A LAMMER kinase gene generates two types of transcripts, the long OsDR11L and the short OsDR11S, which have opposite functions in rice resistance to a bacterial pathogen.
Abstract
Alternative splicing of genes can increase protein diversity and affect mRNA stability. Genome-wide transcriptome sequencing has demonstrated that alternative splicing occurs in a large number of intron-containing genes of different species. However, despite the phenomenon having been known for decades, it is largely unknown how the alternatively spliced transcripts function differently. Here, we report that two alternatively spliced transcripts of the rice (Oryza sativa) LAMMER kinase gene OsDR11, long OsDR11L and short OsDR11S, play opposite roles in rice resistance against Xanthomonas oryzae pv oryzae (Xoo), which causes the most damaging bacterial disease in rice worldwide. Overexpressing OsDR11S or suppressing OsDR11L in rice enhanced resistance to Xoo, which was accompanied by an accumulation of jasmonic acid (JA) and induced expression of JA signaling genes. In contrast, suppressing OsDR11S was associated with increased susceptibility to Xoo, along with decreased levels of JA and expression of JA signaling genes. The OsDR11S and OsDR11L proteins colocalized in the nucleus. OsDR11L showed autophosphorylation activity in vitro, while OsDR11S did not. In the presence of OsDR11S, autophosphorylation of OsDR11L was inhibited, and overexpression of OsDR11S suppressed OsDR11L expression. OsDR11 appeared to contribute to a minor quantitative trait locus against Xoo. These results suggest that OsDR11L is a negative regulator in rice disease resistance, which may be associated with suppression of JA signaling. The results also suggest that OsDR11S may inhibit the function of OsDR11L at both the transcription and protein kinase activity levels, leading to resistance against Xoo.
Bacterial blight caused by Xanthomonas oryzae pv oryzae (Xoo) is the most damaging bacterial disease in rice (Oryza sativa) worldwide. Resistance to Xoo can be classified into two main genetic categories in rice: qualitative resistance regulated by major disease resistance (MR) genes and quantitative resistance regulated by multiple genes or quantitative trait loci (QTLs; Kou and Wang, 2010; Zhang and Wang, 2013). An MR gene against Xoo may be a disease-resistance gene that initiates effector-triggered immunity, a pattern-recognition receptor gene that initiates pathogen-associated molecular pattern-triggered immunity, or a gene that mediates resistance in a manner that cannot be explained by present immunity models (Zhang and Wang, 2013). Several resistance QTLs contributed by defense-responsive genes that function downstream of MR-initiated defense signaling pathways against Xoo have been identified (Kou and Wang, 2012, 2013). One important group of rice defense-responsive genes encode protein kinases that participate in rice-Xoo interactions (Yuan et al., 2007; Shen et al., 2010, 2011; Geng et al., 2013; Chen et al., 2014).
The LAMMER kinases, named after the EHLAMMERILG motif that is important for kinase activity, are conserved in eukaryotes. These kinases have been reported to be involved in various biological activities and physiological processes, including development, cell differentiation, abiotic stress response, metabolism, and reproduction in yeast, insect, animal, and human (Yun et al., 1994; García-Sacristán et al., 2005; James et al., 2009; Rabinow and Samson, 2010; Rodgers et al., 2010). One of the best known biochemical functions of LAMMER kinases is the regulation of alternative splicing via phosphorylation of Ser/Arg-rich (SR) splicing factors and SR-like proteins or other substrates such as cyclin-dependent kinase-inhibitor Rum1 (Kang et al., 2013; Yu et al., 2013; Zhao et al., 2013).
To date, the LAMMER kinases have only been studied in Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum). The tobacco LAMMER kinase PK12 is induced by phytohormone ethylene in vivo, and it phosphorylates both animal and plant proteins in vitro (Sessa et al., 1996; Savaldi-Goldstein et al., 2000). In Arabidopsis, overexpression of PK12 influences development (Savaldi-Goldstein et al., 2003). Arabidopsis has three members of the LAMMER kinase family (Savaldi-Goldstein et al., 2003). AFC1 can complement a yeast mitogen-activated protein kinase mutant (Bender and Fink, 1994). AFC2, but not AFC1, has the ability to phosphorylate SR splicing factors (Golovkin and Reddy, 1999; Reddy and Shad Ali 2011); however, it is unknown whether plant LAMMER kinases are involved in the regulation of host-pathogen interactions.
Alternative splicing of a gene is a widespread posttranscriptional regulatory mechanism that can increase protein diversity and affect mRNA stability. Genome-wide high-throughput transcriptome sequencing demonstrated that alternative splicing occurs in 95% of intron-containing genes in human and 61% of those in Arabidopsis (Pan et al., 2008; Marquez et al., 2012). Extensive alternative splicing events also occur in rice, with 33% to 48% of intron-containing genes being involved (Lu et al., 2010; Zhang et al., 2010). Despite the phenomenon of alternative splicing having been known for decades, however, the biological roles of alternatively spliced transcripts from a single gene are poorly understood.
A previous study revealed that rice cDNA clone EI39C8 (GenBank accession no. BF108323), which corresponds to OsDR11 and encodes a LAMMER kinase-type protein, is a pathogen-induced defense-responsive gene (Zhou et al., 2002). To evaluate whether OsDR11 is involved in rice disease resistance, we generated OsDR11-transgenic plants and analyzed the kinase activity of OsDR11. The results suggest that OsDR11 can generate two types of transcripts, the long OsDR11L and the short OsDR11S, which are differentially expressed in rice-Xoo interaction. OsDR11L has protein kinase activity and suppresses rice disease resistance; OsDR11S does not possess kinase activity and promotes resistance. The ability of OsDR11L to undergo autophosphorylation is influenced by the presence of OsDR11S in vitro. OsDR11 may contribute to a minor Xoo-resistance QTL, which may operate through OsDR11S inhibition of OsDR11L function.
RESULTS
Transcriptional and Structural Characterization of OsDR11
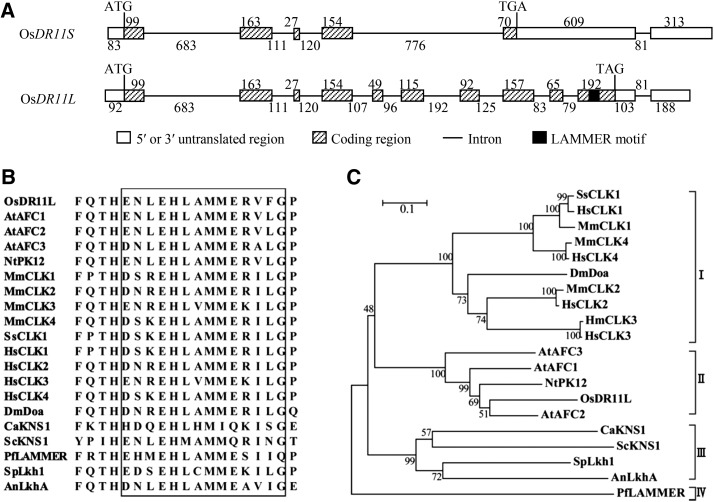
Previous study showed that pathogen infection induced expression of full-length cDNA EI39C8 of OsDR11 from rice variety Minghui 63 (Zhou et al., 2002). EI39C8 is one of the alternatively spliced transcripts of the Os12g27520 locus, according to the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). It contains 1518 nucleotides (nt) and encodes a protein consisting of 170 amino acids. Based on DNA gel blot analysis and genome-wide sequence analysis, Minghui 63, rice variety Zhenshan 97, and genome-sequenced rice variety Nipponbare all harbored a single copy of OsDR11 (Supplemental Fig. S1). We found that the Minghui 63 cDNA library (Chu et al., 2003) also contained another full-length cDNA, EI114F3, from OsDR11. EI114F3 has 1496 nt and encodes a protein consisting of 370 amino acids. In the following text, we refer to EI39C8 and EI114F3 as OsDR11S and OsDR11L, respectively (Fig. 1A). OsDR11L has 11 exons and the translation terminates at exon 10, while OsDR11S has six exons and excludes exons 5, 6, and 7, resulting in a shift in the reading frame and leading to premature termination of translation (Fig. 1A). Both alternative transcripts OsDR11S and OsDR11L were also found in rice varieties Mudanjiang 8 and Zhenshan 97.
Figure 1.
Analysis of OsDR11 and encoded product sequences. A, Structures of OsDR11S and OsDR11L. ATG, Translation start codon; TGA or TAG, translation stop codon. The numbers indicate the nt of each substructure. B, Alignment of multiple amino acid sequences of LAMMER motifs (framed) from different species. Protein accessions: Arabidopsis AtAFC1 (P51566), AtAFC2 (P51567), and AtAFC3 (P51568); N. tabacum NtPK12 (AAC04324); Mus musculus MmCLK1 (NP_001036099), MmCLK2 (NP_001156904), MmCLK3 (NP_031739), and MmCLK4 (NP_031740); Sus scrofa SsCLK1 (EU431337); Homo sapiens HsCLK1 (NP_001155879), HsCLK2 (NP_003984), HsCLK3 (NP_001123500), and HsCLK4 (NP_065717); Drosophila melanogaster DmDOA (P49762); Plasmodium falciparum PfLammer (AAK38173); Candida albicans CaKNS1 (XP_722185); Saccharomyces cerevisiae ScKNS1 (P32350); Schizosaccharomyces pombe SpLkh1 (Q10156); Aspergillus nidulans AnLkhA (XP_658592). C, Phylogenetic relationship of LAMMER family proteins in different species.
The products encoded by OsDR11 were highly homologous with the defined LAMMER kinases. Sequence analysis showed that OsDR11L encodes a LAMMER kinase-type protein based on the conserved LAMMER motif EHLAMMERVFG (Fig. 1B). However, the protein deduced from OsDR11S does not include the entire LAMMER protein kinase domain because of the premature termination of translation. Alternative splicing of LAMMER kinases also exist in other species, such as protein kinase Clk/Sty in mammalian and AFC2 in Arabidopsis, which leads to truncated proteins lacking the LAMMER motif, like OsDR11S (Duncan et al., 1997; Marquez et al., 2012). Sequence comparison of the full-length forms showed that OsDR11L shares 41% to 71% identity with other LAMMER kinases from various organisms. The LAMMER motif is highly conserved in species ranging from yeast to humans and including rice (Fig. 1B). A phylogenic analysis of the LAMMER proteins from different organisms indicated that they form four separate groups (Fig. 1C). All the proteins from mammals and insects are in group I. The proteins from both monocots and dicots fall into group II. OsDR11L is the most closely related to AtAFC2, one of the LAMMER kinases in Arabidopsis (Fig. 1C). Fungal LAMMER proteins are in group III, and group IV only includes LAMMER protein from the malaria parasite Plasmodium falciparum (Fig. 1C).
The results collectively suggest that OsDR11 has at least two alternative transcripts, OsDR11S and OsDR11L, in rice variety Minghui 63. They also suggest that OsDR11L encodes a LAMMER kinase-type protein.
Analysis of the Relationship of OsDR11 and Quantitative Resistance
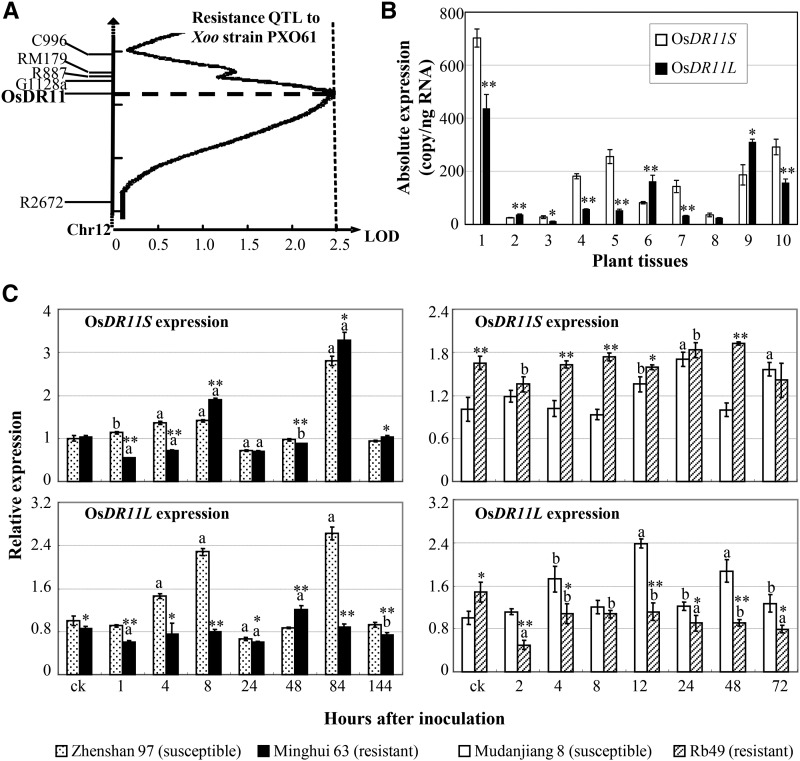
Minghui 63 is resistant to Xoo by both qualitative resistance, conferred by MR genes Xa3/Xa26 and xa25, and quantitative resistance, conferred by resistance QTLs (Sun et al., 2004; Xiang et al., 2006; Hu et al., 2008; Liu et al., 2011). A recombinant inbred line segregation population developed from a cross between susceptible Zhenshan 97 and Minghui 63 has been used to study quantitative disease resistance against Xoo, and several defense-responsive genes contributing to resistance QTLs have been characterized by using this population (Qiu et al., 2007; Ding et al., 2008; Hu et al., 2008; Kou et al., 2010; Deng et al., 2012). We also used this population to comap OsDR11 and resistance QTLs. The mapping showed that OsDR11 was colocalized with the peak of a minor resistance QTL, which had a logarithm of odds of 2.51, on rice chromosome 12 (Fig. 2A; Chen, 2001). This resistance QTL explained 4.82% of the phenotypic variation against Xoo and was contributed by an allele from resistance Minghui 63 in this population.
Figure 2.
The features of OsDR11. A, Colocalization of OsDR11 with a minor disease resistance QTL against Xoo. B, Expression of OsDR11S and OsDR11L during development in rice variety Minghui 63. 1, Callus; 2, shoot at the two-tiller stage; 3, root at the two-tiller stage; 4, leaf at the booting (panicle development) stage; 5, sheath at the booting stage; 6, panicle at the booting stage; 7, flag leaf at the heading stage; 8, panicle at the heading stage; 9, stamen at the flowering stage; 10, pistil at the flowering stage. Asterisks indicate significant difference between two rice lines with the same treatment at **P < 0.01 or *P < 0.05. C, OsDR11S and OsDR11L expression after pathogen infection. Plants were inoculated with Xoo strain PXO61 at the booting stage. The letter “a” or “b” above the bars indicates significant difference between pathogen-infected and noninfected (ck) plants in the same rice line at P < 0.01 or P < 0.05, respectively.
To ascertain the tissue-specific expression characteristics of OsDR11 in Minghui 63, we examined the two transcripts in different tissues collected throughout its life cycle. In general, both OsDR11S and OsDR11R were continuously expressed throughout the life cycle, but they showed different expression patterns (Fig. 2B). OsDR11S expression was higher in callus, root, leaf, sheath, and pistil, while OsDR11L expression was more abundant in shoot, panicle, and stamen (Fig. 2B).
Since the leaf tissue is the major site of Xoo invasion (Kou and Wang, 2013), we next checked the expression of OsDR11S and OsDR11L in leaves of two pairs of rice lines after Xoo infection. The first pair of rice lines was Minghui 63 and Zhenshan 97. Minghui 63 has resistance to some Xoo strains, which is conferred by MR gene Xa3/Xa26, but Zhenshan 97 is highly susceptible to Xoo (Sun et al., 2004; Xiang et al., 2006). The second pair of rice lines was transgenic line Rb49 and japonica rice variety Mudanjiang 8. Rb49, with the genetic background of Mudanjiang 8, carries Xa3/Xa26 and is highly resistant to some Xoo strains, and Mudanjiang 8 is highly susceptible to Xoo (Cao et al., 2007). In susceptible Zhenshan 97, both OsDR11S and OsDR11L had similar expression patterns; they were slightly induced at 4 to 8 h after inoculation, returned to normal level at 48 h, and then reached their peak level at 84 h (Fig. 2C). In contrast, the expression patterns of the two variants in resistant Minghui 63 were totally different. There was an early (8 h) and a late (84 h) increase of OsDR11S expression level after a slight suppression, but no obvious induction peak occurred in OsDR11L expression after Xoo infection (Fig. 2C). The same results were observed in another pair of rice lines, Mudanjiang 8 and Rb49. The two variants had a similar expression pattern in susceptible Mudanjiang 8 after Xoo infection, with expression being slowly induced and reaching a peak at 12 to 24 h (Fig. 2C). In contrast, the two variants had distinct expression patterns in the resistant Rb49. OsDR11S had relatively stable expression in both Mudanjiang 8 and Rb49, but the expression level was significantly higher in the resistant line than in the susceptible line, either with or without pathogen infection. OsDR11L was rapidly suppressed after infection in Rb49, and the level remained significantly lower in the resistant line compared with Mudanjiang 8 after Xoo infection (Fig. 2C).
In summary, quantitative resistance result suggests that OsDR11 may contribute to a minor resistance QTL against Xoo and the distinct expression patterns of OsDR11S and OsDR11L in resistant rice lines after pathogen inoculation suggest that OsDR11S and OsDR11L may be differentially involved in rice-Xoo interaction.
Analysis of the Roles of the Two OsDR11 Variants in Rice-Xoo Interaction
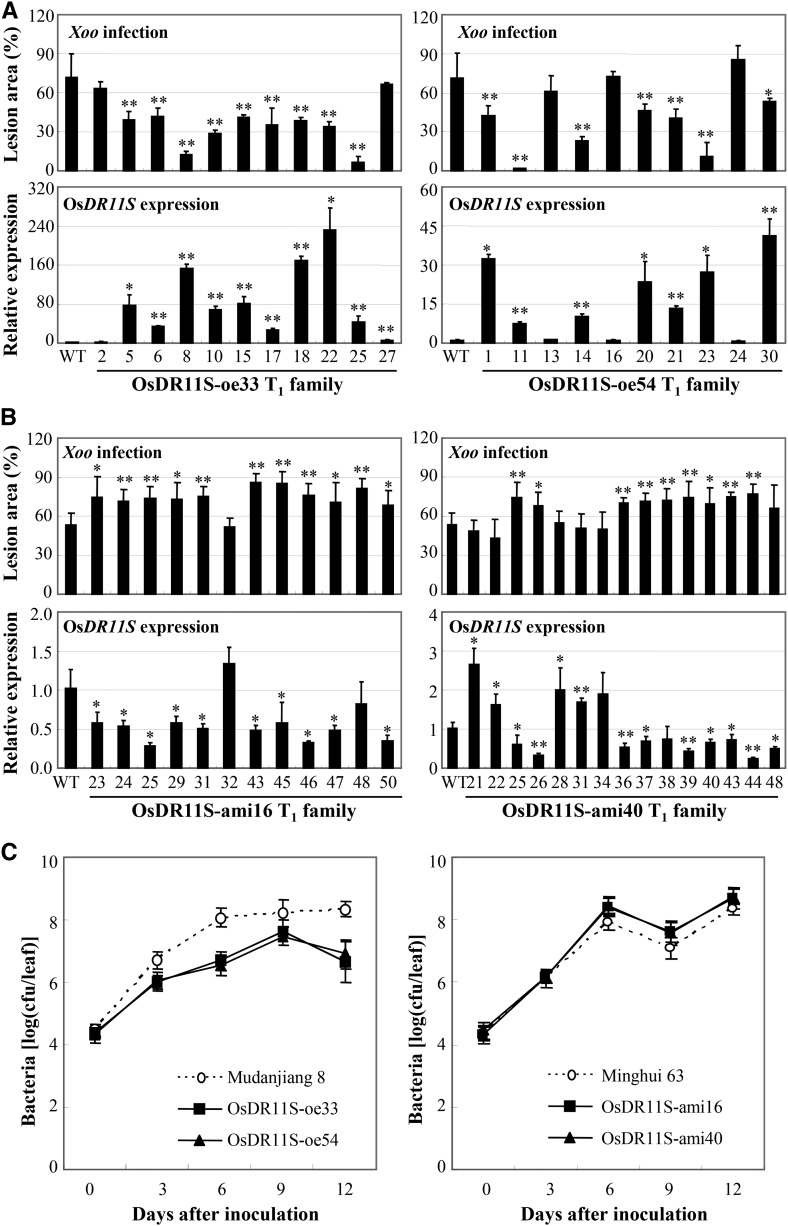
To investigate whether OsDR11S and OsDR11L were involved differently in rice defense to Xoo infection, we manipulated the expression of both OsDR11S and OsDR11L. Fifteen independent positive OsDR11S-overexpressing (oe) plants (also named D35UM8) were obtained. Seven of the 15 T0 OsDR11S-oe plants were significantly more resistant (P < 0.05) to Xoo, with lesion areas ranging from 14% to 39%, compared to 60% for wild-type Mudanjiang 8 (Supplemental Table S1). To confirm that the enhanced resistance of the transgenic plants was due to overexpression of OsDR11S, two T1 families were further analyzed individually for Xoo resistance and their OsDR11S expression level. All T1 plants showing significantly enhanced (P < 0.01) resistance had increased expression of OsDR11S, and wild-type siblings had no significant differences from wild-type plants (Fig. 3A). While the transgenic lines showed increased resistance, it was not directly correlated with the expression levels. Such as the expression levels of plants 18, 22, and 25 did not correlate with resistance levels in the OsDR11S-oe33 T1 family (Fig. 3A), which may suggest that above a certain level of expression of OsDR11S will make no difference. We also generated OsDR11S-Flag-oe plants (D230UM). The phenotypes of these plants were the same as the OsDR11S-oe plants, which did not carry the Flag tag. The reduced lesion area of OsDR11S-Flag-oe plants caused by Xoo significantly negatively correlated with increased expression of OsDR11S (r = −0.606, n = 15, P < 0.05 in OsDR11S-Flag-oe2 family; r = −0.661, n = 14, P < 0.05 in OsDR11S-Flag-oe7 family; Supplemental Fig. S2).
Figure 3.
Modulating OsDR11S expression influenced the resistance to Xoo. Rice plants were inoculated with Xoo strain PXO99 at the booting stage. Data represent mean (three to six replicates from one plant for disease area, three replicates from one plant for gene expression, and three replicates from each type of plant for bacterial growth) ± sd. Asterisks indicate significant difference between transgenic and wild-type (WT) plants at **P < 0.01 or *P < 0.05. A, Enhanced resistance to Xoo was associated with overexpression of OsDR11S in T1 families. WT, Mudanjiang 8. B, Increased susceptibility to Xoo was associated with suppression of OsDR11S in T1 families. WT, Minghui 63. C, Modulating OsDR11S expression influenced the growth of PXO99 in rice leaves. 0 h, Immediately after inoculation; cfu, colony-forming unit.
Fifteen independent positive OsDR11S-suppressing (D205RM) plants were obtained by using an artificial microRNA (ami) strategy. These OsDR11S-ami plants showed a phenotype that was opposite to that of OsDR11S-oe plants. Twelve of the 15 positive T0 plants showed significantly increased susceptibility (P < 0.05) to Xoo, with lesion areas ranging from 62.6% to 78.3%, compared to 56.7% for wild-type Mudanjiang 8 (Supplemental Table S2). Two T1 families were further analyzed for their Xoo resistance and OsDR11S expression level. The increased susceptibility of T1 plants significantly negatively correlated with reduced OsDR11S transcripts (r = −0.651, n = 13, P < 0.05 in OsDR11S-ami16 family; r = −0.852, n = 16, P < 0.001 in OsDR11S-ami40 T1 family; Fig. 3B).
Bacterial growth results reflected the phenotype of the transgenic plants (Fig. 3C). The growth rates of bacteria on the leaves of OsDR11S-oe plants were 7.4- to 20.3-fold lower than on the susceptible wild-type plant leaves at 6 to 12 d after infection. In OsDR11S-ami plants, the rates were 3.7- to 7.7-fold higher than in the wild-type plant at 6 to 12 d after infection (Fig. 3C). These results suggest that OsDR11S may act as a positive regulator in the rice response to Xoo infection.
Nineteen independent positive OsDR11L-oe (D126UM) plants and eight independent positive OsDR11L-Flag-oe (D231UM) plants were generated. However, overexpressing OsDR11L appears having no obvious effect on rice response to Xoo infection (Supplemental Tables S3 and S4; Supplemental Fig. S3).
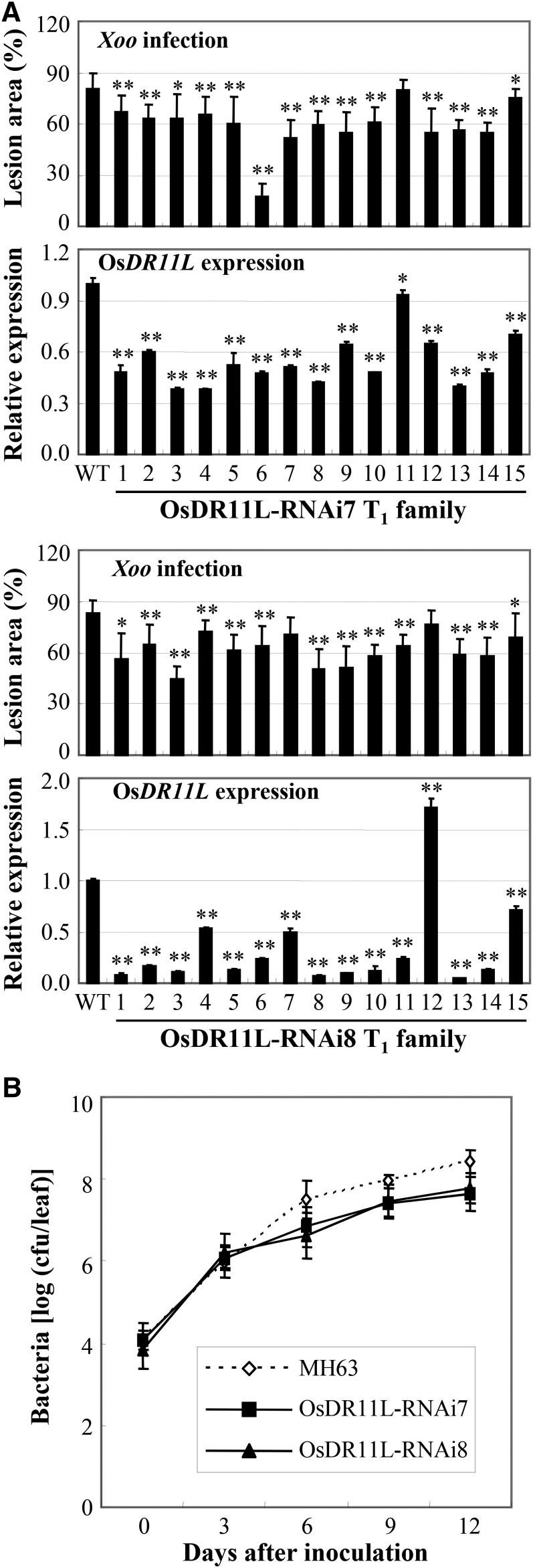
Ten independent positive OsDR11L-suppressing plants (D229RM) were generated in rice variety Minghui 63 by using the RNA interference (RNAi) strategy. Seven of the 10 T0 OsDR11L-RNAi plants were significantly less susceptible to Xoo (P < 0.05), with lesion areas ranging from 64.3% to 74.3%, compared to 97.2% for wild-type Minghui 63 (Supplemental Table S5). Two T1 families were further analyzed for their Xoo resistance and OsDR11L expression level. The reduced susceptibility of T1 plants significantly correlated with reduced OsDR11L transcription (r = 0.528, n = 16, P < 0.05 in OsDR11L-RNAi7 family and r = 0.773, n = 16, P < 0.01 in OsDR11L-RNAi8 family; Fig. 4A). The bacteria growth rate in OsDR11L-RNAi plants was 6.0- to 11.9-fold lower than in wild-type Minghui 63 at 6 to 12 d after infection (Fig. 4B). These results suggest that OsDR11L may act as a negative regulator in the rice response to Xoo infection.
Figure 4.
Suppressing OsDR11L expression enhanced the resistance to Xoo. Rice plants were inoculated with Xoo strain PXO99 at the booting stage. Data represent mean (three to six replicates from one plant for disease area, three replicates from one plant for gene expression, and three replicates from each type of plant for bacterial growth) ± sd. Asterisks indicate significant difference between transgenic and wild-type (WT) plants at **P < 0.01 or *P < 0.05. A, Enhanced resistance to Xoo cosegregated with suppression of OsDR11L in T1 families. B, Suppressing OsDR11L expression reduced the growth of Xoo in rice leaves. 0 h, Immediately after inoculation; cfu, colony-forming unit.
Analysis of OsDR11-Related Defense Pathway in Rice-Xoo Interaction
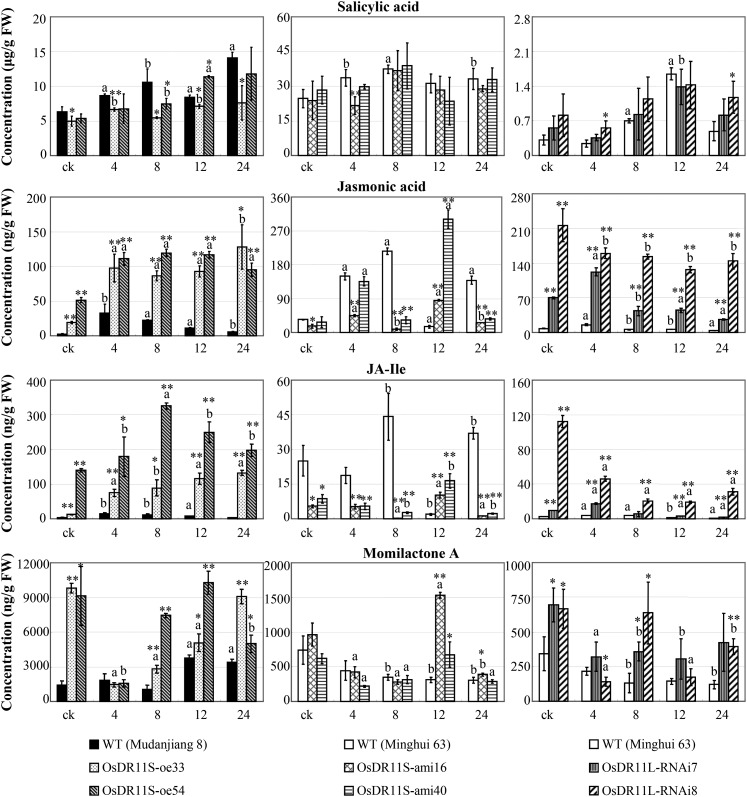
Salicylic acid (SA)-dependent signaling and JA-dependent signaling are important pathways in plant resistance to pathogens. To identify a signaling pathway in which OsDR11S and OsDR11L were possibly involved, we quantified the concentrations of the endogenous phytohormones SA, JA, and JA-Ile, which is an active metabolite of JA and involved in biotic challenges (Liu et al., 2012), in rice leaves. The SA level was slightly lower in OsDR11S-oe plants compared to wild-type plants, and it generally showed no obvious difference in OsDR11S-ami and OsDR11L-RNAi plants compared to their corresponding wild-type plants (Fig. 5). Before or after Xoo infection, the JA and JA-Ile levels were both significantly higher (P < 0.05) in OsDR11S-oe plants and OsDR11L-RNAi plants than in wild-type Mudanjiang 8 and Minghui 63, respectively. In contrast, the JA and JA-Ile levels were significantly lower (P < 0.05) in OsDR11S-ami plants than in wild-type Minghui 63 at most of the time points examined (Fig. 5).
Figure 5.
Modulating OsDR11S and OsDR11L expression influenced the levels of phytohormones and phytoalexin. Rice plants were inoculated with Xoo strain PXO99 at the booting stage. Each data point represents ten replicates of leaf samples from ten different individual plants in one line. Bars represent mean (three replicates) ± sd. The letter “a” or “b” above the bars indicates significant difference between pathogen-infected and noninfected (ck) plants in the same rice line at P < 0.01 or P < 0.05, respectively. Asterisks indicate significant difference between transgenic and wild-type (WT) plants at **P < 0.01 or *P < 0.05.
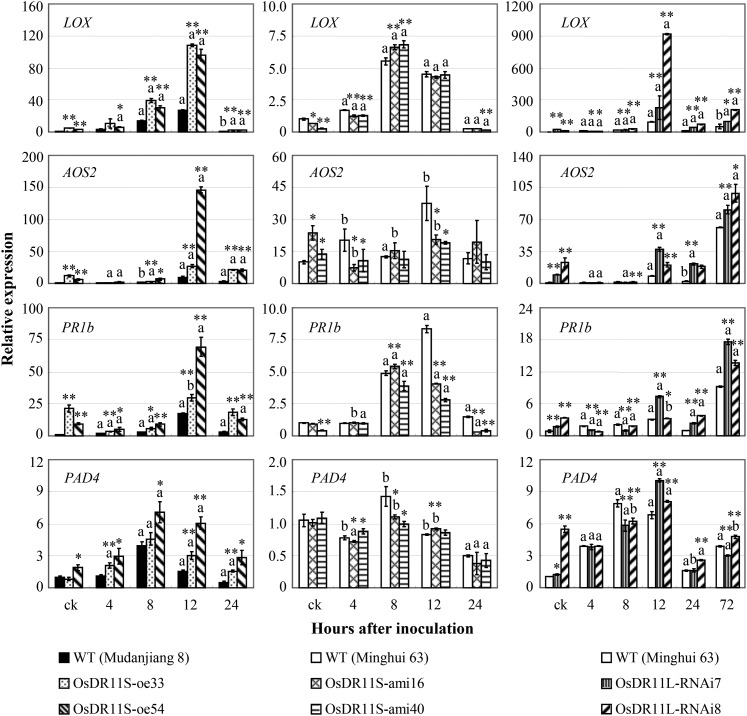
Consistent with the altered concentration of JA and JA-Ile in different transgenic plants, the expression of a set of pathogen-induced defense-responsive genes was also changed in these transgenic plants. Lipoxygenase (D14000) and allene oxide synthase2 (AY062258) are involved in JA synthesis (Lyons et al., 2013), and the expression of basic pathogenesis-related protein1 (U89895) and phytoalexin-deficient4 (AK243523) have been reported to be influenced by both SA and JA signaling (Qiu et al., 2007; Tao et al., 2009; Shen et al., 2011; Ke et al., 2014). Xoo infection influenced the expression of all four genes (Fig. 6). The expression of lipoxygenase, allene oxide synthase2, and basic pathogenesis-related protein1 in OsDR11S-oe and OsDR11L-RNAi plants was significantly increased compared to their corresponding wild-type plants both before and after Xoo infection. In contrast, the expression levels of the three genes were significantly lower or not significantly different in OsDR11S-ami plants compared to wild-type plants at most of the time points examined. The expression level of phytoalexin-deficient4 was slightly higher in OsDR11S-oe and OsDR11L-RNAi plants compared to wild-type plants, but it generally showed no obvious difference between OsDR11S-ami and wild-type plants.
Figure 6.
Modulating the expression of OsDR11S and OsDR11L influenced the expression of other defense-responsive genes analyzed by quantitative reverse transcription PCR. Rice plants were inoculated with Xoo strain PXO99 at the booting stage. Each data point represents ten replicates of leaf samples from ten different individual plants in one line. Bars represent mean (three replicates) ± sd. The letter “a” or “b” above the bars indicates significant difference between pathogen-infected and noninfected (ck) plants in the same rice line at P < 0.01 or P < 0.05, respectively. Asterisks indicate significant difference between transgenic and wild-type (WT) plants at **P < 0.01 or *P < 0.05.
Phytoalexins serve as antibiotics in response to biotic stresses. The terpenoid momilactone A is an important phytoalexin in rice (Liu et al., 2012). The momilactone A was significantly higher in OsDR11S-oe and OsDR11L-RNAi plants both before and after Xoo infection, but its level in OsDR11S-ami plants did not differ from the wild-type control at most of the time points examined (Fig. 5).
These results suggest that OsDR11S-promoted disease resistance may be associated with activation of the JA-dependent pathway and accumulation of phytoalexin momilactone A, while OsDR11L-suppressed disease resistance may be associated with suppression of both JA-dependent signaling and accumulation of momilactone A.
Analysis of the Biochemical Features of the Two Alternatively Spliced Forms of OsDR11
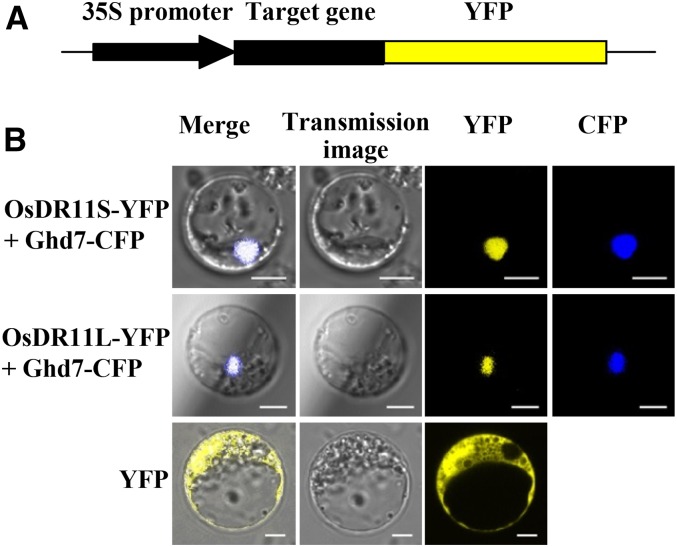
To investigate the possible biochemical function of OsDR11S and OsDR11L proteins, their subcellular localizations were determined by fusing their respective coding regions in frame to the yellow fluorescence protein (YFP) reporter gene under the control of the 35S promoter. The fusion genes were transiently expressed in rice protoplast cells (Fig. 7). Similar to other members of the LAMMER family in other species, the OsDR11 proteins contain a putative nuclear localization signal at the amino terminus. So, we cotransformed the target gene with Ghd7-cyan fluorescence protein (CFP) gene. Rice Ghd7 has been used as a marker since it was reported as a transcription factor localized in the nucleus (Xue et al., 2008). The results showed that the OsDR11S-YFP and OsDR11L-YFP fusion proteins, respectively, colocalized with the marker in nucleus, suggesting that OsDR11S and OsDR11L may function in the nucleus.
Figure 7.
Subcellular localization of OsDR11. Ghd7-CFP, nuclear localization marker. A, The schematic diagram indicates the construct used for protein location. B, OsDR11S and OsDR11L located in rice nuclear.
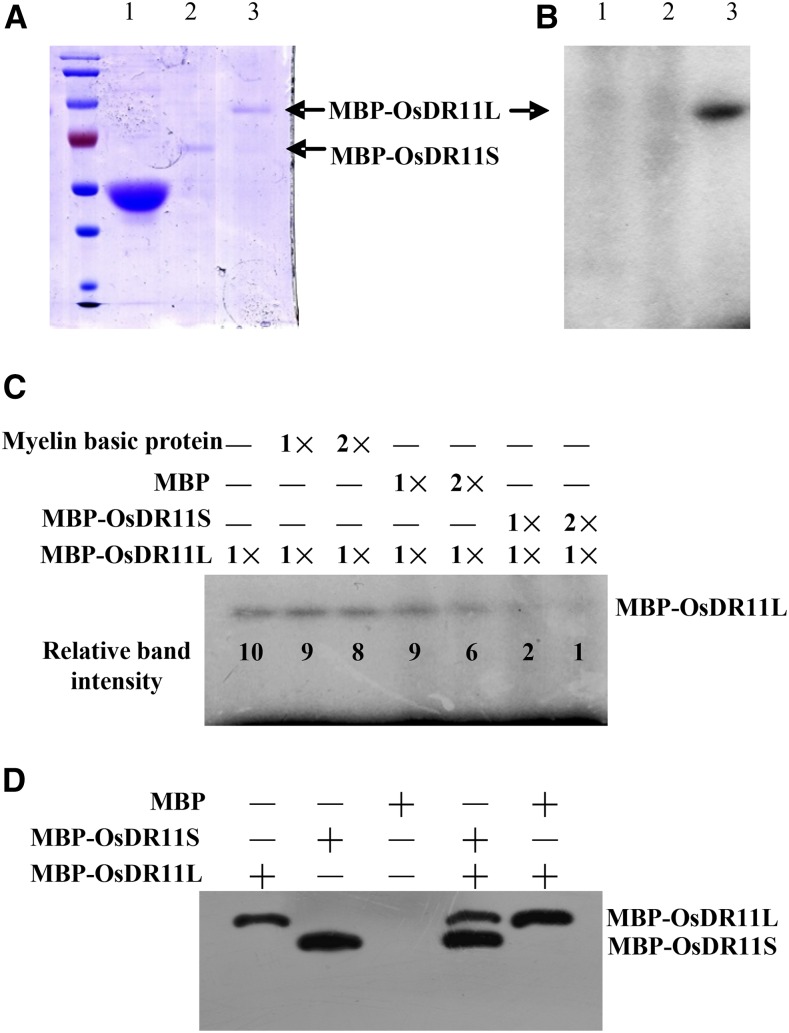
To determine whether OsDR11S and OsDR11L encode functional protein kinases, the coding regions of OsDR11S and OsDR11L were fused in frame to the maltose-binding protein (MBP) and expressed and purified in bacteria (Fig. 8A). The OsDR11S-MBP and OsDR11L-MBP recombination proteins had the expected molecular masses of 60 kD and 80 kD (Fig. 8A). The in vitro kinase assay showed that the OsDR11L-MBP had autophosphorylation activity, while OsDR11S-MBP and MBP did not (Fig. 8B). Both OsDR11S-MBP and OsDR11L-MBP were unable to phosphorylate the myelin basic protein as substrate (Fig. 8C). OsDR11L could not phosphorylate OsDR11S as substrate, but an increased OsDR11S concentration resulted in decreased autophosphorylation of OsDR11L (Fig. 8C). Next, we used western blot analysis to confirm that the inactivation of OsDR11L kinase activity in the presence of OsDR11S was not due to protein degradation (Fig. 8D). These results suggested that OsDR11S can inactivate or inhibit the kinase activity of OsDR11L.
Figure 8.
The ability of OsDR11L to undergo autophosphorylation was influenced by the presence of OsDR11S. A, Purification of proteins. 1, MBP; 2, recombination protein MBP-OsDR11S; 3, recombination protein MBP-OsDR11L. B, Autophosphorylation assay in vitro. 1, MBP; 2, MBP-OsDR11S; 3, MBP-OsDR11L. C, The ability of autophosphorylation of OsDR11L was influenced by the presence of OsDR11S in vitro. The numbers under each band are the relative ratios of the OsDR11L kinase signals. D, The proteins detected by western blotting using anti-OsDR11 antibody.
Analysis of the Influence of OsDR11S on the Transcriptional Expression of OsDR11L
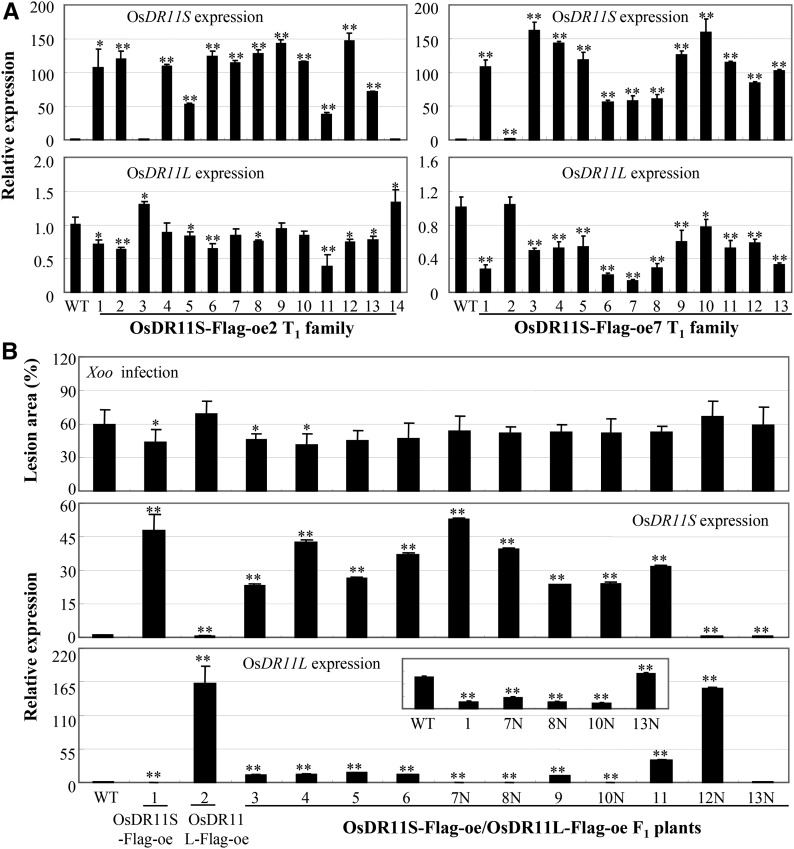
To study whether OsDR11S influenced OsDR11L expression in vivo, we further analyzed the expression level of OsDR11L in OsDR11S-oe plants. Two OsDR11S-oe T1 families were analyzed individually for OsDR11L expression. Most of T1 plants overexpressing OsDR11S showed significantly suppressed expression (P < 0.05) of OsDR11L compared to wild-type plants (Fig. 9A). We further analyzed the expression levels of OsDR11S and OsDR11L in F1 plants that were generated from a cross between the OsDR11S-Flag-oe plants and OsDR11L-Flag-oe plants. The expression levels of OsDR11L in F1 plants that had high expression levels of OsDR11S were lower compared to the levels of OsDR11L in their parent OsDR11L-oe line, and the expression levels of OsDR11S and OsDR11L were significantly negatively correlated in these plants (r = −0.545, n = 14, P < 0.05) (Fig. 9B). The expression levels of OsDR11L in six positive F1 plants (3 to 6, 9, and 11) were only 7% to 23% of those in OsDR11L-Flag-oe plants, while the expression levels of OsDR11S in these positive F1 plants were 49% to 89% of those in OsDR11S-Flag-oe plants. Consistent with the expression features of OsDR11S and OsDR11L, the F1 and F2 plants overexpressing both OsDR11S and OsDR11L had lesion areas similar to those of the OsDR11S-Flag-oe line after Xoo infection (Fig. 9B; Supplemental Fig. S4). The expression levels of OsDR11S and OsDR11L significantly associated with lesion area caused by Xoo (r2 = 0.717, P = 0.001).
Figure 9.
Overexpressing OsDR11S suppressed the expression of OsDR11L. WT, Mudanjiang 8. A, The expression level of OsDR11L in OsDR11S-Flag-oe T1 plants. B, The phenotype to Xoo and expression of OsDR11L and OsDR11S of the F1 plants generated from the cross between OsDR11S-Flag-oe and OsDR11L-Flag-oe lines. 7N, 8N, 10N, 12N, and 13N, Negative F1 plants. Asterisks indicate significant difference between transgenic and wild-type plants at **P < 0.01 or *P < 0.05.
The coding sequences of OsDR11S and OsDR11L were further analyzed. The first 443 nt of OsDR11S were identical to those of OsDR11L, and the last 70 nt of OsDR11S were identical to nt 700–769 of OsDR11L. There seemed to be a dynamic balance between the expression of OsDR11S and OsDR11L. When expression of OsDR11S was high, the transcription of OsDR11L may have been reduced by a negative regulation mechanism. These results suggest that OsDR11S can influence OsDR11L transcript abundance in vivo.
DISCUSSION
Although LAMMER proteins have been well studied in some species, such as in mice, humans, Arabidopsis, and tobacco, and they are reported to have functions in differentiation and development (Nikolakaki et al., 2002; Talevich et al., 2011; Yun et al., 1994), the exact role of LAMMER kinase in rice is poorly understood. Here, we provide evidence that LAMMER protein kinase gene OsDR11 is involved in rice-Xoo interaction.
Two Variants of OsDR11 Play Opposite Roles in Rice-Xoo Interaction
Alternative splicing of mRNAs is an important biological phenomenon of gene transcription, although how the alternatively spliced transcripts function differently in a given genome is largely unknown. Alternative splicing of LAMMER kinase gene has also been reported in Arabidopsis, but the functions of the premature termination codon transcripts generated through alternative splicing are unknown (Marquez et al., 2012). LAMMER kinases appear to play vital roles in various physiological processes, but their roles in plant defense are not yet known. The present results suggest that the two alternative spliced transcripts of OsDR11 are involved in rice-Xoo interaction but function differently.
Many protein kinases function as either positive or negative components in disease resistance against a pathogen species (Matsui et al., 2010; Christiansen et al., 2011; Shen et al., 2011). However, a protein kinase serving as both a positive and a negative regulator in defense responses to the same pathogen species has been reported (Shen et al., 2010). The present results on OsDR11S and OsDR11L in the context of rice-Xoo interaction provide another example that a protein kinase gene can play opposite roles via alternatively spliced transcripts. This hypothesis is supported by the following evidence. First, the OsDR11S expression level was significantly higher in the resistant line than in the susceptible line after Xoo infection, but OsDR11L was more abundant in the susceptible line after Xoo infection (Fig. 2C). Second, overexpressing OsDR11S and suppressing OsDR11L increased rice resistance to Xoo, and suppressing OsDR11S increased susceptibility (Figs. 3 and 4). These results suggest that both variants of OsDR11 have roles in rice-Xoo interaction, with OsDR11S functioning as a positive regulator, while OsDR11L functions as a negative one.
OsDR11-Mediated Disease Resistance May Be Associated with Activation of the JA-Dependent Pathway and Accumulation of Phytoalexin Momilactone A
SA and JA are important phytohormone molecules in plant-pathogen interactions. SA is generally involved in the activation of defense responses against biotrophic and hemibiotrophic pathogens, and JA is usually associated with defense against necrotrophic pathogens (Bari and Jones, 2009). Xoo is a biotrophic pathogen (Li and Wang, 2013). For example, the enhanced Xoo resistance of OsWRKY13-oe rice plants and OsDR10-suppressing rice plants was associated with activation of the SA-dependent pathway and suppression of the JA-dependent pathway (Qiu et al., 2007, 2008; Xiao et al., 2009). However, the present results suggest that OsDR11-mediated rice resistance to Xoo is accompanied by activation of the JA-associated pathway, but not the SA pathway. This inference is supported by the following evidence. First, the enhanced resistance of OsDR11S-oe plants and OsDR11L-suppressing plants was associated with increased endogenous levels of JA and its active metabolite JA-Ile, but not SA (Fig. 5). Second, although Xoo is a biotrophic pathogen, rice resistance against Xoo appears to employ complex regulatory mechanisms to trigger effective defense responses in natural environments. For example, the enhanced Xoo resistance in OsMPK6-oe rice plants and OsEDR1-knockout rice plants was associated with activation of both SA- and JA-associated pathways (Shen et al., 2010, 2011). The Xoo resistance mediated by rice OsWRKY45-2 or C3H12 was associated with JA, but not SA (Tao et al., 2009; Deng et al., 2012). Furthermore, exogenous application of JA can enhance the resistance of rice to Xoo (Yamada et al., 2012; Ke et al., 2014).
Phytoalexins contribute to plant basal immunity and accumulate around infection sites soon after pathogen attack (González-Lamothe et al., 2009). Our results also show that the endogenous accumulation of momilactone A was significantly higher in OsDR11S-oe plants and OsDR11L-suppressing plants before and after Xoo infection, which was associated with resistance against Xoo infection and accumulation of JA (Fig. 5). Previous studies showed that the accumulation level of momilactone A can be induced by infection of rice pathogens such as Xoo and fungal Magnaporthe oryzae (Liu et al., 2012; Riemann et al., 2013). Momilactone A can inhibit the growth of M. oryzae, and a reduced level of momilactone A is associated with compromised resistance against Xoo (Hasegawa et al., 2010; Ke et al., 2014). Exogenous treatment with JA increased accumulation of momilactone A (Nojiri et al., 1996; Kato-Noguchi, 2011). Thus, this study suggests that OsDR11S-mediated rice resistance may also be associated with the accumulation of momilactone A, which may be positively regulated by JA signaling.
OsDR11S May Regulate the Function of OsDR11L
The LAMMER motif of LAMMER kinases is important for kinase activity (Savaldi-Goldstein et al., 2000). This may explain why OsDR11L showed autophosphorylation ability while OsDR11S, which lacks LAMMER motif, did not (Fig. 8). Although OsDR11S does not have autophosphorylation ability, our results suggest that OsDR11S appears to be a dominant and functional variant. This inference is supported by evidence that the F1 and F2 plants overexpressing both OsDR11S and OsDR11L showed the phenotype of OsDR11S-oe plants in response to Xoo infection.
OsDR11L negatively influenced rice resistance to Xoo. The fine balance of OsDR11S with respect to OsDR11L is an important feature in OsDR11 gene regulation. This hypothesis is supported by the following evidence. First, alternative splicing plays a role in the functional complexity of rice, and 59% of the alternative splicing events in rice were organ specific (Lu et al., 2010). In our results, both alternatively spliced variants are continuously expressed throughout the rice life cycle (Fig. 2B) and the expression level of both variants is influenced by Xoo infection (Fig. 2C), which suggest that alternative splicing of OsDR11 is needed throughout rice development and in response to pathogen infection. Second, because of the pretermination codon by alternative splicing, OsDR11S likely regulates the expression level of OsDR11L by influencing the alternative splicing of OsDR11. Increasing evidence shows that alternative splicing coupled to mRNA degradation not only removes aberrant mRNAs but also acts as a mechanism to regulate gene expression, including the expression of genes playing essential roles in plant stress adaptation (Dubrovina et al., 2013). In this study, the expression of OsDR11L was reduced in OsDR11S-oe plants and in F1 plants generated from the cross between OsDR11S-oe and OsDR11L-oe plants (Fig. 9B), which suggests that overexpression of OsDR11S can regulate the alternative splicing of OsDR11. These results were consistent with LAMMER kinase studies in other species. For example, overexpressing Clk/Sty in mammalian cells leads to a switch in the alternative splice site selection of miniature gene constructs (Duncan et al., 1997), and PK12 LAMMER kinase modulates alternative splicing in plants in a dose-dependent manner (Savaldi-Goldstein et al., 2003). However, the mechanism of how overexpressing OsDR11S can reduce expression level of OsDR11L needs further studies.
OsDR11S may also regulate OsDR11L function by influencing its kinase activity. This inference can be supported by the fact that autophosphorylation ability of OsDR11L was inhibited by the presence of OsDR11S, not the degradation of OsDR11L (Fig. 8). Both OsDR11S and OsDR11L were located in the nucleus. This colocalization provides the foundation for OsDR11S being able to affect OsDR11L. However, physical interaction of the two types of proteins was not detected by either coimmunoprecipitation assay in vivo or biomolecular fluorescence complementation assay in rice protoplasts. Thus, further study is needed to elucidate how OsDR11S direct influences the kinase function of OsDR11L.
CONCLUSION
Rice LAMMER kinase gene OsDR11 is alternatively spliced in rice-Xoo interaction. The two transcripts of OsDR11, OsDR11L, and OsDR11S, play opposite roles, as negative and positive regulators, respectively, in rice resistance to Xoo, which is associated with the JA-dependent pathway. OsDR11S appears to suppress the negative function of OsDR11L in rice-Xoo interaction by both transcriptionally suppressing OsDR11L and inhibiting the kinase activity of OsDR11L. In addition, OsDR11 may contribute to a minor resistance QTL against Xoo through the function of OsDR11S. Elucidating the molecular mechanisms of the two alternatively spliced transcripts provide important insights into the regulatory mechanisms of alternatively spliced transcripts in rice.
MATERIALS AND METHODS
Gene Isolation and Structural Analysis
To isolate the OsDR11 gene, two cDNA fragments of OsDR11, EI39C8 and EI114F3, from rice (Oryza sativa) variety Minghui 63 (Zhang et al., 2005) were used to screen the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) using the BLAST program (Altschul et al., 1997). A series of PCR primers (Supplemental Table S6) were designed for sequencing the genomic DNA of OsDR11 from Minghui 63. The analysis indicated that cDNA sequences EI39C8 and EI114F3 from Minghui 63 were highly homologous to gene locus LOC_Os12g27520 and harbored the entire coding sequence. These two variants also contained the 3′ untranslated region of OsDR11. The structures of OsDR11 were determined by comparatively sequencing the genomic DNA and cDNA.
QTL Analysis and DNA Gel Blot Analysis
A rice recombinant inbred line population was used to analyze comapping of OsDR11 and resistance QTLs. This population consisted of 241 lines developed from a cross between susceptible indica Zhenshan 97 (O. sativa ssp. indica) and resistant indica Minghui 63 (O. sativa ssp. indica) by single-seed descent. Minghui 63 carried MR genes Xa3/Xa26 and xa25 for Xoo resistance (Sun et al., 2004; Xiang et al., 2006; Liu et al., 2011). A molecular linkage map containing 221 markers and covering the whole rice genome was developed with this population (Xing et al., 2002). This population had been used to study the quantitative disease resistance for four strains (Chinese strains JL691 and KS-1-21 and Philippines strains PXO61 and PXO339) of Xoo (Chen, 2001; Hu et al., 2008). OsDR11 was mapped into this population using cDNA EI39C8. Mapmaker 3.0 was used for linkage analysis and QTL map for composite interval mapping at a threshold of logarithm of odds 2.5 (Li et al., 2001; Wang et al., 1999).
Rice total DNA was digested with restriction enzymes, separated by electrophoresis on 0.8% agarose gels, and blotted onto nylon membranes. DNA gel blot analysis was conducted as previously described, using the cDNA EI39C8 as probe for hybridization (Xiao et al., 2009).
Pathogen Inoculation
To evaluate bacterial blight disease, plants were inoculated with Xoo at the booting (panicle development) stage by the leaf-clipping method (Chen et al., 2002), and disease was scored by measuring the percentage of the lesion area (lesion length/leaf length) at 11 to 15 d after inoculation. Xoo growth rate in rice leaves was determined by counting colony-forming units (Sun et al., 2004).
Gene Expression Analyses
Two-centimeter leaf fragments next to bacterial infection sites were used for RNA isolation. Quantitative reverse transcription PCR was conducted as described previously (Qiu et al., 2007). To compare the expression levels of OsDR11S and OsDR11L during development, the known concentrations of plasmids EI39C8 and EI114F3 harboring OsDR11S and OsDR11L, respectively, were amplified as standards, and linear regression models were established to calculate the absolute expression of OsDR11S and OsDR11L in rice tissues. To analyze the relative expression of a gene, the expression level of the rice actin gene was used as an internal control, and the expression level relative to the control is presented. The relative expression level of a gene after treatment can be compared only to itself in corresponding control. To identify the two alternative forms of OsDR11, gene-specific primers 83J4-2F/83J4-2R were used to detect the expression of OsDR11L (Supplemental Table S6). For detecting the expression of OsDR11S, primers 39C8-5F/39C8-6R were used in wide-type, OsDR11S-oe, and OsDR11S-suppressing plants, and primers 39C8-6F/39C8-7R were used in OsDR11S-Flag-oe plants (Supplemental Table S6).
Transformation
The overexpression constructs of OsDR11S and OsDR11L were created by cutting the fragment harboring the full-length coding region from cDNA clones EI39C8 and EI114F3 by using restriction enzymes KpnI and BamHI, respectively, and ligating them into the transformation vector pU1301, which contained a maize ubiquitin gene promoter (Cao et al., 2007). To suppress OsDR11S, an artificial miRNA construct was generated according to a previous description (Warthmann et al., 2008). To construct an RNAi vector of OsDR11L, a 256-nt fragment amplified from Minghui 63 cDNA by using gene-specific primers (Supplemental Table S6) was inserted into the pDS1301 vector (Yuan et al., 2007). The constructs were introduced into Agrobacterium tumefaciens EHA105 by electroporation. Agrobacterium-mediated transformation was performed according to a published protocol (Lin and Zhang, 2005). The overexpressing construct was transferred into japonica rice variety Mudanjiang 8, and the suppressing constructs were transferred into indica rice variety Minghui 63. Primers for positive transplant test are listed in Supplemental Table S6.
Quantification of Phytohormones and Phytoalexin
Leaf fragments about 3 cm long next to the inoculation sites were used for analysis. Samples were prepared and quantified using the UFLC-ESI-MS/MS system as reported previously (Liu et al., 2012).
Subcellular Localization
To produce the constructs for assaying subcellular localization, the coding regions of OsDR11S and OsDR11L were amplified by PCR using gene-specific primers (Supplemental Table S6) and inserted into pM999-YFP for fusion with the reporter gene. Rice Ghd7 has been used as a marker since it was reported as a transcription factor localized in the nucleus (Xue et al., 2008). The 35S:Ghd7-CFP was kindly provided by Dr. Lei Wang of Huazhong Agricultural University. The constructs of OsDR11S-YFP or OsDR11L-YFP and Ghd7-CFP were cotransformed and transiently expressed in rice protoplasts according to a previous description (Yang et al., 2012).
Protein Expression and in Vitro Kinase Assay
The entire coding regions of OsDR11S and OsDR11L were amplified from cDNA clones EI39C8 and EI114F3, respectively, by PCR using gene-specific primers (Supplemental Table S6). After BamHI and BamHI/PstI digestion, respectively, the PCR products of OsDR11S and OsDR11L were cloned into the pMAL-c2x vector, which harbors an MBP gene at the 5′ end of the multiple cloning sites (New England Biolabs). The expression plasmid was transformed into Escherichia coli strain BL21 (DE3). The MBP-tagged proteins were purified from E. coli using mylase resin (New England Biolabs).
For the autophosphorylation kinase activity assay, 1.2 μg of purified recombinant proteins, which were quantified using a Bradford assay (2-D Quant Kit; GE Healthcare), was incubated with kinase buffer containing 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm MnCl2, and 0.3 μCi γ-32P-ATP in a final volume of 20 μL. The mixture was incubated for 30 min at room temperature, and the reactions were terminated by the addition of protein sample buffer. Proteins were fractionated on a 12% SDS-PAGE gel and exposed to film. The kinase signals were calculated using Quantity one software (Bio-Rad, version 4.6).
Western blotting was performed as described previously (Yuan et al., 2009). The anti-OsDR11 antibody, which was custom synthesized by Abmart against peptide PRFASPPLRE (60th to 69th amino acid of both OsDR11L and OsDR11S), was used for protein expression analysis.
Statistical Analysis
The significant differences between control and treatment of the samples were analyzed by the pairwise t test in Excel (Microsoft). The correlation analysis between lesion area and gene expression level was performed using the CORREL analysis in Excel. R-squared value of the expression levels of OsDR11S and OsDR11L and lesion areas in OsDR11S-Flag-oe/OsDR11L-Flag-oe F1 plants was analyzed using multiple linear regression analysis in SPSS (IBM SPSS Statistics, Version 19.0; IBM [released 2010]). A two-tailed P < 0.05 was considered statistically significant.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KR107935 (OsDR11).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. DNA gel blot analysis of OsDR11 gene in rice varieties Minghui 63 (MH) and Zhenshan 97 (ZS).
Supplemental Figure S2. Enhanced resistance to Xoo was associated with overexpression of OsDR11S in two T1 families.
Supplemental Figure S3. The response of OsDR11L-oe (A) and OsDR11L-Flag-oe (B) plants to infection by Xoo strain PXO99 was not closely associated with overexpression of OsDR11L, respectively.
Supplemental Figure S4. The F2 plants overexpressing both OsDR11S-Flag and OsDR11L-Flag showed the phenotype of OsDR11S-Flag-oe line in response to Xoo infection.
Supplemental Table S1. The phenotype of OsDR11S-oe T0 plants after inoculation with Xoo strain PXO99.
Supplemental Table S2. The phenotype of OsDR11S-suppressing T0 plants after inoculation with Xoo strain PXO99.
Supplemental Table S3. The phenotype of OsDR11L-oe T0 plants inoculated with Xoo strain PXO99.
Supplemental Table S4. The phenotype of OsDR11L-Flag-oe T0 plants inoculated with Xoo strain PXO99.
Supplemental Table S5. The phenotype of OsDR11L-suppressing T0 plants after inoculation with Xoo strain PXO99.
Supplemental Table S6. Primers used for PCR amplification.
Supplementary Material
Glossary
- JA
jasmonic acid
- QTL
quantitative trait locus
- SR
Ser/Arg-rich
- SA
salicylic acid
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (31330062), and the Special Key Project by the Ministry of Science and Technology of China (2016YFD0100903).
Articles can be viewed without a subscription.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR (1994) AFC1, a LAMMER kinase from Arabidopsis thaliana, activates STE12-dependent processes in yeast. Proc Natl Acad Sci USA 91: 12105–12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S (2007) The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. (2001) Population structure of Pyricularia grisea from central and southern China and comparative mapping of QTL for blast- and bacterial blight-resistance in rice and barley. Doctoral thesis. Huazhong Agriculture University, Wuhan, China
- Chen H, Wang S, Zhang Q (2002) New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology 92: 750–754 [DOI] [PubMed] [Google Scholar]
- Chen X, Zuo S, Schwessinger B, Chern M, Canlas PE, Ruan D, Zhou X, Wang J, Daudi A, Petzold CJ, et al. (2014) An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant 7: 874–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen KM, Gu Y, Rodibaugh N, Innes RW (2011) Negative regulation of defence signalling pathways by the EDR1 protein kinase. Mol Plant Pathol 12: 746–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZH, Peng KM, Zhang LD, Zhou B, Wei J, Wang S (2003) Construction and characterization of a normalized whole-life-cycle cDNA library of rice. Chin Sci Bull 48: 229–235 [Google Scholar]
- Deng H, Liu H, Li X, Xiao J, Wang S (2012) A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol 158: 876–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovina AS, Kiselev KV, Zhuravlev YN (2013) The role of canonical and noncanonical pre-mRNA splicing in plant stress responses. BioMed Res Int 2013: 264314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Bell JC (1997) In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol 17: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sacristán A, Fernández-Nestosa MJ, Hernández P, Schvartzman JB, Krimer DB (2005) Protein kinase clk/STY is differentially regulated during erythroleukemia cell differentiation: a bias toward the skipped splice variant characterizes postcommitment stages. Cell Res 15: 495–503 [DOI] [PubMed] [Google Scholar]
- Geng S, Li A, Tang L, Yin L, Wu L, Lei C, Guo X, Zhang X, Jiang G, Zhai W, et al. (2013) TaCPK2-A, a calcium-dependent protein kinase gene that is required for wheat powdery mildew resistance enhances bacterial blight resistance in transgenic rice. J Exp Bot 64: 3125–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS (1999) An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J Biol Chem 274: 36428–36438 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Mitchell G, Gattuso M, Diarra MS, Malouin F, Bouarab K (2009) Plant antimicrobial agents and their effects on plant and human pathogens. Int J Mol Sci 10: 3400–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Mitsuhara I, Seo S, Imai T, Koga J, Okada K, Yamane H, Ohashi Y (2010) Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol Plant Microbe Interact 23: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Hu KM, Qiu DY, Shen XL, Li XH, Wang SP (2008) Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant 1: 786–793 [DOI] [PubMed] [Google Scholar]
- James BP, Staatz WD, Wilkinson ST, Meuillet E, Powis G (2009) Superoxide dismutase is regulated by LAMMER kinase in Drosophila and human cells. Free Radic Biol Med 46: 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang EH, Kim JA, Oh HW, Park HM (2013) LAMMER kinase LkhA plays multiple roles in the vegetative growth and asexual and sexual development of Aspergillus nidulans. PLoS One 8: e58762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H. (2011) Convergent or parallel molecular evolution of momilactone A and B: potent allelochemicals, momilactones have been found only in rice and the moss Hypnum plumaeforme. J Plant Physiol 168: 1511–1516 [DOI] [PubMed] [Google Scholar]
- Ke Y, Liu H, Li X, Xiao J, Wang S (2014) Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host-pathogen interactions. Plant J 78: 619–631 [DOI] [PubMed] [Google Scholar]
- Kou Y, Li X, Xiao J, Wang S (2010) Identification of genes contributing to quantitative disease resistance in rice. Sci China Life Sci 53: 1263–1273 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13: 181–185 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S (2012) Toward an understanding of the molecular basis of quantitative disease resistance in rice. J Biotechnol 159: 283–290 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S (2013) Bacterial blight resistance in rice. In Varshney R, Tuberosa R, eds, Translational Genomics for Crop Breeding. John Wiley and Sons, Hoboken, NJ, pp 11–30 [Google Scholar]
- Li C, Sun CQ, Mu P, Chen L, Wang XK (2001) QTL analysis of anther length and ratio of stigma exsertion, two key traits of classification for cultivated rice (Oryza sativa L.) and common wild rice (O. rufipogon Griff.). Yi Chuan Xue Bao 28: 746–751 [PubMed] [Google Scholar]
- Li H, Wang S (2013) Disease resistance. In Zhang Q, Wing RA, eds, Genetics and Genomics of Rice, Vol 5 Springer, New York, pp 161–175 [Google Scholar]
- Lin YJ, Zhang Q (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Liu H, Li X, Xiao J, Wang S (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S (2011) A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ 34: 1958–1969 [DOI] [PubMed] [Google Scholar]
- Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, et al. (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20: 1238–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R, Manners JM, Kazan K (2013) Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep 32: 815–827 [DOI] [PubMed] [Google Scholar]
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Miyao A, Takahashi A, Hirochika H (2010) Pdk1 kinase regulates basal disease resistance through the OsOxi1-OsPti1a phosphorylation cascade in rice. Plant Cell Physiol 51: 2082–2091 [DOI] [PubMed] [Google Scholar]
- Nikolakaki E, Du C, Lai J, Giannakouros T, Cantley L, Rabinow L (2002) Phosphorylation by LAMMER protein kinases: determination of a consensus site, identification of in vitro substrates, and implications for substrate preferences. Biochemistry 41: 2055–2066 [DOI] [PubMed] [Google Scholar]
- Nojiri H, Sugimori M, Yamane H, Nishimura Y, Yamada A, Shibuya N, Kodama O, Murofushi N, Omori T (1996) Involvement of jasmonic acid in elicitor-induced phytoalexin production in suspension-cultured rice cells. Plant Physiol 110: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1: 538–551 [DOI] [PubMed] [Google Scholar]
- Rabinow L, Samson ML (2010) The role of the Drosophila LAMMER protein kinase DOA in somatic sex determination. J Genet 89: 271–277 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Shad Ali G (2011) Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip Rev RNA 2: 875–889 [DOI] [PubMed] [Google Scholar]
- Riemann M, Haga K, Shimizu T, Okada K, Ando S, Mochizuki S, Nishizawa Y, Yamanouchi U, Nick P, Yano M, et al. (2013) Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J 74: 226–238 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Haas W, Gygi SP, Puigserver P (2010) Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab 11: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Aviv D, Davydov O, Fluhr R (2003) Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Sessa G, Fluhr R (2000) The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J 21: 91–96 [DOI] [PubMed] [Google Scholar]
- Sessa G, Raz V, Savaldi S, Fluhr R (1996) PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell 8: 2223–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu H, Yuan B, Li X, Xu C, Wang S (2011) OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ 34: 179–191 [DOI] [PubMed] [Google Scholar]
- Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64: 86–99 [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Talevich E, Mirza A, Kannan N (2011) Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol Biol 11: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DL, Zhu J, Li ZKL, Paterson AH (1999) Mapping QTLs with epistatic effects and QTL×environment interactions by mixed linear model approaches. Theor Appl Genet 99: 1255–1264 [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cao Y, Xu C, Li X, Wang S (2006) Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet 113: 1347–1355 [DOI] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4: e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Tan F, Hua P, Sun L, Xu G, Zhang Q (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105: 248–257 [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yamada S, Kano A, Tamaoki D, Miyamoto A, Shishido H, Miyoshi S, Taniguchi S, Akimitsu K, Gomi K (2012) Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol 53: 2060–2072 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhao X, Cheng K, Du H, Ouyang Y, Chen J, Qiu S, Huang J, Jiang Y, Jiang L, et al. (2012) A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Yu EY, Lee JH, Kang WH, Park YH, Kim L, Park HM (2013) Fission yeast LAMMER kinase Lkh1 regulates the cell cycle by phosphorylating the CDK-inhibitor Rum1. Biochem Biophys Res Commun 432: 80–85 [DOI] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S (2009) Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol 50: 947–955 [DOI] [PubMed] [Google Scholar]
- Yuan B, Shen X, Li X, Xu C, Wang S (2007) Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226: 953–960 [DOI] [PubMed] [Google Scholar]
- Yun B, Farkas R, Lee K, Rabinow L (1994) The Doa locus encodes a member of a new protein kinase family and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev 8: 1160–1173 [DOI] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang S (2013) Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr Opin Plant Biol 16: 188–195 [DOI] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42: 772–780 [DOI] [PubMed] [Google Scholar]
- Zhao S, Chen D, Geng Q, Wang Z (2013) The highly conserved LAMMER/CLK2 protein kinases prevent germ cell overproliferation in Drosophila. Dev Biol 376: 163–170 [DOI] [PubMed] [Google Scholar]
- Zhou B, Peng K, Zhaohui C, Wang S, Zhang Q (2002) The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci China C Life Sci 45: 449–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.