Comparative analysis of immunostained metaphase chromosomes at the single cell level with ChIP-seq of individual genes reveals a chromatin basis for biased homoeolog gene expression in polyploids.
Abstract
Histone modifications regulate gene expression in eukaryotes, but their roles in gene expression changes in interspecific hybrids or allotetraploids are poorly understood. Histone modifications can be mapped by immunostaining of metaphase chromosomes at the single cell level and/or by chromatin immunoprecipitation-sequencing (ChIP-seq) for analyzing individual genes. Here, we comparatively analyzed immunostained metaphase chromosomes and ChIP-seq of individual genes, which revealed a chromatin basis for biased homoeologous gene expression in polyploids. We examined H3K4me3 density and transcriptome maps in root-tip cells of allotetraploid cotton (Gossypium hirsutum). The overall H3K4me3 levels were relatively equal between A and D chromosomes, which were consistent with equal numbers of expressed genes between the two subgenomes. However, intensities per chromosomal area were nearly twice as high in the D homeologs as in the A homeologs. Consistent with the cytological observation, ChIP-seq analysis showed that more D homeologs with biased H3K4me3 levels than A homeologs with biased modifications correlated with the greater number of the genes with D-biased expression than that with A-biased expression in most homeologous chromosome pairs. Two chromosomes displayed different expression levels compared with other chromosomes, which correlate with known translocations and may affect the local chromatin structure and expression levels for the genes involved. This example of genome-wide histone modifications that determine expression bias of homeologous genes in allopolyploids provides a molecular basis for the evolution and domestication of polyploid species, including many important crops.
Chromatin structure is a repeating unit of nucleosomes, each consisting of approximately 147 bp of DNA wrapping around a histone octamer with two copies each of H2A, H2B, H3, and H4 core histones (Kornberg, 1974; Luger et al., 1997). The core histone tails can be modified to mediate gene expression, growth, and development in plants and animals (Jenuwein and Allis, 2001; Berger, 2007; Li et al., 2007; Liu et al., 2010; Ha et al., 2011). Some modifications, such as histone H3 Lys K9 acetylation (H3K9ac) and Lys 4 trimethylation (H3K4me3), are euchromatic marks and often associated with active transcription, whereas other modifications, such as H3K9me2 and H3K27me3, are known as heterochromatic marks and related to gene repression (Jenuwein and Allis, 2001; Li et al., 2007).
Changes in chromatin structure and consequently gene expression can occur during development and/or in response to internal (“genome shock”) and external (environmental) signals (McClintock, 1984; Chen, 2007; Chen and Tian, 2007). This genome shock is often associated with interspecific hybridization and polyploidization (McClintock, 1984; Comai, 2005; Chen, 2007). The additional genetic materials in new polyploid species can facilitate adaptation to local environments as well as for crop domestication (Chen, 2007; Soltis et al., 2014). This may explain why the majority of crop plants are of polyploid origin and many other crops are grown as hybrids. The most widely cultivated allotetraploid cotton (Gossypium hirsutum) is an excellent example of polyploidization and domestication from small trees into an annual crop (Wendel and Cronn, 2003; Guan et al., 2014). Genome doubling is predicted to have occurred at 1 to 2 million years ago through hybridization and chromosome doubling between the extant progenitors that are closely related to Gossypium arboreum (A2) or Gossypium herbaceum (A1), originating in the Old World, and Gossypium raimondii (D5), originating in the New World (Wendel, 1989; Wendel and Cronn, 2003). Interestingly, the genome size of G. arboreum (1,746 Mb/1C) is almost twice the size of G. raimondii (885 Mb/1C), which is largely related to the expansion of transposable elements, including long terminal repeat retrotransposons (Hawkins et al., 2006; Wang et al., 2010; Zhang et al., 2015). The transposable element-rich regions often form heterochromatin and are different from genic regions in the euchromatin (Wang et al., 2010; Guan et al., 2014).
Chromatin modifications can be visualized and quantified using immunostaining of metaphase chromosomes that have revealed dramatic regional differences in the distribution of specific histone modifications, particularly in centromeric (constitutive) heterochromatin in plants and animals (Carchilan et al., 2007; Terrenoire et al., 2010). The technique has been applied in meiotic (pachytene) chromosomes in maize to display regional variation of methylated histone isoforms, with distinctive differences between heterochromatic and euchromatic regions (Shi and Dawe, 2006). However, these studies are very limited in plants (Houben and Schubert, 2003; Jin et al., 2008). The differences of histone modifications between homeologous chromosomes and their consequences on expression of homeologous genes in a polyploid species are largely unknown.
In this study, distribution patterns of histone modifications (H3K4me3 and H3K27me2) in heterochromatic and euchromatic regions of homeologous chromosomes in G. hirsutum were quantified and comparatively analyzed with transcriptome data. The results have revealed distinctive patterns of heterochromatin and euchromatin between homeologous chromosomes and an association of transcriptionally active chromatin with gene-rich regions. Although the overall fluorescent intensities of histone marks are equally distributed between A and D homeologous chromosomes, the mean fluorescent intensities per chromosome bin of histone modifications linked to active transcription are significantly different between A and D homeologs. Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis further shows that different numbers of A and D homeologs with biased H3K4me3 levels among homeologous pairs in the allotetraploid cotton correlate with the number and levels of expressed genes between A and D homeologs. Two chromosomes display different expression levels relative to other chromosomes, probably because of existing translocations. These data indicate that both histone modifications and genome organization contribute to the expression bias of homeologous genes in allopolyploids.
RESULTS
H3K4me3 Distributions among A and D Homeologous Chromosomes
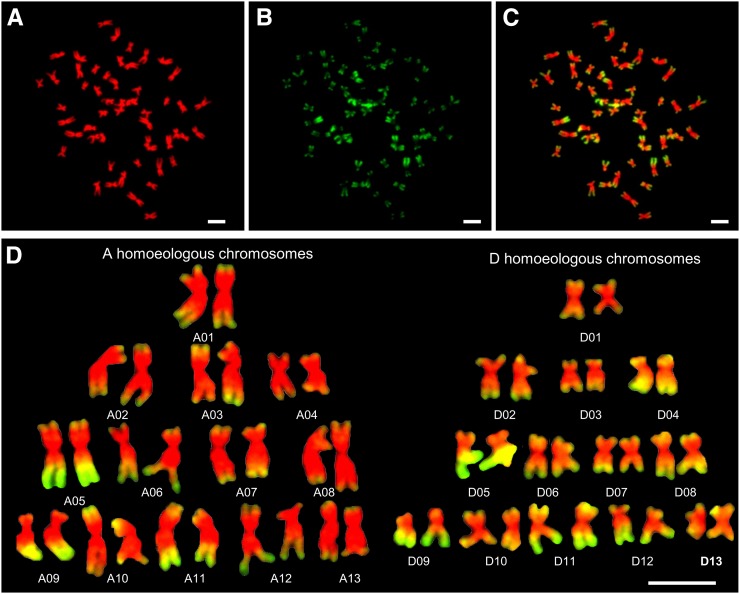
To determine histone modification patterns in allotetraploid cotton, metaphase chromosome spreads in root tip cells (Fig. 1A) of G. hirsutum were immunostained with antibodies against H3K4me3 (Fig. 1B). Centromeric heterochromatin was consistently unstained, while the arms of most chromosomes showed distinct patterns of strongly and weakly stained regions (Fig. 1C). The signals were enriched at the distal end of all chromosomes in G. hirsutum (Fig. 1C; Supplemental Fig. S1A), which were consistent with high levels of expressed genes in the distal regions (Zhang et al., 2015). We distinguished A and D homeologous chromosomes by genomic DNA in situ hybridization (GISH) with genome DNA probe from G. arboreum (Supplemental Fig. S1B). Using a combination of chromosome size, chromosome arm ratio, and H3K4me3 signal patterns (Table I; Supplemental Fig. S1B), we identified all chromosomes and constructed karyotypes based on three reproducible cells, including one shown in Figure 1D. The order of cytological chromosomes was based on the length of each chromosome, which corresponded to the length of sequenced chromosomes (Table I). If no significant difference existed between cytological chromosome lengths (±0.05), correlated coefficients were calculated among H3K4me3 immunostaining intensities and gene densities between each chromosome pair, and the highest correlation coefficient value was chosen for chromosome assignment (see below). Overall, the H3K4me3 signal level was nearly equal between A (8,106,887 ± 1,355,268; total fluorescent luminance intensities per chromosome among all A chromosomes) and D (8,253,784 ± 1,528,778) homeologous chromosomes. The A homeologous chromosomes are twice the size as the D homeologous chromosomes (Zhang et al., 2015). Thus, signal intensities per chromosome area were higher in the D than in the A homeologous chromosomes (Fig. 1D; Supplemental Fig. S1B). The mean intensities per area of D chromosomes (61,144) were approximately 50% higher than the A chromosomes (40,709; t test, P = 2.31E-12; Table I). The H3K4me3 distribution pattern in the A homeologs was similar to that in G. arboreum (Supplemental Fig. S2A), the extant A-genome progenitor, suggesting conservation of H3K4me3 distribution patterns from the progenitor to the allotetraploid cotton. The immunostaining patterns of another active marker, histone H4 Lys 12 acetylation (H4K12ac; Supplemental Fig. S2B), were very similar to those stained by H3K4me3, except for clustered distributions at nucleolar organizer regions, which may contribute to the differential activity of rDNA loci, a phenomenon known as nucleolar dominance (Pikaard, 1999).
Figure 1.
Distinctive immunostaining patterns of antibodies against H3K4me3 fluorescent labels across metaphase chromosomes in allotetraploid cotton. A to C, Metaphase chromosomes from G. hirsutum were counterstained with DAPI (pseudocolored red; A) and immunostained with antibodies against H3K4me3 (Cy3, pseudocolored green; B), and merged (C). D, Immunostained karyotypes constructed from the chromosome spread shown in A to C, bar = 5 μm.
Table I. Chromosome morphology and H3K4me3 intensities in allotetraploid cotton.
| Physical Chromosome | Relative Lengtha | Arm Ratio | Physical Length | Total Signalb | Signalc | Area | Intensity per Aread |
|---|---|---|---|---|---|---|---|
| % | A | Base Pairs | % | µm2 | |||
| A01 | 4.66 ± 0.10 | 1.43 ± 0.15 | 99884700 | 657,674 ± 162,407 | 4.02 ± 0.84 | 16.52 ± 2.06 | 39,923 ± 3,677 |
| A02 | 4.43 ± 0.19 | 1.48 ± 0.15 | 84447906 | 466,557 ± 145,342 | 2.81 ± 0.44 | 15.72 ± 3.28 | 29,611 ± 2,000 |
| A03 | 4.82 ± 0.11 | 1.35 ± 0.14 | 100263045 | 557,364 ± 145,175 | 3.37 ± 0.33 | 14.91 ± 3.46 | 38,735 ± 4,374 |
| A04 | 3.65 ± 0.17 | 1.40 ± 0.13 | 62913722 | 290,055 ± 96,335 | 1.74 ± 0.29 | 11.95 ± 1.82 | 24,189 ± 3,418 |
| A05 | 4.57 ± 0.12 | 1.35 ± 0.18 | 92047023 | 1,054,724 ± 249,696 | 6.4 ± 0.50 | 16.75 ± 1.36 | 63,725 ± 6,371 |
| A06 | 5.13 ± 0.19 | 2.08 ± 0.10 | 103170444 | 589,725 ± 75,662 | 3.62 ± 0.28 | 15.94 ± 1.59 | 37,641 ± 3,572 |
| A07 | 4.34 ± 0.26 | 1.46 ± 0.13 | 78251018 | 655,358 ± 94,698 | 4.01 ± 0.07 | 15.53 ± 5.31 | 41,775 ± 5,902 |
| A08 | 5.17 ± 0.20 | 1.52 ± 0.12 | 103626341 | 466,423 ± 91,853 | 2.84 ± 0.14 | 15.66 ± 2.79 | 29,889 ± 1,446 |
| A09 | 3.76 ± 0.15 | 2.53 ± 0.12 | 74999931 | 673,873 ± 132,876 | 4.12 ± 0.55 | 13.41 ± 0.73 | 50,653 ± 4,187 |
| A10 | 4.98 ± 0.17 | 1.34 ± 0.14 | 100866604 | 691,678 ± 80,475 | 4.25 ± 0.17 | 15.05 ± 3.10 | 46,905 ± 3,083 |
| A11 | 4.59 ± 0.28 | 1.53 ± 0.11 | 93316192 | 776,202 ± 81,460 | 4.77 ± 0.30 | 16.17 ± 2.15 | 48,862 ± 3,418 |
| A12 | 4.51 ± 0.17 | 2.06 ± 0.29 | 87484866 | 847,605 ± 40,103 | 5.24 ± 0.55 | 16.89 ± 2.08 | 50,595 ± 2,477 |
| A13 | 4.37 ± 0.16 | 1.52 ± 0.11 | 79961121 | 379,649 ± 70,493 | 2.31 ± 0.07 | 14.63 ± 2.13 | 26,298 ± 2,094 |
| Subtotal | 59.22 ± 0.18 | 1160232963 | 8,106,887 ± 1,355,268 | 49.51 ± 0.55 | 199.14 ± 25.82 | 40,755 ± 3,892e | |
| D01 | 3.05 ± 0.06 | 1.20 ± 0.21 | 61456009 | 457,157 ± 38,618 | 2.63 ± 0.17 | 9.38 ± 1.23 | 50,545 ± 3,492 |
| D02 | 3.80 ± 0.08 | 1.33 ± 0.16 | 67284553 | 543,146 ± 193,935 | 3.27 ± 0.88 | 10.48 ± 1.49 | 52,736 ± 7,173 |
| D03 | 2.49 ± 0.08 | 1.76 ± 0.05 | 46690656 | 424,859 ± 69,337 | 2.63 ± 0.52 | 9.29 ± 1.30 | 45,909 ± 2,105 |
| D04 | 2.81 ± 0.21 | 2.05 ± 0.16 | 51454130 | 742,481 ± 138,203 | 4.58 ± 0.92 | 9.35 ± 0.61 | 80,571 ± 7,593 |
| D05 | 3.15 ± 0.15 | 2.14 ± 0.11 | 61933047 | 1,114,191 ± 131,179 | 6.86 ± 0.66 | 11.23 ± 0.50 | 99,538 ± 4,848 |
| D06 | 3.18 ± 0.08 | 1.23 ± 0.11 | 64294643 | 550,891 ± 34,863 | 3.40 ± 0.36 | 9.87 ± 0.64 | 55,963 ± 2,085 |
| D07 | 2.86 ± 0.13 | 1.31 ± 0.06 | 55312611 | 584,611 ± 108,816 | 3.57 ± 0.14 | 11.82 ± 1.61 | 50,195 ± 3,894 |
| D08 | 3.38 ± 0.13 | 1.29 ± 0.25 | 65894135 | 603,040 ± 216,770 | 3.61 ± 0.75 | 9.93 ± 1.31 | 59,857 ± 5,890 |
| D09 | 2.61 ± 0.09 | 1.49 ± 0.14 | 50995436 | 618,220 ± 109,011 | 3.77 ± 0.16 | 9.22 ± 0.68 | 67,618 ± 5,151 |
| D10 | 3.17 ± 0.11 | 1.31 ± 0.17 | 63374666 | 560,210 ± 129,677 | 3.04 ± 0.47 | 9.71 ± 0.94 | 57,695 ± 3,669 |
| D11 | 3.78 ± 0.10 | 1.79 ± 0.13 | 66087774 | 968,916 ± 163,002 | 5.93 ± 0.39 | 12.40 ± 1.61 | 78,848 ± 4,944 |
| D12 | 2.90 ± 0.16 | 2.29 ± 0.27 | 59109837 | 688,976 ± 140,019 | 4.22 ± 0.66 | 10.63 ± 1.62 | 65,350 ± 2,746 |
| D13 | 3.04 ± 0.18 | 1.23 ± 0.17 | 60534298 | 485,345 ± 55,347 | 2.99 ± 0.31 | 11.67 ± 1.36 | 41,733 ± 1,419 |
| Subtotal | 40.02 ± 0.18 | 774421795 | 8,253,784 ± 1,528,778 | 50.49 ± 0.55 | 134.99 ± 13.03 | 61,350 ± 5,668e |
Relative chromosomal length.
H3K4me3 fluorescence signals across chromosome surface.
Percent of fluorescence signal of a single chromosome relative to all A and D chromosomes.
Total signal levels per chromosome surface area.
Mean fluorescent intensity per area in the A and D homoeologs.
H3K27me2 Marks Present in Nuclear Peripherals and Devoid in Active rDNA Loci
H3K27me2 marks correlated with heterochromatin and gene silencing in plants and animals (Martin and Zhang, 2005). Immunostaining using antibodies against H3K27me2 showed that the signals were distributed throughout the metaphase chromosomes of G. arboreum (Supplemental Fig. S3A) and G. hirsutum (Supplemental Fig. S3B). Signals in telomeric regions were enriched in some chromosomes. In the interphase, H3K27me2 antibody signals were clustered as speckles around the nuclear peripherals (Supplemental Fig. S3, C and D). There are three 45S rDNA loci, two major and one minor loci, in G. arboreum (in some cases, minor loci could not be detected by fluorescence in situ hybridization (FISH; Hanson et al., 1995; Wang et al., 2008). H3K27me2 antibody signals were colocalized with one of the rDNA loci, but not with the other rDNA loci (Supplemental Fig. S3E). This suggests silencing of one rDNA locus and expression of two other loci that are subject to nucleolar dominance (Pikaard, 1999).
Correlation between Gene Densities and H3K4me3 Intensities
Previous studies have identified a complete set of 26 chromosome pairs by BAC-FISH (Wang et al., 2006). Based on that information, we merged the H3K4me3 intensities with the chromosome morphology map in tetraploid cotton (Fig. 1, C and D). The combination of morphology and H3K4me3 density was used to assign G. hirsutum chromosomes from A01 to A13 and from D01 to D13 (Supplemental Fig. S4; Fig. 1). Seven pairs (A01-D01, A04-D04, A05-D05, A06-D06, A08-D08, A09-D09, and A11-D11) had the highest correlation coefficient values (P < 0.01); five pairs (A02-D02, A03-D03, A10-D10, A12-D12, and A13-D13) had higher (but not the highest) correlation coefficient values with P < 0.01; only one pair (A07-D07) had the value within a low confidence level (Supplemental Table S6). With the chromosomes identified, the maps of antibody intensity and gene density among 13 pairs of A and D homeologous chromosomes were comparatively analyzed.
G. hirsutum genome consisted of similar numbers of genes in A and D homeologous chromosomes but more repetitive sequences in the A homeologs (69.1%) than in the D homeologs (58.0%; Supplemental Data S1; Zhang et al., 2015). These repeats were enriched in the centromeres and gradually lost toward distal regions of chromosomes (Fig. 2A). On the contrary, the distribution of genes showed an opposite trend. Because the D homeologous chromosomes are smaller but essentially with similar numbers of genes (Zhang et al., 2015), the gene density in the D homeologs should be higher.
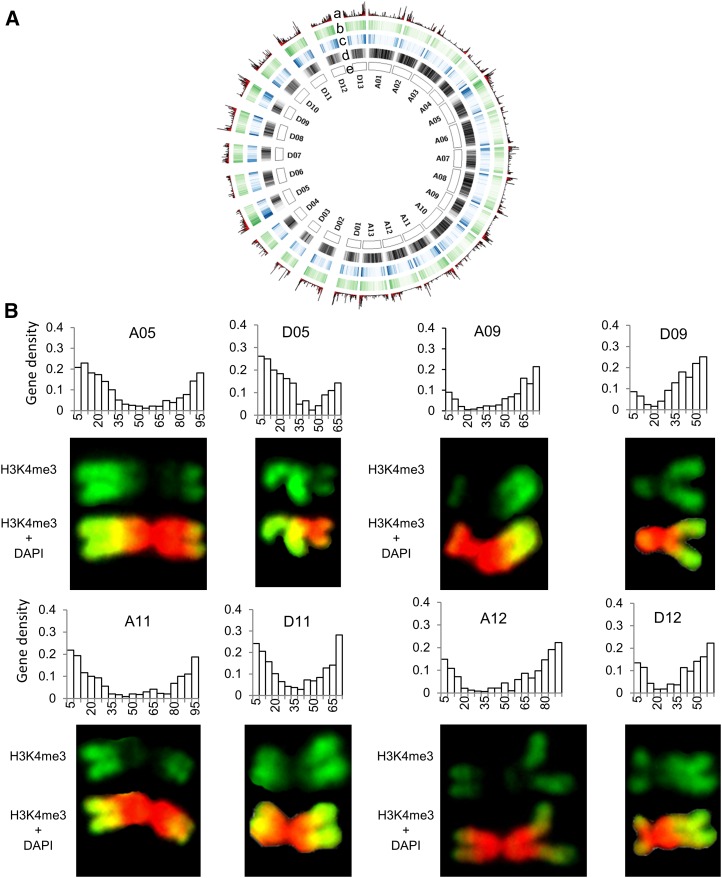
Figure 2.
Correlation of gene density with H3K4me3 levels of A and D homeologous chromosomes in allotetraploid cotton. A, a to d, The nonoverlapping 500-kb windows indicate (a) transcript density (FPKM) of the genes expressed in the root tip cells from high (tall bars) to low (short bars), (b) density of H3K4me3 in the root tip cells from high (dark green) to low (light green), (c) density of the genes expressed in the root tip cells from high (dark blue) to low (light blue), and (d) density of repeats from high (dark gray) to low (light gray). B, H3K4me3 levels are associated with gene-rich regions among homeologous chromosomes in allotetraploid cotton. Examples are A05-D05, A09-D09, A11-D11, and A12-D12 homeologous chromosomes. Histograms showing gene numbers (y axis) relative to 5-Mb windows (x axis) in each chromosome (see Fig. 1).
In humans, the level of histone modifications (e.g. H3K4me3) is directly proportional to the density of genes along each chromosome (Terrenoire et al., 2010). We tested this relationship in G. hirsutum by comparing gene density maps in 5-Mb windows across each chromosome (Supplemental Data S2) with H3K4me3 signal patterns (Table I). The H3K4me3 levels across each chromosome were quantified by Intensity Profile of the Olympus cellSens Dimension, which could generate the intensity profiles over different positions.
The results indicate that gene density and H3K4me3 intensity maps were consistent in chromosome pairs, such as A05-D05, A09-D09, A11-D11, and A12-D12 (Fig. 2B), except for chromosomes that involve translocations (Menzel et al., 1986; Wang et al., 2006; Zhang et al., 2015; see below). The chromosome pairs consistent with the gene and H3K4me3 densities have relatively high gene numbers and evenly distributed gene densities. This is true not only for intensely stained regions (e.g. landmark bands on chromosomes A09q, A05q, D05q, and D09q; p and q denote short and long arms of each chromosome) but also for weakly stained areas (e.g. the signal distribution along chromosomal bands A05p and A12q; Fig. 2B).
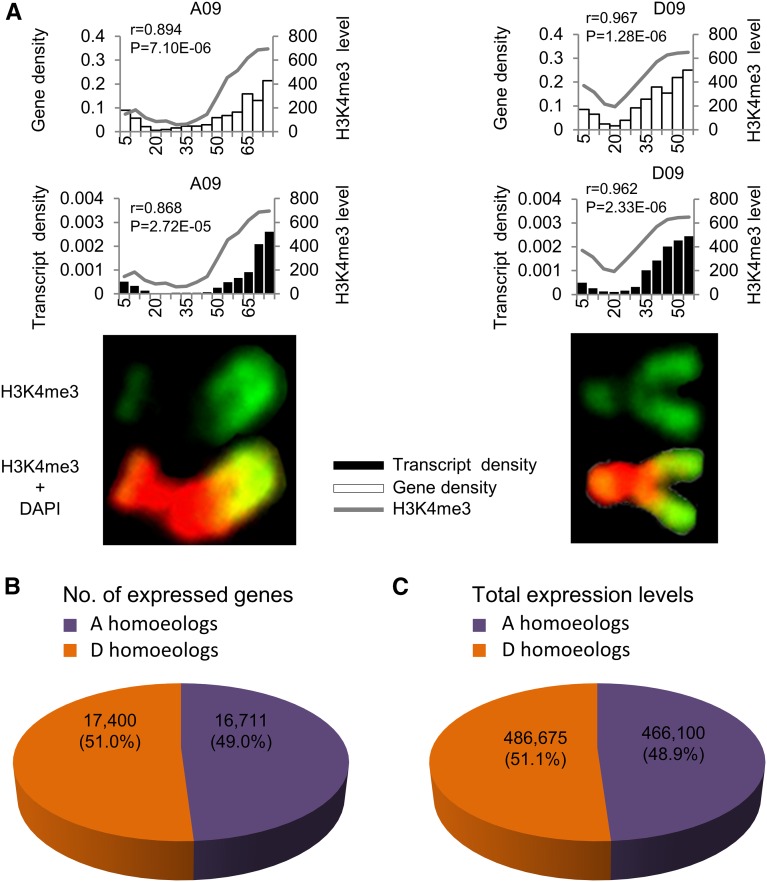
For example, chromosomes A09 and D09 have 75.0 and 51.0 Mb DNA, respectively. Assuming a linear relationship between genomic DNA distribution and physical position on the metaphase chromosome, we divided each chromosome into a number of bins with 5 Mb per bin, resulting in 14 bins on chromosome A09 and 11 bins on chromosome D09. These bins were quantified for their overall fluorescence intensities (Fig. 3A). Correlation coefficients between the gene density and H3K4me3 intensity distributions among bins on each chromosome were 0.894 and 0.967 in A09 and D09, respectively (P < 0.01), suggesting a significant correlation between gene density and H3K4me3 intensity.
Figure 3.
Correlation of gene and transcript densities with H3K4me3 levels between homeologous chromosomes A09 and D09. A, Quantitative analysis of anti-H3K4me3 fluorescent levels between homeologous chromosomes A09 and D09 relative to gene densities and transcript levels in root tip cells. Histograms in the left (A09 chromosome) and right (D09 chromosome) indicate gene densities (top) and transcript levels (bottom) relative to anti-H3K4me3 fluorescent levels (y axis, the opposite). Black, unfilled box, and line, respectively, indicate distribution patterns of transcripts, genes, and anti-H3K4me3 fluorescent levels in 5-Mb windows. P values indicate significant levels of Pearson correlation coefficient (r) between each comparison. H3K4me3 intensity values are shown in Supplemental Data S2. B, Total number of A and D homeologous genes expressed in the root-tip cells. C, Total expression levels (FPKM) of the A and D homeologous genes in the root-tip cells.
Transcriptome Analysis of Root Tip Cells in G. hirsutum Cotton
Different levels of chromatin modifications could result in gene expression differences along the chromosomes. We tested this in tetraploid cotton by RNA-seq analysis using sequence libraries prepared from the root-tip cells, which were the same cells used to prepare for metaphase chromosome immune-staining assays. Three independent mRNA-seq libraries were constructed and sequenced by Illumina paired-end reads. A total of 171 million reads were obtained from three libraries, and 85% to 88% reads were mapped onto A and D homeologs of G. hirsutum sequence (Zhang et al., 2015), among which approximately 49% and 51% reads were mapped onto the A and D homeologs, respectively (Supplemental Table S1). At the whole genome level, an equal number of A (16,711) and D (17,400) homeologous genes was expressed in the root tip cells of allotetraploid cotton (Supplemental Table S1; Fig. 3B), consistent with the results from expression analysis in over 30 tissues (Zhang et al., 2015). The overall expression levels (fragments per kilobase per million [FPKM]) in the A and D homeologs were proportional to the number of expressed genes (Supplemental Tables S1 and S2; Fig. 3, B and C) and also consistent with total H3K4me3 signal intensities in the A and D homeologous chromosomes (Table I; Fig. 2A). For example, transcription levels and H3K4me3 immunostaining intensities on each chromosome were significantly correlated (r = 0.868 for A09 and 0.962 for D09, P < 0.01; Fig. 3A).
Differential Gene Expression between A and D Homeologs
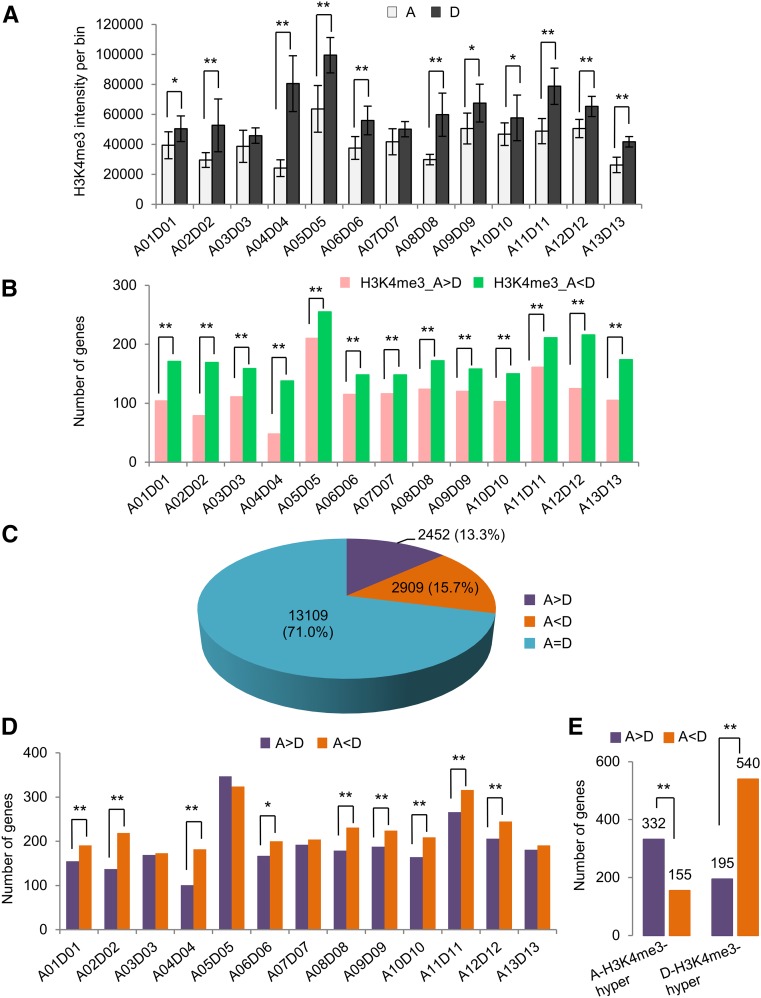
Although the total number of genes did not show expression bias in multiple tissues, as many as 30% of genes can be expressed differently between A and D homeologs in a given tissue (Zhang et al., 2015). The reason for this is unknown. Since we know where each homeologous pair is located in the genome sequence, we can associate expression patterns with H3K4me3 status of immuno-staining chromosomal bins in root-tip cells. We found that the H3K4me3 intensities per surface area in most D homeologous chromosomes, except for A03 versus D03 and A07 versus D07, were higher than those in the A homeologs (Fig. 4A), which could lead to biased expression of the D homeologs. To further test this relationship, we identified 18,470 homeologous gene pairs using reciprocal BLAST hits between A and D homeologs, as previously reported (Zhang et al., 2015), and compared them with ChIP-seq data of H3K4me3 obtained from the same type of root-tip cells (Supplemental Table S5; Supplemental Data S3). Consistent with the cytological data, more D homeologs than the A homeologs had high H3K4me3 levels among all 13 homeologous pairs of chromosomes (Fig. 4B).
Figure 4.
H3K4me3 and gene expression level differences between homeologous chromosomes in the root-tip cells of allotetraploid cotton. A, Anti-H3K4me3 fluorescent intensities in 13 homeologous chromosome pairs from immunostaining cells. Single (*) and double (**) asterisks indicate statistically significant levels of P < 0.05 and P < 0.01, respectively, using Student’s t test. Error bar ± sd. B, ChIP-seq data showed the number of genes with A homeolog bias (A > D) and D homeolog bias (A < D) of H3K4me3 levels among 13 homeologous chromosome pairs. Double (**) asterisks indicate statistically significant levels of P < 0.01 using χ2 test. C, Number and percentage of homeologs showing equal (A = D), A homeolog-biased (A > D), and D homeolog-biased (A < D) expression. D, Number of the genes with A homeolog-biased (A > D) and D homeolog-biased (A < D) expression among 13 homeologous chromosome pairs. Single (*) and double (**) asterisks indicate statistically significant levels of P < 0.05 and P < 0.01, respectively, using χ2 test. E, Number of genes with A homeolog-biased (A > D) and D homeolog-biased (A < D) expression relative to hyper-H3k4me3 levels in A homeologs or D homeologs. Double (**) asterisks indicate statistically significant levels of P < 0.01 using χ2 test.
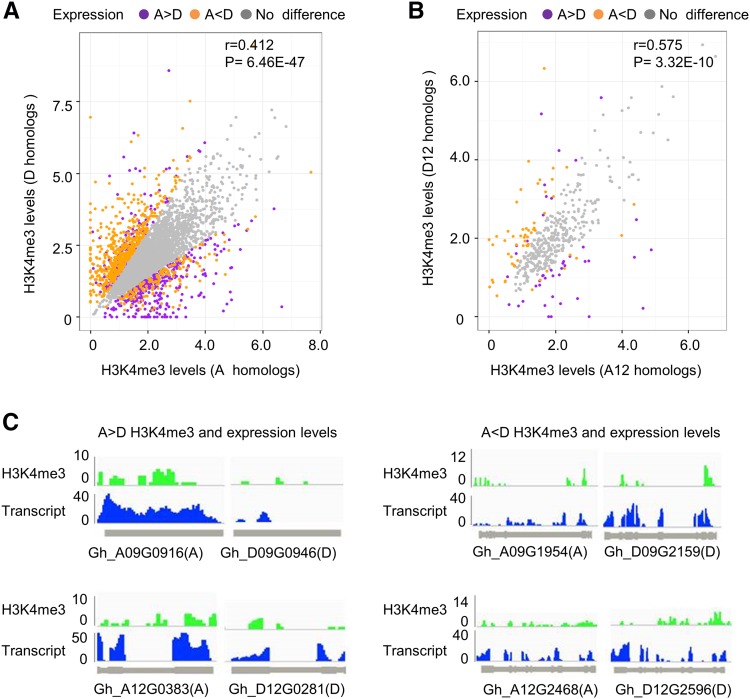
At the level of individual genes, we identified 5,361 (29%) homeologous genes with unequal (biased) expression in the root-tip cells. Among them, 2,452 (13%) were expressed toward A homeologs (A > D), while 2,909 (16%) were expressed toward D homeologs (D > A; Fig. 4C). The number of genes with D homeolog-biased expression was higher than that with A homeolog-biased expression among most homeologous pairs, except for A03 versus D03, A05 versus D05, A07 versus D07, and A13 versus D13 pairs (Supplemental Data S3; Fig. 4D). Among the genes that were hypermethylated at H3K4me3 in the A homeologs relative to the D homeologs, more than 3-fold of A homeologs were expressed at higher levels than the D homeologs. Likewise, among the genes that were hypermethylated at H3K4me3 in the D homeologs relative to the A homeologs, nearly 4-fold of D homeologs were expressed at higher levels than the A homeologs (Fig. 4E). Further analysis using pairwise comparison of A and D homeologs with H3K4me3 levels indicated statistically significant correlation between expression and H3K4 me3 levels of A and D homeologs (r = 0.412, P < 0.01; Fig. 5A). Among hypermethylated H3K4me3 loci, more D homeologs (540) than A homeologs (332) displayed biased expression (P < 0.01, using χ2 test). This biased expression was also observed for individual homeologous chromosome pairs. For example, more D homeologs (49) than A homeologs (31) on A12 and D12 homeologous chromosomes showed biased expression (P < 0.01, using χ2 test; Fig. 5B). This trend was observed at the level of several individual genes examined (Fig. 5C). For example, higher levels of H3K4me3 in Gh_A09G0916 and Gh_A12G0383 correlated with higher levels of expression in these loci relative to their respective homeologs (Gh_D09G0946 and Gh_D12G0281). Likewise, H3K4me3 and expression levels were higher in the D homeologs (Gh_D09G2159 and Gh_D12G2596) than in their corresponding A homeologs (Gh_A09G1954 and Gh_A12G2468). Together, these data suggest that H3K4me3 modifications play an important role in the expression bias of homeologous genes. Overall, more D homeologs were hypermethylated at H3K4me3 and expressed at higher levels than A homeologs.
Figure 5.
Correlation of homeolog-biased expression with H3K4me3 levels. A, Pairwise comparison of H3K4me3 levels (y axis, D homeologs; x axis, A homeologs) for 5,361 pairs of homeologous genes. Purple, orange, and gray dots indicate A-biased (A > D), D-biased (A < D), and equal (A = D or no difference) expression, respectively. P value of Spearman correlation coefficient (r) is shown. B, Pairwise comparison of H3K4me3 levels (y axis, D homeologs; x axis, A homeologs) for homeologous genes from A12 and D12 chromosomes. Dots are shown as in A. P value of Spearman correlation coefficient (r) is shown. C, Examples of hyper-H3K4me3 levels correlating with enhanced expression of the A homeologs (A > D, left) and D homeologs (A < D, right). Colors indicate H3K4me3 (green) and expression (blue) levels.
Effects of Chromosomal Translocation on Homeologous Gene Expression
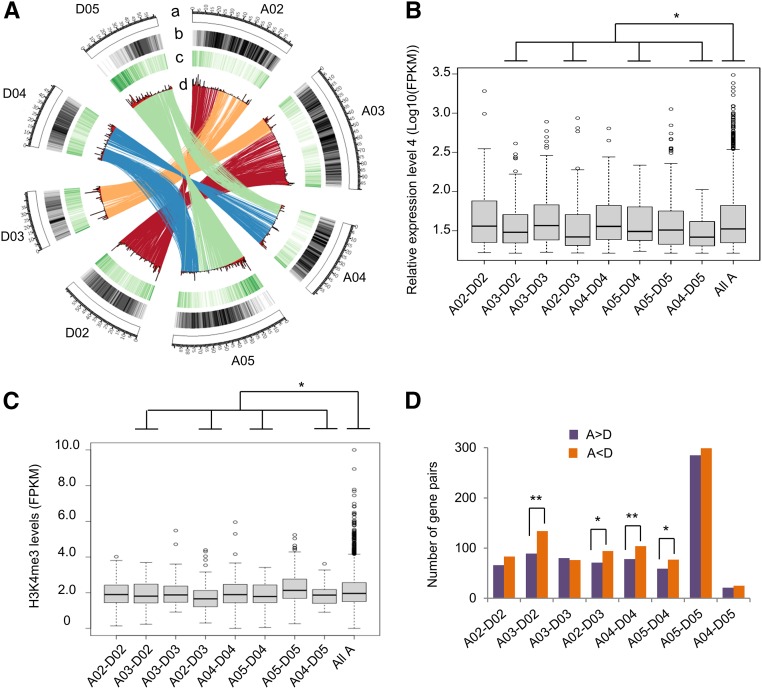
Notably, several homeologous chromosome pairs with an equal number of expression-biased genes involved chromosomal translocations (Fig. 4D). Two translocations have occurred between chromosomes A02 versus A03 and A04 versus A05 based on cytological and sequencing analyses (Menzel et al., 1986; Wang et al., 2006; Zhang et al., 2015; Fig. 6A). As a result, the comparison between putative homeologous pairs did not fully reflect the expected homeologous relationship prior to translocation. Expression levels of the genes between the translocated regions were different from that of all chromosomes (Fig. 6B). This was consistent with the difference of H3K4me3 distribution levels between translocated regions and all chromosomes (Fig. 6C). While the number of expression-biased genes of A02-D02 and A03-D03 gene pairs matched the rest of the genome, the number of genes with D-biased expression was significantly higher in the translocated A02-D03 and the A03-D02 regions (Fig. 6D; χ2 test, P < 0.05). The trend was similar but to a lesser degree in the comparison between A04-D04 versus A05-D05 and A04-D05 versus A05-D04. These translocations could affect the local chromatin structure and hence expression levels for the genes involved.
Figure 6.
Chromosomal translocations and their effects on biased gene expression. A, Diagram of two translocations between chromosomes A02 and A03, and A04 and A05. A, Diagram of chromosomes. B to D, Nonoverlapping 500-kb windows indicate repeat density from high (dark gray) to low (light gray; B), density of the H3K4me3 levels in the root-tip cells from high (dark green) to low (light green; C), and transcript density (FPKM) of the genes expressed in the root-tip cells (D). Color ribbons indicate translocation regions. B, Relative expression levels of the genes among the translocated chromosomes. The gene expression was divided into four levels (from the lowest to the highest). Expression level 4 (the highest) of homeologous genes on translocated A chromosomes was shown. The control was expression level 4 of homeologous genes among all A chromosomes. An asterisk indicates statistical significance of P < 0.05, using Student’s t test. C, H3K4me3 levels of the level-4 genes among the translocated chromosomes. An asterisk indicates statistical significance of P < 0.05, using Student’s t test. D, Distribution of the genes with A homeolog expression bias (A > D) and D homeolog expression bias (A < D) in the translocated regions between A02 and A03 and between A04 and A05 homeologous chromosomes. Single (*) and double (**) asterisks indicate statistically significant levels of P < 0.05 and P < 0.01, respectively, using χ2 test.
Correlation of homeolog-biased expression with H3K4me3 could lead to functional consequences (Supplemental Data S4). Gene Ontology analysis showed enrichments of biased expression of A homeologous genes in plant cell wall and those of biased expression of D homeologous genes in metabolic process in root-tip cells (Supplemental Fig. S6). In other tissues such as cotton ovules and fibers, A homeologous genes with biased expression include fiber-related transcription factor genes, while biased expression of D homeologs is related to stress responses (Zhang et al., 2015).
DISCUSSION
Euchromatic and Heterochromatic Marks in Plant Chromosomes
Histone modifications mediate epigenetic regulation of gene expression, growth, and development in plants and animals (Berger, 2007; Li et al., 2007; Zhang, 2008). Histone marks such as H3K4me3 and H4K12ac are associated with euchromatin and gene activation, while marks like H3K27me2 correlate with heterochromatin and gene repression (Jenuwein and Allis, 2001; Li et al., 2007). In allotetraploid cotton, the H3K4me3 density difference between A and D homeologous chromosomes could be related to the genome size, which is twice as large in the former as in the latter. In addition, retrotransposons including long terminal repeat retrotransposons are more abundant in the A homeologs compared to D homeologs (Zhang et al., 2015). Together, more repetitive sequences and larger size in the A subgenome than in the D subgenome are associated with more heterochromatin per megabase in the A homeologs than in the D homeologs and consequently less euchromatin per megabase in the A homeologs than in the D homeologs. On each chromosome, the distal portions were enriched with H3K4me3 and H4K12ac so the euchromatin is more abundant near chromosome ends than proximal regions, which is consistent with the distribution of genes and genetic recombination along cotton chromosomes (Paterson et al., 2012). This gradient distribution of H3K4me3 across metaphase chromosomes of G. hirsutum is consistent with the notion that H3K4me3 is a conserved euchromatic mark in plant species (Fuchs et al., 2006). H4K12ac has very similar distribution patterns as H3K4me3, except for clustering of H4K12ac near the nucleolar organizing regions, as previously reported in other plant chromosomes (Belyaev et al., 1997; Jasencakova et al., 2000). This indicates a role for H4K12ac in differential expression of uniparental rRNA genes subjected to nucleolar dominance (Lawrence et al., 2004).
H3K27 methylation has been linked to several silencing phenomena in animals, including X chromosome inactivation and genomic imprinting (Martin and Zhang, 2005). H3K27me2 is highly concentrated in the centromeric heterochromatin in Arabidopsis (Arabidopsis thaliana; Lindroth et al., 2004; Mathieu et al., 2005) and maize (Shi and Dawe, 2006). However, this distribution pattern is not conserved in cotton compared to maize or Arabidopsis (Fuchs et al., 2006). H3K27me2 signals are patchy in the metaphase and display some weak bands in interphase cells (Supplemental Fig. S3). Allotetraploid cotton has two major and one minor nucleolar organizing regions (Hanson et al., 1995), and H3K27me2 is more prominent in one of them. This suggests that H3K27me2 could cause silencing of one rDNA locus, while H4K12ac could be associated with active rDNA loci.
Both Chromatin and Genome Organization Mediate Homeologous Gene Expression
After polyploidization, genetic changes such as mutations, sequence elimination, and chromosomal rearrangements as well as epigenetic mechanisms at transcriptional and posttranscriptional levels can contribute to gene expression variation (Chen, 2007; Doyle et al., 2008; Soltis et al., 2014). Polyploidy may induce epigenetic modifications of homeologous chromosomes to reprogram gene expression and developmental patterns of allopolyploids (Song and Chen, 2015), leading to genome-wide dosage-dependent and independent gene expression novelty (Shi et al., 2015). Histone marks such as H3K4me3 are known to correlate with gene expression diversity between species and in Arabidopsis allopolyploids (Ha et al., 2011; Shi et al., 2015). In this study, we associated H3K4me3 levels with expression changes of homeologous genes in allotetraploid cotton. Although the total H3K4me3 levels are relatively equal between A and D homeologous chromosomes, the fluorescent intensity per area of H3K4me3 is significantly higher in the D homeologous chromosomes than in the A homeologs. Consistent with the cytological data, ChIP-seq analysis showed more D homeologs with biased H3K4me3 levels than A homeologs with biased H3K4me3 levels, which could loosen chromatin structure for the D homeologs, allowing genes to be expressed at higher levels. Indeed, there are more genes with D homeolog-biased expression than with A homeolog-biased expression in most homeologous chromosome pairs. This suggests an overall greater effect of H3K4me3 on D homeologs than on A homeologs.
Chromosomal translocations can result in hitherto unforeseen and large-scale changes in gene expression that are the consequence of alterations in normal chromosome territory positioning in human (Harewood et al., 2010). In allotetraploid cotton, homeologous chromosome pairs with chromosomal translocations could possess different chromatin structure and change the number of genes with homeolog-biased expression among the chromosomes involved. Expression levels of the genes between the translocated regions were different from that of all chromosomes and were consistent with the difference of H3K4me3 distribution levels between translocated regions and all chromosomes. Thus, during polyploidization, genome reorganization including translocation may alter chromatin structure that induces gene expression changes and potentially phenotypic variation in polyploids.
It is difficult to determine the exact relationship between an immunostained and sequenced chromosome unless a set of DNA markers specific to each chromosome is used on the same cells where the chromosomes are immunostained. This would be technically challenging and cannot be resolved at the present stage. The current approach using the chromosome length, immunolabeling intensity, and gene density maps has shown to work well in cotton as in mammalian cells (Terrenoire et al., 2010). The ChIP-seq results have confirmed the cytological data. Distributions between H3K4me3 intensities and gene densities are comparable and consistent between most homeologous chromosome pairs. With improvement of technologies, chromatin and expression analysis of single cells or chromosomes would increase the resolution and precision of chromatin regulation. This would be extremely valuable to resolve gene expression changes between homeologous chromosomes in polyploid species for the genetic improvement of important polyploid crops such as wheat (food), cotton (fiber), and canola (oil).
MATERIALS AND METHODS
Immunostaining and FISH
Root tips were harvested from rapidly growing roots of A-genome extant progenitor (Gossypium arboreum L. cv JLZM) and tetraploid cotton (Gossypium hirsutum L. accession TM-1) and treated with N2O for 1 to 3 h and fixed in a phosphate buffer solution (PBS) containing 4% paraformaldehyde. After washing with PBS, the fixed root tips were digested with 4% Cellulase Onozuka R-10 (Yakult Honsha) and 1% Pectinase Y-23 (MP Biomedicals) for 2 h at 37°C. The digested root tips were washed in PBS and water (10 min each) and squashed in a drop of water. The slides were rapidly deep-frozen with liquid nitrogen. After removing coverslips, slides were immediately treated with PBS containing 3% bovine serum albumin and 1% Triton X-100. Commercial antibodies against H4K12Ac (Abcam) and H3K4me3, H3K27me2, and H3K9me2 (Upstate Biotechnology) were diluted into 1:100 in PBS and applied to the slides that were subsequently covered with coverslips. The slides were incubated in a moist chamber at 37°C for overnight. After washing in PBS, the antibodies were detected using the secondary antibodies of Cy3-conjugated antirabbit. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Following histone immunostaining, FISH was performed on chromosome spreads according a previously published protocol (Han et al., 2009). Digoxigenin-labeled probes were detected using fluorescein-conjugated antidigoxigenin (Roche Diagnostics). Chromosomes were counterstained with Vectashield Mounting medium containing DAPI (Vector Laboratories) under a coverslip. To help chromosome identification, we used telomere and GISH for chromosome analysis. Telomeric DNA (42 copies of TTTAGGG) was used as digoxigenin-labeled probe (fluorescein-conjugated antidigoxigenin) for FISH analysis, and genomic DNA from G. arboreum was used as digoxigenin-labeled probe (fluorescein-conjugated antidigoxigenin) for GISH analysis.
Analysis of Chromosome Images and Immunostaining Signal Intensities
Slides were examined using an Olympus BX53 fluorescence microscope (objective UPlanSApo 100× 1.40, oil) at room temperature (Olympus). The images of chromosomes, immunostaining, and FISH signal channels were captured and merged using Olympus cellSens Dimension software V5.1 with an Olympus DP72 CCD camera. To determine H3K4me3 signal levels of A and D chromosomes, only the cells with nonoverlapping chromosomes were analyzed, and the sum of relative signal levels of each chromosome was calculated using Olympus cellSens Dimension. Final image adjustments were processed using Photoshop CS 3.0 (Adobe). For each chromosome, the level of H3K4me3 intensities was estimated using the Intensity Profile of the Olympus cellSens, which measured the signal intensity per 0.0339 μm per unit. The chromosomes were first partitioned into homologous pairs using morphological features (arm length and short/long arm ratios) and H3K4me3 patterns and assigned based on arm length of cytological chromosome (from short to long). For quantitative analysis, we divided each chromosome into bins (5 Mb per bin) using H3K4me3 fluorescent intensities and gene densities. The correlation between H3K4me3 fluorescent intensities and gene densities in each bin on each chromosome pair was estimated using Pearson correlation coefficients and tested for statistical significance (Supplemental Tables S3 and S4). The confidence of homeology assignment was based on Pearson correlation coefficients and P values of immunostaining densities (H3K4me3) at distal regions between A and D homeologous chromosomes (Supplemental Table S6).
RNA Extractions, RNA-Seq Libraries, and Sequencing
RNA samples of root tips in three replicates were extracted using a modified mirvana mirna isolation kit procedure (Ambion), and their quality was evaluated on Nanodrop 2000 (Thermo Fisher Scientific) and Agilent Bioanalyzer (Agilent Technologies). mRNA-seq libraries were constructed following the RNA-seq library prep kit protocol and sequenced by Illumina HiSEquation 2500 machine (Illumina) with 100 base pair-end reads at Nanjing Agricultural University.
RNA-Seq Data Preprocessing and Quality Control
Raw reads were examined and filtered by the following steps: (1) removing the last index of original FASTQ format with perl script, (2) performing quality control of the sequencing data by NGS QC Toolkit V2.3 (Patel and Jain 2012), and (3) discarding the low-quality reads to ensure that >80% of the bases of each retained read had the Illumina quality >20 (q20 indicating 1% sequencing error rate).
Mapping and Analysis of Gene Expression in Homeologous Chromosomes
Clean reads were mapped onto the G. hirsutum genome (Zhang et al., 2015). The mapping was processed by TopHat version 2.0.12 (http://ccb.jhu.edu/software/tophat/index.shtml) utilizing Bowtie2 version 2.23 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Mapping results generated by TopHat were filtered to retain only uniquely mapped reads before being transferred into Cuffdiff for estimating read counts. For each gene, the value of FPKM reads was calculated by in-house scripts based on the count table of Cuffdiff output. Only the genes with mean FPKM of ≥1 were used for further analysis. The reads were used to estimate the number of the genes and expression levels of the each homeolog or chromosome. The same chromosome coordinates were used for visualization and distribution of repeats, genes, and FPKM values in the G. hirsutum genome (Zhang et al., 2015) using the program Circos (Krzywinski et al., 2009). Expression bias of homeologous genes was estimated to identify the genes with differential expression between A and D homeologs using statistical tests (FDR < 0.05) with at least 1.5-fold changes in average expression levels (FPKM) across three biological replicates. The RNA-seq data were deposited at GEO (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE71418.
ChIP and ChIP-seq Library Preparation
ChIP was performed by immunoprecipitating chromatin with antibodies against H3K4me3 (Upstate Biotechnology) with two biological replicates of root tips, as described previously (Saleh et al., 2008). ChIP-seq libraries for immuno-precipitated and input samples from two biological replicates of root tips were constructed and sequenced by Illumina HiSEquation 2500 following the manufacturer’s recommendations (Illumina). The ChIP-seq data has been deposited at GEO (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE83371.
Computational Analysis of ChIP-seq Data
Sequencing reads were mapped onto the G. hirsutum reference genome (Zhang et al., 2015) by SOAP2 version 2.21t software using default parameters (Li et al., 2009). MACS software was used to identify peaks (P < 0.01; Zhang et al., 2008). H3K4me3 distributions of homeologous genes were estimated to identify the genes with differential H3K4me3 levels between A and D homeologs using statistical tests (FDR < 0.05) with at least 1.5-fold changes (FPKM).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GSE83372.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Antibodies against H3K4me3 immunostaining and telomere locations of metaphase chromosomes in allotetraploid cotton.
Supplemental Figure S2. Immunostaining images generated using antibodies against H3K4me3 and H4K12ac, respectively, in metaphase chromosomes of G. arboreum.
Supplemental Figure S3. Immunostaining images of interphase and metaphase chromosomes with antibodies against H3K27me2.
Supplemental Figure S4. Epigenetic chromosome map using antibodies against H3K4me3.
Supplemental Figure S5. Gene Ontology analysis of the genes with homeolog expression bias.
Supplemental Table S1. Number and percentage of sequencing reads mapped onto the allotetraploid cotton (AD) genome sequence.
Supplemental Table S2. Number and expression levels of A and D homeologs.
Supplemental Table S3. Correlation coefficients between immunostaining and gene densities of homeologous A chromosomes in G. hirsutum (TM-1).
Supplemental Table S4. Correlation coefficients between immunostaining and gene densities of homeologous D chromosomes in G. hirsutum (TM-1).
Supplemental Table S5. Number and percentage of sequencing reads of ChIP-seq mapped onto the allotetraploid cotton (AD) genome sequence.
Supplemental Table S6. Correlation coefficients of immunostaining densities (enriched with H3K4me3) in the distal portions between A and D homeologous chromosomes in G. hirsutum (TM-1).
Supplemental Data S1. Gene densities, repeats, and expression levels in allotetraploid cotton
Supplemental Data S2. H3K4me3 immunostaining levels along each chromosome.
Supplemental Data S3. Levels of H3K4me3 and expression bias (A > D and A < D) of homeologous genes.
Supplementary Material
Acknowledgments
We thank Dr. Yan Hu for coordinating sequencing activities and Jingya Yuan for assistance in FISH.
Glossary
- ChIP-seq
chromatin immunoprecipitation-sequencing
- DAPI
4′,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- FPKM
fragments per kilobase per million
- GISH
genomic DNA in situ hybridization
- PBS
phosphate buffer solution
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (31290213 and 91631302), the National Key Research and Development Program (2016YFD0101006), National Science Foundation of the United States (IOS1025947 and IOS1444552), Research and Education Innovation Consortium of Jiangsu Province (2013-2015), Jiangsu Collaborative Innovation Center for Modern Crop Production, and Fundamental Research Funds for the Central Universities (KYRC201204 and KYZ201202-1). The authors declare no competing interest of this work.
Articles can be viewed without a subscription.
References
- Belyaev ND, Houben A, Baranczewski P, Schubert I (1997) Histone H4 acetylation in plant heterochromatin is altered during the cell cycle. Chromosoma 106: 193–197 [DOI] [PubMed] [Google Scholar]
- Berger SL. (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Carchilan M, Delgado M, Ribeiro T, Costa-Nunes P, Caperta A, Morais-Cecílio L, Jones RN, Viegas W, Houben A (2007) Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19: 1738–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Tian L (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci 11: 199–208 [DOI] [PubMed] [Google Scholar]
- Guan X, Song Q, Chen ZJ (2014) Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci 19: 516–528 [DOI] [PubMed] [Google Scholar]
- Ha M, Ng DW, Li WH, Chen ZJ (2011) Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res 21: 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Gao Z, Birchler JA (2009) Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RE, Zwick MS, Choi S, Islam-Faridi MN, McKnight TD, Wing RA, Price HJ, Stelly DM (1995) Fluorescent in situ hybridization of a bacterial artificial chromosome. Genome 38: 646–651 [DOI] [PubMed] [Google Scholar]
- Harewood L, Schütz F, Boyle S, Perry P, Delorenzi M, Bickmore WA, Reymond A (2010) The effect of translocation-induced nuclear reorganization on gene expression. Genome Res 20: 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16: 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Schubert I (2003) DNA and proteins of plant centromeres. Curr Opin Plant Biol 6: 554–560 [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I (2000) Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12: 2087–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jin W, Lamb JC, Zhang W, Kolano B, Birchler JA, Jiang J (2008) Histone modifications associated with both A and B chromosomes of maize. Chromosome Res 16: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Kornberg RD. (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Menzel MY, Hasenkampf CA, Dougherty BJ, Richmond KL, Campbell LB (1986) Characteristics of duplications and deficiencies from chromosome translocations in Gossypium hirsutum. J Hered 77: 189–201 [Google Scholar]
- Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427 [DOI] [PubMed] [Google Scholar]
- Pikaard CS. (1999) Nucleolar dominance and silencing of transcription. Trends Plant Sci 4: 478–483 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Shi J, Dawe RK (2006) Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Zhang C, Ko DK, Chen ZJ (2015) Genome-wide dosage-dependent and -independent regulation contributes to gene expression and evolutionary novelty in plant polyploids. Mol Biol Evol 32: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS (2014) The polyploidy revolution then…and now: Stebbins revisited. Am J Bot 101: 1057–1078 [DOI] [PubMed] [Google Scholar]
- Song Q, Chen ZJ (2015) Epigenetic and developmental regulation in plant polyploids. Curr Opin Plant Biol 24: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrenoire E, McRonald F, Halsall JA, Page P, Illingworth RS, Taylor AM, Davison V, O’Neill LP, Turner BM (2010) Immunostaining of modified histones defines high-level features of the human metaphase epigenome. Genome Biol 11: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Guan B, Guo W, Zhou B, Hu Y, Zhu Y, Zhang T (2008) Completely distinguishing individual A-genome chromosomes and their karyotyping analysis by multiple bacterial artificial chromosome - fluorescence in situ hybridization. Genetics 178: 1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Guo W, Yang Z, Hu Y, Zhang W, Zhou B, Stelly DM, Chen ZJ, Zhang T (2010) Structure and size variations between 12A and 12D homoeologous chromosomes based on high-resolution cytogenetic map in allotetraploid cotton. Chromosoma 119: 255–266 [DOI] [PubMed] [Google Scholar]
- Wang K, Song X, Han Z, Guo W, Yu JZ, Sun J, Pan J, Kohel RJ, Zhang T (2006) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113: 73–80 [DOI] [PubMed] [Google Scholar]
- Wendel JF. (1989) New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86: 4132–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186 [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33: 531–537 [DOI] [PubMed] [Google Scholar]
- Zhang X. (2008) The epigenetic landscape of plants. Science 320: 489–492 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.