GGPP synthase activity essential for chloroplast function and embryo development is provided by two differentially targeted enzyme isoforms encoded by the same gene in Arabidopsis.
Abstract
A wide diversity of isoprenoids is produced in different plant compartments. Most groups of isoprenoids synthesized in plastids, and some produced elsewhere in the plant cell derive from geranylgeranyl diphosphate (GGPP) synthesized by GGPP synthase (GGPPS) enzymes. In Arabidopsis (Arabidopsis thaliana), five genes appear to encode GGPPS isoforms localized in plastids (two), the endoplasmic reticulum (two), and mitochondria (one). However, the loss of function of the plastid-targeted GGPPS11 isoform (referred to as G11) is sufficient to cause lethality. Here, we show that the absence of a strong transcription initiation site in the G11 gene results in the production of transcripts of different lengths. The longer transcripts encode an isoform with a functional plastid import sequence that produces GGPP for the major groups of photosynthesis-related plastidial isoprenoids. However, shorter transcripts are also produced that lack the first translation initiation codon and rely on a second in-frame ATG codon to produce an enzymatically active isoform lacking this N-terminal domain. This short enzyme localizes in the cytosol and is essential for embryo development. Our results confirm that the production of differentially targeted enzyme isoforms from the same gene is a central mechanism to control the biosynthesis of isoprenoid precursors in different plant cell compartments.
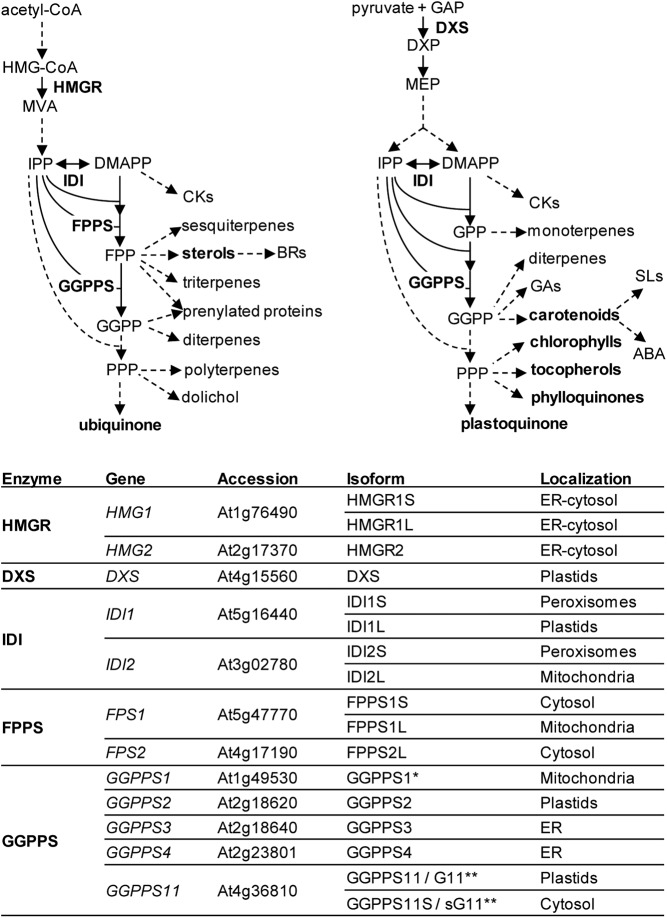
Plants produce tens of thousands of isoprenoid compounds, including some that are essential for respiration, photosynthesis, and regulation of growth and development. Despite their structural and functional diversity, all isoprenoids derive from the same five-carbon precursors, the double-bond isomers isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which can be interconverted by IPP/DMAPP isomerase (IDI) enzymes. Plants use two unrelated pathways to synthesize these units (Fig. 1). The mevalonic acid (MVA) pathway synthesizes IPP in the cytosol, whereas the methylerythritol 4-phosphate (MEP) pathway supplies both IPP and DMAPP in the plastid (Bouvier et al., 2005; Vranová et al., 2013; Rodriguez-Concepción and Boronat, 2015). IPP and DMAPP units can be exchanged between cell compartments to a certain level. For example, MVA-derived IPP can be imported by mitochondria for the biosynthesis of ubiquinone (Lütke-Brinkhaus et al., 1984; Disch et al., 1998). However, this limited exchange of common isoprenoid precursors is not active enough to rescue a genetic or pharmacological blockage of one of the pathways with IPP/DMAPP produced by the noninhibited pathway (Bouvier et al., 2005; Vranová et al., 2013; Rodriguez-Concepción and Boronat, 2015). Addition of IPP units to DMAPP generates longer prenyl diphosphate molecules, including C10 geranyl diphosphate (GPP), C15 farnesyl diphosphate (FPP), and C20 geranylgeranyl diphosphate (GGPP), which are then used in specific downstream pathways to produce particular isoprenoids (Fig. 1). FPP and GGPP pools represent nodes of the major metabolic branch points in the isoprenoid biosynthesis network (Vranová et al., 2011; Vranová et al., 2013). As prenyl diphosphates grow longer, however, their transport between cell compartments becomes increasingly restrained (Bick and Lange, 2003).
Figure 1.
Isoprenoid biosynthetic pathways and enzymes in Arabidopsis. Solid arrows indicate single enzymatic steps, and dashed arrows represent multiple steps. Mevalonic acid (MVA) pathway: HMG-CoA, hydroxymethylglutaryl-CoA. Methylerythritol 4-phosphate (MEP) pathway: GAP, glyceraldehyde 3-phosphate; DXP, deoxyxylulose 5-phosphate. Prenyl diphosphates: IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; PPP, polyprenyl diphosphate. Hormones: CKs, cytokinins; BRs, brassinosteroids; GAs, gibberellins; SLs, strigolactones; ABA, abscisic acid. Enzymes: HMGR, HMG-CoA reductase; DXS, DXP synthase; IDI, IPP/DMAPP isomerase; FPPS, FPP synthase; GGPPS, GGPP synthase. *GGPPS activity unclear. **Isoforms reported in this work.
The two pathways for the production of isoprenoid precursors have been extensively studied in Arabidopsis (Arabidopsis thaliana; Fig. 1). All the MEP pathway enzymes are encoded by nuclear genes and imported into plastids, whereas cytosolic, endoplasmic reticulum (ER), and peroxisomal-associated locations have been found for MVA enzymes (Pulido et al., 2012; Rodriguez-Concepción and Boronat, 2015). The main rate-determining enzymes of the MEP and MVA pathways are deoxyxylulose 5-phosphate synthase (DXS) and hydroxymethylglutaryl-CoA reductase (HMGR), respectively (Fig. 1). Most plants contain small gene families encoding these two enzymes (Rodriguez-Concepción and Boronat, 2015). While several Arabidopsis genes encode proteins with homology to DXS, only one of them produces a functional enzyme with DXS activity (Phillips et al., 2008a). In the case of HMGR, the HMG1 gene produces long and short transcripts encoding two enzyme isoforms (HMGR1L and HMGR1S, respectively) that only differ in their N-terminal region, whereas the HMG2 gene produces only one isoform, HMGR2 (Caelles et al., 1989; Enjuto et al., 1994; Lumbreras et al., 1995). The three HMGR isoforms are primarily attached to the ER and have the same topology in the membrane, with the highly divergent N-terminal region and the highly conserved catalytic domain exposed to the cytosol.
Downstream enzymes such as IDI, FPP synthase (FPPS), and GGPP synthase (GGPPS) are also encoded by small gene families in Arabidopsis and localize to multiple subcellular compartments (Fig. 1). The two genes encoding IDI in Arabidopsis, IDI1 and IDI2, produce long and short transcripts encoding enzyme isoforms that differ in length at their N-terminal ends (Okada et al., 2008; Phillips et al., 2008b; Sapir-Mir et al., 2008). The long IDI1 isoform is targeted to plastids, the long IDI2 isoform is transported to mitochondria, and both short isoforms are sorted to peroxisomes. The two genes encoding FPPS in Arabidopsis produce three isoforms (Cunillera et al., 1997; Manzano et al., 2006; Keim et al., 2012). FPS1 encodes a long isoform targeted to mitochondria (FPP1L) and a short one lacking the N-terminal end that remains in the cytosol, whereas FPS2 only produces a cytosolic enzyme (Fig. 1). Unlike IDI and FPPS, GGPPS paralogs are encoded by a high number of genes in plant genomes, with a particularly large gene family present in Arabidopsis (Lange and Ghassemian, 2003; Coman et al., 2014). From the 12 initially reported genes, however, only four have been conclusively shown to encode true GGPPS enzymes (Nagel et al., 2015; Wang et al., 2016). Two of them, GGPPS3 and GGPPS4, encode proteins sorted to the ER, and the other two, GGPPS2 and GGPPS11, encode plastidial isoforms (Okada et al., 2000; Beck et al., 2013; Coman et al., 2014; Ruiz-Sola et al., 2016). The GGPPS1 gene encodes the only mitochondrial member of the family, but the in vivo activity of the protein is still unclear (Zhu et al., 1997; Okada et al., 2000; Beck et al., 2013; Nagel et al., 2015; Wang et al., 2016). To date, the production of more than one enzyme isoform from a single GGPPS-encoding gene has not been reported.
Despite the presence of at least two GGPPS enzymes in Arabidopsis plastids, GGPPS11 (At4g36810, from herein referred to as G11) is by far the most abundant and ubiquitously expressed isoform (Beck et al., 2013; Ruiz-Sola et al., 2016). G11 is required for the production of all major groups of plastidial isoprenoids, including carotenoids and the side chains of chlorophylls, tocopherols, and prenylated quinones (Ruiz-Sola et al., 2016). Strikingly, several phenotypes have been described for G11-defective mutant alleles (Ruppel et al., 2013; Ruiz-Sola et al., 2016). By carrying out a comprehensive analysis of these alleles, we uncovered here the existence of two differentially targeted G11 enzymes, each of them indispensable for a distinct developmental process likely through the production of a different group of essential isoprenoids.
RESULTS AND DISCUSSION
Different G11-Defective Alleles Show a Range of Phenotypes from Variegation to Embryo Lethality
To better understand the role of G11, the most abundant GGPPS isoform in Arabidopsis, we carefully revised the phenotypes associated to partial or complete loss-of-function mutants (Fig. 2). The ggpps11-1 mutant, originally named ggps1-1 (Ruppel et al., 2013) and here referred to as g11-1, harbors a point mutation that changes a conserved residue (D163R) in G11 (Fig. 2A; Supplemental Fig. S1). The g11-1 allele shows a temperature-dependent variegated phenotype that resembles that of the chs5 mutant (Araki et al., 2000), later renamed dxs-3 (Phillips et al., 2008a), which harbors a D627N mutation in DXS. It is therefore likely that the phenotype of these mutants might be associated to the temperature sensitivity of the corresponding DXS or G11 mutant enzymes, both of them involved in the production of photosynthesis-related isoprenoids (Fig. 1).
Figure 2.
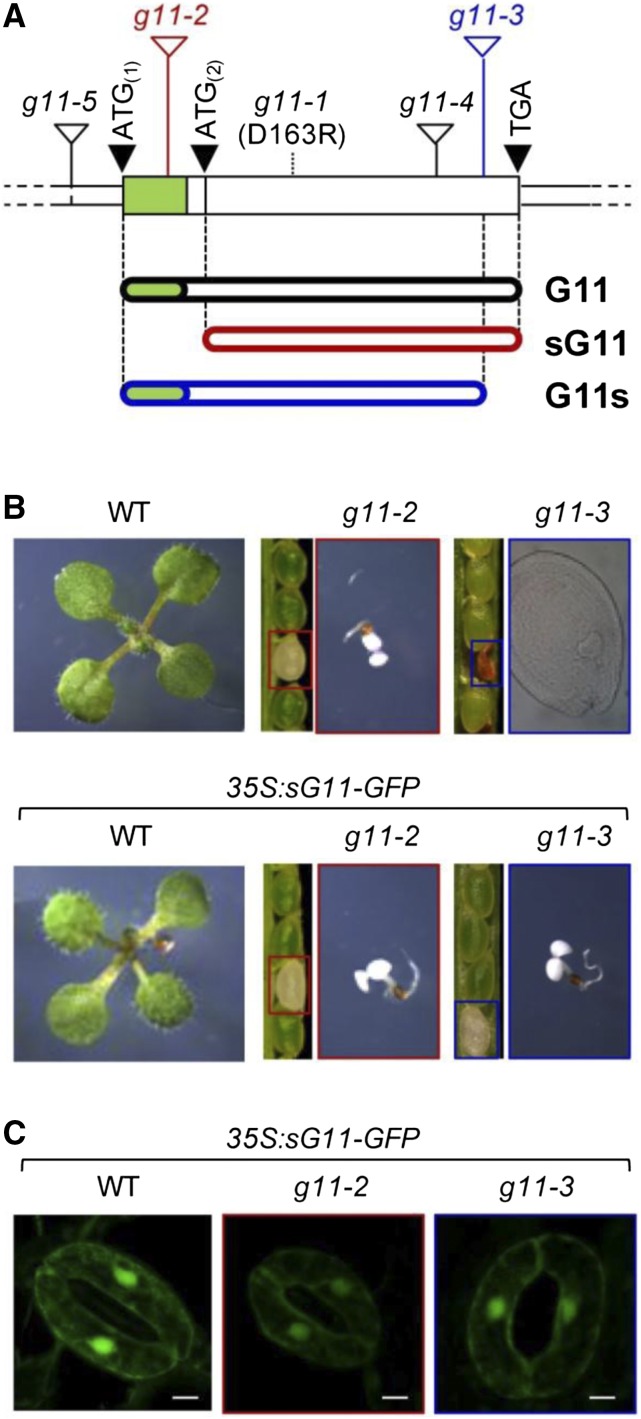
G11 mutant alleles and associated phenotypes. A, G11 gene model according to TAIR v10 annotation. The protein-coding sequence (which lacks introns) is shown as a wider box with a green section corresponding to the predicted plastid targeting sequence. The position of translation start and stop codons is shown with black triangles. The position of T-DNA insertions is represented with white triangles. The position of the point mutation in the g11-1 allele is shown with a dashed line. Lower bars represent encoded proteins. B, Phenotype of G11-defective mutants either producing or not producing a sG11 protein. Representative seedlings of the indicated genotypes grown under long-day conditions for 10 d are shown to the same scale. Segregating populations of seeds in siliques of plants heterozygous for the g11-2 and g11-3 mutations is also shown. Boxed seeds correspond to the homozygous albino mutants represented in the right. Brownish seeds did not produce seedlings due to a blockage in embryo development at the heart stage (as shown in the corresponding picture). C, Cytosolic localization of the sG11-GFP protein. Pictures show GFP fluorescence from the sG11-GFP protein in stomata from 10-d-old seedlings of the indicated genotypes (bars, 5 µm).
A second partial loss-of-function allele, ggpps11-5 (SALK_140601, g11-5), shows a pale phenotype and a developmental delay, likely because a T-DNA insertion upstream of the predicted ATG translation start codon (Fig. 2A; Supplemental Fig. S1) results in a decreased accumulation of G11 transcripts (Ruiz-Sola et al., 2016). In this mutant, lower levels of G11-encoding transcripts are expected to result in an overall reduction in the accumulation of fully active, wild-type G11 protein (Ruiz-Sola et al., 2016). Similarly, a general inhibition of the MEP pathway with sublethal concentrations of the DXS inhibitor clomazone also results in a pale phenotype (Pulido et al., 2013; Perelló et al., 2014). Therefore, the phenotype of g11-1 and g11-5 plants is consistent with the reported role of G11 as the major isoform transforming MEP-derived precursors into GGPP for photosynthesis-related isoprenoid products. Further supporting this conclusion, a seedling-lethal albino phenotype visually identical to that of knockout MEP pathway mutants such as dxs-1, also known as cla1 (Phillips et al., 2008a), was observed in the case of the ggpps11-2 line (SALK_015098, ggps1-2 or g11-2), which harbors a T-DNA insertion in the N-terminal end of the protein coding region of the G11 gene (Fig. 2; Supplemental Fig. S1; Ruppel et al., 2013). By contrast, T-DNA insertions interrupting the C-terminal end of the G11 protein in alleles ggpps11-3 (SALK_085914, ggps1-3 or g11-3) and ggpps11-4 (SAIL_712_D06, g11-4) cause an arrest of embryo development (Fig. 2; Supplemental Fig. S1; Ruppel et al., 2013; Ruiz-Sola et al., 2016). This embryo lethal phenotype has never been observed in MEP pathway mutants (Phillips et al., 2008a).
The Distinct Phenotypes of G11 Alleles Correlate with Differential Subcellular Localization and Activity of the Corresponding Enzymes
To investigate the molecular basis of the puzzling phenotype differences observed between g11-2 (albino, seedling lethal) and g11-3 (embryo lethal) plants (Fig. 2B), we first validated the position of the T-DNA in the mutant genomes by PCR amplification and sequencing of the insertion sites (Supplemental Fig. S1). Insertion of the T-DNA within the predicted plastid targeting sequence in the g11-2 allele (Fig. 2A; Supplemental Fig. S1) is expected to prevent the transcription of a full-length G11 cDNA harboring the first ATG codon (ATG(1)). However, a second in-frame ATG codon (ATG(2)) exists downstream of the T-DNA insertion that could potentially act as an alternative translation initiation point to generate a shorter protein, which we named sG11 (Fig. 2A; Supplemental Fig. S2). We speculated that this shorter protein might not be imported into plastids as it lacked the N-terminal plastid-targeting domain. To test this prediction, a DNA sequence encoding sG11 was generated by removing the ATG(1) codon. The generated sequence was then fused to the N terminus of the GFP reporter in the pCAMBIA1302 vector to obtain the 35S:sG11-GFP construct. As shown in Figure 2C and Supplemental Figure S3, green fluorescence corresponding to the sG11-GFP protein was excluded from plastids and localized in the cytosol of cells from Arabidopsis plants transformed with the construct. By contrast, transgenic plants expressing a similar construct with the wild-type G11 sequence (Ruiz-Sola et al., 2016) showed a predominant association of GFP fluorescence to plastids (Supplemental Fig. S3).
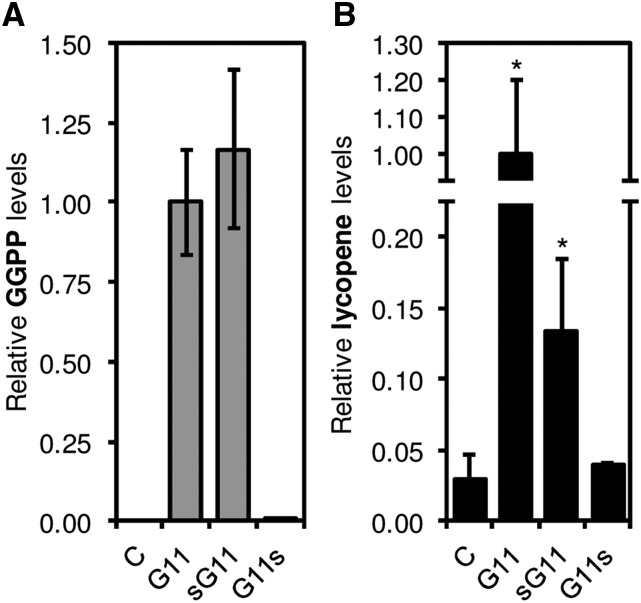
We next evaluated whether sG11 retained the genuine enzymatic activity of the full-length enzyme (Fig. 3). Recent in vitro activity assays followed by analysis of reaction products by liquid chromatography-mass spectrometry (LC-MS; Nagel et al., 2015) or thin-layer chromatography (Wang et al., 2016) confirmed that G11 synthesizes GGPP as the main product. They also revealed that other proteins previously believed to be true GGPPS isoforms actually belong to a novel type of prenyldiphosphate synthases that mainly produce C25 geranylfarnesyl diphosphate (GFPP) or even longer products (Nagel et al., 2015; Wang et al., 2016). The sG11 protein lacks 19 residues predicted to be present in the N-terminal region of the mature G11 enzyme (Fig. 2A; Supplemental Fig. S2). While this N-terminal sequence is not conserved in other GGPPS enzymes and does not include residues determining product length (Supplemental Fig. S2), we aimed to experimentally confirm whether its absence in sG11 had any impact on the ability of the protein to produce GGPP from IPP and DMAPP. In vitro activity assays like those described in Nagel et al. (2015) were carried out using protein extracts from Escherichia coli cells overproducing sG11 or a pseudomature form of G11 lacking the predicted plastid-targeting sequence (Supplemental Fig. S4). Analysis of reaction products by LC-MS detected the production of similar amounts of GGPP in both G11 and sG11 extracts (Fig. 3A). No GPP, FPP, or GFPP were detected in the assays (Supplemental Fig. S4), further indicating that the sG11 protein remains an active GGPPS enzyme. To next determine whether sG11 could also produce GGPP in vivo, we used a heterologous system based on carotenoid (lycopene) production in E. coli strains carrying the pACCRT-BI vector (Beck et al., 2013). Plasmids encoding the full-length G11 or sG11 proteins were used together with pACCRT-BI to cotransform E. coli cells. While cells cotransformed with pACCRT-BI and an empty plasmid control synthesized minor amounts of lycopene due to the presence of only trace levels of GGPP in the bacteria (Vallon et al., 2008), those harboring G11 and sG11 constructs showed significantly increased levels of the carotenoid (Fig. 3B), supporting the conclusion that sG11 synthesizes GGPP in vivo. Together, our results suggest that the lack of the N-terminal region in the sG11 protein produced by g11-2 plants prevents its targeting to plastids but does not override GGPPS activity.
Figure 3.
GGPPS activity assays. A, In vitro assays. Protein extracts from E. coli cells overproducing similar amounts of the indicated proteins or transformed with an empty vector (C) were mixed with IPP and DMAPP, and the production of GGPP was quantified by LC-MS. Levels are represented relative to those in G11 samples. Mean and sd of n = 3 extracts are shown. No GGPP was detected in C samples, and only traces were identified in G11s extracts. B, In vivo assays. Bacterial cells were cotransformed with pACCRT-BI (which lacks a functional GGPPS-encoding gene to produce lycopene) together with a plasmid expressing the indicated G11 isoform or an empty vector (C). Positive transformants were used to measure lycopene levels by normalizing A472 to bacterial growth (OD 600 nm). Levels are represented relative to those in G11 samples. Data correspond to the mean and sd of n = 10 independent transformants. Asterisks mark statistically significant differences from the C control (P < 0.05, one-tailed Student’s t test assuming equal variances).
In the case of the g11-3 mutant, the T-DNA interrupts the sequence encoding the highly conserved C-terminal region of G11 (Supplemental Fig. S2), resulting in a shorter protein that we named G11s (Fig. 2A). Sequencing of the T-DNA insertion site in the g11-3 genome confirmed that the last 21 residues of the wild-type G11 protein are replaced by a single Ser residue in the G11s protein (Supplemental Fig. S1). To test whether loss of the C-terminal region compromised GGPPS activity in the G11s protein, the corresponding DNA sequence was amplified from g11-3 seedlings and cloned into plasmids for E. coli expression. Both in vitro (Fig. 3A) and in vivo (Fig. 3B) activity assays showed that the recombinant G11s protein is unable to produce GGPP. These results suggest that the blockage of embryo development at the heart stage observed in the g11-3 mutant (Fig. 2B; Supplemental Fig. S5) and the g11-4 allele (Ruiz-Sola et al., 2016) might be due to a complete lack of G11 activity. This embryo-lethal phenotype was complemented by expressing a genomic G11 sequence including the promoter and the full protein-coding region (Ruiz-Sola et al., 2016). Most interestingly, embryo development was also rescued by expressing the cytosolic sG11-GFP protein in g11-3 plants (Fig. 2, B and C). Transgenic g11-3 35S:sG11-GFP plants, however, showed a seedling-lethal (albino) phenotype resembling the g11-2 mutant. As expected, the cytosolic sG11-GFP protein was unable to rescue the albino phenotype of the g11-2 mutant (Fig. 2B). We therefore concluded that embryo development beyond the heart stage required the presence of G11 activity in the cytosol (despite two ER-associated GGPPS enzymes exist in Arabidopsis, GGPPS3 and 4), whereas photosynthetic seedling development required the activity of G11 in the plastid (despite a second plastidial enzyme with the same activity, GGPPS2, is found in this plant).
Several Transcription Initiation Sites in the G11 Gene Lead to the Production of Isoforms with Different N-Terminal Ends
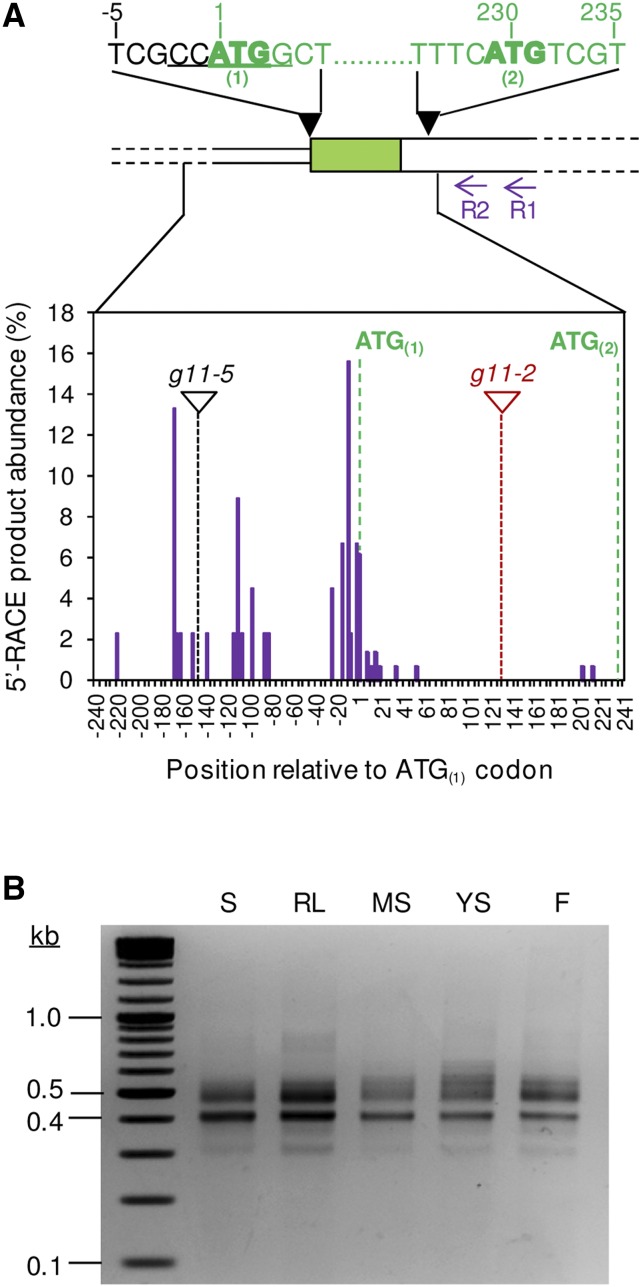
A number of Arabidopsis genes encoding isoprenoid biosynthetic enzymes have been shown to produce transcripts of different lengths encoding isoforms with or without N-terminal transit peptides for plastids and mitochondria (Fig. 1). To determine whether a similar alternative transcription initiation mechanism also occurs for G11, rapid amplification of cDNA 5′ ends (5′-RACE) experiments were performed on RNA extracted from different tissues to assess the length of G11 transcripts in vivo (Fig. 4). The protocol used reverse primers optimized for 5′-RACE (see Fig. 4A; Supplemental Table S1) to amplify gene-specific PCR products while ruling out the possibility of genomic DNA contamination (see “Materials and Methods”). Separation of PCR-amplified products by gel electrophoresis showed the presence of cDNAs of different sizes (Fig. 4B), suggesting that the G11 gene lacks a strictly defined transcription start site. However, amplified fragments could be grouped in two major classes: “long” (approximately 0.5 kb or longer) and “short” (approximately 0.4 kb or shorter). The 5′-RACE products amplified from siliques were cloned and sequenced. Analysis of inserts revealed that all the “long” products included the ATG(1) codon and hence encoded the full-length G11 protein. While most “short” products contained the ATG(1) codon with or without a few nucleotides upstream (up to 25), some lacked ATG(1) and so they can only produce the cytosolic sG11 protein by using the ATG(2) codon (Fig. 4A; Supplemental Fig. S1). The similar pattern of “long” and “short” transcripts detected in seedlings, rosette leaves, and flowers by 5′-RACE experiments (Fig. 4B) suggests that both types of transcripts are likely produced in all tissues. To verify whether the relative abundance of transcripts either lacking or not the ATG(1) codon was indeed similar in different tissues, 5′-RACE products from seedlings were cloned and compared with those from siliques by digestion with NcoI. As shown in Figure 4A, a NcoI target site overlaps the ATG(1) codon, and therefore it can be used to rapidly identify clones lacking this sequence. In both seedlings and siliques, about 10% of the clones could not be cleaved by NcoI, confirming that transcripts exclusively encoding sG11 are low abundant but indeed detectable at similar levels in different tissues.
Figure 4.
Transcription and translation start sites in the G11 gene. A, G11 gene model showing the position of presumed translation start codons ATG(1) and ATG(2) (in bold). The NcoI site in the ATG(1) region is underlined. Numbers indicate the position relative to the first nucleotide of the ATG(1) codon. The position of primers G11-R-R1 and R2 used for 5′-RACE experiments is also shown. Amplified 5′-RACE products (B) were cloned and sequenced to determine the position of transcription initiation sites. The graph represents the percentage of transcripts found to start at the indicated position in siliques based on the analysis of 145 clones (purple columns). The position of ATG(1) and ATG(2) codons and T-DNA insertion sites of the indicated alleles is also shown. B, Agarose gel electrophoresis of 5′-RACE products isolated from seedlings (S), rosette leaves (RL), mature siliques (MS), young siliques (YS), and flowers (F).
G11 Activity Is Essential to Produce Both Plastidial Isoprenoids and Unidentified Extraplastidial Isoprenoids Required for Embryo Development
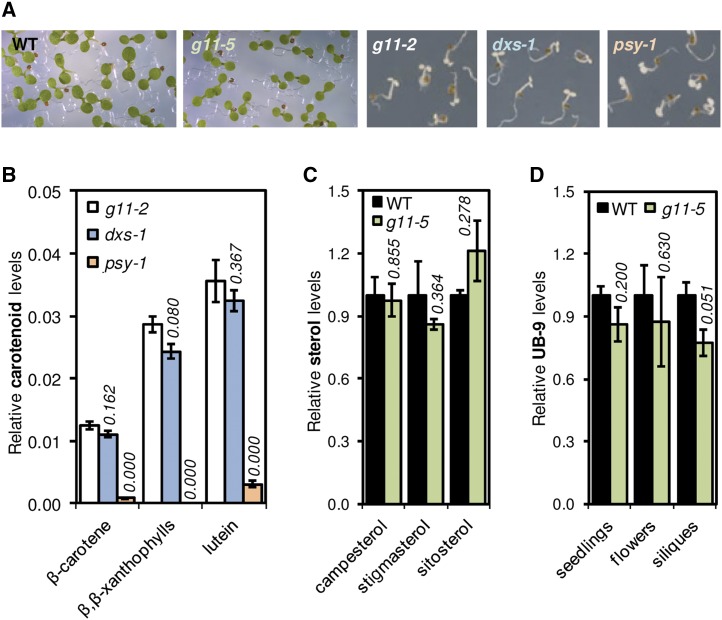
We next aimed to determine the nature of the isoprenoids derived from GGPP produced by plastidial and cytosolic forms of G11. Our previous results with the g11-5 allele showed that the pale phenotype of the mutant was due to a decreased expression of the G11 gene, which caused a reduced accumulation of the major groups of MEP-derived isoprenoids produced from GGPP in the plastid, i.e. carotenoids, chlorophylls, tocopherols, phylloquinones, and plastoquinone (Ruiz-Sola et al., 2016). To verify whether the albino phenotype of g11-2 seedlings was the result of a blockage in the production of plastidial GGPP-derived isoprenoids, we analyzed the levels of carotenoids in the g11-2 mutant (Fig. 5). As controls, we used dxs-1, a MEP pathway null mutant with a complete loss of DXS activity (Phillips et al., 2008a), and psy-1, a knockout mutant in which a specific blockage in the carotenoid pathway results in a similar albino phenotype (Pokhilko et al., 2015; Fig. 5A). Analysis of g11-2 seedlings showed low but detectable levels of carotenoids such as lutein, β-carotene, and β,β-xanthophylls (Fig. 5B). The levels of all these metabolites in g11-2 seedlings were similar to those in the dxs-1 mutants but much higher than the amounts detected in psy-1 seedlings. The results suggest that loss of G11 or DXS activity do not completely block the production of carotenoids in seedlings (as the loss of PSY activity does). The carotenoids detected in g11-2 seedlings might derive from small amounts of GGPP synthesized from MEP-derived precursors by GGPPS2 (the other plastidial GGPPS enzyme found in Arabidopsis). In the case of the dxs-1 mutant, however, the MEP pathway is completely blocked, and hence it is most likely that MVA-derived IPP or DMAPP precursors are transported to the plastid and used to produce GGPP and downstream products. Alternatively, an enhanced import of cytosolic GGPP by nondifferentiated plastids like those found in the albino mutants would make DXS and G11 (but not PSY) dispensable to produce carotenoids. In any case, such a residual production of plastidial isoprenoids is clearly insufficient to support photosynthetic development. We therefore conclude that the albino phenotype of g11-2 seedlings is due to a defective production of GGPP and downstream isoprenoids in the plastid.
Figure 5.
Levels of MEP-derived and MVA-derived isoprenoid products in plants with reduced G11 activity. A, Seedlings of the indicated mutants and the parental Columbia wild-type (WT) grown for 1 week under short-day conditions. These seedlings were used for the metabolite analyses shown in the corresponding graphs. B, Levels of the indicated carotenoids in albino mutants relative to those in wild-type seedlings. C, Levels of major sterols in wild-type and G11-defective g11-5 seedlings. D, Levels of the ubiquinone UB-9 in seedlings, flowers, and young siliques of wild-type and g11 -5 plants. The graphs represent mean and se of at least n = 3 independent samples. Italic numbers above the bars indicate P values (Student’s t test) relative to g11-2 (B) or wild-type (C and D) samples.
To identify the isoprenoid metabolite required for embryo development that is produced from sG11-derived GGPP, it would be necessary to compare the metabolite profile of g11-2 and g11-3 embryos in the transition from globular to heart stage (Fig. 2; Supplemental Fig. S5). Because this is extremely challenging, we followed an alternative approach and evaluated whether the levels of extraplastidial isoprenoids were altered in the g11-5 mutant, which is expected to produce lower amounts not only of plastid-localized G11 but also of cytosolic sG11 enzymes. While MVA-derived precursors are used to produce a wide variety of isoprenoids (Fig. 1), only sterols and ubiquinone appear to be required for proper embryo development (Schrick et al., 2000; Okada et al., 2004). Sterols are not synthesized from GGPP but from FPP (Fig. 1). We therefore expected that reducing sG11 activity (i.e. cytosolic GGPP production) in g11-5 seedlings would not cause a decreased sterol accumulation. Consistently, the levels of sterols (campesterol, stigmasterol, and sitosterol) were found to be similar in wild-type and g11-5 seedlings (Fig. 5C).
In the case of ubiquinone, the predominant form in Arabidopsis (UB-9) contains a C45 solanesyl moiety synthesized by a mitochondrial polyprenyl diphosphate synthase that elongates an initial FPP or GGPP acceptor with IPP units (Hsieh et al., 2011; Ducluzeau et al., 2012). Mitochondria import MVA-derived IPP from the cytosol, as they lack their own biosynthetic pathway (Lütke-Brinkhaus et al., 1984; Disch et al., 1998). Then, specific isoforms of FPPS and GGPPS enzymes targeted to mitochondria (in Arabidopsis, FPPS1L, and GGPPS1; Fig. 1) are presumed to produce FPP and GGPP for ubiquinone synthesis. FPPS1L-defective fps1 mutants only showed a limited decrease in UB-9 levels (Closa et al., 2010), suggesting that the biosynthesis of the ubiquinone solanesyl chain might predominantly rely on the supply of GGPP by GGPPS1, the only GGPPS isoform known to be targeted to mitochondria (Beck et al., 2013; Nagel et al., 2015). However, we found that the T-DNA insertion mutant ggpps1-1 (SAIL_559_G01) contains wild-type levels of UB-9 (Supplemental Fig. S6). Levels of other GGPP-derived isoprenoids were also unaltered in ggpps1-1 seedlings, which showed a wild-type phenotype in terms of plant growth and development (Supplemental Fig. S6). Actually, the role of GGPPS1 as a true GGPPS enzyme still remains controversial as it has not been conclusively established whether its main product is GGPP (Wang et al., 2016) or GFPP (Nagel et al., 2015).
In the absence of a mitochondrial source of GGPP, cytosolic GGPP might be transported to this organelle and used for ubiquinone synthesis. Blockage of this cytosolic source when sG11 activity is lost could actually explain why embryo development is arrested at the same heart stage in mutants defective in ubiquinone synthesis (Okada et al., 2004) and sG11 activity, i.e. g11-3 (Supplemental Fig. S5) and g11-4 (Ruiz-Sola et al., 2016). Analysis of UB-9 contents in seedlings, flowers, and young siliques of the g11-5 mutants detected slightly reduced levels compared to wild-type samples, but the differences were not found to be statistically significant (Fig. 5D). In the case of mutant siliques, however, the observed reduction in ubiquinone levels was close to statistical significance (Student’s t test; P = 0.051). Together, these results suggest two possibilities. First, sG11-derived GGPP might be critical for the biosynthesis of ubiquinone during embryogenesis, allowing embryo development to progress beyond the heart stage. Considering that a ca. 50% reduction of G11 transcript levels in g11-5 seedlings only causes a 20%–30% decrease in plastidial isoprenoid content (Ruiz-Sola et al., 2016), the observed 20% reduction of UB-9 levels in young siliques of the g11-5 mutant might indeed result from partly reduced sG11 activity (and hence GGPP supply) in developing embryos or seeds. Because other GGPPS isoforms are also expressed in these tissues at different stages (Beck et al., 2013), it remains unclear why none of them is able to complement the loss of sG11 activity and hence rescue embryo development in g11-3 and g11-4 mutants. Alternatively, sG11 might produce GGPP for another specific class of unidentified metabolites required for embryogenesis and, perhaps, with roles in other cells and tissues during the plant life cycle, as deduced from the wide distribution of sG11-encoding transcripts (Fig. 4B).
CONCLUSION
Our results support the conclusion that the Arabidopsis G11 gene can produce two enzyme isoforms: one translated from the ATG(1) codon and carrying a plastid-targeting peptide (G11) and a shorter version translated from the downstream ATG(2) codon and lacking the plastid-targeting peptide (sG11). Mechanistically, the production of these two differentially targeted isoforms might rely on both the use of alternative transcription start sites (resulting in the synthesis of mRNAs with or without a 5′-region encoding the plastid transit peptide) or the use of alternative translation start sites (ATG(1) or ATG(2)) in the long transcript. The NetStart algorithm for the prediction of translation start codons in Arabidopsis (http://www.cbs.dtu.dk/services/NetStart/) actually gives a similar score to ATG(1) (0.627) and ATG(2) codon (0.651), suggesting that both could be functional in vivo. It is remarkable that a similar situation has been reported to occur in other Arabidopsis genes encoding key isoprenoid biosynthetic enzymes, including HMGR, IDI, and FPPS. Unlike these other enzymes, however, no redundancy appears to occur in the case of Arabidopsis GGPPS enzymes, as the loss of G11 cannot be rescued by plastidial GGPPS2, ER-localized GGPPS3 and GGPPS4, or mitochondrial GGPPS1. Furthermore, the Met residue encoded by the ATG(2) codon in G11 does not appear to be conserved in the rest of isoforms confirmed to synthesize GGPP (Supplemental Fig. S2), suggesting that G11 might be the only Arabidopsis GGPPS-encoding gene producing more than one isoform.
Whereas G11 activity in the plastid is indispensable for the production of plastidial isoprenoids supporting chloroplast development and photosynthesis, sG11 activity in the cytosol supplies the precursors for an unidentified isoprenoid metabolite that is essential for embryo development. Interestingly, loss of sG11 activity in g11-3 and g11-4 mutants prevents embryo development to proceed beyond the heart stage (Supplemental Fig. S5; Ruiz-Sola et al., 2016), similar to that observed in ubiquinone-defective mutants (Okada et al., 2004), whereas complete loss of FPPS activity in Arabidopsis fps1 fps2 double mutants blocks embryogenesis at the earlier globular stage (Closa et al., 2010). These results suggest that while FPP-derived isoprenoids are needed for the transition of the embryo from globular to heart stages, progression beyond the heart stage requires ubiquinone or/and a different metabolite produced from sG11-supplied precursors. Besides synthesizing GGPP as a homodimer, the plastidial G11 isoform has been found to produce GPP upon heterodimerization with another plastidial GGPPS-like protein (Wang and Dixon, 2009; Chen et al., 2015). Whether the cytosolic sG11 protein unveiled here also has alternative enzyme activities upon association with other proteins remains unknown. In any case, our results support the conclusion that the production of GGPP required for essential functions in different cell compartments relies on the activity of G11 isoforms. Other GGPPS paralogs might be maintained in the Arabidopsis genome for developmental and/or condition-specific subfunctionalization. Future experiments, including the high-resolution analysis of isoprenoid profiles in specialized tissues and organs (e.g. embryos) from wild-type and GGPPS-defective mutants, should provide additional insights on the biological role of specific isoforms and their corresponding downstream GGPP-derived products, further allowing to understand the astounding complexity of the mechanisms used by plants to produce isoprenoids.
MATERIALS AND METHODS
Plant Material and Constructs
All the Arabidopsis (Arabidopsis thaliana) lines used in this work are in the Columbia background. The T-DNA insertion alleles g11-3, g11-4, g11-5, dxs-1, and psy-1 were already available in the lab (Pokhilko et al., 2015; Ruiz-Sola et al., 2016). Homozygous ggpps1-1 mutants were isolated from a segregating population of the SAIL_559_G01 line supplied by the European Arabidopsis Stock Centre (http://arabidopsis.info/). Primers for PCR-based genotyping of the mutants and sequencing of T-DNA insertion sites are indicated in Supplemental Table S1. The 35S:sG11-GFP construct was generated by deleting the ATG(1) codon of the G11 sequence in the pSG11G plasmid (Ruiz-Sola et al., 2016). As shown in Supplemental Table S2, the pSG11G plasmid was digested with NcoI (which has a target sequence overlapping the ATG(1) codon; Fig. 4A) and subsequently treated with Mung Bean nuclease (New England Biolabs) to remove single-stranded extensions and generate blunt ends that were eventually ligated using T4 ligase enzyme (Roche). After Agrobacterium-mediated plant transformation, homozygous lines containing a single T-DNA insertion of the 35S:sG11-GFP construct were selected based on the segregation of the resistance marker (hygromycin). Seeds were surface-sterilized and germinated in petri dishes with solid Murashige and Skoog medium without vitamins or Suc. After stratification for 3 d at 4°C, plates were incubated in a growth chamber at 22°C and illuminated for 16 h (long-day) or 8 h (short-day) a day with fluorescent white light at a photosynthetic photon flux density of 60 µmol m−2 s−1.
GGPPS Activity Assays
Constructs to express different truncated G11 versions were generated as described (Supplemental Table S2). Recombinant proteins were produced in Escherichia coli BL21 pGROE cells using the Overnight Express AutoInduction System 1 (Merck Millipore). After growth for 72 h at 18°C, bacterial cells were recovered by centrifugation, and pellets were resuspended in reaction buffer (25 mm HEPES, pH 7.2, 10 mm MgCl2, 10% v/v glycerol) supplemented with 1 mg/mL lysozyme and one tablet of complete protease inhibitor cocktail (Roche) for every 10 mL of buffer. The resuspended pellet was incubated at 4°C for 10 min, and after a brief sonication (five pulses of 20 s at 30 W), the cell lysate was centrifuged at 19,000g at 4°C for 5 min. The supernatant was used for SDS-PAGE, protein quantification, and GGPP activity assays as described (Nagel et al., 2015). The reaction mix contained 10 µg of total protein from extracts showing similar levels of recombinant protein in 200 µL of reaction buffer, 50 µm IPP, and 50 µm DMAPP. After incubation for 1 h at 30°C, the reaction was stopped by adding 800 µL of methanol. A previously described LC-FTMS system (Henneman et al., 2008) was adapted to detect prenyl diphosphate products. A Luna C18(2) precolumn (2.0 × 4 mm) and an analytical column (2.0 × 150 mm, 100 nm, particle size 3 μm) from Phenomenex were used for chromatographic separation at 40°C, using an Acquity UPLC (H-Class), connected to an LTQ-Orbitrap XL hybrid mass spectrometry system (Waters) operating in negative electrospray ionization mode heated at 300°C with a source voltage of 4.5 kV for full-scan LC-MS in the m/z range 100 to 1300. The injection volume was 10 µL. Compounds were separated by a linear gradient between solution A (20 mm NH4HCO3 with 0.1% triethylamine) and solution B (acetonitrile/H2O, 4:1 with 0.1% triethylamine). The gradient was as follows: 0 to 18 min, 100% A to 80% A; 18 to 23 min, 80% A to 0% A; 23 to 25 min, 0% A; 25 to 30 min, 0% A to 100% A; equilibration with 100% A. Acquisition and visualization of the data were performed using Xcalibur software (Thermo Fischer). GPP, FPP, and GGPP standards were obtained from Sigma and used for quantification.
For in vivo activity assays, E. coli TOP10 cells were cotransformed with both pACCRT-BI (Beck et al., 2013) and plasmids harboring the corresponding G11 versions generated as described (Supplemental Table S2). Transformants containing both plasmids were selected on LB medium containing both ampicillin (100 µg/mL) and chloramphenicol (25 µg/mL). Positive colonies were selected and grown overnight at 37°C in liquid LB. Fresh liquid LB medium supplemented with the appropriate antibiotics was then inoculated with the overnight culture and incubated for 7 more days at 30°C. Aliquots of 10 mL of grown culture were harvested and used for lycopene extraction and quantification as described (Beck et al., 2013).
Microscopy
Subcellular localization of the GFP fusion proteins was analyzed by direct examination of plant tissue samples with an Olympus FV 1000 Confocal Laser Scanning Microscope. Green fluorescence corresponding to the fusion proteins was detected using an argon laser for excitation with blue light at 488 nm and a 500 to 510-nm filter, whereas a 610 to 700-nm filter was used for detection of chlorophyll autofluorescence. Clearing of Arabidopsis seeds and examination of embryo developmental stages was performed as described (Ruiz-Sola et al., 2016).
5′-RACE
For 5′-RACE analysis, total RNA from different organs of wild-type plants was isolated using an RNA purification kit (Sigma-Aldrich) and used for first-strand cDNA synthesis with the SMARTer RACE cDNA amplification kit (Clontech). High-Fidelity AccuPrime Taq DNA Polymerase (Invitrogen) was used with primers provided in the kit and the G11-R-R1 primer (Supplemental Table S1) for 5′-RACE reactions, adding an initial denaturation step of 2 min at 94°C to the recommended PCR program to activate the polymerase, and changing the elongation temperature from 72°C to 68°C. PCR products were cloned into the cloning vector pCRII-TOPO (Invitrogen) for further restriction enzyme and sequencing analysis using the G11-R-R2 primer (Supplemental Table S1).
Analysis of Metabolite Levels
Published methods were used for the extraction, separation, and quantification of photosynthetic pigments (chlorophylls and carotenoids; Rodríguez-Concepción et al., 2004), sterols (Closa et al., 2010), and prenylquinones, including ubiquinone (Martinis et al., 2011).
Accession Numbers
Accessions are deposited in the TAIR database (www.arabidopsis.org): At1g49530, GGPPS1; At4g36810, GGPPS11 (here referred to as G11). See also Figure 1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Arabidopsis G11 gene and mutants.
Supplemental Figure S2. Sequence alignment of Arabidopsis GGPPS isoforms.
Supplemental Figure S3. Subcellular localization of G11-GFP and sG11-GFP fusion proteins in transgenic Arabidopsis plants.
Supplemental Figure S4. In vitro activity of truncated G11 isoforms.
Supplemental Figure S5. Embryogenesis is blocked at the heart stage in the g11-3 mutant.
Supplemental Figure S6. Developmental and metabolic phenotypes of the ggpps1-1 mutant.
Supplemental Table S1. Primers used in this work.
Supplemental Table S2. Constructs and cloning details.
Acknowledgments
We are very grateful to Gaétan Glauser (University of Neuchâtel, Switzerland) for the analysis of prenylquinones. Technical support from M. Rosa Rodríguez-Goberna and members of the CRAG Services is greatly appreciated.
Glossary
- MVA
mevalonic acid
Footnotes
This work was funded by grants from the Ministerio de Economía y Competitividad (BES-2009-025359, BIO2011-23680, BIO2014-59092-P, PCIN-2015-103, BIO2015-71703-REDT, and FPDI-2013-018882), Ministerio de Educación, Cultura y Deporte (FPU14/05142), European Commission H2020 ERA-IB-2 program (BioProMo), and Generalitat de Catalunya (2014SGR-1434 and 2015FI_B 00007).
Articles can be viewed without a subscription.
References
- Araki N, Kusumi K, Masamoto K, Niwa Y, Iba K (2000) Temperature-sensitive Arabidopsis mutant defective in 1-deoxy-D-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiol Plant 108: 19–24 [Google Scholar]
- Beck G, Coman D, Herren E, Ruiz-Sola MA, Rodríguez-Concepción M, Gruissem W, Vranová E (2013) Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol Biol 82: 393–416 [DOI] [PubMed] [Google Scholar]
- Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415: 146–154 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429 [DOI] [PubMed] [Google Scholar]
- Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A (1989) Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Mol Biol 13: 627–638 [DOI] [PubMed] [Google Scholar]
- Chen Q, Fan D, Wang G (2015) Heteromeric geranyl(geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol Plant 8: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Closa M, Vranová E, Bortolotti C, Bigler L, Arró M, Ferrer A, Gruissem W (2010) The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J 63: 512–525 [DOI] [PubMed] [Google Scholar]
- Coman D, Altenhoff A, Zoller S, Gruissem W, Vranová E (2014) Distinct evolutionary strategies in the GGPPS family from plants. Front Plant Sci 5: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388 [DOI] [PubMed] [Google Scholar]
- Disch A, Hemmerlin A, Bach TJ, Rohmer M (1998) Mevalonate-derived isopentenyl diphosphate is the biosynthetic precursor of ubiquinone prenyl side chain in tobacco BY-2 cells. Biochem J 331: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau AL, Wamboldt Y, Elowsky CG, Mackenzie SA, Schuurink RC, Basset GJ (2012) Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J 69: 366–375 [DOI] [PubMed] [Google Scholar]
- Enjuto M, Balcells L, Campos N, Caelles C, Arró M, Boronat A (1994) Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA 91: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneman L, van Cruchten AG, Denis SW, Amolins MW, Placzek AT, Gibbs RA, Kulik W, Waterham HR (2008) Detection of nonsterol isoprenoids by HPLC-MS/MS. Anal Biochem 383: 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FL, Chang TH, Ko TP, Wang AH (2011) Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol 155: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim V, Manzano D, Fernández FJ, Closa M, Andrade P, Caudepón D, Bortolotti C, Vega MC, Arró M, Ferrer A (2012) Characterization of Arabidopsis FPS isozymes and FPS gene expression analysis provide insight into the biosynthesis of isoprenoid precursors in seeds. PLoS One 7: e49109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Campos N, Boronat A (1995) The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J 8: 541–549 [DOI] [PubMed] [Google Scholar]
- Lütke-Brinkhaus F, Liedvogel B, Kleinig H (1984) On the biosynthesis of ubiquinones in plant mitochondria. Eur J Biochem 141: 537–541 [DOI] [PubMed] [Google Scholar]
- Manzano D, Busquets A, Closa M, Hoyerová K, Schaller H, Kamínek M, Arró M, Ferrer A (2006) Overexpression of farnesyl diphosphate synthase in Arabidopsis mitochondria triggers light-dependent lesion formation and alters cytokinin homeostasis. Plant Mol Biol 61: 195–213 [DOI] [PubMed] [Google Scholar]
- Martinis J, Kessler F, Glauser G (2011) A novel method for prenylquinone profiling in plant tissues by ultra-high pressure liquid chromatography-mass spectrometry. Plant Methods 7: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Bernholz C, Vranová E, Košuth J, Bergau N, Ludwig S, Wessjohann L, Gershenzon J, Tissier A, Schmidt A (2015) Arabidopsis thaliana isoprenyl diphosphate synthases produce the C25 intermediate geranylfarnesyl diphosphate. Plant J 84: 847–859 [DOI] [PubMed] [Google Scholar]
- Okada K, Kasahara H, Yamaguchi S, Kawaide H, Kamiya Y, Nojiri H, Yamane H (2008) Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol 49: 604–616 [DOI] [PubMed] [Google Scholar]
- Okada K, Ohara K, Yazaki K, Nozaki K, Uchida N, Kawamukai M, Nojiri H, Yamane H (2004) The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol Biol 55: 567–577 [DOI] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelló C, Rodríguez-Concepción M, Pulido P (2014) Quantification of plant resistance to isoprenoid biosynthesis inhibitors. Methods Mol Biol 1153: 273–283 [DOI] [PubMed] [Google Scholar]
- Phillips MA, D’Auria JC, Gershenzon J, Pichersky E (2008b) The Arabidopsis thaliana type I Isopentenyl Diphosphate Isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell 20: 677–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, León P, Boronat A, Rodríguez-Concepción M (2008a) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci 13: 619–623 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Bou-Torrent J, Pulido P, Rodríguez-Concepción M, Ebenhöh O (2015) Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol 206: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Pulido P, Perello C, Rodriguez-Concepcion M (2012) New insights into plant isoprenoid metabolism. Mol Plant 5: 964–967 [DOI] [PubMed] [Google Scholar]
- Pulido P, Toledo-Ortiz G, Phillips MA, Wright LP, Rodríguez-Concepción M (2013) Arabidopsis J-protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. Plant Cell 25: 4183–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Boronat A (2015) Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr Opin Plant Biol 25: 17–22 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Forés O, Martinez-García JF, González V, Phillips MA, Ferrer A, Boronat A (2004) Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola MA, Coman D, Beck G, Barja MV, Colinas M, Graf A, Welsch R, Rütimann P, Bühlmann P, Bigler L, et al. (2016) Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol 209: 252–264 [DOI] [PubMed] [Google Scholar]
- Ruppel NJ, Kropp KN, Davis PA, Martin AE, Luesse DR, Hangarter RP (2013) Mutations in GERANYLGERANYL DIPHOSPHATE SYNTHASE 1 affect chloroplast development in Arabidopsis thaliana (Brassicaceae). Am J Bot 100: 2074–2084 [DOI] [PubMed] [Google Scholar]
- Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam S, Frydman A, Gidoni D, Eyal Y (2008) Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol 148: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Vallon T, Ghanegaonkar S, Vielhauer O, Müller A, Albermann C, Sprenger G, Reuss M, Lemuth K (2008) Quantitative analysis of isoprenoid diphosphate intermediates in recombinant and wild-type Escherichia coli strains. Appl Microbiol Biotechnol 81: 175–182 [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64: 665–700 [DOI] [PubMed] [Google Scholar]
- Vranová E, Hirsch-Hoffmann M, Gruissem W (2011) AtIPD: a curated database of Arabidopsis isoprenoid pathway models and genes for isoprenoid network analysis. Plant Physiol 156: 1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen Q, Fan D, Li J, Wang G, Zhang P (2016) Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol Plant 9: 195–204 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Suzuki K, Saito T, Okada K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M (1997) Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol Biol 35: 331–341 [DOI] [PubMed] [Google Scholar]