DWARF 14 protein, a strigolactone receptor, is transported through phloem to axillary buds and the transport is required for full function of strigolactones to suppress shoot branching.
Abstract
The cell-to-cell transport of signaling molecules is essential for multicellular organisms to coordinate the action of their cells. Recent studies identified DWARF14 (D14) as a receptor of strigolactones (SLs), molecules that act as plant hormones and inhibit shoot branching. Here, we demonstrate that RAMOSUS3, a pea ortholog of D14, works as a graft-transmissible signal to suppress shoot branching. In addition, we show that D14 protein is contained in phloem sap and transported through the phloem to axillary buds in rice. SLs are not required for the transport of D14 protein. Disruption of D14 transport weakens the suppression of axillary bud outgrowth of rice. Taken together, we conclude that the D14 protein works as an intercellular signaling molecule to fine-tune SL function. Our findings provide evidence that the intercellular transport of a receptor can regulate the action of plant hormones.
Intercellular transport of signaling molecules is critical in cell-to-cell communication and therefore essential for multicellular organisms to coordinate the action of their cells. Plants use two distinct systems, apoplastic and symplastic, for the intercellular transport of signaling molecules (Bloemendal and Kück, 2013). In the apoplastic system, signaling molecules are secreted from cells to the apoplast. After moving through the apoplast, they are then perceived by receptors in other cells. Most plant hormones and peptides are thought to be transported by this system (Bloemendal and Kück, 2013). Various kinds of molecules are also transported in the symplastic system through the plasmodesmata (PD), channels that connect the cytoplasm of plant cells with neighboring cells (Bloemendal and Kück, 2013). PD have a larger diameter than gap junctions, structures for symplastic transport in animals, and are therefore able to transport macromolecules, including proteins (Bloemendal and Kück, 2013). Disruption of PD permeability causes defects in several aspects of growth and development, indicating the importance of symplastic transport in the regulation of plant development (De Storme and Geelen, 2014). For example, the short-distance transport of transcription factors through PD is crucial in tissue patterning and organ formation (Chen et al., 2016; Kawade et al., 2013; Tsukagoshi et al., 2010; Yadav et al., 2011).
Signaling molecules can also be symplastically transported over long distances through the phloem. The phloem is composed of companion cells (CCs), sieve elements (SEs), and parenchyma cells. CCs and SEs are connected via PD. Molecules synthesized in or transported to the CCs are sent to the SEs and then released into the flow of the phloem sap. Various molecules, such as plant hormones, RNAs, and proteins, are present in the phloem sap (Turgeon and Wolf, 2009). For example, Arabidopsis (Arabidopsis thaliana) FT and rice Hd3a, a long-distance florigen signal, are synthesized in the CCs of leaves and transported through the phloem to the shoot apical meristem where flowering is induced (Liu et al., 2013). ELONGATED HYPOCOTYL5, another example of a transported protein, is a transcription factor that regulates photomorphogenesis and is transported from shoot to root through phloem and regulates light-dependent root elongation and nitrate uptake in Arabidopsis (Chen et al., 2016).
Strigolactones (SLs) are a class of plant hormones that regulate many aspects of plant growth and development in order to optimize growth in response to changes in environmental conditions (Brewer et al., 2013; Gomez-Roldan et al., 2008; Seto et al., 2012; Umehara et al., 2008). In rice, three proteins, DWARF3 (D3), DWARF14 (D14), and DWARF53 (D53), an F-box protein, an α/β-fold hydrolase protein, and a Clp ATPase protein, respectively, have been identified as components of the SL signaling pathway (Arite et al., 2009; Ishikawa et al., 2005; Jiang et al., 2013; Zhou et al., 2013). D53 represses the downstream events of SL signaling in the absence of SLs. The receptor of SLs is D14. Perception and hydrolysis of SLs by D14 induces formation of a complex of D14, D3, and D53. Subsequently, D53 is polyubiquitinated by D3 and degraded via the 26S proteasome pathway (Hamiaux et al., 2012; Jiang et al., 2013; Yao et al., 2016; Zhou et al., 2013).
In contrast to the rapid progress in understanding mechanisms of SL signal transduction, information about the sites of SL biosynthesis and function are still fragmental. The shoot branching defects in scions of SL biosynthesis mutants were rescued by grafting to wild-type rootstocks or stem interstocks (Bainbridge et al., 2005; Beveridge, 2000; Booker et al., 2004, 2005; Foo et al., 2001; Morris et al., 2001; Simons et al., 2007). This implies that SLs or intermediates synthesized in rootstocks and the stem interstocks are transported to scions. In petunia, SL transport is regulated by PLEIOTROPIC DRUG RESISTANCE 1 (PDR1), an SL efflux carrier (Kretzschmar et al., 2012; Sasse et al., 2015). It has been shown that SLs are required for the response to phosphate deficiency in shoots and that production and transport of SLs are regulated by phosphate availability (Foo et al., 2013; Kohlen et al., 2011; Kretzschmar et al., 2012; López-Ráez et al., 2008; Sun et al., 2014; Umehara et al., 2010; Yoneyama et al., 2012, 2013).
Grafting experiments are widely used to determine whether hormone mutants may have defects in biosynthesis or signaling (Beveridge et al., 1996; Beveridge, 2000; Beveridge et al., 2005; Booker et al., 2004, 2005; Foo et al., 2001; Johnson et al., 2006; Morris et al., 2001; Simons et al., 2007). In principle, biosynthesis mutants are rescued by grafting with wild-type plants, while signaling mutants are not. Indeed, branching in scions of the pea ramosus1 (rms1) mutant, an SL biosynthesis mutant, was inhibited by wild-type rootstocks (Beveridge, 2000; Foo et al., 2001; Morris et al., 2001). On the other hand, defects in rms4, a D3 mutant of pea, were not rescued by grafting (Beveridge et al., 1996; Beveridge, 2000; Johnson et al., 2006; Morris et al., 2001). Although the increased branching phenotype of pea rms3 was sometimes reduced by grafting with wild-type plants, rms3 was regarded as a signaling mutant because the rescue of the phenotypes was not consistent or only partial (Beveridge et al., 1996, 2000; Morris et al., 2001). Recently, it was shown that RMS3 is a pea ortholog of D14 (de Saint Germain et al., 2016). This, together with the grafting studies, indicates that RMS3 or downstream components of RMS3 may act non-cell-autonomously despite D14 functioning as a receptor in the SL signaling pathway. In Arabidopsis, comparison of expression patterns of AtD14 mRNA and protein suggest short-distance transport of the AtD14 protein (Chevalier et al., 2014). Furthermore, the D14 protein was detected in the phloem sap of rice and Arabidopsis using proteomic analysis (Aki et al., 2008; Batailler et al., 2012). These reports indicate that the D14 protein is transported through phloem.
Here, we demonstrate by further grafting experiments that RMS3 works as a graft-transmissible signal that partially suppresses shoot branching in pea. We show that D14 is present in the phloem sap of rice and transported to the axillary buds. Disruption of D14 transport weakens the function of SLs to suppress the growth of tillers in rice. We propose that the D14 protein functions as an intercellular signaling molecule to fine-tune SL signaling.
RESULTS
RMS3, a Pea Ortholog of D14, Works as a Graft-Transmissible Signal
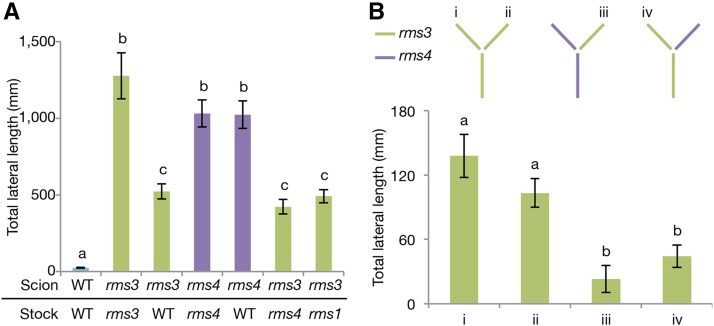
Previous research indicated that RMS3, a pea ortholog of D14, may control a graft-transmissible signal, because branching of the rms3 scion was partially inhibited by grafting to a wild-type rootstock (Beveridge et al., 1996). In this study, we further investigated these results using other rootstocks and two-shoot grafts. Wild-type rootstocks reduced shoot branching of rms3 scions as previously reported (Beveridge et al., 1996; Fig. 1A; Supplemental Fig. S1, A and B), indicating that RMS3 or downstream components of the SL signal work as a mobile signal. As shown in previous studies, the complementation of rms3 branching is much weaker than that of SL biosynthesis mutants grafted onto a wild-type rootstock (Beveridge et al., 1996; Beveridge, 2000; Foo et al., 2001; Morris et al., 2001; Fig. 1; Supplemental Fig. S1).
Figure 1.
RMS3 works as a graft-transmissible signal. A, Total length of lateral branches of wild type (WT), rms3, and rms4 scions grafted with WT, rms3, rms4, and rms1 at their epicotyls 42 d after grafted plants were scored. B, Total length of lateral branches of rms3 grafted with rms3 and rms4 using the two-shoot graft technique 27 d after grafted plants were scored. rms3 scions grafted to rms3 stock (i, ii); rms3 scion grafted with an rms4 scion to rms4 stock (iii); and rms3 scion grafted with an rms4 scion to rms3 stock (iv). Data are means ± se; n = 15 to 20 (A); n = 9 to 12 (B). The different letters denote significant differences at P < 0.05 (one-way ANOVA with Tukey-Kramer test).
RMS4, an ortholog of D3 in pea, is another component of SL signaling (Johnson et al., 2006). In contrast to rms3, branching of rms4 scions was never suppressed by grafting to wild-type rootstocks (Fig. 1A; Supplemental Fig. S1, A and C; Beveridge et al., 1996; Beveridge, 2000; Johnson et al., 2006; Morris et al., 2001). This indicates that RMS4 or downstream components of RMS4 are not graft-transmissible. Interestingly, branching of rms3 scions was also suppressed when grafted onto either rms4 SL-response or rms1 SL-biosynthesis mutant rootstocks (Fig. 1A; Supplemental Fig. S1B). These results indicate that SLs and downstream components of the SL signal are not required for the graft-transmissible function of RMS3. Thus, RMS3 itself, presumably its mRNA or protein, works as a graft-transmissible signal to suppress shoot branching.
The direction of transport of the RMS3 signal was analyzed using two-shoot grafting. As illustrated in Figure 1B, rms3 and rms4 scions were grafted on an rms3 or an rms4 rootstock. While standard grafting experiments showed that the RMS3 signal is transported acropetally (Fig. 1A), the partial suppression of branching in rms3 scions grafted with an rms4 scion and rms3 rootstock indicates that the suppressive signal is able to basipetally move from one shoot to another (Fig. 1B; Supplemental Fig. S1D). These various grafting results indicate that the transport of the RMS3 signal occurs both acropetally and basipetally. The bidirectional transport of the RMS3 signal is in contrast with the unidirectional transport of SLs, as suggested by grafting studies (Foo et al., 2001). These differences might reflect a difference in the transport mechanisms of the RMS3 signal and SLs. It has been shown previously that SL is transported in a polar manner by PDR1 (Sasse et al., 2015). In contrast, the phloem, which moves according to source and sink relationships, can move in both directions (Thieme et al., 2015).
D14 Protein Is Contained in the Phloem Sap of Rice
We used rice for further examination of D14 transport because of the availability of reporter lines. The D14 protein has been detected previously in the phloem sap of rice and Arabidopsis using proteomic analysis (Aki et al., 2008; Batailler et al., 2012). To confirm the presence of the D14 protein in rice phloem sap, we expressed a D14:GFP fusion protein under the control of the native D14 promoter in wild-type rice plants. Since the intensity of GFP fluorescence in phloem was not sufficiently high, localization of the D14:GFP fusion protein was examined by immunostaining with an anti-GFP antibody. The lack of a detectable signal in nontransgenic wild-type plants (Fig. 2A) indicates that the specificity of this antibody is sufficiently high. The signal observed in pD14::D14:GFP plants is most likely derived from the D14:GFP fusion protein rather than from a degraded D14:GFP fusion protein, because free GFP was not detected in western blotting experiments (Supplemental Fig. S2).
Figure 2.
D14 protein is distributed in SEs. Immunostaining of phloem using an anti-GFP antibody in WT (A), pD14::D14:GFP/WT (B) and pD14::D14:3xGFP/WT (C). B, pD14::D14:GFP/WT. C, pD14::D14:3xGFP/WT. Asterisk, SEs; triangle, CCs; bars = 5 μm.
Phloem is composed of CCs, SEs (through which phloem sap is transported), and parenchyma cells. In pD14::D14:GFP lines, the GFP signal was observed in the CCs and SEs (Fig. 2B) confirming that the D14 protein exists in the phloem sap. Since mature SEs are highly specialized cells lacking nuclei and ribosomes, proteins in phloem sap are thought to be synthesized in CCs linked to the SEs or in immature SEs and transported through PD (Turgeon and Wolf, 2009). In general, the increase in the Mr caused by fusion with another protein reduces the PD permeability of the protein (Kawade et al., 2013; Tsukagoshi et al., 2010; Yadav et al., 2011). To confirm that D14 protein is transported from CCs to SEs, we generated pD14::D14:3xGFP lines that produce the D14 protein fused with three repeated copies of the GFP protein. In the pD14::D14:3xGFP lines, the GFP signal was detected in CCs but not in SEs (Fig. 2C). This result supports the hypothesis that the D14 protein is transported from CCs to SEs.
D14 Protein Is Transported to the Axillary Buds in Rice
To understand the significance of intercellular D14 protein movement, we examined D14 protein movement in axillary buds in more detail. We first investigated D14 mRNA expression patterns by in situ hybridization. Our results indicate that D14 mRNA accumulates in vascular bundles and leaf primordia (Fig. 3, A and B). In vascular bundles, D14 mRNA is expressed in CCs and parenchyma cells in phloem and xylem parenchyma cells (Fig. 3C). D14 mRNA was below detectable levels in axillary meristems (Fig. 3B).
Figure 3.
D14 mRNA expression pattern. In situ hybridization analysis using a D14 mRNA anti-sense probe in basal part of rice shoot (A), axillary bud in the axil of second leaf (B) and vascular bundle of a leaf (C). Arrows, a vascular bundles; triangles, axillary meristems; Ph, phloem; Xy, xylem. Bars = 50 μm.
Next, we examined the spatial localization of the D14 protein in wild-type rice plants transformed with pD14::D14:GFP and pD14::D14:3xGFP. We confirmed that GFP mRNA expression patterns in the transgenic lines were indistinguishable from that of endogenous D14 mRNA (Fig. 3B; Supplemental Fig. S3, A and C). In pD14::D14:GFP lines, GFP fluorescence was observed in the axillary meristem where D14:GFP mRNA was not detected (Fig. 4A; Supplemental Fig. S3, A and C). In pD14::D14:3xGFP lines that produce immobile D14:3xGFP fusion protein, while GFP fluorescence was observed in the vascular bundles and the basal part of the meristem, it was undetected in the axillary meristems (Fig. 4B). These results suggest that the GFP fluorescence observed in the axillary meristem of pD14::D14:GFP lines is derived from D14:GFP transported to the axillary meristem.
Figure 4.
D14 protein is transported to axillary buds. Axillary buds in the axil of the second leaf of pD14::D14:GFP/WT (A), pD14::D14:3xGFP/WT (B), pD14::D14:GFP/d10 (C) and pD14::D14:3xGFP/d10 (D). Left, visible light; right, pseudocoloring image of GFP fluorescence intensities; arrows, vascular bundles; triangles, axillary meristems. Bars = 50 μm.
We then tested whether SLs influence D14 protein transport to axillary buds. pD14::D14:GFP was introduced into dwarf10 (d10), an SL biosynthesis mutant (Arite et al., 2007). GFP fluorescence was observed in axillary meristems and leaf primordia in pD14::D14:GFP/d10 plants (Fig. 4, A and C). This indicates that SLs are not required for the transport of the D14 protein to the axillary buds.
pD14::D14:GFP and pD14::D14:3xGFP lines showed different patterns of GFP fluorescence localization in leaf primordia. GFP fluorescence was detected in leaf primordia in pD14::D14:GFP lines but not in pD14::D14:3xGFP lines, despite the fact that both endogenous D14 mRNA and GFP mRNA were transcribed (Figs. 3B and 4B; Supplemental Fig. S3, B and C). This implies that posttranscriptional regulation of D14:3xGFP, such as suppression of translation or promotion of protein degradation, exists in leaf primordia. Since SL-dependent D14 protein degradation has been reported previously (Chevalier et al., 2014), we hypothesized that the absence of GFP florescence in leaf primordia in pD14::D14:3xGFP lines was caused by D14 degradation through the same SL-dependent mechanism. GFP fluorescence, however, was not detected in the leaf primordia in pD14::D14:3xGFP lines in the d10 background (Fig. 4D). This implies that GFP florescence was abolished in the leaf primordia by other mechanisms in pD14::D14:3xGFP lines. Assuming that the same posttranscriptional regulation applies to D14:GFP mRNA expressed in the leaf primordia, the GFP florescence observed in the leaf primordia in pD14::D14:GFP lines should be derived from D14:GFP protein that has been transported to the leaf primordia. Therefore, we can conclude that the D14 protein level in the leaf primordia is regulated by at least two distinct mechanisms. One is the unknown posttranscriptional regulation and the other is likely to be the transport of D14, which would be substantially hindered in pD14::D14:3xGFP.
D14 Transport Is Required to Fully Suppress Tiller Growth in Rice
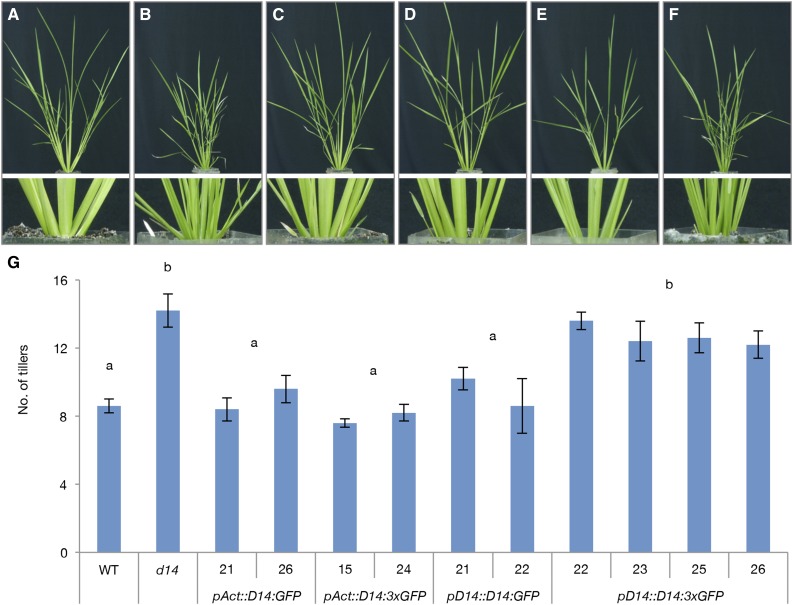
Next, we examined whether the transport of D14 protein is required for its function. Plants were grown in soil for 28 d and their tiller growth was analyzed. Rice ACTIN1 promoter is active in all tissues including axillary buds (Supplemental Fig. S3D). When D14:GFP or D14:3xGFP were expressed constitutively by the rice ACTIN1 promoter (pAct::D14:GFP, pAct::D14:3xGFP) in the d14 mutant, the tillering phenotype of the d14 mutant was rescued (Fig. 5, A–D, G; Supplemental Fig. S4A). This result suggests that both D14:GFP and D14:3xGFP proteins are functional. Introduction of pD14::D14:GFP to the d14 mutant also fully suppressed the tillering, indicating that the D14 promoter used in this experiment contains a region sufficient to control tiller growth (Fig. 5A, B, E, G; Supplemental Fig. S4A). However, the rescue by pD14::D14:3xGFP was much weaker than that in other constructs whether grown on soil (Fig. 5, C–G; Supplemental Fig. S4A) or in hydroponic culture system (Supplemental Fig. S4). These results observed with multiple independent lines suggest that the transport of the D14 protein is required to fully suppress tiller growth.
Figure 5.
D14 transport regulates tiller outgrowth. Twenty-eight-d-old wild type (WT) (A), d14-1 (B), pAct::D14:GFP (C), pAct::D14:3xGFP (D), pD14::D14:GFP (E), and pD14::D14:3xGFP (F) grown in soil. Top, whole shoot; bottom, enlarged view of base part. G, Number of tillers. The different numbers along the x axis denote independent transgenic lines. Data are means ± se, n = 5. Two-way ANOVA with genotype and line as factors revealed a significant effect of genotype (P < 0.001). The different letters denote significant differences between genotypes at P < 0.001 (Tukey-Kramer test).
DISCUSSION
Inhibition of axillary bud outgrowth was the first in planta function of SLs (Gomez-Roldan et al., 2008; Umehara et al., 2008) to be identified. Since this discovery, extensive studies using several species have revealed that SLs have a role in the control of multiple aspects of development, including root development, secondary growth of the stem, leaf senescence, seed germination, and hypocotyl growth (Brewer et al., 2013; Seto et al., 2012). These roles share a common theme, namely, that the function of SLs is to adjust the plant’s growth to its environment and to optimize its growth and reproduction. It has been demonstrated that SL function is finely tuned at various stages, such as at its biosynthesis, transport, perception, and signaling, in order to achieve this unique function (Drummond et al., 2015; Foo et al., 2013; Kohlen et al., 2011; Kretzschmar et al., 2012; López-Ráez et al., 2008; Sun et al., 2014; Umehara et al., 2010; Yoneyama et al., 2012, 2013). In this study, we have demonstrated that D14, a receptor of SL, is transported between cells and that this transport is required for the full function of SLs. Components in plant signaling pathways are regulated at various levels, such as transcription, translation, subcellular localization, stability, and degradation of proteins (Chevalier et al., 2014, Marin et al., 2010; Pérez-Torres et al., 2008; Russinova et al., 2004). In addition to these known types of regulation, our study has shown that the intercellular transport of receptors also functions to modulate plant hormone signaling.
In angiosperms, D14 homologs are classified into three subclades: D14, KAI2/D14Like, and D14like2 (DLK2; Delaux et al., 2012; Waters et al., 2012). Intriguingly, D14 is not the only D14 family member found in the phloem sap by proteomic analysis of rice and Arabidopsis. D14Like and AtDLK2 were also identified in phloem sap of rice (Aki et al., 2008) and Arabidopsis (Batailler et al., 2012), respectively. As these studies revealed a limited number of proteins due to technical limitations, intercellular transport may be a general feature of D14 family proteins. We have also presented evidence that the function of RMS3, as evidenced by graft-transmissible inhibition of branchin but not the downstream components of the SL signaling pathway, has a mobility component. The results of these experiments suggest that D14 transport may be a phenomenon that is conserved in a wide range of plant species. However, in Arabidopsis, despite the fact that short-distance movement of AtD14 protein is observed and that it is present in phloem sap, branching defects in the Atd14 mutant were not suppressed by grafting with a wild-type rootstock (Chevalier et al., 2014). The function of DAD2, the petunia D14 ortholog, was also not transmitted by grafting in petunia (Simons et al., 2007). These results may be caused by the technical limitations of grafting. Otherwise, the mobility of the D14 protein and/or the dependence of its function on its transport may differ between species. Certainly these species are diverse with respect to other aspects of SL biosynthesis and response (Brewer et al., 2015, 2016; Dun et al., 2012; Guan et al., 2012; Mashiguchi et al., 2009; Minakuchi et al., 2010; Shinohara et al., 2013; Yoneyama et al., 2012).
The mechanism of D14 transport is currently unknown. We showed that RMS3 signal is transported bidirectionally in the two-shoot grafting system. This may suggest passive diffusion of RMS3 in the phloem sap. Indeed, many proteins in phloem sap move both acropetally and basipetally (Thieme et al., 2015; Turgeon and Wolf, 2009). On the other hand, phloem loading and unloading of D14/RMS3 might be actively regulated at PD connecting CCs and SEs, as FT loading is controlled by FT-INTERACTING PROTEIN 1 (Liu et al., 2013).
The biological role of D14 transport is still unclear. One hypothesis is that D14 is transported in order to deliver SLs. However, we have shown that D14 can be transported in the absence of SLs in the SL biosynthesis d10 mutant and that SL biosynthesis mutant rms1 rootstocks that do not produce SLs can transport RMS3 signal to rms3 shoots. This implies that SLs are not required for D14 movement. Also, previous grafting experiments have shown that RMS3 is not required for SL transport in pea (Morris et al., 2001), as mutant rms3 rootstocks can inhibit branching in SL biosynthesis mutant scions. These results indicate that the transport of D14 and SLs is independent.
Another hypothesis is that D14 transport is involved in the fine-tuning of shoot branching in response to the environment. The availability of light and nutrients is a major environmental factor that influences the pattern of shoot branching (Drummond et al., 2015; González-Grandío et al., 2013; Henry et al., 2011; Kebrom et al., 2010; Kelly et al., 2012; Kohlen et al., 2011; Mason et al., 2014; Osuna et al., 2007; Rabot et al., 2012; Su et al., 2011; Umehara et al., 2010; Whipple et al., 2011). Therefore, it is crucial for plants to sense light and nutrient conditions accurately and adjust their branching patterns accordingly. Recent studies have shown that SLs may be involved in the regulation of both. It is known that SL biosynthesis is up-regulated in the root in nutrient-deficient conditions (Foo et al., 2013; Kohlen et al., 2011; Kretzschmar et al., 2012; López-Ráez et al., 2008; Sun et al., 2014; Umehara et al., 2010; Yoneyama et al., 2012, 2013). The expression of SL signaling genes is also known to be influenced by carbon source conditions (Osuna et al., 2007) and the quality of light (Drummond et al., 2015). For example, in Arabidopsis, the mRNA level of AtD14 and MAX2, the Arabidopsis D3 ortholog, was decreased by Suc treatment (Osuna et al., 2007) and Suc has been shown to promote branching (Henry et al., 2011; Kelly et al., 2012; Mason et al., 2014; Rabot et al., 2012; Su et al., 2011; Umehara et al., 2010; Whipple et al., 2011). The high ratio of far-red light to red light suppresses shoot branching (González-Grandío et al., 2013; Kelly et al., 2012; Whipple et al., 2011). In petunia, D14 transcription is up-regulated by far-red light and down-regulated by red light (Drummond et al., 2015). In most species, axillary buds are located at the base of a leaf and are enclosed in several young leaves, where it might be difficult to sense photosynthesis conditions and the quality of light. Thus, it seems plausible that D14 transport might contribute to the transmission of information regarding light conditions from the mature leaves to the axillary buds. On the other hand, the amount of SLs transported from the roots reflects the nutrient conditions in the soil. A possible scenario is that the information from both leaves and roots is combined in the axillary bud to allow adjustment of bud outgrowth according to the surrounding environmental conditions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The d10-2 rice mutant was described previously (Umehara et al., 2008). The d14-1 mutant (Arite et al., 2009) was backcrossed to Nipponbare four times and then used in this study. Sterilized seeds were germinated in water for 1 d in the dark. They were grown on solidified media for 7 d and then transferred to hydroponic culture bottles (Umehara et al., 2008; 16 h light, 8 h dark at 25°C). To analyze the tillering phenotype of rice plants grown on soil, plants were grown for 28 d in an incubator (16 h light, 8 h dark at 26°C).
The mutants rms3-2 (K564) and rms4-1 (K164) are derived from the wild-type pea cultivar Torsdag (L107) and have been described previously in Arumingtyas et al., 1992. The mutant rms1-2T was obtained by backcrossing rms1-2 with the wild-type cultivar Torsdag. The mutants rms3-5 (M2T-32) and rms4-3 (M3T-946) are derived from the dwarf wild-type pea cultivar Térèse and have been described previously in Rameau et al., 1997.
Grafting
Grafts were performed using the epicotyl-epicotyl wedge graft technique as described in Beveridge et al., 1994 on 6-d-old seedlings grown at two per 2-L pot in Green Fingers EcoZ Plus potting mix with approximately 2 g of Osmocote (Scotts) per pot under natural photoperiod and 23°C.
Two-shoot grafts were performed using the epicotyl-epicotyl wedge graft technique and two scions grafted on the same rootstock instead of one. Plants were grown in a growth chamber with a 16-h-light (21°C):8-h-dark (16°C) photoperiod at a light intensity of 150 µE m–2 s–1.
Nodes were numbered from the first scale leaf as node 1. The lengths of the lateral branches and buds at each node along the stem(s) were measured using a ruler (branches >10 cm) or digital calipers (branches and buds <10 cm).
Plasmid Construction and Transgenic Rice Production
To construct pD14::D14:GFP, an approximately 4.5-kb region containing the D14 promoter and D14 open reading frame was amplified using the primer set D14 F and D14 R. The PCR fragment was introduced into the pGWB4 expression vector (Nakagawa et al., 2007) using the Gateway system (Invitrogen). To construct pD14::D14:3xGFP, pD14::D14:GFP Δ STOP, and GFP Δ STOP, a GFP region was amplified from pD14::D14:GFP using primer sets D14 F2 and GFP R, GFP F and GFP R, and GFP F and GFP R2, respectively. They were cloned into the pBI 101.2 vector. To construct pAct::D14:GFP, D14:GFP was amplified from pD14::D14:GFP using primer sets D14 F3 and GFP R3 and then cloned into the pAct/Hm2 vector (Inukai et al., 2005). To construct pAct::D14:3xGFP, D14:GFP Δ STOP, and GFP Δ STOP, a GFP region was amplified from pD14::D14:GFP using primer sets D14 F3 and GFP R, GFP F and GFP R, and GFP F and GFP R2, respectively. They were cloned into the pAct/Hm2 vector. To construct pHSP::GFP, the promoter region of OsHSP101 (Chang et al., 2007) was amplified using the primer set OsHSP F and OsHSP R. After subcloning into pBluescript SK, it was cloned into pBI 101.2, together with the GFP fragment. The primer sets used for construction are described in Supplemental Table S1. These constructs were transformed into wild-type rice (cv Nipponbare), the d14 mutant, or the d10-2 mutant. Transformation of rice was performed as described in Nakagawa et al., 2002.
Observation of GFP Fluorescence
The basal parts of rice seedlings were mounted in 7% agarose gel and sectioned at 50-μm thickness using a microslicer. GFP fluorescence was observed with a confocal laser-scanning microscope (FV1000; Olympus). For each construct, except for pD14::D14:3xGFP in a d10 background, more than 10 independent lines were observed at the T0 generation and at least 2 lines were selected among these for more detailed analysis at the T1 generation. For pD14::D14:3xGFP in a d10 background, three independent lines were observed at the T0 generation. The selected lines were then used for the analysis of phenotypes, immunostaining, and mRNA in situ hybridization.
Immunostaining
Immunostaining was performed as described in Yamaji and Ma, 2007. Polyclonal anti-GFP rabbit IgG (Thermo Fisher) and Alexa Fluor 555 goat antirabbit IgG (Molecular Probes) were used as primary and secondary antibodies, respectively. Fluorescence of the secondary antibody and auto-fluorescence of the cell wall were observed with a confocal laser-scanning microscope (LSM700; Carl Zeiss). At least two independent lines were observed for each construct.
In Situ Hybridization
In situ hybridization was performed as described in Kouchi et al., 1995. The full-length D14 cDNA was amplified using the primer set D14 F and D14 R. The PCR fragment was cloned into pENTR/D-TOPO (Thermo Fisher) and then cleaved by Hind III. A 3′ region of the fragment was used as a template to make the D14 antisense probe. A GFP fragment, cloned into pBluescript SK as described above, was used as a template to make the GFP antisense probes. All experiments were carried out with the same concentration of probes and same detection time. At least two independent lines were observed for each construct.
Statistical Analysis
To compare the total length of lateral branches between grafted plants or the number of tillers between plants grown in soil, one-way or two-way ANOVA with the Tukey-Kramer test was performed, respectively. To analyze the effect of genotype on tiller length between plants grown using hydroponic culture, the Kruskal-Wallis test was performed. Subsequently, each genotype was compared using the Steel-Dwass test. These analyses were carried out using R, version 3.1.3.
Supplemental Data
The following supplemental materials are available.
Supplemental Materials and Methods. A list of supplemental materials and methods.
Supplemental Figure S1. RMS3 works as a graft-transmissible signal.
Supplemental Figure S2. Free GFP is not detected in pD14::D14:GFP line.
Supplemental Figure S3. Expression pattern of introduced genes.
Supplemental Figure S4. D14 transport regulates tiller outgrowth.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Jianfeng Ma and Naoki Yamaji for their support in the immunostaining experiments and Tomomitsu Abe for his critical suggestions.
Glossary
- CC
companion cell
- PD
plasmodesmata
- SE
sieve element
- SL
strigolactone
References
- Aki T, Shigyo M, Nakano R, Yoneyama T, Yanagisawa S (2008) Nano scale proteomics revealed the presence of regulatory proteins including three FT-Like proteins in phloem and xylem saps from rice. Plant Cell Physiol 49: 767–790 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Arumingtyas EL, Floyd RS, Gregory MJ, Murfet IC (1992) Branching in Pisum: inheritance and allelism tests with 17 ramosus mutants. Pisum Genet 24: 17–31 [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O (2005) Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J 44: 569–580 [DOI] [PubMed] [Google Scholar]
- Batailler B, Lemaître T, Vilaine F, Sanchez C, Renard D, Cayla T, Beneteau J, Dinant S (2012) Soluble and filamentous proteins in Arabidopsis sieve elements. Plant Cell Environ 35: 1258–1273 [DOI] [PubMed] [Google Scholar]
- Beveridge CA. (2000) Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul 32: 193–203 [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC (1994) Branching mutant rms-2 in Pisum sativum: grafting studies and endogenous indole-3-acetic acid levels. Plant Physiol 104: 953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC (1996) Branching in pea (action of genes Rms3 and Rms4). Plant Physiol 110: 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemendal S, Kück U (2013) Cell-to-cell communication in plants, animals, and fungi: a comparative review. Naturwissenschaften 100: 3–19 [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Gui R, Mason MG, Beveridge CA (2015) Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol 168: 1820–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA (2013) Diverse roles of strigolactones in plant development. Mol Plant 6: 18–28 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F, Meyers E, Scaffidi A, Frickey T, Akiyama K, Seto Y, Dun EA, Cremer JE, et al. (2016) LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 113: 6301–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Huang PS, Lin HR, Lu CH (2007) Transactivation of protein expression by rice HSP101 in planta and using Hsp101 as a selection marker for transformation. Plant Cell Physiol 48: 1098–1107 [DOI] [PubMed] [Google Scholar]
- Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X (2016) Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 26: 640–646 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857–871 [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Geelen D (2014) Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front Plant Sci 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RS, Janssen BJ, Luo Z, Oplaat C, Ledger SE, Wohlers MW, Snowden KC (2015) Environmental control of branching in petunia. Plant Physiol 168: 735–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CG, Beveridge CA (2001) Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol 126: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant 6: 76–87 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR (2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Henry C, Rabot A, Laloi M, Mortreau E, Sigogne M, Leduc N, Lemoine R, Sakr S, Vian A, Pelleschi-Travier S (2011) Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant Cell Environ 34: 1776–1789 [DOI] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, et al. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H (2013) ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 23: 788–792 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA (2010) Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ 33: 48–58 [DOI] [PubMed] [Google Scholar]
- Kelly G, David-Schwartz R, Sade N, Moshelion M, Levi A, Alchanatis V, Granot D (2012) The pitfalls of transgenic selection and new roles of AtHXK1: a high level of AtHXK1 expression uncouples hexokinase1-dependent sugar signaling from exogenous sugar. Plant Physiol 159: 47–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Sekine M, Hata S (1995) Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell cycle. Plant Cell 7: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhu Y, Shen L, Yu H (2013) Emerging insights into florigen transport. Curr Opin Plant Biol 16: 607–613 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73: 2460–2465 [DOI] [PubMed] [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA (2014) Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci USA 111: 6092–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, et al. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Turnbull CG, Murfet IC, Beveridge CA (2001) Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol 126: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot A, Henry C, Ben Baaziz K, Mortreau E, Azri W, Lothier J, Hamama L, Boummaza R, Leduc N, Pelleschi-Travier S, et al. (2012) Insight into the role of sugars in bud burst under light in the rose. Plant Cell Physiol 53: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Rameau C, Bodelin C, Cadier D, Grandjean O, Miard F, Murfet IC (1997) New ramosus mutants at loci Rms1, Rms3 and Rms4 resulting from the mutation breeding program at Versailles. Pisum Genet 29: 7–12 [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J, Simon S, Gübeli C, Liu GW, Cheng X, Friml J, Bouwmeester H, Martinoia E, Borghi L (2015) Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr Biol 25: 647–655 [DOI] [PubMed] [Google Scholar]
- Seto Y, Kameoka H, Yamaguchi S, Kyozuka J (2012) Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol 53: 1843–1853 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC (2007) Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 143: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Abernathy SD, White RH, Finlayson SA (2011) Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant Cell Environ 34: 1986–1998 [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65: 6735–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Turgeon R, Wolf S (2009) Phloem transport: cellular pathways and molecular trafficking. Annu Rev Plant Biol 60: 207–221 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kisugi T, Nomura T, Yoneyama K (2013) Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta 238: 885–894 [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.