The proteasome is required for local and systemic immune responses and is targeted by Pseudomonas type III effectors.
Abstract
Recent evidence suggests that the ubiquitin-proteasome system is involved in several aspects of plant immunity and that a range of plant pathogens subvert the ubiquitin-proteasome system to enhance their virulence. Here, we show that proteasome activity is strongly induced during basal defense in Arabidopsis (Arabidopsis thaliana). Mutant lines of the proteasome subunits RPT2a and RPN12a support increased bacterial growth of virulent Pseudomonas syringae pv tomato DC3000 (Pst) and Pseudomonas syringae pv maculicola ES4326. Both proteasome subunits are required for pathogen-associated molecular pattern-triggered immunity responses. Analysis of bacterial growth after a secondary infection of systemic leaves revealed that the establishment of systemic acquired resistance (SAR) is impaired in proteasome mutants, suggesting that the proteasome also plays an important role in defense priming and SAR. In addition, we show that Pst inhibits proteasome activity in a type III secretion-dependent manner. A screen for type III effector proteins from Pst for their ability to interfere with proteasome activity revealed HopM1, HopAO1, HopA1, and HopG1 as putative proteasome inhibitors. Biochemical characterization of HopM1 by mass spectrometry indicates that HopM1 interacts with several E3 ubiquitin ligases and proteasome subunits. This supports the hypothesis that HopM1 associates with the proteasome, leading to its inhibition. Thus, the proteasome is an essential component of pathogen-associated molecular pattern-triggered immunity and SAR, which is targeted by multiple bacterial effectors.
The ubiquitin-proteasome system (UPS) is one of the main protein degradation systems of eukaryotic cells that not only removes misfolded and defective proteins but also controls various cellular pathways through the selective elimination of short-lived regulatory proteins (Vierstra, 2009). The UPS regulates many fundamental cellular processes, such as protein quality control, DNA repair, and signal transduction (Sadanandom et al., 2012). Selective protein degradation by the UPS proceeds from the ligation of one or more ubiquitin proteins to the ε-amino group of a Lys residue within specific target proteins catalyzed by the consecutive action of E1, E2, and E3 enzymes. The resulting ubiquitinated proteins are then recognized and degraded by the 26S proteasome. The 26S proteasome itself is a 2.5-MD ATP-dependent protease complex composed of 31 subunits divided into two types of subcomplexes, namely the 20S core protease (CP) and the 19S regulatory particles (RPs). While the CP is a broad-spectrum ATP- and ubiquitin-independent protease complex, the RP subcomplex assists in recognizing ubiquitinated target proteins and in opening the channel of the CP to insert the unfolded substrates into the CP chamber for degradation (Smalle and Vierstra, 2004). During the past few years, several studies have revealed that the UPS controls various processes in almost all aspects of plant homeostasis, comprising cell division, plant development, responses to plant hormones, as well as abiotic and biotic stress responses (Sadanandom et al., 2012).
It is becoming increasingly obvious that protein turnover via the UPS controls multiple aspects of plant immunity, including recognition, receptor accumulation, and downstream defense signaling (Marino et al., 2012). Plant immunity relies on a multilayered system to detect and resist attempted pathogen invasion. Cell-surface pattern recognition receptors (PRRs) recognize conserved pathogen-associated molecular patterns (PAMPs) and initiate PAMP-triggered immunity (PTI; Jones and Dangl, 2006). This recognition leads to the production of reactive oxygen species (ROS), the activation of mitogen-activated protein kinases (MAPKs), transcriptional reprogramming, and callose deposition at the cell wall (Boller and Felix, 2009). Adapted plant pathogens are able to overcome PTI by delivering effector proteins into host cells and inducing effector-triggered susceptibility. On the other hand, resistant plants have evolved the ability to monitor the presence or activities of effectors by intracellular immune receptors, commonly referred to as resistance proteins, resulting in effector-triggered immunity (ETI; Jones and Dangl, 2006). ETI is typically accompanied by the hypersensitive response (HR), a form of localized programmed cell death at the primary infection site (Hofius et al., 2007), thereby restricting pathogen spread within infected tissue.
Localized pathogen attack also leads to increased resistance toward secondary infection in uninfected parts of plants. This type of increased resistance is referred to as systemic acquired resistance (SAR; Fu and Dong, 2013). After SAR has been induced, plants are primed (i.e. sensitized) to respond more rapidly and more effectively to a secondary infection. Long-distance signaling between the primary infected leaf and distal leaves is required for the onset of SAR. The defense hormone salicylic acid (SA) is shown to be critical for the establishment of SAR by inducing SAR-related gene expression via the downstream regulator NONEXPRESSER OF PR GENES1 (NPR1), a transcriptional coactivator (Fu and Dong, 2013). However, other signaling metabolites, such as pipecolic acid (Návarová et al., 2012), also have been shown to play an essential role in the establishment of SAR (Bernsdorff et al., 2016).
The intricate molecular processes underlying the cellular changes during PTI, ETI, and SAR require a high degree of proteomic plasticity, likely involving the UPS. For instance, members of the U-box E3 ligase family have been identified as negative regulators of PTI (Trujillo et al., 2008; Stegmann et al., 2012). In addition, many key plant defense signaling components are degraded by the 26S proteasome pathway, including the PAMP receptor FLS2 (Lu et al., 2011), the master regulator of SA-dependent defense NPR1 (Spoel et al., 2009), and the transcription factor WRKY45 (Matsushita et al., 2013). Apart from its function in regulating the turnover of components implicated in plant immunity, several proteasome components have been identified to contribute directly to defense responses such as ROS production and HR formation (Marino et al., 2012). In particular, PBA1, the catalytic subunit of the 20S proteasome, has been proposed to act as a caspase-like enzyme during the induction of programmed cell death in response to avirulent bacterial strains (Hatsugai et al., 2009). Concomitant with the role of 20S subunits in plant immunity, RPN1a, a component of the RP, has been described to be required for resistance against biotrophic fungi (Yao et al., 2012). The latter study also showed that the accumulation of RPN1a is affected by SA and that the rpn1a mutant has defects in SA accumulation upon infection with Pseudomonas syringae pv tomato DC3000 (Pst). However, based on the analysis of additional mutants, it appears that not all proteasome subunits play a similar role in immunity (Yao et al., 2012).
Considering the pivotal role of the UPS in plant defense responses, it is not surprising that pathogens have evolved virulence factors that can manipulate the UPS in an attempt to enhance virulence during plant-pathogen interactions (Dudler, 2013). Gram-negative bacterial pathogens use a type III secretion system to inject so-called type III effector (T3E) proteins into host cells to interfere with host cellular functions and immunity (Macho, 2016). Several T3Es from different genera of plant pathogenic bacteria, such as Pseudomonas and Xanthomonas, were shown to suppress plant defense responses by acting as E3 ligases or by promoting ubiquitination and degradation of target proteins (Nomura et al., 2006; Singer et al., 2013; Üstün and Börnke, 2014, 2015; Banfield, 2015). A more direct way to subvert the UPS is achieved by SylA, a secreted small nonribosomal peptide from Pseudomonas syringae pv syringae, which binds to the catalytic subunits of the 26S proteasome to inhibit its activity and suppress plant immune reactions, including stomatal closure and SA-mediated signaling (Groll et al., 2008; Schellenberg et al., 2010; Misas-Villamil et al., 2013). The first bacterial T3Es identified that directly target the proteasome for defense suppression are XopJ from Xanthomonas campestris pv vesicatoria and HopZ4 from Pseudomonas syringae pv lachrymans. Both closely related T3Es interact with the proteasomal component RPT6, a subunit of the 19S RP, to inhibit proteasome activity (Üstün et al., 2013, 2014; Üstün and Börnke, 2015). In effect, this results in impaired turnover of the SA master regulator NPR1 and the attenuation of SA-dependent defense responses.
Despite these significant advances in the recent years, we still lack knowledge of how the proteasome is involved in defense responses and whether targeting the host proteasome is a general strategy of plant pathogens. To address this question, we analyzed the contribution of the proteasome to plant immunity during local and systemic defense responses of Arabidopsis (Arabidopsis thaliana) against Pst. Here, we show that the proteasome subunits RPT2a and RPN12a are required for PTI events and the establishment of SAR. In turn, through a systematic screen of the T3E repertoire of Pst, we identified the T3Es HopM1, HopAO1, HopA1, and HopG1 as putative inhibitors of proteasome activity. Further biochemical analysis revealed that HopM1 interacts with multiple proteins, including UPS-related proteins. Thus, the proteasome is an essential component of PTI and SAR, which is targeted by multiple bacterial effectors.
RESULTS
The Proteasome Is Required during PTI and Suppressed in a T3E-Dependent Manner
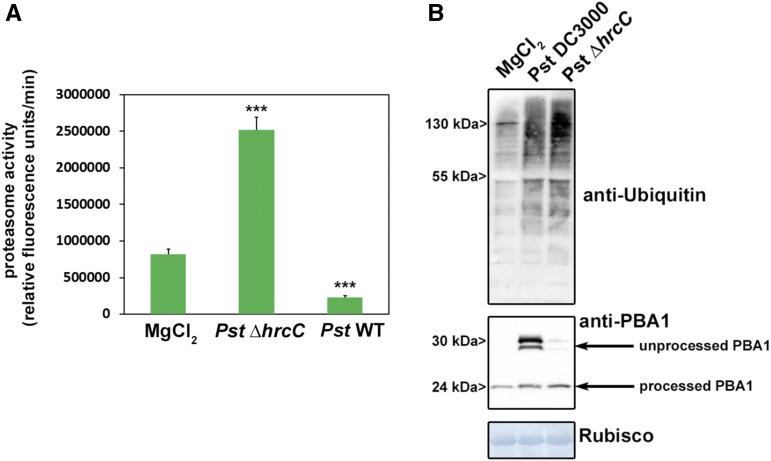
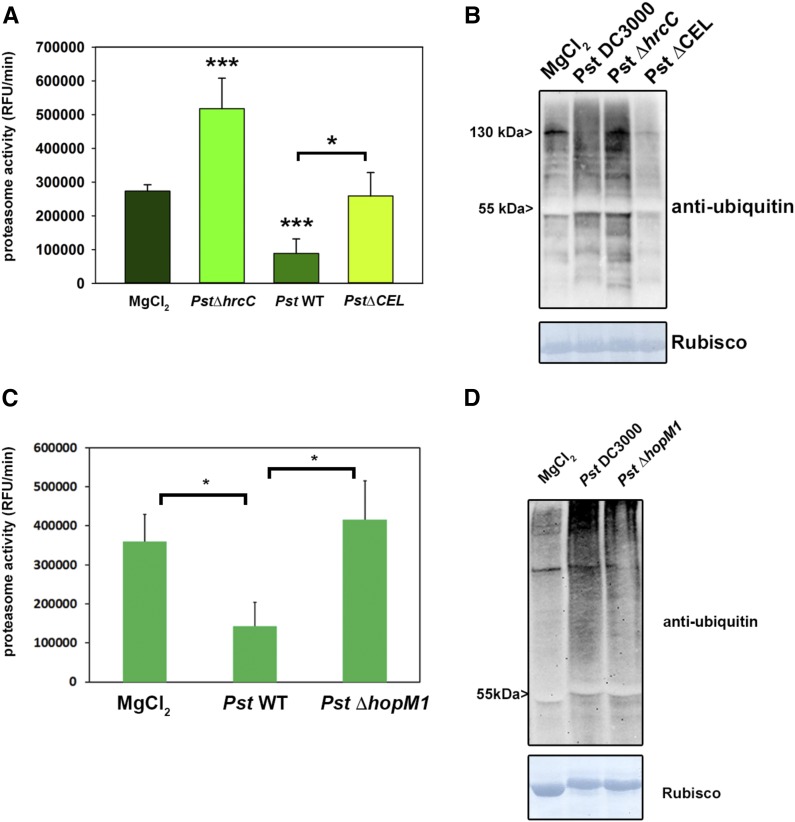
In order to investigate whether proteasome activity is modulated during the interaction of Pst with Arabidopsis, the proteasome activity was monitored using a fluorogenic peptide (Suc-LLVY-AMC). This peptide is a well-established substrate for the chymotrypsin-like activity of the proteasome (Üstün et al., 2013). Proteasome activity was measured in leaves of wild-type Arabidopsis plants inoculated either with a Pst or a Pst ΔhrcC strain, which is not able to deliver T3Es into the host cell. The measurements revealed that proteasome activity is induced significantly in Pst ΔhrcC-infected leaves compared with the mock control (Fig. 1A). In contrast, infection with Pst leads to a significant reduction in proteasome activity. These results indicate that proteasome activity is induced during PTI and that Pst can suppress this induction in a T3E-dependent manner. Immunoblot analysis using an antibody directed against ubiquitin on extracts from Pst- and Pst ΔhrcC-infected leaves revealed the accumulation of ubiquitinated proteins in both cases (Fig. 1B, top). To further confirm proteasome malfunctioning, we probed the same membrane with an antibody directed against the proteasome core subunit PBA1. One essential posttranslational modification is the processing of the N terminus of PBA1. Treatment with the proteasome inhibitor MG132 blocks proteasome maturation, and an unprocessed form of PBA1 accumulates (Book et al., 2010). Similarly, plants infected with Pst also showed the accumulation of unprocessed PBA1 (Fig. 1B, bottom), indicating disturbed proteasome maturation. These findings suggest that proteasome activity is induced as part of the defense response to bacteria and also constitutes a virulence target for Pst effectors.
Figure 1.
Pst prevents the induction of proteasome activity during basal defense in a T3SS-dependent manner. A, Proteasome activity in leaves of Arabidopsis plants infected with either Pst wild-type bacteria or a Pst ∆hrcC strain lacking a functional T3SS. Samples were taken 2 d post inoculation (dpi), and the relative proteasome activity was determined. Each bar represents the mean of three biological replicates ± sd. MgCl2 infiltration serves as a mock control. Asterisks indicate statistical differences according to Student’s t test (***, P < 0.001) in comparison with the mock control. The experiment was repeated three times with similar results. B, Accumulation of ubiquitinated proteins in Arabidopsis leaves after infection with different Pst strains (top) and accumulation of the 20S subunit PBA1 (bottom). All experiments were carried out more than three times with similar results.
Suppression of Proteasome Activity by Pst Is Independent of PTI Inhibition and a Functional SA Signaling Pathway
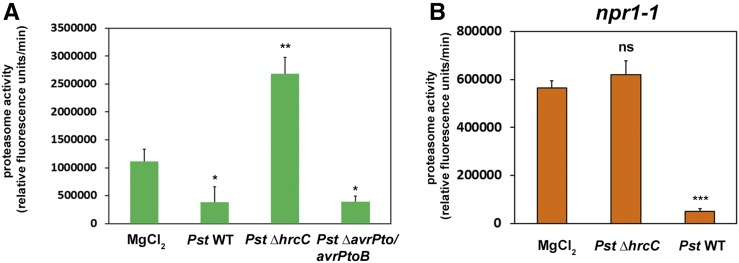
To rule out that the T3E-dependent suppression of early PTI during the Pst-Arabidopsis interaction compromises proteasome function, we assessed whether the inhibition of the proteasome is linked to PTI suppression by T3Es. To this end, we infected plants with a Pst ΔavrPto/avrPtoB deletion strain that is compromised in the suppression of early immune responses (He et al., 2006; Kvitko et al., 2009; Cunnac et al., 2011). In the absence of both T3Es, Pst showed a similar degree of proteasome inhibition than the wild type (Fig. 2A). This excludes the possibility that the inhibition of early events of PTI is responsible for the effect on proteasome function and demonstrates that neither AvrPto nor AvrPtoB is required for proteasome suppression.
Figure 2.
Suppression of proteasome induction by Pst is independent of the ability to suppress PTI and does not require a functional SA signaling pathway. A, Proteasome activity in Arabidopsis leaves infected with a Pst strain impaired in the inhibition of PTI (∆avrPto/avrPtoB) compared with the wild type and a T3SS-deficient strain (Pst ∆hrcC). Samples were taken 2 dpi, and the relative proteasome activity was determined. Each bar represents the mean of three biological replicates ± sd. MgCl2 infiltration serves as a mock control. Asterisks indicate statistical differences according to Student’s t test (**, P < 0.01 and *, P < 0.05). B, Proteasome activity in leaves of npr1-1 Arabidopsis plants 2 dpi with different Pst strains. Each bar represents the mean of three biological replicates ± sd. MgCl2 infiltration serves as a mock control. Asterisks indicate a statistical difference according to Student’s t test (***, P < 0.001); ns, not significant. Both measurements were performed two times with similar results.
Previous results showed that the proteasome is activated by SA treatment and that plants impaired in SA signaling are unable to induce proteasome activity upon infection (Gu et al., 2010; Üstün et al., 2013). In the light of these observations, we investigated whether the inhibitory effect of Pst on the proteasome is due to its ability to dampen SA signaling (DebRoy et al., 2004). Thus, we performed Pst infection assays with npr1-1 mutant lines that lack a functional SA signal transduction (Cao et al., 1997). Pst ΔhrcC was not able to induce proteasome activity in the npr1-1 mutant (Fig. 2B), supporting the notion that proteasome activity seems to be at least partially induced by NPR1-dependent SA signaling. However, Pst was still able to further inactivate the proteasome below the levels detected in the mock control (Fig. 2B). Therefore, we conclude that Pst disables proteasome function independently of its ability to suppress SA signaling. This suggests that Pst directly inhibits proteasome activity, presumably by T3Es.
Proteasome Mutants Are More Susceptible to Pathogenic and Nonpathogenic Pseudomonas spp. Strains
In order to obtain genetic evidence for the involvement of the proteasome in defense responses, a set of Arabidopsis mutant lines carrying defects in different proteasomal subunits was infected with Pst. RPT2a is a subunit of the 19S RP of the proteasome, where it gates the axial channel of the 20S core particle and controls substrate entry and product release. The Arabidopsis rpt2a-2 mutant is a T-DNA insertion with only 25% of the RPT2 protein amount compared with the wild type, due to the expression of the second RPT2 isoform encoded by the RPT2b gene (Lee et al., 2011). The rpt2a-2 plants are affected in root elongation, leaf/organ size, trichome branching, endoreduplication, inflorescence stem fasciation, and flowering time (Lee et al., 2011). RPN12a also is part of the 19S RP, where it is involved in complex assembly, and the Arabidopsis rpn12a-1 mutant was originally created by exon-trap mutagenesis and expresses an RPN12a-NPTII fusion protein whose incorporation into the 26S proteasome complex was proposed to have subtle effects on proteasome function (Smalle et al., 2002). The rpn12a-1 mutant shows decreased rates of leaf formation, reduced root elongation, delayed skotomorphogenesis, and altered growth responses to exogenous cytokinins, suggesting that the mutant has decreased hormone sensitivity (Smalle et al., 2002). Both mutant plants are defective in ubiquitin-dependent proteolysis and exhibit decreased 26S proteasome activity (Kurepa et al., 2008; Lee et al., 2011). We first tested the susceptibility of both mutant genotypes toward virulent Pseudomonas syringae pv maculicola ES4326 (Psm) and monitored bacterial multiplication and symptom development 2 d post infection. As shown in Figure 3A, rpt2a-2 and rpn12a-1 mutants supported bacterial growth to significantly higher levels than the wild-type plants and also showed accelerated symptoms (Fig. 3B). Thus, these data indicate that a fully functional proteasome is required to mount an efficient local defense response against virulent Psm. Infecting plants with the Pst strain also resulted in an enhanced bacterial proliferation in the proteasome mutants (Fig. 3C), which also was reflected by the stronger development of disease symptoms (Fig. 3D). To strengthen our genetic data, bacterial growth measurement of Pst was conducted upon pharmaceutical inhibition of the proteasome by the proteasome inhibitor MG132. In line with our previous observations, inhibition of the proteasome by MG132 also resulted in a significantly enhanced bacterial growth (Supplemental Fig. S1). Prompted by the finding that proteasome activity is highly induced upon Pst ∆hrcC infection, we next tested whether the proteasome mutant lines also show a higher sensitivity toward this strain. Measurement of bacterial growth revealed that the proteasome mutants supported more bacterial growth of the nonpathogenic Pst ∆hrcC (Fig. 3E). Thus, the proteasome seems to play a critical role during PTI and is implicated in early immune responses.
Figure 3.
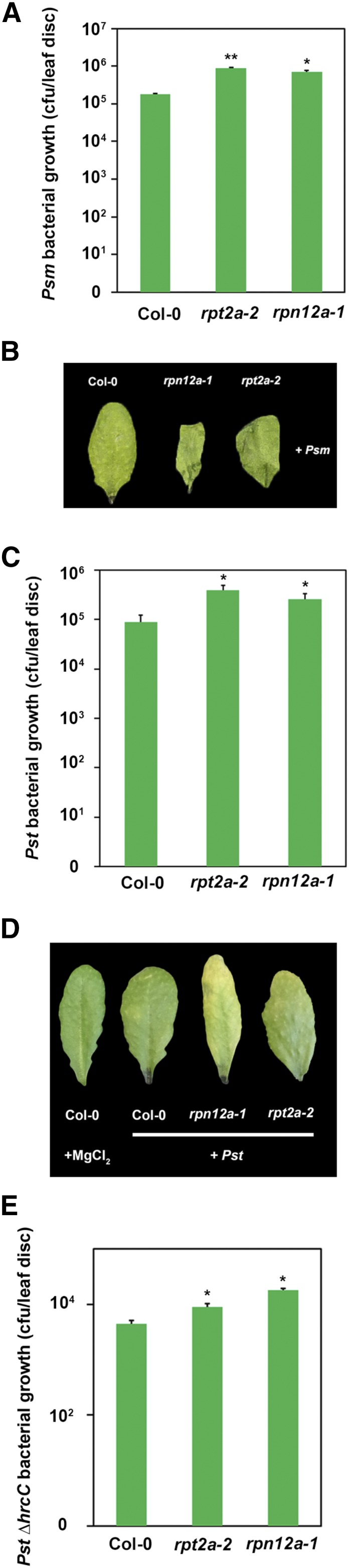
Arabidopsis mutant lines defective in different proteasome subunits display enhanced susceptibility toward infection with Pst and Psm. A, Bacterial density in leaves of different Arabidopsis genotypes infected with Psm. Leaves were syringe infiltrated with 1 × 105 colony-forming units (cfu) mL−1 bacteria, and bacterial multiplication was determined at 2 dpi. Each bar represents the mean of three biological replicates ± sd. Asterisks indicate statistical differences according to Student’s t test (**, P < 0.01 and *, P < 0.05). B, Phenotypes of Psm-infected Arabidopsis leaves 2 dpi. C, Bacterial density in leaves of different Arabidopsis genotypes infected with Pst. Leaves were syringe infiltrated with a bacterial suspension of 1 × 105 cfu mL−1, and in planta bacterial populations were determined 2 dpi. Each bar represents the mean of three biological replicates ± sd. Asterisks indicate statistical differences according to Student’s t test (*, P < 0.05). D, Phenotypes of Psm-infected Arabidopsis leaves 2 dpi. E, The bacterial multiplication of an avirulent Pst ∆hrcC strain is enhanced in leaves of Arabidopsis proteasome mutant lines. Leaves were syringe infiltrated with a bacterial suspension of 1 × 105 cfu mL−1, and bacterial multiplication was determined 2 dpi. Each bar represents the mean of three biological replicates ± sd. Asterisks indicate statistical differences according to Student’s t test (*, P < 0.05). Bacterial growth and infection assays were carried out two times with similar results.
The Proteasome Is Required for PTI
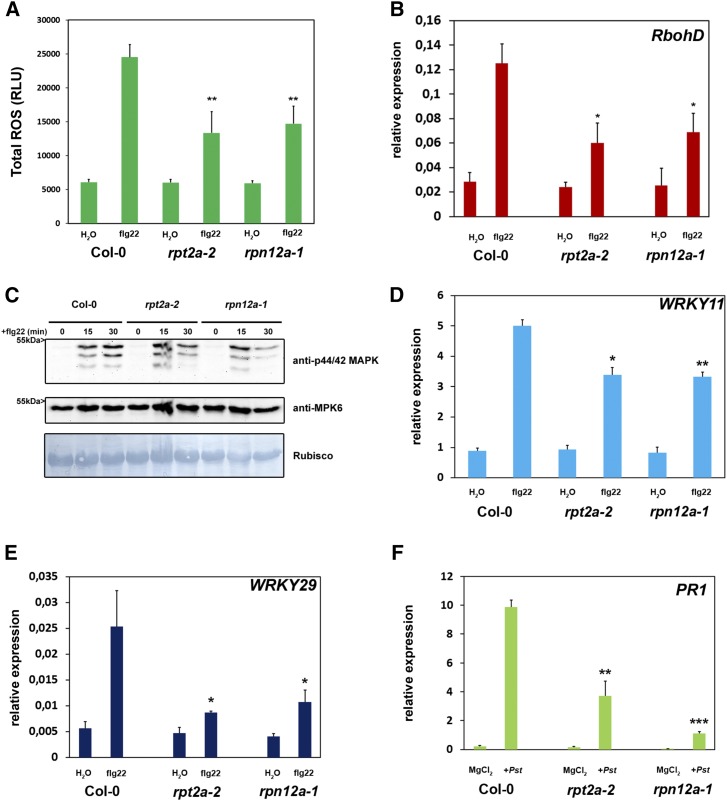
Because the proteasome is involved in local defense responses toward pathogenic and nonpathogenic bacteria, we further analyzed the role of the proteasome during classical PTI responses. Recognition of flg22 by the PRR FLS2 (Gómez-Gómez and Boller, 2000) leads to an oxidative burst, one of the first measurable responses of plants to PAMP perception (Nicaise et al., 2009). ROS production in rpt2a-2 and rpn12a-1 plants was significantly attenuated compared with the Columbia-0 (Col-0) wild-type plants upon flg22 treatment (Fig. 4A), indicating that early PTI responses such as ROS generation are partially dependent on a functional proteasomal turnover. In accordance with the decreased oxidative burst, the transcriptional response of RbohD, encoding an NADPH oxidase that is essential for flg22-triggered ROS production, was dampened in the proteasome mutant lines compared with Col-0 plants (Fig. 4B). Activation of PTI also leads to the activation of MAPK cascades and the subsequent phosphorylation of MPK3, MPK6, and MPK4/11. Thus, we analyzed whether this signaling cascade is altered in the proteasome mutants. Immunoblot analysis using an antibody against phosphorylated MPK3, MPK6, and MPK4/11 revealed that, in comparison with Col-0 plants, both proteasome mutants exhibited impaired kinetics of MAPK activation, as the phosphorylation signal faded out more rapidly in the rpt2a-2 and rpn12a-1 mutants (Fig. 4C).
Figure 4.
The proteasome is required for PAMP-triggered responses. A, Total production of ROS in relative light units (RLU) during treatment with 1 µm flg22 for 45 min. Each bar represents the mean of four biological replicates ± sd. Statistical significance compared with Col-0 plants treated with flg22 is indicated by asterisks (Student’s t test; **, P < 0.01). ROS production was evaluated in at least two independent experiments with similar results. B, Quantitative real-time PCR (RT-PCR) of ROS (RboHD) after flg22 treatment is reduced in proteasome mutants. Plants were treated with 1 µm flg22 or water (control). UBC9 (for ubiquitin carrier protein) was used as a reference gene. Similar results were obtained in three independent experiments. Each bar represents the mean of three biological replicates ± sd. Changes in fold expression are significant for all genes in comparison with the wild type (+flg22) according to Student’s t test (*, P < 0.05) C, MAPK signaling is impaired in proteasome mutant lines. Twelve-day-old seedlings were treated with 1 µm flg22, and samples were collected 0 to 30 min after treatment as indicated. Activated MAPKs were detected by immunoblotting using anti-p44/42 MAPK antibody. Proteins also were detected with anti-AtMPK6 antibody, and Amido Black staining shows equal loading. Experiments were conducted twice with similar results. D and E, PAMP-dependent induction of PTI marker genes is impaired in Arabidopsis proteasome mutant plants. Quantitative RT-PCR is shown for immune response marker genes (WRKY11 and WRKY29) 60 min after flg22 treatment. Plants were treated with 1 µm flg22 or water (control). UBC9 was used as a reference gene. Similar results were obtained in three independent experiments. Each bar represents the mean of three biological replicates ± sd. Changes in fold expression are significant for all genes in comparison with the wild type (+flg22) according to Student’s t test (**, P < 0.01 and *, P < 0.05). F, Relative PR1 expression is decreased in proteasome mutant lines in response to Pst infection. Plants were infiltrated with Pst, and gene expression was analyzed 24 h post inoculation. Each bar represents the mean of three biological replicates ± sd. Changes in gene expression are significant for all genes in comparison with the wild type (+Pst infection) according to Student’s t test (**, P < 0.01 and ***, P < 0.001). All experiments analyzing PTI responses were performed two times with similar results.
In order to assess whether PTI is perturbed at the transcriptional level, we investigated the expression of PTI marker genes, WRKY11 and WRKY29, in rpt2a-2 and rpn12a-1 mutant plants upon flg22 stimulus (Stegmann et al., 2012). Real-time PCR revealed that the transcriptional activation of both PTI marker genes was diminished significantly in the proteasome mutant lines (Fig. 4, D and E), providing further indications for a compromised PTI response in the proteasome mutants. Moreover, activation of PR1 expression, a marker gene for the SA pathway, also was reduced in the proteasome mutants compared with the control (Fig. 4F). Taken together, these data indicate that a fully functional proteasome is required during early and late PTI responses.
Role of the Proteasome in Defense Priming during SAR
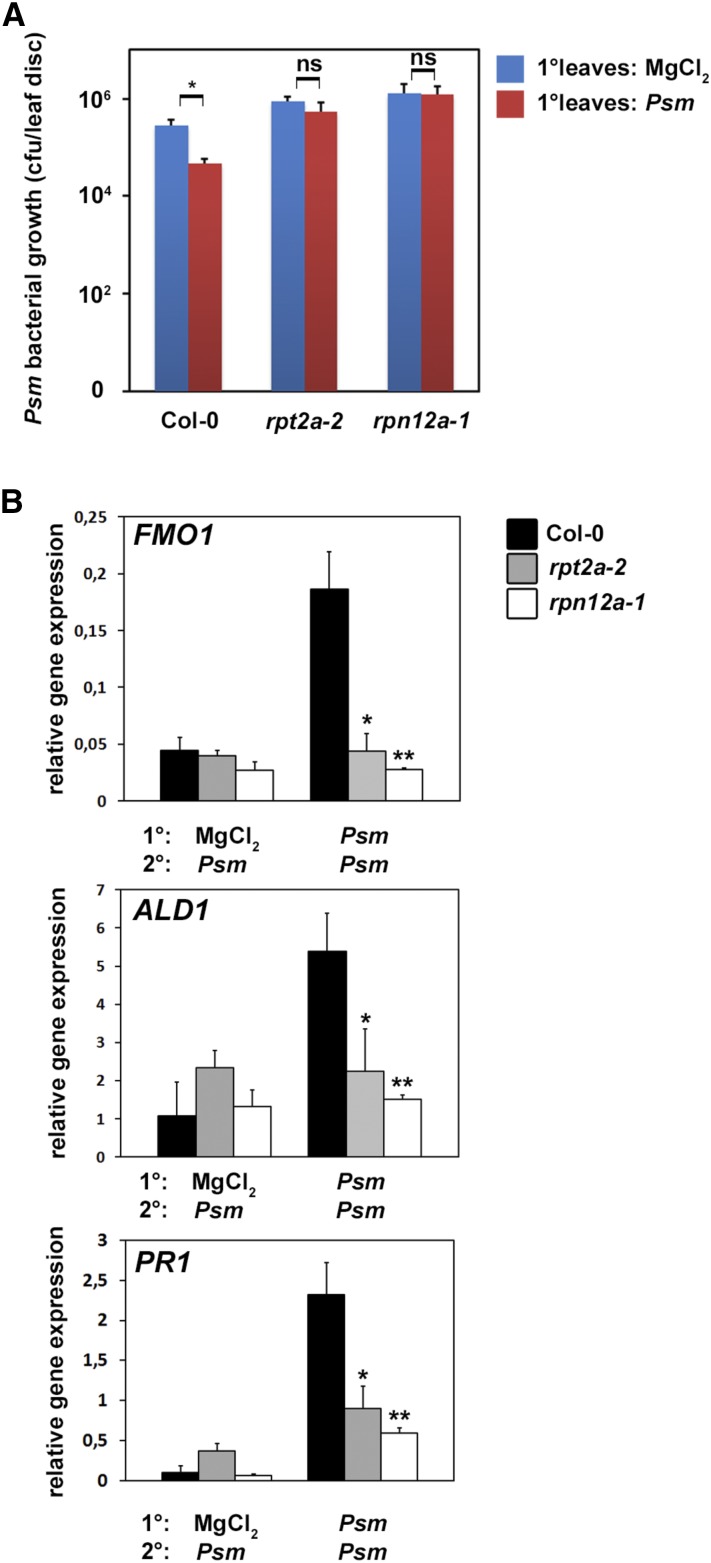
Based on the altered expression of the SA-inducible PR1 gene in the proteasome mutant lines (Fig. 4F), we investigated the role of the proteasome in defense priming during SAR. The Arabidopsis proteasome mutants rpt2a-2 and rpn12a-1 were infected in lower (1°) leaves with the SAR-inducing pathogen Psm (Návarová et al., 2012). Analysis of bacterial growth after a secondary infection of systemic (2°) leaves 2 d after the primary infection revealed that bacterial multiplication was inhibited significantly in systemic leaves of wild-type Col-0 plants (Fig. 5A). This indicates that the primary infection with Psm induced effective SAR and primed the plants for a secondary infection. In contrast, there was no significant difference in bacterial growth in systemic leaves of the proteasome mutants compared with the unprimed mock control. Thus, the establishment of SAR is impaired in proteasome mutants. This suggests that the proteasome plays an important role not only during local defense responses but also in the establishment of systemic defense priming and SAR.
Figure 5.
The proteasome is required for SAR. A, SAR assay in Col-0, rpt2a-2, and rpn12a-1 plants. 1° leaves were infiltrated with either 10 mm MgCl2 or Psm (optical density [OD] = 0.005), and 2 d later, three 2° leaves were challenge infected with Psm (OD = 0.001). Bacterial growth in 2° leaves was assessed 2 d after 2° leaf inoculation. Bars represent means of three biological replicates ± sd. The asterisk denotes a statistically significant difference between the indicated samples (* P < 0.05; Student’s t test); ns, not significant. B, SAR priming of defense gene expression. Relative ALD1, FMO1, and PR1 expression is shown at 10 h after treatment of 2° leaves. Transcript levels were assessed by quantitative real-time PCR analysis from three replicate samples (n = 3). Significant differences in comparison with Col-0 (Psm/Psm infected) were calculated using Student’s t test and are indicated by asterisks: *, P < 0.05 and **, P < 0.01. Priming experiments were performed two times with similar results.
To further corroborate this finding at the molecular level, SAR marker gene expression was analyzed in 2° leaves of plants primed with Psm infection in 1° leaves. FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1) is a critical component of SAR establishment required for SA accumulation in systemic, noninoculated leaves (Mishina and Zeier, 2006), while AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) represents the aminotransferase required for the biosynthesis of the SAR signaling metabolite pipecolic acid (Návarová et al., 2012). As shown in Figure 5B, both FMO1 and ALD1 mRNA levels were strongly induced in 2° leaves of wild-type Arabidopsis plants primed with Psm infection in 1° leaves compared with the unprimed control. On the contrary, rpt2a-2 and rpn12a-1 plants displayed no induction of FMO1 or ALD1 expression in challenge-infected 2° leaves irrespective of the type of treatment of the 1° leaf. Thus, rpt2a-2 and rpn12a-1 plants lack induction of the essential SAR components FMO1 and ALD1 in systemic tissue upon a priming infection with Psm in 1° leaves. In order to analyze gene expression changes downstream of FMO1 and ALD1, we determined the expression of PR1 upon systemic infection of primed plants. PR1 expression in systemic leaves of primed rpt2a-2 and rpn12a-1 plants was reduced significantly and displayed no induction in primed proteasome mutants (Fig. 5B). Taken together, these data indicate that a disturbance of proteasome function has repercussions on SAR gene induction.
Pst T3Es Inhibit the Proteasome
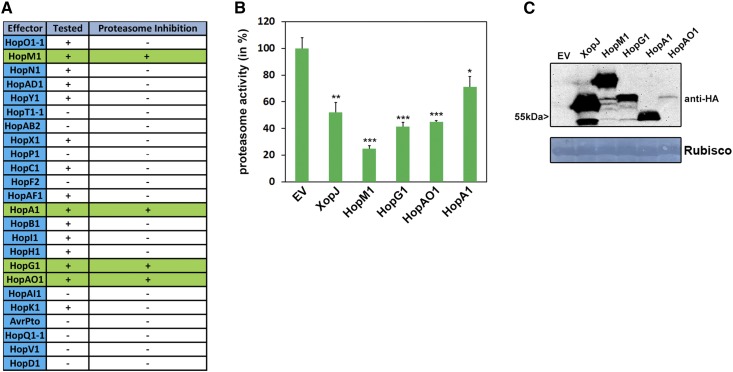
In our initial experiments, Pst suppressed proteasome activity in a type III secretion-dependent manner. In order to identify the effectors suppressing the proteasome activity, we transiently expressed Pst T3Es in Nicotiana benthamiana. In total, we tested 16 Pst T3Es after transient expression in N. benthamiana for their ability to inhibit proteasome activity (Fig. 6A). The effector proteins AvrPto and AvrPtoB were excluded from this analysis, as previous assays using a Pst ΔavrPto/avrPtoB deletion strain showed that this strain was still able to compromise proteasome function (Fig. 2A). This experimental approach identified four T3Es (HopM1, HopG1, HopAO1, and HopA1) whose expression reproducibly led to a significant reduction in proteasome activity in N. benthamiana leaves (Fig. 6B). Notably, the inhibitory effect on the proteasome of HopM1, HopG1, and HopAO1 in transient assays was even stronger than that of XopJ, a T3E from X. campestris, which was shown to inhibit the proteasome by degrading the RP subunit RPT6 (Üstün and Börnke, 2015). The expression of all T3Es tested was verified by immunoblotting using an anti-hemagglutinin (HA) antibody (Fig. 6C). The remaining 12 T3Es from Pst tested were not able to suppress proteasome activity (Supplemental Figs. S2–S7), demonstrating that the interference with proteasome activity is specific for certain T3Es. This suggests that the proteasome represents a virulence target for Pst and that T3Es from Pst either directly or indirectly impede proteasome function.
Figure 6.
T3Es from Pst suppress proteasome activity in N. benthamiana. A, Table of candidate effectors tested for their ability to modulate proteasome function. T3Es tested in proteasome activity measurements are indicated by +, and those not tested are indicated by −. T3Es suppressing proteasome activity are highlighted in green. B, Transient expression of T3Es HopM1, HopG1, HopAO1, and HopA1 in N. benthamiana suppresses proteasome activity. Proteasome activity is shown in N. benthamiana leaves following transient expression of T3Es XopJ, HopM1, HopG1, HopA1, HopAO1, or empty vector control (EV). Relative proteasome activity in total protein extracts was determined by monitoring the breakdown of the fluorogenic peptide Suc-LLVY-AMC at 30°C in a fluorescence spectrophotometer. The empty vector control was set to 100%. Data represent means ± sd (n = 3). Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) determined by Student’s t test (compared with the empty vector control). C, Protein extracts from N. benthamiana leaves transiently expressing T3Es tagged with HA and empty vector at 48 h post inoculation. Equal volumes representing approximately equal protein amounts of each extract were immunoblotted, and proteins were detected using anti-HA antiserum. Amido Black staining served as a loading control. Proteasome activity measurements were carried more than three times with these T3Es.
HopM1 Inhibits Proteasome Activity during the Pst-Arabidopsis Interaction
Because HopM1 inhibits proteasome activity up to 80% and based on its previous association with protein degradation (Nomura et al., 2006), we concentrated our efforts on this candidate T3E. We first infected Arabidopsis plants with a Pst ∆CEL mutant strain that lacks six open reading frames present in the Conserved Effector Locus (CEL), including the core T3Es AvrE and HopM1 (Alfano et al., 2000). The Pst ∆CEL mutant multiplies 200- to 500-fold less than Pst and fails to produce disease symptoms in Col-0 leaves. Complementation analysis suggests that HopM1 is responsible mainly for the Pst ∆CEL mutant phenotype (Nomura et al., 2006). Plants infected with Pst ∆CEL display proteasome activity comparable to untreated plants (Fig. 7A). This result indicates that HopM1 renders Pst unable to inhibit proteasome activity during infection below the basal level but is still able to prevent its induction.
Figure 7.
Pst T3E HopM1 is required for proteasome inhibition during the Pst-Arabidopsis interaction. A, Proteasome activity (in relative fluorescence units [RFU]) in leaves of Arabidopsis plants infected with Pst wild-type bacteria, Pst ∆hrcC, and Pst ∆CEL harboring HopM1. Samples were taken 2 dpi, and the relative proteasome activity was determined. Each bar represents the mean of three biological replicates ± sd. MgCl2 infiltration serves as a mock control. Asterisks indicate statistical differences according to Student’s t test (* P < 0.05 and ***, P < 0.001). The experiment was repeated three times with similar results. B, Accumulation of ubiquitinated proteins in Arabidopsis leaves after infection with different Pst strains determined using an anti-ubiquitin antibody. C, Proteasome activity in leaves of Arabidopsis plants infected with Pst wild-type bacteria, Pst ∆hrcC, and Pst ∆hopM1. Samples were taken 2 dpi, and the relative proteasome activity was determined. Each bar represents the mean of three biological replicates ± sd. MgCl2 infiltration serves as a mock control. Asterisks indicate statistical differences according to Student’s t test (*, P < 0.05). The experiment was repeated two times with similar results. D, Accumulation of ubiquitinated proteins in Arabidopsis leaves after infection with different Pst strains determined using an anti-ubiquitin antibody. All experiments including the Pst ∆CEL and ∆hopM1 strains were carried out three times with similar results.
Consistently, the accumulation of ubiquitinated proteins also was less pronounced compared with Pst wild-type infected leaves (Fig. 7B). Because this conserved effector locus also harbors AvrE, HopAA1-1, and HopN1 besides HopM1 (Kvitko et al., 2009), we decided to test a Pst strain that only carries a deletion in HopM1 (Pst ∆hopM1). Infection of Arabidopsis plants with Pst ∆hopM1 shows that this deletion strain is not able to reduce proteasome activity compared with the wild-type Pst strain (Fig. 7C). This is also partially reflected by a reduced accumulation of ubiquitinated proteins compared with Pst-infected plants (Fig. 7D). Thus, HopM1 is responsible for the inhibition of proteasome activity during the compatible interaction of Pst and Arabidopsis.
HopM1 Interacts with Components of the UPS in Vivo
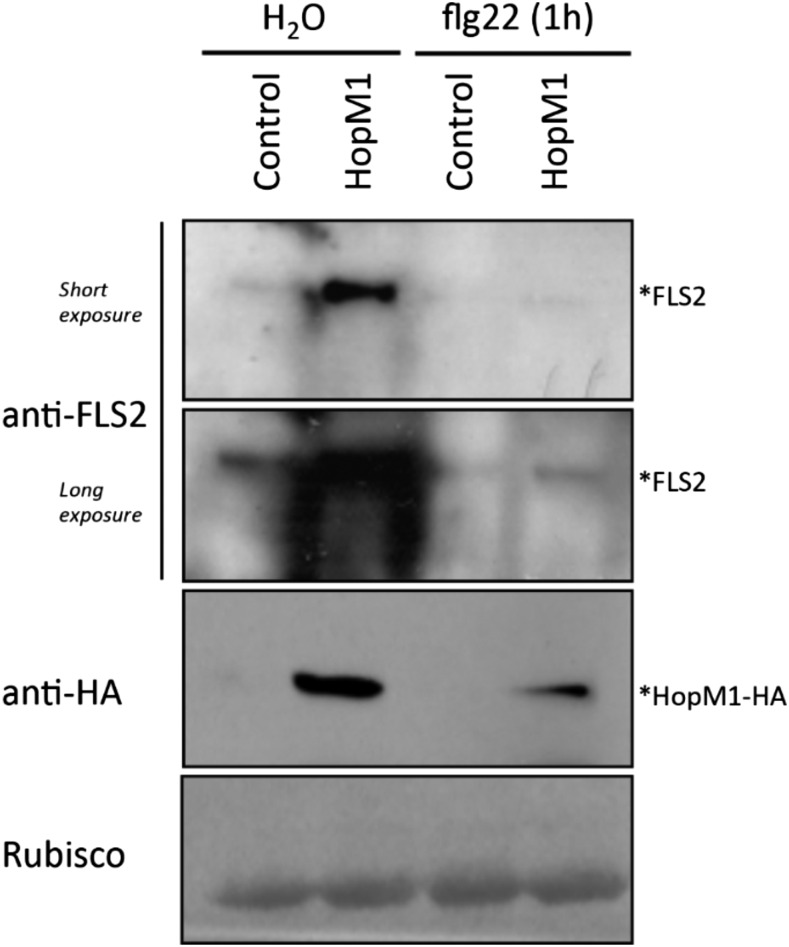
To demonstrate the molecular mechanism by which HopM1 reduces total proteasome activity, an unbiased proteomics-based experiment was performed to find the in vivo interactors of HopM1. After transient expression, HopM1 was immunoprecipitated from leaf extracts and interacting proteins were identified using liquid chromatography-tandem mass spectrometry (MS/MS) analysis. The expression of HopM1 in N. benthamiana was monitored by anti-GFP immunoblotting (Supplemental Fig. S8). In accordance with our previous results, many proteins related to the ubiquitin-proteasome system were identified as HopM1 interactors. The N. benthamiana orthologs of known HopM1 interactors, such as Arabidopsis AtMIN7 (a host ADP ribosylation guanine nucleotide-exchange factor involved in membrane traffic) and AtMIN10 (a 14-3-3 protein), also were detected, reflecting that the interactions were specific to HopM1 (Nomura et al., 2006). After performing Fisher’s exact test (P ≤ 0.05), a number of proteins related to 26S proteasome non-ATPase regulatory subunits 2, 3, 6, 12, and 14 were enriched significantly in the two independent experiments (Table I). The E3 ubiquitin protein ligases UPL1 and UPL3 also were enriched significantly, further suggesting the role of HopM1 in directly perturbing the proteasome activity. However, the most interesting protein detected was the proteasome-associated protein ECM29, which is known to stop protein degradation by inhibiting the proteasomal ATPase activity in yeast (De La Mota-Peynado et al., 2013). The flg22 receptor FLS2 is a known target of direct ubiquitination after PAMP elicitation and subsequent degradation by the proteasome (Lu et al., 2011). To analyze the possible effect of HopM1-mediated inhibition of proteasome activity on PTI signaling, the protein levels of FLS2 were monitored in Arabidopsis protoplasts transfected with HopM1. The FLS2 protein accumulated in HopM1-expressing protoplasts prior to PAMP elicitation. Significantly higher levels of FLS2 also were observed after longer (60-min) treatments with flg22 (Fig. 8).
Table I. List of UPS-related proteins coimmunoprecipitated with HopM1.
Significant values (Fisher’s exact test, P ≤ 0.05) are indicated in boldface. The top two sections list the 26S proteasome- and ubiquitin-related proteins, while the bottom section includes some of the known interacting proteins of HopM1 in Arabidopsis. The experiment was repeated twice as indicated. The full mass spectrometry data set of HopM1 interactions from both repeats was submitted to the PRIDE database and can be accessed at http://www.ebi.ac.uk/pride/archive/. Rep, Repetition.
| Name of the Protein | Accession No. | Mass | Fisher’s Exact Test (P ≤ 0.05) | Total Unique Peptide Count |
Arabidopsis Homolog | |||
|---|---|---|---|---|---|---|---|---|
| GFP Control |
HopM1 |
|||||||
| Rep 1 | Rep 2 | Rep 1 | Rep 2 | |||||
| kD | ||||||||
| 26S proteasome-related proteins | ||||||||
| Cluster of proteasome-associated protein ECM29 | NbS00044170g0008.1_SGN | 198 | <0.00010 | 1 | 0 | 18 | 22 | AT2G26780 (ARM repeat superfamily protein) |
| Cluster of 26S proteasome regulatory subunit | NbS00007460g0008.1_SGN [2] | 142 | 0.023 | 8 | 1 | 14 | 14 | AT2G32730 (26S proteasome regulatory complex, non-ATPase subcomplex, Rpn2/Psmd1 subunit) |
| Cluster of probable 26S proteasome non-ATPase regulatory subunit 3 | NICBE_198688.1_TGAC [3] | 55 | <0.00010 | 4 | 0 | 15 | 22 | AT1G75990 (PAM domain, PCI/PINT-associated module protein) |
| Cluster of 26S protease regulatory subunit 6B homolog | NICBE_268659.1 TGAC [4] | 64 | 0.08 | 6 | 0 | 8 | 7 | AT5G58290 (RPT3, regulatory particle AAA ATPase3) |
| Cluster of 26S protease non-ATPase regulatory subunit 2 1A | NICBE_166405.1_TGAC [2] | 98 | < 0.00010 | 6 | 0 | 19 | 21 | AT2G20580 (RPN1A, 26S proteasome regulatory subunit S2 1A) |
| 26S protease regulatory subunit 7 homolog A | NICBE_154102.1_TGAC (+2) | 48 | 0.0013 | 2 | 0 | 13 | 11 | AT1G53750 (RPT1A, regulatory particle AAA 1A) |
| Cluster of 26S protease regulatory subunit 6A homolog | NICBE_248120.1_TGAC [4] | 53 | 0.018 | 2 | 0 | 12 | 3 | AT3G05530 (ATS6A.2, RPT5A, regulatory particle AAA ATPase5A) |
| Cluster of 26S proteasome non-ATPase regulatory subunit 11 | NbS00016436g0001.1_SGN [4] | 47 | 0.08 | 3 | 0 | 9 | 4 | AT1G29150 (ATS9, non-ATPase subunit 9; RPN6, regulatory particle non-ATPase6) |
| Cluster of 26S proteasome non-ATPase regulatory subunit 2 1A | NICBE_172900.1_TGAC [2] | 98 | 0.0011 | 6 | 0 | 15 | 18 | AT2G20580 (ATRPN1A, RPN1A, 26S proteasome regulatory subunit S2 1A) |
| Cluster of 26S proteasome non-ATPase regulatory subunit 12 | NICBE_244180.1_TGAC [2] | 51 | 0.0069 | 1 | 0 | 10 | 7 | ATSG09900 (EMB2107, embryo-defective2107; MSA, mariposa; RPN5A, regulatory particle non-ATPase subunit 5A) |
| Probable 26S proteasome non-ATPase regulatory subunit 6 | NICBE_167161.1_TGAC (+1) | 44 | 0.033 | 2 | 0 | 7 | 8 | AT4G24820 (RPN7, 26S proteasome regulatory subunit) |
| 26S protease regulatory subunit 8 homolog A | NICBE_417606.1_TGAC (+1) | 47 | 0.077 | 2 | 1 | 8 | 7 | AT5G19990 (ATSUG1, RPT6A, regulatory particle AAA ATPase6A) |
| Cluster of 26S proteasome regulatory subunit S10B homolog B | NICBE_194312.1_TGAC | 44 | 0.13 | 3 | 0 | 7 | 6 | AT1G45000 (AAA-type ATPase family protein) |
| 26S proteasome non-ATPase regulatory subunit 14 | NICBE_364948.l_TGAC (+1) | 45 | 0.039 | 0 | 0 | 6 | 2 | AT5G23540 (Mov34/MPN/PAD-l family protein) |
| 26S proteasome non-ATPase regulatory subunit 7 | NbS00041405g0012.1_SGN (+1) | 35 | 0.48 | 3 | 0 | 2 | 6 | AT5G05780 (AE3, asymmetric leaves enhancer3; ATHMOV34; RPN8A, RP non-ATPase subunit 8A) |
| Ubiquitin-related proteins | ||||||||
| Cluster of ubiquitin protein ligase1 | NbS00002172g0012.1_SGN [6] | 319 | <0.00010 | 2 | 0 | 59 | 23 | AT1G55860 (UPL1, ubiquitin-protein ligase1) |
| Cluster of auxin transport protein BIG | NbS00009154g0017.1_SGN [2] | 537 | <0.00010 | 4 | 0 | 41 | 16 | AT3G02260 (ASA1, attenuated shade avoidance1; BIG; CRM1, corymbosa1; DOC1, dark overexpression of CAB1; LPR1, low phosphate-resistant root1; TIR3, transport inhibitor response3; UMB1, umbrella1) |
| Cluster of E3 ubiquitin protein ligase UPL3 | NbS00031527g0002.1_SGN [3] | 199 | 0.0048 | 1 | 0 | 16 | 1 | AT4G38600 (KAK, kaktus; UPL3, ubiquitin-protein ligase3) |
| Cluster of E3 ubiquitin protein ligase UPL1 | NICBE_298798.1_TGAC [2] | 389 | 0.0016 | 0 | 0 | 7 | 9 | AT1G55860 (UPL1, ubiquitin-protein ligase1) |
| Ubiquitin conjugation factor E4 | NbS00018754g0010.1_SGN | 91 | 0.26 | 3 | 0 | 8 | 3 | AT5G15400 (MUSE3, mutant, SNC1-enhancing3) |
| Cluster of ubiquitin-conjugating enzyme E2 36 | NICBE_251503.1_TGAC [2] | 17 | 0.11 | 1 | 0 | 8 | 0 | AT1G16890 (ATUBC36, UBC36, ubiquitin-conjugating enzyme36) |
| Cluster of E3 ubiquitin protein ligase listerin | NbS00000796g0017.1_SGN | 135 | 0.039 | 0 | 0 | 2 | 6 | AT5G58410 (HEAT/U-box domain-containing protein) |
| Probable ubiquitin conjugation factor E4 | NICBE_244276.1_TGAC | 83 | 0.56 | 3 | 0 | 5 | 2 | AT5G15400 (MUSE3, mutant, SNC1-enhancing3) |
| Known HopM1 interactions | ||||||||
| Cluster of ARF guanine nucleotide-exchange factor 2 | NbS00006288g0001.1_SGN [3] | 207 | <0.00010 | 5 | 0 | 28 | 29 | AT3G43300 (MIN7-like) |
| Cluster of Sec7 guanine nucleotide-exchange factor | NbS00008405g0008.1_SGN [2] | 171 | <0.00010 | 5 | 0 | 35 | 46 | AT3G43300 (MIN7-like) |
| Sec7 guanine nucleotide-exchange factor | NbS00016714g0003.1_SGN | 197 | 0.0052 | 0 | 0 | 9 | 4 | AT3G43300 (M1N7-like) |
| Cluster of 14-3-3-1-like protein 16R | NICBE_421663.1_TGAC [4] | 29 | 0.14 | 7 | 0 | 17 | 7 | AT4G24150 (MIN10-like) |
Figure 8.
HopM1 leads to the accumulation of FLS2 protein. Western blots show protein levels in Arabidopsis Col-0 protoplasts transformed with either control or HopM1-HA using FLS2-specific antibody. The protoplasts were harvested after treating with water as a control or 100 nm flg22 for 1 h. The experiment was repeated at least three times with similar results.
DISCUSSION
Plant immunity has to be tightly regulated to ensure effective immune response activation with minimal negative effects on plant growth and reproduction. One way to regulate certain immune processes is the proteasome-mediated recycling of defense components, highlighting an important role of the plant proteasome in plant immunity. Thus, evidence has accumulated that the 26S proteasome regulates plant defense responses at several layers of the surveillance system and, hence, constitutes a strategic target for plant pathogens (Marino et al., 2012; Dudler, 2013).
In this study, we show that the fully functional proteasome subunits RPT2a and RPN12a are required for the proper execution of PTI events in locally infected leaves and the establishment of SAR. Thus, these subunits are essential for full resistance against Pseudomonas spp. bacteria and also for the growth restriction of virulent bacteria in local and systemic tissue. Furthermore, we show that Pst is able to inhibit proteasome activity and that this phenomenon is dependent on T3E injection. A screen of a Pst T3E collection identified candidate effector proteins for proteasome interference.
In the past, the proteasome was associated mainly with the regulation of plant growth and development, because the UPS plays an essential role in almost all aspects of hormone perception and signaling. Not surprisingly, Arabidopsis proteasome subunit mutants show a range of developmental defects (Kurepa and Smalle, 2008). This is also true for the Arabidopsis rpt2a-2 and rpn12a-1 mutants used in this study, both of them being part of the 19S RP of the proteasome. The rpt2a-2 mutant is affected in root elongation, leaf/organ size, trichome branching, endoreduplication, inflorescence stem fasciation, and flowering time (Lee et al., 2011). The rpn12a-1 mutant shows a decreased rate of leaf formation, reduced root elongation, delayed skotomorphogenesis, and altered growth responses to exogenous cytokinins (Smalle et al., 2002). Both mutants are impaired in ubiquitin-dependent degradation and have reduced proteasome activity, and further studies with rpt2a-2 mutants revealed that the assembly of the 26S proteasome is impaired (Kurepa et al., 2008; Lee et al., 2011).
Beyond their role in the regulation of plant growth and development, certain proteasome subunits also have been implicated in plant immunity. For instance, a mutation in the 19S RP subunit RPN1a was identified as a suppressor of increased resistance to the adapted biotrophic powdery mildew pathogen Golovinomyces cichoracearum in ENHANCED DISEASE RESISTANCE2 (EDR2) loss-of-function mutants (Yao et al., 2012). The Arabidopsis rpn1a single mutant was more susceptible toward local infection with P. syringae strains as well as to G. cichoracearum (Yao et al., 2012). Infected rpn1a plants have reduced late defense responses, such as the accumulation of the defense hormone SA and a reduced expression of the defense marker gene PR1. The observation that the rpt2a and rpn12a mutants show similar enhanced susceptibility phenotypes and reductions in PR1 expression suggests that interference with proteasome function in general leads to compromised immunity. Another report shows that RPT2a is involved in the defense response mediated by a coiled coil-nucleotide-binding site-leucine-rich repeat protein (Chung and Tasaka, 2011). In that case, RPT2a interacts with the resistance protein UNI/uni-1D and a loss of RPT2a in the constitutively active uni-1D mutant represses PR1 gene expression (Igari et al., 2008; Chung and Tasaka, 2011).
From a number of additional Arabidopsis proteasome subunit mutants tested, only those affected in RPT2a and RPN8a function fully suppressed edr2-mediated powdery mildew resistance, indicating that the proteasome subunits have distinct roles in plant defense responses (Yao et al., 2012). Consistent with this scenario, Hatsugai et al. (2009) reported that the proteasome subunit PBA1 functions as a plant CASPASE3-like enzyme. PBA1 RNA interference plants have reduced DEVDase activity besides a decreased overall proteasome activity (Hatsugai et al., 2009). This defect abolished the membrane fusion associated with both disease resistance and HR in response to avirulent bacterial strains but not to a virulent strain. As a consequence of this compromised HR, PBA1 RNA interference plants display enhanced susceptibility toward avirulent Pseudomonas spp. strains, while the growth of virulent Pst is comparable to that in Arabidopsis wild-type plants (Hatsugai et al., 2009).
A possible explanation for the enhanced susceptibility phenotype of rpn1a plants is that a defect in proteasome function interferes with the turnover of a regulator of SA signaling and, thus, prevents the onset of SA-mediated defense (Yao et al., 2012). A similar scenario could at least in part explain the compromised immunity in rpt2a-2 and rpn12a-1 mutants. A possible candidate for proteasomal turnover during defense is the master regulator of SA signaling, NPR1, whose functionality was shown to be dependent on continuous proteasomal degradation (Spoel et al., 2009). Compromised NPR1 function due to reduced proteasomal turnover also would explain the reduced expression of the NPR1-regulated PR1 gene in proteasome mutants.
However, proteasomal degradation also was reported for immune components acting at early stages of PTI (e.g. the receptor kinase FLS2). Previous studies revealed that FLS2 is ubiquitinated by the E3 ligases PUB12 and PUB13, leading to its degradation via the proteasome (Lu et al., 2011). The pub12 and pub13 mutants displayed elevated immune responses to flagellin treatment, indicating that these E3 ligases act as negative regulators of PTI. The observation that rpt2a and rpn12a plants show reduced induction of PTI marker genes (WRKY11 and WRKY29) suggests that the proteasome also can act as a positive regulator of induced immunity. Compromised proteasome function, for instance, could interfere with the turnover of regulators of PTI gene expression. It has been shown in rice (Oryza sativa) that WRKY45 degradation via the proteasome is required for the activity of WRKY45 as a transcriptional activator of immunity (Matsushita et al., 2013). The likely Arabidopsis ortholog of rice WRKY45 is WRKY70. Proteasomal turnover has not been demonstrated to be required for WRKY function in Arabidopsis, but it is conceivable that similar mechanisms are involved in fine-tuning defense gene expression.
The altered kinetics of MAPK phosphorylation supports the hypothesis that upstream PRR signaling, such as FLS2 degradation, is disturbed in the proteasome mutants but does not exclude direct effects of proteasome activity on MAPK signaling.
The generation of ROS also depends on phosphorylation events during the first steps of PAMP recognition. The FLS2-associated kinase BIK1 directly interacts with and phosphorylates RBOHD, which is the NADPH oxidase that generates ROS (Kadota et al., 2014). Continuous proteasome-mediated degradation of BIK1 is required to ensure optimal immune outputs (Monaghan et al., 2014). It is possible that the turnover of BIK1 is affected in the rpt2a-2 and rpn12a-1 mutant plants, supporting our findings that ROS production is perturbed in plants with lowered proteasome activity.
Apart from its function in regulating the turnover of components implicated in ROS production, proteasome components have been identified to directly contribute to ROS-mediated defense. In N. benthamiana, the expression of three genes encoding subunits of the 20S proteasome is induced after treatment with the elicitor cryptogein (Dahan et al., 2001; Suty et al., 2003). N. benthamiana cell lines overexpressing the β1-subunit have decreased levels of NtRbohD gene induction and oxidative burst after cryptogein treatment, indicating that this subunit acts as a negative regulator of early plant responses to cryptogein (Lequeu et al., 2005). Because a loss of RPT2a leads to an accumulation of PBA1 (Lee et al., 2011), the homolog of the N. benthamiana β1-subunit, it is tempting to speculate that ROS signaling is impaired in these plants in a similar manner.
In addition to a reduced local defense response, rpt2a-2 and rpn12a-1 mutants also are compromised in the establishment of SAR. The mounting of SAR requires the generation of signal in locally infected leaves, which is transmitted subsequently to systemic tissue, where it is perceived and confers a primed state that enables a faster and stronger defense response upon a secondary infection (Conrath et al., 2015). The exact nature of the signal(s) involved in this process is currently up for debate; however, it is proposed that several hormonal pathways, such as SA, ethylene, auxin, and jasmonic acid, play roles in SAR (Pastor et al., 2014). Because of its involvement in nearly all aspects of hormonal signaling, it appears reasonable to assume that the proteasome could play a critical role for the execution of the different phases of priming. Based on genetic and physiological evidence, SA plays a pivotal role in SAR (Fu and Dong, 2013). SA signaling strongly depends on NPR1 as a central regulator, and interference with the proteasomal turnover of NPR1 also would affect SAR and defense priming in rpt2a-2 and rpn12a-1 mutants. However, recent evidence suggests that the nonprotein amino acid pipecolic acid is a critical SAR regulator (Návarová et al., 2012). Pipecolic acid elevations are indispensable for SAR and necessary for virtually the whole transcriptional SAR response, although a moderate but significant SA-independent component of SAR activation was revealed recently (Bernsdorff et al., 2016). Future experiments will have to clarify at which step the initiation of SAR is affected by a defect in proteasome function.
Measurement of the overall proteasome activity in leaves infected with a nonpathogenic Pst ∆hrcC strain revealed a significant induction of proteasome activity. Thus, increased proteasome activity appears to be part of the defense response induced by Pst bacteria unable to deliver T3Es into the potential host cell. Consistent with this finding, previous work demonstrated that an X. campestris pv vesicatoria strain lacking a functional T3SS also induced proteasome activity upon infection of pepper (Capsicum annuum) plants (Üstün et al., 2013). This argues for a contribution of early signaling events, such as the recognition of PAMPs and subsequent phosphorylation events, to the induction of the proteasome activity. In accordance with that, the bacterial PAMP flg22 has been shown to activate proteasome peptidase activity upon application, leading to alterations in posttranslational modifications in certain proteasome subunits (Sun et al., 2013). Treatment with flg22 also resulted in an accumulation of ubiquitinated proteins (Sun et al., 2013), which is in line with our observation that massive protein turnover leads to an enhanced ubiquitination of proteins during PTI.
The npr1-1 mutant, which is defective in SA-dependent defense responses, does not display any induction of proteasome activity after infection with nonpathogenic Pst, suggesting that the elevation of proteasomal activity during induced defense depends on the SA signaling pathway. Previous work implies that SA acts at the transcriptional level to up-regulate the expression of certain subunits of the proteasome (Yao et al., 2012; Üstün et al., 2013). In addition, posttranslational modification of subunits (e.g. by phosphorylation) would provide a means to rapidly alter the activity or other biochemical properties of the complex.
Virulent Pst bacteria that inject the full repertoire of T3Es into their host cell are not only able to prevent the induction of proteasome activity but also to suppress it below the basal level detected in the mock-treated control. This suggests that T3Es act to suppress the induction of proteasome activity during defense, which likely occurs at different levels and through different sets of effector proteins. First, the ability to prevent the induction of proteasome activity is consistent with the activity of T3Es acting to suppress SA-mediated defense responses (Kazan and Lyons, 2014). In addition, in npr1-1, wild-type Pst bacteria not only prevent induction of the proteasome but also are able to further inhibit its activity below the basal threshold. This indicates that T3Es also exert a more direct effect on the proteasome to reduce its activity. The ability to prevent the induction of proteasome activity is independent of the capacity to interfere with very early events of pathogen perception, such as the activation of FLS2 by flg22, because a Pst mutant lacking the two effectors AvrPto and AvrPtoB is still able to prevent elevated activity levels. There are several Pseudomonas spp. T3Es that have been suggested to interfere with SA production or signaling. For instance, HopI1 has been shown to interfere with SA synthesis inside chloroplasts, preventing its accumulation (Jelenska et al., 2007, 2010). In addition, HopM1 and AvrE are representatives of T3Es that have the ability to suppress SA-dependent basal immunity and disease necrosis, although the targets of these effectors with respect to SA signaling remain to be discovered (DebRoy et al., 2004). However, the interference with SA synthesis is not sufficient to reduce proteasome activity below basal levels, as transient expression of HopI1 in leaves of N. benthamiana shows no effect on activity.
A direct inhibition of the proteasome through T3Es of Pst targeting its components has not been described so far. However, XopJ, a T3E from X. campestris pv vesicatoria 85-10, was shown to proteolytically degrade the 19S RP subunit RPT6 in order to inhibit proteasome activity and to interfere with SA-mediated defense in susceptible pepper host plants (Üstün et al., 2013; Üstün and Börnke, 2015). A similar mechanism has been proposed for the T3E HopZ4 from P. syringae pv lachrymans (Üstün et al., 2014). Both effectors belong to the YopJ family, which is widespread among animal and plant pathogens but whose members are absent from Pst (Lewis et al., 2011). Also, Pst does not possess SylA, a secreted toxin produced, for instance, by P. syringae pv syringae, which directly targets the catalytic subunits of the 26S proteasome (Groll et al., 2008; Schellenberg et al., 2010; Baltrus et al., 2011; Misas-Villamil et al., 2013). Thus, the suppression of proteasome activity below the basal level by virulent Pst likely involves a previously unidentified effector. We have conducted a screen of a collection of Pst T3Es for their ability to suppress proteasome activity when expressed transiently in leaves of N. benthamiana. The analyses identified four T3Es, namely HopM1, HopG1, HopAO1, and HopA1, that reproducibly inhibited the proteasome in N. benthamiana and thus represent candidates for effectors interfering with proteasome activity during the infection of Arabidopsis with Pst. The functions of HopG1, HopAO1, and HopA1 so far have not been shown to be associated with the UPS. HopG1 was demonstrated to inhibit plant innate immunity associated with its localization to mitochondria (Block et al., 2010), while HopA1 associates with EDS1 to disrupt EDS1-TIR NB LRR disease complexes (Bhattacharjee et al., 2011). As the direct target proteins for both T3Es are still not known, it is possible that both T3Es could directly or indirectly associate with the UPS to modulate proteasome activity. The Tyr phosphatase HopAO1 targets the Arabidopsis receptor kinases EF-TU RECEPTOR and FLS2, reducing their Tyr phosphorylation and thus inhibiting PTI activation (Macho et al., 2014). Because the phosphorylation of certain proteasome subunits is crucial to activate its assembly and also induce its activity (Satoh et al., 2001), it might be possible that HopAO1 could target components of the proteasome to reduce their phosphorylation status. Whether this T3E also is able to interact with multiple target proteins in plants (e.g. with components of the proteasome or other UPS-related proteins) will be the subject of future studies and clarify its role as a proteasome inhibitor.
The identification of HopM1 as a candidate effector protein for suppression of the proteasome is striking. This effect is unlikely to be related to its ability to interfere with SA-dependent defense responses (DebRoy et al., 2004), because a HopM1 deletion strain still prevents the induction of proteasome activity above basal levels. However, in contrast to the wild type, Pst ΔhopM1 bacteria have lost the ability to suppress proteasome activity below the levels of the mock-infected control, suggesting that HopM1 interferes directly with proteasome function. The discovery of HopM1 as a proteasome inhibitor apparently contradicts previous findings, where it has been shown that HopM1 promotes the proteasome-dependent degradation of AtMIN7, a host ADP ribosylation factor guanine nucleotide-exchange factor required for vesicle trafficking during PTI and ETI (Nomura et al., 2006, 2011). However, mass spectrometry analysis suggests that HopM1 resides in a complex with several proteasome subunits. This opens the possibility that HopM1 might directly target proteasome components to interfere with its function. Moreover, using a yeast two-hybrid approach, HopM1 was identified to interact with RAD23, a ubiquitin receptor that delivers ubiquitinated proteins to the 26S proteasome for degradation (Nomura et al., 2006). We speculate that this interaction, on the one hand, mediates the degradation of AtMIN7 but, on the other hand, might affect the recognition of other ubiquitinated proteins by the 26S proteasome. This could result in an inefficient degradation of other substrates and could explain why AtMIN7 is removed by the proteasome while overall proteasome activity is reduced. Besides its function to destabilize AtMIN7 and thereby interfere with vesicle trafficking, HopM1 also has an AtMIN7-independent function: it is able to suppress ROS production and stomatal closure during plant immunity (Lozano-Durán et al., 2014). The increased protein levels of FLS2 observed in HopM1-expressing cells supports the hypothesis that impaired proteasome activity may interfere with the proper recycling of the receptor, thereby dampening the PTI response. It is highly plausible that HopM1 triggers the accumulation of inactive FLS2 receptors in the cells by compromising the recycling of the ubiquitinated receptor post elicitation. This effect on PTI is in line with our observations that the proteasome inhibition by genetic means negatively affects ROS production, defense gene expression, and also MAPK signaling. Thus, these results and previous findings suggest that HopM1 suppresses proteasome activity to dampen ROS generation and suppress stomatal closure to ensure bacterial proliferation during infection.
In conclusion, this work further supports the proposition of a prominent role of the proteasome during early and late defense responses toward Pseudomonas spp. and establishes that Pst possesses T3Es, which directly or indirectly interfere with proteasome activity during infection. These T3Es may affect proteasome function through multiple mechanisms by (1) preventing the SA-dependent induction of activity above basal levels and (2) through direct interaction with proteasomal components to interfere with their function (summarized in Supplemental Fig. S9). Our experiments have provided candidate T3Es for this second group, and future studies will have to clarify the molecular mechanisms of how these effectors target the proteasome.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Nicotiana benthamiana plants were grown in soil in a greenhouse with daily watering and subjected to a 16-h-light/8-h-dark cycle (25°C/21°C) at 300 µmol m−2 s−1 light and 40% relative humidity. Arabidopsis (Arabidopsis thaliana) seeds were sown on Murashige and Skoog (MS) agar (Sigma-Aldrich) plates supplemented with 2% (w/v) Suc and cultivated in tissue culture under a 16-h-light/8-h-dark regime (irradiance of 150 µmol m−2 s−1) at 50% humidity. Arabidopsis seeds germinated on soil were grown under short-day conditions (8 h of light/16 h of dark [23°C/21°C]). The rpt2-2 and rpn12a-1 mutants in the Col-0 background were originally described by Kurepa et al. (2008).
Cultivation of Bacteria
Pseudomonas syringae pv maculicola strain ES4326 obtained from J. Zeier and Pseudomonas syringae pv tomato DC3000 wild-type and deletion strains were grown in King’s B medium containing the appropriate antibiotic (rifampicin) at 28°C.
Assessment of SAR, Defense Priming during SAR, and Local Plant Resistance
The experiments were carried out essentially as detailed by Návarová et al. (2012) with slight modifications. In brief, for SAR induction, plants were infiltrated into three 1° leaves with a suspension of Psm (OD = 0.005). Infiltration with 10 mm MgCl2 served as a control treatment. For SAR growth assays, 2° leaves were inoculated with Psm (OD = 0.001) 2 d after the 1° treatment. Growth of Psm in 2° leaves was scored another 2 d later by homogenizing discs originating from infiltrated areas of three different leaves, plating appropriate dilutions on King’s B medium, and counting colony numbers after incubating the plates at 28°C for 2 d.
For the assessment of defense priming during SAR, 2° leaves were infiltrated with either 10 mm MgCl2 or Psm (OD = 0.005) 2 d after the 1° treatment. The 2° leaves were collected 10 h after the treatment.
For the determination of local defense responses, such as gene expression of PR1, bacterial suspensions of OD = 0.1 were infiltrated and harvested 24 h post inoculation for RNA extraction. For local growth assays, bacterial solutions of OD = 0.002 (Pst and Psm) were infiltrated into three full-grown leaves per plant. Bacterial growth was assessed 2 d after infiltration as described above.
Transient Expression Assays
For the infiltration of N. benthamiana leaves, Agrobacterium tumefaciens C58C1 was infiltrated into the abaxial air space of 4- to 6-week-old plants using a needleless 2-mL syringe. Agrobacteria were cultivated overnight at 28°C in the presence of appropriate antibiotics. The cultures were harvested by centrifugation, and the pellet was resuspended in sterile water to a final OD = 1. The cells were used for the infiltration directly after resuspension.
Immunoblotting
Leaf material was homogenized in SDS-PAGE loading buffer (100 mm Tris-HCl, pH 6.8, 9% β-mercaptoethanol, 40% glycerol, 0.0005% Bromophenol Blue, and 4% SDS) and, after heating for 10 min at 95°C, subjected to electrophoresis. Separated proteins were transferred onto nitrocellulose membranes. Proteins were detected by an anti-HA-peroxidase high-affinity antibody (Roche), anti-ubiquitin antibody (Agrisera), or anti-AtPBA1 (Enzo Life Sciences) via chemiluminescence.
MAPK Assay
Arabidopsis seedlings were grown for 12 d on MS agar plates and then transferred to six-well plates (four to six seedlings per well) on which each well contained 4 mL of liquid medium containing liquid 1× MS medium. Seedlings were treated with 1 µm flg22 peptide, and after 0 to 30 min as indicated, the seedlings were frozen in liquid nitrogen. The frozen seedlings were ground in liquid nitrogen and homogenized in 100 µL of extraction buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 15 mm EGTA, 10 mm MgCl2, 1 mm Na2MoO4·2H2O, 0.5 mm NaVO3, 1 mm NaF, 30 mm β-glycerol phosphate, 0.5 mm phenylmethylsulfonyl fluoride, one tablet per 10 mL of extraction buffer of proteinase inhibitor cocktail [Roche], and phosphatase inhibitor cocktail [Roche]). After centrifugation at 13,000 rpm for 30 min at 4°C, the protein concentration of the supernatants was determined using a Bradford assay. Thirty micrograms of protein was separated on a 12.5% polyacrylamide gel. Immunoblot analysis was performed using anti-phospho-p44/42 MAPK (1:1,000; Cell Signaling Technology) or anti-AtMPK6 (1:2,000; Sigma) as primary antibody and peroxidase-conjugated goat anti-rabbit antibody (1:5,000; Sigma).
ROS Assay
ROS production was monitored using a luminol-based assay modified after Dubiella et al. (2013). In brief, leaf discs (4 mm diameter) from four 4-week-old Arabidopsis plants were sampled using a cork borer and floated overnight on sterile water. The following day, the water was replaced with a solution of 17 mg mL−1 (w/v) luminol (Sigma) and 10 mg mL−1 horseradish peroxidase (Sigma) containing 1 µm flg22. Luminescence was captured over 45 min using the Synergy HT (BioTek Instruments) multiplate reader.
Measurement of Proteasome Activity
Proteasome activity in plant extracts was determined spectrofluorometrically using the fluorogenic substrate Suc-LLVY-NH-AMC (Sigma) according to Üstün et al. (2013). In brief, four leaf discs with a diameter of 0.7 cm each were harvested and frozen in liquid nitrogen. The leaf material was ground in 200 µL of extraction buffer (50 mm HEPES-KOH, pH 7.2, 2 mm ATP, 2 mm dithiothreitol, and 250 mm Suc). After centrifugation, the protein concentration of the supernatant was adjusted to 1 µg µL−1 with extraction buffer. Fifty micrograms of total protein was mixed with 220 µL of proteolysis buffer (100 mm HEPES-KOH, pH 7.8, 5 mm MgCl2, 10 mm KCl, and 2 mm ATP). The reaction was started after 5 min at 30°C by the addition of 0.2 mm Suc-LLVY-AMC. Released amino-methyl-coumarin was measured every 2 min between 0 and 120 min using a fluorescence spectrophotometer with an excitation wavelength of 360 nm and an emission wavelength of 460 nm.
RNA Extraction and Expression Analysis
Total RNA was isolated from leaf material as described and then treated with RNase-free DNase to degrade any remaining DNA. First-strand cDNA synthesis was performed from 2 µg of total RNA using Revert-Aid reverse transcriptase. For quantitative RT-PCR, the cDNAs were amplified using SensiFAST SYBR Lo-ROX Mix (Bioline) in the AriaMx Realtime PCR System (Agilent Technologies). PCR was optimized, and reactions were performed in triplicate. The transcript level was standardized based on cDNA amplification of UBC9 as a reference. Statistical analysis was performed using Student’s t test. Primers are provided in Supplemental Table S1.
Protein Extraction and Mass Spectrometry
Adult N. benthamiana plants were syringe infiltrated with A. tumefaciens either expressing HopM1-GFP (HopM1 cloned in pGWB5 vector) or a vector GFP alone. After 3 d, 10 g of plant tissue was ground in liquid nitrogen, and proteins were isolated in extraction buffer (150 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm EDTA, 2 mm EGTA, 10 mm dithiothreitol, 2% [w/v] polyvinylpyrrolidone, 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich], 50 mm NaF, 10 mm Na2MoO4, 1 mm sodium orthovanadate, and 0.5 mm phenylmethylsulfonyl fluoride). After centrifugation at 20,000g at 4°C for 30 min, the supernatants were filtered through Miracloth (Millipore). Supernatants were incubated for 2 h at 4°C with 200 µL of anti-GFP trap beads (Chromatek). The beads were washed three times with 1 mL of modified extraction buffer (150 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm EDTA, 2 mm EGTA, and 1% protease inhibitor cocktail). Proteins were eluted by adding 50 μL of 5× SDS buffer and then boiled for 5 to 10 min and run on a 10% SDS-PAGE gel. The gel was stained with Coomassie colloidal stain (Invitrogen), and proteins from each lane were trypsin digested and subjected to liquid chromatography-MS/MS analysis. Reverse-phase chromatography was used to separate tryptic peptides prior to mass spectrometric analysis using an Acclaim PepMap µ-precolumn cartridge (300 µm i.d. × 5 mm, 5 μm, 100 Å) and an Acclaim PepMap RSLC (75 µm × 25 cm, 2 µm, 100 Å; Thermo Scientific). Peptides were eluted directly via a Triversa Nanomate nanospray source (Advion Biosciences) into a Thermo Orbitrap Fusion mass spectrometer (Q-OT-qIT; Thermo Scientific). The raw data were processed using MSConvert in the ProteoWizard Toolkit (version 3.0.5759). MS/MS spectra were searched with Mascot engine (Matrix Science; version 2.4.1) against the N. benthamiana database (available on request from Sophien Kamoun at the Sainsbury Laboratory; http://www.tsl.ac.uk/staff/sophien-kamoun/), the P. syringae database (http://www.uniprot.org/), and the common Repository of Adventitious Proteins Database (http://www.thegpm.org/cRAP/index.html). Peptides were generated from a tryptic digestion with up to two missed cleavages, carbamidomethylation of Cys as fixed modifications, and oxidation of Met as variable modifications. Precursor mass tolerance was 5 ppm, and product ions were searched at 0.8-D tolerance. Scaffold (version Scaffold_4.3.2; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95% probability by the Scaffold Local FDR algorithm. The full set of mass spectrometry data is accessible at PRIDE (http://www.ebi.ac.uk/pride/archive/; Vizcaíno et al., 2016) under accession number PXD004883.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Inhibition of the proteasome by MG132 treatment significantly promotes bacterial growth of Pst in Arabidopsis.
Supplemental Figure S2. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopG1, HopH1, HopAO1, or empty vector control.
Supplemental Figure S3. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopAF1, HopX1, XopJ, or empty vector control.
Supplemental Figure S4. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopM1, HopA1, HopB1, or empty vector control.
Supplemental Figure S5. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopN1, HopY1, XopJ, or empty vector control.
Supplemental Figure S6. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopK1, HopI1, HopC1, XopJ, or empty vector control.
Supplemental Figure S7. Proteasome activity in N. benthamiana leaves following transient expression of T3Es HopM1, HopO1-1, HopAD1, XopJ, or empty vector control.
Supplemental Figure S8. HopM1 interacts with proteasome-associated proteins; immunoprecipitation of HopM1 using GFP trap agarose beads.
Supplemental Figure S9. Role of the proteasome during plant immunity.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Susanne Jeserigk for technical assistance, Alan Collmer and Jürgen Zeier for providing Pseudomonas spp. strains, Jan Smalle for providing the rpt2a-2 and rpn12a-1 seeds, and the Proteomic Research Technology Platform at Warwick University.
Glossary
- UPS
ubiquitin-proteasome system
- CP
core protease
- RP
regulatory particles
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- ROS
reactive oxygen species
- ETI
effector-triggered immunity
- HR
hypersensitive response
- SAR
systemic acquired resistance
- SA
salicylic acid
- T3E
type III effector
- Col-0
Columbia-0
- HA
hemagglutinin
- MS/MS
tandem mass spectrometry
- MS
Murashige and Skoog
- OD
optical density
- RT
reverse transcription
- dpi
days post inoculation
Footnotes
This work was supported by the Federation of the European Biochemical Societies (long-term fellowship to S.Ü.), by the Deutsche Forschungsgemeinschaft (grant no. CRC973 to F.B.), by the Royal Society (to V.N.), and by the Biotechnology and Biological Sciences Research Council (grant no. BB/L019345/1 to V.N.).
Articles can be viewed without a subscription.
References
- Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA 97: 4856–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield MJ. (2015) Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell Microbiol 17: 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Döring AC, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Block A, Guo M, Li G, Elowsky C, Clemente TE, Alfano JR (2010) The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol 12: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285: 25554–25569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chung K, Tasaka M (2011) RPT2a, a 26S proteasome AAA-ATPase, is directly involved in Arabidopsis CC-NBS-LRR protein uni-1D-induced signaling pathways. Plant Cell Physiol 52: 1657–1664 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Langenbach CJ, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA 108: 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan J, Etienne P, Petitot AS, Houot V, Blein JP, Suty L (2001) Cryptogein affects expression of alpha3, alpha6 and beta1 20S proteasome subunits encoding genes in tobacco. J Exp Bot 52: 1947–1948 [DOI] [PubMed] [Google Scholar]
- De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, Roelofs J (2013) The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J Biol Chem 288: 29467–29481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101: 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R. (2013) Manipulation of host proteasomes as a virulence mechanism of plant pathogens. Annu Rev Phytopathol 51: 521–542 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Groll M, Schellenberg B, Bachmann AS, Archer CR, Huber R, Powell TK, Lindow S, Kaiser M, Dudler R (2008) A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature 452: 755–758 [DOI] [PubMed] [Google Scholar]
- Gu C, Kolodziejek I, Misas-Villamil J, Shindo T, Colby T, Verdoes M, Richau KH, Schmidt J, Overkleeft HS, van der Hoorn RA (2010) Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J 62: 160–170 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- Hofius D, Tsitsigiannis DI, Jones JD, Mundy J (2007) Inducible cell death in plant immunity. Semin Cancer Biol 17: 166–187 [DOI] [PubMed] [Google Scholar]
- Igari K, Endo S, Hibara K, Aida M, Sakakibara H, Kawasaki T, Tasaka M (2008) Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J 55: 14–27 [DOI] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT (2010) Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci USA 107: 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr Biol 17: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90: 324–335 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Toh-E A, Smalle JA (2008) 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J 53: 102–114 [DOI] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Minami A, Marshall RS, Book AJ, Farmer LM, Walker JM, Vierstra RD (2011) The RPT2 subunit of the 26S proteasome directs complex assembly, histone dynamics, and gametophyte and sporophyte development in Arabidopsis. Plant Cell 23: 4298–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequeu J, Simon-Plas F, Fromentin J, Etienne P, Petitot AS, Blein JP, Suty L (2005) Proteasome comprising a beta1 inducible subunit acts as a negative regulator of NADPH oxidase during elicitation of plant defense reactions. FEBS Lett 579: 4879–4886 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lee A, Ma W, Zhou H, Guttman DS, Desveaux D (2011) The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol 12: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Bourdais G, He SY, Robatzek S (2014) The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol 202: 259–269 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP. (2016) Subversion of plant cellular functions by bacterial type-III effectors: beyond suppression of immunity. New Phytol 210: 51–57 [DOI] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh MH, Sklenar J, Derbyshire P, et al. (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S (2012) Ubiquitination during plant immune signaling. Plant Physiol 160: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2013) The nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73: 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil JC, Kolodziejek I, Crabill E, Kaschani F, Niessen S, Shindo T, Kaiser M, Alfano JR, van der Hoorn RA (2013) Pseudomonas syringae pv. syringae uses proteasome inhibitor syringolin A to colonize from wound infection sites. PLoS Pathog 9: e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C (2009) Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 150: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, He SY (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY (2011) Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor V, Balmer A, Gamir J, Flors V, Mauch-Mani B (2014) Preparing to fight back: generation and storage of priming compounds. Front Plant Sci 5: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H (2001) Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40: 314–319 [DOI] [PubMed] [Google Scholar]
- Schellenberg B, Ramel C, Dudler R (2010) Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol Plant Microbe Interact 23: 1287–1293 [DOI] [PubMed] [Google Scholar]
- Singer AU, Schulze S, Skarina T, Xu X, Cui H, Eschen-Lippold L, Egler M, Srikumar T, Raught B, Lee J, et al. (2013) A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog 9: e1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HH, Fukao Y, Ishida S, Yamamoto H, Maekawa S, Fujiwara M, Sato T, Yamaguchi J (2013) Proteomics analysis reveals a highly heterogeneous proteasome composition and the post-translational regulation of peptidase activity under pathogen signaling in plants. J Proteome Res 12: 5084–5095 [DOI] [PubMed] [Google Scholar]
- Suty L, Lequeu J, Lançon A, Etienne P, Petitot AS, Blein JP (2003) Preferential induction of 20S proteasome subunits during elicitation of plant defense reactions: towards the characterization of “plant defense proteasomes.” Int J Biochem Cell Biol 35: 637–650 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Üstün S, Bartetzko V, Börnke F (2013) The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog 9: e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Börnke F (2014) Interactions of Xanthomonas type-III effector proteins with the plant ubiquitin and ubiquitin-like pathways. Front Plant Sci 5: 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Börnke F (2015) The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol 168: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, König P, Guttman DS, Börnke F (2014) HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol Plant Microbe Interact 27: 611–623 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Wu Y, Nie H, Tang D (2012) RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J 71: 1015–1028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.