Characterization of Arabidopsis hpe1 mutants revealed a novel strategy to optimize light-harvesting pigments that improved photosynthetic efficiency and biomass production.
Abstract
Maximizing light capture by light-harvesting pigment optimization represents an attractive but challenging strategy to improve photosynthetic efficiency. Here, we report that loss of a previously uncharacterized gene, HIGH PHOTOSYNTHETIC EFFICIENCY1 (HPE1), optimizes light-harvesting pigments, leading to improved photosynthetic efficiency and biomass production. Arabidopsis (Arabidopsis thaliana) hpe1 mutants show faster electron transport and increased contents of carbohydrates. HPE1 encodes a chloroplast protein containing an RNA recognition motif that directly associates with and regulates the splicing of target RNAs of plastid genes. HPE1 also interacts with other plastid RNA-splicing factors, including CAF1 and OTP51, which share common targets with HPE1. Deficiency of HPE1 alters the expression of nucleus-encoded chlorophyll-related genes, probably through plastid-to-nucleus signaling, causing decreased total content of chlorophyll (a+b) in a limited range but increased chlorophyll a/b ratio. Interestingly, this adjustment of light-harvesting pigment reduces antenna size, improves light capture, decreases energy loss, mitigates photodamage, and enhances photosynthetic quantum yield during photosynthesis. Our findings suggest a novel strategy to optimize light-harvesting pigments that improves photosynthetic efficiency and biomass production in higher plants.
The tremendous increase in world population and environmental deterioration pose serious challenges to agricultural production and food security (Ray et al., 2013). To meet this challenge, crops with high yield potential need to be developed (Long et al., 2015). However, the yield traits that have played key roles during the green revolution have had their potential nearly exhausted; thus, new strategies are needed. Photosynthesis, the unique biological process responsible for the conversion of light energy to chemical forms, is the ultimate basis of crop yield (Zhu et al., 2010). Theoretically, enhancing photosynthetic efficiency should be an excellent strategy to increase crop yield. However, the improvement of photosynthetic efficiency has played only a minor role in the remarkable crop productivity improvement achieved in the last half-century (Zhu et al., 2010; Ort et al., 2015).
In the light reactions of photosynthesis, light energy is used by chlorophyll and associated pigments, water is split, and electron transport on the chloroplast membrane reduces NADP, resulting in a proton gradient that powers the phosphorylation of ADP. NADPH and ATP power the Calvin cycle, which assimilates and reduces carbon dioxide to carbohydrate (Ort et al., 2015). Strategies to improve photosynthesis mainly include the optimization of light capture, light energy conversion in the light reaction, and carbon capture and conversion in the dark reaction (Ort et al., 2015). Previous research focused mainly on the optimization of dark reactions through the improvement of carbon capture and conversion to directly increase biomass (Miyagawa et al., 2001; Kebeish et al., 2007; Lin et al., 2014; Ort et al., 2015). However, less effort has been spent to optimize light capture and light energy conversion in the light reactions to improve the whole photosynthetic efficiency (Ort et al., 2015).
Maximizing light capture by the adjustment of antenna size can optimize light capture and light energy conversion, but it is difficult to achieve (Blankenship and Chen, 2013). Antenna in photosynthetic systems typically consist of pigments specifically bound to membrane-associated proteins. These antenna pigment-protein complexes closely associate with the reaction center complexes and deliver absorbed energy to the reaction centers, where some of the energy originally in the photon is captured by electron-transfer processes (Blankenship, 2002; Green and Parson, 2003). However, light saturation could take place at intensities much lower than would be expected if every chlorophyll was able to carry out photosynthesis by itself (Blankenship, 2002). The light saturation problem also has been addressed from the antenna perspective, and many efforts are under way to truncate the antenna system in photosynthetic microorganisms. A smaller antenna associated with each reaction center will, in principle, also shift the light-response curve, so that light saturation sets in at higher intensities, thereby reducing excess light and increasing productive light. While the concept of increased efficiency due to reduced antenna size is simple, reaching this goal has not yet been achieved (Blankenship and Chen, 2013). In green algae, the reduction of light-harvesting pigments by decreasing the expression of the chlorophyll a oxygenase gene, which is responsible for the synthesis of chlorophyll b via the oxidation of chlorophyll a (Czarnecki and Grimm, 2012), led to efficient photosynthesis due to the balance between captured light and photochemical reactions (Perrine et al., 2012). However, there is still no success in higher plants.
In this study, we performed a large-scale genetic screen using the model organism Arabidopsis (Arabidopsis thaliana) and identified two independent alleles of an uncharacterized gene that we named HIGH PHOTOSYNTHETIC EFFICIENCY1 (HPE1), whose mutation confers improved photosynthetic efficiency by optimizing light-harvesting pigment. A deficiency of HPE1 shows higher light reaction activity of photosynthesis, more efficient carbon fixation, and significantly increased biomass production. Interestingly, HPE1 encodes a chloroplast protein containing an RNA recognition motif and regulates the splicing of RNAs of plastid genes by directly associating with target RNAs. HPE1 mutation results in a splicing deficiency of plastid genes that may alter the expression of chlorophyll-related genes, probably through plastid-to-nucleus signaling. Altered expression of chlorophyll-related genes changes the content of light-harvesting pigments and optimizes the light-harvesting system. Our characterization of HPE1 mutants suggests a novel strategy to optimize light harvesting and improve photosynthetic efficiency in higher plants.
RESULTS
Loss of HPE1 Confers Improved Light Reaction Activity of Photosynthesis
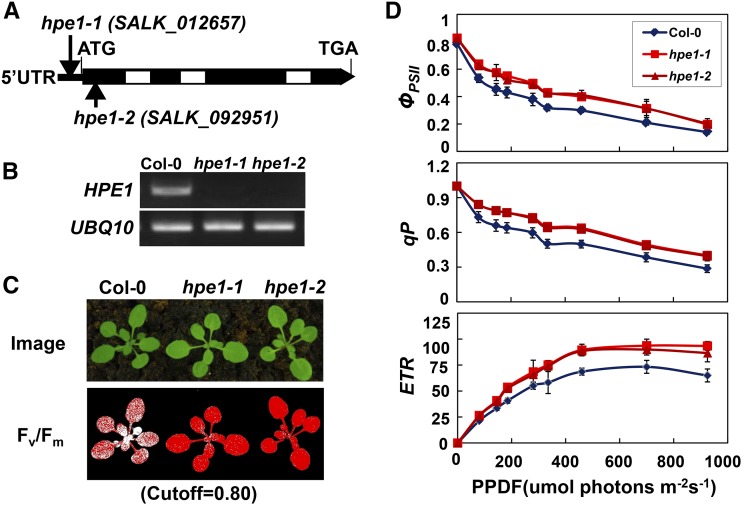
To better understand the regulatory mechanism of photosynthesis and identify mutants with increased photosynthetic efficiency, we screened many Arabidopsis mutant pools by using a chlorophyll fluorescence video imaging system (Jin et al., 2014). We identified hpe mutants that exhibit high maximum photochemical efficiency of PSII (Fv/Fm) under growth light. One hpe1 mutant (SALK_012657), At1g70200, showed higher photosynthetic efficiency than wild-type plants (Fig. 1, A–C). This allele contains a T-DNA element inserted into the 5′ untranslated region of the HPE1 gene (Fig. 1A). Interestingly, we also obtained another independent hpe1 allele (SALK_092951) that harbors a T-DNA element inserted into the first exon of the HPE1 gene and exhibits a phenotype identical to that of hpe1 mutants (Fig. 1, A–C). These results were confirmed with twice-backcrossed mutants and imply that a deficiency of HPE1 results in higher light reaction activity during photosynthesis. We subsequently named SALK_012657 and SALK_092951 as hpe1-1 and hpe1-2, respectively. Next, to further precisely determine the light reaction activity of photosynthesis in hpe1 mutants, we analyzed the light intensity dependence of three chlorophyll fluorescence parameters: light-response curves of PSII quantum yield, photochemical quenching, and electron transport rate. PSII quantum yield, photochemical quenching, and electron transport rate were much higher in the hpe1 mutants than in the wild type (Fig. 1D), together with higher Fv/Fm in the hpe1 mutants (Fig. 1C), which confirms that the light reaction activity of photosynthesis is higher in hpe1 mutants than in the wild type.
Figure 1.
Isolation of hpe1 Arabidopsis mutant plants. A, Schematic diagram of the HPE1 gene inferred using DNA sequence analysis. Exons (black boxes) and introns (lines) are indicated. Positions of the T-DNA insertions corresponding to hpe1-1 and hpe1-2 are shown. The ATG start codon and TGA stop codon are shown. UTR, Untranslated region. B, RT-PCR analysis of HPE1 transcription in the wild type (Columbia-0 [Col-0]) and hpe1 mutants. C, False-color images representing Fv/Fm under growth light conditions in 3-week-old wild-type and hpe1 mutant plants. Red pixels indicate that Fv/Fm is above the cutoff value (0.8). D, Light-response curves of PSII quantum yield (ΦPSII), photochemical quenching (qP), and electron transport rate (ETR) in the wild type and hpe1 mutants. Measurements were performed at the following light intensities: 0, 81, 145, 186, 281, 335, 461, 701, and 926 μmol photons m−2 s−1. PPDF, Photosynthetic photon flux density. Each data point represents at least 20 independent plants.
The Content of Carbohydrates and Biomass Production Are Increased in hpe1 Mutants
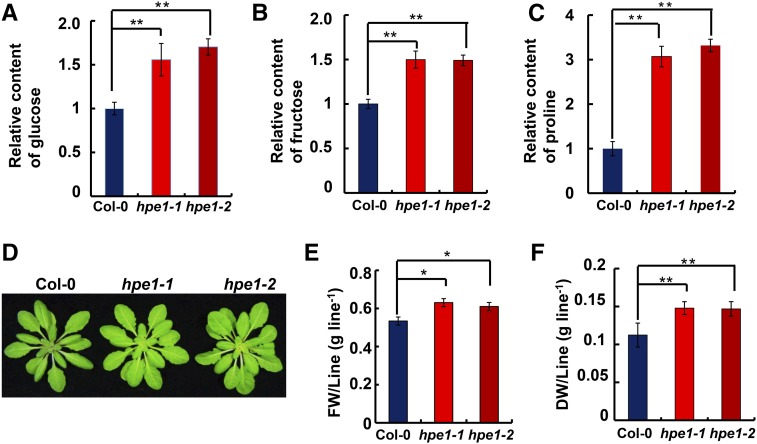
Because NADPH and ATP from the light reaction are used to power the Calvin cycle, we speculated that improved light reaction performance can promote carbon fixation during the dark reaction of photosynthesis in the hpe1 mutants. To test this, we first determined the contents of representative carbohydrates in leaves using gas chromatography-mass spectrometry (GC-MS). The hpe1 mutant plants accumulated higher contents of carbohydrates per unit of leaf area, including Glc and Fru. The contents of Glc and Fru were approximately 51.1% and 49.9% higher in the hpe1 mutants than in the wild-type plants, respectively (Fig. 2, A and B). To determine whether the greater accumulation of carbohydrates in hpe1 mutants is due to the decrease in carbohydrate turnover, we measured the degradation rate of wild-type and mutant plants after inhibiting the photosynthetic light reactions using 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) treatment. Interestingly, after a 5-h DCMU treatment, the content of Glc was decreased by approximately 39.9% in wild-type, 40.3% in hpe1-1, and 42.5% in hpe1-2 plants. The content of Fru was decreased by approximately 38.4% in wild-type, 39.1% in hpe1-1, and 41.2% in hpe1-2 plants (Supplemental Fig. S1). These results indicate that the turnover rate of carbohydrates in hpe1 mutants is comparable to that in wild-type plants, implying that the greater accumulation of carbohydrates is due to the more efficient carbon fixation in hpe1 mutants. More interestingly, the content of Pro, the only proteinogenic secondary amino acid that responds to metabolic stress and participates in metabolic signaling (Phang et al., 2010), was significantly higher in the hpe1 mutants than in the wild-type plants (Fig. 2C; Supplemental Fig. S1), implying that the hpe1 mutants may be resistant to adverse stress.
Figure 2.
Metabolite and biomass production of hpe1 Arabidopsis mutant plants. A to C, Content analysis of Glc (A), Fru (B), and Pro (C) in the wild-type and hpe1 mutant plants by GC-MS. D, Representative photographs of selected 5-week-old wild-type and hpe1 mutant plants. E, Fresh weights (FW) of 5-week-old wild-type and hpe1 mutant plants. F, Dry weights (DW) of 5-week-old wild-type and hpe1 mutant plants. Each data point represents at least 20 independent plants. Significant differences were identified at 5% (*) and 1% (**) probability levels using Student’s t test.
To evaluate whether more efficient photosynthesis could result in increased biomass, we compared the growth phenotype of the hpe1 mutants and the wild type. Interestingly, 5-week-old mature hpe1 mutants showed a substantial increase in biomass (Fig. 2D), including both greater fresh weight (Fig. 2E) and dry weight (Fig. 2F), although the hpe1 mutants and wild-type controls showed no obvious differences at the young plant stage. Rosette size and leaf number of the hpe1 mutants and the wild type showed no obvious differences (Supplemental Fig. S2), which indicates that the increased biomass may be due to the more fleshy leaves of the hpe1 mutants than the wild-type controls. Together, these results suggest that carbon fixation is more efficient and biomass production is increased in hpe1 mutants.
HPE1 Is Localized Specifically to Chloroplast and Affects RNA Splicing of Plastid Genes
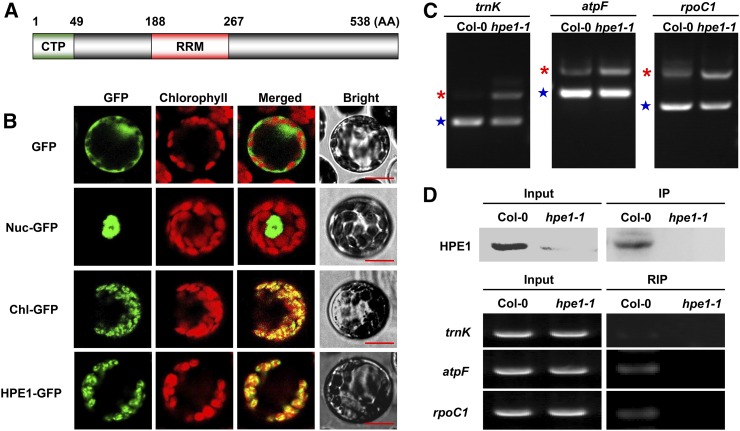
To elucidate the mechanism underlying the improved photosynthetic efficiency and biomass of the hpe1 mutants, we analyzed the molecular functions of HPE1. HPE1 encodes a protein of 538 amino acids of unknown function according to The Arabidopsis Information Resource (www.arabidopsis.org). HPE1 protein contains a predicted N-terminal chloroplast transit peptide (1–49; Fig. 3A), based on TargetP prediction (Emanuelsson et al., 2000). Analysis of the subcellular localization of HPE1-GFP fusion proteins by confocal laser scanning microscopy revealed that HPE1 is localized specifically to the chloroplast (Fig. 3B). Notably, HPE1 contains an RRM (188–267; Fig. 3A), also known as an RNA-binding domain or ribonucleoprotein domain (RNP), which is one of the most abundant protein domains involved in RNA binding and is found in all life kingdoms (Maris et al., 2005), suggesting that HPE1 may be involved in the regulation of plastid RNA metabolism. To test this, we first examined the transcription of plastid genes using real-time PCR. The transcription of plastid genes regulated by nucleus-encoded RNA polymerase, plastid-encoded RNA polymerase, or both was comparable in the hpe1 mutants and the wild type (Supplemental Fig. S3). Furthermore, we analyzed plastid RNA processing in the hpe1 mutants and found that no obvious differences in RNA editing or stability were detected between the hpe1 mutants and wild-type controls. Finally, we determined the RNA splicing efficiency of the intron-containing plastid genes. Interestingly, we found increased accumulation of unspliced precursors of Lys tRNA (trnK), ATPase F subunit (atpF), and RNA polymerase β-subunit-1 (rpoC1) but reduced accumulation of spliced RNAs of trnK, atpF, and rpoC1 in the hpe1 mutants in varying degrees (Fig. 3C; Supplemental Fig. S4B). Notably, the group II intron in the trnK gene of land plants encodes a conserved protein called MatK, which shares sequence similarity with canonical group II maturases in domains that also may assist the splicing of its own and other chloroplast group II introns (Stern et al., 2010). However, there are no significant effects of HPE1 deficiency on the splicing of other plastid genes that contain introns (Supplemental Fig. S4A). These results suggest that HPE1 only regulates the splicing of some chloroplast RNAs.
Figure 3.
Confirmation of the subcellular location and molecular function of HPE1. A, Schematic diagram of the HPE1 protein, including the chloroplast transit peptide (CTP) and RNA recognition motif (RRM). B, Subcellular localization of HPE1 within the chloroplast using the GFP assay. GFP, Control with empty vector; Nuc-GFP, nuclear control; Chl-GFP, chloroplast control; HPE1-GFP, HPE1-GFP fusion. Bars = 10 μm. C, Analysis of splicing defects in the hpe1 mutants using RT-PCR. Products of RT-PCR were isolated and sequenced. Asterisks indicate unspliced mRNA precursors, and stars indicate spliced mature mRNA. D, Confirmation of the association of HPE1 with target RNA using RNA immunoprecipitation (RIP). The top gels show western blots of proteins present in crude leaf extracts derived from Col-0 and hpe1 mutant plants and proteins immunoprecipitated (IP) with the anti-HPE1 antibody. In the bottom gels, RT-PCR was used to detect the association of trnK, rpoC1, and atpF introns with HPE1. Five additional independent biological replicates were performed, and similar results were obtained.
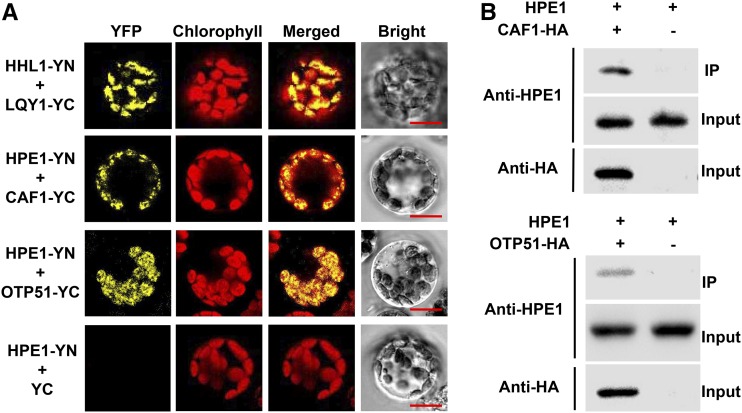
We further assessed whether HPE1 is associated with target RNAs using RNA immunoprecipitation, an analysis that detects the presence of the corresponding RNA in the protein immunoprecipitates by reverse transcription (RT)-PCR. The immunoprecipitated HPE1 complexes were found to specifically coprecipitate the RNAs of trnK, atpF, and rpoC1, as revealed by comparative RT-PCR analysis of the corresponding samples derived from the hpe1 mutants and wild-type plants (Fig. 3D). These results thus indicate that HPE1 associates in vivo with target RNA, including trnK, atpF, and rpoC1, which suggests that HPE1 is a novel chloroplast RNA-splicing factor. Introns and multiple splicing factors assemble into RNP complexes in land plants and in Chlamydomonas reinhardtii to regulate RNA splicing (Perron et al., 2004; Stern et al., 2010). Previous studies reported that RNA splicing of trnK, atpF, and rpoC1 also is regulated by CAF1 (Asakura and Barkan, 2006), OTP51 (de Longevialle et al., 2008), OTP70 (Chateigner-Boutin et al., 2011), or WTF1 (Kroeger et al., 2009). To determine whether HPE1 assembles into RNP complexes with other RNA-splicing factors in chloroplasts, we analyzed the interaction between HPE1 and CAF1, OTP51, OTP70, or WTF1. The interaction of HPE1 with CAF1 or OTP51 was detected by bimolecular fluorescence complementation (BiFC) analysis (Fig. 4A), which was further confirmed by coimmunoprecipitation assays (Fig. 4B). However, the interaction of HPE1 with OTP70 or WTF1 was not detected (Supplemental Fig. S6). These observations indicate that HPE1 forms a complex specifically with CAF1 and OTP51 to coregulate chloroplast RNA splicing.
Figure 4.
Interaction analysis of HPE1 with other RNA-splicing factors of plastid genes. A, Interaction of HPE1 with other splicing factors involved in the splicing of trnK, rpoC1, and atpF introns using BiFC analysis. HPE1 fused with the N terminus of YFP (YN) and CAF1 and OTP51 fused with the C terminus of YFP (YC) were cotransfected into protoplasts and visualized using confocal microscopy. As a positive control, both HYPERSENSITIVE TO HIGH LIGHT1 (HHL1) fused with YN and LOW QUANTUM YIELD OF PHOTOSYSTEM II1 (LQY1) fused with YC were cotransfected into protoplasts. As a negative control, HPE1 was fused with YN and empty vector YC. Bars = 10 μm. B, Confirmation of the interaction between HPE1 and chloroplast splicing factors CAF1 and OTP51 using the coimmunoprecipitation assay. Fusion proteins of CAF1-hemagglutinin (HA) and OTP51-HA were expressed in Arabidopsis protoplasts and precipitated with anti-HA-coupled agarose. The immunoprecipitates (IP) were probed with anti-HPE1 antibodies, and protoplasts expressing the empty HA vector were used as controls. All experiments were repeated three times with similar results.
Light-Harvesting Pigments Are Optimized through Down-Regulation of the Expression of Nucleus-Encoded Chlorophyll-Related Genes in hpe1 Mutants
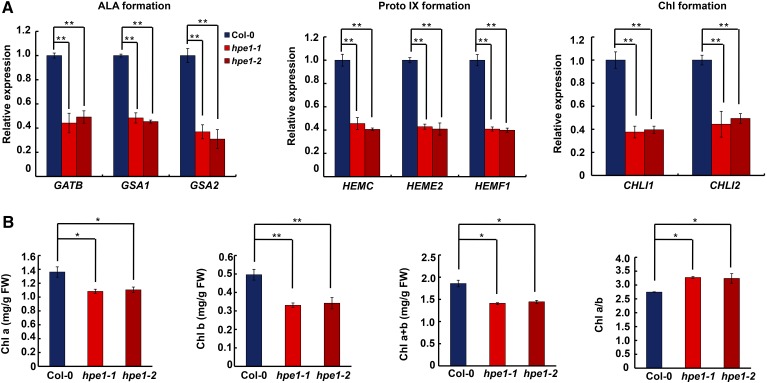
Biochemical analysis indicated that HPE1 is involved in the posttranscriptional regulation of gene expression in plastids, but it is still unclear how the loss of HPE1 improved photosynthetic efficiency and biomass production. Interestingly, we found that the leaves of the hpe1 mutants had a chlorophyll-deficient phenotype (Fig. 2D). Notably, the content of chlorophyll also was decreased in mutants of other chloroplast RNA-splicing factors, including CAF1 and OTP51 (Asakura and Barkan, 2006; de Longevialle et al., 2008). Both chlorophyll a and chlorophyll b contents were decreased in the hpe1 mutants (Fig. 5B; Supplemental Fig. S9), consistent with other chloroplast RNA-splicing mutants (Asakura and Barkan, 2006; de Longevialle et al., 2008; Kroeger et al., 2009; Chateigner-Boutin et al., 2011). Interestingly, different from mutants of other chloroplast RNA-splicing regulators, the extent of chlorophyll reduction in hpe1 mutants was smaller, which may be related to the smaller contribution of HPE1 to plastid RNA splicing. The reduced chlorophyll is probably still able to meet the need of light capture but meanwhile reduces the light-harvesting antenna size, thus ameliorating the loss of light energy. In addition, the reduction in chlorophyll b content was greater than the reduction in chlorophyll a content (Fig. 5B; Supplemental Fig. S9), resulting in a higher chlorophyll a/b ratio in the hpe1 mutants than in the wild type (Fig. 5B; Supplemental Fig. S9). Taken together, these results suggest that the down-regulation of chlorophyll content may be a common theme in response to impaired plastid RNA splicing.
Figure 5.
Analysis of chlorophyll-related gene expression and chlorophyll contents. A, Effects of HPE1 deficiency on the expression of chlorophyll-related genes in three different steps. The left graph indicates 5-aminolevulinic acid (ALA) formation-related genes; the middle graph indicates protoporphyrin IX (Proto IX) formation-related genes; and the right graph indicates chlorophyll (Chl) formation-related genes. B, Analysis of chlorophyll a (Chl a) content, chlorophyll b (Chl b) content, total chlorophyll a and chlorophyll b (Chl a+b) content, and the ratio of chlorophyll a to chlorophyll b (Chl a/b). FW, Fresh weight. All experiments were repeated three times with similar results. Significant differences were identified at 5% (*) and 1% (**) probability levels using Student’s t test.
The plastid-encoded gene trnE-UUC encodes tRNAGlu, which functions in plastid protein synthesis. Apart from this function, tRNA-Glu is a precursor for the biosynthesis of tetrapyrroles, including chlorophyll in plants, archaea, and most bacteria (Levicán et al., 2005). However, the trnE-UUC gene does not contain an intron; thus, its expression should not be affected in hpe1 mutants. Consistently, quantitative RT-PCR assay indicated that the expression of trnE-UUC was indeed not affected in hpe1 mutants (Supplemental Fig. S5). Thus, the decreased chlorophyll content in hpe1 mutants should not be due to alteration in the expression of trnE-UUC, which is required for the biosynthesis of chlorophyll.
Chlorophyll synthesis is carried out by enzymes encoded by nuclear genes (Czarnecki and Grimm, 2012). Real-time PCR showed that multiple key genes in the chlorophyll synthesis pathway, including 5-aminolevulinic acid formation-related genes, protoporphyrin IX formation-related genes, and chlorophyll formation-related genes, were all down-regulated 3- to 4-fold in the hpe1 mutants compared with the wild type (Fig. 5A), suggesting that defects in chlorophyll accumulation in the hpe1 mutants may be due to the down-regulation of chlorophyll synthesis-related genes. Given the role of HPE1 in plastid RNA splicing, we wondered whether mutants of other regulators of plastid RNA splicing also show decreased expression of nucleus-encoded chlorophyll-related genes. Therefore, we analyzed the mutants of CAF1 and OTP51 (Supplemental Fig. S7). The results showed that the expression of nucleus-encoded chlorophyll synthesis-related gene mutants also was repressed in caf1 and otp51. Notably, the down-regulation of these nucleus-encoded genes was more significant in caf1 and otp51 mutants than in hpe1 mutants, probably due to more severe plastid RNA splicing deficiency in caf1 and otp51 mutants (Supplemental Fig. S7). These results indicate that decreased plastid RNA splicing can repress the expression of nucleus-encoded chlorophyll-related genes, but how this regulation is achieved remains unclear.
One possibility is that a decrease in plastid RNA splicing elicits a retrograde plastid-to-nucleus signaling to repress the expression of nucleus-encoded chlorophyll-related genes. There are multiple distinct putative plastid retrograde signaling pathways based on the sources of the signals, and plastid gene expression is a key signal in plastid-to-nuclear signaling (Chi et al., 2013), which is mediated by the PPR motif-containing protein GENOMES UNCOUPLED1 (GUN1; Cottage et al., 2007). ABA INSENSITIVE4 (ABI4), an APETALA2-type transcription factor, is common to all retrograde signaling pathways. ABI4 binds the promoter of a retrograde-regulated gene through a conserved motif found in close proximity to a light-regulatory element to regulate gene expression, such as light-harvesting complex subunit b (Lhcb; Koussevitzky et al., 2007). However, Lhcb4, which is regulated by the GUN1 pathway, was unaffected before and after lincomycin treatment (Supplemental Fig. S8), implying that the down-regulation of chlorophyll-related genes in hpe1 mutants is independent of the GUN1-ABI4 pathway. Thus, further investigation is warranted to understand how impaired splicing of plastid RNA leads to the down-regulation of nucleus-encoded chlorophyll-related genes.
The Coordination of Light Capture with Conversion Is Improved in hpe1 Mutants
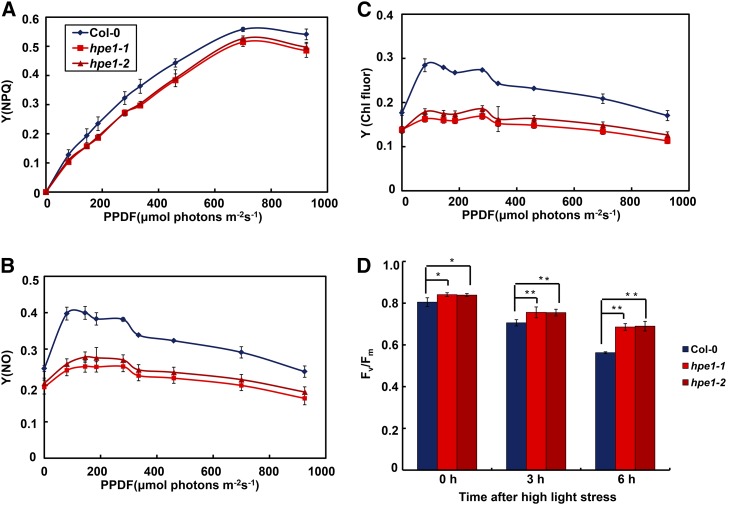
We further compared the utility and loss of light energy during the light reactions in the hpe1 mutants. Heat, including regulatable nonphotochemical quenching yield (Fig. 6A) and nonregulatable nonphotochemical quenching yield (Fig. 6B), and chlorophyll fluorescence (Fig. 6C) were reduced significantly in the hpe1 mutants compared with the wild type, together with faster photochemical quenching and electron transport rate (Fig. 1D). These results suggest that the optimized chlorophyll decreases the loss of light energy in hpe1 mutants. The preponderance of light-harvesting pigment optimization should be more significant under excess light (Blankenship and Chen, 2013). We found that hpe1 mutants still show higher photosynthetic efficiency under high-light stress (Fig. 6D), consistent with the greater accumulation of Pro in the hpe1 mutants (Fig. 2C), suggesting that loss of light energy and photodamage were reduced in hpe1 mutants under high light.
Figure 6.
Loss of the light energy of photosynthesis in the hpe1 mutants. A to C, Loss of light energy in the hpe1 mutants. Light-response curves of regulatable nonphotochemical quenching yield [Y(NPQ)] (A), nonregulatable nonphotochemical quenching yield [Y(NO)] (B), and chlorophyll fluorescence yield [Y(Chl fluor)] (C). PPDF, Photosynthetic photon flux density. Each data point represents at least 20 independent plants. D, Analysis of tolerance to high-light stress of the hpe1 mutants. Plants were exposed to high light at 1,200 μmol photons m–2 s–1 at 0, 3, and 6 h. The fraction of active PSII (Fv/Fm) was measured after dark incubation for 30 min. Data represent means ± se (n = 20). Significant differences were identified at 5% (*) and 1% (**) probability levels using Student’s t test. Differences between the hpe1 mutants and wild-type plants were more significant after high-light treatment.
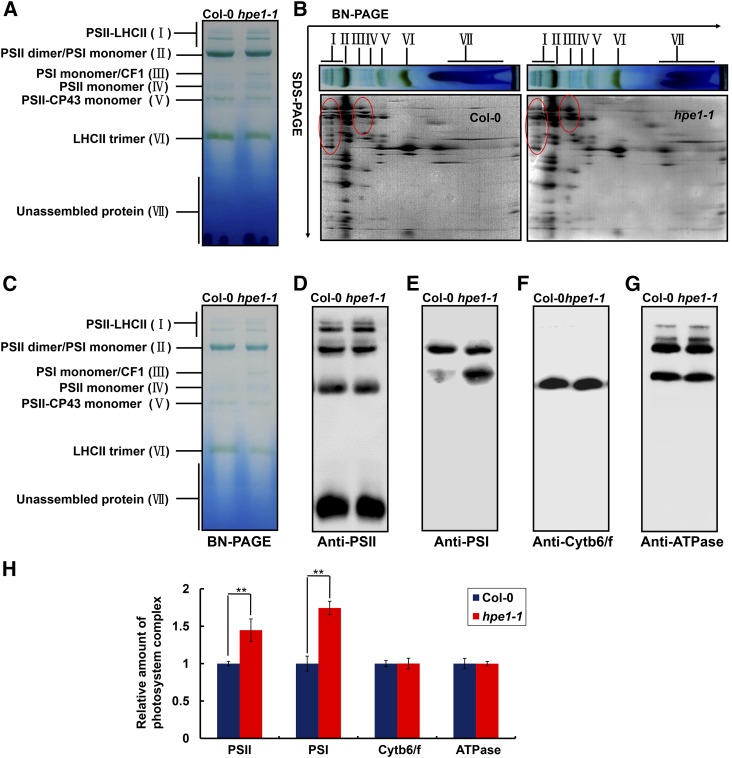
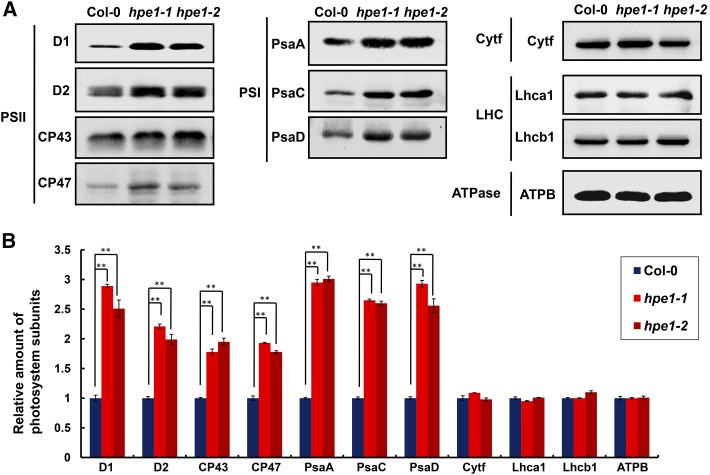
To examine the effect of the optimized light-harvesting system on the reaction center in hpe1 mutants, we determined the levels of photosystem complexes located in the specialized thylakoid membrane through blue native (BN)-PAGE analysis. Interestingly, on an equal chlorophyll basis, the abundance of the photosystem complexes that specifically associate with light-harvesting pigments, namely PSI and PSII, was approximately 44.7% and 74.4% greater in the hpe1 mutants than in the wild-type plants (Fig. 7). In particular, the abundance of the PSII core subunits D1, D2, CP43, and CP47 was approximately 120.9%, 71.1%, 67.2%, and 50.9% higher in the hpe1 mutants than in the wild-type plants, respectively. PSI subunits PsaA, PsaC, and PsaD were approximately 168.9%, 120.1%, and 120.4% higher in hpe1 mutants than in the wild-type plants, respectively (Fig. 8). However, the amounts of cytochrome b6/f and ATPase complex, which are not associated with light-harvesting pigments, showed no obvious difference between the hpe1 mutants and wild-type plants (Figs. 7 and 8).
Figure 7.
Analysis of photosystem complexes from the wild-type and hpe1 mutant plants. A, BN-PAGE analysis of chlorophyll-protein complexes. Equal thylakoid membranes (10 μg of chlorophyll) from the leaves of the wild type and hpe1-1 mutants were solubilized by treatment with 1% (w/v) dodecyl β-d-maltoside and separated by BN-PAGE. The assignments of the macromolecular protein complexes of thylakoid membranes indicated at left were identified according to Jin et al. (2014). B, Two-dimensional (2D) BN/SDS-PAGE fractionation of thylakoid membrane protein complexes. After separation in the first dimension on a nondenaturing gel, the protein lanes were applied to a denaturing 2D gel followed by Coomassie Blue staining. The proteins with different abundance between hpe1 mutants and wild-type plants are circled in red. The PSII and PSI proteins, which appeared more in hpe1 mutants, are circled in red. C, BN-PAGE for immunoblot analysis (3 μg of chlorophyll). D, A representative immunoblot with anti-CP43 antiserum used to probe the PSII complex. E, A representative immunoblot with anti-PsaA antiserum used to probe the PSI complex. F, A representative immunoblot with anti-cytochrome f antiserum used to probe the cytochrome b6/f (Cytb6/f) complex. G, A representative immunoblot with anti-ATPB antiserum used to probe the ATP synthase (ATPase) complex. Three independent biological replicates for all experiments were performed, and a representative one is shown. H, Proteins immunodetected from D to G were analyzed with Phoretix 1D Software (Phoretix International). Values (means ± se; n = 3 independent biological replicates) are given as ratios to protein amounts of the wild type (Col-0) and hpe1-1 mutants. **, P < 0.01, by Student’s t test.
Figure 8.
Analysis of thylakoid membrane protein accumulation in the wild type and hpe1 mutants. A, Thylakoid membrane proteins from the wild type (Col-0) and hpe1 mutants were separated by 15% SDS-urea-PAGE, transferred onto polyvinylidene difluoride membranes, and probed with antibody against known thylakoid membrane proteins obtained from Agrisera. Samples were loaded on an equal chlorophyll basis. Cytf, Cytochrome b6/f complex; LHC, light-harvesting complex; ATPase, ATP synthase complex. Three independent biological replicates for all experiments were performed, and a representative one is shown. B, Proteins immunodetected from A were analyzed with Phoretix 1D Software (Phoretix International). Values (means ± se; n = 3 independent biological replicates) are given as ratios to protein amounts of the wild type (Col-0) and hpe1 mutants. **, P < 0.01, by Student’s t test.
In addition, considering the lower chlorophyll content in the hpe1 mutants, we calculated the relative abundance of PSII complexes and proteins on an equal leaf area or fresh weight basis. On an equal fresh weight basis, the abundance of PSII and PSI complexes was still approximately 15.8% and 39.5% greater in hpe1 mutants than in the wild-type plants, respectively (Supplemental Fig. S10A). The abundance of PSII subunits D1, D2, CP43, and CP47 was approximately 100.8%, 59.2%, 56%, and 42.4% greater in hpe1 mutants than in the wild-type plants, respectively. The abundance of PSI subunits PsaA, PsaC, and PsaD was approximately 140.8%, 100.1%, and 100.3% higher in hpe1 mutants than in the wild-type plants, respectively (Supplemental Fig. S10B). However, the abundance of cytochrome b6/f and ATPase complexes was approximately 20.9% and 17.6% lower in hpe1 mutants than in the wild-type plants, respectively (Supplemental Fig. S10A). The abundance of cytochrome f, Lhca1, Lhcb1, and ATPB was approximately 12.8%, 24.1%, 20%, and 19.8% lower in hpe1 mutants than in the wild-type plants, respectively (Supplemental Fig. S10B). Thus, the increase in chlorophyll-associated photosystem complexes including both PSII and PSI may be due to photodamage mitigation in hpe1, which suggests that the balance between captured light and photochemical reactions may be improved in these mutants.
DISCUSSION
Maximizing light capture by optimizing light-harvesting pigments to improve photosynthetic efficiency, although very appealing, is also very challenging. While the principle of increasing efficiency by reducing antenna size is simple, this goal has not yet been achieved (Blankenship and Chen, 2013). In this study, we revealed that the chloroplast RNA-splicing factor HPE1 provides a means to improve photosynthetic efficiency through the optimization of light-harvesting pigments in higher plants.
Photosynthetic Efficiency Can be Improved through the Optimization of Light-Harvesting Pigment in Higher Plants
There are several lines of evidence supporting that the loss of HPE1 confers improved photosynthetic efficiency. First, two independent mutants of the hpe1 gene show greater Fv/Fm and actual photochemical efficiency of PSII (Fig. 1C), more PSII quantum yield, and faster electron transport rates (Fig. 1D), indicating that the activity of the light reaction was higher in hpe1 mutants than in wild-type plants. Second, the significant reduction in chlorophyll fluorescence (Fig. 6C) and heat, including regulatable nonphotochemical quenching yield (Fig. 6A) and nonregulatable nonphotochemical quenching yield (Fig. 6B) in the hpe1 mutants, indicates that the energy loss is mitigated in hpe1 mutants. Third, the content of carbohydrates, including Glc and Fru, was higher in hpe1 mutants (Fig. 2, A and B; Supplemental Fig. S1), implying that improved light-reaction performance can promote carbon fixation. Finally, increased fresh weight and dry weight indicated that efficient photosynthesis leads to improved biomass production (Fig. 2, D–F). These results suggest that light capture and light energy conversion in the light reaction are optimized in hpe1 mutants.
The antennae of photosynthetic systems consist of pigments specifically bound to membrane-associated proteins and are responsible for photon absorption, while excitation transfer delivers energy to the reaction centers, where some of the energy in the photon is captured by electron transfer processes (Blankenship, 2002; Green and Parson, 2003). Maximizing light capture by reducing the antenna size represents another strategy to optimize light capture and light energy conversion (Blankenship and Chen, 2013). In algae, decreasing light-harvesting pigments is an optional approach to reduce antenna size (Perrine et al., 2012). We found that levels of both chlorophyll a and chlorophyll b were reduced in hpe1 mutants (Fig. 5B). Moreover, the decrease of chlorophyll b content was more significant than that in chlorophyll a content, resulting in a higher chlorophyll a/b ratio in the hpe1 mutants than in the wild type (Fig. 5B), which is reversely related to antenna size (Kirst et al., 2012; Perrine et al., 2012; Blankenship and Chen, 2013). In addition, the level of another light-harvesting pigment, carotenoids, also was decreased slightly (Supplemental Fig. S9), together with less light-harvesting complex II, as revealed using the BN gel in hpe1 mutants (Fig. 7A), implying that the antenna size also is reduced in hpe1 mutants. Moreover, the adjustment of light-harvesting pigments optimizes light capture (Fig. 1C), decreases energy loss (Fig. 6, A–C), ameliorates photodamage of the photosystem (Figs. 6D, 7, and 8), and improves carbon fixation during photosynthesis. These results suggest that photosynthetic efficiency can be improved through the optimization of light-harvesting pigments in higher plants.
The Regulation of Light-Harvesting Pigment
In plants, light-harvesting pigments are regulated both transcriptionally and posttranslationally. Most of the genes encoding enzymes involved in chlorophyll biosynthesis have been identified in plants, and these catalyze the formation of 5-aminolevulinic acid, protoporphyrin IX, and mature chlorophyll. The transcriptional regulation of these genes remains unclear, although posttranslational regulation, including enzyme activity control, is well understood (Czarnecki and Grimm, 2012). To explore the possible cause underlying the optimization of light-harvesting pigment in hpe1 mutants, we first determined that HPE1 is a chloroplast protein (Fig. 3B), consistent with the predicted chloroplast-targeting signal in this protein (Fig. 3A). In addition, HPE1 also contains an RRM (Fig. 3A). The RRM motif is one of the most abundant protein domains in RNA-binding proteins and is found in all life kingdoms (Maris et al., 2005), suggesting that HPE1 is involved in the regulation of chloroplast RNA processing. RT-PCR and sequencing analysis showed that the unspliced pre-mRNAs, including trnK, ropC1, and atpF, are all increased in hpe1 mutants (Fig. 3C; Supplemental Fig. S4), suggesting that HPE1 regulates plastid RNA splicing. RNA immunoprecipitation analysis indicates that HPE1 associates with target RNAs directly (Fig. 3D). Together, these results suggest that HPE1 is a novel chloroplast RNA-splicing regulator. In addition, BiFC and coimmunoprecipitation analyses showed that HPE1 interacts with other plastid RNA-splicing factors, including CAF1 and OTP51 (Fig. 4), which share common target RNAs with HPE1 (Asakura and Barkan, 2006; de Longevialle et al., 2008), suggesting that HPE1 may form a complex specifically with CAF1 and OTP51 to coregulate chloroplast group II intron splicing.
Interestingly, the chlorophyll contents in mutants of chloroplast RNA-splicing regulators, including CAF1 and OTP51 (Asakura and Barkan, 2006; de Longevialle et al., 2008), are decreased. The contents of chlorophyll also are decreased in hpe1 mutants, very similar to caf1 and otp51 mutants. However, the degree of chlorophyll decrease is different in mutants of different chloroplast RNA-splicing regulators. Deficiency in some chloroplast RNA-splicing events results in albinism, but some also result in yellow or light-green leaves (Kroeger et al., 2009). The leaves of hpe1 mutants are light green and show less of a decrease in chlorophyll compared with mutants of other chloroplast RNA-splicing factors (Figs. 2D and 5B; Supplemental Fig. S9). The slight chlorophyll decrease in hpe1 mutants may optimize the light-harvesting antenna. The plastid-encoded gene trnE-UUC encodes tRNAGlu, whose amino acyl form tRNA-Glu is a precursor for the biosynthesis of tetrapyrroles (e.g. heme and chlorophyll) in plants, archaea, and most bacteria (Levicán et al., 2005). Quantitative RT-PCR assay indicates that the transcript of trnE-UUC is not affected in hpe1 mutants (Supplemental Fig. S5), suggesting that the down-regulation of chlorophyll content in hpe1 mutants may not be due to the alteration of trnE-UUC expression.
We found that multiple nucleus-encoded key genes of the chlorophyll synthesis pathway were markedly down-regulated in hpe1 mutants (Fig. 5A), suggesting that defects in chlorophyll accumulation may be caused by the down-regulation of chlorophyll synthesis-related genes. The down-regulation of chlorophyll-related genes reduces the size of the light-harvesting complex, ensuring optimized light capture and energy conversion during the light reaction. Finally, optimization of light reactions improves the efficiency of carbon fixation, which leads to greater photosynthetic efficiency and biomass production (Supplemental Fig. S11). It is seemingly perplexing that the loss of a plastid RNA-splicing regulator affects the expression of nuclear genes. One probability is that the loss of hpe1 elicits plastid-to-nucleus signaling, leading to the down-regulation of these nuclear genes. A significant consequence of the hpe1 mutation is the accumulation of unspliced RNAs; however, the spliced RNAs and the encoded proteins are only slightly reduced (Supplemental Figs. S4B and S10). Thus, we speculate that such plastid-to-nucleus signaling is more likely triggered by an accumulation of unspliced mRNAs (Fig. 5A; Supplemental Fig. S7), which is reminiscent of the well-known unfolded protein response that is triggered by an accumulation of unfolded proteins in the endoplasmic reticulum (Bernales et al., 2006; Howell, 2013; Popp and Maquat, 2013). However, we cannot introduce exogenous unspliced pre-mRNAs to plastids to directly test this hypothesis due to technological limits, and we are not able to exclude the possibility that such plastid-to-nucleus signaling is triggered by the down-regulation of plastid proteins.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
T-DNA Arabidopsis (Arabidopsis thaliana) mutants used in this study were in the Col-0 background. The hpe1-1 and hpe1-2 mutants were obtained from the Arabidopsis Biological Resource Center (stock nos. SALK_012657C and SALK_092951C). Arabidopsis was grown in the soil in a growth chamber (100 μmol photons m−2 s−1, 12-h/12-h photoperiod, 21°C, and 60% relative humidity). Plants for the chlorophyll fluorescence assays and protein analysis were 3 and 5 weeks of age, respectively. To study the effects of high light, plants were placed in a high-light growth chamber (1,200–1,500 μmol photons m−2 s−1).
Determination of Carbohydrate and Amino Acid Levels
Plant leaf tissues of the same area were placed into liquid N2 immediately and then ground using a pestle. Quantification of monosaccharides (Glc and Fru) and amino acids was performed using GC-MS-based methods as described (Lisec et al., 2006). The degradation of carbohydrate was analyzed after 5 h of DCMU treatment according to methods described previously (Kowallik and Schatzle, 1980).
Pigment Analysis
Chlorophyll from 3-week-old plants was extracted with 80% acetone in 2.5 mm HEPES-KOH (pH 7.5), and the amount of chlorophyll was determined as described previously (Wellburn, 1994). Carotenoids were extracted and analyzed as described previously using spectrofluorometry (Yang et al., 2012) and HPLC (Pogson et al., 1996; Li et al., 2009). Pigments were identified by comparing retention times with reference standards.
Chlorophyll Fluorescence
Chlorophyll fluorescence parameters were measured with the MAXI version of the IMAGING-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments). Plants were dark adapted for 30 min before measurements were made, and light-response curves were determined as described (Lu et al., 2011).
Isolation of Thylakoid Membranes
Thylakoid membranes were prepared as described previously (Robinson and Yocum, 1980). Isolated thylakoid membranes were quantified on the basis of total chlorophyll as described (Porra et al., 1989).
Production of Anti-HPE1 Polyclonal Antibodies
Affinity-purified anti-HPE1 polyclonal antibodies were prepared by GenScript. A 15-amino acid peptide (corresponding to amino acids 467–481 of HPE1) with an additional N-terminal Cys residue, CFDKPEAKPARVEGK, was synthesized, conjugated with keyhole limpet hemocyanin, and used to induce antibodies against HPE1.
RT-PCR and Quantitative Real-Time RT-PCR
Total RNA was extracted from Arabidopsis rosette leaves using the RNeasy Plant Mini Kit (Qiagen). The RNA samples were reverse transcribed into first-strand cDNA using the PrimeScript RT Reagent Kit (TaKaRa). Both random and oligo(dT)-containing mix primers were used to reverse transcribe the first-strand cDNA. For RT-PCR, UBIQUITIN10 was used as the control gene. Quantitative real-time RT-PCR was performed using gene-specific primers and SYBR Premix ExTaq reagent (TaKaRa) on a real-time RT-PCR System (Roche; LC480) according to the manufacturer’s instructions. Reactions were performed in triplicate for each sample, and expression levels were normalized against ACTIN and UBIQUITIN4.
BN-PAGE and Immunoblot Analyses
BN-PAGE was performed as described (Schägger et al., 1994) with modifications (Peng et al., 2006). For the quantification of thylakoid proteins, gels were loaded on an equivalent chlorophyll basis in amounts ensuring that immunodetection was in the linear range. Primary antibodies and antisera were induced in rabbits. Antisera against photosynthetic proteins were purchased from Agrisera.
Subcellular Localization of GFP Fusions and BiFC
Subcellular localization of GFP fusion proteins and BiFC were performed as described previously (Zhang et al., 2011).
Immunoprecipitation
Immunoprecipitation was performed as described (Zhang et al., 2015) with minor modifications. Four milliliters of Arabidopsis protoplasts was transfected with 400 μg of plasmid of 35S:CAF1-HA or 35S:OTP51-HA. Total proteins were extracted with 500 μL of protein extraction buffer, and 20 μL of protein extract was used as input. The total cell extracts were further incubated with 30 μL of anti-HA affinity matrix (Roche) for 6 h at 4°C with rotation. After washing five times with ice-cold phosphate-buffered saline buffer (pH 7.4), the bound proteins were eluted by boiling the gel using 30 μL of SDS-PAGE sample buffer without β-mercaptoethanol and loaded onto an SDS-PAGE device for immunoblotting.
Analysis of RNA Splicing
Analysis of RNA splicing by RT-PCR was performed as described (Valkov et al., 2009). The RT-PCR products of unspliced and spliced RNA were identified by sequencing. Quantitative RT-PCR analysis of RNA splicing was performed as described (de Longevialle et al., 2008). The primers used are listed in Supplemental Table S1.
RNA Immunoprecipitation
RNA immunoprecipitation assays were performed (García-Andrade et al., 2013) as described with minor revisions. Two grams of leaf tissue from Arabidopsis plants (4 weeks of age) was ground to a fine powder with a mortar and pestle in liquid N2 and homogenized in 12.5 mL g−1 lysis buffer (50 mm Tris-HCl, pH 7.4, 2.5 mm MgCl2, 100 mm KCl, 0.1% Nonidet P-40, 1 μg mL−1 leupeptin, 1 μg mL−1 aprotonin, 0.5 mm phenylmethylsulfonyl fluoride, one tablet of Complete proteinase inhibitor [Roche], and 50 units mL−1 RNase OUT [Invitrogen]). Cell debris were pelleted by centrifugation for 5 min at 12,000g at 4°C. Clarified lysates were incubated with 4 μg mL−1 anti-HPE1 antibody for 15 min at 4°C and then with 100 μL mL−1 Protein A agarose (Roche) for 30 min at 4°C. Beads were washed six times for 10 min with lysis buffer at 4°C and then divided for protein and RNA analyses. RNAs were recovered by incubating the beads in 0.5 volumes of proteinase K buffer (0.1 m Tris-HCl, pH 7.4, 10 mm EDTA, 300 mm NaCl, 2% SDS, and 1 μg μL−1 proteinase K [Roche]) for 15 min at 65°C, extracted with saturated phenol, phenol:chloroform:isoamyl alcohol, and chloroform, and precipitated with ethanol. For RT-PCR assays, 1 μg of total RNA was used for the input fraction, and 20% of the RNA immunoprecipitate was used for immunoprecipitation.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Degradation of metabolite production in hpe1 mutant and wild-type plants.
Supplemental Figure S2. Growth parameters of wild-type and hpe1 mutant plants.
Supplemental Figure S3. Transcript levels of representative genes that encode chloroplast proteins were analyzed using quantitative real-time RT-PCR.
Supplemental Figure S4. RNA splicing analysis in hpe1 mutants and wild-type plants.
Supplemental Figure S5. The level of TrnE transcript was analyzed using quantitative real-time RT-PCR.
Supplemental Figure S6. Interaction analysis of HPE1 with other splicing factors involved in the splicing of trnK, rpoC1, and atpF introns using BiFC.
Supplemental Figure S7. Analysis of the expression of chlorophyll-related genes in other plastid splicing mutants.
Supplemental Figure S8. Analysis of the expression of plastid-to-nucleus signaling-related genes.
Supplemental Figure S9. HPLC analysis of pigments in the wild type (Col-0), hpe1-1, and hpe1-2 plants.
Supplemental Figure S10. Protein amounts of the photosystems were calculated based on fresh weight.
Supplemental Figure S11. Presumed working model of HPE1 in the regulation of photosynthesis.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Yonggang Zheng (Johns Hopkins University) for critical reading of the article and the Arabidopsis Biological Resource Center for providing plant materials.
Glossary
- GC-MS
gas chromatography-mass spectrometry
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- RRM
RNA recognition motif
- RNP
ribonucleoprotein domain
- RT
reverse transcription
- BiFC
bimolecular fluorescence complementation
- BN
blue native
- Col-0
Columbia-0
Footnotes
This work was supported by the grants from the National Science and Technology Major Project Foundation of China (grant no. 2016ZX08009003-005-005), the National Natural Science Foundation of China (grant nos. 31425003 and 31500195), the Natural Science Foundation of Guangdong Province, PR China (grant no. 2014A030310491), the China postdoctoral Science Foundation (grant nos. 2015M572399 and 2016T90808), and the Fundamental Research Funds for the Central Universities.
Articles can be viewed without a subscription.
References
- Asakura Y, Barkan A (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22: 487–508 [DOI] [PubMed] [Google Scholar]
- Blankenship RE. (2002) Molecular Mechanisms of Photosynthesis. Blackwell Science, Oxford [Google Scholar]
- Blankenship RE, Chen M (2013) Spectral expansion and antenna reduction can enhance photosynthesis for energy production. Curr Opin Chem Biol 17: 457–461 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- Cottage AJ, Mott EK, Wang JH, Sullivan JA, MacLean D, Tran L, Choy MK, Newell CA, Kavanagh TA, Aspinall S, et al. (2007) GUN1 (GENOMES UNCOUPLED1) encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. Photosynth Res 91: 276 [Google Scholar]
- Czarnecki O, Grimm B (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot 63: 1675–1687 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- García-Andrade J, Ramírez V, López A, Vera P (2013) Mediated plastid RNA editing in plant immunity. PLoS Pathog 9: e1003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR, Parson WW (2003) Light-Harvesting Antennas in Photosynthesis. Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Jin H, Liu B, Luo L, Feng D, Wang P, Liu J, Da Q, He Y, Qi K, Wang J, et al. (2014) HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26: 1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stäbler N, Schönfeld B, Kreuzaler F, Peterhänsel C (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Kirst H, García-Cerdán JG, Zurbriggen A, Melis A (2012) Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol 158: 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kowallik W, Schatzle S (1980) Enhancement of carbohydrate degradation by blue light. In Senger H, ed, The Blue Light Syndrome. Springer, Berlin, pp 344–360 [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levicán G, Katz A, Valenzuela P, Söll D, Orellana O (2005) A tRNA(Glu) that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett 579: 6383–6387 [DOI] [PubMed] [Google Scholar]
- Li Z, Ahn TK, Avenson TJ, Ballottari M, Cruz JA, Kramer DM, Bassi R, Fleming GR, Keasling JD, Niyogi KK (2009) Lutein accumulation in the absence of zeaxanthin restores nonphotochemical quenching in the Arabidopsis thaliana npq1 mutant. Plant Cell 21: 1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR (2014) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG (2015) Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66 [DOI] [PubMed] [Google Scholar]
- Lu Y, Hall DA, Last RL (2011) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S (2001) Overexpression of a cyanobacterial fructose-1,6-sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19: 965–969 [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine Z, Negi S, Sayre RT (2012) Optimization of photosynthetic light energy utilization by microalgae. Algal Research 1: 134–142 [Google Scholar]
- Perron K, Goldschmidt-Clermont M, Rochaix JD (2004) A multiprotein complex involved in chloroplast group II intron splicing. RNA 10: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang JM, Liu W, Zabirnyk O (2010) Proline metabolism and microenvironmental stress. Annu Rev Nutr 30: 441–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D (1996) Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8: 1627–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE (2013) Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson HH, Yocum CF (1980) Cyclic photophosphorylation reactions catalyzed by ferredoxin, methyl viologen and anthraquinone sulfonate: use of photochemical reactions to optimize redox poising. Biochim Biophys Acta 590: 97–106 [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, Bock R, Cardi T (2009) Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol 150: 2030–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. (1994) The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Yang S, Zeng X, Li T, Liu M, Zhang S, Gao S, Wang Y, Peng C, Li L, Yang C (2012) AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis. J Exp Bot 63: 3959–3973 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.