. Ecology 65: 1105–1112 [Google Scholar]
- Stuppy WH, Maisano JA, Colbert MW, Rudall PJ, Rowe TB (2003) Three-dimensional analysis of plant structure using high-resolution x-ray computed tomography. Trends Plant Sci 8: 2–6 [DOI] [PubMed] [Google Scholar]
- Tanabata T, Shibaya T, Hori K, Ebana K, Yano M (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol 160: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboven P, Herremans E, Borisjuk L, Helfen L, Ho QT, Tschiersch H, Fuchs J, Nicolaï BM, Rolletschek H (2013) Void space inside the developing seed of Brassica napus and the modelling of its function. New Phytol 199: 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Mott R (2009) The 1001 Genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windt CW, Blümler P (2015) A portable NMR sensor to measure dynamic changes in the amount of water in living stems or fruit and its potential to measure sap flow. Tree Physiol 35: 366–375 [DOI] [PubMed] [Google Scholar]
- Yamauchi D, Tamaoki D, Hayami M, Uesugi K, Takeuchi A, Suzuki Y, Karahara I, Mineyuki Y, Momose A, Yashiro W (2012) Extracting tissue and cell outlines of Arabidopsis seeds using refraction contrast x-ray CT at the SPring-8 facility. AIP Conf Proc 1466: 237 [Google Scholar]
- Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, et al. (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5: 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Jalink H, Denkert R, Reaney M (2006) Factors affecting the density of Brassica napus seeds. Seed Sci Technol 34: 633–645 [Google Scholar]
- Young LW, Parham C, Zhong Z, Chapman D, Reaney MJT (2007) Non-destructive diffraction enhanced imaging of seeds. J Exp Bot 58: 2513–2523 [DOI] [PubMed] [Google Scholar]
- Zhang X, Sun J, Cao X, Song X (2015) Epigenetic mutation of RAV6 affects leaf angle and seed size in rice. Plant Physiol 169: 2118–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.