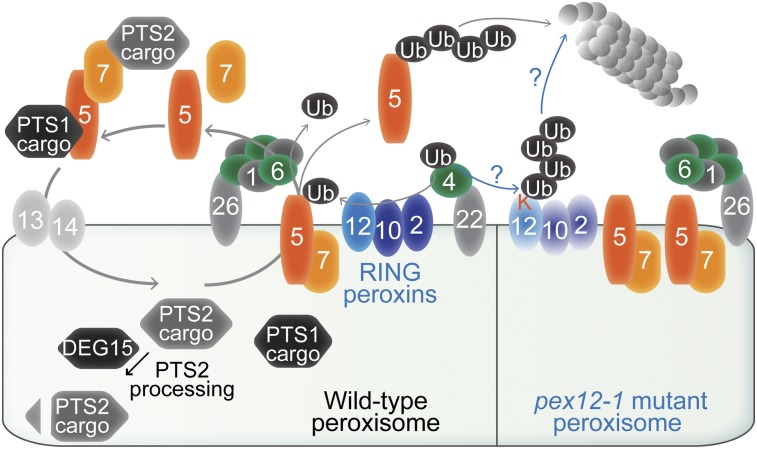
A pex12-1 mutation creating an ectopic lysine causes degradation of the RING peroxin complex and reveals PEX12 involvement in retrotranslocating peroxisome matrix protein receptors.
Abstract
Most eukaryotic cells require peroxisomes, organelles housing fatty acid β-oxidation and other critical metabolic reactions. Peroxisomal matrix proteins carry peroxisome-targeting signals that are recognized by one of two receptors, PEX5 or PEX7, in the cytosol. After delivering the matrix proteins to the organelle, these receptors are removed from the peroxisomal membrane or matrix. Receptor retrotranslocation not only facilitates further rounds of matrix protein import but also prevents deleterious PEX5 retention in the membrane. Three peroxisome-associated ubiquitin-protein ligases in the Really Interesting New Gene (RING) family, PEX2, PEX10, and PEX12, facilitate PEX5 retrotranslocation. However, the detailed mechanism of receptor retrotranslocation remains unclear in plants. We identified an Arabidopsis (Arabidopsis thaliana) pex12 Glu-to-Lys missense allele that conferred severe peroxisomal defects, including impaired β-oxidation, inefficient matrix protein import, and decreased growth. We compared this pex12-1 mutant to other peroxisome-associated ubiquitination-related mutants and found that RING peroxin mutants displayed elevated PEX5 and PEX7 levels, supporting the involvement of RING peroxins in receptor ubiquitination in Arabidopsis. Also, we observed that disruption of any Arabidopsis RING peroxin led to decreased PEX10 levels, as seen in yeast and mammals. Peroxisomal defects were exacerbated in RING peroxin double mutants, suggesting distinct roles of individual RING peroxins. Finally, reducing function of the peroxisome-associated ubiquitin-conjugating enzyme PEX4 restored PEX10 levels and partially ameliorated the other molecular and physiological defects of the pex12-1 mutant. Future biochemical analyses will be needed to determine whether destabilization of the RING peroxin complex observed in pex12-1 stems from PEX4-dependent ubiquitination on the pex12-1 ectopic Lys residue.
Oilseed plants obtain energy for germination and early development by utilizing stored fatty acids (Graham, 2008). This β-oxidation of fatty acids to acetyl-CoA occurs in peroxisomes, organelles that also house other important metabolic reactions, including the glyoxylate cycle, several steps in photorespiration, and phytohormone production (Hu et al., 2012). For example, indole-3-butyric acid (IBA) is β-oxidized into the active auxin indole-3-acetic acid (IAA) in peroxisomes (Zolman et al., 2000, 2007, 2008; Strader et al., 2010; Strader and Bartel, 2011). Many peroxisomal metabolic pathways generate reactive oxygen species (Inestrosa et al., 1979; Hu et al., 2012), and peroxisomes also house antioxidative enzymes, like catalase and ascorbate peroxidase, to detoxify hydrogen peroxide (Wang et al., 1999; Mhamdi et al., 2012).
Peroxisomes can divide by fission or be synthesized de novo from the endoplasmic reticulum (ER). Preperoxisomes with peroxisomal membrane proteins bud from the ER and fuse, allowing matrix proteins to be imported to form mature peroxisomes (van der Zand et al., 2012; Mayerhofer, 2016). Peroxin (PEX) proteins facilitate peroxisome biogenesis and matrix protein import. Most peroxins are involved in importing proteins destined for the peroxisome matrix, which are imported after recognition of a type 1 or type 2 peroxisome-targeting signal (PTS). The PTS1 is a tripeptide located at the C terminus of most peroxisome-bound proteins (Gould et al., 1989; Chowdhary et al., 2012). The less common PTS2 is a nonapeptide usually located near the N terminus (Swinkels et al., 1991; Reumann, 2004). PTS1 proteins are recognized by PEX5 (van der Leij et al., 1993; Zolman et al., 2000), PTS2 proteins are recognized by PEX7 (Marzioch et al., 1994; Braverman et al., 1997; Woodward and Bartel, 2005), and PEX7 binds to PEX5 to allow matrix protein delivery in plants and mammals (Otera et al., 1998; Hayashi et al., 2005; Woodward and Bartel, 2005). The cargo-receptor complex docks with the membrane peroxins PEX13 and PEX14 (Urquhart et al., 2000; Otera et al., 2002; Woodward et al., 2014), and PEX5 assists cargo translocation into the peroxisomal matrix (Meinecke et al., 2010) before dissociating from its cargo (Freitas et al., 2011).
After cargo delivery, PEX5 is recycled to enable further rounds of cargo recruitment (Thoms and Erdmann, 2006). This process requires a set of peroxins that is implicated in ubiquitinating PEX5 so that it can be retrotranslocated back to the cytosol. PEX5 ubiquitination is best understood in yeast. In Saccharomyces cerevisiae, Pex5 is monoubiquitinated through the action of the peroxisome-tethered ubiquitin-conjugating enzyme Pex4 and the peroxisomal ubiquitin-protein ligase Pex12 (Platta et al., 2009) and returned to the cytosol with the assistance of a peroxisome-tethered ATPase complex containing Pex1 and Pex6 (Grimm et al., 2012). S. cerevisiae Pex5 also can be polyubiquitinated and targeted for proteasomal degradation (Kiel et al., 2005). The cytosolic ubiquitin-conjugating enzyme Ubc4 cooperates with the peroxisomal ubiquitin-protein ligase Pex2 to polyubiquitinate Pex5 (Platta et al., 2009). Pex10 has ubiquitin-protein ligase activity (Williams et al., 2008; Platta et al., 2009; El Magraoui et al., 2012), but whether Pex10 directly ubiquitinates Pex5 is controversial. Pex10 promotes Ubc4-dependent Pex5 polyubiquitination when Pex4 is absent (Williams et al., 2008); however, Pex10 is not essential for Pex5 mono- or polyubiquitination (Platta et al., 2009), but rather enhances both Pex4/Pex12- and Ubc4/Pex2-mediated ubiquitination (El Magraoui et al., 2012). Recycling of the PTS2 receptor PEX7 is less understood, although the Pex5 recycling pathways are implicated in shuttling and degrading Pex7 in Pichia pastoris (Hagstrom et al., 2014).
Although PEX5 ubiquitination has not been directly demonstrated in plants, the implicated peroxins are conserved in Arabidopsis, and several have been connected to PEX5 retrotranslocation. The PEX4 ubiquitin-conjugating enzyme binds to PEX22, which is predicted to be a peroxisomal membrane protein based on ability to restore peroxisome function to yeast mutants (Zolman et al., 2005). The pex4-1 mutant displays increased membrane-associated PEX5 (Ratzel et al., 2011; Kao and Bartel, 2015), suggesting that ubiquitin supplied by PEX4 promotes PEX5 retrotranslocation. PEX1 and PEX6 are members of the ATPases associated with diverse cellular activities (AAA) family and are tethered to peroxisomes by the peroxisomal membrane protein PEX26 (Goto et al., 2011; Li et al., 2014). The pex6-1 mutant displays PTS1 import defects and decreased PEX5 levels (Zolman and Bartel, 2004), suggesting that impaired PEX5 recycling can lead to increased PEX5 degradation. Indeed, pex4-1 restores PEX5 levels in the pex6-1 mutant (Ratzel et al., 2011), suggesting that Arabidopsis PEX4 also is involved in PEX5 ubiquitination and degradation when retrotranslocation is impeded.
In addition to allowing for further rounds of PTS1 cargo import, several lines of evidence suggest that in the absence of efficient retrotranslocation, PEX5 retention in the peroxisomal membrane impairs peroxisome function. Slightly reducing levels of the PEX13 docking peroxin ameliorates the physiological defects of pex4-1 without restoring matrix protein import (Ratzel et al., 2011), presumably because decreasing PEX5 docking reduces its accumulation in the peroxisomal membrane. In addition, overexpressing PEX5 exacerbates rather than ameliorates the peroxisomal defects of pex4-1 (Kao and Bartel, 2015), suggesting that pex4-1 defects are linked to excessive PEX5 lingering in the peroxisome membrane rather than a lack of PEX5 available for import.
The three Really Interesting New Gene (RING) peroxins (PEX2, PEX10, and PEX12) from Arabidopsis each possesses in vitro ubiquitin-protein ligase activity (Kaur et al., 2013). Null mutations in the RING peroxin genes confer embryo lethality in Arabidopsis (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005; Prestele et al., 2010), necessitating other approaches to study the in vivo functions of these peroxins. Expressing RING peroxins with mutations in the C-terminal zinc-binding RING domains (ΔZn) confers matrix protein import defects for PEX2-ΔZn and photorespiration defects for PEX10-ΔZn but no apparent defects for PEX12-ΔZn (Prestele et al., 2010). Targeting individual RING peroxins using RNAi confers β-oxidation deficiencies and impairs PTS1 cargo import (Fan et al., 2005; Nito et al., 2007). A screen for delayed matrix protein degradation (Burkhart et al., 2013) uncovered a missense pex2-1 mutant and a splicing pex10-2 mutant that both display PTS1 import defects (Burkhart et al., 2014), suggesting roles in regulating the PTS1 receptor, PEX5. A missense pex12 mutant (aberrant peroxisome morphology 4, apm4) has defects in β-oxidation and PTS1 import and increased membrane-associated PEX5 (Mano et al., 2006). These findings highlight the essential roles of the RING peroxins in Arabidopsis development and peroxisomal functions, but the RING peroxin interactions and the individual roles of the RING peroxins in PEX5 retrotranslocation remain incompletely understood.
In this study, we describe a missense pex12-1 mutant recovered from a forward genetic screen for β-oxidation deficient mutants. The pex12-1 mutant displayed severe peroxisomal defects, including reduced growth, β-oxidation deficiencies, matrix protein import defects, and inefficient processing of PTS2 proteins. Comparing single and double mutants with impaired RING peroxins revealed that each RING peroxin contributes to complex stability and influences PEX5 accumulation. Furthermore, decreasing PEX4 function ameliorated pex12-1 defects, suggesting that the Glu-to-Lys substitution in pex12-1 lures ubiquitination, perhaps by pex12-1 itself, leading to PEX4-dependent degradation of the mutant protein.
RESULTS
Isolation of a Mutant Defective in PEX12
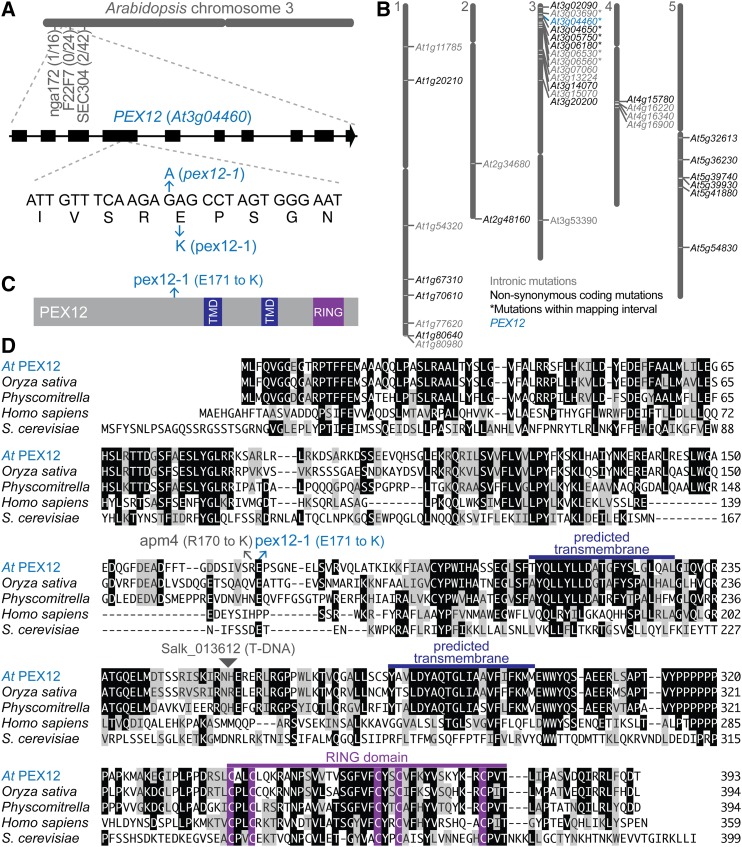
To identify genes important for peroxisome function, we screened the progeny of ethyl methanesulfonate (EMS)-mutagenized seeds for mutants displaying elongated IBA-resistant hypocotyls (HRs) in the dark (Strader et al., 2011) and assessed the resultant putative mutants for incomplete processing of PTS2 proteins to enrich for mutants defective in peroxins. We mapped the causal lesion in one such mutant (HR390) to a region on the top of chromosome 3 (Fig. 1A). Sequencing genomic DNA prepared from back-crossed mutant lines identified seven homozygous EMS-consistent intronic or nonsynonymous coding-sequence mutations in the mapping interval (Fig. 1B), including a mutation in PEX12 (At3g04460) that we named pex12-1. This missense mutation in exon 4 would cause a Glu-to-Lys substitution at position 171 in the pex12-1 protein (Fig. 1, C and D). Although Glu171 is not in a highly conserved region of the PEX12 protein, it is intriguing that the previously described apm4 mutant (Mano et al., 2006) carries an Arg-to-Lys substitution just one residue upstream (Fig. 1D), indicating that an ectopic Lys residue is not tolerated in this region of PEX12.
Figure 1.
Identification of pex12-1. A, The causal mutation in an IBA-resistant mutant was mapped to an interval on the top of chromosome 3 that included PEX12 (At3g04460). The number of recombinants at each marker over the number of chromosomes assayed is indicated. A schematic of the PEX12 gene with exons indicated as rectangles and introns as lines is shown above the portion of exon 4 containing the pex12-1 mutation. B, Whole-genome sequencing identified a missense mutation in PEX12. Gene identifiers are shown at their positions on the five Arabidopsis chromosomes for genes containing homozygous mutations consistent with EMS mutagenesis in introns (gray text) or in exons and resulting in nonsynonymous amino acid changes (black or blue text). The seven genes with mutations within the chromosome 3 mapping interval defined in (A) are indicated with asterisks. C, PEX12 protein diagram with predicted transmembrane domains in dark blue, the RING domain in purple, and the position of the pex12-1 Glu171-to-Lys substitution in light blue. D, PEX12 amino acid alignment with the positions of the pex12-1, apm4 (Mano et al., 2006), and a T-DNA insertion line (Salk_013612) conferring embryo lethality (Fan et al., 2005). The MegAlign program (Clustal W; DNASTAR) was used to compare Arabidopsis (At) PEX12 with orthologs from Oryza sativa (Os10g0467200; NP_001064809.1), Physcomitrella patens (XP_001757098.1), Homo sapiens (O00623.1), and Saccharomyces cerevisiae (NP_013739.1). Identical residues in three or more sequences are boxed in black; chemically similar residues are highlighted in gray. The predicted transmembrane domains are marked in dark blue, and the RING domain is highlighted in purple. TMD, Transmembrane domain.
The pex12-1 Mutant Displays Severe Peroxisome Defects
We compared the phenotypes of pex12-1 to mutants defective in other peroxins implicated in recycling or degrading the PTS1 cargo receptor, PEX5. PEX12 is part of a peroxisomal membrane complex of three RING-family ubiquitin-protein ligases (PEX2, PEX10, PEX12; Fan et al., 2005; Mano et al., 2006; Kaur et al., 2013; Burkhart et al., 2014) that together with the PEX4 ubiquitin-conjugating enzyme are thought to be involved in the PEX5 ubiquitination that allows retrotranslocation by the PEX1 and PEX6 ATPases. Mutations in PEX2, PEX4, PEX6, PEX10, or PEX12 can cause peroxisomal defects ascribed to impaired PEX5 recycling and consequent defects in matrix protein import (Zolman and Bartel, 2004; Zolman et al., 2005; Mano et al., 2006; Burkhart et al., 2013).
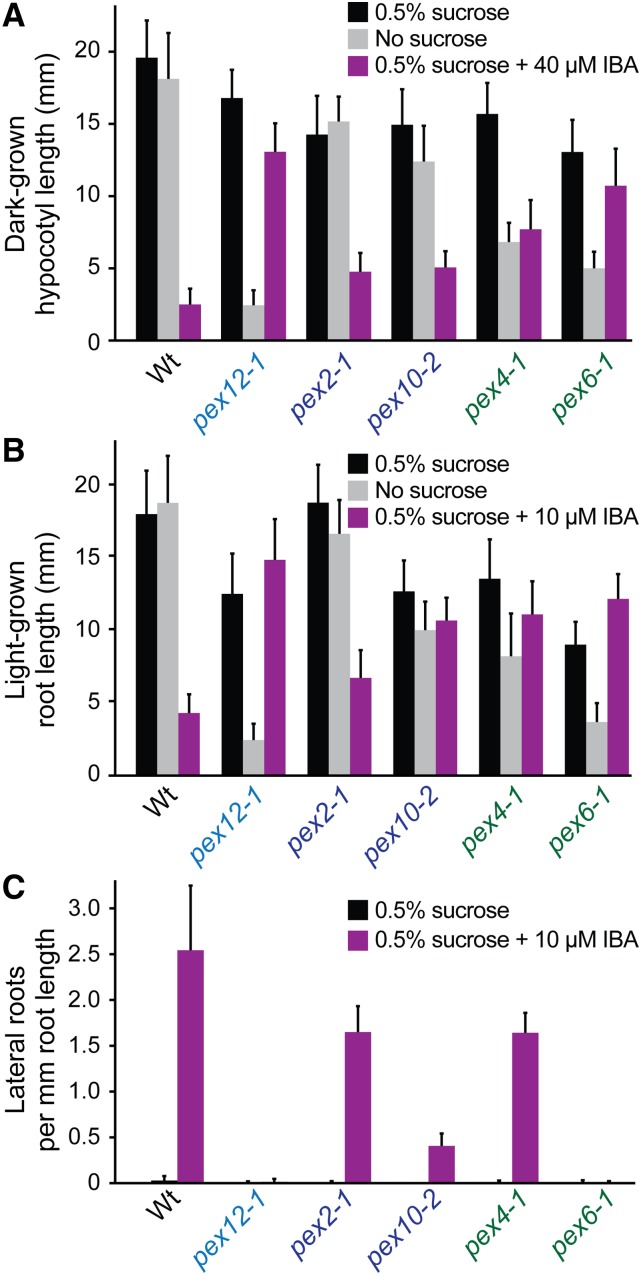
Wild-type seedling growth is fueled by peroxisomal β-oxidation of seed storage lipids (Graham, 2008) and is similar with or without an exogenous carbon source such as Suc (Fig. 2, A and B). Mutants with defective peroxisomes that inefficiently metabolize fatty acids display reduced growth that can be partially ameliorated by providing Suc (Hayashi et al., 1998; Zolman et al., 2000, 2001). As previously reported (Zolman and Bartel, 2004; Zolman et al., 2005; Burkhart et al., 2014), some mutants defective in peroxisomal ubiquitin-dependent machinery, pex4-1 and pex6-1, displayed growth defects without Suc whereas pex2-1 and pex10-2 resembled wild type in this assay (Fig. 2, A and B). We found that pex12-1 displayed more prominent growth defects without Suc than other available mutants in the peroxisomal ubiquitination machinery (Fig. 2, A and B).
Figure 2.
The pex12-1 seedlings display severe peroxisome-related physiological defects. A, The pex12-1 seedlings are resistant to the inhibitory effects of IBA on dark-grown hypocotyl elongation. Seedlings were grown under yellow-filtered light for 1 d followed by 5 d in the dark with the indicated supplements. Mean hypocotyl lengths and sd of the means are shown (n ≥ 15). B, The pex12-1 seedlings are resistant to the inhibitory effects of IBA on light-grown primary root growth. Seedlings were grown under yellow-filtered light for 7 d with the indicated supplements. Mean root lengths and sd of the means are shown (n ≥ 15). C, The pex12-1 seedlings are resistant to the stimulatory effects of IBA on light-grown lateral root formation. Five-day-old light-grown seedlings were moved to 0.5% Suc-supplemented plant nutrient media with or without IBA and grown for 4 d. Mean lateral roots per millimeter root length and sd of the means are shown (n ≥ 13).
Peroxisomes also house phytohormone β-oxidation, providing a second assay to evaluate peroxisome function (Zolman et al., 2000). In wild type, IBA treatment inhibits cell elongation (Strader et al., 2011) because IBA is β-oxidized to IAA (Strader et al., 2010), resulting in short dark-grown hypocotyls (Fig. 2A) and light-grown roots (Fig. 2B). Mutants with impaired peroxisomes have slowed IBA-to-IAA conversion (Strader et al., 2010) and display resistance to the inhibitory effects of IBA on root and hypocotyl elongation (Zolman et al., 2000; Strader et al., 2011). We found that pex12-1 exhibited nearly complete IBA resistance in these assays, similar to pex6-1 (Fig. 2, A and B; Zolman and Bartel, 2004). In contrast, pex10-2 and pex4-1 roots were moderately IBA resistant, and pex2-1 showed only slight IBA resistance (Fig. 2, A and B; Zolman et al., 2005; Burkhart et al., 2014). IBA-derived IAA also stimulates lateral root formation in wild type (Zolman et al., 2000; Strader et al., 2011). Like pex6-1, pex12-1 was completely resistant to IBA-induced lateral root production whereas pex2-1, pex10-2, and pex4-1 showed intermediate resistance in this assay (Fig. 2C).
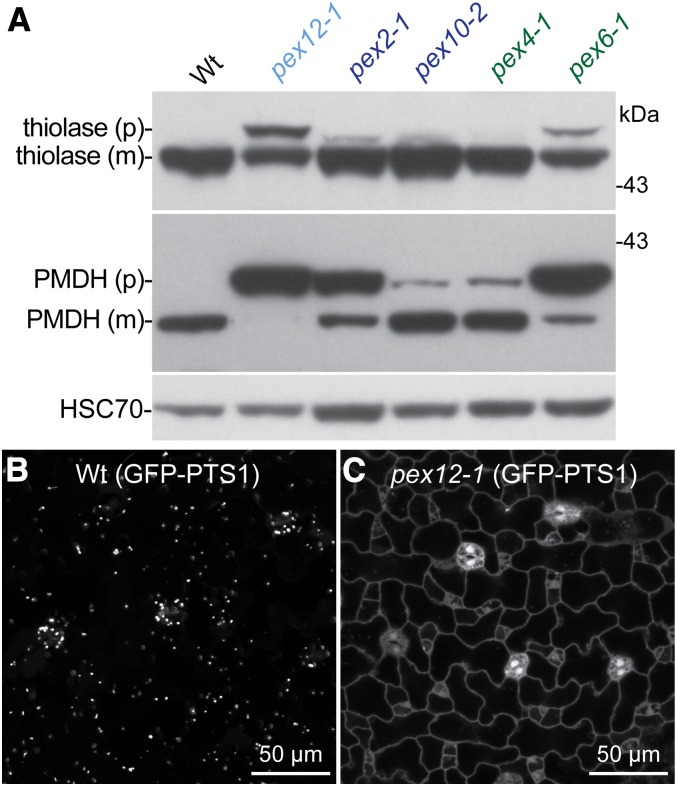
Peroxisome-related physiological defects are often associated with defects in matrix protein import, which can be evaluated by immunoblotting or microscopy. After import into the organelle, PTS2 proteins are proteolytically cleaved by the DEG15 protease to remove the N-terminal PTS2-containing region (Helm et al., 2007; Schuhmann et al., 2008). The molecular mass shift between full-length and cleaved PTS2 cargo can be monitored by immunoblotting as a proxy for matrix protein import (Fig. 3; Woodward and Bartel, 2005). Because DEG15 is a PTS1 protein (Reumann, 2004) and because the two cargo receptors are interdependent in plants (Hayashi et al., 2005; Woodward and Bartel, 2005; Ramón and Bartel, 2010), robust PTS2 processing requires both PTS1 and PTS2 import. Unlike wild-type seedlings, which fully process thiolase and peroxisomal malate dehydrogenase (PMDH), mutants defective in components of the peroxisomal ubiquitin-dependent machinery display various degrees of impaired processing (Zolman et al., 2005; Mano et al., 2006; Ratzel et al., 2011; Burkhart et al., 2014); pex2-1 and pex6-1 accumulated high levels of PMDH precursor and detectable thiolase precursor whereas pex10-2 and pex4-1 displayed minor defects in this assay (Fig. 3A). We found that pex12-1 seedlings also accumulated PTS2-containing precursor proteins, displaying more severe defects than the previously characterized mutants defective in peroxisomal ubiquitination machinery (Fig. 3A). For all mutants assayed, PMDH processing appeared to be more severely impaired than thiolase processing (Fig. 3A). This difference may reflect varying import efficiencies of different PTS2 cargo proteins or varying cytosolic stabilities of mislocalized PTS2-containing proteins.
Figure 3.
The pex12-1 seedlings inefficiently process PTS2-containing proteins and display matrix protein import defects. A, Protein extracts of 7-d-old light-grown seedlings were processed for immunoblotting with the indicated antibodies. Thiolase and PMDH are synthesized as precursors (p); the N-terminal PTS2-containing region is cleaved to give the mature (m) form after import into peroxisomes. HSC70 was used to monitor protein loading. The positions of molecular mass markers (in kDa) are indicated on the right. B and C, A transgene expressing GFP-PTS1 was introduced into wild type (B) and pex12-1 (C). Cotyledon epidermal cells from 4-d-old light-grown seedlings were imaged for GFP using confocal microscopy. Scale bar = 50 μm.
To directly visualize matrix protein import in pex12-1, we used a peroxisomally targeted GFP (GFP-PTS1). Unlike the typical punctate GFP-PTS1 fluorescence observed in wild type (Fig. 3B), we detected cytosolic GFP-PTS1 fluorescence in cotyledon epidermal cells in pex12-1 seedlings (Fig. 3C), suggesting a nearly complete block of PTS1 cargo import into peroxisomes. These extreme defects in seedling growth, IBA responsiveness, PTS2 processing, and PTS1 cargo import (Figs. 2 and 3) indicate that the pex12-1 mutation severely impairs peroxisome function.
pex12-1 Defects Are Rescued by PEX12-CFP
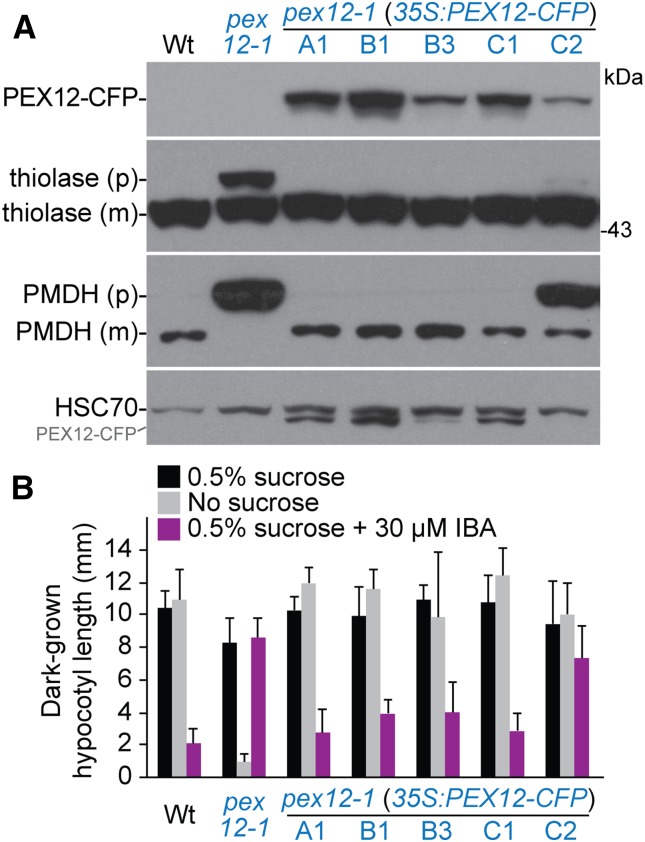
To confirm that the mutation we identified in pex12-1 was responsible for the observed defects, we transformed a construct expressing PEX12-CFP driven by the cauliflower mosaic virus 35S promoter (Fan et al., 2005) into pex12-1 and characterized physiological and molecular phenotypes of the transformed lines. We generated five independent pex12-1 35S:PEX12-CFP lines and found that PTS2-processing of PMDH and thiolase were fully restored in four lines (Fig. 4A). These lines also were Suc independent and displayed substantially restored IBA responsiveness (Fig. 4B). The C2 line, which accumulated less PEX12-CFP compared to other transformants, incompletely rescued PTS2 processing (Fig. 4A) and IBA responsiveness (Fig. 4B), suggesting that the threshold level of PEX12-CFP necessary for function was not achieved in this line. This successful complementation confirmed that the pex12-1 mutation caused the peroxisome-related defects that we observed.
Figure 4.
Molecular and physiological defects of pex12-1 are rescued by a 35S:PEX12-CFP transgene. A, Protein extracts of 8-d-old wild type, pex12-1, and pex12-1 35S:PEX12-CFP T3 lines were processed for immunoblotting with antibodies to GFP (to detect PEX12-CFP), thiolase, and PMDH (to monitor PTS2 processing), and HSC70 (to monitor loading). PEX12-CFP reactivity from a previous probing remains visible in the HSC70 panel. B, Wild-type, pex12-1, and pex12-1 35S:PEX12-CFP T3 lines were grown under yellow-filtered light for 1 d followed by 4 d in the dark on the indicated media. Mean hypocotyl lengths and sd of the means are shown (n ≥ 10).
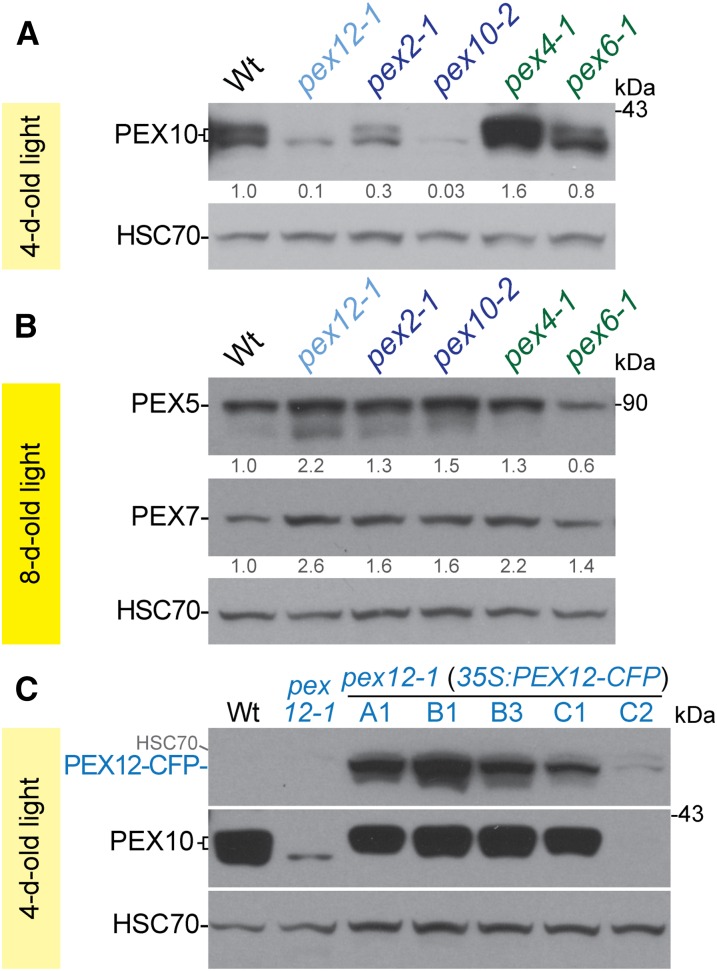
RING Peroxin Defects Destabilize PEX10 and Elevate Peroxisomal Cargo Receptor Levels
In yeast and mammals, the three RING peroxins interact with each other (Okumoto et al., 2000; Hazra et al., 2002), and disruption of any one RING peroxin can decrease the stability of the RING peroxin complex (Hazra et al., 2002; Agne et al., 2003; Okumoto et al., 2014). However, this phenomenon has not been demonstrated in plants. We were unable to monitor PEX12 or PEX2 levels in pex12-1 because we lack a PEX12 antibody and because an antibody recognizing PEX2 (Sparkes et al., 2005) detects overexpressed PEX2 but not endogenous PEX2 in wild-type seedlings (Burkhart et al., 2014). However, we were able to monitor PEX10 levels in the various mutants to assess whether the plant RING peroxins exhibit interdependence. We found that not only pex10-2 (Burkhart et al., 2014) but also pex12-1 and pex2-1 seedlings had low PEX10 levels, with the pex12-1 mutant showing several-fold more marked PEX10 reduction than pex2-1 (Fig. 5A). PEX10 levels were restored in our complementing pex12-1 35S:PEX12-CFP lines (Fig. 5C), indicating that the reduced PEX10 levels in the pex12-1 mutant were attributable to the pex12-1 mutation. We concluded that both PEX12 and PEX2 are important for PEX10 accumulation in Arabidopsis. In contrast to the reduced PEX10 levels in the RING peroxin mutants, we found nearly wild-type PEX10 levels in the pex6-1 mutant and slightly elevated PEX10 levels in the pex4-1 mutant (Fig. 5A), suggesting that PEX4 might directly or indirectly contribute to PEX10 turnover.
Figure 5.
Mutations in RING peroxins result in PEX10 destabilization and elevated levels of the receptor peroxins PEX5 and PEX7. A, Protein extracts of 4-d-old light-grown seedlings were processed for immunoblotting with antibodies recognizing PEX10 or HSC70 (to monitor loading). The positions of molecular mass markers (in kDa) are indicated on the right. Numbers below bands indicate PEX10/HSC70 ratios normalized to the wild-type ratio. Light yellow banner indicates samples from 4-d-old light-grown seedlings. B, Protein extracts of 8-d-old light-grown seedlings were processed for immunoblotting with antibodies recognizing PEX5, PEX7, or HSC70 (to monitor loading). Numbers below bands indicate PEX5/HSC70 or PEX7/HSC70 ratios normalized to the wild-type ratio. Yellow banner indicates samples from 8-d-old light-grown seedlings. C, Protein extracts of 4-d-old light-grown seedlings were processed for immunoblotting with antibodies recognizing GFP (to monitor PEX12-CFP), PEX10, or HSC70 (to monitor loading). HSC70 reactivity from a previous probing remains visible in the PEX12-CFP panel.
Although RING peroxins interact with each other, they have distinct functions (Okumoto et al., 2000; Prestele et al., 2010; Burkhart et al., 2014). S. cerevisiae Pex12, assisted by the Pex4 ubiquitin-conjugating enzyme, is implicated in monoubiquitinating Pex5 to allow Pex5 recycling back to the cytosol for further rounds of cargo delivery (Platta et al., 2009). S. cerevisiae Pex2 coupled with Ubc4 is involved in polyubiquitinating Pex5, which targets Pex5 for proteasomal degradation (Platta et al., 2009). S. cerevisiae Pex10 appears to assist both Pex2 and Pex12 in these roles (El Magraoui et al., 2012). In Arabidopsis pex4, pex6, and pex12 mutants, PEX5 is retained in the peroxisomal membrane, indicating that PEX5 recycling is defective (Mano et al., 2006; Ratzel et al., 2011; Kao and Bartel, 2015). This defect in PEX5 retrotranslocation is accompanied by increased PEX5 degradation in pex6-1 (Zolman and Bartel, 2004; Ratzel et al., 2011; Kao and Bartel, 2015). We used immunoblotting to evaluate PEX5 levels and found that the RING peroxin mutants, especially pex12-1, displayed slightly elevated PEX5 levels (1.3-fold to 2.2-fold; Fig. 5B), suggesting that the Arabidopsis RING peroxin complex normally contributes to PEX5 degradation.
Pex7 degradation is promoted by Pex5 recycling in P. pastoris (Hagstrom et al., 2014). We found that mutations in peroxisomal ubiquitin-dependent machinery resulted in slightly elevated PEX7 levels (1.4-fold to 2.6-fold; Fig. 5B). This elevation might reflect reduced RING complex-mediated PEX7 degradation or might be an indirect consequence of elevated PEX5 levels in most of these mutants, as PEX5 and PEX7 are interdependent in plants (Hayashi et al., 2005; Woodward and Bartel, 2005; Ramón and Bartel, 2010).
Genetic Interactions between RING Peroxins
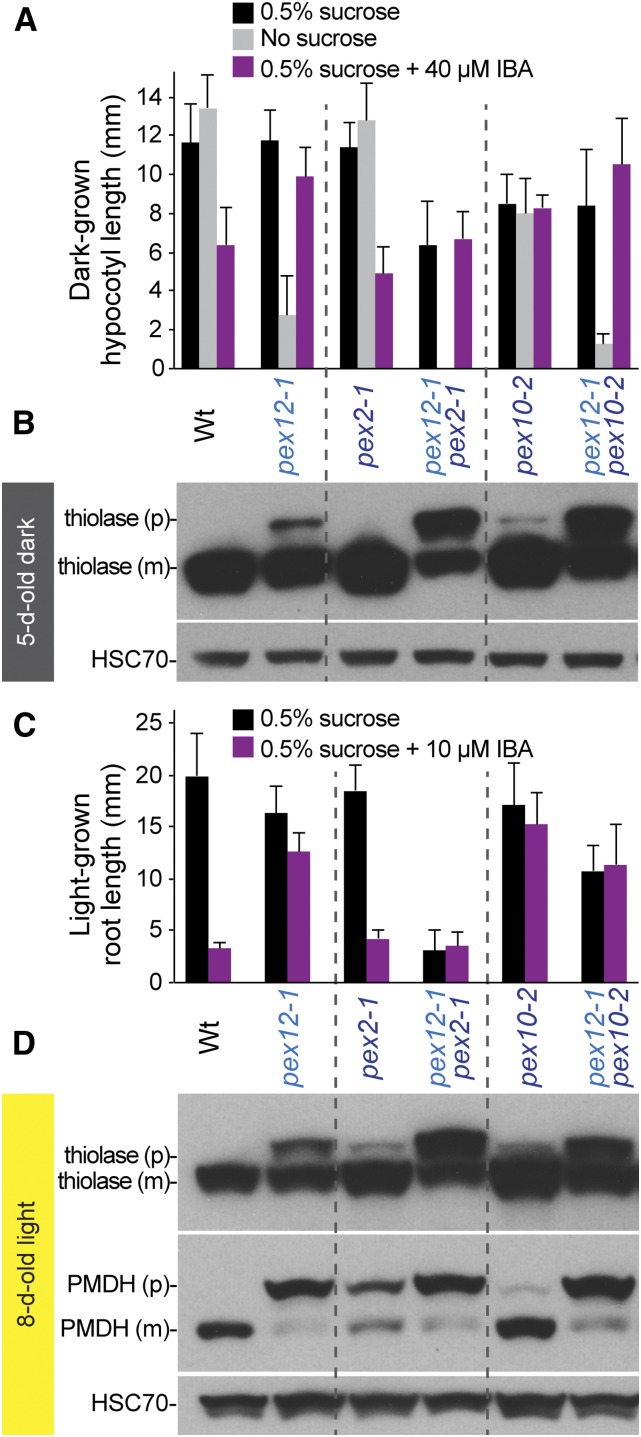
To explore the genetic interactions between RING peroxins, we constructed double mutants of pex12-1 with pex2-1 and pex10-2. Although pex2-1 resembles wild type in most physiological assays (Fig. 2, A and B; Burkhart et al., 2014), pex2-1 markedly exacerbated pex12-1 defects (Figs. 6 and 7). Elongation of dark-grown hypocotyls (Fig. 6A) and light-grown roots (Figs. 6C and 7A) were much further impaired in the pex12-1 pex2-1 double mutant compared to either single mutant even when Suc was provided. In addition, the pex12-1 pex2-1 double mutant failed to germinate without Suc supplementation, and IBA at the concentrations assayed did not inhibit elongation of pex12-1 pex2-1 hypocotyls or roots in the dark or in the light, respectively (Fig. 6, A and C).
Figure 6.
Combining pex12-1 with mutations in other RING peroxins exacerbates peroxisomal defects. A, Seedlings were grown with the indicated supplements under yellow-filtered light for 1 d followed by 4 d in the dark. Mean hypocotyl lengths and sd of the means are shown (n ≥ 14). B, Protein extracts of 5-d-old dark-grown seedlings were processed for immunoblotting with antibodies to thiolase (to monitor PTS2 processing) and HSC70 (to monitor loading). The dark gray banner indicates samples from 5-d-old dark-grown seedlings. C, Seedlings were grown with the indicated supplements under yellow-filtered light for 8 d. Mean root lengths and sd of the means are shown (n ≥ 16). D, Protein extracts of 8-d-old light-grown seedlings were processed for immunoblotting with antibodies to thiolase and PMDH (to monitor PTS2 processing) and HSC70 (to monitor loading).
Figure 7.
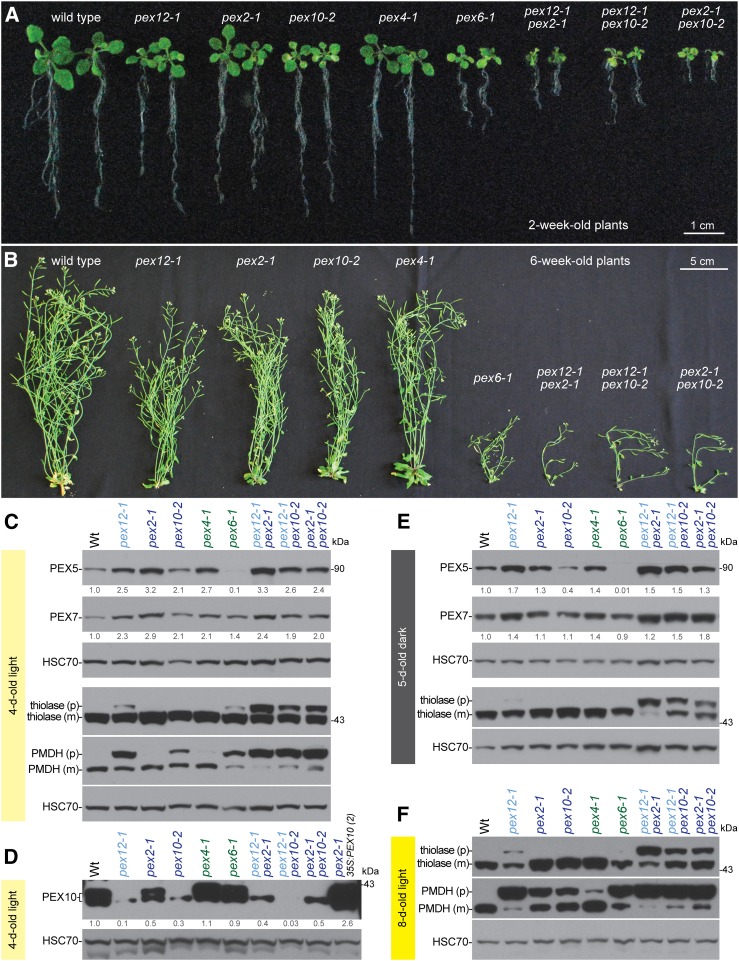
Mutations in two RING peroxin genes result in severe growth and molecular defects. A, RING peroxin double mutants showed synergistic growth defects as 2-week-old plants. Seedlings were grown on plant nutrient media with 0.5% Suc. Representative 2-week-old individuals were selected for imaging. Scale bar = 1 cm. B, RING peroxin double mutants showed synergistic growth defects as 6-week-old plants. Two-week-old plants were moved to soil and grown for additional 4 weeks. Plants were grown in constant light and watered twice a week. Representative 6-week-old individuals were selected for imaging. Scale bar = 5 cm. C to F, Single RING peroxin mutants had elevated PEX5 levels, especially in pex12-1 and pex2-1, and double mutants showed higher PEX5 levels compared to single mutants. Thiolase and PMDH PTS2-processing defects were worsened in RING peroxin double mutants. Protein extracts of 4-d-old light-grown seedlings were processed for immunoblotting (C and D). The pex2-1 35S:PEX10 (2) was used as an overexpressing PEX10 control (Burkhart et al., 2014). Numbers below bands indicate PEX5/HSC70, PEX7/HSC70, or PEX10/HSC70 ratios normalized to the wild-type ratio. Protein extracts of 5-d-old dark-grown seedlings were processed for immunoblotting (E). Protein extracts of 8-d-old light-grown seedlings were processed for immunoblotting (F).
The pex10-2 mutation also exacerbated pex12-1 physiological defects, reducing IBA responsiveness and hypocotyl growth without Suc in the dark (Fig. 6A) and reducing light-grown root growth with Suc (Figs. 6C and 7A). Even though the pex10-2 single mutant displays more apparent physiological defects than pex2-1 (Figs. 2 and 6, A and C; Burkhart et al., 2014), pex12-1 pex10-2 seedling growth was less impaired than growth of pex12-1 pex2-1 seedlings (Figs. 6, A and C, and 7A).
When grown for 2 weeks on Suc-supplemented media, the pex12-1, pex2-1, pex10-2, and pex4-1 single mutants displayed smaller rosettes than wild type but relatively normal root elongation (Fig. 7A) whereas the pex6-1 mutant displayed smaller and paler rosettes (Zolman and Bartel, 2004) as well as reduced root growth (Fig. 7A). The three double mutants, including the previously characterized pex2-1 pex10-2 double mutant (Burkhart et al., 2014), all exhibited synergistically reduced rosette sizes and root growth (Fig. 7A). Similar trends were observed in 6-week-old plants; despite the severe seedling peroxisomal defects (Figs. 2 and 3), mature pex12-1 plants resembled pex2-1 and pex10-2 plants, displaying only slightly reduced plant size, whereas pex6-1 was more impaired (Fig. 7B). Unlike the corresponding single mutants, all three RING peroxin double mutants were quite small as mature plants (Fig. 7B) and produced limited numbers of seeds.
We examined whether the exacerbated physiological defects in the double mutants were accompanied by worsened PTS2 protein processing. Indeed, both pex12-1 pex2-1 and pex12-1 pex10-2 accumulated more unprocessed precursor thiolase proteins than the single mutants when grown in the dark (Figs. 6B and 7E) or the light (Figs. 6D and 7, C and F). PMDH was already almost fully unprocessed in 8-d-old light-grown pex12-1 seedlings (Figs. 6D and 7F), but 4-d-old light-grown pex12-1 seedlings displayed an incomplete defect, allowing us to detect worsened PMDH processing defects in the pex12-1 pex2-1 and pex12-1 pex10-2 double mutants (Fig. 7C). As with the physiological defects, pex2-1 more dramatically worsened the pex12-1 PTS2-processing defects than pex10-2 (Fig. 7, C and F). Although the pex2-1 pex10-2 double mutant displayed heightened defects in processing thiolase (Fig. 7, C, E, and F) and PMDH (Fig. 7, C and F) compared to the pex2-1 or pex10-2 single mutants, these defects were not as extreme as the pex12-1 pex2-1 double mutant (Fig. 7, C, E, and F).
Because the RING peroxin single mutants displayed elevated PEX5 and PEX7 levels (Fig. 5B), we compared levels of these peroxins in the double and single mutants. Similar to 8-d-old light-grown seedlings (Fig. 5B), PEX5 levels were elevated 2-fold to 3-fold in 4-d-old light-grown pex4-1 and RING peroxin mutants, but markedly diminished in pex6-1 (Fig. 7C). PEX5 levels in the three double mutants were elevated similarly to the single mutants (Fig. 7C). In 5-d-old dark-grown seedlings, PEX5 levels were slightly elevated in pex12-1 and pex2-1 and in the three double mutants, but were not elevated in the pex10-2 single mutant (Fig. 7E). PEX7 levels generally mirrored PEX5 levels: in mutants with elevated PEX5, PEX7 levels were elevated as well (Figs. 5B and 7, C and E). This elevation suggests that PEX7, like PEX5, may be subject to ubiquitin-dependent degradation upon recycling. However, even though PEX5 levels were markedly reduced in the pex6-1 mutant, PEX7 levels were not reduced (Figs. 5B and 7, C and E), indicating that PEX7 is not necessarily subject to excessive degradation in conditions that trigger PEX5 degradation.
We also examined PEX10 levels in the double mutants. The low levels of PEX10 in the pex12-1 and pex10-2 single mutants were further reduced in the pex12-1 pex10-2 double mutant (Fig. 7D), suggesting that the residual pex10-2 protein was destabilized when PEX12 function was reduced. In contrast, combining pex12-1 or pex10-2 with pex2-1 did not appear to reduce PEX10 levels below levels in the pex12-1 or pex10-2 single mutants (Fig. 7D).
Reducing PEX4 Function Ameliorates pex12-1 Defects
The three Arabidopsis RING peroxins are ubiquitin-protein ligases with autoubiquitination capabilities in vitro (Kaur et al., 2013). Because the Glu171Lys mutation causing pex12-1 defects was in a relatively nonconserved region of the protein lacking Lys residues (Fig. 1D), we speculated that the ectopic Lys in the mutant protein might be subject to ubiquitination, triggering pex12-1 degradation and consequent peroxisome-related defects. Because Pex4 assists Pex12 in monoubiquitinating Pex5 in S. cerevisiae (Platta et al., 2009), we hypothesized that Arabidopsis PEX4 might provide the ubiquitin to the responsible ligase. To test this hypothesis, we used two pex4 mutants, a missense pex4-1 allele with moderate peroxisome-related defects (Zolman et al., 2005), and an intronic pex4-2 allele with minor peroxisome-related defects and reduced PEX4 protein levels (Kao and Bartel, 2015).
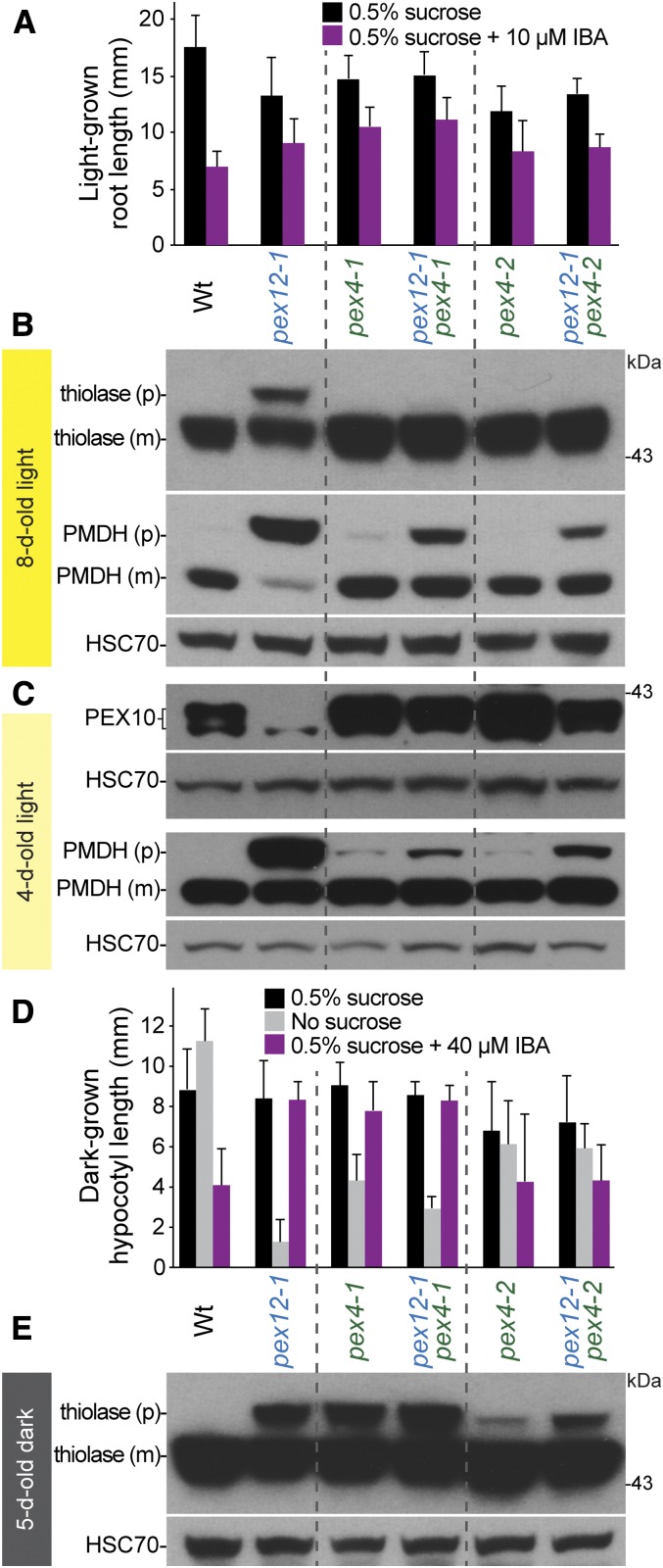
We found that introducing the pex4-1 or pex4-2 mutants into pex12-1 allowed longer hypocotyl growth without Suc (Fig. 8D), suggesting that pex12-1 fatty acid β-oxidation efficiency improved when PEX4 function was reduced. Although the pex12-1 pex4-1 double mutant showed similar IBA responsiveness as pex12-1 and pex4-1 (Fig. 8, A and D), the pex12-1 pex4-2 double mutant displayed only slight IBA resistance in the dark, similar to pex4-2 (Fig. 8D), suggesting that slightly reducing PEX4 function improved IBA β-oxidation in pex12-1. Similarly, pex4-2 improved PTS2 processing of thiolase in dark-grown pex12-1 seedlings (Fig. 8E), and both pex4-1 and pex4-2 markedly improved PTS2 processing of both thiolase and PMDH in light-grown pex12-1 seedlings (Fig. 8, B and C). Finally, we found that both pex4 mutants dramatically increased PEX10 levels in pex12-1 (Fig. 8C). The partial rescue of pex12-1 physiological and PTS2-processing defects that accompanied impairment of a ubiquitin-conjugating enzyme implies that excessive ubiquitination contributes to pex12-1 phenotypic defects.
Figure 8.
The pex12-1 defects are suppressed by impairing the PEX4 ubiquitin-conjugating enzyme. A, Seedlings were grown with the indicated supplements under yellow-filtered light for 8 d. Mean root lengths and sd of the means are shown (n ≥ 22). B, The pex4 mutations improve thiolase and PMDH PTS2 processing in light-grown pex12-1 seedlings. Protein extracts of 8-d-old light-grown seedlings were processed for immunoblotting. C, The pex4 mutations restore PEX10 levels in pex12-1. Protein extracts of 4-d-old light-grown seedlings were processed for immunoblotting. D, The pex4 mutations improve pex12-1 growth on medium lacking Suc. Seedlings were grown under yellow-filtered light for 1 d followed by 4 d in the dark. Mean hypocotyl lengths and sd of the means are shown (n ≥ 22). E, pex4-2 improves thiolase PTS2 processing in dark-grown pex12-1 seedlings. Protein extracts of 5-d-old dark-grown seedlings were processed for immunoblotting.
DISCUSSION
Peroxisome-associated ubiquitination machinery is involved in peroxisomal matrix protein receptor retrotranslocation and maintaining peroxisome functions in yeast (Platta et al., 2014). In plants, however, the genetic interactions among RING peroxins and how RING peroxins influence cargo receptor translocation are incompletely understood. We identified an Arabidopsis pex12-1 mutant in a forward-genetic screen for mutants with elongated IBA-resistant hypocotyls in the dark (Strader et al., 2011). Severe peroxisomal defects were conferred by the pex12-1 mutation, including impaired fatty acid and IBA β-oxidation (Fig. 2), inefficient matrix protein import (Fig. 3), and reduced growth (Fig. 7, A and B). Similarly, the previously described apm4 allele of pex12 displays reduced growth that is ameliorated by Suc, resistance to the IBA analog 2,4,-dichlorophenoxybutyric acid, and inefficient import of PTS1 and PTS2 proteins into peroxisomes (Mano et al., 2006).
We combined pex12-1 with mutations in the other two RING peroxins, pex2-1 and pex10-2 (Burkhart et al., 2014), to investigate the genetic interactions among RING peroxins. We found that double mutants defective in any two RING peroxins displayed worsened peroxisomal defects compared to the single mutants (Figs. 6 and 7). This result is consistent with distinct functions for each RING peroxin, which also is suggested by previous findings that neither overexpressing PEX2 in the pex10-2 mutant nor overexpressing PEX10 in the pex2-1 mutant ameliorates peroxisomal defects (Burkhart et al., 2014). Interestingly, the pex12-1 pex2-1 double mutant displayed more severe defects than the pex12-1 pex10-2 double mutant, even though the pex2-1 single mutant is less impaired than the pex10-2 single mutant. If as in other systems, Arabidopsis PEX12 monoubiquitinates PEX5 for recycling and PEX2 polyubiquitinates PEX5 for degradation, then the synergistic pex12-1 pex2-1 double mutant phenotypes imply that a defect in PEX5 recycling is particularly deleterious in the absence of a mechanism for PEX5 degradation and vice versa. This synergy also implies that despite very low PEX10 levels (Fig. 7D), more RING peroxin complex function remains in pex12-1 pex10-2 than in the pex12-1 pex2-1 mutant. PEX10 function in PEX5 ubiquitination is controversial; S. cerevisiae Pex10 enhances activities of the other two RING peroxins (El Magraoui et al., 2012), whereas mammalian PEX10 displays ubiquitin-protein ligase activity and is needed for PEX5 ubiquitination (Okumoto et al., 2014). Our results suggest that as in S. cerevisiae, Arabidopsis PEX10 assists in but is not the predominant ubiquitin-protein ligase involved in PEX5 ubiquitination.
Intriguingly, pex12-1 and apm4 (Mano et al., 2006) carry Glu-to-Lys or Arg-to-Lys substitutions, respectively, in adjacent residues in a relatively nonconserved region of PEX12 that is N-terminal to the first predicted transmembrane domain (Fig. 1D). The residues mutated in pex12-1 and apm4 are in a 49-residue stretch lacking Lys residues (Fig. 1D). The ectopic Lys residues in the pex12-1 and apm4 proteins would be on the cytosolic surface of the peroxisome along with the catalytic RING domains (Kaur et al., 2013). We hypothesize that pex12-1 (and apm4) is a self-destructive protein. Ubiquitin provided by PEX4 might allow pex12-1 autoubiquitination on the ectopic Lys residue, which might prompt pex12-1 degradation, destabilization of the RING complex, and degradation of other RING peroxins (Fig. 9). An Arabidopsis PEX12 antibody would be useful for testing this hypothesis directly. Because we lack such an antibody, we explored the downstream predictions of this hypothesis using a PEX10 antibody. Indeed, we found reduced PEX10 levels in all three single RING peroxin mutants (Figs. 5A and 7D), indicating that disrupting PEX12 or PEX2 can lead to PEX10 degradation, like in yeast and mammals (Hazra et al., 2002; Agne et al., 2003; Okumoto et al., 2014). We found even lower PEX10 levels in the pex12-1 pex10-2 double mutant, further validating that functional PEX12 promotes PEX10 accumulation. Unlike the low PEX10 levels observed in RING peroxin mutants, PEX10 levels were increased in pex4 mutants (Figs. 5A, 7D, and 8C), and pex4 mutants fully restored PEX10 levels in the pex12-1 mutant (Fig. 8C). This PEX4-dependent RING peroxin degradation suggests the presence of a quality-control system on the peroxisomal membrane to recognize and eliminate dysfunctional peroxisomal components. The restored PEX10 levels in pex12-1 pex4 might be an indirect consequence of stabilizing pex12-1 protein, which promotes PEX10 stability. Alternatively or in addition, PEX4 could play a direct role in destabilizing other RING peroxin complex members when one member is absent or dysfunctional. The development of specific antibodies to PEX12 and PEX2 will be necessary to distinguish among these possibilities and to test whether the similar apm4 mutation also encodes a self-destructive ligase.
Figure 9.
A working model of peroxisome-associated ubiquitination machinery in wild type (left) and the pex12-1 mutant (right). Peroxins are numbered ovals. In wild type, the PEX5-PEX7-cargo complex docks at the peroxisome and delivers cargo into the matrix. The PEX4 ubiquitin-conjugating enzyme is suggested to work with the PEX2, PEX10, and PEX12 RING peroxins to ubiquitinate PEX5, which allows PEX5 (and PEX7) retrotranslocation by the PEX1-PEX6 ATPase complex for further rounds of cargo recruitment or proteasomal degradation. Although ubiquitination of PEX5 has not been directly shown in Arabidopsis, it is demonstrated in yeast (for review, see Platta et al., 2014) and is inferred from the low PEX5 levels in the pex6-1 mutant (Zolman and Bartel, 2004) that can be restored by treatment with a proteasome inhibitor (Kao and Bartel, 2015) and in pex6-1 pex4-1 (Ratzel et al., 2011) and pex6-1 pex2-1 (Burkhart et al., 2014) double mutants. The elevated PEX5 levels in pex2, pex10, and pex12 mutants (this work) also support the hypothesis that PEX5 is a substrate of these ubiquitin-protein ligases. Decreasing PEX4 function partially ameliorates pex12-1 defects, consistent with the possibility that the pex12-1 ectopic Lys substitution (K) is ubiquitinated, leading to pex12-1 degradation, RING peroxin complex destabilization, peroxisomal retention of receptors, and severe matrix protein import defects.
Ubiquitin-protein ligases might be prone to self-destructive alleles. Intriguingly, the pex2-1 mutation, like pex12-1 and apm4, results in an ectopic Lys residue (Arg161Lys) just N-terminal to the first predicted transmembrane domain (Burkhart et al., 2014). It would be interesting to learn whether PEX2 levels are impacted by this mutation and whether reducing PEX4 activity also restores peroxisome function to the pex2-1 mutant. This similarity of ectopic Lys residues of pex12-1 and pex2-1 raises another possibility to explain the synergistic defects observed in the pex12-1 pex2-1 double mutant; perhaps the pex12-1 and pex2-1 proteins effectively compete with PEX5 as ubiquitination substrates, limiting the amount of PEX4-provided ubiquitin available to modify PEX5.
The topology and composition of the peroxisome-associated ubiquitination machinery resembles the ER-associated degradation machinery (Gabaldón et al., 2006; Schlüter et al., 2006; Schliebs et al., 2010). Unlike ER-associated degradation, which degrades a variety of misfolded ER proteins, PEX5 was until recently the only validated substrate of the peroxisome-associated ubiquitination machinery. However, PEX2 can target mammalian peroxisomes for autophagy by ubiquitinating not only PEX5, but also a peroxisomal membrane protein (Sargent et al., 2016). Intriguingly, the Arabidopsis pex2-1 and pex10-2 mutants were identified in a forward genetic screen selecting for delayed degradation of the peroxisomal matrix protein isocitrate lyase (Burkhart et al., 2013, 2014). Moreover, ubiquitinated isocitrate lyase and several other peroxisomal matrix proteins were found in a proteomic study (Kim et al., 2013), consistent with the possibility that the peroxisome-associated ubiquitination machinery might also retrotranslocate peroxisomal matrix substrates. The single and double RING peroxin mutants described here could be useful in learning whether any of the peroxisome-associated ubiquitin-protein ligases ubiquitinate isocitrate lyase or other peroxisomal proteins.
S. cerevisiae Pex12 is involved in monoubiquitinating Pex5 for recycling (Platta et al., 2009). PEX5 membrane association is heightened in the apm4 allele of pex12 (Mano et al., 2006), similarly implicating Arabidopsis PEX12 in PEX5 retrotranslocation. We found elevated PEX5 levels in the Arabidopsis pex12-1 mutant (Figs. 5B, 7C, and 7E), indicating that PEX12 also contributes to PEX5 degradation. This contribution could be direct or could be an indirect consequence of a PEX12 role in stabilizing PEX10 (Figs. 5A, 7D, and 8C) or PEX2, as we also detected elevated PEX5 levels in pex10-2 and pex2-1 mutants (Figs. 5B and 7C). Alternatively, the polyubiquitination that directs PEX5 to the proteasome could require initial monoubiquitination provided by PEX12.
Although a recycling-defective GFP-PEX7 fusion promotes degradation of native PEX7 in Arabidopsis (Cui et al., 2013), little else is known about PEX7 degradation in plants. We found that mutants with elevated PEX5 levels (pex12-1, pex2-1, pex10-2, and pex4-1) also displayed increased PEX7 levels (Figs. 5B and 7, C and E), suggesting that impairing PEX5 recycling can slow PEX7 degradation. Mammalian PEX7 recycling requires PEX5 and the ATP-dependent PEX5 recycling machinery (Rodrigues et al., 2015), and Pex7 degradation is promoted by Pex5 recycling in P. pastoris (Hagstrom et al., 2014). Interestingly, PEX7 levels were not decreased in the pex6-1 mutant (Figs. 5B and 7, C and E), which displays decreased PEX5 levels that can be partially restored by inhibiting the proteasome (Kao and Bartel, 2015) or by reducing PEX2 (Burkhart et al., 2014) or PEX4 (Ratzel et al., 2011) function. Although the elevated PEX7 levels in RING peroxin mutants hint at a role for ubiquitination in PEX7 degradation, the mismatch between PEX5 and PEX7 levels in the pex6-1 mutant (Figs. 5B and 7, C and E) suggests that PEX7, unlike PEX5, is not subject to ubiquitin-dependent degradation when PEX5 retrotranslocation is stalled in pex6-1. It is possible that the topology of PEX5 and PEX7 during matrix protein docking and translocation explains this difference; the N-terminal domain of mammalian PEX5 remains exposed to the cytosol where it can be subject to ubiquitination (Gouveia et al., 2003) but at least part of mammalian PEX7 is protease protected in the peroxisomal matrix (Rodrigues et al., 2014).
Interestingly, we detected slightly different PEX5 and PEX7 accumulation in various mutants depending on the growth conditions. In 4-d-old light-grown seedlings, pex2-1 accumulated more PEX5 than pex12-1 (Fig. 7C) whereas in 5-d-old dark-grown seedlings, pex12-1 had more PEX5 than pex2-1 (Fig. 7E). Perhaps PEX2-assisted PEX5 degradation is more prevalent in light- than in dark-grown plants. Light-dependent differences in PEX5 levels are also described for pex7 mutants, which show reduced PEX5 levels that are more apparent in light-grown seedlings (Ramón and Bartel, 2010). Our findings do not explain the low PEX5 levels in pex7 mutants (Ramón and Bartel, 2010) because low PEX5 levels in pex7-1 are not restored in the pex2-1 pex7-1 double mutant (Burkhart et al., 2014). Hence, the low PEX5 levels in pex7 mutants may reflect impacts of light on cytosolic PEX5 rather than the direct involvement of RING peroxin-mediated ubiquitination of PEX5 in the peroxisomal membrane.
Defective peroxisomes or β-oxidation underlie human peroxisome biogenesis disorders. Affected patients display developmental delays and neurological defects, and there is no cure (Braverman et al., 2016). PEX12, PEX2, and PEX10 mutations are found in approximately 4% to 9%, 15% to 4%, and 3% to 5% of peroxisome biogenesis disorder patients, respectively (Steinberg et al., 1993; Ebberink et al., 2011). Unlike Arabidopsis PEX12 (Kaur et al., 2013), mammalian PEX12 does not appear to have ubiquitin ligase activity but promotes PEX10 activity (Okumoto et al., 2014). However, the elevated PEX5 levels seen in our Arabidopsis pex12-1, pex10-2, and pex2-1 mutants (Figs. 5B and 7, C and E) also are observed in fibroblasts from patients with RING peroxin mutations, including complementation group 3 (CG3; pex12), CG7 (pex10), and CG10 (pex2; Dodt and Gould, 1996). Isolation of genetic or chemical suppressors of the Arabidopsis RING peroxin mutants described here might be informative for further understanding of the roles of these peroxins in plants as well as the etiology of these disorders.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild type and mutants were in the Columbia-0 accession. The pex2-1 (Burkhart et al., 2014), pex4-1 (Zolman et al., 2005), pex4-2 (Kao and Bartel, 2015), pex6-1 (Zolman and Bartel, 2004), pex10-2 (Burkhart et al., 2014), pex2-1 pex10-2 (Burkhart et al., 2014), and wild type-carrying GFP-PTS1 (Zolman and Bartel, 2004) were as previously described. Double mutants including pex12-1 pex2-1, pex12-1 pex4-1, pex12-1 pex4-2, pex12-1 pex10-2, and pex12-1 GFP-PTS1 were selected from progeny of the corresponding crosses by PCR-based genotyping. Genotyping markers are listed in Supplemental Table S1. We used the dCAPS Web site (http://helix.wustl.edu/dcaps/dcaps.html) to design a genotyping marker for pex12-1 (Neff et al., 2002). pex12-1 was back crossed to wild type at least once before phenotypic assays.
The pPZP221-PEX12-CFP plasmid (Fan et al., 2005) was transformed into Agrobacterium tumefaciens strain GV3101 (pMP90; Koncz et al., 1992), which was used to transform pex12-1 plants by floral dipping (Clough and Bent, 1998). T1 seeds were selected on plant nutrient media supplemented with 0.5% Suc and 90 μg/mL gentamycin. Gentamycin-resistant T1 plants were moved to soil and genotyped by PCR for the presence of the PEX12 cDNA and the genomic pex12-1 mutation (Supplemental Table S1). Homozygous lines were identified in the T3 generation and characterized.
For physiological assays and immunoblotting, seeds were surface-sterilized using 3% NaOCl and 0.1% Triton X-100, then washed, suspended in 0.1% (w/v) agar (Cat. no. 90000-762; VWR), and stratified at 4°C in the dark for 1 d to 3 d. Seeds were then plated on plant nutrient medium (Haughn and Somerville, 1986) solidified with 0.6% (w/v) agar with indicated concentrations of Suc and with or without IBA. The 100 mM IBA stock solution was dissolved in ethanol. Equal volumes of ethanol were added to mock plates for normalizing ethanol effects, although we did not observe growth effects at the tested ethanol concentrations (0.01–0.03% [v/v]). For physiological assays involving IBA, light was filtered through yellow long-pass filters to reduce photochemical breakdown of auxin (Stasinopoulos and Hangarter, 1990). Plates for dark-grown hypocotyl assays and immunoblotting experiments were incubated at 22°C in constant yellow light for 1 d to promote germination and then wrapped in two layers of aluminum foil for an additional 4 d or 5 d. Plates for light-grown root assays and indicated immunoblotting experiments were incubated at 22°C in constant yellow light for 7 or 8 d. Plates for immunoblotting experiments using the PEX10 antibody were incubated for only 4 d because the PEX10 antibody did not reliably detect PEX10 protein in older seedlings. For light-grown lateral root assays, seeds were sown on 0.5% (w/v) Suc-supplemented plant nutrient media and incubated at 22°C for 5 d, after which seedlings were transferred to plates with or without IBA for additional 4 d in constant yellow light. Physiological assays and immunoblotting experiments were repeated at least twice with similar results; representative results are shown. For propagation, 2-week-old plants were moved to soil (Sungro Metro-Mix 366, Cat. no. TX366; BWI) and grown at 22°C in constant white light for 12 to 16 weeks with watering twice a week.
Recombinant Mapping and Whole-Genome Sequencing
Wild-type seeds were mutagenized with 0.24% (v/v) EMS (Normanly et al., 1997). The progeny of the mutagenized seeds were screened for elongated IBA-resistant dark-grown hypocotyls as described in Strader et al. (2011). HR390 (pex12-1) was outcrossed to Landsberg erecta for recombination mapping. A mapping population was selected that had IBA-resistant root elongation coupled with PMDH PTS2-processing defects. Recombination mapping markers are listed in Supplemental Table S1.
Once-back-crossed HR390 (pex12-1) was used for whole-genome sequencing. Approximately 2000 F3 seeds pooled from two backcrossed lines were plated on 0.5% (w/v) Suc-supplemented plant nutrient media covered with sterile filter paper. Genomic DNA was purified from 5-d-old light-grown seedlings as described in Thole et al. (2014) and sequenced at the Washington University (St. Louis) Genome Technology Access Center using HiSeq 2000 (Illumina). Sequencing data processed for homozygous EMS-consistent lesions in introns and coding sequences as described in Farmer et al. (2013) are listed in Supplemental Table S2.
Immunoblotting
Protein extracts from seedlings grown as described above were processed for immunoblotting as described in Kao and Bartel (2015). Membranes were incubated overnight with rabbit primary antibodies against PEX5 (1:100, Zolman and Bartel, 2004); PEX7 (1:800, Ramón and Bartel, 2010); PEX10 (1:500, Burkhart et al., 2014); PMDH2 (1:2000–5000, Pracharoenwattana et al., 2007); or thiolase (1:5000–10,000, Lingard et al., 2009) followed by horseradish peroxidase-linked goat anti-rabbit secondary antibody (1:5000, Cat. no. sc-2030; Santa Cruz Biotechnology). Mouse primary antibodies against GFP (1:100, Cat. no. sc-9996; Santa Cruz Biotechnology) and HSC70 (1:100,000, Cat. no. HPA-817; Stressgen) were paired with horseradish peroxidase-linked goat anti-mouse secondary antibody (1:5000, Cat. no. sc-2031; Santa Cruz Biotechnology). Antibodies were visualized by Western Bright ECL substrate (Cat. no. K-12045; Advansta) and exposed on autoradiography film (Cat. no. 30-810C; Genesee). Representative films were scanned, and band intensity was analyzed using ImageJ (National Institutes of Health).
Confocal Microscopy
Cotyledons of 4-d-old light-grown seedlings of wild type and pex12-1 carrying GFP-PTS1 were mounted in water under a cover glass (Cat. no. 12-544-E; Thermo Fisher Scientific). GFP fluorescence in epidermal cells was detected using an LSM710 laser scanning confocal microscope (Carl Zeiss) equipped with a spectral detector (META). Tissues were imaged using a 40× oil immersion objective. GFP was excited with a 488 nm argon laser, and GFP emission was collected between 493 nm and 530 nm.
Accession Numbers
Sequence information for PEX2 (At1g79810), PEX4 (At5g25760), PEX6 (At1g03000), PEX10 (At2g26350), and PEX12 (At3g04460) can be found in the Arabidopsis Information Resource or GenBank.
Supplemental Material
The following supplemental materials are available.
Supplemental Table S1. Genotyping and recombinant mapping markers used in this study.
Supplemental Table S2. Genes from whole-genome sequencing of pex12-1 with homozygous mutations consistent with EMS mutagenesis in introns or in exons resulting in nonsynonymous amino acid changes.
Supplementary Material
Acknowledgments
We thank Jianping Hu for providing the pPZP221-PEX12-CFP plasmid, Steven Smith for the PMDH2 antibody, Lucia Strader for developing the IBA-resistant hypocotyl screen, and Savina Venkova for assistance with rescreening putative mutants. We are grateful to Joseph Faust, Kim Gonzalez, Roxanna Llinas, Mauro Rinaldi, Andrew Woodward, Zachary Wright, Pierce Young, and two anonymous reviewers for critical comments on the article.
Footnotes
This research was supported by the National Institutes of Health (under grant no. R01GM079177) and the Robert A. Welch Foundation (under grant no. C-1309). Y.-T.K. was partially supported by the Studying Abroad Scholarship from the Ministry of Education, Taiwan. M.J.V. was partially supported by a Howard Hughes Medical Institute Professors Grant (grant no. 52005717 to B.B.). Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the National Institutes of Health (grant no. S10RR026399). Whole-genome sequencing at the Genome Technology Access Center at Washington University in St. Louis was supported by the National Institutes of Health National Center for Research Resources (under grant no. UL1 RR024992).
Articles can be viewed without a subscription.
References
- Agne B, Meindl NM, Niederhoff K, Einwächter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell 11: 635–646 [DOI] [PubMed] [Google Scholar]
- Braverman NE, Raymond GV, Rizzo WB, Moser AB, Wilkinson ME, Stone EM, Steinberg SJ, Wangler MF, Rush ET, Hacia JG, Bose M (2016) Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 117: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman NE, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D (1997) Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet 15: 369–376 [DOI] [PubMed] [Google Scholar]
- Burkhart SE, Kao YT, Bartel B (2014) Peroxisomal ubiquitin-protein ligases peroxin2 and peroxin10 have distinct but synergistic roles in matrix protein import and peroxin5 retrotranslocation in Arabidopsis. Plant Physiol 166: 1329–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart SE, Lingard MJ, Bartel B (2013) Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193: 125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary G, Kataya AR, Lingner T, Reumann S (2012) Non-canonical peroxisome targeting signals: identification of novel PTS1 tripeptides and characterization of enhancer elements by computational permutation analysis. BMC Plant Biol 12: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui S, Fukao Y, Mano S, Yamada K, Hayashi M, Nishimura M (2013) Proteomic analysis reveals that the Rab GTPase RabE1c is involved in the degradation of the peroxisomal protein receptor PEX7 (peroxin 7). J Biol Chem 288: 6014–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebberink MS, Mooijer PA, Gootjes J, Koster J, Wanders RJ, Waterham HR (2011) Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum Mutat 32: 59–69 [DOI] [PubMed] [Google Scholar]
- El Magraoui F, Bäumer BE, Platta HW, Baumann JS, Girzalsky W, Erdmann R (2012) The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J 279: 2060–2070 [DOI] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B (2013) Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 peroxisomal protease in matrix protein degradation. Plant Cell 25: 4085–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas MO, Francisco T, Rodrigues TA, Alencastre IS, Pinto MP, Grou CP, Carvalho AF, Fransen M, Sá-Miranda C, Azevedo JE (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J Biol Chem 286: 40509–40519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (2006) Origin and evolution of the peroxisomal proteome. Biol Direct 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Mano S, Nakamori C, Nishimura M (2011) Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S (1989) A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108: 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia AM, Guimaraes CP, Oliveira ME, Reguenga C, Sa-Miranda C, Azevedo JE (2003) Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J Biol Chem 278: 226–232 [DOI] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Grimm I, Saffian D, Platta HW, Erdmann R (2012) The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta 1823: 150–158 [DOI] [PubMed] [Google Scholar]
- Hagstrom D, Ma C, Guha-Polley S, Subramani S (2014) The unique degradation pathway of the PTS2 receptor, Pex7, is dependent on the PTS receptor/coreceptor, Pex5 and Pex20. Mol Biol Cell 25: 2634–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Hazra PP, Suriapranata I, Snyder WB, Subramani S (2002) Peroxisome remnants in pex3Δ cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3: 560–574 [DOI] [PubMed] [Google Scholar]
- Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, Arnold GJ, Adamska I, Görg A, Lottspeich F, Gietl C (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa NC, Bronfman M, Leighton F (1979) Detection of peroxisomal fatty acyl-coenzyme A oxidase activity. Biochem J 182: 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YT, Bartel B (2015) Elevated growth temperature decreases levels of the PEX5 peroxisome-targeting signal receptor and ameliorates defects of Arabidopsis mutants with an impaired PEX4 ubiquitin-conjugating enzyme. BMC Plant Biol 15: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Zhao Q, Xie Q, Hu J (2013) Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins. J Integr Plant Biol 55: 108–120 [DOI] [PubMed] [Google Scholar]
- Kiel JA, Emmrich K, Meyer HE, Kunau WH (2005) Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem 280: 1921–1930 [DOI] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J, Rédei GP (1992) T-DNA transformation and insertion mutagenesis. In Koncz C, Chua N-H, Schell J, eds, Methods in Arabidopsis Research. World Scientific, Singapore, pp 224–273 [Google Scholar]
- Li XR, Li HJ, Yuan L, Liu M, Shi DQ, Liu J, Yang WC (2014) Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell 26: 619–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J 47: 604–618 [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J 13: 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer PU. (2016) Targeting and insertion of peroxisomal membrane proteins: ER trafficking versus direct delivery to peroxisomes. Biochim Biophys Acta 1863: 870–880 [DOI] [PubMed] [Google Scholar]
- Meinecke M, Cizmowski C, Schliebs W, Krüger V, Beck S, Wagner R, Erdmann R (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol 12: 273–277 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Noctor G, Baker A (2012) Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys 525: 181–194 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M (2007) Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol 48: 763–774 [DOI] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B (1997) Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K, Abe I, Fujiki Y (2000) Molecular anatomy of the peroxin Pex12p: ring finger domain is essential for Pex12p function and interacts with the peroxisome-targeting signal type 1-receptor Pex5p and a ring peroxin, Pex10p. J Biol Chem 275: 25700–25710 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Noda H, Fujiki Y (2014) Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J Biol Chem 289: 14089–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Okumoto K, Tateishi K, Ikoma Y, Matsuda E, Nishimura M, Tsukamoto T, Osumi T, Ohashi K, Higuchi O, Fujiki Y (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol Cell Biol 18: 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Setoguchi K, Hamasaki M, Kumashiro T, Shimizu N, Fujiki Y (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol Cell Biol 22: 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R (2009) Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Hagen S, Reidick C, Erdmann R (2014) The peroxisomal receptor dislocation pathway: to the exportomer and beyond. Biochimie 98: 16–28 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2007) Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Prestele J, Hierl G, Scherling C, Hetkamp S, Schwechheimer C, Isono E, Weckwerth W, Wanner G, Gietl C (2010) Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc Natl Acad Sci USA 107: 14915–14920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón NM, Bartel B (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel SE, Lingard MJ, Woodward AW, Bartel B (2011) Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TA, Alencastre IS, Francisco T, Brites P, Fransen M, Grou CP, Azevedo JE (2014) A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway. Mol Cell Biol 34: 2917–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TA, Grou CP, Azevedo JE (2015) Revisiting the intraperoxisomal pathway of mammalian PEX7. Sci Rep 5: 11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent G, van Zutphen T, Shatseva T, Zhang L, Di Giovanni V, Bandsma R, Kim PK (2016) PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol 214: 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliebs W, Girzalsky W, Erdmann R (2010) Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol 11: 885–890 [DOI] [PubMed] [Google Scholar]
- Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A (2006) The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol 23: 838–845 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I (2008) The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol 148: 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Hawes C, Baker A (2005) AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol 139: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SJ, Raymond GV, Braverman NE, Moser AB (1993) Peroxisome biogenesis disorders, Zellweger syndrome spectrum. In Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds, GeneReviews, Seattle, WA [Google Scholar]
- Strader LC, Bartel B (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Beisner ER, Liu J, Venkova SV, Strader LC (2014) Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 (Bethesda) 4: 1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Erdmann R (2006) Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta 1763: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Urquhart AJ, Kennedy D, Gould SJ, Crane DI (2000) Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J Biol Chem 275: 4127–4136 [DOI] [PubMed] [Google Scholar]
- van der Leij I, Franse MM, Elgersma Y, Distel B, Tabak HF (1993) PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 90: 11782–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A, Gent J, Braakman I, Tabak HF (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang H, Allen RD (1999) Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol 40: 725–732 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Geers E, Distel B (2008) Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun 374: 620–624 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Fleming WA, Burkhart SE, Ratzel SE, Bjornson M, Bartel B (2014) A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol Biol 86: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.