A tomato vacuolar invertase inhibitor regulated by the global fruit ripening regulator RIPENING INHIBITOR is responsible for sucrose metabolism and fruit ripening.
Abstract
Fruit ripening is a complex process that involves a series of physiological and biochemical changes that ultimately influence fruit quality traits, such as color and flavor. Sugar metabolism is an important factor in ripening, and there is evidence that it influences various aspects of ripening, although the associated mechanism is not well understood. In this study, we identified and analyzed the expression of 36 genes involved in Suc metabolism in ripening tomato (Solanum lycopersicum) fruit. Chromatin immunoprecipitation and gel mobility shift assays indicated that SlVIF, which encodes a vacuolar invertase inhibitor, and SlVI, encoding a vacuolar invertase, are directly regulated by the global fruit ripening regulator RIPENING INHIBITOR (RIN). Moreover, we showed that SlVIF physically interacts with SlVI to control Suc metabolism. Repression of SlVIF by RNA interference delayed tomato fruit ripening, while overexpression of SlVIF accelerated ripening, with concomitant changes in lycopene production and ethylene biosynthesis. An isobaric tags for relative and absolute quantification-based quantitative proteomic analysis further indicated that the abundance of a set of proteins involved in fruit ripening was altered by suppressing SlVIF expression, including proteins associated with lycopene generation and ethylene synthesis. These findings provide evidence for the role of Suc in promoting fruit ripening and establish that SlVIF contributes to fruit quality and the RIN-mediated ripening regulatory mechanisms, which are of significant agricultural value.
Fleshy fruit ripening is a complex process involving a series of physiological and biochemical changes that influence fruit characteristics such as color, flavor, aroma, and texture (Giovannoni, 2004). Considerable progress has been made in elucidating the biochemical and molecular basis of fruit ripening, including the discovery and characterization of associated transcription factor networks and phytohormone signaling pathways (Alba et al., 2005; Giovannoni, 2007; Klee and Giovannoni, 2011). As an example, the MADS box transcription factor, RIPENING INHIBITOR (RIN) has been defined as a global regulator of ripening and has been shown to play a key role in modulating ethylene biosynthesis, carbohydrate metabolism, and aroma compound production (Vrebalov et al., 2002; Ito et al., 2008; Qin et al., 2012). Much current research into ripening focuses on the identification of new candidate genes that affect fruit quality traits (Fujisawa et al., 2013; Zhong et al., 2013), an important example of which is flavor.
In general, fruit ripening is characterized by a substantial accumulation of carbohydrates (Fraser et al., 1994) that provide energy for fruit development and contribute to its flavor (Rolland et al., 2002; Zhu et al., 2013), thereby promoting consumption and seed dispersal. Indeed, the content and composition of sugars largely determine fruit sweetness and nutritional quality (Borsani et al., 2009). Although the regulatory effects of sugars on photosynthetic activity and plant metabolism have long been recognized (Godt and Roitsch, 1997; Koch, 2004; Kocal et al., 2008), the role of sugars as signaling molecules in fruit is less understood. In many plant species, the major form of carbohydrate transported from the leaves to the fruit is Suc, which acts as an important form of energy storage/translocation and plays a central role in growth and development. There is also increasing evidence that Suc may play a nonnutritive role as a regulator of cellular metabolism, possibly by altering gene expression (Huber and Huber, 1996; Rolland et al., 2002; Vaughn et al., 2002; Ruan et al., 2010; Eveland and Jackson, 2012). Suc imported into fruits is degraded into hexoses (Glc and Fru) for various metabolic and biosynthetic processes and resynthesized in the cytosol, vacuole, and apoplast (Koch, 2004). Suc turnover in vivo is catalyzed by Suc synthase (SS) and invertase (Ruan et al., 2010). SS converts Suc into UDP-Glc and Fru in the presence of UDP, whereas invertase hydrolyzes Suc into Glc and Fru in an irreversible reaction (Klann et al., 1996; Ruan et al., 2010).
Invertases play a major role in plant development and in responses to biotic and abiotic stresses and have been suggested to have important regulatory functions in carbon metabolism and development in fruit (Jin et al., 2009). According to their subcellular locations, invertases are classified as cell wall invertase (CWI), vacuolar invertase (VI), and cytoplasmic invertase (CI) or neutral invertase (NI) forms (Sturm, 1999). In tomato fruit, CWI activity is tightly linked with fruit sugar levels (Fridman et al., 2004) and is thought to hydrolyze extracellular Suc when the apoplastic pathway is active (Jin et al., 2009). Since the vacuole is a storage organelle for sugars, VI is believed to modulate the Suc/hexose ratio in fruits. Antisense suppression of TIV1, a VI gene, in tomato was reported to result in reduced hexose accumulation during fruit ripening (Klann et al., 1996). Similar findings were reported in other fruits, such as grape (Vitis vinifera) berry (Davies and Robinson, 1996) and muskmelon (Cucumis melo; Yu et al., 2008), suggesting that VI controls the sugar composition in broad range of fleshy fruit. Given the critical role of VI in fruit development and ripening, its activity is likely to be tightly regulated in vivo. Studies have shown that the invertase is regulated by a range of signals at the transcriptional level (Long et al., 2002; Proels and Roitsch, 2009) and that invertase activity is controlled posttranslationally by inhibitor proteins (Hothorn et al., 2004).
Invertase inhibitors bind to invertases to form inactive complexes. Vacuolar invertase inhibitors (VIFs) have been isolated from tobacco (Nicotiana tabacum), Arabidopsis (Arabidopsis thaliana), and potato (Solanum tuberosum; Greiner et al., 1998; Link et al., 2004; Brummell et al., 2011). Moreover, ectopic expression of Nt-inhh, a tobacco VIF, in potato tubers strongly reduced VI activity and blocked hexose accumulation (Greiner et al., 1999), while heterologous expression of recombinant potato VIF (INH2) suppressed potato VI activity in vitro (Brummell et al., 2011). However, to our knowledge nothing has been reported regarding the action or significance of fruit VIFs, which, if present, may be important for regulating VI activity and hence the sugar composition of fruits.
Previous studies suggested a link between sugars and pigment metabolism in fleshy fruit: In vitro Suc supplementation promotes color changes in citrus (Citrus unshiu) fruit epicarp (Iglesias et al., 2001) and in tomato (Solanum lycopersicum) fruit pericarp discs (Télef et al., 2006). However, evidence that the internal quality trait (sugar levels) and external quality trait (color) of fruits are linked in vivo is lacking. Moreover, much remains to be learned about how sugars affect fruit quality traits and regulate ripening remains. In this study, we identified a set of genes involved in Suc synthesis and degradation, including VI and VIF, in tomato as direct targets of the ripening regulator RIN. Our findings indicate a role for Suc in promoting pigment biosynthesis and fruit ripening in vivo and establish VIF as a novel genetic tool for regulating fruit ripening.
RESULTS
Expression Analysis of Genes Involved in Suc Metabolism during Tomato Fruit Ripening
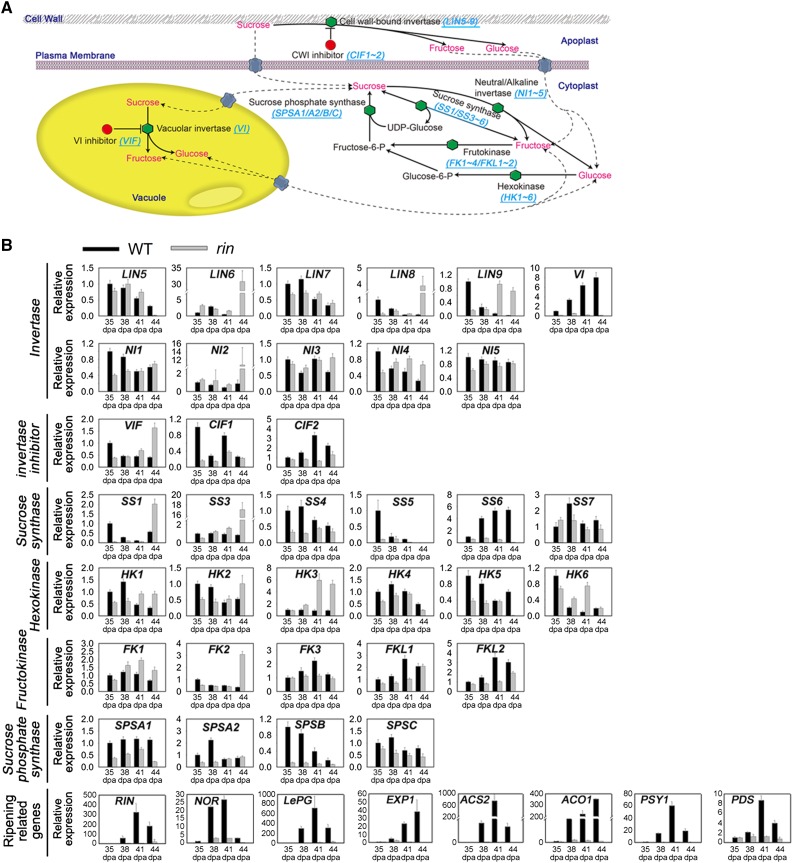
The Suc metabolic pathway is well established; however, the expression profiles of genes involved in this process during fruit ripening have not been well characterized. Suc metabolism includes the degradation and resynthesis of Suc in the cytosol, vacuole, and apoplast. Several gene families are involved in these processes, including those encoding invertase, invertase inhibitor, Suc synthase, hexokinase, fructokinase, and Suc phosphate synthase (Carrari and Fernie, 2006; Fig. 1A). We identified the orthologs of these genes in tomato in the SOL Genomics Network (SGN; http://solgenomics.net/) databases using the reported gene sequence of each gene family from tomato or another Solanaceous species as a query. A total of 36 genes were identified (Supplemental Table S1), of which 20 have previously been reported (Klann et al., 1992; Elliott et al., 1993; Wang et al., 1993; Kanayama et al., 1997; Menu et al., 2001; Fridman and Zamir, 2003; Lunn and MacRae, 2003; German et al., 2004; Kandel-Kfir et al., 2006; Jin et al., 2009; Goren et al., 2011; Li et al., 2012). The 16 unreported genes were named numerically or on the basis of the results of phylogenetic analysis. The expression of these 36 genes during ripening in wild-type tomato or in fruits at an equivalent stage, based on the number of days after anthesis, in the rin mutant during fruit ripening was analyzed by real-time quantitative reverse transcription (qRT)-PCR, using gene specific primers (Supplemental Table S2). To understand the temporal expression of these Suc metabolism-related genes in the overall context of fruit ripening, the expression of ripening marker genes (RIN, NOR, LePG, EXP1, ACS2, ACO1, PSY1, PDS; Supplemental Table S3) was also determined.
Figure 1.
Expression of genes involved in Suc metabolism during fruit ripening. A, Diagram of Suc metabolism in tomato. LIN, Cell wall invertase; VI, vacuolar invertase; NI, cytoplasmic invertase; VIF, vacuolar invertase inhibitor; CIF, cell wall invertase inhibitor; SS, Suc synthase; HK, hexokinase; FK, fructokinase; SPS, Suc phosphate synthase. B, Expression analyses of genes involved in Suc metabolism during fruit ripening in the wild type (WT) and the rin mutant by qRT-PCR. The stages of fruit ripening include 35 DPA, 38 DPA, 41 DPA, and 44 DPA. The gene transcript levels are normalized against the ACTIN gene, followed by normalization against the wild type at 35 DPA. Values are means ± sd of three independent experiments. The expression of ripening marker genes is shown. RIN, Ripening inhibitor; NOR, nonripening; LePG, polygalacturonase A; EXP1, expansin 1; ACS2, 1-aminocyclopropane-1-carboxylate synthase 2; ACO1, 1-aminocyclopropane-1-carboxylate oxidase; PSY1, phytoene synthase 1; PDS, phytoene desaturase.
Eleven invertase genes were identified in the tomato genome, of which five are predicted to encode CWI proteins (LIN5–9); one encodes a VI (VI), and five encode CI (NI1–5). The expression of these genes had different patterns from each other in the rin mutant during ripening (Fig. 1B). Notably, LIN8, LIN9, NI2, and NI4 showed higher expression levels in rin, whereas VI expression was significantly lower in the mutant throughout ripening.
The activity of invertase has been shown to be controlled posttranslationally by binding to inhibitor proteins (Hothorn et al., 2004), forming an inactive complex. Although no inhibitor proteins were identified in the tomato genome for CI, one gene encoding an inhibitory protein, VIF, was found for VI, and two inhibitors, CIF1 and CIF2, were identified for CWI. The expression of VIF was higher in the fruit of rin mutant than in the wild type, while the opposite expression patterns were found for CIF1 and CIF2 (Fig. 1B). These results indicated that RIN promotes the expression of the VI gene and suppresses the expression of its inhibitor. Conversely, RIN suppresses the expression of the CWI gene and promotes the expression of its inhibitor.
Six Suc synthase genes (SS1, SS3, SS4, SS5, SS6, and SS7) were identified in the tomato genome, which showed different patterns of gene expression from each other in the rin mutant at equivalent days after anthesis, and the expression of SS6 was notably lower in the mutant throughout ripening (Fig. 1B). In addition, six hexokinase genes (HK1–6) and six fructokinase genes (FK1–FKL2) were identified in the tomato genome. Their expression also showed different patterns from each other in the rin mutant during ripening (Fig. 1B), with the exception of the fructokinase gene, FK4, whose expression has been specifically linked to stamen development (German et al., 2002) and was not detected in this study. Lastly, four Suc phosphate synthase genes (SPSA1–SPSC) were identified, and their expression was shown to be generally lower in rin mutant compared with the wild type (Fig. 1B).
ChIP-qPCR Analysis Reveals That RIN Directly Binds to the Promoters of Genes Involved in Suc Metabolism
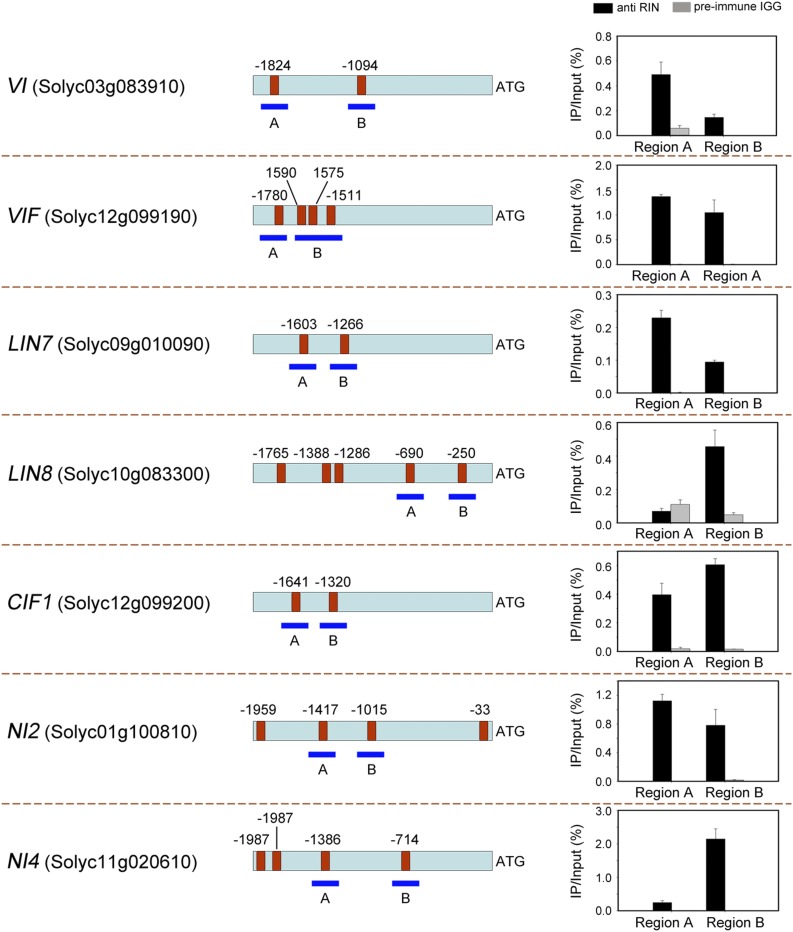
RIN regulates the expression of ripening-related genes, either directly or indirectly (Fujisawa et al., 2013). To examine whether genes involved in Suc metabolism are also directly regulated by RIN, a chromatin immunoprecipitation (ChIP) assay was carried out. An analysis of the promoter regions of the genes analyzed in this study identified varying numbers of CArG-box binding motifs (Supplemental Table S1), which are elements that are commonly found in MADS-box transcription factor binding sites (Ito et al., 2008). The cross-linked DNA-protein complexes were immunoprecipitated with anti-RIN polyclonal antibody that was prepared using a bacterial expressed recombinant RIN protein. To amplify the promoter sequences surrounding the CArG-box binding sites from the immunoprecipitated DNA, specific primers were designed (Supplemental Table S4), and the binding of RIN to the promoter of ACC synthase 2 (ACS2), a known RIN-target gene (Ito et al., 2008), served as a positive control (Supplemental Fig. S1).
ChIP screening using affinity-purified RIN antibodies, in combination with quantitative PCR (ChIP-qPCR), resulted in an enrichment in the promoter regions of 16 genes compared with when nonspecific antibodies (preimmune rabbit IgG) were used (Fig. 2; Supplemental Fig. S2). Notably, RIN directly bound to the promoter of VI and VIF (Fig. 2). Taken together with the gene expression analysis showing that VI expression was lower in the rin mutant than in the wild type, while VIF expression was higher (Fig. 1B), these data suggest that RIN modulates Suc degradation in the vacuole during fruit ripening. A similar result was found for genes encoding CWIs, specifically LIN7 and LIN8, and the gene encoding the inhibitory protein of CWI, CIF1 (Fig. 2). However, while the expression of LIN7 and LIN8 was up-regulated, the expression of CIF1 was down-regulated in the rin mutant during fruit ripening (Fig. 1B). This indicates that Suc degradation associated with CWI is inhibited by the RIN protein during fruit ripening. The ChIP-qPCR analysis also showed that RIN directly bound to the promoter of NI2 and NI4 (Fig. 2), which encodes CI. Lastly, RIN was shown to bind directly to the promoter of several other genes involved in Suc metabolism, e.g. SS3, HK3, and SPSA1 (Supplemental Fig. S2).
Figure 2.
Chromatin immunoprecipitation reveals direct binding of RIN to the promoters of genes involved in Suc metabolism. The promoter regions of the target genes are indicated. Red boxes represent CArG-box elements, and numbers above the box indicate the position of these motifs relative to the translational start site. The blue fragments with uppercase letters represent the regions used for ChIP-qPCR. Values are the percentage of DNA fragments that were coimmunoprecipitated with specific anti-RIN antibodies or nonspecific antibodies (preimmune rabbit IgG) relative to the input DNA. Error bars represent the sd of three independent experiments. VI, Vacuolar invertase; VIF, vacuolar invertase inhibitor; LIN, cell wall invertase; CIF, cell wall invertase inhibitor; NI, cytoplasmic invertase.
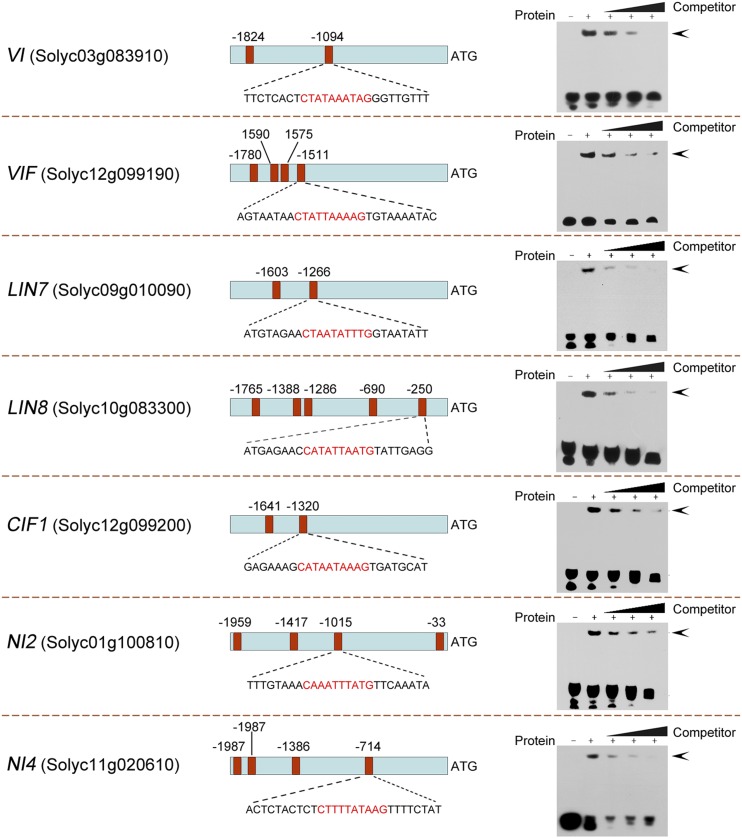
Gel Mobility Shift Assay Confirms That RIN Binds to the Promoters of Suc Metabolic Genes
In order to confirm the binding ability of RIN to the promoter regions of genes identified in the ChIP assay, an electrophoretic mobility shift assay (EMSA) was performed with purified recombinant RIN protein. Specifically we examined the ability of RIN to bind to double-stranded and biotin-labeled probes (Supplemental Table S5) for the genes shown in Figure 2 (VI, VIF, LIN7, LIN8, CIF1, NI2, and NI4), which contained CArG-box motifs. For each gene, a band shift was observed when the purified RIN protein was mixed with the biotin-labeled probe (Fig. 3). Formation of the DNA-protein complexes could be prevented by addition of an excessive amount of the corresponding unlabeled probe, indicating specific binding of RIN to the biotin-labeled probe. Based on the different extent of competition by the unlabeled DNA fragment that was observed among these genes, we concluded that RIN had different binding affinities for the promoters of the analyzed genes.
Figure 3.
RIN binding to the regulatory regions of target genes revealed by EMSA. The promoter regions of the target genes are indicated. Red boxes indicate CArG-box elements in the promoter region, and numbers above represent the position of these motifs relative to the translational start site. The probe sequences used for EMSA for each target gene are shown, with red letters representing the CArG box. The protein-DNA complexes were separated on 6% native polyacrylamide gels. Triangles indicate the increasing amounts (10×, 100×, or 1000×) of unlabeled probes used for competition. The specific complexes formed are indicated by arrowheads. VI, Vacuolar invertase; VIF, vacuolar invertase inhibitor; LIN, cell wall invertase; CIF, cell wall invertase inhibitor; NI, cytoplasmic invertase.
Identification of SlVIF and Assessment of Its Physical Interaction with VI
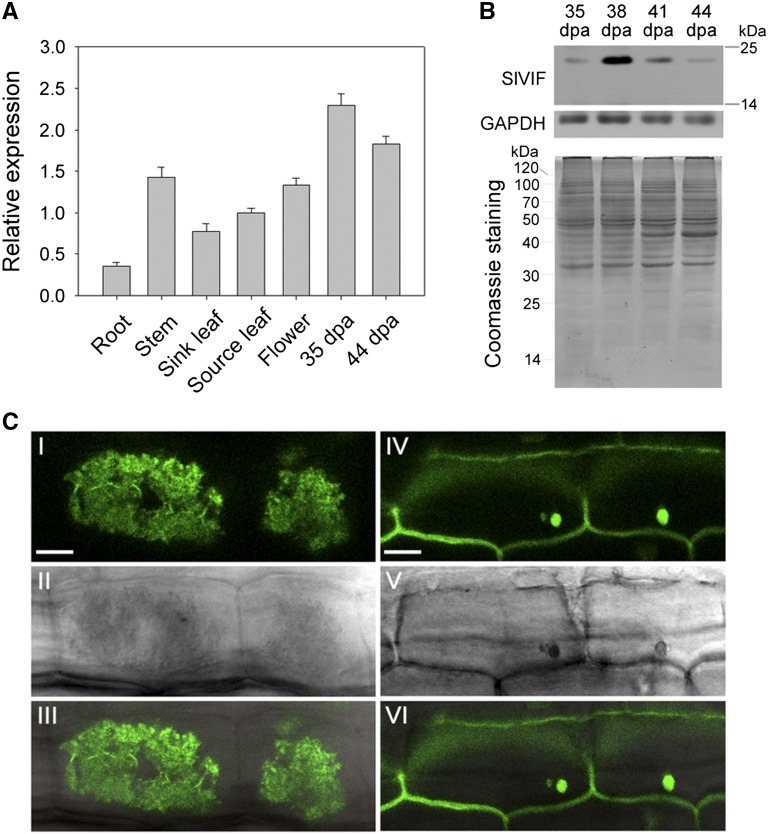
Having demonstrated that VI and VIF were directly regulated by RIN, the putative VI inhibitor gene, VIF, was chosen for further functional analysis. VIF genes have been cloned from Arabidopsis, tobacco, and potato, and reported to modulate Suc metabolism (Link et al., 2004); however, little is known about the expression of VIF genes in tomato fruit or their roles in ripening. Tomato VIF (GenBank accession no. KC007445; hereafter referred to as SlVIF) was cloned based on sequence similarity to the N. tabacum VIF (AY145781). SlVIF showed 81% and 71% amino acid identity with NtVIF (Y12806) and StVIF (FJ810206), respectively (Supplemental Fig. S3). All deduced proteins contained the four conserved Cys residues that are a hallmark of all known plant invertase inhibitors (Rausch and Greiner, 2004). We observed that SlVIF was expressed in both vegetative and reproductive organs, but at different levels (Fig. 4A). Notably, SlVIF mRNA levels were high in fruit, and SlVIF protein levels peaked at 38 d post anthesis (DPA) before dropping (Fig. 4B), consistent with SlVIF transcript abundance.
Figure 4.
Characterization of tomato SlVIF. A, qRT-PCR analyses of SlVIF in vegetative and reproductive tomato organs. The ACTIN gene was used as an internal control. Values are the means of three biological replicates. Bars represent standard deviations. B, Western-blot analysis of SlVIF protein abundance during fruit ripening. Proteins were isolated from fruit at 35 DPA, 38 DPA, 41 DPA, and 44 DPA. An anti-glyceraldehyde-3-phosphate dehydrogenase immunoblot was used as a protein control. In addition, Coomassie Brilliant Blue staining was used as a loading control. C, Subcellular localization of GFP:SlVIF fusion protein. I, to III, Stable expression of the GFP:SlVIF fusion protein in tomato root cells, showing fluorescent signal in the vacuole. IV to VI, Stable expression of GFP alone in tomato root cells, showing fluorescent signals throughout the cell. I and IV show fluorescent images; II and V are the same images viewed using bright-field microscopy. III and VI are overlays of fluorescent and bright-field images. Bar = 10 μm.
To confirm that SlVIF indeed localizes in the vacuole, we transformed tomato with a construct encoding SlVIF fused to a GFP marker, GFP:SlVIF, as well as with a GFP-only construct. When we imaged transgenic tomato root cells, we observed that the GFP:SlVIF fusion protein produced a strong signal in the vacuole (Fig. 4C), while the GFP-only control displayed fluorescence throughout the cell (Fig. 4C).
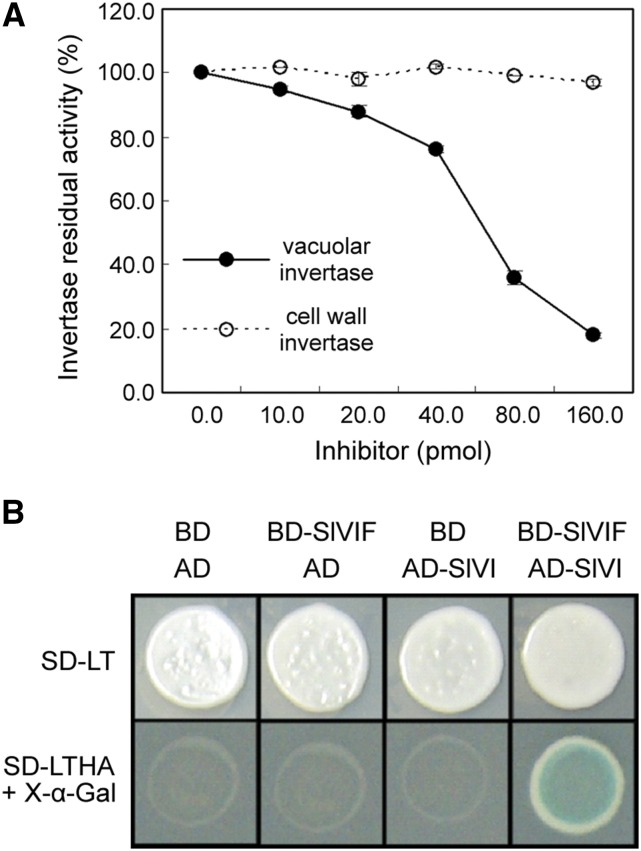
Recombinant SlVIF protein was purified and tested for its inhibitory activity against VI extracted from 44 DPA tomato fruit to confirm whether SlVIF indeed acts as a SlVI inhibitor. As shown in Figure 5A, a decrease in the VI activity was observed upon increasing the SlVIF concentration, while SlVIF had no inhibitory effects on CWI activity. To investigate whether SlVIF physically interacted with SlVI, a yeast two-hybrid (Y2H) assay was performed. The cDNA fragments of SlVIF and SlVI were cloned into pGBKT7 (BD) and pGADT7 (AD) vectors, respectively. The resulted plasmids, BD-SlVIF and AD-SlVI, were cotransformed into yeast strains. Yeasts cotransformed with BD and AD, BD-SlVIF and AD, and AD-SlVI and BD were used as controls. As shown in Figure 5B, yeast cotransformed with BD-SlVIF and AD-SlVI displayed normal growth and blue color on the selective SD/-Leu/-Trp/-His/-Ade medium containing X-α-Gal, whereas the controls showed no growth. Taken together, these results indicate that SlVIF interacts with SlVI to suppress SlVI activity.
Figure 5.
Interactions between SlVIF and SlVI. A, Inhibitory effects of recombinant SlVIF on tomato vacuolar invertase and cell wall invertase. Residual invertase activity was measured after preincubation with 10 to 160 pmol of recombinant SlVIF. Suc concentration in the assay was 20 mM. B, Interaction between SlVIF and SlVI in yeast. BD and AD represent plasmids pGBKT7 and pGADT7, respectively. Transformants were grown on selective medium SD-LT (SD/-Leu/-Trp) and SD-LTHA (SD/-Leu/-Trp/-His/-Ade) containing X-α-Gal for blue color development.
SlVIF Influences Suc Metabolism and Fruit Ripening
To examine the effects of SlVIF on sugar metabolism and fruit ripening, a 35S:SlVIF overexpression construct and a SlVIF RNAi construct were separately introduced into tomato. Three independent transgenic lines corresponding to each construct and containing single-copy transgenes were identified by qRT-PCR analyses (Mason et al., 2002). The SlVIF overexpression line 2 (OE2) and SlVIF silenced line 4 (S4) lines with the highest and lowest transcript level of SlVIF, respectively, were selected for further study.
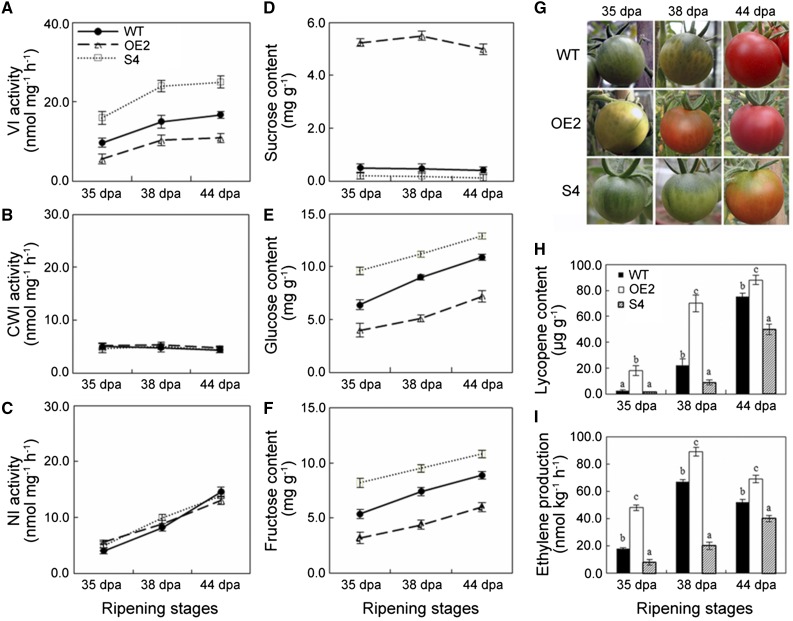
Enzyme activity assays revealed that the VI activity was suppressed in fruit of the OE2 line but elevated in fruit of the S4 line, compared with wild-type fruit at different ripening stages (Fig. 6A). CWI and NI activities were not affected by presence of the transgenes (Fig. 6, B and C), confirming that SlVIF specifically inhibited VI activity. To investigate whether alteration of the SlVIF activity affected fruit sugar metabolism, levels of Suc, Glc, and Fru were measured. Overexpression of SlVIF resulted in 10-fold increase in Suc levels and a 40% decrease in hexose levels in 38-DPA OE2 fruits. In contrast, silencing SlVIF led to lower Suc levels and higher hexose levels than in the wild-type control fruit (Fig. 6, D–F). These results demonstrated that SlVIF influenced sugar metabolism and that altering its expression caused a substantial change in Suc levels in fruits.
Figure 6.
Function of tomato SlVIF in Suc metabolism and fruit ripening. Invertase activity, sugar content, color change phenotype, lycopene content, and ethylene production in fruit of SlVIF overexpressing line 2 (OE2), SlVIF-silenced line 4 (S4), and the wild type (WT) were examined during fruit ripening. A, Vacuolar invertase (VI) activity. B, Cell wall invertase (CWI) activity. C, Neutral invertase (NI) activity. D, Suc content. E, Glc content. F, Fru content. G, Photos of fruit from transgenic lines and wild-type control. H, Lycopene content. I, Ethylene production. Bars represent standard deviations of three biological replicates.
We noted that alteration of Suc levels in the transgenic lines was accompanied by changes in the onset of fruit ripening (Fig. 6G). Fruit color changed from 35 DPA to 44 DPA in wild-type fruit, but the color transition from green to red was faster in the OE2 line and slower in the S4 line (Fig. 6G). Moreover, we determined that while the red colored carotenoid pigment, lycopene, was detectable from 38 DPA in the wild-type fruit, it started to accumulate in the OE2 line at 35 DPA and continued to increase over time, whereas no significant changes in lycopene levels were observed from 35 DPA to 38 DPA in fruit of the S4 line (Fig. 6H). We concluded that altering sugar content affected carotenoid pigment biosynthesis, which in turn affected the timing of the onset of ripening. We also observed that overexpression of SlVIF led to a 60% increase of ethylene production at 35 DPA compared with the control, and repression of SlVIF expression resulted in significant reduction in ethylene production throughout the ripening process (Fig. 6I), suggesting that the altered sugar levels also affected phytohormone production to coordinately regulate fruit ripening.
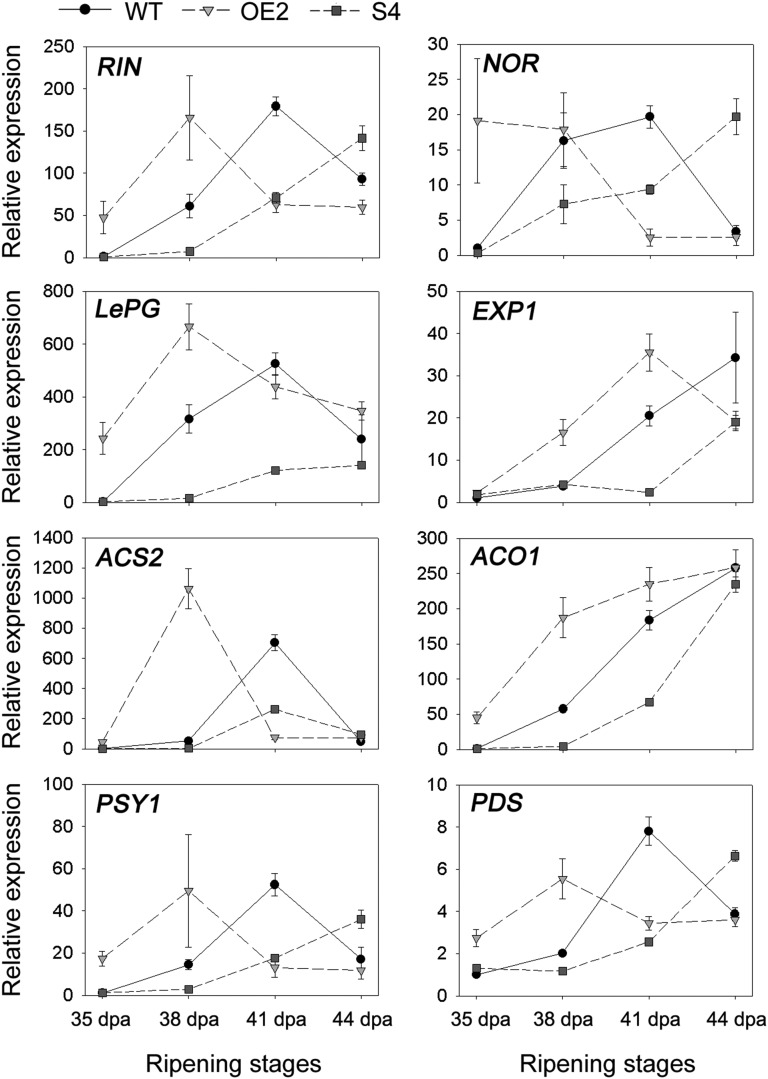
To better understand the mechanism of action of SlVIF in ripening fruit, the transcript levels of a set of ripening-related marker genes (RIN, NOR, LePG, EXP1, ACS2, ACO1, PSY1, and PDS) were evaluated in SlVIF-overexpressing and -silenced tomato fruits by qRT-PCR (Fig. 7). The results indicated that the expression levels of these ripening marker genes were significantly influenced by SlVIF. Notably, the ripening regulators RIN and NOR were repressed in the SlVIF RNAi fruits at 38 DPA and 41 DPA but exhibited higher transcript levels at 44 DPA compared with the wild type. This suggests that gene expression of RIN and NOR was delayed in the SlVIF RNAi tomatoes during fruit ripening.
Figure 7.
Expression of ripening marker genes in SlVIF-overexpressing and -silenced tomato fruits. The gene transcript levels were determined by qRT-PCR. Total RNA was extracted from pericarp tissues of SlVIF-overexpressing line 2 (OE2), SlVIF-silenced line 4 (S4), and the wild type (WT) at 35 DPA, 38 DPA, 41 DPA, and 44 DPA. The gene transcript levels are normalized against the ACTIN gene, followed by normalization against the wild type at 35 DPA. Values are means ± sd of three independent experiments. RIN, Ripening inhibitor; NOR, nonripening; LePG, polygalacturonase A; EXP1, expansin 1; ACS2, 1-aminocyclopropane-1-carboxylate synthase 2; ACO1, 1-aminocyclopropane-1-carboxylate oxidase; PSY1, phytoene synthase 1; PDS, phytoene desaturase.
SlVIF Modulates the Expression of Ripening-Related Proteins
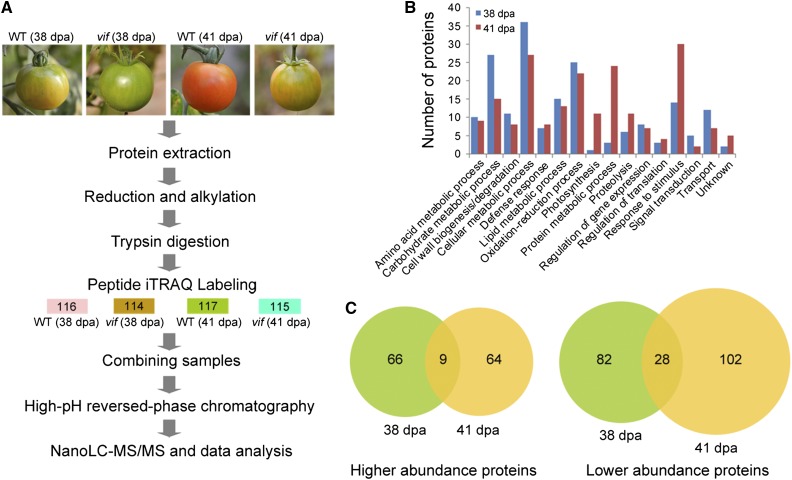
To further elucidate how SlVIF coordinately regulated fruit ripening, we performed an isobaric tags for relative and absolute quantification (iTRAQ)-based quantitative proteomic analysis of wild-type and SlVIF RNAi fruit. Proteins were extracted from the frozen pericarp tissue of wild-type and SlVIF RNAi fruits at 38 DPA and 41 DPA and labeled with 4-plex iTRAQ reagents (Fig. 8A). We analyzed three independent biological replicates, with a replicate corresponding to tissue pooled from fruits from five different plants for each stage and each genotype. Using a S. lycopersicum protein database (ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/), a total of 4240, 4225, and 4381 proteins were identified in both wild type and transgenic line in biological replicates 1, 2, and 3, respectively, with a global false discovery rate (FDR) <1% in each. A 2-fold cutoff led to the identification of 185 and 203 proteins with significantly altered levels in the SlVIF silenced fruits at 38 DPA and 41 DPA, respectively (Supplemental Table S6).
Figure 8.
Identification of differentially expressed proteins in SlVIF silenced tomato fruits by iTRAQ-based quantitative proteomic analysis. A, Schematic diagram of the workflow used in this study. Three sets of biological replicate samples from wild-type (WT) and SlVIF-silenced fruit (vif) at 38 DPA and 41 DPA were analyzed by iTRAQ, using the NanoLC-MS/MS workflow for examining proteome changes. B, GO categories of differentially expressed proteins at 38 DPA or 41 DPA. C, Venn diagram of differentially expressed proteins identified at 38 DPA and 41 DPA. The numbers of differentially abundant proteins at each ripening stage are shown.
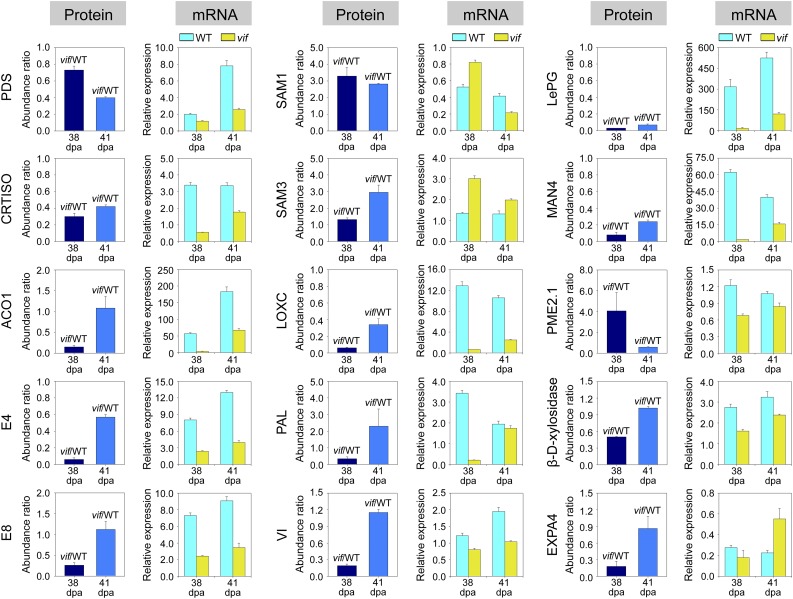
The differentially expressed proteins identified at 38 DPA and 41 DPA were categorized using Blast2go (https://www.blast2go.com/) into fourteen functional categories (Fig. 8B). Overall 139 of them were more abundant and 212 were less abundant, respectively, in the transgenic fruit than in wide type (Fig. 8C). We concluded that a set of ripening related proteins were significantly affected by SlVIF expression (Fig. 9). These included proteins involved in carotenoid biosynthesis, such as phytoene desaturase (PDS), in ethylene synthesis, such as ACC oxidase 1 (ACO1) and peptide Met sulfoxide reductase msrA (E4), and in cell wall degradation, such as polygalacturonase A (LePG) and pectinesterase (PME). To determine whether the protein expression patterns correlated with transcript levels, qRT-PCR was carried out for the corresponding genes using gene specific primers (Supplemental Table S7). Of the 15 genes analyzed, the expression of 12, including PDS, CRTISO, ACO1, E4, and LePG, were consistent with the protein abundance variations (Fig. 9). These results further indicated that SlVIF affected the expression of ripening-related genes.
Figure 9.
Quantitative proteomic analysis showing that ripening-related genes are affected by SlVIF expression. The changes in protein abundance in the SlVIF-silenced fruits (vif) at 38 DPA and 41 DPA are shown as the ratio between vif and the wild type (WT). The mRNA expression levels were measured by qRT-PCR. Total RNA was extracted from pericarp tissues of SlVIF silenced fruit (vif) and the wild type at 35 DPA, 38 DPA, 41 DPA, and 44 DPA. The gene transcript levels were normalized against the ACTIN gene, followed by normalization against the wild type at 35 DPA. The changes in transcript levels at 38 DPA and 41 DPA are shown. The results for both protein and mRNA expression are means ± sd from three independent experiments. PDS, Phytoene desaturase; CRTISO, carotenoid isomerase; ACO1, 1-aminocyclopropane-1-carboxylate oxidase; E4, peptide Met sulfoxide reductase; E8, 1-aminocyclopropane-1-carboxylate oxidase-like protein; SAM1, S-adenosyl-Met synthase 1; SAM3, S-adenosyl-Met synthase 3; LoxC, lipoxygenase; PAL, Phe ammonia-lyase; VI, vacuolar invertase; LePG, polygalacturonase A; MAN4, mannan endo-1,4-β-mannosidase; PME2.1, pectinesterase; EXPA4, expansin.
DISCUSSION
RIN Regulates Expression of Genes Involved in Suc Metabolism
Given the importance of the ripening process in determining the quality, taste, and aroma of fruit, considerable effort had been focused on understanding its control (Klee and Giovannoni, 2011; Ecker, 2013). The MADS-box transcription factor, RIN, has been shown to be a global regulator of fruit ripening (Vrebalov et al., 2002; Martel et al., 2011) and to be critical for the production of characteristic tomato aromas derived from the LOX pathway (Qin et al., 2012). However, the particular genes related to sugar metabolism and their specific modulation by RIN have not been well defined. The VI, VIF, SS, HK, FK, and SPS gene families have been shown to be involved in Suc metabolism by regulating the degradation and resynthesis of Suc (Carrari and Fernie, 2006). Here, we identified a total of 36 genes involved in Suc metabolism in tomato (Supplemental Table S1) and analyzed their expression in the wild type and the rin mutant by qRT-PCR using gene specific primers (Fig. 1). Many of these genes were differentially expressed in the rin mutant compared with the wild type at equivalent DPA, including 11 invertase genes (LIN5–9, VI, NI1–5), a VI inhibitor gene (VIF), six Suc synthase genes (SS1, SS3, SS4, SS5, SS6, and SS7), four Suc phosphate synthase genes (SPSA1–SPSC), and six hexokinase genes (HK1–6). The data suggest that RIN regulates Suc metabolism during ripening by modulating the expression of a specific set of genes that play crucial roles in Suc degradation and resynthesis.
Sugars have important hormone-like functions as primary messengers in signal transduction in addition to their essential roles as substrates in the carbon and energy metabolism and in polymer biosynthesis (Rolland et al., 2002). Following Suc degradation by SS and VI, the resulting hexoses undergo phosphorylation by FK and HK for further metabolism (Claeyssen and Rivoal, 2007), suggesting that FK and HK are important in hexose metabolism, hexose sensing, and signaling (Eveland and Jackson, 2012). In this study, we showed that RIN governs sugar metabolism via the regulation of a number of genes, including SlVI and SlVIF, in the Suc synthesis and degradation pathway (Fig. 1B). SlVI expression was lower in the rin mutant while that of SlVIF was higher, suggesting that RIN can modulate Suc degradation during fruit ripening via promoting SlVI expression and suppressing its inhibitor SlVIF. Interestingly, the expression of RIN changed substantially in the SlVIF RNAi tomatoes during fruit ripening (Fig. 7), suggesting that SlVIF affects RIN expression via a positive feed-back loop.
SlVIF Functions on Suc Metabolism and Lycopene Synthesis
The deduced SlVIF sequence was shown to share a high homology with vacuolar invertase inhibitors from tobacco and potato (Supplemental Figure S3). Four conserved Cys residues were identified that, in a potato vacuolar invertase inhibitor, have been shown to form disulfide bridges to stabilize the protein (Brummell et al., 2011). Gene expression analysis showed that SlVIF was expressed in both vegetative and reproductive organs (Fig. 4A) and that its expression increased in fruit from 35 DPA to 44 DPA (Fig. 4B), indicating that it is involved in fruit ripening. Greiner et al. (1999) suggested that the homologous protein from tobacco, NtVIF, is vacuolar, because overexpression of NtVIF in potato strongly inhibited VI but not CWI. This is consistent with our observation that recombinant GFP-SlVIF fusion protein accumulated in the vacuole of tomato root cells (Fig. 4C). Overexpression of SlVIF in tomato reduced the activity of VI, whereas silencing its expression resulted in increased VI activity (Fig. 6A), indicating that invertase activity is regulated by its endogenous inhibitor. Importantly, altered SlVIF expression did not affect the activities of CWI or NI (Fig. 6, B and C), demonstrating the specificity of SlVIF for SlVI. These results were further confirmed by Y2H analysis (Fig. 5B). Previous reports indicated a role for invertases and invertase inhibitors in mediating sugar levels in plants (Greiner et al., 1999). Induction of the VIF expression in potato tubers contributes to cold-induced sweetening resistance due to its inhibition of VI and resulting decrease in the accumulation of reducing sugars (Brummell et al., 2011). Consistent with its predicted role, overexpression of SlVIF in tomato fruits significantly inhibited Suc hydrolysis, leading to an increase in Suc content and a reduction in hexose levels, while suppression of SlVIF promoted Suc hydrolysis and enhanced hexose levels (Fig. 6, D–F).
Posttranslational regulation of enzymes involved in the sugar metabolic pathway represents an important mechanism for regulating sugar levels and composition in plants (Koch, 2004). Moreover, posttranslational elevation of CWI activity by silencing of its inhibitor has been shown to result in increased hexose accumulation in tomato fruits (Jin et al., 2009), and a number of studies have shown that VI activity is correlated with hexose levels in hexose-accumulating species of tomato (Miron and Schaffer 1991; Klann et al., 1996). External Suc supplementation was previously reported to promote a color break in citrus (C. unshiu) and tomato fruit (Iglesias et al., 2001; Télef N, et al., 2006), indicating a link between sugars and pigments. Color transition in tomato fruits is due to the degradation of chlorophylls and the simultaneous accumulation of carotenoids, mainly lycopene, which is affected by several factors (Giovannoni, 2004). However, evidence supporting a role for sugars in pigment synthesis and consequently fruit ripening in vivo is still lacking. In this study, overexpression of SlVIF in tomato decreased hexose content during ripening, in contrast to fruits from wild-type and SlVIF RNAi plants (Fig. 6, E and F), suggesting a connection between VI activity and hexose accumulation. Interestingly, altering Suc levels by modification of SlVIF expression resulted in a clear change in the timing of the onset of fruit ripening. Specifically, the red transition was faster in fruit of the SlVIF-overexpressing line, but slower in the SlVIF RNAi line compared with the wild-type fruit (Fig. 6G), indicating that SlVIF plays a role in the regulation of Suc metabolism and pigment biosynthesis.
SlVIF Modulates Ethylene Production and the Expression of Ripening-Related Genes
Ethylene is thought to be required for the Suc-induced modulation of carotenoid accumulation in both climacteric and nonclimacteric fruit (Iglesias et al., 2001; Télef et al., 2006), and ethylene production is concomitant with color change and lycopene biosynthesis, which is an indicator of ripening in many climacteric fruit (Giovannoni, 2007). Suppression of acid invertase expression can stimulate ethylene production in tomato and muskmelon fruit (Klann et al., 1996; Yu et al., 2008), and we saw that overexpression of SlVIF in tomato increased ethylene production (Fig. 6I) and led to a precocious color shift, due to elevated lycopene accumulation (Fig. 6H). In contrast, ethylene production was decreased, and fruit coloring was delayed by silencing SlVIF in tomato (Fig. 6G). This suggests that SlVIF is involved in the regulation of ethylene production and fruit ripening. Using iTRAQ-based quantitative proteomic analysis, we identified 185 and 203 proteins whose abundance was significantly different in SlVIF-silenced fruit compared with wild-type fruit at 38 DPA and 41 DPA, respectively (Fig. 8). Of these, 139 were more abundant in the transgenic fruit and 212 were less abundant (Fig. 8C). Suppression of SlVIF expression affected the abundance of many ripening-related proteins (Fig. 9), including PDS, ACO1, E4, and LePG. qRT-PCR analysis indicated that the expression levels of several ripening-related genes (PDS, ACO1, E4, and LePG) were consistent with the corresponding protein abundance variations (Fig. 9). Altering sugar content and/or SlVIF expression also affected the abundance of other ripening-related genes, suggesting a potential role for SlVIF in the regulation of fruit ripening. Finally, ChIP-qPCR and EMSA assays showed that RIN directly bound to the promoters of SlVI and SlVIF, along with those of 14 other genes involved in Suc metabolism (Figs. 2 and 3; Supplemental Fig. S2). These results indicate that RIN controls sugar metabolism by modulating the expression of genes involved in Suc synthesis and degradation.
In conclusion, our study highlights an important role for SlVIF in fruit ripening and establishes a link between Suc metabolism, lycopene synthesis, and ripening in tomato fruit. Our results further suggest that SlVIF has potential value as a genetic tool to regulate tomato fruit ripening through manipulating the interaction of VI and its inhibitor.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum cv Ailsa Craig) plants were grown at 25°C with a 16-h photoperiod. The flowers were tagged at anthesis to determine ripening stages. For plants grown in vitro, seeds were germinated on Murashige and Skoog medium (Murashige and Skoog, 1962) with 16 h of light at 25°C and 8 h of darkness at 22°C. Seeds of the tomato mutant rin in the ‘Ailsa Craig’ background were kindly provided by Dr. James J. Giovannoni (Boyce Thompson Institute for Plant Research, Cornell University, Ithaca, NY). Fruit ripening stages used in this study were 35, 38, 41, and 44 DPA. Fruits of rin or transgenic lines were collected at the equivalent ripening stages determined by the number of DPA.
Identification of Tomato Genes Involved in Suc Metabolism
In order to identify tomato orthologs of genes involved in Suc metabolism (i.e. genes encoding invertases, invertase inhibitors, Suc synthases, Suc phosphate synthases, fructokinases, and hexokinases), BLAST searches were carried out against the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and SGN (http://solgenomics.net/) databases using the reported gene sequence as a query. The uniqueness of the identified genes was manually verified to remove redundant sequences. GENSCAN (http://genes.mit.edu/GENSCAN.html) was used to predict the putative open reading frames and protein sequences.
qRT-PCR
Total RNA (2 μg) was extracted from pericarp tissues as described by Moore et al. (2005). The extracted RNA was treated with DNase (Promega) and reverse-transcribed using an oligo(dT)18 primer with Moloney murine leukemia virus reverse transcriptase (Promega) to synthesize cDNA. qRT-PCR was performed with Takara SYBR Green Master Mix and gene-specific primers in a volume of 20 μL using an Mx3000P system (Stratagene). The following condition was applied for PCR amplification: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. ACTIN (BT012695) was used as an internal control, and the relative expression was calculated using the comparative 2(−ΔCt) method (Schmittgen and Livak, 2008). Each experiment had three biological repeats, each with three technical replicates.
ChIP Assay
ChIP assays were performed as described previously (Qin et al., 2012). Pericarp tissue from 41 DPA fruit was collected and submerged in 1% formaldehyde under a vacuum to cross-link genomic DNA and protein. Nuclei were enriched and then sonicated in nuclei lysing buffer containing 50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS, 0.1 mm phenylmethylsulfonyl fluoride, and proteinase inhibitors. The conditions for sonication were optimized to shear DNA to an average size of 500 ∼ 1000 bp. A small aliquot of sonicated chromatin was reversely cross-linked and served as the input DNA control. The remaining chromatin sample was centrifuged at 16,000g for 5 min at 4°C, and the supernatant was diluted 10-fold in ChIP dilution buffer (1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, 167 mm NaCl, 0.1 mm phenylmethylsulfonyl fluoride, and proteinase inhibitors). Protein A-agarose/salmon sperm DNA beads (Millipore) were used to preclear the chromatin solution for 1 h at 4°C. Immunoprecipitation was carried out with affinity-purified RIN polyclonal antibody (see below), and the tube containing the samples were incubated on a rotator with gentle agitation (25 rpm) for 12 h at 4°C. Reactions with preimmune serum IgG and without antibody were used as negative and mock controls, respectively. Protein-chromatin immunocomplexes were captured on Protein A-agarose beads by incubating at 4°C for 1 h. The beads were collected by centrifugation at 16,000g for 2 min at 4°C, and the bead-bound immunocomplexes were eluted with elution buffer (1% SDS, 0.1 m NaHCO3) by gently rotating for 15 min at 65°C. The beads were collected and the supernatant (eluate) was transferred to a fresh tube. The cross linking of immunoprecipitated DNA was then reversed by incubation of the eluate in 0.2 m NaCl at 65°C overnight. The immunoprecipitated DNA was purified after Proteinase K (Invitrogen) treatment and eluted in TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA) before being used for PCR using the same conditions as for qRT-PCR analysis. The RIN binding sites (CArG box) in the promoters of selected genes were analyzed using PLACE Web Signal Scan (http://www.dna.affrc.go.jp/PLACE/signalup.html). Primers used for PCR amplification are listed in Supplemental Table S4.
EMSA
Recombinant His-tagged RIN protein was expressed in Escherichia coli and purified as described previously (Qin et al., 2012). The binding activity of RIN to specific DNA sequences was assayed using a Lightshift Chemiluminescent EMSA kit (Thermo Scientific). Briefly, purified RIN protein (1 μg) in a binding buffer containing 10 mm Tris-HCl, pH 7.2, 50 mm KCl, 1 mm dithiothreitol 2.5% glycerol, 0.05% NP-40, and 50 ng μL−1 polydeoxy (inosinate-cytidylate) was incubated for 20 min at room temperature in the presence or absence of unlabeled (double-stranded) competitor probes. The 3′ biotin end-labeled dsDNA probes, which were constructed by annealing complementary oligonucleotides, were then added, and the incubation continued for 20 min. The sequences of the biotin-labeled probes, are listed in Supplemental Table S5. Protein-DNA complexes were separated on 6% native polyacrylamide gels, and the biotin-labeled probes were detected according to the instructions provided with the EMSA kit.
Recombinant Protein Expression and Antibody Preparation
Recombinant RIN protein was prepared as described previously (Qin et al., 2012). For recombinant SlVIF protein preparation, a SlVIF gene fragment without the predicted putative vacuolar sorting sequence (SignalP 4.0) was amplified from tomato cDNA using primers SlVIF-F (5′-GGTACCAACAACAACAACAACATCAT-3′) and SlVIF-R (5′-GAGCTCTCATAATAACATTCTAATTA-3′). The fragments were digested using KpnI and SacI and inserted into the pET30a vector (Merck). The resulting plasmid was transformed into E. coli BL21 (Lys) cells, and expression of the recombinant protein was induced by 1 mm of isopropyl-β-d-thiogalacto-pyranoside. Recombinant His-tagged proteins were purified using Ni-NTA His-Bind resin (Merck).
Antibodies were raised against the recombinant RIN and SlVIF proteins in New Zealand white rabbits by AbMax Biotechnology. Polyclonal antibodies were affinity purified from antisera using AminoLink Plus coupling resin, according to the manufacturer’s instructions (Thermo Scientific).
Western Blot Analysis
Proteins were extracted from tomato fruit harvested at different ripening stages as in Bate et al. (2004), fractionated on a 12% SDS-PAGE gel and electrotransferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore). The membranes were blocked at 20°C for 2 h with 5% bovine serum albumin in PBS-Tween buffer (137 mm NaCl, 2.7 mm KCl, 8.1 mm NaH2PO4, 1.5 mm KH2PO4, and 0.1% Tween 20, pH 7.4). Immunoblots were performed at 4°C overnight with affinity-purified rabbit polyclonal anti-SlVIF as described by Wang et al. (2014). The membranes were washed with PBS-Tween (3 × 10 min) and treated with the corresponding secondary antibodies (Abmart; 1:5000 dilutions) conjugated to horseradish peroxidase. Immunoreactive bands were visualized using a chemiluminescence detection kit (SuperSignal, Pierce Biotechnology). As a protein control, an anti-glyceraldehyde-3-phosphate dehydrogenase (Abmart) immunoblot was used. Additionally, Coomassie Brilliant Blue staining was used as a loading control.
Plasmid Construction and Plant Transformation
To construct the 35S:GFP:SlVIF vector, the full-length SlVIF cDNA was inserted into a GFP expression vector pCAMBIA2300. The SlVIF overexpression construct was made by cloning the full-length SlVIF cDNA into the pBI121 vector (kindly provided by Dr. Jing Bo Jin from Institute of Botany, the Chinese Academy of Sciences, Beijing) downstream of the 35S promoter.
To construct the SlVIF RNAi plasmid, a 285-bp SlVIF fragment was amplified from tomato cDNA (prepared from fruit pericarp at 35 DPA as described above) by the primer pair 5′-TATAACACCGTCCTACGAGCCG-3′ and 5′-TCTTTATATCTGGTTCACGTAACG-3′. The resulting product was cloned into the PCR8/GW/TOPO Gateway entry vector (Invitrogen). The cloned fragment was subsequently transferred into the destination vector pK7GWIWG2 (Karimi et al., 2002) using the LR Clonase II enzyme (Invitrogen) according to the manufacturer’s instructions.
The above constructs were individually transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Tomato transformation was performed according to Fillatti et al. (1987). The presence of transgenes in kanamycin-resistant plants was confirmed by PCR using a forward primer corresponding to the 35S promoter and a reverse primer specific for SlVIF: 5′-CGGAAACCTCCTCGGATTCCATT-3′ and 5′-GGCTACCGTTACATCGGCTCGTA-3′.
Invertase Activity and Inhibition Assays
The activities of VI, CWI, and NI were assayed according to Miron and Schaffer (1991) and Klann et al. (1993). Approximately 2 g of pericarp was homogenized in 10 mL of extraction buffer containing 50 mm HEPES-NaOH, pH 7.5, 5 mm MgC12, 1 mm EDTA, 2.5 mm dithiothreitol,10 mm ascorbic acid, and 5% polyvinylpolypyrrolidone. After centrifugation at 20,000g for 30 min at 4°C, the supernatants were dialyzed for 16 h against 25 mm HEPES-NaOH, pH 7.5, and 0.5 mm EDTA and used as a crude enzyme extract for VI and NI activity assays. The insoluble pellet was washed three times with extraction buffer and extracted for 1 h with 10 mL of extraction buffer containing 1 m NaCl. After centrifugation at 20,000g for 30 min at 4°C, the supernatants were dialyzed overnight and used as crude enzyme extract for CWI assays. For VI and CWI activity assays, 0.2 mL of enzyme extract was incubated for 30 min at 37°C with 0.6 mL of 100 mm sodium acetate, pH 4.8, and 0.2 mL of 100 mm Suc. Reactions were stopped by placing the reaction tubes in boiling water for 5 min and reducing sugars were measured using dinitrosalicylic acid (Sumner, 1921). NI activity was assayed in 50 mm HEPES-NaOH, pH 7.5, instead of sodium acetate, pH 4.8.
For inhibition studies, VI or CWI enzyme preparations were mixed with recombinant SlVIF protein and incubated in the assay buffer for 30 min at 37°C prior to addition of Suc and measurement of enzyme activity.
Y2H Assay
To generate the vector system for the Y2H analysis, the SlVIF cDNA fragment encoding the mature protein and the full-length cDNA of SlVI (GenBank accession no. M81081) were cloned into the pGBKT7 (BD) and pGADT7 (AD) vectors (Clontech), using specific primers (SlVIF: 5′-CGGAATTCAACAACAACAACAACATCAT-3′ and 5′-AACTGCAGTCATAATAACATTCTAATTATG-3′; SlVI: 5′-CGGAATTCATGGCCACTCAGTGTTATGA-3′ and 5′-CCGCTCGAGTTACAAGTCTTGCAAAGGGA-3′), resulting in the BD-SlVIF and AD-SlVI plasmids. The two plasmids were cotransformed into the yeast strain Saccharomyces cerevisiae AH109 according to the manufacturer’s instructions (Clontech) and dripped on synthetic dropout nutrient medium SD-LT (SD/-Leu/-Trp) and SD-LTHA (SD/-Leu/-Trp/-His/-Ade) containing X-α-Gal. As a control, BD and AD, BD-SlVIF and AD, and BD and AD-SlVI were also cotransformed and analyzed, respectively.
Sugar, Lycopene, and Ethylene Measurements
Sugars were extracted and determined according to Zhu et al. (2013).
Pericarp lycopene content was measured as described by Choi and Huber (2008). Tomato pericarp tissue samples (5 g) were homogenized for 1 min in 50 mL of hexane-acetone-ethanol (2:1:1, v/v) wrapped in aluminum foil to exclude light, then 15 mL of water was added and the samples were vortexed for 10 s. After allowing phase separation on ice, the lycopene concentration was determined by measuring the absorbance of the organic phase (hexane) at 503 nm. The lycopene content was calculated using the molar extinction coefficient of 17.2 L mol−1 m−1 and expressed as μg g−1 fresh weight. Three independent samples derived from five fruits at each ripening stage were used for lycopene measurements.
Ethylene production was determined according to Zhu et al. (2010). Fruit were harvested at different ripening stages, weighed, and transferred to 1 L gas-tight jars. The jars were sealed and incubated at 25°C for 2 h, then 1 mL of gas sample was withdrawn from the headspace with a syringe and injected into a gas chromatograph (SQ-206, Beifen-Ruili Analytical Instrument Company), equipped with an activated alumina column and a flame ionization detector. Three independent samples derived from five fruits at each ripening stage were used for the ethylene measurements.
Protein Isolation, iTRAQ Labeling, and NanoLC-Tandem Mass Spectrometry Analysis
Fruits were harvested at 38 DPA and 41 DPA, and total cellular protein was extracted from fruit pericarp tissue as described by Saravanan and Rose (2004). Proteins were solubilized in a protein buffer consisting of 500 mm triethylammonium bicarbonate and 0.6% (w/v) SDS, pH 8.5, and the protein concentrations were determined using the Bradford method (Bradford, 1976) with bovine serum albumin as a standard. Proteins from each sample were reduced with 10 mm tris-(2-carboxyethyl)phosphine, alkylated with 50 mm methyl methanethiosulfonate, and digested with 10 ng μL−1 trypsin using the filter-aided sample preparation method (Wiśniewski et al., 2009). The tryptic peptides were labeled using the iTRAQ Reagents 4-plex kit (Applied Biosystems) according to the manufacturer’s protocol. Samples taken from wild-type and SlVIF-silenced tomatoes at 38 DPA were labeled with iTRAQ tags 116 and 114, respectively, while samples from wild-type and SlVIF-silenced tomatoes at 41 DPA were labeled with iTRAQ tags 117 and 115, respectively. Three independent biological replicates were analyzed. The iTRAQ-labeled samples were combined and then fractionated with high-pH reversed-phase chromatography using the Agilent Technologies 1290 UPLC system. Briefly, the pooled iTRAQ-labeled peptides were reconstituted using buffer A (20 mm ammonium formate, pH 10, in water) and loaded onto a 4.6 × 250 mm, 150 Å size Durashell C18 (L) column containing 5 μm particles (Agela Technologies). The peptides was eluted at a flow rate of 0.8 ml min−1 with a gradient of 2% buffer B (20 mm ammonium formate in 80% acetonitrile, pH 10) for 5 min, 2% to 30% buffer B for 25 min, and 30% to 90% buffer B for 10 min. The system was maintained in 90% buffer B for 10 min and then equilibrated with 2% buffer B for 10 min. The elution was monitored by measuring UV A210,, and fractions were collected every 1 min. A total of 48 fractions were collected and pooled into a total of six fractions. After reconstitution in formic acid, the labeled peptides were desalted and submitted for NanoLC-tandem mass spectrometry (MS/MS) analysis.
The MS analysis was carried out using a NanoLC system (NanoLC-2D Ultra Plus, Eksigent) equipped with a Triple TOF 5600 Plus mass spectrometer (AB SCIEX). The iTRAQ-labeled peptide mixtures were desalted on a 100 μm × 20 mm trap column and eluted on an analytical 75 μm × 150 mm column. Both the trap column and the analytical column were filled with the Magic C18-AQ 5 μm 200 Å phase (Michrom Bioresources). Peptides were separated by a gradient formed by 0.1% formic acid (mobile phase A) and 100% acetonitrile, 0.1% formic acid (mobile phase B), from 5% to 30% of mobile phase B over 75 min at a flow rate of 300 nL/min. The precursor ions were selected across the mass range of 350 to 1500 m/z. High-resolution mode (>30,000) was applied using 250 ms accumulation time per spectrum. A maximum of 25 precursors per cycle from each MS spectrum were chosen for fragmentation with 100 ms minimum accumulation time for each precursor and dynamic exclusion for 18 s. Tandem mass spectra were recorded in high-sensitivity mode (resolution >15,000) with rolling collision energy and iTRAQ reagent collision energy adjustment on.
Protein identification and quantification were performed using ProteinPilot 4.5 software (AB SCIEX), and database searches were carried out using the S. lycopersicum protein database ITAG2.4_proteins_full_desc.fasta (ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/). The following parameters was applied: (1) sample type: iTRAQ 4-plex (peptide labeled); (2) Cys alkylation: MMTS; (3) digestion: trypsin; (4) instrument: TripleTOF 5600; (5) species: none; (6) quantitate: yes; (7) bias correction: yes; (8) background correction: yes; (9) search effort: thorough; and (10) FDR analysis: yes. The peptide for quantification was automatically chosen by the Pro Group algorithm (AB SCIEX) to calculate the reporter peak area. The reverse database search method (Elias and Gygi, 2007) was used to estimate the global FDR for peptide identification. Proteins identified below the 1% global FDR were used to calculate the meaningful cutoff value with the experimental replicate method (Gan et al., 2007).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KC007445.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The binding ability of RIN to the promoter of ACS2 as revealed by chromatin immunoprecipitation.
Supplemental Figure S2. Chromatin immunoprecipitation reveals the direct binding of RIN to the promoters of genes involved in Suc metabolism.
Supplemental Figure S3. Alignment of amino acid sequences of invertase inhibitors from various plants.
Supplemental Table S1. Genes involved in Suc metabolism.
Supplemental Table S2. Primers used in qRT-PCR analysis of Suc metabolism genes.
Supplemental Table S3. Ripening marker genes used for qRT-PCR analysis in this study.
Supplemental Table S4. Gene-specific primers used in ChIP-qPCR assay.
Supplemental Table S5. Primers used for probes of EMSA.
Supplemental Table S6. Identification of the differentially expressed proteins in the SlVIF silenced tomato fruit (vif) using iTRAQ-based quantitative proteomic analysis.
Supplemental Table S7. Primers for qRT-PCR analysis of selected genes identified in iTRAQ.
Supplementary Material
Acknowledgments
We are grateful to Dr. James J. Giovannoni (Boyce Thompson Institute for Plant Research, Cornell University) for providing the tomato seeds of the wild type (‘Ailsa Craig’) and mutant rin used in the study. We thank Dr. Jing Bo Jin (Institute of Botany, the Chinese Academy of Sciences) for providing pBI121 vector. We also thank Dr. Zhuang Lu for analysis of MS/MS.
Glossary
- iTRAQ
isobaric tags for relative and absolute quantification
- FDR
false discovery rate
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31530057 to S.T.), the National Basic Research Program of China (973 Program; grant no. 2011CB100604 to S.T.), and the Youth Innovation Promotion Association CAS (to G.Q.).
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Niu X, Wang Y, Reimann KS, Helentjaris TG (2004) An invertase inhibitor from maize localizes to the embryo surrounding region during early kernel development. Plant Physiol 134: 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani J, Budde CO, Porrini L, Lauxmann MA, Lombardo VA, Murray R, Andreo CS, Drincovich MF, Lara MV (2009) Carbon metabolism of peach fruit after harvest: changes in enzymes involved in organic acid and sugar level modifications. J Exp Bot 60: 1823–1837 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Chen RK, Harris JC, Zhang H, Hamiaux C, Kralicek AV, McKenzie MJ (2011) Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J Exp Bot 62: 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Choi ST, Huber DJ (2008) Influence of aqueous 1-methylcyclopropene concentration, immersion duration, and solution longevity on the postharvest ripening of breaker-turning tomato (Solanum lycopersicum L.) fruit. Postharvest Biol Technol 49: 147–154 [Google Scholar]
- Claeyssen E, Rivoal J (2007) Isozymes of plant hexokinase: occurrence, properties and functions. Phytochemistry 68: 709–731 [DOI] [PubMed] [Google Scholar]
- Davies C, Robinson SP (1996) Sugar accumulation in grape berries. Cloning of two putative vacuolar invertase cDNAs and their expression in grapevine tissues. Plant Physiol 111: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR. (2013) Epigenetic trigger for tomato ripening. Nat Biotechnol 31: 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Elliott KJ, Butler WO, Dickinson CD, Konno Y, Vedvick TS, Fitzmaurice L, Mirkov TE (1993) Isolation and characterization of fruit vacuolar invertase genes from two tomato species and temporal differences in mRNA levels during fruit ripening. Plant Mol Biol 21: 515–524 [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP (2012) Sugars, signalling, and plant development. J Exp Bot 63: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat Biotechnol 5: 726–730 [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fridman E, Zamir D (2003) Functional divergence of a syntenic invertase gene family in tomato, potato, and Arabidopsis. Plant Physiol 131: 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan CS, Chong PK, Pham TK, Wright PC (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J Proteome Res 6: 821–827 [DOI] [PubMed] [Google Scholar]
- German MA, Asher I, Petreikov M, Dai N, Schaffer AA, Granot D (2004) Cloning, expression and characterization of LeFRK3, the fourth tomato (Lycopersicon esculentum Mill.) gene encoding fructokinase. Plant Sci 166: 285–291 [Google Scholar]
- German MA, Dai N, Chmelnitsky I, Sobolev I, Salts Y, Barg R, Schaffer AA, Granot D (2002) LeFRK4, a novel tomato (Lycopersicon esculentum Mill.) fructokinase specifically expressed in stamens. Plant Sci 163: 607–613 [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Godt DE, Roitsch T (1997) Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol 115: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren S, Huber SC, Granot D (2011) Comparison of a novel tomato sucrose synthase, SlSUS4, with previously described SlSUS isoforms reveals distinct sequence features and differential expression patterns in association with stem maturation. Planta 233: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Greiner S, Krausgrill S, Rausch T (1998) Cloning of a tobacco apoplasmic invertase inhibitor. Proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol 116: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Rausch T, Sonnewald U, Herbers K (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol 17: 708–711 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16: 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 431–444 [DOI] [PubMed] [Google Scholar]
- Iglesias DJ, Tadeo FR, Legaz F, Primo-Millo E, Talon M (2001) In vivo sucrose stimulation of colour change in citrus fruit epicarps: interactions between nutritional and hormonal signals. Physiol Plant 112: 244–250 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL (2009) Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 21: 2072–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama Y, Dai N, Granot D, Petreikov M, Schaffer A, Bennett AB (1997) Divergent fructokinase genes are differentially expressed in tomato. Plant Physiol 113: 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel-Kfir M, Damari-Weissler H, German MA, Gidoni D, Mett A, Belausov E, Petreikov M, Adir N, Granot D (2006) Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization. Planta 224: 1341–1352 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klann EM, Chetelat RT, Bennett AB (1993) Expression of acid invertase gene controls sugar composition in tomato (Lycopersicon) fruit. Plant Physiol 103: 863–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann EM, Hall B, Bennett AB (1996) Antisense acid invertase (TIV1) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol 112: 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E, Yelle S, Bennett AB (1992) Tomato fruit acid invertase complementary DNA: nucleotide and deduced amino acid sequences. Plant Physiol 99: 351–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kocal N, Sonnewald U, Sonnewald S (2008) Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol 148: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Li Z, Palmer WM, Martin AP, Wang R, Rainsford F, Jin Y, Patrick JW, Yang Y, Ruan YL (2012) High invertase activity in tomato reproductive organs correlates with enhanced sucrose import into, and heat tolerance of, young fruit. J Exp Bot 63: 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link M, Rausch T, Greiner S (2004) In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Lett 573: 105–109 [DOI] [PubMed] [Google Scholar]
- Long JC, Zhao W, Rashotte AM, Muday GK, Huber SC (2002) Gravity-stimulated changes in auxin and invertase gene expression in maize pulvinal cells. Plant Physiol 128: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, MacRae E (2003) New complexities in the synthesis of sucrose. Curr Opin Plant Biol 6: 208–214 [DOI] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason G, Provero P, Vaira AM, Accotto GP (2002) Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menu T, Rothan C, Dai N, Petreikov M, Etienne C, Destrac-Irvine A, Schaffer A, Granot D, Ricard B (2001) Cloning and characterization of a cDNA encoding hexokinase from tomato. Plant Sci 160: 209–218 [DOI] [PubMed] [Google Scholar]
- Miron D, Schaffer AA (1991) Sucrose phosphate synthase, sucrose synthase, and invertase activities in developing fruit of Lycopersicon esculentum Mill. and the sucrose accumulating Lycopersicon hirsutum Humb. and Bonpl. Plant Physiol 95: 623–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Payton P, Wright M, Tanksley S, Giovannoni J (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56: 2885–2895 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Proels RK, Roitsch T (2009) Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. J Exp Bot 60: 1555–1567 [DOI] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S (2012) Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Rausch T, Greiner S (2004) Plant protein inhibitors of invertases. Biochim Biophys Acta 1696: 253–261 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS (2010) Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant 3: 942–955 [DOI] [PubMed] [Google Scholar]
- Saravanan RS, Rose JKC (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4: 2522–2532 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Sturm A. (1999) Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner JB. (1921) Dinitrosalicylic acid: a reagent for the estimation of sugar in normal and diabetic urine. J Biol Chem 47: 5–9 [Google Scholar]
- Télef N, Stammitti-Bert L, Mortain-Bertrand A, Maucourt M, Carde JP, Rolin D, Gallusci P (2006) Sucrose deficiency delays lycopene accumulation in tomato fruit pericarp discs. Plant Mol Biol 62: 453–469 [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99: 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang F, Smith AG, Brenner ML (1993) Isolation and sequencing of tomato fruit sucrose synthase cDNA. Plant Physiol 103: 1463–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S (2014) Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol 15: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yu X, Wang X, Zhang W, Qian T, Tang G, Guo Y, Zheng C (2008) Antisense suppression of an acid invertase gene (MAI1) in muskmelon alters plant growth and fruit development. J Exp Bot 59: 2969–2977 [DOI] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Liu R, Li B, Tian S (2013) Characterisation of genes encoding key enzymes involved in sugar metabolism of apple fruit in controlled atmosphere storage. Food Chem 141: 3323–3328 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhang ZQ, Qin GZ, Tian SP (2010) Effects of brassinosteroids on postharvest disease and senescence of jujube fruit in storage. Postharvest Biol Technol 56: 50–55 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.