Abstract
The stepwise progression of common endoderm progenitors into differentiated liver and pancreas organs is regulated by a dynamic array of signals that are not well understood. The nuclear receptor subfamily 5, group A, member 2 gene nr5a2, also known as Liver receptor homolog-1 (Lrh-1) is expressed in several tissues including the developing liver and pancreas. Here, we interrogate the role of Nr5a2 at multiple developmental stages using genetic and chemical approaches and uncover novel pleiotropic requirements during zebrafish liver and pancreas development. Zygotic loss of nr5a2 in a targeted genetic null mutant disrupted the development of the exocrine pancreas and liver, while leaving the endocrine pancreas intact. Loss of nr5a2 abrogated exocrine pancreas markers such as trypsin, while pancreas progenitors marked by ptf1a or pdx1 remained unaffected, suggesting a role for Nr5a2 in regulating pancreatic acinar cell differentiation. In the developing liver, Nr5a2 regulates hepatic progenitor outgrowth and differentiation, as nr5a2 mutants exhibited reduced hepatoblast markers hnf4α and prox1 as well as differentiated hepatocyte marker fabp10a. Through the first in vivo use of Nr5a2 chemical antagonist Cpd3, the iterative requirement for Nr5a2 for exocrine pancreas and liver differentiation was temporally elucidated: chemical inhibition of Nr5a2 function during hepatopancreas progenitor specification was sufficient to disrupt exocrine pancreas formation and enhance the size of the embryonic liver, suggesting that Nr5a2 regulates hepatic versus pancreatic progenitor fate choice. Chemical inhibition of Nr5a2 at a later time during pancreas and liver differentiation was sufficient to block the formation of mature acinar cells and hepatocytes. These findings define critical iterative and pleiotropic roles for Nr5a2 at distinct stages of pancreas and liver organogenesis, and provide novel perspectives for interpreting the role of Nr5a2 in disease.
Keywords: nuclear receptor, endoderm development, hepatopancreas progenitors, exocrine pancreas, liver
1. Introduction
The liver and pancreas develop from common progenitors in the embryonic gut endoderm through a complex series of developmental events, including the specification of progenitor cells, epithelial budding and outgrowth, and differentiation of precursors into mature cells with specialized function. How these stepwise events are regulated is not well understood. An emerging theme is that these two endodermally-derived organs share many regulatory signals during their development, including fibroblast growth factor 10, bone morphogenetic proteins (Bmps), Wnt, and prostaglandin E2 (PGE2) (Chung et al., 2008; Dong et al., 2007; Goessling et al., 2008; Nissim et al., 2014; Wandzioch and Zaret, 2009). Moreover, the same signals are often used iteratively to regulate different events in the progression from shared gut endoderm into diverse cell types (Goessling et al., 2008; McLin et al., 2007; Ober et al., 2006). In this work, we define multiple roles of the nuclear receptor Nr5a2 in specific stages of pancreas and liver organogenesis.
Pancreas and liver progenitors develop as distinct sub-populations of the endoderm gut tube, distinguishable by gene expression patterns prior to organ bud formation. In zebrafish, pancreas progenitors are marked as early as 14 hpf by pancreatic and duodenal homeobox 1 (pdx1) and liver progenitors are marked later by prospero homeobox 1 (prox1) and hematopoietically expressed homeobox (hhex) at 24 hpf (Biemar et al., 2001; Field et al., 2003c; North and Goessling, 2011; Ober et al., 2003). A subset of pancreas progenitors acquires an endocrine fate, manifested by expression of insulin or other endocrine markers. Progenitors of the pancreas or liver subsequently emerge as epithelial buds from the endoderm tube. Cells of the endocrine pancreas lineage emerge at 24 hpf as the posterodorsal pancreatic bud containing a “principal islet” (Field et al., 2003a). Anterior to this bud, the remaining pancreas progenitors in the gut activate expression of the pancreas specific transcription factor 1a (ptf1a) by 32 hpf and subsequently emerge as the anteroventral pancreatic bud visible by 40 hpf, engulfing the principal islet by 48 hpf (Field et al., 2003a; Lin et al., 2004; Zecchin et al., 2004). Likewise, liver progenitors begin budding by 28 hpf, activate expression of the hepatocyte nuclear transcription factor 4 alpha (hnf4α) by 30–34 hpf, and by 46 hpf they completely detach from the gut tube (Field et al., 2003c). Lastly, pancreas and liver progenitor cells differentiate into mature cell types with specialized function. The ptf1a-expressing pancreas progenitors mainly give rise to acinar cells, the functional cells of the exocrine pancreas that produce and secrete digestive enzymes, and also to pancreatic ductal epithelial cells, which contribute to the epithelial lining of the tubes that transport digestive enzymes from the acinar cells into the duodenum (Grapin-Botton, 2005). Acinar cells are marked by expression of trypsin at 48 hpf while pancreatic ductal epithelial cells are marked by expression of cytokeratins at 48 hpf (Yee et al., 2005). Likewise, markers of differentiated hepatocytes, including liver fatty acid binding protein 10a (fabp10a) are visible by approximately 48hpf (Her et al., 2003). After 48 hpf, the nascent pancreas and liver organs expand laterally in opposite directions from the midline (Field et al., 2003c).
In addition to these similar developmental milestones in organogenesis, the developing pancreas and liver are also often regulated by the same signaling pathways. Early in development, these signaling pathways act on a common pool of endoderm progenitors to specify them to liver or pancreas fate, and hence consequences on the pancreas and liver may be opposing. For example, between 12–20 hpf, Wnt or Prostaglandin E2 (PGE2) activity has opposite effects, resulting in a smaller exocrine pancreas but larger liver at 72 hpf (Goessling et al., 2008; Nissim et al., 2014). Later in development after pancreas and liver progenitors have been specified, Wnt or PGE2 activity stimulates both the exocrine pancreas and liver enlargement (Ober et al., 2006; Goessling et al., 2008; Murtaugh, 2008; Nissim et al., 2014). These effects can be due to signals affecting fate decisions of hepatopancreatic progenitors or differential impact of these signaling pathways on the pancreas and liver progenitors depending on the developmental stage during which they act. These examples highlight that a single signaling pathway can be repeatedly used over time to direct distinct developmental events.
A number of extracellular signals such as Bmps, Wnts, fibroblast growth factors (Fgfs), and PGE2 have been demonstrated to have pleiotropic roles in pancreas and liver development (Deutsch et al., 2001; Goessling et al., 2008; McLin et al., 2007; Nissim et al., 2014; Ober et al., 2006; Rossi et al., 2001; Wandzioch and Zaret, 2009; Zaret and Grompe, 2008). In contrast, assigning multiple sequential, developmentally distinct roles to transcription factors has been more challenging due to limitations in temporal control over transcription factor activity, partially from a lack of specific inhibitors. One transcription factor, the orphan nuclear receptor NR5A2 is a candidate transcription factor that may have diverse roles in both pancreas and liver organogenesis (Fayard et al., 2004; Pare et al., 2001).
Nr5a2 is primarily expressed in the developing and mature gastrointestinal endoderm, including liver hepatocytes and exocrine pancreas cells (Bookout et al., 2006; Pare et al., 2004; Rausa et al., 1999). The Nr5a2 promoter contains binding sites for a number of genes that regulate early endoderm development, including the GATA factors (Fayard et al., 2004; Pare et al., 2001). Further evidence suggests that Nr5a2 expression may be regulated by transcription factors expressed in pancreas progenitors, including Pancreatic-duodenal Homeobox 1 (PDX1). In endoderm progenitor cells, the NR5A2 protein has been shown to promote the expression of genes involved in hepatopancreas maturation, including the hepatocyte nuclear factors (HNFs) (Pare et al., 2001). In the mature liver and pancreas, NR5A2 also regulates transcriptional networks responsible for cholesterol and bile acid homeostasis and the production of digestive enzymes (Chong et al., 2012; Fayard et al., 2004; Hale et al., 2014; Holmstrom et al., 2011). Given established pathway connections, NR5A2 is thought to function as a signal linking early endoderm development and endoderm differentiation (Fayard et al., 2004; Pare et al., 2001).
Investigations into the role of NR5A2 in endoderm organogenesis have been complicated because global loss of NR5A2 is lethal in early mouse embryogenesis at E6.5–7.5 due to gastrulation defects, before development of the liver or pancreas (Gu et al., 2005; Labelle-Dumais et al., 2006; Pare et al., 2004). However, lineage-specific knockout of Nr5a2 has been accomplished in mouse models: selective loss of NR5A2 in already specified pancreas progenitor cells by a Pdx1early-Cre results in severe reduction in exocrine and endocrine pancreas cell number, supporting a post-specification role for NR5A2 in pancreas cell fate (Fayard et al., 2004; Hale et al., 2014). Additionally, loss of Nr5a2 in differentiated mouse hepatocytes, as driven by an Albumin-Cre, results in no apparent effects (Lee et al., 2008). Due to the relatively late expression of Albumin-Cre in differentiated hepatocytes, the requirements of NR5A2 for specification and outgrowth of hepatic progenitors and hepatic differentiation have not been elucidated. These data indicate the need for a detailed investigation of the role of NR5A2 at all stages of hepatopancreatic development.
While NR5A2 has specific functions in development and metabolism of liver and pancreas, it may also have more general functions in self-renewal and cell cycle progression of stem and progenitor cells. NR5A2 acts as a regulator of embryonic stem cell (ESC) pluripotency by maintaining the expression of ESC identity genes, including Oct4 and Nanog, and is further capable of replacing OCT4 in the reprogramming of adult differentiated fibroblasts into induced pluripotent stem cells (Gu et al., 2005; Heng et al., 2010b). In mouse ESCs and intestinal crypt stem cells, NR5A2 also regulates genes involved in self-renewal and growth, including c-Myc, n-Myc, Cyclin D1, and Cyclin E1, often by functioning as a coactivator of β-catenin/Tcf4 (Botrugno et al., 2004; Heng et al., 2010a; Wagner et al., 2010). Consistent with a general role in cell cycle progression, NR5A2 also regulates the proliferation of breast cancer and pancreatic cancer cell lines (Annicotte et al., 2005; Benod et al., 2011). Reduced NR5A2 activity in Estrogen Receptor-α+ (ERα+) breast cancer cell lines can abrogate estradiol-induced proliferation (Annicotte et al., 2005). Furthermore, blocking NR5A2 activity in pancreatic cancer cell lines can inhibit proliferation in vitro through the down-regulation of c-Myc, Cyclin D1, and Cyclin E1 (Benod et al., 2011). The relevance of NR5A2 activity to cancer has been strengthened by the repeated association of NR5A2 with pancreatic ductal adenocarcinoma in genome wide association studies (GWAS) (Benod et al., 2011; Petersen et al., 2010; Ueno et al., 2015).
We have previously harnessed the zebrafish model as a tool for interrogating the iterative roles of signaling pathways at specific events in pancreas or liver development (Goessling et al., 2008; Nissim et al., 2014). To study the role of Nr5a2 during distinct developmental time windows in zebrafish, an important recent advance has been the identification of a pharmacologic antagonist (Benod et al., 2013). This molecule, named “Compound 3” (Cpd3), directly binds to the NR5A2 ligand binding domain and inhibits transcriptional activation of downstream target genes and proliferation of human cancer cell lines (Benod et al., 2013). Prior to the current study, the impact of Cpd3-mediated NR5A2 inhibition has not been investigated in vivo.
In this study, we use genetic and pharmacologic approaches to show that Nr5a2 is repeatedly used to regulate both hepatopancreatic specification and differentiation. In zebrafish, nr5a2 is expressed in the anteroventral pancreas and liver buds during outgrowth from the gut endoderm tube. Zebrafish nr5a2 null mutants exhibit abrogated exocrine pancreas and liver development, while endocrine pancreas development is largely unaffected. In the pancreas, markers of differentiated acinar cells are lost, while exocrine pancreas progenitor markers ptf1a and pdx1 appear unchanged, consistent with a role for nr5a2 in regulating progenitor cell differentiation. Additionally, prox1 expression is reduced in a subset of pancreatic progenitors whose contribution to pancreas development is largely undefined. A loss of the prox1 expressing progenitors in the pancreatic region of the nr5a2oz3 mutants may have an impact on pancreas development. In the liver, nr5a2 regulates both progenitor outgrowth and differentiation as mutants demonstrate a reduction in hepatoblast and differentiated hepatocyte markers.
Importantly, our study is the first in vivo study of Nr5a2 pharmacologic antagonism with Cpd3, defining distinct temporal requirements for Nr5a2 activity in both the pancreas and liver. First, we demonstrate that Nr5a2 regulates the commitment of bipotent hepatopancreas progenitor cells to distinct hepatic or pancreatic lineages. The loss of Nr5a2 activity during the specification of hepatopancreas progenitor cells is sufficient to disrupt the formation of the mature exocrine pancreas and expand the mature liver, potentially through the preferential commitment of progenitors to the hepatic lineage at the expense of the pancreatic lineage. Second, we demonstrate that Nr5a2 is necessary for the expansion and differentiation of the liver and pancreas. Inhibition of Nr5a2 activity with chemical antagonist during differentiation reduces the size of both organs. Taken together, our results reveal that Nr5a2 is a transcription factor with multiple sequential and developmentally distinct roles in hepatopancreatic development.
2. Materials and Methods
2.1 Zebrafish husbandry
Zebrafish were maintained according to Institutional Animal Care and Use Committee protocols (HMS 04626) and in compliance with NIH guidelines. Transgenic lines Tg(fabp10a:GFP), Tg(elastase:GFP), Tg(fabp10a:DsRed), Tg(prox1a:Citrine), Tg(sox17:eGFP), and Tg(trypsin:GFP) were previously described (Bagnat et al., 2007; Bussmann and Schulte-Merker, 2011; Her et al., 2003; Nissim et al., 2014; Wan et al., 2006).
2.2 Generation of nr5a2+/oz3 mutants
Custom TALEN constructs were designed to target the first constitutively present exon of nr5a2 and constructed by the University of Utah mutagenesis core facility (Cermak et al., 2011). Linearized plasmids were transcribed with mMessage mMachine Sp6 transcription kit for injection of 150 pg each. Mutagenesis was confirmed by reduced BsrI cleavage within the spacer between TALEN binding sites. Founder mutants were identified using high resolution melt analysis by progeny with melt curves that deflect from WT, using primers (F: 5′ ACTCTTATGTTTTCAGCCCCACAGTTT 3′) and (R: 5′ TCACAGGTCAGCAACCCATAGTGAT 3′) (Dahlem et al., 2012). PCR fragments from deflecting progeny were subsequently cloned into pCRII-TOPO vectors; inserts were Sanger sequenced for confirmation. Heterozygous progeny were raised to adulthood and genotyped prior to subsequent matings. nr5a2+/oz3 adult heterozygotes were identified by PCR amplification using primers (F: 5′ ACGAACCTCATAACACATGACAGCCA 3′), (R: 5′ AGCTCTCACAGGTCAGCAACCCATA 3′) and subsequent BsrI restriction enzyme digestion. Mutants were identified by the destruction of a BsrI cut site. The nr5a2oz3 carriers were outcrossed for at least two generations prior to analysis.
2.3 Embryonic zebrafish expression studies
In situ hybridization was conducted on paraformaldehyde-fixed embryos using standard protocols (http://zfin.org/ZFIN/SD/ThisseProtocol.html) and established probes (Nissim et al., 2014). nr5a2 probe was generated from zebrafish cDNA using the following primers: 5′ TGTAAGGGCTTCTTCAAGCGC 3′ (forward) and 5′ GGAGAACAGTGTCTGGTCAGCC 3′ (reverse). hnf4α probe was generated from zebrafish cDNA using the following primers: 5′ CGCAGTGTACGCAAAAACCA 3′ (forward) and 5′ GGTGAGCGTGAGGTGCTTCATT 3′ (reverse). wnt2 probe was generated from zebrafish cDNA using the following primers: 5′ ATGAACTTTTTGCCAAATGGAA 3′ (forward) and 5′ TCAGGACTGGGTTTTGCAGG 3′ (reverse). Changes in trypsin, fabp10a, pdx1, ptf1a, prox1, and hnf4α expression were scored in images using ImageJ to quantify pancreas, liver, endoderm, or progenitor population size. Volumetric analysis was performed using 3D reconstruction of confocal stacks in ImageJ.
2.4 Morpholino Injections
Morpholino (GeneTools) knockdown was performed as previously described (North et al., 2007), utilizing 2 nanoliters of 100 μM nr5a2 morpholino (5′ TCACTCTCAAAACTACTGGACATTT 3′).
2.5 Chemical Treatments
Zebrafish embryos were exposed to 100 μM (or 200 μM when specified) pharmacologic Nr5a2 antagonist, 1-(3′-(1-(2-(4-Morpholinyl)ethyl)-1H-pyrazol-3-yl)-3-biphenylyl)ethanone (Cpd3, ChemBridge) (Benod et al., 2013) at the specified time points. Embryo water containing 100 μM Cpd3 was replaced at 24-hour intervals. DMSO carrier content for 100 μM treatments was 0.04%, and 0.08% for 200 μM treatments.
2.6 Fluorescence activated cell sorting (FACS)
For hepatocyte cell isolations, Tg(fabp10a:GFP) embryos (72 hpf) were incubated in 50μg/mL Liberase™ (Roche) at 37ºC for 1.5 hours, manually dissociated with a p1000 pipette, strained through a 30μm mesh filter, and suspended in 1% fetal bovine serum. SYTOX Red Dead Cell Stain (ThermoFisher) was added to the cell suspension to detect nonviable cells. Cell suspensions were separated into GFP+ and GFP− fractions using a BD FACSAria II SORP flow cytometer (Goessling et al., 2008). For elastase:GFP cell counting, whole fluorescent embryos were manually dissociated in 0.25% trypsin for 20 minutes and analyzed for GFP fluorescence by flow cytometry (> 100,000 cells were analyzed per sample; n = 10–20 per treatment).
2.7 Total RNA isolation and production of antisense RNA
Cells isolated by FACS (approximately 200,000) were processed using the RNAqueous-Micro Total RNA Isolation Kit (ThermoFisher) according to the manufacturer’s protocol. DNase I (Ambion) processing was performed on total cellular RNA according to manufacturer’s protocol. Amplified cDNA was prepared using the Ovation Pico WTA System V2 (NuGEN) in the Molecular Genetics Core Facility at Beth Israel Deaconess Medical Center. RNA was isolated from whole embryos using TRIzol® Reagent and treated with the TURBO DNA-free DNAse kit (Life Technologies); cDNA was generated using iScript cDNA synthesis reagents (Bio-Rad).
2.6 qRT-PCR
qRT-PCR was performed on amplified cDNA libraries using the SYBR Green Supermix (Bio-Rad) and relative expression levels were calculated using the ΔΔCt method. Expression was normalized to ef1α. See Supplementary Table 1 for primer sequences.
2.8 TUNEL staining
TUNEL staining was performed on paraformaldehyde fixed embryos using the ApopTag® Peroxidase In Situ Apoptosis Detection Kit (Chemicon International). Apoptosis signal was developed using the ImmPACT DAB Peroxidase (HRP) Substrate Kit (Vector Laboratories).
2.9 Wholemount immunohistochemistry
Embryos were fixed overnight in 4% paraformaldehyde at 4ºC and permeabilized in acetone at −20ºC. Embryos were incubated with Anti-phospho-histone H3 (Ser10) Antibody (EMD Millipore) overnight and again with secondary antibody Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 488) (Abcam).
3. Results
3.1 nr5a2 is expressed in the developing endoderm of zebrafish embryos
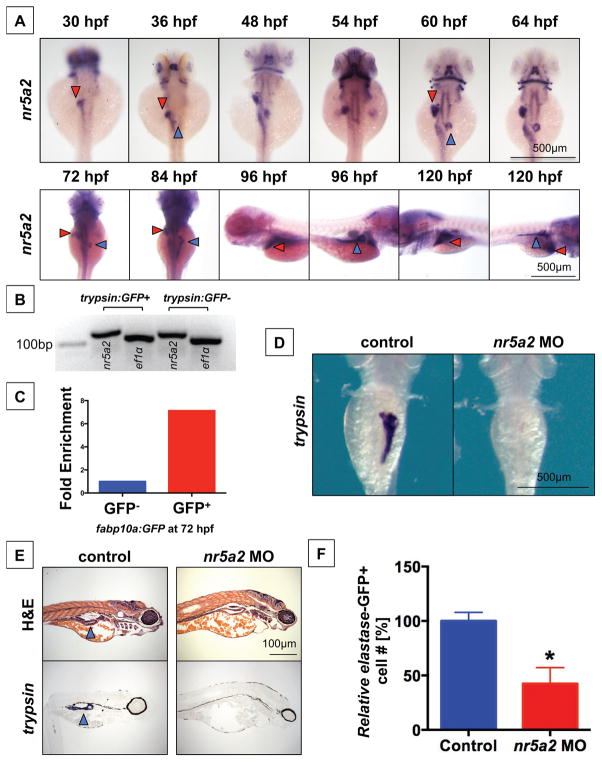
Previous work has shown that nr5a2 is expressed in the developing digestive organs (Lin et al., 2000), and we demonstrate here that transcripts also appear to be maternally provided and are present in embryonic blastomeres and their derivatives until the end of gastrulation (Figure S1A, Figure 1A). Later in the endoderm, nr5a2 transcripts are first detected in the liver bud and anteroventral pancreas bud by 30 hpf and 36 hpf, respectively (Figure 1A) (Bertrand et al., 2007; Lin et al., 2000). Expression of nr5a2 was detected in the liver bud during budding stages II and III when the liver bud emerges and separates from the gut tube, and persisted through hepatocyte differentiation, which begins at approximately 44 hpf. nr5a2 expression also appeared during the outgrowth of the developing pancreas; however, nr5a2 expression remained restricted to a portion of differentiated pancreas acinar cells (60–72 hpf) until approximately 84 hpf, when it was expressed in most exocrine pancreas cells (Figure 1A). Fluorescence activated cell sorting (FACS)-isolated GFP+ pancreatic acinar cells and hepatocytes from the Tg(trypsin:GFP) and Tg(fabp10a:GFP) reporter lines, respectively, confirms that nr5a2 is expressed in isolated acinar cells and hepatocytes, and that nr5a2 expression is specifically enriched in GFP+ hepatocytes relative to GFP- cells isolated from Tg(fabp10a:GFP) embryos (Figure 1B–C). These results demonstrate that nr5a2 is expressed in gut endoderm during hepatopancreatic development, and that nr5a2 is present in the endoderm during critical stages of hepatopancreatic progenitor specification and differentiation.
Figure 1. nr5a2 is expressed in the developing endoderm and is required for exocrine pancreas formation.
A) Expression pattern of nr5a2 in the developing endoderm by in situ hybridization. nr5a2 appears in the developing liver bud (red arrow) by 30 hpf and the pancreas bud (blue arrow) by 36 hpf. After 84 hpf, it is expressed in the mature liver (red arrow) and exocrine pancreas (blue arrow).
B) Expression of nr5a2 in FACS isolated acinar cells from the Tg(trypsin:GFP) line at 72 hpf by qRT-PCR.
C) Expression of nr5a2 in FACS isolated hepatocytes from pooled Tg(fabp10a:GFP) embryos at 72 hpf by qRT-PCR (fold enrichment displayed as an average of technical triplicates).
D) Splice-site morpholino (MO) knockdown of Nr5a2 reduces the size of the exocrine pancreas by in situ hybridization for trypsin at 96 hpf.
E) nr5a2 MO injected embryos (96 hpf) have a reduced exocrine pancreas size (blue arrow) by histology and section in situ hybridization for trypsin (n = 5).
F) FACS quantification of GFP+ cells (% of control) from elastase:GFP reporter fish at 96 hpf demonstrates that MO knockdown of nr5a2 reduces the number of exocrine pancreas cells (n = 20 per treatment; p < 0.05; unpaired t-test).
3.2 nr5a2 is required for exocrine pancreas development
The requirement of nr5a2 in zebrafish pancreas development was initially assessed by morpholino-mediated knockdown. nr5a2 morphants exhibited impaired exocrine pancreas formation, as assessed by in situ hybridization for trypsin at 96 hpf (Figure 1D). Liver formation was also inhibited by morpholino-mediated knockdown, as assessed by fluorescence microscopy of the Tg(fabp10a:DsRed) reporter line (Figure S1B). Histological analysis of controls and nr5a2 morphant embryos confirmed the absence of exocrine pancreas structure (Figure 1E). Furthermore, section in situ hybridization for trypsin in control and nr5a2 MO injected embryos revealed that nr5a2 morphants lack mature acinar cells (Figure 1E). To further confirm that nr5a2 is required for the formation of exocrine pancreas cells, nr5a2 MO was injected into elastase:GFP reporter embryos followed by FACS analysis to quantify the number of GFP-labeled exocrine pancreas cells present in pooled embryo groups. elastase is expressed in the exocrine pancreas following trypsin expression, and is a marker of mature acinar cells (Mudumana et al., 2004; Wan et al., 2006). nr5a2 morphants contained a significantly reduced quantity of elastase GFP+ cells (Figure 1F). These findings suggest a conserved impact of nr5a2 function in exocrine pancreas formation.
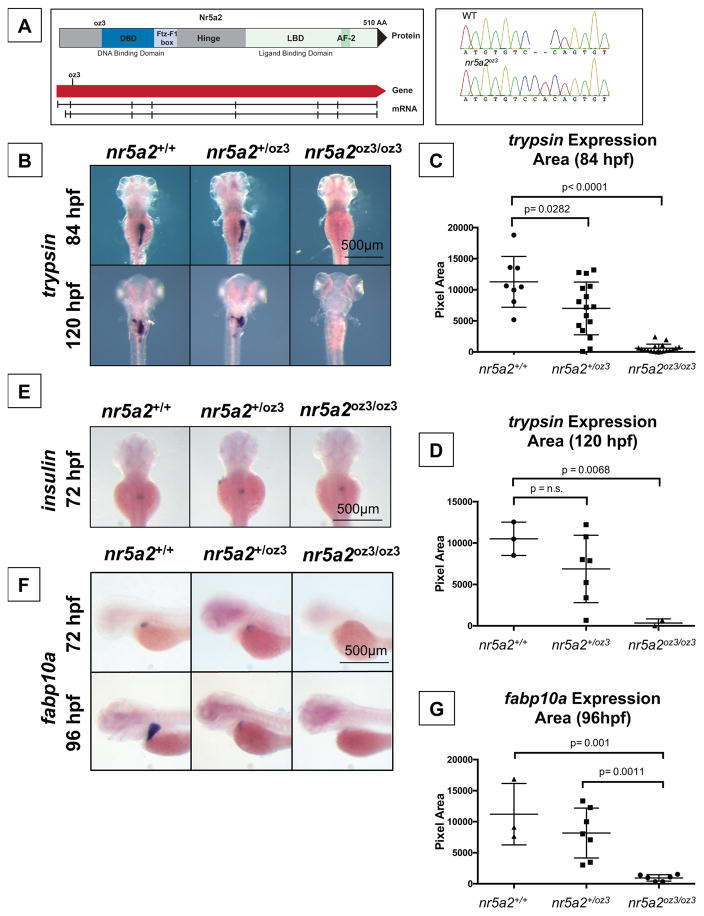
To genetically test whether nr5a2 is required for the formation and maturation of the endoderm, we generated zebrafish mutants using TALEN-based genome editing (Cermak et al., 2011; Sander et al., 2011) resulting in an early frameshift due to a 2-basepair insertion within exon 3 (Figure 2A). The mutation disrupts the protein sequence prior to major functional domains (DNA-binding domain and ligand binding domains), thereby generating a likely null allele (Figure 2A) (Lin et al., 2000). Heterozygous nr5a2+/oz3 fish were viable and fertile through adulthood. nr5a2oz3/oz3 homozygous mutants were embryonic lethal at 8–9 days post fertilization (dpf). Homozygous nr5a2oz3/oz3 and heterozygous nr5a2+/oz3 embryos are grossly morphologically indistinguishable from wildtypes during early embryogenesis. The earliest visible phenotype appears upon examination of endoderm development by in situ hybridization or fluorescent reporter lines, as described below.
Figure 2. Genetic modulation of Nr5a2 disrupts the formation of the mature exocrine pancreas and liver.
A) The oz3 genetic loss of function mutation is a two base pair insertion within exon 3 of the nr5a2 transcript, before the DNA-binding domain and ligand-binding domain.
B) trypsin in situ hybridization at 84 and 120 hpf reveals that nr5a2oz3/oz3 embryos do not develop a mature exocrine pancreas.
C) Quantification of exocrine pancreas size in nr5a2+/+, nr5a2+/oz3, and nr5a2oz3/oz3 embryos by trypsin area calculations. nr5a2oz3/oz3 and nr5a2+/oz3 embryos have a significantly reduced exocrine pancreas size relative to nr5a2+/+ siblings at 84 hpf (n > 8 per group; for nr5a2+/+ vs. nr5a2+/oz3 (p = 0.0282; for nr5a2+/+ vs. nr5a2oz3/oz3, p < 0.0001; unpaired t-test).
D) Quantification of exocrine pancreas size in nr5a2+/+, nr5a2+/oz3, and nr5a2oz3/oz3 embryos by trypsin area calculations. At 120 hpf, nr5a2oz3/oz3 have a significantly reduced exocrine pancreas size relative to nr5a2+/+ siblings (n > 2 per group; p = 0.0068; unpaired t-test).
E) In situ hybridization for insulin demonstrates that nr5a2+/+, nr5a2+/oz3, and nr5a2oz3/oz3 embryos develop an endocrine pancreas.
F) In situ hybridization for fabp10a reveals that nr5a2oz3/oz3 embryos have a reduced liver size at 72 and 96 hpf.
G) Quantification of liver size by fabp10a area calculations demonstrates that nr5a2oz3/oz3 embryos have a significantly reduced liver size relative to nr5a2+/+ and nr5a2+/oz3 categories (n > 3 per group; p = 0.001; unpaired t-test).
Expression of markers for the mature exocrine (trypsin) and endocrine (insulin) pancreas in wildtype, nr5a2+/oz3 and nr5a2oz3/oz3 embryos revealed that genetic loss of nr5a2 completely disrupted the development of the mature exocrine pancreas at 72 and 84 hpf, while leaving the endocrine pancreas largely intact (Figure 2B–E). Quantification of exocrine pancreas size by area of trypsin expression (84 and 120 hpf) demonstrated that nr5a2oz3/oz3 embryos have a significantly smaller exocrine pancreas than nr5a2+/+ siblings (Figure 2C–D). Additionally, heterozygous nr5a2+/oz3 embryos have a variably penetrant, but statistically significant, reduced exocrine pancreas phenotype, which ranges from completely normal to fully absent (Figure 2C,D). The heterozygous exocrine pancreas phenotypes suggest that nr5a2 is haploinsufficient for reliable patterning.
The loss of mature exocrine pancreas markers was not specific to the trypsin gene, as mutants also exhibited carboxypeptidase A (carbA) expression deficits in the exocrine pancreas with similarly variable penetrance in nr5a2+/oz3 embryos (Figure S1C). In situ hybridization for trypsin expression in the newly differentiated exocrine pancreas confirms that trypsin+ cells fail to form in the nr5a2oz3/oz3 embryos all together (Figure S1D). Cell death may be an unlikely explanation for the nr5a2 mutant phenotype, because TUNEL labeling appears normal through development in these mutants and morphants, and proliferation is also largely unchanged in the early embryo (Figure S2A–D). Further, the endocrine pancreatic lineages may remain unaffected in nr5a2oz3/oz3 mutants, and consistent with this, nr5a2 is not co-expressed in endocrine β-cells (Figure S3A). These results demonstrate the requirement of nr5a2 for normal exocrine pancreas development and reveal that optimal exocrine pancreas development is sensitive to nr5a2 gene dose.
3.3 Nr5a2 is required for liver development
Given our previous findings that the same developmental pathways co-regulate pancreas and liver formation (Goessling et al., 2008; Nissim et al., 2014), we examined whether liver development is affected in nr5a2 mutants. nr5a2oz3/oz3 homozygous mutants displayed severely impaired expression of the mature hepatocyte marker fabp10a. Liver size, calculated by fabp10a expression area, at 72 and 96 hpf was significantly reduced, with a subset of nr5a2+/oz3 heterozygous embryos also exhibiting reduced liver size (Figure 2F–G). Given that nr5a2 regulates aspects of cholesterol and bile acid homeostasis (Paré et al., 2004), and may therefore impact the expression of genes involved in lipid transport, we confirmed that a reduction in liver size was not specific to the fabp10a hepatocyte marker. Expression of an additional hepatocyte marker, transferrin-a, was also reduced in the nr5a2oz3/oz3 embryos, as assessed by in situ hybridization (Figure S3B). Furthermore, in situ hybridization for fabp10a at earlier stages of liver differentiation revealed that fabp10a+ hepatocytes fail to form in nr5a2oz3/oz3 embryos (Figure S3C). These results demonstrate that nr5a2 is required for the development of differentiated hepatocytes.
3.4 Expression of prox1, but not ptf1a and pdx1, is reduced in pancreas progenitors by nr5a2oz3/oz3
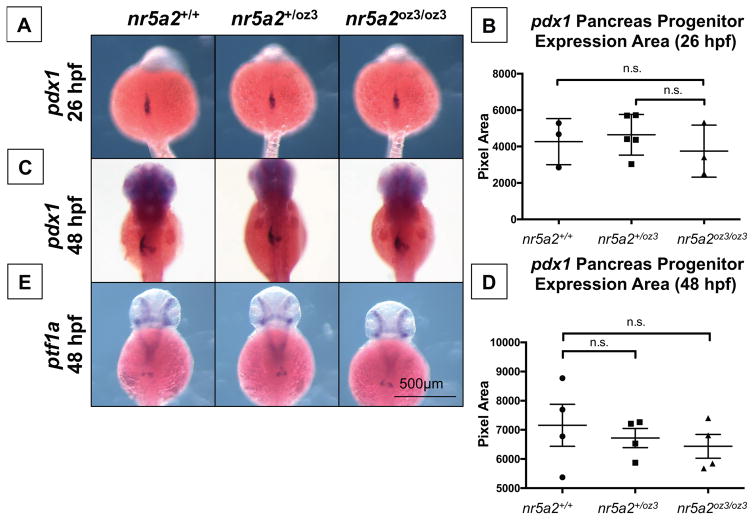
The observed reduction in exocrine pancreas size may result from defects in pancreas progenitor cell formation or from defects in differentiation. We therefore sought to determine whether standard exocrine pancreas progenitors marked by pdx1 and ptf1a (Tiso et al., 2009) were impacted in nr5a2oz3/oz3 mutants. pdx1 expression appeared normal in pancreas progenitors during early morphogenesis (26 hpf), and this progenitor expression also appeared relatively normal by 48 hpf (Figure 3A–D); similarly, ptf1a expressing progenitors were also largely unaffected at 48 hpf (Figure 3E).
Figure 3. Nr5a2 is not required for the formation of pdx1 and ptf1a pancreas progenitors.
A) The area of pdx1 pancreas progenitors is similar across nr5a2+/+, nr5a2+/oz3, and nr5a2oz3/oz3 embryos at 26 hpf.
B) No significant differences in pdx1 progenitor population size identified by a quantification of pdx1 expression area by genotype (p > 3 per group; for nr5a2+/+ vs. nr5a2+/oz3, p = 0.6746; for nr5a2+/+ vs. nr5a2oz3/oz3, p = 0.6606; unpaired t-test).
C) pdx1 pancreas progenitors are formed at 48 hpf in nr5a2+/oz3 and nr5a2oz3/oz3 embryos; however, subtle difference in pdx1 progenitor budding is noted with variable penetrance across the nr5a2oz3/oz3 population.
D) No significant differences in the size of the pdx1 pancreas progenitor pool were observed by quantification of expression area at 48 hpf (n > 3 per group; for nr5a2+/+ vs. nr5a2+/oz3, p = 0.6024; for nr5a2+/+ vs. nr5a2oz3/oz3, p = 0.4178; unpaired t-test).
E) ptf1a progenitors are formed in nr5a2+/oz3 and nr5a2oz3/oz3 embryos by 48 hpf.
We also examined the expression of prox1, a less commonly used hepatopancreas progenitor marker, thought to regulate the differentiation of pancreas progenitors and the expansion of the exocrine pancreas (Wang et al., 2005; Westmoreland et al., 2012). Expression of prox1 is reduced in the pancreas bud of nr5a2oz3/oz3 and nr5a2+/oz3 embryos at 48 hpf (Figure 4A). However, at 30 hpf, prox1 expression in the pancreas bud of nr5a2oz3/oz3 embryos appears normal, suggesting that Nr5a2 regulates prox1 expression only during specific stages of bud formation or progenitor differentiation (Figure S3D). Furthermore, the expression of prox1 in the pancreas does not recover in the mutants by the 96 hpf time point (Figure S4A–B). These results indicate that while nr5a2 is not required for the expression of conventional progenitor markers pdx1 and ptf1a, nr5a2 is required for the maintenance, but not the initiation, of prox1 expression in the pancreas bud.
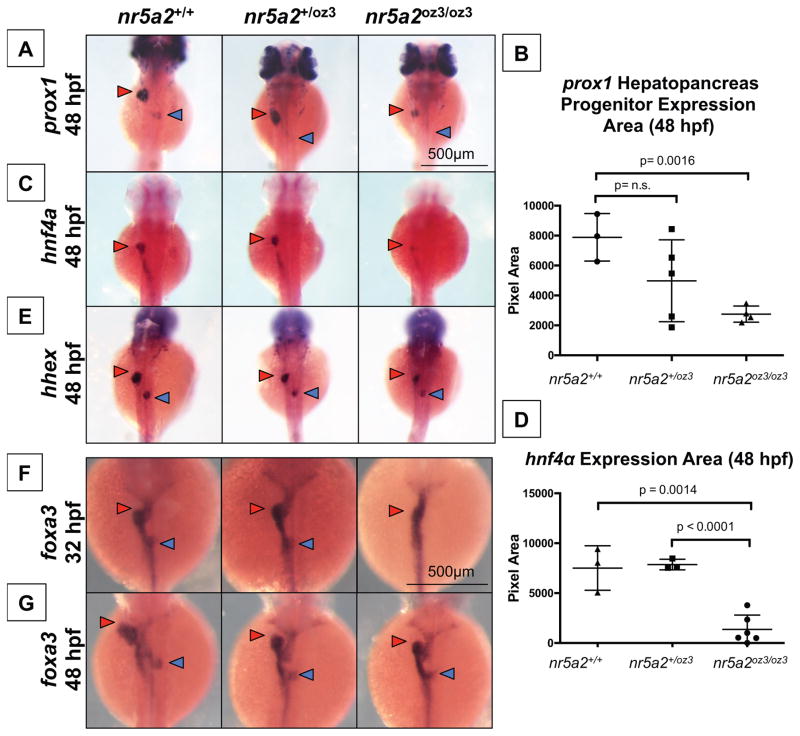
Figure 4. A subset of hepatopancreas progenitors require nr5a2 for specification.
A) nr5a2oz3/oz3 mutants have reduced prox1 expression in the liver (red arrow) and pancreas (blue arrow) buds relative to nr5a2+/+ controls at 48 hpf.
B) Quantification of prox1 expression area in the combined liver and pancreas buds (n > 3 per group; for nr5a2+/+ vs. nr5a2+/oz3, p = 0.1502; for nr5a2+/+ vs. nr5a2+/oz3, p = 0.0016; unpaired t-test).
C) In situ hybridization reveals consistent decreased hnf4α expression in the hepatoblasts (red arrow) of nr5a2oz3/oz3 embryos.
D) Quantification of hnf4α expression area in the intestine and liver buds shows significant expression reduction in nr5a2oz3/oz3 embryos relative to nr5a2+/+ and nr5a2+/oz3 groups (n > 3 per group; for nr5a2+/+ vs. nr5a2oz3/oz3, p = 0.0014; for nr5a2+/oz3 vs. nr5a2oz3/oz3, p < 0.0001; unpaired t-test).
E) The size of the hhex liver bud (red arrow) is reduced in the nr5a2oz3/oz3 embryos compared to nr5a2+/+ siblings, while the pancreas bud (blue arrow) is largely unaffected.
F) In situ hybridization for foxa3 endoderm marker consistently demonstrates delayed gut looping, liver (red arrow), and pancreas (blue arrow) budding in the nr5a2oz3/oz3 embryos at 32 hpf.
G) The sizes of the foxa3 liver (red arrow) and pancreas (blue arrow) buds are consistently reduced in the nr5a2+/oz3 and nr5a2oz3/oz3 mutant populations at 48 hpf.
3.5 Nr5a2 is required for emergence of an hnf4α and prox1 expressing liver bud
Given the observed impact of loss of nr5a2 on differentiated hepatocytes, we examined hepatoblast marker expression in nr5a2 mutants to assess the role of nr5a2 in hepatic progenitor formation. nr5a2 is known to transcriptionally activate the hepatoblast marker hnf4α in vitro, suggesting that hnf4α expression would be reduced in nr5a2oz3/oz3 homozygotes (Kyrmizi et al., 2006; Pare et al., 2001). nr5a2oz3/oz3 embryos had a reduced expression size of hepatic progenitor markers hnf4α and prox1 in the liver bud at 48 hpf (Figure 4A–D). We next examined whether other hepatoblast markers were affected in the nr5a2oz3 mutants, including hhex. We observed that the size of hhex expression in the liver bud was also reduced, though less substantially, in the nr5a2oz3/oz3 mutants (Figure 4E). These results demonstrate that nr5a2 is required for the formation of a liver bud that contains an appropriately sized population of hnf4α and prox1 expressing hepatocyte progenitors, and further indicate that nr5a2 may be required for the formation of the subsets of hepatic progenitors labeled by these markers.
Reduced expression of prox1 and hnf4α, as well as subtle differences in budding (as revealed by pdx1 expression) in nr5a2oz3/oz3 embryos by 48 hpf, suggest that defects in liver and pancreas budding may underlie the mature liver and pancreas phenotypes observed in nr5a2oz3/oz3 mutants. To evaluate this possibility, we examined the expression of foxa3, a global marker for the developing gut tube, liver bud, and pancreas buds (Field et al., 2003c). Homozygous nr5a2oz3/oz3 mutants exhibited altered gut looping and disrupted liver and pancreas budding at 32 hpf (Figure 4F). At 48 hpf, the outgrowth of the liver bud and the fusion of the pancreas buds were delayed in both nr5a2oz3/oz3 homozygotes and nr5a2+/oz3 heterozygous embryos (Figure 4G). These findings point to a role for nr5a2 in gut morphogenesis, including gut looping and the outgrowth of the liver and pancreas buds.
3.6 Nr5a2 chemical antagonist disrupts exocrine pancreas development in vivo
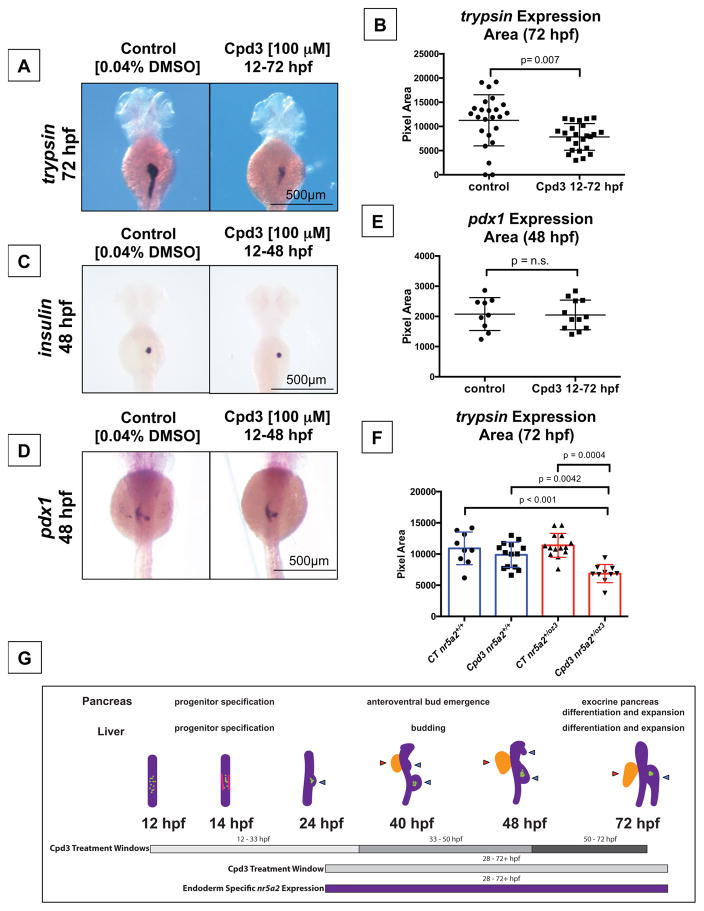
Cpd3 is a known chemical antagonist of human NR5A2 that directly binds to the NR5A2 ligand binding domain and renders NR5A2 transcriptionally inactive in vitro (Benod et al., 2013). We exposed zebrafish embryos to Cpd3 to determine whether Cpd3 has the ability to disrupt Nr5a2-mediated developmental events and produce phenotypes consistent with the nr5a2+/oz3 or nr5a2oz3/oz3 mutants. Treatment of developing embryos after gastrulation (starting at 12 hpf) with 100 μM Cpd3 significantly disrupted the formation of the mature exocrine pancreas, as assessed by trypsin expression area at 72 hpf (Figure 5A–B). Cpd3 reduced the average pancreas size, but did not ablate exocrine pancreas formation as severely as observed in the mutants, suggesting partial loss of Nr5a2 at doses utilized, consistent with the effects observed for nr5a2oz3 haploinsufficiency. Further corroborating the nr5a2oz3/oz3 mutants, the Cpd3-treated embryos formed insulin-expressing endocrine pancreas cells normally (Figure 5C). The islet1 endocrine pancreas lineages, including β-cells (insulin) and δ-cells (somatostatin), were not visibly affected by chemical Nr5a2 inhibition (Figure 5C, Figure S5A–B). Similar to the nr5a2oz3/oz3 mutant, pdx1-expressing pancreas progenitors were also unaffected by Cpd3 treatment (Figure 5D–E). Lastly, we assessed whether heterozygote nr5a2+/oz3 fish were sensitized to the Nr5a2 chemical antagonist Cpd3. Heterozygotes were more sensitive to the chemical antagonist and showed a statistically significant reduction in pancreas size in response to Cpd3 treatment relative to nr5a2+/oz3 controls and Cpd3 treated nr5a2+/+ embryos (Figure 5F, Figure S5C). Collectively, these data demonstrate that chemical inhibition of Nr5a2 over the duration of pancreas development results in similar phenotypes to those observed in heterozygous nr5a2 mutants and suggests specificity of the inhibitor in vivo.
Figure 5. Nr5a2 chemical antagonist Cpd3 impacts exocrine pancreas development in vivo.
A) The Nr5a2 antagonist (Cpd3) produces reduced exocrine pancreas sizes that mimic the nr5a2+/oz3 heterozygotes when treated from 12–72 hpf.
B) Quantification of exocrine pancreas size by area of trypsin expression at 72 hpf. Cpd3 significantly reduces exocrine pancreas size compared to vehicle (0.04% DMSO) treated controls (n = 24 per group; p = 0.007; unpaired t-test).
C) Loss of Nr5a2 activity by Cpd3 treatment does not visibly impact the formation of the insulin expressing β-cells.
D) Cpd3 treatment does not impact the size of the pdx1 pancreas progenitor pool at 48 hpf.
E) Quantification of pdx1 pancreas progenitor expression area at 48 hpf in control and Cpd3 treated groups (n > 9 per group; p = 0.9089; unpaired t-test).
F) Quantification of exocrine pancreas size by area of trypsin expression at 72 hpf. Heterozygote nr5a2+/oz3 embryos show heightened sensitivity to 100 μM Cpd3 treatment relative to the nr5a2+/+ sibling controls (n > 10 per group; for control nr5a2+/+ vs. Cpd3 treated nr5a2+/oz3, p < 0.001; for control nr5a2+/oz3 vs. Cpd3 treated nr5a2+/oz3, p = 0.0004; for Cpd3 treated nr5a2+/+ vs. Cpd3 treated nr5a2+/oz3, p = 0.0042; ordinary one-way ANOVA with Tukey’s multiple comparisons test).
G) Drug treatment windows to examine the impact of Nr5a2 loss of function on distinct stages of pancreas and liver development, including progenitor specification, bud emergence, and organ differentiation and expansion. At 12 hpf, insulin expressing β-cells form within the gut tube (green dots). By 14 hpf, pdx1 expressing pancreas progenitors appear in two stripes (pink lines) An endocrine cell cluster (green) forms (beginning at approximately 24 hpf) and the dorsoventral pancreas bud emerges from the gut tube. By 40 hpf, the dorsoventral and anteroventral buds (blue arrows) have emerged from the gut tube, along with the liver bud (red arrow). Post 50 hpf, the liver and pancreas buds differentiate and expand. Figure adapted (Tiso et al., 2009).
3.7 Nr5a2 is required at multiple stages of pancreas and liver development
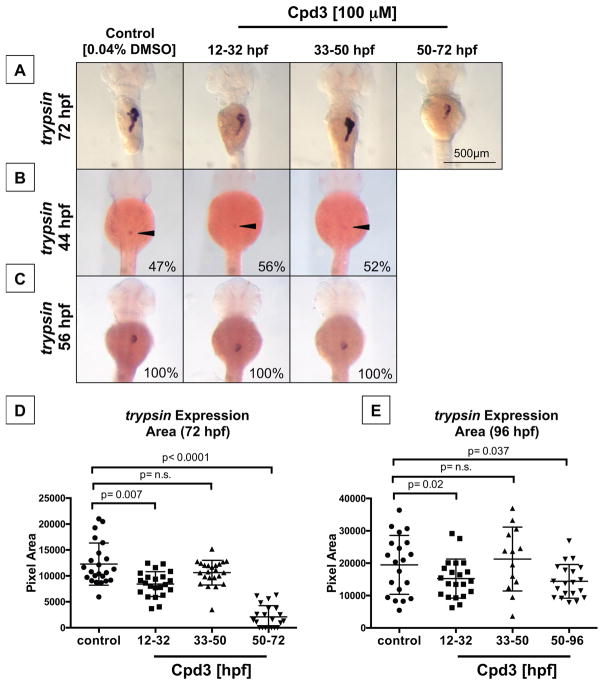
In order to determine when Nr5a2 is temporally required for hepatopancreas development, embryos from the Tüpfel long fin (TL) background were exposed to Cpd3 during time windows that correspond to distinct pancreas and liver developmental events (Figure 5G). For the pancreas studies, we performed Nr5a2 antagonist treatments over the periods of posterodorsal and ventral bud specification (12 – 32 hpf), ventral bud formation and emergence (33 – 50 hpf), exocrine pancreas differentiation and expansion (50 – 96 hpf), and the period of endoderm-specific nr5a2 expression (28 – 96 hpf) (Figure 5G). Antagonism of Nr5a2 activity in the 12 – 32 hpf and 50 – 96 hpf windows, but not the 33 – 50 hpf window, reduced the size of the exocrine pancreas (Figure 6A, Figure S6A). Despite presenting with a reduced exocrine pancreas size, the embryos in the 12 – 32 hpf treatment group initiated exocrine differentiation along a standard timeline (Figure 6B–C). At 44 hpf, approximately 50% of fish in the control and treatment groups had trypsin-expressing exocrine pancreas cells (Figure 6B). By 56 hpf, the size of the trypsin bud was also consistent across the control, 12 – 32 hpf, and 33 – 50 hpf treatment groups (Figure 6C). Furthermore, 12 – 32 hpf treatment did not alter the size of the pancreas progenitor pool marked by pdx1 at 36 hpf (Figure S6).
Figure 6. Two temporally defined roles for Nr5a2 during pancreas development.
A) Timed Cpd3 treatment reduces trypsin expression at 72 hpf for the 12 – 32 hpf and 50 – 72 hpf treatment windows, but not during the 33 – 50 hpf window.
B) Control (n = 22/46), 12–32 hpf (n = 19/36), and 33–50 hpf (n = 14/25) treated fish all have the same percentage of fish with differentiated exocrine pancreas cells at 44 hpf, indicating that drug treatments do not alter the timing of initial exocrine cell differentiation.
C) Control (n = 12/12), 12–32 hpf (n = 11/11), and 33–50 hpf (n = 14/14) treated fish all have similarly sized trypsin cell clusters at 56 hpf, demonstrating that initial exocrine pancreas differentiation is unaffected by previous nr5a2 inactivation.
D) Quantification of exocrine pancreas size (by area of trypsin expression at 72 hpf) demonstrates that the 12 – 32 hpf and 50 – 96 hpf Cpd3 treatment groups have significantly reduced exocrine pancreas sizes relative to controls. Exocrine pancreas size in the 33 – 50 hpf treatment group was not significantly altered by treatment (n > 21 per group; for control vs. 12 – 32 hpf, p = 0.007; for control vs. 33 – 50 hpf, p = 0.09; for control vs. 50 – 72 hpf, p < 0.0001; unpaired t-test).
E) Quantification of exocrine pancreas size by area of trypsin expression demonstrates that antagonist-induced exocrine pancreas reduction persists through 96 hpf for the 12 – 32 hpf and 50 – 96 hpf treatment categories. The size of the pancreas in 33 – 50 hpf treatment categories remains unchanged relative to the vehicle treated controls (n > 13 per group; for control vs. 12 – 32 hpf, p = 0.02; for control vs. 33 – 50 hpf, p = 0.5929; for control vs. 50 – 72 hpf, p = 0.037; unpaired t-test).
Quantification of the exocrine pancreas size across all treatment groups at 72 and 96 hpf confirmed that a reduction in Nr5a2 activity during 12 – 32 hpf and 50 – 96 hpf was sufficient to reduce the size of the trypsin-expressing exocrine pancreas (Figure 6D–E). Importantly, loss of Nr5a2 during ventral bud formation and emergence (33 – 50 hpf) did not impact acinar cell expansion or differentiation (Figure 6A,D–E). We hypothesize that the loss of Nr5a2 activity between 12 – 32 hpf reduces the competence of the total exocrine pancreas progenitor pool, thereby impacting exocrine pancreas expansion and differentiation beyond the 56 hpf time point. Furthermore, reduced exocrine pancreas size in the 50 – 96 hpf treatment group reveals a separate and distinct role for Nr5a2 during exocrine pancreas differentiation or expansion.
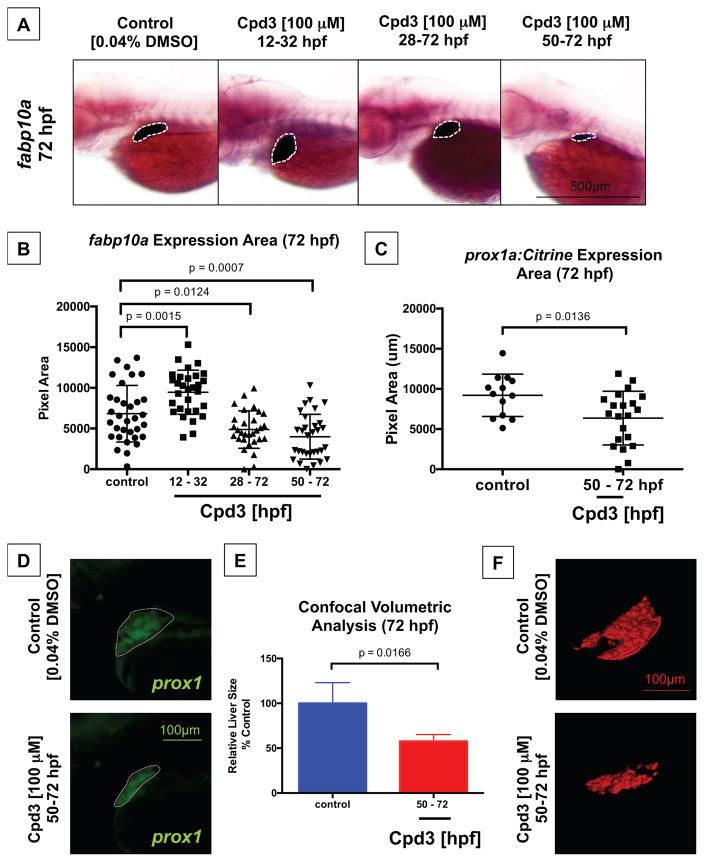
Next, we performed Cpd3 exposures to examine critical windows of Nr5a2 function for liver formation. Timed drug experiments were employed to reduce Nr5a2 activity during important stages of liver development, including liver progenitor and bud specification (12 – 32 hpf) as well as liver differentiation and outgrowth (50 – 72 hpf). Unexpectedly, area measurements performed on fabp10a expression demonstrated that loss of Nr5a2 during liver specification (12 – 32 hpf) produced a larger fabp10a-expressing liver when measured at 72 hpf (Figure 7B). Given that loss of Nr5a2 activity from 12 – 32 hpf reduces the exocrine pancreas size and expands the size of the liver, Nr5a2 may be implicated in hepatic-versus-pancreatic commitment or the specification of bipotent hepatopancreas progenitor populations. To evaluate this possibility, we examined the expression of hepatoblast markers at 36 hpf following 12 – 32 hpf Cpd3 treatment using in situ hybridization. Similar to the pancreas progenitor findings following 12 – 32 treatment, there were no notable differences in the size of the prox1 or hnf4α hepatoblast populations (Figure S6B). Furthermore, the size of the liver bud by confocal analysis of the Tg(sox17:eGFP) line in control and 12 – 32 hpf treated embryos was largely unchanged at 36 hpf (Figure S6C). Although no obvious differences in the size of the hepatoblast population or liver bud were observed, progenitors may have an intrinsically enhanced ability to respond to inductive cues, possibly through epigenetic mechanisms. Additionally, Nr5a2 may regulate proliferation in hepatoblast and pancreas progenitor cell populations, thereby contributing to increased liver and reduced pancreas size.
Figure 7. Liver differentiation and outgrowth depends on Nr5a2 activity.
A) Cpd3 treatment reduces the size and alters the position of the fabp10a liver.
B) Quantification of liver size by area of fabp10a expression at 72 hpf. Cpd3 treatment from 12 – 32 hpf enhances the size of the mature liver relative to vehicle (DMSO) treated controls (n > 27; p = 0.0015; unpaired t-test). Treatment from 28 – 72 hpf and 50 – 72 hpf reduces the median liver size relative to vehicle (DMSO) treated controls (n > 27; p = 0.0124; p = 0.0007; unpaired t-test).
C) Quantification of liver bud size by area of fluorescence expression in the Tg(prox1a:Citrine) reporter line at 72 hpf. Cpd3 treatment from 50–72 hpf significantly reduces the size of the prox1+ liver (n > 13; p = 0.0136).
D) Fluorescent microscopy images of the embryonic liver in Tg(prox1a:Citrine) reporters following vehicle (DMSO) or Cpd3 treatment from 50 – 72 hpf.
E) Confocal volumetric analysis of liver volume in control (DMSO) and Cpd3 treated (50 – 72 hpf) Tg(fabp10a:GFP) lines at 72 hpf demonstrates that Cpd3 treatment significantly reduces the volume of the liver (n > 3; p = 0.0166).
F) Representative 3D renderings of 72 hpf GFP+ livers from the Tg(fabp10a:GFP) line that were exposed to control DMSO or Cpd3 treatment from 50 – 72 hpf. Cpd3 (50 – 72 hpf) livers are visibly smaller than the control livers.
Similar to the nr5a2+/oz3 heterozygote embryos, the 28 – 72 hpf and 12 – 72 hpf treated embryos had a significantly reduced liver size relative to their control counterparts. These findings suggested a second and later role for Nr5a2 in the differentiation of specified progenitors (Figure 7A–B). As expected, reduced Nr5a2 activity during the subsequent stages of liver differentiation and outgrowth (50 – 72 hpf) significantly reduced the size of the fabp10a liver at 72 hpf, demonstrating that Nr5a2 activity during these stages is required for hepatic maturation (Figure 7A–B). We performed Cpd3 treatment (50 – 72 hpf) on Tg(prox1a:Citrine) reporter embryos and observed a similarly significant reduction in the size of the prox1+ liver bud in Cpd3 treated fish (Figure 7C–D). Additionally, confocal volumetric analysis and 3D volumetric reconstruction of livers from the Tg(fabp10a:GFP) reporter fish line demonstrate that the Cpd3 treatment reduced the total volume of the embryonic liver at 72 hpf (Figure 7E–F). These experiments further confirm a separate stage-dependent role for Nr5a2 during liver differentiation and outgrowth.
Wnt/β-catenin signaling has a role in multiple stages of hepatic development, including the specification, proliferation, and differentiation of hepatic progenitors (Decaens et al., 2008; Goessling et al., 2008; McLin et al., 2007; Ober et al., 2006; Poulain and Ober, 2011; Tan et al., 2008). In particular, the mesodermal-derived wnt2 signal has been shown to regulate hepatoblast proliferation (Poulain and Ober, 2011). We assessed expression of wnt2 in nr5a2oz3 mutants and found no changes compared to wildtype embryos (Figure S7A). Furthermore, treatment of embryos with Cpd3 at 50 – 72 hpf did not impact the expression of Wnt/β-catenin target genes assayed by qPCR (Figure S7B). These data suggest that the impact of nr5a2 on hepatic development occurs through a mechanism independent of Wnt/β-catenin signaling.
4. Discussion
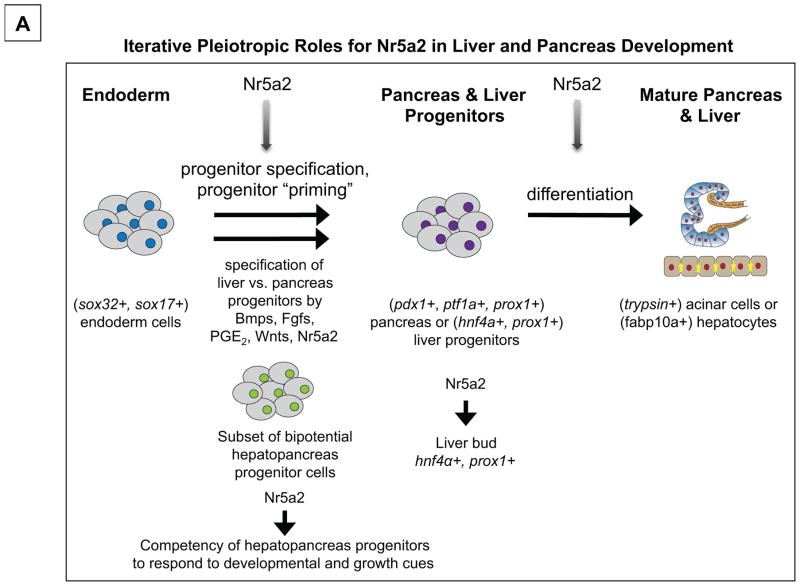
In this study, we characterize the requirement of Nr5a2 activity in zebrafish pancreas and liver development. Zygotic loss of nr5a2 disrupted the formation of the mature exocrine pancreas and liver. Analysis of hepatopancreas progenitor marker expression in nr5a2 null mutants reveals that Nr5a2 is required for the expansion and organization of endoderm progenitors as well as expression of hepatoblast markers (Figure 8). Timed inhibition of Nr5a2 using a chemical antagonist also reveals that Nr5a2 is required for the differentiation of both the liver and pancreas, as well as for the specification of the hepatopancreas progenitors (Figure 8). This study is the first to demonstrate that Nr5a2 is employed at multiple stages throughout hepatopancreas development to achieve distinct developmental milestones. We present evidence of several novel roles for Nr5a2 in endoderm development, including in the regulation of liver development and the specification of common endoderm progenitors to the hepatic or pancreatic fate. Furthermore, we accomplish the first in vivo inhibition of Nr5a2 with the chemical antagonist Cpd3 and demonstrate that it can be used to modify the specification and differentiation of hepatopancreas progenitors.
Figure 8. Pleiotropic roles for Nr5a2 in liver and pancreas development.
A) Model for Nr5a2 function during multiple stages of hepatopancreas specification and differentiation. We postulate that Nr5a2 initially functions during stages of hepatopancreas progenitor specification, possibly acting to prime progenitors to competently receive differentiation cues, or to regulate their commitment to the hepatic and pancreatic lineages. Later, Nr5a2 is required for the expression of hepatoblast markers, including hnf4α and prox1, which may be crucial for liver bud outgrowth and differentiation. Finally, Nr5a2 is required for the differentiation of pancreas and liver progenitors (following 50 hpf) into mature acinar cells and hepatocytes, respectively.
4.1 Nr5a2 regulates hepatopancreas morphogenesis and hepatopancreas progenitor specification upstream of pancreas and liver differentiation
It has been demonstrated that NR5A2 is required for exocrine pancreas differentiation in mice; however, roles for NR5A2 in hepatopancreas morphogenesis and progenitor specification have been unexplored due to the embryonic lethality of Nr5a2−/− mice (Hale et al., 2014; Labelle-Dumais et al., 2006; Paré et al., 2004). The nr5a2oz3/oz3 global knockout fish that we have generated survive through stages of hepatopancreas progenitor formation and differentiation, enabling the evaluation of novel roles for Nr5a2 in these stages of endoderm development. The nr5a2 mutant zebrafish displays defects in several events prior to hepatopancreas differentiation, including disrupted expression of hepatic and pancreas progenitor markers such as prox1 and hnf4α, and delayed liver and pancreas budding.
First, delayed morphogenesis of the endoderm was identified by examination of the pan-endoderm marker foxa3 in the nr5a2oz3/oz3 mutants. The pancreas and liver buds failed to emerge from the gut tube and expand on a normal developmental timescale in the homozygous mutants. We propose that altered morphogenesis of the liver and pancreas buds could contribute to the pancreas and liver defects observed in the nr5a2oz3/oz3 mutant. For example, liver bud outgrowth in nr5a2oz3/oz3 embryos is likely required for the emergence of the hnf4α- and hhex-expressing hepatoblasts. Therefore, when loss of Nr5a2 function reduces the size of the foxa3 expressing liver bud, the total population of hepatoblast cells may be similarly diminished. Future studies should seek to clarify the impact of a loss of nr5a2 on the migration and outgrowth of pancreas and liver progenitors that give rise to the mature endodermal organs, with attention to the impact of nr5a2 on proliferation in these populations.
Although Nr5a2 was not required for initial expression of standard pancreas progenitors markers pdx1 and ptf1a, prox1 expression in the pancreas progenitors was largely absent at 48 hpf. Additionally, prox1 and hnf4α had reduced expression in the liver bud. The loss of progenitor marker expression in the nr5a2oz3/oz3 mutants during liver and pancreas budding may be intricately coupled to the hepatopancreas morphogenesis defects. PROX1 is thought to control the migration of hepatoblast cells in the liver bud, and genetic ablation of Prox1 in the mouse leads to a failure of liver bud outgrowth (North and Goessling, 2011; Seth et al., 2014; Sosa-Pineda et al., 2000). Furthermore, loss of Prox1 in mice disrupts pancreas morphology, morphogenesis, and growth through control of pancreatic branching and tip progenitor differentiation (Wang et al., 2005; Westmoreland et al., 2012). We therefore posit that prox1 deficiency visible at 48 hpf in the hepatopancreas progenitors of nr5a2oz3/oz3 embryos may be partially responsible for the defects in liver and pancreas outgrowth, as well as the reduced size of the mature pancreas and liver structures. Loss of hnf4α expression in the liver bud may similarly underlie failed hepatocyte differentiation.
Importantly, our study identifies a general need to evaluate the role of hepatopancreas progenitor markers in the formation of the mature pancreas and liver. Although markers such as prox1, ptf1a, pdx1, hhex, and hnf4α identify populations of hepatopancreas progenitor cells, delineation of the functional roles of these genes in endoderm organ development is incomplete. Identifying the functions of prox1 and hnf4α in zebrafish, for example, will provide a greater understanding of the mechanisms by which genetic nr5a2 inactivation leads to a diminished liver size.
4.2 Nr5a2 controls hepatopancreas progenitor specification and priming
Timed drug studies with the Nr5a2 antagonist Cpd3 discovered a previously unidentified role for Nr5a2 in pancreas versus liver progenitor specification. Inactivation of Nr5a2 during early pancreas progenitor specification was sufficient to disrupt downstream exocrine pancreas formation and promote excess liver formation. There are several possible mechanisms that could explain the reduced exocrine pancreas and expanded liver size following Cpd3 treatment during hepatopancreas progenitor specification. One possibility we propose is that Nr5a2 activity could restrict liver expansion or progenitor differentiation during the 12 – 32 hpf specification window, while simultaneously promoting pancreas commitment and expansion. In a normal developmental context, Nr5a2 is expressed primarily in the liver bud prior to 32 hpf, where it may regulate the speed of liver growth and differentiation. Further studies are needed to explore the possibility that temporally restricted loss of Nr5a2 activity can differentially impact proliferation or differentiation rate in the developing liver and pancreas.
Another possibility is that Nr5a2 may be directly involved in the priming of hepatopancreas progenitors. Inhibition of Nr5a2 activity between the 12–32 hpf window significantly reduced the size of the exocrine pancreas at 72 and 96 hpf, despite unaltered expression of progenitor markers pdx1 or ptf1a and normal timing of exocrine pancreas differentiation. In our proposed progenitor priming model, the presence of Nr5a2 in pancreas or hepatic progenitor cells would establish epigenetic landscapes or transcriptional programs that enable progenitors to respond to future developmental cues (Figure 8). This model could partially explain why pancreas progenitors form, yet fail to respond to differentiation cues. Recent studies suggest that pancreatic progenitors adopt poised enhancer states during gut tube formation, and that these poised states can pre-program cells to activate pancreas-specific transcriptional programs in response to pancreas induction cues (Wang et al., 2015). Smaller exocrine pancreas size following Cpd3 treatment from 12 – 32 hpf may result because the pancreas progenitors have an altered developmental potential and a reduction in their developmental competence to become mature pancreatic acinar cells. Similarly, we hypothesize that the expanded liver following Cpd3 treatment from 12 – 32 hpf could occur because shared hepatopancreas progenitors develop a heightened competence to differentiate into hepatic lineages. Specifically, we hypothesize that Nr5a2 normally acts as a repressor of hepatic fate in a subset of shared hepatopancreas progenitors during specification stages by restricting the response to differentiation cues, and that a loss of Nr5a2 is sufficient to release this repression. Further studies are needed in order to elucidate the role of Nr5a2 in hepatopancreas progenitor priming and specification events.
4.3 Novel developmental roles for Nr5a2 inform studies on Nr5a2-dependent disease
In addition to its function in embryonic development, NR5A2 also impacts human and murine cancer development. A genome wide association study (GWAS) has implicated NR5A2 as a risk modulator for pancreatic ductal adenocarcinoma (PDAC) (Murtaugh, 2014; Petersen et al., 2010). Interestingly, acinar-to-ductal metaplasia, in which acinar cells lose their mature markers and transdifferentiate into ductal-like pancreas cells, is thought to be a critical step in PDAC formation (Murtaugh, 2014; von Figura et al., 2014). A loss of Nr5a2 in the mature exocrine pancreas using lineage-specific knockout disrupts acinar cell identity and can accelerate Kras-mediated PDAC precursor lesions (von Figura et al., 2014). Given the role of NR5A2 in regulating hepatopancreas progenitor development and acinar cell differentiation, we speculate that understanding the developmental roles of NR5A2 may elucidate the association of NR5A2 in cellular transformation and oncogenesis. Importantly, as a nuclear receptor that can bind ligands that modify its function, NR5A2 may offer a viable therapeutic target for cancers, including PDAC. Our study demonstrates that Cpd3 treatment blocks exocrine pancreas and hepatocyte differentiation, indicating that the drug can be used to modulate involvement of NR5A2 in cellular identity in vivo. Pharmacologic modulation of NR5A2 in vivo through antagonism or agonism may therefore have broad application for the treatment of endoderm-derived cancers and other disease processes impacted by NR5A2.
Supplementary Material
Zygotic loss of nr5a2 disrupts development of exocrine pancreas.
Nr5a2 regulates acinar differentiation downstream of progenitor cell formation.
Nr5a2 regulates hnf4α and prox1 expression in hepatic progenitors.
Nr5a2 is required for the formation of differentiated hepatocytes.
Nr5a2 antagonism reveals roles at multiple stages of hepatopancreas development.
Acknowledgments
We thank Chad Walesky from the Goessling Laboratory for providing the hnf4α probe. We also extend our gratitude to Eric Ortlund at Emory University for helpful discussions, and staff of the Beth Israel Deaconess Medical Center, Ohio State University, and Brigham and Women’s Hospital zebrafish facilities for zebrafish husbandry. This work was supported by the NIH K08 DK105326 (S.N.), NIH T32 GM007226 (O.W.), R01DK090311, R24OD017870 (W.G.), R01GM088041 (S.L.A.), NINDS T32 NS077984 (J.C.T.), the National Pancreas Foundation (S.N.), the Harvard Digestive Diseases Center (P30 DK034854; S.N., W.G.) and the Anna Fuller Fund (W.G.). J.C.T. is a recipient of a Pelotonia Postoctoral Fellowship. O.W. is the recipient of a Sternlicht Director’s Fund Award for Graduate Students in Diabetes Research at the Harvard Stem Cell Institute. S.N. is a recipient of the Burroughs Wellcome Fund Career Award for Medical Scientists. W.G. is a Pew Scholar in the Biomedical Sciences. There are no conflicts of interest to report.
Footnotes
Author Contributions
S.N., O.W., and W.G. conceived and designed the experiments. S.N., O.W., J.H., and W.G. analyzed the resulting data. J.C.T. generated the nr5a2 TALEN mutant. O.W. and J.H. propagated mutant lines and validated the endoderm phenotypes in the nr5a2oz3 background. O.W., S.N., and J.H. performed embryonic drug experiments. S.N., O.W., and J.W. performed morpholino experiments. S.S.B., M.C., O.W., S.N., and J.W. conducted FACS sorting and qRT-PCR. K.A. performed in situ hybridizations to assess Wnt signaling in nr5a2oz3 mutants. S.N. and J.W. performed morpholino injections and morpholino analysis. S.N., O.W., and W.G. wrote the manuscript. All authors reviewed and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annicotte JS, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F, Maudelonde T, Lazennec G, Cavailles V, Fajas L. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Cheung ID, Mostov KE, Stainier DYR. Genetic control of single lumen formation in the zebrafish gut. Nature Cell Biology. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- Benod C, Carlsson J, Uthayaruban R, Hwang P, Irwin JJ, Doak AK, Shoichet BK, Sablin EP, Fletterick RJ. Structure-based discovery of antagonists of nuclear receptor LRH-1. The Journal of Biological Chemistry. 2013;288:19830–19844. doi: 10.1074/jbc.M112.411686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benod C, Vinogradova MV, Jouravel N, Kim GE, Fletterick RJ, Sablin EP. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proceedings of the National Academy of Sciences. 2011;108:16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PLL, Escrivà H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genetics. 2007;3:2085–2100. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Developmental Biology. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botrugno OA, Fayard E, Annicotte JSS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Molecular Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:1–11. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HK, Biesinger J, Seo Y-KK, Xie X, Osborne TF. Genome-wide analysis of hepatic LRH-1 reveals a promoter binding preference and suggests a role in regulating genes of lipid metabolism in concert with FXR. BMC Genomics. 2012:13. doi: 10.1186/1471-2164-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WSS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Developmental Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaens T, Godard C, de Reyniès A, Rickman DS, Tronche F, Couty JPP, Perret C, Colnot S. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology (Baltimore, Md) 2008;47:247–258. doi: 10.1002/hep.21952. [DOI] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lora J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nature Genetics. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends in Cell Biology. 2004;14:250260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Developmental Biology. 2003a;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DYR. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Developmental Biology. 2003c:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis A-P, Puder M, Clevers H, Moon RT, Zon LI. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Developmental Biology. 2008:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A. Ductal cells of the pancreas. The International Journal of Biochemistry & Cell Biology. 2005;37:504–510. doi: 10.1016/j.biocel.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Molecular and Cellular Biology. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MA, Swift GH, Hoang CQ, Deering TG. The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development (Cambridge, England) 2014;141:3123–3133. doi: 10.1242/dev.109405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. The Nuclear Receptor Nr5a2 Can Replace Oct4 in the Reprogramming of Murine Somatic Cells to Pluripotent Cells. Cell Stem Cell. 2010a;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010b;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Her GM, Chiang CCC, Chen WYY, Wu JLL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio) FEBS letters. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- Holmstrom SR, Deering T, Swift GH, Poelwijk FJ, Mangelsdorf DJ, Kliewer SA, MacDonald RJ. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes & Development. 2011:1674–1679. doi: 10.1101/gad.16860911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes & Development. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle-Dumais C, Jacob-Wagner M, Pare JF, Belanger L, Dufort D. Nuclear receptor NR5A2 is required for proper primitive streak morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:3359–3369. doi: 10.1002/dvdy.20996. [DOI] [PubMed] [Google Scholar]
- Labelle-Dumais C, Jacob-Wagner M, Paré JF, Bélanger L, Dufort D. Nuclear receptor NR5A2 is required for proper primitive streak morphogenesis. Developmental Dynamics. 2006;235:3359–3369. doi: 10.1002/dvdy.20996. [DOI] [PubMed] [Google Scholar]
- Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Molecular endocrinology. 2008;22:1345–1356. doi: 10.1210/me.2007-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Developmental Biology. 2004;274:491–503. doi: 10.1016/j.ydbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Lin W, Wang HW, Sum C, Liu D, Hew CL, Chung B. Zebrafish ftz-f1 gene has two promoters, is alternatively spliced, and is expressed in digestive organs. The Biochemical Journal. 2000;348(Pt 2):439–446. [PMC free article] [PubMed] [Google Scholar]
- McLin VAA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development (Cambridge, England) 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Developmental Dynamics. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC. The what, where, when and how of Wnt/beta-catenin signaling in pancreas development. Organogenesis. 2008;4:81–86. doi: 10.4161/org.4.2.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC. Putting GWAS to the functional test: NR5A2 and pancreatic cancer risk. Gut. 2014;63:535–536. doi: 10.1136/gutjnl-2013-305030. [DOI] [PubMed] [Google Scholar]
- Nissim S, Sherwood RI, Wucherpfennig J, Saunders D, Harris JM, Esain V, Carroll KJ, Frechette GM, Kim AJ, Hwang KL, Cutting CC, Elledge S, North TE, Goessling W. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Developmental Cell. 2014;28:423–437. doi: 10.1016/j.devcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W. Chapter 10 – Endoderm Specification, Liver Development, and Regeneration. Methods in Cell Biology. 2011;101:205–223. doi: 10.1016/B978-0-12-387036-0.00010-4. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, Field HA, Stainier DYR. From endoderm formation to liver and pancreas development in zebrafish. Mechanisms of Development. 2003:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Paré JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Bélanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. Journal of Biological Chemistry. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- Paré JF, Roy S, Galarneau L, Belanger L. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the Hnf3beta, Hnf4alpha, and Hnf1alpha gene promoters. The Journal of Biological Chemistry. 2001;276:13136–13144. doi: 10.1074/jbc.M010737200. [DOI] [PubMed] [Google Scholar]
- Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M, Helzlsouer K, Holly EA, Jacobs EJ, Klein AP, LaCroix A, Li D, Mandelson MT, Olson SH, Risch HA, Zheng W, Albanes D, Bamlet WR, Berg CD, Boutron-Ruault MCC, Buring JE, Bracci PM, Canzian F, Clipp S, Cotterchio M, de Andrade M, Duell EJ, Gaziano JM, Giovannucci EL, Goggins M, Hallmans G, Hankinson SE, Hassan M, Howard B, Hunter DJ, Hutchinson A, Jenab M, Kaaks R, Kooperberg C, Krogh V, Kurtz RC, Lynch SM, McWilliams RR, Mendelsohn JB, Michaud DS, Parikh H, Patel AV, Peeters PH, Rajkovic A, Riboli E, Rodriguez L, Seminara D, Shu XOO, Thomas G, Tjønneland A, Tobias GS, Trichopoulos D, Van Den Eeden SK, Virtamo J, Wactawski-Wende J, Wang Z, Wolpin BM, Yu H, Yu K, Zeleniuch-Jacquotte A, Fraumeni JF, Hoover RN, Hartge P, Chanock SJ. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nature Genetics. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausa FM, Galarneau L, Belanger L, Costa RH. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3beta in the developing murine liver, intestine and pancreas. Mechanisms of Development. 1999;89:185–188. doi: 10.1016/s0925-4773(99)00209-9. [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes and Development. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JRJR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Ye J, Yu N, Guez F, Bedford DC, Neale GA, Cordi S, Brindle PK, Lemaigre FP, Kaestner KH, Sosa-Pineda B. Prox1 ablation in hepatic progenitors causes defective hepatocyte specification and increases biliary cell commitment. Development (Cambridge, England) 2014;141:538–547. doi: 10.1242/dev.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nature Genetics. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, Michalopoulos GK, Kaestner KH, Monga SP. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology (Baltimore, Md) 2008;47:1667–1679. doi: 10.1002/hep.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso N, Moro E, Argenton F. Zebrafish pancreas development. Molecular and Cellular Endocrinology. 2009;312:24–30. doi: 10.1016/j.mce.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Ueno M, Ohkawa S, Morimoto M, Ishii H. Genome-wide association study-identified SNPs (rs3790844, rs3790843) in the NR5A2 gene and risk of pancreatic cancer in Japanese. Nature Scientific Reports. 2015;5 doi: 10.1038/srep17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Morris JP, Wright CV, Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2014;63:656–664. doi: 10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RT, Xu X, Yi F, Merrill BJ, Cooney AJ. Canonical Wnt/catenin regulation of liver receptor homolog 1 mediates pluripotency gene expression. Stem Cells. 2010;28:1794–1804. doi: 10.1002/stem.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Korzh S, Li Z, Mudumana SP, Korzh V, Jiang YJJ, Lin S, Gong Z. Analyses of pancreas development by generation of gfp transgenic zebrafish using an exocrine pancreas-specific elastaseA gene promoter. Experimental Cell Research. 2006;312:1526–1539. doi: 10.1016/j.yexcr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, Lam DK, Raum JC, Stoffers DA, Ren B, Sander M. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kilic G, Aydin M, Burke Z, Oliver G. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Developmental Biology. 2005:182–194. doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Westmoreland JJ, Kilic G, Sartain C, Sirma S, Blain J. Pancreas-specific deletion of Prox1 affects development and disrupts homeostasis of the exocrine pancreas. Gastroenterology. 2012;142:999–1009. doi: 10.1053/j.gastro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Developmental Biology. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Developmental Biology. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.