A Vigna umbellata oxalyl-CoA synthetase, VuAAE3, confers tolerance to Al stress by regulating cytosolic oxalate metabolism.
Abstract
Acyl Activating Enzyme3 (AAE3) was identified to be involved in the catabolism of oxalate, which is critical for seed development and defense against fungal pathogens. However, the role of AAE3 protein in abiotic stress responses is unknown. Here, we investigated the role of rice bean (Vigna umbellata) VuAAE3 in Al tolerance. Recombinant VuAAE3 protein has specific activity against oxalate, with Km = 121 ± 8.2 µm and Vmax of 7.7 ± 0.88 µmol min−1 mg−1 protein, indicating it functions as an oxalyl-CoA synthetase. VuAAE3-GFP localization suggested that this enzyme is a soluble protein with no specific subcellular localization. Quantitative reverse transcription-PCR and VuAAE3 promoter-GUS reporter analysis showed that the expression induction of VuAAE3 is mainly confined to rice bean root tips. Accumulation of oxalate was induced rapidly by Al stress in rice bean root tips, and exogenous application of oxalate resulted in the inhibition of root elongation and VuAAE3 expression induction, suggesting that oxalate accumulation is involved in Al-induced root growth inhibition. Furthermore, overexpression of VuAAE3 in tobacco (Nicotiana tabacum) resulted in the increase of Al tolerance, which was associated with the decrease of oxalate accumulation. In addition, NtMATE and NtALS3 expression showed no difference between transgenic lines and wild-type plants. Taken together, our results suggest that VuAAE3-dependent turnover of oxalate plays a critical role in Al tolerance mechanisms.
Oxalic acid is the simplest of the dicarboxylic acids with pKa values of 1.23 and 3.83, thus existing as anion, i.e. oxalate, in the cellular environment. Oxalate in plants generally exists in two forms of soluble salts and insoluble calcium oxalate crystal. Many crop plants and pasture weeds produce and accumulate oxalate (Libert and Franceschi, 1987). Being a relatively strong acid, reducing agent, and a very strong chelating agent, it has long been recognized that oxalate must have pivotal roles in biological and metabolic processes. For example, oxalate was supposed to be implicated in a metabolically plausible pH-stat (Davies and Asker, 1983) and as an important counterion to inorganic cations such as sodium and potassium (Osmond, 1963). Both physiological and biochemical studies have suggested a role for oxalate in the regulation of calcium concentrations by balancing soluble and insoluble forms of oxalate (Nakata, 2012). In addition, there is evidence that oxalate functions in both abiotic and biotic stress response in plants. In rice (Oryza sativa), Yang et al. (2000) reported that lead (Pb) toxicity induced more root oxalate secretion from tolerant varieties than from sensitive varieties, and oxalate was able to detoxify Pb by chelating it as less-toxic complex. Zhu et al. (2011) demonstrated that root oxalate secretion is involved in differential Cd tolerance in two tomato (Lycopersicon esculentum) cultivars. Furthermore, oxalate has been demonstrated to be involved in protection against herbivory through regulation of calcium oxalate crystal formation (Franceschi and Nakata, 2005; Nakata, 2012).
Oxalate has also been reported to be implicated in the tolerance of some plant species to Al toxicity, which is a major factor limiting plant growth and development on acid soils (Kochian et al., 2004). One strategy for plants to utilize oxalate against Al toxicity relies on the secretion of oxalate from Al-stressed roots, thereby chelating Al to form nontoxic complex. For example, secretion of oxalate from plant roots in response to Al stress has been reported in taro (Colocasia esculenta; Ma and Miyasaka, 1998), common buckwheat (Fagopyrum esculentum; Zheng et al., 1998), tartary buckwheat (Fagopyrum tataricum; Yang et al., 2011b), tomato (Yang et al., 2011a), spinach (Spinacia oleracea; Yang et al., 2005), Polygonum species (You et al., 2005), tea (Camellia sinensis; Morita et al., 2011), and grain amaranth (Amaranthus hypochondriacus; Fan et al., 2016). Our previous study found that all oxalate accumulators including eight cultivars within four species were able to secrete oxalate in response to Al stress (Yang et al., 2008). The second strategy involves chelation of Al with oxalate and sequestration of this nontoxic complex into vacuoles. For example, Al was found to be present as Al-oxalate complex in leaves of Melastoma malabathrichum and buckwheat, two Al-accumulating plant species (Watanabe et al., 1998; Shen et al., 2002). Plants such as buckwheat even use both mechanisms of Al detoxification (Ma et al., 1997). Thus far, the utilization of oxalate as a mechanism to tolerate Al seems to be limited to those plant species that accumulate oxalate. However, the role of oxalate in Al tolerance in plant species that in general do not accumulate oxalate remains unknown.
Although oxalate in plant cells provides many benefits, the metabolism of oxalate must be tightly controlled to maintain cellular functions. For example, exposure of renal epithelial cells to this strong organic acid can result in a series of deleterious effects such as disruption of membrane integrity and mitochondrial metabolism, metal precipitation, and free radical formation (Scheid et al., 1996). Some pathogens also secrete oxalate to aid in colonization by stimulating stomatal opening, interfering with cell wall, and acting as an elicitor of programmed cell death (Bateman and Beer, 1965; Guimarães and Stotz, 2004; Kim et al., 2008; Williams et al., 2011). Recently, Nakata (2015) reported that transgenic Arabidopsis (Arabidopsis thaliana) plants overexpressing a bacterial gene encoding an oxalic acid biosynthesis enzyme displayed retarded growth and development.
To regulate oxalate level, plants have evolved two pathways to degrade oxalate. In one pathway, oxalate is oxidized into CO2 and H2O2 by oxalate oxidase, an enzyme belongs to germin protein family. The proposition of another pathway for oxalate catabolism can date back to 1961 (Giovanelli and Tobin, 1961), but the enzyme has not been discovered until recently. Foster et al. (2012) characterized Arabidopsis ACYL-ACTIVATING ENZYME3 (AAE3) gene encoding an oxalyl-CoA synthetase that is capable of catalyzing the first step in the CoA-dependent pathway of oxalate catabolism. To date, oxalate oxidase was found only in monocots but seems absent in dicots, but oxalyl-CoA synthetase was reported to exist in Arabidopsis (Foster et al., 2012), Medicago truncatula (Foster et al., 2016), and yeast (Saccharomyces cerevisiae; Foster and Nakata, 2014). While oxalate oxidase was found to be localized to the cell wall and oxalyl-CoA synthetase was localized in cytoplasm, both play a role in defending plants against oxalate-secreting fungal phytopathogens.
Catabolism of oxalate by oxalyl-CoA synthetase results in the formation of oxalyl-CoA, followed by formyl-CoA, formate, and finally CO2. In this catabolic pathway, the enzymes involved in oxidation of formate into CO2 have been well documented as formate dehydrogenase. Intriguingly, we recently demonstrated that Al-induced formate accumulation represents an early event related to Al-induced root growth inhibition in rice bean, and Vigna umbellata Formate Dehydrogenase (VuFDH) encoding a formate dehydrogenase is involved in Al tolerance through oxidation of formate to form NADH and CO2 (Lou et al., 2016). However, how Al stress resulted in the accumulation of formate remains unclear. The potential of formate formation from oxalate catabolism led us to hypothesize whether this pathway exist in rice bean. More interestingly, a gene previously named as peroxisomal-coenzyme A synthetase (PCAS) was found to be up-regulated significantly in root apices of rice bean, and actually PCAS has high amino acid sequence homology with Arabidopsis AAE3 (Fan et al., 2014). Thus, it is likely that one possible route to formate accumulation originates from degradation of oxalate mediated by PCAS.
In this study, we isolated a full-length VuAAE3 cDNA and found that the expression of VuAAE3 in rice bean root tips is enhanced greatly by Al stress. We also showed that Al stress causes rapid accumulation of oxalate in rice bean root tips, a process that may contribute to Al-induced root elongation inhibition. In accord with this, overexpression of VuAAE3 in tobacco resulted in increased Al tolerance and decreased oxalate production. Therefore, our results contribute to not only a further understanding of Al toxicity mechanisms in higher plants, i.e. disorder of oxalate metabolism, but may provide a novel approach to improve plant Al tolerance through biotechnology.
RESULTS
Cloning and Sequence Analysis of VuAAE3 from Vigna umbellata
On the basis of rice bean root tip Al stress-responsive suppression subtractive hybridization libraries (Fan et al., 2014), we obtained a full-length VuAAE3 cDNA via rapid amplification of cDNA ends (RACE)-PCR method (GenBank accession no. KX354978; Supplemental Fig. S1). The VuAAE3 coding region is 1560 bp in length, and encodes a protein of 519 amino acids. A BLAST search of orthologs in Arabidopsis revealed that VuAAE3 is an ortholog of AtAAE3 (At3g48990), displaying 69% identity and 79% similarity with AtAAE3 (Fig. 1). Recently, AtAAE3 has been functionally characterized as an enzyme involved in the ligation of oxalate with CoA to form oxalyl-CoA (Foster et al., 2012). Further BLAST analysis using VuAAE3 protein sequence found that orthologs are present in both dicots such as soybean (Glycine max) and tomato and monocots such as rice and maize (Zea mays), suggesting the evolution of AAE3 protein before divergence of monocots and dicots. Similar to AtAAE3, VuAAE3 is characterized by having an AMP binding domain and acetyl-CoA synthetase domain (Fig. 1). Phylogenetic relationship analysis indicated that VuAAE3 was most closely clustered with GmAAE3 from soybean, and AAE3 proteins from monocots and dicots are separated into two distinct clades (Supplemental Fig. S2).
Figure 1.
Amino acid sequence alignment of AAE3 proteins from rice bean (VuAAE3; KX354978), Arabidopsis (AtAAE3; At3g48990), and M. truncatula (MtAAE3; XP_003599555.1). The conserved AMP binding domain and acetyl-CoA synthetase domain are indicated.
VuAAE3 Encodes an Oxalyl-CoA Synthetase
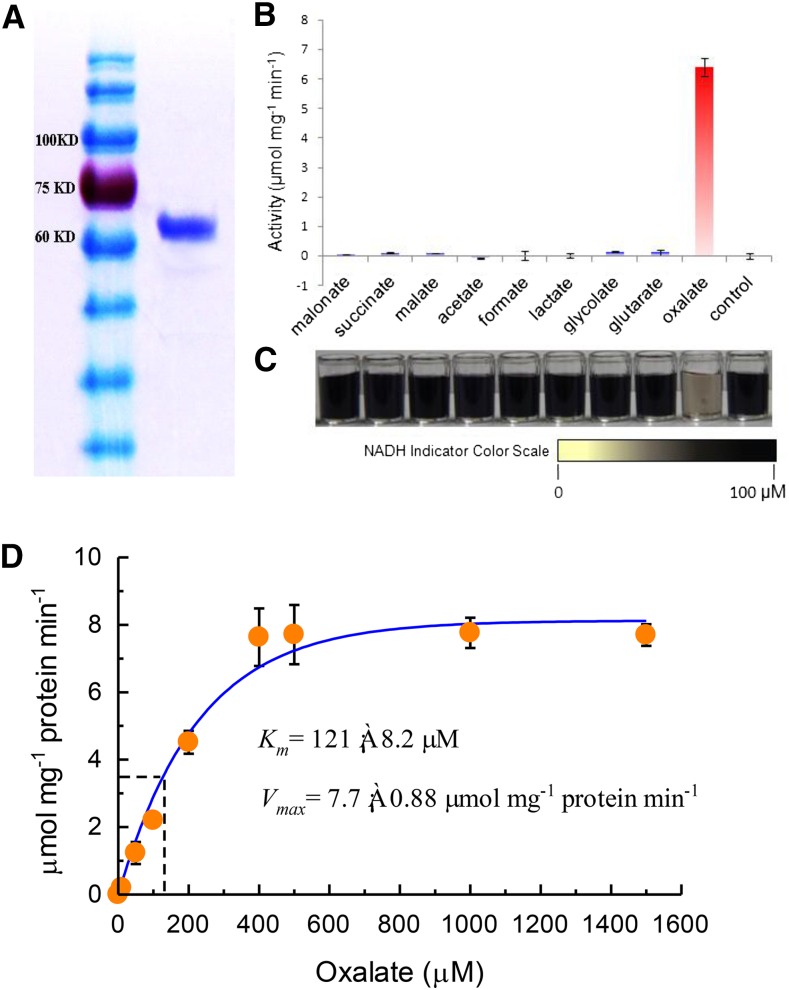
VuAAE3 is supposed to function as an Oxalyl-CoA synthetase as indicated by the BLAST result. To investigate its natural organic acid substrate, the histidine (HIS)-tagged fusion of VuAAE3 was constructed, expressed in Escherichia coli, and purified by nickel-affinity chromatography. The recombinant protein was supposed to be >90% pure as visualized by Coomassie Brilliant Blue staining on the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Fig. 2A). Then, eight organic acids were used as potential substrates for VuAAE3 using a coupled-enzyme assay (Chen et al., 2011; Foster et al., 2012). The recombinant protein displayed higher enzyme activity toward oxalate, while negligible activities were found against the other seven organic acids (Fig. 2B). We further developed a color-visualizing method to test the specificity of VuAAE3 protein to oxalate. A highly water-soluble disulfonated tetrazolium salt was used as a chromogenic indicator for NADH, showing faint yellow to black proportional to NADH concentration (Ishiyama et al., 1997). If organic acid tested could be the substrate for VuAAE3, the consumption of NADH results in color change from black toward faint yellow. As shown in Figure 2C, only when oxalate was used as substrate was the color changed accordingly. We also tested a range of other short chain fatty acids and some organic acids involved in carbonate metabolism, but no significant activity was found (data not shown). Thus, VuAAE3 is an oxalyl-CoA synthetase specifically involved in oxalate degradation.
Figure 2.
Biochemical analysis of VuAAE3. A, SDS-PAGE gel of purified His-VuAAE3 protein (right) and molecular weight markers (left). B, Reaction rate against different organic anions. The reaction mixture contains 0.5μg/mL recombinant protein and 375 μm each of the tested substrates; the mixture was incubated for 1 h at room temperature. bars indicate ± se (n = 3). C, Visual inspection of NADH residue level indicated by nitroblue tetrazolium and l-Methoxy-5-methylphenazinium methosulfate. D, Kinetic analysis of VuAAE3 using a range of oxalate concentrations. Km and Vmax were determined from nonlinear regression to the Michaelis-Menten equation for concentrations up to 1500 µm oxalate.
We next performed kinetic analysis of VuAAE3 using a range of oxalate concentrations. At saturating concentration of CoA (0.5 mm) and ATP (5 mm), the enzyme displayed Michaelis-Menten kinetics with respect to oxalate concentration up to 400 µm (Fig. 2D). Using this data, a Vmax of 7.7 ± 0.88 µmol min−1 mg−1 protein and a Km of 121 ± 8.2 µm was calculated. The Vmax for VuAAE3 is lower than the 11.4 ± 1.0 and 19 ± 0.9 µmol min−1 mg−1 protein reported for AtAAE3 (Foster et al., 2012) and MtAAE3 (Foster et al., 2016), respectively. However, the Km for VuAAE3 is smaller than AtAAE3 (149.0 ± 12.7 µm) but larger than MtAAE3 (81.0 ± 8.1).
VuAAE3 Is Localized to Both Cytoplasm and Nucleus
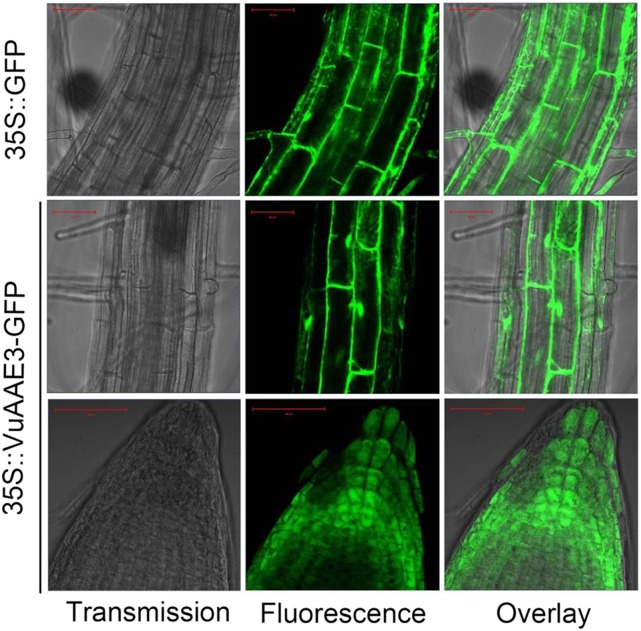
Both AtAAE3 and MtAAE3 are reported to be localized to cytoplasm (Foster et al., 2012; 2016). In order to investigate the subcellular localization of VuAAE3, we constructed transgenic Arabidopsis plants overexpressing a VuAAE3-GFP fusion protein or a GFP protein alone under the control of Cauliflower mosaic virus 35S promoter. In both root tip and mature root region, a strong GFP signal was observed in cytoplasm and nucleus in 35S::VuAAE3::GFP lines similar to that seen in the 35S::GFP control (Fig. 3). Thus, our result indicates that rice bean VuAAE3 is a soluble oxalyl-CoA synthetase with no specific subcellular localization.
Figure 3.
Subcelluar localization of VuAAE3 in transgenic plants. Both 35S::GFP and 35S::VuAAE3-GFP constructs were introduced individually into Arabidopsis. The homozygous seedlings of T4 generation were used to observe GFP fluorescence. Bar = 50 μm.
Expression Pattern of VuAAE3
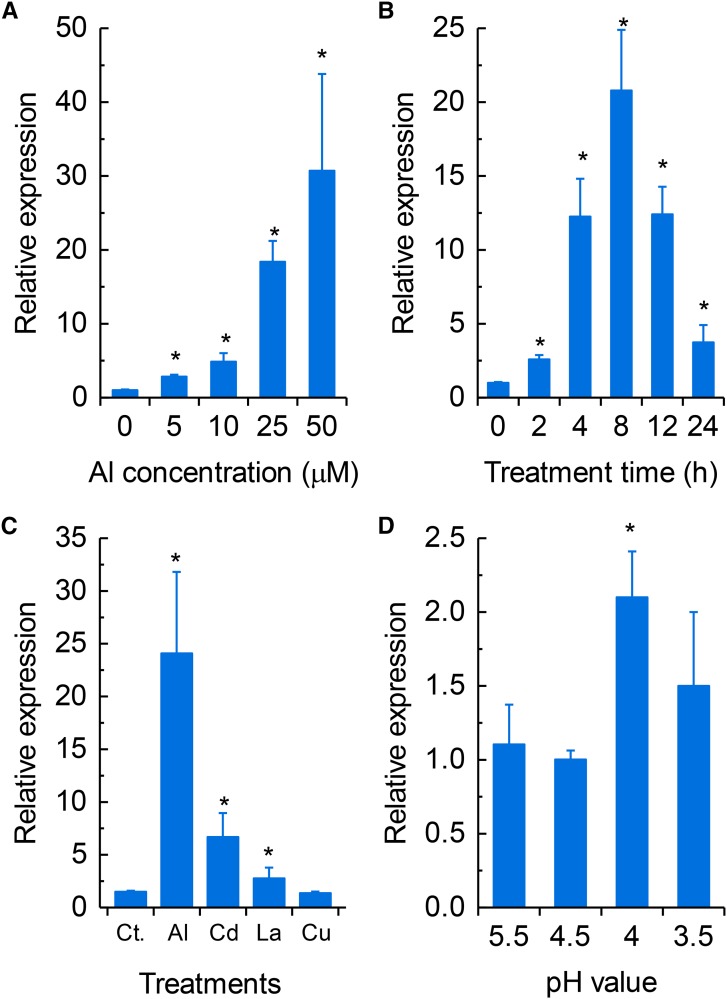
We have previously found that VuAAE3 expression was up-regulated by Al stress (Fan et al., 2014). To characterize the expression of VuAAE3 comprehensively, we used quantitative real-time PCR (qRT-PCR) and in planta VuAAE3 promoter-GUS reporter assay to investigate the expression pattern of VuAAE3. In a dose-response experiment, as shown in Figure 4A, the expression of VuAAE3 increased with increasing external Al concentrations after 8 h of exposure. In addition, in a time-course experiment, the expression of VuAAE3 was found to have increased within 2 h of exposure to 25 µm Al and to be dramatically increased as exposure time prolonged, although this increase had fallen after 8 h of exposure (Fig. 4B).
Figure 4.
Rice bean VuAAE3 expression analysis. A, Dose-dependent VuAAE3 expression in rice bean root tips (0–1 cm). The roots were exposed to various concentrations of Al for 8 h. B, Time-dependent VuAAE3 expression in rice bean root tips (0–1 cm). The roots were exposed to 25 µm Al for various times. C, Metal-dependent VuAAE3 expression in rice bean root tips (0–1 cm). The seedlings were subjected to nutrient solution as control (Ct.) or the same nutrient solution containing Al (25 µm), CdCl2 (20 µm), LaCl3 (10 µm), and CuCl2 (0.5 µm) for 8 h. D, pH-dependent VuAAE3 expression in rice bean root tips (0–1 cm). Seedlings were grown in nutrient solution with different pH values for 8 h. All data were normalized relative to VuAAE3 expression in the absence of Al at pH 4.5. The expression was determined by real-time RT-PCR and 18S rRNA was used as an internal control. Values are expressed as means ± sd (n = 3). The asterisk indicates significant differences between treatment and control (pH 4.5 without Al stress).
The expression of VuAAE3 responses to other stressors, e.g. heavy metals and low pH, was also examined (Fig. 4C). Exposure of rice bean roots to Al, Cd, and La induced the expression of VuAAE3, but the expression induction was significantly lower in Cd and La than in Al. Exposure to Cu, however, had no effect on the expression of VuAAE3. Previously, we have found that several Al-responsive genes, e.g. VuSTOP1, VuMATE1, and VuFDH are also responsive to low pH regulation (Fan et al., 2014; 2015; Lou et al., 2016). In order to examine whether expression of VuAAE3 is also sensitive to low pH, we investigated the effect of low pH stress on VuAAE3 expression. In a pH range of 5.5 to 3.5, the expression of VuAAE3 was induced only slightly by pH 4.0, but no significant difference among other pH conditions was detected (Fig. 4D).
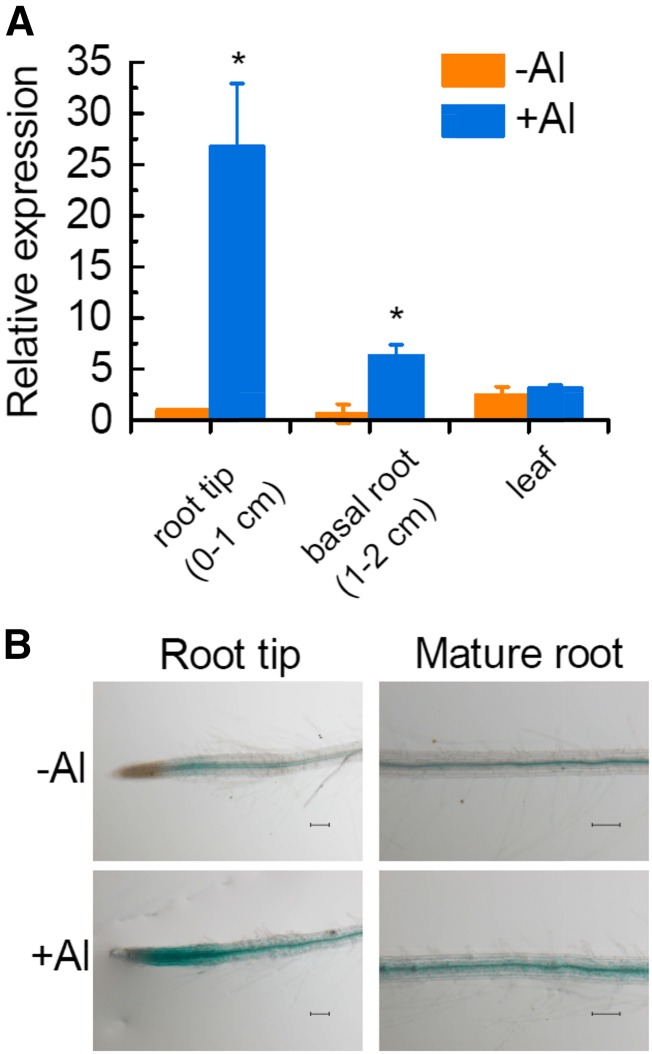
We also investigated the spatial patterning of VuAAE3 expression in either the presence or absence of Al. In the absence of Al stress, VuAAE3 is expressed in root tip, basal root, and leaf, but the expression is more abundant in leaf than in root (Fig. 5A). In the presence of Al stress, however, the expression of VuAAE3 was up-regulated in both root tip and basal root but not in leaf (Fig. 5A). The expression induction was much greater in root tip than that in basal root (Fig. 5A). To further investigate the tissue-specific localization of VuAAE3 expression, a 1456-bp DNA sequence upstream of the translation start codon was isolated (Supplemental Fig. S3). This promoter fragment was fused to a GUS reporter gene and transformed into Arabidopsis wild-type plants. As shown in Figure 5B, GUS activity was observed in the root tip and mature root region, both confining to the vascular cylinder in the absence of Al stress. Al stress resulted in the increase of GUS activity in both root tip and mature root region, but the intensity and location are different. In root tip, Al stress caused GUS activity to extend from vascular cylinder to the whole root tip (including cortex and epidermis). However, in mature root region, the enhancement of GUS activity seems to be limited to vascular cylinder.
Figure 5.
Tissue-specific expression of VuAAE3 in response to Al stress. A, Seedlings of rice bean (3 d old) were exposed to 0 or 25 µm Al for 8 h. Root tip (0–1 cm), basal root (1–2 cm), and leaves were sampled for RNA extraction. The expression was determined by real-time RT-PCR and 18S rRNA was used as an internal control. Values are expressed as means ± sd (n = 3). The asterisk indicates significant differences between treatments. B, Seedlings (7 d old) of transgenic Arabidopsis lines carrying PVuAAE3::GUS construct were subject to 1:30 strength Hoagland nutrient solution at pH 5.0 with or without 4 µm Al for 6 h. Bar = 100 µm.
Oxalate Accumulation Contributes to Al-Induced Inhibition of Root Growth
Since VuAAE3 catalyzes oxalate to form oxalyl-CoA (Fig. 2) and Al stress rapidly induced VuAAE3 expression (Fig. 4B), we wonder whether oxalate accumulated in rice bean root tip when exposed to Al stress and the accumulation of oxalate is actually harmful to root growth. To test this hypothesis, we first measured the internal oxalate content in response to Al stress. As shown in Figure 6, the internal oxalate content of rice bean root apices is relatively constant in the absence of Al. However, Al stress causes a significant increase in oxalate content of rice bean root tip, and this increase occurs rapidly (within 2 h). Further, it appears that the longer the exposure time, the more the increase in oxalate content within 8 h of the onset of exposure. In a parallel experiment, our results showed that oxalate secretion rate from excised root tip decreased over time, irrespective of being treated with Al or not (Supplemental Fig. S4). Although the secretion rate in Al-stressed root tip seems greater than that in Al-free control, they are not statistically different, and the secreted oxalate only represents at most 2% of internal content. Thus, the secretion of oxalate and the difference in oxalate secretion between Al-stressed and Al-free root tip are more likely due to leakage from cut damage and passive efflux from root tip rather than an active Al-dependent process. These findings suggest that rice bean root tip oxalate accumulation is a relatively early event in response to Al stress.
Figure 6.
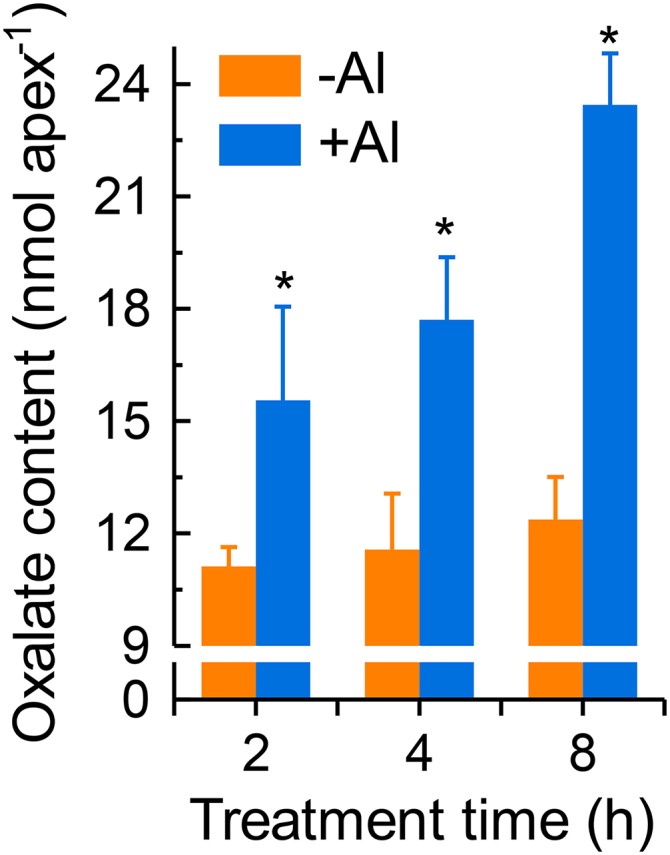
The effect of Al stress on rice bean root tip oxalate content. Seedlings were exposed to nutrient solution containing 0 or 25 µm Al for different times. After treatment, the root tips were homogenized thoroughly in deionized water for oxalate analysis. Data are means ± sd (n = 3). Asterisk indicate significant difference between wild-type and transgenic lines within treatment at P < 0.05.
We next investigated the relationship among oxalate content, root growth inhibition, and VuAAE3 expression. At growth condition of pH 4.5, exogenous application of oxalate resulted in a dose-dependent inhibition of primary root elongation, although oxalate concentration of 0.25 mm had no significant negative effect on root elongation (Fig. 7A). However, only a several-fold increase of VuAAE3 expression was observed after external oxalate supply (Fig. 7B), an induction significantly lower than that caused by Al stress (Fig. 4). Unfortunately, an effort to analyze internal oxalate accumulation after external oxalate incubation was unsuccessful, mainly due to the contamination of internal oxalate with apoplastic oxalate (data not shown). To further support oxalate being the substrate responsible for VuAAE3 expression induction, we analyzed VuFDH expression which functions downstream of VuAAE3 to catalyze formate oxidative degradation. Similar to VuAAE3, the expression of VuFDH could be induced by exogenous oxalate in a dose-dependent manner (Supplemental Fig. S5). Because the primary and secondary dissociation constant of oxalic acid are 1.23 and 3.83, respectively, almost all the oxalate molecules are present as anions at present study conditions (pH 4.5). Without the aid of specific transporters, oxalate anion is very hard to pass across plasma membrane against the electrochemical potential. Thus, it seems possible that only a moderate induction of both VuAAE3 and VuFDH was observed when exogenous oxalate was applied.
Figure 7.
The effect of exogenous oxalate on rice bean root growth and VuAAE3 expression. A, The effect of oxalate on rice bean root elongation. Seedlings were exposed to nutrient solution (pH 4.5) containing different concentrations of exogenous oxalate for 24 h. Root elongation was measured with a ruler before and after treatment (n = 12). Different letters indicate significant differences between treatments at P < 0.05. B, VuAAE3 expression in response to exogenous oxalate. After treatment, root tips (0–1 cm) were excised for RNA extraction and qRT-PCR analysis of VuAAE3 (n = 3). Different letters indicate significant differences between treatments at P < 0.05. C, VuAAE3 promoter activity assay of transgenic Arabidopsis lines carrying GUS reporter gene under the control of VuAAE3 promoter. Roots and leaves were excised from 3-week-old transgenic seedlings and subject to water or oxalate (1 mm) for 2 h. Bars represent 100 µm in roots and 1 mm in leaves.
In order to further test that the expression of VuAAE3 is really induced by oxalate, transgenic Arabidopsis lines carrying GUS reporter genes under the control of VuAAE3 promoter were used. The roots and leaves of GUS-reporter lines were excised and incubated with 1 mm oxalate or water as control. As shown in Figure 7C, oxalate could induce the GUS activity in both root apex and leaf of Arabidopsis. Overall, these results support a role for VuAAE3 in Al-induced oxalate degradation.
Overexpression of VuAAE3 in Transgenic Tobacco Confers Al Tolerance
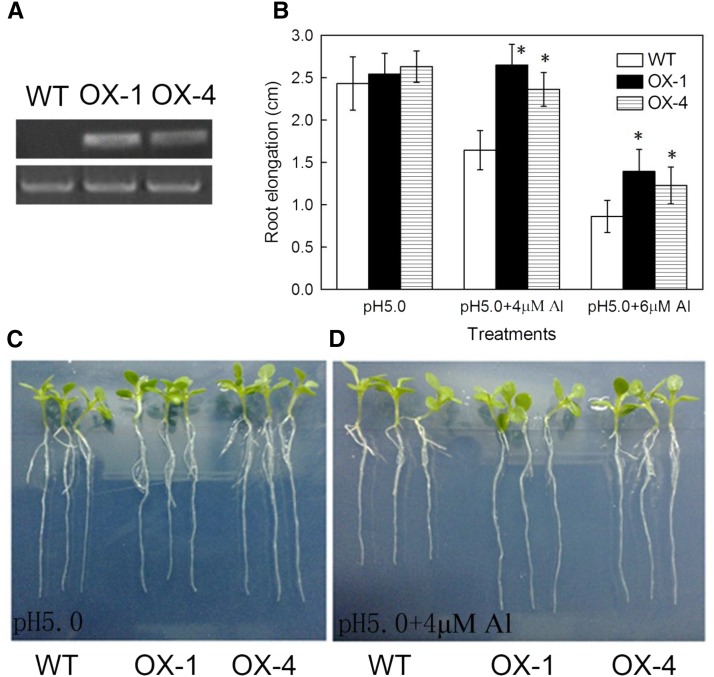
To further characterize the role of VuAAE3 in Al stress response, a 35S::VuAAE3 construct was introduced into tobacco plants, and two independent homozygous T2 transgenic lines (OX-1 and OX-4) were selected for further phenotypic and physiological analysis. Semi-RT-PCR analyses showed that VuAAE3 was highly expressed in the roots of both transgenic lines, but not in wild-type plants (Fig. 8A).
Figure 8.
Overexpression of VuAAE3 enhances Al tolerance in tobacco. A, Detection of expression of VuAAE3 in the wild-type and VuAAE3 overexpression lines. RT-PCR analysis was performed to detect the mRNA expression of VuAAE3 (32 cycles) and the internal control NtACTIN (29 cycles). B, Root elongation of wild-type (WT) and the transgenic lines overexpressing VuAAE3 (OX-1 and OX-4). Data are expressed as means ± sd (n = 15). Asterisk indicates significant difference between wild-type and transgenic lines within treatment at P < 0.05. C, Representative seedlings of wild-type and overexpression lines grown in the 1:30 strength Hoagland nutrient solution at pH 5.0 for 6 d. D, Representative seedlings showing difference in Al sensitivity between wild-type and overexpression lines. Seedlings were grown in the 1:30 Hoagland nutrient solution containing 4 µm Al for 6 d.
In a test of Al tolerance, both wild-type and transgenic plants were grown hydroponically either in the presence (at different concentrations) or absence of Al. After transferring seedlings to nutrient solution for 6 d, there were no significant differences in root growth between wild-type and transgenic lines in the absence of Al (Fig. 8, B and C). While the root elongation of wild type was inhibited by 33% after 6 h of exposure to 4 µm Al, the root elongation of OX-4 was not inhibited, and that of OX-1 was only inhibited by 11% (Fig. 8, B and D). Increase of the Al concentration to 6 µm resulted in inhibition of the root elongation of wild type by 65%, while root elongation of both OX-1 and OX-4 was inhibited by approximately 50% (Fig. 8B). These observations suggest that overexpression of VuAAE3 in tobacco confers increased Al tolerance.
To examine if the increased Al tolerance of transgenic tobacco plants overexpressing VuAAE3 is associated with a decrease in oxalate accumulation, we compared the oxalate content in transgenic lines to that in wild-type plants either in the presence or absence of Al stress conditions. In the absence of Al, the oxalate content was similar in two transgenic tobacco lines to that in wild-type plants. However, while Al stress resulted in a significant increase (about one-third) in the oxalate content, the increase was not observed in both transgenic lines (Fig. 9).
Figure 9.
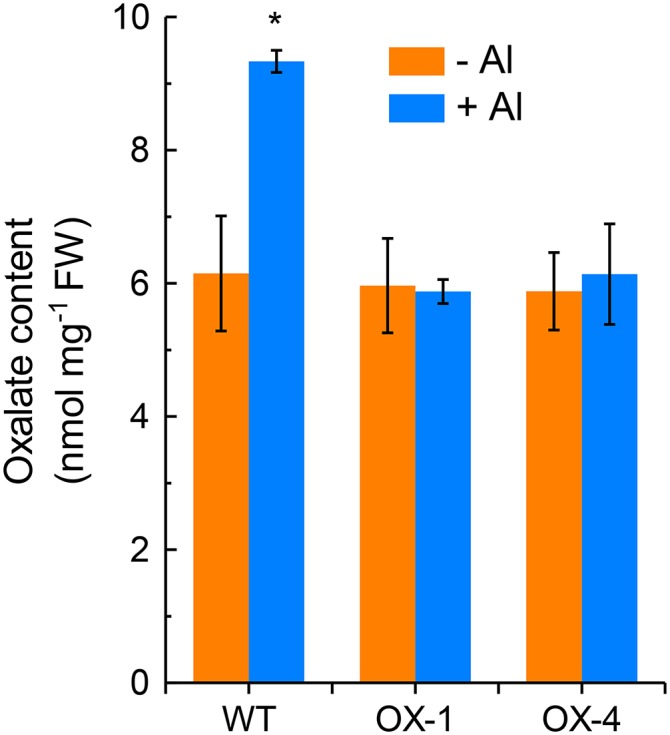
The effect of Al stress on oxalate content in wild-type and overexpression tobacco lines. The plants of wild-type and two independent transgenic lines were exposed to 1:30 strength Hoagland nutrient solution with 0 (−Al) or 4 µm Al (+Al) for 24 h. After treatment, root tips were homogenized thoroughly in deionized water for oxalate content analysis. Data are means ± sd (n = 3). Asterisk indicates significant differences between treatments at P < 0.05.
In tobacco, the induction of two genes, i.e. NtMATE and NtALS3, was reported to be involved in Al tolerance mechanism (Ohyama et al., 2013). To determine whether the increased Al tolerance of VuAAE3 overexpressing lines is associated with changes of their expression, we also compared the expression of both genes in OX-1 to that of wild-type plants. There was no difference between the expressions of either of these genes in wild type versus OX-1, although the expression of both was induced by Al (Supplemental Fig. S6). Taken together, these results suggest that the increased Al tolerance in transgenic lines was related to the decrease of oxalate content, but not via effects on the expression of NtMATE and NtALS3.
DISCUSSION
In this study, we demonstrated that rice bean VuAAE3 encodes an oxalyl-CoA synthetase. The strong evidence for this is provided by the fact that VuAAE3 protein is able to specifically catalyze the conversion of oxalate and CoA into oxalyl-CoA (Fig. 2). Furthermore, bioinformatic analysis revealed that VuAAE3 displays high sequence similarity with Arabidopsis AtAAE3 and M. truncatula MtAAE3, which have been identified as oxalyl-CoA synthetases (Fig. 1; Foster et al., 2012, 2016). In addition, both qRT-PCR analysis and in planta promoter activity assay suggested that the expression of VuAAE3 was responsive to exogenous oxalate (Fig. 7).
Being the intermediates or end products of plant cell metabolism, organic acids are ubiquitous and play pivotal roles in biochemical and physiological processes. Oxalate has been reported to be involved in various biotic and abiotic stresses, such as metal toxicity, pathogen invasion, and insect chewing (Park et al., 2009; Korth et al., 2006; Franceschi and Nakata, 2005; Guo et al., 2005; Nakata and McConn, 2000). One of the best documented roles for oxalate in abiotic stresses is in Al tolerance, because oxalate has strong chelate ability to Al3+ (Ma et al., 1997). At first thought, oxalate biodegradation should play a negative role in Al tolerance, since less oxalate is available to detoxify excessive Al. However, we here suggest that oxalate accumulation is one of the early events leading to plant root growth arrest in response to Al stress. This conclusion is supported by the following lines of evidence. First, exogenous oxalate inhibits root elongation, and such inhibition is actually accompanied by the expression induction of VuAAE3 (Fig. 7). Second, the enhanced Al tolerance of transgenic tobacco plants is associated with the decrease of oxalate accumulation (Fig. 9).
In accord with our findings, other reports also support the view that oxalate accumulation is harmful to plant growth and development. For instance, transgenic Arabidopsis plants overexpressing a bacterial oxalic acid biosynthetic enzyme gene displayed not only significant increase in oxalate content, but also a reduction in plant stature as well as a pronounced delay in bolting and seed set (Nakata, 2015). Unlike Arabidopsis, which is an oxalate nonaccumulating plant, the oxalate-accumulating plant M. truncatula has also evolved CoA-dependent pathway of oxalate biodegradation (Foster et al., 2016), suggesting that oxalate content must be tightly controlled even in oxalate-accumulating plant species. The BLAST search has also revealed that orthologs of AtAAE3 and VuAAE3 present in genomes of rice and tomato, both of which are oxalate accumulating species (Supplemental Fig. S2; Yu et al., 2010; Yang et al., 2011). Buckwheat is another species that accumulates oxalate (Ma et al., 1998). Interestingly, analysis of buckwheat root tip Al-responsive genes through RNA sequence revealed that a gene highly homologous to AtAAE3 was found to be induced by Al stress (data not shown). Although the biochemical and physiological role of these AAE3 proteins has to be identified, these findings imply that oxalate must be strictly regulated in plants.
Previously we have demonstrated the accumulation of formate in response to either Al or low pH stress, but the mechanisms of formate production remain unclear (Lou et al., 2016). It has been proposed that formate may originate from photorespiration, glycolysis, and cell wall synthesis or degradation (Igamberdiev et al., 1999; Hanson et al., 2000). Recently, a novel pathway of oxalate degradation that proceeds from oxalate to oxalyl-CoA to formyl-CoA to formate and eventually CO2 has been suggested in plants (Foster et al., 2012, 2016). Here, we also found that oxalate degradation contributes to formate production in rice bean. First, exogenous application of oxalate induced expression of not only VuAAE3, but VuFDH (Fig. 7; Supplemental Fig. S5). Because VuFDH is specifically involved in the degradation of formate, this result suggests that formate was produced when oxalate was degraded by VuAAE3. Second, oxalyl-CoA synthetase catalyzes the first step toward oxalate degradation, followed by oxalyl-CoA decarboxylase, formyl-CoA hydrolase, and formate dehydrogenase. We presented evidence to support the existence of oxalyl-CoA synthetase, namely VuAAE3 in this study, and formate dehydrogenase (VuFDH) in a previous study (Lou et al., 2016). In a further support, we have isolated a gene homologous to Arabidopsis oxalyl-CoA decarboxylase which is responsible for catalyzing the second step in this pathway (data not shown).
It is worth to note that the expression of VuFDH was greatly induced by low pH stress, but VuAAE3 was almost not, although both displayed similar expression pattern to Al stress (Fig. 4; Lou et al., 2016). In fact, the expression pattern of VuFDH and VuAAE3 also differed in response to other metals (Fig. 4; Lou et al., 2016). These results suggest that while oxalyl-CoA synthetase pathway is conserved for Al tolerance mechanism in plants, it may be not essential for other abiotic stresses. Alternatively, while formate may mainly be produced from oxalate degradation under Al stress, oxalate may originate from other pathways under low pH stress, which contributes to the differential expression pattern between VuAAE3 and VuFDH under Al and low pH stress conditions.
Since the identification and characterization of Arabidopsis AtAAE3 that encodes an oxalyl-CoA synthetase specifically involved in the process of oxalate degradation, homologs of AtAAE3 have been isolated from S. cerevisiae and M. truncatula (Foster and Nakata, 2014; Foster et al., 2016). So far, the role of AAE3 proteins has been ascribed to defense against fungal plant pathogens (Foster et al., 2012, 2016), but the role of AAE3 proteins in abiotic stress is unclear. Therefore, to the best of our knowledge, this is the first report to show that AAE3 plays an important role in tolerance to Al toxicity, the major abiotic stress in acid soils (Kochian et al., 2004). Given VuAAE3 also plays a pivotal role in defense against pathogen invasion the same as AtAAE3 and MtAAE3, this study clearly points to the convergence of biotic and abiotic stress responses on VuAAE3. It is not surprising that there is cross talk between biotic and abiotic stress at transcriptional levels. There is ample evidence that Al may act as an elicitor of pathogenesis-related transduction pathways. For example, in wheat, most Al-responsive genes share homologies with genes induced by pathogens (Hamel et al., 1998). In a previous study, a pathogenesis-related gene was also found to be up-regulated by Al stress in rice bean (Fan et al., 2014). In addition, our recent study suggests that rice bean VuFDH has dual roles both in abiotic and biotic stress tolerance (Lou et al., 2016). More compelling evidence comes from Arabidopsis AtALMT1, which has been well-documented to be involved in both Al tolerance and pathogen resistance (Rudrappa et al., 2008; Lakshmanan et al., 2012; Kobayashi et al., 2013).
In Arabidopsis, loss-of-function mutant of AtAAE3 exhibited an increase of oxalate content in the normal growth conditions (Foster et al., 2012). However, in this study, we found that overexpression of VuAAE3 in tobacco had no measurable influence on oxalate content in normal growth conditions (Fig. 9). The discrepancy may lie in the fact that oxalate is mainly compartmentalized in vacuoles, since it is a strong acid and chelator (Franceschi and Nakata, 2005). Thus, cytoplasmic oxalate content must be low enough to avoid undesired reaction between oxalate and other essential elements such as Ca2+. Obvious cytosolic presence of VuAAE3 (Fig. 3) and oxalyl-CoA synthetase proteins from Arabidopsis and M. truncatula reinforced the possibility that cytoplasmic oxalate must be tightly controlled. We deduce that the biodegradation of cytoplasmic oxalate is attributed to the affinity of VuAAE3 to oxalate, but not to the amount of VuAAE3 proteins. This may also be the reason of why oxalate content was not changed in two transgenic tobacco lines overexpressing VuAAE3 in the absence of Al stress (Fig. 9). However, when oxalate accumulates in response to Al stress, overexpression of VuAAE3 in two transgenic tobacco lines is able to react with excessive oxalate, which results in oxalate content stable in transgenic lines under Al stress (Fig. 9).
Transgenic approach is a compelling solution to increase the Al tolerance of plants in acid soils (Ryan et al., 2011; Kochian et al., 2015). To date, a number of genes involved in different biological processes have been adopted to genetically modify Al tolerance (Ryan et al., 2011). Here, we demonstrated that manipulation of oxalate metabolism via overexpression of VuAAE3 could increase Al tolerance, provided more chances for genetic modification of crop Al tolerance. More interestingly, oxalate is one of the known antinutritional factors, which has negative health concerns, when we consume crops containing high amount of oxalate, such as tomato, spinach, soybean and grass pea (Lathyrus sativus). Chakraborty et al. (2013) found that the fruits of transgenic tomato plants expressing a fungus Flammulina velutipes oxalate decarboxylase under the control of fruit-specific promoter have significant less oxalate, but pronounced more nutrients such as Fe and Ca. Constitutive or seed-specific expression of Flammulina velutipes oxalate decarboxylase has also been demonstrated to be not only helpful for improving seed nutritional quality but also tolerance to the fungal pathogen (Kumar et al., 2016). Thus, it will be interesting to examine in the near future whether overexpression of VuAAE3 in crops could have multiple advantages not only in stress tolerance but also in human nutrition.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice bean (Vigna umbellata) seeds were soaked in deionized water overnight and germinated at 26°C in the dark. The germinated seeds were cultured in 0.5 mm CaCl2 (pH 4.5) solution for 3 d. The solution was renewed daily. Seedlings of similar size were transplanted into nutrient solution of the following composition (μm): CaSO4 (200), CaCl2 (200), MgSO4 (100), KNO3 (400), NH4NO3 (300), NaH2PO4 (5), H3BO3 (3), MnCl2 (0.5), ZnSO4 (0.4), CuSO4 (0.2), Fe-EDTA (10), and (NH4)6Mo7O24 (1). The solution was adjusted to pH 4.5 with HCl, and renewed daily. After 2 d of culture, the plants were subjected to the following treatments. The nutrient solution was used as the control treatment solution. For the time course experiment, seedlings were exposed to 25 μm AlCl3 for 0, 2, 4, 8, 12, or 24 h. For the Al concentration-dependence experiment, seedlings were exposed to 0, 5, 10, 25, or 50 μm AlCl3 for 12 h. For other treatments, the seedlings were exposed to nutrient solution (pH 4.5) containing 25 μm AlCl3, 20 μm CdCl2, 10 μm LaCl3, or 0.5 μm CuCl2 or in different pH conditions for 12 h. For the exogenous oxalate experiment, the seedlings were exposed to nutrient solution (pH 4.5) containing 0, 0.25, 0.5, or 1.0 mm sodium oxalate for 24 h. All experiments were performed in an environmentally controlled growth room with a 12 h/30°C day and a 12 h/22°C night regime, a light intensity of 300 to 350 μmol photons m−2 s−1 and a relative humidity of 60%.
Cloning and Sequence Analysis
The full-length VuAAE3 cDNA was amplified by a RACE method using the SMART RACE cDNA Amplification kit (Clontech). The gene-specific primers for 3′-RACE and 5′-RACE amplification were listed in Supplemental Table S1. Amplified cDNA fragments were cloned into MD-18T clone vector (Takara) and sequenced. The full-length cDNA sequence was assembled with the help of ClustalW2 software, and the open reading frame was verified by RT-PCR. The deduced amino acid sequence was predicted by Open Reading Frame Finder (http://www.ncbi.nlm.nih.gov/orffinder/). Amino acid sequence alignment and phylogenetic analysis was performed using DNAMAN and Mega 6.0 software, respectively.
A 1456 bp of DNA sequence upstream of VuAAE3 was amplified from rice bean genome walker libraries (Lou et al., 2016) using gene-specific primers (Supplemental Table S1). Amplified fragments were cloned into the pMD18-T vector (Takara). The sequences that extended upstream of the cDNA clones were isolated as the 5′ upstream regions of the gene. Putative cis-elements of 5′-flanking region was analyzed using software PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
RNA Isolation and Gene Expression Analysis
Total RNA was isolated from eight root tips (0–1 or 1–2 cm) or 100 mg leaf tissue using the RNeasymini kit (Tiangen). First-strand cDNA was synthesized from 1 μg of total RNA using SuperScript reverse transcriptase (Takara). One microliter (100 ng μL−1) of cDNA in 10 μL solution systems was used for quantitative analysis of gene expression performed with SYBR Premix Ex Taq (Takara) on a LightCycler 480 machine (Roche Diagnostics). The primers of genes were listed in Supplemental Table S1, and the PCR amplification conditions were as follows: 94°C for 5 min; 45 cycles of 94°C for 10 s, 55°C for 15 s, and 72°C for 15 s. For each gene, expression data were normalized with expression level of internal control (18S rRNA for rice bean and NtACTIN for tobacco [Nicotiana tabacum]) and calculated by equation 2−ΔΔCp. The experiment was performed with three biological and technical replications.
Subcellular Localization and GUS Analysis of VuAAE3
The full-length gene fragment and promoter region of VuAAE3 was PCR cloned and ligated into the binary vector 35S::GFP (modified from pCAMBIA1300) and pCAMBIA1301 (fusion to the GUS gene) vector, respectively (Supplemental Table S1). The resultant 35S::VuAAE3::GFP and VuAAE3p::GUS plasmid was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis wild-type (Col-0) plants by Agrobacterium-mediated transformation. Homozygous T3 plants were used. GFP fluorescence was observed via confocal laser scanning microscopy (LSM710: Karl Zeiss). GUS staining was performed according to Jefferson et al. (1987).
Overexpression of VuAAE3 in Tobacco and Al Tolerance Evaluation
The VuAAE3 coding region, carrying its stop codon, was amplified by PCR using primer pairs (Supplemental Table S1) and ligated into a modified pCAMBIA1300 vector under the control of the Cauliflower mosaic virus 35S promoter, then transformed into A. tumefaciens (strain GV1301). Tobacco plants were transformed as described by Horsch et al. (1985). Transgenic lines carrying VuAAE3were selected by PCR using the primers described above. For evaluating Al tolerance of VuAAE3 overexpression of tobacco, seeds from T2 homozygous and wild-type lines were first surface-sterilized with 50% sodium hypochlorite for 5 min and then washed four times with deionized water. Then, seeds were sown onto Murashige and Skoog plates containing 3% (w/v) Suc and 0.8% (w/v) agar (pH 5.7). Following incubation in a refrigerator at 4°C for 3 d, the seeds were then placed in a growth chamber in 12- h light/12-h dark conditions at 23°C. When the length of the primary root had reached ∼1 cm, the seedlings were transferred to the one-thirtieth-strength Hoagland nutrient solution (without NH4H2PO4 and with 1 mm CaCl2) containing 0, 4, or 6 μm AlCl3 at pH 5.0 for 6 d. The solution was renewed every 2 d. Al sensitivity was evaluated by relative root elongation expressed as (root elongation with Al treatment/root elongation without Al) × 100.
Extraction of Oxalate and Determination
For tissue oxalate content, plant materials were ground into fine powder in liquid nitrogen, extracted by deionized water three times, and combined. For oxalate in root exudates, the treatment solution bathing the roots were directly collected. The collected solution was allowed to pass first through a cation exchange column (16 mm × 14 cm) filled with 5 g Amerlite IR-120B resin (H+ form, Muromachi Chemical) and then through an anion-exchange column filled with 1.5 g Dowex 1 × 8 resin (100–200 mesh, formate form). HCl (1 m) was used as eluent to collect oxalate retained in the anion-exchange resin. The eluent was concentrated to dryness using a rotary evaporator at 40°C and redissolved in deionized water. Oxalate was detected by ion chromatography (ICS 3000; Dionex) equipped with an IonPac AS11 anion-exchange analytical column (4 × 250 mm) and a guard column (4 × 50 mm). The mobile phase was 30 mm NaOH at flow rate of 0.6 mL min−1.
Recombinant Protein Purification and Enzyme Activity Assay
Full-length cDNA of VuAAE3 was obtained by PCR amplification using a pair of specific primers (Supplemental Table S1) and cloned into the pET-28a (+) vector. The vector was introduced into the Escherichia coli line BL21 (DE3) for protein expression. The expression of the recombinant protein was induced by 1 mm of IPTG at 28°C for 4 h, and the cells was collected and resuspended with binding buffer (20 mm sodium phosphate, 0.5 m sodium chloride, and 40 mm imidazole pH 7.4). The recombinant enzyme was collected using HisTrap FF column (GE Healthcare) under the manufacturer’s instructions and purified by Ultra filtration (30 kD Mr cutoff; Millipore) and equilibrated by 0.1 m Tris-HCl, pH 7.5. The purity of the His-tagged protein was determined by SDS-PAGE followed by Coomassie Brilliant Blue staining.
The assay buffer contains 0.7 mg of purified recombinant protein, 0.1 m Tris-HCl (pH 8.0), or 0.1 m NaPO4 (pH 8.0), 2 mm DTT, 5 mm ATP, 10 mm MgCl2, 0.5 mm CoA, 0.4 mm NADH, 1 mm phosphoenolpyruvate, and 10 units each of myokinase, pyruvate kinase, and lactate dehydrogenase and the carboxylic acid substrate, in a final volume of 1 mL. In the substrate assay, 0.4 μm of candidate substrate was used, and up to 1.5 mm oxalate was used for the enzyme kinetic assay. The mixture was incubated at room temperature for 1 h, and the reaction rate was measured by the NADH concentration at 340 nm spectrophotometrically.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession number: V. umbellata VuAAE3 (KX354978).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of rice bean VuAAE3 cDNA.
Supplemental Figure S2. Phylogram of AAE3 proteins.
Supplemental Figure S3. The sequence and cis-element analysis of VuAAE3 promoter.
Supplemental Figure S4. The effect of Al stress on oxalate secretion from root tips of rice bean.
Supplemental Figure S5. The effect of exogenous oxalate on rice bean VuFDH expression.
Supplemental Figure S6. The effect of Al stress on the expression of Al-tolerance gene expression in tobacco.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Footnotes
This work was supported by the 973 Project (2014CB441002), the Natural Science Foundation of China (31222049, 31071849, and 31501827), and the Fundamental Research Funds for the Central Universities.
Articles can be viewed without a subscription.
References
- Bateman DF, Beer SV (1965) Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotiumrolfsii. Phytopathology 55: 204–211 [PubMed] [Google Scholar]
- Chakraborty N, Ghosh R, Ghosh S, Narula K, Tayal R, Datta A, Chakraborty S (2013) Reduction of oxalate levels in tomato fruit and consequent metabolic remodeling following overexpression of a fungal oxalate decarboxylase. Plant Physiol 162: 364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kim HU, Weng H, Browse J (2011) Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell 23: 2247–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DD, Asker H (1983) Synthesis of oxalic Acid by enzymes from lettuce leaves. Plant Physiol 72: 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Lou HQ, Gong YL, Liu MY, Cao MJ, Liu Y, Yang JL, Zheng SJ (2015) Characterization of an inducible C2 H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol 208: 456–468 [DOI] [PubMed] [Google Scholar]
- Fan W, Lou HQ, Gong YL, Liu MY, Wang ZQ, Yang JL, Zheng SJ (2014) Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ 37: 1586–1597 [DOI] [PubMed] [Google Scholar]
- Fan W, Xu JM, Lou HQ, Xiao C, Chen WW, Yang JL (2016) Physiological and molecular analysis of aluminium-induced organic acid anion secretion from grain amaranth (Amaranthus hypochondriacus L.) roots. Int J Mol Sci 17: E608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Kim HU, Nakata PA, Browse J (2012) A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Luo B, Nakata PA (2016) An oxalyl-CoA dependent pathway of oxalate catabolism plays a role in regulating calcium oxalate crystal accumulation and defending against oxalate-secreting phytopathogens in Medicago trungatula. PLoS One 11: e0149850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Nakata PA (2014) An oxalyl-CoA synthetase is important for oxalate metabolism in Saccharomyces cerevisiae. FEBS Lett 588: 160–166 [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Nakata PA (2005) Calcium oxalate in plants: formation and function. Annu Rev Plant Biol 56: 41–71 [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Tobin NF (1961) Adenosine triphosphate- and coenzyme A-dependent decarboxylation of oxalate by extracts of peas. Nature 190: 1006–1007 [DOI] [PubMed] [Google Scholar]
- Guimarães RL, Stotz HU (2004) Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol 136: 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Tan H, Zhu Z, Lu S, Zhou B (2005) Effect of intermediates on ascorbic acid and oxalate biosynthesis of rice and in relation to its stress resistance. Plant Physiol Biochem 43: 955–962 [DOI] [PubMed] [Google Scholar]
- Hamel F, Breton C, Houde M (1998) Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta 205: 531–538 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Gage DA, Shachar-Hill Y (2000) Plant one-carbon metabolism and its engineering. Trends Plant Sci 5: 206–213 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Bykova NV, Kleczkowski LA (1999) Origins and metabolism of formate in higher plants. Plant Physiol Biochem 37: 503–513 [Google Scholar]
- Ishiyama M, Miyazono Y, Sasamoto K, Ohkura Y, Ueno K (1997) A highly water-soluble disulfonated tetrazolium salt as a chromogenic indicator for NADH as well as cell viability. Talanta 44: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Min J-Y, Dickman MB (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol Plant Microbe Interact 21: 605–612 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi Y, Sugimoto M, Lakshmanan V, Iuchi S, Kobayashi M, Bais HP, Koyama H (2013) Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol 162: 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Kochian LV, Piñeros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66: 571–598 [DOI] [PubMed] [Google Scholar]
- Korth KL, Doege SJ, Park SH, Goggin FL, Wang Q, Gomez SK, Liu G, Jia L, Nakata PA (2006) Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiol 141: 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Chattopadhyay A, Ghosh S, Irfan M, Chakraborty N, Chakraborty S, Datta A (2016) Improving nutritional quality and fungal tolerance in soya bean and grass pea by expressing an oxalate decarboxylase. Plant Biotechnol J 14: 1394–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert B, Franceschi VR (1987) Oxalate in crop plants. J Agric Food Chem 35: 926–938 [Google Scholar]
- Lou HQ, Gong YL, Fan W, Xu JM, Liu Y, Cao MJ, Wang M-H, Yang JL, Zheng SJ (2016) A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol 171: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Hiradate S, Matsumoto H (1998) High aluminum resistance in buckwheat. Ii. Oxalic acid detoxifies aluminum internally. Plant Physiol 117: 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Miyasaka SC (1998) Oxalate exudation by taro in response to Al. Plant Physiol 118: 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Zheng SJ, Matsumoto H, Hiradate S (1997) Detoxifying aluminium with buckwheat. Nature 390: 569–5709403684 [Google Scholar]
- Morita A, Yanagisawa O, Maeda S, Takatsu S, Ikka T (2011) Tea plant (Camellia sinensis L.) roots secrete oxalic acid and caffeine into medium containing aluminum. Soil Sci Plant Nutr 57: 796–802 [Google Scholar]
- Nakata PA. (2012) Plant calcium oxalate crystal formation, function, and its impact on human health. Front Biol 7: 254–266 [Google Scholar]
- Nakata PA. (2015) An assessment of engineered calcium oxalate crystal formation on plant growth and development as a step toward evaluating its use to enhance plant defense. PLoS One 10: e0141982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata PA, McConn MM (2000) Isolation of Medicago truncatula mutants defective in calcium oxalate crystal formation. Plant Physiol 124: 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama Y, Ito H, Kobayashi Y, Ikka T, Morita A, Kobayashi M, Imaizumi R, Aoki T, Komatsu K, Sakata Y, et al. (2013) Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol 162: 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond B. (1963) Oxalates and ionic equilibria in Australian saltbushes (Atriplex). Nature 198: 503–504 [Google Scholar]
- Park S-H, Doege SJ, Nakata PA, Korth KL (2009) Medicago truncatula-derived calcium oxalate crystals have a negative impact on chewing insect performance via their physical properties. Entomol Exp Appl 131: 208–215 [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62: 9–20 [DOI] [PubMed] [Google Scholar]
- Scheid C, Koul H, Hill WA, Luber-Narod J, Kennington L, Honeyman T, Jonassen J, Menon M (1996) Oxalate toxicity in LLC-PK1 cells: role of free radicals. Kidney Int 49: 413–419 [DOI] [PubMed] [Google Scholar]
- Shen R, Ma JF, Kyo M, Iwashita T (2002) Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 215: 394–398 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Osaki M, Yoshihara T, Tadano T (1998) Distribution and chemical speciation of aluminum in the Al accumulator plant, Melastoma malabathrichum L. Plant Soil 201: 165–173 [Google Scholar]
- Williams B, Kabbage M, Kim H-J, Britt R, Dickman MB (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7: e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Jung JY, Song WY, Suh HS, Lee Y (2000) Identification of rice varieties with high tolerance or sensitivity to lead and characterization of the mechanism of tolerance. Plant Physiol 124: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Zhang L, Zheng SJ (2008) Aluminum-activated oxalate secretion does not associate with internal content among some oxalate accumulators. J Integr Plant Biol 50: 1103–1107 [DOI] [PubMed] [Google Scholar]
- Yang JL, Zheng SJ, He YF, Matsumoto H (2005) Aluminium resistance requires resistance to acid stress: a case study with spinach that exudes oxalate rapidly when exposed to Al stress. J Exp Bot 56: 1197–1203 [DOI] [PubMed] [Google Scholar]
- Yang JL, Zhu XF, Peng YX, Zheng C, Ming F, Zheng SJ (2011a) Aluminum regulates oxalate secretion and plasma membrane H+-ATPase activity independently in tomato roots. Planta 234: 281–291 [DOI] [PubMed] [Google Scholar]
- Yang JL, Zhu XF, Zheng C, Zhang YJ, Zheng SJ (2011b) Genotypic differences in Al resistance and the role of cell-wall pectin in Al exclusion from the root apex in Fagopyrum tataricum. Ann Bot (Lond) 107: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JF, He YF, Yang JL, Zheng SJ (2005) A comparison between aluminum resistance among Polygonum species originating on strongly acidic and neutral soils. Plant Soil 276: 143–151 [Google Scholar]
- Yu L, Jiang J, Zhang C, Jiang L, Ye N, Lu Y, Yang G, Liu E, Peng C, He Z, et al. (2010) Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J Exp Bot 61: 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Ma JF, Matsumoto H (1998) High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol 117: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Zheng C, Hu YT, Jiang T, Liu Y, Dong NY, Yang JL, Zheng SJ (2011) Cadmium-induced oxalate secretion from root apex is associated with cadmium exclusion and resistance in Lycopersicon esulentum. Plant Cell Environ 34: 1055–1064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.