OsDEX1 binds Ca2+ and plays a conserved role in the development of tapetal cells and pollen formation in rice.
Abstract
In flowering plants, successful male reproduction requires the sophisticated interaction between somatic anther wall layers and reproductive cells. Timely degradation of the innermost tissue of the anther wall layer, the tapetal layer, is critical for pollen development. Ca2+ is a well-known stimulus for plant development, but whether it plays a role in affecting male reproduction remains elusive. Here we report a role of Defective in Exine Formation 1 (OsDEX1) in rice (Oryza sativa), a Ca2+ binding protein, in regulating rice tapetal cell degradation and pollen formation. In osdex1 anthers, tapetal cell degeneration is delayed and degradation of the callose wall surrounding the microspores is compromised, leading to aborted pollen formation and complete male sterility. OsDEX1 is expressed in tapetal cells and microspores during early anther development. Recombinant OsDEX1 is able to bind Ca2+ and regulate Ca2+ homeostasis in vitro, and osdex1 exhibited disturbed Ca2+ homeostasis in tapetal cells. Phylogenetic analysis suggested that OsDEX1 may have a conserved function in binding Ca2+ in flowering plants, and genetic complementation of pollen wall defects of an Arabidopsis (Arabidopsis thaliana) dex1 mutant confirmed its evolutionary conservation in pollen development. Collectively, these findings suggest that OsDEX1 plays a fundamental role in the development of tapetal cells and pollen formation, possibly via modulating the Ca2+ homeostasis during pollen development.
Higher plants alternate their life cycle between sporophytic and gametophytic generations that result from two sequential processes: sporogenesis and gametogenesis (Goldberg et al., 1993). Formation of the male gametophyte is a complex process that starts with anther morphogenesis, followed by microspore formation via meiosis and mitosis (Ma, 2005; Gómez et al., 2015; Zhang and Liang, 2016). The somatic cell layers surrounding the microsporocytes include the epidermis, endothecium, middle layer, and tapetum, which are required for normal pollen development. The differentiation of the innermost tapetal cell layer is crucial for pollen formation (Kelliher and Walbot, 2012; Fu et al., 2014; Zhang and Yang, 2014). After the formation of tapetal cells, subsequent tapetal cell and callose degradation, as well as primexine formation, are vital for pollen development (Ma, 2005; Li et al., 2006; Wu and Cheun, 2000; Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Li et al., 2011a; Chang et al., 2012; Ji et al., 2013; Niu et al., 2013; Sun et al., 2013).
During tapetum development, regulated tapetal cell death is of vital importance for primexine formation, sporopollenin synthesis, and exine formation (Shi et al., 2015; Zhang and Liang 2016). Until now, several transcription factors and their associated targets have been reported to play role in tapetal cell death (Sorensen et al., 2003; Jung et al., 2005; Li et al., 2006; Aya et al., 2009; Xu et al., 2010; Li et al., 2011a; Niu et al., 2013; Ji et al., 2013; Fu et al., 2014; Ko et al., 2014). In rice (Oryza sativa), mutations in the basic helix-loop-helix (bHLH) transcription factor TDR Interacting Protein2 (TIP2), also called bHLH142, show compromised inner anther wall layer differentiation and defects in microspore development (Fu et al., 2014; Ko et al., 2014). A mutant of the bHLH transcription factor, Undeveloped Tapetum1(UDT1), displays abnormal development and degeneration of the tapetum and middle layer (Jung et al., 2005). gamyb, which encodes a MYB transcription factor, shows aborted tapetum degradation (Aya et al., 2009). Another bHLH transcription factor, Eternal Tapetum1 (EAT1), also called DTD, as well as one PHD finger protein, Persistent Tapetal Cell1 (PTC1), regulate microspore development via controlling tapetal cell death (Li et al., 2011a; Ji et al., 2013; Niu et al., 2013). TDR and EAT1 directly regulate the expression of proteases, which are regarded as executors for programmed cell death (PCD) in animals (Li et al., 2006; Niu et al., 2013; Woltering, 2010). For instance, EAT1 regulates the two aspartic proteases OsAP25 and OsAP37, which trigger PCD in both yeast and plants (Niu et al., 2013). As a positive tapetal PCD determinant, TDR regulates the expression of OsCP1, a Cys protease that is involved in tapetal degradation (Lee et al., 2004; Li et al., 2006; Niu et al., 2013). In addition, OsCP1 is also regulated by two ATP-dependent RNA helicases, AIP1 and AIP2 (Li et al., 2011b).
Tapetal cell death is also associated with the degradation of callose and the formation of primexine on the surface of the microspore, which is the first step of the pollen cell wall formation (Ariizumi and Toriyama, 2011; Shi et al., 2015). Mutants defective in callose degradation in rice also have defects in exine formation and pollen development (Wan et al., 2011). In Arabidopsis (Arabidopsis thaliana), Defective in Exine Formation1 (DEX1), No Exine Formation1 (NEF1), Ruptured Pollen Grain1 (RPG1), Ruptured Pollen Grain2 (RPG2), and No Primexine and Plasma Membrane Undulation (NPU) are required for primexine formation, and their corresponding mutants are defective in exine formation (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Chang et al., 2012; Sun et al., 2013). After the formation of the primexine, sporopollenin was synthesized in the tapetum and transported to the microspore. Several lipid metabolism-related genes, such as Defective Pollen Wall (DPW), CYP704B2, and CYP703A3 have been reported to be essential for rice exine formation (Shi et al., 2011; Li et al., 2010; Yang et al., 2014b; Shi et al., 2015; Zhang et al., 2016). Additionally, sporopollenin precursor transport-related genes, such as Postmeiotic Deficient Anther1 (OsABCG15/PDA1), OsABCG26, and OsC6 (Zhang et al., 2010; Qin et al., 2013; Zhu et al., 2013; Zhang and Li 2014; Zhao et al., 2015), are also required for pollen exine formation (Supplemental Fig. S1).
Up to now, the progress made toward understanding tapetal cell death is mainly restricted to the level of transcription factors and their related regulatory effects (Sorensen et al., 2003; Li et al., 2006; Aya et al., 2009; Xu et al., 2010; Li et al., 2011a; Niu et al., 2013; Ji et al., 2013; Fu et al., 2014; Ko et al., 2014). Consequently, further approaches are needed to reveal additional details of tapetal cell death regulation. Ca2+ is an essential component regulating a wide range of biological processes including cell division, differentiation, motility, and PCD (Poovaiah and Reddy, 1993; Trewavas and Malhó, 1998; Zielinski, 1998; Reddy, 2001; Lopez-Fernandez et al., 2015). Ca2+ affects cell death, including apoptosis, autophagy, and autolysis (Groover and Jones, 1999; Giorgi et al., 2008) by triggering the increase of cytosolic Ca2+([Ca2+]cyt; McConkey and Orrenius, 1997; Ferrari et al., 2002; Orrenius et al., 2003; Smaili et al., 2003; Giorgi et al., 2008). The endoplasmic reticulum (ER) is considered as an important Ca2+-rich organelle, and ER-localized proteins have been reported to influence Ca2+ homeostasis in cells (Lam et al., 1994; Foyouzi-Youssefi et al., 2000; Kowaltowski et al., 2000; Pinton et al., 2000; Vanden Abeele et al., 2002). In Arabidopsis, BCL2-Associated X protein inhibitor1 acts as an inducer of ER stress-mediated PCD, which may play a role in affecting Ca2+ homeostasis on ER stress-mediated PCD in plants (Watanabe and Lam, 2008). Overexpression of ER-localized Bcl-2 protein promoted Ca2+ leakage from the ER and suppressed Ca2+ reuptake by the ER, causing the consequent increase of [Ca2+]cyt and subsequent apoptosis (Hofer et al., 1998; Foyouzi-Youssefi et al., 2000; Vanden Abeele et al., 2002; Ferrari et al., 2002).
Ca2+ signal is sensed by a series of Ca2+ binding proteins, calmodulin (CaM), calcineurin B-like proteins, and Ca2+-dependent protein kinases, and activates a number of transcriptional regulators by a kinase cascade or by direct interaction of CaM in a Ca2+-dependent manner (Hiraga et al., 1993; Corneliussen et al., 1994; Enslen et al., 1995; Tokumitsu et al., 1995; Shi et al., 1999). Many Ca2+ sensors contain domain E and F (EF)-hand domains for Ca2+ binding (Snedden and Fromm, 1998). The typical EF-hand is a helix-loop-helix structure, in which the residues with +X*+Y*+Z*-Y*-X**-Z are associated with Ca2+ binding (Day et al., 2002; Derbyshire et al., 2007; Rigden et al., 2011). In addition, other motifs such as helix-loop-strand, helix-loop-turn, strand-loop-helix, strand-loop-strand, and several structural contexts without regular secondary structure elements either before or after the DxDxDG-containing loop can provide affinity for binding Ca2+ (Rigden and Galperin, 2004).
In this work, we characterized a Ca2+ binding protein, OsDEX1, which is required for tapetal function and pollen development in rice. OsDEX1 is conserved in flowering plants, and it is able to complement the mutant of its Arabidopsis counterpart. These findings provides the first evidence for an essential role of the Ca2+ homeostasis in plant pollen development.
RESULTS
Phenotypic Analysis of osdex1
To identify genes that are required for rice male reproduction, we isolated the defective in exine formation1 in rice (osdex1) mutant (see below) from a rice mutant library generated in ssp. japonica cv 9522 background by 60Co γ-ray irradiation (Chen et al., 2006), which showed a complete male sterility phenotype. Compared with wild-type plants, osdex1 mutant displayed morphologically normal vegetative and nonreproductive floral organs, but smaller and pale yellow anthers lacking normal mature pollen grain, leading to complete male sterility (Fig. 1). All of the F1 progeny between wild type and osdex1 were fertile, and the segregation rate of F2 generation was approximately 1:3 (sterility/fertility = 24:81), suggesting osdex1 in caused by a single recessive mutation.
Figure 1.
Phenotypic comparison between the wild type and the osdex1 mutant. A, Wild-type plant (left) and the osdex1 mutant plant (right) after heading. B, Part of the wild-type panicle showing the dehisced anther (left) and part of the osdex1 panicle (right) showing a smaller anther at the pollination stage. C, Wild-type (left) and osdex1 (right) flower organs after removal of the palea and lemma. D, Wild-type (left) and osdex1 (right) flowers before anthesis. E, Wild-type stained pollen at stage 13. Bars = 10 cm (A), 2 cm (B), 5 mm (C and D), and 100 μm (E).
OsDEX1 Affects Tapetal Cell Death and Primexine Formation
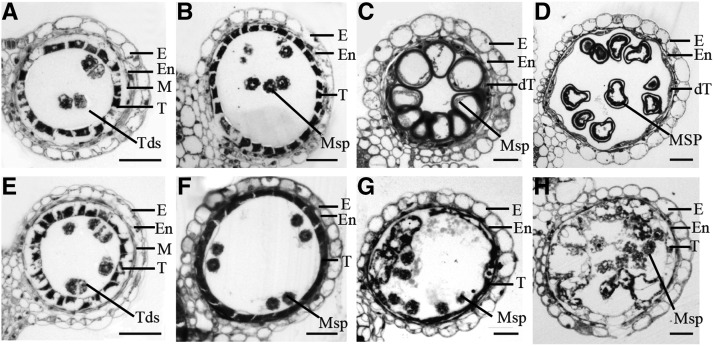
Transverse sectioning was used to investigate the cellular morphological alterations of osdex1 during pollen development, which was delineated based on a previous report (Zhang et al., 2011). No detectable morphological defect was observed in osdex1 anthers until stage 8b when the microspore mother cells of both wild type and osdex1 had undergone the second meiosis to produce ellipsoidal-shaped dyads, and tapetal cells were darkly stained with a vacuolated shape (Fig. 2, A and E). At stage 9, the wild-type microspores had separated from the tetrads and were distributed in anther lobes (Fig. 2B), while osdex1 microspores were not separated (Fig. 2F). At stage 10, the wild-type microspores were covered in a thick layer of exine, displayed a round shape due to vacuolization, and the tapetum were degenerated into a thin layer (Fig. 2C). In contrast, osdex1 microspores were smaller, lacked a clear layer of exine compared with the wild type, and the tapetal cells were not degenerated but appeared expanded in some regions (Fig. 2G). At stage 11, the wild-type tapetal cells were completely degraded and sickle-shape microspores exhibited storage starch accumulation (Fig. 2D). However, osdex1 displayed a persistent tapetal layer, and the microspores showed a similar morphology to previous stages (Fig. 2H). These observations indicate that osdex1 exhibits delayed tapetal degradation and aborted exine formation.
Figure 2.
Bright field microscopy of transverse sections showing anther and microspore development in wild type and osdex1. Locules from the anther section of the wild type (A–D) and osdex1 (E–H) from stage 8 to stage 11. dT, degenerated tapetal layer; E, Epidermis; En, endothecium; M, middle layer; Msp, microspores; T, tapetal layer; Tds, tetrads. Bars = 15 µm. A and E, Stage 8b. B and F, Stage 9. C and G, Stage 10. D and I, Early stage 11.
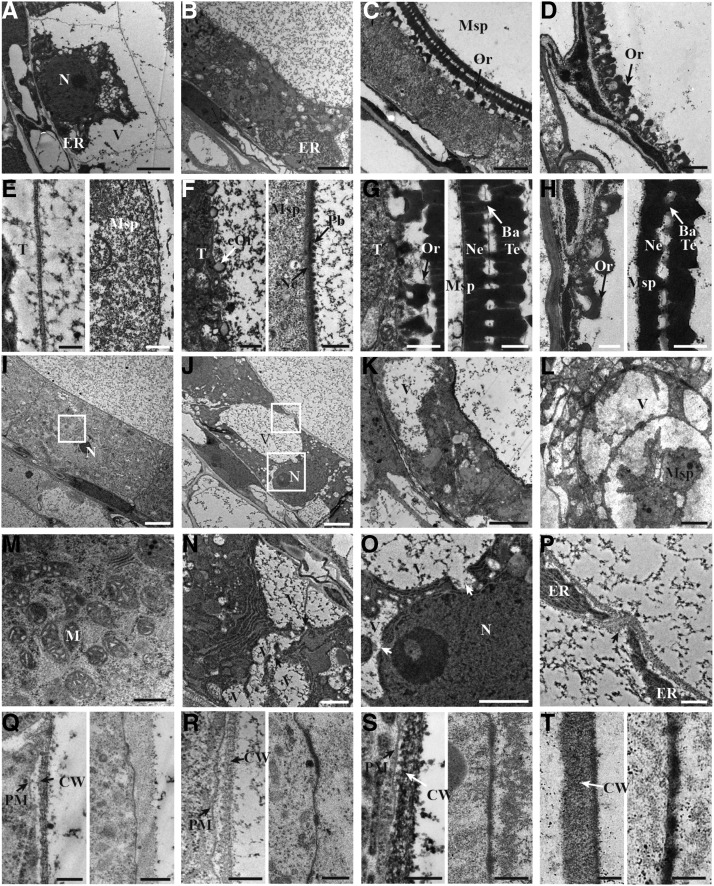
Transmission electron microscopy (TEM) was used to further investigate the defects in osdex1 pollen formation and tapetum degeneration. Consistent with the observations by light microscopy, no visible morphological differences were observed between the wild type and osdex1 (Supplemental Fig. S2) until stage 8a. At stage 8b, wild-type tapetal cells became largely vacuolated with a thin layer of cell wall (Fig. 3, A and E), and the plasma membrane (PM) of wild-type microspore showed regular undulations and primexine deposition (Fig. 3E). In osdex1 tapetal cells, where vacuoles were reabsorbed, their cytoplasm appeared condensed with a large number of mitochondria and they displayed a thicker cell wall (Fig. 3, I, M, and Q). PM undulation was not observed in mutant microspores and primexine deposition did not occur (Fig. 3Q). At stage 9, wild-type tapetal cells displayed a waved shape accompanied by an enrichment of organelles, such as mitochondria and ER, and small bubbles that likely represented the early stages of orbicule development (Fig. 3, B and F). Correspondingly, a thin layer of exine (including probacula and nexine) was present on the wild-type microspore surface (Fig. 3F). Consistent with this observation, sporopollenin precursor synthesis and transport-related genes such as DPW, CYP703A3, CYP704B2, OsC6, and PDA were highly expressed in wild-type anthers (Supplemental Fig. S3; Shi et al., 2011; Yang et al., 2014b; Li et al., 2010; Zhu et al., 2013; Zhang et al., 2010). By contrast, in osdex1, at the same stage, tapetal cells were flat with a thicker cell wall and contained a larger number of vacuoles with variable size compared to wild type (Fig. 3, J and R). Notably, the vacuoles in the mutant tapetal cells were of irregular shape and were attached to each other exhibiting membrane ablation (Fig. 3N). Furthermore, the structures of nucleus and ER seemed disrupted in the mutant tapetal cells (Fig. 3, O and P). No intact exine structure was formed on the osdex1 microspore surface, which only showed fragmented primexine deposition (Fig. 3R). Furthermore, no obvious expression of DPW, CYP703A3, CYP704B2, OsC6, and PDA was detected in the mutant, suggesting the defective synthesis and transport of sporopollenin precursors (Supplemental Fig. S3). At stage 10, wild-type tapetal cells were thin and contained a large number of mature orbicules on the inner surface (Fig. 3, C and G). As a result, wild-type microspores possessed a thick layer of exine on the surface (Fig. 3G). By contrast, osdex1 tapetal cells were expended in several regions because of large vacuoles (Fig. 3K). The persistent tapetal cells were covered by a thick cell wall without any orbicules (Fig. 3S). Moreover, mutant microspores were covered by a thin layer of darkly stained material, lacking the normal structure of exine (Fig. 3S). At stage 11, the tapetal layer in wild-type anthers was almost completely degenerated and orbicules were present around microspores forming a thick exine (Fig. 3, D and H), while osdex1 exhibited expanded tapetal cells with large vacuoles, and extremely thick cell walls (Fig. 3, L and T) without sculptured exine on the microspores (Fig. 3T). These results suggest that OsDEX1 affects the normal tapetal cell death, synthesis of sporopollenin precursors, and the formation of pollen wall ranging from the primexine to multiple-layer exine during male development.
Figure 3.
TEM images of the anthers from the wild type and osdex1. A to D, TEM observation showing tapetal cells of wild-type anthers at stage 8b (A), stage 9 (B), stage 10 (C), and stage11 (D). E to H, Wild-type tapetal cell wall (left) and microspore cell wall (right) at stage 8b (E), stage 9 (F), stage 10 (G), and stage11 (H). I to L, TEM observation showing tapetal cells of osdex1 anthers at stage 8b (I), stage 9 (J), stage 10 (K), and stage 11 (L). M, Higher magnification of the highlighted region in (I) showing details in tapetal cells. N, Vacuoles fusion in osdex1 tapetal cells at stage 9. Black arrows show the attachment of vacuoles. O and P, Higher magnification of the highlighted region in (J) showing details in tapetal cells. White arrows show the breakage of vacuoles. Black arrow shows the breakage of ER. Q to T, osdex1 tapetal cell wall (left) and microspore cell wall (right) at stage 8b (Q), stage 9 (R), stage 10 (S), and stage 11 (T). Ba, Bacula; CW, cell wall; eOr, early stage of orbicules; Pb, probacula; M, mitochondria; N, nucleus; Ne, nexine; Or, orbicules; T, tapetal cells; Te, tectum; V, vacuoles. Bars = 2 μm (A–C and I–K), 1 μm (D and N), 10 μm (L), and 0.5 μm (E–H, M, O–P, and Q–T).
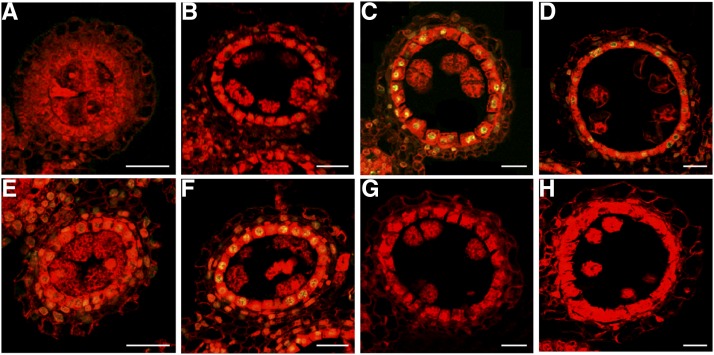
The persistence of tapetal cells in osdex1 mutants was investigated using the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in which the fluorescein-12-dUTP-labeled DNA is catalytically incorporated into fragmented DNA, and can be visualized by confocal laser scanning microscope (Li et al., 2006). The wild-type tapetum displayed DNA fragmentation from stage 8b until stage 9, and there was no DNA fragmentation at stage 7 and stage 8a. In contrast, the DNA fragmentation in osdex1 tapetal cells could be observed starting from stage 7 until stage 8a, and declined at stage 8b and stage 9 (Fig. 4). Therefore, DNA fragmentation appears to be activated early in osdex1 tapetal cells and disrupted at the stages when wild-type tapetal cells show DNA fragmentation. To further assess tapetal cell death, the expression pattern of tapetal cell death-related genes was determined. OsAP25, OsAP37 and OsCP1 were not induced in osdex1 anthers compared to wild-type anthers undergoing cell death (Supplemental Fig. S4), suggesting that the normal protein degradation pathway was disrupted in the osdex1 mutant during tapetum development.
Figure 4.
DNA fragmentation is initiated earlier and subsequently blocked in osdex1 mutant. A to H, DNA fragment signal at stage 7, stage 8a, stage 8b, and stage 9 in wild type (A–D) and osdex1 mutant (E–H). The red fluorescence shows the propidium iodide staining of anther cells using confocal laser scanning microscope; the yellow fluorescence shows the TUNEL-positive nuclei staining in confocal laser scanning microscope overlays of fluorescein staining and propidium iodide staining. Bars = 15 μm.
Our observations using TUNEL and TEM suggest abnormal tapetal cell degeneration in osdex1. Consistent with the earlier induction of DNA fragmentation in osdex1 tapetal cells, in osdex1 we observed up-regulation of five endonuclease-encoding genes (LOC_07g45100, LOC_01g03740, LOC_04g58850, LOC_02g50040, and LOC_08g29700) that are down-regulated in the tapetal cell death-deficient mutants gamyb and tip2 at stage 8 (Aya et al., 2009; Fu et al., 2014). Similarly at stage 7, the genes were either up-regulated or showed the opposite expression pattern compared to gamyb and tip2. These findings are in agreement with the opposite cell death phenotype. At stage 9 when the DNA fragmentation signal was declined, the expression of these genes was down-regulated (Supplemental Fig. S4).
OsDEX1 Affects Callose Degradation
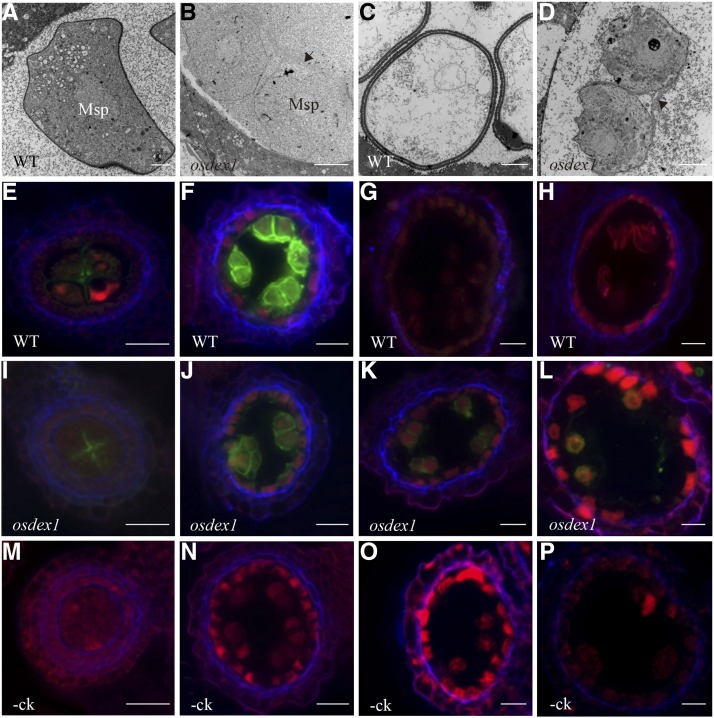
The TEM analysis indicated that osdex1 microspores were covered in an electro-dense matrix at later stages (Fig. 5, A to D), likely indicative of abnormal callose degradation. To further characterize the role of OsDEX1 in callose dynamics during microspore development, aniline blue staining and immunolabling with a callose antibody were performed (Figs. 5, E to L; S5, E to L). At stage 7, both wild-type and osdex1 tetrads were labeled with aniline blue, indicating the relatively normal synthesis of callose in osdex1 (Supplemental Fig. S5, E and I). A similar pattern was observed using callose immunolabeling, although in this case staining was weaker around osdex1 tetrads than in wild type at stage 8b (Fig. 5, F and J). This is supported by the TEM observations showing a looser electron-dense matrix around osdex1 tetrads (Supplemental Fig. S5, A and B). Also at stage 8b, both aniline blue and callose antibody stained cell-wall material in the center and the border region surrounding the wild-type tetrads (Figs. 5, F and J; S5, F and J). Remarkably, in osdex1, aniline blue staining was restricted mainly to the inner tetrad walls, suggesting some changes in cell-wall composition of the outer walls, possibly because of precocious yet incomplete callose degradation (Supplemental Fig. S5F). Specific callose labeling was detected by aniline blue and callose antibody directly adjoining osdex1 microspores, and weakly in the diffuse halos surrounding them, until stage 10 (Figs. 5, K and L; S5, K and L) when staining was absent around the wild-type microspores. These results suggest that OsDEX1 affects callose degradation during microspore development.
Figure 5.
Callose degradation is retarded in osdex1. A to D, TEM of extracellular materials from the wild type and osdex1 at stage 9 (A and B), and stage 10 (C and D). Black arrows show the site of callose deposition. E to L, Immunolabeling of wild-type (E–H) and osdex1 (I–L) anther sections from stage 7 to stage 10 observed by epifluorescence microscopy. M to P, Negative controls of immunolabeling. In (E) to (P), the green channel shows immunostaining with callose antibody; blue counterstaining shows 1,4- and 1,3;1,4-glucan polymers stained with 0.01% calcofluor white; and red staining shows background autofluorescence. Bars = 2 μm (A), 5 μm (B–D), and 15 μm (E–P).
Cloning and Expression Analysis of OsDEX1
To identify the gene responsible for the osdex1 phenotype, we employed a map-based cloning approach. OsDEX1 was mapped to chromosome 3 between the markers of Os315 and Os315-2, which incorporated a 26 kb DNA fragment and seven putative genes (Supplemental Fig. S6A). After sequencing anther-expressed candidate genes within this region, a 1 bp deletion was identified in the coding region of a gene corresponding to LOC_Os03g61050 (http://www.gramene.org/, also annotated as Os03g0825700 by http://rapdb.dna.affrc.go.jp/), which deletion resulted in a frame shift and subsequently an earlier termination of the translation of this gene (Supplemental Fig. S6B). Moreover, three additional alleles of OsDEX1 were identified, and we re-named osdex1 as osdex1-1, and the additional three alleles were designated as osdex1-2, osdex1-3, and osdex1-4 (Supplemental Fig. S6B). osdex1-2 has a one-nucleotide G to A substitution at an intron boundary (position 4296), causing the alternative splicing of OsDEX1. osdex1-3 had a 374 bp deletion in the 10th exon leading to a 286 bp deletion of the mRNA. osdex1-4 had a 4 bp deletion in the sixth exon leading to an earlier termination of OsDEX1. All of the mutants exhibited similar male sterile phenotype and expanded tapetal cells (Supplemental Fig S7). Allelic tests confirmed that the four mutants were indeed alleles of a single locus. The OsDEX1 transcript is 3241 bp in length and contains a 2556 bp coding sequence with 12 introns, a 180 bp 5′ untranslated region, and a 505 bp 3′ untranslated region (Supplemental Fig. S6B). The predicted OsDEX1 protein encompasses 851 amino acids and harbors one putative N-terminal signal peptide (amino acids 1 to 23), one integrin α-N-terminal domain (amino acids 69 to 582) containing two phenylalanyl-glycyl-glycyl-alanyl-prolyl (FG-GAP) domains (amino acids 460 to 486 and 554 to 579; PF01839; http://pfam.xfam.org), and one transmembrane domain (amino acids 816 to 826; Supplemental Fig. S6C). The integrin α-N-terminal domain is predicted to mediate cell adhesion, and the FG-GAP domain has been shown to be important for ligand binding (Loftus et al., 1994).
To gain insights into the function of OsDEX1, quantitative RT-PCR (qRT-PCR) analysis was used to further investigate the spatio-temporal expression pattern of OsDEX1. In the wild type, OsDEX1 expression was detectable in roots, shoots, and leaves, with a preferable expression in the anther, starting from stage 6, reaching an expression maximum at stage 8 and stage 9. OsDEX1 expression showed a dramatic reduction in osdex1-1, particularly in anthers (Supplemental Fig. S6D). Although the OsDEX1 expression was high in wild-type leaves and much reduced in the mutant, no visible phenotypic change was observed in mutant leaves under normal growth conditions, suggesting no obvious role of OsDEX1 or redundant gene function during leaf development. Furthermore, in situ hybridization also showed that OsDEX1 was expressed in tapetal cells and microspores from stage 8a to stage 9, and low level at stage 10 (Supplemental Fig. S6, E to L).
OsDEX1 Has a Conserved Function in Pollen Development
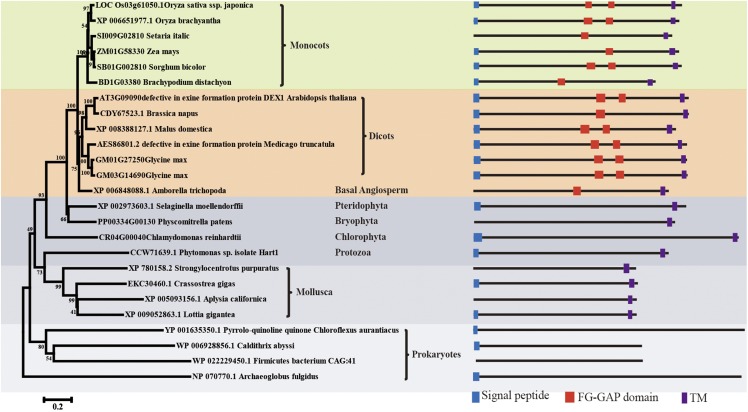
To elucidate the evolutionary conservation and distribution of OsDEX1, the full-length OsDEX1 protein was used as the query to search for its closest relatives in public databases, including the National Center for Biotechnology Information, The Arabidopsis Information Resource, Phytozome (http://www.phytozome.net/), and PLAZA (http://bioinformatics.psb.ugent.be/plaza/), which yielded 25 sequences from 24 different species, and all of which were then included in the phylogenetic analysis (Fig. 6). Among the 24 species, besides rice, there are monocots such as Oryza brachyantha, dicots such as Arabidopsis, basal angiosperm such as Amborella trichopoda, Pteridophyta such as Selaginella moellendorffii, Bryophyta such as Physcomitrella patens, Chlorophyta such as Chlamydomonas reinhardtii, Protozoa such as Phytomonas sp. isolate Hart, Mollusca such as Strongylocentrotus purpuratus, and Prokaryotes such as Firmicutes bacterium. These data suggested that OsDEX1 belongs to an ancient protein subfamily, present in both prokaryotes and eukaryotes, which was designated as DEX1 (PTHR21419:SF23) by the panther database (http://www.pantherdb.org/). Notably, proteins in this family only exist in lower animals, while they are present in both higher plants and lower plants. OsDEX1 clustered together with other monocot DEX1 sequences in the same clade, while proteins from dicots, including the Arabidopsis homolog DEX1, clustered in a separate clade. In addition, the homologous eukaryotic sequences all contained a transmembrane domain, suggesting their similar membrane localization in the cell. The FG-GAP domain was only detectable in the sequences of flowering plants, which has been shown to be important for ligand binding (Loftus et al., 1994). These results suggest that OsDEX1 might have an evolutionarily conserved conserved ancient function in male gametophyte development that, however, may have obtained the ability ligand binding during the formation of angiosperms.
Figure 6.
Phylogenetic analysis of OsDEX1 and its related proteins. Neighboring-joint analysis was performed using MEGA 6.1 (see “Materials and Methods”) based on the alignment given in Supplemental Data Set S1 online of OsDEX1 with the most similar OsDEX1 sequences from the species shown. The species were classified by evolutionary relationship.
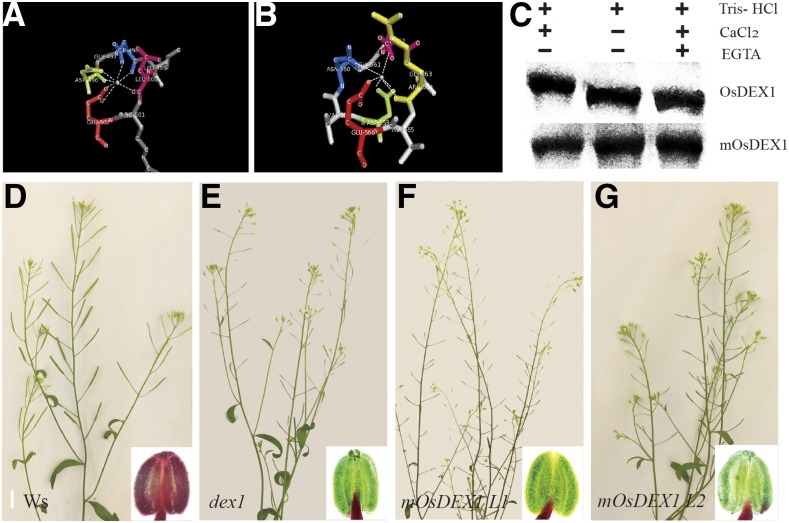
Based on the phylogenetic analysis, OsDEX1 was identified as a putative ortholog of Arabidopsis DEX1. dex1 was also reported as a male sterile line due to its delayed primexine formation, and aborted pollen wall development, but lacked characterization of its tapetal and biochemical function (Paxson-Sowders et al., 2001). The amino acid sequence of OsDEX1 was overall 66.5% identical to that of DEX1 (Supplemental Fig. S8), suggesting that they may have similar function. To test this hypothesis, OsDEX1 cDNA driven by DEX1 promoter (1.7 Kb) was introduced into dex1 heterozygous plants. More than 100 T1 transformants were identified, and among the dex homozygous lines containing the transgene, 42% displayed full fertility and 25% showed partial fertility. Notably, Alexander staining showed that in the fully fertile lines, all the pollen grains in the anther were stained red, indicating that these pollen grains are viable (Fig. 7C). In the partially fertile lines, some anthers showed all pollen grains stained in red, while others showed a reduced amount of red-stained pollen grains (Fig. 7D). Altogether, these results demonstrate that OsDEX1 fulfills a conserved function in pollen development.
Figure 7.
A to D, Transgenic lines containing Pro:DEX1:OsDEX1 in the Arabidopsis dex1 mutant display rescued male fertility. Insets show the pollen tested by Alexander staining using bright field microscopy. Ws, wild-type plant of Wassileskija ecotype. Bar = 10 mm.
OsDEX1 Is a Calcium Binding Protein
The phylogenetic analysis suggested the importance of the FG-GAP domain in flowering plants, and FG-GAP domain has also been also reported to mediate calcium binding (Tuckwell et al., 1992; Loftus et al., 1994; Springer, 1997). To investigate whether the conserved FG-GAP domain of OsDEX1 can indeed bind to Ca2+, a three-dimensional (3D) structure prediction of OsDEX1 was conducted using SWISSMODEL (http://www.swissmodel.expasy.org/), which revealed that the 3D structure of OsDEX1 was rich in β-sheet linked by loops (Supplemental Fig. S9), implying its potential ability to bind Ca2+. Structural analysis showed the oxygen atoms from the side chains of the first, third, and fifth residues in the EF-hand motif that may form a pocket suitable for calcium atom binding (Fig. 8, A and B).
Figure 8.
Recombinant OsDEX1 has Ca2+ binding activity. A, 3D structure of EF-hand motif of YesW. B, 3D structure of EF-hand motif of OsDEX1. C, In vitro Ca2+ binding assay shows that OsDEX1 has Ca2+ binding activity. D to G, Failure in complementation by OsDEX1 with the mutated Ca2+ binding sites in dex1. Insets show pollen tested by Alexander staining using bright field microscopy. Bar = 10 mm.
An in vitro Ca2+ binding assay (Gregersen et al., 1990; Ling and Zielinski, 1993; Libich and Harauz, 2002) showed that the truncated protein of OsDEX1 (amino acids 336 to 634), containing 3 EF-hands motifs, was shifted in an acrylamide-SDS gel after incubation with 1 mm CaCl2. However, when Ca2+ was chelated by adding EGTA, no shift in fragment migration was detected. In addition, when an isoform of OsDEX1 the first, third, fourth, fifth, and sixth residues of the conserved calcium binding domain were muted was incubated with CaCl2, no shift was observed (Fig. 8C). Importantly, when the mutated OsDEX1 cDNA driven by DEX1 promoter (1.7 Kb) was introduced into the Arabidopsis dex1 mutant, the transgenic plants homozygous for dex1 failed to produce viable pollen grains (Fig. 8, D to G). These results identify OsDEX1 as a Ca2+ binding protein and provide evidence for a pivotal role of Ca2+ binding in the function of OsDEX1.
OsDEX1 Is Required for Adequate Ca2+ Homeostasis in Tapetum
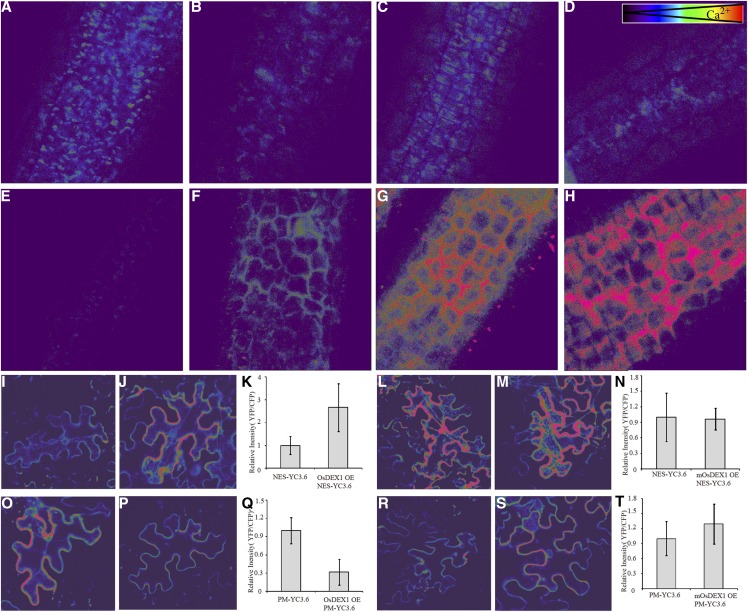
The abnormal tapetal cell death phenotype and the alteration of the Ca2+ binding ability of a truncated OsDEX1 protein suggested that OsDEX1 might regulate tapetal cell death by affecting Ca2+ distribution in the tapetal cells, as suggested by previous reports (Wyllie, 1980; Cohen and Duke, 1984). To test this, yellow cameleons 3.6 (YC3.6) expressed under the control of the constitutive UBI10 promoter, a powerful tool monitoring the spatio-temporal dynamics of Ca2+ fluxes in different tissues of Arabidopsis and rice (Krebs et al., 2012; Nehera et al., 2015), was used to monitor Ca2+ homeostasis in rice tapetum. There are three detectable somatic cell layers in the rice anther at stage 9, i.e. the epidermis, endothecium, and tapetum. In Figure 9, the anther images show the morphology of tapetal cells, which is consistent with a previous report (Zhao et al., 2015). PM-YC3.6, which is a plasma membrane targeted version of YC3.6, is able to distinguish the faint Ca2+ signal at the cytosolic side of the plasma membrane from the strong autofluorescence of anthers. During anther development from stage 8 to stage 10, only a faint Ca2+ signal was observed in the wild-type anthers. In sharp contrast, in the osdex1 mutant, the plasma membrane-localized Ca2+ reporter indicated a dramatic increase of intracellular Ca2+ concentration. in the tapetal cells specifically initiating from stage 9, that was even further enhanced at stage 10 (Fig. 9, A to H). These results identify rapid and strong elevations of intracellular Ca2+ concentration that occur stage specific only in osdex1 and the coincide with the establishment of tapetal phenotypes in this mutant. These findings support the conclusion that the increase in [Ca2+] specifically observed in osdex1 represents an important facet of the cell death ongoing in these cells and there by attribute a crucial function for maintaining appropriate cellular [Ca2+] homeostasis to OsDEX1.
Figure 9.
The Ca2+ concentration calculated by emission ratios between YFP and CFP intensities using confocal laser scanning microscope in rice anthers and tobacco epidermal cells. A to H, Ca2+ concentration on plasma membrane in tapetal cells of wild type (A–D) and osdex1 (E and F) from stage 8 to stage 11. I to T, Comparison of cytosolic (I to N) and plasma membrane (O–T) Ca2+ concentration in tobacco epidermal cells overexpressed with OsDEX1 (J and P) or mOsDEX1 (M and S). I, L, O, and R, Control of YC3.6 overexpression. K, N, Q, and T, Statistical analysis of ratios between YFP and CFP intensities in the cells (K and N) and on the plasma membrane (Q and T).
In another assay for cytosolic [Ca2+] homeostasis, cytoplasmic NES-YC3.6 or plasma membrane targeted PM-YC3.6 were expressed alone or coexpressed together with either OsDEX1 protein or non-Ca2+ binding mOsDEX1 protein transiently in tobacco epidermal cells. Consistent with the results in rice tapetal cells, [Ca2+]PM was lower when PM-YC3.6 was coexpressed with OsDEX1 compared to cell that did only express PM-YC3.6 while [Ca2+]cyt was elevated when NES-YC3.6 was coexpressed with OsDEX1 compared to cells that only expressed NES-YC3.6. Importantly, the Ca2+ concentration was not affected when PM-YC3.6 or NES-YC3.6 were coexpressed with mOsDEX1, which lacks Ca2+ binding activity (Fig. 9). These results reveal that the expression of OsDEX1 modulates the Ca2+ homeostasis in the cells, which is considered as a key mechanism for cell death regulation (Giorgi et al., 2008).
To determine the subcellular localization of OsDEX1 at which this protein impacts on cellular [Ca2+] dynamics, the full-length OsDEX1 coding region was fused with the GFP at its C terminus, and transiently expressed in tobacco epidermal cells. The OsDEX1-GFP signal was detected on the ER (Supplemental Fig. S10), which is in agreement with previous reports suggesting that the ER is a main storage site for Ca2+ in the cell (Pozzan et al., 1994). Considering the results described above, we therefore hypothesize that OsDEX1 may regulate tapetal cell death via modulating Ca2+ homeostasis between the ER and other cellular compartments.
DISCUSSION
Rice is one of the most important foods for the global population. Rice yield improvement is largely dependent on hybrid breeding that employs male sterile lines. Although much progress has been made toward understanding the molecular mechanisms underlying plant male development, the role of Ca2+ during anther development has remained elusive. In this study, we have demonstrated that an FG-GAP domain-containing protein, OsDEX1, is required for pollen wall development and tapetum function in a way has not been elucidated in previous descriptions of the Arabidopsis dex1 mutant (Paxson-Sowders et al., 2001). The evolutionary importance of the OsDEX1-associated pathway is supported by the fertility restoration of dex1 by OsDEX1.
OsDEX1 Affects Tapetal Function and Anther Development
The primexine functions as a template for sporopollenin deposition, which is critical for pollen development. Several genes have been identified that influence primexine formation in Arabidopsis (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Chang et al., 2012; Sun et al., 2013), while no gene has been identified in rice. In this study, we identified the ortholog of Arabidopsis DEX1 in rice, which shares a similar function with Arabidopsis DEX1 in regulating primexine formation and male fertility (Paxson-Sowders et al., 2001). Similar to dex1, osdex1 showed defects in plasma membrane undulation as well as primexine formation, resulting the failure in exine formation (Fig. 3). Therefore, DEX1 appears to play an evolutionarily conserved role in male reproduction in model dicot Arabidopsis and monocot rice plants.
Primexine is a structure located at the periphery of the haploid microspores; however, it is believed that primexine formation is controlled by sporophytic cells. Evidence for this comes from reported primexine defective mutants, such as dex1, nef, hkm, and rpg1, whose heterozygous mutants showed normal primexine formation (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Chang et al., 2012; Sun et al., 2013). Tapetal cell death is of vital importance for exine formation (Sorensen et al.,2003; Li et al., 2006; Zhang et al., 2008; Aya et al., 2009; Xu et al., 2010; Niu et al., 2013; Ji et al.,2013). Although its role in primexine formation has not been described previously, we agree that tapetal cell death is also required for primexine formation. In a previous report of Arabidopsis DEX1 (Paxson-Sowers et al., 2001), mutant defects in microspore release from the tetrad, tapetal function as well as a biochemical characterization of DEX1 have not been reported. In this study, we show that the newly produced microspores within the osdex1 tetrad are covered by an electron-dense matrix, and that microspore release into the lobe is inhibited until at least stage 9. Immunolabeling and aniline blue assays indicate that degradation of microspore callose is abnormal in osdex1 (Figs. 5 and S5), which has not been reported in Arabidopsis mutants such as dex1, npg1, hkm, rpg1 rpg2, npu, and nef (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Ariizumi et al., 2005; Guan et al., 2008; Chang et al., 2012, Sun et al., 2013). In addition, diffuse halos that surround osdex1 microspores at stage 10 only contain very low levels of callose, suggesting that some other polymers are aberrantly deposited onto the osdex1 microspores. Taken together, the weaker staining of callose by callose antibody and aniline blue at stage 8b, the less compact matrix detected by TEM observation around osdex1 tetrads, and the persistence of callose until stages 9 and 10 suggest that callose degradation may initiate early in osdex1, but then fails to complete due to defects in tapetal development.
As a nutritive tissue, tapetal function such as proper cell death and the biosynthesis of sporollenin precursors is essential for pollen wall formation. Either premature or delayed tapetal degradation frequently causes reduced male sterility (Balk and Leaver, 2001; Ku et al., 2003; Lee et al., 2004; Jung et al., 2005; Luo et al., 2006; Kawanabe et al., 2006; Li et al., 2006; Oshino et al., 2007; Li et al., 2011; Aya et al.,2009; Tan et al., 2012; Niu et al.,2013; Ji et al., 2013; Fu et al., 2014; Ko et al., 2014; Yang et al., 2014; Zhang et al., 2014). We observed a persistent and partially expanded tapetal layer in osdex1 until stage 11. However, compared to wild-type tapetal cells that are rich in organelles at stage 9, osdex1 tapetal cells form abnormal vacuoles. The vacuoles expand and encapsulate organelles, leading to degradation of the inclusions in the tapetal cells. These observations suggest that expanding vacuoles may contribute to an alternative form of tapetal cell death in osdex1, distinct from normal tapetal cell death (Li et al., 2006; Niu et al., 2013). Consistent with this, the tapetal cell death executors OsAP25, OsAP37, and OsCP1 fail to be induced in osdex1 (Supplemental Fig. S4), suggesting that the normal protein degradation pathway involved in the tapetal cell death is not activated in osdex1. Furthermore, compared with the cell death-deficient mutant tdr whose tapetum expansion is uniform (Li et al., 2006), the expansion of different tapetal cells in osdex1 varies (Fig. 3), suggesting a different molecular mechanism of OsDEX1 on regulating tapetal cell death compared with TDR.
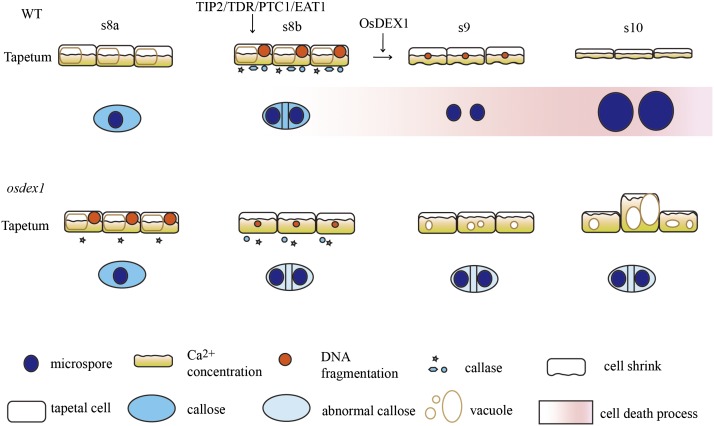
Tapetal cell death is characterized by changes such as DNA fragmentation, protease activation, and cell shrinkage (Li et al., 2006; Aya et al., 2009; Xu et al., 2010; Phan et al., 2011; Li et al., 2011a; Niu et al., 2013; Fu et al., 2014; Fig. 10). Notably, DNA fragmentation is observed from stage 8b, while tapetal cell shrinkage and protease induction are observed from stage 9 in the wild-type anthers. The link between DNA fragmentation and cell death remains to be elucidated. DNA fragmentation is observed in the osdex1 mutant (Fig. 4), but no cell shrinkage or protease induction could be observed, suggesting that OsDEX1 may act as a component required for the tapetal cell death signal transduction (Fig. 10). The early appearance of DNA fragmentation in osdex1 could be explained by the earlier induction of endonuclease genes (Supplemental Fig. S4). How OsDEX1 regulates the expression of these genes requires further investigation.
Figure 10.
Model for OsDEX1 in anther development. Transcription factors such as TIP2, TDR, PTC1, and EAT1 function as master valves to switch cell death signaling on or off; OsDEX1 buffers the Ca2+ concentration in the cells to function as a component of cell death signaling.
OsDEX1 Is a Ca2+ Binding Protein
Ca2+ concentration in the cytoplasm, organelles, and cell wall is maintained within a certain range (Knight, 2000; White, 2000). Ca2+ binding proteins are required for the decoding of transient [Ca2+]cyt (Enslen et al., 1995; Snedden and Fromm, 1998; Day et al., 2002; Kudla et al., 2010; Steinhorst and Kudla, 2013), and usually contain EF-hand motifs that are rich in negative amino acid for the coordination with Ca2+. EF-hand usually form a helix-loop-helix structure (Day et al., 2002; Derbyshire et al., 2007; Rigden et al., 2011). However, there are some β-propeller structure proteins shown to possess Ca2+ binding activity (Cioci et al., 2006). It has been reported that an eight-bladed β-propeller structure protein, RG lyase YesW from saprophytic Bacillus subtilis, had the ability for Ca2+ binding, despite lacking the canonical EF-hand motif (Ochiai et al., 2007). 3D-structure predictions show that OsDEX1 harbors a β-sheet-rich region linked by loops of negatively amino acids that form a pocket for Ca atom, similar to YesW (Supplemental Figs. S9 and 8). The in vitro Ca2+ binding assay indicates that truncated OsDEX1 has the Ca2+ binding activity, and the genetic complementation using the mutated OsDEX1 suggests that the Ca2+ binding activity is required for pollen development in vivo (Fig. 8). Furthermore, OsDEX1 is an ER-localized protein (Supplemental Fig. S10), and osdex1 loss-of-function mutant accumulated Ca2+ on the plasma membrane, while overexpressing OsDEX1 prevented the Ca2+ accumulation on the plasma membrane (Fig. 9). These results suggest a role for OsDEX1 in affecting the level of Ca2+ on the plasma membrane (Fig. 10).
OsDEX1 Has a Conserved Function during Plant Male Reproduction
The pollen wall is essential for pollen grains resistant to various biotic and abiotic stresses in flowering plants. In this study, we identified and analyzed 25 DEX1-like proteins from flowering plants, lower plants, lower animals, fungi, and bacteria. Their phylogenetic relationship suggests that these genes probably share a common ancestor and have a conserved function. Notably, the proteins in flowering plants share one or two FG-GAP domains that are required for the Ca2+ binding (Fig. 6). The protein structure similarity in flowering plants highlights the conserved and essential role of OsDEX1 and its homologs in pollen wall formation in flowering plants. To our knowledge, OsDEX1 is the first member of the DEX1 family that has been identified as having Ca2+ binding activity.
In summary, we have demonstrated that the FG-GAP domain protein OsDEX1 is able to bind Ca2+ to modulate cellular Ca2+ homeostasis, and regulates tapetal degradation and pollen wall formation. A conserved role for DEX1-like proteins was established by the genetic complementation of Arabidopsis dex1 by OsDEX1. This finding provides important insights into the role of Ca2+in male reproduction in plants.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Molecular Cloning of OsDEX1
Rice (Oryza sativa ssp. japonica, 9522) plants were grown in the paddy field of Shanghai JiaoTong University. Male sterile plants in the F2 progenies generated by a cross between wild-type GuangLuAi species (indica) and the osdex1 mutant (japonica) were selected for mapping. Twenty-four pairs of InDel molecular markers were designed based on the polymorphism between japonica and indica for mapping. Further fine-mapping of OsDEX1was performed using the previously published method from Li et al. (2006).
Characterization of Mutant Plant Phenotypes
Anthers from different developmental stages, as defined in Zhang et al. (2011), were collected based on the comparison and analysis of wild-type and osdex1 plants on glume length and morphology. Observation of anther development by semithin section analysis, TEM, and callose staining were performed according to previous studies (Fu et al., 2014; Aditya et al., 2015). For callose immunolabeling, sections were first deparaffinated using xylene before labeling with anticallose antibody (Meikle et al., 1991). Images were captured on a Zeiss A1 AxioImager using a black and white camera and ZEN2012 software. Identical exposure times were used to enable comparisons between stages and samples. Green immunostaining (Alexafluor-488) was viewed using Zeiss Filter Set 38; blue counterstaining (0.01% calcofluor white) was viewed using Zeiss filter set 49, and red staining (autofluorescence) was viewed using Zeiss filter set 43.
TUNEL Assay
Samples from wild-type and osdex1 anthers at different developmental stages were collected. The TUNEL assay was performed as reported in Fu et al. (2014). The samples were analyzed under a fluorescence confocal scanner microscope (Leica SP5II system). Images were recorded using a HCX PL APO CS 20*0.7 DRY objective. The imaging parameters were as follows: image dimension (1024*1024); pinhole (2.19 airy unit); scanning speed (100 Hz); line average (3). The fluorescein signal was excited using the 488 nm laser line of the Argon laser (30%). The image parameters for this channel were as follows: 488 nm 48%; smart gain (817.0); offset (−2.5%). The propidium iodide signal was excited using the 543 nm laser line of the HeNe 543. The image parameters for this channel were as follows: 543 nm 33%; smart gain (654.0); offset (−2.5%). The overlays of fluorescein signal and propidium iodide signal were shown as the TUNEL-positive signal. All pictures were taken in the same setting.
Cloning of OsDEX1 and Plant Transformation
A 2.5 kb cDNA sequence of OsDEX1 was amplified from the cDNA reverse transcripted by the RNA extracted from anthers of 9522. A 1.7 kb upstream sequence of Arabidopsis DEX1 was amplified from the genomic DNA of Col-0. The cDNA of mutated EF-hand motif 1 was amplified by the primers mEF1 F1 and mEF1 R1, and mEF1 F2 and mEF1 R2. The PCR products of the amplification were taken as the template for a second PCR, with the primers mEF1 F1 and mEF1 R2. The cDNA of mutated EF-hand motif 2 was amplified by the primers mEF2 F1 and mEF2 R1, and mEF2 F2 and mEF2 R2. The PCR products of the amplification were taken as the template for a second PCR, with the primers mEF2 F1 and mEF2 R2. The two fragments were ligated together by NcoI and XbaI, which existed in the primer and the pBlueScript SK, and then ligated with the 5′ end of the cDNA to a full-length cDNA. Taking the full-length wild-type and mutated full-length cDNA as the templates, the amplified fragments were cloned into the binary vector pCAMBIA1301. Plasmids were then transferred into Agrobacterium tumefaciens GV3101 and Arabidopsis heterozygous dex1 plants. The transformants were screened for the presence of transgene on hygromycin medium. Over 100 positive transgenic plants in each transformation were obtained and genotyped for the homozygous dex1 mutant background (primers used are listed in Supplemental Table S1). The PM-YC3.6 (Krebs et al., 2012) were transferred into A. tumefaciens EHA105. Calli induced from young panicles of the wild type 9522 and osdex1 mutant were used for transformation (Li et al., 2006). For transgenic plants, at least three independent lines were obtained for each construct.
qRT-PCR and In Situ Hybridization
Total RNA from corresponding tissues was isolated using TRI reagent from rice tissues. Ninety milligrams of RNA was used to synthesis cDNA in each sample using the Primescript RT reagent kit with genomic DNA eraser (Takara). qRT-PCR was performed with the lightCycler system (Roche) using SYBR Premix Ex Taq GC (Takara) according to the manufacturer’s instructions. Amplification was conducted following this protocol: 95°C for 2 min; 40 cycles of 95°C for 5 s; and 55°C for 30 s; 72°C for 30 s. ACTIN (Supplemental Table S1) was used as an internal control, and a relative quantization method (D cycle threshold) was used to quantify the relative expression level of target genes. Three biological replicates with three technique replicates each were included in producing the statistical analysis and the error range analysis. In situ hybridization was performed as described in Fu et al. (2014). Two OsDEX1 cDNA fragments generated by PCR were used for preparing antisense and sense probes.
Phylogenetic Analysis
Multiple alignments were performed using Muscal 3.6 (http://www.ebi.ac.uk/Tools/msa/muscle/). A phylogenetic tree was constructed with the aligned sequences from the full-length region of OsDEX1. MEGA (version 6.0; http://www.megasoftware.net/index.html) and the Neighbor-Jointing methods were used with p-distance model and pairwise deletion and bootstrap (1000 replicates; random seed). The Max Parsimony method of MEGA was also employed to support the Neighbor-Jointing tree, using the default parameter.
Heterologous Expression and In Vitro Ca2+ Binding Assay
Wild-type and EF-hands-mutated OsDEX1 cDNA was amplified by primers cDNA F and cDNA R fused with GST, expressed in BL21 DE3 Escherichia coli cells. Proteins were purified using amylose and GSSH resin, respectively. The protein extraction was incubated with 1 mM CaCl2 or 1 mM CaCl2, together with 1 mM EGTA at 4°C overnight. The mobility was detected in 10% SDS-PAGE.
Calcium Imaging
FRET-based Ca2+ imaging was performed as described in Krebs et al. (2012). Anthers at different stages from YC3.6 transgenic lines in wild type or osdex1 were used for analyses. The samples were analyzed using a fluorescence confocal scanner microscope (Leica SP5II system). The objective, the imaging dimension, and scanning speed were same as above. The pinhole was 2.32 airy unit. The CFP signal was excited using the 458 nm laser line of the Argon laser (30%). The image parameters for the YFP channel were as follows: smart gain (1028.1); offset (−16.1%). The image parameters for this channel were as follows: smart gain (763.0); offset (−12.7%). The ratios between CFP and YFP signal intensity, subtracting the background fluorescence in a region outside the anthers, were calculated. For the ratio calculation, the parameters for image scaling were as follows: Min (0); Max (2).
For the Ca2+ imaging in tobacco cells, the epidermis cells transformed with corresponding plasmids for 2 d were observed. The objective, imaging dimension, and scanning speeds were same as above. The pinhole was 1.00 airy unit. The CFP signal was excited using the 458 nm laser line of the Argon laser (50%). The image parameters for this channel were as follows: 458 nm 33%; smart gain (761.0); offset (0.1%). The image parameters for YFP channel were as follows: smart gain (531.0); offset (−1.3%). For the statistical analysis of [Ca2+]cyt, the ratio of fluorescence intensity in channel CFP and YFP in the whole region of the cell was measured and calculated as described (Behera et al., 2015). For the statistical analysis of [Ca2+]PM, the ratios of fluorescence intensity in channels CFP and YFP around the plasma membrane were measured (n > 15 each).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XP_015631235; XP_006651977; XP_008666953; XP_003563417; XP_004981208; XP_002466172; ERN09669; NP_566343; CDY67523; XP_008388127; XP_006573337; XP_006576567; XP_003604604; XP_001782562; XP_002973603; XP_001703106; CCW71639; XP_780158; XP_005093156; EKC30460; XP_009052863; WP_012257615; WP_022229450; NP_070770.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The model of anther development from stage 5 to stage 9.
Supplemental Figure S2. Transmission electron micrographs of the anthers from the wild type (A) and the osdex1 mutant (B) at stage 8a. Bars = 20 μm.
Supplemental Figure S3. Expression of genes associated with sporopollenin synthesis and transport genes in the wild type and osdex1 during pollen development. Bars = SE.
Supplemental Figure S4. Protease genes and endonuclease genes expression in osdex1 and tapetum cell death deficient mutants.
Supplemental Figure S5. Callose degradation is abnormal in osdex1.
Supplemental Figure S6. Molecular identification and expression pattern of OsDEX1.
Supplemental Figure S7. Phenotype of osdex1-2 and osdex1-4. A, Wild-type flower (left) and osdex1-2 mutant flower (right) before anthesis.
Supplemental Figure S8. Alignment of OsDEX1 and DEX1.
Supplemental Figure S9. Structure prediction of OsDEX1 and DEX1.
Supplemental Figure S10. Sublocalization analysis using confocal laser scanning microscope of OsDEX1 in tobacco epidermal cell.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set 1. Protein sequences used for phylogenetic tree.
Acknowledgments
We thank Lu Zhu, Jie Xu, and Wanwan Zhu for TEM observation at the Instrumental Analysis Center of Shanghai Jiao Tong University. We also thank Lisa O’Donovan from the University of Adelaide for assistance with callose immunolabeling.
Footnotes
This research was supported by the National Key Technologies Research and Development Program of China, Ministry of Science and Technology (grant no. 2016YFD 0100804); the National Natural Science Foundation of China (grants no. 31322040 and no. 31271698); the National Key Basic Research Developments Program of the Ministry of Science and Technology, China (grant no. 2013 CB126902); the Innovative Research Team, Ministry of Education; the 111 Project (grant no. B14016); the Science and Technology Commission of Shanghai Municipality (grant no. 13JC1408200); and the National Transgenic Major Program (grant no. 2016ZX08009003-003-007).
Articles can be viewed without a subscription.
References
- Abeele FV, Skryma R, Shuba Y, Van Coppenolle F, Slomianny C, Roudbaraki M, Mauroy B, Wuytack F, Prevarskaya N (2002) Bcl-2-dependent modulation of Ca2+ homeostasis and store-operated channels in prostate cancer cells. Cancer Cell 1: 169–179 [DOI] [PubMed] [Google Scholar]
- Aditya J, Lewis J, Shirley NJ, Tan HT, Henderson M, Fincher GB, Burton RA, Mather DE, Tucker MR (2015) The dynamics of cereal cyst nematode infection differ between susceptible and resistant barley cultivars and lead to changes in (1,3;1,4)-β-glucan levels and HvCslF gene transcript abundance. New Phytol 207: 135–147 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Wang N, Zhang CX, Schmitz-Thom I, Strohkamp S, Schultke S, Hashimoto K, Xiong LZ, Kudla J (2015) Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytol 206: 751–760 [DOI] [PubMed] [Google Scholar]
- Chang HS, Zhang C, Chang YH, Zhu J, Xu XF, Shi ZH, Zhang XL, Xu L, Huang H, Zhang S, Yang ZN (2012) No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol 158: 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chu HW, Yuan Z, Pan AH, Liang WQ, Huang H, Shen MS, Zhang DB (2006) Isolation and genetic analysis for rice mutants treated with 60Co γ-ray. J Xiamen Univ Nat Sci 45: 82–85 [Google Scholar]
- Cioci G, Mitchell EP, Chazalet V, Debray H, Oscarson S, Lahmann M, Gautier C, Breton C, Perez S, Imberty A (2006) Beta-propeller crystal structure of Psathyrella velutina lectin: an integrin-like fungal protein interacting with monosaccharides and calcium. J Mol Biol 357: 1575–1591 [DOI] [PubMed] [Google Scholar]
- Cohen JJ, Duke RC (1984) Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J Immunol 132: 38–42 [PubMed] [Google Scholar]
- Corneliussen B, Holm M, Waltersson Y, Onions J, Hallberg B, Thornell A, Grundström T (1994) Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factor domains. Nature 368: 760–764 [DOI] [PubMed] [Google Scholar]
- Day IS, Reddy VS, Shad Ali G, Reddy AS (2002) Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 3: H0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, McCann MC, Roberts K (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslen H, Tokumitsu H, Soderling TR (1995) Phosphorylation of CREB by CaM-kinase IV activated by CaM-kinase IV kinase. Biochem Biophys Res Commun 207: 1038–1043 [DOI] [PubMed] [Google Scholar]
- Ferrari D, Pinton P, Szabadkai G, Chami M, Campanella M, Pozzan T, Rizzuto R (2002) Endoplasmic reticulum, Bcl-2 and Ca2+ handling in apoptosis. Cell Calcium 32: 413–420 [DOI] [PubMed] [Google Scholar]
- Foyouzi-Youssefi R, Arnaudeau S, Borner C, Kelley WL, Tschopp J, Lew DP, Demaurex N, Krause KH (2000) Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc Natl Acad Sci USA 97: 5723–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Romagnoli A, Pinton P, Rizzuto R (2008) Ca2+ signaling, mitochondria and cell death. Curr Mol Med 8: 119–130 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JF, Talle B, Wilson ZA (2015) Anther and pollen development: a conserved developmental pathway. J. Integr. Plant Biol. 57: 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen HJ, Heizmann CW, Kaegi U, Celio MR (1990) Ca2+-dependent mobility shift of parvalbumin in one- and two-dimensional gel-electrophoresis. Adv Exp Med Biol 269: 89–91 [DOI] [PubMed] [Google Scholar]
- Groover A, Jones AM (1999) Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol 119: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T (1993) Identification and characterization of nuclear calmodulin-binding proteins of Saccharomyces cerevisiae. Biochim Biophys Acta 1177: 25–30 [DOI] [PubMed] [Google Scholar]
- Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant 6: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T, Ariizumi T, Kawai-Yamada M, Uchimiya H, Toriyama K (2006) Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol 47: 784–787 [DOI] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H. (2000) Calcium signaling during abiotic stress in plants. Int Rev Cytol 195: 269–324 [DOI] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Sun-Ben Ku M, Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC, Chan MT (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaltowski AJ, Vercesi AE, Fiskum G (2000) Bcl-2 prevents mitochondrial permeability transition and cytochrome c release via maintenance of reduced pyridine nucleotides. Cell Death Differ 7: 903–910 [DOI] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Ku S, Yoon H, Suh HS, Chung YY (2003) Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217: 559–565 [DOI] [PubMed] [Google Scholar]
- Lam M, Dubyak G, Chen L, Nuñez G, Miesfeld RL, Distelhorst CW (1994) Evidence that BCL-2 represses apoptosis by regulating endoplasmic reticulum-associated Ca2+ fluxes. Proc Natl Acad Sci USA 91: 6569–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jung KH, An G, Chung YY (2004) Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol Biol 54: 755–765 [DOI] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, Liang W, Zhang D (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D (2011a) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang Q, Wu C (2011b) Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23: 1416–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Harauz G (2002) Interactions of the 18.5-kDa isoform of myelin basic protein with Ca2+-calmodulin: in vitro studies using fluorescence microscopy and spectroscopy. Biochem Cell Biol 80: 395–406 [DOI] [PubMed] [Google Scholar]
- Ling V, Zielinski RE (1993) Isolation of an Arabidopsis cDNA sequence encoding a 22 kDa calcium-binding protein (CaBP-22) related to calmodulin. Plant Mol Biol 22: 207–214 [DOI] [PubMed] [Google Scholar]
- Loftus JC, Smith JW, Ginsberg MH (1994) Integrin-mediated cell adhesion: the extracellular face. J Biol Chem 269: 25235–25238 [PubMed] [Google Scholar]
- Lopez-Fernandez MP, Maldonado S (2015) Programmed cell death in seeds of angiosperms. J Integr Plant Biol 57: 996–1002 [DOI] [PubMed] [Google Scholar]
- Luo XD, Dai LF, Wang SB, Wolukau JN, Jahn M, Chen JF (2006) Male gamete development and early tapetal degeneration in cytoplasmic male-sterile pepper investigated by meiotic, anatomical and ultrastructural analyses. Plant Breeding 125: 395–399 [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- McConkey DJ, Orrenius S (1997) The role of calcium in the regulation of apoptosis. Biochem Biophys Res Commun 239: 357–366 [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Bonig I, Hoogenraad NJ, Clarke AE, Stone BA (1991) The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta 185: 1–8 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Orrenius S, Zhivotovsky B, Nicotera P (2003) Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol 4: 552–565 [DOI] [PubMed] [Google Scholar]
- Oshino T, Abiko M, Saito R, Ichiishi E, Endo M, Kawagishi-Kobayashi M, Higashitani A (2007) Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol Genet Genomics 278: 31–42 [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P, Ferrari D, Magalhães P, Schulze-Osthoff K, Di Virgilio F, Pozzan T, Rizzuto R (2000) Reduced loading of intracellular Ca2+ stores and downregulation of capacitative Ca2+ influx in Bcl-2-overexpressing cells. J Cell Biol 148: 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Reddy AS (1993) Calcium and signal transduction in plants. CRC Crit Rev Plant Sci 12: 185–211 [DOI] [PubMed] [Google Scholar]
- Pozzan T, Rizzuto R, Volpe P, Meldolesi J (1994) Molecular and cellular physiology of intracellular calcium stores. Physiol Rev 74: 595–636 [DOI] [PubMed] [Google Scholar]
- Qin P, Tu B, Wang Y, Deng L, Quilichini TD, Li T, Wang H, Ma B, Li S (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol 54: 138–154 [DOI] [PubMed] [Google Scholar]
- Reddy ASN. (2001) Calcium: silver bullet in signaling. Plant Sci 160: 381–404 [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Galperin MY (2004) The DxDxDG motif for calcium binding: multiple structural contexts and implications for evolution. J Mol Biol 343: 971–984 [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Woodhead DD, Wong PW, Galperin MY (2011) New structural and functional contexts of the Dx[DN]xDG linear motif: insights into evolution of calcium-binding proteins. PLoS One 6: e21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, Shanklin J, Ma H, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaili SS, Hsu YT, Carvalho AC, Rosenstock TR, Sharpe JC, Youle RJ (2003) Mitochondria, calcium and pro-apoptotic proteins as mediators in cell death signaling. Braz J Med Biol Res 36: 183–190 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (1998) Calmodulin, calmodulin-related proteins and plant responses to the environment. Trends Plant Sci 3: 299–304 [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Springer TA. (1997) Folding of the N-terminal, ligand-binding region of integrin α-subunits into a β-propeller domain. Proc Natl Acad Sci USA 94: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MX, Huang XY, Yang J, Guan YF, Yang ZN (2013) Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod 26: 83–91 [DOI] [PubMed] [Google Scholar]
- Tan H, Liang WQ, Hu JP, Zhang DB (2012) MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev Cell 20: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Enslen H, Soderling TR (1995) Characterization of a Ca2+/Calmodulin-dependent protein dinase cascade. Molecular cloning and expression of calcium/calmodulin-dependent protein kinase kinase. J Biol Chem 270: 19320–19324 [DOI] [PubMed] [Google Scholar]
- Trewavas AJ, Malhó R (1998) Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol 1: 428–433 [DOI] [PubMed] [Google Scholar]
- Tuckwell DS, Brass A, Humphries MJ (1992) Homology modelling of integrin EF-hands. Evidence for widespread use of a conserved cation-binding site. Biochem J 285: 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Abeele F, Skryma R, Shuba Y, van Coppenolle F, Slomianny C, Roudbaraki M, Mauroy B, Wuytack F, Prevarskaya N (2002) Bcl-2-dependent modulation of Ca2+ homeostasis and store-operated channels in prostate cancer cells. Cancer Cell 1: 169–179 [DOI] [PubMed] [Google Scholar]
- Wan L, Zha W, Cheng X, Liu C, Lv L, Liu C, Wang Z, Du B, Chen R, Zhu L, He G (2011) A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233: 309–323 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2008) BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem 283: 3200–3210 [DOI] [PubMed] [Google Scholar]
- White PJ. (2000) Calcium channels in higher plants. Biochim Biophys Acta 1465: 171–189 [DOI] [PubMed] [Google Scholar]
- Woltering EJ. (2010) Death proteases: alive and kicking. Trends Plant Sci 15: 185–188 [DOI] [PubMed] [Google Scholar]
- Wu HM, Cheun AY (2000) Programmed cell death in plant reproduction. Plant Mol Biol 44: 267–281 [DOI] [PubMed] [Google Scholar]
- Wyllie AH. (1980) Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang YX, Dong CH, Yu JY, Shi L, Tong CB, Li ZB, Huang JY, Liu SY (2014a) Cysteine Protease 51 (CP51), an anther-specific cysteine protease gene, is essential for pollen exine formation in Arabidopsis. Plant Cell Tissue Organ Cult 119: 383–397 [Google Scholar]
- Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, Liang W, Zhang D (2014b) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56: 979–994 [DOI] [PubMed] [Google Scholar]
- Zhang D and Liang W (2016). Pushing the Boundaries of Scientific Research: 120 Years of Addressing Global Issues. Science/AAAS, Washington, DC, pp. 45–48. doi/. 351.6278.1223-c. [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H (2014) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang D, Shi J, Yang X (2016) Role of Lipid Metabolism in Plant Pollen Exine Development. Springer International Publishing, Cham, Switzerland: [DOI] [PubMed] [Google Scholar]
- Zhang D, Yang L (2014) Specification of tapetum and microsporocyte cells within the anther. Curr Opin Plant Biol 17: 49–55 [DOI] [PubMed] [Google Scholar]
- Zhang DB, Li H (2014). Exine export in pollen. In Plant ABC Transporters. Springer International Publishing, Cham, Switzerland [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D (2015) Two ATP Binding Cassette G transporters, rice ATP Binding Cassette G26 and ATP Binding Cassette G15, collaboratively regulate rice male reproduction. Plant Physiol 169: 2064–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi JX, Zhao GC, Zhang DB, Liang WQ (2013) Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J Plant Biol 56: 59–68 [Google Scholar]
- Zielinski RE. (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–725 [DOI] [PubMed] [Google Scholar]